Abstract
Lactobacillus paraplantarum BGCG11, a putative probiotic strain isolated from a soft, white, artisanal cheese, produces a high-molecular-weight heteropolysaccharide, exopolysaccharide (EPS)-CG11, responsible for the ropy phenotype and immunomodulatory activity of the strain. In this study, a 26.4-kb region originating from the pCG1 plasmid, previously shown to be responsible for the production of EPS-CG11 and a ropy phenotype, was cloned, sequenced, and functionally characterized. In this region 16 putative open reading frames (ORFs), encoding enzymes for the production of EPS-CG11, were organized in specific loci involved in the biosynthesis of the repeat unit, polymerization, export, regulation, and chain length determination. Interestingly, downstream of the eps gene cluster, a putative transposase gene was identified, followed by an additional rfb gene cluster containing the rfbACBD genes, the ones most probably responsible for dTDP-l-rhamnose biosynthesis. The functional analysis showed that the production of the high-molecular-weight fraction of EPS-CG11 was absent in two knockout mutants, one in the eps and the other in the rfb gene cluster, as confirmed by size exclusion chromatography analysis. Therefore, both eps and rfb genes clusters are prerequisites for the production of high-molecular-weight EPS-CG11 and for the ropy phenotype of strain L. paraplantarum BGCG11.
INTRODUCTION
A “functional starter” is defined as “cultures that possess properties which contribute to food safety and/or offer one or more organoleptic, technological, nutritional, or health advantages” (1). Accordingly, some strains of lactic acid bacteria (LAB) that produce exopolysaccharides (EPSs) meet both technological and health aspect criteria. Recently, it has been shown that EPS-producing LAB strains might be capable of improving the rheological characteristics of fermented milk products, as well as having immunomodulatory potential (2, 3).
Bacterial exopolysaccharides (EPSs) represent a large group of carbohydrate polymers which can be either covalently associated with the cell surface, forming a capsule, or loosely attached or even totally secreted into the environment of the cell (2). EPSs are classified as homo- or heteropolysaccharides, depending on whether they are composed, respectively, of one or more than one type of sugar. EPSs are widely distributed among bacteria. They can have a protective function in the natural environment against phagocytosis and predation by protozoa, phage attack, antibiotics or toxic compounds, and osmotic stress. Moreover, EPSs also have a role in cell recognition, adhesion to surfaces, and biofilm formation (4). EPSs can interfere with adhesion of pathogens and probiotics to intestinal mucus or have an immunomodulatory effect (3, 5–9). Furthermore, EPSs can have the ability to lower the cholesterol and triglyceride levels in the serum and liver of rats (10). For that reason, detailed characterization of the genes involved in EPS production is of great importance.
The genetic information for heteropolymer-type EPS production is organized into gene clusters, located in chromosomal or plasmid DNA. Sometimes more than one EPS operon can be found in the genome of bacteria (11, 12). In the eps operons several genes with specific functions are present: genes involved in the assembling of repeating units, chain length determination, polymerization, and export and those involved in regulation (13). Heteropolysaccharides are made by polymerizing repeating-unit precursors formed in the cytoplasm. The sugar nucleotides, such as UDP-glucose, UDP-galactose, and dTDP-rhamnose, play an essential role in heteropolysaccharide biosynthesis due to their role in the sugar activation necessary for monosaccharide polymerization (13). It has been shown previously that a 9-fold increase in the number of EPS plasmid copies can lead to an almost 3-fold increase in the eps expression level, which results in an almost 4-fold increase in the EPS production level in Lactococcus lactis NIZO B40. In the same study the authors indicated that the maximum amount of NIZO B40 EPS production is limited by the level of the expression products of the eps gene cluster rather than by the level of EPS precursors (14).
In lactobacilli, eps gene clusters have been identified in Lactobacillus helveticus NCC2745, Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus rhamnosus, and Lactobacillus plantarum (15–18), among others. In Lactobacillus paraplantarum BGCG11, formerly classified as Lactobacillus casei CG11, isolated from a soft, white, artisanal cheese, the assumption of plasmid localization of the eps gene cluster was proposed after plasmid-curing experiments when some derivatives lacking the pCG1 plasmid had lost their EPS-CG11 production and ropy phenotype (19). In previous work it was shown that when L. paraplantarum BGCG11 was grown in minimal medium supplemented with glucose as the only sugar source, EPS-CG11 was composed of glucose (75.7%), rhamnose (20.5%), galactose (2.1%), and mannose (1.7%). In the same medium, novobiocin (NB) derivatives which had lost the ropy phenotype produced residual EPS in an amount almost 10 times lower and with a different composition of sugars: glucose (86.6%), mannose (6.2%), galactose (4.1%), and rhamnose (3.1%) (20). The biosynthesis of more than one EPS polymer by one strain was noticed previously (21–24). Recently, it has been shown that the EPS-CG11 polymer is responsible for in vitro anti-inflammatory or immunosuppressive effects (3). Taking these findings into consideration, together with the fact that there have been no data on functional eps genes clusters in L. paraplantarum until now, the aim of this study was to decipher the genetic organization of the eps gene cluster in the strain L. paraplantarum BGCG11.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. paraplantarum BGCG11 and its derivative NB1 were grown at 30°C in deMan-Rogosa-Sharpe (MRS) broth (Merck, Darmstadt, Germany). Escherichia coli DH5α cells, used as cloning hosts, were grown in LB medium with aeration at 37°C. When appropriate, erythromycin (Em) (Serva, Heidelberg, Germany) was added at 300 μg ml−1 for E. coli.
DNA manipulations.
Plasmid DNA from E. coli cells was isolated using a Qiagen Plasmid Mini-Prep kit (Qiagen, Hilden, Germany). Total DNA from L. paraplantarum BGCG11 and NB1 was extracted as previously described (25) with a slight modification (cells were treated with lysozyme before lysis with SDS). Plasmid DNA was isolated and purified as previously described (26). Restriction enzymes (Fermentas UAB, Vilnius, Lithuania) were used as recommended by the manufacturer. The plasmids and DNA fragments were analyzed by horizontal electrophoresis on 1% (wt/vol) agarose gels, in 1× TAE (40 mM Tris-acetate, 1 mM EDTA [pH 8.0]) buffer with ethidium bromide (0.5 μg ml−1), at a constant voltage of 2.5 V cm−1 and visualized using a charge-coupled-device (CCD) camera (Biometra BDR2/5/6; Bio Doc Analyze GmbH, Göttingen, Germany). GeneRuler DNA ladder mix (Fermentas UAB) was used for comparison of the sizes of obtained DNA fragments.
Southern blot hybridization.
Hybridization experiments were carried out essentially as previously described (27). DNA probes were labeled following the instructions of the manufacturer using a digoxigenin (DIG) DNA labeling and detection kit (Roche Applied Science, Mannheim, Germany). Hybridizations were carried out at 65°C.
Plasmid construction.
