Abstract
Agr family includes three groups of genes, Ag1, Agr2 and Agr3, which encode the thioredoxin domain-containing secreted proteins and have been shown recently to participate in regeneration of the amputated body appendages in amphibians. By contrast, higher vertebrates have only Agr2 and Agr3, but lack Ag1, and have low ability to regenerate the body appendages. Thus, one may hypothesize that loss of Ag1 in evolution could be an important event that led to a decline of the regenerative capacity in higher vertebrates. To test this, we have studied now the expression and role of Ag1 in the regeneration of fins of a representative of another large group of lower vertebrates, the fish Danio rerio. As a result, we have demonstrated that amputation of the Danio fins, like amputation of the body appendages in amphibians, elicits an increase of Ag1 expression in cells of the stump. Furthermore, down-regulation of DAg1 by injections of Vivo-morpholino antisense oligonucleotides resulted in a retardation of the fin regeneration. These data are in a good agreement with the assumption that the loss of Ag1 in higher vertebrates ancestors could lead to the reduction of the regenerative capacity in their modern descendants.
Agr proteins belong to the superfamily of the thioredoxin domain-containing protein disulphide isomerases, but differ from most of the latter due to their ability to be secreted out of the cell1. Because of this property, Agrs can operate as non-cell-autonomous factors, regulating such processes as cell growth and differentiation during the embryonic development, tumorogenesis and regeneration2,3,4,5,6,7,8,9. In particular, it have been shown recently that all three sub-families of Agrs, Ag1, Agr2 and Agr3, are involved in regulation of the limb and tail regeneration in the Xenopus laevis tadpoles (Ivanova et al., 2013). Interestingly, as we have established in this work, higher vertebrates that demonstrate low ability to regenerate the body appendages, including reptiles, birds and mammals, have no genes of Ag1 subfamily, which were apparently lost by their ancestors. In connection with this, an attractive hypothesis might be formulated, according to which just the extinction of Ag1 during evolution could be one of the events that resulted in a reduced regenerative capacity of modern higher vertebrates. Accordingly, this hypothesis predicts that Ag1 could be also involved in the process of appendages regeneration in other lower vertebrates, which, by contrast to higher vertebrates, have this gene and demonstrate high regenerative capacity.
To test this prediction, we have investigated now the involvement of Ag1 in the process of fins regeneration in zebrafish (Danio rerio). By contrast to amphibians, Agr family in bony fishes, including Danio, is represented by only two genes: DAg1, that is lost in higher vertebrates, and DAgr2, which is a common ancestor of both Agr2 and Agr3 in amphibians and higher vertebrates5. The expression pattern of DAgr2 in Danio embryos and in adults was recently described10. As far as we know, however, there is no literature data on DAg1 expression, neither in embryogenesis, nor during regeneration of the amputated fins. Therefore in the present work, we first of all cloned DAg1 cDNA and investigated its expression by the whole mount in situ hybridization and qRT-PCR in embryonic development. In addition, we analyzed by the same methods the dynamics of DAg1 expression during regeneration of the amputated caudal, pectoral and pelvic fins in adult fishes. As a result, we have found out that the expression of DAg1 is well correlated with the regeneration process. Unlike the Xenopus Xag1/2, however, DAg1 is intensively expressed already in the epithelium of the intact fins, but the level of its expression drops down at the 1 day post amputation (1 dpa). After that, the expression of DAg1 begins to gradually increase, even exceeding by 5 dpa the initial level in the intact fins. Given that the expression of another fish's representative of Agr family, DAgr2, was not yet analyzed during fins regeneration, we performed in addition in the present work such an analysis, which yielded results very resembling those obtained for DAg1. Finally, by down-regulating DAg1 functioning by Vivo-morpholino antisense oligonucleotides, we demonstrate that this protein is essential for the fin regeneration. Thus, our data demonstrate tight involvement of both Agr genes in the process of fins regeneration, which in turn is in a good agreement with the hypothesis implying that the loss of Ag1 in higher vertebrates' ancestors could be a cause of the reduction of regenerative capacity in their modern descendants.
Results
DAg1 expression in embryonic development
We have shown recently that Danio rerio has, like other bony fishes, Agr genes of two sub-families: Ag1 (DAg1), that disappeared in higher vertebrates, and Agr2 (DAgr2)5. The expression of one of these genes, DAgr2, has been already studied (Shih et al., 2007). As far as we know, however, there are no reports describing the expression of DAg1. Therefore, we have analyzed the expression pattern of this gene in the Danio rerio embryonic development.
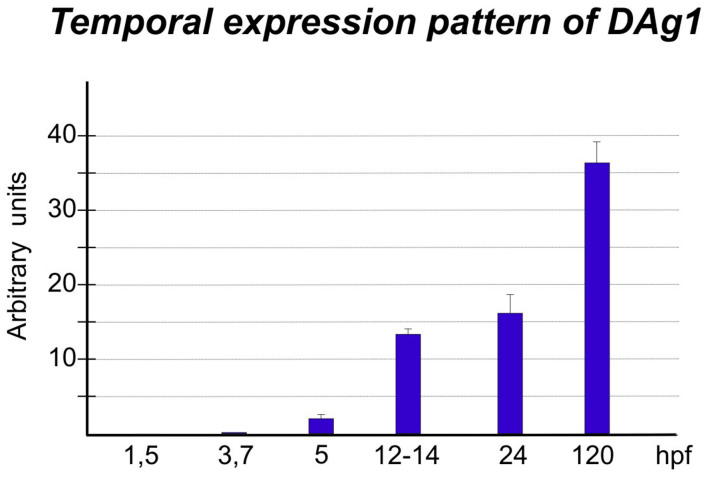
First of all, we have studied the temporal expression pattern of DAg1 by making qRT-PCR with specific primers at the following development stages: stage 8–16-cells, (1,5 hours post fertilization, cleavage period), oblong stage (3,7 hpf, blastula period), 40–50% epiboly stage (5 hpf, gastrula period), 6–12 somite stage (12–14 hpf, segmentation period), prim-5 stage (24 hpf, pharyngula period) and 120 hpf (larval period). As a result, we have found out that DAg1 expression starts during the blastula to gastrula transition period and then gradually increases till larval period (Fig. 1).
