Abstract
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.Multiple recent publications on RNA sequencing (RNA-seq) have demonstrated the power of next-generation sequencing technologies in whole-transcriptome analysis. Vendor-specific protocols used for RNA library construction often require at least 100 ng total RNA. However, under certain conditions, much less RNA is available for library construction. In these cases, effective transcriptome profiling requires amplification of subnanogram amounts of RNA. Several commercial RNA amplification kits are available for amplification prior to library construction for next-generation sequencing, but these kits have not been comprehensively field evaluated for accuracy and performance of RNA-seq for picogram amounts of RNA. To address this, 4 types of amplification kits were tested with 3 different concentrations, from 5 ng to 50 pg, of a commercially available RNA. Kits were tested at multiple sites to assess reproducibility and ease of use. The human total reference RNA used was spiked with a control pool of RNA molecules in order to further evaluate quantitative recovery of input material. Additional control data sets were generated from libraries constructed following polyA selection or ribosomal depletion using established kits and protocols. cDNA was collected from the different sites, and libraries were synthesized at a single site using established protocols. Sequencing runs were carried out on the Illumina platform. Numerous metrics were compared among the kits and dilutions used. Overall, no single kit appeared to meet all the challenges of small input material. However, it is encouraging that excellent data can be recovered with even the 50 pg input total RNA.
Keywords: cDNA synthesis kits, polyA, ribodepletion
INTRODUCTION
For over a decade, hybridization-based approaches (microarrays) have been the method of choice for comprehensive assessment of RNA expression. The advent of next-generation sequencing (NGS) allowed a potentially more comprehensive way to measure expression level,1–3 identify alternative RNA processing,4,5 and discover new genes6 by allowing RNA analysis through sequencing of cDNA at massively parallel scales. This procedure is denoted RNA sequencing (RNA-seq).
RNA-seq has been widely used for whole-transcriptome analysis, yet often faces the limitation of requiring at least 100 ng total RNA for library construction. Sometimes, it is not possible to obtain that much RNA, for example, if one is working with single cells or small clinical samples. Methods for constructing RNA libraries from low starting amounts of RNA in a high-throughput and reproducible manner are thus required. For instance, Tang et al.7, 8 used a modified protocol that has been initially developed for single-cell microarray analysis to perform transcriptome analysis using individual mouse oocytes and early embryonic cells. To further address this need, several vendors have recently developed RNA amplification kits designed to use low starting input amounts. Results from one such kit (SMARTer; Clontech, Mountain View, CA, USA) performing RNA-seq from individual circulating tumor cells and single neurons have been reported.9,10 More recently, in-house developed protocols have been used to examine gene expression from single cells in developing embryos.11 Vendors have also optimized or retooled RNA amplification kits that were initially developed for microarray analysis. Tariq et al.12 applied the Ovation RNA-Seq System (NuGEN Technologies Incorporated, San Carlos, CA, USA) to do whole-transcriptome RNA-seq analysis from minute amounts of total RNA isolated from mouse tissue.
In response to the increased interest in working with small amounts of input material, numerous protocols have been developed, tested, and compared in different laboratories.11,13–15 One limitation of the comparison experiments is that they are all done in the same lab, sometimes the lab that developed a procedure, and it can be difficult to extrapolate those results to a broader group of researchers.
This paper reports the results of a field evaluation of low-input RNA amplification kits carried out by the DNA Sequence Research Group (DSRG) of the Association of Biomolecular Resource Facilities (ABRF). One novel aspect of this study is that the work is done in many different core laboratories, allowing comparison of the output material across different generation sites.
The ABRF DSRG compared RNA-seq results from standardized RNA samples using 4 different kits: Ovation RNA amplification kit, SMARTer amplification kit, TransPLEX-WTA2-SEQ kit (Sigma-Aldrich, St. Louis, MO, USA), and SuperAMP kit (Miltenyi Biotech, San Diego, CA, USA). To assess how the kits worked across different amounts of starting RNA, we amplified and sequenced 3 dilutions (5 ng, 500 pg, and 50 pg) of a standard commercial RNA (Clontech) spiked with External RNA Control Consortium (ERCC) control RNAs (Ambion, Life Technologies, Carlsbad, CA, USA). The reproducibility of these 4 kits was tested by performing amplification with each kit type at 3 core facilities, for a total of 12 participating sites. RNA and amplification kits were distributed by the DSRG to the participating core facilities to conduct amplification. Amplified cDNA samples were then collected by the DSRG for library construction at a single site (to reduce variability not associated with the amplification itself) with the Illumina TruSeq RNA kit (Illumina Incorporated, San Diego, CA, USA). Illumina TruSeq mRNA libraries were also prepared using 5 ng, 100 ng, and 1 μg total RNA spiked with ERCC control Mix 1 and 100 ng and 1 μg ribosomal-depleted RNA spiked with ERCC control Mix 1 as standards. All libraries were pooled and sequenced on the Illumina HiSeq 2000 platform. The data obtained from all samples were used to assess the effects of RNA abundance on the amplification kits under study.
MATERIALS AND METHODS
Input Material
The human universal reference total RNA [(part number) 636538; Clontech] spiked in with ERCC control mix (part number 4456740) was used in this study. The quality of the RNA (RNA integrity number 8.2) was determined on the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) as per the manufacturer’s instructions.
Control “Standard” RNA-Seq Library Construction
To construct libraries for unamplified control suitable for Illumina sequencing, the total RNA sample was processed using both the polyA-based TruSeq V2 as well as a ribo-depletion-based mRNA enrichment method (Epicentre, Madison, WI, USA).
The polyA-based TruSeq V2 RNA sample preparation kit was used following the low-throughput modified protocol according to the manufacturer’s instructions. Because neither of the kits for the control data sets is configured to use the low starting amounts used for the other kits, input amounts of 5 ng, 100 ng, and 1 μg total RNA were tested. It should be noted that 5 ng is 20-fold lower than the recommended minimum for the TruSeq kit.
For the ribo-depletion method, 100 ng and 1 μg total RNA were used for ribosomal depletions using the Ribo-Zero Gold Kit for human, mouse, or rat (part number MRZG12324; Epicentre) according to the manufacturer’s instructions.