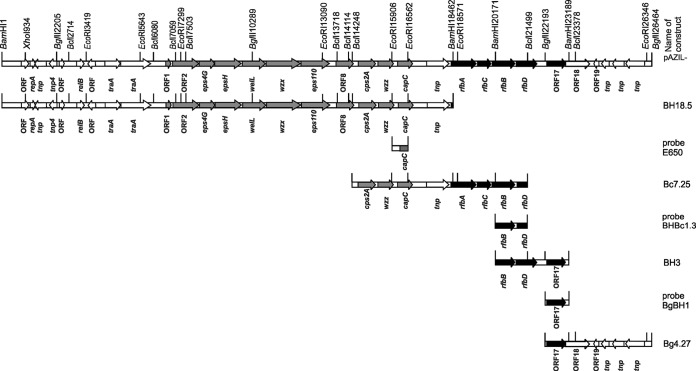
The plasmids and constructs used in the study are listed in Table 1. In order to obtain the nucleotide sequence(s) of the eps gene cluster, total plasmids from L. paraplantarum BGCG11, as well as from the BGCG11 EPS-negative (EPS−) derivative NB1 (for comparison), were subjected to BamHI restriction enzyme digestion. The DNA fragments obtained were ligated into pAZIL vector (28) predigested with BamHI, using T4 DNA ligase (Promega, Madison, USA) in the buffer as recommended by the manufacturer. Nucleotide sequencing of the 18,466-bp BamHI fragment revealed that it contained part of the eps gene cluster. In order to search the neighboring DNA region and to complete the cloning of the entire eps gene cluster, the next step was performed by constructing new plasmid libraries in the pAZIL vector using BclI and BglII restriction enzymes. Overlapping the BclI clone with the previously cloned 18,466-bp BamHI fragment was done by Southern blot hybridization using an E650 DNA probe to map in the overlapping region at the end of the pAZIL-BH18.5 clone, resulting in the clone pAZIL-Bc7.25. The downstream part of the eps gene cluster was selected by screening the already constructed BamHI plasmid library using the BHBc1.3 probe, resulting in the overlapping clone pAZIL-BH3. To complete the cloning of the eps gene cluster located in the pCG1 plasmid, an additional clone, pAZIL-Bg4.27, was selected using the probe BgBH1 from the constructed BglII plasmid library. The complete eps and rfb gene clusters were assembled by selecting overlapping clones obtained from three independent plasmid libraries (constructed by BamHI, BclI, and BglII restriction enzymes) of the pCG1 plasmid. The construct pAZIL-BH18.5 carrying the BamHI fragment (18,466 bp), was selected for further work. The BamHI fragment (18,466 bp) was originally from the plasmid pCG1, which is present only in the parental strain BGCG11. That was confirmed by Southern blot DNA-DNA hybridization using a 656-bp EcoRI DNA fragment subcloned from the pAZIL-BH18.5 into the pAZIL vector (named pAZIL-E650) as a probe. The total plasmids pCG11 and pNB1, digested with several restriction enzymes (see Fig. S1a in the supplemental material), were used for hybridization (see Fig. S1b). It was confirmed that pAZIL-BH18.5 contains a DNA fragment from the plasmid not present in the derivative NB1. Based on the nucleotide sequence analysis of the DNA fragment in the pAZIL-E650 construct (98% identity with the part of the sequence belonging to the polysaccharide biosynthesis protein [GenBank accession number YP_008122774.1] from L. plantarum 16), the whole DNA fragment in pAZIL-BH18.5 (Fig. 1) was analyzed. Since an 18,466-bp BamHI fragment in pAZIL-BH18.5 did not contain the complete information for the eps gene cluster, additional cloning of neighboring DNA regions from pCG1 was performed (Fig. 1). The continuation of the genetic information for EPS-CG11 production was mapped on the construct pAZIL-Bc7.25, selected from the BclI plasmid library. In the next two steps, overlapping fragments with additional DNA were selected, first from the BamHI (the clone pAZIL-BH3) and then from the BglII (the clone pAZIL-Bg4.27) plasmid libraries. Complete information for EPS-CG11 production, consisting of two gene clusters, was cloned by combining clones from the three constructed plasmid libraries of the pCG1 plasmid.
TABLE 1.
Plasmid and constructs used in this study
| Name | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| pAZIL | Shuttle cloning vector, 7,109 bp, Emr | 28 |
| pAZIL-BH18.5 | 18,466-bp BamHI fragment from pCG1 cloned in BamHI site of pAZIL, Emr | This work |
| pAZIL-BH3 | 3,018-bp BamHI fragment from pCG1 cloned in BamHI site of pAZIL, Emr | This work |
| pAZIL-Bg4.27 | 4,266-bp BglII fragment from pCG1 cloned in BglII site of pAZIL, Emr | This work |
| pAZIL-E650 | 656-bp EcoRI fragment from pCG1 cloned in EcoRI site of pAZIL, Emr | This work |
| pAZIL-BHBc1.3 | 1,328-bp BamHI/BclI fragment from pCG1 cloned in BamHI/BclI site of pAZIL, Emr | This work |
| pAZIL-BgBH1 | 996-bp BglII/BamHI fragment from pCG1 cloned in BglII/BamHI site of pAZIL, Emr | This work |
| pGhost9 | Emr, vector for insertional mutagenesis with a temperature-sensitive origin of replication | 31 |
| pGhost/31 | pGhost9 derivative carrying part of the orf2 gene | This work |
| pGhost/32 | pGhost9 derivative carrying part of the rfbB gene | This work |
| p-GEM-T-Easy | 3,015 bp, Ampr, PCR cloning vector | Promega |
Emr, resistance to erythromycin; Ampr, resistance to ampicillin.
FIG 1.
Linear gene map of the pCG1 region carrying eps gene clusters of 26,464 bp and the scheme of construction of overlapping clones used for sequencing and probes for Southern blot hybridization. Positions of relevant restriction sites are indicated. The sizes and orientations of predicted ORFs are indicated by arrows. Genes representing the eps gene cluster are indicated by gray arrows; those for specific rfb gene clusters are indicated by black arrows.
Sequencing and annotation of the putative EPS biosynthetic cluster.
DNA sequencing reactions were performed by Macrogen Europe (Amsterdam, The Netherlands), a nucleotide sequencing service. The obtained nucleotide sequences were assembled into a ca. 17-kb contig containing the complete nucleotide sequence of the putative eps gene clusters of L. paraplantarum BGCG11. The NCBI Entrez nucleotide database was searched for similar sequences using BLASTx, and alignment of overlapping fragments was performed with BLASTn (29). Membrane-spanning regions of translated gene products were predicted using the Phobius program (http://phobius.sbc.su.se/). Additionally, the Pfam database (30) was searched for conserved motifs in the translated gene products.
Construction of the knockout mutants.
