Abstract
Clock circadian regulator (CLOCK)/brain and muscle arnt-like protein-1 (BMAL1) complex governs the regulation of circadian rhythm through triggering periodic alterations of gene expression. However, the underlying mechanism of circadian clock disruption in hepatocellular carcinoma (HCC) remains unclear. Here, we report that a long noncoding RNA (lncRNA), highly upregulated in liver cancer (HULC), contributes to the perturbations in circadian rhythm of hepatoma cells. Our observations showed that HULC was able to heighten the expression levels of CLOCK and its downstream circadian oscillators, such as period circadian clock 1 and cryptochrome circadian clock 1, in hepatoma cells. Strikingly, HULC altered the expression pattern and prolonged the periodic expression of CLOCK in hepatoma cells. Mechanistically, the complementary base pairing between HULC and the 5' untranslated region of CLOCK mRNA underlay the HULC-modulated expression of CLOCK, and the mutants in the complementary region failed to achieve the event. Moreover, immunohistochemistry staining and quantitative real-time polymerase chain reaction validated that the levels of CLOCK were elevated in HCC tissues, and the expression levels of HULC were positively associated with those of CLOCK in clinical HCC samples. In functional experiments, our data exhibited that CLOCK was implicated in the HULC-accelerated proliferation of hepatoma cells in vitro and in vivo. Taken together, our data show that an lncRNA, HULC, is responsible for the perturbations in circadian rhythm through upregulating circadian oscillator CLOCK in hepatoma cells, resulting in the promotion of hepatocarcinogenesis. Thus, our finding provides new insights into the mechanism by which lncRNA accelerates hepatocarcinogenesis through disturbing circadian rhythm of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the major histologic subtype of primary liver cancer, accounting for 70% to 85% of cases in most countries, and a leading cause of cancer deaths every year worldwide [1], [2]. It has been reported that the chronic exposure to toxic chemicals, chronic infections with hepatitis B virus (HBV) or hepatitis C virus, as well as hepatosteatosis are the major risk factors for HCC [3], [4]. A recent study describes that disturbance of circadian gene expression is exhibited in HCC [5], indicating that the development of HCC impacts the orchestrated circadian rhythm of liver cells. However, the major cause and the underlying mechanism of circadian rhythm disorders of HCC remain a mystery. In response to the daily 24-hour light–dark cycle, living organisms have developed an evolutionarily conserved program that coordinates body systems and synchronizes the internal milieu with the environment [6], [7]. At the cellular level, these rhythms are controlled by transcriptional feedback loops that produce fluctuations in gene expression, a process associated with circadian changes in chromatin architecture, messenger RNA (mRNA) processing, protein activity, and protein turnover [8]. In mammals, the transcription factors clock circadian regulator (CLOCK) and brain and muscle arnt-like protein-1 (BMAL1) drive the central oscillator within the hypothalamus and even in peripheral tissue [9]. At the core, CLOCK and BMAL1, the basic helix–loop–helix (bHLH)-PAS domain transcription factors, form a heterodimer to drive rhythmic expression of genes harboring E-box elements in their promoter [10], [11]. Notably, as a peripheral tissue, liver itself is a biological clock capable of actually generating diurnal rhythms [12]. Approximately, 10% of genes are rhythmic in the liver, driven by the circadian clock. More recent RNA-sequencing data have shown that, of these rhythmic genes, only about a fifth is driven by de novo transcription, indicating that posttranscriptional regulation plays crucial roles in this event [13]. Heretofore, the role of circadian oscillators, such as CLOCK, in hepatocarcinogenesis has not been well documented.
The non–protein-coding portion of the mammalian genome is transcribed into a vast array of RNA species [14]. Among them, long noncoding RNAs (lncRNAs)—defined as transcribed RNA molecules larger than 200 nt in length—are poorly conserved expression by diverse mechanisms [15], [16], [17]. Many of the identified lncRNAs show spatial- and temporal-specific patterns of expression, indicating that lncRNA expression is strongly regulated. Alterations in the primary structure, secondary structure, and expression levels of lncRNAs as well as their cognate RNA-binding proteins underlie diseases ranging from neurodegeneration to cancer [18], [19], suggesting that lncRNAs emerge as vital modulators in physiologic and pathologic states. Regulation of gene expression by lncRNAs can be mediated at transcriptional step and posttranscriptional step [20]. Nevertheless, whether lncRNAs participate in the modification of circadian rhythm of hepatoma cells remains largely unknown.
In the current article, we are interested in the role of lncRNAs in circadian rhythm disorders of HCC. We show that HULC, an lncRNA overexpressed in HCC, enhances the hepatocarcinogenesis through disturbing the circadian rhythm of hepatoma cells. HULC alters the expression pattern and prolongs the periodic expression of circadian oscillator CLOCK in hepatoma cells. Our finding provides new insights into the mechanism by which lncRNA accelerates hepatocarcinogenesis through perturbing circadian rhythm of HCC.
Materials and Methods
Patient Samples
Twenty HCC tissues and their corresponding nearby nontumorous liver tissues and 10 HCC tissues used in this study were obtained from Tianjin First Center Hospital (Tianjin, China) after surgical resection. Written consents approving the use of their tissues for research purposes were obtained from patients. The study protocol was approved by the Institute Research Ethics Committee at the Nankai University.
Cell Lines and Cell Culture
The human immortalized liver L-O2 cell line and L-O2-X cell line were cultured in PRMI-1640 medium (Gibco, CA). The human hepatoma cell lines, HepG2 and HepG2.2.15 (a hepatoma HepG2 cell line integrated full-length HBV DNA), were maintained in Dulbecco’s modified Eagle’s medium (Gibco). All of these cell lines were supplemented with heat-inactivated 10% fetal bovine serum (Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin in 5% CO2 at 37°C.