Both the polyA and rRNA-depleted RNAs were subjected to thermal fragmentation using the elute, prime, and fragment mix from the TruSeq RNA sample preparation kit. RNA fragments were then converted to double-stranded (ds) cDNA using reverse transcriptase and random primers provided in the TruSeq RNA library preparation kit. The end repair, dA-tailing, ligation of bar-coded adaptors (PN. 514104; Bioo Scientific, Austin, TX, USA) to cDNA fragments for Illumina sequencing and AMPure purification steps (Beckman Coulter Genomics, Brea, CA, USA) were done on the Tecan Freedom EVO Liquid Handling Robot (Tecan Group, Männedorf, Switzerland).
Generation of cDNA Using RNA Amplification Kits
The same batch of RNA and ERCC control mix was shipped to 12 different laboratories on dry ice. For comparison studies between amplification kits, each participant was instructed to combine 1 μl total RNA (1 μg/μl) with 2 μl of 1:100 dilution of ERCC spike-in mix 1 and 7 μl water. Serial dilutions (5 ng, 500 pg, and 50 pg) of the initial RNA cocktail (100 ng/μl) were used as substrates for amplification reactions using the manufacturer’s recommended protocols.
There were 4 different RNA amplification kits used in this study: SMARTer Ultra Low input RNA (part number 634935); SuperAmp (R&D Systems, Minneapolis, MN, USA); Ovation RNA-Seq System V2 (part number 7102; NuGEN Technologies Incorporated); and SeqPlex RNA (Sigma-Aldrich; R&D Systems). Kits were generously provided by the suppliers.
SMARTer cDNA Synthesis
For cDNA synthesis using the SMARTer Ultra Low RNA-Seq System, RNA samples were primed with modified Oligo-dT primer (SMART CDS primer II A 5′-AAGCAGTGGTATCAACGCAGAGTACT(30) N-1N-3′, where N = A, C, G, or T, and N-1 = A, G, or C), and first-strand synthesis was carried out using SMARTScribe (Clontech) reverse transcriptase (Supplemental Fig. 1 (418.9KB, docx) ). Once the reverse-transcriptase reaction reaches the 5′ end of an RNA molecule, the terminal transferase activity of Moloney murine leukemia virus adds a few nontemplated bases to the 3′ end of cDNA. The SMARTer II A Oligonucleotide (5′-AAGCAGTGGTATCAACGCAGAGTACXXXXX-3′, where X = undisclosed bases) then base pairs with the nontemplate nucleotide stretch, creating an extended template. The SMARTScribe then switches template and continues transcribing to the end of oligonucleotide. The resulting full-length, single-stranded (ss) cDNA contains the complete 5′ end of the mRNA, as well as anchor sequences that are complementary to the SMARTer Oligonucleotide for second-strand synthesis. The ds cDNA was purified using Agencourt AMPure beads (Beckman Coulter Genomics). The purified cDNA was then amplified by long-distance PCR using the Advantage 2 PCR Kit (part number 639206; Clontech) and the included PCR primer (5′-AAGCAGTGGTATCAACGCAGAGT-3′) for 12 PCR cycles (5 ng), 14 PCR cycles (500 pg), and 17 cycles (50 pg) in 50 μl reaction according to the manufacturer’s instructions.
SuperAMP cDNA Synthesis
RNA samples were combined with 5 μl supermagnetic MACS MicroBeads (Miltenyi Biotec) for mRNA isolation (Supplemental Fig. 2 (418.9KB, docx) ). The bead-RNA mixture was applied to a column and rinsed to remove rRNA and other contaminants. First-strand cDNA mix was added, then in-column cDNA synthesis was carried out at 42°C for 45 min. After rinsing, the 3′ tailing mix was applied on top of the column matrix and incubated at 37°C for 1 h for 3′ tailing of ss cDNA. After washing steps, the column was removed from the thermoMACS separator (Miltenyi Biotec) and placed into a 0.2 ml PCR tube. Resuspended PCR mix (New England Biolabs, Ipswich, MA, USA) was applied to elute material from the magnetic beads. A total of 2 μl New England Biolabs Phusion High-Fidelity DNA polymerase was added to the eluate and amplified ss cDNA by running SuperAMP PCR for 40 cycles. The ds cDNA was then purified using the High Pure PCR Product Purification Kit (Roche, Basel, Switzerland) following the manufacturer’s instructions. This Miltenyi Biotec product was a prerelease item provided by Miltenyi Biotec for the purposes of this project. Following discussions between Miltenyi Biotec scientists and participating researchers, they have further improved their protocol by optimizing the bead composition and reducing the final number of PCR cycles. These changes have not been evaluated in this study.
Ovation cDNA Synthesis
For cDNA synthesis with the Ovation RNA-Seq kit, RNA samples were reverse transcribed by using a combination of random hexamers and poly-T RNA/DNA chimeric primer mix (Supplemental Fig. 3 (418.9KB, docx) ). The ds cDNA with a unique DNA/RNA heteroduplex at one end was then generated by fragmentation of the mRNA using RNA-dependent DNA polymerase. The resulting cDNA was then purified using RNAClean XP purification beads (Beckman Coulter, Brea, CA, USA). The purified cDNA was then subjected to linear amplification by the so-called single primer isothermal amplification (SPIA) process. In this protocol, DNA/RNA chimeric SPIA primer binding, DNA replication, strand displacement, and RNA cleavage are repeated for multiple rounds, resulting in accumulation of SPIA cDNA. Random hexamer priming was then used to convert ss-SPIA cDNA to ds cDNA.
SeqPlex cDNA Synthesis
With the SeqPlex RNA amplification kit, the RNA samples were reverse transcribed with random primers having semidegenerate 3′ ends and defined universal 5′ ends (Supplemental Fig. 4 (418.9KB, docx) ). The displaced single strands generated during the process serve as a template for primer annealing and second-strand cDNA synthesis. The resultant ds cDNA flanked by a single universal primer was amplified by 17–19 cycles of PCR. The cDNA was then purified using the GenElute PCR Clean-Up Kit (catalog number NA1020; Sigma-Aldrich) or AMPure beads (part number A63881; Beckman Coulter Genomics). Finally, the primer at both ends was removed using primer removal enzyme followed by purification using the GenElute PCR Clean-Up Kit or AMPure beads (Agencourt).