The genes orf2 (GenBank accession number HG316787, position 6949 to 7917) and rfbB (GenBank accession number HG316787, position 19902 to 20930) were inactivated in pCG1 by homologous integration using a protocol described by Maguin et al. (31). For insertional inactivation, a part of the orf2 gene and a part of the rfbB gene were amplified using the primer pair ORF2F (5′-CTAAGACTAGTAAGGATGC-3′) and ORF2R (5′-GCTTAGCTCCCAAAATCCAGG-3′) and the pair RFBBF (5′-GGTAATCGGGCCAACTTGG-3′) and RFBBR (5′-CGTAAATCATGGCCGGGACG-3′), respectively. KapaTaq DNA polymerase (Kapa Biosystems, INC., Boston, MA, USA) was used to amplify DNA fragments by PCR using a GeneAmp PCR System 2700 thermal cycler (Applied Biosystems, Foster City, CA, USA). PCR products were purified with a QiaQuick PCR purification kit (Qiagen) according to the protocol of the supplier. The commercial vector p-GEM-T-Easy (Promega) was used for cloning the PCR products. Standard heat shock transformation was used for plasmid transfer into E. coli. For plasmid isolation from E. coli, a QIAprep Spin Miniprep kit was used according to the manufacturer's recommendations (Qiagen, Hilden, Germany). A fragment from the orf2 gene (NcoI-NcoI, 674 bp, position 7070 to 7744) was cloned into the pGhost9 vector, resulting in the construct pGhost/31. A fragment from the rfbB gene (NcoI-NcoI, 742 bp, position 20019 to 20761) was cloned into the pGhost9 vector, resulting in the construct pGhost/32. Plasmid constructs were sequenced by the Macrogen Sequencing Service (Macrogen, Netherlands). In the next step, strain BGCG11 was transformed with pGhost/31 and pGhost/32, individually, and grown on MRS plates containing 7.5 μg/ml of erythromycin at 28°C for 48 h. Obtained transformants were transferred to 37°C and incubated for 48 h, resulting in the knockout mutants MUT1 for the orf2 gene and MUT2 for the rfbB gene.
EPS extraction, purification, and analysis by high performance liquid chromatography (HPLC).
EPS was isolated and purified from BGCG11, its derivative NB1, and both knockout mutants (MUT1 and MUT2) from cultures grown in basal minimum medium (BMM). The purified EPS was analyzed by means of size exclusion chromatography (SEC) coupled with a multiangle laser light scattering (MALLS) detector. These procedures were done as described by Nikolic et al. (3). In short, EPS fractions were separated in size exclusion columns (TSKgel G3000 PWXL plus TSKgel G5000 PWXL protected with a TSKgel guard column; Supelco-Sigma) at 40°C with a flow rate 0.45 ml min−1 using 0.1 M NaNO3 as the mobile phase. The chromatographic equipment (Waters, Milford, MA) was coupled to a photodiode array (PDA) 996 detector, set at 280 nm for protein detection, and to a 410 refraction index (RI) detector, which allows the quantification of each EPS fraction by using calibration equations made with dextran standards. The Empower software (Waters) was used for analysis. Additionally, a third static MALLS detector (Dawn Heleos II; Wyatt Europe GmbH, Dembach, Germany) was coupled in series to analyze the molar mass distribution of the EPS fractions using the software Astra, version 3.5 (Wyatt Europe GmbH).
Nucleotide sequence accession number.
The complete eps and rfb gene clusters with surrounding genes were submitted to the European Nucleotide Archive (ENA) and GenBank under accession number HG316787.
RESULTS AND DISCUSSION
Exopolysaccharides produced by LAB display a wide variety of composition, structure, and size of molecules (22). Although these macromolecules are synthesized in representatives of different LAB, such as Streptococcus, Lactococcus, Lactobacillus, Leuconostoc, and Pediococcus, organization of the genes involved in EPS synthesis is conserved constitutively. Until now, a large number of operons for EPS production have been characterized, and it has been noticed that eps clusters in LAB have a conserved functional structure, with most genes found in the same position (except for in L. rhamnosus) (17, 22). In this work we have identified, sequenced, and functionally characterized a 26,464-bp region from the pCG1 plasmid present in L. paraplantarum BGCG11 containing 16 open reading frames (ORFs) assumed to be involved in EPS-CG11 biosynthesis.
Cloning of the EPS-CG11 gene clusters.
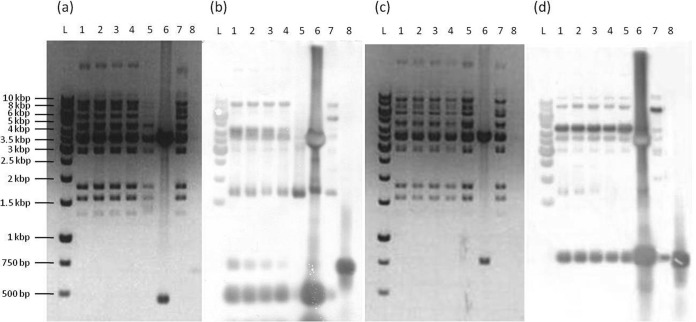
Total plasmids isolated from L. paraplantarum BGCG11 (pCG11) and its EPS-CG11-negative derivative NB1 (pNB1) were subjected to BamHI digestion in order to distinguish the DNA fragments belonging to the pCG1 plasmid (responsible for the EPS-CG11 production), present in BGCG11 but not in the NB1 derivative (Fig. 2). It was noticed that the resulting BamHI restriction profile of the pCG11 plasmid showed extra BamHI plasmid bands originating from the pCG1 plasmid. A pCG1 plasmid library consisting of BamHI fragments from the pCG11 plasmid cloned into the pAZIL vector was constructed as described in Materials and Methods. Afterwards, restriction analysis of the obtained clones was performed with the same enzyme, as described above.
FIG 2.
The comparison of the total plasmid profile of L. paraplantarum BGCG11 (pCG11) and the EPS-CG11-negative derivative NB1 (pNB1). Lane L, GeneRuler DNA ladder mix; lane 1, pCG11; lane 2, BamHI restriction digestion of total pCG11; lane 3, BamHI restriction digestion of total pNB1. The arrows indicate plasmid bands present only in BGCG11.
Identification of the eps and rfb gene clusters and putative functions of predicted ORFs.
It was found that the eps gene cluster, located in a 17-kb region of the pCG1 plasmid as proposed previously (19), has a unique structure and is divided into two gene clusters by a transposase gene. The functional structure of the eps gene cluster was similar to that of previously known operons for EPS production as it has genes for enzymes responsible for regulation, polymerization, export, assembling repeating units, and chain length determination (32). The complete sequence of 26,464 bp consists of 31 predicted ORFs, 16 of which represent the gene cluster responsible for EPS-CG11 production (Table 2 and Fig. 3). The other genes are involved in plasmid replication, mobilization, and transposition.
Table 2.