Plasmid Construction
The full-length HULC was amplified from cDNA of HepG2 cells and then cloned into pcDNA3.1 vector. The resulting vector was sequenced and named pcDNA3.1-HULC. The HULC containing the mutant in (or out of) complementary region between HULC and CLOCK 5′ untranslated region (UTR) was constructed (termed as HULC-mut-in or HULC-mut-out). All primers are listed in Supplementary Table S1.
One ~ 150–base pair fragment of CLOCK 5′UTR was subcloned into pGL3-control vector (Promega, Madison, WI) immediately upstream of the start codon of the luciferase gene to generate pGL3-CL-5′UTR. Mutant construct of complementary region of CLOCK 5′UTR (named pGL3-CL-5′UTR-mut), carrying a substitution of 16 nucleotides within the complementary region between HULC and CLOCK 5′UTR, was conducted by using overlapping extension polymerase chain reaction (PCR). All primers are listed in Supplementary Table S1.
Cell Transfection
The cells were cultured in a 6-well or 24-well plate for 24 hours and then were transfected with plasmids or siRNAs. All transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. siRNA oligonucleotides, including targeting HULC (or CLOCK) and a nonspecific scrambled control (si-Ctrl), were synthesized by RiboBio (Guangzhou, China). The siRNA duplexes sequences are all listed in Supplementary Table S1.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from the cells (or tissues) using Trizol (Invitrogen) according to the manufacturer’s protocol. Reverse transcription was performed using poly(A)-tailed total RNA and reverse transcription primer with ImPro-IIReverse Transcriptase (Promega) according to the manufacturer’s instructions. The qRT-PCR was performed as described in the method of Fast Start Universal SYBR Green Master (Rox) (Roche Diagnostics GmbH, Mannheim, Germany). GAPDH was used as the control for qRT-PCR. All primers are listed in Supplementary Table S1.
Luciferase Reporter Gene Assays
Luciferase reporter gene assays were performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Briefly, cells were cultured in 24-well plates at 3 × 104 cells per well. After 24 hours, the cells were transiently co-transfected with the pRL-TK plasmid (Promega) containing the Renilla luciferase gene, which is used for internal normalization, and with various constructs containing different sequence of CLOCK 5′UTR (or pGL3-control) or various constructs of wild and mutant HULC. All experiments were performed at least three times.
Western Blot Analysis
Western blot analysis protocol was described previously [21]. The primary antibodies used were mouse anti–β-actin (Sigma, St Louis, MO) and rabbit anti-CLOCK (Proteintech Group, Chicago, IL). All experiments were repeated at least three times.
Immunohistochemistry Staining (IHC)
The HCC tissue and peritumor liver tissue microarrays were obtained from the Xi'an Aomei Biotechnology Co, Ltd (Xi'an, China). These microarrays were composed of 33 HCC tissues and 4 peritumor liver tissues, which included duplicate core biopsies (1 mm in diameter) from fixed, paraffin-embedded tumors. Immunohistochemical staining of samples was performed as previously reported [22], and the primary antibody of rabbit anti-CLOCK (Proteintech Group) or primary antibody of rabbit anti-Ki-67 (Cell Signaling Technology, Beverly, MA) was used. The extents of cytosolic staining were considered in the scoring. The percentage of immunoreactivity in tumor cells or hepatocytes was graded as: 0 (< 10%); 1, low (10% to 30%); 2, intermediate (30% to 50%); 3, high (> 50%). Categorization of immunostaining intensity was performed by three independent observers. The medical records of the patients were listed in Supplementary Table S2.
3-(4,5-Dimethylthiazol-2-yl)-2,5 Diphenyltetrazolium Bromide (MTT) Assays
HepG2 cells were seeded onto 96-well plates (1000 cells/well) for 24 hours before transfection, and MTT assays were used to assess cell proliferation every day from the first day until the third day after transfection. The protocol was described previously [23].
Flow Cytometry Analysis
Forty-eight hours after transfection, the cells (1 × 106) were harvested and washed with cold PBS twice. Washed cells were fixed in 75% ethanol at 4°C overnight. The fixed cells were rinsed twice with PBS and treated with propidium iodine solution including 50 μg/ml propidium iodine (Sigma) and 50 μg/ml RNaseA (Sigma) at 37°C for 60 minutes. Stained cells were analyzed by a FACScan flow cytometer (Becton Dickinson, Bedford, MA).
Cloning Formation Assays
For clonogenicity analysis, 48 hours after transfection, 500 viable transfected cells were placed in six-well plates and maintained in complete medium for 2 weeks. Colonies were fixed with methanol and stained with methylene blue.
Soft Agar Assays
Anchorage-independent cell growth was assessed by the soft agar assays. Briefly, 2 ml of 0.5% agar was added to each well of a 6-well plate. Detached LO-2 (LO-2-HULC) cells were mixed with an agarose suspension (0.3% final concentration) and then layered onto the 0.5% agarose underlay. Culture medium was changed every 3 days, and the number of cell foci ≥ 100 mm in diameter was counted after 21 days.
