Abstract
Objectives
Autism spectrum disorders (ASD) are diagnosed based on early-manifesting clinical symptoms, including markedly impaired social communication. We assessed the viability of resting-state functional MRI (rs-fMRI) connectivity measures as diagnostic biomarkers for ASD and investigated which connectivity features are predictive of a diagnosis.
Methods
Rs-fMRI scans from 59 high functioning males with ASD and 59 age- and IQ-matched typically developing (TD) males were used to build a series of machine learning classifiers. Classification features were obtained using 3 sets of brain regions. Another set of classifiers was built from participants' scores on behavioral metrics. An additional age and IQ-matched cohort of 178 individuals (89 ASD; 89 TD) from the Autism Brain Imaging Data Exchange (ABIDE) open-access dataset (http://fcon_1000.projects.nitrc.org/indi/abide/) were included for replication.
Results
High classification accuracy was achieved through several rs-fMRI methods (peak accuracy 76.67%). However, classification via behavioral measures consistently surpassed rs-fMRI classifiers (peak accuracy 95.19%). The class probability estimates, P(ASD|fMRI data), from brain-based classifiers significantly correlated with scores on a measure of social functioning, the Social Responsiveness Scale (SRS), as did the most informative features from 2 of the 3 sets of brain-based features. The most informative connections predominantly originated from regions strongly associated with social functioning.
Conclusions
While individuals can be classified as having ASD with statistically significant accuracy from their rs-fMRI scans alone, this method falls short of biomarker standards. Classification methods provided further evidence that ASD functional connectivity is characterized by dysfunction of large-scale functional networks, particularly those involved in social information processing.
Keywords: Autism, Biomarkers, Machine learning classification, Social brain
Highlights
-
•
We distinguish rs-fMRI scans from ASD and TD individuals with high accuracy.
-
•
ASD versus TD classification using behavioral metrics was much more accurate.
-
•
Highly predictive brain features largely originated from the canonical social brain.
-
•
High performing brain features also correlated with individual symptom severity.
1. Introduction
Autism spectrum disorders (ASD) are clinically characterized by marked social and communication impairments as well as restricted interests and repetitive behaviors. Diagnosis is typically made in early childhood based on clinical interviews and observation of behavior. There is significant need for biomarkers to improve diagnostic precision when behavioral symptoms are equivocal and to identify infants or young children who might be at risk for ASD before reliable behavioral symptoms manifest (Yerys and Pennington, 2011).
Recent studies applied multivariate classification techniques to neuroimaging data to characterize ASD using features that are predictive of a diagnosis on the level of individuals. These classifier studies achieved relatively high classification accuracy (~60–85%) using multiple imaging modalities including structural MRI (Sato et al., 2013; Ecker et al., 2010), diffusion tensor MRI (DTI) (Ingalhalikar et al. 2012; Lange et al., 2010), magnetoencephalography (Roberts et al., 2011) and resting-state functional MRI (rs-fMRI; which measures “functional connectivity”, correlations between spontaneous BOLD signal fluctuations in different brain regions) (Uddin et al., 2013; Nielsen and Zielinski, 2013; Anderson et al., 2011). Rs-fMRI is a particularly interesting technique as it can investigate, in a task-independent manner, the hypothesis that ASD involves the disruption of large-scale brain networks (Castelli et al., 2002; Belmonte et al., 2004). These multivariate techniques have provided convergent evidence about brain differences that underlie ASD and unveiled additional informative brain features.
Given the recent success of these neuroimaging methods, it is tempting to cite these findings as grounds for establishing a neuroimaging-based diagnostic biomarker for ASD. However, several benchmarks must be met to fulfill the promise of neuroimaging-based biomarkers including: establishing standard analytic techniques, as such methodological factors influence connectivity measures (Jo et al., 2013; Gotts et al. 2013; Power et al., 2014); demonstrating biomarkers' robustness to variability across larger numbers of individuals and sites—to date, only one multisite classifier study exists (Nielsen, and Zielinski, 2013); and addressing the diagnostic potential of brain-based biomarkers by comparing their diagnostic or prognostic accuracy to that of simpler, more easily obtained ratings of behavior. The present study examines each of these issues.
In this study, we determined the best methods for performing classification of ASD vs. TD participants using rs-fMRI data by applying several popular classification techniques to three separate sets of brain-based features. We also addressed classifier generalizability by including a large in-house cohort of high-functioning ASD individuals and typically developing (TD) individuals (118 total participants) and a replication cohort obtained from the ABIDE dataset (178 individuals). Given similar accuracies achieved using different methods in previous rs-fMRI ASD classification studies (Uddin et al., 2013; Nielsen, and Zielinski, 2013; Anderson et al., 2011) we expected that there would be little effect of classifier method or brain region set.
Second, to determine the upper bounds of diagnostic performance using machine learning classification, we determined whether classification algorithms based on rs-fMRI data perform comparably to classifiers based on questionnaire data from the Social Responsiveness Scale (SRS) (Constantino, and Gruber, 2005). This questionnaire was expected to be highly predictive of ASD diagnoses as it is a measure of social functioning, the hallmark deficit in ASD. While SRS has been validated relative to “gold standard” interview and observation schedules, this measure is independent of the actual diagnosis criteria (Lord et al., 1994; Lord et al., 2000). The action of classifying participants as having a disorder characterized by social functioning deficits based on a measure of social functioning may be somewhat circular in its logic; however, the simplicity of the SRS and the ease of its administration make it an important benchmark of diagnostic utility for rs-fMRI based classification. In addition, such a behavioral classifier provides a more realistic ceiling of classifier performance that is tailored to the dataset in question. It is important to clarify that the SRS cannot be a biomarker as it is a clinical measure of social impairment designed to interrogate autistic symptoms. Performing classification on these measures simply gives an estimate of how well these individuals can be distinguished using a continuous measure of behavior that is independent of the diagnosis itself.
Finally, we investigated which connectivity features and brain networks are most predictive of ASD and further, which connections track individual symptom expression. We identified a disperse set of connections throughout the brain that were highly predictive of an ASD diagnosis. Classification accuracy increased by including regions beyond those seen in meta-analyses of task-based fMRI studies.
2. Methods and materials
2.1. Participants
2.1.1. NIMH
Fifty-nine typically developing (TD) male participants (mean age ± standard deviation (SD) = 18.3 ± 3.05) and 59 high-functioning participants with an autism spectrum disorder (ASD, mean age ± SD = 17.66 ± 2.72) took part in the study, including 29 ASD and 28 TD participants previously described (Gotts et al., 2012). Participants with ASD were recruited from the Washington, DC, metropolitan area and met Diagnostic and Statistical Manual-IV diagnostic criteria as assessed by an experienced clinician. Scores on the SRS (Constantino, and Gruber, 2005), an informant-based rating scale used to assess social and communication traits quantitatively, were obtained from parents for all ASD participants and 45 TD participants. Participant groups did not differ in terms of full-scale IQ or age (Table 1). Informed assent and consent were obtained from all participants and/or their parent/guardian when appropriate in accordance with the National Institutes of Health Institutional Review Board approved protocol. See Appendix A.1 and Table 1 for further details.
Table 1.
Demographic characteristics of in-house cohort.
| TD (N = 59) |
ASD (N = 59) |
|||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Age | 18.3 | 3.05 | 17.66 | 2.72 |
| IQ | 115.76 | 11.70 | 111.02 | 15.87 |
| ADOS: soc + comm | 11.69 | 4.16 | ||
| SRS | 19.82 | 11.54 | 91.75 | 30.20 |
| Whole brain tSNR | 324.59 | 53.66 | 314.37 | 36.94 |
| Average head movement (per TR) | 0.047 | .0019 | 0.069 | .042 |
2.1.2. ABIDE
The ABIDE dataset is an open-access multi-site image repository comprising structural and rs-fMRI scans from ASD and TD individuals (Di Martino et al., 2014). Acquisition parameters and protocol information can be found at http://fcon_1000.projects.nitrc.org/indi/abide/. Data from three of the five sites with the most subjects that met the following criteria were included in our analyses: males with a full-scale IQ > 80 and age within one standard deviation of the range of our in-house sample. Other sites were excluded due to excessive difficulties with anatomical FreeSurfer parcellation. The included sites were New York University (NYU), University of Utah School of Medicine (USM), and University of California Los Angeles 1 (UCLA_1). Participants were included if their scans met quality assurance standards (see Appendix A.2). These inclusion criteria and an additional step for matching ASD and TD prevalence resulted in a cohort of 178 individuals (89 TD; 89 ASD). Participant demographic and clinical data are provided in Inline Supplementary Table S1.
Inline Supplementary Table S1.
