Abstract
Proteolytic cleavage of the amyloid precursor protein (APP) by the two proteases α- and β-secretases controls the generation of the amyloid β peptide (Aβ), a key player in Alzheimer’s disease pathogenesis. The α-secretase ADAM10 and the β-secretase BACE1 have opposite effects on Aβ generation and are assumed to compete for APP as a substrate, such that their cleavages are inversely coupled. This concept was mainly demonstrated in studies using activation or overexpression of α- and β-secretases. Here, we report that this inverse coupling is not seen to the same extent upon inhibition of the endogenous proteases. Genetic and pharmacological inhibition of ADAM10 and BACE1 revealed that the endogenous, constitutive α-secretase cleavage of APP is largely uncoupled from β-secretase cleavage and Aβ generation in neuroglioma H4 cells and in neuronally differentiated SH-SY5Y cells. In contrast, inverse coupling was observed in primary cortical neurons. However, this coupling was not bidirectional. Inhibition of BACE1 increased ADAM10 cleavage of APP, but a reduction of ADAM10 activity did not increase the BACE1 cleavage of APP in the neurons. Our analysis shows that the inverse coupling of the endogenous α- and β-secretase cleavages depends on the cellular model and suggests that a reduction of ADAM10 activity is unlikely to increase the AD risk through increased β-secretase cleavage.
Keywords: Alzheimer’s disease, Alpha-secretase, Beta-secretase, Amyloid precursor protein
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder which causes progressive cognitive decline. A central player in AD pathogenesis is the amyloid β peptide (Aβ), which forms toxic oligomeric aggregates leading to neurodegeneration (for review see Hardy and Selkoe, 2002; Haass and Selkoe, 2007; Laferla et al., 2007). The Aβ peptide derives from proteolytic processing of the type I membrane protein amyloid precursor protein (APP) by two protease activities referred to as β- and γ-secretases. β-secretase, which is the aspartyl protease BACE1 (Vassar et al., 1999), cleaves in the ectodomain of APP at the N-terminus of the Aβ domain. As a result the large APP ectodomain (APPsβ) is released from cells. A short C-terminal fragment of APP (C99) remains in the membrane and undergoes subsequent processing by the γ-secretase protease complex (De Strooper et al., 1998). This cleavage occurs at the C-terminus of Aβ and leads to Aβ secretion.
A third protease activity, called α-secretase, cleaves APP within the Aβ domain, and thus, has the potential to prevent Aβ generation (for a review see Lichtenthaler, 2011a). Similar to BACE1-cleavage, α-cleavage occurs within the ectodomain of APP close to the membrane and leads to the secretion of the APP ectodomain (APPsα). α-secretase cleavage occurs constitutively and is mediated by the metalloprotease ADAM10 (Jorissen et al., 2010; Kuhn et al., 2010; Postina et al., 2004). Additionally, the α-secretase cleavage can be pharmacologically increased above its constitutive level, which is referred to as regulated α-secretase cleavage. In contrast to the constitutive α-cleavage, the regulated process can be mediated by different proteases, including ADAM10 and ADAM17 (for review see Lichtenthaler, 2011b).
Several previous studies showed that a pharmacological activation of α-secretase, in particular by phorbol esters and muscarinic agonists, increased the α-secretase cleavage and reduced Aβ levels (e.g. Amtul et al, 2010; Caccamo et al., 2006; Donmez et al., 2010; Fu et al., 2009; Hung et al., 1993; Kim et al., 2009; Nitsch et al., 1992; Skovronsky et al., 2000; Tachida et al., 2008), suggesting that α- and β-secretase cleavages are inversely coupled. Likewise, overexpression of ADAM10 in a mouse model of AD reduced Aβ levels and brain pathology (Postina et al., 2004), making the enhancement of the α-secretase cleavage a promising therapeutic approach for Aβ reduction in AD (for review see Fahrenholz, 2007). Furthermore, overexpression of BACE1 in cells or mice reduced APP α-secretase cleavage (Kuhn et al., 2010; Sala Frigerio et al., 2010; Vassar et al., 1999; Willem et al., 2004). Together, these findings led to the concept that α- and β-secretases compete for APP as a substrate and that their cleavages are inversely coupled. This concept, however, mainly relied on the activation or overexpression of α- and β-secretases. It is much less clear whether an inverse coupling also exists for the endogenous α- and β-secretases and their inhibition. In fact, pharmacological inhibition of α-secretase may not increase APPsβ and Aβ levels (Kim et al., 2008; Kuhn et al., 2010; Marcello et al., 2007). Likewise, a BACE1 inhibitor increased APPsα levels in one study (May et al., 2011), but not in another (Kim et al., 2008). The reasons for the discrepancies in the previous studies are not clear, but may result from experimental differences, such as the cell line used or the expression level of APP (endogenous versus overexpressed).
Resolving whether endogenous α- and β-secretase cleavages are coupled is essential for better understanding AD pathogenesis and for evaluating the therapeutic potential of both proteases. For example, a reduction of the constitutive α-cleavage may increase the risk for AD by leading to enhanced β-secretase cleavage and Aβ levels in certain families (Kim et al., 2009). Likewise, reduced APPsα levels have been observed in the CSF of AD patients (Colciaghi et al., 2002), indicating that a reduced α-cleavage may contribute to AD pathogenesis. Conversely, an inhibition of ADAM10 is considered for different forms of cancer, where high ADAM10 expression is observed (Crawford et al., 2009). Thus, a side effect of ADAM10 inhibition in cancer may be an increased AD risk.
To resolve whether endogenous α- and β-secretase cleavages are indeed inversely coupled, we systematically investigated whether a loss-of-cleavage of APP by ADAM10 or BACE1 leads to a compensatory increase in the other cleavage product. To use experimental conditions as physiological as possible, we analyzed only endogenously expressed proteins, comparing a commonly used tumoral cell line with a neuronally differentiated cell line and with primary neuronal cultures. To further support our findings and to exclude any possible technical artifacts, we compared the genetic knock-out and knock-down to pharmacological inhibition of ADAM10 and BACE1.
Our analysis clearly shows that the inverse coupling of ADAM10 and BACE1 depends on the cellular model. Inhibition of the secretases in the cell lines did not show a major coupling of APP α-secretase cleavage with β-secretase cleavage and Aβ generation. In contrast, coupling was observed in primary cortical neurons. However, this coupling was not bidirectional. Inhibition of BACE1 activity increased ADAM10 cleavage of APP, but a reduction of ADAM10 activity did not increase the BACE1 cleavage of APP.
Materials and methods
Cell culture
The neuroglioma cell line H4 was cultured in Optimem medium plus Glutamax (Gibco) containing 10% fetal bovine serum (FBS, Sigma Aldrich) and 1% penicillin/streptomycin (P/S, Gibco). The neuroblastoma SH-SY5Y cells were cultured in F12/DMEM (Lonza) supplemented with 15% FBS, 1% P/S and non essential amino acids (NAA, Gibco) (Hogl et al., 2011). To induce neuronal differentiation, SH-SY5Y cells were cultured in the presence of 10 μM all-trans retinoic acid (Sigma Aldrich) at low serum concentration (1%) for three days (adapted from Encinas et al., 2000). All the experiments on differentiated SH-SY5Y cells were performed in regular serum condition (15%). HEK293 EBNA and HEK293T cells were cultured in DMEM/glutamax (Gibco) supplemented with 10% FBS and 1% P/S, while HCT8 (a kind gift of Olivier Gires) cells were cultured in RPMI-1640 medium supplemented with 10% FBS and 1% P/S.
Embryonic primary cortical neurons were prepared as described (Colombo et al., 2007; Kuhn et al., 2010; Mitterreiter et al., 2010). Briefly, cortex samples from E16.5 wild-type C57-Bl6J or ADAM10 conditional KO embryos (Gibb et al., 2010) were collected and dissociated in DMEM plus 200 U of papain (Sigma Aldrich). Neurons were plated in 6 well plates precoated with poly D lysine (1.5 × 106 cells/ well). Plating medium was B27/Neurobasal (Gibco) supplemented with 0.5 mM glutamine and 1% P/S. All experimental procedures on animals were performed in accordance with the European Communities Council Directive (86/609/EEC).
Primary cultures have been characterized by immunofluorescent staining specific for neuronal and glial cells. 8DIV neurons were washed twice with PBS and then fixed in 4% paraformaldehyde (PFA)/sucrose for 10 min at room temperature (RT). Cells were permeabilized with PBS/0.1% Triton X-100 and incubated for 1 h at RT with blocking solution (PBS/5% normal goat serum). Primary antibodies were incubated over night at 4 °C: β3 tubulin (specific for neuronal cells, Cell Signaling) and GFAP (specific for glial cells, Millipore). Fixed neurons were washed three times with PBS and then incubated for 1 h at RT with anti-mouse 546-Alexa Fluor and anti-rabbit 488-Alexa Fluor secondary antibodies (Invitrogen) for neuronal and glial cell detection, respectively. Cells were washed twice and nuclei were stained using TOPRO intercalating agent according to the manufacturer’s protocol (Invitrogen). Laser confocal analysis was performed using a Zeiss LSM 510 Meta inverted confocal microscope, equipped with Zeiss LSM software and a Plan Apochromat 10× lens.
