Abstract
Humans have coevolved with their microbes over thousands of years, but this relationship, is now being dramatically affected by shifts in the collective human microbiome resulting from changes in the environment and societal norms. Resulting perturbations of intestinal host-microbe interactions can lead to miscues and altered host responses that increase the risk of pathogenic processes and promote “western” disorders such as inflammatory bowel diseases, cancers, obesity, diabetes, autism, and asthma. Given the current challenges and limitations in gene therapy, approaches that can reshape the gut microbiome represent a reasonable strategy for restoring the balance between host and microbes. In this review and commentary, we highlight recent progress in our understanding of the intestinal microbiome in the context of health and diseases, focusing on mechanistic concepts that underlie the complex relationships between host and microbes. Despite these gains, many challenges lie ahead that make it difficult to close the gap between the basic sciences and clinical application. We will discuss the potential therapeutic strategies that can be used to manipulate the gut microbiota, recognizing that the promise of pharmabiotics (“bugs to drugs”) is unlikely to be completely fulfilled without a greater understanding of enteric microbiota and its impact on mammalian physiology. By leveraging the knowledge gained through these studies, we will be prepared to enter the era of personalized medicine where clinical inventions can be custom-tailored to individual patients to achieve better outcomes.
Keywords: Cancer, Dysbiosis, Gut–brain axis, Inflammation, Obesity, SCFA, Secondary bile acid, Vitamin D receptor
Introduction
Through co-evolution, hosts and microbes have forged a mutually beneficial or tolerant relationship, which is manifested in virtually all life forms. In humans and mammals, the acquisition of gut microbes does not occur randomly and is highly dependent on host factors, environmental cues, and self-assembly rules exert by microbes themselves. Once fully developed, the gut microbiome becomes an “essential” acquired organ that provides many vital functions to the host. However, the fundamental nature and stability of this evolutionarily determined relationship between host and microbe is now being threatened by drastic changes in the environment, diet, and life style over the past 50–100 years which have almost certainly reshaped the collective human gut microbiome. Corresponding and adaptive changes in the collective human genome, on the other hand, cannot proceed with such rapidity. Resulting mismatches in host-microbe relationships can then lead to homeostatic chaos, possibly explaining the increased incidence and prevalence of many disorders that have merged with alarming frequency in the modern age.
In this review and commentary, we highlight recent progress in understanding host-microbe interactions in the context of health and disease. In doing so, we provide specific examples where mechanistic insights into host-microbe relationships have transformed our conceptual thinking in this area. At the same time, we also bring up the many limitations and daunting challenges ahead of us that must be overcome to move the field forward. With the era of personalized medicine upon us, new knowledge will create opportunities to maintain health, effectively treat illness, and achieve better clinical outcomes.
The microbial organ: acquire and essential for health
The human gut microbiome is dominated by four phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. Firmicutes and Bacteroidetes are generally the most abundant of the gut microbiota, followed by Proteobacteria and Actinobacteria, with minor contributors, including Verrucomicrobia and Fusobacteria.1 Bacteroides and Ruminococcus are consistent with enriched intake of animal sources, while a plant-based diet favors Prevotella.2 The ratio of Prevotella to Bacteroides constitutes a potentially useful index for clinical diagnosis. Butyrate-producing bacteria, including Clostridium groups IV (Faecalibacterium prausnitzii) and XIVa, Roseburia spp., Butyricicoccus, and lactic acid bacteria (LAB), mainly Lactobacillus and Bifidobacterium, are believed to benefit the host through anti-inflammatory, anti-tumorigenic, and pathogen exclusion properties that they possess.3, 4, 5 There are also interactions between lactic acid- and butyrate-producing bacteria, which involve the ability of the latter to feed on lactate.1 Dysbiosis caused by a variety of perturbations can increase the risk of disease directly or indirectly when the delicate balance in bacterial community and host and microbiota are perturbed.
As an acquired and essential organ of the body, the gut microbiota provide a wide variety of beneficial functions, including: i) gleaning indigestible ingredients from food and synthesizing nutritional factors, such as vitamins; ii) detoxifying the deleterious xenobiotics and affecting the host metabotypes; iii) development of a robust systemic and intestinal immune system; vi) providing signals for epithelial renewal and maintaining gut integrity; and iv) secretion of anti-microbial products, which negatively select against pathogenic bacteria through the development of colonization resistance.6, 7 These functions are vital, because in the absence of gut microbiota or with its ablation with broad spectrum antibiotics, significant consequences can happen, e.g. improper development of the gut immune system and the development of Clostridium difficile antibiotic-associated colitis, respectively.