In Vivo Tumorigenicity Assays
Nude mice were housed and treated according to guidelines established by the National Institutes of Health Guide for the Care and Use of Laboratory Animals. We conducted the animal transplantation according to the Declaration of Helsinki. Briefly, pretreated HepG2 cells (treated with pcDNA3.1, pcDNA3.1-HULC, or pcDNA3.1-HULC and CLOCK siRNA) were harvested and resuspended at 2 × 107 per ml with sterile phosphate-buffered saline. Three groups of 4-week-old male BALB/c athymic nude mice (Experiment Animal Center of Peking, China) (each group, n= 6) were subcutaneously injected at the shoulder with 0.2 ml of the cell suspensions. Tumor growth was measured after 5 days from injection and then every 5 days. Tumor volume (V) was monitored by measuring the length (L) and width (W) with calipers and calculated with the formula (L × W2) × 0.5. After 25 days, tumor-bearing mice and controls were sacrificed, and the tumors were excised and measured.
Statistical Analysis
Each experiment was repeated at least three times. Statistical significance was assessed by comparing mean values (6 SD) using a Student’s t test for independent groups and was assumed for *P < .05, **P < .01, and ***P < .001, no significant (NS). Pearson correlation coefficient was used to determine the correlation between the expressions of HULC and CLOCK in tumorous tissues. CLOCK expression in tumor tissues and matched adjacent nontumor tissues was compared using Wilcoxon signed-rank test.
Results
HULC Upregulates CLOCK and Perturbs Its Rhythmical Expression in Hepatoma Cells
Given that the heterodimer of CLOCK/BMAL1 is essential for the modulation of circadian rhythm [9], we evaluated the endogenous expression levels of CLOCK and BMAL1 in L-O2 and HepG2 cells. qRT-PCR assays manifested that the endogenous levels of BMAL1 were much higher than those of CLOCK in the cells (Supplementary Figure S1A), suggesting that CLOCK might be the limitative component in the formation of the heterodimer. Moreover, the transient transfection of HULC resulted in the upregulation of CLOCK but failed to affect the expression of BMAL1 in hepatoma cells (Supplementary Figure S1B). Therefore, we focused on the investigation of CLOCK in the cells. qRT-PCR and Western blotting showed that HULC was able to heighten the expression levels of CLOCK after 32 hours of transfection in L-O2 and HepG2 cells in a dose-dependent manner (Supplementary Figure S1C; Figure 1, A and B). Because HepG2.2.15 cells contained high levels of endogenous HULC [24], we observed that HULC siRNA dose-dependently lessened the mRNA and protein levels of CLOCK in the cells (Supplementary Figure S1B, Figure 1C). Meanwhile, the transfection (or interference) efficiency of HULC (or HULC siRNA) was verified by qRT-PCR in the cells (Figure 1, A–C). To further validate above outcomes, we examined the effect of HULC on the downstream factors of CLOCK in hepatoma cells. It has been reported that period circadian clock 1 (PER1) and cryptochrome circadian clock 1 (CRY1) are clock genes regulated by CLOCK/BMAL1 [25], [26], [27]. As expected, qRT-PCR assays exhibited that HULC upregulated the expression of PER1 and CRY1 in L-O2 and HepG2 cells (Figure 1D), indicating that HULC is able to upregulate CLOCK in hepatoma cells.
Figure 1.
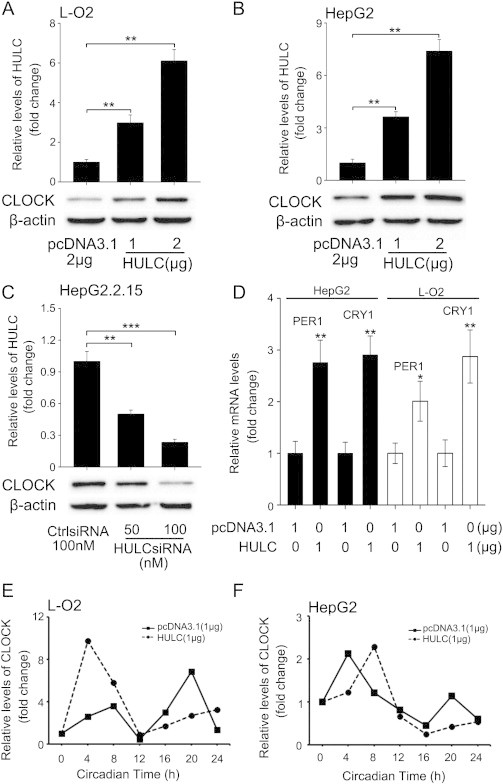
HULC upregulates CLOCK and perturbs its rhythmical expression in hepatoma cells. (A and B) The protein levels of CLOCK were examined by Western blotting in L-O2 and HepG2 cells transfected with pcDNA3.1-HULC (termed as HULC), respectively. Meanwhile, the transfection efficiency of HULC was detected by qRT-PCR. (C) The protein levels of CLOCK were examined by Western blotting in HepG2.2.15 cells transfected with CLOCK siRNA. The interference efficiency of CLOCK siRNA was detected by qRT-PCR in the cells. (D) The effect of HULC on the expression of downstream target genes of CLOCK, PER1 and CRY1, was measured by qRT-PCR in HepG2 and L-O2 cells. (E and F) The effect of HULC on rhythmical expression of CLOCK was assessed by qRT-PCR in L-O2 and HepG2 cells during 24 hours, respectively. Statistically significant differences are indicated: **P < .01, ***P < .001; Student’s t test.