Construction of Libraries from Amplified cDNA
The yield of all the amplified products was measured using a Qubit (Invitrogen, Carlsbad, CA, USA). The fragment size of the purified ds cDNA was assessed on the Agilent 2100 Bioanalyzer. Details of the amount of cDNA recovered, fragment sizes of amplified cDNA for each dilution, and difficulty level of the kits are included in Supplemental Table 1 (364.3KB, xlsx) .
All kits except the Sigma-Aldrich SeqPlex RNA amplification kit generated cDNA that required fragmentation prior to library generation. Fragmentation was carried out with a Covaris S220 AFA Ultrasonicator (Woburn, MA, USA) using the recommended conditions for generating 200–500 bp fragments. When possible, 500 ng cDNA was used for subsequent manipulations, though frequently the entire sample was used (Supplemental Table 1 (364.3KB, xlsx) ). All the libraries were constructed on the Tecan Freedom EVO300 Liquid Handling Robot using the TruSeq DNA Sample Preparation kit reagents at a single site (Stowers Institute, Kansas City, MO, USA). All adapter ligation mixes were amplified for 12 cycles. Finished libraries were purified from free adaptor product using AMPure beads (Agencourt). The resulting purified libraries were quantitated using a Qubit, and the size range of the products was confirmed by Caliper’s LabChip GX (Supplemental Table 2 (364.3KB, xlsx) ).
Sequencing
The indexed 35 cDNA libraries (30 from amplified and 5 from nonamplified cDNA) were combined in equimolar ratios into 1 pool and loaded into 7 lanes of the Illumina HiSeq 2000 for a 50-cycle sequencing run, with the Illumina PhiX sample used as control. A new pool was created for underrepresented libraries (based on the data generated using the first pool) and subjected to sequencing on another 5 lanes of HiSeq 2000. Image analysis and base calling were performed using Illumina pipeline version 1.5.15.1. Average number of total reads is shown in Supplemental Table 3 (364.3KB, xlsx) .
Alignments
Reads were aligned with TopHat 2.0.6 (Bowtie 2.0.2): bt2-very-sensitive setting, Ensembl 66 Gene Transfer Format (GTF), to University of California, Santa Cruz (UCSC) hg19. The Ensembl GTF was modified to reflect UCSC chromosome names. Read hits to genomic features were quantitated using intersectBed.
There were 2 additional alignment versions made: 1 without multireads (Unique), and another with no multireads and with only 1 read per unique start position in the genome (Unique-Rmdup), for purposes of comparison. Library complexity was examined using read uniqueness, as (N unique reads) / (N total reads), for each sample.
ERCC Controls
Unaligned reads were aligned to ERCC92 sequences (http://tools.invitrogen.com/downloads/ERCC92.fa) with Bowtie 2.0.2: very fast setting and quantitated with samtools idxstats. Observed relative spike concentrations were correlated to expected (http://tools.invitrogen.com/content/sfs/manuals/cms_095046.txt) and plotted. Sample similarities were measured by Pearson correlation of all ERCC values.
Reads per Kilobase of Exon per Million Reads
Exon- and junction-aligning reads were converted into reads per kilobase of exon per million reads (RPKMs) per sample, where M is the total number of exon- and junction-aligned reads for all genes per sample. Sample similarities were measured by Pearson correlation of all gene RPKMs.
Gene Detection
All read sets were randomly down sampled to 10 million reads to avoid biases in gene detection due to differential number of recovered reads. Stochasticity in detections was addressed in 2 ways: 1) down sampling was repeated 5 times; and 2) gene detectability was calculated using 1, 2, or 5 reads. Detectability of each Ensembl gene biotype was evaluated for all methods.
Gene Coverage
For each gene and sample, base-by-base read depth values were extracted from the whole-genome BedGraph file. Intronic positions were removed, coverage vectors for minus-strand genes were reversed, and remaining values divided into 100 bins and averaged to produce a uniform 100-column matrix of gene coverage’s for each sample. Genes shorter than 100 bp were dropped. Global coverage bias per sample was graphed by converting rows of the matrix to Z scores and plotting the column sums. Gene-wise trends in coverage bias and variability were investigated by taking the coefficient of variance of the original coverage vector per gene and also by taking the ratio of the first-quarter mean depth (5′ coverage) to fourth-quarter mean depth (3′ coverage) on the gene body.
RESULTS
Library Generation
One of the primary goals in many ABRF Research Group studies is the assessment of site-to-site variability in experimental manipulations. For the current low-input study, 12 sites participated in the comparison of 4 kits, with 3 sites per kit. There were 2 sites unable to generate material resulting in sequenceable libraries. Because reagents for these studies are donated by the manufacturers, it is not always possible to repeat compromised experiments. Thus, data series for 2 sites are unavailable.
Control universal human reference RNA, representing a pool of total RNA from different human tissues, was distributed to the sites. As described in MATERIALS AND METHODS, all kit manufacturers’ protocols were faithfully followed to generate cDNA. The resulting output cDNA was quantified by Qubit and further characterized by bioanalyzer. The different kits produced patterns and sizes of cDNAs stereotypical for each kit but distinct from each other. Representative trace files are shown in Supplemental Table 1 (364.3KB, xlsx) . The cDNAs from all sites were sent to The Stowers Institute for library production and sequencing on the Illumina HiSeq platform; details of the syntheses are provided in Supplemental Table 1 (364.3KB, xlsx) . The control libraries were also produced at Stowers Institute. Because all libraries were synthesized simultaneously, using automation throughout, it is expected that variances at the construction step will be as controlled for as possible. Libraries were evaluated by Bioanalyzer analysis prior to sequencing (Supplemental Table 2 (364.3KB, xlsx) ).
Despite similarities in the bioanalyzer appearance of the libraries, they exhibited different behaviors on the sequencer. The Clontech samples showed the strong presence of a particular sequence derived from the SMART adapters used during amplification. This is a known contaminant with this system (Clontech, personal communication). The Miltenyi Biotec samples demonstrated more rapid decline in base quality over the course of the run, though the cause is not known. Because these are from samples prepared at different sites, it seems probable that they represent common outcomes from these kits (Fastqc results not shown).