Results of BLASTx comparison of predicted ORFs from eps and rfb gene clusters with the NCBI Entrez nucleotide database
| ORF (no. of aa)a | Position (nt)b | Protein(s) with highest homology (GenBank accession no.) | Predicted domain(s) or superfamily in encoded ORF (identification no.)c | Organism | Amino acid identity (%) |
|---|---|---|---|---|---|
| orf1 (62) | 6746–6934 | Putative priming glycosyltransferase (YP_008122775.1) | Bacterial sugar transferase (Pfam 02397); exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase (TIGR03025) | Lactobacillus plantarum 16 | 77 |
| orf2 (322) | 6949–7917 | Glycosyltransferase family 8 (WP_003663531) | Glycosyltransferase family 8 (Pfam 01501) | Lactobacillus reuteri | 36 |
| eps4G (232) | 7956–8654 | Putative ribitol phosphotransferase (CAI34193.1) | CDP-alcohol phosphatidyltransferase (cl00453) | Streptococcus pneumoniae | 51 |
| Eps4G (AAN63683) | Streptococcus thermophilus | 48 | |||
| epsH (341) | 8680–9705 | EpsH (NP_053026) | Glycosyltransferase family 11 (Pfam 01531) | Lactococcus lactis subsp. cremoris | 33 |
| welL (323) | 9744–10715 | Putative glycosyltransferase (WP_003566665) | Glycosyltransferase family A (cl11394) | Lactobacillus casei | 35 |
| WelL (CCC15461) | Lactobacillus pentosus IG1 | 34 | |||
| wzx (463) | 10778–12169 | Putative polysaccharide biosynthesis protein (WP_003676723) | Wzx, a subfamily of the multidrug and toxic compound extrusion (MATE)-like proteins (cd13128); polysaccharide biosynthesis protein (pfam01943), RfbX (COG2244) | Lactobacillus reuteri | 36 |
| Flippase (WP_002911476) | Streptococcus sanguinis | 27 | |||
| eps11O (369) | 12195–13304 | Eps11O, putative polysaccharide polymerase (AAN63798) | No domains | Streptococcus thermophilus | 27 |
| orf8 (226) | 13596–14276 | UDP-d-galactose:(glucosyl)lipopolysaccharide-1, 6-d-galactosyltransferase (YP_001844233) | Glycosyltransferase_GTB_type (cl10013) | Lactobacillus fermentum IFO 3956 | 42 |
| cps2A (256) | 14522–15292 | Exopolysaccharide biosynthesis protein (YP_008122772); | Chain length determinant protein (pfam02706); capsular polysaccharide biosynthesis protein (COG3944) | Lactobacillus plantarum 16 | 95 |
| Exopolysaccharide biosynthesis protein Cps2A | Lactobacillus plantarum WCFS1 | 92 | |||
| wzz (239) | 15304–16023 | Exopolysaccharide biosynthesis protein, chain length determinant Wzz (CCC15467) | Capsular exopolysaccharide family (TIGR01007); chain length determinant protein tyrosine kinase EpsG (TIGR03029); exopolysaccharide transport protein family (TIGR01005); exopolysaccharide/PEP-CTERM locus tyrosine autokinase (TIGR03018) | Lactobacillus pentosus IG1 | 97 |
| capC (257) | 16010–16783 | Capsular polysaccharide biosynthesis protein (CCC15468) | Capsular polysaccharide biosynthesis protein CapC (COG4464) | Lactobacillus pentosus IG1 | 91 |
| Polysaccharide biosynthesis protein, phosphotyrosine-protein phosphatase (YP_008122774) | Lactobacillus plantarum 16 | 94 | |||
| tnp (309) | 17355–18284 | IS30 family transposase (YP_795436) | Transposase and inactivated derivatives, IS30 family (COG2826) | Lactobacillus brevis ATCC 367 | 99 |
| rfbA (289) | 18438–19307 | Glucose-1-phosphate thymidylyltransferase | Glucose-1-phosphate thymidylyltransferase, short form, RmlA (TIGR01207); dTDP-glucose pyrophosphorylase RfbA (COG1209) | Lactobacillus plantarum | 99 |
| rfbC (193) | 19311–19892 | dTDP-4-dehydrorhamnose 3,5-epimerase (YP_003062617) | dTDP-4-dehydrorhamnose 3,5-epimerase, RmlC (TIGR01221); RfbC (COG1898) | Lactobacillus plantarum JDM1 | 99 |
| rfbB (342) | 19902–20930 | dTDP-glucose 4,6-dehydratase (YP_004889090) | dTDP-d-glucose 4,6-dehydratase, RfbB (COG1088) | Lactobacillus plantarum WCFS1 | 99 |
| rfbD (280) | 20963–21805 | dTDP-4-dehydrorhamnose reductase (WP_003646252) | RmlD substrate binding domain (Pfam 04321); dTDP-4-dehydrorhamnose reductase RfbD (COG1091) | Lactobacillus plantarum | 95 |
| orf17 (233) | 22271–22972 | 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase (YP_003063113) | Glycosyltransferase family A (cl11394), UDP-N-acetylglucosamine diphosphorylase/glucosamine-1-phosphate N-acetyltransferase (TIGR01173); glucose-1-phosphate thymidylyltransferase, long form (TIGR01208) | Lactobacillus plantarum JDM1 | 100 |
| orf18 (341) | 22974–23999 | Ribitol-5-phosphate 2-dehydrogenase (YP_003063114) | Medium chain reductase/dehydrogenase (MDR)/zinc-dependent alcohol dehydrogenase-like family (cl16912); threonine dehydrogenase and related Zn-dependent dehydrogenases (COG1063) | Lactobacillus plantarum JDM1 | 100 |
The number of amino acids (aa) encoded.
The position from the beginning of the sequence of pCG1 plasmid (from the BamHI restriction site). nt, nucleotides.
Identification numbers are from databases from Pfam protein families, clusters of orthologous groups of proteins (COG), The Institute for Genomic Research (TIGR), and NCBI Conserved Domains Database.
FIG 3.

Schematic genetic organization of polysaccharide biosynthesis of eps gene clusters in L. paraplantarum BGCG11. ORF1 is a putative priming glycosyltransferase (black arrow). Other glycosyltransferases are indicated with dark gray arrows (ORF2, eps4G, epsH, welL, and ORF8); lighter-gray arrows indicate genes putatively involved in polymerization and the export process in EPS-CG11 biosynthesis (wzx and eps11O); arrows with hatched lines indicate genes putatively involved in the regulation of EPS-CG11 biosynthesis (cps2A, wzz, and capC). The transposase tnp separates the above-mentioned genes from the rfbACBD gene cluster (indicated by arrows with horizontal stripes) putatively involved in the synthesis of dTDP-rhamnose.
The ORFs thought to be involved in the glycosyltransferase (GTF) function are located at the beginning of the eps gene cluster. EPS-CG11 biosynthesis starts with a priming gtf gene, orf1, at the beginning of eps gene cluster. ORF1 has 77% identity at the amino acid level with a putative priming glycosyltransferase (p-GTF) of L. plantarum 16 (Table 2). The product of this gene has a role in the first (priming) step in the synthesis of an oligosaccharide “block” since it is a member of the family of EPS biosynthesis polyprenyl glycosylphosphotransferases which transfer the sugar from, for instance, UDP-glucose or UDP-galactose to a lipid carrier, such as undecaprenyl phosphate. This gene is highly evolutionarily conserved (33). In comparison to previously described eps gene clusters, the GTF that initiates EPS biosynthesis could be located at the beginning of the eps gene cluster, as in Lactobacillus helveticus NCC2745 (15), downstream from the other genes in the eps gene cluster, in the central region of the eps clusters, as in L. lactis NIZO B40 (34), Streptococcus thermophilus strains (35, 36), and L. delbrueckii subsp. bulgaricus Lfi5 (16), or even near the end of 3′ region, as in the case of L. rhamnosus (17). Downstream of orf1 there are five genes assigned as GTFs, and their function should be in transferring the other sugars of the EPS subunit in an ordered way. Among them, the genes orf2, epsH, and welL encode proteins belonging to different GTF superfamilies (families 8, 11, and A, respectively), while eps4G encodes a putative ribitol phosphotransferase. Among other GTFs encoded by the eps gene cluster, Eps4G showed more than 50% amino acid identity with a putative ribitol phosphotransferase in Streptococcus pneumoniae, while others (ORF2, EpsH, and WelL) share less than 40% amino acid identity with GTF gene products. ORF8, as a putative 1,6-d-galactosyltransferase, might have a role in the addition of galactose to the EPS-CG11 polymer. ORF8 has 42% identity at the amino acid level with UDP-d-galactose, for example, the (glucosyl) lipopolysaccharide-1,6-d-galactosyltransferase from Lactobacillus fermentum IFO 3956 (Table 2). These GTF gene products are involved in the synthesis of repeating-unit blocks that are linked to the lipid carrier at the inner side of the cytoplasmic membrane.
