Abstract
The rate of biological invasions is expected to increase as the effects of climate change on biological communities become widespread. Climate change enhances habitat disturbance which facilitates the establishment of invasive species, which in turn provides opportunities for hybridization and introgression. These effects influence local biodiversity that can be tracked through genetic and genomic approaches. Metabarcoding and metagenomic approaches provide a way of monitoring some types of communities under climate change for the appearance of invasives. Introgression and hybridization can be followed by the analysis of entire genomes so that rapidly changing areas of the genome are identified and instances of genetic pollution monitored. Genomic markers enable accurate tracking of invasive species’ geographic origin well beyond what was previously possible. New genomic tools are promoting fresh insights into classic questions about invading organisms under climate change, such as the role of genetic variation, local adaptation and climate pre-adaptation in successful invasions. These tools are providing managers with often more effective means to identify potential threats, improve surveillance and assess impacts on communities. We provide a framework for the application of genomic techniques within a management context and also indicate some important limitations in what can be achieved.
Keywords: adaptation, admixture, climate change, decision framework, genomics, hybridization, invasive species, management
Introduction
Biological invasions constitute a major environmental change driver, affecting conservation, agriculture and human health. Invasive alien species (IAS) impacts include, alterations of ecosystem features such as hydrology, fire regimes, food webs, soil nutrients and nutrient cycling (Hänel and Chown 1998; Asner and Vitousek 2005; Van Wilgen 2009; Veldtman et al. 2011); negative effects on populations (Blackburn et al. 2004; Butchart et al. 2010; Ziska et al. 2011); facilitation of further invasion and its associated impacts (O'Dowd et al. 2003); changes to evolutionary trajectories, such as by hybridization (McDonald et al. 2008); and evolutionary shifts in species responding to the new introductions (Strauss et al. 2006). Much attention has therefore been given to understanding the invasion process. A broadly accepted perspective on biological invasions now exists that distinguishes the stages of invasion and the mechanisms that are involved in each stage (Blackburn et al. 2011).
Although the field has grown rapidly (Mooney and Hobbs 2000; Sax et al. 2005; Richardson 2011; McGeoch et al. 2012; Spear et al. 2013), several significant challenges remain. Foremost among these is limited understanding of the mechanisms underlying each of the key transitions in the invasion process, reflected by periodic calls for additional work in these areas (e.g. Puth and Post 2005; Hulme et al. 2008). Most recently, attention has turned to the quantification and forecasting of IAS impacts. Several reviews have concluded that current knowledge and capability in this area are inadequate, rendering management responses either ineffective or inefficient (Pyšek et al. 2012; Hulme et al. 2013; Ricciardi et al. 2013; Simberloff et al. 2013).
Further challenges are presented by the simultaneous increase in the impacts of other environmental change drivers. Biological invasions (e.g. Lambdon et al. 2008; Chown et al. 2012; Richardson and Ricciardi 2013) are not the only forms of change increasing in frequency and extent. Climate change, as a consequence of anthropogenic greenhouse gas emissions, is continuing, with indications that the rate of change may be increasing owing to a rise in CO2 emissions (Rignot et al. 2011). Similarly, landscapes continue to change through human interventions (Barnosky et al. 2012). Exploitation is ongoing with apparently little abatement, resulting in substantial population declines, even within protected areas (Chown 2010; Craigie et al. 2010; Jackson 2010; Laurance et al. 2012). While some forms of pollution have declined notably, others have come to replace these (Sutherland et al. 2010, 2012).
Although the form of the interactions between biological invasions and these other environmental change drivers has yet to be fully generalized (Darling and Côté 2008; Bradley et al. 2010; Treasure and Chown 2014), the current view is that interactions are likely most often to be synergistic (Brook et al. 2008; Walther et al. 2009). Moreover, the simultaneous effects of environmental change drivers on indigenous species, and human responses to counter at least some of the effects of environmental change, such as through managed relocation (Schwartz et al. 2012), are likely to make management decisions about biological invasions much more difficult than in the past (Webber and Scott 2012).
The field of invasion science has recognized many of these challenges and research directions are clearly changing to meet them (McGeoch et al. 2012; Ricciardi et al. 2013; Simberloff et al. 2013). Much of this new work remains ecological in nature (at the organismal, population, species or ecosystem levels), including work that aims to guide management interventions to address the impacts of biological invasions (Richardson and Pyšek 2012; Blackburn et al. 2014). Nonetheless, from early on in the development of the field, several other approaches have been considered. Among these, the utility of genetics and the significance of evolutionary processes have long been recognized (e.g. Baker 1974). However, it is only recently that they have seen a resurgence of interest (Huey et al. 2000; Callaway and Maron 2006; Dormontt et al. 2011; Lawson Handley et al. 2011). Indeed, the significance of genetic approaches and an evolutionary perspective are now recognized as important not only for understanding the ability of species to progress along the stages of invasion (e.g. Pandit et al. 2011; Richardson and Pyšek 2012), but also for improving management interventions that might reduce the rates and impacts of invasions (Lawson Handley et al. 2011; Prentis and Pavasovic 2013). Examples include better genotypic matching of biocontrol agents and their hosts, and understanding the coevolutionary dynamics of invasive species and those with which they interact (Phillips and Shine 2006; McDonald et al. 2008; Gaskin et al. 2011). Nonetheless, additional scope exists for modern genetic approaches, and notably genomics, to contribute to the understanding and forecasting that is required to limit the rates and impacts of biological invasions, especially given the expectation of interactions with other global change drivers.
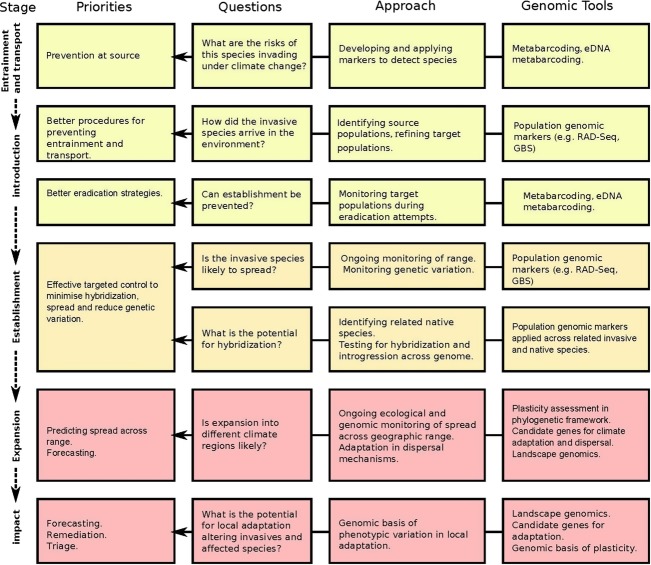
Here, we focus on the use of genomics to understand the changing form of invasion with climate change. We briefly review the contributions that genetic and evolutionary approaches have already made, discuss recent technological developments and then highlight further potential of this approach. We base our overview on the unified framework for biological invasions (Blackburn et al. 2011), recognizing that each of the stages in the invasion process can be mitigated by an accompanying management response (Fig.1). In addition, although various views exist about how invasive species should be defined (Pyšek et al. 2004; Valéry et al. 2008), here, to avoid uncertainty (McGeoch et al. 2012), we adopt the definition provided by Richardson et al. (2011). In consequence, we are concerned with the processes by which species are transported outside their natural range through direct or indirect anthropogenic intervention and become established, proliferate and interact with the biota in their new ranges.
Figure 1.
Genomic tools that can add to capacity for understanding risk and monitoring management actions at each stage of the invasion process. Tools can assist in the initial detection of an invasion, monitoring its spread after the initial detection and understanding the potential for invasions to expand and impact native species following evolutionary adaptation. Management priorities and questions relevant to each stage are also shown.
Genetic and evolutionary studies of invasive organisms
An increasing number of studies have documented evolutionary changes in invasive populations, typically over ecological timescales (Lee 2002; Prentis et al. 2008; Whitney and Gabler 2008). These studies have shown that evolutionary adaptation can be rapid in a broad range of invasive organisms. Examples include copepods adapting to different salinity levels (Lee and Petersen 2002), soapberry bugs adapting to new host plants (Carroll et al. 2001), and invasive weeds adapting through mimicry to evade eradication (Barrett 1983). Moreover, several of the documented examples of rapid evolutionary adaptation involve responses to climate change over a few decades. These include the evolution of Drosophila body size and inversion frequencies following invasion across continents with latitudinal climate variation (Huey et al. 2000), the evolution of rabbits along a heat gradient following invasion into Australia (Williams and Moore 1989), and changes in flowering time in Solidago goldenrods following introduction into Europe (Weber and Schmid 1998).
Recent work has continued to reinforce the notion that evolutionary genetic changes in invasive species can be rapid and often involve responses to climate. For instance, purple loosestrife, Lythrum salicaria, has invaded wetlands across an introduced range that covers a latitudinal gradient extending in eastern North America from 39°N to 48.5°N. Along this gradient, Colautti and Barrett (2013) showed an adaptive cline in flowering time, with earlier flowering at the northern invasion front, where it increases survival and fruit set, but with selection against earlier flowering in the south where it decreases vegetative growth. Climate adaptation involving changes in flowering traits has been recorded in several other invasive plant species (Moran and Alexander 2014). Evolution of traits in response to climate in invasive species has also been found for a range of animals. In Drosophila subobscura, introduced into Chile in the late 1970s, evolutionary divergence in thermal preferences among populations from different climates has weakened clines in chromosomal inversion (Castañeda et al. 2013). Likewise, physiological tolerances appear to have diverged in response to climate in the mite Halotydeus destructor (Hill et al. 2013). Several researchers have argued that evolution in invasive species can provide general answers to questions of whether species will be able to adapt to rapid climate change (Moran and Alexander 2014), including those about the speed of evolutionary change, the specific nature of the change that has occurred and factors that might limit evolutionary change.
Apart from their use to investigate adaptive changes, genetic tools have also been used to understand the dynamic nature of invasive processes. In particular, genetic markers have been widely used to detect invasive organisms, investigate patterns of historical movement in invasive species and examine the role of genetic variation in ensuring the success of invasions (Armstrong and Ball 2005; Lawson Handley et al. 2011; Blanchet 2012). Most past studies have focussed on markers thought to be selectively neutral, initially allozymes and then later microsatellites, AFLPs and other DNA markers. These markers can indicate the status of invasive species, origin of invasive populations and the levels of genetic variation in invading populations when compared to the populations of origin. Recent examples include tracking invasions of the winter annual Geranium carolinianum in China from multiple origins (Shirk et al. 2014), identifying a key translocation event in the invasion by a coregonid fish of a major Scandinavian watershed (Præbel et al. 2013), the use of mtDNA markers and microsatellites to trace the invasion history of eastern mosquitofish (Gambusia holbrooki) into Australia (Ayres et al. 2012) and determining multiple origins of invasive acacias in South Africa and South Australia (Le Roux et al. 2011; Millar et al. 2012). Genetic markers have also provided much information on the biology of invasive species, such as modes of reproduction (Weeks et al. 1995; Ali et al. 2014; Molins et al. 2014), movement patterns and rates (Bronnenhuber et al. 2011; Kirk et al. 2011; Berthouly-Salazar et al. 2013) and predator–prey relationships (Kasper et al. 2004; Blanchet 2012; Valdez-Moreno et al. 2012).
These types of studies are now being supplemented with a range of genomic and transcriptomic approaches that are providing answers to questions about the origins of invasive populations at a new level of resolution, providing new ways of monitoring for invasive organisms, and leading to an understanding of the nature of the changes that underlie adaptive shifts. We consider the promise provided by these studies within the context of detecting and managing biological invasions as climate change proceeds, and within a framework of the different steps involved in detecting and responding to an invader, understanding its source, and then tracking its impact on the surroundings (Fig.1).
Technology
Model and nonmodel organisms
The genomes of several invasive species have now been sequenced and assembled as well as annotated to differing extents. These include the tunicate Ciona intestinalis (Dehal et al. 2002), dengue mosquito vector Aedes aegypti (Nene et al. 2007), argentine ant Linepithema humile (Smith et al. 2011), fire ant Solenopsis invicta, (Wurm et al. 2011), diamondback moth Plutella xylostella (You et al. 2013), domestic cat Felis catus (Pontius et al. 2007) and mallard Anas platyrhynchos (Huang et al. 2013). Then, there are the genomes of model species that are also invasive, which include Drosophila species such as D. melanogaster and D. suzukii (Adams et al. 2000; Chiu et al. 2013), rats and mice (Waterston et al. 2002; Gibbs et al. 2004), and some weedy plants, such as the castor bean (Chan et al. 2010). Some of these species are useful for climatic studies, based on evidence for geographic variation in climate responses in species such as the drosophilids (Hoffmann et al. 2003) and C. intestinalis (Dybern 1965).
Many genomic approaches can be applied to population genetic studies of nonmodel species that lack a reference genome assembly. The use of sequence tagging and ‘Pool-Seq’ technologies, together with relatively inexpensive RNA-Seq, genotyping-by-sequencing (GBS) and other reduced-representation library methods, means that data on many thousands of single nucleotide polymorphisms (SNPs) can be collected at moderate cost on multiple samples from a population without a reference genome (Box 1) (e.g. Hohenlohe et al. 2010; Elshire et al. 2011; Bi et al. 2013; Neves et al. 2013; Yeaman et al. 2014). However, even draft reference genomes or genomes from related species can be useful when implementing these approaches to identify variants. The expansion of reference genomes across a wider array of species will facilitate the study of invasion genomics. Several impediments to genome construction still exist (Ellegren 2014), although new technologies are emerging to overcome them and the cost of short-read sequencing is declining. Nonetheless, many plants and animals have large genomes, and the common occurrence of higher ploidy, especially in invasive plants, adds significant difficulty and expense. Higher ploidy also complicates other approaches such as SNP identification and analyses (see below).
Box 1: Sequencing approaches for ecological genomics.
Whole-genome resequencing is cost-prohibitive and unnecessary in many cases. As an alternative, several methods of genome reduction make it possible for many individuals to be combined in one sequencing run while maintaining high coverage. This results in the reliable identification of SNPs and genotypes for each individual at a subset of regions throughout the genome.
Restriction-enzyme-based reduced-representation sequencing: Genomic DNA is digested with specific restriction enzymes, followed by ligation of adaptors, amplification and sequencing. Several methods employ this strategy, including genotyping-by-sequencing (GBS; Elshire et al. 2011) and RAD-Seq (Hohenlohe et al. 2010). Repetitive regions of genomes can be avoided using methylation-sensitive enzymes (Elshire et al. 2011). This approach is relatively inexpensive and easy to use in nonmodel organisms, but can suffer from biases such as those associated with the loss of restriction sites in some individuals (Gautier et al. 2013).
RNA-Seq: Sequencing mRNA from the genes that are expressed in the whole organism or specific sampled tissues at a particular point in time and development and under specific conditions (the ‘transcriptome’ of a whole organism or specific tissue; Wang et al. 2009). This method targets the expressed portion of the genome (protein-coding sequences and untranslated regions of genes). Gene expression and sequence information is simultaneously identified, as it is possible to compare counts of reads that map to genes and identify variants given sufficient expression of that gene in each individual.
Targeted sequence capture by hybridization: This approach uses oligoprobes to enrich specific regions of the genome for subsequent next-generation sequencing. The scale of the capture can range from a handful to several thousand targeted loci, making it appropriate for wide range of projects. However, prior genomic knowledge is essential for the development of the probes (e.g. through RAD-Seq or RNA-Seq projects). The development of noncommercial protocols and PCR-based probes (Peñalba et al. 2014) is making this approach more attractive for smaller-scale sequencing projects in nonmodel species. Several alternative methods are available that enrich target regions prior to sequencing (for review, see Mamanova et al. 2010).
Gene-space sequencing: Many genomes are replete with repetitive regions along with genes present in high copy number (e.g. chloroplast genes), and whole-genome sequencing without reducing their quantity can be a substantial waste of sequencing space. Consequently, some approaches seek to deplete these regions rather than enrich specific portions of the genome (e.g. Matvienko et al. 2013).