Gut microbial interactions are complex and fluid, capable of adjusting to physiological perturbations that are encountered on a daily basis. However, large or selective shifts in the gut microbiota as a consequence of host pathobiology, alterations of diet, medications, and other environmental triggers can upset critical inter-microbe as well as host-microbe relationships to initiate pathophysiological processes leading to disease. Two examples of this are the loss of beneficial microbes and their products and the emergence of disease-promoting microbes that produce microbial metabolites and proinflammatory mediators that negatively impact the intestine and other organ systems.
Short-chain fatty acids (SCFA), such as acetate, butyrate and propionate, are major fermentation products of microorganisms in gut. SCFA are the main energy source for colonocytes that also provide a number of other beneficial effects in maintaining intestinal homeostasis. For example, butyrate-producing bacteria have recently gained attention because they are important for a healthy colon and when altered contribute to emerging diseases, such as IBD.8 Butyrate can be produced directly by certain groups of bacteria: Butyricicoccus pullicaecorum; Faecalibacterium prausnitzii, Rosebuia and indirectly by cross-feeding some butyrate producers with lactate, as in the case of Eubacterium ballii, Anaerostipes caccae, and Escherichia coli.3 While butyrate is produced in colon, it can affect distal organs. Human serum butyrate, for instance, is in the range of 4 μM in British adults, and 29 μM in the hepatic portal vein which brings fats and other nutrients from the digestive tract to the liver.4 Shifts in butyrate-producing bacteria caused by bioavailability of substrate or changes in gut microbial membership and abundance can drastically change the production and amount of SCFAs that is delivered to the gut and distal organs.
Gut microbiota also produce a host of other metabolites that include many as yet unidentified or incompletely characterized natural products,5 as well as compounds well known to us. Among the latter are conjugated linoleic acids, vitamins (e.g. folate, riboflavin), and secondary bile acids, all having local and systemic effects.9 Conjugated linoleic acid, derived from bacterial metabolism of dietary linoleic acid, has many putative effects on host functions, including being anti-inflammatory and in regulating metabolic pathways.9 About 5% of secreted bile acid escape reabsorption in the ileum and enter the colon, where, because of their biophysical properties, they can dramatically affect the microbial landscape by suppressing many commensal gut microbes that are bile-intolerant while promoting others that are bile-tolerant. Most of the primary bile acids are rapidly converted to secondary bile acids through 7-alpha dehydroxylation by bacteria. Secondary bile acids (SBA) activate the nuclear farnesoid X receptor (FXR) and thereby protect against muscle fat deposition.6 SBA lithocholic acid (LCA) also bind to the vitamin D receptor (VDR) to promote detoxifying mechanisms that protect host cells against injury and inflammation.7, 10, 11 At physiological levels, SBA may contribute to the regulation of mucosal barrier function, cell renewal, and immune function. At higher, non-physiological levels, they can be cytotoxic, genotoxic, and proinflammatory, contributing to the development of mucosal inflammation and carcinogenesis.
There is increasing evidence that the reach of gut microbes extends beyond the intestine, affecting systemic processes, such as metabolism and organ functions of brain, cardiovascular system, liver, and others. Several metabolomic studies have identified hundreds of compounds in blood that are specifically derived or dependent on the presence of gut microbes.12 These findings have enlarged our thinking about the impact of the gut microbiome, particularly in influencing developmental processes and in the physiological regulation of a vast array of tissue and cell functions in the body.