Interestingly, orf8 is separated from the ORFs encoding other GTFs by two genes, wzx and the eps11O. The flippase (Wzx) is potentially involved in translocation of the EPS subunits across the cytoplasmic membrane while the other protein, assigned as Eps11O, is a polysaccharide polymerase and could be involved in the polymerization process of EPS into long polysaccharides on the outer side of the cytoplasmic membrane, as similarly described for L. rhamnosus GG (22). The wzx gene of BGCG11 is presumed to encode a polysaccharide biosynthesis protein (36% amino acid identity with Lactobacillus reuteri), a member of a family of integral membrane proteins, and shows 27% amino acid identity to the flippase from Streptococcus sanguinis (Table 2). Both Wzx and Eps11O from the eps gene cluster could be integral membrane proteins since the prediction (http://phobius.sbc.su.se/) showed 13 transmembrane domains for Wzx and 10 transmembrane domains for Eps11O (data not shown). The supposed function of these two proteins is the polymerization and export process in EPS-CG11 biosynthesis since the proteins show similar homology to S. thermophilus and the wzx and the wzy genes in L. rhamnosus RW-9595M (17).
With high (95%) identity at the amino acid level with the EPS biosynthesis protein from L. plantarum 16, Cps2A belongs to a family of chain length determinant proteins (or Wzz protein) with a region of bacterial tyrosine kinases (Table 2). The product of the gene wzz showed 97% amino acid identity with the Lactobacillus pentosus IG1 chain length determiner Wzz, a member of the family of the chain length determinant protein-tyrosine kinase EpsG and also a member of the exopolysaccharide/PEP-CTERM locus tyrosine autokinase, with C-terminal tyrosine residues (likely to be autophosphorylation sites). The next gene product, CapC, has 94% identity at the amino acid level with a polysaccharide biosynthesis protein, phosphotyrosine-protein phosphatase, from L. plantarum 16 and 91% identity at the amino acid level with a capsular polysaccharide biosynthesis protein from L. pentosus. Gene products of cps2A, wzz, and capC have more than 90% identity at the amino acid level with previously described proteins involved in the regulation of EPS biosynthesis, strongly indicating their role in the regulation of the EPS-CG11 biosynthesis process. Cps2A can be located in the membrane since it has two transmembrane domains on the N and C termini of the predicted protein. With this characteristic, Cps2A resembles export proteins (Wzx, Wzy, Wzz, and homologues to ExoP-like proteins) that consist of a transmembrane domain containing two putative transmembrane helices and a domain containing a nucleotide-binding motif (32). In the previously described models for EPS biosynthesis in L. rhamnosus GG (22) and strains of S. pneumoniae (37), Wzz and Wzb (phosphotyrosine-protein phosphatase) are thought to be involved in the regulation of EPS biosynthesis. Taking this into consideration, Wzz and CapC from the eps gene cluster might be part of the phosphorylation complex. Wzz is a member of the family of the chain length determinant protein-tyrosine kinase, with C-terminal tyrosine residues, which could be autophosphorylation sites, while CapC is a phosphotyrosine-protein phosphatase. The organization of the previously described eps gene clusters is similar to that of the cps gene cluster cps1A-I in L. plantarum WCFS1 (18).
Following the mentioned genes there is a tnp gene, and its product showed 99% amino acid identity with an IS30 family transposase from Lactobacillus brevis ATCC 367. The tnp gene separates the above-mentioned preceding genes in the eps gene cluster from the four genes putatively involved in dTDP-l-rhamnose biosynthesis (rfbA, rfbC, rfbB, and rfbD). Using the IS Finder reference database for insertion sequences (IS) (https://www-is.biotoul.fr//), the presence of complete IS30 sequences was detected at positions between 17256 and 17305 nucleotides (left end) and between 18253 and 18298 nucleotides (right end). Also, partial IS sequences were found, as follows: a transposase of the IS30 family (position 24475 to 24771) had only the left end of the IS sequence at position 24461 to 24500; a transposase of the IS3 family (position 25456 to 26223) had only the left end of the insertion sequence at the position between 25424 and 25452 nucleotides; and partial IS sequences for transposases of the IS5 family were found for ISLho2 at the position between nucleotides 1207 and 1229 and for ISAba15 at the position between nucleotides 1841 and 1868. However, the tnp gene is not involved in EPS-CG11 biosynthesis. Interestingly, in the strain L. plantarum WCFS1 there are four functional cps gene clusters, but only after cps1A-I are there rmlACBD genes responsible for the presence of rhamnose in the polysaccharide polymer.
The rfbACBD genes represent a specific rfb gene cluster encoding proteins with a very high percentage of amino acid identity (over 95%) with the ones in the L. plantarum strains, indicating that they are highly conserved, as was shown previously in L. rhamnosus (22). RfbA, with 99% amino acid identity to glucose-1-phosphate thymidylyltransferase from L. plantarum, is the first of four enzymes thought to be involved in the biosynthesis of dTDP-l-rhamnose. The next, RfbC, with 99% amino acid identity to dTDP-4-dehydrorhamnose 3,5-epimerase from L. plantarum JDM1, is a member of a family which catalyzes the isomerization of dTDP-4-dehydro-6-deoxy-d-glucose with dTDP-4-dehydro-6-deoxy-l-mannose. The RfbB showed 99% identity at the amino acid level with dTDP-glucose 4,6-dehydratase from L. plantarum WCFS1 and most likely functions as the second of four steps in the dTDP-l-rhamnose pathway (the dehydration of dTDP-d-glucose to dTDP-4-keto-6-deoxy–d-glucose) in the synthesis of l-rhamnose. The fourth, RfbD, with 95% amino acid identity with the dTDP-4-dehydrorhamnose reductase from L. plantarum, is assumed to synthesize dTDP-l-rhamnose from alpha-d-glucose-1-phosphate. This protein also has an RmlD substrate binding domain, responsible for binding a sugar nucleotide. A similar rfb gene cluster separated by insertional elements was also found in Lactobacillus gasseri ATCC 33323T and Lactobacillus johnsonii NCC533 and NCC2767 strains, suggesting that the eps clusters could be a hot spot of recombination events, which under selective pressure result in diverse EPS structures (38).
Since rhamnose represents more than 20% of the total monosaccharide composition in EPS-CG11, it could be expected that the rfb gene cluster has its own promoter upstream from rfbA. According to the literature on four L. rhamnosus strains, genes involved in biosynthesis of the sugar nucleotide precursor dTDP-l-rhamnose (designated rmlACBD) are expressed from their own promoters, as well as cotranscribed with the upstream eps genes (17). The presence of the rfbACBD genes in the rfb gene cluster could be responsible for the specific composition, and are possibly involved in the physicochemical characteristics, of the EPS-CG11 polymer.