Figure 1. Temporal pattern of DAg1 expression in Danio rerio embryogenesis.
QRT-PCR of total RNA from embryos at the indicated stages with primers to cDNAs of DAg1. The data were normalized by using results of qRT-PCR with primers to transcripts of two housekeeping genes: Ef1-alpha and ODC (see Materials and Methods for details).
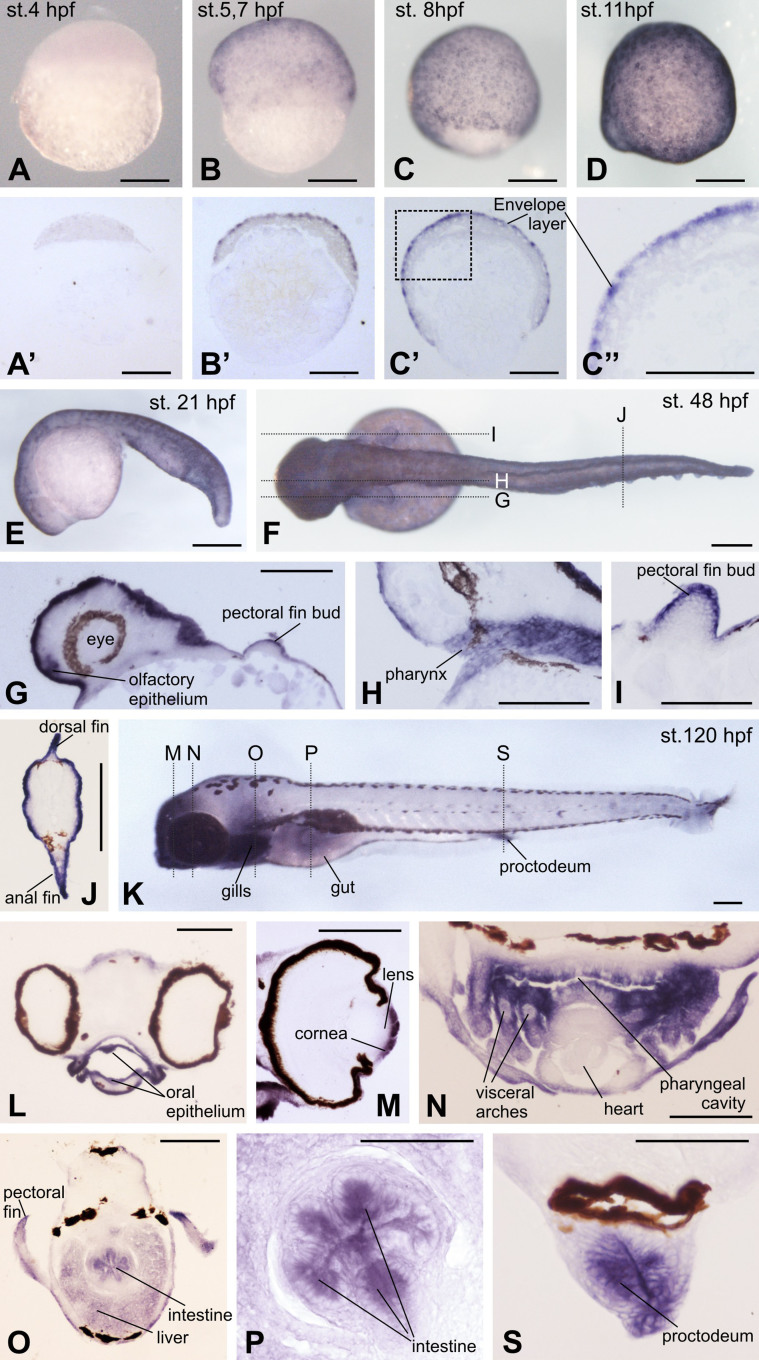
To study the spatial expression pattern of DAg1 during Danio rerio embryogenesis, we have used the method of the whole-mount in situ hybridization with the digoxygenin-labeled anti-sense RNA probe prepared on the base of the previously cloned cDNA template (see Materials and Methods). As a result, we have confirmed the data obtained by qRT-PCR by revealing a uniform expression of DAg1 in all cells of the surface layer of the embryonic ectoderm at stage 30–40% of epiboly (Fig. 2A and B) and then by observing progressive increase of the signal in these cells at later stages (Fig. 2A–F). To reveal possible sites of DAg1 expression in the inner structures, these embryos were embedded into paraplast and sectioned onto histological slices. This has revealed that at least before the pharyngula period, the expression of DAg1 could be seen exclusively in the outermost enveloping layer of the embryo (Fig. 2A′–C″). At hatching and larval period, however, the expression was revealed, besides the surface epithelium, in the pectoral fin buds, olfactory epithelium of the nasal pit, in the gill epithelium and in the epithelial cells lining the pharynx, the intestine and the proctodeum (Fig. 2G–J and K–S). Also, DAg1 is expressed at this period in such derivatives of the embryonic ectodermal epithelia as eye cornea (Fig. 2M).
Figure 2. Spatial expression pattern of DAg1 in Danio rerio embryos as revealed by in situ hybridization.
