Abstract
Drosophila species lack most hallmarks of adaptive immunity yet are highly successful against an array of natural microbial pathogens and metazoan enemies. When attacked by figitid parasitoid wasps, fruit flies deploy robust, multi-faceted innate immune responses and overcome many attackers. In turn, parasitoids have evolved immunosuppressive strategies to match, and more frequently to overcome, their hosts. We present methods to examine the evolutionary dynamics underlying anti-parasitoid host defense by teasing apart the specialized immune-modulating venoms of figitid parasitoids and, in turn, possibly delineating the roles of individual venom molecules. This combination of genetic, phylogenomic, and "functional venomics" methods in the Drosophila-parasitoid model should allow entomologists and immunologists to tackle important outstanding questions with implications across disciplines and to pioneer translational applications in agriculture and medicine.
Introduction
Flies in the genus Drosophila act as hosts to a number of pathogens and parasites, including many species of endoparasitoid wasps [1,2*]. Parasitoids represent a unique class of venomous organisms that physically threaten their hosts, yet keep them alive to support their developing progeny. To meet developmental needs and protect their offspring, parasitoid venoms have been tailored to their hosts through dynamic co-evolutionary pressures [2*]. Although Drosophila spp. possess versatile innate immune responses [1,3], their parasitoid wasps have evolved powerful and complex immune-suppressive strategies that can partially compromise or completely incapacitate host defenses [4–6]. Immune suppression is mediated through the cellular and biochemical actions of venom components such as virus-like particles (VLPs, Fig. 1) and venom proteins [7–10,11*,12*,13*]. The effects of wasps of the Leptopilina and Ganaspis spp. on host immunity and development are specific and potent.
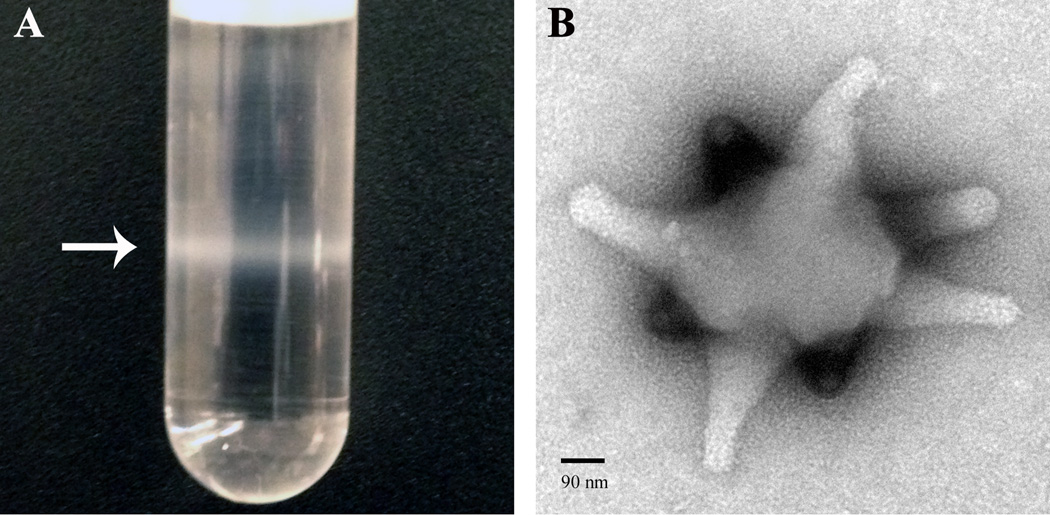
Figure 1. Virulence factors from Leptopilina species.
(A) An opaque band of concentrated purified VLPs (arrow) is visualized after ultracentrifugation of venom gland extract in a nycodenz gradient. (B) A uranyl acetate stained VLP, visualized by the negative stain transmission electron microscope (Zeiss 902) method. Downward-facing extensions from VLP body (i.e., “spikes”) are in a different focal plane and stain more intensely than upward-facing ones. VLP spikes appear to widen slightly at their termini. Spiked VLPs have been reported from L. heterotoma, L, victoriae, and L. boulardi [7,8,10]. Abundant proteins from VLPs of the L. heterotoma and L. victoriae sister species are produced in secretory cells of the wasp’s venom gland. VLP proteins and precursors transit through an extensive and conserved canal system within the venom gland, localize to specific VLP regions, and enter host hemocytes post infection [9,61]. The biological nature of VLPs (i.e., whether they are true viruses, virus-like, or simply secretions of wasp’s venom glands), their macromolecular constituents, and precise modes of action on the host’s immune system and development remain a significant challenge.
Sources estimate that parasitoid wasps comprise up to 20% of all insect species and 70% of Hymenoptera [14]. Members of this large and diverse group of arthropods are considered keystone species within their native ecosystems, acting as important regulators of complex food-web dynamics and overall ecosystem stability [15,16]. Due to their roles in maintaining herbivorous insect populations and their impressive host specificity, the presence or absence of hymenopteran parasitoids acts as a useful bio-indicator of ecosystem health and diversity [17,18].The importance of these host-parasite interactions in efficacious agricultural and horticultural pest control, and in forest conservation, has also been recognized for decades [19–22]. The fruit fly Drosophila suzukii poses an emerging agricultural threat and is already blamed for 500 million dollars in damage annually. Drosophila endoparasitoids are ideal candidates for biocontrol [23] and potentially pose minimal ecological disruption of other established biological networks.