To dissect the influence of HULC in circadian rhythm of hepatoma cells, we observed the effect of HULC on CLOCK during the period of 24 hours in L-O2 and HepG2 cells according to the previous report of measuring time [9]. We transfected HULC into L-O2 and HepG2 cells and extracted the mRNA at 4-hour intervals. Intriguingly, the expression of CLOCK displayed two peaks during 24 hours in L-O2 and HepG2 cells. The expression peaks of CLOCK were present at 8 hours (peak I) and 20 hours (peak II) in L-O2 cells, whereas HepG2 cells showed peak I at 4 hours and peak II at 20 hours. Importantly, we observed that the overexpression of HULC advanced the expression peak I of CLOCK from 8 to 4 hours in L-O2 cells and delayed the peak II over 4 hours in the cells; however, in HepG2 cells, the overexpression of HULC delayed the expression of peaks I and II of CLOCK over 4 hours (Figure 1, E and F), suggesting that the HULC-disturbed rhythm is different in hepatoma cells and normal liver cells. In addition, HULC prolonged the expression period of CLOCK in L-O2 and HepG2 cells (Figure 1, E and F). Thus, we conclude that HULC perturbs the rhythmical expression of CLOCK in hepatoma cells and modifies the CLOCK expression pattern of hepatic cells.
HULC Targets the 5'UTR of CLOCK mRNA via Complementary Base Pairing in Hepatoma Cells
Next, we undertook to nail the underlying mechanism of HULC eliciting CLOCK. A previous study described that lncRNA interacted with and stabilized mRNA [28], and RNA structure in the UTRs of mRNAs influences posttranscriptional regulation of gene expression [29]. Therefore, we are interested in whether HULC governs the expression of CLOCK at posttranscriptional step through interacting with the mRNA of CLOCK. Thus, we screened the sequence of HULC and CLOCK using bioinformatics (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Notably, we obtained a complementary area about 16 nt between HULC and the 5'UTR of CLOCK mRNA. Accordingly, we conjectured that the complementary region might underlie the modulation of CLOCK by HULC.
To verify the assumption, we constructed two mutants of HULC into pcDNA3.1. One mutant was changed in the sequence of complementary region (termed HULC-mut-in); the other was altered in the sequence nearby the complementary region (termed HULC-mut-out) (Figure 2A). Interestingly, our data revealed that HULC-mut-in lost the function in L-O2 and HepG2 cells relative to wild-type HULC after 32-hour transfection (Figure 2, B and D). However, the HULC-mut-out still heightened the expression of CLOCK in the cells in a dose-dependent manner (Figure 2, C and E). Moreover, we cloned the 5'UTR of CLOCK mRNA containing the complementary region and the relative mutant into pGL3-control (termed pGL3-CL-5'UTR and pGL3-CL-5'UTR-mut) individually (Figure 3A). Luciferase reporter gene assays indicated that HULC was able to increase the activities of pGL3-CL-5'UTR in a dose-dependent manner but unaltered those of pGL3-CL-5'UTR-mut in L-O2 and HepG2 cells (Figure 3, B and D). Inversely, the activities of pGL3-CL-5'UTR were petered out by treatment with HULC siRNA in HBV X gene stably transfected L-O2-X (expressing a high level of endogenous HULC) [24] and HepG2.2.15 cells, whereas HULC siRNA failed to influence the activity of pGL3-CL-5'UTR-mut in the system (Figure 3, C and E). In addition, the downstream target genes of CLOCK, such as PER1 and CRY1 [30], were upregulated by HULC (or HULC-mut-out) but not by HULC-mut-in in HepG2 cells after 32-hour transfection (Supplementary Figure S2, A and B). Together, we conclude that the complementary region between HULC and the 5'UTR of CLOCK mRNA is required for the modulation of CLOCK mediated by HULC.
Figure 2.
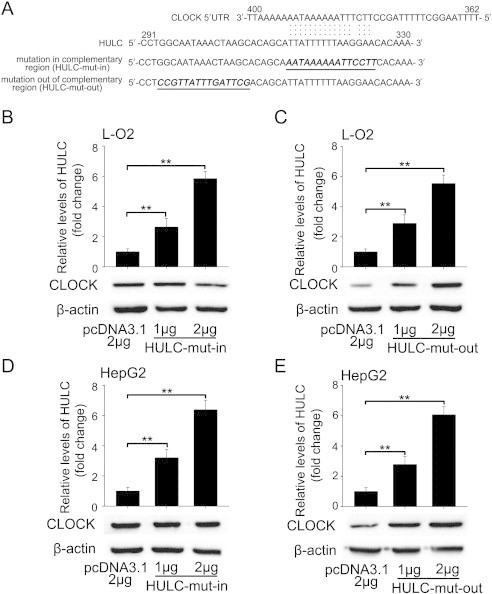
HULC interacts with the 5'UTR of CLOCK mRNA via complementary base pairing in hepatoma cells. (A) A model shows the complementary region between HULC and CLOCK 5'UTR by bioinformatics prediction, in which the generated mutant sites in (or out of) the complementary region between HULC and CLOCK 5'UTR were marked. (B and C) The protein levels of CLOCK were measured by Western blotting in L-O2 cells transfected with HULC-mut-in (or HULC-mut-out), respectively. The transfection efficiency of HULC-mut-in (or HULC-mut-out) was detected by qRT-PCR in the cells. (D and E) The experiments above were repeated in HepG2 cells. Statistically significant differences are indicated: **P < .01; Student’s t test.
Figure 3.