(A–D and E). Up to 21 hpf, DAg1 is expressed only in the superficial enveloping layer. (A′–C′). Histological sections of embryos shown on A–C. (C″). Enlarged fragment framed on C′ by dotted line. (F–J). Dorsal view of the embryo at hatching stage (F) and histological sections (G–J) of sibling embryos corresponding to the planes indicated by dotted lines on F. (K–S). Left side view of embryo at 5 days stage (K) and histological sections (L–S)of sibling embryos corresponding to the planes indicated by dotted lines on K. Scale bar everywhere is 200 μm.
To control specificity of in situ hybridization with DAg1 probe, we arranged two types of the control experiments. First, we made in situ hybridization on embryos preliminary cut in halves. As a result, no additional sites of expression, besides those observed during hybridization of the whole embryos, were revealed (see Supplementary Fig. S1A). Second, we performed in situ hybridization with sense probe. In this case, no signal was seen at all (see Supplementary Fig. S1B and C). Thus, both these controls confirm specificity of the revealed pattern of DAg1 expression.
Expression of DAg1 and DAgr2 during fins regeneration
Due to its ability to easily repair amputated fins, Danio rerio is a suitable model for the investigation of the regeneration mechanisms. Given this, and taking in mind recently revealed involvement of the amphibians Agr genes in processes of the tail and the hindlimb bud regeneration, we have studied the expression of DAg1 during regeneration of different fins of the adult fishes. In addition, the expression of another fish representative of Agr family, DAgr2, has been analyzed, since it was not yet investigated during regeneration elsewhere. As an internal control, we have also tested in these experiments the expression of two regeneration markers, Igf2b and Fgf20a, which were shown before to be strongly activated after the caudal fin amputation11,12. To be able to compare the target genes expression levels in different experiments, the geometric mean of expression levels of two reference housekeeping genes, ornithine decarboxylase (ODC) and elongation factor 1alpa (EF-1alpha), was determined in each of experiments for normalization of the data obtained.
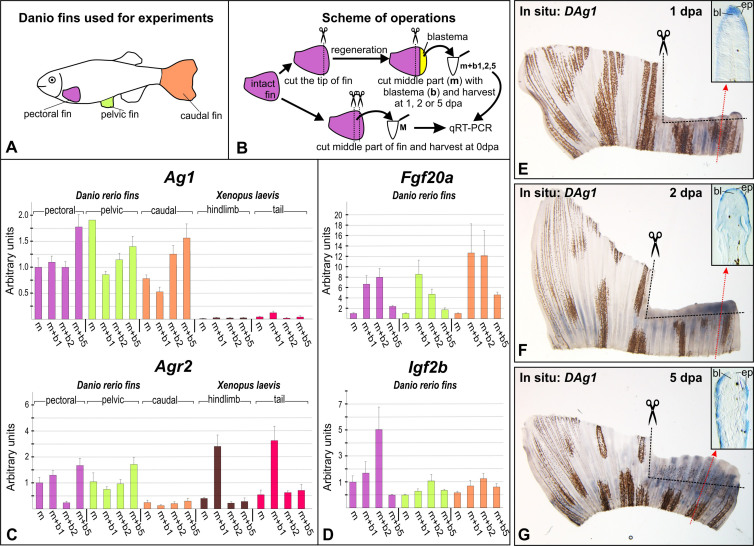
A common feature of the post-amputation expression dynamics revealed for DAg1 and DAgr2 in the distal parts of stumps of all types of the tested fins was some drop of their expression levels at the first day post-amputation 1 dpa (Fig. 3C, m + b1). Meanwhile, the expression of both these genes has started to be gradually increased at later stages, such that by 5 dpa it has exceeded the expression levels in the intact fins (Fig. 3C, m and m + b5).
Figure 3. Analysis of Ag1, Agr2, Fgf20a and IGF2b expression in the regenerating fins.
(A and B). Schemes of experiments. Tissue pieces of hindlimbs and tails of stage 52 Xenopus laevis tadpoles were collected as was previously described5. Drawings for these figures were done by MBT and AGZ. (C and D). QRT-PCR analysis of Ag1 and Agr2 (C) and Fgf20a and Igf2b (D) expression in the intact and regenerating of fins of adult Danio rerio and in hindlimbs and tails of stage 52 Xenopus laevis tadpoles. All graphs represent means of triplicate experiments. Bars indicate standard deviations. The geometric mean of expression of Danio rerio and Xenopus laevis ornithine decarboxylase (ODC) and elongation factor 1alpa (EF-1alpha) was used for normalization of experimental values (see Materials and Methods for details). The value of normalized PCR signal in "m" sample taken from pectoral fin was taken as an arbitrary unit in each series. (E–G). In situ hybridization of regenerating caudal fins with the probe to DAg1 at 1, 2 and 5 dpa. Photos on insets demonstrate histological sections of regenerating fins at levels whose approximate positions are indicated by red dotted arrowed lines. Black dashed lines indicate places of amputations. Abbreviations: bl - blastema, ep - epidermis.
By contrast, the expression of Fgf20a and Igf2b sharply increased already at 1 dpa and then decreased by 5 dpa to the intact level (Fig. 3D). Such temporal expression patterns of these two genes correspond well to the literature data, thus validating other results obtained in our experiments.
The revealed dynamics of Agr expression in the amputated Danio fins obviously differs from that recently revealed for same genes during regeneration of the Xenopus tadpoles tails and hindlimb buds5. By contrast to the initially high concentrations of Agr transcripts in Danio fins and dropping of these concentrations at 1 dpa, we observed low concentrations of XAg1 and XAgr2 transcripts in the intact tadpoles' tails and hindlimb buds and an increase of their concentrations after amputation.