Table S1.
Demographic characteristics of ABIDE cohort.
| TD (N=89) |
ASD (N=89) |
|||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Age | 17.58 | 5.66 | 16.81 | 5.56 |
| IQ | 110.24 | 12.68 | 104.3 | 12.89 |
| ADOS: soc + comm | 11.89 | 3.34 | ||
| SRS | 17.62 | 13.34 | 93.48 | 32.62 |
| Whole brain tSNR | 355.58 | 129.64 | 366.75 | 188.36 |
| Average head movement (per TR) | 0.07 | .031 | 0.077 | .035 |
2.2. fMRI acquisition
Functional MRI data were collected using a GE, Signa 3T whole-body MRI scanner at the NIH Clinical Center NMR Research Facility. For each participant, a high-resolution T1-weighted anatomical image (MPRAGE) was obtained (124 axial slices, 1.2 mm slice thickness, field of view = 24 cm, 224 × 224 acquisition matrix). Spontaneous brain activity was measured during functional MRI using a gradient-echo echo-planar series with whole-brain coverage while participants maintained fixation on a central cross and were instructed to lie still and rest quietly (repetition time = 3500 ms, echo time = 27 ms, flip angle = 90°, 42 axial interleaved slices per volume, 3.0 mm slice thickness, field of view = 22 cm, 128 × 128 acquisition matrix, single-voxel volume = 1.7 × 1.7 × 3.0 mm). Each resting scan lasted 8 min, 10 s for a total of 140 consecutive whole-brain volumes. A GE 8-channel send–receive head coil was used for all scans, with a SENSE factor of 2 used to reduce gradient coil heating during the session.
2.3. fMRI preprocessing
fMRI data were preprocessed using AFNI software package (Cox, 1996) in accordance with pipelines recommended by Jo et al. (2013) with one exception: we did not employ cardiac and respiratory denoising so that a common preprocessing pipeline could be used on ABIDE data that lacked physiological measures. See Appendix A.2 for further details.
2.4. Connectivity measures and feature matrices
Three sets of regions of interest (ROIs) were used to create three separate fMRI timecourse correlation matrices for subjects' processed EPI time series. These ROI sets included one set of 49 spherical regions (5 mm radius) derived from coordinates in Di Martino et al. (2009), one set of 264 spherical regions (5 mm radius) from Power et al. (2011) and one set of 162 cortical and subcortical ROIs from each subject's FreeSurfer Destrieux atlas anatomical segmentation. Timecourses were extracted and averaged within each region. Linear correlations were computed between the average timecourses of each region in a ROI set and Fisher transformed. For each ROI set, this process yielded a Ns × Nf feature matrix, F, for use in classification, where Ns = number of subjects and Nf = number of features (Fisher transformed correlation values). F has an associated label vector, L, containing the diagnoses of the participants (ASD or TD) coded as a binary variable.
Subjects' clinical and demographic data (age, IQ, and scores from SRS sub-scales for: social awareness, social cognition, social communication, social motivation, and autism mannerisms) were used to create an additional feature matrix for the NIMH cohort. TD participants who did not have an SRS score were excluded from this classifier.
2.5. Classification of region × region correlation matrices
Classification algorithms were implemented using Scikit-learn (Pedregosa et al., 2011). Leave-one-out (LOO) cross-validation was performed on each F using the following classification algorithms: Random forest (RF), K-Nearest Neighbor (KNN), Linear Support Vector Machines (L-SVM), Gaussian kernel support vector machines (rbf-SVM), L1-regularized logistic regression (L1LR), L2-regularized logistic regression (L2LR), Elastic-net-regularized logistic regression (ENLR), Gaussian Naïve Bayes (GNB), and Linear Discriminant Analysis (LDA). For each algorithm we report accuracy, sensitivity (proportion of ASD individuals correctly classified), specificity (proportion of TD individuals correctly classified), positive predictive value (PPV), and negative predictive value (NPV). Statistical significance was estimated using permutation tests. For the highest performing classifiers, cross-validation was repeated using stratified-3-fold and stratified-10-fold techniques. See Appendixes A.3 and A.4 for descriptions of cross-validation, hyperparameter tuning, and permutation testing.
3. Results
3.1. Comparison of classifier and feature set performance
In order to determine the best methods for classifying of ASD vs. TD participants using rs-fMRI, we performed LOO cross-validation on our in-house cohort using nine popular classification algorithms and features derived from three different ROI sets. High LOO cross-validation accuracy was achieved through several machine-learning algorithms across all three ROI sets (Table 2). However, two classification algorithms, L2LR (average accuracy 73.33%) and L-SVMs (average accuracy 73.89%), consistently performed the best. Additionally, the two larger ROI sets, the Power (264 regions) and Destrieux (162 regions) sets, yielded higher performing classifiers than the DiMartino (49 regions) ROI set.
Table 2.
Cross-validation performance for the in-house cohort using the DiMartino, Power, and Destrieux ROI sets as well as a behavioral classifier.
| Classifier type | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| LOO-cross-validation | |||||
| DiMartino ROI set | |||||
| RF | 66.67 | 71.67 | 64.67 | 65.15 | 68.52 |
| KNN | 60.83 | 70.00 | 51.67 | 59.15 | 63.27 |
| L-SVM | 69.17 | 71.67 | 66.67 | 68.25 | 70.18 |
| RBF-SVM | 66.67 | 76.67 | 56.67 | 63.89 | 70.83 |
| GNB | 60.83 | 73.33 | 48.33 | 58.67 | 64.44 |
| LDA | 66.67 | 68.33 | 65.00 | 66.13 | 67.24 |
| L1LR | 61.67 | 58.33 | 65.00 | 62.50 | 60.94 |
| L2LR | 67.50 | 66.67 | 68.33 | 67.80 | 67.21 |
| ENLR | 72.50 | 70.00 | 75.00 | 73.68 | 71.43 |
| Destrieux Atlas | |||||
| RF | 66.67 | 70.00 | 63.33 | 65.63 | 67.86 |
| KNN | 68.33 | 65.00 | 71.67 | 69.64 | 67.19 |
| L-SVM | 74.58 | 69.49 | 79.66 | 77.36 | 72.31 |
| RBF-SVM | 76.67 | 70.00 | 83.33 | 80.77 | 73.53 |
| GNB | 60.00 | 68.33 | 51.67 | 58.57 | 62.00 |
| LDA | 74.17 | 71.67 | 76.67 | 75.44 | 73.02 |
| L1LR | 70.83 | 75.00 | 66.67 | 69.23 | 72.72 |
| L2LR | 76.67 | 75.00 | 78.33 | 77.59 | 75.81 |
| ENLR | 72.50 | 71.67 | 73.33 | 72.88 | 72.13 |
| Power ROI set | |||||
| RF | 65.00 | 65.00 | 65.00 | 65.00 | 65.00 |
| KNN | 65.00 | 73.33 | 56.67 | 62.86 | 68.00 |
| L-SVM | 75.83 | 75.00 | 76.67 | 76.27 | 75.41 |
| RBF-SVM | 70.83 | 73.33 | 68.33 | 69.84 | 71.93 |
| GNB | 60.83 | 70.00 | 51.67 | 59.15 | 63.27 |
| LDA | 69.17 | 73.33 | 65.00 | 67.69 | 70.91 |
| L1LR | 65.83 | 70.00 | 61.67 | 64.62 | 67.27 |
| L2LR | 75.83 | 75.00 | 76.67 | 76.27 | 75.41 |
| ENLR | 72.50 | 75.00 | 70.00 | 71.43 | 73.68 |
| Behavior | |||||
| RF | 91.35 | 91.53 | 91.11 | 93.10 | 89.13 |
| KNN | 93.26 | 91.53 | 95.56 | 96.43 | 89.58 |
| L-SVM | 90.38 | 83.05 | 100.00 | 100.00 | 81.82 |
| RBF-SVM | 91.35 | 84.75 | 100.00 | 100.00 | 83.33 |
| GNB | 95.19 | 93.22 | 97.78 | 98.21 | 91.67 |
| LDA | 88.46 | 81.36 | 97.78 | 97.96 | 80.00 |
| L1LR | 93.27 | 93.22 | 93.33 | 94.83 | 91.30 |
| L2LR | 94.23 | 93.22 | 95.56 | 96.49 | 91.49 |
| ENLR | 95.19 | 94.92 | 95.56 | 96.55 | 93.48 |
| Stratified-10-fold cross-validation | |||||
| DiMartino ROI set | |||||
| L-SVM | 69.39 | 66.33 | 73.00 | 73.57 | 68.24 |
| L2LR | 67.65 | 67.67 | 67.33 | 66.95 | 69.52 |
| Destrieux atlas | |||||
| L-SVM | 74.55 | 71.67 | 77.00 | 81.51 | 75.12 |
| L2LR | 79.09 | 73.33 | 85.00 | 83.33 | 77.38 |
| Power ROI set | |||||
| L-SVM | 75.30 | 73.00 | 78.00 | 79.55 | 76.26 |
| L2LR | 73.56 | 72.67 | 74.00 | 77.67 | 76.07 |
| Behavior | |||||
| L-SVM | 91.36 | 84.67 | 100.00 | 100.00 | 84.81 |
| L2LR | 94.27 | 93.33 | 95.50 | 96.90 | 92.33 |
| Stratified-3-fold cross-validation | |||||
| DiMartino ROI set | |||||
| L-SVM | 72.05 | 79.56 | 64.56 | 69.19 | 75.98 |
| L2LR | 65.34 | 67.72 | 62.54 | 66.18 | 65.94 |
| Destrieux atlas | |||||
| L-SVM | 72.93 | 71.40 | 74.65 | 73.70 | 73.00 |
| L2LR | 73.68 | 69.47 | 77.98 | 75.69 | 72.85 |
| Power ROI set | |||||
| L-SVM | 76.26 | 76.14 | 76.32 | 77.01 | 77.34 |
| L2LR | 74.57 | 78.07 | 71.23 | 74.17 | 77.25 |
| Behavior | |||||
| L-SVM | 94.26 | 93.33 | 95.56 | 96.75 | 92.98 |
| L2LR | 94.23 | 94.91 | 93.33 | 94.91 | 93.33 |
RF = Random Forests, KNN = K-Nearest Neighbor, L-SVM = Linear Support Vector Machine, RBF-SVM = Gaussian Kernel Support Vector Machine, GNB = Gaussian Naïve Bayes, LDA = Linear Discriminant Analysis, L1LR = L1 Logistic Regression, L2LR=L2 Logistic Regression, ENLR = Elastic-net Logistic Regression.
