Abstract
Flavonoids comprise a large and diverse group of polyphenolic plant secondary metabolites. In plants, flavonoids play important roles in many biological processes such as pigmentation of flowers, fruits and vegetables, plant-pathogen interactions, fertility and protection against UV light. Being natural plant compounds, flavonoids are an integral part of the human diet and there is increasing evidence that dietary polyphenols are likely candidates for the observed beneficial effects of a diet rich in fruits and vegetables on the prevention of several chronic diseases. Within the plant kingdom, and even within a single plant species, there is a large variation in the levels and composition of flavonoids. This variation is often due to specific mutations in flavonoid-related genes leading to quantitative and qualitative differences in metabolic profiles. The use of such specific flavonoid mutants with easily scorable, visible phenotypes has led to the isolation and characterisation of many structural and regulatory genes involved in the flavonoid biosynthetic pathway from different plant species. These genes have been used to engineer the flavonoid biosynthetic pathway in both model and crop plant species, not only from a fundamental perspective, but also in order to alter important agronomic traits, such as flower and fruit colour, resistance, nutritional value. This review describes the advances made in engineering the flavonoid pathway in tomato (Solanum lycopersicum). Three different approaches will be described; (I) Increasing endogenous tomato flavonoids using structural or regulatory genes; (II) Blocking specific steps in the flavonoid pathway by RNA interference strategies; and (III) Production of novel tomato flavonoids by introducing novel branches of the flavonoid pathway. Metabolite profiling is an essential tool to analyse the effects of pathway engineering approaches, not only to analyse the effect on the flavonoid composition itself, but also on other related or unrelated metabolic pathways. Metabolomics will therefore play an increasingly important role in revealing a more complete picture of metabolic perturbation and will provide additional novel insights into the effect of the introduced genes and the role of flavonoids in plant physiology and development.
Keywords: GC/MS, LC/MS, Metabolic engineering, Metabolomics, Tomato
Introduction
Flavonoids represent a large family of low molecular weight polyphenolic secondary metabolites that are widespread throughout the plant kingdom (Koes et al. 1994). They are involved in a diverse range of biological processes, for example (I) in providing pigmentation to flowers, fruits and seeds to attract pollinators and seed dispersers, (II) in protection against ultraviolet light, (III) in plant defence against pathogenic micro organisms, (IV) in plant fertility and germination of pollen, and (V) in acting as signal molecules in plant-microbe interactions (Koes et al. 1994; Dixon and Paiva 1995; Dooner and Robbins 1991).
To date, more than 6,000 different flavonoids have been described and the number is still increasing (Harborne and Williams 2000). Flavonoids are defined by their basic skeleton, the flavan-nucleus, consisting of two aromatic rings with six carbon atoms (ring A and B) interconnected by a hetero cycle including three carbon atoms (ring C). According to the modifications of the central C-ring they can be divided into different structural classes like flavanones, isoflavones, flavones, flavonols, flavanols and anthocyanins (Fig. 1). The huge diversity in flavonoid structures is due to modifications of the basic skeleton by enzymes such as glycosyl transferases, methyl transferases and acyl transferases. In a single plant species, dozens of different flavonoids may be present and most of these are conjugated to various sugar moieties (Forkmann and Heller 1999).
Fig. 1.
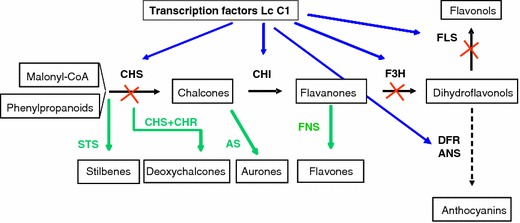
Schematic overview of flavonoid pathway engineering in tomato. Solid black arrows represent the active pathway in tomato fruit peel. Anthocyanin production is occasionally found in vegetative tissues (dotted arrow). Increased flavonol biosynthesis was obtained by upregulating the pathway in fruit flesh using the transcription factor genes Lc and C1, or by relieving the block in the pathway in fruit peel using the petunia CHI gene (blue arrows). An RNAi approach was used to block specific steps in the pathway (red crosses). Newly introduced flavonoid branches are depicted in green
The flavonoid biosynthetic pathway in plants has been almost completely elucidated. Many of the structural and some of the regulatory genes have been cloned from several model plants, including maize, Antirrhinum, tobacco, Petunia and Arabidopsis (Holton and Cornisch 1995) and have been expressed in genetically modified model plants and micro-organisms (Dixon and Steele 1999; Forkmann and Martens 2001; Winkel-Shirley 2001; Hwang et al. 2003). Today, standard molecular tools are available to genetically modify plants including several global important crops such as maize, potato, tomato, sugar beet and wheat (Sévenier et al. 2002).
Although some plants, such as onions (flavonols), blueberries (anthocyanins) and soybean (isoflavonoids) contain high levels of certain flavonoids, in other species the composition of these secondary metabolites is ‘sub-optimal’. Therefore, genetic engineering strategies have regularly been looked upon as possible means to modify flavonoid biosynthesis in order to influence, for example, flower pigmentation in ornamental plants (Van der Krol et al. 1988; Courtney-Gutterson et al. 1994; Deroles et al. 1998; Tanaka et al. 1998; Davies et al. 1998; Mol et al. 1999; Aida et al. 2000; Suzuki et al. 2002; Zuker et al. 2002; Fukui et al. 2003) or to improve for resistance against pathogens (Yu et al. 2003; Fischer et al. 1997; Jeandet et al. 2002).
In the past decade it has become increasingly clear that the composition of secondary metabolites greatly influences the quality and health potential of food and food products (Stobiecki et al. 2002). In particular, flavonoids have been suggested to protect against oxidative stress, coronary heart disease, certain cancers, and other age related diseases (Kuo 1997; Yang et al. 2001; Ross and Kasum 2002; Rein et al. 2006). At least part of these presumed health promoting properties of flavonoids can be attributed to the well-documented antioxidant properties of these compounds (Rice-Evans et al. 1995, 1997). Besides antioxidant activity, the inhibitory effect of flavonoids on enzymatic activities (Castelluccio et al. 1995; Rice-Evans et al. 1997; Pietta 2000) and their interaction with signal transduction pathways, leading to changes in the expression of genes involved in cell survival, cell proliferation and apoptosis (Yang et al. 2001; Sarkar and Li 2003; O’prey et al. 2003; Van Dross et al. 2003) may also contribute to their health-promoting properties. For this reason, there is currently a growing interest in the development of agronomically important food crops with optimized levels and composition of flavonoids and for many plant-based products ‘enhanced anti-oxidant content’ is now exploited in their marketing strategies. An excellent candidate for such an approach is tomato, one of the most important vegetable crops world-wide. In tomato fruit, flavonoids accumulate mainly in the peel, whereas in the flesh, which comprises >95% of total fruit weight, only traces of flavonoids can be found. The main flavonoids in tomato peel are naringenin-chalcone and rutin (quercetin-rutinoside). In the model cultivar Moneymaker, the levels of these compounds can reach up to 50 and 10 mg/kg FW, respectively. These levels are low compared to levels found in, for example, yellow onions, which contain up to 300 mg quercetin/kg FW (Hertog et al. 1993; Crozier et al. 1997). This suggests that there is considerable room for improvement of the flavonoid composition in tomato fruit.
In this paper we give an overview of different strategies used to alter the levels and composition of flavonoids in tomato fruit by engineering the flavonoid biosynthesis pathway or by screening the natural biodiversity and specific mutants using metabolomics approaches.
The flavonoid biosynthesis pathway in tomato fruit
Two classes of genes can be distinguished within the flavonoid pathway: structural genes encoding enzymes that directly participate in the formation of flavonoids, and regulatory genes that control the expression of the structural genes. The precursors for the synthesis of most flavonoids are malonyl-CoA and p-coumaroyl-CoA, which are derived from carbohydrate metabolism and the phenylpropanoid pathway, respectively (Forkmann and Heller 1999).
In tomato fruit, flavonoids are mainly produced in the peel. In the flesh, the flavonoid pathway is inactive due to a lack of expression of flavonoid biosynthesis genes. The major flavonoids in tomato fruit are a chalcone and a flavonol: naringenin chalcone and rutin (quercetin-rutinoside) respectively. The pathway leading to the production of these tomato flavonoids will be described in more detail below and is illustrated in Fig. 1.
