Abstract
The eukaryotic RNA exosome processes and degrades RNA by directing substrates to the distributive or processive 3′ to 5′ exoribonuclease activities of Rrp6 or Rrp44, respectively. The non-catalytic nine-subunit exosome core (Exo9) features a prominent central channel. While RNA can pass through the channel to engage Rrp44, it is not clear how RNA is directed to Rrp6 or if Rrp6 utilizes the central channel. Here, we report a 3.3 Å crystal structure of a ten-subunit RNA exosome complex from Saccharomyces cerevisiae composed of the Exo9 core and Rrp6 bound to single-stranded polyA RNA. The Rrp6 catalytic domain rests atop the Exo9 S1/KH ring above the central channel, the RNA 3′ end is anchored in the Rrp6 active site, and the remaining RNA traverses the S1/KH ring in an opposite orientation to that observed in a structure of a Rrp44-containing exosome complex. Solution studies with human and yeast RNA exosome complexes suggest that the RNA path to Rrp6 is conserved and dependent on the integrity of the S1/KH ring. While selection to Rrp6 or Rrp44 is stochastic in vitro, the fate of a particular RNA may be determined in vivo by the manner in which co-factors present RNA to the RNA exosome.
The eukaryotic RNA exosome core is an essential nine-subunit complex formed by two stacked rings of six RNase PH-like proteins on the bottom and three S1/KH domain “cap” proteins on the top. Exo9 associates with the endo- and 3′ to 5′ processive exoribonuclease Rrp44, and the 3′ to 5′ distributive exoribonuclease Rrp6 (ref. 1). In Saccharomyces cerevisiae, the cytoplasmic exosome includes Exo9 and Rrp44 (Exo10Rrp44), the nuclear exosome contains Exo9, Rrp44 and Rrp6 (Exo11Rrp44/Rrp6)2, and a nucleolar exosome with Exo9 and Rrp6 (Exo10Rrp6) has been posited in human3. Each exosome complex may be uniquely equipped to target different RNA substrates, in conjunction with protein cofactors, to catalyze RNA turnover, quality control, or processing in the context of their respective cellular compartments4.
Rrp6 is a RNase D family member5, and is proposed to hydrolyze RNA via two metal ion catalysis6,7. It includes an N-terminal PMC2NT domain that associates with a cofactor Rrp47 (ref. 8), an exoribonuclease domain (EXO), an HRDC domain, and a C-terminal domain (CTD) that associates with the Exo9 core9. Structural studies revealed that the EXO and HRDC domains constitute the catalytic module10,11 while a Exo10Rrp44+Rrp6Cterm structure revealed how Rrp6 CTD residues interact with Exo9 (ref. 12). No structures yet exist for Rrp6 in complex with Exo9 or RNA.
We proposed that Rrp44 and Rrp6 utilize an overlapping channel within the S1/KH ring to engage RNA, that Rrp6 activities are modulated by Exo9, and that Rrp6 stimulates Rrp44 in binding and degradation of ssRNA in the context of Exo11Rrp44/Rrp6 (ref. 13). To provide a structural basis for these observations, we crystallized Rrp6 with Exo9 and a 24-nt single-stranded polyA RNA (polyA24 RNA).
Global architecture of Exo10Rrp6-polyA RNA
The 3.3 Å structure of Exo10Rrp6 was obtained in the presence of a 24-nucleotide (nt) single-stranded polyA RNA using Rrp6 (128-685) that lacked exoribonuclease activity (D238N)13,14, the PMC2NT domain (1-127)13 and its last 48 C-terminal residues (Extended Data Fig. 1; Extended Data Table 1). Rrp6 is positioned atop the Exo9 S1/KH ring with Exo9 subunits resembling those of human Exo9 (ref. 15) and yeast Exo9 (ref. 12) in Exo10Rrp44+Rrp6Cterm (Fig. 1a, b). PolyA24 RNA is coordinated within the S1/KH ring with Rrp6 active site residues contacting the RNA 3′ end (Fig. 1c). The Rrp6 EXO domain contacts two of the three S1/KH ring proteins, Rrp4 and Rrp40, and the HRDC domain is proximal to Rrp4, although no direct contacts are apparent. Consistent with previous results12, the Rrp6 CTD wraps around the N-terminal domain (NTD) of Csl4 and PH-like subunit, Rrp43, before emerging at the top of the Exo9 core (Fig. 1a; Extended Data Fig. 2a,b). No electron density was observed for CTD residues 629-684 and for residues (517-524) that link the HRDC domain and CTD. The location of the Rrp6 N-terminus (Met128) places the Rrp6 PMC2NT domain over the Exo9 central channel in an ideal position to interact with Rrp47 to facilitate substrate recruitment (Fig. 1a). Alignment of exosome-associated Rrp6 to the yeast Rrp6 catalytic module (EXO-HRDC) reveals few differences (Extended Data Fig. 2c).
Figure 1. Overall structure of a RNA bound Rrp6 exosome.
a,b,c, Cartoon and surface representation of S. cerevisiae Exo10Rrp6 bound to polyA24 RNA. Views from the (a) sides, (b) top, and (c) close-up of RNA engaged by the Rrp6 EXO domain and S1 domains of Rrp40, Rrp4, and Csl4. RNase PH-like ring as transparent surfaces, S1/KH ring and Rrp6 as cartoons, RNA as spheres.
The NTD of Rrp4 and S1 domains of Rrp4 and Rrp40 interact with the Rrp6 EXO domain burying 2230 Å2 of surface area in the complex. Rrp6 interactions with the Rrp4 NTD (Region 2) and S1 domain (Region 3) are more extensive (1750 Å2) while interactions with the Rrp40 S1 domain and Rrp4 S1 domain encompass a region proximal to the RNA binding site (Region 1). These surfaces are highly conserved (Fig. 2a,b) in comparison to the Rrp6 CTD (Extended Data Fig. 2a), which is important for Rrp6 interaction with the exosome core as illustrated by analytical gel filtration studies showing the CTDs of both yeast and human Rrp6 are required for association with the exosome (Extended Data Fig. 3).
Figure 2. Rrp6 contacts the S1/KH ring and RNA.
a,b, Rrp6 EXO domain interface with Rrp4 and Rrp40 as yellow buried surfaces (a) and in atomic detail (b). a, Rrp6 (teal; left), Rrp4 (pale green; right) and Rrp40 (magenta; right) in open book representation (160°). Below, surfaces colored by sequence conservation from red (conserved) to blue (variable). b, Contacts between Rrp6, Rrp4 and Rrp40 in stick representation colored as in (a). c, Contacts between RNA (orange), magnesium (green) and Rrp6 active site residues (teal) in Exo10Rrp6-RNA. A catalytic aspartate, D238, is mutated (D238N; red). Potential hydrogen bonds depicted as dashed lines.