Glossary of genetic terms
| Genetic architecture: | the genetic underpinnings of phenotypic traits, including the number loci, their effect sizes and location in the genome. |
| Linkage disequilibrium: | the nonrandom association between alleles at two or more loci. |
| Quantitative trait locus (QTL): | a genomic area associated with variation in a quantitative trait in the progeny of a genetic cross. |
| Standing genetic variation: | existing variation in a population as opposed to variation that results from new mutations. |
| Wright fixation index (FST): | the proportion of the total genetic variability that occurs among populations. It is a measure of the level of population genetic differentiation. |
An important feature of invasive species for evolutionary studies is that the introduction and spread of the invader is often documented (Baker 1974), and in many cases, this includes historic collections. Genomic studies are benefiting from increased progress in obtaining information from specimens stored in herbariums, museum collections or recovered from natural repositories such as sediment layers in lake bottoms (Krehenwinkel and Tautz 2013; Martin et al. 2014). Such studies are particularly applicable to invasive organisms that are often represented well in collections with longitudinal sampling. These specimen records often cover periods over which populations have been exposed to relatively rapid climate change (such as between 1970 and 2000) (Hansen et al. 2013). More formal approaches are also now being applied to secure the preservation of material across time, such as the resurrection initiative for plants (Franks et al. 2008).
In future, methods can be employed that maximize the genomic information obtained from such limited material, rather than focusing on a few anonymous markers. Whole-genome resequencing has been used successfully for historic samples (Rowe et al. 2011), although contamination by nontarget DNA can make this expensive proposition even more cost-prohibitive. Sequence capture, which utilizes probes to enrich target sequences (e.g. Bi et al. 2013) (Box 1), offers a compromise between minimizing sequencing costs and maximizing sequence information from target regions of the study organism. Pooling individual samples together prior to either whole-genome (e.g. Futschik and Schlotterer 2010) or reduced-representation (e.g. Vandepitte et al. 2012) sequencing is another approach that can greatly reduce costs, while still providing useful information about the genomic extent and location of variation and divergence. Genome-wide analysis of historic DNA in invasive species will provide exciting opportunities to assess the temporal dynamics of evolutionary change during colonization and spread, and over periods of rapid climate change. It can also provide evidence of past changes in population size across time through coalescence methods (do Amaral et al. 2013) and indicate parts of the genome that have been affected by selection and population processes (Excoffier et al. 2009). Such data will improve inference of invasion history and aid in the identification of the genetic basis of recent adaptation.
Top-down and bottom-up approaches
Ecological and landscape genomics are approaches that incorporate information about the phenotype, genotype and the local environment to make inferences about the loci involved in local adaptation. ‘Bottom-up’ approaches use the genomic signatures of selection to identify loci likely important for adaptation and do not rely on phenotypic information (Wright and Gaut 2005). Several different methods are used, such as tests examining the site frequency spectrum or the extent of linkage disequilibrium, which can indicate the location of a recent selective sweep (Smith and Haigh 2007; Thornton et al. 2007). The popular FST outlier method identifies regions of the genome under divergent selection by determining which sites have strong differentiation among populations relative to the entire genome or a set of putatively neutrally evolving loci (Lewontin and Krakauer 1973; Beaumont and Nichols 1996; Foll and Gaggiotti 2008; Steane et al. 2014). Another approach involves identifying associations between alleles and the local environment, while controlling for population structure, to make inferences about local adaptation (e.g. Coop et al. 2010; Günther and Coop 2013). Many of these tests of selection could be applied to invasive species to identify the genes associated with adaptation during invasion and in responses to climate change. These methods are appealing because genes under selection during invasion can be identified without a priori identification of specific functional traits and do not require common garden experiments. However, the loci identified may only be in linkage disequilibrium with the functional site(s). Moreover, population structure can hinder the ability of these tests to correctly identify adaptive loci, and certain demographic scenarios, particularly those involving nonequilibrium conditions, have been shown to generate a large number of false positives (Lotterhos and Whitlock 2014). A critical assessment of how complicated invasion histories, including founder events and genetic admixture, might impact the ability of these methods to detect loci under very recent divergent selection has not yet been conducted, but these processes could represent an impediment to the successful implementation of these tests in some invasive species.
‘Top-down’ methods offer a complementary approach for identifying the genetic basis of adaptation (Wright and Gaut 2005; Barrett and Hoekstra 2011). Here, the genetic basis of specific traits under selection during invasion can be dissected by looking for the cosegregation of these traits and genetic markers. Quantitative trait loci (QTL) mapping uses pedigreed populations descended from known parents. Linkage disequilibrium between genetic markers and causal loci enables the identification of genomic regions segregating with the phenotypes of interest. This method limits the identification of loci to those present in the initial cross, and the genomic resolution is low due to high linkage disequilibrium in the mapping population. Association mapping studies use unpedigreed mapping to look for associations between traits and markers, either using a candidate gene approach or genome-wide markers (GWAS studies) (Box 1). Such studies can be prone to false positives through spurious associations brought about by population structure, although there are statistical means to control for these effects (Tian et al. 2008). The approaches offer greater resolution than QTL studies due to the lower linkage disequilibrium found in natural populations relative to pedigreed mapping populations.
Top-down and bottom-up approaches each have their own advantages and disadvantages (Wright and Gaut 2005; Barrett and Hoekstra 2011; Le Corre and Kremer 2012; Sork et al. 2013), suggesting that tackling the problem from both ends will often yield the most useful results. However, there are biases common to all these methods that may obscure the genetic architecture of adaptation during invasion, or to climate change signals (Moran and Alexander 2014). For example, detecting small effect loci underlying polygenic traits has been a major challenge regardless of the method applied (Pritchard et al. 2010; Le Corre and Kremer 2012). Well-powered QTL and GWAS studies appear to be an effective solution to this problem (Bloom et al. 2013), and new analytical methods that attempt to integrate information from GWAS with allele frequency data from many populations are providing a powerful way to identify the signal of local adaptation in polygenic traits (Turchin et al. 2012; Berg and Coop 2014).
Although there have been few studies that have applied these approaches to identify genes important for adaptation during invasion, the growth of genomic data in nonmodel organisms suggest that these methods will soon be commonplace for invasive species. As strong selection will be required to see evolutionary changes over short time scales, theory suggests that large-effect loci (Yeaman and Whitlock 2011) or perhaps clusters of adaptive loci may be more likely to underlie a rapid response to selection. However, genetic changes from standing variation as opposed to new mutations are likely to be essential for adaptation during invasion, due to the waiting time required for new mutations (Barrett and Schluter 2008; Prentis et al. 2008). This suggests that polygenic adaptation from standing variation present in the original introduction is likely, so adaptation through multiple loci of small effect may also play an important role in the invasion. Both empirical and theoretical studies are needed to determine which features of the genetic architecture of rapid adaptation, if any, might distinguish it from longer-term adaptive evolution and if differences in genetic architecture could impact the propensity of organisms to become invasive and/or rapidly adapt to continuing climate change.
The identification of candidate ‘invasion’ genes using these top-down and bottom-up approaches is only the first step. Comparisons across species will be essential for determining whether there are similar functional groups, genes or genetic pathways that frequently evolve during invasion and in responses to changing environments further precipitated by local climate change. This might be expected, for example, if invading species commonly evolve along similar fitness trade-offs in response to changes in the biotic and abiotic environment in the introduced range (Blossey and Notzold 1995; Bossdorf et al. 2005; He et al. 2010). The context dependence and sometimes idiosyncratic nature of invasion and adaptation may, however, make such generalities unlikely (though see also Richardson and Pyšek 2006, 2012). Experimental studies, where possible, will be important to assess fitness effects of these candidate loci in nature, while controlling for genetic background (Barrett and Hoekstra 2011). The development of more model invasive species amenable to reverse genetics will be key for dissecting the functional role of candidate loci and will be a step forward for invasion genomics (Stewart et al. 2009).
The initial stages – entrainment risk and transport
The introduction of alien species is either intentional, usually for economic benefit of some form, or unintentional, owing to the fact that species move around in all kinds of ways and include human vectors and human-mediated pathways in their dispersal portfolio (Hulme et al. 2008). For intentional introductions, understanding entrainment risk amounts to investigating the socio-political motivations for introducing new species. These are many and diverse and, through time, have ranged from agriculture and horticulture, which are ongoing (Dehnen-Schmutz 2011), to the activities of acclimatization societies (Cassey et al. 2004). In the nineteenth century, acclimatization societies were established to ‘improve’ what were considered impoverished biotas in newly colonized regions (e.g. North America, New Zealand and Australia) through the importation of plants, birds and mammals, typically from Europe, but also from other regions. Investigations of survival during transport belong in the realm either of plant propagation and animal husbandry, or maintenance of biocontrol agents (e.g. Teshler et al. 2004). While these areas may have much to inform the investigation of biological invasions and their evolutionary dynamics (e.g. Vorsino et al. 2012), we will not consider them here. Rather, our emphasis is on unintentional introductions.
Much of the understanding required to reduce the rates of unintentional introductions is concerned with improving knowledge of the pathways and vectors of invasion (Hulme 2009). Preventing an invasion is the most cost-effective way of dealing with it (Simberloff et al. 2013). These pathways and vectors are now becoming much better understood across a range of activities in different areas, and for different organisms (e.g. Drake and Lodge 2004; Tatem and Hay 2007; Huiskes et al. 2014). Changing human traffic patterns in tandem with changing climates (including their variability) is likely to change substantially the sites of entrainment risk for any given location (Tatem 2009). In consequence, although something of an invasion cycle exists, where disturbed entrainment areas tend to harbour similar suites of species (Lee and Chown 2009), forecasting which species and areas to prioritize for surveillance may be difficult because of rapidly changing environments and species distributions. Ongoing invasion of source areas (see e.g. Roy et al. 2011) may likewise mean changing risk profiles. Genomic approaches offer a powerful tool to help manage these risks.
At their most straightforward, DNA barcoding approaches can identify alien species at a given location (Darling and Blum 2007). They are now proving useful for a range of taxa. Mostly, the aim has been to characterize the diversity of an assemblage and identify alien species among its members in a given recipient area, or to verify whether given populations are from species thought to be nonindigenous (e.g. Scheffer et al. 2006; Smith and Fisher 2009; Porco et al. 2012, 2013; Fernández-Álvarez and Machordom 2013; Zhang et al. 2013a). Such approaches nonetheless are subject to the same kinds of problems as those facing DNA barcoding generally (see review by Taylor and Harris 2012). These include substantial error rates in some taxonomic groups (e.g. Hickerson et al. 2006; Meier et al. 2006; Zhang et al. 2013b) and lack of clarity about the questions being posed or rigorous independent assessment of the hypotheses being tested (Collins and Cruickshank 2013).
As DNA barcoding approaches were developed, it has now become possible to screen simultaneously samples of many species. Such metabarcoding provides relatively reliable estimates of community composition and turnover (Yoccoz et al. 2012; Yu et al. 2012; Ji et al. 2013). The approach has also been broadened to include what may be termed environmental metabarcoding or eDNA metabarcoding (Taberlet et al. 2012a; Bohmann et al. 2014). Here, rather than the organisms being sequenced directly, DNA is typically extracted from soil or water (Collins et al. 2013; Porco et al. 2013; Piaggio et al. 2014), but may also come from other sources such as the gut contents of flies (Calvignac-Spencer et al. 2013). The terms metagenomics and ecogenomics have also been applied to this approach, although typically metagenomics involves analysis of functional characteristics and assembly of whole genomes for microbes (Taberlet et al. 2012a). Considerable success has been had in detecting invasive alien species using this approach, with the method surpassing traditional survey approaches both in terms of sampling effort and sensitivity. Notable demonstrations thereof are for carp in waterways linking to the Laurentian Great Lakes (Jerde et al. 2011), the American bullfrog in France (Ficetola et al. 2008; Dejean et al. 2012) and several other aquatic species (Bohmann et al. 2014; Rees et al. 2014). Much interest exists in the application of eDNA barcoding to surveillance of ballast water in ships, a substantial source of aquatic invasions, with novel tools being developed to enable rapid screening by nonspecialists (Mahon et al. 2012).
A further area where eDNA metabarcoding is proving useful is in understanding the extent, ecology and ecosystem functioning consequences of microbial invasions (Litchman 2010; Shade et al. 2012). Microbial invasions are significant across the globe with substantial impacts on a range of ecosystems and on human and animal health (Van der Putten et al. 2007; Jones et al. 2008; Vellinga et al. 2009; Keesing et al. 2010; Cowan et al. 2011; Carroll et al. 2014). Because metagenomics has deep roots in microbiology, the approach enables sophisticated portraits to be painted of the identity of the players involved in a given system, the extent to which invasions are significant and the functional roles that key members of the community play (Litchman 2010; Cowan et al. 2011; Shade et al. 2012; Adriaennsens and Cowan 2014; Ramirez et al. 2014).
Metabarcoding and eDNA metabarcoding approaches are not without their problems, including challenges to do both with resolving taxa (see above and Taberlet et al. 2012b; Brodin et al. 2013; Pyšek et al. 2013), and with the skills, methods and databases required to ensure that these approaches deliver their full potential (Yoccoz 2012). Nonetheless, the field is developing rapidly (e.g. Liu et al. 2013; Zhang et al. 2013b; Bohmann et al. 2014), and management risks posed by differentiating among increasing numbers of species introductions and range shifts precipitated by changing climates (Walther et al. 2009), could readily be reduced using these approaches.
Although barcoding and metabarcoding approaches are increasingly being applied to the detection of invasives and have been used in several successful detections (e.g. Dejean et al. 2012; Collins et al. 2013; Egan et al. 2013; Takahara et al. 2013), they could also be used to reduce risks of introduction by understanding the composition of species in source hot spots. While doing so might at first seem impracticable, reductions in costs of the technical methods (Lemmon et al. 2012; Rocha et al. 2013), and routine surveillance of areas for incoming species (e.g. Bourlat et al. 2013), which are often the same sources for outgoing ones (Lee and Chown 2009), mean that the data may become increasingly available for species detection. The main barriers to overcome are making sure that sequence information, biological records and distributional data are available in linked and readily useable forms [although their downstream use also faces a variety of challenges (Amano and Sutherland 2013)], achieving interoperability among the various approaches and information databases, and estimating the extent of errors of omission and commission for various taxa (Collins and Cruickshank 2013). While these barriers are high, they are being reduced by increasing discussions among major data providers and ongoing work to improve interoperability. Although this provider approach is important, good practice by investigators (such as coherence in data submitted to major providers) is probably the most significant way to reduce barriers. DNA barcoding approaches may be especially helpful where changing climates encourage change in human agricultural and business practices, creating rapidly changing invasion scenarios.
Identifying source populations
Although genetic markers have been used for some time and are now used almost routinely for identifying source populations of species, genomic markers promise a much higher level of resolution and they are being increasingly applied. A recent example is provided by the dengue vector mosquito, Aedes aegypti, whose source populations have traditionally been identified using mtDNA markers coupled with microsatellites (Endersby et al. 2011; Brown et al. 2014). These provide some indication of population sources and are currently used in countries such as Australia to locate source populations. However, a much higher level of resolution for identifying source populations can be obtained from SNP nuclear markers that can now be scored in the thousands; this was demonstrated in the extent to which geographic samples of Ae. aegypti could be completely resolved based on 18 000 SNP markers compared to 10–15 other markers (Rasic et al. 2014) (Fig.2).
Figure 2.

Comparison of population structure of Aedes aegypti mosquitoes as determined from microsatellite markers versus SNP markers. Based on a discriminant analysis, the SNP markers provide a far higher level of resolution of populations including the invaded (non-Asian) range of this species (from Rasic et al. 2014).
SNP markers are now being developed for other invasive species including invasive plants (De Kort et al. 2014) and mammals (White et al. 2013) as well as insects. These markers promise to revolutionize risk assessment and quarantine control because they can reveal both contemporary and historical population processes. The potential of this approach in detecting and understanding past patterns based on whole-genome sequencing is evident from recent human studies on invasions and contractions of different cultural groups that provide new insights into patterns of relatedness and the spread of cultural practices (Sikora et al. 2014).
Understanding source populations and spread dynamics may benefit from using nuclear and extranuclear genomes. For instance, in the Russian wheat aphid, Zhang et al. (2014) tested patterns of invasion by characterizing more than 500 clones for genetic markers in nuclear DNA, mtDNA and endosymbionts. They were able to show that Turkey and Syria were the most likely sources of invasion to Kenya and South Africa respectively, as well as establishing patterns of invasion into the New World. Having the different sources of genomic information was essential to this study as the recent introduction limited the genetic resolution of the aphid markers used. Combining nuclear DNA and organellar markers has traditionally improved the understanding of invasive processes, and it is becoming clear that including vertically transmitted endosymbiont or parasitic organisms can provide new insight into the origins of invasive species as well as the diseases they carry (Gupta et al. 2014; Mobegi et al. 2014). The addition of mtDNA provides information on introgression patterns between diverged populations or subspecies because mtDNA genomes track maternal lineages (McCormick et al. 2014).