The first enzyme in the flavonoid pathway is chalcone synthase (CHS), which catalyses the step-wise condensation of three acetate units starting from malonyl-CoA and the phenylpropanoid p-coumaroyl-CoA to yield the yellow-coloured naringenin chalcone (4,2′,4′,6′-tetrahydroxychalcone), the most abundant flavonoid in red tomato fruits (Holton and Cornish 1995; Tanaka 1998; Muir et al. 2001). Genomic and cDNA sequences encoding CHS have been isolated and characterised from many plant species, including tomato, and the expression of endogenous CHS genes has been studied in detail. Unlike tomato, most plants do not accumulate chalcones and after its formation, naringenin chalcone is rapidly isomerized by the enzyme chalcone isomerase (CHI) to form the flavanone naringenin. Even in the absence of CHI, naringenin chalcone may spontaneously isomerise to form naringenin (Holton and Cornish 1995). This compound is subsequently hydroxylated at position C-3 to form the dihydroflavonol dihydrokeampferol.The reaction is carried out by flavanone-3-hydroxylase (F3H), a member of the 2-oxoglutarate-dependent dioxygenase family which is highly conserved among widely divergent plant species (Britsch et al. 1993). Dihydrokaempferol (DHK) can be further hydroxylated, either at the 3′ position or at both the 3′ and 5′ positions of the B-ring. The former reaction leads to the formation of dihydroquercetin (DHQ), carried out by the P450 hydroxylase flavonoid 3′-hydroxylase (F3′H). The latter hydroxylation steps are catalysed by the P450 enzyme flavonoid 3′,5′-hydroxylase (F3′5′H), responsible for the conversion of DHK into dihydromyricetin (DHM). Dihydrokaempferol and dihydroquercetin are substrates for the enzyme flavonol synthase (FLS), a 2-oxoglutarate-dependent dioxygenase, leading to the production of the flavonols kaempferol and quercetin, respectively. The latter compound is a major flavonoid in tomato fruit peel, in addition to naringenin chalcone. In tomato, dihydromyricitin is the preferred substrate of the enzyme dihydroflavonol reductase (DFR), the first step in the branch leading to delphinidin-type anthocyanins, which are usually found only in vegetative tissues of tomato.
Modifying the flavonoid pathway using regulatory genes
The coordinate transcriptional control of structural biosynthetic genes has evolved as a major mechanism determining the final assembly of secondary metabolites in plants. This regulation is achieved by specific transcription factors which interact with promoter regions of target genes and thereby modulate the rate of initiation of mRNA synthesis (Ranish and Hahn 1996). Transcription factors controlling pigmentation pattern and intensity through regulating the expression of several flavonoid-anthocyanin structural genes have been identified in many plants (Holton and Cornish 1995). The control of these regulatory genes appeared to be highly dependent on tissue type, internal signals (e.g. hormones), and external signals (e.g. microbial elicitors, UV radiation (Memelink et al. 2000; Martin et al. 2001; Vom Endt et al. 2002).
In particular three plant species have been important for elucidating the transcriptional control of the anthocyanin biosynthetic pathway: maize, Antirrhinum and Petunia. More recently, regulatory genes involved in flavonoid biosynthesis have been found in Arabidopsis, tomato and apple. In general, these regulatory genes can be divided into two transcription factor families; one with sequence homology to a regulatory protein encoded by the vertebrate proto-oncogene c-MYB, and the other with similarity to the vertebrate basic-Helix-Loop-Helix (bHLH) protein encoded by the proto-oncogene c-MYC (Mol et al. 1998).
In various plant species tissue-specific regulation of the structural genes involved in flavonoid biosynthesis is controlled by a combination of regulators from these two transcription factor families. Homologous sets of transcription factors are able to control different sets of structural genes, thus allowing regulatory diversity in the flavonoid pathway in different plant species as well as different tissues (Koes et al. 1994; Quattrochio et al. 1993). Transcription factors from different plant species, however, often show a remarkable sequence homology, indicating that they are derived from a common ancestor. Moreover, ectopic expression of transcription factor genes in various plant species has confirmed that these regulatory genes are functionally conserved among different plant species and thus could provide a useful tool for genetic modification of crop plants in order to modify their final metabolite composition. However, the final quantity and class of flavonoid-derived metabolites that are produced are dependent on a number of parameters such as: the binding affinity of the transcription factor to specific promoter sites of their target structural genes; the ability of the introduced regulators to cooperate with endogenous transcription factors; the functionality of these endogenous transcription factors (Quattrocchio et al. 1993; Grotewold 2006). As a result, ectopic expression of flavonoid regulatory genes of different origin shows a highly variable profile of flavonoids in transgenic plants.
Amongst the most well characterized flavonoid regulatory plant genes are the maize leaf colour (LC) gene belonging to the MYC type R gene family and the MYB type C1 (colourless) gene. As early as 15 years ago, activation of the anthocyanin production has been achieved in Arabidopsis and tobacco plants by the introduction of the maize regulator genes R and C1. In both plant species, expression of the R regulatory gene alone was sufficient to enhance anthocyanin pigmentation in tissues that originally produce anthocyanins. In contrast, the expression of the C1 gene alone had no visible effect. Interestingly, accumulation of anthocyanins in tissues that normally do not contain any of these pigments was observed in transgenic Arabidopsis plants expressing both C1 and LC genes (Lloyd et al. 1992). Also in LC over-expressing cherry tomato plants, anthocyanins accumulated in leaves, stems, sepals, petals, veins and, to a lesser extent, in developing green fruits (Goldsbrough et al. 1996).
Ectopic expression of LC and C1 does not only result in enhanced anthocyanin production. Other flavonoid classes have been reported to accumulate when both LC and C1 are expressed. For example, in red ripe tomato fruits, the introduction and co-ordinate expression of the regulatory genes LC and C1 was sufficient to induce the flavonoid biosynthesis in fruit flesh, a tissue that normally does not produce flavonoids (Bovy et al. 2002; Fig. 1). The main compounds accumulating in these transgenic fruits were glycosides of the flavonol kaempferol. In addition, a more modest increase in glycosides of naringenin was observed. Taken together, the total flavonol content of ripe transgenic tomatoes over-expressing LC/C1 was about 20-fold higher compared to wild type fruits (Bovy et al. 2002; Le Gall et al. 2003; Table 1). Remarkably, these fruits did not accumulate any anthocyanins, although all structural flavonoid genes required for the production of kaempferol-type flavonols and pelargonidin-type anthocyanins were strongly induced due to the introduced LC/C1 transcription factors. Combined biochemical and transcriptional analysis of the transgenic lines indicated that the lack of anthocyanins could be explained by a low, LC/C1 independent expression of the gene encoding flavanone-3′,5′-hydroxylase in tomato fruit, together with a strong preference of the tomato dihydroflavonol reductase (DFR) enzyme to use the F3′5′H reaction product dihydromyricetin as substrate for the production of delphinidin-type anthocyanins (Martens et al. 2002). In contrast to fruits, light-stressed LC/C1 seedlings, as well as leaves and nodes of some LC/C1 tomato plants, clearly accumulated these delphinidin-type anthocyanins. When compared to fruits, expression of the F3′5′H gene appeared to be at least ten-fold higher in the purple-coloured LC/C1 leaves (Bovy et al. 2002).
Table 1.
Flavonoid content (mg/kg FW) in red fruits of several transgenic tomato lines
| Line | Control | CHI | FNS | FNS + CHI | CHS-as | STS | AS | Lc/C1 |
|---|---|---|---|---|---|---|---|---|
| Naringenin chalcone | 25 | 1 | 25 | 3.3 | 0.1 | 12.5 | 25 | 25 |
| Quercetin | 5 | 300 | 2 | 200 | 0.03 | 3.5 | 5 | 5 |
| Kaempferol | 1 | 70 | 70 | 1 | 70 | |||
| Luteolin | 5 | 100 | ||||||
| Resveratrol | 1.6 | |||||||
| Aureusidin | 2 |
For all transgenic lines except Lc/C1, the flavonoids are predominantly present in the fruit peel and likewise, appr. ten-fold higher levels can be obtained by specifically analysing peel instead of whole fruit samples
Homologs of the maize flavonoid transcription factor genes LC and C1 have also been isolated from dicot species. Petunia, Arabidopsis, tomato, Antirrhinum and apple contain genes with sequence homology to transcription factors that regulate transcription of structural anthocyanin biosynthesis genes (Cone et al. 1986; Goodrich et al. 1992; Grotewold et al. 1994; Quattrocchio et al. 1998; Ramsay et al. 2003; Takos et al. 2006). In contrast to maize, where LC and C1 regulate all genes of the pathway from CHS until 3GT, it has been shown that, in dicots, distinct sets of MYB/MYC transcription factors are responsible for regulating different parts the pathway (early: CHS up to F3H, or late: DFR to 3GT). For example, in Antirrhinum, anthocyanin production is regulated by three regulatory genes—Delila, Eluta and Rosea. The Antirrhinum Delila gene (DEL), a MYC (bHLH) homologue, is required for pigmentation of the flower tube. The first two steps of the flavonoid pathway, CHS and CHI, show negligible regulation by Delila but subsequent steps (F3H, DFR, 3GT) have an absolute prerequisite for the Delila gene product and show quantitative regulation by Eluta and Rosea (Martin et al. 1991).
Over-expression of Delila in tobacco and tomato resulted in enhanced pigmentation of flowers in tobacco, whereas vegetative tissues were affected in tomato. In both plant species this was at least partly due to increased expression of the DFR gene. A ten-fold increase of DFR mRNA levels was observed in tomato and a four-fold increase in tobacco when DEL was over-expressed. Transcript levels of CHS were only slightly increased, two and three-fold, for tobacco and tomato, respectively (Mooney et al. 1995).