Down-regulation of gene expression by RNAi or knock-out
To induce the knock-down of ADAM10 and BACE1, H4 and differentiated SH-SY5Y cells were transfected with 5 nM of siGenome pool (Dharmacon) targeting the specific gene and RNAiMax reagent (Invitrogen) in accordance to the manufacturer’s protocol. The same amount of non-targeting siRNAs (C2 pool) was used as control. 48 h after transfection, conditioned media were collected for a further 24 h and cells lysed on ice in lysis buffer (150 mM NaCl, 50 mM Tris pH 7.5, 1% Triton X-100, protease inhibitor cocktail from Roche). To down-regulate ADAM10 in primary neurons, we prepared primary cultures from ADAM10 conditional knock-out mice (ADAM10 cKO, Gibb et al., 2010) which contain two flox sites flanking the ADAM10 gene. After neuronal infection with a lentiviral vector expressing the CRE recombinase, we were able to induce the excision of the ADAM10 gene. An empty viral vector was used as control.
Briefly, Cre was amplified with iCre-NHE1 fw (GATCGCTAGCATGG TGCCCAAGAAGAAGAGG) and iCre-EcoR1 rev (GATCGAATTCTCAATCG CCCTCGAGCAGCCTCACC) and subcloned into FU2Δ-Zeo vector digested with Nhe1 and EcoR1. Cre-expressing lentiviral particles were generated as previously described (Kuhn et al., 2010). Briefly, lentiviruses were generated by transient cotransfection of HEK293T cells with the plasmids psPAX2, pCDNA3.1 (−)-VSV-G and as transfer vector pLKO2mod-EGFP-WPRE using Lipofectamine 2000 (Invitrogen). Lentiviral particles for infection of murine primary cortical neurons were concentrated and purified by ultracentrifugation. Lentiviral stocks were stored at −80 °C until use. To knock-down BACE1 in primary neurons, specific short hairpin RNAs (shRNAs; Sh1 (2498) aagcctacactggta caaag; Sh2 (2920) ggtcttcccagcataggttca) were expressed in neuronal cells using lentiviral vectors as described (Kuhn et al., 2010). A viral vector to express a scramble shRNA sequence was used as control. Plated neurons were infected with lentivirus at 3DIV. At 7DIV culture medium was completely changed to collect shedding products for 24 h. At 8DIV media were collected and cells lysed in lysis buffer.
Pharmacological inhibition
To inhibit ADAM10 and BACE1, cells were cultured for 24 h with the broad-spectrum metalloprotease inhibitor TAPI1 (50 μM, International Peptides) (Darlak et al., 1990) or with the specific BACE1 inhibitor C3 (2 μM, Calbiochem, β-secretase inhibitor IV) (Stachel et al., 2004), respectively. After this incubation period, media were collected and cells lysed on ice in lysis buffer. H4 and differentiated SH-SY5Y cells were used at 80% confluence, while primary neurons were treated at 7DIV and samples collected at 8DIV.
Antibodies
Mouse mAb APPC 2C11 (IgG2a, APP C-terminus, for full length APP in lysates, generated by immunization against CKMQQNGYENPTYK FFEQMQN-COOH peptide), mouse mAb 22C11 (APP N-terminal for total soluble APP ectodomain in culture media, provided by K. Beyreuther), rat mAb A16M 14D6 (IgG2a, specific for human APPsα in culture media, generated by immunization against CYEVHHQ-COOH peptide), rat mAb A16M 5G11 (IgG2c, specific for murine APPsα in culture media, generated by immunization against DAEF GHDSGFEVRHQKC-COOH peptide), pAb 192wt (specific for APPsβ in culture media, provided by D. Schenk), pAb ADAM10 (Calbiochem), mAb 3D5 (specific for BACE1, kind gift of R. Vassar, Zhao et al., 2007), tubulin (Santa Cruz Biotechnology). To detect APPsβ in H4 conditioned media, 500 μl of conditioned medium was immunoprecipitated with protein A sepharose beads and mouse mAb BAWT antibody (specific for APPsβ, Kuhn et al., 2010) for 2 h at 4 °C.
Western blot analysis
Cell lysate protein concentration was measured using the Bradford method according to the manufacturer’s protocol (BioRad). Media loading was normalized to lysate protein concentration. After protein quantification, samples were directly loaded on 8% acrylamide gels for Western blot analysis and detection of the specific APP fragments. Membranes were incubated over night at 4 °C. Blots were developed using horseradish peroxidase-conjugated secondary antibodies and the ECL chemiluminescence system. Quantification of Western blots was carried out using Fuji Las-4000 software (Fuji Film inc.) and was based on at least 6 independent replicates.
mAb 5G11antibody characterization
HEK293EBNA cells were transiently transfected with Peak12-linker (Lichtenthaler et al., 2003), Peak12-mAPP695, Peak12-mAPPsα, Peak12-mAPPsβ′ and Peak12-mAPPsβ (all murine sequences) expressing vectors and Lipofectamine 2000. 48 h after transfection, media were collected for a 24 h period and cells lysed in lysis buffer. Samples were analyzed by Western blot using 8% acrylamide gels.
ELISA measurements
Aβx-40 (Aβ40) peptides were quantified in culture media by a Meso Scale Discovery (MSD) sandwich immunoassay using the MSD Sector Imager 2400. 96-well Multi-SPOT plates pre-coated with anti-Aβ40 C-terminus capture antibody were incubated in block buffer (1% BSA, 0.1% Tween in PBS) for 1 h at room temperature and then washed twice in wash buffer (0.1% Tween in PBS). Samples and Aβ peptide standards (MSD) were added together with either ruthenylated 6E10 or 4 G8 antibodies (1 μg/ml in Block buffer) for the detection of human or mouse Aβ, respectively. Plates were covered and incubated at room temperature for 2 h before washing three times with wash buffer. For detection, 150 μl of 2× MSD Read buffer was added and the light emission at 620 nm after electrochemical stimulation was measured using the MSD Sector Imager 2400 reader. The corresponding concentrations of Aβ peptides were calculated using the MSD Discovery Workbench software.
To specifically measure Aβ peptides starting at amino acid 1 (BACE1 cleavage site, Aβ1-x) in cell culture supernatants of H4 and differentiated SH-SY5Y cells, a 96-well plate coated with Anti-Human Aβ (N) monoclonal antibody (clone 82E1) was used according to the manufacturer’s protocol (IBL, Japan). This assay is specific to human Aβ (Horikoshi et al., 2004).
Despite the relatively higher BACE1 expression in differentiated SH-SY5Y cells compared to H4 cells (see also Fig. 1), APPsβ levels in the SH-SY5Y cells were below the detection limit of our immunoblot assay and were measured using the Meso Scale Discovery (MSD) sandwich immunoassay kit specific for APPsβ detection. Differentiation of SH-SY5Y cells is known to mildly increase BACE1 expression, but more strongly ADAM10 expression, suggesting increased α-secretase cleavage in SH-SY5Y cells compared to H4 cells (Endres et al., 2005; Holback et al., 2008).
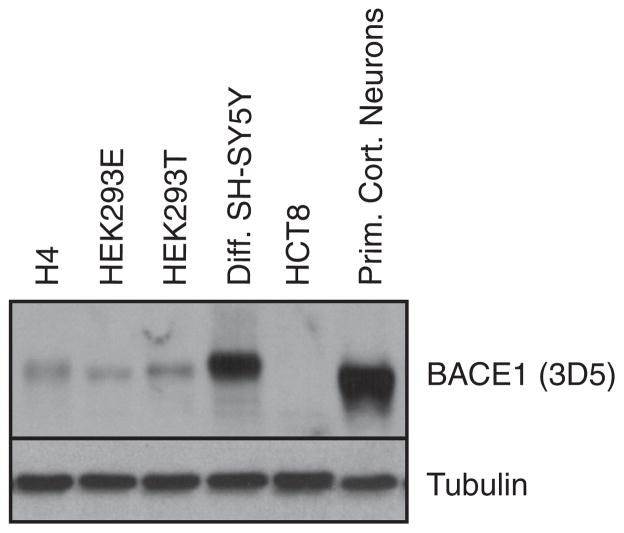
Fig. 1.
BACE1 protein level in different cell types. Western blot analysis of BACE1 protein level in H4, HEK293E (EBNA), HEK293T, differentiated SH-SY5Y, HCT8 and primary cortical neuronal cells. Cellular lysates were blotted for BACE1 using mAb 3D5. Loading control: tubulin.
BACE1 protein level analysis
H4, HEK293 EBNA, HEK293T, ileocecal colorectal adenocarcinoma HCT8 cells, differentiated SH-SY5Y and primary cortical neuronal cells were lysed in lysis buffer (as above). A comparative analysis of the BACE1 protein level among the different lines was performed by Western blot using 8% acrylamide gels and the 3D5 antibody.
Statistical analysis
All experiments were repeated at least three times in duplicates (at least n=6) using independent culture preparations. Quantitative data were statistically analyzed by two-tailed type 3 (unequal variance) t-test. A p-value<0.05 was considered significant.
Results
Neuroglioma cell line H4
To analyze whether the constitutive α- and β-secretase cleavages are coupled, α- or β-secretase was silenced by RNA interference or inhibited pharmacologically. The effect of these treatments on the processing of the endogenous APP was measured in H4 human neuroglioma cells, which are frequently used for studying APP processing (for example Colombo et al., 2009), in neuronally differentiated human SH-SY5Y neuroblastoma cells and in primary murine neurons. The three cell types differ in their BACE1 expression levels, with H4 cells having the lowest BACE1 level, the differentiated SH-SY5Y cells an intermediate level and the primary neurons the highest BACE1 level (Fig. 1).
