Abstract
Proteinase 3 (PR3) and myeloperoxidase (MPO) are two major autoantigens in patients with vasculitis with ANCA. The genes encoding these autoantigens are abnormally expressed in peripheral granulocytes of patients with active ANCA-associated vasculitis. This study provides evidence that this transcriptional dysregulation results in a variety of mRNA processing events from the PRTN3 gene locus. In addition to elevated levels of PR3 message, leukocyte RNA from patients contained PR3 transcripts with an alternative 3′ untranslated region. Furthermore, we detected usage of an alternative transcription start site within intron 1 of the PRTN3 gene locus that coincided with active disease (odds ratio, 3.3; 95% confidence interval, 1.3 to 8.4; P=0.01). This promoter may be developmentally regulated, because it was active in normal human bone marrow, multiple leukemia cell lines, MCF-7 cells, and subjects after GM-CSF treatment but not subjects with a neutrophil left shift. This transcript, which lacks exon 1 of PRTN3, encodes a 24-kD protein (p24PR3/MBN) with a sequence similar to that previously described for myeloblastin. Notably, PR3, p24PR3/MBN, and MPO were synthesized in cultured neutrophils from patients with active ANCA-associated vasculitis, indicating that increased transcription results in newly synthesized autoantigens in peripheral neutrophils of patients. The synthesis of p24PR3/MBN seems to expand the autoantigen repertoire, because immunoblots showed that sera from patients recognized p24PR3/MBN. These findings emphasize the importance of transcriptional dysregulation of the autoantigen in autoimmune disease.
Keywords: ANCA, GN, transcription regulation
Altered gene expression profiles are often associated with disease. In autoimmune diseases, dysregulated expression can be a salient characteristic. Leukocyte gene expression signatures differ among patients with rheumatoid arthritis, SLE, and ANCA-associated vasculitis (AAV).1,2 Specifically, patients with AAV frequently have increased expression of the two principle autoantigen genes proteinase 3 (PR3) and myeloperoxidase (MPO),3,4 and this elevated expression is associated with active disease.4 Fundamental to the pathogenesis of AAV, ANCA binds these autoantigens on the surface of neutrophils, causing neutrophil activation with degranulation of inflammatory agents that injure neighboring vascular endothelium.5–8 Although antibodies to PR3 and MPO play a pivotal role in AAV, understanding the regulation and consequences of dysregulation of the genes encoding PR3 and MPO may provide unique insight into the disease process.
PR3 and MPO genes are normally silenced in mature circulating neutrophils, but the observation that they are expressed in peripheral leukocytes of patients with ANCA suggested that gene silencing is disrupted. Indeed, histone modifications involved in transcriptional repression were depleted at these loci, showing an association between increased transcription of PR3 and MPO and lack of epigenetic silencing in patients with ANCA.9 Although this observation implicates epigenetic processes in the regulation of autoantigen genes, upstream events that disrupt epigenetic silencing or act directly to modulate expression remain ill-defined. In addition, the consequence of transcriptional dysregulation, beyond increased or persistent expression of PR3 and MPO transcripts, is an open question.
The studies here focus on consequences of transcriptionally active loci in AAV. Aberrant and alternative transcripts have been detected in disease settings, such as cancer10 and SLE.11,12 The presence of alternative transcripts from the autoantigen gene loci would be especially informative in the setting of an autoimmune disease, because alternative transcripts could be templates for protein isoforms of autoantigens other than their archetypal form. Interestingly, a prior report indicates that the PRTN3 locus encodes two highly homologous proteins, PR3 and myeloblastin (MBN).13 These two proteins may be functionally different, with PR3 having antimicrobial functions14 and MBN promoting myeloid proliferation,15 and structurally distinct on the basis of differences in their N-terminal amino acid sequence.16,17 The studies here characterize aberrantly expressed transcripts and investigate whether they are translated in peripheral neutrophils, which implies that the presence of increased PR3 and MPO message would result in newly expressed autoantigens.
Results
Northern Blot Analysis Indicates That PR3 Transcripts of Abnormal Size Are Expressed in Patients with AAV
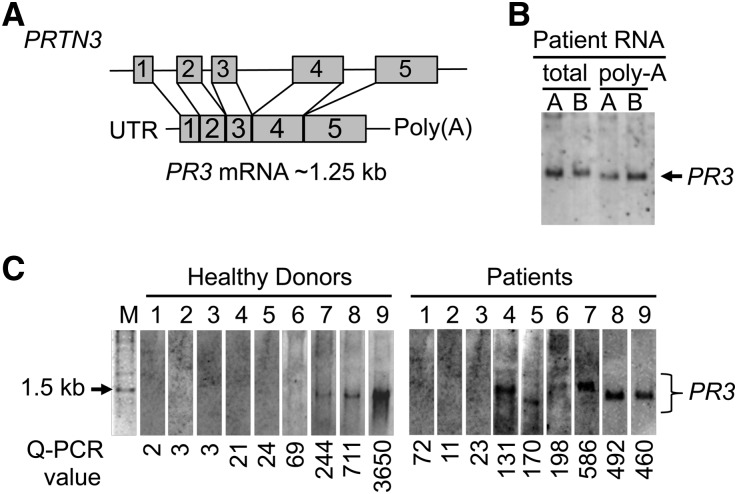
Northern blotting was performed to examine whether increased expression of PR3 mRNA in disease represented the canonical transcript of predicted size [1023 nucleotides plus poly(A) tail] (Figure 1A) or alternate forms, including splice variants. Data indicated that four of nine patients expressed at least one irregular PR3 transcript, and of four patients, three patients expressed a variant larger (100–400 additional nucleotides) than the currently annotated transcript (Figure 1C). The robust bands corresponding to PR3 mRNA by Northern blot were coincident with high TaqMan quantitative PCR values (Figure 1C). Unexpectedly, high TaqMan values for PR3 were measured in three of nine age-matched healthy donors’ samples, and Northern analysis of these samples detected a single band corresponding to the PR3 transcript. Importantly, even with extremely elevated levels of total PR3 mRNA, the PR3 transcript in healthy donors migrated at the expected approximately 1.25-kb size, suggesting that increased expression alone is not sufficient to result in alternative forms of PR3 message (Figure 1C).