Activation tagging in a tomato line accumulating anthocyanins has led to the identification of the ANT1 gene, a transcriptional regulator of the anthocyanin biosynthesis pathway encoding a myb transcription factor (Mathews et al. 2003) with strong similarity to Petunia AN2. This ANT1 gene appeared to be responsible for the intense purple coloured vegetative tissue and purple spots in the fruit epidermis of the tomato mutant. The tomato cultivar Micro-Tom, constitutively over-expressing a single genomic ANT1 gene, demonstrated phenotypes varying from weak to strong anthocyanin accumulation. In seedlings of transgenic ANT1 tomato plants the anthocyanin levels were increased up to 3.5 mg/g fresh weight—an almost 500-fold increase compared to untransformed seedlings. LC-MS analysis revealed that nine discrete anthocyanins were responsible for the pigmentation found in ANT1 transenic seedlings. These consisted of the 3-rutinoside-5-glucosides of delphinidin, petunidin and malvidin (all delphinidin-type anthocyanins). Further acylation of these anthocyanins with caffeic acid or coumaric acid resulted in six additional pigment molecules. Over-expression of ANT1 resulted in the up-regulation of early (CHS, CHI-like) as well as late (DFR) genes of anthocyanin biosynthesis. In addition flavonoid modifying genes (i.e. 3-O-glucosyltransferase and 5-O-glucosyltransferase) and genes encoding proteins involved in flavonoid transport (HD-GL2, permease and GST) were increased as well.
Besides transcription factors which increase the activity of the flavonoid pathway, also negative regulators of flavonoid biosynthesis have been described. For example, mutation of the DE-ETIOLATED1 gene (DET1) has been shown to result in high pigmented phenotypes (hp-2) in tomato. The darker fruits of these mutants are due to elevated levels of both flavonoids and carotenoids (Bino et al. 2005). Fruit specific suppression of the regulatory gene DET-1 resulted in increased levels of both secondary metabolite groups. Flavonoid levels were increased up to 3.5 fold, lycopene content was two-fold higher and β-carotene levels accumulated up to ten-fold compared to wild type fruits (Davuluri et al. 2005).
Modifying the flavonoid pathway using structural genes
The major flavonoid accumulating in tomato fruit peel is naringenin chalcone, the product of CHS. Naringenin chalcone accumulates during ripening concommittant with an increase in CHS gene expression. In addition to naringenin chalcone, the flavonol quercetin rutinoside, or rutin, also accumulates in tomato fruit peel. Biochemical and molecular analysis of peel and flesh samples of WT tomato fruits revealed that, in the flesh, all flavonoid genes show very low expression levels, in line with the lack of flavonoids in fruit flesh. In peel, all the genes required for the production of rutin were strongly induced upon ripening, except for the gene encoding chalcone isomerase (CHI), which demonstrated low expression throughout ripening (Muir et al. 2001; Bovy et al. 2002). Chalcone isomerase is required to convert naringenin chalcone into naringenin and both the accumulation of naringenin chalcone and the low CHI expression suggest that CHI is a rate-limiting step in the pathway leading to rutin. Ectopic expression of the PetuniaCHI gene in tomato fruits indeed relieved the block in the pathway (Fig. 1), leading to an up to 70-fold increase in tomato fruit peel of total flavonols, consisting mainly of the flavonols rutin (quercetin 3-rutinoside) and isoquercetin (quercetin-3-glucoside), and to a smaller but still substantial extent of kaempferol glycosides (Table 1). In these high-flavonol transformants, naringenin chalcone levels were strongly reduced, suggesting that the Petunia CHI enzyme utilizes the natural naringenin chalcone pool as substrate (Muir et al. 2001; Verhoeyen et al. 2002).
To enhance the levels of flavonols in the fruit flesh, a four-gene construct was introduced into tomato, leading to concomitant ectopic expression of PetuniaCHS, CHI, F3H and FLS in tomato fruit. This resulted in increased levels of flavonols in both peel (primarily quercetin glycosides) and flesh (primarily kaempferol glycosides) (Colliver et al. 2002). When expressed separately, none of these four genes was sufficient to lead to flavonol production in fruit flesh: CHS over-expression resulted in accumulation of naringenin in the flesh, CHI only affected flavonol levels in the peel, and F3H and FLS showed no effects on flavonoid levels, neither in peel, nor in flesh. Crossing experiments with single gene transgenic lines revealed that concomitant expression of both CHS and FLS was sufficient to lead the pathway to the accumulation of glycosides of naringenin as well as kaempferol in tomato flesh (Colliver et al. 2002; Verhoeyen et al. 2002). In addition, these results suggested that, in tomato flesh, ectopic expression of CHI and F3H is not required for flavonol production. This could be due to the presence of sufficient endogenous CHI enzyme or spontaneous conversion of naringenin chalcone to naringenin. In addition, in vitro experiments revealed that FLS has the capacity to convert naringenin into kaempferol, suggesting that the FLS enzyme harbours an intrinsic F3H-like activity as well (Martens et al. 2003; Lukačin et al. 2003).
Blocking specific steps in the flavonoid pathway by RNAi
In order to block specific metabolic conversions in the endogenous tomato flavonoid biosynthesis pathway, RNA interference has been used to down-regulate the expression of specific structural flavonoid genes. Expression analysis of the endogenous tomato flavonoid genes CHS, CHI, F3H and FLS revealed that CHS, F3H and FLS were expressed in peel tissue during all stages of fruit development, peaking at the turning stage. In contrast, the CHI transcript levels remained below detection levels (Muir et al. 2001). Based on these gene expression data we decided to block the flavonoid pathway leading to flavonols at CHS, F3H and FLS using RNAi-mediated gene silencing (Fig. 1).
RNAi inhibition of the tomato CHS1 gene resulted in a strong reduction of total flavonoid levels (naringenin chalcone and quercetin rutinoside) (Schijlen et al. 2007). Gene expression analysis showed that both Chs1 and Chs2 expression levels were reduced. As a result, CHS activity was also dramatically reduced in these lines, finally resulting in a 99% reduction of the total flavonoid levels relative to wild type. Interestingly, these RNAi tomato fruits displayed several phenotypic alterations of which parthenocarpic fruit development appeared to be the most severe. The occurrence of parthenocarpic fruits has also been reported in tomato overexpressing stilbene synthase (Giovinazzo et al. 2005; Schijlen et al. 2006), suggesting a role for flavonoids in fertilization, seed and fruit development.
A clear reduction of flavonols was obtained by introducing an FLS RNAi construct. Biochemical analysis revealed that in FLS RNAi tomato peel, quercetin-3-rutinoside levels were strongly decreased (1.2 mg/kg FW) when compared to wild type (78 mg/kg FW; Figs. 2 and 3). Although these fruits were indistinguishable from wild type tomatoes, vegetative tissues for example leaves, stem and flower buds clearly accumulated anthocyanins due to the decreased FLS activity. This suggests that dihydroflavonols, which are the natural substrates for both FLS and DFR, were thus more efficiently converted into anthocyanins. Due to decreased FLS activity there appears to be less competition between the flavonol and anthocyanin branches in vegetative tissue of FLS RNAi tomatoes, thereby improving the metabolic flux towards anthocyanin end products (Fig. 4).
Fig. 2.
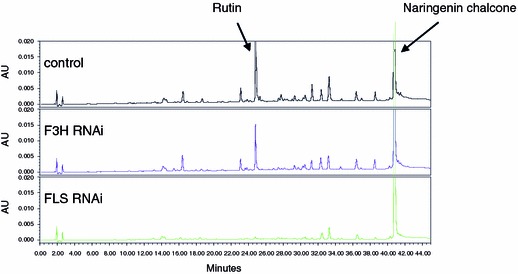
Reduced flavonoid metabolites in RNAi tomato fruit; Upper panel Wild type, middle panel F3H RNAi, lower panel FLS RNAi
Fig. 3.
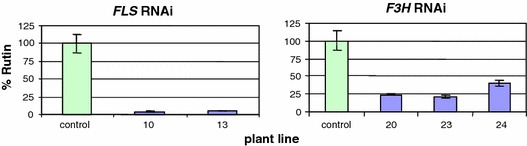
Biochemical analysis of primary transformants of FLS and F3H RNAi plants. Analyses were carried out using three individual cuttings per plant line (three fruits pooled from each plant). Rutin levels were expressed relative to the levels in control plants
Fig. 4.

Anthocyanin accumulation in vegetative tissues of FLS RNAi tomato plants (left and middle panel: FLS RNAi; right panel: wild type)
By using an RNAi construct for F3H, tomato flavonoid biosynthesis could be reduced beyond this step as well, although not as effectively as for Chs and Fls (Fig. 2). In F3H RNAi the rutin levels were decreased to approximately 20% of wild type values (Fig. 3). This relatively low decrease may have resulted either from a partial F3H down-regulation or is due to an intrinsic F3H-like activity of FLS, thereby using naringenin to form flavonols (Martens et al. 2003; Lukačin et al. 2003) This is in line with the observation that simultaneous overexpression of CHS and FLS was sufficient to accumulate flavonols in the fruit flesh, despite the very low expression of endogenous CHI and F3H genes in this tissue (Colliver et al. 2002).
