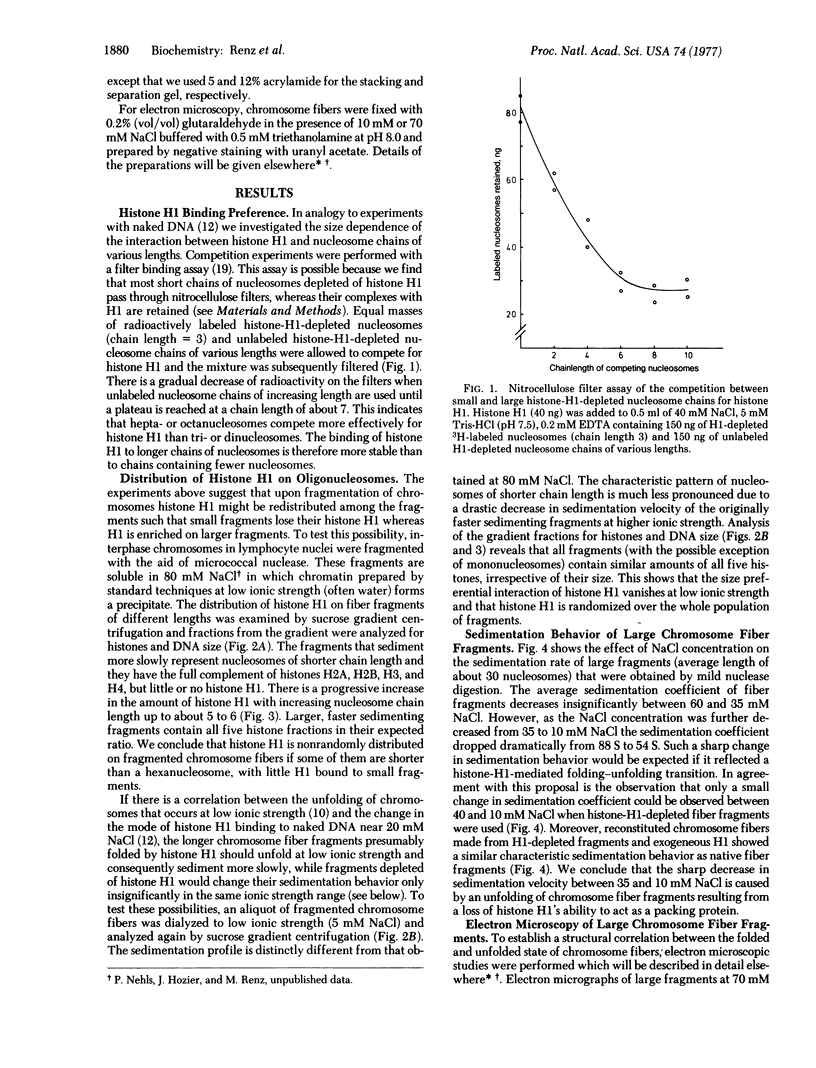
Abstract
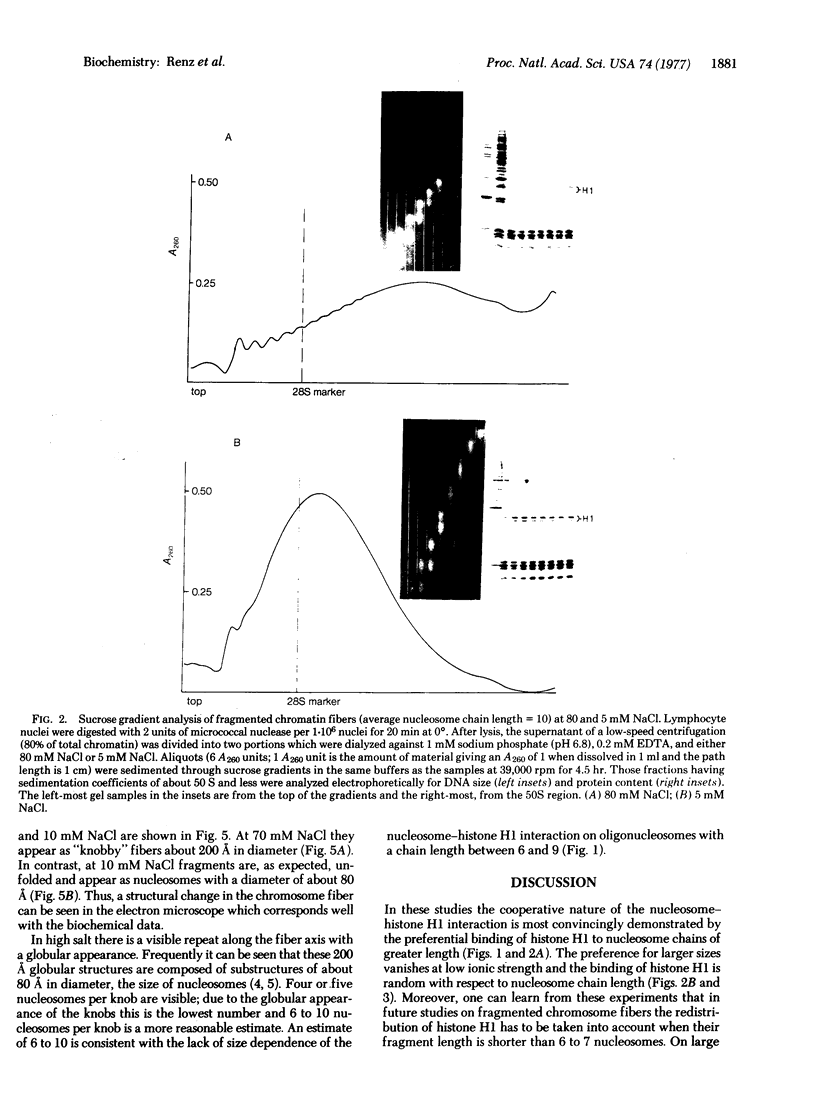
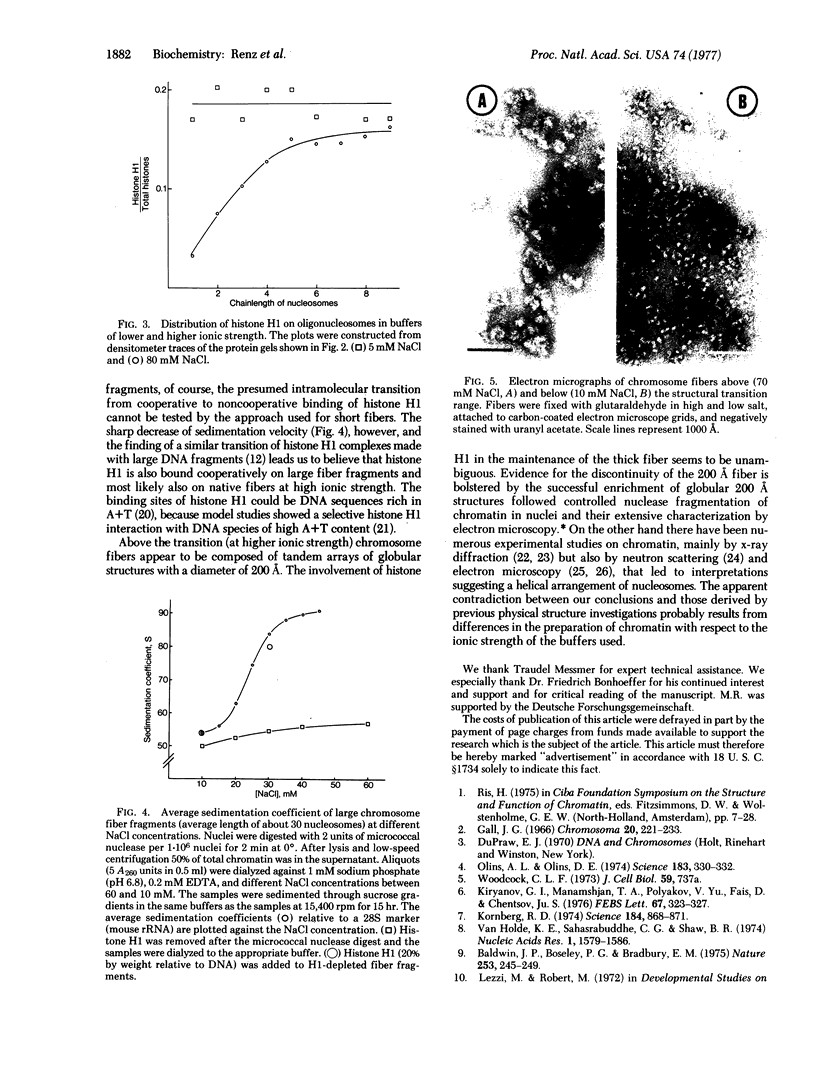
At high ionic strength (e.g., physiological salt concentrations) chromosome fibers are 200 A in diameter and composed of discrete globular structures that are held together by histone H1. At low ionic strength the fibers unfold and appear as the familiar chains of nucleosomes (80 A in diameter). The unfolding of chromosome fibers occurs within a narrow salt range. It results from a change in the mode of the interaction between histone H1 and the chromosome fiber and is very likely the consequence of a change from cooperative binding between histone H1 and DNA to a noncooperative binding. In the noncooperative binding state histone H1 molecules are randomly redistributed along the chromosome fiber.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin J. P., Boseley P. G., Bradbury E. M., Ibel K. The subunit structure of the eukaryotic chromosome. Nature. 1975 Jan 24;253(5489):245–249. doi: 10.1038/253245a0. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Danby S. E., Rattle H. W., Giancotti V. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. Histone H1 in chromatin and in H1 - DNA complexes. Eur J Biochem. 1975 Sep 1;57(1):97–105. doi: 10.1111/j.1432-1033.1975.tb02280.x. [DOI] [PubMed] [Google Scholar]