The structure of Exo10Rrp6 bound to polyA24 RNA reveals how an RNase D family member interacts with RNA (Fig. 2c). The Rrp6 catalytic domain and active site bind A21A22A23A24 with the 2′-OH and 3′-OH of the terminal A24 coordinated via two main chain hydrogen bonds to the backbone amide and carbonyl oxygen of His241 and Glu240 side chain carboxylate, similar to that observed for AMP in the Rrp6 catalytic domain10. An additional contact is observed to A24 between the N7 adenine atom and Gln345 side chain amide. The scissile phosphate of A24 and A23 is coordinated by a single magnesium ion that bridges the phosphate and the side chain carbonyl of Asn238 (Asp in wild-type enzyme). The scissile phosphate is within hydrogen bond distance of the invariant side chain hydroxyl of Tyr361 (Extended Data Fig. 4a,c). Consistent with the proposed reaction mechanism7, two metal ions coordinated by conserved side chain carboxylates in the yeast Rrp6-AMP complex are ideally positioned to contact the scissile phosphate (Extended Data Fig. 4c,d). Several contacts between metal ions and RNA are missing in our structure due to the D238N substitution.
The remaining contacts to polyA24 RNA reveal several non-specific interactions, consistent with the ability of Rrp6 to degrade RNA of any sequence. A23 2′-OH is within hydrogen bonding distance to the side chain carboxylate of Asp296 while the A23 phosphate is within hydrogen bonding distance of the Leu328 backbone amide. The A23 base is situated over the A24 base with van der Waals contacts provided by side chains of Trp299, Met295 and Phe294. Phe294 is positioned between the A23 and A22 bases, presumably disrupting base stacking interactions. The A22 base and ribose are cradled by van der Waals contacts to Phe294, Gly292, His291 and Tyr315 while its phosphate is within hydrogen bonding distance with the backbone amide of His291. Lys319, His326 and Tyr315 contribute the remaining contacts to the A21 base. Importantly, none of the aforementioned contacts appear to interrogate the identity of the base. Sixteen of eighteen residues observed in direct contact to the RNA or metal cofactors are conserved in human Rrp6 (Extended Data Fig. 4a,b)10,11.
RNA passes through the S1/KH ring
Four of six nucleotides of polyA24 RNA observed in electron density (5′-A19-A24-3′) are coordinated by Rrp6 placing the 5′ end (A19) proximal to conserved, basic side chains from Rrp4 (Lys122 and Arg123), while Rrp40 Lys108 points toward A24 and Arg110 is proximal to the phosphate of A21 and ribose of A20 (Fig. 3a). Additional densities with spacing consistent with the RNA phosphate backbone, modeled as PO4a and PO4b, are observed within the S1/KH ring nearest to Arg150 and proximal to Arg145 and Arg202 of Csl4 (Extended Data Fig. 1a-c). The aforementioned Csl4, Rrp4 and Rrp40 residues are highly conserved15. No additional density consistent with RNA is observed within the PH-like ring.
Figure 3. Rrp6 activities are dependent on the S1/KH ring.
a,b, Partially overlapping paths for RNA in complex with (a) Exo10Rrp6 and (b) Exo10Rrp44+Rrp6Cterm within the S1/KH ring. RNA and S1/KH side chains depicted as sticks, Rrp6 (a) as a surface, the stem loop (b) in Exo10Rrp44+Rrp6Cterm as a ribbon. Color-coded labels adjacent to subunits. c, RNA decay activities for yeast Exo10Rrp6 and Exo11Rrp44/Rrp6 with mutations in the S1/KH ring or a channel occluding insertion lower in the PH-like ring (Rrp45Large). d, RNA decay activities for human Exo10RRP6 with insertions at the top (RRP41Large) or middle of the PH-like ring (RRP45Large). RNA decay assays were conducted at least three times for each of the samples with typical results depicted in (c) and (d).
Rrp6 and Rrp44 utilize a shared portion of the central channel to engage RNA substrates13, and alignment of Exo10Rrp6 and Exo10Rrp44+Rrp6Cterm reveals overlapping paths for RNA within the S1/KH central channel (Fig 3a,b), albeit in opposing directions. A subset of side chains from the S1/KH cap proteins contact RNA in both structures, but none make contacts that would enforce directionality. To determine the importance of the S1/KH ring for RNA degradation, yeast Exo10Rrp6 and Exo11Rrp44/Rrp6 were reconstituted with Csl4 and Rrp40 containing amino acid substitutions for Arg145, Arg150, Arg202; and Lys107, Lys108, and Arg110, respectively. These mutations severely diminish Rrp6 activity in Exo10Rrp6 and Exo11Rrp44/Rrp6 as well as Rrp44 activity in Exo11Rrp44/Rrp6 (Fig. 3c; Extended Data Fig. 5a). In contrast, Rrp6 activity is not inhibited by an insertion in Rrp45 that occludes the PH-like ring channel below the S1/KH ring13 although this mutation inhibits Rrp44 activity. Reconstituted human Exo10Rrp6 (Extended Data Fig. 3b) reveals similar dependencies as a channel occluding insertion in Rrp41 near the S1/KH ring impaired Rrp6 activity while a channel occluding insertion in Rrp45 well below the S1/KH ring failed to inhibit Rrp6 (Fig. 3d; Extended Data Fig. 5b). Combining insertions inhibits Rrp6 to greater degree, likely because the Rrp45 insertion restricts movement of the Rrp41 insertion. These data suggest that the RNA path to Rrp6 depends on the integrity of the S1/KH ring but does not require the full extent of the PH-like ring central channel. In contrast, at least for yeast Exo11Rrp44/Rrp6, the RNA path to Rrp44 relies on the integrity of central channel throughout both S1/KH and PH-like rings. These data are consistent with contacts observed to RNA in structures of Exo10Rrp6 and Exo10Rrp44+Rrp6Cterm.