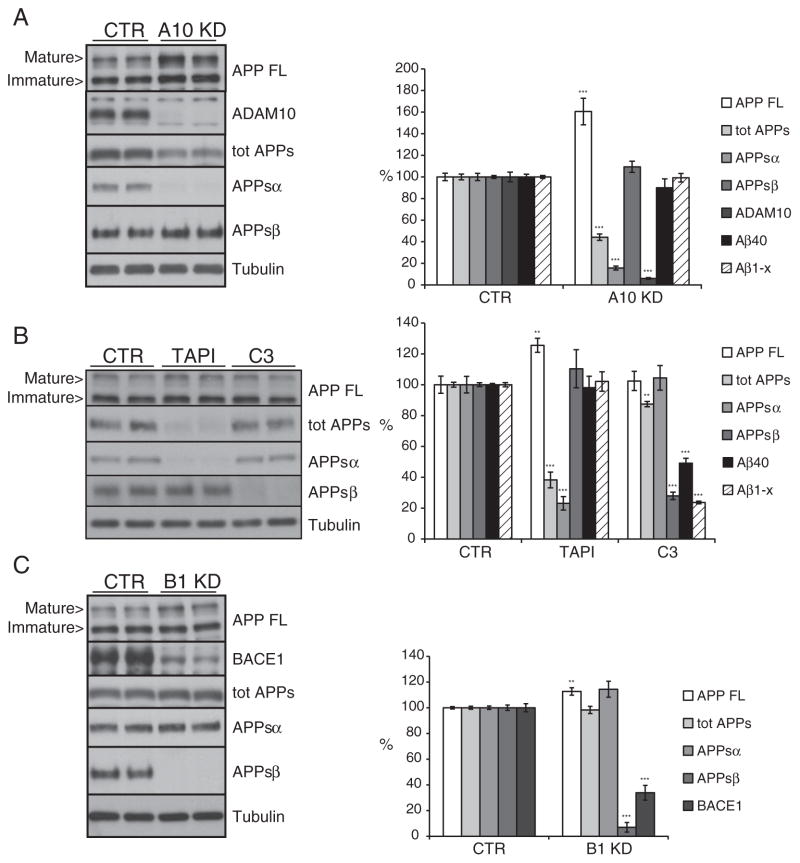
In H4 cells ADAM10 was knocked down with siRNA pools shown to be specific for ADAM10 (Kuhn et al., 2010). This reduced cellular ADAM10 protein levels to 6%, as measured by quantitative immunoblot analysis (Fig. 2A, p<0.001). Levels of the α-secretase cleavage product APPsα were reduced to 15% (Fig. 2A, p<0.001), demonstrating that ADAM10 is essential for constitutive α-secretase cleavage in H4 cells. Total APPs levels were reduced to 44% (Fig. 2A, p<0.001), similar to our previous findings in HEK293 cells (Kuhn et al., 2010). Surprisingly, however, in the culture media APPsβ together with the Aβ40 levels were not significantly changed (Fig. 2A). Because Aβ peptides may show heterogeneity at their N-terminus, we also specifically measured Aβ peptides starting at amino acid 1 (Aβ1-x). Similar to Aβ40, no significant changes were seen for Aβ1-x (Fig. 2A). Full length APP protein accumulated in the cells (Fig. 2A, 160%, p<0.001). Taken together, these results show that in H4 cells the loss of α-cleavage is not coupled to an increase of the BACE1 cleavage products Aβ and APPsβ.
Fig. 2.
ADAM10 and BACE1 are not coupled in neuroglioma H4 cells. Shown are immunoblots and the densitometric quantification of the indicated proteins and peptides. (A) Cells were transfected with a pool of siRNA specific for ADAM10 (A10 KD). (B) Cells were treated for 24 h with the metalloprotease inhibitor TAPI1 (50 μM) or the BACE1 inhibitor C3 (2 μM). (C) Cells were transfected with a pool of siRNA specific for BACE1 (B1 KD). CTR: pool of control, non-targeting siRNA. Cellular lysates and media were blotted for full length APP (APP FL, mAb 2C11), total secreted APP ectodomain (tot APPs, mAb 22C11), soluble APPα ectodomain (APPsα, mAb 14D6), soluble APPβ ectodomain (APPsβ, pAb 192wt), ADAM10 and BACE1 (mAb 3D5). Loading control: tubulin. Aβ40 level in culture media detected by Meso Scale Discovery (MSD) sandwich immunoassay. Quantifications show mean value of at least six independent experiments (± SEM), *=p<0.05, **=p<0.01 and ***=p<0.001 (compared to CTR).
The pharmacological inhibition of the α-secretase activity with TAPI1 produced similar results compared to the ADAM10 knock-down. Mild accumulation of full length APP in the cell lysate (Fig. 2B, 125%, p<0.01) correlated with a strong reduction of total secreted APP (Fig. 2B, 38%, p<0.001) and of the APPsα level (Fig. 2B, 23%, p<0.001). As for the ADAM10 knock-down experiment, Aβ and APPsβ levels were not significantly affected by TAPI1 treatment (Fig. 2B).
Next, BACE1 was knocked down in H4 cells, reducing BACE1 protein levels to 33% of control (Fig. 2C, p<0.001). As expected APPsβ levels were also strongly reduced (Fig. 2C, 7%, p<0.001). In contrast to the ADAM10 knock-down, however, full length APP only slightly accumulated (Fig. 2C, 112%, p<0.01), while total APPs levels were not significantly reduced. This shows that in H4 cells BACE1 only contributes to a small extent to total APP shedding. Importantly the nearly complete inhibition of APPsβ generation did not yield a significant increase in the APP α-cleavage product APPsα.
The pharmacological inhibition of BACE1 achieved through the specific inhibitor C3 (Stachel et al., 2004) confirmed the observations found with BACE1 knock-down. The reduced APPsβ level (Fig. 2B, 27%, p<0.001) did not produce a compensatory increase in the α–pathway as indicated by the unchanged APPsα level (Fig. 2B). The total secreted ectodomain level was only slightly decreased (Fig. 2B, 87%, p<0.01).
As a whole, our data show that in H4 cells there is no major inverse coupling of the constitutive α-secretase cleavage of APP with the endogenous β-secretase cleavage and Aβ generation.
Differentiated SH-SY5Y cells
Next, we analyzed whether cells with more neuronal like properties would show a more pronounced inverse coupling of α- and β-secretase cleavage compared to the neuroglioma H4 cells. To this aim, human neuroblastoma SH-SY5Y cells were differentiated into neuron-like cells (Encinas et al., 2000). Afterwards ADAM10 or BACE1 were knocked-down or pharmacologically inhibited as done with the H4 cells.
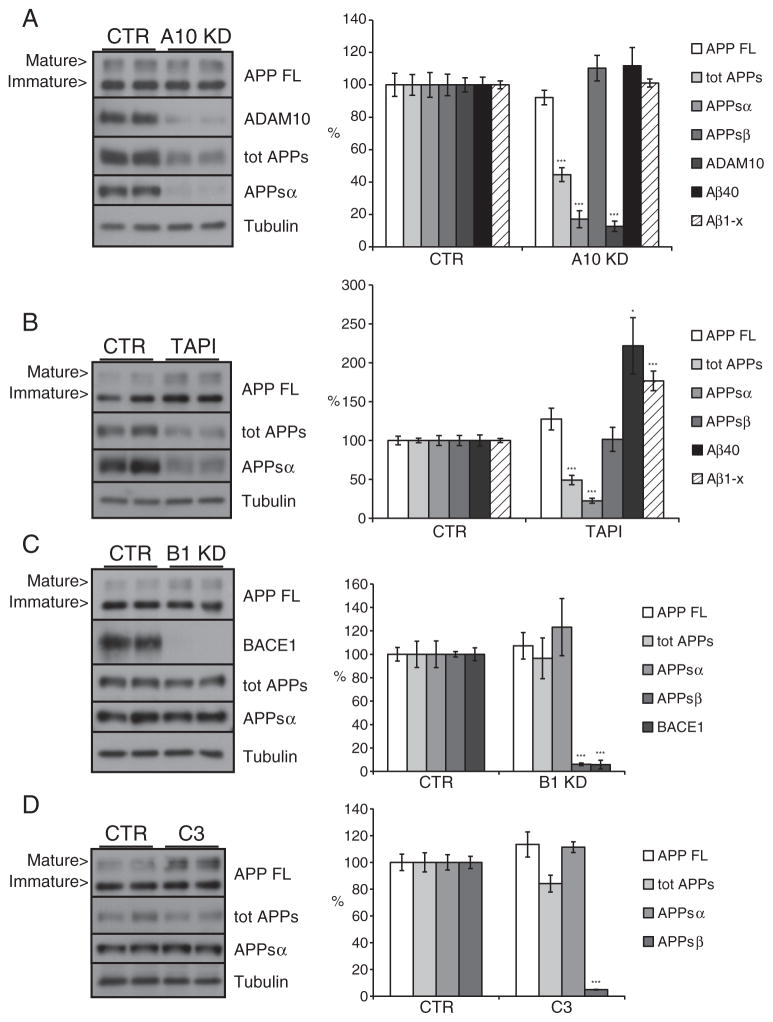
ADAM10 knock-down (Fig. 3A, 12%, p<0.001) produced no major changes in the APP full length protein levels in the lysate (Fig. 3A), but reduced total secreted APP to 45% (Fig. 3A, p<0.001). APPsα levels were efficiently reduced (Fig. 3A, 17%, p<0.001), but no significant changes in APPsβ were observed (Fig. 3A). In these cells, APPsβ levels were below the detection limit of our immunoblot assay (see Materials and methods section) and were measured with a sandwich immuno assay. The uncoupling between α- and β-secretase cleavages was also confirmed by the unaffected levels of Aβ40 and Aβ1-x in ADAM10 knock-down culture media (Fig. 3A).
