Abstract
End-stage renal disease (ESRD) presenting in a familial autosomal dominant pattern points to an underlying monogenic cause. Nail-patella syndrome (NPS) is an autosomal dominant disorder that may lead to ESRD caused by mutations in the transcription factor LMX1B. Renal-limited forms of this disease, termed nail-patella-like renal disease (NPLRD), and LMX1B nephropathy have recently been described. We report a large family, from the North East of England, with seven affected members with varying phenotypes of renal disease, ranging from ESRD at 28 years of age to microscopic haematuria and proteinuria and relatively preserved renal function. In this family, there were no extra-renal manifestations to suggest NPS. Genome-wide linkage studies and inheritance by descent (IBD) suggested disease loci on Chromosome 1 and 9. Whole exome sequencing (WES) analysis identified a novel sequence variant (p.R249Q) in the LMX1B gene in each of the three samples submitted, which was confirmed using Sanger sequencing. The variant segregated with the disease in all affected individuals. In silico modelling revealed that R249 is putatively located in close proximity to the DNA phosphoskeleton, supporting a role for this residue in the interaction between the LMX1B homeodomain and its target DNA. WES and analysis of potential target genes, including CD2AP, NPHS2, COL4A3, COL4A4 and COL4A5, did not reveal any co-inherited pathogenic variants. In conclusion, we confirm a novel LMX1B mutation in a large family with an autosomal dominant pattern of nephropathy. This report confirms that LMX1B mutations may cause a glomerulopathy without extra-renal manifestations. A molecular genetic diagnosis of LMX1B nephropathy thus provides a definitive diagnosis, prevents the need for renal biopsies and allows at risk family members to be screened.
Keywords: end-stage renal disease, LMX1B, podocytopathy, proteinuria, transcription factor
Introduction
Nail-patella syndrome (NPS, OMIM 161200) is a rare cause of autosomal dominant end-stage renal disease (ESRD). Typically, patients with NPS will exhibit extra-renal manifestations including absent/hypoplastic finger/toe nails and patellae, elbow dysplasia and iliac horns [1]. Renal manifestations occur in 30–50% of patients, and typically include haematuria and proteinuria and sometimes nephrotic syndrome. ESRD occurs in 5–10% of cases [2, 3]. Mutations in the LIM homeodomain transcription factor (LMX1B) underlie the molecular basis of NPS [4]. Antenatally, LMX1B is important for dorsal–ventral patterning, and is also expressed in the developing glomerulus, within S-shaped bodies. Murine data suggest that, post-natally, LMX1B is necessary for podocyte maintenance, and regulates expression of CD2AP, NPHS2 [5], COL4A3 and COL4A4 [6]. It has been suggested that intra-familial variation in renal failure seen in NPS may relate to heterozygous variants (which may segregate independently) in LMX1B-regulated genes [2].
Recently, LMX1B mutations have been identified in renal limited forms of NPS, termed nail-patella-like renal disease (NPLRD). Previous reports include three families with an autosomal dominant pattern of focal and segmental glomerular sclerosis (FSGS) in whom missense mutations (two with R246Q and one with R246P) in LMX1B were identified [7]. Another report detailed an R246Q missense mutation in LMX1B, which was associated with childhood-onset microscopic haematuria and subnephrotic proteinuria but lacked any extra-renal manifestations [8]. Thus, LMX1B is a candidate gene for autosomal dominant nephropathies, including familial cases of FSGS and unexplained proteinuria.
We describe a large family where affected individuals had ESRD, chronic kidney disease or haematuria and proteinuria in an autosomal dominant pattern. Renal biopsies were unhelpful and failed to show glomerular or basement membrane defects consistent with an inherited glomerulopathy, and therefore we pursued a possible underlying genetic cause for a unifying diagnosis. Using a combination of traditional linkage studies and inheritance by descent (IBD) approaches with whole exome sequencing (WES) strategies, we identified a novel heterozygous mutation in LMX1B (p.R249Q) which segregated with disease.