Figure 1.
Northern blot analysis for PR3 transcripts in leukocyte RNA from healthy donors and patients with ANCA. (A) Schematic of PRTN3 locus with 5 exons and spliced PR3 mRNA with predicted size: 1.25 kb including an approximately 250-nucleotide polyA tail. (B) PR3 probe, amplified from nucleotide 336 to nucleotide 627 of PR3 cDNA, was tested on total RNA and polyA mRNA from patient leukocytes. (C) Total leukocyte RNA (15 µg) was probed for PR3-specific transcripts from healthy donors (lanes 1–9 in left panel) and patients with AAV (lanes 1–9 in right panel). C is a composite of four separate experiments. Lanes 4, 6, and 7 in right panel contain transcripts larger than the expected size for PR3 message. Relative levels of PR3 message as determined by TaqMan quantitative RT-PCR (Q-PCR) are below each lane. Values were on the basis of a standard curve generated from values of high PR3-expressing THP-1 cells versus PR3-null Jurkat cells. Sequences of primer and probe are listed in Supplemental Table 1. Forward primer resides in exon 4, and reverse primer resides in exon 5. M, 1 kb DNA molecular weight marker.
Secondary Distal Polyadenylation Site Used at the PRTN3 Locus
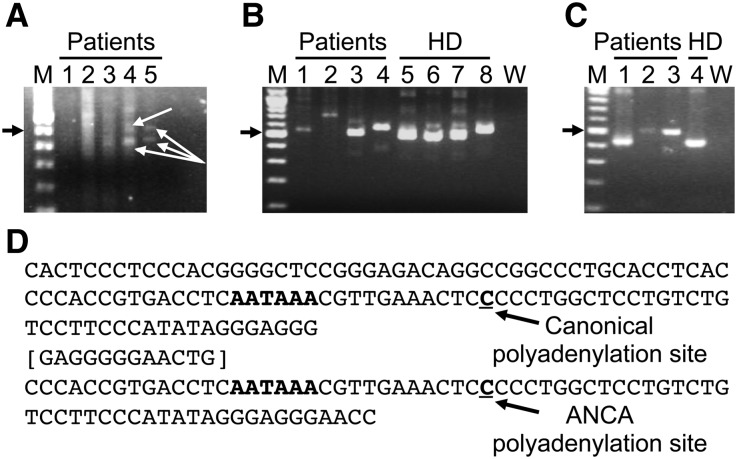
We used 3′ rapid amplification of cDNA ends (RACE) to explore whether different sized PR3 transcripts resulted from differences in the 3′ untranslated region (UTR) for PR3. Data from multiple sets of forward primers found variations in 3′UTR length in patients with AAV (Figure 2). Two isoforms of PR3 mRNA were identified and sequenced from two of three patients with ANCA (Figure 2A). Nested PCR confirmed the presence of the larger 3′UTR in three other patients (Figure 2B, lane 4 and C, lanes 2 and 3) and one healthy donor (Figure 2B, lane 8). Sequence of the lower band aligned with the published PR3 transcript 3′UTR. The larger PCR amplicon contained a 68-bp repeat with a unique 12-bp spacer (Figure 2D) that aligned with the downstream genomic sequence of PRTN3. Inclusion of this genomic sequence provided a distal polyadenylation site used in mRNA processing in patients with AAV. Changes in the 3′UTR of the PR3 mRNA may alter binding of crucial cis-acting proteins or microRNAs that affect mRNA half-life and ultimately, protein output.
Figure 2.
3′RACE amplification of PRTN3 transcripts from leukocyte RNA of patients with AAV and healthy donors. (A) Leukocyte RNA from three different patients with AAV (patient 1 at two different times in disease [lanes 1 and 2], patient 2 at two different times in disease [lanes 3 and 4], and patient 3 [lane 5]) was analyzed by 3′RACE, and PCR was performed with the following primers (listed in Supplemental Table 1): first round used PR3 634 exon 5 FOR and Qo, and second round used PR3 634 exon 5 FOR and Qi. Arrows indicate bands that were sequenced from patients 2 and 3. (B) 3′RACE was performed on leukocyte RNA from four patients with AAV (lanes 1–4) and four age-matched healthy donors (HDs; lanes 5–8). PCR was performed with the following primers: first round used PR3 374 exon 3 FOR and Qo, and second round used PR3 547 exon 4 FOR and Qi. PCR products were cloned and sequenced; no sequences were obtained from patient samples used in lanes 1 and 2. (C) The extended 3′UTR of the PRTN3 transcript was confirmed in an additional 3′RACE experiment on three patients with AAV (lanes 1–3) and one healthy donor (lane 4) with the following primers: first round used PR3 547 exon 4 FOR and Qo, and second round used PR3 617 exon 5 FOR and Qi. PCR products were cloned and sequenced. (D) Sequence of the 3′UTR of PRTN3 transcripts used either the canonical polyadenylation site or a downstream polyadenylation site that is within a 68-nucleotide repeat separated by a 12-nucleotide spacer (brackets). The sequence obtained from the larger band, corresponding to the extended 3′UTR of the PRTN3 transcript, indicates an alternative polyadenylation site in 5 of 10 patients with AAV (A, lanes 4 and 5; B, lane 4; and C, lanes 2 and 3) and 1 of 4 healthy donors (B, lane 8). Solid arrow indicates the 500-bp band. M, 100 base pair molecular weight marker. W, water control.