The role of gut dybiosis in causing and sustaining disease states
The development of gut dysbiosis can set into play processes that activate the host immune and inflammation response, disturb intestinal homeostasis, and cause metabolic abnormalities. As an example, many microbes are selected by an inflammatory milieu because of their ability to survive the hostile inflammatory milieu, in contrast to many commensal microorganisms that cannot tolerate this type of harsh environment. In turn, it is to their benefit to maintain the inflammatory process to prevent the return of competing commensal microorganisms, i.e. creating a vicious cycle that leads to chronic disease. Their production of pathogen-associated molecular patterns (PAMPs) that include agents like flagellin, peptidoglycans, and lipopolysaccharide (LPS), further fuels the inflammatory process and contribute to the extent, severity, and duration of mucosal injury. Inflammation-induced intestinal barrier dysfunction and frank ulceration can also promote systemic entry of PAMPs that can affect many distant organs. For example, increased LPS translocation, has been proposed as driver of inflammation associated with obesity-related metabolic disorders13 and type 2 diabetes.13, 14 LPS, when subcutaneously infused into mice fed a normal diet, can also induce chronic inflammation that promotes the development of obesity and conditions of insulin resistance.14
Fei et al in fact demonstrated a causal relationship between endotoxin producers in the gut and obesity/insulin resistance outcomes, which can be tracked by changes in gut permeability, serum endotoxin, and inflammatory biomarkers.15
Dysbiosis associated with various disorders can also be characterized by lower community diversity. Resulting alterations in both structural (membership) and functional profiles of the gut microbiota in these circumstances are believed to be major contributors to the etiopathogenesis of complex immune, infectious, metabolic, and cancerous disorders, including inflammatory bowel disease (IBD),16 neonatal necrotizing enterocolitis,11, 12 gastrointestinal (GI) cancers, asthma,17 allergy, and infectious diseases. Even in organs that used to be considered sterile, such as esophagus and lungs, microbiota have been found that may contribute to the pathogenesis and progression of disease.13, 14 Other studies have also demonstrated widespread systemic effects of the gut microbiome determine various physiological states, such as cardiac size, hepatic gene expression, central nervous system function, and behavioral patterns.18 Thus, gut dysbiosis can disrupt host-microbe homeostasis and cause and/or contribute to many human diseases beyond the digestive system.19
Alterations of the gut microbiota have been associated with host metabolic disorders, including metabolic syndrome, type 2 diabetes, and obesity. One study demonstrated that richness of the gut microbiome correlated with certain metabolic markers.16 Akkermansia muciniphila, a mucin-degrading microbe that resides in the mucus layer, has been reported to prevent high-fat diet-induced metabolic disorders, including fat-mass gain, metabolic endotoxemia, adipose tissue inflammation, and insulin resistance.17 These effects appeared to be mediated by enhanced intestinal levels of endocannabinoids that control inflammation, the gut barrier, and gut peptide secretion. The results provide a rationale for the development of a treatment that uses this human mucus colonizer for the prevention or treatment of obesity and its associated metabolic disorders. Two recent reviews have explored the microbiota in relation to metabolic phenotype and disease risk.18, 19
In other recent studies by Hazen's group, the gut microbiota have been shown to promote atherosclerosis through metabolism of dietary carnitine and phosphatidylcholine.20, 21 Both are sources of dietary choline which is positively correlated with Bacteroides that is often associated with a Western diets. Choline is metabolized gut microbiota to trimethylamine which further undergoes hepatic metabolism to form trimethylamine oxide, the active agent that promotes atherosclerosis through its proinflammatory properties.20
The gut microbiota also appears to affect the central nervous system. A recent study showed that the gut microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders, using a mouse model of autism spectrum disorder (ASD).21 Another study demonstrated that the leakiness of the intestine appears to be important in the development of Parkinson's disease,22 promoting systemic exposure to intestinal bacteria and their toxins. Finally, the gastrointestinal tract is sensitive to stress and stress mediators, including catecholamines. Irritable bowel syndrome (IBS) is a common disorder through the gut–brain axis that might be triggered by gut bacterial imbalance.23 The brain–gut axis allows bidirectional communication between the central nervous system and the enteric nervous system, linking emotional and cognitive centers of the brain with intestinal functions.23 An association between dysbiosis and stress and depressive disorder has also been proposed.22, 24
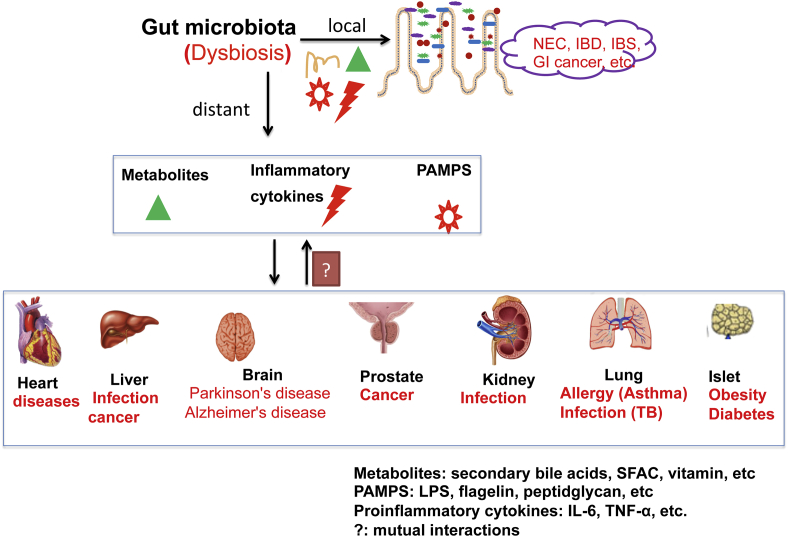
In summary, host-microbe interactions can have health and disease promoting effects in the gut and also in distal organs (Fig. 1). The effects can be mediated by a plethora of microbe-derived effector molecules that include metabolites (SBA, SCFA), immune and inflammatory modulators PAMPs, MAMPs (microbe-associated molecular pattern), and secreted small natural products. Perturbations of physiological host-microbe interactions can have significant consequences to immune and metabolic homeostasis at both the local and systemic level. The new “steady-state” created by these events can become chronic and difficult to break because of self-reinforcing host and microbial processes that are set into play.