Materials and Methods
Clinical and genetic investigations
Clinical data and historical renal biopsies were reviewed where available. Following informed consent, DNA was obtained from all affected patients and their unaffected relatives where available. This study was approved by the Northern and Yorkshire Regional Ethics Committee. Genomic DNA was extracted from blood samples collected in EDTA tubes using the QIAGEN Blood and Cell Culture DNA kit according to the manufacturer's instructions.
Genotyping and linkage studies
We carried out a genome-wide linkage search using Affymetrix GeneChip® Human Mapping 250K Sty Arrays according to the manufacturer's protocol (http://www.affymetrix.com) in six affected members and two unaffected members of the family. Genehunter software was used to calculate a multipoint parametric logarithm of odds (LOD) score, assuming an autosomal dominant mode of inheritance. IBD across the genome was estimated using Combinatorial Conflicting Homozygosity (CCH) [9].
Whole exome sequencing
WES was performed in three affected individuals from the family using genomic DNA by AROS Applied Biotechnology AS, Denmark. The reads were processed and analysed using a comprehensive bioinformatics workflow to identify variants. The quality of the reads was first checked with FastQC [10]. Poly-N tails were trimmed off from reads with an in-house Perl script. The 13 bp on the 5′ of all reads was clipped off with Seqtk [11] to remove biased sequencing reads caused by random hexamer priming [12]. Low-quality bases (Q ≤ 20) and standard Illumina (Illumina, Inc., CA, USA) paired-end sequencing adaptors on 3′ ends of reads were trimmed off using Trim Galore [13], and only those that were at least 20 bp in length after trimming were kept. High-quality reads were then mapped to the human reference genome hg19 with Burrows-Wheeler Aligner [14]. The alignments were then refined with tools of the GATK suite [15]. Variants for the samples were called according to the GATK Best Practice recommendations [16, 17]. This included recalibration. Non-synonymous exonic variants were subsequently filtered by the minor-allele frequency (MAF); as reported in 1000 Genomes 2011 release, ESP5400 and [18]. Variants with a MAF of above 0.05 were excluded. ANNOVAR [19] was used for annotations and prediction of functional consequences. In parallel, causal variants in this study were identified through the use of QIAGEN's Ingenuity® Variant Analysis™ software (www.qiagen.com/ingenuity) from QIAGEN Redwood City.
Sanger sequencing was used to confirm whether variants segregated with disease phenotypes. All coding regions of the LMX1B gene were amplified and sequenced directly using Sanger sequencing (primer sequences available on request), and segregation analysis was performed using DNA samples from affected and unaffected members of the family.
Homology modelling LMX1B
HHPred and Modeller were used to model the homeodomain of LMX1B (NP_002307) against the crystal structure of the NKX2.5 homeodomain bound to ANF-242-DNA (PDB code 3RKQ) [20]. A similar modelling strategy has been recently reported [7]. The homology model was visualized using PyMOL (http://www.pymol.org/).
Results
We report a large four generation family from the North East of England, in whom there was an autosomal dominant pattern of ESRD of unknown cause. The index case (II:3) shown in Figure 1A presented at the age of 15 years with dipstix proteinuria during a routine assessment (Table 1). Subsequently, she developed nephrotic-range proteinuria and progressive renal impairment leading to ESRD by the age of 28 years. A renal biopsy was performed which failed to provide a specific diagnosis. Her sibling (II:5) initially presented with proteinuria during her first pregnancy (aged 16 years), which was persistent but non-progressive. However, during her third pregnancy, aged 29, she developed hypertension and nephrotic-range proteinuria. A renal biopsy (performed at the age of 30 years) demonstrated mesangial proliferative glomerulonephritis with moderate granular deposits of C3 in the mesangium. Electron microscopy was unable to demonstrate glomerular basement membrane (GBM) changes due to poor tissue preservation. ESRD was reached at the age of 40 years. Proteinuria in another sibling (II:4) was identified at the age of 53 years during her workup as a potential living-related kidney donor for II:3. Her renal function was preserved (serum creatinine 79 µmol/L and eGFR >90). Given this identification of three siblings with proteinuria, a screen of additional family members was performed. The clinical data are presented in Table 1. Clinical examination of affected family members did not reveal any extra-renal manifestations that would suggest NPS. Two unaffected family members remained well with no proteinuria. II:6 successfully donated a kidney to his sibling II:3 and remains well with no evidence of proteinuria.