Identification of a Surrogate Promoter within Intron 1 of PRTN3
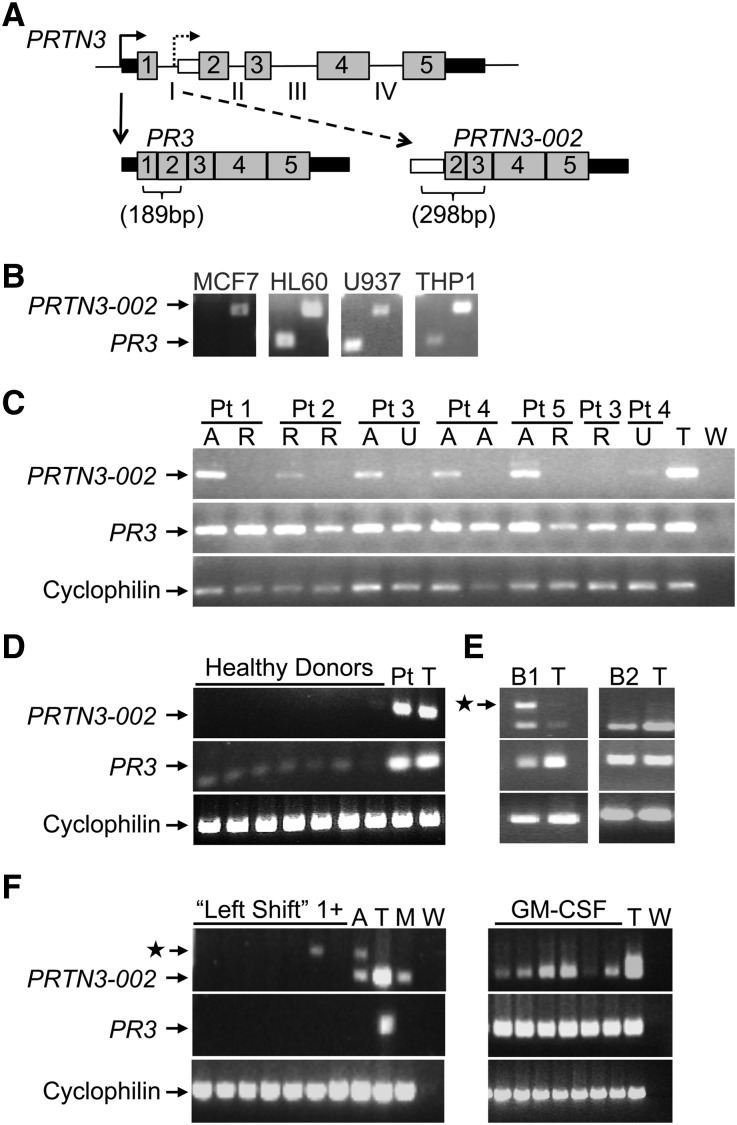
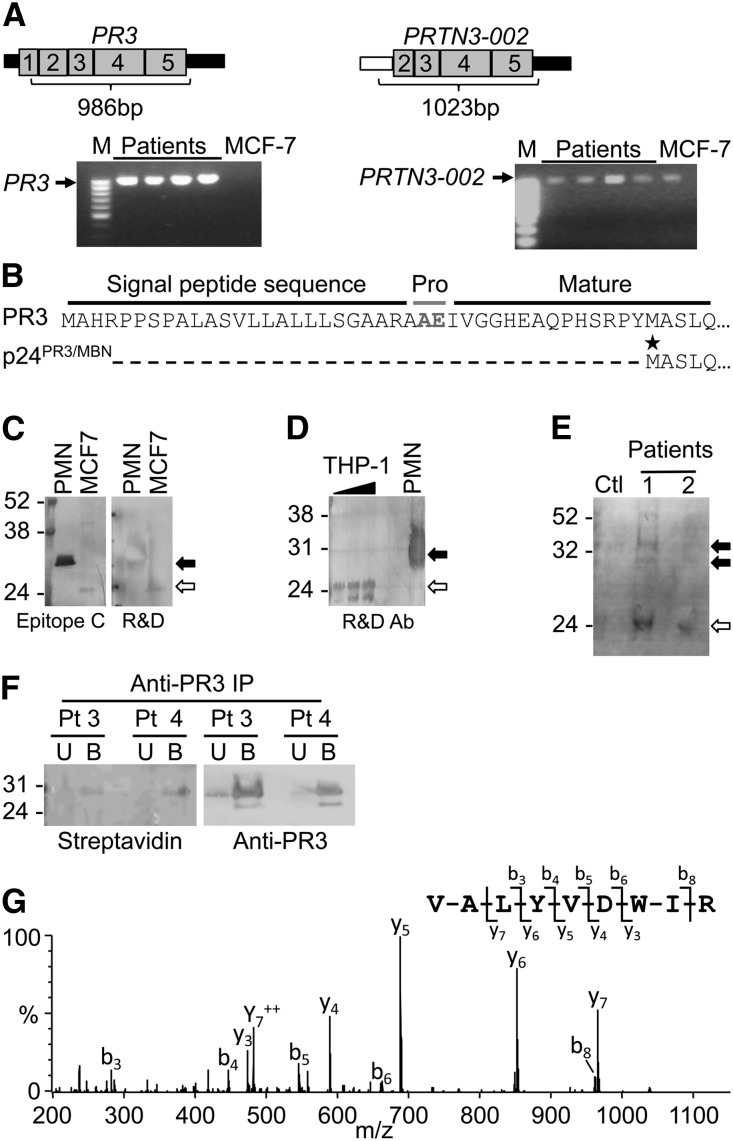
After identifying PR3 mRNA with an extended 3′UTR, we questioned whether alternative forms of PR3 transcripts had been annotated in published databases. Expressed sequence tags with homology to PR3 mRNA but lacking exon 1 of PRTN3 were identified in MCF-7 cells, and data from the publicly available Burge laboratory RNA sequencing project revealed sequence reads that aligned only with exons 2–5 of PRTN3 (Supplemental Figure 4). PCR primers designed to distinguish between the alternative (PRTN3–002) and canonical (PR3) transcripts (Figure 3A) were validated using MCF-7 cells negative for PR3 but positive for PRTN3–002 and monocytic cell lines HL-60, THP-1, and U937 positive for both transcripts (Figure 3B). Paired samples from five patients with AAV were tested for PR3 and PRTN3–002 transcripts (Figure 3C). All samples were positive for the PR3 message. The presence of PRTN3–002 transcript fluctuates during disease course, and it seems to be present more frequently during active disease. Healthy donors were negative for PRTN3–002, although slightly positive for PR3 transcripts (Figure 3D). Both PRTN3–002 and PR3 were detected in human bone marrow and subjects who underwent GM-CSF treatment (Figure 3, E and F). The serum levels of GM-CSF, measured by ELISA and listed in Supplemental Table 1, were not different between patients positive or negative for the PRTN3–002 transcript. Subjects with a clinically diagnosed left shift were negative (Figure 3F). These data indicate that active transcription of PRTN3–002 occurs in neutrophil progenitors but not normal peripheral neutrophils or band cells.