Figure 1.
Host-bacterial interactions that could potentially mediate the gut microbiota human diseases in local intestine and distant organs. Gut microbiota influences amino acid bioavailability, is a source of metabolites (SBA, SCFA, PAMPs). Dysbiosis is associated with dysfunction of intestinal barriers and enhances proinflammatory cytokines (TNF-α, IFN-γ, IL-1β, and IL-8). All these factors could potentially influence pathogenesis and progression of human diseases.
Reshaping the gut microbiota to restore host-microbial balance
The therapies for many disorders that have a microbial component in their pathogenesis are mostly focused on the host side (infectious diseases being the exception). Now, a lot of thought is being given to manipulating the microbial side of the equation to restore host-microbial balance. There are various approaches to shape the gut microbiota, including personalized probiotic, prebiotics (fiber), vitamin/mineral supplementation, dietary, fecal microbiota transplantation (FMT), and/or the use of antibiotics. Prebiotics largely comprise a group of carbohydrates that cannot be degraded by the host but can otherwise promote the growth, fitness and functional properties of beneficial bacteria. Probiotics are live microbes that bestow the host with health advantages, whether it is anti-inflammatory, immunomodulatory, or trophic to the gut mucosa. They may also provide benefit in digestion and absorption of many dietary nutrients and minerals. Because prebiotics and probiotics differ in their properties and mechanisms of action, their efficacy for treating many disorders is often unpredictable. Patients take these agents empirically and physician who prescribe them do so without consideration of patient factors, the nature of the disease, and clear endpoints. In addition, these agents are marketed as nutraceuticals and, as such, are not subject to quality control or proof of efficacy. Finally, the notion that these agents can reshape the endogenous gut microbiome in a consistent and predictable way is probably untenable. The gut microbiota in most conditions has a substantial degree of resilience that would preclude fitness and colonization by non-indigenous probiotic microbes.
FMT is currently receiving a great deal of attention, having the theoretical advantage of being a diverse microbial community preselected under conditions of health. FMT has been found to be relatively safe and effective for the treatment of refractory C. difficile infection.25 It is also being considered the treatment of IBD, but these studies are currently under FDA review. Issues regarding safety and standardization of FMT have to be considered, particularly since many of these patients may be immunocompromised. We feel that FMT is not likely to be effective in moderate or severe cases of IBD in absence of other therapies directed against the host inflammatory and immune dysregulation. Even then, the question remains how long the membership, diversity, and function of the transferred microbiota can be sustained under different set of conditions presented by the new host's genetic, environmental, and physiological factors.
Several clinical and experimental studies have shown that diet is one of the most consistent and predictable ways of reshaping the gut microbiome. As shown by David, et al,26 different diets can cause very rapid shifts in gut microbial composition and function in healthy human subjects. Shifts in microbial assemblage induced by diet can have consequences for intestinal health, as was demonstrated in a study of genetically susceptible IL-10 deficient mice fed a diet rich in saturated milk fat. This diet promoted a bloom of sulfite reducing Proteobacteria (Bilophila wadsworthia), which increased the incidence and severity of spontaneous colitis in these animals.27 Long-term patterns of dietary consumption are associated with development of specific enterotypes that can have consequences for host immune function and disease risk. A recent study, for example, demonstrated that restricting life-long food intake by 30 per cent below what is needed to maintain body weight in mice can significantly change the composition of the gut microbiota.28 This calorie restricted diet promoted beneficial bacteria, such as Lactobacillus, and reduced harmful bacteria. Another examples is that of vitamin/mineral supplementation used for prevention and treatment of diseases. Vitamin D and its receptor VDR were shown to mitigate the dysbiosis associated with intestinal inflammation,29 possibly by restoring immune homeostasis, but also by direct effects on the gut microbiota.