Fig. 3.
ADAM10 and BACE1 are not coupled in differentiated neuroblastoma SH-SY5Y cells. SH-SY5Y cells were differentiated for three days into neuron-like cells and then treated as indicated. Shown are immunoblots and the densitometric quantification of the indicated proteins and peptides. (A) Cells were transfected with a pool of siRNA specific for ADAM10 (A10 KD). (B) Cells were treated for 24 h with the metalloprotease inhibitor TAPI1 (50 μM). (C) Cells were transfected with a pool of siRNA specific for BACE1 (B1 KD) or (D) treated with the BACE1 inhibitor C3 (2 μM). CTR: pool of non-targeting siRNA. Cellular lysates and media were blotted for full length APP (APP FL, mAb 2C11), total secreted APP ectodomain (tot APPs, mAb 22C11), α soluble APP ectodomain (APPsα, mAb 14D6), ADAM10 and BACE1 (mAb 3D5). β soluble APP ectodomain level (APPsβ) in culture media was detected by specific ELISA assay. Loading control: tubulin. Aβ40 level in culture media detected by Meso Scale Discovery (MSD) sandwich immunoassay. Quantifications were from at least six independent replicates (±SEM), *=p<0.05 **=p<0.01 and ***=p<0.001 (compared to CTR).
Pharmacological inhibition through TAPI1 treatment validated the data obtained with the ADAM10 knock-down. Full length APP showed a trend to a slight accumulation (Fig. 3B, 125%, ns), while total secreted APP was reduced to 50% in culture media (Fig. 3B, p<0.001). The APPsα level was reduced to 20% (Fig. 3B, p<0.001) but this reduction was not coupled to an increase in the APPsβ level. In contrast to what was observed in the ADAM10 knock-down experiments, the Aβ40 and the Aβ1-x levels in the conditioned medium surprisingly were increased in response to the TAPI1 treatment (Fig. 3B, 221%, p<0.05 and 176%, p<0.001 respectively). Because APPsβ levels were not altered, it is unlikely that the increase in Aβ is a direct consequence of the reduced ADAM10 cleavage. It may rather reflect that TAPI1 is a broad-spectrum metalloprotease inhibitor, which potentially also blocks Aβ-degrading enzymes or other steps in Aβ metabolism (see Discussion).
Overall our analysis shows that, similar to H4 cells, in neuronally differentiated SH-SY5Y cells the reduction of ADAM10 activity does not produce any major increase in the β-cleavage of APP.
Next, BACE1 was knocked down in the differentiated SH-SY5Y cells (Fig. 3C, 5%, p<0.001). As expected, this nearly completely abolished APPsβ levels (Fig. 3C, 5%, p<0.001), while the levels of the full length protein, the total secreted APP and the APPsα fragment remained unchanged (Fig. 3C). Likewise, the pharmacological inhibition of BACE1 with C3 strongly inhibited APPsβ (Fig. 3D, 5%, p<0.001), while all other analyzed APP species were unaffected. This demonstrates that the amount of α-secretase cleavage is not influenced by a reduction of BACE1 cleavage.
Thus we conclude that similar to the H4 cells the α- and the β-secretase cleavages of APP are uncoupled in differentiated SH-SY5Y cells and that the α-secretase ADAM10 is the major APP sheddase, whereas BACE1 is a minor APP sheddase in these cells.
Embryonic primary cortical neurons
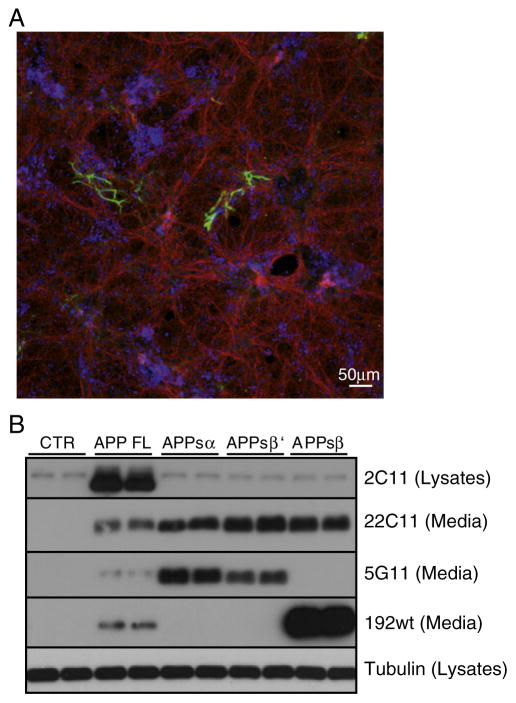
Next, the potential inverse coupling between α- and β-secretase cleavages was investigated in primary murine neurons prepared at embryonic day 16.5 and analyzed after 8 days in vitro. At this time point the cortical neuron culture consists mostly of neurons with very little contamination of glial cells, as revealed by immunocytochemical analysis with markers specific for neuronal or glial cells (Fig. 4A). To detect the endogenous α-secretase cleaved murine APPsα in neurons, we generated the monoclonal antibody 5G11. To validate its specificity HEK293 cells were transfected with different constructs encoding murine full length APP695 or one of its soluble ectodomain fragments arising through cleavage at the α-, β′- or β-cleavage sites (APPsα, APPsβ ′ and APPsβ). As a control HEK293 cells were mock transfected. 5G11 detected APPsα and to a lesser extent APPsβ ′, but not APPsβ (Fig. 4B).
Fig. 4.
Characterization of embryonic neuronal cortical cultures and antibody 5G11 specificity. (A) Primary cortical neuronal cultures were fixed at 8DIV and immunostained for neuronal cells (β3 tubulin in red) and glial cells (GFAP in green). Nuclei were stained with the TOPRO reagent in blue. Staining was repeated with at least 3 independent neuronal preparations. (B) Western blot analysis of cell lysates and media from HEK293 cells overexpressing murine full length APP695 (mAPP695), the α soluble ectodomain of APP (APPsα), the β ′ soluble ectodomain (APPsβ ′) or the β ectodomain (APPsβ). CTR: wild type HEK293E cells. Cellular lysates were blotted for full length APP (APP FL, mAb 2C11), while culture media for total secreted APP ectodomain (tot APPs, mAb 22C11), α soluble APP ectodomain (APPsα, mAb 5G11), β soluble APP ectodomain (APPsβ, pAb 192wt). Loading control: tubulin.
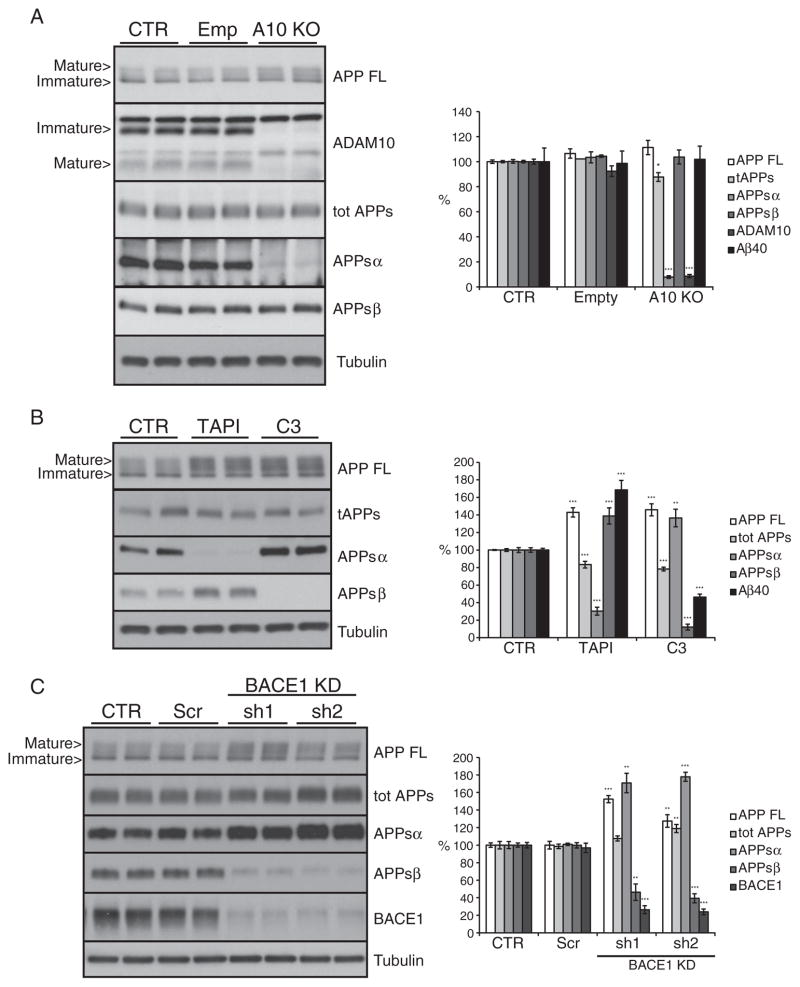
In order to block α-secretase cleavage and measure its influence on β-secretase cleavage, we used primary embryonic neurons from a conditional, floxed ADAM10 knock-out mouse (Gibb et al., 2010). Mice with a regular ADAM10 knock-out (not conditional) die at embryonic day 9.5 (Hartmann et al., 2002), excluding the possibility to prepare primary neurons. To achieve the ADAM10 knock-out in the conditional knock-out neurons, the neurons were transduced with a lentivirus expressing the Cre recombinase gene, which resulted in efficient knock-out of ADAM10, with only a minor percentage of cells still expressing ADAM10 (Fig. 5A, 8%, p<0.001). As a control, a lentivirus containing an empty vector (Emp) was used. Non-transduced neurons (CTR) were used as an additional control. Knock-out of ADAM10 nearly completely abolished APPsα levels (Fig. 5A, 7%, p<0.001), which agreed well with the decrease in ADAM10 levels. Antibody 5G11 detects APPsα and not APPsβ, but to a low degree also APPsβ ′ as described above (Fig. 4B). APPsβ ′ ends with amino acid 10 of the Aβ sequence. However, given that the ADAM10 knock-out nearly completely abolished the 5G11 signal, the β ′-cleavage does not appear to have a major role in shedding of the endogenous APP in primary neurons. Total secreted APP was only slightly decreased (Fig. 5A, 87%, p<0.05) and full length APP slightly accumulated in the lysates (Fig. 5A, 111%, not significant), demonstrating that α-secretase cleavage only contributes to a small extent to total APP shedding in primary neurons. Importantly, APPsβ and Aβ40 levels (Fig. 5A) were unaffected by the ADAM10 knock out, demonstrating that β-secretase cleavage is not significantly increased when ADAM10 cleavage is blocked.