Figure 3.
RT-PCR detection of an alternative transcript from the PRTN3 locus that initiates transcription at an intron 1 promoter. (A) Introns (lines; roman numerals), exons (numbered; gray boxes), and UTRs (black boxes) of the PRTN3 gene are depicted with the structure of two alternative transcripts, PR3 and PRTN3–002, shown below. White box marks the 5′UTR of PRTN3–002, which is encoded by intron 1. Primers for PCR to discriminate between PR3 and PRTN3–002 mRNA: forward primer resides in exon 1 for PR3 and intron 1 for PRTN3–002. (B) Primer specificity was verified using MCF-7 cells and monocytic cell lines HL60, U937, and THP-1. (C) Five patients (Pts) with AAV were screened by RT-PCR for PR3 and PRTN3–002 transcripts at two different times. Two patients (3 and 4) were screened a third time. Disease status of each sample was active (A), remission (R), or unclear (U). Patients labeled 1–5 correspond to patients 80, 46, 16, 1, and 20 listed in Supplemental Table 1. Cyclophilin was used as a control for RNA integrity. (D) Seven healthy donors were screened by RT-PCR for PR3 and PRTN3–002 transcripts. RNA from a patient with AAV positive for both PR3 and PRTN3–002 transcripts and THP-1 cells (T) was included as a positive control. (E) Two lots of purchased, pooled human bone marrow RNA (B1 and B2) were screened for PRTN3–002 and PR3 mRNA. Control was THP-1 RNA (T). (F) The presence of PRTN3–002 and PR3 mRNA was tested in banked RNA from patients with left shift; controls included one acute monocytic leukemia sample (A) and THP-1 (T) and MCF-7 (M) cells. Star indicates an RT-PCR product that retains intron 2, which was verified by sequence analysis (data not shown). In a separate reaction, samples from donors treated with GM-CSF were screened for PRTN3–002 and PR3 mRNA. W, water control.
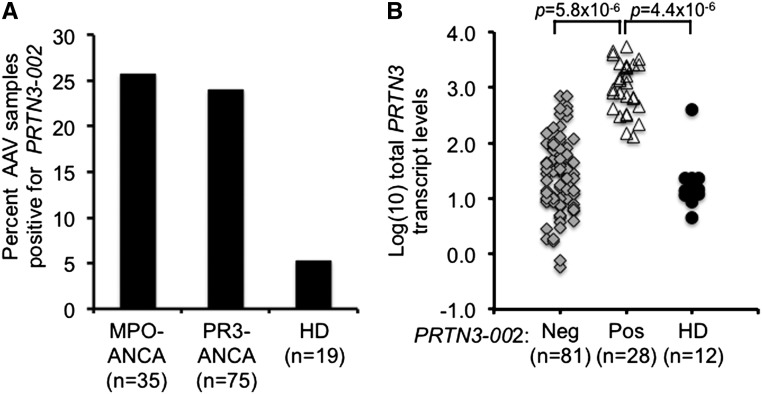
Using the transcript-specific RT-PCR primers, the presence of the PRTN3–002 transcript was tested in a larger group of patients with AAV and healthy donors. PRTN3–002 was not detected in healthy donors (n=18), except for one individual who unexpectedly expressed PR3 mRNA at levels similar to patients (discussed below). Among the patient samples tested, PRTN3–002 was detected in 25%. This fraction was evenly divided on the basis of ANCA serotype, with 24% of PR3-ANCA samples (n=75) and 26% of MPO-ANCA (n=35) samples positive for PRTN3–002 (Figure 4A). Instead of an association with ANCA serotypes, PRTN3–002 message may be related to disease activity. Patients expressing the PRTN3–002 transcript were more likely to have a Birmhingham Vasculitis Activity Score>1 compared with patients negative for the PRTN3–002 message (odds ratio, 3.3; 95% confidence interval, 1.32 to 8.42; P=0.01) (Supplemental Table 1).
Figure 4.