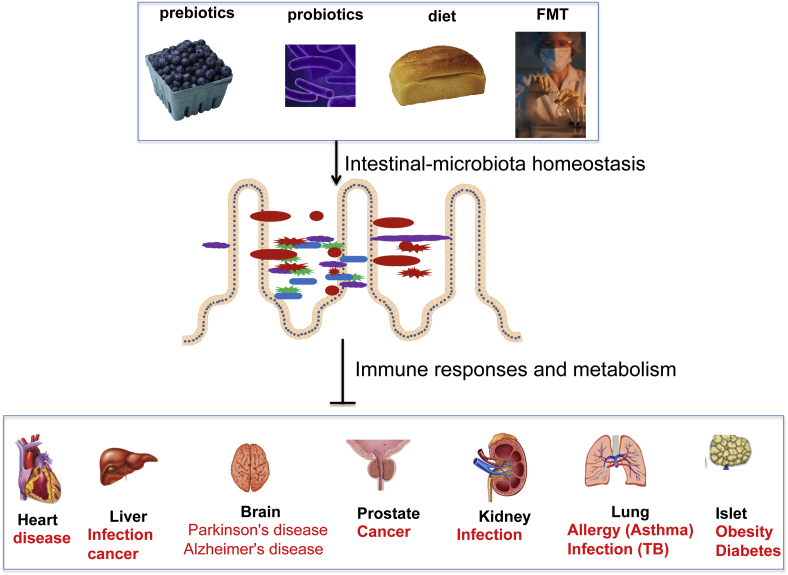
In summary, targeting the gut microbiota is a promising strategy for the prevention and treatment of human diseases believed to be affected by the development of dysbiosis. Restoring the healthy host-microbial interactions by personalized prebiotics/probotics, FMT, and dietary may be useful in achieving better clinical outcomes (Fig. 2).
Figure 2.
Targeting the gut microbiota in prevention and treatment of human diseases. Prebiotics are non-digestible carbohydrates fermented in gut that selectively stimulate the growth and/or activity of a limited number of bacteria and thereby, confer health benefits on the host. Probiotics are live microorganisms, which, when administered in adequate amount, confer a health benefit on the host. Personalized dietary and fecal microbiota transplantation (FMT)/of healthy donor feces to patients are used to prevent or treat diseases through restoring healthy host-bacterial interactions.
Challenges in the field
The study of the gut microbiome has advanced rapidly with new developments in technology (both cultivation-dependent and -independent) and bioinformatic tools for assessing community structure, function, and potential interrelationships among represented microorganisms. As a direct results, we've come to realize that the gut microbiome can be quite heterogeneious among different populations, influenced by changes in external factors (e.g. environmental and dietary cues), states of health, and intermicrobial and host assembly rules. An enormous amount of data has been generated by the Human Microbiome Project and other consortiums, but most of this information remains descriptive and inferential, limited in many cases by the lack of supporting experimental and clinical data, incomplete development and vetting of ‘omic’ technologies, and still evolving bioinformatics platforms for analyzing and integrating large datasets. These data potentially represent a treasure trove of information, not only for studies of bacteria, but also other microbes including viruses, fungi and Archea. However, many formidable challenges impede efforts to move the field forward. Waiting for new technologies and bioinformatics tools represents only part of the solution. Understanding how these data relate to human physiology and disease will require a closer partnership between the clinical and basic sciences so that information is no longer viewed out of context of clinical metadata and outcomes.
Limitations of technology and bioinformatics tools
Despite rapid advances in technology and analysis of large datasets, our ability to study the functional profiles of gut microbiota and their impact on host remains a major challenge. Taxonomic information provided by the study of 16S rRNA gene sets does not provide much information of community function, although attempts have been made by using reference genomes of highly represented microbiota to infer function.27 This approach remains unproven, largely because the genome inventories of microbial functional genes are still limited and incompletely curated. This may be helped by new methodologies in cultivation of single strains that may either be poorly represented in the gut microbiota or difficult to grow.30 These approaches will lead to the development of more complete microbial reference genomes. Even then, the conclusions made from bioinformatics analyses has to be vetted by experimental and clinical studies to determine if these approaches are meritorious. Shotgun sequencing of sample DNA to obtain metagenomic profiles is another approach to gain insights into community function. Again, the interpretation of data is limited by incomplete functional gene inventories, the fact that these data may not reflect true gene expression, and biases introduced by small sample biomass requiring gene amplification and host DNA contamination. Metatranscriptomes in theory provide a much more useful measure of gut microbial community function, but, even then, this approach currently requires fairly substantial sample biomass (restricting studies to mostly luminal samples) because yield is compromised by attempts to remove structural RNA to enrich for mRNA. The interpretation of these data, as with metagenomes, is further limited by the incomplete inventories of microbial functional genes. Combining metagenomic and metatranscriptome information, however, may provide greater confidence and ability to interpret this collective information, by confirming and potentially identifying the origin of encoded transcripts.