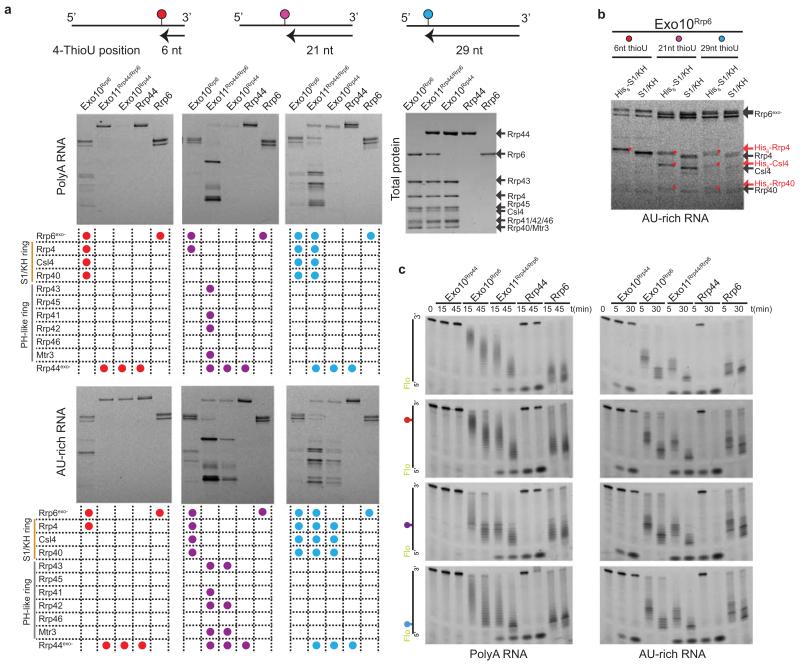
To map RNA interactions through Exo9 to Rrp6 and Rrp44, catalytically dead yeast exosome complexes were reconstituted and subjected to long wavelength UV crosslinking using 36-nt polyA or AU-rich RNA substrates bearing single 4-thioU substitutions 6, 21, or 29 nt from the 3′ end (Fig. 4a, Extended Data Fig. 6a-c). As the 4-thioU probe is moved 3′ to 5′, AU-rich RNA crosslinks to Rrp44, followed by the PH-like ring, and finally to the S1/KH cap in Exo10Rrp44 and Exo11Rrp44/Rrp6, in agreement with previous studies12,13,16,17. In contrast, crosslinks are only observed to Rrp44 in Exo10Rrp44 with polyA36 RNA; however, addition of Rrp6 in Exo11Rrp44/Rrp6 results in crosslinks that progress from Rrp44 through the PH-like ring to the S1/KH ring and Rrp6 as the 4-thioU probe is moved 3′ to 5′. The results with polyA36 RNA are in line with previous biochemistry showing that Rrp6 stimulates Rrp44 binding to polyA RNA in Exo11Rrp44/Rrp6 (ref 13).
Figure 4. RNA paths to Rrp6 and Rrp44.
a, RNA crosslinking pattern schematic for yeast exosomes (Extended Data Fig. 6a). Subunits labeled, background colored with Rrp6 (blue), S1/KH ring (orange), PH-like ring (grey) and Rrp44 (pink). 4-thioU positions indicated by colored circles at 6, 21 or 29 nt from the 3′ end. b, RNA crosslinking pattern schematic for human Exo10RRP6 as in (a). Crosslinks (left); proteins (right) (Extended Data Fig. 7). c, Exo11Rrp44/Rrp6 cartoon. Subunits and rings colored and labeled. RNA paths depicted by arrows, arrowhead at 3′ end with 4-thioU positions indicated as in (a). d, RNA degradation at 5-fold enzyme excess with Rrp6 and Rrp44 products (left) and time course for crosslinking to 4-thioU RNA 6 nt from 3′ end (right). e, Exo11Rrp44/Rrp6 model indicating RNA paths to Rrp6 (left) or Rrp44 (right). Subunits colored as in Fig. 1 next to surfaces colored according to electrostatic potential27. Visible subunits and RNA (yellow) labeled in left panels. Three putative paths past the S1/KH ring to Rrp6 denoted by yellow arrows and labels (P1, P2, P3) (Extended Data Fig. 8a). Side view shows an RNA path to Rrp44 (yellow arrow; 5′ and 3′ ends labeled) indicating that RNA could pass by Rrp6 (Extended Data Fig. 8b). RNA from PDB 4IFD was edited to remove the stem loop after nt 36. Assays in (a,b,d) conducted at least three times with typical results depicted.
Crosslinking to the Exo10Rrp6 complex reveals RNA contacts to Rrp6 and Rrp4 when 4-thioU is 6 nt from the 3′ end. Although no structural impediment exists to prevent RNA from entering the PH-like ring, polyA36 and AU-rich RNA crosslinks are only observed to Rrp4, Csl4 and Rrp40 even when the 4-thioU is positioned 21 or 29 nt from the 3′ end (Fig. 4c; Extended Data Fig. 6). This pattern contrasts with that observed for Exo10Rrp44 and Exo11Rrp44/Rrp6, where crosslinks are observed to PH-like ring subunits when 4-thioU is placed 21 nt from the 3′ end. Crosslinking patterns to human Exo10Rrp6 utilized a 36-nt AU-rich substrate as reported previously13. Similar to results obtained for yeast, UV-induced crosslinks are observed to the three S1/KH ring proteins and RRP45, but to none of the other five PH-like proteins (Fig. 4b). UV crosslinking to complexes with loop insertions mirror results observed in decay assays (Fig. 3d) with diminished crosslinking when a loop insertion is placed proximal to the S1/KH ring (Extended Data Fig. 7). These data suggest that the integrity of the S1/KH ring central channel is important for Rrp6 activity in both human and yeast systems.
The results are consistent with a model in which distinct but overlapping paths guide RNA to Rrp6 or to Rrp44 (Fig. 4c). So how is a path selected? Path selection appears stochastic in vitro because degradation products of Rrp6 and Rrp44 are observed under conditions of limiting enzyme (Fig. 3c) or limiting substrate (Fig. 4d). The distributive mechanism underlying Rrp6 activity suggests repeated substrate binding and release while the processive mechanism utilized by Rrp44 suggests that it binds and holds onto substrates until completely degraded. Thus at steady state, binding and UV crosslinking likely reflect the stable interaction with Rrp44 even when Rrp6 is present13 (Extended Data Fig. 6a). Additional evidence for stochastic sampling of the two paths is evident by UV crosslinking under conditions of slight enzyme excess (Fig. 4d). As predicted based on the distributive and processive mechanisms of Rrp6 and Rrp44, respectively, crosslinked products are observed to Rrp44, Rrp6 and the S1/KH ring proteins at the earliest times, and this pattern is lost once most of the RNA finds its way to the Rrp44 active site.