By utilizing a systematic program that we call functional venomics, we seek to gain unique insights into the evolution, conservation, and mechanisms of parasite virulence and host immunity. We present experimental approaches to uncover the individual roles of the highly specialized, immune modulating venom factors of the natural hymenopteran parasites of Drosophila melanogaster that are central to the evolutionary arms race between these organisms.
The Drosophila-parasitoid model of the evolutionary arms race
More hymenopteran parasitoids are catalogued every year [24]. However, our understanding of the cellular and molecular processes that underlie the tightly interwoven interspecies relationships between parasitoids and their prey is limited to only a small number of species. Into this gap steps the genetically flexible model organism D. melanogaster. Much of our understanding of innate immunity is derived from this natural host of several hymenopteran parasitoids [3,23]. Drosophila-parasitoid pairs are bound within an “evolutionary arms race,” a phrase originating from the Red Queen Hypothesis, which posits that competing organisms must constantly adapt to meet the challenges of their obligate, antagonistic relationships [25].
Flies in the genus Drosophila support the development of endoparasitoids such as the figitid (Leptopilina, Ganaspis), braconid (Asobara, Aphaereta), diapriid (Trichopria), and pteromalid (Pachycrepoideus) [1,2*,26] wasps. Smaller than fruit flies, they are conveniently cultured on fly larvae or pupae, on standard media [26]. Generation times are relatively short at 3–4 weeks, their development closely tied to their hosts [27]. These wasps follow the haplo-diploid mechanism of sex determination, making them ideal for classical genetic mapping. The genome sizes and karyotypes for some endoparasitoids of Drosophila species are known [28].
D. melanogaster’s differential immune responses to braconid [29] and figitid (e.g., L. boulardi, L. heterotoma, L. victoriae, Ganaspis xanthopoda) [30–32] wasps include, for example, variable levels of cytokine-based hemocyte activation and wasp egg encapsulation. Wasps that parasitize D. melanogaster also attack other drosophilids, such as Zaprionus indianus, yet induce unique anti-wasp defenses in the latter because of differences in immune cell types [33]. Thus, drosophilids and their parasitoids present a powerful model for unraveling the dual mysteries of comparative molecular host immunity and specialized parasitoid virulence that underlie such diverse host-parasite interactions.
Parasitoid attack activates inflammatory signaling and multiple immune responses
Drosophila species have developed several local (cell migration and wound healing at the site of oviposition [26]), and systemic, cellular and humoral immune responses [1,3,5,26]. Two experimental approaches have been instrumental in clarifying anti-wasp reactions in D. melanogaster: (a) microarray-based transcriptomics followed by experimental verification of major gene expression trends; and (b) conventional genetic analysis of larval hematopoiesis and immune competence against wasp attack. When interpreted in the larger context of the well-characterized and phylogenetically-conserved signaling modules and networks that control immunity, development, cell division, and differentiation, results from the simple fly model are highly relevant to other systems, from insects to mammals.
The examination of genome-wide host responses against A. tabida, L. boulardi, and G. xanthopoda has pointed to the transcriptional activation and involvement of the Toll/NF-κB, JAK-STAT, and the pro-phenol oxidase cascade pathways [29–31]. These molecular findings support genetic experiments in which animals mutant in either Toll/NF-κB or JAK-STAT pathway components are unable to successfully encapsulate wasp eggs [34].
These immune pathways protect the host against parasitoids in two ways. First, the Toll/NF-κB signaling controls hemocyte load [34,35]. Evidence for hemocyte load as a key determinant of host success comes from direct correlation between hemocyte concentration and encapsulation capacity in larvae of laboratory strains [5,34] as well as in natural populations of species of the D. melanogaster subgroup [36]. Hemocyte deficit compromises encapsulation ability, whereas an abundance of hemocytes contributes to high resistance.
Successful encapsulation also requires the presence of appropriate and functional hemocyte lineages. L. boulardi attack, for example, promotes limited mitosis and lamellocyte differentiation among hematopoietic progenitors of the larval lymph gland, while impeding crystal cell development [37,38]. The molecular mechanisms underlying these changes are not entirely understood, although requirements for the NF-κB proteins Dorsal and Dif, in basal versus wasp-activated hematopoiesis in the larval lymph gland, have recently been identified [39**]. Notch and reactive oxygen species (ROS) signaling also contribute to the mechanisms of lamellocyte differentiation in the lymph gland [40,41].