Analysis
Various sequencing metric and data comparisons were carried out to assess the output of the different kits. In some cases, the analyses utilized the entire data set generated from a particular kit, whereas other analyses were normalized to the same number of reads in an attempt to emphasize the validity of the comparisons (Supplemental Table 3 (364.3KB, xlsx) ). Control data sets were generated from libraries using established commercial reagents and suggested protocols for construction. Although providing an essential analytic comparison, data sets from these TruSeq polyA-based and Ribo-Zero ribo-depleted libraries were generated with much more starting material, i.e., under less-challenging conditions. Therefore, these libraries would be expected to perform better. Another important caveat is that the lowest RNA concentration of 50 pg was out of specifications for the Sigma-Aldrich and NuGEN Technologies Incorporated kits, whereas the lowest amount of RNA used in the TruSeq V2 kit, 5 ng, was also out of the manufacturer-recommended range of starting material.
Read Mapping and Library Complexity
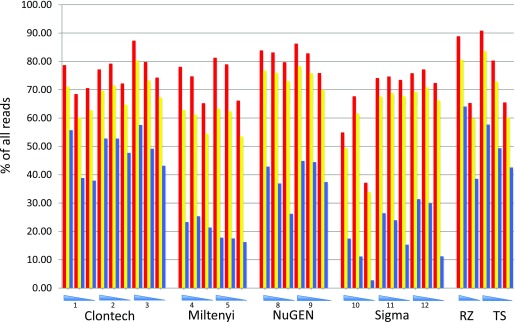
Initial analyses focused on the basic parameters of mapped reads and read compositions. All the reads from each kit and concentration were used for these calculations. Figure 1 shows the percent distributions of reads into those that aligned, reads that aligned and uniquely mapped, and uniquely mapping reads from which duplicate reads have been removed (Supplemental Table 3 (364.3KB, xlsx) shows the data from which these figures were derived). Several trends are observable. One is that the percent distribution of aligned reads for a given kit is not greatly affected by input concentrations. Another observation is that site-to-site variability using different kits appears small. Finally, whereas all methods produced predominantly uniquely mapping reads, the percentage of duplicate reads (those beginning with the same chromosomal coordinates) varied substantially between kits and across the concentration ranges detected, with the lowest input amounts showing the largest percentage of putative PCR duplicates (Fig. 1 and Supplemental Table 3 (364.3KB, xlsx) ). The results revealed that the highest percentage of unique reads was generated by Clontech, which showed unique read values very close to those generated by the control libraries. The Miltenyi Biotec and Sigma-Aldrich kits produced the lowest percentage of unique sequences.
Figure 1.
Read composition comparison. The percentage of all recovered reads falling into particular categories is shown: red indicates all aligning reads, yellow represents all aligning reads mapping to a unique genomic location, and blue shows the uniquely mapping reads with putative PCR duplicates (i.e., reads starting at the same coordinate) removed. The numbers represent the different sites providing the starting cDNA from the indicated kits; RNA concentrations from high to low are represented by the blue triangles. RZ, Ribo-Zero; TS, TruSeq.
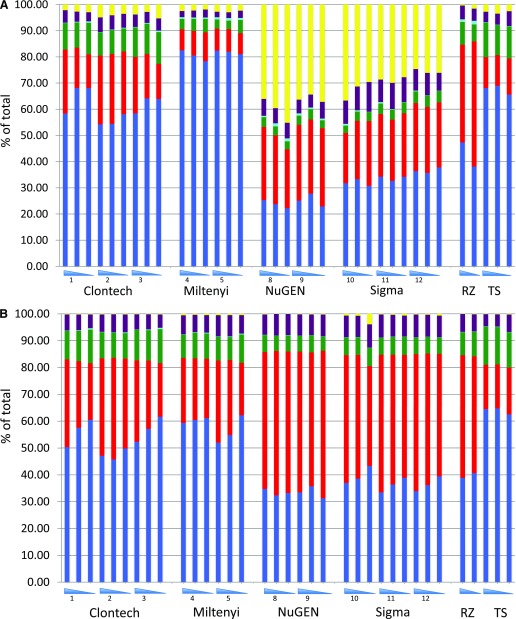
The distribution of reads across several broad biologic categories, i.e., read composition, is shown in Fig. 2. These comparisons are shown with duplicate reads included (Fig. 2A) and removed (Fig. 2B). The most noticeable and important trend here is that polyA selection-based methods (Clontech, Miltenyi Biotec, and “TS” for Illumina TruSeq kit) show a substantially higher proportion of exonic reads than the ribo-depletion-based methods (NuGEN Technologies Incorporated, Sigma-Aldrich, and “RZ” for Ribo-Zero). This difference can be up to 4-fold in certain pairwise comparisons, an important factor when deciding the number of reads required to match a specific experimental goal. The loss of reads in the exonic compartment was primarily due to the increased number of rRNA reads (∼35% for NuGEN Technologies Incorporated and ∼29% for Sigma-Aldrich) and intronic reads (representing unprocessed or partially processed transcripts). Although NuGEN Technologies Incorporated and Sigma-Aldrich had many more rRNA reads than Ribo-Zero-depleted material, the intronic fraction was higher in the latter. Removal of putative PCR duplicates (i.e., reads starting at the same nucleotide) had varying effects on read representation. For NuGEN Technologies Incorporated and Sigma-Aldrich, whereas the proportion of exonic reads increased overall, the proportion of intronic reads increased more. For Miltenyi Biotec, Clontech, and both control libraries, the exonic read proportion actually decreased.
Figure 2.
(A) Comparison of RNA category percentages. The percent composition of several broad categories of RNAs is displayed; 100% represents the total number of uniquely aligning reads for each sample set. Blue indicates exonic reads, red shows intronic, yellow represents ribosomal, purple indicates intergenic, aqua shows novel splice junctions, and green indicates known splice junctions. The kits used and participating sites are indicated, with RNA input amounts from highest to lowest represented by blue triangles. (B) Comparison of RNA category percentages. Designations shown are the same as those in (A), except the putative PCR duplicates have been removed from these data sets prior to percent category calculations.