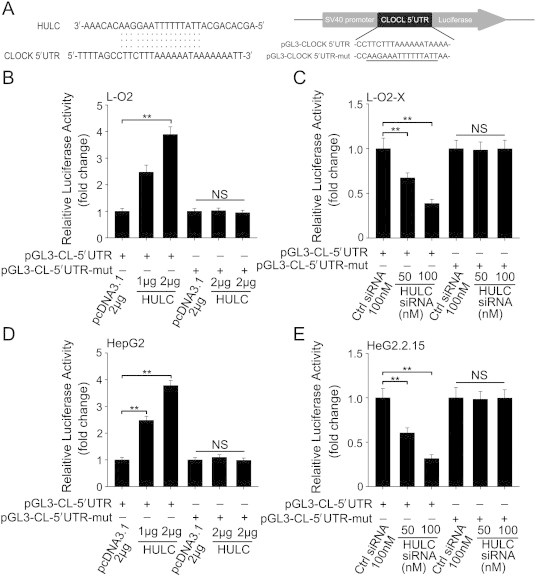
The 5'UTR of CLOCK mRNA interacts with HULC via complementary base pairing in hepatoma cells. (A) A model shows the complementary region between HULC and CLOCK 5'UTR. Schematic diagram exhibits the generated mutant site at the CLOCK 5'UTR in complementary region binding to HULC, which is inserted in pGL3-control vector. (B) The effect of HULC on reporters of pGL3-CL-5'UTR (or pGL3-CL-5'UTR-mut) was examined by luciferase reporter gene assays in L-O2 cells. (C) The effect of HULC siRNA on reporters of pGL3-CL-5'UTR (or pGL3-CL-5'UTR-mut) was measured by luciferase reporter gene assays in HBx-elevated HULC-highly-expressing L-O2-X cells. (D and E) The experiments above were repeated in HepG2 and HepG2.2.15 cells. Statistically significant differences are indicated: **P < .01, no significance (NS); Student’s t test.
The Expression Levels of HULC Are Positively Correlated with Those of CLOCK in Clinical HCC Tissues
Next, we assessed the expression relationships between HULC and CLOCK in clinical HCC tissues. Noticeably, immunohistochemical staining manifested that the positive rate of CLOCK was 75.76% (25/33) in HCC tissues using tissue microarrays. One out of four was positive in the peritumor liver tissues (Figure 4A). Meanwhile, qRT-PCR showed that the mRNA levels of CLOCK were higher in HCC tissues compared with those in their adjacent nontumorous liver tissues in 20 paired clinical samples (Figure 4B). The levels of HULC were positively related to those of CLOCK in 30 cases of clinical HCC tissues (Figure 4C). Thus, we conclude that the expression of HULC is positively related to that of CLOCK in clinical liver cancer tissues.
Figure 4.
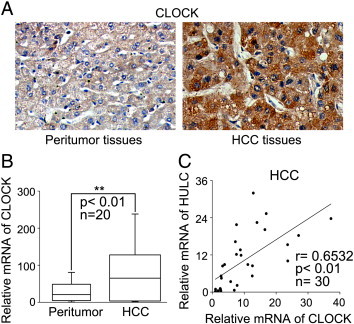
The expression levels of HULC are positively correlated with those of CLOCK in clinical HCC tissues. (A) The expression of CLOCK was examined by IHC staining in clinical HCC tissues and peritumor tissues using tissue microarray. Representative example of IHC observed in peritumor tissues and HCC tissues using rabbit anti-CLOCK Ab. (B) The relative mRNA levels of CLOCK were assessed by qRT-PCR in 20 pairs of clinical HCC tissues and corresponding nontumorous tissues (**P < .01; Wilcoxon signed-rank test). (C) The correlation of HULC mRNA levels with CLOCK mRNA levels was examined by qRT-PCR in 30 cases of clinical HCC tissues (**P < .01; Pearson correlation coefficient, r= 0.6532).
CLOCK Is Required for the Proliferation of Hepatoma Cells Mediated by HULC In Vitro and In Vivo
Given that dysregulation of the circadian rhythm is implicated in many types of cancers [31], we are interested in whether CLOCK is involved in the proliferation of hepatoma cells mediated by HULC. Luciferase reporter gene assays illustrated that HULC-mut-in was unable to affect the activity of pGL3-CL-5'UTR, whereas HULC-mut-out worked well in HepG2 cells in a dose-dependent manner (Figure 5A). Moreover, MTT assays corroborated that CLOCK siRNA was capable of blocking the HULC-enhanced proliferation of hepatoma cells (Figure 5B). In addition, cell cycle analysis by flow cytometry showed that HULC resulted in the increase of S-phase HepG2 cells from 15.75% to 35.94%. Notably, CLOCK siRNA inhibited the effect of HULC on the S-phase HepG2 cells, decreasing from 35.94% to 22.15% (Figure 5C). Clone formation assay revealed that HULC siRNA was able to block the proliferation of HepG2 cells. Moreover, CLOCK siRNA potently abolished HULC-strengthened proliferation of HepG2 cells (Figure 5D). Meanwhile, the interference efficiency of CLOCK siRNA (or CLOCK siRNA*) was validated by Western blotting in the cells (Supplementary Figure S3A). Moreover, soft agar assays exhibited that the efficiency of LO-2 cells to generate colonies by anchorage-independent growth in a semisolid medium was remarkably heightened by overexpressing HULC (Figure 5E). Together, our observations suggest that CLOCK is implicated in the proliferation of hepatoma cells in vitro.
Figure 5.