Data from both genetic and microarray-based transcriptomic experimental approaches point to the existence of a homeostatic set point, governed by positive and negative regulators (and potential direct targets) of Toll/NF-κB signaling, in the activation (via Spatzle-processing enzyme (SPE)) and resolution (via Cactus/IκB) of the encapsulation response [32]. Both SPE and Spatzle (Spz, the ligand for the Toll receptor) are secreted proteins, and either their ectopic expression, or loss-of-function mutations in cactus, leads to chronic hemocyte overproliferation, lamellocyte differentiation, aggregation, and melanization. Genetic studies of hematopoietic tumor phenotypes have identified cytokine-mediated anti-wasp inflammatory circuitry encoded in the fly’s genome, paralleling conserved pathways found in mammals. The mutant, dysregulated signaling cycles that produce hematopoietic tumors and the pathways that lead to wasp encapsulation are different in two respects: (a) in mutants, activation is not induced, but is constitutive; and (b) mutants lack the ability to terminate pathway activation and return to homeostasis [32,42].
Direct correlations have been reported between anti-parasitoid defenses of D. melanogaster [43] and D. paramelanica [44] and reactive oxygen and nitric oxide species (NO). D. paramelanica, a species within the Drosophila subgroup, lacks the hemocyte-mediated encapsulation response of D. melanogaster and other arthropods, but demonstrates a NO-dependent protection against infection [44]. Intriguingly D. paramelanica is not overcome by L. heterotoma attack, even though this endoparasitoid demonstrates wide-spread virulence and is a generalist wasp of the Drosophila subgroup [30]. Recent experiments suggest a role for ROS in lamellocyte differentiation [40,41]. Whether the ROS and NO cytoprotective mechanisms tie into the major humoral and cellular inflammatory immune signaling cascades, or represent independent and robust arms of anti-wasp immunity, remains to be determined.
Genetic screens and pathway analyses in D. melanogaster continue to provide rich insights into the complexity and conservation of basal and wasp-activated hematopoietic development [45,46,47*,48–51]. These findings, easily transferrable to other flies and insects, will propel phylogenomic discoveries of the major and minor immune mechanisms across and beyond the drosophilids and their diverse macroparasitoids.
Hybrid “omics” approaches predict molecular virulence effectors
Recent de novo figitid wasp (L. boulardi, L. heterotoma, and Ganaspis sp.) transcriptomes, from pertinent tissues, have initiated the characterization of venom and VLP proteins in the absence of genomic sequencing. These high-throughput RNA-seq analyses of Ganaspis and Leptopilina spp. wasps [12,52] or cDNA-based venom gland libraries [11,13], in conjunction with proteomics, have identified several specific venom molecules. Similarity-based analyses of these molecules revealed shared ancestry between the venom gland products of Leptopilina spp. and aculeate hymenopterans. However, a large proportion of figitid venom sequences are novel, or homologous only to other unannotated proteins [13*]. While unknown sequences present significant experimental hurdles, bioinformatic tools can frequently predict sequence-based function in phylogenomic contexts for newly discovered venom molecules (Fig 2). Gene ontology enrichments can sometimes identify shared activities within complex venom mixtures, providing snapshot profiles for comparing one species’ venom to that from another species [11*,12*]. Enzymatic pathway annotations (e.g., EC, KEGG, BioSystems) can clarify putative protein identifications, suggesting both general bioactivities and specific metabolic or developmental pathways that may be targeted by particular parasitoid venoms [13*].