If to assume that concentrations of housekeeping EF-1alpha and ODC transcripts are approximately similar in the fish fins and in the body appendages of the Xenopus tadpoles, then, knowing PCR effectiveness of all the compared templates, one may compare the expression levels of both Agr transcripts in these species. By this way, we have found out that while DAgr2 concentration in the intact Danio fins was in average in 3–4 times higher than the concentration of its ortholog Xagr2 in the Xenopus body appendages, the concentration of DAg1 exceeded that of XAg1 more than in 15–20 times (see PCR effectiveness for the Danio rerio and Xenopus laevis Ag1, Agr2, EF-1alfa and ODC in Materials and Methods and in Table 1). Importantly, the concentration of DAg1 transcripts after its dropping at 1 dpa in the amputated fins still remained higher than the increased concentration of XAg1 in the amputated body appendages of the Xenopus tadpoles (Fig. 3C). Thus, if to suppose that the increased concentration of XAg1 transcripts in the amputated tadpole appendages at 1 dpa is critical to ensure necessary level of the activity of this protein for the regeneration process, then the concentration of DAg1 transcripts detected at 1 dpa in the amputated fins could be also sufficient for the same function.
Table 1. PCR efficiency of analyzed cDNA.
| Xag1 | DAg1 | Xagr2 | Dagr2 | XEFalfa | DEFalfa | XODC | DODC | |
|---|---|---|---|---|---|---|---|---|
| PCR Efficiency | 82% | 83% | 82% | 80% | 90% | 92% | 93% | 90% |
The results of qRT-PCR experiments were confirmed by in situ hybridization with DAg1 probe (Fig. 3E–G). Interestingly, at 1 and 2 dpa the revealed signal was higher in more rapidly growing lateral parts of the amputated fins (Fig. 3E–F). As we have shown by histological sectioning, DAg1 is expressed in these regenerating parts of fins mainly in epidermis and in cells of the regenerating blastema, located just beneath the wound epithelium. Noteworthy, the expression of DAg1 in the blastema cells was especially evident in the 1 dpa fins. In general, this expression pattern of DAg1 very resembles the expression of its orthologs, Xag1/2, which we observed earlier in the regenerating tails and limb buds of the Xenopus laevis tadpoles5.
DAg1 is essential for regeneration of the caudal fin
To test directly if DAg1 is actually involved in the mechanism of the fin regeneration, we sought to inhibit translation of its mRNA by injecting into the stumps of the amputated caudal fins of antisense Vivo-morpholino oligonucleotides (Vivo-MO). The efficiency of this MO was tested and confirmed in experiments in which synthetic mRNA encoding for the Myc-tagged DAg1 was injected into the Xenopus laevis embryos either in a mixture with DAg1 VivoMO or with control-misDAg1 VivoMO (see Supplementary Fig. S2).
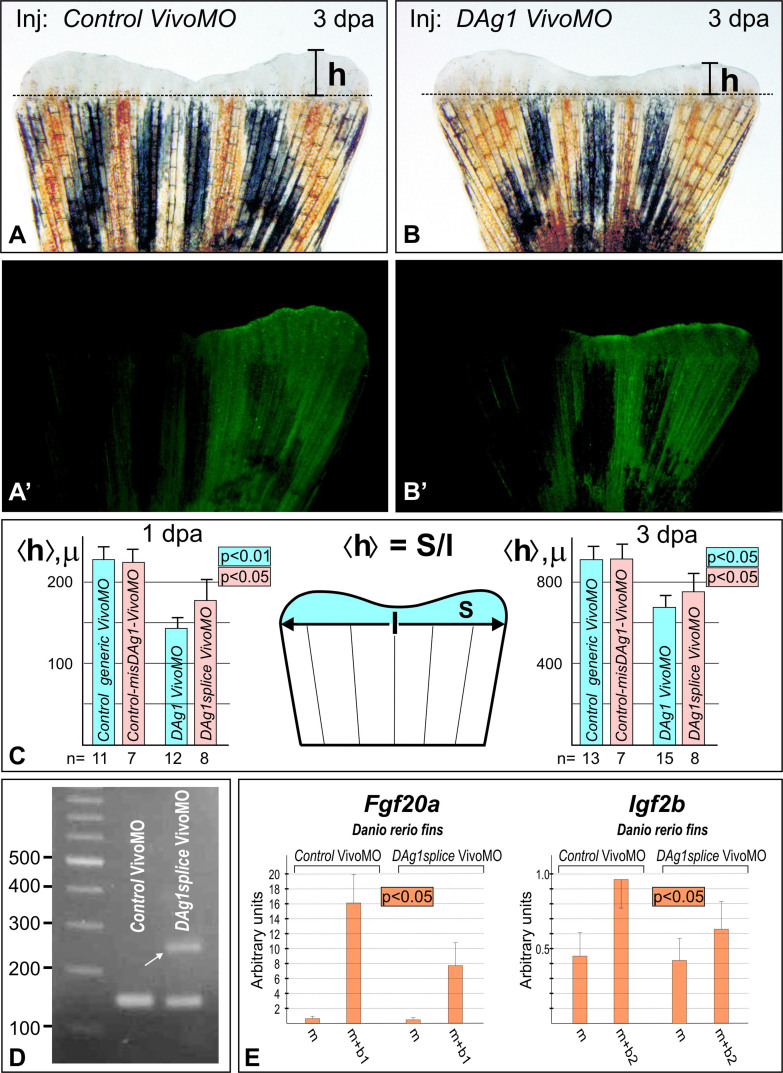
As a result of injections of the amputated fins with DAg1Vivo MO, we observed statistically significant retardation of the fin regeneration on the injected side (Fig. 4 and Supplementary Fig. S3). Importantly, no retardation of regeneration was seen in fins injected with any of two types of the control MO: control-generic VivoMO or control-misDAg1 VivoMO ( Fig. 4 A and C, Supplementary Fig. S3A and C).