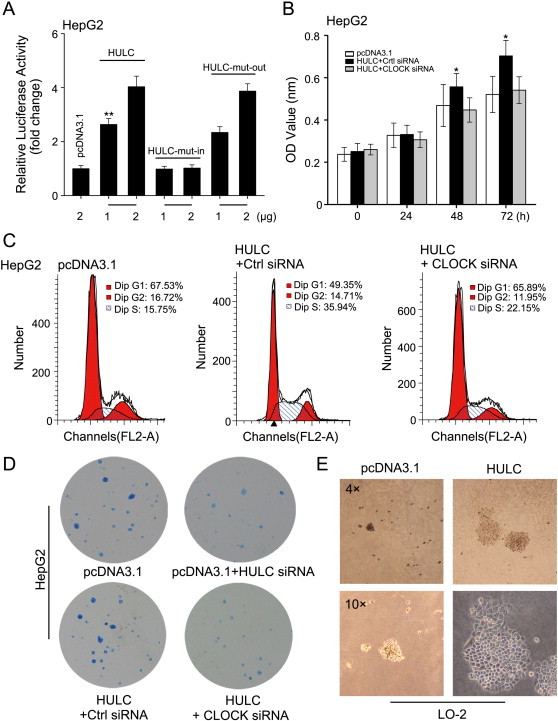
CLOCK is required for HULC-accelerated proliferation of hepatoma cells in vitro. (A) The effect of HULC-mut-in (or HULC-mut-out) on reporters of pGL3-CL-5'UTR was assessed by luciferase reporter gene assays in HepG2 cells. (B–D) The cell proliferation was measured by MTT assays, flow cytometry, and cloning formation in HepG2 cells transfected with HULC (or HULC/CLOCK siRNA) individually. (E) The effect of HULC on the neoplastic transformation in LO-2 cells was detected by soft agar assays. Statistically significant differences are indicated: *P < .05, **P < .01, NS; Student’s t test.
Next, we evaluated the effect of CLOCK on the tumor growth mediated by HULC in mouse models. Notably, we observed that the volume and weight of tumors were remarkably elevated in the group of HULC pretreated HepG2 cells relative to control group. Strikingly, our data exhibited that CLOCK siRNA markedly eliminated the tumor growth enhanced by HULC in nude mice (Figure 6, A–C). Moreover, IHC further confirmed that the expression of Ki-67, a marker of proliferation, in the tumor tissues was reconciled with the tumor growth (Supplementary Figure S3B). Meanwhile, our data validated that the levels of HULC and CLOCK were corresponding to the treatment in the tumor tissues from mice (Figure 6, D and E). Thereby, we conclude that CLOCK is required for the proliferation of hepatoma cells mediated by HULC in vitro and in vivo.
Figure 6.
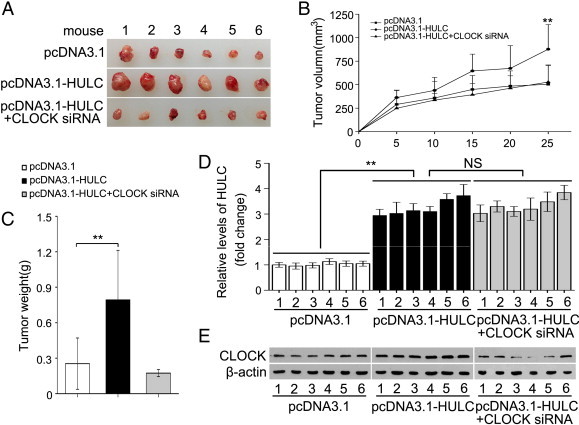
CLOCK is required for HULC-accelerated proliferation of hepatoma cells in vivo. (A and B) Photographs of dissected tumors from nude mice and their growth curve, respectively. (C) The average weight of tumors from mice. (D and E) Expression levels of HULC and CLOCK were detected by qRT-PCR and Western blotting in the tumor tissues from mice, respectively. Statistically significant differences are indicated: **P < .01, NS; Student’s t test.
Discussion
The altered circadian rhythm accompanies stages of cancer progression. However, the role of a circadian oscillator, CLOCK, in liver cancer remains poorly understood. A spot of characterized human lncRNAs, which emerged as regulatory RNAs, has been associated with a spectrum of biological functions, and the disruption of these functions has critical roles in the development of cancer [32]. In this study, we are interested in whether lncRNA perturbs the circadian rhythm of HCC.
Previous studies pointed out that HULC, as an lncRNA, was overexpressed in HCC tissues and was governed by an RNA-binding protein IGF2BPs [33], [34]. Our group has reported that HULC is able to accelerate the growth of HCC through downregulating its neighbor gene p18 [24]. Here, we were curious whether HULC participates in the circadian rhythm disturbance in liver cancer. Strikingly, we found that HULC was able to heighten the expression levels of CLOCK and prolong the periodic expression of CLOCK in hepatoma cells. Importantly, HULC altered the expression pattern and prolonged the periodic expression of CLOCK in the cells. The expression patterns of CLOCK were different in LO-2 and HepG2 overexpressing HULC, however, suggesting that the cell context might influence the cell behavior to the stimulation. Previous studies described that lncRNAs were implicated in the development and differentiation of human cells [35]; their inaccurate expression could lead to disease [36], [37]. In the present article, we observed that the disturbance of circadian rhythm in LO-2 cells mediated by HULC led to the promotion of malignance abilities of liver cells in vitro. Collectively, we first provide evidence that an lncRNA HULC disturbs the circadian rhythm of hepatocellular carcinima through upregulating oscillator CLOCK.
LncRNAs serve as modulators to control the physiologic and pathologic progresses. Given that lncRNAs are poorly conserved and control the expression of genes in vatious manners, such as posttranscriptional gene regulation through controlling processes like protein synthesis, RNA maturation and transport, and, very recently, in transcriptional gene silencing through regulating the chromatin structure [16]. In addition, lncRNAs act as cis- or trans-regulators, scaffolds, precursors for small RNAs, and structural RNAs to govern the gene expression [38], [39]. LncRNA interacts with mRNA to increase its stability as well [28]. As a regulatory RNA, HULC is able to sponge miR-372 [40]. Accordingly, we supposed that HULC might upregulate CLOCK in a manner of direct interacting with CLOCK mRNA. To test out hypothesis, we screened the sequences of HULC and CLOCK. Intriguingly, we obtained a complementary region between HULC and the 5'UTR of CLOCK mRNA, hinting that HULC has potential to target CLOCK mRNA. Not surprisingly, our experiments validated that the complementary base pairing between HULC and 5'UTR of CLOCK mRNA was responsible for the modulation of CLOCK mediated by HULC. Moreover, we revealed that the levels of HULC were positively correlated with those of CLOCK in clinical HCC tissues.