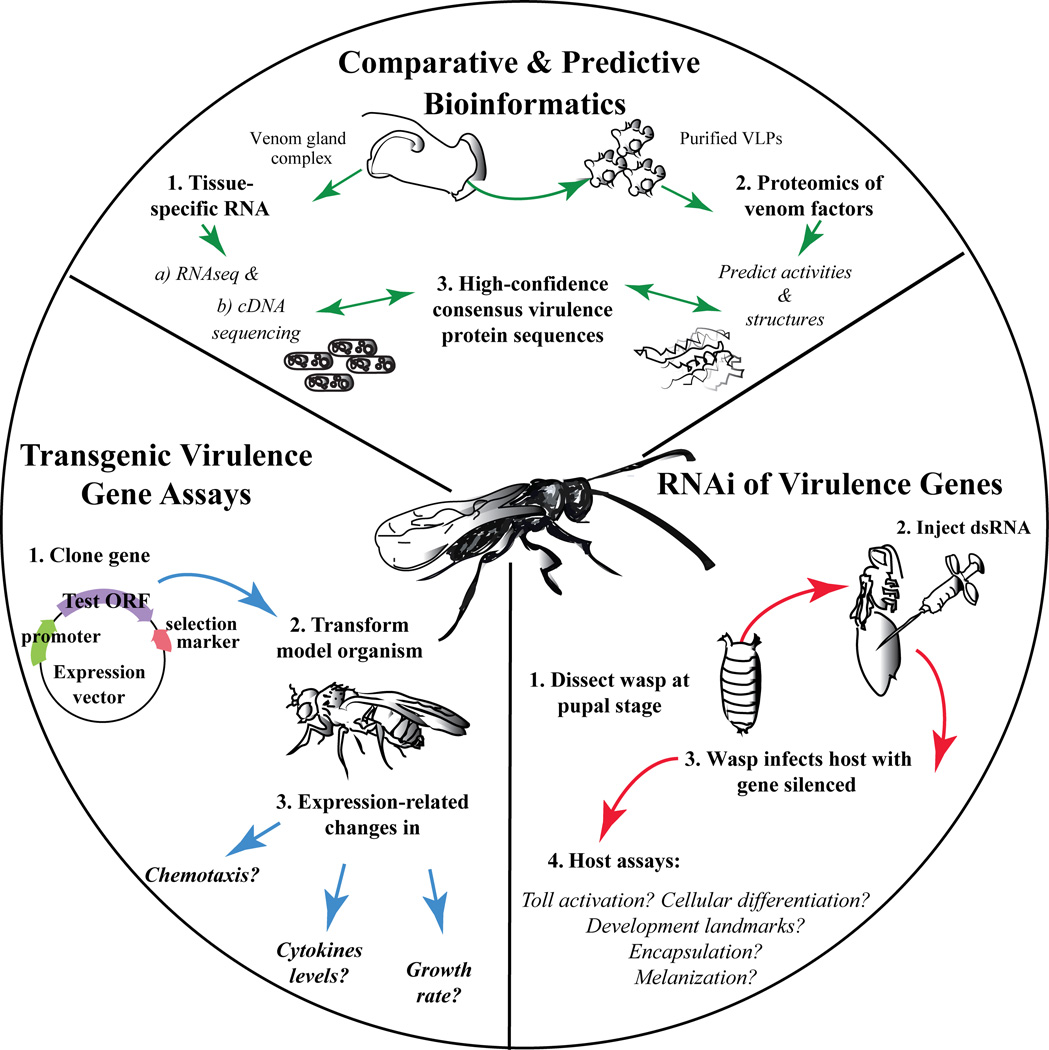
Figure 2. Structural and functional parasitoid venomics.
(Top sector) Comparative and predictive bioinformatics: Either total RNA or mRNA from venom glands is sequenced. In addition, proteomes of purified immune-suppressive factors (e.g., from venom fluid or purified VLPs) are matched with RNA and genomic sequences, if available. High-confidence matches and consensus sequences are fed into structural bioinformatics algorithms. Structural folds, domain architectures, catalytic motifs, etc., are predicted to provide functional insight, especially for novel and un-annotated proteins. (Right bottom sector) RNAi of virulence genes: For high- or medium-throughput analysis of virulence proteins of interest, newly demonstrated RNAi approaches would be appropriate. Blocking translation of single virulence proteins can provide specific biological activity information given non-redundant functions in hosts. (Left bottom sector) Transgenic virulence gene assays: For proteins with clear-cut functions in blocking host immunity, spatially and temporally controlled in vivo transgenic virulence, development, and other assays can be used in Drosophila and in other model systems to assess molecular and signaling targets.
Comparative transcriptomics of isogenized Drosophila hosts infected by L. heterotoma-14 or L. boulardi-17 provide clear evidence of distinct host-parasite specificities that relate to attack arsenals and resistance response [30]. While L. boulardi-17 infection affects the expression of more than 400 genes, only a small subset are differentially regulated after L. heterotoma-14 attack. This analysis and other inter-specific comparisons suggest that host-parasite resistance-virulence success interdependencies correlate to wasp venom compositions [11*,12*,53]. Intra-specific comparisons indicate that L. boulardi (Lb) strains differ in the expression of LbGAP alleles. LbGAP encodes a RhoGAP domain-containing venom protein that significantly affects host hemocyte shapes and function [11*,53,54]. Lbm, the more widely successful strain of Lb, expresses higher levels of LbGAP RNA than the less successful strain Lby, suggesting allelic differences in cis-regulation as the basis for variation in virulence [53]. Early molecular insights into the venom and VLP compositions present clear experimental challenges (Fig. 1), but hint at the richness of information that is yet to be uncovered about host-specificity of immunosuppressive venom components and their mechanisms of control over host development and immune physiology.
The dawn of functional venomics in parasitoid insects
Since inflammatory reactions (i.e., hemocyte recruitment, activation, migration, and adhesion; cytokine secretion; and gene expression changes in immune tissues) represent a major anti-wasp host defense mechanism [32], it is not surprising that suppression of inflammation is a shared property among parasitoids of Drosophila spp. and potentially other endoparasitoids with broadly similar life histories. This fundamental function is realized in a variety of scenarios. For example, L. heterotoma and L. victoriae VLPs kill hemocytes (Fig. 1), [9,55] and L. boulardi venom modulates hemocyte cell shape [53], whereas Ganaspis SERCA suppresses the intracellular calcium burst that normally accompanies hemocyte activation/migration [52**]. PDV Vankyrin proteins of ichneumonid wasps, when expressed in D. melanogaster cells, selectively block immune signaling, hemocyte proliferation and differentiation, and embryonic development [39**]. Additional validation has come from recent studies in which anti-inflammatory effects of N. vitripennis venom were demonstrated in both macrophage and fibrosarcoma cell lines, making the transition to mammalian systems a reality [56**].