Fig. 5.
α and β activity coupling in primary cortical neurons. Shown are immunoblots and the densitometric quantification of the indicated proteins and peptides. (A) ADAM10 conditional knock-out neurons transducted with a lentiviral vector for the CRE recombinase expression to induce gene knock out (A10 KO). (B) cells treated for 24 h with metalloproteinase family inhibitor (TAPI1, 50 μM) and BACE1 inhibitor (C3, 2 μM) and (C) wild type neurons infected with lentiviral vectors to express specific shRNAs against BACE1 sequence (sh1 and sh2 — BACE1 KD). CTR = not transducted neurons. Empty = empty viral vector; Scr = scramble shRNA. Cellular lysates and media were blotted for full length APP (APP FL, mAb 2C11), total secreted APP ectodomain (tot APPs, mAb 22C11), α soluble APP ectodomain (APPsα, mAb 5G11), β soluble APP ectodomain (APPsβ, pAb 192wt), ADAM10 and BACE1 (mAb 3D5). Loading control: tubulin. Aβ40 level in culture media detected by Meso Scale Discovery (MSD) sandwich immunoassay. Quantifications were from at least six independent replicates (±SEM), *=p<0.05 **=p<0.01, ***=p<0.001 (compared to CTR).
Next, α-secretase cleavage was pharmacologically blocked in wild-type cortical neurons with TAPI1. This metalloprotease inhibitor strongly reduced APPsα level in the culture media (Fig. 5B, 30%, p<0.001), while total secreted APP was only mildly reduced (Fig. 5B, 83%, p<0.001%), in agreement with the results obtained in the ADAM10 knock-out neurons. Surprisingly, however, the APPsβ level was significantly increased by TAPI1 treatment (Fig. 5B, 139%; p<0.001). This discrepancy between the ADAM10 knock-out and the broad-spectrum metalloprotease inhibitor was also confirmed by ELISA measurement of secreted Aβ40, showing a significant increase only in response to TAPI1 treatment (Fig. 5B, 168%, p<0.001). Potentially, TAPI1 also blocks Aβ and APPsβ degrading enzymes. Alternatively, it may inhibit APP cleavage or degradation by other metalloproteases besides ADAM10, which in turn may yield more APP available for β-secretase cleavage, resulting in the observed increase in APPsβ and Aβ (see Discussion). In fact, full length APP significantly accumulated in the cells upon TAPI1 treatment (Fig. 5B, 142%; p<0.001), in contrast to the only mild increase upon ADAM10 knock-out in the neurons.
From this data set we conclude that the specific down-regulation of ADAM10 does not affect APP β-secretase cleavage, while inhibition with the broad-spectrum inhibitor TAPI1 can raise the levels of the β-secretase cleavage products, presumably acting on other metalloproteases than ADAM10.
Next, BACE1 was blocked in wild-type neurons by lentiviral RNAi-mediated knock-down using two distinct shRNA sequences. As a control, a lentiviral vector for a scrambled RNA sequence (Scr) and non-transduced neurons (CTR) were used. Compared to the controls, BACE1 knock-down efficiently reduced BACE1 protein levels to 26% and 23% (Fig. 5C, both p<0.001) with the sh1 and the sh2-sequences respectively. Full length APP accumulated in cell lysates after the knock down (Fig. 5C, 150%–130%, p<0.001 and p<0.01 respectively), while total secreted APP showed only a mild increase (Fig. 5C, 110% for sh1, ns and 120% for sh2, p<0.01). Likewise APPsβ level was clearly reduced (Fig. 5C, 46% and 39%, p<0.01 and p<0.001 respectively). In contrast to the H4 and differentiated SH-SY5Y cells, the reduced APPsβ levels were accompanied by a strong increase in APPsα levels (Fig. 5C, 170%–180%, p<0.01 and p<0.001 respectively), indicative of an inverse coupling of α- and β-secretase cleavages under conditions when BACE1 is inhibited. The pharmacological inhibition of BACE1 by C3 confirmed the results obtained with BACE1 knock-down. The APPsβ was reduced by 90% (Fig. 5B, p<0.001) and this decrease was coupled to an increase in the APPsα level (Fig. 5B, 136%, p<0.01). Full length APP accumulated in the C3 treated cells (Fig. 5B, 146%, p<0.001) while the total shed APP was reduced to78% in comparison to controls (Fig. 5B, p<0.001).
Taken together, both knock-down and inhibition of BACE1 clearly show that in primary cortical neurons the loss of BACE1 cleavage is coupled to an increase in the APP α-secretase cleavage.
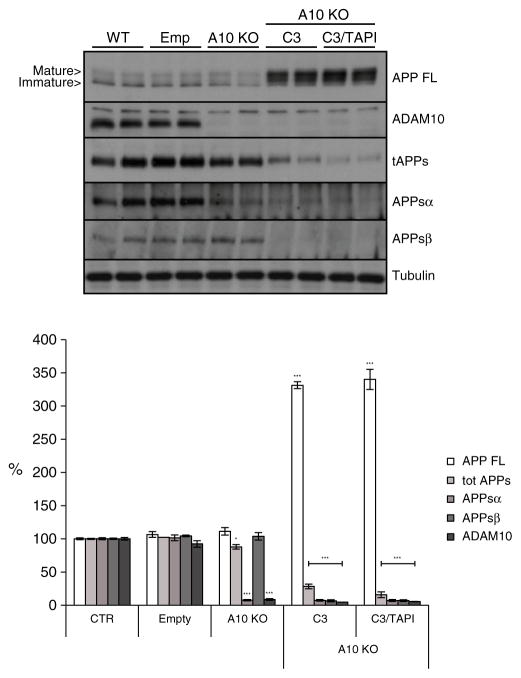
ADAM10 mediates the increase of α-cleavage upon BACE1 inhibition
Next, we investigated whether the increased α-secretase cleavage upon BACE1 inhibition was mediated by the constitutive α-secretase ADAM10. To this aim, CRE-transduced ADAM10 knock-out neurons were treated with or without C3 for 24 h. BACE1 inhibition nearly completely suppressed APPsβ (Fig. 6, 7%, p<0.001). In contrast to C3-treated wild-type neurons (Fig. 5B) the simultaneous knock-out of ADAM10 completely prevented the compensatory increase in APPsα, demonstrating that ADAM10 is responsible for the increased α-cleavage in response to BACE1 inhibition in the wild-type neurons. Total APPs levels were also strongly reduced (Fig. 6, 28%, p<0.001), while full length APP levels were dramatically increased (Fig. 6, 330%, p<0.001). This demonstrates that ADAM10 and BACE1 together are responsible for most of APP shedding in neurons. Addition of TAPI1 to the C3-treated ADAM10 knock-out neurons led to a slight further reduction of total APP shedding (Fig. 6, 15%, p<0.001), suggesting that a metalloprotease other than ADAM10 and not cleaving at the α-clevage site contributes to APP shedding to a minor extent.
Fig. 6.
ADAM10 compensates for BACE1 inhibition in primary cortical neurons. Western blot analysis and densitometric quantification of ADAM10 knock-out primary cortical neurons (A10 KO) treated for 24 h with TAPI1 (50 μM) and C3 (2 μM). CTR: non-transduced neurons. Emp: Empty viral vector. Cellular lysates and media were blotted for full length APP (APP FL, mAb 2C11), total secreted APP ectodomain (tot APPs, mAb 22C11), α soluble APP ectodomain (APPsα, mAb 5G11), β soluble APP ectodomain (APPsβ, pAb 192wt) and ADAM10. Loading control = tubulin. Quantifications were from at least six independent replicates (±SEM), *=p<0.05 **=p<0.01, ***=p<0.001 (compared to CTR).