In order to ensure that there is no other rfb gene cluster on the chromosome (since the presence of 3% rhamnose was revealed in the low-molecular-weight residual EPS in the NB1 derivative), Southern blot hybridization using chromosomal DNA with a probe complementary to the rfb gene was performed (see Fig. S1c and S1d in the supplemental material). The results clearly showed that only the BGCG11 wild-type strain gave specific hybridization signals with this probe. This result strongly indicates that the obtained signal corresponds to the pCG1 plasmid present only in BGCG11 and not to a gene present at the bacterial chromosome.
The last two genes in the rfb gene cluster are orf17 and orf18, encoding proteins with 100% amino acids identity with two proteins from L. plantarum JDM1. The former gene, orf17, encodes a protein thought to be involved in UDP-N-acetylglucosamine and peptidoglycan biosynthesis and shows identity with 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase, a member of GTF family A. It has predicted domains of UDP-N-acetylglucosamine diphosphorylase/glucosamine-1-phosphate N-acetyltransferase and possibly a bifunctional enzyme, GlmU, which catalyzes the last two reactions in the four-step pathway of UDP-N-acetylglucosamine biosynthesis from fructose-6-phosphate. Hence, orf17 might be involved in the biosynthesis and degradation of surface polysaccharides. The product of the latter gene, orf18, is a putative ribitol-5-phosphate 2-dehydrogenase, part of a medium-chain reductase/dehydrogenase (MDR)/zinc-dependent alcohol dehydrogenase-like family, and has predicted domains of threonine dehydrogenase and related Zn-dependent dehydrogenases. ORF18 might not be directly related to EPS-CG11 biosynthesis since, as a putative ribitol-5-phosphate 2-dehydrogenase, it participates in pentose and glucuronate interconversions (39).
Subsequently, downstream from the eps gene cluster, a hypothetical protein and three putative transposases in opposite directions are present in the pCG1 plasmid. This has been noticed previously in other eps operons, where transposases and insertion elements are also present, indicating that possible horizontal gene transfers and homologous recombination events contribute to the ubiquity and diversity of EPSs among bacteria (22, 40).
Insertional inactivation of the orf2 and the rfbB genes.
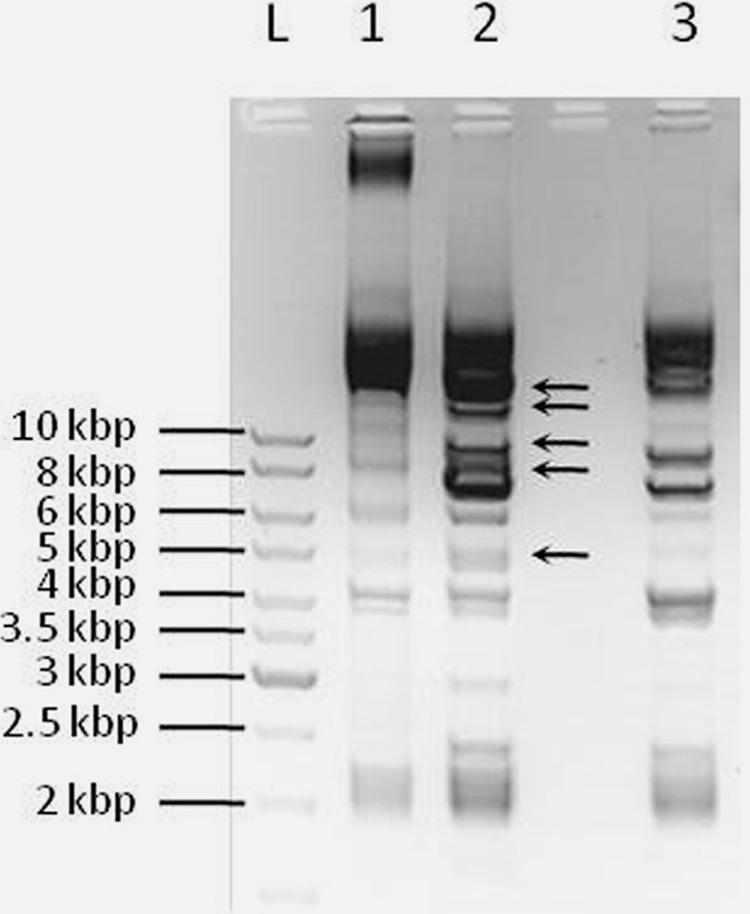
In order to test the functionality of the eps and rfb gene clusters and their role in the production of high-molecular-weight (HMW) polymer, the MUT1 mutant with an inactivated glycosyltransferase (the orf2 gene) and the MUT2 mutant with an inactivated rfbB gene from the rfb mutant gene cluster were constructed as described in Materials and Methods. The insertional inactivation was checked by Southern blotting hybridization (Fig. 4). Both mutants have lost the ropy phenotype (observed visually in comparison to the BGCG11 wild-type strain in Fig. S2 in the supplemental material).
FIG 4.
The electrophoresis of total plasmid DNA of the strain L. paraplantarum BGCG11 (pCG11) and its MUT1 (a) or MUT2 (c) mutant digested with EcoRI restriction enzyme. (b) Southern blot hybridization with MUT1. Lane L, GeneRuler 1-kb DNA ladder; lanes 1, 2, 3, 4, 5, and 7, digested pCG11 (different isolations); lane 6, digested MUT1; lane 8, DNA probe from the orf2 gene. (d) Southern blot hybridization with MUT2. Lane L, GeneRuler 1-kb DNA ladder; lanes 1, 2, 3, 4, 5, and 7, digested pCG11 (different isolations); lane 6, digested MUT2; lane 8, DNA probe from the rfbB gene.