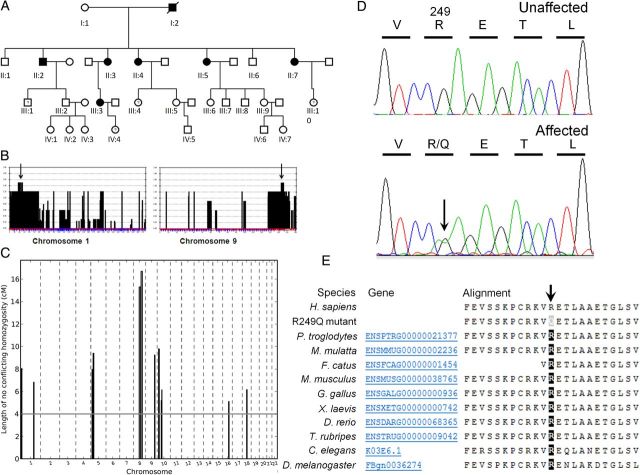
Fig. 1.
(A) Pedigree demonstrating an autosomal dominant pattern of disease. Circles represent females, and squares represent males. Shaded, affected. (B) Selected LOD plots from genome-wide linkage studies showing maximal LOD scores (1.505) on Chromosome 1 and 9 (arrowed). (C) IBD analysis using a CCH algorithm identified 12 loci in which there are >500 consecutive SNPs consistent with IBD. (D) Sequence chromatograms showing c.746G>A, p.R249Q heterozygous change in LMX1B. (E) Amino acid alignment confirming evolutionary conservation of the arginine residue (R) at position 249 across different species (reference sequence NM_002316.3). The R249Q missense mutation is predicted to be ‘probably damaging’ with a score of 1.000 (sensitivity: 0.00 and specificity: 1.00) using PolyPhen-2.
Table 1.
Clinical features of affected and unaffected family members
| Family ID | Age at presentation | Renal phenotype | ESRD | Comments |
|---|---|---|---|---|
| I:2 | Unknown | Haematuria/nephrotic-range proteinuria. Renal biopsy non-diagnostic | No | Died aged 45 years of myocardial infarction |
| II:1 | Unaffected | |||
| II:2 | Age 27 years with proteinuria | CKD stage 2 aged 60, biopsy non-diagnostic | No | |
| II:3 | Age 15 years with proteinuria | Progressive proteinuria and renal impairment | Age 28 years | Renal transplant X3 (graft loss secondary to chronic allograft nephropathy X2 and transplant pyelonephritis) |
| II:4 | Age 53 years with proteinuria | Minimal proteinuria, preserved renal function | No | |
| II:5 | Age 16 years with proteinuria | Progressive nephrotic-range proteinuria, exacerbated by pregnancy. Renal biopsy (aged 30 years) showed early mesangial proliferative glomerulonephritis |
Age 40 years | Renal transplant X2 (graft loss secondary to acute cellular rejection and chronic allograft nephropathy) |
| II:6 | Unaffected | Living-related kidney donor to II:3 | ||
| II:7 | Age 23 years with proteinuria | Progressive nephrotic-range proteinuria and renal impairment. Renal biopsy inconclusive |
Age 44 years | |
| III:3 | Age 36 years with proteinuria | CKD stage 2 aged 41 years. Subnephrotic proteinuria |
No |
Initial genetic studies were performed using a genome-wide linkage approach. Affymetrix GeneChip® Human Mapping 250K arrays were used to genotype eight family members, including six affected members (II:2, II:3, II:4, II:5, II:7 and III:3) and two unaffected members (II:1 and II:6). LOD scores were calculated using Genehunter. Putative disease loci were identified in Chromosome 1 (LOD 1.505) and Chromosome 9 (LOD 1.505; Figure 1B). In parallel, IBD analysis was performed using a CCH algorithm. This identified 12 loci in which there were >500 consecutive SNPs consistent with IBD, including the putative loci on Chromosome 1 and Chromosome 9 (Figure 1C). The Chromosome 1 region (Chr1: 184,113,318-184,118,523) was gene poor and contained no candidate genes, whereas the Chromosome 9 region (Chr9: 125,077,344-131,145,855) was 6 Mb in length and included the candidate gene LMX1B.