LOO cross-validation has high variance in its estimates of a classifier's true prediction error (Hastie et al., 2009), so we additionally performed stratified-10-fold and stratified-3-fold cross-validation for L2LR and L-SVM. Classifier performance remained stable (Table 1). All L2LR and L-SVM cross-validation accuracies were significant for each methodological variant (p < .001; chance accuracy 47.27–51.24%).
Behavioral features alone outperformed fMRI classifiers across all learning algorithms (peak accuracy 95.19%; Table 2). Once again, classification accuracy was highly significant (p < 0.001; chance accuracy 50.50–51.84%).
It is possible that the lower accuracies seen with fMRI-based classifiers were due to insufficient data to train the classifiers (“underfitting”). To address this problem, we repeated the analysis with a combined data pool of our in-house cohort and a matched cohort from the ABIDE dataset (combined Ns = 296). Cross-validation accuracy did not improve (Inline Supplementary Table S2). Cross-site differences, in either true underlying correlations or in nuisance variables, caused problems for these classifiers (see Inline Supplementary Fig. S1). Removing linear effects of site identity from correlation matrices or correcting for global level of correlation prior to cross-validation did not improve performance. There were no differences in temporal signal-to-noise ratio (tSNR) between ASD and TD scans in either the NIMH or the ABIDE cohorts. Cross-validation performance on the ABIDE dataset was markedly lower than on the combined datasets or in-house dataset (Inline Supplementary Table S3). This suggests that sample size may not be the sole factor limiting accuracy; across all methods rs-fMRI-based classifiers plateaued around ~ 75% accuracy.
Inline Supplementary Table S2.
Table S2.
Cross-validation performance for the combined in-house and ABIDE cohort using the DiMartino, Power, and Destrieux ROI set.
|
LOO cross-validation | |||||
|---|---|---|---|---|---|
|
Classifier type |
Accuracy |
Sensitivity |
Specificity |
PPV |
NPV |
| DiMartino ROI set | |||||
| RF | 58.11 | 61.33 | 54.79 | 58.23 | 57.97 |
| KNN | 61.82 | 49.33 | 74.66 | 66.67 | 58.92 |
| L-SVM | 64.53 | 66.67 | 62.33 | 64.52 | 64.54 |
| RBF-SVM | 65.88 | 65.33 | 66.44 | 66.67 | 65.10 |
| GNB | 53.04 | 56.67 | 49.32 | 53.46 | 52.55 |
| LDA | 69.59 | 89.33 | 49.32 | 64.42 | 81.82 |
| L1LR | 67.23 | 65.33 | 69.18 | 68.53 | 66.01 |
| L2LR | 66.22 | 66.00 | 66.44 | 66.89 | 65.54 |
| ENLR | 66.55 | 66.67 | 66.44 | 67.11 | 65.99 |
| Power ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| RF | 55.41 | 53.33 | 57.53 | 56.34 | 54.55 |
| KNN | 63.85 | 54.00 | 73.97 | 68.07 | 61.02 |
| L-SVM | 73.65 | 75.33 | 71.92 | 73.38 | 73.94 |
| RBF-SVM | 73.31 | 74.80 | 71.77 | 73.08 | 73.55 |
| GNB | 53.16 | 56.00 | 49.32 | 52.17 | |
| LDA | 69.59 | 88.00 | 50.68 | 64.71 | 80.43 |
| L1LR | 72.30 | 68.67 | 76.03 | 74.64 | 70.25 |
| L2LR | 69.26 | 67.33 | 71.23 | 70.63 | 67.97 |
| ENLR | 67.22 | 65.33 | 69.18 | 68.53 | 66.01 |
| Destrieux atlas | Accuracy | Sensitivity | Specificity | PPV | NPV |
| RF | 63.51 | 65.33 | 61.64 | 63.64 | 63.38 |
| KNN | 58.45 | 56.67 | 60.27 | 59.44 | 63.38 |
| L-SVM | 73.65 | 74.00 | 73.29 | 74.00 | 73.29 |
| RBF-SVM | 70.61 | 68.00 | 73.29 | 72.34 | 69.03 |
| GNB | 50.68 | 60.67 | 40.41 | 51.12 | 50.00 |
| LDA | 72.97 | 92.00 | 53.42 | 66.90 | 86.67 |
| L1LR | 73.69 | 70.67 | 76.71 | 75.71 | 71.79 |
| L2LR | 75.00 | 73.33 | 76.71 | 76.39 | 73.68 |
| ENLR | 69.26 | 66.67 | 71.92 | 70.92 | 67.77 |
| Stratified -10-fold cross-validation | |||||
| DiMartino ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 68.62 | 69.33 | 67.90 | 68.75 | 68.79 |
| L2LR | 65.90 | 63.33 | 68.48 | 69.27 | 64.19 |
| Power ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 69.62 | 69.33 | 69.90 | 70.91 | 69.98 |
| L2LR | 68.55 | 70.00 | 67.14 | 69.73 | 68.62 |
| Destrieux atlas | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 69.87 | 70.00 | 69.90 | 70.73 | 70.26 |
| L2LR | 73.25 | 70.67 | 75.95 | 76.01 | 72.66 |
| Stratified-3-fold cross-validation | |||||
| DiMartino ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 66.22 | 63.33 | 67.11 | 67.03 | 65.77 |
| L2LR | 67.23 | 6.00 | 68.52 | 68.35 | 66.35 |
| Power ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 65.16 | 66.67 | 63.61 | 65.74 | 64.78 |
| L2LR | 67.57 | 65.33 | 69.88 | 69.10 | 66.31 |
| Destrieux atlas | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 67.21 | 62.00 | 72.56 | 71.05 | 65.11 |
| L2LR | 71.27 | 68.67 | 73.92 | 73.21 | 69.90 |
Inline Supplementary Fig. S1.
Fig. S1.
Connectivity matrices from the two subject cohorts show significant whole-brain differences during ASD vs. TD comparisons. A series of thresholded t-tests (p < .005) on the connectivity matrices from the DiMartino (left column), Power (middle column), and Destrieux (right column) ROI sets are shown. Matrices show t-values from the comparisons. Comparing all of the NIMH data to all of the ABIDE data (top row), regardless of ASD or TD distinction, reveals whole-brain connectivity differences in all three ROI sets. The t-test of ASD-TD reveals incongruent group differences in the NIMH cohort (middle row) and ABIDE cohort (bottom row).
Inline Supplementary Table S3.
Table S3.