In the future, genomic information will likely prove useful in identifying source risk and predicted outcomes of invasions when they are detected. For instance, in ragweed, it appears, based on microsatellite and chloroplast markers, that invasive populations have developed in the native range of this species associated with human habitation, and these populations are then likely to form the basis of invasive populations into other areas (Martin et al. 2014). Predictions are that invasions will grow in number and be increasingly difficult to distinguish from range shifts (Walther et al. 2009). Genomic techniques that can facilitate rapid identification of source populations will prove especially useful for addressing this problem (for an application in identifying the basis of range expansion, see Krehenwinkel and Tautz 2013).
Colonization and establishment: genetic diversity
Multiple founders can facilitate population establishment in two ways. First, propagule pressure mitigates the impact of demographic stochasticity; second, increased genetic and phenotypic variation improves the chances of successful colonization (Colautti et al. 2006; Simberloff 2009; Rius and Darling 2014). A recent meta-analysis, which controlled for the effects of population size, found strong evidence that higher levels of genotypic and phenotypic diversity in founder groups increased establishment success (Forsman 2014). Genetic variation can improve the likelihood of establishment for many reasons (Zhu et al. 2000; Willi et al. 2006; Gamfeldt and Källström 2007; Dlugosch and Parker 2008; Prentis et al. 2008; Thompson et al. 2012; Berthouly-Salazar et al. 2013), and evidence is mounting that a large proportion of invasive species have experienced multiple introductions (e.g. Bossdorf et al. 2005; Roman and Darling 2007; Thompson et al. 2012; Rius and Darling 2014).
The role of genetic variation in successful invasions has proven contentious though, with authors often taking opposing views depending on the nature of the system that they study (Rius and Darling 2014). Indeed, many species have spread successfully with low genetic diversity (Myburgh et al. 2007; Darling et al. 2008a; Richards et al. 2012). The success of invaders with low genetic variability may depend on the nature of the environment into which they invade; weedy species moving into agricultural landscapes may require less genetic variability because they enter a relatively homogeneous landscape, whereas for species invading natural environments, a high level of variability may be required (Moran and Alexander 2014).
Genomic approaches provide a means to help resolve the apparent paradox. For example, success with limited genetic diversity may be due to the expression of phenotypic plasticity, aided by epigenetic responses to the environment (Richards et al. 2012) that can be identified with genomic approaches. Epigenetic mechanisms may be particularly important in these situations because they generate heritable changes in response to environmental cues, although their ecological significance remains unclear (Bossdorf et al. 2008). Alternatively, functional traits may remain variable even when genetic variation is low, as the loss of rare alleles will not greatly affect the quantitative trait distribution, and nonadditive genetic variance may either aid in fitness directly, or be converted to additive variance due to allele frequency shifts (Dlugosch and Parker 2008; Dawson et al. 2012; Le Roux et al. 2013). Genomic tools provide a way of identifying variation in functional traits (see next section). They also provide new means to understand the nature of introductions of invasive species. As mentioned in the previous section, genomic markers provide a much higher level of resolution when identifying source populations, and they can therefore clearly indicate the number of introductions from different environmental regions involved in invasions.
Colonization and establishment: adaptive loci
Identifying risk factors associated with establishment would be assisted by more information on the genetic basis of adaptive changes in colonizing species. If these changes were known and repeatable, it might help predict when colonizations occur. One of the best examples is the evolutionary changes involved in the repeated independent colonization of freshwater environments by sticklebacks (summary in Bell and Aguirre 2013). These colonization events have been associated with repeatable changes in lateral plate morphology across populations involving the ectodysplasin (EDA) locus. Many loci in different genomic regions also appear to be involved, but nevertheless there appear to be changes across the same set of genomic regions in different, isolated freshwater populations of sticklebacks following invasion (Jones et al. 2012). The stickleback situation may be unusual because there has been a repeated process of invasions that have been occurring over 10 million years (Bell and Aguirre 2013), but more cases need to be examined. Because invasive species often occupy similar habitats in the introduced range to those in the native range (see Guisan et al. 2014 for a recent overview and perspective), there is an opportunity to determine whether the same genomic regions contribute to adaptations in the native and introduced ranges across many invasive species.
While there is abundant evidence for changes in neutral markers following invasion, characterization of adaptive molecular differences, such as those established in stickleback populations, remains relatively scarce. Transcriptome comparisons of plants in common garden experiments suggest that invasive populations of ragweed show altered patterns of gene expression that are distinct from most populations from the native range, and genes showing altered patterns of expression appear to be linked to stress and biotic responses (Hodgins et al. 2013). A similar pattern has since been identified in Canada thistle (Guggisberg et al. 2013), but whether changes to expression during invasion primarily involve certain functional groups, as indicated by these studies, will require more examples across a broader array of organisms.
One of the challenges in finding the genetic basis of adaptive change is that causal connections between genetic changes and traits can be difficult to identify. Often researchers are faced with the problem of locating small but crucial changes within a mass of genomic data. However, the implementation of population genomic analysis of SNP variation across the genome has enabled the identification of putatively selected loci (or those linked to the causal loci). This approach has already proven useful in several cases. Pyrenean rocket (Sisymbrium austriacum subsp. chrysanthum) is a small colonizing herb of rocky soils that has recently invaded Belgium and the Netherlands, and Vandepitte et al. (2014) used a RAD-Seq approach to identify outlier loci in comparisons between invaded and native populations. Multiple native and invaded populations were investigated including those represented by herbarium specimens covering ca. 100 years in the invasive range. Several outlier loci that could underlie adaptive differences were discovered, and the sampling enabled a comparison of outliers across space as well as time. Some of the SNPs were located in genes known to affect flowering time in the closely related Arabidopsis thaliana. These findings point to likely adaptive shifts rather than changes associated with demographic processes. This study demonstrates the utility of phenotype–genotype approaches for identifying candidate adaptive loci in invasive species coupled with functional information from model species. As the genomes of invasive species become better defined and genetic tools are developed, it should be increasingly possible to also pursue bottom-up approaches in a range of invasive species particularly because these can often be relatively easily grown under controlled conditions.
Colonization and establishment: hybridization and polyploidy
Hybridization can act as a stimulus to invasion (Ellstrand and Schierenbeck 2000; Schierenbeck and Ellstrand 2009) and occurs on a continuum from the combining of largely reproductively isolated genomes to the mixing of distinct populations within a species. Hybridization offers many potential advantages during invasion. It may increase adaptive potential through the injection of genetic variation and the formation of beneficial gene combinations (Anderson 1948). Hybridization is known to result in transgressive segregation, where phenotypic variation in the hybrids exceeds that of the parents (Rieseberg et al. 2003). In sunflowers and Louisiana irises, there are particularly compelling cases for hybridization facilitating the colonization of novel habitats (Rieseberg et al. 2003, 2007; Arnold et al. 2012).
Over the short term, heterosis (improved performance of hybrid offspring) could be important for overcoming demographic stochasticity associated with initial establishment (Drake 2006; Keller and Taylor 2010; Rius and Darling 2014). Although heterosis is transitory in most cases (Hochholdinger and Hoecker 2007), fixed heterozygosity can be maintained across generations in allopolyploids (assuming disomic inheritance occurs) and asexually reproducing species (Ellstrand and Schierenbeck 2000; Rieseberg et al. 2007). In addition, if dominance is the primary cause of heterosis, the advantage of hybridization may continue in later generations and facilitate the purging of genetic load (Burke and Arnold 2001). Alternative mildly deleterious alleles can become fixed within diverging populations due to the effects of genetic drift. Hybridization between these lineages could produce descendants with an intrinsic fitness advantage over their parents that is maintained if the combined effects of recombination and natural selection erode the frequency of these deleterious alleles (Ellstrand and Schierenbeck 2000; Burke and Arnold 2001).
Genomic tools can help in identifying the consequences of these processes because they can be followed across different parts of the genome. This applies particularly to plants where invasive taxa often arise as hybrids (Moody and Les 2007; Ainouche et al. 2009; Schierenbeck and Ellstrand 2009). One famous case is of the damaging riparian invasive Tamarix species in the USA. The main invasive lineage arose from hybridization between two introduced species, T. chinensis and T. ramosissima, which have largely separate ranges with some overlap in Asia, but do not appear to hybridize there (Gaskin and Schaal 2002). Genome-wide neutral markers revealed that hybrids dominate the invasive range, and the level of introgression is strongly correlated with latitude (Gaskin and Kazmer 2009). A corresponding latitudinal cline in cold hardiness suggests that hybridization may have contributed to the rapid adaptation of Tamarix to climatic extremes in North America (Friedman et al. 2008). In reed canarygrass, multiple introductions into North America of genetic material from different European regions have mitigated the impact of genetic bottlenecks (Lavergne and Molofsky 2007). This species has higher genetic diversity and heritable phenotypic variation in its invasive range relative to its native range. Comparisons of neutral differentiation (FST) to differentiation in quantitative traits among populations (QST) provide evidence for rapid selection of genotypes with greater vegetative colonization ability and phenotypic plasticity in the introduced range.
Hybridization is expected to be important from the perspective of invasiveness under climate change for two reasons. First, evidence is growing that climate change is increasing rates of hybridization, as species that were previously geographically isolated come into contact with each other (Garroway et al. 2010; Muhlfeld et al. 2014). Second, evidence from genomic studies is revealing that genes associated with climate change adaptation can be of hybrid origin (Becker et al. 2013; De La Torre et al. 2014). Thus, when hybrids are formed, they may contribute to rapid adaptation under climate change. Although most information on hybridization comes from plants, hybridization is also likely to play an important role in the adaptation of animal lineages to climate change. Genomic comparisons have indicated introgression of invasive genes across lineage boundaries affected by climate in birds (Taylor et al. 2014) and stick insects (Nosil et al. 2012). Introgressed genotypes can be favoured under changing climatic conditions as evident in hybridization events in swallowtail butterflies (Scriber 2011) and in Anopheles mosquitoes (Besansky et al. 2003). Genome scans across multiple related species along climate gradients are likely to provide many other examples of candidate genomic regions associated with climate adaptation and exchanged through hybridization and introgression.
Polyploidy has occurred frequently in the evolutionary history of angiosperms, often associated with hybridization events (Wood et al. 2009). There are several examples of recently derived invasive polyploids (Ainouche et al. 2009; Schierenbeck and Ellstrand 2009; te Beest et al. 2012), and polyploidy is over-represented among alien plant taxa (e.g. Pandit et al. 2011). Polyploidy is associated with several advantages (reviewed in Otto and Whitton 2000; Comai 2005; Otto 2007; te Beest et al. 2012) that could aid in invasion, including pre-adapting species to the environmental conditions in the new range (e.g. Henery et al. 2010), masking deleterious alleles (Otto and Whitton 2000; Otto 2007), restoring fertility through chromosome doubling (e.g. Ainouche et al. 2009) or through changes to the reproductive system to that contribute to colonization ability (e.g. Robertson et al. 2011). In addition, allopolyploids benefit from fixed heterozygosity due to the combining of divergent parental genomes. This feature may have contributed to the evolutionary success of polyploids over diploids in the Arctic enabling them to survive dramatic shifts in climate (Brochmann et al. 2004).
One of the best examples of polyploidy contributing to invasion success is tetraploid spotted knapweed, Centaurea stoebe. Although both diploids and tetraploids were introduced into the USA from Europe, invasive populations are dominated by tetraploids (Treier et al. 2009). Tetraploids are pre-adapted to a wider range of climates compared to diploids, and following introduction, they may have further adapted to their introduced environment (Treier et al. 2009; Henery et al. 2010). A classic example of hybridization and polyploidy coinciding with invasion is Spartina anglica. This species is a recently formed allopolyploid (12×) that is highly invasive compared to its parental species, is able to colonize a broader range of habitats and exhibits substantial phenotypic plasticity (Thompson 1991; Ainouche et al. 2009), despite a strong genetic bottleneck that occurred during its formation (Baumel et al. 2001). Neo-allopolyploids often undergo extensive alterations to their genome and transcriptome, and genomic studies of S. anglica are dissecting the genetic, epigenetic and expression changes following hybridization and genome duplication (Salmon et al. 2005; Ainouche et al. 2009; Chelaifa et al. 2010). However, the mechanism by which S. anglica has become invasive remains elusive, and it is not known whether the success of this species reflects fixed heterosis and restored fertility of the hybrid following genome duplication, or phenotypic novelty due to epigenetic and transcriptome remodelling.
Whole-genome and reduced-representation genomic approaches are invaluable for detecting hybridization, and more powerful than any other method to quantify the extent of introgression of genomic material and (potentially) functional genes involved in environmental tolerance and invasiveness. They have been used to estimate rates and genomic extent of hybridization (Larson et al. 2013; Parchman et al. 2013) and identify the potential for genetic swamping, or hybrid incompatibilities that may be deleterious to native species (Fitzpatrick et al. 2010). Applying genetic and genomic methods to recent polyploids poses extra challenges due to the presence of multiple genome and gene copies; this can complicate short-read alignment and de novo assembly of genomes, but can also be useful in establishing patterns of relatedness among species and populations (Griffin et al. 2011). Reference genomes are especially valuable tools in these cases and are helping to reveal within-species diversity and hybrid origins of polyploids (e.g. Evans et al. 2014; Marcussen et al. 2014), and reduced-representation library methods can now be applied even without a reference genome (Lu et al. 2013). Genomic studies of several plant groups are pointing to porous species barriers across thousands of years where ongoing hybridization and gene flow between species is present despite species maintaining their morphological integrity (e.g. De La Torre et al. 2014; Griffin and Hoffmann 2014). Such a dynamic process of hybridization and polyploidy may also occur among different chromosomal races within a species as in the case of buffel grass, which is weedy and invasive in some areas of its current range and consists of different polyploids that vary in their environmental distributions and invasiveness (Kharrat-Souissi et al. 2014).
Colonization and establishment: phenotypic plasticity
Although the link between phenotypic plasticity and distribution has not been comprehensively supported (Dawson et al. 2012), phenotypic plasticity clearly contributes to environmental tolerance in various ways (e.g. Ghalambor et al. 2007; Des Marais et al. 2013; Schilthuizen and Kellermann 2014). Moreover, at least under conditions of resource abundance, it appears that invasive species do show greater plasticity than their indigenous counterparts (Davidson et al. 2011), although the way in which such comparisons should be made has been the subject of much recent discussion (Van Kleunen et al. 2010; Leffler et al. 2014).
Phenotypic plasticity plays a significant role in the responses of both plants and animals to environmental change (De Witt and Scheiner 2004; Ellers and Stuefer 2010; Nicotra et al. 2010). The extent to which plasticity promotes longer-term adaptation may vary, however, ranging from cases where it promotes adaptation through to those where it might inhibit adaptation. The outcome depends on a range of circumstances, including the extent and form of environmental predictability, the extent of dispersal and the genetic variance in new environments (Chown and Terblanche 2007; Ghalambor et al. 2007; Scoville and Pfrender 2010; Chevin et al. 2013). Nonetheless, currently, it appears that phenotypic plasticity can play a major role in species responses to climate change, such as in changing the timing of breeding in birds (Gienapp et al. 2008) and responses of plants along climate gradients (Matesanz and Valladares 2014; McLean et al. 2014). Some evidence also exists suggesting that invasive species have advantages over indigenous ones as a consequence either of the extent or form of their plasticity (Stachowicz et al. 2002; Chown et al. 2007; Kleynhans et al. 2014). Given that at least some evidence supports the idea that phenotypic plasticity may enhance the success of nonindigenous species as climates change, an understanding of the genomic basis of plasticity may improve forecasts of where such advantage should be expected and what form it should take.
The genomic and transcriptomic basis of variation in phenotypic plasticity is still poorly understood. Several gene expression studies have now been undertaken across related species or populations exposed to different environmental conditions to assess whether these might provide clues about the mechanistic basis of variability in plastic responses (Smith et al. 2013; Dunning et al. 2014; Meier et al. 2014). A few of them have considered populations from native and invaded parts of their range to assess whether there are differences in plastic responses to the different environmental conditions encountered in invasive situations. For instance, in sticklebacks, the invasion of freshwater environments by marine populations has involved an increase in expression plasticity, including some genes that are thought to be involved in adaptation to these new habitats (Morris et al. 2014), while in D. melanogaster populations from warm environments, there was an enrichment of down-regulated genes when flies were raised in conditions that were not commonly encountered in their native range (Levine et al. 2011).