Concurrent with traditional genetic approaches, WES analysis was performed in three affected family members (II:3, II:5 and II:7). Variant analysis identified 908 potentially pathogenic variants. Variants were filtered for associated kidney disease/expression within kidney tissue. Potential disease variants were found in four candidate genes, namely CDH6 (on Chromosome 5), MYH9 (on Chromosome 22), LMX1B (on Chromosome 9) and CD9 (on Chromosome 12). The variant in LMX1B was the only variant that was compatible with an IBD hypothesis within the identified putative disease loci. Sanger sequencing of these genes was undertaken in all affected and three unaffected family members. As expected from linkage and IBD studies, the only variant which segregated completely with disease was the c.746G>A; p.R249Q LMX1B variant. Importantly, the LMX1B variant had been identified using WES in all three of the patients analysed, and Sanger sequencing confirmed its presence in all affected family members (Figure 1D). There were no additional variant alleles identified following sequencing of the whole coding region of LMX1B. The variant was absent from unaffected family members. The mutation c.746G>A; p.R249Q (Reference sequence NM_002316.3) is novel and is not present in databases including 1000 Genomes (http://www.1000genomes.org/), Exome Variant Server (http://evs.gs.washington.edu/EVS/) or the Exome Aggregation Consortium” (ExAC) data set with exome sequence data obtained from 63 352 individuals (http://exac.broadinstitute.org/). There are, however, two previous reports of pathogenic missense mutations at the identical amino acid position occurring in NPS patients, namely c.745C>G; p.R249G [reported as c.676C>G; p.R226G, using a shorter isoform of LMX1B (NM_001174147.1) as reference [21]] and c.746G>C; p.R249P [reported as c.677G>C; p.R226P again using the shorter isoform of LMX1B (NM_001174147.1) as reference [22]].
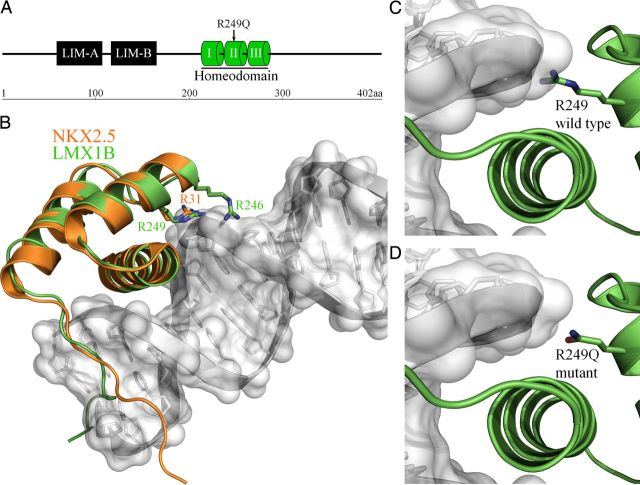
The arginine residue at position 249 in LMX1B is highly conserved in the DNA-binding motif of homeodomain-containing proteins [23] and across various species (Figure 1E). The LMX1B protein consists of two amino-terminal zinc-binding LIM domains and a homeodomain, which functions to regulate target gene transcription (Figure 2A). In silico modelling of LMX1B revealed that R249 is putatively located in close proximity to the backbone of the DNA strand (Figure 2B), where the positively charged side-chain is well placed to interact with the negatively charged DNA phosphoskeleton. Mutation of R249 to proline (reported as R226P [22]) was shown to impair DNA binding in vitro using an electrophoretic mobility shift assay and reduced transactivation by >90% in an in vitro luciferase reporter assay [24]. The novel R249Q mutation we have identified is also predicted to affect the homeodomain–DNA interaction (PolyPhen-2 score, 1.000).