Cross-validation performance for the ABIDE cohort using the DiMartino, Power, and Destrieux ROI sets.
|
LOO cross-validation | |||||
|---|---|---|---|---|---|
|
Classifier type |
Accuracy |
Sensitivity |
Specificity |
PPV |
NPV |
| DiMartino ROI set | |||||
| RF | 63.48 | 67.03 | 59.77 | 63.54 | 63.41 |
| KNN | 53.93 | 34.07 | 74.71 | 58.49 | 52.00 |
| L-SVM | 67.98 | 72.53 | 63.22 | 67.35 | 68.75 |
| RBF-SVM | 70.22 | 71.43 | 68.97 | 70.65 | 69.77 |
| GNB | 58.43 | 65.93 | 50.57 | 58.25 | 68.67 |
| LDA | 65.17 | 64.84 | 65.52 | 66.29 | 64.04 |
| L1LR | 66.29 | 61.54 | 71.26 | 69.14 | 63.92 |
| L2LR | 69.10 | 73.63 | 64.37 | 68.37 | 70.00 |
| ENLR | 65.17 | 65.93 | 64.37 | 65.93 | 64.37 |
| Power ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| RF | 56.18 | 58.24 | 54.02 | 56.99 | 55.29 |
| KNN | 52.25 | 35.16 | 70.11 | 55.17 | 50.83 |
| L-SVM | 69.10 | 70.33 | 67.82 | 69.57 | 68.60 |
| RBF-SVM | 59.55 | 60.44 | 58.62 | 60.44 | 58.62 |
| GNB | 55.06 | 67.03 | 42.53 | 54.95 | 55.22 |
| LDA | 57.87 | 62.64 | 52.87 | 58.16 | 57.50 |
| L1LR | 56.74 | 57.14 | 56.32 | 57.78 | 55.68 |
| L2LR | 65.17 | 61.54 | 68.97 | 67.47 | 63.16 |
| ENLR | 52.81 | 56.04 | 49.43 | 53.68 | 51.81 |
| Destrieux atlas | Accuracy | Sensitivity | Specificity | PPV | NPV |
| RF | 48.31 | 49.45 | 47.13 | 49.45 | 47.13 |
| KNN | 56.74 | 36.26 | 78.16 | 63.46 | 53.97 |
| L-SVM | 70.79 | 71.43 | 70.11 | 71.43 | 70.11 |
| RBF-SVM | 66.85 | 68.13 | 65.52 | 67.39 | 66.28 |
| GNB | 53.93 | 64.84 | 42.53 | 54.13 | 53.62 |
| LDA | 59.55 | 63.74 | 55.17 | 59.80 | 59.26 |
| L1LR | 68.54 | 69.23 | 67.82 | 69.23 | 67.82 |
| L2LR | 71.35 | 70.33 | 72.41 | 72.73 | 70.00 |
| ENLR | 65.17 | 63.74 | 66.67 | 66.67 | 63.74 |
| Stratified-10-fold cross-validation | |||||
| DiMartino ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 65.13 | 64.67 | 65.14 | 66.13 | 65.13 |
| L2LR | 69.71 | 72.44 | 66.67 | 69.76 | 70.72 |
| Power ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 62.48 | 64.89 | 60.28 | 64.33 | 63.48 |
| L2LR | 62.39 | 58.44 | 66.81 | 65.52 | 60.74 |
| Destrieux atlas | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 66.90 | 68.11 | 65.69 | 67.18 | 67.96 |
| L2LR | 67.94 | 66.11 | 70 | 69.92 | 67.65 |
| Stratified-3-fold cross-validation | |||||
| DiMartino ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 61.24 | 63.87 | 58.62 | 61.59 | 62.44 |
| L2LR | 64.04 | 53.84 | 74.71 | 69.36 | 61.26 |
| Power ROI set | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 60.69 | 61.47 | 59.77 | 62.11 | 59.57 |
| L2LR | 59.00 | 52.76 | 65.52 | 61.27 | 57.20 |
| Destrieux atlas | Accuracy | Sensitivity | Specificity | PPV | NPV |
| L-SVM | 64.06 | 63.84 | 64.37 | 65.13 | 63.37 |
| L2LR | 62.37 | 58.28 | 66.67 | 64.59 | 60.49 |
Another possible explanation for the plateaued performance of the rs-fMRI classifiers is the high number of uninformative features included in the model. However, neither dimensionality reduction of the feature matrices (principal components analysis) nor univariate feature selection (t-test filters) markedly improved cross-validation accuracy (Inline Supplementary Table S4, Inline Supplementary Table S5).
Inline Supplementary Table S4.
Table S4.
Stratified-10-fold cross-validation accuracy using principal components analysis to reduce the dimensionality of the feature set.1
| Number of principal components |
|||||
|---|---|---|---|---|---|
| Classifier type | 1 | 5 | 10 | 50 | 100 |
| NIH dataset: | |||||
| Dimartino ROI set | |||||
| L-SVM | 51.59 | 60.23 | 62.50 | 70.30 | 69.24 |
| L2LR | 58.41 | 68.48 | 68.79 | 70.38 | 69.47 |
| Power ROI set | |||||
| L-SVM | 52.42 | 65.61 | 68.79 | 72.95 | 74.62 |
| L2LR | 59.47 | 67.80 | 72.12 | 74.77 | 77.58 |
| Destrieux ROI set | |||||
| L-SVM | 51.59 | 71.21 | 71.97 | 76.36 | 75.61 |
| L2LR | 59.47 | 71.36 | 71.97 | 76.36 | 78.79 |
| NIH & ABIDE dataset: | |||||
| Dimartino ROI set | |||||
| L-SVM | 50.69 | 50.36 | 55.47 | 62.78 | 64.23 |
| L2LR | 48.00 | 51.66 | 59.78 | 61.18 | 66.93 |
| Power ROI set | |||||
| L-SVM | 50.69 | 56.43 | 57.70 | 63.86 | 70.25 |
| L2LR | 47.30 | 58.09 | 58.10 | 64.46 | 65.17 |
| Destrieux ROI set | |||||
| L-SVM | 50.69 | 51.37 | 59.09 | 66.22 | 66.53 |
| L2LR | 46.99 | 57.43 | 61.52 | 67.26 | 66.87 |
During each fold of cross-validation principal components analysis is performed on the training data and the testing data is projected onto the resulting components.
Inline Supplementary Table S5.
Table S5.
Stratified-10-fold cross-validation accuracy using t-test filters to select features during training.1
| Percent of total features kept in model |
||||||||
|---|---|---|---|---|---|---|---|---|
| Classifier type | 0.01% | 0.10% | 1% | 5% | 10% | 25% | 50% | 75% |
| NIH dataset: | ||||||||
| Dimartino ROI set | ||||||||
| L-SVM | 47.42 | 47.42 | 58.26 | 60.76 | 60.76 | 65.91 | 68.63 | 70.30 |
| L2LR | 50.00 | 50.98 | 50.15 | 61.06 | 65.38 | 70.30 | 69.62 | 70.23 |
| Power ROI set | ||||||||
| L-SVM | 59.09 | 58.26 | 67.05 | 67.88 | 69.39 | 74.39 | 73.63 | 74.70 |
| L2LR | 61.74 | 68.64 | 66.89 | 72.20 | 72.05 | 72.20 | 74.70 | 73.86 |
| Destrieux ROI set | ||||||||
| L-SVM | 54.92 | 60.76 | 74.47 | 72.88 | 69.39 | 70.98 | 75.53 | 75.15 |
| L2LR | 55.00 | 66.06 | 69.62 | 71.06 | 73.71 | 79.70 | 73.81 | 77.12 |
| NIH & ABIDE dataset: | ||||||||
| Dimartino ROI set | ||||||||
| L-SVM | 50.69 | 50.69 | 50.69 | 50.69 | 50.69 | 64.18 | 61.92 | 63.79 |
| L2LR | 50.00 | 54.72 | 61.89 | 63.17 | 60.02 | 65.24 | 66.83 | 66.28 |
| Power ROI set | ||||||||
| L-SVM | 50.69 | 50.69 | 65.13 | 65.17 | 63.83 | 68.21 | 67.59 | 68.91 |
| L2LR | 56.08 | 64.16 | 61.13 | 69.28 | 66.90 | 67.16 | 69.91 | 71.61 |
| Destrieux ROI set | ||||||||
| L-SVM | 50.69 | 50.69 | 61.57 | 67.60 | 66.31 | 69.31 | 69.55 | 68.29 |
| L2LR | 59.11 | 61.77 | 64.53 | 70.64 | 72.64 | 71.63 | 73.32 | 73.61 |
During each fold of cross-validation the training data is filtered to keep only the features that show differences between the ASD and TD scans (t-statistics). The classifier is only tested on these features as well.