A challenge is to link these expression changes to adaptive variation in plasticity; population differences in gene expression under different conditions may indicate adaptive changes in plasticity, but expression changes have not yet been clearly linked mechanistically to such changes. One problem in making such connections is that there is a level of complexity in organism's transcriptome that largely remains to be explored even in model organisms. For instance, in D. melanogaster, the transcriptome can be modified qualitatively in a variety of ways, such as through alternative splicing and expression of alternative isoforms, that may ultimately have more impact on plastic responses than quantitative changes in gene expression. These sources of variation for climate adaptation are only just starting to be explored (Telonis-Scott et al. 2013).
Another challenge is to consider plastic changes across generations as well as within them. Epigenetic modifications such as DNA methylation and histone modifications can be induced by environment and influence gene expression and transposable element activity, with direct effects on the phenotype (Glastad et al. 2011; Herrera et al. 2012; Zhang et al. 2013c). In plants, methylation can also be inherited through multiple generations, thus partly acting like heritable genetic variation. There are several examples of invasive species where epigenetic variation far exceeds standing genetic variation (Richards et al. 2012; Liebl et al. 2013). Genomics is integral to the field of ecological epigenetics. Whole-genome bisulphite sequencing can reconstruct entire methylation profiles (Schmitz et al. 2013; Wang et al. 2013) while reduced-representation genomic methods and genetic methods such as MS-AFLP can enable quantification of the extent of epigenetic compared to genetic variation within and among populations (Pérez et al. 2006). The hypothesis that epigenetic variation is important to invasion in some cases is intriguing, and a handful of studies exist directly linking epigenotype to phenotypic means, plasticities and environmental tolerance (Herrera et al. 2012; Zhang et al. 2013c), but in general, few data have been collected that examine adaptive and heritable epigenetic effects (Furrow and Feldman 2014).
Spread
Understanding the spread/expansion phase of an invasion has similarities with species range projections in climate change biology (Caplat et al. 2013). To expand its range into new environments, a species requires appropriate dispersal traits and tolerance of a range of environments or capacity to experience the new environment as consistently receptive. Dispersal traits are a key characteristic of invasive species, and dispersal occurs through direct movement (in the case of animals) or indirect movement, by seed or vegetative dispersal (in the case of plants), dispersal through soil or water (e.g. the plant pathogen Phytophthora cinnamomi) or human-mediated dispersal (e.g. weedy species along roadsides, cane toads in transport vehicles and vertebrates on ships).
Genomic studies on populations can be used to test ecological hypotheses about dispersal patterns in invasive species once these have become established in a new area and started to spread, including the widely documented lag phase before populations disperse widely (Richardson et al. 2011). For instance, Rohfritsch et al. (2013) used genomic data to map the spread of an invasive oyster across Europe following its introduction from Asia. This approach is particularly powerful when combined with ecological models that predict the extent to which populations are expected to occupy new space across time or maintain refugia under a changing climate, both of which should be reflected in patterns of genetic variation (Fordham et al. 2014). For example, genomic approaches have been used to understand patterns of spread in Australian Acacia species that have subsequently invaded southern Africa. These include estimating the impacts of human-assisted dispersal in the native range prior to introduction (Le Roux et al. 2013) and better forecasting of the effects of climate change by accounting for genotypic variation (see Millar et al. 2011; Thompson et al. 2011).
Population genomic data can also be used to understand the impact of spreading invaders through hybridization and introgression. As discussed above, invaders spreading into non-native habitat, particularly under climate change, are expected to hybridize with local species where reproductive isolation is incomplete. This can lead to the incursion of genes from the invaders into native species. As a consequence, local species can become genetically ‘polluted’, resulting in the loss of species integrity. Genomic markers provide a way of detecting such pollution (e.g. Yamazaki et al. 2005; Sampson and Byrne 2008; Millar et al. 2012).
Mechanisms of dispersal can evolve, as in the case of leg length in the cane toad (Phillips et al. 2006), and seed dispersal in Abronia umbellata (Darling et al. 2008b). In these cases, genomic studies on quantitative traits can provide information on the genes and pathways involved. Dispersal evolution may involve traits directly involved in movement, such as cane toad leg length, or more subtle changes that nevertheless enhance dispersal potential, such as altered timing of reproduction that increases seed dispersal. Genomic data can identify the nature of these evolved changes in mechanisms that alter the potential of populations to spread. For instance, QTLs have been isolated that control the formation of rhizomes in perennial wild rye that represents an important trait facilitating colonization in weedy ryegrass species (Yun et al. 2014). Understanding the genomic basis of traits involved in non-human-mediated dispersal may aid in predicting the invasive potential of suspect species before introduction or at the establishment stage (Pérez et al. 2006), as well as projecting their future spread in cases where dispersal traits can evolve.
Management implications
The broad consensus in the literature is that climate change is likely to interact synergistically with biological invasions, leading to increasing economic and biodiversity impact (Walther et al. 2009; Scholes et al. 2014). Much evidence exists for recent changes in pest and disease distributions (Bebber et al. 2013), although substantial variation exists among taxa and geographic regions in the extent to which an overall increase in the burden of invasives under climate change might be expected (Bellard et al. 2013). A complicating factor is that human activity in the form of trade, transport and adaptation to climate change is likely to increase too. This will complicate decisions about range shifts relative to anthropogenic introductions and thus potentially confounding management decisions that are required to limit the overall burden of invasive species impacts. Even in relatively remote areas, these problems are playing out (Chown et al. 2012). Overall, the prospects are for increased impacts of invasion as climate change proceeds (Scholes et al. 2014).
From a management perspective, prevention, usually involving some form of risk assessment, is clearly the most cost-effective solution to invasions (Simberloff et al. 2013). Therefore, considerable focus should be on prevention including the identification of the source and receiving areas for introductions (such as ports). Internationally, risk assessment is accepted as an essential policy instrument for managing biological risk. Risk is quantified by examining the invasiveness, impacts and potential distribution of species, areas which are all being improved by the adoption of genomic approaches. Thereafter (i.e. in a postborder context), substantial focus on interventions across the stages of invasion is critical for management success both in relation to typical population processes (such as those involved in emergence from lag phases) (Kueffer et al. 2013; Ricciardi et al. 2013; Simberloff et al. 2013) and to changes in the environment associated with climate change and human adaptation to such change. Likewise, understanding when invasion success may be driven by factors other than climate change, such as local adaptation to climates, evolution of increased competitive ability and enemy release (Blossey and Notzold 1995; Chun et al. 2010; Van Kleunen et al. 2010; Hill et al. 2013), is also important for managing invasions. Prioritization of management actions to species of greatest risk (Randall et al. 2008; Byrne et al. 2011; Forsyth et al. 2012) is critical given the requirement for effective outcomes with investment of limited resources. As hybridization has been demonstrated to facilitate invasiveness, assessment of genetic risk is an important management tool (Byrne et al. 2011), particularly in the context of managed relocation as a climate change adaptation strategy (Schwartz et al. 2012).
Throughout this review, we have demonstrated how genomics approaches can improve understanding, and in many cases forecasting, at each stage of invasion (Fig.1). These include better understanding of source areas and the identity of introduced species. In this sense, genomic approaches are typically improvements on genetic tools that have been available for sometime; they allow source populations to be identified more accurately, provide a clearer picture regarding multiple interactions, including the role of hybridization, and lead to better estimates of population size, gene flow and other processes. NGS techniques can provide ways of detecting invasive organisms that have previously not been available, and identifications become more accurate as additional sequence information is added. Microsatellite and mtDNA markers have been widely used to understand the population dynamics of agricultural pests and disease vectors in the past and thereby assisting in programs aimed at controlling pests and/or limiting their impact. However, the new genomic tools provide an unprecedented view of past and present population processes (e.g. Brown et al. 2014; Rasic et al. 2014). With invasion frequency increasing under climate change, these are all important tools in improving an understanding of invasion processes.
Where control of invasive species is being considered, genomic approaches can help in defining units for eradication and risks of reinvasion once eradication is achieved (Fig.1). At present, markers such as microsatellites are often used for defining movement patterns among populations of invasive pest mammals (e.g. Hampton et al. 2004; Abdelkrim et al. 2005; Berry et al. 2012; Adams et al. 2014). With the use of genomics approaches, managers should become much more confident of likely movement patterns and reinvasion potential as local movement patterns can be tracked much more accurately than in the past.
However, genomics tools promise much more than ways of simply improving detection and an understanding of spread. High-density markers provide a way of tracking changes in different parts of the genome, essential for understanding hybridization and introgression as well as adaptation under climate change including the application of quantitative genomic approaches to identify regions with candidate genes (Fig.1). This can be used to understand the roles of various evolutionary processes at key steps of the invasion pathway, and the ways in which climate change will interact with the latter. Key areas include investigation of evolutionary change underpinning increased competitive ability and enemy release, especially in a biological control context. Genomics methods can also help redirect resources by improving understanding of the burden of invasion, an especially important role as that burden increases (e.g. Pfenninger et al. 2014). They can assist with rehabilitation in the case where substantial invasive impacts result in widespread ecosystem changes, by understanding patterns of resistance and identifying resistant genotypes that can persist into the future. For example, where trees are affected by introduced pathogens, there is the possibility of identifying tree genotypes that survive outbreaks (Stukely and Crane 1994; McKinney et al. 2014). As information becomes available on adaptive changes in traits controlling the spread and climate change adaptation, it should also be possible to refine models about the non-native range of an invasive species. For instance, Kearney et al. (2009) modelled the expected range of Aedes aegypti in northern Australia based on anticipated evolutionary changes in egg desiccation tolerance under climate change and showed the potential threat posed by this species around a population centre.
In summary, genomic tools are proving to be extremely useful for managing invasive species. While they may be seen as costly for less well-resourced nations, several studies have demonstrated that they are more cost-effective than traditional surveillance tools and other methods. Moreover, technology is now advancing to the stage where users do not need to be familiar with its intricacies (in much the same way, most mobile phone users could not build one), and substantial programmes of technology assistance are in place through various international agencies (such as the Consultative Group on International Agricultural Research, www.cgiar.org). In consequence, the myriad ways genomic tools can be utilized will enable society to contain the economic cost of biological invasions accentuated by climate change, and prevent a world dominated by weedy species.
Acknowledgments
Melodie McGeoch and two anonymous reviewers provided helpful comments on the manuscript. This work arose from a workshop sponsored by CSIRO's Cutting Edge Symposium series and is linked to research being undertaken under the Science Industry Endowment Fund. SLC is supported by Australian Research Council Grant DP140102815.
Literature cited
- Abdelkrim J, Pascal M, Calmet C. Samadi S. Importance of assessing population genetic structure before eradication of invasive species: examples from insular Norway rat populations. Conservation Biology. 2005;19:1509–1518. [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Adams AL, van Heezik Y, Dickinson KJM. Robertson BC. Identifying eradication units in an invasive mammalian pest species. Biological Invasions. 2014;16:1481–1496. [Google Scholar]
- Adriaennsens EM. Cowan DA. Using signature genes as tools to address environmental viral ecology and diversity. Applied and Environmental Microbiology. 2014 doi: 10.1128/AEM.00878-14. doi: 10.1128/AEM.00878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainouche ML, Fortune PM, Salmon A, Parisod C, Grandbastien MA, Fukunaga K, Ricou M. Misset MT. Hybridization, polyploidy and invasion: lessons from Spartina (Poaceae) Biological Invasions. 2009;11:1159–1173. [Google Scholar]
- Ali S, Gladieux P, Rahman H, Saqib MS, Fiaz M, Ahmad H, Leconte M, et al. Inferring the contribution of sexual reproduction, migration and off-season survival to the temporal maintenance of microbial populations: a case study on the wheat fungal pathogen Puccinia striiformis f.sp tritici. Molecular Ecology. 2014;23:603–617. doi: 10.1111/mec.12629. [DOI] [PubMed] [Google Scholar]
- Amano T. Sutherland WJ. Four barriers to the global understanding of biodiversity conservation: wealth, language, geographical location and security. Proceedings of the Royal Society B. 2013;280:20122649. doi: 10.1098/rspb.2012.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Amaral FR, Albers PK, Edwards SV. Miyaki CY. Multilocus tests of Pleistocene refugia and ancient divergence in a pair of Atlantic Forest antbirds (Myrmeciza. Molecular Ecology. 2013;22:3996–4013. doi: 10.1111/mec.12361. [DOI] [PubMed] [Google Scholar]
- Anderson E. Hybridization of the habitat. Evolution. 1948;2:1–9. [Google Scholar]
- Armstrong KF. Ball SL. DNA barcodes for biosecurity: invasive species identification. Philosophical Transactions of the Royal Society B. 2005;360:1813–1823. doi: 10.1098/rstb.2005.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold ML, Ballerini ES. Brothers AN. Hybrid fitness, adaptation and evolutionary diversification: lessons learned from Louisiana Irises. Heredity. 2012;108:159–166. doi: 10.1038/hdy.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asner GP. Vitousek PM. Remote analysis of biological invasion and biogeochemical change. Proceedings of the National Academy of Sciences of the USA. 2005;102:4383–4386. doi: 10.1073/pnas.0500823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres RM, Pettigrove VJ. Hoffmann AA. Genetic structure and diversity of introduced eastern mosquitofish (Gambusia holbrooki) in south-eastern Australia. Marine and Freshwater Research. 2012;63:1206–1214. [Google Scholar]
- Baker HG. The evolution of weeds. Annual Review of Ecology and Systematics. 1974;5:1–24. [Google Scholar]
- Barnosky AD, Hadly EA, Bascompte J, Berlow EL, Brown JH, Fortelius M, Getz WM, et al. Approaching a state shift in Earth's biosphere. Nature. 2012;486:52–58. doi: 10.1038/nature11018. [DOI] [PubMed] [Google Scholar]
- Barrett SCH. Crop mimicry in weeds. Economic Botany. 1983;37:255–282. [Google Scholar]