Frequency of PRTN3–002 mRNA and relationship to total PRTN3 transcripts in peripheral leukocytes. (A) Patients with AAV (n=112) and healthy donors (n=19) were screened for PRTN3–002 mRNA by RT-PCR. Percentage of patients positive for PRTN3–002 mRNA is shown for two serotypes: MPO-ANCA and PR3-ANCA. An ANCA-negative sample and a dual positive ANCA sample were excluded. (B) Total PRTN3 transcript levels determined by TaqMan quantitative RT-PCR, which uses primers in exons 4 and 5, were compared from patients that were negative (Neg; gray diamonds; n=81) or positive (Pos; open triangles; n=28) by RT-PCR for PRTN3–002 mRNA and healthy donors (HDs; black circles; n=12). There were no quantitative RT-PCR data for three patient samples and seven healthy donor samples. P values were calculated by t test.
Increased transcriptional activity from the PRTN3 locus could favor initiation of transcription from the intron 1 promoter and explain the presence of this alternative transcript. If it were the case, one prediction is that there would be a quantitative difference in the amount of total PR3 mRNA in patients positive for PRTN3–002. Total PR3 transcript levels were evaluated by quantitative RT-PCR with primers in exons 4 (forward primer) and 5 (reverse primer), which will amplify both PR3 and PRTN3–002 transcripts. Patients with AAV positive for PRTN3–002 had significantly greater levels of total PR3 mRNA than patients negative for PRTN3–002 and healthy donors (Figure 4B). One sample from a healthy donor was noted as high for PR3 levels (Figure 4B), and RNA from this healthy donor was positive for PRTN3–002 by RT-PCR. Detecting PRTN3–002 in samples with high levels of total PR3 message is consistent with the idea that increased transcriptional activity from the PRTN3 gene is either permissive for or reflects alternative promoter usage.
Peripheral Neutrophils from Patients with AAV Actively Translate Expressed mRNAs
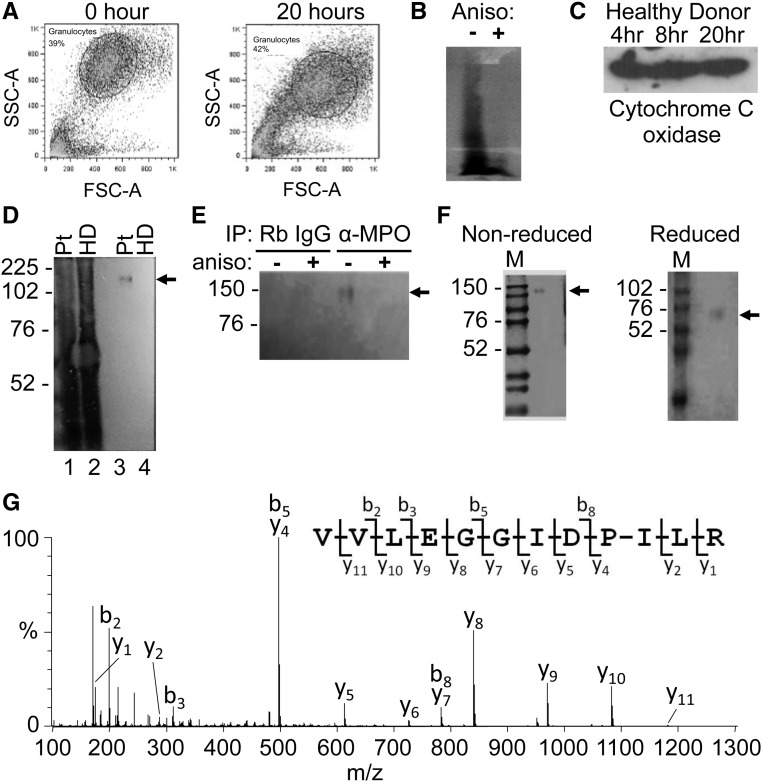
Of pivotal importance is whether aberrantly expressed transcripts in peripheral neutrophils are translated into protein. New protein synthesis was detected with the Click-It labeling reaction, which chemically attaches biotin to a metabolically incorporated methionine analog. Metabolic labeling was tested on neutrophils isolated from a healthy donor. Flow cytometry analysis revealed that a high percentage of purified neutrophils maintained a characteristic forward scatter and side scatter distribution for granulocytes after 20 hours in culture (Figure 5A). A Western blot probed with streptavidin indicated that the analog and attached biotin were incorporated into newly synthesized proteins, and loss of signal from neutrophils cultured in the presence of the protein synthesis inhibitor anisomycin verified that protein labeling was, indeed, a result of protein synthesis (Figure 5B). Immunoblot analysis for cytochrome C oxidase, the terminal enzyme of the mitochondrial respiratory chain, confirmed that protein synthesis was sustained during the culture period (Figure 5C). Next, the procedure was applied to neutrophils from a patient with active AAV. PR3 protein contains only 3 methionine residues, whereas MPO contains 28 methionine residues. To increase chances for success and because levels of PR3 and MPO transcripts are highly correlated,4 we first tested for MPO protein synthesis. General protein synthesis in neutrophils from a patient with active disease was confirmed, and MPO containing the methionine analog was detected in the labeled-protein pool by immunoprecipitation (Figure 5D). Additional controls established specificity of the immunoprecipitation and authenticity of MPO synthesis in cultured neutrophils (Figure 5E). For additional validation, affinity-purified proteins were separated by SDS-PAGE (Figure 5F), and bands were excised and subjected to in-gel digestion. Peptide mixtures were analyzed by tandem mass spectrometry (MS). MS/MS spectra were assigned to MPO with statistically significant scores (Figure 5G).
Figure 5.