Gene Abundance and Biotype Comparisons
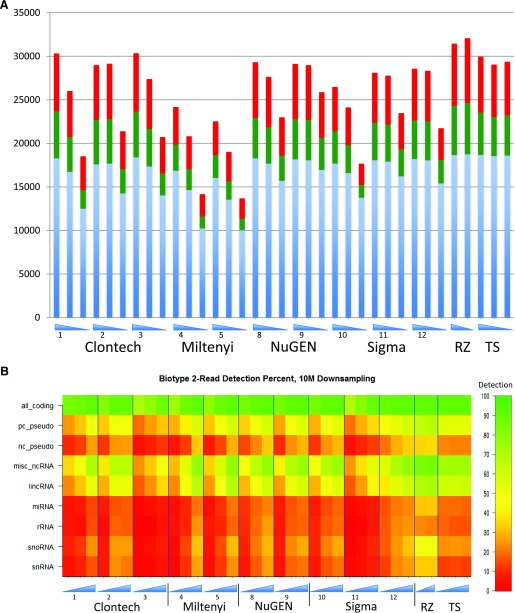
The sensitivity of gene detection was assessed for each input amount of RNA and kit. This comparison, based on a detection threshold of 2 reads, is shown in Fig. 3. The genes used are the GRCh37 Ensembl version 66 compilations of 53,598 genes. To normalize detection for the variable read numbers recovered, 5 random samplings of 10 million reads each were carried out per library (the lowest read count for any library was 13.3 million). Detected genes are broken down into coding, noncoding, and pseudogenes. The composition of detected gene biotypes showed variability across protocols and dilutions. However, the number of genes detected reduced with lower amounts of input RNA. This trend was evident for all the low-input protocols tested and at most of the sites. The unamplified control, tested using 3 different amounts of input RNA (5 ng, 100 ng, and 1 μg), was more robust. The results revealed that the 5 ng Clontech and NuGEN Technologies Incorporated samples performed nearly as well as the control data sets, with Sigma-Aldrich very close. The Miltenyi Biotec protocol yielded the fewest number of detected genes, which may be a consequence of the large number of amplification cycles used in this protocol (see DISCUSSION).
Figure 3.
(A) Detection sensitivity. The number of genes represented by 2 alignment hits is shown for the different construction kits and protocols; coding genes are shown in blue, ncRNAs in green, and pseudogenes in red. RNA concentrations going from high to low are represented by blue triangles, and the site numbers are indicated. The queried set is the Ensembl version 66 compilation of 53,599 genes. (B) Sensitivity of gene detection by gene type. The different biotypes are indicated on the left-hand side. For each library, detected gene percentages represent the mean from 5 different 10 million read down samplings. Heatmap values are detected genes as percent detectable, detection requires ≥2 reads, and “detectable” genes are those with ≥2 reads in the 1 μg Ribo-Zero control. Kit types and dilutions are indicated as in the other figures.
Because both polyA and non-polyA-based kits were used, the proportion of gene biotypes recovered would be expected to differ substantially. To assess this, the detectability of different classes of noncoding and protein-coding RNAs was calculated and plotted on the heatmap in Fig. 3B and Supplemental Table 4 (364.3KB, xlsx) . For each sample and biotype, the number of genes detected by at least 2 reads in that sample is shown as a percentage of detectable genes in that biotype. The values for detectable genes in a particular biotype were determined experimentally: a gene was considered “detectable” if it had at least 2 reads in the 1 μg Ribo-Zero control. Thus, by definition, Ribo-Zero detected the most genes in every biotype. The 50 pg libraries had substantial sensitivity loss in all biotypes; noncoding RNAs (ncRNAs) in particular showed up to 60% loss versus their 5 ng counterparts. Minimal detectability of small housekeeping ncRNAs like mi-, sn-, and snoRNAs, as well as their pseudogenes, is to be expected and may be due more to library size selection than to amplification bias. Performance of ribo-depleted libraries was more robust than polyA libraries across lowered input amounts: comparing detected genes at 5 ng and 50 pg for each kit, the ribo-depletion-based kits had average losses of 14 and 37% for protein-coding and ncRNAs, respectively, whereas the polyA-based kits had average losses of 30 and 52%.
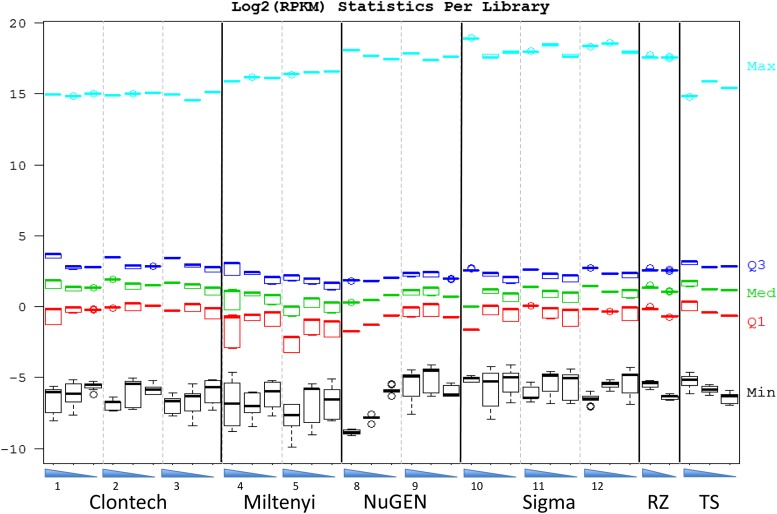
To determine if any potential biases in expression level are generated during amplification, RPKM values per gene were calculated for each kit. RPKMs showed the least variation among kits, sites, and dilutions. RPKMs were calculated using bedTools to count reads on exons. The distributions of 5 RPKM percentiles per library (0, 25, 50, 75, and 100%) are shown in Fig. 4. A consistent trend was that the maximum RPKMs are higher in the ribo-depleted libraries than polyA based; this is caused by the mitochondrial rRNAs and 1 somatic rRNA pseudogene.
Figure 4.
RPKM measurements of detected reads. Gene abundance was quantified and divided into quantiles based on their relative levels. RNA concentrations going from high to low are represented by blue triangles, and the site numbers are indicated.