With an initial molecular description of select wasp venom proteins and some understanding of possible effects of these proteins on insect immune responses and development in hand, the parasitoid wasp venomics field is poised for great strides. For proteins whose structures allow clear functional predictions, a combination of logical and rigorous in vitro [57] and in vivo approaches [39**] can be used (Fig. 2). In addition to the fly model system, cultured Drosophila S2 cells have clear-cut phenotypes with abundant functional genomics resources [58*] and can be utilized for new high-throughput functional bioassays, protein expression, and structural biology studies. In addition to Drosophila S2 and mammalian cell lines, simpler model organisms such as yeast or C. elegans can also serve significant roles in the functional venomics toolkit (Fig 2). Classical techniques in Saccharomyces cerevisae, for example, rely on inducible promoters appropriate for potentially toxic effects of venom proteins, or can provide diagnostic phenotypes in genetic screens (Fig 2). The successful use of RNAi to silence of Figitidae venom genes has recently been demonstrated [59**], adding to a list of insect species amenable to this technique. This reverse genetic approach will facilitate direct correlations between host immunological modulation and venom protein function (Fig. 2).
Conclusions and outlook
The ultimate goal of the larger community of researchers studying the parasitic wasps of Drosophila spp. is to use the extensive and incisive genetic tools to obtain deeper insights into the molecular basis of the host-parasite arms race. Flies utilize multiple local and systemic, cellular and humoral mechanisms to respond to wasp attack. The wasp’s molecular armament has co-evolved to antagonize these Drosophila anti-macroparasitoid immune mechanisms. Whether wasp immune-suppressive effectors target just a single immune arm to overcome host defenses, or wasps use a multipronged strategy to inactivate multiple facets of host immunity, remains to be determined. Powerful computational and experimental tools are now available to systematically probe critical aspects of host-parasite immune physiology, especially those aspects that do not fit neatly into either the Imd or Toll responses [3]. The genetic regulation of hematopoietic stem-cell niche maintenance, effector cell differentiation, and activation of cellular defenses through conserved NF-κB, JAK-STAT, Notch, and JNK pathways [50,60] will be extrapolated to other Drosophila species to explore the diverse anti-parasitoid responses of insects. Functional venomics should therefore integrate and build upon the findings of molecular insect immunity. Genomic sequencing of the Leptopilina, Ganaspis and Asobara spp. experimental models will drive translational applications, with wide-reaching impacts in agriculture and medicine.
HIGHLIGHTS.
Parasitoids induce humoral and cellular immune signaling in Drosophila.
“Omics” approaches are integrating insect immunity and functional venomics.
Bioinformatics of predicted venom proteins should direct future experimentation.
Model systems genetics will facilitate testing of parasitoid venom function.
ACKNOWLEDGEMENTS
We are grateful to past and present members of the Govind lab for their contributions. Many thanks to Zoe Papadopol for fly stock maintenance and J. Ramroop and Dr. S. Hoskins for advice and editing. This review was produced with funding from National Science Foundation 1121817.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST STATEMENT
The authors declare no conflicts of interest with this work.
AUTHORS’ CONTRIBUTIONS
All authors contributed to the preparation of the manuscript.
REFERENCES
- 1.Govind S. Innate immunity in Drosophila : Pathogens and pathways. Insect Sci. 2008;15:29–43. doi: 10.1111/j.1744-7917.2008.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Keebaugh ES, Schlenke TA. Insights from natural host-parasite interactions: the Drosophila model. Dev Comp Immunol. 2014;42:111–123. doi: 10.1016/j.dci.2013.06.001. Disscusses the benefits of harnessing the natural host-pathogen model of Drosophila to uncover highly specific and novel host defense strategies
- 3.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Annu. rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 4.Dubuffet A, Colinet D, Anselme C, Dupas S, Carton Y, Poirie M. Variation of Leptopilina boulardi success in Drosophila hosts: What is inside the black box? In: P G, editor. Advances in Parasitology. Vol. 70. Academic Press; 2009. pp. 147–188. [DOI] [PubMed] [Google Scholar]
- 5.Lee M, Kalamarz M, Paddibhatla I, Small C, Rajwani R, Govind S. Virulence factors and strategies of Leptopilina spp.: Selective responses in Drosophila hosts. In: Prevost G, editor. Advances in Parasitology. Vol. 70. Academic Press; 2009. pp. 123–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nappi A, Poirie M, Carton Y. The role of melanization and cytotoxic by-products in the cellular immune responses of Drosophila against parasitic wasps. In: Prevost G, editor. Advances in Parasitology. Vol. 70. Academic Press; 2009. pp. 99–121. [DOI] [PubMed] [Google Scholar]