Figure 4. Blocking of DAg1 mRNA translation by Vivo-morpholino injections results in retardation of the caudal fin regeneration.
(A and A'). Caudal fin injected by the control-generic Vivo-morpholino mixed with FLD in the right side. No retardation of regeneration is seen on the injected side. Dashed line indicates the level of amputation. Scale bar 1 mm. (B and B'). Caudal fin injected by the DAg1 Vivo-morpholino mixed with FLD in the right side. Note retardation of regeneration of the injected side. Scale bar 1 mm. (C). Quantification of the mean height of the regenerating part of caudal fins injected by control-generic Vivo MO or DAg1 Vivo MO at 1 dpa (left) and 3 dpa (right). Numbers of the injected fins are indicated by n. The schema in the middle demonstrates how mean height <h> was calculated. Drawing on this figure was done by AGZ. (D). RT-PCR analysis of DAg1 transcripts in 1 dpa regenerating caudal fins injected with either control-generic Vivo MO or DAg1splice Vivo MO. White arrow indicates additional band generated by DAg1 un-spliced transcript. (E). QRT-PCR analysis of Fgf20a and Igf2b expression in the regenerating caudal fin at 1 dpa and 2 dpa respectively (at these days the expression levels of Fgf20a and Igf2b reach maximal values, see Fig. 3D) maximal expression levels of these genes injected with either control-generic Vivo MO or DAg1splice Vivo MO. For scheme of experiments and abbreviations see picture D and legends on Fig. 3).
To test specificity of DAg1 Vivo-MO effect, we used another Vivo-MO, designed to a region that included donor splicing site at the border of exon 2 and intron 2–3 of DAg1 gene (Supplementary Fig. S4). Blocking of this splicing site should potentially result in the translation frame-shifting, accompanied by an extension of RNA template on additional 104 nucleotides in comparison with the normal spliced mRNA. When this DAg1splice Vivo-MO was injected, we did observe a retardation of the caudal fin regeneration, while less expressed than in case of DAg1 Vivo-MO injections. Importantly, the longer form of DAg1 transcript was detected by RT-PCR when 2 dpa tissue samples of regenerating fins were analyzed, whereas no such form of DAg1 transcripts was revealed in the control fins (Fig. 4D). Obviously, this result confirms that DAg1splice Vivo-MO indeed penetrate even into the nucleus and block splicing of DAg1 transcripts. However, this blocking was not full, probably because relatively small amount of MO was able to pass, beginning from the intercellular space, through all the cytoplasm and to reach its targets within cell nuclei.
To further test specificity of DAg1splice Vivo-MO effects, we analyzed by qRT-PCR the expression of two regeneration markers, Fgf20 and IGF, in the control and DAg1splice Vivo-MO-injected regenerating fins. As a result, a moderate but statistically significant inhibition of the expression of both genes by DAg1splice Vivo-MO was observed (Fig. 4E).
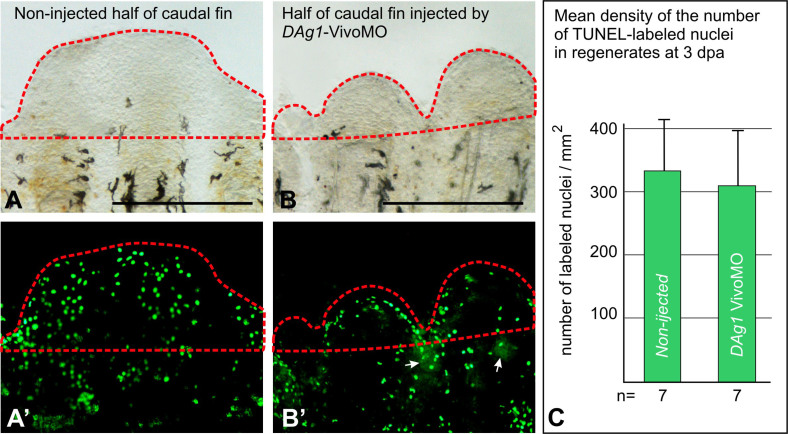
Finally, to verify if the retardation of regeneration observed in the case of DAg1-VivoMO could be caused by cell death, which is a non-specific effect seen with some MOs, we investigated cell apoptosis in regenerating fins injected with DAg1 VivoMO by means of TUNEL assay. As a result, no statistically significant difference in average concentration of apoptotic cells in regenerates was revealed between normally regenerating non-injected parts of the fins and their retarded parts that were injected with DAg1 VivoMO (Fig. 5). Thus, we concluded that the observed effect of the retardation of the fin regeneration was not the result of cell death.
Figure 5. TUNEL assay of apoptosis in the regenerating caudal fins injected with Vivo MO.
(A and A′). Revealing by TUNEL assay of apoptotic nuclei in regenerating part (framed by red dashed line) of the non-injected side of the caudal fin at 3 dpa. Scale bar 500 μm. (B and B′). Revealing by TUNEL assay of apoptotic nuclei in regenerating part (framed by red dashed line) of the side of caudal fin injected by DAg1Vivo MO at 3 dpa. White arrows indicate traces of FLD co-injected with MO. Scale bar 500 μm. (C). Quantification of TUNEL-labeled nuclei in the regenerating parts (regenerates) of caudal fins non-injected and injected by DAg1 Vivo MO.
The retardation of regeneration of the amputated fins injected by DAg1 Vivo-MO, compared with the fins injected by control Vivo-MOs, was observed within 3–5 days after injection, during which the fins were repeatedly injected by Vivo-MO solution. After the cessation of repeated injections, the difference with the control fins gradually disappeared within a few days. In sum, these data confirm an essential role of DAg1 for the fin regeneration.
Discussion
In the present work, we have cloned for the first time cDNA of the Danio rerio ortholog of the Xenopus XAg1 gene, DAg1, studied its expression pattern in early embryogenesis and demonstrated its role in the process of the caudal fin regeneration.