Structural analysis of the Exo10Rrp6 polyA complex suggests at least four potential paths past the S1/KH ring to Rrp6 although paths 1 and 2 appear most likely with respect to electrostatics and conservation (Fig 4e; Extended Data Fig. 8a). Importantly, these paths are available in Exo11Rrp44/Rrp6 as they do not involve surfaces from the PH-like ring or Rrp44. Modeling Rrp6 onto the Exo10Rrp44 RNA complex shows that the central channel is still accessible and that RNA paths to Rrp6 and Rrp44 are available in Exo11Rrp44/Rrp6 (Fig 4e; Extended Data Fig. 8b).
A wider channel in Exo10Rrp6
Rrp6 can stimulate Rrp44 binding and decay activities13, a phenomenon readily apparent in crosslinking to polyA RNA (Fig. 4a; Extended Data Fig. 6a). These results suggest that Rrp6 enhances RNA access to the PH-like ring and central channel. Structures of Exo10Rrp6 and Exo10Rrp44+Rrp6Cterm were compared to query differences that might account for this activity. While the global architecture of the PH-like ring does not differ (Fig. 5d), the Exo9 channel widens in Exo10Rrp6 through movement of Rrp4, Rrp40 and Csl4 away from the central channel, widening the gap between Rrp4 and Rrp40 S1 domains by ~4 Å (Fig. 5a-c). Although Exo10Rrp6 and Exo10Rrp44+Rrp6Cterm structures are bound to RNA, the increase in channel width in Exo10Rrp6 might account for Rrp6-mediated stimulation of Rrp44, especially since Rrp44 and Rrp6 do not physically interact. It remains unclear how Rrp6 exerts this change or if an Exo10Rrp44 apo structure differs from its RNA-bound configuration, but it is clear that Rrp6 EXO and CTD are both required for this activity as addition of either element alone is not sufficient to stimulate Exo10Rrp44 when added in trans (Fig. 5e). These data suggest that the Rrp6 CTD is required to bring the catalytic module in proximity to the S1/KH ring, perhaps eliciting channel widening through EXO domain interaction with Rrp4 and Rrp40. Further data will be required to determine if channel widening is a regulated feature of Exo11Rrp44/Rrp6 or if additional cytoplasmic factors elicit channel widening of Exo10Rrp44.
Figure 5. S1/KH ring widening in Exo10Rrp6.
a,b, Close-up of the central channel looking down the S1/KH ring. Rrp4 (green), Rrp40 (magenta) and Csl4 (light blue) in cartoon with transparent surfaces from (a) Exo10Rrp6 and (b) Exo10Rrp44+Rrp6Cterm. Distances indicated between respective labeled amino acids. c, Channel dimensions in Exo10Rrp6 (red) and Exo10Rrp44+6Cterm (black) calculated using HOLE28. d, Superposition of PH-like rings for Exo10Rrp6 (red) and Exo10Rrp44+6Cterm (grey). e, Rrp6exo- (128-733), and not Rrp6CTD (518-733) or Rrp6CAT (128-518), is required to stimulate Rrp44 activity. RNA decay assays conducted at least three times with typical results depicted.
Conclusions
The structure of Exo10Rrp6 shows Rrp6 positioned above the Exo9 S1/KH ring while the Exo10Rrp44+Rrp6Cterm structure shows Rrp44 below the PH-like ring. It is notable that Rrp6 activity is altered and becomes dependent on the S1/KH ring when associated with Exo9 (ref. 13). Although shorter RNAs may be directed to Rrp44 via a channel-independent “direct access” route18, Rrp44 remains highly dependent on the integrity of the central channel throughout both S1/KH and PH-like rings. The dependency on the Exo9 core is remarkable given that both Rrp44 and Rrp6 active sites are exposed to solvent in Exo10Rrp6 and Exo10Rrp44+Rrp6Cterm complexes.
That a similar segment of the S1/KH ring is used to engage RNA in opposing directions suggests that overlapping paths to Rrp6 and Rrp44 may serve to commit the exosome to distributive or processive degradation depending on how a particular RNA substrate is delivered to the exosome. Because the paths overlap, the exosome would be unable to interact with another substrate until completing the task at hand. While path choice appears stochastic in vitro (Fig. 4d), the nuclear cofactors Mpp6 (ref. 19), Rrp47 (ref. 8), and TRAMP20 may bias selection of a particular path to facilitate transitions between editing, processing or degradation activities of the exosome.
Rrp6 can stimulate Rrp44 activities when associated with the Exo9 core, perhaps through widening of the S1/KH ring. Channel gating mechanisms as a point of regulation have been described in many systems including, but not limited to, the proteasome21-25. While many degradation complexes place their catalytic activities in a protected compartment that is accessed by gating a substrate responsive channel, the RNA exosome appears inside out, with its catalytic subunits located on the periphery of the exosome core. While the functional relevance of this architecture is not yet clear, it is evident that the exosome core can modulate the activities of its catalytic subunits by requiring RNA to pass through distinct elements of the central channel before being processed or degraded.
Methods
Yeast exosome reconstitution and purification
Expression, purification, and reconstitution of recombinant exosome subunits and complexes have been previously described in detail13,26. In brief, all subunits were cloned as N-terminal 6× histidine Smt3 fusions in pRSFDuet-1 expression vectors (Novagen), and expressed in the E. coli expression strain BL21 (DE3) RIL (Stratagene) either as heterodimers (Smt3-Rrp41/Rrp45, Smt3-Rrp42/Mtr3, Smt3-Rrp46/Rrp43) or as distinct subunits (Smt3-Csl4, Smt3-Rrp4, Smt3-Rrp40, Smt3-Rrp6, Smt3-Rrp44). Generation of the Rrp40 and Csl4 point mutants was performed by PCR-mediated site-directed mutagenesis. For the mutant subunits, cells were grown in shaker flasks at 37°C in Superbroth (Teknova), induced by cold shock on ice, addition of ethanol to a final concentration of 2% and 0.05 mM isopropyl-β-D-thiogalactoside (IPTG), and grown at 18°C overnight. For all other proteins, cells were grown in shaker flasks at 37°C in Superbroth and induced by cold shock on ice and addition of 0.4 mM IPTG, and then subjected to shaking at 18°C overnight. Cells were harvested and lysed as described previously. After high speed centrifugation to pellet the insoluble material, the supernatant was loaded on a nickel-NTA column (Qiagen) and allowed to flow by gravity. After washing with 350 mM NaCl, 20 mM Tris pH 8.0, 1 mM BME, and 20 mM imidazole, the column was further washed with 350 mM NaCl, 50 mM KCl, 20 mM Tris pH 8.0, 1 mM BME, 2 mM ATP, 10 mM MgSO4 to displace chaperone impurities. Protein was eluted with 350 mM NaCl, 20 mM Tris pH 8.0, 1 mM BME, and 250 mM imidazole and then purified by size exclusion chromatography. All subunits were purified on the Superdex 200 (GE) with the exception of Smt3-Csl4 and Smt3-Rrp40, which were purified on a Superdex 75 (GE). Wild-type and mutant subunits eluted at the same volumes on gel filtration. Only Smt3-Rrp6, Smt3-Rrp44, and Smt3-Rrp42/Mtr3 were subjected to overnight cleavage by the SUMO protease Ulp1 (ref. 29) with another step of purification on the Superdex 200 to remove the 6x histidine Smt3 tags; Smt3-Rrp42/Mtr3 was cleaved by Ulp1 prior to formation of Exo9 as inclusion of this tag interferes with reconstitution. At this stage, subunits were concentrated to 6 to 12 mg/mL and stored at −80°C. For reconstitution, Exo9 subunits were mixed together as Smt3 fusions, incubated on ice for 30 minutes, followed by addition of Ulp1 for another 30 minutes prior to overnight dialysis. After purification, exosomes were concentrated to 12-14 mg/mL and stored at −80°C in a final buffer of 100 mM NaCl, 20 mM Tris pH 8.0, 0.1 mM MgCl2, 1 mM TCEP.