Metabolic labeling for new protein synthesis in human neutrophils purified from peripheral circulation. (A) Flow cytometry was used to assess cell viability of purified neutrophils immediately after isolation (left panel) or after 20 hours in culture (right panel). FSC-A, forward scatter-area; SSC, side scatter-area. (B) Streptavidin Western blot on neutrophil extracts to determine protein synthesis activity of metabolically labeled neutrophils in the absence (−) or presence (+) of anisomycin (aniso), a protein synthesis inhibitor. (C) Labeled neutrophil extracts were precipitated with streptavidin and Western blot with Cytochrome C oxidase antibody to measure translation during in culture period. (D) Streptavidin Western blot on unpurified, labeled neutrophil extracts from a patient (Pt) with AAV (lane 1) and healthy donor (HD; lane 2) shows new protein synthesis. To detect synthesis of MPO in cultured neutrophils, streptavidin Western blot was performed on labeled extracts from a patient with ANCA (lane 3) and a healthy donor (lane 4) immunopurified with anti-MPO antibody. (E) Specificity of detection of newly synthesized MPO was tested with streptavidin Western blot on labeled neutrophil extracts cultured in the absence (−) or presence (+) of anisomycin and immunopurified with normal rabbit (Rb) IgG or anti-MPO antibody. IP, immunoprecipitation. (F) Labeled extracts immunopurified with anti-MPO antibody were subjected to SDS-PAGE. Bands that were detected by Coomassie Blue staining of the gel were excised, digested enzymatically, and analyzed by tandem MS. (G) The MS/MS spectrum acquired from a parent ion at m/z 640.9 (+2) confirmed that MPO was present in the bands. The most abundant peptide fragment ions extending from the N and C termini are denoted b and y, respectively. Neutrophils from patients 6, 41, and 60 listed in Supplemental Table 1 were used for metabolic labeling to detect synthesis of MPO. M, protein molecular weight marker. Molecular mass in kilodaltons is shown to the left of the images. Solid arrows in D–F denote position of MPO.
Translation of PR3 and PRTN3–002 Transcripts
Full-length PR3 and PRTN3–002 transcripts were detected in leukocyte RNA of patients with ANCA (Figure 6A). The PR3 transcript codes for 32- and 29-kD proteins corresponding to prepro-PR3 and processed PR3, respectively. The PRTN3–002 transcript codes for a protein that lacks the prepro sequence of PR3, the signal peptide sequence (Supplemental Figure 6), and the dipeptide (alanine-glutamate) plus an additional 14 amino acids coded by exon 2 and has a predicted size of 24 kD (Figure 6B). Interestingly, the predicted amino acid sequence encoded by PRTN3–002 is identical to the amino acid sequence reported for MBN,15 and to distinguish it from PR3, we will refer to the protein translated from PRTN3–002 as p24PR3/MBN. Appreciating that the tertiary structure of these two proteins may be different, commercially available antibodies were screened for reactivity against PR3 and/or p24PR3/MBN. By Western blot (Supplemental Figure 6), three antibodies of six antibodies tested on lysates from polymorphonuclear leukocytes recognized both PR3 and a 24-kD protein consistent with the size of p24PR3/MBN expressed in MCF-7 cells18 and THP-1 cells (Figure 6, C and D). However, attempts to use these antibodies to immunopurify p24PR3/MBN from MCF-7 and THP-1 cells were unsuccessful (data not shown).
Figure 6.
Translation of PR3 and PRTN3–002 mRNA in neutrophils from patients with ANCA disease. (A) RT-PCR using transcript-specific primers indicated that both PR3 and PRTN3–002 mRNAs were the expected full length. (B) Predicted protein sequences for PR3 versus p24PR3/MBN. The star indicates the first methinonine of p24PR3/MBN, which is in frame with PR3. (C) Available anti-PR3 antibodies were screened for crossreactivity against p24PR3/MBN by Western blot (Supplemental Figure 6). PMN, polymorphonuclear leukocytes; R&D, R&D Systems. (D) Anti-PR3 antibody reacted with a 24-kD band in THP-1 protein lysates. (E) Metabolically labeled protein lysates from neutrophils of patients with AAV (n=2; patients 56 and 79 listed in Supplemental Table 1) and one disease control (Ctl) were processed for detection of newly synthesized proteins. Anti-PR3 immunoblot showed reactivity with three bands in lysates from patient 1. The position corresponding to prepro-PR3 (upper band in E) and processed PR3 (C–E) is marked by solid arrows. The position corresponding to p24PR3/MBN in C–E is marked by open arrows. (F) Neutrophils from two patients (Pts; 6 and 26 in Supplemental Table 1) were metabolically labeled, and extracts were immunoprecipitated (IP) with anti-PR3 antibody. Unbound (U) and bound (B) fractions were analyzed by Western blot. Bands testing positive for both streptavidin probe (left panel) and anti-PR3 antibody (right panel) were excised, digested enzymatically, and analyzed by tandem MS. (G) The MS/MS spectrum acquired from a parent ion of 567.9 (+2) confirmed that PR3 was present in the bands. The most abundant peptide fragment ions extending from the N and C termini are denoted b and y, respectively. Neutrophils from patients 6, 26, 56, and 79 listed in Supplemental Table 1 and one disease control was used for metabolic labeling to detect synthesis of PR3. M, protein molecular weight marker. Molecular mass in kilodaltons is shown to the left of the images.
Purified neutrophils from three patients were cultured with the methionine analog. By Western blot (Figure 6E), one of three samples contained biotinylated proteins reactive with anti-PR3 antibody migrating at approximately 32, approximately 29, and approximately 24 kD, consistent with prepro-PR3 (32 kD), processed PR3 (29 kD), and p24PR3/MBN (24 kD). To further validate metabolic labeling of PR3, affinity-purified proteins were separated by SDS-PAGE, and bands were excised and subjected to in-gel digestion. Peptide mixtures were analyzed by tandem MS. One MS/MS spectrum was assigned to PR3 with a statistically significant score (Figure 6, F and G).