As we have revealed, the expression of this gene is localized in the embryonic epitheliums, thus, resembling in that the expression of DAg1 ortholog in Xenopus laevis, which is also activated for the first time in the ectodermal epithelium beginning from the late blastula stage4,13. However, in contrast to its Xenopus counterpart, we were unable to reveal any signs of the anterior to posterior gradient of DAg1 expression. Instead, this gene is expressed quite uniformly in all cells of the superficial ectodermal layer of the embryo till larval stages. Similarly to XAg1 in Xenopus, DAg1 is also expressed at the hatching stage in cells of other tissues of epithelium origin, including, olfactory pits, gills, pharynx, intestine, proctodeum and cornea.
In the last decade, more and more data is accumulated, which demonstrate involvement of Agr proteins in regulation of many processes, including cell proliferation, cell differentiation and cancerogenesis6,8,14. As we have shown recently, the Xenopus ortholog of DAg1 is intensively expressed in the anterior non-neural subdomain of the ectodermal epithelium, adjacent to cells of the telencephalic primordium located at the anterior border of the neural plate, and regulates development of the telencephalon by promoting the Fgf8 signaling4. A similar location of the surface ectoderm cells expressing DAg1, adjacent to cells of the presumptive telencephalon, was revealed now in Danio embryos. In connection with this, it would be interesting to verify in the future experiments whether DAg1 can also regulate the telencephalic development in Danio, like its ortholog XAg1 does this in Xenopus.
As we have demonstrated in the present work, the expression of DAg1, like the expression of its Xenopus ortholog Xag1, increases after amputation of the body appendages (fins). Unlike the hindlimbs and tails of the Xenopus tadpoles, however, a significantly higher expression level of DAg1 is revealed already in the intact fins before amputation. Given our data demonstrating an essential role of DAg1 for the regeneration process, one may hypothesized that such a high basal level of its expression is necessary to ensure timely participation of DAg1 in the repairing of the adult fish fins, which might be more frequent and critical for survival of the population than damage of limbs and tails of the Xenopus tadpoles.
It was shown that the expression of one of Agr proteins, Agr2, is activated after amputation of the newt limb, and Agr2 functioning is critical for the process of the limb regeneration3,7. In consistent with this, we have revealed recently the activation of expression of both Ag1 and Agr2 after amputation of the tail or hindlimb bud in the Xenopus laevis tadpoles5. Importantly, we have also shown in the cited work that Ag1 had disappeared somewhere during evolutionary transition from the amphibians to higher vertebrates. Given all these data, along with the results of the present work which demonstrate an essential role of DAg1 for the fin regeneration in Danio, one may hypothesizes that disappearance of Ag1 family genes in evolution could be one of the critical events, which resulted in a reduced regenerative capacity observed in extant higher vertebrates.
Methods
Animal experiments were performed in accordance with guidelines approved by the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry (Moscow, Russia) Animal Committee and handled in accordance with the Animals (Scientific Procedures) Act 1986 and Helsinki Declaration.
Cloning of DAg1 cDNA and dig-probe synthesis
For generating the template for synthesis of the DAg1 probe, DNA fragment corresponding to the protein coding region of DAg1 cDNA was obtained by RT-PCR from the first strand obtained from the total zebrafish embryos (48 hpf ) RNA with the following primers:
DAg1 forward: 5′-ATGAATTCTTATGAAATTCTGAAGACA,
DAg1 reverse: 5′-GAGTCGACGATCAGAGTTCAGTCTGC.
The obtained DNA fragment was cloned in pGEM-T (Promega) vector and correct clone was selected as a result of sequencing of five random clones. The resulting Dig-labeled RNA antisense probe for the whole-mount in situ hybridization was synthesized by T7 polymerase (mMessageMachine) from the PCR-product obtained with DAg1 forward and M13 reverse primers from DAg1pGEM-T plasmid construct.
To construct templates for testing the efficiency of DAg1 VivoMo, DAg1 cDNA was obtained by PCR with the following primers (restriction sites of BamHI and NcoI are underlined, start- and stop-codones are in frames):
Forward-DAg1- 5'- and Reverse-DAg1- 5'-
and Reverse-DAg1- 5'-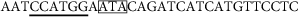 .
.
The obtained cDNA was sub-cloned into pCS2-Myc-tag by BamHI and NcoI, upstream and in frame with the last Myc-tag. Capped synthetic mRNA encoding for DAg1-Myc was synthesized with SP6 Message Machine Kit (Ambion) using the obtained plasmids cut by NotI.
Animals handling, samples fixation, whole-mount in situ hybridization and hystological sections
Danio rerio embryos were staged according to standard staging table (Westerfield, 2000).
In regeneration experiments adult fishes were anesthetized in 0.1% tricaine (Sigma, Aldrich). Xenopus laevis tadpoles were obtained as we described previously5. Fin amputations were performed in adult fishes with razor blades or with microscissor. The latter was used when a half of fin should be amputated. Animals were allowed to regenerate for various time periods at 29,5°C and the fins were cut off again and fixed for qRT-PCR or in situ hybridization. Tails and hind limb buds of the Xenopus laevis tadpoles at stage 52 were amputated by microscissors and were further processed for RT-PCR analysis at different days postamputation as we previously described5. For in situ hybridization, Danio rerio embryos or caudal fins were fixed in MEMFA (3.7% paraformaldehyde, 2мМ EGTA, 1мМ MgSO4х7H2O, 0.1мМ MOPS, pH 7.4) during 24 hours, dehydrated by ethanol and processed for in situ hybridization. The in situ hybridization was performed as described5. For histological sectioning, embryos or fins were embedded in paraplast and cut into 15 μm sections. To control the penetrability of embryos for DAg1 probe, some embryos were cut as previously described before the hybridization procedure by a microknife4. Also, to control specificity, some embryos were hybridized with sense probe to DAg1.