Human exosome reconstitution and purification
For human, expression, purification and reconstitution strategies of the Exo9 core were identical except Smt3-RRP43/MTR3/RRP42 was expressed as a trimer, Smt3-RRP46 was expressed alone (as described previously15,26), and RRP451-302/RRP41 was purified with a non-cleavable N-terminal hexa-histidine tag on RRP45. RRP6 (EXOSC10, PM/SCL-100) residues 180 to 804 were PCR-amplified from a previously generated full-length expression construct11, cloned into pET28a-Smt3 and transformed into BL21 (DE3) RIL. For expression, cells were grown in shaker flasks to ~0.6 OD600, and induced overnight at 18°C with 0.25 mM IPTG in the presence of 2% ethanol. Subsequent purification of human Smt3-RRP6180-804 and reconstitution of HsExo10Rrp6 is identical to that described for the yeast system. The purification method for the catalytic region, RRP6180-606, used for core interaction studies has been described previously11. The channel occluding RRP41 and RRP45 mutants were engineered based on those described for S. cerevisiae13, and were generated by PCR to encode electrostatic and steric loop insertions with the primary sequence GTGESEGESES between amino residues Gly93 and Arg94 of RRP45 and between Arg62 and Ala63 of RRP41.
Crystallization and structure determination
Exosome samples were mixed with polyA24 RNA (Invitrogen) in a 1:1.1 molar ratio and incubated on ice for 1 hour prior to crystallization. Crystals grew at 18°C by vapor diffusion in either sitting (Greiner Bio One, Crystalquick) or hanging (Hampton Research, VDX) drop formats, in 7-11% PEG3350, 100 mM MES pH 6.7, 4-15% MPD, typically taking 3 to 5 days to appear. Crystals were harvested within 2 weeks. Partial degradation of RNA (20-24 nt) was observed in washed crystals (Extended Data Fig. 1d). For cryoprotection, well solution was replaced with crystallization buffer augmented with 25-30% MPD and crystals were incubated for three days before harvesting; one day before harvest, trays were transferred to 4°C. A light polarizer was used to exclude multiple crystals and to identify crystals with single regions, which were broken off and flash frozen in liquid nitrogen for data collection. X-ray diffraction data were collected at the Advanced Photon Source 24-ID-E and 24-ID-C beam lines, and the National Synchrotron Light Source X29 beam line. Data was obtained from a single crystal diffracted at NSLS X29 at a wavelength of 1.075 Å at 100 K. Data were processed using HKL200030 and statistics reported in Extended Data Table 1 were obtained using Phenix31 including CC1/2 (%) and CC* (%) values of 99.9 (33.8) and 100.0 (71.1) for data between 50-3.3 (3.42-3.3) Å. The structure solved by molecular replacement using Phaser32 and coordinates of human apo Exo9 (PDB: 2NN6) and yeast Rrp6CAT (PDB: 2HBL) as search models followed by docking yeast Exo10Rrp44+6Cterm (PDB: 4IFD) (Extended Data Table 1). The structure is refined to R/Rfree values of 0.227/0.265. The final model includes six nucleotides of the polyA 24mer, and two phosphate ions that likely represent the RNA backbone (Extended Data Fig. 1a-c), and 2751 of the 3156 amino acids present in the crystal (Extended Data Table 2). The asymmetric unit contains one complex. Iterative rounds of refinement were accomplished using Phenix31. RNA and side chains were manually built using O33 and Coot34. The model was initially refined using secondary structure restraints in conjunction with positional refinement followed by individual B-factor refinement. Secondary structure restraints were released and a final round of positional and B-factor refinement was performed followed by refinement of TLS parameters that resulted in a further decrease in R and Rfree values. Figures depicting the structure were prepared with Pymol27. Surface conservation was calculated using ConSurf35. Structure quality was assessed using MolProbity36 indicating the model has excellent geometry with 95.7% in favored and 100% in allowed regions of Ramachandran space. The structure also scored in the 100th percentile for the Clash and MolProbity scores.