- 7.Rizki RM, Rizki TM. Parasitoid virus-like particles destroy Drosophila cellular immunity. Proc Natl Acad Sci USA. 1990;87:8388–8392. doi: 10.1073/pnas.87.21.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morales J, Chiu H, Oo T, Plaza R, Hoskins S, Govind S. Biogenesis, structure, and immune-suppressive effects of virus-like particles of a Drosophila parasitoid, Leptopilina victoriae. J Insect Physiol. 2005;51:181–195. doi: 10.1016/j.jinsphys.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Chiu H, Morales J, Govind S. Identification and immuno-electron microscopy localization of p40, a protein component of immunosuppressive virus-like particles from Leptopilina heterotoma a virulent parasitoid wasp of Drosophila. J Gen Virol. 2006;87:461–470. doi: 10.1099/vir.0.81474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gueguen G, Rajwani R, Paddibhatla I, Morales J, Govind S. VLPs of Leptopilina boulardi share biogenesis and overall stellate morphology with VLPs of the heterotoma clade. Virus Res. 2011;160:159–165. doi: 10.1016/j.virusres.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colinet D, Deleury E, Anselme C, Cazes D, Poulain J, Azema-Dossat C, Belghazi M, Gatti J-L, Poirie M. Extensive inter- and intraspecific vemon variation in closely related parasites targeting the same host: The case of Leptopilina parasitoids of Drosophila. Insect Biochem and Mol Biol. 2013;43:601–611. doi: 10.1016/j.ibmb.2013.03.010. By utilizing a combined transcriptomic-proteomic approach, the main secreted venom proteins of three closely related Drosophila endoparasitoid wasps were identified and revealed both inter- and intra-specific variations in venom protein composition
- 12. Goecks J, Mortimer NT, Mobley JA, Bowersock GJ, Taylor J, Schlenke TA. Integrative approach reveals composition of endoparasitoid wasp venoms. PLoS One. 2013;8:e64125. doi: 10.1371/journal.pone.0064125. Describes a transcriptomic-proteomic approach for identifying the venom composition of two Drosophila endoparasitoid wasps
- 13. Heavner ME, Gueguen G, Rajwani R, Pagan PE, Small C, Govind S. Partial venom gland transcriptome of a Drosophila parasitoid wasp, Leptopilina heterotoma reveals novel and shared bioactive profiles with stinging Hymenoptera. Gene. 2013;526:195–204. doi: 10.1016/j.gene.2013.04.080. The partial venom transcriptome of L. heterotoma identifies potential molecular strategies associated with host immune suppresion and parasitoid success
- 14.Bonet A. Parasitoid wasps, natural enemies of insects. In: Del Claro K, Oliveira PS, Rico-Gray V, editors. Tropical Biology and Conservation Management. VII. Paris, France: Eolss Publishers; 2009. [Google Scholar]
- 15.Barbosa P, Castellanos I. Ecology of predator-prey interactions. Oxford: Oxford University Press; 2005. [Google Scholar]
- 16.Van Veen FJ, Muller CB, Pell JK, Godfray HC. Food web structure of three guilds of natural enemies: predators, parasitoids and pathogens of aphids. J Anim Ecol. 2008;77:191–200. doi: 10.1111/j.1365-2656.2007.01325.x. [DOI] [PubMed] [Google Scholar]
- 17.Barbosa P. Conservation biological control. San Diego: Academic Press; 1998. [Google Scholar]
- 18.Anderson A, McCormack S, Helden A, Sheridan H, Kinsella A, G P. The potential of parasitoid Hymenoptera as bioindicators of arthropod diversity in agricultural grasslands. Journal of Applied Ecology. 2011;48:382–390. [Google Scholar]
- 19.Clausen CP. Biological control of insect pests in the continental United States. Washington DC: USDA; 1956. [Google Scholar]
- 20.Clausen CP, editor. Introduced Parasites and Predators of Arthropod Pests and Weeds: A World Review. Washington DC: USDA Agricultural Research Service; 1978. [Google Scholar]
- 21.Gilstrap FE, Hart WG. Biological control of the Mediterranean fruit fly in the United States and Central America. USDA ARS. 1987;56:1–64. [Google Scholar]
- 22.van Driesche R, Hoddle M, Center T. Control of Pests and Weeds by Natural Enemies: An Introduction to Biological Control. Wiley-Blackwell; 2008. [Google Scholar]
- 23.Kacsoh BZ, Schlenke TA. High hemocyte load is associated with increased resistance against parasitoids in Drosophila suzukii a relative of D. melanogaster. PLoS One. 2012;7:e34721. doi: 10.1371/journal.pone.0034721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Triana JL, Whitfield JB, Rodriguez JJ, Smith MA, Janzen DH, Hallwachs WD, Hajibabaei M, Burns JM, Solis MA, Brown J, et al. : Review of Apanteles sensu stricto (Hymenoptera, Braconidae, Microgastrinae) from Area de Conservacion Guanacaste, northwestern Costa Rica, with keys to all described species from Mesoamerica. Zookeys. 2014;383:1–565. doi: 10.3897/zookeys.383.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;11:1–30. [Google Scholar]