QRT-PCR
The total RNA from whole embryos or amputated pieces of fins (see scheme of operation on Fig 3A and B) was extracted by RNA isolation KIT (MASHEREY-NAGEL) according to the manufacturer's protocol. For each type of samples shown on Fig. 3B, three replicates, each containing RNA from five amputated pieces of the same type, were prepared. The concentration of the extracted RNA was measured by means of Qubit® fluorometer (Invitrogene), while RNA integrity was checked visually by gel electrophoresis. 250 ng of total RNA extracted from each sample was reverse transcribed in 20 μl of final volume by M-MLV reverse transcriptase (Promega) in presence of 10 pmol of oligo-dT primer (Evrogen), according to the manufacturer's guidelines (Promega) (+RT sample). In parallel, same reaction was assembled in each case without adding of M-MLV reverse transcriptase (-RT control). For qPCR reaction, which was performed on a ANK-32 (Syntol), 2 μl of + RT and -RT solutions of each type were mixed in parallel tubes with qPCRmix-HS SYBR (x5, Evrogen), corresponding primers (5 pmol each) and milli-Q water till the final volume 25 μl. A standard 40-cycle program with hot start was used; the annealing temperature was 59°C, elongation −72°C and melting 95°C, all lasted for 25 seconds. The PCR data were imported into Microsoft Excel and analyzed by using the ΔΔCt method. The geometric mean of expression of two reference housekeeping genes: ornithine decarboxylase (ODC)15 and elongation factor 1alpa (EF-1alpha) was used for normalization of the target genes expression levels. The following pairs of primers for Danio rerio genes were used:
ODC forward 5′- CTCCACCTTCAATGGCTTCCAG;
ODC reverse 5′- AGTGGGATGGCACGTTTCCAG;
EF-1alpha forward 5′- AAGAACGTGTCAGTCAAGGACAT;
EF-1alpha reverse 5′ - CGTAACCCTGAGAGATCTGACCA;
Fgf20a forward 5′- AAGGGCGAACTGTACGGAT;
Fgf20a reverse 5′- TTGAGGGCTACATAATAAT;
Igf2b forward 5′ - GCAGGTCATTCCAGTGATGC;
Igf2b reverse 5′- TCTGAGCAGCCTTTCTTTGC;
DAgr2 forward 5′- AACCACAGAGCGTGTCAGT;
DAgr2 reverse 5′- ACAGTGCTAATGCTTTCATTG;
DAg1 forward 5′- CACTGGCCGCTCTGTATACTT;
DAg1 reverse 5′- CTCTTGAGAGAGTTTGGACTGT;
DAg1 reverse-2 primer for revealing by qRT-PCR of the unspliced form of DAg1 transcript in experiments with DAg1splice VivoMO and used in pair with DAg1 forward primer 5′-AGCCAGTCCCTCCTCATACG.
Primers for the Xenopus laevis Xag2, Xagr2, ODC and EF-1alpha were the same as in5.
PCR efficiency (PE) for Xag1, DAg1, Xagr2, Dagr2, XEFalfa, DEFalfa, XODC and DODC were calculated by making qPCR form progressive dilutions of the 1st strand of cDNA obtained on the base of total RNA samples extracted from Xenopus laevis and Danio rerio midneurula stage embryos, building standard curves and using the on-line qPCR efficiency calculator (http://www.thermoscientificbio.com/webtools/qpcrefficiency). As a result, the following PE values were obtained (see Table 1). As one may see, PEs for orthologous Agrs as well as for orthologous housekeeping genes appear to be very similar. This fact justifies the comparison of concentrations of the Xenopus and Danio Agrs templates normalized in relation to concentrations of housekeeping templates.
For visualization of additional band generated by un-spliced DAg1 nuclear transcript in experiments with DAg1splice VivoMO, the PCR mixture after completion of qRT-PCR was resolved in 2% agarose gel and photographed.
Vivo-morpholino injections and fin length measurements
To test effects of DAg1 functioning down-regulation during fin regeneration, the 0,4 mM solutions of vivo-porter morpholino oligonucleotides (Gene-Tools) mixed with the tracer FLD (fluorescein lysinated dextran, Invitrogen, 40 kDa, 5 μg/μl)) were used for injections into caudal fin tissues. The following Vivo-MOs were used:
DAg1 VivoMO 5′ ACAGAGCGGCCAGTGCTGCATGATT;
DAg1splice VivoMO 5' AATTTGCTGATACCTCTTGAGAGAG (sequence complementary to the 2nd exon is underlined; for genomic sequence see Ensembl Acc.: ENSDARG00000060682);
control-generic VivoMO 5′ TCTgTggATgTCTTgCTCTTCCAgg;
control-misDAg1 VivoMO 5' ACAGATCGGCAAGTTCTGCATTATT.
The day before amputation, caudal fins of fishes anesthetized by Tricain were pre-injected with the use of Eppendorf FemtoJet microinjector by Vivo-MOs mixed with FLD into the area between bony rays along the line of the subsequent plane of amputation (4 nl per one injection, 12–15 injections per fin) in order to pre-inhibit the strong basal DAg1 expression. Amputations were performed with razor blades along the FLD-marked line under the fluorescent stereomicroscope Leica M205. Immediately after amputation, stumps of the fins were injected with vivo-MOs once again, and then injections were repeated at 24 hpa and 48 hpa. The fins were photographed by using Leica M205 stereomicroscope and Leica camera (DC 400F). To calculate the mean of the regenerate length of the caudal fins (from the amputation plane to the distal tip of the fin) injected by the control or anti-DAg1 vivo-MOs, the following formula was used: S/L, where L was the fin width and S was the square of the regenerate, both measured by using ImageJ software (http://rsb.info.nih.gov/ij). Statistical significance was determined with the Student's test and the significance was set at P < 0,01 or P < 0,05.