RNA biochemistry
For yeast exosomes, unless otherwise noted, exoribonuclease assays were performed under multiple turnover conditions as described previously13. In brief, 10 nM synthetic 49-nt RNAs (Invitrogen) bearing 5′ fluorescein labels were incubated at 30°C with 1 nM RNA exosome for various time points, and RNA degradation was monitored by resolving reaction intermediates by denaturing TBE-PAGE (Invitrogen), and detected using a Fuji FLA-5000 scanner (FITC filter). A similar protocol was followed for human exosome complexes, the exception being that these assays were performed at 37°C, with 5 nM of the 49-nt RNA and 50 nM HsExo10Rrp6. For mix-in experiments (Fig. 5e), 2-fold molar excess of Rrp6 protein was incubated with Exo10Rrp44 on ice for 1 hour prior to initiating RNA decay by addition of RNA. For steady state UV-RNA crosslinking to 4-thioU-bearing RNAs, 500 nM of exosome complexes with mutations in the exoribonuclease active sites (D238N for Rrp6, D551N for Rrp44) were incubated for 20 minutes on ice with 50 nM 36-nt 5′ fluorescein RNAs with a single internal 4-thioU (Thermo Scientific) in a 70 μL reaction volume. Binding buffer includes 50 mM KCl, 20 mM Tris pH 8.0, 10 mM DTT, 0.5 mM MgCl2. Crosslinking was performed by subjecting the RNA-exosome mixture to long-range UV (365 nm) for 15 minutes in the dark using a 4W handheld lamp. For the time course described in Fig. 4d, 350 nM of exosome was incubated with 150 nM of RNA, and the binding reactions allowed to proceed for indicated times before initiating crosslinking for 10 minutes. 15 μL was quenched with LDS loading buffer, and the crosslinked products were separated by SDS-PAGE (Invitrogen) and visualized with a Fuji FLA-5000 scanner (FITC filter). 4-thioU RNA included the following sequences, each of which contained a 5′-Fluorescein: 5′-Fl-AAU UAU 4thioUUA UUA UUU AUU UAU UAU UUA UUU AUU UAA; 5′-Fl-AAU UAU UUA UUA UU4thioU AUU UAU UAU UUA UUU AUU UAA; 5′-Fl-AAU UAU UUA UUA UUU AUU UAU UAU UUA UU4thioU AUU UAA; 5′-Fl-AAA AAA 4thioUAA AAA AAA AAA AAA AAA AAA AAA AAA AAA; 5′-Fl-AAA AAA AAA AAA AA4thioU AAA AAA AAA AAA AAA AAA AAA; 5′-Fl-AAA AAA AAA AAA AAA AAA AAA AAA AAA AA4thioU AAA AAA. For UV crosslinking to human exosome complexes, samples were incubated with 250 nM of the AU-rich RNA and protein in binding buffer for 60 minutes on ice, and then placed in a UV-Stratalinker (Stragene) and subjected to 300,000 μJ of short-wave (254 nm) UV radiation. Samples were quenched with LDS loading buffer, and crosslinked products were separated and visualized as described above.
Extended Data
Extended Data Figure 1. View highlighting electron densities covering RNA in the Exo10Rrp6-polyA structure.
a, Simulated annealing 2Fo-Fc omit map contoured at 1.0 σ. b, Simulated annealing Fo-Fc omit map contoured at 2.0 σ. c, Final 2Fo-Fc omit map contoured at 1.0 σ. RNA and phosphate oxygen, nitrogen, carbon and phosphate atoms are colored red, blue, orange and deep orange, respectively. Electron density maps shown as wire mesh colored blue for 2Fo-Fc maps and green for the Fo-Fc map. Rrp6, Rrp40, Rrp4 and Csl4 are labeled and shown in cartoon representation in teal, magenta, green and light blue, respectively. d, 20 to 24 nucleotides of polyA RNA are present in crystals of Exo10Rrp6. Crystals of Exo10Rrp6 bound to polyA24 RNA were first washed by two rounds of transfer to 1 μL well solution, then dissolved in water and TBE-urea sample buffer, and analyzed by 15% TBE-urea PAGE. A drop without crystals was run as a control. RNA was stained by Sybr Gold.
Extended Data Figure 2. Rrp6 features within Exo10Rrp6.
a, The primary sequence of the catalytic module of Rrp6 (EXO and HRDC) is highly conserved, whereas sequence conservation within the CTD varies. Rrp6 in Exo10Rrp6 is represented as a surface, and colored according to sequence conservation as calculated by ConSurf35, colored from red (highly conserved) to blue (variable). The Exo9 core is depicted as a transparent cartoon, with the S1/KH ring in orange, and the PH-like ring in grey. b, Superposed structures of the Rrp6 CTD from Exo10Rrp6 (residues 525-628; teal) and Exo10Rrp44+Rrp6Cterm (residues 532-557, 565-619; magenta; PDB 4IFD) reveal overall similarity for residues within the respective models. c, The conserved linker between the EXO and HRDC domain of Rrp6 adopts an α-helix when associated with the exosome core. Superposition of the Rrp6 catalytic domains from the structures of Exo10Rrp6 and Rrp6-AMP (PDB: 2HBL) shows residues Pro424 to Asn433 rearrange from a loop in the AMP-bound structure to a helix that is adjacent to a conserved albeit disordered loop in the S1 domain of Rrp4. These residues were previously thought to comprise a protein interaction module10. Rrp6 is represented as cartoons, with Rrp6 from the Exo10Rrp6 structure in teal, and the Rrp6-AMP structure in grey. Rrp4 is in light green and depicted as a cartoon with a transparent surface, with the S1 loop representing residues Gly145 to Gln158 shown as a dashed line.
Extended Data Figure 3. The Rrp6 CTD is required for interaction with human and yeast RNA exosome cores.
a, Schematic representation of Rrp6 including from N- to C-terminus the PMC2NT, EXO, HRDC and C-terminal (CTD) domains. EXO and HRDC comprise Rrp6CAT. Below are lines representing individual elements used in gel filtration analysis in c and d along with amino acid numbering specific for S. cerevisiae (Sc) Rrp6 and H. sapiens (Hs) RRP6. b, Gel filtration profile for reconstituted human Exo10Rrp6 (left) with SDS-PAGE analysis of the peak fractions (right). Subunits labeled in capital letters corresponding to yeast nomenclature for clarity. Apparent molecular weights denoted in kilodaltons. c, Interaction studies of human RRP6 with the HsExo9. The human Rrp6 catalytic domain (residues 180-606) does not interact well with human Exo9; however addition of Rrp6 180-804 suggests that CTD residues 607-804 are important for stable interaction with Exo9. Reconstitution experiments on mixtures of HsExo9, HsExo9 with 2-fold excess of RRP6180-606 (the catalytic region), HsExo9 with 2-fold excess of RRP6180-804 (the catalytic region with a portion of the carboxy terminus), and RRP6180-606. Gel filtration analysis on RRP6180-804 in isolation was not possible as this construct is unstable in the absence of the SMT3 tag. Mixtures (0.15 ml at ~2 mg/ml) were dialyzed overnight at 4°C against reconstitution buffer (50 mM NaCl, 20 mM Tris-HCl [pH 8.0], and 10 mM DTT). Mixtures were applied to a Superose 6 10/300 GL column (G.E. Health Sciences) equilibrated with reconstitution buffer with the resulting UV traces (upper panels) and analyzed fractions (lower panel). d, Interaction studies of budding yeast Rrp6 with ScExo10Rrp44. Rrp6 (residues 128-733) and Rrp6CTD, but not Rrp6CAT, interact with Exo10Rrp44. Rrp6 (residues 128-733), Rrp6CAT (residues 128-518), the Rrp6CTD (residues 518-733), or both were incubated with Exo10Rrp44 for one hour on ice before analysis by size exclusion chromatography (Superose 6) in reconstitution buffer. Rrp6 and Rrp6CAT were added in 1.5-fold molar excess to Exo10Rrp44, while Rrp6CTD was added in 3-fold molar excess. For panel c and d, fractions were analyzed by SDS-PAGE and protein detected with Sypro Ruby.