Production of novel tomato flavonoids by introducing new branches of the flavonoid pathway
Production of flavonoid related stilbenes
Nowadays, there is a lot of interest on the putative health effects of the flavonoid related stilbenes that are rarely found in the plant kingdom. Therefore, several attempts have been made to produce these phenolic compounds in a more common dietary source such as tomato. The introduction of a grape STS cDNA in tomato resulted in the accumulation of stilbenes in fruits (Giovinazzo et al. 2005; Schijlen et al. 2006; Fig. 1; Table 1)) Of these stilbenes, the major compounds were trans-resveratrol and its glucoside piceid. Whereas flavonoids normally do not accumulate in fruit flesh, significant levels of stilbenes were also detected in this tissue. As for flavonoids, it was found that the content of the new metabolites strongly varied during fruit maturation. In addition, a five fold difference in stilbene accumulation was found between the two research groups, probably also reflecting differences in growing conditions. On a whole fruit basis, total stilbene accumulation was reported up to 53 mg/kg FW (Giovinazzo et al. 2005) and 10 mg/kg FW (Schijlen et al. 2006).
Since the introduced stilbene synthase competes with endogenous chalcone synthase for their common substrate 4-coumaroyl CoA (Schröder and Schröder 1990), STS overexpression might be expected to lead to a reduced level of endogenous flavonoids. Although Giovinazzo et al. (2005) did not find any alterations in flavonoid levels in tomatoes overexpressing stilbene synthase, Schijlen et al. (2006) reported significant changes in the levels of naringenin chalcone. These differences are most likely due to differences in ‘background’ levels of for example endogenous phenylpropanoid and flavonoid biosynthesis as a result of environmental and/or genetic differences. Nevertheless, overexpression of resveratrol synthase appeared to be a successful approach to enable stilbenes to be produced in tomato. Interestingly, the stilbene levels in these transgenic tomatoes are significantly higher than those found in different red wines (ranging from 0.5 to 10 mg/l), currently the most common source of resveratrol (Celotti et al. 1996).
Production of deoxychalcones
In the plant kingdom, the presence of deoxychalcones is mainly restricted to the Leguminoseae, where production of 6′-deoxychalcones results from a combination of chalcone reductase and chalcone synthase activity (Davies et al. 1998). Overexpression of both CHS and CHR in tomato resulted in the accumulation of deoxychalcones (Schijlen et al. 2006; Fig. 1; Table 1). In these fruits, deoxychalcones accumulates up to 265 mg/kg FW fruit peel, with butein and isoliquiritigenin as major new metabolites (up to 89 mg/kg FW and 176 mg/kg FW respectively). Because no other deoxychalcone-related flavonoid classes accumulated in these tomatoes it can be concluded that the 6′-deoxychalcones synthesized in CHS/CHR tomato fruits, were not incorporated into the subsequent 5-deoxy(iso)flavonoid pathway. This can be explained by the lack of endogenous type II CHI activity in tomato, necessary for the conversion of isoliquiritigenin into the 5-deoxy flavonoid liquiritigenin, the substrate for IFS and thus precursor for iso-flavonoid biosynthesis which normally only occurs in leguminous plants (Heller and Forkmann 1993). A similar accumulation of deoxychalcones,was described for transgenic Petunia flowers overexpressing CHR. (Davies et al. 1998) Although the accumulation of deoxychalcones in our tomato lines was less than reported in CHR Petunia, in both plant species the same metabolic efficiency rate, i.e. ratio between hydroxy flavonoids and deoxychalcones (3:2) was obtained. The higher deoxychalcone accumulation in Petunia flowers is likely due to an approximately ten-fold higher flavonoid background level of these flowers compared to tomato fruit peel. In agreement with results obtained in CHR-overexpressing Petunia and tobacco plants (Joung et al. 2003), overexpression of the CHR gene in tomato resulted in strong competition for common substrates between the endogenous hydroxy-flavonoid and the introduced deoxyflavonoid pathway. As a consequence, a clear loss of 6-hydroxyflavonoids (reduced to one third of wild type values) accompanied the accumulation of deoxychalcones in CHR tomato fruit peel.
Production of flavones and aurones
Although flavones are ubiquitous in plants, they are not commonly found in crops. Major food sources of flavones are celery and parsley. In an attempt to produce flavones in tomato, heterologous expression of a FNS-II gene derived from Gerbera has been used (Fig. 1). Although this appeared to be an effective way to introduce new flavone-derived metabolites in tomato fruit, high amounts of flavones could only be obtained after simultaneous enhancement of the endogenous flavonoid biosynthesis. Therefore, heterologous expression of FNS to create a new flavone branch was combined with overexpression of CHI to stimulate the metabolic flux through the bottleneck at the CHI-step in tomato fruit peel (Schijlen et al. 2006). The tomatoes obtained accumulated high levels of flavones in their peel, mainly as luteolin aglycone and luteolin 7-glucoside (up to 340 and 150 mg/kg FW respectively). In addition to flavones, flavonol levels in peel tissue of these transgenic fruits were also strongly increased compared to the wild-type (Table 1). These flavonols were identified as quercetin-3,7-trisaccharide, quercetin-3-trisaccharide, rutin, isoquercetin, quercetin-3-rhamnoside, kaempferol-3-rutinoside, kaempferol-3-glucoside and quercetin aglycone. The accumulation of isoquercetin (quercetin-3-glucoside) and quercetin aglycone, both precursors of rutin, as well as luteolin aglycon, suggests that both rhamnosyl transferase required for the production of rutin (=quercetin-3-rutinoside) and glucosyl transferase become rate limiting in the peel of CHI/FNS transgenic fruits.
Flavonol accumulation (quercetin up to 67 mg/kg FW; rutin up to 900 mg/kg FW) in fruit peel of CHI/FNS overexpressing tomatoes was up to 16-fold higher compared to the wild type. Analagous to tomato plants overexpressing CHI alone (Muir et al. 2001; Verhoeyen et al. 2002), naringenin chalcone levels in fruit peel were strongly reduced in CHI/FNS plants (Table 1). This indicates that CHI/FNS overexpression indeed leads to an increased flux through the pathway towards flavones and flavonols at the expense of the CHI substrate naringenin chalcone.
Aurones are a minor subclass of plant flavonoid pigments that provide the typical bright yellow color of some popular ornamental plant species including Antirrhinum majus. We isolated the complete cDNA encoding Aureusidin synthase (AmAs1, a polyphenol oxidase homologue (Nakayama et al. 2000) from yellow snapdragon petals and used this for constitutive expression in tomato. The resulting transgenic tomato plants produced aureusidin aglycon as well as three different aureusidin glycosides in the fruit peel up to approximately 21 mg/kg fresh weight total aurones (expressed as aureusidin equivalents) (unpublished results).
Natural variation in flavonoids
Although the previous attempts to enhance the flavonoid biosynthesis in tomato focused on transgenic strategies, non-transgenic approaches can also be applied for metabolic engineering to develop tomato with altered flavonoid composition. Among the different factors that may influence the polyphenol content of tomatoes, genotype is certainly among the most relevant. Therefore, the evaluation of flavonoid gene expression and polyphenol patterns of wild tomato species, as well as mutant tomato lines, can be a first step on a successful path to increase the nutritional value of tomato fruit.
The obtained molecular knowledge about the cultivated tomato can be utilized for subsequent screening of available genetic resources. For example, expression analysis of the flavonoid biosynthetic pathway in the cultivated tomato fruit peel suggests that the lack of flavonoid accumulation may be due to a mutation leading to a loss of CHI expression but not the rest of the pathway. This raised the possibility of screening wild tomato species for enhanced CHI expression. It has been found that only a few wild Lycopersicon accessions express CHI in tomato fruit peel and that even a smaller subset of these also express genes of the flavonol biosynthetic pathway, including CHI, in the fruit flesh (Willits et al. 2005). Lycopersicon pennellii v. puberulum (LA1926) expressed high levels of all five biosynthetic genes (CHS-a, CHS-b, CHI, F3H, and FLS) in both fruit peel and fruit flesh and was therefore selected as a candidate for breeding.
Fruits of L. esculentum typically accumulate moderately low levels of quercetin-rutinoside (3 μg/mg in peel and 0.1 μg/mg Dry Weight in flesh). In addition the peel tissue accumulates significant amounts of naringenin chalcone. In contrast, L. pennellii v. puberulum accumulates slightly higher levels of flavonols (4.7 μg/mg Dry Weight in the peel and 1.2 μg/mg DW in the flesh) and no naringenin chalcone. In order to enhance the flavonoid biosynthesis in cultivated tomato, an inter-specific cross was made between a cultivar of L. esculentum. and the wild accession L. pennellii. Fruits from the F1 hybrid plants showed a ∼12 fold increase in flavonoid levels over L. esculentum (Willits et al. 2005). The results show that a wild accession can be successfully used to backcross a high flavonol trait into a cultivated tomato.