- Barrett RDH. Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Barrett RDH. Schluter D. Adaptation from standing genetic variation. Trends in Ecology and Evolution. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Baumel A, Ainouche ML. Levasseur JE. Molecular investigations in populations of Spartina anglica C.E. Hubbard (Poaceae) invading coastal Brittany (France. Molecular Ecology. 2001;10:1689–1701. doi: 10.1046/j.1365-294x.2001.01299.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA. Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society B. 1996;263:1619–1626. [Google Scholar]
- Bebber DP, Ramatowski MAT. Gurr SJ. Crop pests and pathogens move polewards in a warming world. Nature Climate Change. 2013;3:985–988. [Google Scholar]
- Becker M, Gruenheit N, Steel M, Voelckel C, Deusch O, Heenan PB, McLenachan PA, et al. Hybridization may facilitate in situ survival of endemic species through periods of climate change. Nature Climate Change. 2013;3:1039–1043. [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M. Pyšek P. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell MA. Aguirre WE. Contemporary evolution, allelic recycling, and adaptive radiation of the threespine stickleback. Evolutionary Ecology Research. 2013;15:377–411. [Google Scholar]
- Bellard C, Thuiller W, Leroy B, Genovesi P, Bakkenes M. Courchamp F. Will climate change promote future invasions? Global Change Biology. 2013;19:3740–3748. doi: 10.1111/gcb.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. Coop G. The population genetic signature of polygenic local adaptation. Plos Genetics. 2014;8:e1004412. doi: 10.1371/journal.pgen.1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry O, Algar D, Angus J, Hamilton N, Hilmer S. Sutherland D. Genetic tagging reveals a significant impact of poison baiting on an invasive species. Journal of Wildlife Management. 2012;76:729–739. [Google Scholar]
- Berthouly-Salazar C, Hui C, Blackburn TM, Gaboriaud C, van Rensburg BJ, van Vuuren BJ. Le Roux JJ. Long-distance dispersal maximizes evolutionary potential during rapid geographic range expansion. Molecular Ecology. 2013;22:5793–5804. doi: 10.1111/mec.12538. [DOI] [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, et al. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proceedings of the National Academy of Sciences of the USA. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi K, Linderoth T, Vanderpool D, Good JM, Nielsen R. Moritz C. Unlocking the vault: next-generation museum population genomics. Molecular Ecology. 2013;22:6018–6032. doi: 10.1111/mec.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn TM, Cassey P, Duncan RP, Evans KL. Gaston KJ. Avian extinction and mammalian introductions on oceanic islands. Science. 2004;305:1955–1958. doi: 10.1126/science.1101617. [DOI] [PubMed] [Google Scholar]
- Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošík V, Wilson JRU. Richardson DM. A proposed unified framework for biological invasions. Trends in Ecology and Evolution. 2011;26:333–339. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Blackburn TM, Essl F, Evans TA, Hulme PE, Jeschke JM, Kühn I, Kumschick S, et al. A unified classification of alien species based on the magnitude of their environmental impacts. PLoS Biology. 2014;12:e1001850. doi: 10.1371/journal.pbio.1001850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet S. The use of molecular tools in invasion biology: an emphasis on freshwater ecosystems. Fisheries Management and Ecology. 2012;19:120–132. [Google Scholar]
- Bloom JS, Ehrenreich IM, Loo WT, Thuy-Lan Vo L. Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blossey B. Notzold R. Evolution of increased competitive ability in invasive nonindigenous plants - a hypothesis. Journal of Ecology. 1995;83:887–889. [Google Scholar]
- Bohmann K, Evans A, Gilbert MT, Carvalho GR, Creer S, Knapp M, Yu DW, et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology and Evolution. 2014;29:358–367. doi: 10.1016/j.tree.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers WE, Siemann E. Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Richards CL. Pigliucci M. Epigenetics for ecologists. Ecology Letters. 2008;11:106–115. doi: 10.1111/j.1461-0248.2007.01130.x. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Borja A, Gilbert J, Taylor MI, Davies N, Weisberg SB, Griffith JF, et al. Genomics in marine monitoring: new opportunities for assessing marine health status. Marine Pollution Bulletin. 2013;74:19–31. doi: 10.1016/j.marpolbul.2013.05.042. [DOI] [PubMed] [Google Scholar]
- Bradley BA, Blumenthal DM, Wilcove DS. Ziska LH. Predicting plant invasions in an era of global change. Trends in Ecology and Evolution. 2010;25:310–318. doi: 10.1016/j.tree.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen AC. Elven R. Polyploidy in arctic plants. Biological Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- Brodin Y, Ejdung G, Strandberg J. Lyrholm T. Improving environmental and biodiversity monitoring in the Baltic Sea using DNA barcoding of Chironomidae (Diptera) Molecular Ecology Resources. 2013;13:996–1004. doi: 10.1111/1755-0998.12053. [DOI] [PubMed] [Google Scholar]
- Bronnenhuber JE, Dufour BA, Higgs DM. Heath DD. Dispersal strategies, secondary range expansion and invasion genetics of the nonindigenous round goby, Neogobius melanostomus, in Great Lakes tributaries. Molecular Ecology. 2011;20:1845–1859. doi: 10.1111/j.1365-294X.2011.05030.x. [DOI] [PubMed] [Google Scholar]
- Brook BW, Sodhi NS. Bradshaw CJA. Synergies among extinction drivers under global change. Trends in Ecology and Evolution. 2008;23:453–460. doi: 10.1016/j.tree.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, Zhao HY, Caccone A. Powell JR. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 2014;68:514–525. doi: 10.1111/evo.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JM. Arnold ML. Genetics and the fitness of hybrids. Annual Review of Genetics. 2001;35:31–52. doi: 10.1146/annurev.genet.35.102401.085719. [DOI] [PubMed] [Google Scholar]
- Butchart SHM, Walpole M, Collen B, van Strien A, Scharlemann JPW, Almond REA, Baillie JEM, et al. Global biodiversity: indicators of recent declines. Science. 2010;328:1164–1168. doi: 10.1126/science.1187512. [DOI] [PubMed] [Google Scholar]
- Byrne M, Stone L. Millar MA. Assessing genetic risk in revegetation. Journal of Applied Ecology. 2011;48:1365–1373. [Google Scholar]
- Callaway RM. Maron JL. What have exotic plant invasions taught us over the past 20 years? Trends in Ecology and Evolution. 2006;21:369–374. doi: 10.1016/j.tree.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Calvignac-Spencer S, Merkel K, Kutzner N, Kuhl H, Boesch C, Kappeler PM, Metzger S, et al. Carrion fly-derived DNA as a tool for comprehensive and cost-effective assessment of mammalian biodiversity. Molecular Ecology. 2013;22:915–924. doi: 10.1111/mec.12183. [DOI] [PubMed] [Google Scholar]
- Caplat P, Cheptou PO, Diez J, Guisan A, Larson BMH, Macdougall AS, Peltzer DA, et al. Movement, impacts and management of plant distributions in response to climate change: insights from invasions. Oikos. 2013;122:1265–1274. [Google Scholar]
- Carroll SP, Dingle H, Famula TR. Fox CW. Genetic architecture of adaptive differentiation in evolving host races of the soapberry bug, Jadera haematoloma. Genetica. 2001;112:257–272. [PubMed] [Google Scholar]
- Carroll SP, Jørgensen PS, Kinnison MT, Bergstrom CT, Denison RF, Gluckman P, Smith TB, et al. Applying evolutionary biology to address global challenges. Science. 2014 doi: 10.1126/science.1245993. doi: 10.1126/science.1245993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassey P, Blackburn TM, Sol D, Duncan RP. Lockwood JL. Global patterns of introduction effort and establishment success in birds. Proceedings of the Royal Society B. 2004;271(Suppl 6):S405–S408. doi: 10.1098/rsbl.2004.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda LE, Balanyá J, Rezende EL. Santos M. Vanishing chromosomal inversion clines in Drosophila subobscura from Chile: is behavioral thermoregulation to blame? American Naturalist. 2013;182:249–259. doi: 10.1086/671057. [DOI] [PubMed] [Google Scholar]
- Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, et al. Draft genome sequence of the oilseed species Ricinus communis. Nature Biotechnology. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H, Monnier A. Ainouche M. Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina x townsendii and Spartina anglica (Poaceae) New Phytologist. 2010;186:161–174. doi: 10.1111/j.1469-8137.2010.03179.x. [DOI] [PubMed] [Google Scholar]
- Chevin L-M, Collins S. Lefèvre F. Phenotypic plasticity and evolutionary demographic responses to climate change: taking theory out to the field. Functional Ecology. 2013;27:967–979. [Google Scholar]
- Chiu JC, Jiang XT, Zhao L, Hamm CA, Cridland JM, Saelao P, Hamby KA, et al. Genome of Drosophila suzukii, the Spotted Wing Drosophila. G3-Genes Genomes. Genetics. 2013;3:2257–2271. doi: 10.1534/g3.113.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL. Temporal biodiversity change in transformed landscapes: a southern African perspective. Philosophical Transactions of the Royal Society B. 2010;365:3729–3742. doi: 10.1098/rstb.2010.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL. Terblanche JS. Physiological diversity in insects: ecological and evolutionary contexts. Advances in Insect Physiology. 2007;33:50–152. doi: 10.1016/S0065-2806(06)33002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL, Slabber S, McGeoch MA, Janion C. Leinaas HP. Phenotypic plasticity mediates climate change responses among invasive and indigenous arthropods. Proceedings of the Royal Society B. 2007;274:2661–2667. doi: 10.1098/rspb.2007.0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chown SL, Huiskes AHL, Gremmen NJM, Lee JE, Terauds A, Crosbie K, Frenot Y, et al. Continent-wide risk assessment for the establishment of nonindigenous species in Antarctica. Proceedings of the National Academy of Sciences of the USA. 2012;109:4938–4943. doi: 10.1073/pnas.1119787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun YJ, van Kleunen M. Dawson W. The role of enemy release, tolerance and resistance in plant invasions: linking damage to performance. Ecology Letters. 2010;13:937–946. doi: 10.1111/j.1461-0248.2010.01498.x. [DOI] [PubMed] [Google Scholar]
- Colautti RI. Barrett SCH. Rapid adaptation to climate facilitates range expansion of an invasive plant. Science. 2013;342:364–366. doi: 10.1126/science.1242121. [DOI] [PubMed] [Google Scholar]
- Colautti RI, Grigorovich IA. MacIsaac HJ. Propagule pressure: A null model for biological invasions. Biological Invasions. 2006;8:1023–1037. [Google Scholar]
- Collins RA. Cruickshank RH. The seven deadly sins of DNA barcoding. Molecular Ecology Resources. 2013;13:969–975. doi: 10.1111/1755-0998.12046. [DOI] [PubMed] [Google Scholar]
- Collins RA, Armstrong KF, Holyoake AJ. Keeling S. Something in the water: biosecurity monitoring of ornamental fish imports using environmental DNA. Biological Invasions. 2013;15:1209–1215. [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nature Reviews Genetics. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A. Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan D, Chown SL, Convey P, Tuffin IM, Hughes KE, Pointing S. Vincent WF. Non-indigenous microorganisms in the Antarctic: assessing the risks. Trends in Microbiology. 2011;19:540–548. doi: 10.1016/j.tim.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Craigie ID, Baillie JEM, Balmford A, Carbone C, Collen B, Green RE. Hutton JM. Large mammal population declines in Africa's protected areas. Biological Conservation. 2010;143:2221–2228. [Google Scholar]
- Darling JA. Blum MJ. DNA-based methods for monitoring invasive species: a review and prospectus. Biological Invasions. 2007;9:751–765. [Google Scholar]
- Darling ES. Côté IM. Quantifying the evidence for ecological synergies. Ecology Letters. 2008;11:1278–1286. doi: 10.1111/j.1461-0248.2008.01243.x. [DOI] [PubMed] [Google Scholar]
- Darling JA, Bagley MJ, Roman J, Tepolt CK. Geller JB. Genetic patterns across multiple introductions of the globally invasive crab genus Carcinus. Molecular Ecology. 2008a;17:4992–5007. doi: 10.1111/j.1365-294X.2008.03978.x. [DOI] [PubMed] [Google Scholar]
- Darling E, Samis KE. Eckert CG. Increased seed dispersal potential towards geographic range limits in a Pacific coast dune plant. New Phytologist. 2008b;178:424–435. doi: 10.1111/j.1469-8137.2007.02349.x. [DOI] [PubMed] [Google Scholar]
- Davidson AM, Jennions M. Nicotra AB. Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecology Letters. 2011;14:419–431. doi: 10.1111/j.1461-0248.2011.01596.x. [DOI] [PubMed] [Google Scholar]
- Dawson W, Rohr RP, van Kleunen M. Fischer M. Alien plant species with a wider global distribution are better able to capitalize on increased resource availability. New Phytologist. 2012;194:859–867. doi: 10.1111/j.1469-8137.2012.04104.x. [DOI] [PubMed] [Google Scholar]
- De Kort H, Vandepitte K, Mergeay J. Honnay O. Isolation, characterization and genotyping of single nucleotide polymorphisms in the non-model tree species Frangula alnus (Rhamnaceae) Conservation Genetics Resources. 2014;6:267–269. [Google Scholar]
- De La Torre AR, Roberts DR. Aitken SN. Genome-wide admixture and ecological niche modelling reveal the maintenance of species boundaries despite long history of interspecific gene flow. Molecular Ecology. 2014;23:2046–2059. doi: 10.1111/mec.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witt TJ. Scheiner SM. Phenotypic Plasticity. Functional and Conceptual Approaches. Oxford: Oxford University Press; 2004. [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, et al. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Dehnen-Schmutz K. Determining non-invasiveness in ornamental plants to build green lists. Journal of Applied Ecology. 2011;48:1374–1380. [Google Scholar]
- Dejean T, Valentini A, Miquel C, Taberlet P, Bellemain E. Miaud C. Improved detection of an alien invasive species through environmental DNA barcoding: the example of the American bullfrog Lithobates catesbeianus. Journal of Applied Ecology. 2012;49:953–959. [Google Scholar]
- Des Marais DL, Hernandez KM. Juenger TE. Genotype-by-environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annual Review of Ecology, Evolution, and Systematics. 2013;44:5–29. [Google Scholar]
- Dlugosch KM. Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Molecular Ecology. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Dormontt EE, Lowe AJ. Prentis PJ. Is rapid evolution important in successful invasions? In: Richardson DM, editor; Fifty years of invasion ecology. The legacy of Charles Elton. Oxford: Wiley-Blackwell; 2011. pp. 175–193. [Google Scholar]
- Drake JM. Heterosis, the catapult effect and establishment success of a colonizing bird. Biology Letters. 2006;2:304–307. doi: 10.1098/rsbl.2006.0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JM. Lodge DM. Global hot spots of biological invasions: evaluating options for ballast-water management. Proceedings of the Royal Society of London B. 2004;271:575–580. doi: 10.1098/rspb.2003.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning LT, Dennis AB, Sinclair BJ, Newcomb RD. Buckley TR. Divergent transcriptional responses to low temperature among populations of alpine and lowland species of New Zealand stick insects (Micrarchus. Molecular Ecology. 2014;23:2712–2726. doi: 10.1111/mec.12767. [DOI] [PubMed] [Google Scholar]
- Dybern BI. The life cycle of Ciona intestinalis (L.) f. typica in the relation to environmental temperature. Oikos. 1965;16:109–131. [Google Scholar]
- Egan SP, Barnes MA, Hwang C-T, Mahon AR, Feder JL, Ruggiero ST, Tanner CE. Lodge DM. Rapid invasive species detection by combining environmental DNA with light transmission spectroscopy. Conservation Letters. 2013;6:402–409. [Google Scholar]
- Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends in Ecology and Evolution. 2014;29:51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Ellers J. Stuefer JF. Frontiers in phenotypic plasticity research: new questions about mechanisms, induced responses and ecological impacts. Evolutionary Ecology. 2010;24:523–526. [Google Scholar]
- Ellstrand NC. Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants? Proceedings of the National Academy of Sciences of the USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES. Mitchell SE. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endersby NM, Hoffmann AA, White VL, Ritchie SA, Johnson PH. Weeks AR. Changes in the genetic structure of Aedes aegypti (Diptera: Culicidae) populations in Queensland, Australia, across two seasons: implications for potential mosquito releases. Journal of Medical Entomology. 2011;48:999–1007. doi: 10.1603/me10264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J, Kim J, Childs KL, Vaillancourt B, Crisovan E, Nandety A, Gerhardt DJ, et al. Nucleotide polymorphism and copy number variant detection using exome capture and next-generation sequencing in the polyploid grass Panicum virgatum. The Plant Journal. 2014;79:993–1008. doi: 10.1111/tpj.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Hofer T. Foll M. Detecting loci under selection in a hierarchically structured population. Heredity. 2009;103:285–298. doi: 10.1038/hdy.2009.74. [DOI] [PubMed] [Google Scholar]