Fig. 2.
(A) Schematic representation of the predicted domain structure of LMX1B including LIM domains and a DNA-binding homeodomain. The position of the R249Q mutation in helix II of the homeodomain is indicated. (B) The homeodomain of LMX1B (shown in green) was modelled against the crystal structure of the NKX2.5 homeodomain (shown in orange; PDB code 3RKQ). The NKX2.5 homeodomain structure was determined in complex with DNA (shown in grey) from the proximal promoter region of the atrial natriuretic factor (ANF) gene [20]. The position of the LMX1B residues mutated in NPS, R246 and R249 (homologous to R31 in NKX2.5) are highlighted. Modelling of wild-type (C) and R249Q mutant (D) LMX1B homeodomain. The shorter and uncharged side-chain of glutamine in the R249Q mutant is predicted to disrupt DNA binding.
Analysis of variants in transcriptional target genes was undertaken in the three affected family members who had undergone WES using QIAGEN's Ingenuity® Variant Analysis™ software. In this analysis, no additional pathogenic variants associated with proteinuria or podocytopathies were detected.
Discussion
This case and others recently described make it clear that mutations in LMX1B may lead to renal-limited disease, and we support the suggested terminology of LMX1B nephropathy [8]. Although important in developmental processes, including limb formation, it is now clear that LXM1B is also important for post-natal podocyte function. Where there are no extra-renal features of classical NPS, a renal biopsy is often performed, given the renal-limited phenotype of proteinuria and cystic kidney disease (CKD)/ESRD of unknown cause. In NPS, renal histological features may be subtle. Light microscopy may not reveal any glomerular abnormalities, and direct immunofluorescence is usually negative. Electron microscopy may reveal podocyte foot process effacement and irregular thickening of the GBM [25, 26]. Often the GBM is noted to have a moth-eaten appearance [8].
Recently, LMX1B mutations have been associated with a histological picture of FSGS, without extra-renal features [7]. Boyer et al. [7] describe a large family, where two index cases presented with nephrotic-range proteinuria and FSGS on renal biopsy. Electron microscopy performed on one of the biopsy samples, even after reprocessing and reanalysis, failed to show GBM changes typical of NPS. LMX1B mutations were also identified in two additional families who had an autosomal dominant pattern of nephrotic syndrome, whose histology was consistent with FSGS and minimal change disease, respectively [7]. Interestingly, detailed immunohistological analysis of a renal biopsy of a 6-year-old child with LMX1B nephropathy (where light microscopy showed normal glomeruli and no abnormalities of the GBM) revealed a cytoplasmic granular staining pattern for CD2AP, rather than a linear staining pattern seen in control samples [8]. In light of these reports, LMX1B, therefore, becomes a candidate gene for non-syndromic autosomal dominant FSGS [27] and autosomal-dominant forms of minimal change disease.
The pathogenesis of the renal phenotype has been investigated in murine studies. Lmx1b is expressed in murine glomeruli [28]. In Lmx1b-null mice, podocyte foot processes are reduced in number and are dysplastic [5]. In vitro, LMX1B binds to and regulates the transcription of CD2AP and NPHS2 [5], thus regulating key podocyte genes critical for podocyte function. Reduced Col4a3 and Col4a4 transcript expression has also been noted in Lmx1b-null mice, leading to reduced expression of their encoded type IV collagen proteins [6].