Patients with PR3-ANCA Disease Have Antibodies to p24PR3/MBN
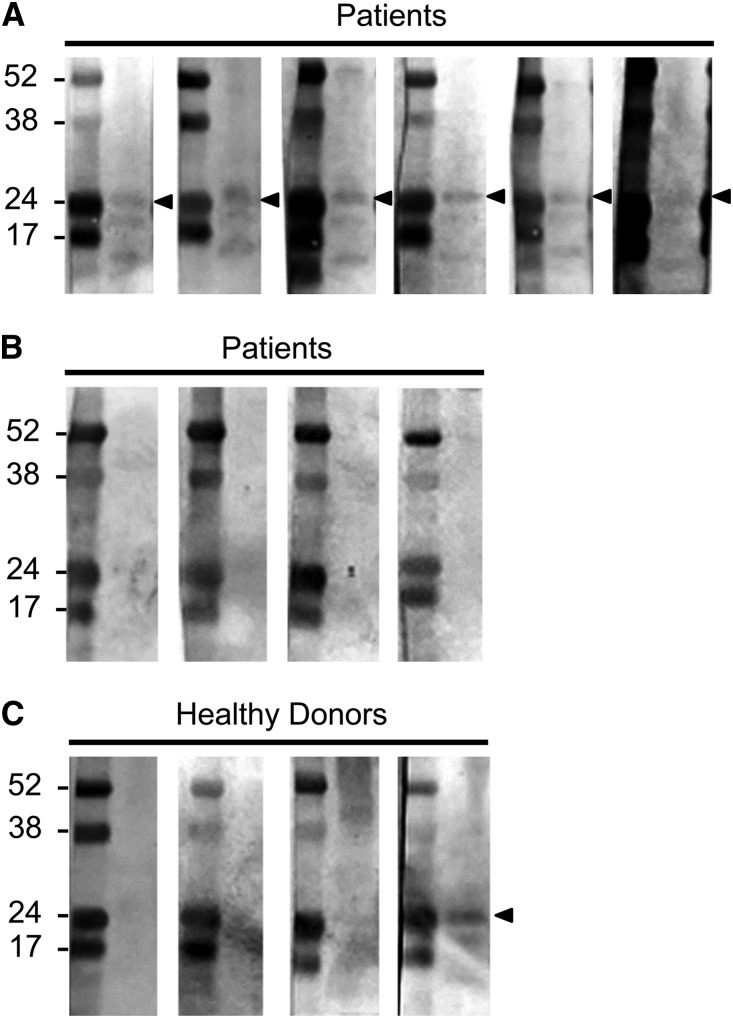
Because p24PR3/MBN was detected by Western blot of cultured neutrophils, we tested whether patients’ sera contained antibodies that recognize p24PR3/MBN. Extracts from MCF-7 cells, which only produce the PRTN3–002 transcript, were processed for immunoblot analysis and probed with sera from 10 patients with PR3-ANCA disease who were either positive or negative for the PRTN3–002 transcript and sera from four healthy donors. Figure 7 shows that sera from six patients and one healthy donor detected a 24-kD protein, similar to the monoclonal PR3 antibody used in Figure 6C. The reactivity to p24PR3/MBN with serum from a healthy donor is consistent with previous reports of naturally occurring antibodies to PR3 in normal individuals.19 Titers of PR3 antibodies were similar among the patients’ sera that reacted to p24PR3/MBN (145.8±37.4) and patients’ sera that failed to react (146.4±26.0), indicating that a high antibody titer does not explain reactivity to p24PR3/MBN. Instead, the reactivity to PR3 among patients with PR3-ANCA disease indicates that p24PR3/MBN may be antigenic.
Figure 7.
Immunoblots with sera from patients with PR3-ANCA disease to p24PR3/MBN. Whole-cell extracts from MCF-7 cells were probed with sera from 10 patients with PR3-ANCA disease and sera from 4 healthy donors. Patient sera samples (n=6) that reacted to a 24-kD protein (denoted with arrowheads) are shown in A. Patient sera samples (n=4) that failed to detect a 24-kD protein are shown in B. Immunoblot with sera from four healthy donors is shown in C, with one serum from one healthy donor recognizing a 24-kD protein. Molecular mass in kilodaltons is shown to the left of the images.
Discussion
We report that transcriptional dysregulation in leukocytes from patients with AAV results in not only increased levels of autoantigen message but also, variant transcripts. Specifically, from the PRTN3 locus, we detected transcripts with different sizes compared with the canonical PR3 transcript, transcripts with an extended 3′UTR containing an additional polyadenylation signal, and transcripts that initiated from a surrogate promoter in intron 1. A significant consequence of the transcriptional activity at the PRTN3 and MPO loci is de novo protein synthesis of PR3, p24PR3/MBN, and MPO in peripheral neutrophils of patients with AAV. The discovery of reactivity to p24PR3/MBN among patients with PR3-ANCA disease suggests a more complex autoantigen picture.