Altered fruit color is a widely used visual marker to identify tomato lines that could be subjected to metabolic profiling in order to investigate the differences in carotenoid, phenolic and flavonoid composition. Clear differences in polyphenol pattern and quantity were found on evaluation of 25 tomato lines that were selected based on their fruit color, together with five more cultivars suitable for processing by food industry (Minoggio et al. 2003). Moreover, naringenin, naringenin-chalcone and rutin with its analogues were the main polyphenols in the examined genotypes. Although the individual polyphenol levels (mg/100 g FW) showed a broad range (from 0.04 to 4.90 for naringenin plus its chalcone, 0.07–2.35 for rutin and 0.03–1.38 for rutin-pentoside) the total polyphenol content difference was up to six fold (ranged from 4.43 to 25.84 mg/100 g; mean ± s.e. 13.15 ± 1.15). In addition, phenolic acids were mainly represented by chlorogenic acid (range 0.03−0.58 mg/100 g) followed by caffeic acid (range 0.0–0.1 mg/100 g), chlorogenic acid derivative (range 0.00–0.07 mg/100 g) and ferulic acid (0.00–0.01 mg/100 g). The total antioxidant activity of these tomatoes was also determined and compared to the total phenol and carotenoid concentrations. Interestingly, within this subset almost all low carotenoid fruits accumulated high amounts of polyphenols and correlated (R2 = 0.79, P < 0.0001) with the most powerful antioxidant potential (Minoggio et al. 2003).
Since the products of both pathways produce antioxidants, it is important to understand how manipulation of one may affect the other and the extent of any cross-talk between the two. Therefore a panel of transgenic and mutant tomato lines has been subjected to metabolite profiling and compared with wild type Ailsa Craig for both carotenoids and phenolics (Long et al. 2006). This was also encouraged by their finding that the rr mutant of tomato is devoid of carotenoids, but still has a yellow colour due to 50% higher levels of naringenin compared to WT. Interestingly, although the authors unfortunately only used technical replicates, all mutants altered in structural genes for carotenoid biosynthesis generally revealed no significant alterations in total phenolic or flavonoid content, even when devoid of carotenoids. In contrast, mutants defective in light perception for example the hp-1 and LA3771 lines, showed increased fruit carotenoid content, as well as elevated phenylpropanoids and flavonoids (chlorogenic acid and rutin respectively).
Among tomato mutants with an altered fruit color, the high pigment mutants (hp-1 and hp-2) and the anthocyanin fruit mutant (Af or Aft) have gained considerable interest for metabolic profiling and subsequent to use these mutations in commercial varieties to increase nutritional value. Both hp mutants contain elevated fruit levels of flavonoids as well as carotenoids (Yen et al. 1997; Mustilli et al. 1999; Long et al. 2006; Bino et al. 2005). Bino et al. (2005) compared the overall metabolic modifications between fruits of high pigment (hp) tomato mutant and isogenic nonmutant (wt) control plants. Targeted high-performance liquid chromatography with photodiode array detection (HPLC–PDA) metabolite analysis showed higher levels of isoprenoids and phenolic compounds in hp-2dg fruit. Levels of the three flavonoids (in μg g−1 FW) were higher in mutant compared to control fruits: at the green stage the ratio hp-2dg compared to control was 7.39 for rutin and 4.07 for quercetin- 3-trisaccharide, representing increased flavonoid levels but slightly less than the thirteen fold reported for hp-1 (Yen et al. 1997). In red hp-2dg mutant fruits the total quercetin content (measured as aglycon level) was approximately three-fold of the wild type fruits. In addition, a three-fold increase of naringenin chalcone was observed in hp-2dg fruits at breaker stage when compared to control. However this difference was less pronounced in red fruits (Bino et al. 2005).
Tomato fruit are not usually reported to contain anthocyanins. However, one exception for this is the LA1996 mutant line with the Anthocyanin fruit (Aft) gene. This mutant has dark green foliage, elevated anthocyanin expression in the hypocotyls of seedlings, and interestingly also anthocyanins in the fruit. Unlike that of the phytochrome high pigment mutants, unripe fruits of LA1996 are not dark green and ripe fruits do not contain increased carotenoid levels (Jones et al. 2003). Anthocyanin expression was strongest in the skin and pericarp tissues beneath the skin composed by petunidin, followed by malvidin and delphinidinin as the principal anthocyanidins in Anthocyanin fruit. The total monomeric anthocyanin concentration of pigment-rich tissues separated from whole LA1996 fruits was estimated to be 20.6 mg/100 g FW in the pericarp tissue and 66.5 g/100 g FW in the skin expressed as petunidin 3- (p-coumaryl-rutinoside)-5-glucoside.
Several crossings of hp-1, hp-2, atv and Aft have been made to obtain a collection of single and double mutants with increased anthocyanin levels in their fruits. These mutants were used for targeted and untargeted fruit metabolite analysis (Van Tuinen et al. 2006).
Whereas the Af mutant contained anthocyanins in peel of fruits and the atv mutant accumulated anthocyanins in vegetative tissues, the double mutant Af atv had unripe and ripe fruits with high levels of anthocyanins resulting in deep purple mature fruits.
Also the Af hp-1w double mutant showed a strong synergistic effect on pigmentation, resulting in high levels of anthocyanins and increased levels of several phenolics compared to the single mutant parents. Moreover, whereas the hp-1w mutant fruit is dark green and does not contain anthocyanins and the Af mutant fruit contained some anthocyanins, rutin and some (yet) unidentified phenylpropanoids were much higher in Af hp-1w tomatoes. These include several antioxidants, either new or at enhanced levels, in the double mutant compared to single mutant parents (Af and hp-1w). Comparable increased anthocyanins were obtained by double mutation of the Af and hp-2j tomato (Van Tuinen et al. 2006).
The results of these mutant analyses show how metabolic profiling can be used to identify individual metabolites altered in particular mutants or breeding lines and thereby can provide a selection method to improve nutritional value in tomato.
The potential of metabolomics
All the examples detailed or referred to above clearly highlight the great interest in understanding the molecular mechanisms behind the control of phenylpropanoid metabolism in plants. Tomato has been a particular favorite plant target, acting often as a model species. Consequently, with our knowledge which is continually expanding, we are becoming more sophisticated in the approaches and tools we have at our disposal for targeted modifications of specific pathway branches in order to achieve a particular desired metabolic profile in ripe fruit. However, knowledge is still limited and much of the ongoing tomato work and work on other contrasting species, continues to reveal just how complex plant metabolism is. Few, if any, pathways ‘stand alone’ but rather, form a single route in a much broader, highly integrated and interactive metabolic network. As such, targeted perturbations such as the introduction of transgenes or directed mutagenesis, may have much more far-reaching effects than anticipated or, alternatively, may have no effect at all due to the ability of such interactive networks to compensate for localized changes. Next to the more targeted analyses, optimized for specific compound groups of interest, metabolomics, through its untargeted nature, offers an additional opportunity for identifying potentially unexpected metabolic effects.
Tomato has already been a popular subject for the development and application of metabolomics technologies. Both HPLC-MS approaches for semi-polar compounds (Minoggio et al. 2003; Bino et al. 2005; Moco et al. 2006; Verhoeven et al. 2006; van Tuinen et al. 2006) and GC-MS approaches for both primary metabolites (Schauer et al. 2005a, b; Urbanczyk-Wochniak and Fernie 2005) and natural volatiles (Tikunov et al. 2005, 2006) have been widely applied. In addition, alternative technologies such as Flow Injection—MS (Overy et al. 2005), FT-IR (Johnson et al. 2003) and MALDI-MS (Fraser et al. 2007) have also been employed for specific projects. Diverse changes in tomato metabolite profiles have been observed in these investigations, involving chemically contrasting compound groups, and following contrasting reasons for biological variation such as nitrogen supplementation (Urbanczyk-Wochniak and Fernie 2005), genomic introgression from another species (Overy et al. 2005), salt stress (Johnson et al. 2003) and transgenesis (Long et al. 2006, Fraser et al. 2007). Many of these authors emphasize the importance of using a non-targeted approach to establish the range of chemical perturbation, to identify changes previously unexpected and to follow the simultaneous effects of multiple gene changes (Levin et al. 2004). Furthermore, the biochemical and bioinformatics tools now typically being employed for metabolomics analyses can often allow the identification of subtle differences which would otherwise have been overlooked (Johnson et al. 2003, Overy et al. 2005) and also for example for making large-scale investigations such as genome-wide metabolic profiling (Schauer et al. 2005b). Of course the topic of ‘unintended effects’ is of most potential relevance regarding the application of transgene approaches to crop plants (Rischer and Oksman-Caldentey 2006). However, the delineation between unintended and unexpected is quickly becoming a grey area considering the wealth of knowledge that is already being generated using metabolomics approaches. The full impact of metabolomics on tomato research has therefore yet to be experienced.
Conclusions
All four transgenic approaches described above have been very effective in introducing novel flavonoid-derived metabolites, normally absent in tomato fruit. Building up and exploiting prior knowledge of pathway control mechanisms opens up new possibilities for metabolic engineering of the tomato flavonoid pathway. Next to fundamental science, this area of research is gaining a major boost through the growing interest in for example anti-oxidant content in food and more generally in plant-based, health-related compounds. The examples given here describe the accumulation of stilbenes, deoxychalcones and flavones and the levels achieved are favourably comparable or even higher than are currently present in natural food sources such as red wine, celery and onions (USDA 2003; Haytowitz et al. 2003). Knowledge of the pathways involved is extensive but, nevertheless, is still incomplete. Metabolomics is predicted to play an important future role in furthering our understanding of the flavonoid pathway and specifically, how it operates in the context of the much broader metabolic network in which it is positioned.