- Bram S., Ris H. On the structure of nucleohistone. J Mol Biol. 1971 Feb 14;55(3):325–336. doi: 10.1016/0022-2836(71)90321-4. [DOI] [PubMed] [Google Scholar]
- CHAUVEAU J., MOULE Y., ROUILLER C. Isolation of pure and unaltered liver nuclei morphology and biochemical composition. Exp Cell Res. 1956 Aug;11(2):317–321. doi: 10.1016/0014-4827(56)90107-0. [DOI] [PubMed] [Google Scholar]
- Carpenter B. G., Baldwin J. P., Bradbury E. M., Ibel K. Organisation of subunits in chromatin. Nucleic Acids Res. 1976 Jul;3(7):1739–1746. doi: 10.1093/nar/3.7.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H. G., Haynes M. E. Electron-microscope observations on cell nuclei in various tissues of a teleost fish: the nucleolus-associated monolayer of chromatin structural units. J Cell Sci. 1976 Jul;21(2):315–327. doi: 10.1242/jcs.21.2.315. [DOI] [PubMed] [Google Scholar]
- Evenson D. P., Mego W. A., Taylor J. H. Subunits of chromosomal DNA. I. Electron microscopic analysis of partially denatured DNA. Chromosoma. 1972;39(3):225–235. doi: 10.1007/BF00290785. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Klug A. Solenoidal model for superstructure in chromatin. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G. Chromosome fibers studied by a spreading technique. Chromosoma. 1966;20(2):221–233. doi: 10.1007/BF00335209. [DOI] [PubMed] [Google Scholar]
- Hozier J. C., Kaus R. Subunit structure of chromosomes in mitotic nuclei of physarum polycephalum. Chromosoma. 1976 Aug 4;57(1):95–102. doi: 10.1007/BF00292952. [DOI] [PubMed] [Google Scholar]