Moreover, this analysis demonstrates that over 50% of total APP shedding was mediated by BACE1 in the neurons, whereas the ADAM10 knock-out alone only mildly reduced APP shedding by about 10%. Thus, in contrast to the cell lines, BACE1 is the major APP sheddase in neurons, whereas ADAM10 is only the minor one. Taken together our data clearly show that ADAM10 is responsible for the increased APPsα levels triggered by BACE1 inhibition.
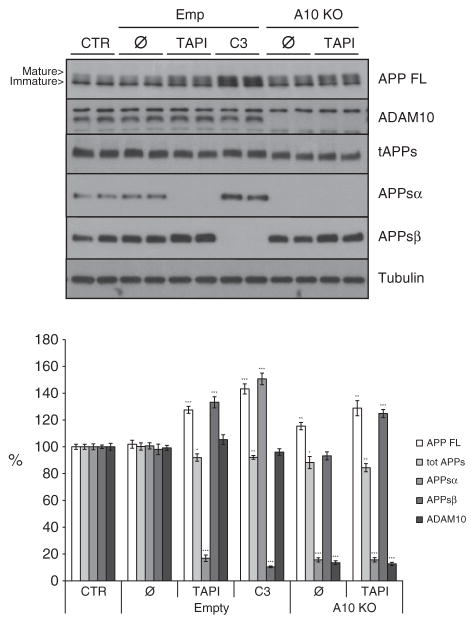
As described above (Fig. 5A and B), TAPI but not ADAM10 knock-out led to increased APPsβ levels. Interestingly, the increase in APPsβ is also seen in TAPI-treated ADAM10 knock-out neurons (Fig. 7, 125%, p<0.001), demonstrating that this increase is ADAM10-independent and likely to be mediated by another TAPI-sensitive metalloprotease.
Fig. 7.
ADAM10 is not required for TAPI1-induced increase in APPsβ level. Western blot analysis and densitometric quantification of primary cortical wild-type (WT) neurons and conditional floxed ADAM10 knock-out neurons transduced with a control virus (Empty (Emp), still expressing ADAM10) or with CRE-virus (A10 KO, ADAM10 knock-out) treated for 24 h with TAPI1 (50 μM) or C3 (2 μM). CTR: non-transduced neurons. Emp: empty viral vector. Cellular lysates and media were blotted for full length APP (APP FL, mAb 2C11), total secreted APP ectodomain (tot APPs, mAb 22C11), α soluble APP ectodomain (APPsα, mAb 5 G11), β soluble APP ectodomain (APPsβ, pAb 192wt) and ADAM10. Loading control = tubulin. Quantifications were from at least six independent replicates (±SEM), *=p<0.05 **= p<0.01, ***=p<0.001 (compared to control CTR).
Taken together our data clearly show that in embryonic cortical primary neurons ADAM10 and BACE1 are coupled. However, this coupling is not bidirectional since only BACE1 inhibition increased ADAM10-cleavage of APP, whereas the knock-out of ADAM10 did not raise β-secretase cleavage of APP or Aβ generation. Only the use of a broad-spectrum metalloprotease inhibitor was able to increase APPsβ and Aβ levels, presumably by acting on additional metalloproteases besides ADAM10, which may be involved in APP metabolism.
Discussion
The concept that α- and β-secretases compete for APP as a substrate and that their cleavages are inversely coupled, was demonstrated in many studies using the activation or overexpression of α- and β-secretases. Here, we report that this inverse coupling is not seen to the same extent upon inhibition of the endogenous proteases. Using genetic and pharmacological inhibition of the α-secretase ADAM10 and the β-secretase BACE1 in two cell lines and in primary neurons, our study demonstrates that the endogenous, constitutive α- and β-secretase cleavages of APP are largely uncoupled and that an inverse coupling only occurs in a cell-type specific manner and is observed in particular in neurons upon BACE1 down-regulation, but not upon knock-out of ADAM10.
Specific reduction of ADAM10 activity by knock-out or knock-down was not coupled to a major increase in BACE1-mediated APPsβ generation or Aβ levels in all three cell types. Similar results were obtained upon pharmacological inhibition of ADAM10 with TAPI1 in H4 cells and in two previous studies upon knock-down of ADAM10 in HEK293 cells (Kuhn et al., 2010) or using a metalloprotease inhibitor unrelated to TAPI1 in APP-transfected CHO cells and primary neurons (Kim et al., 2008). However, for TAPI1 we obtained different results in neuronally differentiated SH-SY5Y cells and in primary neurons. In the SH-SY5Y cells, TAPI1 increased Aβ levels without a concomitant increase in APPsβ. We speculate that this selective effect on Aβ is likely due to the inhibition of Aβ degrading enzymes by the broad spectrum metalloprotease inhibitor TAPI1. In fact, several known Aβ degrading enzymes are metalloproteases, such as insulin-degrading enzyme, neprilysin and matrix metalloproteases (for review Miners et al., 2008). They are inhibited by hydroxamate-based metalloprotease inhibitors similar to TAPI1, and this was reported to decrease Aβ degradation leading to increased Aβ levels (Leissring et al., 2010; Liao and Van Nostrand, 2010; Marcotte et al., 1999; Saghatelian et al., 2004; Yin et al., 2006). We did not observe an increase in the Aβ level in the H4 cells upon TAPI1 treatment, but we speculate that the enzyme pattern expressed by the neuroglioma cell line is different from that expressed by differentiated SH-SY5Y cells with a neuronal phenotype. In the primary neurons, TAPI1 increased both APPsβ and Aβ, while the specific ADAM10 knock-out did not raise the levels of both BACE1 cleavage products. TAPI1 even increased APPsβ in ADAM10 knock-out neurons. Because TAPI1 also increased full length APP levels in the lysate of the neurons, it is possible that other TAPI1-sensitive metalloproteases besides ADAM10 contribute to APP turnover, such that their inhibition makes more APP available for BACE1 cleavage, resulting in the higher Aβ and APPsβ levels.
Interestingly, a recent study reported that ADAM10 knock-out in neurons did not only decrease APPsα, but in parallel also APPsβ and Aβ (Jorissen et al., 2010). Although the reasons for this discrepancy with our data is not yet clear, it may result from the different time points, at which ADAM10 was knocked-out. The other study used primary neurons from mice where the ADAM10 knock-out started at embryonic day E9, and was accompanied by Notch signaling-dependent premature neuronal differentiation and defects in neuronal migration. In our study neurons were prepared at embryonic day E16 and Cre-mediated knock-out started after 3 days in vitro. Thus, it appears possible that the timing, when ADAM10 is switched-off during development, influences the coupling of α- and β-secretase cleavages of APP.
Knock-down or pharmacological inhibition of BACE1 in H4 and differentiated SH-SY5Y cells blocked β-secretase cleavage and did not result in an increased APP cleavage by α-secretase. A similar result was obtained using a BACE1 inhibitor in CHO and HEK293 cells (Kim et al., 2008; Kuhn et al., 2010) and with a novel APP mutation (expressed in HEK293 cells), which blocks BACE1 cleavage and reduces the risk for Alzheimer’s disease (Jonsson et al., 2012). Together, these studies demonstrate that in the investigated cell lines a reduction of BACE1-mediated cleavage is not coupled to increased α-secretase cleavage. In contrast to the cell lines, the inhibition of BACE1 in primary neurons, both genetically and pharmacologically, was coupled to a clearly increased APP cleavage by the α-secretase ADAM10. A similar compensatory increase of α-secretase cleavage in neurons was reported in BACE1 knock-out mice and the CSF of human volunteers treated with a BACE1 inhibitor (May et al., 2011; Sala Frigerio et al., 2010).
The finding that the constitutive ADAM10 and BACE1 cleavages of APP are partially coupled in neurons, but largely uncoupled in the cell lines, is consistent with different mechanistic scenarios. The uncoupling may reflect that α- and β-secretase cleavages occur in different cellular compartments, such that a reduced α-secretase cleavage at the plasma membrane would not necessarily increase the endosomal APP pool available for β-secretase cleavage. Alternatively, the cellular APP levels may not be rate limiting for α- and β-secretase cleavages, such that a reduction of one cleavage does not increase the other cleavage. If however, one of the proteases is overexpressed or strongly activated, as done in numerous studies (e.g. Caccamo et al., 2006; Kim et al., 2008, 2009; Postina et al., 2004; Sala Frigerio et al., 2010; Skovronsky et al., 2000), APP substrate levels may become rate-limiting, leading to the observed inverse coupling of α- and β-secretase cleavages. Interestingly, BACE1 expression is high in neurons, but low in peripheral tissue (Vassar et al., 1999). Accordingly, we found BACE1 to be the major APP sheddase in neurons (mediating>50% of APP shedding), but only the minor one in the cell lines (mediating ~10% of total APP shedding). In this regard, neurons may resemble BACE1 overexpressing cell lines, in which the APP level is rate-limiting for proteolysis and inverse coupling of the secretases is observed. Thus, an inhibition of BACE1 in neurons, but not in the cell lines, would make more APP available for an increased α-cleavage.