3.2. Selection of most informative features
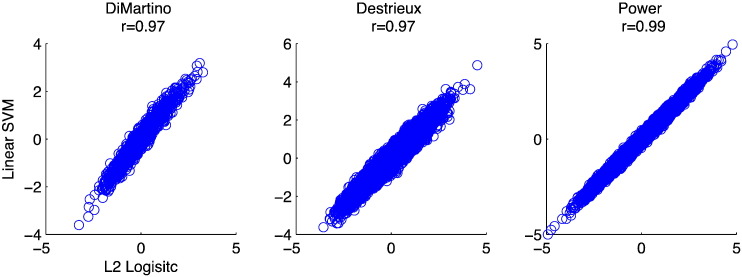
In order to determine which rs-fMRI features were most predictive of ASD or TD classification, we performed Recursive Feature Elimination (RFE) with stratified-10-fold cross-validation for the top performing classifiers (see Appendix A.5 for further details). L2LR and L-SVM performed comparably in cross-validation (Table 2, Inline Supplementary Table S2, Inline Supplementary Table S3). Feature weights for these two algorithms were also highly correlated (r > .97, Appendix B and Inline Supplementary Fig. S2) allowing us to choose one method for feature selection. As the variance in feature weights across cross-validation folds was smaller for L-SVM (4.68 × 10- 5) than for L2LR (1.47 × 10- 2), we only show feature weights associated with L-SVM (Fig. 1). The Power ROI set features (57 features, 0.164% of Nf; 85.10% accuracy) and Destrieux ROI set features (42 features, 0.322% of Nf ; 87.71% accuracy) showed the highest discriminability. We did not use RFE to boost classification accuracy, thus accuracies in this section simply measure the optimal feature subset’s ability to discriminate the two classes; they do not reflect the overall cross-validation accuracy. This measure is more akin to classifier training accuracy and does not directly estimate how predictive these features would be in a novel dataset. For the behavioral classifiers, we found that SRS social motivation, social cognition, and autism mannerisms sub-scales scores were the most predictive across all learning algorithms that weight features (Inline Supplementary Table S6). In addition, demographic features (age and IQ) were not predictive of diagnosis. In the next section, we characterize these highly informative regions and features on the level of interacting large-scale networks.
Inline Supplementary Fig. S2.
Fig. S2.
Feature weights from the top performing linear classifiers are highly correlated. The Spearman correlation coefficient for each pair comparison is shown above the scatter plots (p < 10− 15).
Fig. 1.
Optimal feature subsets chosen via recursive feature elimination (RFE) for each ROI set by L-SVM. The feature weights shown are the average weights from LOO cross-validation. Spheres are centered at the ROI's center of mass, and sphere radius represents the number of features coincident on that region. Comparing sphere radii across ROI sets is not advised due to the difference in the number of regions, in the number of features chosen, and in the cross-validation accuracy stated in the text. Edge thickness indicates absolute value of feature weight in the L-SVM, and color indicates the sign of the feature. ‘Hotter’ edges indicate stronger connectivity in ASD individuals while ‘cooler’ edges represent indicate stronger connectivity in TD individuals.
Inline Supplementary Table S6.
Table S6.
Magnitude of L2-normalized mean feature weights from LOO cross-validation for behavioral classifiers.1
| SRS |
|||||||
|---|---|---|---|---|---|---|---|
| Classifier type | Age | IQ | Social awareness | Social cognition | Social communication | Social motivation | Autism mannerisms |
| RF | .045 | .051 | .165 | .498 | .394 | .567 | .494 |
| L-SVM | .031 | .006 | .324 | .433 | .431 | .488 | .532 |
| L1LR | .092 | .000 | .354 | .562 | .287 | .543 | .416 |
| L2LR | .072 | .022 | .223 | .573 | .257 | .573 | .471 |
| EN | .046 | .005 | .117 | .594 | .280 | .598 | .441 |
| Mean | .057 | .017 | .237 | .532 | .330 | .554 | .471 |
For each classifier mean magnitude of feature weights are computed across LOO cross-validation folds and then scaled by the L2-norm of the resultant vector of feature weights.
3.3. Functional networks associated with top features
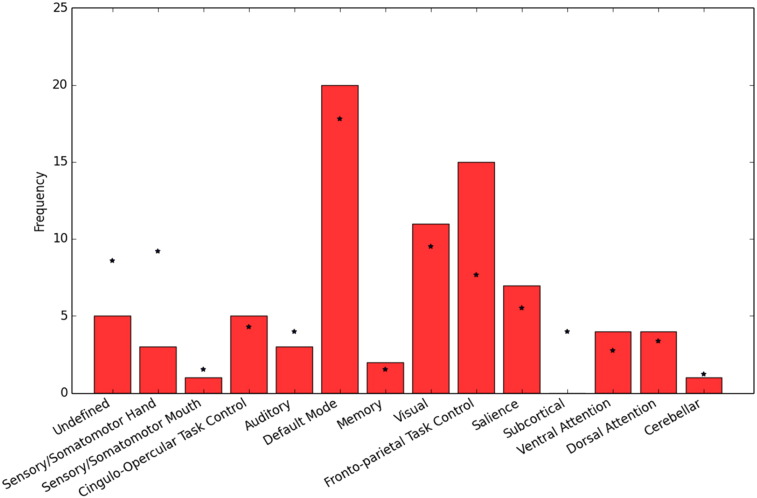
In the DiMartino and Power ROI sets but not the Destrieux ROI set, each ROI is associated with a functional network or task label. DiMartino et. al. labeled ROIs as being derived from either social or non-social tasks and by direction of group differences, yielding 4 possible labels for each region. Power et al labeled ROIs as belonging to 1 of 13 functional networks. In order to test whether functional networks, or connections between networks, were overrepresented in the optimal subset of features, we ran a χ2 test with permutations testing (see Appendix A.6 for further details). No regions or connections across regions were significantly overrepresented in the DiMartino feature set. For the Power ROI set, regions designated as the ‘default-mode network’ and the ‘frontal-parietal control network’ were more prevalent in the optimal subset of features (p < .01) (Inline Supplementary Fig. S3). See Table 3 for a list of the most diagnostic ROIs from the Power ROI set.
Inline Supplementary Fig. S3.
Fig. S3.
Default mode network and frontal–parietal control network regions are highly prevalent in the most important features from the Power ROI set. The frequency of regions within particular functional networks among the top ranked features (red bars) chosen by RFECV are plotted along with the expected frequency of these functional networks if features were chosen at random (*s). The observed frequency of regions was significantly different from the expected frequency (p < .01).
Table 3.
Ranking of regions from the Power ROI set based on the sum of the absolute value of the feature weights coincident on that region chosen by RFE.