- Fernández-Álvarez FA. Machordom A. DNA barcoding reveals a cryptic nemertean invasion in Atlantic and Mediterranean waters. Helgoland Marine Research. 2013;67:599–605. [Google Scholar]
- Ficetola GF, Miaud F, Pompanon F. Taberlet P. Species detection using environmental DNA from water samples. Biology Letters. 2008;4:423–425. doi: 10.1098/rsbl.2008.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick BM, Johnson JR, Kump DK, Smith JJ, Voss SR. Shaffer HB. Rapid spread of invasive genes into a threatened native species. Proceedings of the National Academy of Sciences of the USA. 2010;107:3606–3610. doi: 10.1073/pnas.0911802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foll M. Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham DA, Brook BW, Moritz C. Nogues-Bravo D. Better forecasts of range dynamics using genetic data. Trends in Ecology and Evolution. 2014;29:436–443. doi: 10.1016/j.tree.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Forsman A. Effects of genotypic and phenotypic variation on establishment are important for conservation, invasion, and infection biology. Proceedings of the National Academy of Sciences of the USA. 2014;111:302–307. doi: 10.1073/pnas.1317745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth GG, Le Maitre DC, O'Farrell PJ. van Wilgen BW. The prioritisation of invasive alien plant control projects using a multi-criteria decision model informed by stakeholder input and spatial data. Journal of Environmental Management. 2012;103:51–57. doi: 10.1016/j.jenvman.2012.01.034. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Avise JC, Bradshaw WE, Conner JK, Etterson JR, Mazer SJ, Shaw RG. Weis AE. The Resurrection Initiative: storing ancestral genotypes to capture evolution in action. BioScience. 2008;58:870–873. [Google Scholar]
- Friedman JM, Roelle JE, Gaskin JF, Pepper AE. Manhart JR. Latitudinal variation in cold hardiness in introduced Tamarix and native Populus. Evolutionary Applications. 2008;1:598–607. doi: 10.1111/j.1752-4571.2008.00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrow RE. Feldman MW. Genetic variation and the evolution of epigenetic regulation. Evolution. 2014;68:673–683. doi: 10.1111/evo.12225. [DOI] [PubMed] [Google Scholar]
- Futschik A. Schlotterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamfeldt L. Källström B. Increasing intraspecific diversity increases predictability in population survival in the face of perturbations. Oikos. 2007;116:700–705. [Google Scholar]
- Garroway CJ, Bowman J, Cascaden TJ, Holloway GL, Mahan CG, Malcolm JR, Steele MA, et al. Climate change induced hybridization in flying squirrels. Global Change Biology. 2010;16:113–121. [Google Scholar]
- Gaskin JF. Kazmer DJ. Introgression between invasive saltcedars (Tamarix chinensis and T. ramosissima) in the USA. Biological Invasions. 2009;11:1121–1130. [Google Scholar]
- Gaskin JF. Schaal BA. Hybrid Tamarix widespread in US invasion and undetected in native Asian range. Proceedings of the National Academy of Sciences of the USA. 2002;99:11256–11259. doi: 10.1073/pnas.132403299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskin JF, Bon M-C, Cock MJW, Cristofaro M, Biase AD, De Clerck-Floate R, Ellison CA, et al. Applying molecular-based approaches to classical biological control of weeds. Biological Control. 2011;58:1–21. [Google Scholar]
- Gautier M, Gharbi K, Cezard T, Foucaud J, Kerdelhue C, Pudlo P, Cornuet J-M. Estoup A. The effect of RAD allele dropout on the estimation of genetic variation within and between populations. Molecular Ecology. 2013;22:3165–3178. doi: 10.1111/mec.12089. [DOI] [PubMed] [Google Scholar]
- Ghalambor CK, McKay JK, Carroll SP. Reznick DN. Adaptive versus non-adaptive phenotypic plasticity and the potential for contemporary adaptation in new environments. Functional Ecology. 2007;21:394–407. [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Gienapp P, Teplitsky C, Alho JS, Mills JA. Merila J. Climate change and evolution: disentangling environmental and genetic responses. Molecular Ecology. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- Glastad KM, Hunt BG, Yi SV. Goodisman MAD. DNA methylation in insects: on the brink of the epigenomic era. Insect Molecular Biology. 2011;20:553–565. doi: 10.1111/j.1365-2583.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- Griffin PC. Hoffmann AA. Limited genetic divergence among Australian alpine Poa tussock grasses coupled with regional structuring points to ongoing gene flow and taxonomic challenges. Annals of Botany. 2014;113:953–965. doi: 10.1093/aob/mcu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin PC, Robin C. Hoffmann AA. A next-generation sequencing method for overcoming the multiple gene copy problem in polyploid phylogenetics, applied to Poa grasses. BMC Biology. 2011;9:18. doi: 10.1186/1741-7007-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggisberg A, Lai Z, Huang J. Rieseberg LH. Transcriptome divergence between introduced and native populations of Canada thistle, Cirsium arvense. New Phytologist. 2013;199:595–608. doi: 10.1111/nph.12258. [DOI] [PubMed] [Google Scholar]
- Guisan A, Petitpierre B, Broennimann O, Daehler C. Kueffer C. Unifying niche shift studies: insights from biological invasions. Trends in Ecology and Evolution. 2014;29:260–269. doi: 10.1016/j.tree.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Günther T. Coop G. Robust identification of local adaptation from allele frequencies. Genetics. 2013;195:205–220. doi: 10.1534/genetics.113.152462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SK, Singh S, Nischal A, Pant KK. Seth PK. Molecular-based identification and phylogeny of genomic and proteomic sequences of mosquito-borne flavivirus. Genes & Genomics. 2014;36:31–43. [Google Scholar]
- Hampton JO, Spencer PBS, Alpers DL, Twigg LE, Woolnough AP, Doust J, Higgs T. Pluske J. Molecular techniques, wildlife management and the importance of genetic population structure and dispersal: a case study with feral pigs. Journal of Applied Ecology. 2004;41:735–743. [Google Scholar]
- Hänel C. Chown SL. The impact of a small, alien invertebrate on a sub-Antarctic terrestrial ecosystem: Limnophyes minimus (Diptera, Chironomidae) at Marion Island. Polar Biology. 1998;20:99–106. [Google Scholar]
- Hansen J, Kharecha P, Sato M, Masson-Delmotte V, Ackerman F, Beerling DJ, Hearty PJ, et al. Assessing “dangerous climate change”: required reduction of carbon emissions to protect young people, future generations and nature. PLoS ONE. 2013;8:e81648. doi: 10.1371/journal.pone.0081648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W-M, Thelen GC, Ridenour WM. Callaway RM. Is there a risk to living large? Large size correlates with reduced growth when stressed for knapweed populations. Biological Invasions. 2010;12:3591–3598. [Google Scholar]
- Henery ML, Bowman G, Mraz P, Treier UA, Gex-Fabry E, Schaffner U. Mueller-Schaerer H. Evidence for a combination of pre-adapted traits and rapid adaptive change in the invasive plant Centaurea stoebe. Journal of Ecology. 2010;98:800–813. [Google Scholar]
- Herrera CM, Pozo MI. Bazaga P. Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower-living yeast. Molecular Ecology. 2012;21:2602–2616. doi: 10.1111/j.1365-294X.2011.05402.x. [DOI] [PubMed] [Google Scholar]
- Hickerson MJ, Meyer CP. Moritz C. DNA barcoding will often fail to discover new animal species over broad parameter space. Systematic Biology. 2006;55:729–739. doi: 10.1080/10635150600969898. [DOI] [PubMed] [Google Scholar]
- Hill MP, Chown SL. Hoffmann AA. A predicted niche shift corresponds with increased thermal resistance in an invasive mite, Halotydeus destructor. Global Ecology and Biogeography. 2013;22:942–951. [Google Scholar]
- Hochholdinger F. Hoecker N. Towards the molecular basis of heterosis. Trends in Plant Science. 2007;12:427–432. doi: 10.1016/j.tplants.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Hodgins KA, Lai Z, Nurkowski K, Huang J. Rieseberg LH. The molecular basis of invasiveness: differences in gene expression of native and introduced common ragweed (Ambrosia artemisiifolia) in stressful and benign environments. Molecular Ecology. 2013;22:2496–2510. doi: 10.1111/mec.12179. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sørensen JG. Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. Journal of Thermal Biology. 2003;28:175–216. [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA. Cresko WA. Population genomics of parallel adaptation in threespine stickleback using Sequenced RAD Tags. PloS Genetics. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li Y, Burt DW, Chen H, Zhang Y, Qian W, Kim H, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nature Genetics. 2013;45:776–784. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D. Serra L. Rapid evolution of a geographic cline in size in an introduced fly. Science. 2000;287:308–309. doi: 10.1126/science.287.5451.308. [DOI] [PubMed] [Google Scholar]
- Huiskes AHL, Gremmen NJM, Bergstrom DM, Frenot Y, Hughes KA, Imura S, Kiefer K, et al. Aliens in Antarctica: assessing transfer of plant propagules by human visitors to reduce invasion risk. Biological Conservation. 2014;171:278–284. [Google Scholar]
- Hulme PE. Trade, transport and trouble: managing invasive species pathways in an era of globalization. Journal of Applied Ecology. 2009;46:10–18. [Google Scholar]
- Hulme PE, Bacher S, Kenis M, Klotz S, Kühn I, Minchin D, Nentwig W, et al. Grasping at the routes of biological invasions: a framework for integrating pathways into policy. Journal of Applied Ecology. 2008;45:403–414. [Google Scholar]
- Hulme PE, Pyšek P, Jarošik V, Pergl J, Schaffner U. Vilà M. Bias and error in understanding plant invasion impacts. Trends in Ecology and Evolution. 2013;28:212–218. doi: 10.1016/j.tree.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Jackson JB. The future of the oceans past. Philosophical Transactions of the Royal Society B. 2010;365:3765–3778. doi: 10.1098/rstb.2010.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerde JL, Mahon AR, Chadderton WL. Lodge DM. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conservation Letters. 2011;4:150–157. [Google Scholar]
- Ji Y, Ashton L, Pedley SM, Edwards DP, Tang Y, Nakamura A, Kitching R, et al. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecology Letters. 2013;16:1245–1257. doi: 10.1111/ele.12162. [DOI] [PubMed] [Google Scholar]
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL. Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Chan YF, Schmutz J, Grimwood J, Brady SD, Southwick AM, Absher DM, et al. A Genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Current Biology. 2012;22:83–90. doi: 10.1016/j.cub.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper ML, Reeson AF, Cooper SJ, Perry KD. Austin AD. Assessment of prey overlap between a native (Polistes humilis) and an introduced (Vespula germanica) social wasp using morphology and phylogenetic analyses of 16S rDNA. Molecular Ecology. 2004;13:2037–2048. doi: 10.1111/j.1365-294X.2004.02193.x. [DOI] [PubMed] [Google Scholar]
- Kearney M, Porter WP, Williams C, Ritchie S, et al. Integrating biophysical models and evolutionary theory to predict climatic impacts on species’ ranges: the dengue mosquito Aedes aegypti in Australia. Functional Ecology. 2009;23:528–538. [Google Scholar]
- Keesing F, Belden LK, Daszak P, Dobson AP, Harvell CD, Holt RD, Hudson P, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468:647–652. doi: 10.1038/nature09575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SR. Taylor DR. Genomic admixture increases fitness during a biological invasion. Journal of Evolutionary Biology. 2010;23:1720–1731. doi: 10.1111/j.1420-9101.2010.02037.x. [DOI] [PubMed] [Google Scholar]
- Kharrat-Souissi A, Siljak-Yakovlev S, Brown SC, Baumel A, Torre F. Chaieb M. The polyploid nature of Cenchrus ciliaris L. (Poaceae) has been overlooked: new insights for the conservation and invasion biology of this species - a review. Rangeland Journal. 2014;36:11–23. [Google Scholar]
- Kirk H, Paul J, Straka J. Freeland JR. Long-distance dispersal and high genetic diversity are implicated in the invasive spread of the common reed, Phragmites australis (Poaceae), in northeastern North America. American Journal of Botany. 2011;98:1180–1190. doi: 10.3732/ajb.1000278. [DOI] [PubMed] [Google Scholar]
- Kleynhans E, Mitchell KA, Conlong DE. Terblanche JS. Evolved variation in cold tolerance among populations of Eldana saccharina (Lepidoptera: Pyralidae) in South Africa. Journal of Evolutionary Biology. 2014;27:1149–1159. doi: 10.1111/jeb.12390. [DOI] [PubMed] [Google Scholar]
- Krehenwinkel H. Tautz D. Northern range expansion of European populations of the wasp spider Argiope bruennichi is associated with global warming-correlated genetic admixture and population-specific temperature adaptations. Molecular Ecology. 2013;22:2232–2248. doi: 10.1111/mec.12223. [DOI] [PubMed] [Google Scholar]
- Kueffer C, Pyšek P. Richardson DM. Integrative invasion science: model systems, multi-site studies, focused meta-analysis and invasion syndromes. New Phytologist. 2013;200:615–633. doi: 10.1111/nph.12415. [DOI] [PubMed] [Google Scholar]
- Lambdon PW, Pyšek P, Basnou C, Hejda M, Arianoutsou M, Essl F, Jarošík V, et al. Alien flora of Europe: species diversity, temporal trends, geographical patterns and research needs. Preslia. 2008;80:101–149. [Google Scholar]
- Larson EL, Andres JA, Bogdanowicz SM. Harrison RG. Differential introgression in a mosaic hybrid zone reveals candidate barrier genes. Evolution. 2013;67:3653–3661. doi: 10.1111/evo.12205. [DOI] [PubMed] [Google Scholar]
- Laurance WF, Useche DC, Rendeiro J, Kalka M, Bradshaw CJ, Sloan SP, Laurance SG, et al. Averting biodiversity collapse in tropical forest protected areas. Nature. 2012;489:290–294. doi: 10.1038/nature11318. [DOI] [PubMed] [Google Scholar]
- Lavergne S. Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proceedings of the National Academy of Sciences of the USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson Handley LJ, Estoup A, Evans DM, Thomas CE, Lombaert E, Facon B, Aebi A. Roy HE. Ecological genetics of invasive alien species. BioControl. 2011;56:409–428. [Google Scholar]
- Le Corre V. Kremer A. The genetic differentiation at quantitative trait loci under local adaptation. Molecular Ecology. 2012;21:1548–1566. doi: 10.1111/j.1365-294X.2012.05479.x. [DOI] [PubMed] [Google Scholar]
- Le Roux JJ, Brown GK, Byrne M, Ndlovu J, Richardson DM, Thompson GD. Wilson JRU. Phylogeographic consequences of different introduction histories of invasive Australian Acacia species and Paraserianthes lophantha (Fabaceae) in South Africa. Diversity and Distributions. 2011;17:861–871. [Google Scholar]
- Le Roux JJ, Richardson DM, Wilson JRU. Ndlovu J. Human usage in the native range may determine future genetic structure of an invasion: insights from Acacia pycnantha. BMC Ecology. 2013;13:8. doi: 10.1186/1472-6785-13-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends in Ecology & Evolution. 2002;17:386–391. [Google Scholar]
- Lee JE. Chown SL. Breaching the dispersal barrier to invasion: quantification and management. Ecological Applications. 2009;19:1944–1959. doi: 10.1890/08-2157.1. [DOI] [PubMed] [Google Scholar]
- Lee CE. Petersen CH. Genotype-by-environment interaction for salinity tolerance in the freshwater-invading copepod Eurytemora affinis. Physiological and Biochemical Zoology. 2002;75:335–344. doi: 10.1086/343138. [DOI] [PubMed] [Google Scholar]
- Leffler JA, James JJ, Monaco TA. Sheley RL. A new perspective on trait differences between native and invasive exotic plants. Ecology. 2014;95:298–305. doi: 10.1890/13-0102.1. [DOI] [PubMed] [Google Scholar]
- Lemmon AR, Emme SA. Lemmon EM. Anchored hybrid enrichment for massively high-throughput phylogenomics. Systematic Biology. 2012;61:727–744. doi: 10.1093/sysbio/sys049. [DOI] [PubMed] [Google Scholar]
- Levine MT, Eckert ML. Begun DJ. Whole-genome expression plasticity across tropical and temperate Drosophila melanogaster populations from eastern Australia. Molecular Biology and Evolution. 2011;28:249–256. doi: 10.1093/molbev/msq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC. Krakauer J. Distribution of gene frequency as a test of theory of selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl AL, Schrey AW, Richards CL. Martin LB. Patterns of DNA methylation throughout a range expansion of an introduced songbird. Integrative and Comparative Biology. 2013;53:351–358. doi: 10.1093/icb/ict007. [DOI] [PubMed] [Google Scholar]
- Litchman E. Invisible invaders: non-pathogenic invasive microbes in aquatic and terrestrial systems. Ecology Letters. 2010;13:1560–1572. doi: 10.1111/j.1461-0248.2010.01544.x. [DOI] [PubMed] [Google Scholar]
- Liu S, Li Y, Lu J, Su X, Tang M, Zhang R, Zhou L, et al. SOAPBarcode: revealing arthropod biodiversity through assembly of Illumina shotgun sequences of PCR amplicons. Methods in Ecology and Evolution. 2013;4:1142–1150. [Google Scholar]