As another mechanistic scenario we consider the possibility that a reduction of BACE1 cleavage is always coupled to an increase in α-secretase cleavage. While this was clearly observed in neurons, it was not seen in the cell lines. However, because BACE1 is the minor sheddase in the cell lines, its inhibition may not lead to a significant increase in α-secretase cleavage, as measured by immunoblot analysis.
As a last scenario we speculate that additional proteases besides α- and β-secretases may cleave APP and contribute to its shedding and degradation, in particular when α- or β-secretase is inhibited. In fact, metalloproteases such as meprin β and membrane-type matrix metalloproteases can cleave APP (Ahmad et al., 2006; Jefferson et al., 2011). Thus, inhibition of α-secretase (the major sheddase) in the cell lines may be compensated for through increased cleavage by another metalloprotease. This would explain that neither increased β-secretase cleavage was observed nor accumulation of full length APP in the cell lysate. Such a protease would be expected to cleave APP at peptide bonds different from the α- and β-secretase cleavage sites, because inhibition of α- and β-secretases led to a nearly complete inhibition of the corresponding cleavage products (APPsα and APPsβ), as determined with cleavage-site specific antibodies. In fact, one of the potential proteases, meprin β, was previously shown to cleave APP within the N-terminal half of the ectodomain at a larger distance from the Aβ domain (Jefferson et al., 2011).
Our study provides evidence that indeed additional proteases besides ADAM10 and BACE1 contribute to total APP shedding. In the primary neurons the combination of the ADAM10 knock-out with the specific BACE1 inhibitor reduced total APP shedding to 30%. The addition of the broad-spectrum metalloprotease inhibitor TAPI1 induced a further reduction of APP shedding to 15%, suggesting that – at least under conditions of BACE1 and ADAM10 inhibition – an additional TAPI1-sensitive metalloprotease contributes to total APP shedding. We speculate that the remaining 15% must come from a protease that is neither sensitive to TAPI1 nor to the BACE1 inhibitor C3 or that this remaining cleavage is due to incomplete inhibition of a metalloprotease. This may be the metalloprotease meprin β, which cleaves APP, but is not efficiently inhibited by inhibitors of the TAPI family (Kruse et al., 2004).
Finally, the partial coupling of APP cleavage by ADAM10 and BACE1 in neurons has implications for drug development. BACE1 inhibition does not only lower Aβ, but also increases the release of the neuroprotective fragment APPsα from neurons, thereby potentially promoting an autocrine activity to boost neuronal survival (Furukawa et al., 1996; Meziane et al., 1998; Stein et al., 2004). Conversely, an inhibition of ADAM10 is pursued in different forms of cancer (Crawford et al., 2009; Gibb et al., 2010; Saftig and Reiss, 2011). Given that ADAM10 knock-out does not increase β-secretase cleavage or Aβ levels in neurons, this therapeutic approach may not increase the risk of AD. However, ADAM10 has additional substrates, which may contribute to pathogenesis. For example, neuronal overexpression of a dominant-negative ADAM10 mutant in an AD mouse model aggravated amyloid pathology in the brain (Postina et al., 2004). This may be due to substrates other than APP, because Aβ and APPsβ levels only showed a trend to a mild increase, which however, was not significant. However, if inhibitors are tested, which are not specific to ADAM10, but also affect other metalloproteases, it is likely that they will increase Aβ levels, as observed here with TAPI1.
Taken together, our study unequivocally demonstrates that the choice of the in vitro model plays a pivotal role in the analysis of the pathways involved in APP processing. Most of the tumor cell lines commonly used in the laboratory provide strong advantages over neurons in routine handling. However, several aspects of APP processing, such as coupling of α- and β-secretases and the expression level of BACE1 shows crucial differences between cell lines and neurons.
Acknowledgments
We thank Mara Taverna, Sebastian Hogl, Bastian Dislich and Katrin Moschke for helpful comments on the manuscript. We thank the following institutions for financial support: the Deutsche Forschungsgemeinschaft for SFB596 project B12 to SFL and project Z2 to EK, the competence network degenerative dementias (BMBF) to SFL and the Chinese Scholarship Council to HW.
References
- Ahmad M, Takino T, Miyamori H, Yoshizaki T, Furukawa M, Sato H. Cleavage of amyloid-beta precursor protein (APP) by membrane-type matrix metalloproteinases. J Biochem. 2006 Mar;139(3):517–526. doi: 10.1093/jb/mvj054. [DOI] [PubMed] [Google Scholar]
- Amtul Z, Wang L, Westaway D, Rozmahel RF. Neuroprotective mechanism conferred by 17beta-estradiol on the biochemical basis of Alzheimer’s disease. Neuroscience. 2010 Aug 25;169(2):781–786. doi: 10.1016/j.neuroscience.2010.05.031. Epub 2010 May 21. [DOI] [PubMed] [Google Scholar]
- Caccamo A, Oddo S, Billings LM, Green KN, Martinez-Coria H, Fisher A, LaFerla FM. Neuron. 2006 Mar 2;49(5):671–682. doi: 10.1016/j.neuron.2006.01.020. [DOI] [PubMed] [Google Scholar]
- Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, Padovani A, Di Luca M. [alpha]-Secretase ADAM10 as well as [alpha]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol Med. 2002 Feb;8(2):67–74. [PMC free article] [PubMed] [Google Scholar]
- Colombo A, Repici M, Pesaresi M, Santambrogio S, Forloni G, Borsello T. The TAT-JNK inhibitor peptide interferes with beta amyloid protein stability. Cell Death Differ. 2007 Oct;14(10):1845–1848. doi: 10.1038/sj.cdd.4402202. [DOI] [PubMed] [Google Scholar]
- Colombo A, Bastone A, Ploia C, Sclip A, Salmona M, Forloni G, Borsello T. JNK regulates APP cleavage and degradation in a model of Alzheimer’s disease. Neurobiol Dis. 2009 Mar;33(3):518–525. doi: 10.1016/j.nbd.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML. ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des. 2009;15(20):2288–2299. doi: 10.2174/138161209788682442. [DOI] [PubMed] [Google Scholar]
- Darlak K, Miller RB, Stack MS, Spatola AF, Gray RD. Thiol-based inhibitors of mammalian collagenase. Substituted amide and peptide derivatives of the leucine analogue, 2-[(R, S)-mercaptomethyl]-4-methylpentanoic acid. J Biol Chem. 1990 Mar 25;265(9):5199–5205. [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010 Jul 23;142(2):320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Ceña V, Gallego C, Comella JX. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000 Sep;75(3):991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- Endres K, Postina R, Schroeder A, Mueller U, Fahrenholz F. Shedding of the amyloid precursor protein-like protein APLP2 by disintegrin-metalloproteinases. FEBS J. 2005 Nov;272(22):5808–5820. doi: 10.1111/j.1742-4658.2005.04976.x. [DOI] [PubMed] [Google Scholar]
- Fahrenholz F. Alpha-secretase as a therapeutic target. Curr Alzheimer Res. 2007 Sep;4(4):412–417. doi: 10.2174/156720507781788837. [DOI] [PubMed] [Google Scholar]
- Fu H, Dou J, Li W, Cui W, Mak S, Hu Q, Luo J, Lam CS, Pang Y, Youdim MB, Han Y. Eur. J Pharmacol. 2009 Nov 25;623(1–3):14–21. doi: 10.1016/j.ejphar.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Sopher BL, Rydel RE, Begley JG, Pham DG, Martin GM, Fox M, Mattson MP. Increased activity-regulating and neuroprotective efficacy of alpha-secretase-derived secreted amyloid precursor protein conferred by a C-terminal heparin-binding domain. J Neurochem. 1996 Nov;67(5):1882–1896. doi: 10.1046/j.1471-4159.1996.67051882.x. [DOI] [PubMed] [Google Scholar]
- Gibb DR, El Shikh M, Kang DJ, Rowe WJ, El Sayed R, Cichy J, Yagita H, Tew JG, Dempsey PJ, Crawford HC, Conrad DH. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010 Mar 15;207(3):623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002 Jul 19;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lübke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/ metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002 Oct 1;11(21):2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- Hogl S, Kuhn PH, Colombo A, Lichtenthaler SF. Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and γ-secretase. PLoS One. 2011;6(6):e21337. doi: 10.1371/journal.pone.0021337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holback S, Adlerz L, Gatsinzi T, Jacobsen KT, Iverfeldt K. PI3-K- and PKC-dependent up-regulation of APP processing enzymes by retinoic acid. Biochem Biophys Res Commun. 2008;365:298–303. doi: 10.1016/j.bbrc.2007.10.167. [DOI] [PubMed] [Google Scholar]
- Horikoshi Y, Sakaguchi G, Becker AG, Gray AJ, Duff K, Aisen PS, Yamaguchi H, Maeda M, Kinoshita N, Matsuoka Y. Development of Abeta terminal end-specific antibodies and sensitive ELISA for Abeta variant. Biochem Biophys Res Commun. 2004 Jul 2;319(3):733–737. doi: 10.1016/j.bbrc.2004.05.051. [DOI] [PubMed] [Google Scholar]