| Region rank | Talairach coordinates |
Region label | (103) ∑ |RFE feature weights| | Sign of feature weights to regiona | ||
|---|---|---|---|---|---|---|
| x | y | z | ||||
| 1 | −46 | −60 | 16 | L posterior STS/TPJ | 31.61 | − |
| 2 | 36 | 10 | 1 | R Insula | 18.77 | − |
| 3 | −42 | 44 | 1 | L IFG | 17.23 | + |
| 4 | 31 | 31 | 25 | R MFG | 15.58 | + |
| 5 | 10 | −20 | 67 | R SMA | 13.97 | − |
| 6 | 13 | −4 | 64 | R SMA | 13.71 | − |
| 7 | −44 | 0 | 42 | L precentral gyrus | 13.35 | + |
| 8 | −51 | −61 | 2 | L posterior MTG | 11.94 | − |
| 9 | −42 | 36 | 21 | L MFG | 11.92 | − |
| 10 | 5 | 21 | 35 | R ACC | 11.85 | + |
| 11 | 33 | −53 | 38 | R IPS | 11.44 | ± |
| 12 | 54 | −45 | 32 | R supramarginal gyrus | 11.44 | + |
| 13 | −57 | −29 | −5 | L STS | 11.33 | − |
| 14 | 36 | 21 | 4 | R anterior insula | 11.03 | − |
| 15 | 27 | −35 | −13 | R parahippocampal gyrus | 10.91 | − |
| 16 | 37 | −82 | 8 | R MOG | 10.79 | − |
| 17 | −11 | −55 | 12 | L PCC | 10.52 | ± |
| 18 | −46 | 31 | −9 | L IFG | 10.32 | − |
| 19 | −3 | 23 | 42 | L dorsomedial prefrontal gyrus | 10.32 | + |
| 20 | −3 | 40 | 17 | L ACC | 10.21 | + |
| 21 | 6 | 65 | 0 | R ventromedial prefrontal cortex | 9.34 | + |
| 22 | −52 | −49 | 37 | L supramarginal gyrus | 9.07 | − |
| 23 | 11 | −66 | 35 | R precuneus | 8.96 | + |
| 24 | 43 | −71 | 22 | R MOG | 8.78 | ± |
| 25 | 46 | 17 | −24 | R temporal pole | 8.13 | − |
| 26 | −28 | 49 | 22 | L SFG | 8.05 | + |
| 27 | −20 | 42 | 38 | L SFG | 8.05 | + |
| 28 | 55 | −45 | 8 | R posterior STS | 7.42 | − |
| 29 | 57 | −51 | −14 | R posterior ITG | 6.80 | − |
| 30 | 44 | −54 | 41 | R IPL | 6.62 | + |
| 31 | 6 | 61 | 23 | R SFG | 6.36 | + |
| 32 | 22 | −65 | 41 | R superior parietal lobe | 6.28 | − |
| 33 | 22 | 36 | 38 | R MFG | 6.17 | + |
| 34 | −35 | 17 | 48 | L SFG | 6.10 | + |
| 35 | 51 | −1 | −14 | R MTG | 6.04 | − |
| 36 | −55 | −49 | 7 | L MTG | 5.91 | − |
| 37 | 65 | −9 | 23 | R precentral gyrus | 5.83 | − |
| 38 | 27 | −57 | −10 | R fusiform gyrus | 5.78 | − |
| 39 | −20 | 61 | 21 | L MFG | 5.75 | + |
| 40 | 38 | 41 | 16 | R MFG | 5.64 | + |
| 41 | −15 | −69 | −10 | L lingual gyrus | 5.60 | + |
| 42 | −33 | −76 | −15 | L fusiform gyrus | 5.60 | + |
| 43 | −17 | −60 | 56 | L superior parietal lobe | 5.56 | + |
| 44 | 46 | −57 | 1 | R posterior MTG | 5.45 | − |
| 45 | −8 | 45 | 23 | L medial frontal gyrus | 5.34 | + |
| 46 | 49 | 8 | 0 | R Insula | 5.30 | − |
| 47 | 48 | 21 | 10 | R IFG | 5.30 | − |
| 48 | 46 | −45 | −17 | R fusiform gyrus | 5.28 | − |
| 49 | 37 | −65 | 34 | R angular gyrus | 5.21 | + |
| 50 | 52 | 32 | 3 | R IFG | 5.21 | + |
| 51 | 53 | −43 | 18 | R TPJ | 5.18 | − |
| 52 | 24 | 44 | −10 | R lateral orbitofrontal cortex | 5.17 | + |
| 53 | 48 | 23 | 26 | R MFG | 5.15 | − |
| 54 | −42 | −55 | 39 | L IPL | 5.07 | + |
| 55 | −16 | −76 | 28 | L cuneus | 5.07 | + |
| 56 | 26 | 47 | 27 | R SFG | 5.03 | − |
| 57 | −38 | −33 | 14 | L posterior insula | 4.89 | − |
| 58 | 51 | 8 | −25 | R temporal lobe | 4.89 | − |
| 59 | 29 | −20 | 64 | R precentral gyrus | 4.78 | − |
| 60 | 24 | −85 | 18 | R MOG | 4.77 | − |
| 61 | −2 | −35 | 27 | L PCC | 4.73 | + |
| 62 | −41 | 4 | 31 | L inferior frontal junction | 4.73 | + |
| 63 | 8 | 47 | −10 | R ventromedial prefrontal gyrus | 4.71 | − |
| 64 | −39 | −75 | 37 | L angular gyrus | 4.70 | + |
| 65 | −53 | −24 | 38 | L postcentral gyrus | 4.67 | + |
| 66 | 64 | −22 | −17 | R ITG | 4.67 | + |
| 67 | −47 | −73 | −12 | L fusiform | 4.65 | + |
| 68 | −40 | −85 | −9 | L inferior occipital gyrus | 4.65 | + |
| 69 | −59 | −25 | 12 | L STG | 4.60 | + |
| 70 | 29 | −76 | 19 | R MOG | 4.50 | − |
| 71 | −42 | 23 | 29 | L MFG | 4.48 | + |
| 72 | 53 | −29 | 30 | R IPL | 4.47 | − |
| 73 | −10 | 51 | 39 | L SFG | 4.47 | − |
| 74 | 17 | −76 | −32 | R cerebellum lobule VIIa | 4.41 | − |
| 75 | −50 | −34 | 22 | L IPL | 4.36 | + |
| 76 | 34 | 37 | −8 | R lateral orbitofrontal cortex | 4.36 | − |
| 77 | 17 | −88 | −16 | R occipital pole | 4.22 | + |
| 78 | 22 | −55 | −22 | R cerebellum lobule VI | 4.22 | + |
| 79 | 6 | −71 | 19 | R cuneus | 3.81 | − |
| 80 | 8 | −70 | 7 | R cuneus | 3.81 | − |
| 81 | −3 | −1 | 49 | L SMA | 3.79 | − |
IFG = inferior frontal gyrus, MFG = middle frontal gyrus, SFG = superior frontal gyrus, ITG = inferior temporal gyrus, MTG = middle temporal gyrus, STG = superior temporal gyrus, SMA = supplementary motor area, ACC = anterior cingulate cortex, PCC = posterior cingulate cortex, IPS = intraparietal sulcus, IPL = inferior parietal lobule, MOG = middle occipital gyrus.
While the majority of regions were associated with feature weights of the same sign (indicated by + or −), some regions were associated with both positive and negative feature weights (indicated by ±).
3.4. Classifier Results Correlate with Behavioral Ratings
In this final section, we test whether classifier techniques are sensitive to symptom expression beyond a binary ASD vs. TD decision by assessing whether feature weights correlated with participants’ SRS total sum scores.
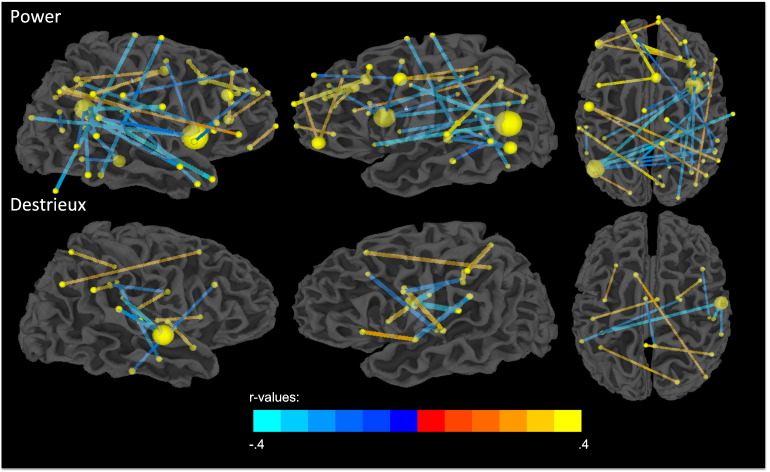
The probability estimate of a scan coming from an ASD or TD individual from the fMRI connectivity data alone derived from LOO cross-validation, P(ASD|fMRI data), was highly correlated subjects’ SRS sum scores (Table 4). L-SVMs trained and tested on the Power ROI set achieved the highest correlation with the SRS sum score (r = .51, p = 2.79 × 10- 8). Many top predictive features from the Power and Destrieux ROI sets also strongly correlated with SRS scores (Fig. 2). Correlation significance values were corrected for multiple comparisons (FDR25 < .05). The Power ROI set yielded a higher proportion of predictive features that were relevant to subject behavior (56.14% of most predictive features) than did the Destrieux ROI set (28.57% of most predictive features).
3.4. Classifier results correlate with behavioral ratings
In this final section, we test whether classifier techniques are sensitive to symptom expression beyond a binary ASD vs. TD decision by assessing whether feature weights correlated with participants' SRS total sum scores.
The probability estimate of a scan coming from an ASD or TD individual from the fMRI connectivity data alone derived from LOO cross-validation, P(ASD|fMRI data), was highly correlated with the subjects' SRS sum scores (Table 4). L-SVMs trained and tested on the Power ROI set achieved the highest correlation with the SRS sum score (r = .51, p = 2.79 × 10−8). Many top predictive features from the Power and Destrieux ROI sets also strongly correlated with SRS scores (Fig. 2). Correlation significance values were corrected for multiple comparisons (FDR (Benjamini, and Hochberg, 1995) < .05). The Power ROI set yielded a higher proportion of predictive features that were relevant to subject behavior (56.14% of most predictive features) than did the Destrieux ROI set (28.57% of most predictive features).
Table 4.
Spearman's correlation of classifier's probability of labeling a scan as ASD (P(ASD|fMRI data)) and participants' SRS scores.
| ROI set |
L2LR |
L-SVM |
||
|---|---|---|---|---|
| r | p | r | p | |
| DiMartino | 0.3195 | <.001 | 0.3169 | <.001 |
| Power | 0.4775 | ≪.001 | 0.5119 | ≪.001 |
| Destrieux | 0.4187 | ≪.001 | 0.4928 | ≪.001 |
Fig. 2.