Hypermethylation in the CLOCK promoter reduced the risk of breast cancer, and lower levels of CLOCK expression were documented in healthy controls relative to normal or tumor tissue from patients with breast cancer [41]. However, the role of CLOCK in hepatocarcinogenesis remains mysterious. On the basis of our observations, we are inquisitive whether CLOCK is involved in the HULC-enhanced development of HCC. Our results exhibited that CLOCK siRNA effectively blocked the proliferation of hepatoma cells mediated by HULC in vitro. Strikingly, CLOCK siRNA was able to remarkably abrogate the HULC-facilitated tumor growth in nude mice as well, indicating that CLOCK might be implicated in the HULC-promoted tumor growth. A wave of studies reports that CLOCK participates in the glucose and lipid homeostasis [42], [43] and governs circadian autophagy rhythm to impair the energy metabolism [44]. In addition, CLOCK is a positive modulator of nuclear factor-κB–mediated transcription to elicit the inflammatory process and regulates the epigenome [11], [45]. It is well known that the above phenomena are pervasive in the development of tumorigenesis. Thereby, it suggests that HULC might be involved in above functions in the development of HCC through eliciting the expression of CLOCK. Potentially, HULC may serve as a novel therapeutic target for HCC.
In aggregate, here we report that the HULC-modulated disturbance of the circadian rhythm contributes to the hepatocarcinogenesis. Briefly, HULC heightens the expression of circadian oscillator CLOCK, in which the complementary base pairing between HULC and the 5'UTR of CLOCK mRNA underlies the event. Notably, we observe that the overexpression of HULC results in disordering the expression pattern and prolonging the periodic expression of CLOCK in hepatoma cells. Moreover, we first provide evidence that CLOCK is involved in the HULC-enhanced proliferation of hepatoma cell in vitro and in vivo. Thus, our finding provides new insights into the mechanism by which lncRNAs modulate hepatocarcinogenesis through perturbing circadian rhythm.
The following are the Supplementary data related to this article.
List of Primers and siRNA Used in This Paper
Clinical Characteristics of Liver Cancer and Normal Tissue Microarray Samples
HULC upregulates CLOCK and perturbs its rhythmical expression in hepatoma cells, related to Figure 1. (A) The endogenous expression levels of CLOCK and BMAL1 were examined by qRT-PCR in L-O2 and HepG2 cell, respectively. (B) The effect of HULC on the expression of circadian oscillators CLOCK and BMAL1 was assessed by qRT-PCR in HepG2 cells. (C) The mRNA levels of CLOCK were examined by qRT-PCR in L-O2 (HepG2) or HepG2.2.15 cells transfected with HULC or HULC siRNA, respectively. Statistically significant differences are indicated: **P < .01, ***P < .001, no significance (NS); Student’s t test.
HULC targets the 5'UTR of CLOCK mRNA via complementary base pairing in hepatoma cells, related to Figure 2, Figure 3. (A and B) The effect of HULC (or HULC-mut-in, HULC-mut-out) on the expression levels of PER1 and CRY1, downstream factors of CLOCK, was measured by qRT-PCT in HepG2 cells. Statistically significant differences are indicated: *P < .05, **P < .01; Student’s t test.
CLOCK is required for the proliferation of hepatoma cells mediated by HULC in vitro and in vivo, related to Figure 5, Figure 6. (A) Interference efficiency of CLOCK siRNA* and CLOCK siRNA was examined by Western blotting in HepG2 cells. (B) The levels of Ki-67 were assessed by IHC in the tumor tissues from mice.
Footnotes
Financial support: This work was supported by grants from the National Basic Research Program of China (973 Program, no. 2015CB553703), National Natural Science Foundation of China (no. 31470756, no. 81272218), and Project of Prevention and Treatment of Key Infectious Diseases (no. 2014ZX0002002-005).