One solution to the vexing problem of assessing microbial function may come from new developments in ‘omic’ technologies and other functional assays. Significant advances have been made in proteomics, lipidomics, and metabolomics that have increased the value of information as well as cost for performing the analysis. For studies of the gut microbiome, these approaches are still under development and will require some form of experimental vetting to substantiate their informational value, but they provide promise because they are more direct measures of microbial function. Other measures of microbial community function are also under development, including candidate gene analysis (e.g. Butyrate transferase and butyrate kinase, dsrA and Biolog™ profiling) which provides an affordable, reliable, and informative metabolic signature of microbial samples. As will be mentioned below, the data from these types of studies will be particularly useful when viewed in the context of clinical and experimental information. In line with this, the second phase of the NIH Human Microbiome Project (HMP 2), large databases, computational systems to rapidly analyze and integrate data sets, and methodologies for functional profiling of human microbiomes are being developed for eventual use by end users.
Clinical challenges
The idiom “garbage in, garbage out” is highly relevant to the study the human gut microbiomes, particularly with regard to the inattention to study design, clinical context, and the type, acquisition, and processing of clinical samples. Most studies, particularly relating to IBD, have been cross-sectional and without recognition that the study populations are heterogeneous and that chronic diseases often have a transitional natural history. IBD, for instance, has traditionally been thought of as being two diseases, Crohn's Disease27 and ulcerative colitis (UC). In actuality, these designations are clinical phenotypes based on clinical presentation and histopathology. Over 160 genetic polymorphisms have now been identified through genome-wide association studies that are associated with increased risk for IBD, many shared between CD and UC. These data indicate that IBD are many diseases having different etiopathogenic mechanisms. Other confounding factors muddy the analysis of the gut microbiome in IBD. Most studies are conducted after the onset of colitis when the immune and inflammatory processes set into play can independently create dysbiosis that negatively selects against commensal microbes in favor of microorganisms that can survive the harsh conditions of chronic inflammation. Whether the observed dysbiosis is causative cannot be determined. In addition, the confounding effects of medications (antibiotics, immunosuppressive agents), changes in diet and daily lifestyle, and introduction of other environmental factors (hospital settings) are usually not taken into consideration. Chronic complex immune disorders like IBD are also transitional diseases, i.e. the initiating disease processes are often quite different from those that present later in the course of disease. For example, the development of maldigestion and malabsorption in CD at later stages of disease, caused by inflammation, anatomic alterations (e.g. fistula, stenosis), and surgery, can dramatically affect the composition and function of the gut microbiota. Consequently, cross-sectional studies collecting samples indiscriminately and failing to recognize these issues are unlikely to yield meaningful information that would help to understand cause-effect relationships or be translatable to the prevention or treatment of IBD.
Almost all human microbiome studies of the GI tract are based on the collection of stool samples, which can potentially be limiting and misleading. First, the gut microbiota are not uniformly distributed throughout the GI tract, but exhibit regional heterogeneity. Their assemblage in these areas is determined by ambient conditions and available nutrients that are provided by their host as well as partnering microbes. It is also known that the mucosa-associated microbiota are quite distinct from those that are found in the lumen, the former being more stable and particularly adept in living with the host. Because both UC and CD are anatomically distinct diseases, local factors, especially gut microbes, are likely to play a key role in their pathogenesis. CD can involve any part of the GI tract, typically starting as a discrete ulceration surrounded by normal mucosa, and eventually penetrating into and sometimes through the bowel wall. UC, on the other hand, only involves the colon, always starts in the rectum, and, in most cases, proceeds proximally as a contiguous front of inflammation. Based on these considerations, stool collection as the sole means to assess the gut microbiota in IBD can be inadequate. Finally, there is currently little standardization in the way samples are collected and processed (e.g. brush, biopsy, how to store samples, optimization of extract techniques, etc). All these nuances are likely to affect the results and analysis, making it difficult to compare results to identify true relationships.