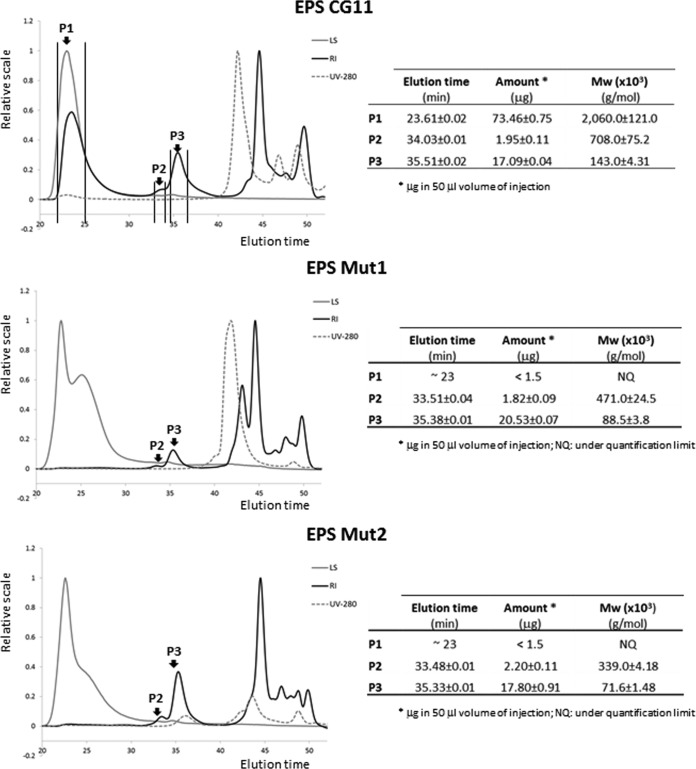
In the case of BGCG11, mutations in the eps mutant gene cluster (MUT1 strain) as well as in the rfb gene cluster (MUT2 strain) lead to the loss of the ropiness in comparison with BGCG11 cultivated in medium with glucose as the sugar source. Similarly, the NB1 strain, derived from BGCG11 without the plasmid pCG1, also lacked the ropy phenotype (3). It has been reported that the ropy phenotype could be related to the size (molar mass) of the polymer (41). In this regard, the EPS fractions purified from the parental and the two mutant strains were characterized by SEC-MALLS (Fig. 5). The results revealed that peak 1, corresponding to EPS-CG11 with the highest molecular mass (∼ 2 × 106 g/mol), was present in the wild-type BGCG11, whereas it was under the quantification level in the mutants MUT1 and MUT2. However, the other two fractions of smaller molar mass (peaks 2 and 3) were produced in similar amounts by the three strains. Thus, it seems that both mutants, MUT1 and MUT2, have lost the capability to synthesize the HMW EPS fraction detected in the wild type and, therefore, the ropy phenotype. In this regard, it was shown that the Δcps1A-I strain, a derivative of L. plantarum WCFS1, produced surface polysaccharides in amounts equal to those of the wild-type strain although the molar mass was reduced and there is a lack of rhamnose (18). The molar mass and the composition of polysaccharides were not affected by cps2A-J, cps3A-J, or cps4A-J mutants, but the genes produced decreased levels of surface polysaccharides. In the quadruple mutant, the amount of surface polysaccharides was strongly reduced (18).
FIG 5.
Analysis of the EPSs from BGCG11, NB1, MUT1, and MUT2 strains by size exclusion chromatography (SEC) coupled to multiangle laser light scattering (MALLS) detection. LS, light scattering detector set at 90°; RI, refraction index; UV-280, PDA detector set at 280 nm; Mw, weighted average molar mass; P, peak.
Although we cannot undoubtedly conclude which genes are responsible for the ropiness, it was shown that the mutant in which the rfb gene cluster is inactivated does not retain the ropy phenotype. The ropiness is lost in both mutants obtained in this work by insertional inactivation (inactivation of the orf2, glycosyltransferase family 8, and the rfbB genes) in which the production of EPS-CG11 high-molecular-weight polymer is disrupted. This ropy phenotype is affected by the presence of sugar in the growth medium; e.g., in medium supplemented with fructose as the only sugar source, there is no visible ropiness of BGCG11 colonies (unpublished data). The influence of using fructose as the sole sugar source on the lack of EPS production of high molecular weight was also noticed in L. delbrueckii subsp. bulgaricus NCFM 2772, but this strain retained the production of EPS of low molecular weight (21). Similarly, L. pentosus LPS26 lost the production of high-molecular-weight EPS A in medium with fructose as the sole sugar source (24). It is possible that if fructose is present in the medium as the sole sugar, there is no available glucose-1-phosphate for the beginning of dTDP-l-rhamnose biosynthesis (42).
Interestingly, none of the sequenced genes was annotated as a transcriptional regulator, indicating that the transcriptional regulatory elements for EPS-CG11 biosynthesis are probably located at the chromosome, and this will be the subject of our further research.
In conclusion, we have described here complete gene information with respect to two gene clusters for high-molecular-weight EPS (EPS-CG11) biosynthesis in the L. paraplantarum BGCG11 strain, which was previously reported to have immunomodulatory effects (3). In the last 10 years EPS operons from different lactobacilli were sequenced, and only a few were partially functionally characterized. Our future work will be focused on the role of BGCG11 and its EPS in the beneficial effects in treatment or prevention of intestinal infections or other diseases where its immunomodulatory effect can be beneficial.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the Ministry of Education, Science and the Technological Development, Republic of Serbia (grant no. 173019). The short stay of Patricia Ruas Madiedo at Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, was covered by the bilateral collaboration project AIB2010SE-00386 between Spain and the Republic of Serbia.
We are grateful to Nathaniel Aaron Sprinkle, the native-English-speaking editor, for the proofreading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03028-14.
REFERENCES
- 1.Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for food fermentation industry. Trends Food Sci Technol 15:67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 2.Ruas-Madiedo P, Abraham A, Mozzi F, de los Reyes-Gavilán CG. 2008. Functionality of exopolysaccharides produced by lactic acid bacteria, p 137–166. Mayo B, López P, Pérez-Martínez G (ed), Molecular aspects of lactic acid bacteria for traditional and new applications. Research Signpost, Kerala, India. [Google Scholar]
- 3.Nikolic M, López P, Strahinic I, Suárez A, Kojic M, Fernández-García M, Topisirovic L, Golic N, Ruas-Madiedo P. 2012. Characterisation of the exopolysaccharide (EPS)-producing Lactobacillus paraplantarum BGCG11 and its non-EPS producing derivative strains as potential probiotics. Int J Food Microbiol 158:155–162. doi: 10.1016/j.ijfoodmicro.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland IW. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9. [DOI] [PubMed] [Google Scholar]
- 5.Kitazawa H, Itoh T, Tomioka Y, Mizugaki M, Yamaguchi T. 1996. Induction of IFN-γ and IL-1α production in macrophages stimulated with phosphopolysaccharide produced by Lactococcus lactis ssp. cremoris. Int J Food Microbiol 31:99–106. [DOI] [PubMed] [Google Scholar]
- 6.Kitazawa H, Harata T, Uemura J, Saito T, Kaneko T, Itoh T. 1998. Phosphate group requirement for mitogenic activation of lymphocytes by an extracellular phosphopolysaccharide from Lactobacillus delbrueckii ssp. bulgaricus. Int J Food Microbiol 40:169–175. doi: 10.1016/S0168-1605(98)00030-0. [DOI] [PubMed] [Google Scholar]
- 7.Ruas-Madiedo P, Gueimonde M, Margolles A, de los Reyes-Gavilán CG, Salminen S. 2006. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J Food Prot 69:2011–2015. [DOI] [PubMed] [Google Scholar]
- 8.Makino S, Ikegami S, Kano H, Sashihara T, Sugano H, Horiuchi H, Saito T, Oda M. 2006. Immunomodulatory effects of polysaccharides produced by Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J Dairy Sci 89:2873–2881. doi: 10.3168/jds.S0022-0302(06)72560-7. [DOI] [PubMed] [Google Scholar]
- 9.Bleau C, Monges A, Rashidan K, Lverdure JP, Lacroix M, van Clasteren MR, Millette M, Savard R, Lamontagne L. 2010. Intermediate chains of exopolysaccharides from Lactobacillus rhamnosus RW-9595M increase IL-10 production by macrophages. J Appl Microbiol 108:666–675. doi: 10.1111/j.1365-2672.2009.04450.x. [DOI] [PubMed] [Google Scholar]
- 10.Maeda H, Zhu X, Suzuki S, Suzuki K, Kitamura S. 2004. Structural characterization and biological activities of an exopolysaccharide kefiran produced by Lactobacillus kefiranofaciens WT-2BT. J Agric Food Chem 52:5533–5538. doi: 10.1021/jf049617g. [DOI] [PubMed] [Google Scholar]