- 26.Small C, Paddibhatla I, Rajwani R, Govind S. An introduction to parasitic wasps of Drosophila and the antiparasite immune response. J Vis Exp. 2012;63:e3347. doi: 10.3791/3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melk JP, Govind S. Developmental analysis of Ganaspis xanthopoda : a larval parasitoid of Drosophila. J Exp Biol. 1999;202:1885–1896. doi: 10.1242/jeb.202.14.1885. [DOI] [PubMed] [Google Scholar]
- 28.Gokhman V, Johnston J, Small C, Rajwani R, Hanrahan S, Govind S. Genomic and karyotypic variation in Drosophilia parasitoids (Hymenoptera, Cynipoidea, Figitidae) Comp Cytogen. 2011;5:211–221. doi: 10.3897/CompCytogen.v5i3.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wertheim B, Kraaijeveld AR, Schuster E, Blanc E, Hopkins M, Pletcher SD, Strand MR, Partridge L, Godfray HC. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 2005;6:R94. doi: 10.1186/gb-2005-6-11-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlenke T, Morales J, Govind S, Clark A. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 2007;26:1486–1501. doi: 10.1371/journal.ppat.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MJ, Mondal A, Small C, Paddibhatla I, Kawaguchi A, Govind S. A database for the analysis of immunity genes in Drosophila : PADMA database. Fly (Austin) 2011;5:155–161. doi: 10.4161/fly.5.2.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paddibhatla I, Lee MJ, Kalamarz ME, Ferrarese R, Govind S. Role for Sumoylation in Systemic Inflammation and Immune Homeostasis in Drosophila Larvae. PLoS Pathog. 2010;6:e1001234. doi: 10.1371/journal.ppat.1001234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kacsoh BZ, Bozler J, Schlenke TA. A role for nematocytes in the cellular immune response of the Drosophilid Zaprionus indianus. Parasitology. 2014;141:697–715. doi: 10.1017/S0031182013001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorrentino RP, Melk JP, Govind S. Genetic analysis of contributions of dorsal group and JAK-Stat92E pathway genes to larval hemocyte concentration and the egg encapsulation response in Drosophila. Genetics. 2004;166:1343–1356. doi: 10.1534/genetics.166.3.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu P, Pan PC, Govind S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development. 1998;125:1909–1920. doi: 10.1242/dev.125.10.1909. [DOI] [PubMed] [Google Scholar]
- 36.Prevost G, Eslin P. Hemocyte load and immune resistance to Asobara tabida are correlated in species of the Drosophila melanogaster subgroup. J Insect Physiol. 1998;44:807–816. doi: 10.1016/s0022-1910(98)00013-4. [DOI] [PubMed] [Google Scholar]
- 37.Krzemien J, Oyallon J, Crozatier M, Vincent A. Hematopoietic progenitors and hemocyte lineages in the Drosophila lymph gland. Dev Biol. 2010;346:310–319. doi: 10.1016/j.ydbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Sorrentino RP, Carton Y, Govind S. Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev Biol. 2002;243:65–80. doi: 10.1006/dbio.2001.0542. [DOI] [PubMed] [Google Scholar]
- 39. Gueguen G, Kalamarz ME, Ramroop J, Uribe J, Govind S. Polydnaviral ankyrin proteins aid parasitic wasp survival by coordinate and selective inhibition of hematopoietic and immune NF-kappa B signaling in insect hosts. PLoS Pathog. 2013;9(8):e1003580. doi: 10.1371/journal.ppat.1003580. A heterologous in vivo Drosophila study demonstrates utility of this genetic system to uncover structure-based functions of immune-suppressive proteins in host immunity and development
- 40.Small C, Ramroop J, Otazo M, Huang LH, Saleque S, Govind S. An Unexpected Link Between Notch Signaling and ROS in Restricting the Differentiation of Hematopoietic Progenitors in Drosophila. Genetics. 2013;197:471–483. doi: 10.1534/genetics.113.159210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinenko SA, Shim J, Banerjee U. Oxidative stress in the haematopoietic niche regulates the cellular immune response in Drosophila. EMBO Rep. 2012;13:83–89. doi: 10.1038/embor.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalamarz ME, Paddibhatla I, Nadar C, Govind S. Sumoylation is tumor-suppressive and confers proliferative quiescence to hematopoietic progenitors in Drosophila melanogaster larvae. Biol Open. 2012;1:161–172. doi: 10.1242/bio.2012043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nappi AJ, Vass E, Frey F, Carton Y. Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur J Cell Biol. 1995;68:450–456. [PubMed] [Google Scholar]
- 44.Carton Y, Frey F, Nappi AJ. Parasite-induced changes in nitric oxide levels in Drosophila paramelanica. J Parasitol. 2009;95:1134–1141. doi: 10.1645/GE-2091.1. [DOI] [PubMed] [Google Scholar]