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Schmieder R, Puehler F, Neuhaus R, Kissel M, Adjei AA, Miner JN, Mumberg D, Ziegelbauer K, Scholz A. Allosteric MEK1/2 inhibitor refametinib (BAY 86-9766) in combination with sorafenib exhibits antitumor activity in preclinical murine and rat models of hepatocellular carcinoma. Neoplasia. 2013;15:1161–1171. doi: 10.1593/neo.13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su JC, Tseng PH, Wu SH, Hsu CY, Tai WT, Li YS, Chen IT, Liu CY, Chen KF, Shiau CW. SC-2001 overcomes STAT3-mediated sorafenib resistance through RFX-1/SHP-1 activation in hepatocellular carcinoma. Neoplasia. 2014 doi: 10.1016/j.neo.2014.06.005. [pii: S1476-5586(14)00079-7, Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakurai T, Kudo M, Umemura A, He G, Elsharkawy AM, Seki E, Karin M. p38alpha inhibits liver fibrogenesis and consequent hepatocarcinogenesis by curtailing accumulation of reactive oxygen species. Cancer Res. 2013;73:215–224. doi: 10.1158/0008-5472.CAN-12-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, Gao Y, Zhang S, Lu Z, Ye L. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15:1282–1291. doi: 10.1593/neo.131362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin YM, Chang JH, Yeh KT, Yang MY, Liu TC, Lin SF, Su WW, Chang JG. Disturbance of circadian gene expression in hepatocellular carcinoma. Mol Carcinog. 2008;47:925–933. doi: 10.1002/mc.20446. [DOI] [PubMed] [Google Scholar]
- 6.Chang HC, Guarente L. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell. 2013;153:1448–1460. doi: 10.1016/j.cell.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337:839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, Gitlin II, Artemicheva NM, Deluca KA, Gudkov AV. Core circadian protein CLOCK is a positive regulator of NF-kappaB–mediated transcription. Proc Natl Acad Sci U S A. 2012;109:E2457–E2465. doi: 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turek FW, Allada R. Liver has rhythm. Hepatology. 2002;35:743–745. doi: 10.1053/jhep.2002.32873. [DOI] [PubMed] [Google Scholar]
- 13.Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I. RNA-methylation–dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241. doi: 10.1002/hep.25895. [DOI] [PubMed] [Google Scholar]
- 16.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aguilo F, Zhou MM, Walsh MJ. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 2011;71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012;45:279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S, Shan C, Kong G, Du Y, Ye L, Zhang X. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-kappaB–inducing kinase (NIK) Oncogene. 2012;31:3607–3620. doi: 10.1038/onc.2011.523. [DOI] [PubMed] [Google Scholar]
- 22.You X, Liu F, Zhang T, Lv N, Liu Q, Shan C, Du Y, Kong G, Wang T, Ye L. Hepatitis B virus X protein upregulates Lin28A/Lin28B through Sp-1/c-Myc to enhance the proliferation of hepatoma cells. Oncogene. 2014;33:449–460. doi: 10.1038/onc.2012.618. [DOI] [PubMed] [Google Scholar]
- 23.Shan C, Xu F, Zhang S, You J, You X, Qiu L, Zheng J, Ye L, Zhang X. Hepatitis B virus X protein promotes liver cell proliferation via a positive cascade loop involving arachidonic acid metabolism and p-ERK1/2. Cell Res. 2010;20:563–575. doi: 10.1038/cr.2010.49. [DOI] [PubMed] [Google Scholar]
- 24.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St John PC, Hirota T, Kay SA, Doyle FJ., III Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2014;111:2040–2045. doi: 10.1073/pnas.1323618111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science. 2012;337:599–602. doi: 10.1126/science.1221592. [DOI] [PubMed] [Google Scholar]
- 27.Hatori M, Hirota T, Iitsuka M, Kurabayashi N, Haraguchi S, Kokame K, Sato R, Nakai A, Miyata T, Tsutsui K. Light-dependent and circadian clock–regulated activation of sterol regulatory element-binding protein, X-box–binding protein 1, and heat shock factor pathways. Proc Natl Acad Sci U S A. 2011;108:4864–4869. doi: 10.1073/pnas.1015959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25(5):666–681. doi: 10.1016/j.ccr.2014.03.010. [Epub 2014 Apr 24] [DOI] [PubMed] [Google Scholar]
- 29.Bugaut A, Balasubramanian S. 5'-UTR RNA G-quadruplexes: translation regulation and targeting. Nucleic Acids Res. 2012;40:4727–4741. doi: 10.1093/nar/gks068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X. O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 2013;17:303–310. doi: 10.1016/j.cmet.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Hammerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, Skawran B, Geffers R, Longerich T, Breuhahn K. Posttranscriptional destabilization of the liver-specific long noncoding RNA HULC by the IGF2 mRNA-binding protein 1 (IGF2BP1) Hepatology. 2013;58:1703–1712. doi: 10.1002/hep.26537. [DOI] [PubMed] [Google Scholar]
- 35.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 37.Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, Holford TR, Hansen J, Paulson J, Zhu Y. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 2010;70:1459–1468. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma D, Li S, Molusky MM, Lin JD. Circadian autophagy rhythm: a link between clock and metabolism? Trends Endocrinol Metab. 2012;23:319–325. doi: 10.1016/j.tem.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of Primers and siRNA Used in This Paper
Clinical Characteristics of Liver Cancer and Normal Tissue Microarray Samples
HULC upregulates CLOCK and perturbs its rhythmical expression in hepatoma cells, related to Figure 1. (A) The endogenous expression levels of CLOCK and BMAL1 were examined by qRT-PCR in L-O2 and HepG2 cell, respectively. (B) The effect of HULC on the expression of circadian oscillators CLOCK and BMAL1 was assessed by qRT-PCR in HepG2 cells. (C) The mRNA levels of CLOCK were examined by qRT-PCR in L-O2 (HepG2) or HepG2.2.15 cells transfected with HULC or HULC siRNA, respectively. Statistically significant differences are indicated: **P < .01, ***P < .001, no significance (NS); Student’s t test.
HULC targets the 5'UTR of CLOCK mRNA via complementary base pairing in hepatoma cells, related to Figure 2, Figure 3. (A and B) The effect of HULC (or HULC-mut-in, HULC-mut-out) on the expression levels of PER1 and CRY1, downstream factors of CLOCK, was measured by qRT-PCT in HepG2 cells. Statistically significant differences are indicated: *P < .05, **P < .01; Student’s t test.
CLOCK is required for the proliferation of hepatoma cells mediated by HULC in vitro and in vivo, related to Figure 5, Figure 6. (A) Interference efficiency of CLOCK siRNA* and CLOCK siRNA was examined by Western blotting in HepG2 cells. (B) The levels of Ki-67 were assessed by IHC in the tumor tissues from mice.