Acknowledgements
AB and RDH acknowledge financial support from the Centre for BioSystems Genomics, an activity carried out under the auspices of the Netherlands Genomics Initiative (NGI). AB acknowledges financial support from the EU project FLAVO (FOOD-CT-2005-513960) and RDH acknowledges financial support from the EU projects FLORA (FOOD-CT-2005-01730) and META-PHOR (FOOD-CT-2006-036220) all of which are focused on the application of metabolomics technologies to plant materials.
Contributor Information
Arnaud Bovy, Email: arnaud.bovy@wur.nl.
Robert D. Hall, Phone: +31-317-477230, FAX: +31-317-418094, Email: robert.hall@wur.nl
References
- Aida R., Kishimoto S., Tanaka Y., Shibata M. Modification of flower color in torenia (Torenia Fournieri Lind.) by genetic transformation. Plant Science. 2000;153:33–42. doi: 10.1016/S0168-9452(99)00239-3. [DOI] [Google Scholar]
- Bino R. J., de Vos C. H. R., Lieberman M., Hall R. D., Bovy A. G., Jonker H. H., Tikunov Y., Lommen A., Moco S., Levin I. The light hyper-responsive high-pigment 2dg mutation of tomato: Alterations in the fruit metabolome. New Phytologist. 2005;166:427–438. doi: 10.1111/j.1469-8137.2005.01362.x. [DOI] [PubMed] [Google Scholar]
- Bovy A., de Vos R., Kemper M., Schijlen E., Almenar Pertejo M., Muir S., Collins G., Robinson S., Verhoeyen M., Hughes S., Santos-Buelga C., van Tunen A. High-flavonol tomatoes resulting from heterologous expression of the maize transcription factor gene LC and C1. Plant Cell. 2002;14:2509–2526. doi: 10.1105/tpc.004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britsch L., Dedio J., Saedler H., Forkmann G. Molecular characterization of flavanone 3 beta-hydroxylases. Consensus sequence, comparison with related enzymes and the role of conserved histidine residues. European Journal of Biochemistry. 1993;217:745–754. doi: 10.1111/j.1432-1033.1993.tb18301.x. [DOI] [PubMed] [Google Scholar]
- Castelluccio C., Paganga G., Melikian N., Bolwell P. G., Pridham J., Sampson J., Rice-Evans C. Antioxidant potential of intermediates in phenylpropanoid metabolism in higher plants. FEBS Letters. 1995;368:188–192. doi: 10.1016/0014-5793(95)00639-Q. [DOI] [PubMed] [Google Scholar]
- Celotti E., Ferrarini R., Zironi R., Conte L. S. Resveratrol content of some wines obtained from dried Valpolicella grapes: Recioto and Amarone. Journal of Chromatography A. 1996;730:47–52. doi: 10.1016/0021-9673(95)00962-0. [DOI] [PubMed] [Google Scholar]
- Colliver S., Bovy A., Collins G., Muir S., Robinson S., de Vos C. H. R., Verhoeyen M. E. Improving the nutritional content of tomatoes through reprogramming their flavonoid biosynthetic pathway. Phytochemical Review. 2002;1:113–123. doi: 10.1023/A:1015848724102. [DOI] [Google Scholar]
- Cone K. C., Burr F. A., Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proceedings of the National Academy of Sciences. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney-Gutterson N., Napoli C., Lemieux C., Morgan A., Firoozabady E., Robinson K. E. Modification of flower color in florist’s chrysanthemum: production of a white-flowering variety through molecular genetics. Biotechnology. 1994;12:268–271. doi: 10.1038/nbt0394-268. [DOI] [PubMed] [Google Scholar]
- Crozier A., Lean M. E. J., McDonald M. S., Black C. Quantitative analysis of the flavonoid content of commercial tomatoes, onions, lettuce, and celery. Journal of Agricultural and Food Chemistry. 1997;45:590–595. doi: 10.1021/jf960339y. [DOI] [Google Scholar]
- Davies K. M., Bloor S. J., Spiller G. B., Deroles S. C. Production of yellow colour in flowers: Redirection of flavonoid biosynthesis in Petunia. Plant Journal. 1998;13:259–266. doi: 10.1046/j.1365-313X.1998.00029.x. [DOI] [Google Scholar]
- Davuluri G. R., van Tuinen A., Fraser P. D., Manfredonia A., Newman R., Burgess D., Brummell D. A., King S. R., Palys J., Uhlich J., Bramley P. M., Pennings H. M. J., Bowler C. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nature Biotechnology. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroles S. C., Bradley J. M., Schwinn K. E., Markham K. R., Bloor S., Manson D. G., Davies K. M. An antisense chalcone synthase cDNA leads to novel colour patterns in lisianthus (Eustoma grandiflorum) flowers. Molecular Breeding. 1998;4:59–66. doi: 10.1023/A:1009621903402. [DOI] [Google Scholar]
- Dixon R. A., Paiva N. L. Stress-induced phenylpropanoid metabolism. Plant Cell. 1995;7:1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Steele C. L. Flavonoids and isoflavonoids —a gold mine for metabolic engineering. Trends in Plant Science. 1999;4:394–400. doi: 10.1016/S1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- Dooner H. K., Robbins T. P., Jorgensen R. A. Genetic and developmental control of anthocyanin biosynthesis. Annual Review of Genetics. 1991;25:173–199. doi: 10.1146/annurev.ge.25.120191.001133. [DOI] [PubMed] [Google Scholar]
- Fischer R., Budde I., Hain R. Stilbene synthase gene expression causes changes in flower colour and male sterility in tobacco. Plant Journal. 1997;11:489–498. doi: 10.1046/j.1365-313X.1997.11030489.x. [DOI] [Google Scholar]
- Forkmann G., Martens S. Metabolic engineering and applications of flavonoids. Current Opinions in Biotechnology. 2001;12:155–160. doi: 10.1016/S0958-1669(00)00192-0. [DOI] [PubMed] [Google Scholar]
- Forkmann G., Heller W. Biosynthesis of flavonoids. In: Barton D., Nakanishi K., Meth-Cohn O., editors. Comprehensive natural products chemistry. Amsterdam: Elsevier; 1999. pp. 713–748. [Google Scholar]
- Fraser P. D., Enfissi E. M. A., Goodfellow M., Eguchi T., Bramley P. M. Metabolite profiling of plant carotenoids using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Plant Journal. 2007;49:552–564. doi: 10.1111/j.1365-313X.2006.02949.x. [DOI] [PubMed] [Google Scholar]
- Fukui Y., Tanaka Y., Kusumi T., Iwashita T., Nomoto K. A rationale for the shift in colour towards blue in transgenic carnation flowers expressing the flavonoid 3′,5′-hydroxylase gene. Phytochemistry. 2003;63:15–23. doi: 10.1016/S0031-9422(02)00684-2. [DOI] [PubMed] [Google Scholar]
- Giovinazzo G., D’Amico L., Paradiso A., Bollino R., Sparvoli F., DeGara L. Antioxidant metabolite profiles in tomato fruit constitutively expressing the grapevine stilbene synthase gene. Plant Biotechnology Journal. 2005;3:57–69. doi: 10.1111/j.1467-7652.2004.00099.x. [DOI] [PubMed] [Google Scholar]
- Goldsbrough A. P., Tong Y., Yoder J. I. LC as a non-destructive visual reporter and transposition excision marker gene for tomato. Plant Journal. 1996;9:927–933. doi: 10.1046/j.1365-313X.1996.9060927.x. [DOI] [Google Scholar]
- Goodrich J., Carpenter R., Coen E. S. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68:955–64. doi: 10.1016/0092-8674(92)90038-E. [DOI] [PubMed] [Google Scholar]
- Grotewold E., Drummond B. J., Bowen B., Peterson T. The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- Grotewold E. The science of flavonoids. New York, NY: Springer Science+Business Media, Inc; 2006. [Google Scholar]
- Harborne J. B., Williams C. A. Advances in flavonoid research since 1992. Phytochemistry. 2000;55:481–504. doi: 10.1016/S0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Haytowitz, D. B., Eldridge, A. L., Bhagwat, S., Gebhardt, S. E., Holden, J. M., Beecher, G. R., Peterson, J., & Dwyer, J. (2003). International Research Conference on Food, Nutrition and Cancer. July 17–18, Washington DC.