- Lotterhos KE. Whitlock MC. Evaluation of demographic history and neutral parameterization on the performance of F-ST outlier tests. Molecular Ecology. 2014;23:2178–2192. doi: 10.1111/mec.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES. Costich DE. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PloS Genetics. 2013;9:e1003215. doi: 10.1371/journal.pgen.1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon AR, Barnes MA, Li F, Egan SP, Tanner CE, Ruggiero ST, Feder JL, et al. DNA-based species detection capabilities using laser transmission spectroscopy. Journal of the Royal Society Interface. 2012;10:20120637. doi: 10.1098/rsif.2012.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, et al. Target-enrichment strategies for next-generation sequencing. Nature Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- Marcussen T, Sandve SR, Heier L, Spannagl M. Pfeifer M The International Wheat Genome Sequencing Consortium. Jakobsen KS, et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345:1251788. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- Martin MD, Zimmer EA, Olsen MT, Foote AD, Gilbert MTP. Brush GS. Herbarium specimens reveal a historical shift in phylogeographic structure of common ragweed during native range disturbance. Molecular Ecology. 2014;23:1701–1716. doi: 10.1111/mec.12675. [DOI] [PubMed] [Google Scholar]
- Matesanz S. Valladares F. Ecological and evolutionary responses of Mediterranean plants to global change. Environmental and Experimental Botany. 2014;103:53–67. [Google Scholar]
- Matvienko M, Kozik A, Froenicke L, Lavelle D, Martineau B, Perroud B. Michelmore R. Consequences of normalizing transcriptomic and genomic libraries of plant genomes using a duplex-specific nuclease and tetramethylammonium chloride. PLoS ONE. 2013;8:e55913. doi: 10.1371/journal.pone.0055913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick H, Cursons R, Wilkins RJ. King CM. Location of a contact zone between Mus musculus domesticus and M. m. domesticus with M. m. castaneus mtDNA in southern New Zealand. Mammalian Biology. 2014;79:297–305. [Google Scholar]
- McDonald DB, Parchman TL, Bower MR, Hubert WA. Rahel FJ. An introduced and a native vertebrate hybridize to form a genetic bridge to a second native species. Proceedings of the National Academy of Sciences of the USA. 2008;105:10837–10842. doi: 10.1073/pnas.0712002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch MA, Spear D, Kleynhans EJ. Marais E. Uncertainty in invasive alien species listing. Ecological Applications. 2012;22:959–971. doi: 10.1890/11-1252.1. [DOI] [PubMed] [Google Scholar]
- McKinney LV, Nielsen LR, Collinge DB, Thomsen IM, Hansen JK. Kjaer ED. The ash dieback crisis: genetic variation in resistance can prove a long-term solution. Plant Pathology. 2014;63:485–499. [Google Scholar]
- McLean EH, Prober SM, Stock WD, Steane DA, Potts BM, Vaillancourt RE. Byrne M. Plasticity of functional traits varies clinally along a rainfall gradient in Eucalyptus tricarpa. Plant Cell and Environment. 2014;37:1440–1451. doi: 10.1111/pce.12251. [DOI] [PubMed] [Google Scholar]
- Meier R, Shiyang K, Vaidya G. Ng PK. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Systematic Biology. 2006;55:715–728. doi: 10.1080/10635150600969864. [DOI] [PubMed] [Google Scholar]
- Meier K, Hansen MM, Normandeau E, Mensberg KLD, Frydenberg J, Larsen PF, Bekkevold D. Bernatchez L. Local adaptation at the transcriptome level in brown trout: evidence from early life history temperature genomic reaction norms. PLoS ONE. 2014;9:13. doi: 10.1371/journal.pone.0085171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar MA, Byrne M. O'Sullivan W. Defining entities in the Acacia saligna (Fabaceae) species complex using a population genetics approach. Australian Journal of Botany. 2011;59:137–148. [Google Scholar]
- Millar MA, Byrne M, Nuberg IK. Sedgley M. High levels of genetic contamination in remnant populations of Acacia saligna from a genetically divergent planted stand. Restoration Ecology. 2012;20:260–267. [Google Scholar]
- Mobegi VA, Duffy CW, Amambua-Ngwa A, Loua KM, Laman E, Nwakanma DC, MacInnis B, et al. Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Molecular Biology and Evolution. 2014;31:1490–1499. doi: 10.1093/molbev/msu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molins MP, Corral JM, Aliyu OM, Koch MA, Betzin A, Maron JL. Sharbel TF. Biogeographic variation in genetic variability, apomixis expression and ploidy of St. John's wort (Hypericum perforatum) across its native and introduced range. Annals of Botany. 2014;113:417–427. doi: 10.1093/aob/mct268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody ML. Les DH. Geographic distribution and genotypic composition of invasive hybrid watermilfoil (Myriophyllum spicatum x M. sibiricum) populations in North America. Biological Invasions. 2007;9:559–570. [Google Scholar]
- Mooney HA. Hobbs RJ. Invasive Species in a Changing World. Washington, D. C: Island Press; 2000. [Google Scholar]
- Moran EV. Alexander JM. Evolutionary responses to global change: lessons from invasive species. Ecology Letters. 2014;17:637–649. doi: 10.1111/ele.12262. [DOI] [PubMed] [Google Scholar]
- Morris MRJ, Richard R, Leder EH, Barrett RDH, Aubin-Horth N. Rogers SM. Gene expression plasticity evolves in response to colonization of freshwater lakes in threespine stickleback. Molecular Ecology. 2014;23:3226–3240. doi: 10.1111/mec.12820. [DOI] [PubMed] [Google Scholar]
- Muhlfeld CC, Kovach RP, Jones LA, Al-Chokhachy R, Boyer MC, Leary RF, Lowe WH, et al. Invasive hybridization in a threatened species is accelerated by climate change. Nature Climate Change. 2014;4:620–624. [Google Scholar]
- Myburgh M, Chown SL, Daniels SR. Jansen van Vuuren B. Population structure, propagule pressure, and conservation biogeography in the sub-Antarctic: lessons from indigenous and invasive springtails. Diversity and Distributions. 2007;13:143–154. [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z, Loftus B, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves LG, Davis JM, Barbazuk WB. Kirst M. Whole-exome targeted sequencing of the uncharacterized pine genome. Plant Journal. 2013;75:146–156. doi: 10.1111/tpj.12193. [DOI] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, et al. Plant phenotypic plasticity in a changing climate. Trends in Plant Science. 2010;15:684–692. doi: 10.1016/j.tplants.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Nosil P, Gompert Z, Farkas TE, Comeault AA, Feder JL, Buerkle CA. Parchman TL. Genomic consequences of multiple speciation processes in a stick insect. Proceedings of the Royal Society B-Biological Sciences. 2012;279:5058–5065. doi: 10.1098/rspb.2012.0813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dowd DJ, Green PT. Lake PS. Invasional ‘meltdown’ on an oceanic island. Ecology Letters. 2003;6:812–817. [Google Scholar]
- Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Otto SP. Whitton J. Polyploid incidence and evolution. Annual Review of Genetics. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Pandit MK, Pocock MJO. Kunin WE. Ploidy influences rarity and invasiveness in plants. Journal of Ecology. 2011;99:1108–1115. [Google Scholar]
- Parchman TL, Gompert Z, Braun MJ, Brumfield RT, McDonald DB, Uy JAC, Zhang G, et al. The genomic consequences of adaptive divergence and reproductive isolation between species of manakins. Molecular Ecology. 2013;22:3304–3317. doi: 10.1111/mec.12201. [DOI] [PubMed] [Google Scholar]
- Peñalba JV, Smith LL, Tonione MA, Sass C, Hykin SM, Skipwith PL, McGuire JA, et al. Sequence capture using PCR-generated probes: a cost-effective method of targeted high-throughput sequencing for nonmodel organisms. Molecular Ecology Resources. 2014;14:1000–1010. doi: 10.1111/1755-0998.12249. [DOI] [PubMed] [Google Scholar]
- Pérez JE, Nirchio M, Alfonsi C. Muñoz C. The biology of invasions: The genetic adaptation paradox. Biological Invasions. 2006;8:1115–1121. [Google Scholar]
- Pfenninger M, Weigand A, Balint M. Klussmann-Kolb A. Misperceived invasion: the Lusitanian slug (Arion lusitanicus auct. non-Mabille or Arion vulgaris Moquin-Tandon 1855) is native to Central Europe. Evolutionary Applications. 2014;7:702–713. doi: 10.1111/eva.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BL. Shine R. An invasive species induces rapid adaptive change in a native predator: cane toads and black snakes in Australia. Proceedings of the Royal Society B. 2006;273:1545–1550. doi: 10.1098/rspb.2006.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BL, Brown GP, Webb JK. Shine R. Invasion and the evolution of speed in toads. Nature. 2006;439:803. doi: 10.1038/439803a. [DOI] [PubMed] [Google Scholar]
- Piaggio AJ, Engeman RM, Hopken MW, Humphrey JS, Keacher KL, Bruce WE. Avery ML. Detecting an elusive invasive species: a diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Molecular Ecology Resources. 2014;14:374–380. doi: 10.1111/1755-0998.12180. [DOI] [PubMed] [Google Scholar]
- Pontius JU, Mullikin JC, Smith DR, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, et al. Initial sequence and comparative analysis of the cat genome. Genome Research. 2007;17:1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porco D, Bedos A, Greenslade P, Janion C, Skarzynski D, Stevens MI, Jansen van Vuuren B. Deharveng L. Challenging species delimitation in Collembola: cryptic diversity among common springtails unveiled by DNA barcoding. Invertebrate Systematics. 2012;26:470477. [Google Scholar]
- Porco D, Decaëns T, Deharveng L, James SW, Skarzynski D, Erséus C, Butt KR, et al. Biological invasions in soil: DNA barcoding as a monitoring tool in a multiple taxa survey targeting European earthworms and springtails in North America. Biological Invasions. 2013;15:899–910. [Google Scholar]
- Præbel K, Gjelland KO, Salonen E. Amundsen PA. Invasion genetics of vendace (Coregonus albula (L.)) in the Inari-Pasvik watercourse: revealing the origin and expansion pattern of a rapid colonization event. Ecology and Evolution. 2013;3:1400–1412. doi: 10.1002/ece3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentis PJ. Pavasovic A. Understanding the genetic basis of invasiveness. Molecular Ecology. 2013;22:2366–2368. doi: 10.1111/mec.12277. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Wilson JRU, Dormontt EE, Richardson DM. Lowe AJ. Adaptive evolution in invasive species. Trends in Plant Science. 2008;13:288–294. doi: 10.1016/j.tplants.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Pickrell JK. Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Current Biology. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puth LM. Post DM. Studying invasion: have we missed the boat? Ecology Letters. 2005;8:715–721. [Google Scholar]
- Pyšek P, Richardson DM, Rejmánek M, Webster GL, Williamson M. Kirschner J. Alien plants in checklists and floras: towards better communication between taxonomists and ecologists. Taxon. 2004;53:131–143. [Google Scholar]
- Pyšek P, Jarošík V, Hulme PE, Pergl J, Hejda M, Schaffner U. Vilà M. A global assessment of invasive plant impacts on resident species, communities and ecosystems: the interaction of impact measures, invading species’ traits and environment. Global Change Biology. 2012;18:1725–1737. [Google Scholar]
- Pyšek P, Hulme PE, Meyerson LA, Smith GF, Boatwright JS, Crouch NR, Figueiredo E, et al. Hitting the right target: taxonomic challenges for, and of, plant invasions. AOB Plants. 2013;5:plt042. [Google Scholar]
- Ramirez KS, Leff JW, Barberán A, Bates ST, Betley J, Crowther TW, Kelly EF, et al. Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proceedings of the Royal Society B. 2014;281:20141988. doi: 10.1098/rspb.2014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall JM, Morse LE, Benton N, Hiebert R, Lu S. Killeffer T. The invasive species assessment protocol: a tool for creating regional and national lists of invasive nonnative plants that negatively impact biodiversity. Invasive Plant Science and Management. 2008;1:36–49. [Google Scholar]
- Rasic G, Filipovic I, Weeks AR. Hoffmann AA. Genome-wide SNPs lead to strong signals of geographic structure and relatedness patterns in the major arbovirus vector, Aedes aegypti. BMC Genomics. 2014;15:12. doi: 10.1186/1471-2164-15-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HC, Maddison BC, Middleditch DJ, Patmore JRM. Gough KC. The detection of aquatic animal species using environmental DNA – a review of eDNA as a survey tool in ecology. Journal of Applied Ecology. 2014;51:1450–1459. [Google Scholar]
- Ricciardi A, Hoopes MF, Marchetti MP. Lockwood JL. Progress toward understanding the ecological impacts of nonnative species. Ecological Monographs. 2013;83:263–282. [Google Scholar]
- Richards CL, Schrey AW. Pigliucci M. Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecology Letters. 2012;15:1016–1025. doi: 10.1111/j.1461-0248.2012.01824.x. [DOI] [PubMed] [Google Scholar]
- Richardson DM. Fifty Years of Invasion Ecology. The Legacy of Charles Elton. Oxford: Wiley-Blackwell; 2011. [Google Scholar]
- Richardson DM. Pyšek P. Plant invasions: merging the concepts of species invasiveness and community invasibility. Progress in Physical Geography. 2006;30:409–431. [Google Scholar]
- Richardson DM. Pyšek P. Naturalization of introduced plants: ecological drivers of biogeographical patterns. New Phytologist. 2012;196:383–396. doi: 10.1111/j.1469-8137.2012.04292.x. [DOI] [PubMed] [Google Scholar]
- Richardson DM. Ricciardi A. Misleading criticisms of invasion science: a field guide. Diversity and Distributions. 2013;19:1461–1467. [Google Scholar]
- Richardson DM, Pyšek P. Carlton JT. A compendium of essential concepts and terminology in invasion ecology. In: Richardson DM, editor; Fifty years of invasion ecology. The legacy of Charles Elton. Oxford: Wiley-Blackwell; 2011. pp. 409–420. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, et al. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Kim S-C, Randell RA, Whitney KD, Gross BL, Lexer C. Clay K. Hybridization and the colonization of novel habitats by annual sunflowers. Genetica. 2007;129:149–165. doi: 10.1007/s10709-006-9011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rignot E, Velicogna I, van den Broeke MR, Monaghan A. Lenaerts JTM. Acceleration of the contribution of the Greenland and Antarctic ice sheets to sea level rise. Geophysical Research Letters. 2011;38:L05503. [Google Scholar]
- Rius M. Darling JA. How important is intraspecific genetic admixture to the success of colonising populations? Trends in Ecology & Evolution. 2014;29:233–242. doi: 10.1016/j.tree.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Robertson K, Goldberg EE. Igic B. Comparative evidence for the correlated evolution of polyploidy and self-compatibility in Solanaceae. Evolution. 2011;65:139–155. doi: 10.1111/j.1558-5646.2010.01099.x. [DOI] [PubMed] [Google Scholar]
- Rocha LA, Bernal MA, Gaither MR. Alfaro ME. Massively parallel DNA sequencing: the new frontier in biogeography. Frontiers of Biogeography. 2013;5:67–77. [Google Scholar]
- Rohfritsch A, Bierne N, Boudry P, Heurtebise S, Cornette F. Lapegue S. Population genomics shed light on the demographic and adaptive histories of European invasion in the Pacific oyster, Crassostrea gigas. Evolutionary Applications. 2013;6:1064–1078. doi: 10.1111/eva.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman J. Darling JA. Paradox lost: genetic diversity and the success of aquatic invasions. Trends in Ecology and Evolution. 2007;22:454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Rowe KC, Singhal S, Macmanes MD, Ayroles JF, Morelli TL, Rubidge EM, Bi K. Moritz CC. Museum genomics: low-cost and high-accuracy genetic data from historical specimens. Molecular Ecology Resources. 2011;11:1082–1092. doi: 10.1111/j.1755-0998.2011.03052.x. [DOI] [PubMed] [Google Scholar]
- Roy HE, Roy DB. Roques A. Inventory of terrestrial alien arthropod predators and parasites established in Europe. BioControl. 2011;56:477–504. [Google Scholar]
- Salmon A, Ainouche ML. Wendel JF. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Molecular Ecology. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- Sampson JF. Byrne M. Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba. Molecular Ecology. 2008;17:2769–2781. doi: 10.1111/j.1365-294X.2008.03779.x. [DOI] [PubMed] [Google Scholar]
- Sax DF, Stachowicz JJ. Gaines SD. Sinauer Associates Inc. Massachusetts: Publishers; 2005. Species invasions. Insights into ecology, evolution, and biogeography. [Google Scholar]
- Scheffer SJ, Lewis ML. Joshi RC. DNA barcoding applied to invasive leafminers (Diptera:Agromyzidae) in the Philippines. Annals of the Entomological Society of America. 2006;99:204–210. [Google Scholar]
- Schierenbeck KA. Ellstrand NC. Hybridization and the evolution of invasiveness in plants and other organisms. Biological Invasions. 2009;11:1093–1105. [Google Scholar]