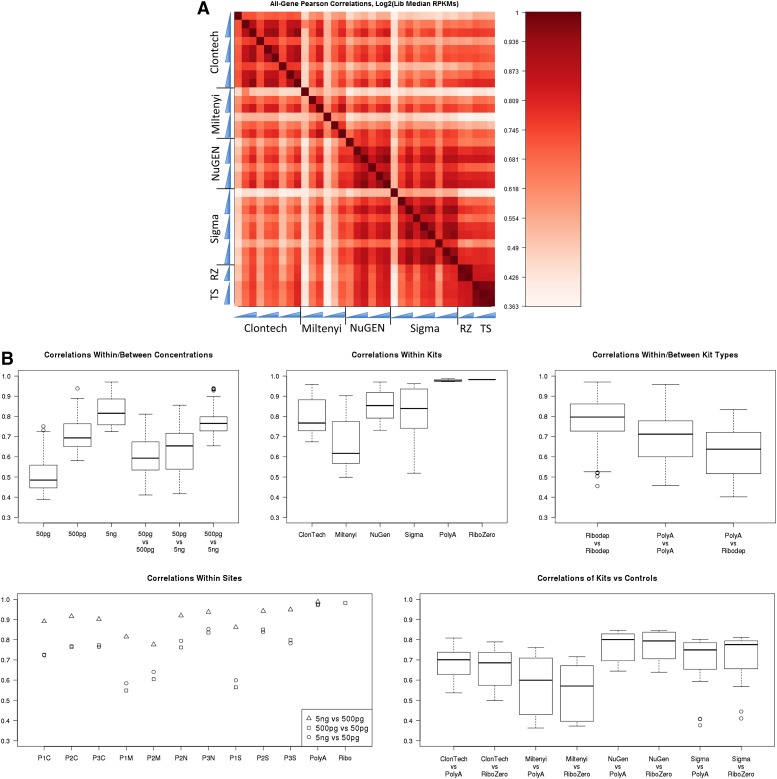
Because the goal of many RNA-seq experiments is to analyze differential gene expression among samples, it was of interest to quantify expression similarities between kits and dilutions. Expression profiles for all expressed genes were compared by Pearson correlation, using log2 (median RPKM), and displayed in a heatmap matrix shown in Fig. 5A. Specific sets of R values were extracted from the matrix and box plotted in Fig. 5B. In accordance with earlier performance metrics, the 50 pg samples had low correlations with the control libraries, whereas 500 pg and 5 ng tended to behave similarly. Not surprisingly, at the higher input amounts, all kits had more similar expression profiles. Also, gene expression values in the ribo-depletion kits (NuGEN Technologies Incorporated and Sigma-Aldrich) were more similar to each other than were the polyA kits (Clontech and Miltenyi Biotec). The greater consistency in RPKMs for ribo-depleting kits parallels their increased gene detection. The NuGEN Technologies Incorporated kit appeared to have the least expression bias because it correlated best with both control libraries and also had highest intrakit R values. Furthermore, it appears that intersite variability is not substantially different from intrasite variability. For example, the correlations between the different input amounts within a site vary from 0.55 to ∼0.9, about the same range as the correlations within and between the concentrations among all sites.
Figure 5.
(A) Gene expression comparisons. Pearson correlations among kits and across dilutions were graphed based on gene expression values. These values were derived from the Log2(Median Library RPKM) per gene. (B) Pairwise correlations of gene expression measurements. Values are displayed as box plots when sufficient data points are used, otherwise individual values are displayed.
Coverage Uniformity
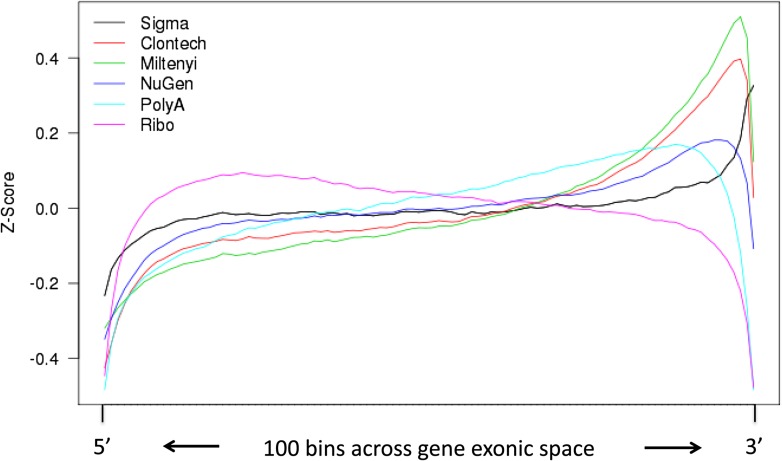
As discussed, the amplification kits and controls used here employ widely varying strategies for generating cDNA. It was therefore of interest to examine the uniformity of gene body coverage with the different kits. This is visualized in Fig. 6. For this figure, the coverage data from a particular kit were pooled for all RNA concentrations and across all sites. This was done because gene body coverage was not substantially affected by dilution or sites (data not shown). Analysis of read coverage across the exonic regions of genes demonstrated that all amplification kits showed 3′ biases in comparison to Ribo-Zero control sample. The Miltenyi Biotec and Smart kits’ polyA-based enrichment methods showed significant 3′ bias, followed by TruSeq and then NuGEN Technologies Incorporated, probably because NuGEN Technologies Incorporated uses both Oligo-dT and random primers during first-step cDNA synthesis. The Sigma-Aldrich kit displayed more even coverage after a pronounced but short 3′ bias. Variation in 3′ bias per input concentration was negligible except for Miltenyi Biotec, which moderately increased in bias with increasing input. Our results also show a slight 5′ bias in Ribo-Zero samples.
Figure 6.
Gene body coverage. The mean coverage from all reporting genes and all samples from the different kits is displayed (i.e., all input concentrations and site data were pooled for a given kit). The figure shows the Z score across each bin, going from 5′ to 3′ left to right. Exonic reads within detected genes were used for the calculations.