The most predictive features from the Power and Destrieux ROI sets correlate with subjects' SRS sum scores. Spheres are centered at each ROI's center of mass, and sphere radius represents the number of significantly correlated features coincident on that region. Edge thickness indicates absolute value of the r-statistic. Edge color indicates the sign and magnitude of the r-statistic. Cooler colors indicate a negative correlation while warmer colors indicate a positive correlation. All correlations are significant at FDR < .05.
4. Discussion
In this study, we addressed the impact of classifier algorithm and ROI set on classification accuracy of rs-fMRI applied to ASD. Contrary to our prediction that there would be little effect of ROI or classifier choice on classification performance, these factors substantially impacted classification accuracy. L2LR and L-SVMs were the most successful in classifying ASD individuals from TD individuals. Further, restricting analysis to loci of group differences in task-based studies of ASD populations greatly impaired accuracy.
We also found that accuracy achieved using simple behavior metrics, the SRS, far exceeded accuracy achieved with rs-fMRI. Nonetheless, classification techniques applied to rs-fMRI data still had demonstrable utility. These data-driven methods identified aberrant connections in ASD participants, revealing connections within specific networks that were predictive of an ASD diagnosis and correlated with symptom expression.
4.1. Implications for rs-fMRI as a diagnostic ASD biomarker
Despite promising results using rs-fMRI data to classify ASD and TD individuals' scans, we exercise substantial caution in heralding this technique as a potential ASD biomarker at this time for several reasons. The first reason is that rs-fMRI classification lacks the sensitivity and specificity of simple behavioral metrics (Table 1). We do not conclude that the SRS is a better “biomarker” than rs-fMRI metrics because SRS is in fact a behavioral measure of social impairment validated against “gold-standard” ASD clinical measures. By comparing rs-fMRI and behavioral classifiers, we show that a set of highly informative features independent of the diagnosis criteria can distinguish the individuals with high accuracy using the same statistical techniques. This procedure provides a classification benchmark more realistic than 100% accuracy. Thus, there is a gap in classification performance between the brain and behavior for which we must account. This gap remains when attempting to reduce the rs-fMRI classifiers to only the most relevant features (Fig. 1 and Inline Supplementary Table S5). The observed discrepancy between performance of brain-based and behavior-based classifiers does not appear to be specific to our cohort or methods. Our best-performing brain-based classifier (peak accuracy = 77%) performed on par with the two single-site rs-fMRI classifiers in the literature (peak accuracy (Anderson et al., 2011) = 78%; peak accuracy (Castelli et al., 2002) = 79%). This advantage of behavioral metrics may be due to many methodological and measurement factors including thermal scanner noise, stubborn head motion artifacts, non-uniform signal quality across the brain, and classifier overfitting. More intriguingly, however, the behavioral advantage may reflect neurobiological heterogeneity of ASD. The wide-range of symptom expression profiles seen in an ASD diagnosis may be due to neurobiological changes that are as variable or even more variable than the disorder itself, thus yielding an extremely difficult classification problem.
The second reason for our qualified excitement is the decline in classifier accuracy when algorithms are trained on data from multiple sites. Though our multi-site classifier (peak accuracy = 75%) substantially outperformed a previous multi-site classifier (peak accuracy (Belmonte et al., 2004) = 60%) (Inline Supplementary Table S2, Inline Supplementary Table S3), large univariate differences in connectivity strength exist between sites (Inline Supplementary Fig. S1). Further work is needed to isolate causes of site effects (e.g., hardware, acquisition parameter, or cohort effects). The third reason is that rs-fMRI classification studies with autism fail to perform on the same level as ASD classification studies using other modalities (i.e. behavior (Williams et al., 2013) structural and diffusion MRI, (Ecker et al., 2010; Uddin et al., 2011) and molecular and genetic screens (Hu, and Lai, 2013)). Finally, prior studies and ours have focused on individuals who were older than the age of typical diagnosis (~2–6 years (Kleinman et al., 2008)) or the age at which reliable behavioral symptoms are detectable (~1–3 years (Bolton et al., 2012)). Presently, it is unclear whether existing classifiers are merely detecting circuit-level consequences of living with ASD for many years. The possible age-dependence of rs-fMRI based classifiers may hinder the generalizability of a given classifier across age and underscores the need to evaluate developmental disorders in a developmental context (Dosenbach et al., 2010; Karmiloff-Smith, 2013). An ideal biomarker would be measurable prior to behavioral symptom onset (Yerys, and Pennington, 2011) and able to distinguish developmental disorders of different etiologies.
4.2. Functional networks disrupted in ASD individuals
Irrespective of the utility of rs-fMRI as an ASD diagnosis biomarker, classifier methods remain valuable tools for investigating brain circuitry in ASD. We found that a disperse set of connections was highly predictive of an ASD diagnosis. Including regions beyond those seen in meta-analyses of task-based fMRI studies increased classification accuracy (Table 1). This accuracy increase with increasing coverage is consistent with the idea that ASD involves disruptions of interacting large-scale brain networks (Gotts et al., 2012).
While both anatomically-defined (Destrieux) and functionally-defined (Power) whole-brain ROI sets performed similarly with regard to classification, the latter resulted in a higher proportion of features that correlated strongly with behavioral measures (Table 2 & Fig. 2). Functionally defined brain regions may be essential for characterizing functional connectivity differences in ASD individuals. Network labels assigned to regions from the Power ROI set enabled us to investigate canonical functional networks that showed atypical connectivity in ASD individuals. Fortuitously, the two best performing algorithms (L2LR and L-SVM) allow for the ready interpretation of feature weights, something that was not previously straightforward due to either feature selection methods (Nielsen, and Zielinski, 2013; Anderson et al., 2011) or choice of classifier algorithm. (Uddin et al., 2013)
Connections involving the putative ‘default mode’ and ‘frontal-parietal task control’ networks were most predictive of ASD. These results are consistent with group-level functional connectivity analyses of high-functioning ASD populations documenting reduced correlation in ASD among regions of these networks (Just et al., 2012; Schipul et al., 2012; Kana et al., 2006) many of which are known to be involved in multiple aspects of social functioning (Frith, and Frith, 2007; Adolphs, 2009). Specifically, regions identified in the present study as most predictive of ASD included the insula, ventromedial prefrontal cortex, anterior, middle, and posterior regions of cingulate cortex, supplementary motor cortex, anterior temporal lobes, posterior aspects of the fusiform gyrus, posterior superior temporal sulcus, temporal parietal junction, intraparietal sulcus, and inferior and middle frontal gyri, bilaterally. These results also converge with other imaging and histological investigations of ASD. Structural MRI studies found cortical thickness and volume differences between ASD and TD individuals in several posterior temporal and parietal regions of these two networks (Wallace et al., 2010). A recent post-mortem study also found laminar disorganization in the posterior superior temporal cortex of children and adolescents with ASD (Stoner et al., 2014). Finally, atypical activity during theory of mind and mentalizing tasks has been observed in many of these regions (Castelli et al., 2002; Lombardo et al., 2010). Thus, the general pattern of findings is consistent with the hypothesis that ASD involves a fractionation of circuits that underlie social processing from other functional networks (Gotts et al., 2012).
4.3. Future directions
The aforementioned limitations of rs-fMRI as a diagnostic biomarker of ASD should not preclude further exploration of this imaging technique and machine-learning methods. This approach may be well suited for predicting the trajectory of the disorder, identifying clinical subgroups on the basis of a functional connectivity phenotype, or identifying individuals who may be more responsive to treatment. Autism is a heterogeneous developmental disorder with a range of symptom expression profiles, and rs-fMRI may have a role in explicating the root of this heterogeneity. Autism studies using other modalities have already had success using techniques that may accomplish this task (Ingalhalikar et al. 2012; Hu, and Lai, 2013). We anticipate that basic and clinical advances will result from studying functional brain networks with multivariate techniques.
Acknowledgments
Thanks go to Steve Gotts, PhD for helpful discussion regarding methods and statistics. The authors declare no competing interests. This work was supported by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation (KAB) and by the Intramural Research Program at NIMH (AM). Ethics approval for this study was granted by the NIH Combined Neuroscience Institutional Review Board under protocol number 10-M-0027. The clinical trial number (clinicaltrials.gov) for this protocol is NCT01031407.
Previous presentation: This work is also to be presented as a poster at Society for Neuroscience, Washington, D.C., Nov. 15–19, 2014.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.nicl.2014.12.013.