To test efficiency and specificity of DAg1 Vivo MO, DAg1-Myc mRNA (see above) was injected into animal pole of 4-cells Xenopus laevis embryos (100 pg/blastomere) either alone or in a mixture with mis-DAg1 VivoMo MO or DAg1 VivoMo (6–8 nl of 0,2 mM water solution). The injected embryos were collected at the gastrula stage and analyzed for the presence of DAg1-Myc by Western blotting with Sigma Anti-Myc antibody (cat. # M 4439) as described previously16. As a result, no inhibition of DAg1-Myc mRNA translation was observed in embryos co-injected with this mRNA and mis-DAg1 VivoMo MO. By contrast, a strong suppression of DAg1-Myc mRNA translation was observed in embryos co-injected with this mRNA and DAg1-Vivo MO (Fig. S2). These results confirm the efficiency and specificity of the latter MO.
TUNEL assay
For this assay, caudal amputated caudal fins were injected in one side as described above. The only difference is that DAg1-Vivo MO was mixed with a decreased amount of FLD (1 μg/μl). This trick allowed us to mark the injected side and simultaneously to reveal FITC-labeled nuclei on the low diffuse signal emitted by FLD. TUNEL assay was done on the whole-mounted caudal fins fixed in MEMFA. To reveal apoptotic cells, DeadEnd™ Fluorometric TUNEL System (Promega) was used in accordance with the manufacturer's recommendations. The number of labeled nuclei and their density (number of labeled nuclei/mm2 of the planar projection of regenerate) were calculated in the MO-injected and non-injected part of each fin by using ImageJ software.
Author Contributions
A.S.I. and I.M.S. performed cloning, qRT-PCR, in situ hybridization and M.O. injections. G.V.E. arranged and performed TUNEL experiments. A.A.M. collected and fixed embryos. M.B.T. and A.G.Z. designed experiments, drew and composed figures and wrote the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Eugenii Gasanov for providing Danio rerio fishes. This work was supported by RFBR grants N 13-04-01516, 13-04-40194-KOMFI to AGZ and grant from Dynasty Foundation to MBT. The experiment on VivoMO injections was supported by 14-14-00557 RSCF grant.
References
- Persson S. et al. Diversity of the protein disulfide isomerase family: identification of breast tumor induced Hag2 and Hag3 as novel members of the protein family. Mol. Phylogenet. Evol 36, 734–740 (2005). [DOI] [PubMed] [Google Scholar]
- Salmans M. L., Zhao F. & Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast Cancer Res 15, 204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A. & Brockes J. P. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science (New York, N.Y 318, 772–777 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tereshina M. B., Ermakova G. V., Ivanova A. S. & Zaraisky A. G. Ras-dva1 small GTPase regulates telencephalon development in Xenopus laevis embryos by controlling Fgf8 and Agr signaling at the anterior border of the neural plate. Biol. Open 3, 192–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A. S., Tereshina M. B., Ermakova G. V., Belousov V. V. & Zaraisky A. G. Agr genes, missing in amniotes, are involved in the body appendages regeneration in frog tadpoles. Sci. Rep 3, 1279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. A. et al. Anterior Gradient-3: a novel biomarker for ovarian cancer that mediates cisplatin resistance in xenograft models. J. Immunol. Methods 378, 20–32 (2012). [DOI] [PubMed] [Google Scholar]
- Kumar A. et al. The aneurogenic limb identifies developmental cell interactions underlying vertebrate limb regeneration. Proc. Natl. Acad. Sci. USA 108, 13588–13593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Rudland P. S., Sibson D. R., Platt-Higgins A. & Barraclough R. Human homologue of cement gland protein, a novel metastasis inducer associated with breast carcinomas. Cancer Res 65, 3796–3805 (2005). [DOI] [PubMed] [Google Scholar]
- Vanderlaag K. E. et al. Anterior gradient-2 plays a critical role in breast cancer cell growth and survival by modulating cyclin D1, estrogen receptor-alpha and survivin. Breast Cancer Res 12, R32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih L. J. et al. Characterization of the agr2 gene, a homologue of X. laevis anterior gradient 2, from the zebrafish, Danio rerio. Gene Expr. Patterns 7, 452–460 (2007). [DOI] [PubMed] [Google Scholar]
- Chablais F. & Jazwinska A. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development (Cambridge, England) 137, 871–879 (2010). [DOI] [PubMed] [Google Scholar]
- Whitehead G. G., Makino S., Lien C. L. & Keating M. T. fgf20 is essential for initiating zebrafish fin regeneration. Science (New York, N.Y 310, 1957–1960 (2005). [DOI] [PubMed] [Google Scholar]
- Sive H. L., Hattori K. & Weintraub H. Progressive determination during formation of the anteroposterior axis in Xenopus laevis. Cell 58, 171–180 (1989). [DOI] [PubMed] [Google Scholar]
- Aberger F., Weidinger G., Grunz H. & Richter K. Anterior specification of embryonic ectoderm: the role of the Xenopus cement gland-specific gene XAG-2. Mech. Dev 72, 115–130 (1998). [DOI] [PubMed] [Google Scholar]
- Xanthos J. B. et al. The roles of three signaling pathways in the formation and function of the Spemann Organizer. Development (Cambridge, England) 129, 4027–4043 (2002). [DOI] [PubMed] [Google Scholar]
- Bayramov A. V. et al. Novel functions of Noggin proteins: inhibition of Activin/Nodal and Wnt signaling. Development (Cambridge, England) 138, 5345–5356 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information