- Heller W., Forkmann G. Biosynthesis of flavonoids. In: Harborn J. B., editor. The flavonoids: Advances in research since 1986. London: Chapman and Hall; 1993. pp. 399–425. [Google Scholar]
- Hertog M. G. L., Hollman P. C., Katan M. B., Kromhout D. Intake of potentially anticarcinogenic flavonoids and their determinants in adults in the Netherlands. Nutrition and Cancer. 1993;20:21–29. doi: 10.1080/01635589309514267. [DOI] [PubMed] [Google Scholar]
- Holton T. A., Cornish E. C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E. I., Kaneko M., Ohnishi Y., Horinouchi S. Production of plant specific flavanones by Escherichia coli containing an artificial gene cluster. Applied and Environmental Microbiology. 2003;69:2699–2706. doi: 10.1128/AEM.69.5.2699-2706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeandet P., Douillet-Breuil A.-C., Bessis R., Debord S., Sbaghi M., Adrian M. Phytoalexins from the Vitaceae: biosynthesis, phytoalexins gene expression in transgenic plants, antifungal activity and metabolism. Journal of Agricultural and Food Chemistry. 2002;50:2731–2741. doi: 10.1021/jf011429s. [DOI] [PubMed] [Google Scholar]
- Johnson H. E., Broadhurst D., Goodacre R., Smith A. R. Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry. 2003;62:919–928. doi: 10.1016/S0031-9422(02)00722-7. [DOI] [PubMed] [Google Scholar]
- Jones C. M., Mes P., Myers J. R. Characterization and Inheritance of the Anthocyanin fruit (Aft) tomato. Journal of Heredity. 2003;94:449–456. doi: 10.1093/jhered/esg093. [DOI] [PubMed] [Google Scholar]
- Joung J.-Y., Kasthuri M., Park J.-Y., Kang W.-J., Kim H.-S., Yoon B.-S., Joung H., Jeon J.-H. An overexpression of chalcone reductase of Pueraria montana var. lobata alters biosynthesis of anthocyanin and 5′-deoxyflavonoids in transgenic tobacco. Biochemical and Biophysical Research Communications. 2003;3003:326–331. doi: 10.1016/S0006-291X(03)00344-9. [DOI] [PubMed] [Google Scholar]
- Koes R. E., Quattrocchio F., Mol J. N. M. The flavonoid biosynthetic pathway in plants: Function and evolution. BioEssays. 1994;16:123–132. doi: 10.1002/bies.950160209. [DOI] [Google Scholar]
- Kuo S.-M. Dietary flavonoids and cancer prevention: evidence and potential mechanism. Critical reviews in Oncogenesis. 1997;8:47–69. doi: 10.1615/critrevoncog.v8.i1.30. [DOI] [PubMed] [Google Scholar]
- Le Gall G., DuPont M. S., Mellon F. A., Davis A. L., Collins G. J., Verhoeyen M. E., Colquhoun I. J. Characterization and content of flavonoid glycosides in genetically modified tomato (Lycopersicon esculentum) fruits. Journal of Agricultural Food and Chemistry. 2003;51:2438–2446. doi: 10.1021/jf025995e. [DOI] [PubMed] [Google Scholar]
- Levin I., lalazar A., Bar M., Schaffer A. A. Non GMO fruit factories:strategies for modulating metabolic pathways in the tomato fruit. Industrial Crops and Products. 2004;20:29–36. doi: 10.1016/j.indcrop.2003.12.010. [DOI] [Google Scholar]
- Lloyd A. M., Walbot V., Davis R. W. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science. 1992;258:1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- Long M., Millar D. J., Kimura Y., Donovan G., Rees J., Fraser P. D., Bramley P. M., Bolwell G. P. Metabolite profiling of carotenoid and phenolic pathways in mutant andtransgenic lines of tomato: Identification of a high antioxidant fruit line. Phytochemistry. 2006;67:1750–1757. doi: 10.1016/j.phytochem.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Lukačin R., Wellmann F., Britsch L., Martens S., Matern U. Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry. 2003;62:287–292. doi: 10.1016/S0031-9422(02)00567-8. [DOI] [PubMed] [Google Scholar]
- Martens S., Teeri T., Forkmann G. Heterologous expression of dihydroflavonol 4-reductases from various plants. FEBS Letters. 2002;531:453–458. doi: 10.1016/S0014-5793(02)03583-4. [DOI] [PubMed] [Google Scholar]
- Martens S., Forkmann G., Britsch L., Wellmann F., Matern U., Lukačin R. Divergent evolution of flavonoid 2-oxoglutarate-dependent dioxygenase in parsley. FEBS Letters. 2003;544:93–98. doi: 10.1016/S0014-5793(03)00479-4. [DOI] [PubMed] [Google Scholar]
- Martin C., Prescott A., Mackay S., Bartlett J., Vrijlandt E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant Journal. 1991;1:37–49. doi: 10.1111/j.1365-313X.1991.00037.x. [DOI] [PubMed] [Google Scholar]
- Martin C., Jin H., Schwinn K. Mechanisms and applications of transcriptional control of phenylpropanoid metabolism. In: Romeo J. T., Saunders J. A., Matthews B. F., editors. Regulation of phytochemicals by molecular techniques. Oxford: Elsevier Science Ltd; 2001. pp. 179–195. [Google Scholar]
- Mathews H., Clendennen S. K., Caldwell C. G., Liu X. L., Connors K., Matheis N., Schuster D. K., Menasco D. J., Wagoner W., Lightner J., Ry Wagner D. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification and transport. Plant Cell. 2003;15:1689–1703. doi: 10.1105/tpc.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memelink J., Menke F. L. H., van der Fits L., Kijne J. W. Transcriptional regulators to modify secondary metabolism. In: Verpoorte R., Alfermann W., editors. Metabolic engineering of plant secondary metabolism. Dordrecht: Kluwer Academic Publishers; 2000. pp. 111–125. [Google Scholar]
- Minoggio M., Bramati L., Simonetti P., Gardana C., Iemoli L., Santahgelo E., Mauri P. L., Spigno P., Soressi G. P., Pietta P. G. Polyphenol pattern and antioxidant activity of different tomato lines and cultivars. Annals of Nutrition and Metabolism. 2003;47:64–69. doi: 10.1159/000069277. [DOI] [PubMed] [Google Scholar]
- Moco S., Bino R. J., Vorst O., Verhoeven H. A., de Groot J., van Beek T. A., Vervoort J., de Vos C. H. R. A liquid chromatography—mass spectrometry—based metabolome database for tomato. Plant Physiology. 2006;11:1205–1218. doi: 10.1104/pp.106.078428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol J., Grotewold E., Koes R. How genes paint flowers and seeds. Trends in Plant Science. 1998;3:212–217. doi: 10.1016/S1360-1385(98)01242-4. [DOI] [Google Scholar]
- Mol J., Cornish E., Mason J., Koes R. Novel coloured flowers. Current Opinion in Biotechnology. 1999;10:198–201. doi: 10.1016/S0958-1669(99)80035-4. [DOI] [PubMed] [Google Scholar]
- Mooney M., Desnos T., Harrison K., Jones J., Carpenter R., Coen E. Altered regulation of tomato and tobacco pigmentation genes caused by the Delila gene of Antirrhinum. Plant Journal. 1995;7:333–339. doi: 10.1046/j.1365-313X.1995.7020333.x. [DOI] [Google Scholar]
- Muir S. R., Collins G. J., Robinson S., Hughes S., Bovy S., De Vos C. H., van Tunen A. J., Verhoeyen M. E. Overexpression of petunia chalcone isomerase in tomato results in fruit containing increased levels of flavonols. Nature Biotechnology. 2001;19:470–474. doi: 10.1038/88150. [DOI] [PubMed] [Google Scholar]
- Mustilli A. C., Fenzi F., Ciliento R., Alfano F., Bowler C. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. The Plant Cell. 1999;11:145–157. doi: 10.2307/3870847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Yonekura-Sakakibara K., Sato T., Kikuchi S., Fukui Y., Fukuchi-Mizutani M., Ueda T., Nakao M., Tanaka Y., Kusumi T., Nishino T. Aureusidin synthase: A polyphenol oxidase homolog responsible for flower coloration. Science. 2000;290:1163–1166. doi: 10.1126/science.290.5494.1163. [DOI] [PubMed] [Google Scholar]
- O’Prey J., Brown J., Fleming J., Harrison P. R. Effects of dietary flavonoids on major signal transduction pathways in human epithelial cells. Biochemistry and Pharmacology. 2003;66:2075–2088. doi: 10.1016/j.bcp.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Overy S. A., Walker H. J., Malone S., Howard T. P., Baxter C. J., Sweetlove L. J., Hill S. A., Quick W. P. Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. Journal of Experimental Botany. 2005;56:287–296. doi: 10.1093/jxb/eri070. [DOI] [PubMed] [Google Scholar]
- Pietta P.-G. Flavonoids as antioxidants. Journal of Natural Products. 2000;63:1035–1042. doi: 10.1021/np9904509. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J. F., Leppen H., Mol J., Koes R. E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell. 1993;5:1497–1512. doi: 10.1105/tpc.5.11.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F., Wing J. F., van der Woude K., Mol J. N. M., Koes R. Analysis of bHLH and MYB-domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant Journal. 1998;13:475–488. doi: 10.1046/j.1365-313X.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Ramsay N. A., Walker A. R., Mooney M., Gray J. C. Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Molecular Biology. 2003;52:679–688. doi: 10.1023/A:1024852021124. [DOI] [PubMed] [Google Scholar]