Overcoming the challenges and moving forward
How can we resolve these issues? First, we should be more circumspect about how to design our studies, collect samples, and analyze data. Upto now, most studies have been technologically driven. As a result, a lot of data has been generated and analyzed in isolation of other metadata. Quality has been sacrificed for quantity and, in the end, the studies have ended up with unsubstantiated associations and questionable conclusions. Ideally, studies aimed at identifying potential causal relationships should be performed prospectively, using each subject as their own control and collecting critical information before the onset of disease to determine how they ultimately related to clinical outcomes. For complex immune disorders, this is difficult largely because subjects at risk who eventual develop the disease cannot be easily identified. Nevertheless, there are a few conditions where this is feasible. In Type I diabetes, “pre-diabetic” subjects can now be identified from whom, stool samples for microbiota analysis are being collected prospectively.31, 32 In IBD, studies of UC patients who undergo total colectomy with Ileal pouch anal anastomosis (IPAA) are being followed prospectively to determine if changes in gut microbiota and/or host response (e.g. transcriptomes, cytokine profiles) to predict who will develop an inflammatory condition called pouchitis.33 Non-UC patients (e.g. those with familial adenomatous polyposis) undergoing the same surgical procedure rarely develop this condition, suggesting this condition is a recapitulation of some of the pathogenic processes that originally caused the UC. IPAA-UC patients are also ideal subjects for study because they are generally no longer on medication, can be serially sampled endoscopically, and over half will develop pouchitis within 12–15 months. Therefore, the incidence and time course of disease make if feasible to collect and analyze corresponding datasets to potentially understand the factors that lead to pouchitis which, in turn, could provide insights into the fundamental cause of UC.
The identification of more homogeneous subsets of subjects is essential for identifying key associations between gut microbiota and clinical outcomes. In this regard, many groups have focused on twin studies where variations in genetics can be minimized.34 Similarly, studies of more homogeneous patient populations (Amish, Hutterites, Ashkenazum, African) have been increased the yield and impact of these types of studies where, in addition to common ancestral genetic backgrounds, factors such as environment, diet, life style, etc can be more easily controlled and studied. Even in a heterogeneous group, such as IBD patients, information collected longitudinally and carefully analyzed in the context of clinical stage and other metadata can provide important insights into the role of gut microbes in disease remission or relapse.
Despite our best efforts to design the most optimal human subjects based study, establishing true causality and defining disease pathogenesis remain difficult. Humans are so individual and the ability to rigorously control clinical parameters and variables is often beyond what is ethically and technically possible. The utilization of experimental approaches and models is therefore an essential counterpart to human-based research. For the study of host-microbe interactions, in vitro and in vivo models have been extremely useful in defining important relationship that could not otherwise be achieved through clinical studies. These types of studies can serve the additional purpose of vetting many of the modeling and bioinformatic approaches used by investigators to draw conclusions from large clinical datasets. One of the limitations of the first phase of the Human Microbiome Project was that studies focused solely on the human microbiota. As a consequence, opportunity was lost in gaining insights of evaluating the significance of findings in the context of host responses and experimental models where study parameters and genetics can be carefully controlled. Having said this, the caveat to the experimental approach that they don't always recapitulate the biology and pathobiology of humans. Thus, studies of gut microbiota should be multi-pronged and the approach should be iterative between humans and experimental systems. Standardizing approaches for sample acquisition and processing are also needed.
Finally, many of us have come to realize that reaching for the high hanging fruit where discovery lies requires a multi-disciplinary team effort, involving basic, translational, and clinical investigators who each bring something to the table. The next phase of research investigation of the gut microbiome should be guided by specific biological questions relevant to the clinical aspects and natural history of the disease, utilizing the full spectrum of ‘omic’ technologies, bioinformatic analysis, and experimental models.
Conflict of interest
There is no conflict of interest.
Acknowledgment
This work was supported by the Swim Across America Research Award to Jun Sun and NIDDK DK42086 (DDRCC), DK097268, and DK47722 to Eugene B Chang.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Jun Sun, Email: jun_sun@rush.edu, junsun7@gmail.com.
Eugene B. Chang, Email: echang@medicine.bsd.uchicago.edu.