- 11.Boels IC, Ramos A, Kleerebezem M, de Vos WM. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl Environ Microbiol 67:3033–3040. doi: 10.1128/AEM.67.7.3033-3040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoshaug EP, Ahlgren JA, Trempy JE. 2007. Exopolysaccharide expression in Lactococcus lactis subsp. cremoris Ropy352: evidence for novel gene organization. Appl Environ Microbiol 73:897–905. doi: 10.1128/AEM.01945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Vuyst L, Degeest B. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev 23:153–177. doi: 10.1111/j.1574-6976.1999.tb00395.x. [DOI] [PubMed] [Google Scholar]
- 14.Boels IC, van Kranenburg R, Kanning MW, Chong BF, de Vos WM, Kleerebezem M. 2003. Increased exopolysaccharide production in Lactococcus lactis due to increased levels of expression of the NIZO B40 eps gene cluster. Appl Environ Microbiol 69:5029–5031. doi: 10.1128/AEM.69.8.5029-5031.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jolly L, Newell J, Porcelli I, Vincent SJ, Stingele F. 2002. Lactobacillus helveticus glycosyltransferases: from genes to carbohydrate synthesis. Glycobiology 12:319–327. doi: 10.1093/glycob/12.5.319. [DOI] [PubMed] [Google Scholar]
- 16.Lamothe GT, Jolly L, Mollet B, Stingele F. 2002. Genetic and biochemical characterization of exopolysaccharide biosynthesis by Lactobacillus delbrueckii subsp. bulgaricus. Arch Microbiol 178:218–228. doi: 10.1007/s00203-002-0447-x. [DOI] [PubMed] [Google Scholar]
- 17.Péant B, LaPointe G, Gilbert C, Atlan D, Ward P, Roy D. 2005. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 151:1839–1851. doi: 10.1099/mic.0.27852-0. [DOI] [PubMed] [Google Scholar]
- 18.Remus DM, van Kranenburg R, van Swam II, Taverne N, Bongers RS, Wels M, Wells JM, Bron PA, Kleerebezem M. 2012. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb Cell Fact 11:149–158. doi: 10.1186/1475-2859-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojic M, Vujcic M, Banina A, Cocconcelli P, Cerning J, Topisirovic L. 1992. Analysis of exopolysaccharide production by Lactobacillus casei CG11, isolated from cheese. Appl Environ Microbiol 58:4086–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cerning J, Renard CMGC, Thibault JF, Bouillanne C, Landon M, Desmazeaud M, Topisirovic L. 1994. Carbon source requirements for exopolysaccharide production by Lactobacillus casei CG11 and partial structure analysis of the polymer. Appl Environ Microbiol 60:3914–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grobben GJ, van Casteren WHM, Schols HA, Oosterveld A, Sala G, Smith MR, Sikkema J, de Bont JAM. 1997. Analysis of the exopolysaccharides produced by Lactobacillus delbrueckii subsp. bulgaricus NCFB 2772 grown in continuous culture on glucose and fructose. Appl Microbiol Biotechnol 48:516–521. doi: 10.1007/s002530051089. [DOI] [Google Scholar]
- 22.Lebeer S, Verhoeven TL, Francius G, Schoofs G, Lambrichts I, Dufrêne Y, Vanderleyden J, De Keersmaecker SC. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol 75:3554–3563. doi: 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tallon R, Bressollier P, Urdaci MC. 2003. Isolation and characterization of two exopolysaccharides produced by Lactobacillus plantarum EP56. Res Microbiol 154:705–712. doi: 10.1016/j.resmic.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Sánchez JI, Martínez B, Guillén R, Jiménez-Díaz R, Rodríguez A. 2006. Culture conditions determine the balance between two different exopolysaccharides produced by Lactobacillus pentosus LPS26. Appl Environ Microbiol 72:7495–7502. doi: 10.1128/AEM.01078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopwood DA, Bibb JM, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate KM, Smith CP, Ward JM, Schrempf H. 1985. Genetic manipulation of Streptomyces: laboratory manual. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 26.Anderson DG, McKay LL. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol 46:549–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojic M, Fira D, Banina A, Topisirovic L. 1991. Characterization of the cell wall-bound proteinase of Lactobacillus casei HN14. Appl Environ Microbiol 57:1753–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojic M, Jovcic B, Strahinic I, Begovic J, Lozo J, Veljovic K, Topisirovic L. 2011. Cloning and expression of a novel lactococcal aggregation factor from Lactococcus lactis subsp. lactis BGKP1. BMC Microbiol 11:265–274. doi: 10.1186/1471-2180-11-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. 2006. Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178:931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hidalgo-Cantabrana C, Sánchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. 2014. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol 80:9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jolly L, Stingele F. 2001. Molecular organization and functionality of exopolysaccharide gene clusters in lactic acid bacteria. Int Dairy J 11:733–745. doi: 10.1016/S0958-6946(01)00117-0. [DOI] [Google Scholar]
- 34.Kleerebezem M, van Kranenburg R, Tuiner R, Boels IC, Looijesteijn E, Hugenholtz J, de Wos WM. 1999. Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved reological properties? Antonie Van Leeuwenhoek 76:357–365. doi: 10.1023/A:1002084822851. [DOI] [PubMed] [Google Scholar]
- 35.Stingele F, Neeser JR, Mollet B. 1996. Identification and characterization of the eps (exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol 178:1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almiron-Roig E, Mulholland F, Gasson MJ, Griffin AM. 2000. The complete cps gene cluster from Streptococcus thermophilus NCFB 2392 involved in the biosynthesis of a new exopolysaccharide. Microbiology 146:2793–2802. [DOI] [PubMed] [Google Scholar]
- 37.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger B, Pridmore RD, Barretto C, Delmas-Julien F, Schreiber K, Arigoni F, Brus̈sow H. 2007. Similarity and differences in the Lactobacillus acidophilus group identified by polyphasic analysis and comparative genomics. J Bacteriol 189:1311–1321. doi: 10.1128/JB.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glaser L. 1963. Ribitol-5-phosphate dehydrogenase from Lactobacillus plantarum. Biochim Biophys Acta 67:525–530. doi: 10.1016/0926-6569(63)90274-8. [DOI] [PubMed] [Google Scholar]
- 40.Mavroidi A, Godoy D, Aanensen DM, Robinson DA, Hollingshead SK, Spratt BG. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J Bacteriol 186:8181–8192. doi: 10.1128/JB.186.24.8181-8192.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruas-Madiedo P, de los Reyes-Gavilán CG. 2005. Methods for the screening, isolation, and characterization of exopolysaccharides produced by lactic acid bacteria. J Dairy Sci 88:843–856. doi: 10.3168/jds.S0022-0302(05)72750-8. [DOI] [PubMed] [Google Scholar]
- 42.Boels IC. 2002. Metabolic engineering of exopolysaccharide production in Lactococcus lactis. Ph.D. thesis Wageningen University, Wageningen, The Netherlands. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.