It has been suggested that variants in genes under the transcriptional control of LMX1B may account for the intra-familial variability of renal dysfunction seen in NPS [2]. In the family we report here, there were extremes of phenotypes, with proteinuria presenting in teenage years leading to ESRD before 30 years of age contrasting with minimal proteinuria and preserved renal function at the age of 53 years. Given that three affected members had undergone WES, we were able to examine for pathogenic variants in genes associated with podocytopathies, but we found no predicted disease-causing variants within these genes, including CD2AP, NHPS2, COL4A3, COL4A4 and COL4A5.
There are no specific treatments for LMX1B nephropathy. Treatment of proteinuria and hypertension with ACE inhibitors/angiotensin receptor blockers is recommended, but it is not known whether prophylactic treatment with these agents is helpful to prevent disease progression. Successful treatment of significant proteinuria in a 6-year-old child with NPS was achieved with combination of enalapril and losartan therapy [29], and that observation certainly argues the case for titration of drugs to achieve effective blocking of the renin–angiotensin–aldosterone system.
As seen in the family presented, proteinuria and renal impairment may be exacerbated during pregnancy. The clinical course is unpredictable, again as demonstrated here, with some patients progressing rapidly to ESRD and others having many years of asymptomatic proteinuria.
ESRD occurs in 5–10% of NPS patients [2, 3] at a mean age of 33 years [30]. Renal transplantation is usually successful in this condition, with no disease recurrence in the renal allograft, and these patients do not develop anti-GBM antibodies.
Missense mutations within the homeodomain are likely to disrupt DNA binding [24, 31], and there is evidence that patients with mutations located in the homeodomain have more proteinuria than those with LIM domain mutations [32]. Homology modelling of LMX1B using the crystal structure of the NKX2.5 homeodomain [20] supports a role for R249 in DNA interaction for subsequent transcriptional activity (Figure 2B). The Nε atom of the homologous arginine residue in NKX2.5 (R31) forms a salt bridge with a phosphate group of the DNA backbone to provide anchor for the DNA recognition helix [23]. The R249Q mutation reported here is predicted to impair the LMX1B–DNA interaction, likely through loss of the positively charged side-chain. Mutation of R249 to proline (reported as R226P) reduced DNA binding by the LMX1B homeodomain in vitro [22]. A similar effect would also be anticipated for the NPS-linked R249G mutation (reported as R226G) [22]. The functional importance of this region of the LMX1B homeodomain is further highlighted by homology modelling of the R246 residue (Figure 2B). Several NPS-linked mutations of R246 have been reported [7, 8], and LMX1B–R246Q exhibited partially impaired transcriptional activity in vitro [8].
Increasingly in patients in whom we suspect an inherited renal disease, genetic investigations may be more definitive and diagnostic than renal biopsies [33]. Historically, a genetic diagnosis in kidney disease has been limited to patients and families, where clinical or histological data display distinctive features that suggest one or a small number of candidate genes that can be sequenced individually. However, with the advent of massively parallel (next-generation) sequencing, a large number of genes can now be investigated in a single patient at a cost that can reasonably be borne by healthcare providers. The benefit of this approach is that it can allow a precise molecular diagnosis to be made even in patients where clinical data are lacking or non-specific, and can sometimes obviate the need for invasive tests such as a kidney biopsy.
However, because a large number of genes are tested simultaneously, the prior probability that any one of the genes or variants identified is responsible for disease is low. While variants known to be common in the healthy population can usually be excluded from consideration, it is not always possible to determine whether a rare or novel variant identified in a patient is actually pathogenic. This is because, even where they change the amino acid sequence of a protein, not all genetic variants change protein function or cause disease. Typically several lines of evidence (including linkage data, co-segregation analysis, comparison with mutation databases and functional and structural data about the protein) are needed to make a diagnosis securely. Over time, as sequence databases, incorporating data from larger numbers of patients and healthy individuals, become available to clinicians, the power of this approach is likely to increase.