- Ranish J. A., Hahn S. Transcription: Basal factors and activation. Current Opinions Genes and Development. 1996;6:151–158. doi: 10.1016/S0959-437X(96)80044-X. [DOI] [PubMed] [Google Scholar]
- Rein D., Schijlen E., Kooistra T., Herbers K., Verschuren L., Hall R., Sonnewald U., Bovy A., Kleeman R. Transgenic flavonoid tomato intake reduces C-reactive protein in human C-reactive protein transgenic mice more than wild-type tomato. Journal of Nutrition. 2006;136:2331–2337. doi: 10.1093/jn/136.9.2331. [DOI] [PubMed] [Google Scholar]
- Rischer H., Oksman-Caldentey K.-M. Unintended effects in genetically modified crops: Revealed by metabolomics? Trends in Biotechnology. 2006;24:102–104. doi: 10.1016/j.tibtech.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Rice-Evans C. A., Millier N. J., Paganga G. Structure-antioxidant activity relationships of flavonoids and phenolic acids. Free Radical Biology & Medicine. 1995;20:933–956. doi: 10.1016/0891-5849(95)02227-9. [DOI] [PubMed] [Google Scholar]
- Rice-Evans C. A., Miller N. J., Paganga G. Antioxidant properties of phenolic compounds. Trends in Plant Science. 1997;2:2152–2159. [Google Scholar]
- Ross J. A., Kasum C. M. Dietary flavonoids: Bioavailability, metabolic effects, and safety. Annual Reviews in Nutrition. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- Sarkar F. H., Li Y. Soy isoflavones and cancer prevention. Cancer Investigation. 2003;21:817–818. doi: 10.1081/CNV-120023773. [DOI] [PubMed] [Google Scholar]
- Schauer N., Zamir D., Fernie A. R. Metabolic profiling of leaves and fruit of wild species tomato: A survey of the Solanum lycopersicum complex. Journal of Experimental Botany. 2005;56:297–307. doi: 10.1093/jxb/eri057. [DOI] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Roessner U., Gur A., Balbo I., Carrari F., Pleban T., Perez-Melis A., Bruedigam C., Kopka J., Willmitzer L., Zamir D., Fernie A. R. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nature Biotechnology. 2005;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Schijlen E., de Vos C. H. R, Jonker H., van den Broeck H., Molthoff J., van Tunen A., Martens S., Bovy A. Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnology Journal. 2006;4:433–444. doi: 10.1111/j.1467-7652.2006.00192.x. [DOI] [PubMed] [Google Scholar]
- Schijlen E. G. W. M., de Vos C. H. R., Martens S., Jonker H. H., Rosin F. M., Molthoff J. W., Tikunov Y. M., Angenent G. C., van Tunen A. J., Bovy A. G. RNAi silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiology. 2007;144:1520–1530. doi: 10.1104/pp.107.100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder J., Schröder G. Stilbene and chalcone synthases: related enzymes with key functions in plant-specific pathways. Zeitschrift Naturforschung. 1990;45:1–8. doi: 10.1515/znc-1990-1-202. [DOI] [PubMed] [Google Scholar]
- Sévenier R., van der Meer I. M., Bino R., Koops A. J. Increased production of nutriments by genetically engineered crops. Jouranl of American College of Nutrition. 2002;21:199S–204S. doi: 10.1080/07315724.2002.10719266. [DOI] [PubMed] [Google Scholar]
- Stobiecki M., Matysiak-Kata I., Frański R., Skała J., Szopa J. Monitoring changes in anthocyanin and steroid alkaloid glycoside content in lines of transgenic potato plants using liquid chromatography/mass spectrometry. Phytochemistry. 2002;62:959–969. doi: 10.1016/S0031-9422(02)00720-3. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Nakayama T., Yonekura-Sakakibara K., Fukui Y., Nakamura N., Yamaguchi M-A., Tanaka Y., Kusumi T., Nishino T. cDNA cloning, heterologous expression, and fuctional characterization of malonyl-Coenzyme A: Anthocyanidin 3-O-Glucoside-6′-O-malonyltransferase from Dahlia flowers. Plant Physiology. 2002;130:2142–2151. doi: 10.1104/pp.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., & Walker, A. R. (2006). Light induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol Preview. DOI:10.1104, 106.088104. [DOI] [PMC free article] [PubMed]
- Tanaka Y., Tsuda S., Kusumi T. Metabolic engineering to modify flower color. Plant and Cell Physiology. 1998;39:1119–1126. [Google Scholar]
- Tikunov Y., Lommen A., de Vos C. H. R., Verhoeven H. A., Bino R. J., Hall R. D., Bovy A. G. A novel approach for non-targeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiology. 2005;139:1125–1137. doi: 10.1104/pp.105.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y., Verstappen F., & Hall, R. D. (2006). A metabolic profiling of natural volatiles: Head space trapping GC/MS. In W. Weckwerth (Ed.), Plant metabolomics: Methods and protocols, methods in molecular biology, Vol. 358 (pp. 39–53). Humana Press: Totowa, USA. [DOI] [PubMed]
- Urbanczyk-Wochniak E., Fernie A. R. Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. Journal of Experimental Botany. 2005;56:309–321. doi: 10.1093/jxb/eri059. [DOI] [PubMed] [Google Scholar]
- USDA Database for the Flavonoid Content of Selected Foods. Agricultural Research Service, Beltsville Human Nutrition Research Center, US Department of Agriculture, Beltsville, MD, March 2003. Avaliable from: www.nal.usda.gov/fnic/foodcomp/Data/Flav/flav pdf. 2003 Mar.
- Van der Krol A. R., Lenting P. E., Veenstra J., Van der Meer I. M., Koes R. E., Gerats A. G. M., Mol J. N. M., Stuitje A. R. An anti-sense chalcone synthase gene in transgenic plants inhibits flower pigmentation. Nature. 1988;333:866–869. doi: 10.1038/333866a0. [DOI] [Google Scholar]
- Van Dross R., Xue Y., Knudson A., Pelling J. C. The chemopreventive bioflavonoid Apigenin modulates signal transduction pathways in keratinocyte and colon carcinoma cell lines. Journal of Nutrition. 2003;133:3800S–3804S. doi: 10.1093/jn/133.11.3800S. [DOI] [PubMed] [Google Scholar]
- Van Tuinen, A., de Vos, C. H. R., Hall, R. D., van der Plas, L. H. W., Bowler, C., & Bino, R. J. (2006). Use of metabolomics for identification of tomato genotypes with enhanced nutritional value derived from natural light-hyperresponsive mutants. In P. W. Jaiwal (Ed.), Plant genetic engineering and molecular farming, Vol. 7 (pp. 339–356). Houston: Studium Press.
- Verhoeven, H. A., De Vos, C. H. R., Bino, R. J., & Hall, R. D. (2006). Plant metabolomics strategies based upon qaudrupole time of flight mass spectrometry (QTOF-MS). In K. Saito, R. A. Dixon, & L. Willmitzer (Eds.), Biotechnology in agriculture and forestry, Vol. 57 (pp. 33–48). Germany: Springer.
- Verhoeyen M. E., Bovy A., Collins G., Muir S., Robinson S., de Vos C. H. R., Colliver S. Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. Jouranl of Experimental Botany. 2002;53:2099–2106. doi: 10.1093/jxb/erf044. [DOI] [PubMed] [Google Scholar]
- Vom Endt D., Kijne J. W., Memelink J. Transcription factors controlling plant secondary metabolism: What regulates the regulators? Phytochemistry. 2002;61:107–114. doi: 10.1016/S0031-9422(02)00185-1. [DOI] [PubMed] [Google Scholar]
- Willits M. G., Kramer C. M., Prata R. T. N., De Luca V., Potter B. G., Steffens J. C., Graser G. Utilization of the genetic resources of wild species to create a nontransgenic high flavonoid tomato. Jouranl of Agricultural and Food Chemistry. 2005;53:1231–1236. doi: 10.1021/jf049355i. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiology. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. S., Landau J. M., Huang M. T., Newmark H. L. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annual Review of Nutrition. 2001;21:381–406. doi: 10.1146/annurev.nutr.21.1.381. [DOI] [PubMed] [Google Scholar]
- Yen H. C., Shelton B. A., Howard L. R., Lee S., Vrebalov J., Giovannoni J. J. The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theoretical and Applied Genetics. 1997;95:1069–1079. doi: 10.1007/s001220050664. [DOI] [Google Scholar]
- Yu O., Shi J., Hession A. O., Maxwell C. A., McGonigle B., Odell J. T. Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry. 2003;63:753–763. doi: 10.1016/S0031-9422(03)00345-5. [DOI] [PubMed] [Google Scholar]
- Zuker A., Tzfira T., Ben-Meir H., Ovadis M., Shklarman E., Itzhaki H., Forkmann G., Martens S., Neta-Sharir I., Weiss D., Vainstein A. Modification of flower color and fragrance by antisense suppression of the flavanone 3-hydroxylase gene. Molecular Breeding. 2002;9:33–41. doi: 10.1023/A:1019204531262. [DOI] [Google Scholar]