- Schilthuizen M. Kellermann V. Contemporary climate change and terrestrial invertebrates: evolutionary versus plastic changes. Evolutionary Applications. 2014;7:56–67. doi: 10.1111/eva.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, Alix A, et al. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes RJ, Settele J, Betts R, Bunn S, Leadley P, Nepstad D, Overpeck J, et al. IPCC Working Group II Assessment Report 5 Chapter 4 Terrestrial and Inland Water Systems. Geneva: Intergovernmental Panel on Climate Change; 2014. [Google Scholar]
- Schwartz MA, Hellmann JJ, McLachlan JM, Sax DF, Borevitz JO, Brennan J, Camacho AE, et al. Managed relocation: integrating the scientific, regulatory, and ethical challenges. BioScience. 2012;62:732–743. [Google Scholar]
- Scoville AG. Pfrender ME. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proceedings of the National Academy of Sciences of the USA. 2010;107:4260–4263. doi: 10.1073/pnas.0912748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scriber JM. Impacts of climate warming on hybrid zone movement: geographically diffuse and biologically porous “species borders”. Insect Science. 2011;18:121–159. [Google Scholar]
- Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, et al. Fundamentals of microbial community resistance and resilience. Frontiers in Microbiology. 2012;3:47. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirk RY, Hamrick JL, Zhang C. Qiang S. Patterns of genetic diversity reveal multiple introductions and recurrent founder effects during range expansion in invasive populations of Geranium carolinianum (Geraniaceae) Heredity. 2014;112:497–507. doi: 10.1038/hdy.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora M, Carpenter ML, Moreno-Estrada A, Henn BM, Underhill PA, Sanchez-Quinto F, Zara I, et al. Population genomic analysis of ancient and modern genomes yields new insights into the genetic ancestry of the Tyrolean iceman and the genetic structure of Europe. PloS Genetics. 2014;10:12. doi: 10.1371/journal.pgen.1004353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simberloff D. The role of propagule pressure in biological invasions. Annual Review of Ecology Evolution and Systematics. 2009;40:81–102. [Google Scholar]
- Simberloff D, Martin JL, Genovesi P, Maris V, Wardle DA, Aronson J, Courchamp F, et al. Impacts of biological invasions: what's what and the way forward. Trends in Ecology and Evolution. 2013;28:58–66. doi: 10.1016/j.tree.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Smith MA. Fisher BL. Invasions, DNA barcodes, and rapid biodiversity assessment using ants of Mauritius. Frontiers in Zoology. 2009;6:31. doi: 10.1186/1742-9994-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. Haigh J. The hitch-hiking effect of a favourable gene. Genetics Research. 2007;89:391–403. doi: 10.1017/S0016672308009579. [DOI] [PubMed] [Google Scholar]
- Smith CD, Zimin A, Holt C, Abouheif E, Benton R, Cash E, Croset V, et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile. Proceedings of the National Academy of Sciences of the USA. 2011;108:5673–5678. doi: 10.1073/pnas.1008617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Bernatchez L. Beheregaray LB. RNA-seq analysis reveals extensive transcriptional plasticity to temperature stress in a freshwater fish species. BMC Genomics. 2013;14:12. doi: 10.1186/1471-2164-14-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sork VL, Aitken SN, Dyer RJ, Eckert AJ, Legendre P. Neale DB. Putting the landscape into the genomics of trees: approaches for understanding local adaptation and population responses to changing climate. Tree Genetics and Genomes. 2013;9:901–911. [Google Scholar]
- Spear D, Foxcroft LC, Bezuidenhout H. McGeoch MA. Human population density explains alien species richness in protected areas. Biological Conservation. 2013;159:137–147. [Google Scholar]
- Stachowicz JJ, Terwin JR, Whitlatch RB. Osman RW. Linking climate change and biological invasions: ocean warming facilitates nonindigenous species invasions. Proceedings of the National Academy of Sciences of the USA. 2002;99:15497–15500. doi: 10.1073/pnas.242437499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steane DA, Potts BM, McLean E, Prober SM, Stock WD, Vaillancourt RE. Byrne M. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Molecular Ecology. 2014;23:2500–2513. doi: 10.1111/mec.12751. [DOI] [PubMed] [Google Scholar]
- Stewart CN, Jr, Tranel PJ, Horvath DP, Anderson JV, Rieseberg LH, Westwood JH, Mallory-Smith CA, et al. Evolution of weediness and invasiveness: charting the course for weed genomics. Weed Science. 2009;57:451–462. [Google Scholar]
- Strauss SY, Lau JA. Carroll SP. Evolutionary responses of natives to introduced species: what do introductions tell us about natural communities? Ecology Letters. 2006;9:354–371. doi: 10.1111/j.1461-0248.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- Stukely MJC. Crane CE. Genetically based resistance of Eucalyptus marginata to Phytophthora cinnamomi. Phytopathology. 1994;84:650–656. [Google Scholar]
- Sutherland WJ, Clout M, Côté IM, Daszak P, Depledge MH, Fellman L, Fleishman E, et al. A horizon scan of global conservation issues for 2010. Trends in Ecology and Evolution. 2010;25:1–7. doi: 10.1016/j.tree.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sutherland WJ, Aveling R, Bennun L, Chapman E, Clout M, Côté IM, Depledge MH, et al. A horizon scan of global conservation issues for 2012. Trends in Ecology and Evolution. 2012;27:12–18. doi: 10.1016/j.tree.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Coissac E, Hajibabaei M. Rieseberg LH. Environmental DNA. Molecular Ecology. 2012a;21:1789–1793. doi: 10.1111/j.1365-294X.2012.05542.x. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Coissac E, Pompanon F, Brochmann C. Willerslev E. Towards next-generation biodiversity assessment using DNA-metabarcoding. Molecular Ecology. 2012b;21:2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x. [DOI] [PubMed] [Google Scholar]
- Takahara T, Minamoto T. Doi H. Using environmental DNA to estimate distribution of an invasive fish species in ponds. PLoS ONE. 2013;8:e56584. doi: 10.1371/journal.pone.0056584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatem AJ. The worldwide airline network and the dispersal of exotic species: 2007-2010. Ecography. 2009;32:94–102. doi: 10.1111/j.1600-0587.2008.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatem AJ. Hay SI. Climatic similarity and biological exchange in the worldwide airline transportation network. Proceedings of the Royal Society of London B. 2007;274:1489–1496. doi: 10.1098/rspb.2007.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HR. Harris WE. An emergent science on the brink of irrelevance: a review of the past 8 years of DNA barcoding. Molecular Ecology Resources. 2012;12:377–388. doi: 10.1111/j.1755-0998.2012.03119.x. [DOI] [PubMed] [Google Scholar]
- Taylor SA, White TA, Hochachka WM, Ferretti V, Curry RL. Lovette I. Climate-mediated movement of an avian hybrid zone. Current Biology. 2014;24:671–676. doi: 10.1016/j.cub.2014.01.069. [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, van Heerwaarden B, Johnson TK, Hoffmann AA. Sgrò CM. New levels of transcriptome complexity at upper thermal limits in wild Drosophila revealed by exon expression analysis. Genetics. 2013;195:809–830. doi: 10.1534/genetics.113.156224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshler MP, Dernovici SA, Ditommaso A, Coderre D. Watson AK. A novel device for the collection, storage, transport, and delivery of beneficial insects, and its application to Ophraella communa (Coleoptera: Chrysomelidae) Biocontrol Science and Technology. 2004;14:347–357. [Google Scholar]
- Thompson JD. The biology of an invasive plant - what makes Spartina anglica so successful. BioScience. 1991;41:393–401. [Google Scholar]
- Thompson GD, Robertson MP, Webber BL, Richardson DM, Le Roux JJ. Wilson JRU. Predicting the subspecific identity of invasive species using distribution models: Acacia saligna as an example. Diversity and Distributions. 2011;17:1001–1014. [Google Scholar]
- Thompson GD, Bellstedt DU, Byrne M, Millar MA, Richardson DM, Wilson JRU. Le Roux JJ. Cultivation shapes genetic novelty in a globally important invader. Molecular Ecology. 2012;21:3187–3199. doi: 10.1111/j.1365-294X.2012.05601.x. [DOI] [PubMed] [Google Scholar]
- Thornton KR, Jensen JD, Becquet C. Andolfatto P. Progress and prospects in mapping recent selection in the genome. Heredity. 2007;98:340–348. doi: 10.1038/sj.hdy.6800967. [DOI] [PubMed] [Google Scholar]
- Tian C, Gregersen PK. Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Human Molecular Genetics. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treasure AM. Chown SL. Antagonistic effects of biological invasion and temperature change on body size of island ectotherms. Diversity and Distributions. 2014;20:202–213. [Google Scholar]
- Treier UA, Broennimann O, Normand S, Guisan A, Schaffner U, Steinger T. Mueller-Schaerer H. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology. 2009;90:1366–1377. doi: 10.1890/08-0420.1. [DOI] [PubMed] [Google Scholar]
- Turchin MC, Chiang CWK, Palmer CD, Sankararaman S, Reich D. Hirschborn JN GIANT Consortium. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nature Genetics. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez-Moreno M, Quintal-Lazama C, Gómez-Lazano R. García-Rivas MdC. Monitoring of an alien invasion: DNA barcoding and the identification of Lionfish and their prey on coral reefs of the Mexican Caribbean. PLoS ONE. 2012;7:e36636. doi: 10.1371/journal.pone.0036636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valéry L, Fritz H, Lefeuvre J-C. Simberloff D. In search of a real definition of the biological invasion phenomenon itself. Biological Invasions. 2008;10:1345–1351. [Google Scholar]
- Van der Putten WH, Klirononos JN. Wardle DA. Microbial ecology of biological invasions. The ISME Journal. 2007;1:28–37. doi: 10.1038/ismej.2007.9. [DOI] [PubMed] [Google Scholar]
- Van Kleunen M, Dawson W, Schlaepfer D, Jeschke JM. Fischer M. Are invaders different? A conceptual framework of comparative approaches for assessing determinants of invasiveness. Ecology Letters. 2010;13:947–958. doi: 10.1111/j.1461-0248.2010.01503.x. [DOI] [PubMed] [Google Scholar]
- Van Wilgen BW. The evolution of fire and invasive alien plant management practices in fynbos. South African Journal of Science. 2009;105:335–342. [Google Scholar]
- Vandepitte K, Honnay O, Mergeay J, Breyne P, Roldán-Ruiz I. De Meyer T. SNP discovery using Paired-End RAD-tag sequencing on pooled genomic DNA of Sisymbrium austriacum (Brassicaceae) Molecular Ecology Resources. 2012;13:269–275. doi: 10.1111/1755-0998.12039. [DOI] [PubMed] [Google Scholar]
- Vandepitte K, De Meyer T, Helsen K, Van Acker K, Roldan-Ruiz I, Mergeay J. Honnay O. Rapid genetic adaptation precedes the spread of an exotic plant species. Molecular Ecology. 2014;23:2157–2164. doi: 10.1111/mec.12683. [DOI] [PubMed] [Google Scholar]
- Veldtman R, Lado TF, Botes A, Proches S, Timm AE, Geertsema H. Chown SL. Creating novel food webs on introduced Australian acacias: indirect effects of galling biological control agents. Diversity and Distributions. 2011;17:958–967. [Google Scholar]
- Vellinga EC, Wolfe BE. Pringle A. Global patterns of ectomycorrhizal introductions. New Phytologist. 2009;181:960–973. doi: 10.1111/j.1469-8137.2008.02728.x. [DOI] [PubMed] [Google Scholar]
- Vorsino AE, Wieczorek AM, Wright MG. Messing RH. Using evolutionary tools to facilitate the prediction and prevention of host-based differentiation in biological control: a review and perspective. Annals of Applied Biology. 2012;160:204–216. [Google Scholar]
- Walther G-R, Roques A, Hulme PE, Sykes MT, Pyšek P, Kühn I, Zobel M, et al. Alien species in a warmer world: risks and opportunities. Trends in Ecology and Evolution. 2009;24:686–693. doi: 10.1016/j.tree.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M. Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colbourne JK, Clark AG. Werren JH. Function and evolution of DNA methylation in Nasonia vitripennis. PloS Genetics. 2013;9:e1003872. doi: 10.1371/journal.pgen.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Webber BL. Scott JK. Rapid global change: implications for defining natives and aliens. Global Ecology and Biogeography. 2012;21:305–311. [Google Scholar]
- Weber E. Schmid B. Latitudinal population differentiation in two species of Solidago (Asteraceae) introduced into Europe. American Journal of Botany. 1998;85:1110–1121. [PubMed] [Google Scholar]
- Weeks AR, Fripp YJ. Hoffmann AA. Genetic structure of Halotydeus destructor and Penthaleus major populations in Victoria (Acari: Penthaleidae) Experimental and Applied Acarology. 1995;19:633–646. [Google Scholar]
- White TA, Perkins SE, Heckel G. Searle JB. Adaptive evolution during an ongoing range expansion: the invasive bank vole (Myodes glareolus) in Ireland. Molecular Ecology. 2013;22:2971–2985. doi: 10.1111/mec.12343. [DOI] [PubMed] [Google Scholar]
- Whitney KD. Gabler CA. Rapid evolution in introduced species, ‘invasive traits’ and recipient communities: challenges for predicting invasive potential. Diversity and Distributions. 2008;14:569–580. [Google Scholar]
- Willi Y, Van Buskirk J. Hoffmann AA. Limits to the adaptive potential of small populations. Annual Review of Ecology, Evolution and Systematics. 2006;37:433–458. [Google Scholar]
- Williams CK. Moore RJ. Phenotypic adaptation and natural selection in the wild rabbit, Oryctolagus cuniculus, in Australia. Journal of Animal Ecology. 1989;58:495–507. [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB. Rieseberg LH. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences of the USA. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI. Gaut BS. Molecular population genetics and the search for adaptive evolution in plants. Molecular Biology and Evolution. 2005;22:506–519. doi: 10.1093/molbev/msi035. [DOI] [PubMed] [Google Scholar]
- Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, Ingram KK, et al. The genome of the fire ant Solenopsis invicta. Proceedings of the National Academy of Sciences of the USA. 2011;108:5679–5684. doi: 10.1073/pnas.1009690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Shimada N. Tago Y. Detection of hybrids between masu salmon Oncorhynchus masou masou and amago salmon O. m. ishikawae occurred in the Jinzu River using a random amplified polymorphic DNA technique. Fisheries Science. 2005;71:320–326. [Google Scholar]
- Yeaman S. Whitlock MC. The genetic architecture of adaptation under migration-selection balance. Evolution. 2011;65:1897–1911. doi: 10.1111/j.1558-5646.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Yeaman S, Hodgins KA, Suren H, Nurkowski KA, Rieseberg LH, Holliday JA. Aitken SN. Conservation and divergence of gene expression plasticity following c. 140 million years of evolution in lodgepole pine (Pinus contorta) and interior spruce (Picea glauca x Picea engelmannii. New Phytologist. 2014;203:578–591. doi: 10.1111/nph.12819. [DOI] [PubMed] [Google Scholar]
- Yoccoz N. The future of environmental DNA in ecology. Molecular Ecology. 2012;21:2031–2038. doi: 10.1111/j.1365-294X.2012.05505.x. [DOI] [PubMed] [Google Scholar]
- Yoccoz NG, Brathen KA, Gielly L, Haile J, Edwards ME, Goslar T, Von Stedingk H, et al. DNA from soil mirrors plant taxonomic and growth form diversity. Molecular Ecology. 2012;21:3647–3655. doi: 10.1111/j.1365-294X.2012.05545.x. [DOI] [PubMed] [Google Scholar]
- You M, Yue Z, He W, Yang X, Yang G, Xie M, Zhan D, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nature Genetics. 2013;45:220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- Yu DW, Ji Y, Emerson BC, Wang X, Ye C, Yang C. Ding Z. Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods in Ecology and Evolution. 2012;3:613–623. [Google Scholar]
- Yun L, Larson SR, Mott IW, Jensen KB. Staub JE. Genetic control of rhizomes and genomic localization of a major-effect growth habit QTL in perennial wildrye. Molecular Genetics and Genomics. 2014;289:383–397. doi: 10.1007/s00438-014-0817-5. [DOI] [PubMed] [Google Scholar]
- Zhang W, Fan X, Zhu S, Zhao H. Fu L. Species-specific identification from incomplete sampling: applying DNA barcodes to monitoring invasive Solanum plants. PLoS ONE. 2013a;8:e55927. doi: 10.1371/journal.pone.0055927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kapli P, Pavlidis P. Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013b;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-Y, Fischer M, Colot V. Bossdorf O. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytologist. 2013c;197:314–322. doi: 10.1111/nph.12010. [DOI] [PubMed] [Google Scholar]
- Zhang B, Edwards O, Kang L. Fuller S. A multi-genome analysis approach enables tracking of the invasion of a single Russian wheat aphid (Diuraphis noxia) clone throughout the New World. Molecular Ecology. 2014;23:1940–1951. doi: 10.1111/mec.12714. [DOI] [PubMed] [Google Scholar]
- Zhu YY, Chen HR, Fan JH, Wang YY, Li Y, Chen JB, Fan JX, et al. Genetic diversity and disease control in rice. Nature. 2000;406:718–722. doi: 10.1038/35021046. [DOI] [PubMed] [Google Scholar]
- Ziska LH, Blumenthal DM, Runion GB, Hunt ER. Diaz-Soltero H. Invasive species and climate change: an agronomic perspective. Climatic Change. 2011;105:13–42. [Google Scholar]