We suggest that in all cases of ESRD of uncertain/unknown cause, a family history is sought. If an autosomal dominant pattern of CKD/ESRD is identified, then an underlying genetic disorder should be considered. There are a growing number of genetic causes of autosomal dominantly inherited renal failure, such that a candidate gene approach would be too costly and time consuming. We anticipate, however, that next-generation sequencing (NGS) approaches, and even whole genome sequencing approaches (WGS), will in the future allow a fast, cost effective and definitive genetic diagnosis to be made. Clearly, the advantages of an early diagnosis of an inherited renal disease allow therapies to be instituted, directed towards reducing blood pressure and proteinuria and perhaps the disease itself. Plans regarding ESRD, transplantation and risk of recurrent disease can be more easily discussed and relatives at risk of the disease can be screened, thereby avoiding invasive renal biopsies and multiple other investigations.
In conclusion, using genetic investigations in a large family affected by proteinuria, CKD and ESRD in whom renal biopsies had been uninformative, we have identified a novel mutation in LMX1B. The mutation segregated with the disease phenotype of LMX1B nephropathy. Genetic investigations of families with similar phenotypes should be a priority for nephrologists, and the advent of NGS and WGS will make this both practical and cost effective.
Conflict of interest statement
The results presented in this paper have not been published previously in whole or part.
Acknowledgements
We sincerely thank patients and family members for their support. This work was supported by the Northern Counties Kidney Research Fund. S.J.R. is supported by a MRC PhD studentship.
References
- 1.Turner JW. A hereditary arthrodysplasia associated with hereditary dystrophy of the nails. J Am Med Assoc. 1933;100:882. [Google Scholar]
- 2.Lemley KV. Kidney disease in nail-patella syndrome. Pediatr Nephrol. 2009;24:2345–2854. doi: 10.1007/s00467-008-0836-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sweeney E, Fryer A, Mountford R, et al. Nail patella syndrome: a review of the phenotype aided by developmental biology. J Med Genet. 2003;40:153–162. doi: 10.1136/jmg.40.3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreyer SD, Zhou G, Baldini A, et al. Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet. 1998;19:47–50. doi: 10.1038/ng0598-47. [DOI] [PubMed] [Google Scholar]
- 5.Miner JH, Morello R, Andrews KL, et al. Transcriptional induction of slit diaphragm genes by Lmx1b is required in podocyte differentiation. J Clin Invest. 2002;109:1065–1072. doi: 10.1172/JCI13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morello R, Zhou G, Dreyer SD, et al. Regulation of glomerular basement membrane collagen expression by LMX1B contributes to renal disease in nail patella syndrome. Nat Genet. 2001;27:205–208. doi: 10.1038/84853. [DOI] [PubMed] [Google Scholar]
- 7.Boyer O, Woerner S, Yang F, et al. LMX1B mutations cause hereditary FSGS without extra-renal involvement. J Am Soc Nephrol. 2013;24:1216–1222. doi: 10.1681/ASN.2013020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isojima T, Harita Y, Furuyama M, et al. LMX1B mutation with residual transcriptional activity as a cause of isolated glomerulopathy. Nephrol Dial Transplant. 2014;29:81–88. doi: 10.1093/ndt/gft359. [DOI] [PubMed] [Google Scholar]
- 9.Levine AP, Connor TM, Oygar DD, et al. doi: 10.1186/s12864-015-1360-4. Combinatorial Conflicting Homozygosity (CCH) analysis enables the rapid identification of shared genomic regions in the presence of multiple phenocopies. BMC Genomics 2014 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews S. FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 11.Li H. Seqtk. https://github.com/lh3/seqtk .