- Hung AY, Haass C, Nitsch RM, Qiu WQ, Citron M, Wurtman RJ, Growdon JH, Selkoe DJ. Activation of protein kinase C inhibits cellular production of the amyloid beta-protein. J Biol Chem. 1993 Nov 5;268(31):22959–22962. [PubMed] [Google Scholar]
- Jefferson T, Čaušević M, auf dem Keller U, Schilling O, Isbert S, Geyer R, Maier W, Tschickardt S, Jumpertz T, Weggen S, Bond JS, Overall CM, Pietrzik CU, Becker-Pauly C. Metalloprotease meprin beta generates nontoxic N-terminal amyloid precursor protein fragments in vivo. J Biol Chem. 2011 Aug 5;286(31):27741–27750. doi: 10.1074/jbc.M111.252718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jönsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012 Jul 11; doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, Bartsch U, Weskamp G, Blobel CP, Glatzel M, De Strooper B, Saftig P. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010 Apr 7;30(14):4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, Norton D, Tesco G, Elliott K, Wagner SL, Moir RD, Becker KD, Tanzi RE. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet. 2009 Oct 15;18(20):3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ML, Zhang B, Mills IP, Milla ME, Brunden KR, Lee VM. Effects of TNFalpha-converting enzyme inhibition on amyloid beta production and APP processing in vitro and in vivo. J Neurosci. 2008 Nov 12;28(46):12052–12061. doi: 10.1523/JNEUROSCI.2913-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse MN, Becker C, Lottaz D, Köhler D, Yiallouros I, Krell HW, Sterchi EE, Stöcker W. Human meprin alpha and beta homo-oligomers: cleavage of basement membrane proteins and sensitivity to metalloprotease inhibitors. Biochem J. 2004 Mar 1;378(Pt 2):383–389. doi: 10.1042/BJ20031163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010 Sep 1;29(17):3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferla FM, Green KN, Oddo S. Intracellular amyloid-beta in Alzheimer’s disease. Nat Rev Neurosci. 2007;8:499–509. doi: 10.1038/nrn2168. [DOI] [PubMed] [Google Scholar]
- Leissring MA, Malito E, Hedouin S, Reinstatler L, Sahara T, Abdul-Hay SO, Choudhry S, Maharvi GM, Fauq AH, Huzarska M, May PS, Choi S, Logan TP, Turk BE, Cantley LC, Manolopoulou M, Tang WJ, Stein RL, Cuny GD, Selkoe DJ. Designed inhibitors of insulin-degrading enzyme regulate the catabolism and activity of insulin. PLoS One. 2010 May 7;5(5):e10504. doi: 10.1371/journal.pone.0010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao MC, Van Nostrand WE. Degradation of soluble and fibrillar amyloid beta-protein by matrix metalloproteinase (MT1-MMP) in vitro. Biochemistry. 2010 Feb 16;49(6):1127–1136. doi: 10.1021/bi901994d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF, Dominguez DI, Westmeyer GG, Reiss K, Haass C, Saftig P, De Strooper B, Seed B. The cell adhesion protein P-selectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J Biol Chem. 2003 Dec 5;278(49):48713–48719. doi: 10.1074/jbc.M303861200. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF. Alpha-secretase in Alzheimer’s disease: molecular identity, regulation and therapeutic potential. J Neurochem. 2011a Jan;116(1):10–21. doi: 10.1111/j.1471-4159.2010.07081.x. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler SF. Alpha-secretase cleavage of the amyloid precursor protein: proteolysis regulated by signaling pathways and protein trafficking. Curr Alzheimer Res. 2011b May 23;9(2):165–177. doi: 10.2174/156720512799361655. [DOI] [PubMed] [Google Scholar]
- Marcello E, Gardoni F, Mauceri D, Romorini S, Jeromin A, Epis R, Borroni B, Cattabeni F, Sala C, Padovani A, Di Luca M. Synapse-associated protein-97 mediates alpha-secretase ADAM10 trafficking and promotes its activity. J Neurosci. 2007 Feb 14;27(7):1682–1691. doi: 10.1523/JNEUROSCI.3439-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte PA, Elmore IN, Guan Z, Magoc TJ, Albert DH, Morgand DW, Curtin ML, Garland RB, Guo Y, Heyman HR, Holms JH, Sheppard GS, Steinman DH, Wada CK, Davidsen SK. Evaluation of the inhibition of other metalloproteinases by matrix metalloproteinase inhibitors. J Enzyme Inhib. 1999;14(6):425–435. doi: 10.3109/14756369909030333. [DOI] [PubMed] [Google Scholar]
- May PC, Dean RA, Lowe SL, Martenyi F, Sheehan SM, Boggs LN, Monk SA, Mathes BM, Mergott DJ, Watson BM, Stout SL, Timm DE, Smith Labell E, Gonzales CR, Nakano M, Jhee SS, Yen M, Ereshefsky L, Lindstrom TD, Calligaro DO, Cocke PJ, Greg Hall D, Friedrich S, Citron M, Audia JE. Robust central reduction of amyloid-β in humans with an orally available, non-peptidic β-secretase inhibitor. J Neurosci. 2011 Nov 16;31(46):16507–16516. doi: 10.1523/JNEUROSCI.3647-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane H, Dodart JC, Mathis C, Little S, Clemens J, Paul SM, Ungerer A. Memory-enhancing effects of secreted forms of the beta-amyloid precursor protein in normal and amnestic mice. Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12683–12688. doi: 10.1073/pnas.95.21.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, Love S. Abeta-degrading enzymes in Alzheimer’s disease. Brain Pathol. 2008 Apr;18(2):240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitterreiter S, Page RM, Kamp F, Hopson J, Winkler E, Ha HR, Hamid R, Herms J, Mayer TU, Nelson DJ, Steiner H, Stahl T, Zeitschel U, Rossner S, Haass C, Lichtenthaler SF. Bepridil and amiodarone simultaneously target the Alzheimer’s disease beta- and gamma-secretase via distinct mechanisms. J Neurosci. 2010 Jun 30;30(26):8974–8983. doi: 10.1523/JNEUROSCI.1199-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsch RM, Slack BE, Wurtman RJ, Growdon JH. Release of Alzheimer amyloid precursor derivatives stimulated by activation of muscarinic acetylcholine receptors. Science. 1992 Oct 9;258(5080):304–307. doi: 10.1126/science.1411529. [DOI] [PubMed] [Google Scholar]
- Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, Flamez P, Dequenne A, Godaux E, van Leuven F, Fahrenholz F. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004 May;113(10):1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, Reiss K. The “a disintegrin and metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol. 2011 Jun-Jul;90(6–7):527–535. doi: 10.1016/j.ejcb.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004 Jul 6;101(27):10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio C, Fadeeva JV, Minogue AM, Citron M, Van Leuven F, Staufenbiel M, Paganetti P, Selkoe DJ, Walsh DM. beta-Secretase cleavage is not required for generation of the intracellular C-terminal domain of the amyloid precursor family of proteins. FEBS J. 2010 Mar;277(6):1503–1518. doi: 10.1111/j.1742-4658.2010.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovronsky DM, Moore DB, Milla ME, Doms RW, Lee VM. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J Biol Chem. 2000 Jan 28;275(4):2568–2575. doi: 10.1074/jbc.275.4.2568. [DOI] [PubMed] [Google Scholar]
- Stachel SJ, Coburn CA, Steele TG, Jones KG, Loutzenhiser EF, Gregro AR, Rajapakse HA, Lai MT, Crouthamel MC, Xu M, Tugusheva K, Lineberger JE, Pietrak BL, Espeseth AS, Shi XP, Chen-Dodson E, Holloway MK, Munshi S, Simon AJ, Kuo L, Vacca JP. Structure-based design of potent and selective cell-permeable inhibitors of human beta-secretase (BACE-1) J Med Chem. 2004 Dec 16;47(26):6447–6450. doi: 10.1021/jm049379g. [DOI] [PubMed] [Google Scholar]
- Stein TD, Anders NJ, DeCarli C, Chan SL, Mattson MP, Johnson JA. Neutralization of transthyretin reverses the neuroprotective effects of secreted amyloid precursor protein (APP) in APPSW mice resulting in tau phosphorylation and loss of hippocampal neurons: support for the amyloid hypothesis. J Neurosci. 2004 Sep 1;24(35):7707–7717. doi: 10.1523/JNEUROSCI.2211-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachida Y, Nakagawa K, Saito T, Saido TC, Honda T, Saito Y, Murayama S, Endo T, Sakaguchi G, Kato A, Kitazume S, Hashimoto Y. Interleukin-1 beta up-regulates TACE to enhance alpha-cleavage of APP in neurons: resulting decrease in Abeta production. J Neurochem. 2008 Mar;104(5):1387–1393. doi: 10.1111/j.1471-4159.2007.05127.x. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999 Oct 22;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Willem M, Dewachter I, Smyth N, Van Dooren T, Borghgraef P, Haass C, Van Leuven F. beta-site amyloid precursor protein cleaving enzyme 1 increases amyloid deposition in brain parenchyma but reduces cerebrovascular amyloid angiopathy in aging BACE×APP[V717I] double-transgenic mice. Am J Pathol. 2004 Nov;165(5):1621–1631. doi: 10.1016/s0002-9440(10)63419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin KJ, Cirrito JR, Yan P, Hu X, Xiao Q, Pan X, Bateman R, Song H, Hsu FF, Turk J, Xu J, Hsu CY, Mills JC, Holtzman DM, Lee JM. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006 Oct 25;26(43):10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Fu Y, Yasvoina M, Shao P, Hitt B, O’Connor T, Logan S, Maus E, Citron M, Berry R, Binder L, Vassar R. Beta-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer’s disease pathogenesis. J Neurosci. 2007 Apr 4;27(14):3639–3649. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]