ERCC Controls
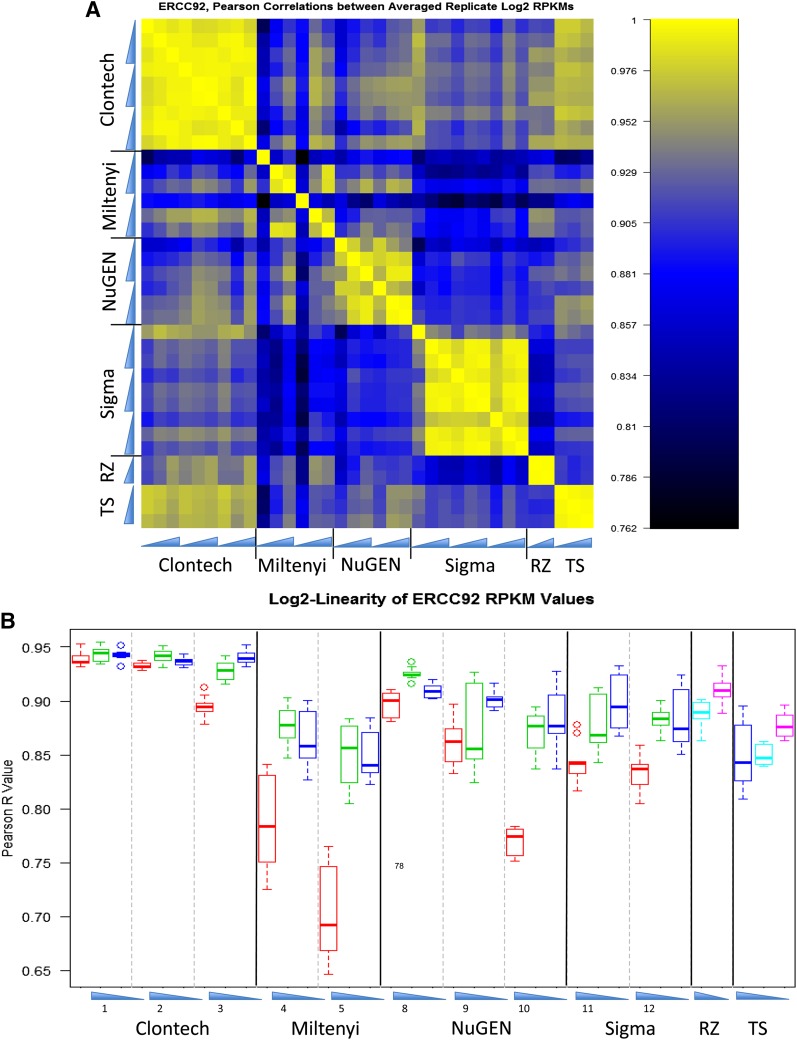
The universal human reference RNA sample distributed to each site was spiked with an ERCC control prior to dilution. ERCC controls are synthetic polyadenylated transcript pools with each transcript present at known concentration. Comparison of these known concentrations with the values recovered in the experiment can provide a quantitative measure of sequence loss or bias during library preparation. ERCC analyses are shown in Fig. 7, the Pearson correlation of ERCC RPKMs between samples displayed as a heatmap matrix is shown in Fig. 7A, and Pearson correlations of observed versus expected concentrations are shown in Fig. 7B. Despite the apparent spread in values in Fig. 7A, the lowest correlations measured (excluding Miltenyi Biotec) were >0.76. These results indicate that ERCC recovery was broadly consistent across treatments and concentrations. Similar to the other metrics displayed, intersite variability was low, and again the lowest dilution, 50 pg, was apparent through compromised performance. However, the degree of falloff seemed to be kit dependent, with the Clontech protocol showing the most consistent results across the dilution series. Figure 7B shows the log linearity of ERCC RPKMs because spike-in transcript concentrations have a logarithmic distribution. If the recovered concentrations of spike-ins equaled the expected concentrations, this value would be 1.0. Particularly at the 2 higher concentrations, all kits and sites performed similarly and were able to approach the expected level of expression. The Clontech kit did the best job of approaching the theoretic perfect value.
Figure 7.
(A) ERCC standards expression. Pearson correlations among kits and across dilutions were graphed based on the recovered RPKM values of ERCC standards. Blue triangles indicate dilution series. (B) Log linearity of ERCC RPKM values. Box plots indicate expression levels relative to the known concentrations of the original standards present. A value of 1.0 indicates equivalence between the published standard values and the recovered values.
DISCUSSION
The ability to elucidate the transcriptome from a single or small number of cells will provide a high-resolution examination of basic processes like differentiation, carcinogenesis, and circulating cell physiology. A number of kits are currently available to assist this effort. These kits differ in their basic approaches, generally splitting into polyA-based versus ribo-depleted protocols. Suggested input concentrations can also vary. Two of the kits tested, NuGEN Technologies Incorporated and Sigma-Aldrich, were challenged with starting material amounts below the manufacturer’s recommendations.
It was of interest to see how the read composition and biotype distribution varied from kit to kit. These comparisons were made between kits but also between the low-input kits and data sets generated from libraries made using commonly utilized higher-input kits. The chief findings here are that the kits showed substantial differences in both the complexity of the libraries generated, as indicated by the percentage of duplicate reads (Fig. 1), and the distribution of recovered reads in different transcript compartments (Fig. 2). These latter differences in compartmentalization were to be expected because ribo-depletion-based kits (here, Sigma-Aldrich and NuGEN Technologies Incorporated) are known to generate substantially more reads in intronic regions and more rRNA reads. The proportion of rRNA reads, at least for the NuGEN Technologies Incorporated kit, was higher than expected based on a previously published study12 and manufacturer’s reports (http://www.nugeninc.com/nugen/index.cfm/products/cs/ngs/rna-seq-v2/), but the fact that the values reported here were generated across all dilutions at different sites suggests that technical issues within the testing laboratories were not involved. The proportion of exonic reads was highest using the Miltenyi Biotec kit, but this metric alone is misleading because these libraries are among the least complex. Both this and the Sigma-Aldrich kits generated many more PCR duplicates than the other kits. The 50 pg libraries, whereas in general are less complex for all kits, controls, and sites, were particularly compromised for Miltenyi Biotec and Sigma-Aldrich, both probably being the prereleased kits (Miltenyi Biotec has since modified its protocol to include fewer PCR cycles, which is expected to improve this). A recent publication13 covering some of the library preparation conditions tested here showed similar overall patterns.