- 12.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krueger F. Trim Galore! http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics. Hoboken, USA: John Wiley & Sons, Inc.; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blazer S, Zimmer EZ, Blumenfeld Z, et al. Natural history of fetal simple renal cysts detected in early pregnancy. J Urol. 1999;162:812–814. doi: 10.1097/00005392-199909010-00066. 3 Pt 1. [DOI] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pradhan L, Genis C, Scone P, et al. Crystal structure of the human NKX2.5 homeodomain in complex with DNA target. Biochemistry. 2012;51:6312–6319. doi: 10.1021/bi300849c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunston JA, Hamlington JD, Zaveri J, et al. The human LMX1B gene: transcription unit, promoter, and pathogenic mutations. Genomics. 2004;84:565–576. doi: 10.1016/j.ygeno.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 22.McIntosh I, Dreyer SD, Clough MV, et al. Mutation analysis of LMX1B gene in nail-patella syndrome patients. Am J Hum Genet. 1998;63:1651–1658. doi: 10.1086/302165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi YI. Homeodomain revisited: a lesson from disease-causing mutations. Hum Genet. 2005;116:433–444. doi: 10.1007/s00439-004-1252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreyer SD, Morello R, German MS, et al. LMX1B transactivation and expression in nail-patella syndrome. Hum Mol Genet. 2000;9:1067–1074. doi: 10.1093/hmg/9.7.1067. [DOI] [PubMed] [Google Scholar]
- 25.Del Pozo E, Lapp H. Ultrastructure of the kidney in the nephropathy of the nail—patella syndrome. Am J Clin Pathol. 1970;54:845–851. doi: 10.1093/ajcp/54.6.845. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Bassat M, Cohen L, Rosenfeld J. The glomerular basement membrane in the nail-patella syndrome. Arch Pathol. 1971;92:350–355. [PubMed] [Google Scholar]
- 27.Kopp JB. An expanding universe of FSGS genes and phenotypes: LMX1B mutations cause familial autosomal dominant FSGS lacking extra-renal manifestations. J Am Soc Nephrol. 2013;24:1183–1185. doi: 10.1681/ASN.2013060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suleiman H, Heudobler D, Raschta AS, et al. The podocyte-specific inactivation of Lmx1b, Ldb1 and E2a yields new insight into a transcriptional network in podocytes. Dev Biol. 2007;304:701–712. doi: 10.1016/j.ydbio.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Proesmans W, Van Dyck M, Devriendt K. Nail-patella syndrome, infantile nephrotic syndrome: complete remission with antiproteinuric treatment. Nephrol Dial Transplant. 2009;24:1335–1338. doi: 10.1093/ndt/gfn725. [DOI] [PubMed] [Google Scholar]
- 30.Looij BJ, Jr, te Slaa RL, Hogewind BL, et al. Genetic counselling in hereditary osteo-onychodysplasia (HOOD, nail-patella syndrome) with nephropathy. J Med Genet. 1988;25:682–686. doi: 10.1136/jmg.25.10.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bongers EM, Gubler MC, Knoers NV. Nail-patella syndrome. Overview on clinical and molecular findings. Pediatr Nephrol. 2002;17:703–712. doi: 10.1007/s00467-002-0911-5. [DOI] [PubMed] [Google Scholar]
- 32.Bongers EM, Huysmans FT, Levtchenko E, et al. Genotype-phenotype studies in nail-patella syndrome show that LMX1B mutation location is involved in the risk of developing nephropathy. Eur J Hum Genet. 2005;13:935–946. doi: 10.1038/sj.ejhg.5201446. [DOI] [PubMed] [Google Scholar]
- 33.Adam J, Connor TM, Wood K, et al. Genetic testing can resolve diagnostic confusion in Alport syndrome. Clin Kidney J. 2014;7:197–200. doi: 10.1093/ckj/sft144. [DOI] [PMC free article] [PubMed] [Google Scholar]