- 45.Fossett N. Signal transduction pathways, intrinsic regulators, and the control of cell fate choice. Biochim Biophys Acta. 2013;1830:2375–2384. doi: 10.1016/j.bbagen.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honti V, Csordas G, Kurucz E, Markus R, Ando I. The cell-mediated immunity of Drosophila melanogaster : hemocyte lineages, immune compartments, microanatomy and regulation. Dev Comp Immunol. 2014;42:47–56. doi: 10.1016/j.dci.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 47. Evans CJ, Liu T, Banerjee U. Drosophila hematopoiesis: Markers and methods for molecular genetic analysis. Methods. 2014;68:242–251. doi: 10.1016/j.ymeth.2014.02.038. This review summarizes the Drosophila hematopoietic system and describes the many useful molecular genetic tools and analytical methods available to researchers, including new molecular markers for blood cell types and lineages
- 48.Morin-Poulard I, Vincent A, Crozatier M. The Drosophila JAK-STAT pathway in blood cell formation and immunity. JAKSTAT. 2013;2:e25700. doi: 10.4161/jkst.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tokusumi Y, Tokusumi T, Shoue DA, Schulz RA. Gene regulatory networks controlling hematopoietic progenitor niche cell production and differentiation in the Drosophila lymph gland. PLoS One. 2012;7:e41604. doi: 10.1371/journal.pone.0041604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zettervall CJ, Anderl I, Williams MJ, Palmer R, Kurucz E, Ando I, Hultmark D. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci USA. 2004;101:14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan KL, Goh SC, Minakhina S. Genetic screen for regulators of lymph gland homeostasis and hemocyte maturation in Drosophila. G3 (Bethesda) 2012;2:393–405. doi: 10.1534/g3.111.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mortimer NT, Goecks J, Kacsoh BZ, Mobley JA, Bowersock GJ, Taylor J, Schlenke TA. Parasitoid wasp venom SERCA regulates Drosophila calcium levels and inhibits cellular immunity. Proc Natl Acad Sci USA. 2013;110:9427–9432. doi: 10.1073/pnas.1222351110. A joint transcriptomic-proteomic approach characterizes the venom of Ganaspis sp.1. The role of one venom protein, a SERCA pump, is then explored in the context of parasitic wasp success
- 53.Colinet D, Schmitz A, Cazes D, Gatti J-L, Poirie M. The origin of intraspecific variation of virulence in an eukaryotic immune suppressive parasite. PLoS Pathog. 2010;6:e1001206. doi: 10.1371/journal.ppat.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Labrosse C, Stasiak K, Lesobre J, Grangeia A, Huguet E. A RhoGAP protein as a main immune suppressive factor in the Leptopilina boulardi (Hymenoptera, Figitidae)-Drosophila melanogaster interaction. Insect. Biochem. Mol. Biol. 2005;35:92–103. doi: 10.1016/j.ibmb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Chiu H, Govind S. Natural infection of D. melanogaster by virulent parasitic wasps induces apoptotic depletion of hematopoietic precursors. Cell Death Differ. 2002;9:1379–1381. doi: 10.1038/sj.cdd.4401134. [DOI] [PubMed] [Google Scholar]
- 56. Danneels EL, Gerlo S, Heyninck K, Van Craenenbroeck K, De Bosscher K, Haegeman G, de Graaf DC. How the Venom from the Ectoparasitoid Wasp Nasonia vitripennis Exhibits Anti-Inflammatory Properties on Mammalian Cell Lines. PLoS One. 2014;9:e96825. doi: 10.1371/journal.pone.0096825. Through the use of NF-κB reporter gene assays in mammalian cell lines, the venom of the ectoparasitoid wasp Nasonia vitripennis is shown to decrease NF-κB signaling in a dose-dependent fashion
- 57.Bitra K, Suderman R, Strand M. Polydnavirus Ank proteins bind NF-kB homodimers and inhibit processing of relish. PLoS Pathog. 2012;8:e1002722. doi: 10.1371/journal.ppat.1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohr SE, Hu Y, Kim K, Housden BE, Perrimon N. Resources for functional genomics studies in Drosophila melanogaster. Genetics. 2014;197:1–18. doi: 10.1534/genetics.113.154344. This review discusses the large variety of software tools and online resources available for functional genomics studies in Drosophila including how such resources can be effectively integrated into a research workflow
- 59. Colinet D, Kremmer L, Lemauf S, Rebuf C, Gatti JL, Poirie M. Development of RNAi in a Drosophila endoparasitoid wasp and demonstration of its efficiency in impairing venom protein production. J Insect Physiol. 2014;63:56–61. doi: 10.1016/j.jinsphys.2014.02.011. A proof-of-principle paper that describes the RNAi-mediated silencing of a venom-protein coding gene in an endoparasitoid wasp. This work also outlines a method for the in vitro rearing of late-stage wasp larvae
- 60.Tokusumi T, Sorrentino RP, Russell M, Ferrarese R, Govind S, Schulz RA. Characterization of a lamellocyte transcriptional enhancer located within the misshapen gene of Drosophila melanogaster. PLoS ONE. 2009;4:e6429. doi: 10.1371/journal.pone.0006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferrarese R, Morales J, Fimiarz D, Webb B, Govind S. A supracellular system of actin-lined canals controls biogenesis and release of virulence factors in parasitoid venom glands. J Exp Biol. 2009;212:2261–2268. doi: 10.1242/jeb.025718. [DOI] [PMC free article] [PubMed] [Google Scholar]