An important consideration with constructing libraries using low amounts of input material is the sensitivity of gene detection. This was assessed by examining the read occurrence of the known set of GRCh37 genes in the sample-specific data sets. Figure 3A shows detection sensitivity for 3 broad classes of genes: protein coding, pseudogenes, and ncRNAs. Interestingly, there were not substantial differences between the polyA-based and ribo-depletion kits for this particular analysis. For the test kit samples, the least sensitive libraries were those constructed with the lowest amount of material. A more detailed breakdown of the biotypes, shown in Fig. 3B, indicates more subtle distinctions among the tested conditions. The heatmap indicates that the Ribo-Zero versus TruSeq controls in particular show noticeable differences for ncRNA biotype detection. However, overall, there are surprisingly minor differences between the kits with regard to the percentages of gene types detected. The main effect observed is that increasing dilution leads to decreased sensitivity at all the amounts tested. This behavior is not unexpected, but the difference between 5 ng and 500 pg seems a little more substantial than for many other metrics tested.
A frequently desired goal of RNA-seq experiments is the assessment of differential gene expression. Comparison of recovered expression values should indicate the suitability of the different kits and dilutions for these experiments. Figure 4 shows that for the detected reads, the RPKM values did not show substantial variability. The “Min” values, representing genes with the lowest RPKMs, had more spread, yet the median values were similar across all conditions and close to control values. This suggests that whereas all the kits might not identify the same spectrum of genes, expression values for the detected genes are accurate. Gene expression comparisons assessed by Pearson correlations further suggest the suitability of the different kits for quantitative comparisons, at least under certain conditions (Fig. 5A). The 50 pg sample again showed the greatest divergence between kits and between the other dilutions with the same kit. Comparison of the correlations (Fig. 5B) reveals additional interesting data. Ribo-depletion approaches appear to have higher correlations and less spread than polyA protocols. The NuGEN Technologies Incorporated kit was better correlated to the controls. Somewhat surprisingly, given Clontech’s excellence in other metrics, this kit showed less correlation with the controls than NuGEN Technologies Incorporated or Sigma-Aldrich. Correlations within kits also showed that NuGEN Technologies Incorporated and Sigma-Aldrich were tied for lowest variability. These results suggest that despite yielding a lower proportion of exonic reads, ribo-depletion approaches provide equivalent or superior quantitative expression data compared to the tested polyA approaches. In general, it would appear that cross kit, method, and dilution correlations outweigh intersite variability. This is an important component assessed in ABRF studies, but not in others in which multiple kits are tested in the same laboratory. The main source of variability did seem to be the success with which the kits were utilized—2 of the sites with 2 different kits (Miltenyi Biotec and NuGEN Technologies Incorporated) were unable to generate cDNA suitable for subsequent library generation.
Analysis of the expression level of the ERCC RNA standards is shown in Fig. 7. Pearson correlations displayed by heatmap in Fig. 7A show the outcome observed in most other metric assessments, namely that the 50 pg samples behave notably poorer than the others in most pairwise combinations, whereas the 500 pg and 5 ng samples closely resemble each other. Nevertheless, all kits performed reasonably well, with Pearson correlation values rarely going below 0.8. The log-linearity plots in Fig. 7B emphasize this conclusion because all kits performed about as well as the controls (except for the 50 pg samples). Clontech actually exceeded the control values and came closest to recapitulating the actual amounts of the added ERCC standards.
The main conclusions that emerge are that expression analyses using small input amounts of RNA, whereas challenging, are relatively consistent across laboratories and that several available kits work reasonably well. Both polyA and ribo-depleted protocols can do a good job recapitulating the transcriptome across a wide range of input concentrations. The middle input concentration of 500 pg behaved nearly identically to 5 ng in this and many other important sequencing metrics. At the lowest tested level of 50 pg input RNA, supposedly corresponding to 5 cells, nearly every sequencing quality metric was clearly compromised (though as noted for the Sigma-Aldrich and NuGEN Technologies Incorporated kits, this amount was outside recommendations). A reasonable assumption is that 5-fold lower input amounts (i.e., single cell) would lead to even more variability and compromised performance, at least with the kits tested here. Other studies, notably those using actual single cells and not diluted control RNAs, have borne this out.10 This study helps push down the lower input limit for standardized library performance.
Another interesting finding is that kit protocols are often robust enough to perform well outside of the manufacturer’s recommendations. With proper controls in place, such as ERCC spike-ins, a good strategy for labs would be to expand the usable parameters for the kits of choice. In general, the specific needs of the user and characteristics of the starting material will often dictate the choice of kits.
An additional conclusion emerging from this study, though it was not directly addressed, is that determining how many reads are needed for an experiment is a complicated question. As is clear in Fig. 1, exonic read proportions can vary widely depending on the kit and protocol used. Whether or not duplicate removal is required for a given analysis will also substantially influence the number of initial reads required. At these very small amounts, library complexity and sampling issues become important and need to be considered.
Low-input RNA-seq analysis using different amplification kits has demonstrated some promising performance, but further development toward single-cell analysis is needed before it can be used as a routine application for basic and clinical research.
Supplementary Material
Acknowledgments
The authors thank Clontech, Sigma-Aldrich, Miltenyi Biotech, and NuGEN Technologies Incorporated for providing reagents, kits, and technical support for the study. The authors thank Illumina for providing library preparation kits and sequencing reagents. The authors also thank the ABRF Executive Board for providing support for this study. Authors declare no competing financial interests.
REFERENCES
- 1.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621–628. [DOI] [PubMed] [Google Scholar]
- 3.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 2011; 12: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008;456:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 2009;25:1026–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6:377–382. [DOI] [PubMed] [Google Scholar]
- 8.Tang F, Barbacioru C, Bao S, Lee C, Nordman E, Wang X, Lao K, Surani MA. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-seq analysis. Cell Stem Cell 2010;6:468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu S, Luo S, Evgrafov O, Li R, Schroth GP, Levitt P, Knowles JA, Wang K. Single-neuron RNA-Seq: technical feasibility and reproducibility. Front Genet 2012;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, Horvath S, Fan G. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013;500:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res 2011;39:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adiconis X, Borges-Rivera D, Satija R, DeLuca DS, Busby MA, Berlin AM, Sivachenko A, Thompson DA, Wysoker A, Fennell T, Gnirke A, Pochet N, Regev A, Levin JZ. Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods 2013;10:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods 2014;11:41–46. [DOI] [PMC free article] [PubMed]
- 15.Bhargava V, Head SR, Ordoukhanian P, Mercola M, Subramaniam S. Technical variations in low-input RNA-seq methodologies. Sci Rep 2014;4:3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.