Appendix A and B. Supplementary data
Supplementary material.
References
- Adolphs R. The social brain: neural basis of social knowledge. Annu. Rev. Psychol. 2009;60:693–716. doi: 10.1146/annurev.psych.60.110707.163514. 18771388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.S., Nielsen J.A., Froehlich A.L. Functional connectivity magnetic resonance imaging classification of autism. Brain. 2011;134(12):3742–3754. doi: 10.1093/brain/awr263. 22006979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte M.K., Allen G., Beckel-Mitchener A., Boulanger L.M., Carper R.A., Webb S.J. Autism and abnormal development of brain connectivity. J. Neurosci. 2004;24(42):9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. 15496656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Bolton P.F., Golding J., Emond A., Steer C.D. Autism spectrum disorder and autistic traits in the Avon Longitudinal Study of Parents and Children: precursors and early signs. J. Am. Acad. Child Adolesc. Psychiatry. 2012;51(3):249–260. doi: 10.1016/j.jaac.2011.12.009. 22365461 [DOI] [PubMed] [Google Scholar]
- Castelli F., Frith C., Happe F., Frith U. Autism, Asperger syndrome and brain mechanisms for the attribution of mental states to animated shapes. Brain. 2002;125(8):1839–1849. doi: 10.1093/brain/awf189. [DOI] [PubMed] [Google Scholar]
- Constantino J., Gruber C. Social Responsiveness Scale (SRS): Manual. West Psychol Serv; Los Angeles, CA: 2005. [Google Scholar]
- Cox R.W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996;29(3):162–173. doi: 10.1006/cbmr.1996.0014. 8812068 [DOI] [PubMed] [Google Scholar]
- Di Martino A., Ross K., Uddin L.Q., Sklar A.B., Castellanos F.X., Milham M.P. Functional brain correlates of social and nonsocial processes in autism spectrum disorders: an activation likelihood estimation meta-analysis. Biol. Psychiatry. 2009;65(1):63–74. doi: 10.1016/j.biopsych.2008.09.022. 18996505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A., Yan C.-G., Li Q. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry. 2014;19(6):659–667. doi: 10.1038/mp.2013.78. 23774715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach N.U., Nardos B., Cohen A.L. Prediction of individual brain maturity using fMRI. Science. 2010;329(5997):1358–1361. doi: 10.1126/science.1194144. 20829489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C., Rocha-Rego V., Johnston P. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage. 2010;49(1):44–56. doi: 10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Frith C.D., Frith U. Social cognition in humans. Curr. Biol. 2007;17(16):R724–R732. doi: 10.1016/j.cub.2007.05.068. 17714666 [DOI] [PubMed] [Google Scholar]
- Gotts S.J., Saad Z.S., Jo H.J., Wallace G.L., Cox R.W., Martin A. The perils of global signal regression for group comparisons: a case study of autism spectrum disorders. Front. Hum. Neurosci. 2013;7:356. doi: 10.3389/fnhum.2013.00356. 23874279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotts S.J., Simmons W.K., Milbury L.A., Wallace G.L., Cox R.W., Martin A. Fractionation of social brain circuits in autism spectrum disorders. Brain. 2012;135(9):2711–2725. doi: 10.1093/brain/aws160. 22791801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T., Tibshirani R., Friedman J. Vol. 763. 2009. (The Elements of Statistical Learning: Data Mining, Inference and Prediction). [Google Scholar]
- Hu V.W., Lai Y. Developing a predictive gene classifier for autism spectrum disorders based upon differential gene expression profiles of phenotypic subgroups. N. Am. J. Med. Sci. (Boston) 2013;6(3) doi: 10.7156/najms.2013.0603107. 24363828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M., Smith A.R., Bloy L., Gur R., Roberts T.P., Verma R. Identifying sub-populations via unsupervised cluster analysis on multi-edge similarity graphs. Med Image Comput Comput Assist Interv. 2012;15:254–261. doi: 10.1007/978-3-642-33418-4_32. 23286056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H.J., Gotts S.J., Reynolds R.C. Effective preprocessing procedures virtually eliminate distance-dependent motion artifacts in resting state FMRI. J. Appl. Math. 2013;2013 doi: 10.1155/2013/935154. 24415902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just M.A., Keller T.A., Malave V.L., Kana R.K., Varma S. Autism as a neural systems disorder: a theory of frontal-posterior underconnectivity. Neurosci. Biobehav. Rev. 2012;36(4):1292–1313. doi: 10.1016/j.neubiorev.2012.02.007. 22353426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kana R.K., Keller T.A., Cherkassky V.L., Minshew N.J., Just M.A. Sentence comprehension in autism: thinking in pictures with decreased functional connectivity. Brain. 2006;129(9):2484–2493. doi: 10.1093/brain/awl164. 16835247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmiloff-Smith A. Challenging the use of adult neuropsychological models for explaining neurodevelopmental disorders: developed versus developing brainsQ J Exp Psychol (Hove) 2013;66(1):1–14. doi: 10.1080/17470218.2012.744424. 23173948 [DOI] [PubMed] [Google Scholar]
- Kleinman J.M., Ventola P.E., Pandey J. Diagnostic stability in very young children with autism spectrum disorders. J. Autism Dev. Disord. 2008;38(4):606–615. doi: 10.1007/s10803-007-0427-8. 17924183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange N., Dubray M.B., Lee J.E. Atypical diffusion tensor hemispheric asymmetry in autism. Autism Res. 2010;3(6):350–358. doi: 10.1002/aur.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo M.V., Chakrabarti B., Bullmore E.T. Atypical neural self-representation in autism. Brain. 2010;133(2):611–624. doi: 10.1093/brain/awp306. 20008375 [DOI] [PubMed] [Google Scholar]
- Lord C., Risi S., Lambrecht L. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000;30(3):205–223. [PubMed] [Google Scholar]
- Lord C., Rutter M., Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J. Autism Dev. Disord. 1994;24(5):659–685. doi: 10.1007/BF02172145. 7814313 [DOI] [PubMed] [Google Scholar]
- Nielsen J.A., Zielinski B.A. Multisite functional connectivity MRI classification of autism: ABIDE results. Front. Hum. Neurosci. 2013;7(September):599. doi: 10.3389/fnhum.2013.00599. 24093016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F., Weiss R., Brucher M. Scikit-Learn.: Machine Learning in Python. 2011;12:2825–2830. [Google Scholar]
- Power J.D., Cohen A.L., Nelson S.M. Functional network organization of the human brain. Neuron. 2011;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. 22099467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J.D., Mitra A., Laumann T.O., Snyder A.Z., Schlaggar B.L., Petersen S.E. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. 23994314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.P., Cannon K.M., Tavabi K. Auditory magnetic mismatch field latency: a biomarker for language impairment in autism. Biol. Psychiatry. 2011;70(3):263–269. doi: 10.1016/j.biopsych.2011.01.015. 21392733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato J.R., Hoexter M.Q., Oliveira P.P.dM., Brammer M.J., Murphy D., Ecker C. Inter-regional cortical thickness correlations are associated with autistic symptoms: a machine-learning approach. J. Psychiatr. Res. 2013;47(4):453–459. doi: 10.1016/j.jpsychires.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Schipul S.E., Williams D.L., Keller T.A., Minshew N.J., Just M.A. Distinctive neural processes during learning in autism. Cereb. Cortex. 2012;22(4):937–950. doi: 10.1093/cercor/bhr162. 21725037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R., Chow M.L., Boyle M.P. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 2014;370(13):1209–1219. doi: 10.1056/NEJMoa1307491. 24670167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin L.Q., Menon V., Young C.B. Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biol. Psychiatry. 2011;70(9):833–841. doi: 10.1016/j.biopsych.2011.07.014. 21890111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin L.Q., Supekar K., Lynch C.J. Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry. 2013;70(8):869–879. doi: 10.1001/jamapsychiatry.2013.104. 23803651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace G.L., Dankner N., Kenworthy L., Giedd J.N., Martin A. Age-related temporal and parietal cortical thinning in autism spectrum disorders. Brain. 2010;133(12):3745–3754. doi: 10.1093/brain/awq279. 20926367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Perkins D., Wheeler D., Hayen A., Bayl V. Can questions about social interaction correctly identify preschool aged children with autism? J. Paediatr. Child. Health. 2013;49(2):E167–E174. doi: 10.1111/jpc.12092. 23350819 [DOI] [PubMed] [Google Scholar]
- Yerys B.E., Pennington B.F. How do we establish a biological marker for a behaviorally defined disorder? Autism as a test case. Autism Res. 2011;4(4):239–241. doi: 10.1002/aur.204. 21710504 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.