References
- 1.Belenguer A., Holtrop G., Duncan S.H. Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol Ecol. Jul 2011;77(1):107–119. doi: 10.1111/j.1574-6941.2011.01086.x. [DOI] [PubMed] [Google Scholar]
- 2.Damman C.J., Miller S.I., Surawicz C.M., Zisman T.L. The microbiome and inflammatory bowel disease: is there a therapeutic role for fecal microbiota transplantation. Am J Gastroenterol. 2012;107(10):1452–1459. doi: 10.1038/ajg.2012.93. [DOI] [PubMed] [Google Scholar]
- 3.Marteau P. Butyrate-producing bacteria as pharmabiotics for inflammatory bowel disease. Gut. Dec 2013;62(12):1673. doi: 10.1136/gutjnl-2012-304240. [DOI] [PubMed] [Google Scholar]
- 4.Cummings J.H., Pomare E.W., Branch W.J., Naylor C.P., Macfarlane G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. Oct 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischbach M.A., Walsh C.T., Clardy J. The evolution of gene collectives: how natural selection drives chemical innovation. Proc Natl Acad Sci U S A. Mar 25 2008;105(12):4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cipriani S., Mencarelli A., Palladino G., Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J Lipid Res. Apr 2010;51(4):771–784. doi: 10.1194/jlr.M001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makishima M., Lu T.T., Xie W. Vitamin D receptor as an intestinal bile acid sensor. Science. May 17 2002;296(5571):1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 8.Machiels K., Joossens M., Sabino J. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. Sep 10 2013 doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 9.Delzenne N.M., Cani P.D. Interaction between obesity and the gut microbiota: relevance in nutrition. Annu Rev Nutr. Aug 21 2011;31:15–31. doi: 10.1146/annurev-nutr-072610-145146. [DOI] [PubMed] [Google Scholar]
- 10.Sun J., Mustafi R., Cerda S. Lithocholic acid down-regulation of NF-kappaB activity through vitamin D receptor in colonic cancer cells. J Steroid Biochem Mol Biol. Jul 2008;111(1–2):37–40. doi: 10.1016/j.jsbmb.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurutka P.W., Thompson P.D., Whitfield G.K. Molecular and functional comparison of 1,25-dihydroxyvitamin D(3) and the novel vitamin D receptor ligand, lithocholic acid, in activating transcription of cytochrome P450 3A4. J Cell Biochem. Apr 1 2005;94(5):917–943. doi: 10.1002/jcb.20359. [DOI] [PubMed] [Google Scholar]
- 12.Swann J.R., Want E.J., Geier F.M. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. Mar 15 2011;108(suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao S., Fei N., Pang X. A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol Ecol. 2014 Feb;87(2):357–367. doi: 10.1111/1574-6941.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cani P.D., Amar J., Iglesias M.A. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. Jul 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 15.Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. Apr 2013;7(4):880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaur N., Chen C.C., Luther J., Kao J.Y. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes. Jul–Aug 2011;2(4):211–216. doi: 10.4161/gmic.2.4.17863. [DOI] [PubMed] [Google Scholar]
- 17.Couzin-Frankel J. Bacteria and asthma: untangling the links. Science. Nov 26 2010;330(6008):1168–1169. doi: 10.1126/science.330.6008.1168. [DOI] [PubMed] [Google Scholar]
- 18.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amirian E.S., Petrosino J.F., Ajami N.J., Liu Y., Mims M.P., Scheurer M.E. Potential role of gastrointestinal microbiota composition in prostate cancer risk. Infect Agent Cancer. 2013;8(1):42. doi: 10.1186/1750-9378-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koeth R.A., Wang Z., Levison B.S. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. May 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartiala J., Bennett B.J., Tang W.H. Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and L-carnitine. Arterioscler Thromb Vasc Biol. Jun 2014;34(6):1307–1313. doi: 10.1161/ATVBAHA.114.303252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey M.T., Dowd S.E., Galley J.D., Hufnagle A.R., Allen R.G., Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun. Mar 2011;25(3):397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saulnier D.M., Ringel Y., Heyman M.B. The intestinal microbiome, probiotics and prebiotics in neurogastroenterology. Gut Microbes. Jan–Feb 2013;4(1):17–27. doi: 10.4161/gmic.22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Palma G., Collins S.M., Bercik P., Verdu E.F. The Microbiota-Gut-Brain axis in gastrointestinal disorders: stressed bugs, stressed brain or both? J Physiol. Apr 22 2014;592(Pt 14):2989–2997. doi: 10.1113/jphysiol.2014.273995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrof E.O., Khoruts A. From stool transplants to next-generation microbiota therapeutics. Gastroenterology. May 2014;146(6):1573–1582. doi: 10.1053/j.gastro.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.David L.A., Maurice C.F., Carmody R.N. Diet rapidly and reproducibly alters the human gut microbiome. Nature. Jan 23 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huttenhower C., Gevers D., Knight R. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang C., Li S., Yang L. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun. 2013;4:2163. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaoping Wu R.L., Zhang Yongguo, Xia Yinglin. Intestinal vitamin D receptor deletion leads to defective autophagy. Gut. 2014 doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Ma J.K., Hatzenpichler Roland, Karymov Mikhail A. Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in human microbiome project's most wanted taxa. PNAS. 2014 doi: 10.1073/pnas.1404753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown C.T., Davis-Richardson A.G., Giongo A. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giongo A., Gano K.A., Crabb D.B. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. Jan 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young V.B., Raffals L.H., Huse S.M. Multiphasic analysis of the temporal development of the distal gut microbiota in patients following ileal pouch anal anastomosis. Microbiome. 2013;1(1):9. doi: 10.1186/2049-2618-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith M.I., Yatsunenko T., Manary M.J. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. Feb 1 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]