- Ilyin Y. V., Varshavsky A. Y., Mickelsaar U. N., Georgiev G. P. Studies on deoxyribonucleoprotein structure. Redistribution of proteins in mixtures of deoxyribonucleoproteins, DNA and RNA. Eur J Biochem. 1971 Sep 24;22(2):235–245. doi: 10.1111/j.1432-1033.1971.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Kiryanov G. I., Manamshjan T. A., Polyakov V. Y., Fais D., Chentsov J. S. Levels of granular organization of chromatin fibres. FEBS Lett. 1976 Sep 1;67(3):323–327. doi: 10.1016/0014-5793(76)80557-1. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Wilkins M. H. A super-coil model for nucleohistone. J Mol Biol. 1972 Jul 14;68(1):115–124. doi: 10.1016/0022-2836(72)90267-7. [DOI] [PubMed] [Google Scholar]
- Renz M., Day L. A. Transition from noncooperative to cooperative and selective binding of histone H1 to DNA. Biochemistry. 1976 Jul 27;15(15):3220–3228. doi: 10.1021/bi00660a010. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Schaller H. Mapping and characterization of promoters in bacteriophages fd, f1 and m13. J Mol Biol. 1975 Feb 25;92(2):261–277. doi: 10.1016/0022-2836(75)90226-0. [DOI] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de NOOIJ E., WESTENBRINK H. G. Isolation of a homogeneous lysine-rich histone from calf thymus. Biochim Biophys Acta. 1962 Aug 27;62:608–609. doi: 10.1016/0006-3002(62)90254-8. [DOI] [PubMed] [Google Scholar]