Extended Data Figure 4. RNA interactions within the Rrp6 active site.
a,b, View of RNA nucleotides 24 to 20 from the 3′ end in the active site of S. cerevisiae Rrp6 from the structure of Exo10Rrp6 (a) and the same RNA modeled into the active site of H. Sapiens Rrp6 (PDB: 3SAF) based on superposing the respective EXO domains (b). Residues involved in RNA contacts in the structure of yeast Exo10Rrp6 are shown as teal sticks in (a); the corresponding residues in human are shown in (b). In both structures, a catalytic aspartate is mutated to asparagine and labeled in red (N238 in yeast; N313 in human). RNA is shown as orange sticks, and magnesium ions are represented as green spheres. c, Structure of the yeast Rrp6 catalytic domain in complex with zinc (Zn; blue sphere) and manganese (Mn; yellow sphere) in complex with AMP (PDB 2HBL). Active site residues are depicted as in a,b. The conserved Tyr361 side chain was mutated to alanine in this structure (labeled Y361A in red). d, Model of the Rrp6 active site constructed by superposing the 3′ nucleotide in the Exo10Rrp6 polyA structure to the AMP ligand from PDB 2HBL. Amino acid side chains are labeled and putative interactions between metal ligands, active site residues and RNA are depicted by dashed lines.
Extended Data Figure 5. Central channel mutations and their impact on Rrp6 degradation of AU-rich RNA.
a, 1 nM of reconstituted wild-type and mutant yeast Exo10Rrp6 and Exo11Rrp44/Rrp6 were incubated with 10 nM 49-nt 5′ fluorescein AU-rich RNA at 30°C. Reaction intermediates were analyzed after indicated time points by denaturing PAGE and imaged with a fluoroimager. Rrp6 activity decreases in the presence of the Rrp40 K107E/K108E/R110D and Csl4 R145A/R150A/R202D mutations in both Exo10Rrp6 and Exo11Rrp44/Rrp6 while is mostly unaffected by the channel occlusion in the Rrp45Large exosome. Rrp44 exoribounclease activity is attenuated by mutations in both the S1/KH and PH-like ring in Exo11Rrp44/Rrp6. b, Exoribonuclease activities of wild-type and mutant forms of human Exo10RRP6 were performed in a 100-μL reaction mixture with 5 nM 49-nt 5′ fluorescein AU-rich RNA and 50 nM protein at 37°C. Reaction intermediates were analyzed after indicated time points as described in (a). Mutation studies reveal that the human RRP6 exoribonucleolytic activity decreases in the presence of a channel occlusion near the S1/KH ring (RRP41Large), while a channel occlusion deeper in the central channel (RRP45Large) of HsExo10RRP6 has little impact on activity.
Extended Data Figure 6. UV crosslinking with 36-nt 4-thioU RNAs for yeast exosome complexes.
a, Raw data used to generate Fig. 4a in main text for polyA and AU-rich RNA. See Methods for crosslinking details. Presence or absence of crosslinks to given subunits is summarized in the table and denoted by color-coded dots, with red corresponding to 4-thioU 6 nt from the 3′ end; purple for 4-thioU 21 nt from the 3′ end; and blue for 4-thioU 29 nt from the 3′ end. b, RNA-protein adducts observed in UV crosslinking to Exo10Rrp6 correspond to the S1/KH cap proteins. Exo10Rrp6 bearing N-terminal hexahistine tags on the S1/KH cap proteins Rrp4, Rrp40, and Csl4 (ref. 13) or tag free variants were crosslinked to the three 4-thioU RNAs described above, and resolved by SDS-PAGE and imaged with a flouorimager. In comparing the His6-tagged and tagless complexes, mobility shifts are apparent in crosslinks corresponding to the S1/KH cap proteins. c, 4-thioU RNAs are bona fide decay substrates for exosome complexes and catalytic subunits, Rrp6 and Rrp44. Conditions using substrate excess (10 nM RNA, 1 nM enzyme), 4-thioU polyA RNAs (left) and AU-rich RNAs (right) are comparably degraded to corresponding 36-nt RNAs lacking 4-thioU. Reactions were performed at 30°C and stopped after indicated time points and resolved by denaturing PAGE and imaged with a fluoroimager. 4-thioU RNAs are color-coded as in (a) and (b).
Extended Data Figure 7. UV crosslinking to AU-rich RNA to components of human Exo10RRP6.
a, UV crosslinking of RNA to HsExo10RRP6 with open and occluded channels. Human RRP6 (Smt3-RRP6180-804), HsExo9, and wild type as well as three different pore occlusion mutants of HsExo10RRP6 were incubated and UV crosslinked to a 24-nt fluorescein-labeled AU-rich RNA substrate. Samples were analyzed for the presence of RNA-protein crosslinks (left panel) or integrity of protein via Sypro staining (right panel, Bio-Rad). Mutant forms of HsExo10RRP6 with insertion loops in RRP41-L and RRP45-L are labeled with red asterisks. b, Identification of UV-induced crosslinks. Identification of RNA-protein crosslinks between RNA to cap components of the human Exo9 core (RRP4, RRP40, and CSL4) was facilitated by reconstituting His6-tagged versions of these subunits into HsExo9 complexes, performing crosslinking assays with a 36-nt fluorescein-labeled AU-rich RNA, and detecting slower mobility for the His6-tagged crosslinked adducts. Similarly, identification of RRP45-RNA adducts was based on changes in adduct mobility when using isoforms with wild-type carboxy termini (RRP451-456) and the long insertions (RRP45-L1-302). RRP6-RNA adducts were determined by crosslinking of an Smt3-RRP6180-804 fusion and comparison of 9- and 10-subunit exosomes (in a).
Extended Data Figure 8. RNA paths to Rrp6 and Rrp44.
Structure of an Exo11Rrp44/Rrp6 model derived by superposing the PH-like ring subunits in Exo10Rrp6 and Exo10Rrp44+6Cterm (PDB 4IFD). a, RNA paths to the Rrp6 active site. RNA (yellow) shown as derived from the Exo10Rrp6 polyA complex. Three views are provided from the top, side and backside of the complex with subunits colored as in Fig. 1 in the main text (left), surfaces colored according to conservation as calculated by ConSurf35 (middle) from red (highly conserved) to blue (variable), and surfaces colored according to electrostatic potential as calculated by Pymol27 (right). Subunits that are visible are labeled in the left panel as is the RNA. Three putative paths that traverse the S1/KH ring into the Rrp6 active site are denoted by yellow arrows and labels (P1, P2, P3) in the top and side views and a fourth path (P4) under the Rrp6 catalytic domain is denoted in the backside view. Based on conservation, electrostatics and crosslinking we deem P1 or P2 as the most likely path of ingress for the incoming RNA substrate. The top and side views utilize cutaway surfaces to illustrate the putative paths and surface properties. b, RNA paths to the Rrp44 active site. Subunits and surfaces depicted as in (a) with the exception that the RNA is now derived from the structure of Exo10Rrp44+6Cterm (PDB 4IFD) with the RNA edited to remove the stem loop after nucleotide 36. Top, side and bottom views are depicted with the RNA path indicated by a yellow arrow with 5′ and 3′ ends labeled. The position of the central channel is also labeled and indicated by a yellow arrow in the top and bottom views. The top and side views show that RNA could pass by Rrp6 to penetrate the S1/KH and PH-like ring central channel.
Extended Data Table 1. Data collection and refinement statistics.
| Exo10Rrp6/polyA | |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 193.3, 200.1, 97.4 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50-3.3 (3.42-3.3) * |
| R merge | 6.1 (65.3) |
| I/σI | 15.5 (1.4) |
| Completeness (%) | 97.7 (96.9) |
| Redundancy | 5.2 (3.1) |
| Refinement | |
| Resolution (Å) | 50-3.3 |
| No. reflections | 56332 |
| Rwork/ Rfree | 22.7/26.5 |
| No. atoms | 21595 |
| Protein | 21450 |
| Ligand/ion | 145 |
| Water | 0 |
| B-factors | |
| Protein | 125 |
| Ligand/ion | 156 |
| Water | n/a |
| R.m.s deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.47 |
One crystal was used.
Highest resolution shell is shown in parenthesis.
Extended Data Table 2. Amino acid and RNA residues.
| Subunit | Residues | Non-native N-terminal residues | Residues observed in structure |
|---|---|---|---|
| Rrp45 | 305 | None | 3-206, 213-301 |
| Rrp41 | 246 | GDPH | 4-242 |
| Rrp43 | 394 | None | 6-101,121-180, 184-191, 209-249, 271-309, 327-394 |
| Rrp46 | 223 | GS | 1-221 |
| Rrp42 | 265 | GDPH | 2-161, 170-264 |
| Mtr3 | 250 | None | 5-20, 43-147, 163-248 |
| Rrp40 | 240 | GDPH | 2-45, 54-235 |
| Rrp4 | 359 | GDPH | 5-14, 50-145, 158-249, 277-357 |
| Csl4 | 292 | GDPH | 5-70, 105-113, 128-162, 186-291 |
| Rrp6 (128-685) | 558 | SL | 128-516, 525-628 |
| RNA | 24 | n.a. | 19-24 |
Acknowledgements
We thank NE-CAT beamlines (Advanced Photon Source) supported by RR-15301 (NIH NCRR). APS is supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. Beamline X29 (National Synchrotron Light Source) supported by the US Department of Energy, the Office of Basic Energy Sciences and P41RR012408 (NIH NCRR). Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers F31GM097910 (E.V.W) and R01GM079196 (C.D.L). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.D.L. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions E.V.W. and C.D.L. designed and E.V.W. performed experiments for the S. cerevisiae exosomes. E.V.W. and C.D.L. determined the structure. K.J. and C.D.L. designed and K.J. performed the experiments for the H. sapiens exosome. E.V.W. and C.D.L. wrote the manuscript.
Author Information Atomic coordinates and structure factors are deposited in the RCSB with accession code 4OO1. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Wasmuth EV, Lima CD. Structure and activities of the eukaryotic RNA exosome. In: Chanfreau GF, Tamanoi F, editors. The Enzymes. Vol. 31. Elsevier; San Diego: 2012. pp. 53–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ to 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 3.Tomecki R, et al. The human core exosome interacts with differentially localized processive RNases: hDis3 and hDis3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg+2 ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stead JA, Costello JL, Livingstone MJ, Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Midtgaard SF, et al. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl. Acad. Sci. USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Januszyk K, Liu Q, Lima CD. Activities of human RRP6 and structure of the human RRP6 catalytic domain. RNA. 2011;17:1566–1577. doi: 10.1261/rna.2763111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makino DL, Baumgärtner M, Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- 13.Wasmuth EV, Lima CD. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell. 2012;48:133–144. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assenholt J, et al. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA. 2008;14:2305–2313. doi: 10.1261/rna.1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 16.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 17.Drążkowsak K, et al. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41:3845–3858. doi: 10.1093/nar/gkt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JJ, et al. Visualization of distinct substrate-recruitment pathways in the yeast exosome by EM. Nature Struct. Mol. Biol. 2013 doi: 10.1038/nsmb.2736. doi:10.1038/nsmb.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milligan L, et al. A yeast exosome cofactor, Mpp6, functions in RNA surveillance and in the degradation of noncoding RNA transcripts. Mol. Cell. Biol. 2008;28:5446–5457. doi: 10.1128/MCB.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 21.Makino DL, Halbach F, Conti E. The RNA exosome and proteasome: common principles of degradation control. Nat. Rev. Mol. Cell. Biol. 2013;14:654–660. doi: 10.1038/nrm3657. [DOI] [PubMed] [Google Scholar]
- 22.Kish-Trier E, Hill CP. Structural biology of the proteasome. Annu. Rev. Biophys. 2013;43:29–49. doi: 10.1146/annurev-biophys-083012-130417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sen M, et al. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Effantin G, Maurizi MR, Steven AC. Binding of the ClpA unfoldase opens the axial gate of ClpP peptidase. J. Biol. Chem. 2010;285:14834–14840. doi: 10.1074/jbc.M109.090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha SS, et al. Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber. EMBO J. 2010;29:3520–3530. doi: 10.1038/emboj.2010.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greimann JC, Lima CD. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The PyMOL Molecular Graphics System . Version 1.5.0.4 Schrödinger. LLC; [Google Scholar]
- 28.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–60. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 29.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter CW Jr., Sweet RM, editors. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–581. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashkenazy H, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]