Promiscuous transcription at the PRTN3 locus was evident by an alternative transcript, PRTN3–002, which used a surrogate promoter residing in intron 1. Although PRTN3–002 is an annotated transcript expressed in MCF-7 cells, we detected this mRNA in cell lines (HL60, THP-1, and U937), bone marrow, and GM-CSF–treated individuals, which suggests a role in myeloid progenitors. PRTN3–002 may play a specific role in patients with AAV on the basis of its appearance during the course of disease. PRTN3–002 was detected in patients with active disease but not all active patients. Instead, patients positive for PRTN3–002 were less likely to be in remission than patients negative for PRTN3–002. This finding suggests that the presence of PRTN3–002 is more specific for disease status than sensitive to disease status. Also, the association of PRTN3–002 with high levels of expression likely contributes to the overall increase in PR3 transcript levels reported earlier.4 Our observation that these transcripts are translated in neutrophils from patients with ANCA emphasizes the importance of understanding the potential protein translated from PRTN3–002. Unfortunately, our study was limited by the availability of antibodies that recognize p24PR3/MBN. Three anti-PR3 antibodies crossreacted with the 24-kD protein by Western blot; however, we were unsuccessful in identifying one that would immunopurify p24PR3/MBN, hindering our ability to sequence the protein by MS.
Is PRTN3–002 the transcript coding for MBN? MBN was named for its proliferative function in HL60 cells.15 The published protein sequence for MBN is identical to the predicted sequence for p24PR3/MBN; however, the published cDNA sequence for MBN diverges from the sequence for the PRTN3–002 message. The cDNA sequence reported for MBN contains a portion of exon 1 and no intron 1 sequence from the PRTN3 gene, indicating that the originally reported MBN cDNA corresponds to the PR3 message and not the PRTN3–002 message. The proliferative function assigned to MBN was on the basis of experiments designed to inhibit MBN production using an antisense oligonucleotide. The antisense oligonucleotide targets the identical sequence in the PR3 message and the MBN message and would inhibit production of both PR3 and MBN. The putative proliferative function of MBN could equally be ascribed to PR3. Mechanistically, PR3 could have a proliferative function, because data indicate that proteolytically active PR3 cleaves p21 (cyclin-dependent kinase inhibitor).20,21 However, Sköld et al.22 described an S-phase inhibitory function for the secreted proform of PR3 that is distinct from p24PR3/MBN, because it contains N-terminal amino acids not encoded by the PRTN3–002 transcript. These data suggest that the PRTN3 gene encodes proteins with either proliferative or inhibitory activities, the latter determined by the proform of PR3 and the former ascribed to MBN. Before definitively naming the protein encoded by the PRTN3-002 transcript, functional experiments, such as overexpression or knockdown, are needed to show if p24PR3/MBN is responsible for the proliferative activity and indeed, MBN.
Amino acid sequence differences between PR3 and p24PR3/MBN suggest differences in post-translational processing. Given that p24PR3/MBN lacks the signal peptide responsible for targeting to the Golgi for post-translational processing, it is reasonable to assume that processing, packaging, folding, and localization of p24PR3/MBN will be different from that of PR3. The fact that anti-PR3 antibodies immunopurified PR3 but not p24PR3/MBN substantiates that there is a significant difference in surface-exposed epitopes. Importantly, these potential structural differences seem to be antigenic, because we observed reactivity to p24PR3/MBN in sera from patients with PR3-ANCA disease.
Alternative promoters of mammalian protein-coding genes are predicted to exist in over 58% of transcriptional units.23,24 An intronic promoter was recently identified in murine Prtn3 that drives expression of an alternative mRNA initiated in intron 2 rather than intron 1.25 Murine Prtn3 diverges from human PRTN3 gene and lacks the alternative promoter in intron 1 and an orthologous translational start codon in exon 2. Murine and human PR3 also differ in the amino acid sequence and biophysical properties.26 The differences in the murine Prtn3 gene, alternative transcript, and Pr3 protein compared with the human counterpart may contribute to the difficulty in developing a mouse model for PR3-ANCA disease that is as robust as the MPO-ANCA mouse model.27 p24PR3/MBN, encoded by PRTN3–002, could have a unique role in neutrophil activation or proliferation. Alternatively, p24PR3/MBN could be antigenic, which is consistent with the Western blots on extracts from MCF-7 cells showing that some patients with AAV have antibodies that recognize this protein.
Future studies are necessary to determine what role p24PR3/MBN may play in the disease process; however, it is tempting to draw parallels to reports of autoantigen conformeropathy.28 AAV has provided a model for discovery of characteristic features of autoimmune diseases. Investigations exemplify how environmentally triggered effects29–32 might alter epigenetic mechanisms33 that result in a deviation in expression and availability of the actual autoantigens recognized in disease.
Concise Methods
Patient Cohort
Patients with ANCA small-vessel vasculitis were categorized by the Chapel Hill Consensus Conference nomenclature34 and identified through the Glomerular Disease Collaborative Network. Study protocols were compliant with and approved by the University of North Carolina Institutional Review Board. Blood donations were obtained from informed and consenting individuals. ANCA samples (n=119) were collected from 79 patients (25 patients donated more than one sample). Supplemental Table 1 summarizes the characteristics of patients participating in this study. Individuals with infection provided left-shift samples (n=7), mean age was 53.1 years, and absolute neutrophil count averaged 13.4×106/ml. Anonymous samples included one patient with acute monocytic leukemia and six GM-CSF–treated patients (n=6). Healthy donors (n=19) had a mean age of 46.3 years (range=20–86); 17 donors were white, 0 donors were black, and 2 donors were other. There were 6 men and 13 women.
Purified neutrophils from six patients with active AAV were included in protein studies (only four patients had a corresponding RNA sample). Mean age was 38.8 years (range=18–64); five patients were white, and one patient was black. There were three men and three women. Absolute neutrophil count averaged 4.45×106/ml (range=3.1–5.8); mean serum creatinine was 3.57 mg/dl (range=2.18–4.96).
Northern Blot Analyses
RNA was isolated as previously described.9 RNA (15 µg) was electrophoresed in 1×3-(N-morpholino)propanesulfonic acid on a 1.2% denaturing agarose gel containing 1×3-(N-morpholino)propanesulfonic acid and 8% 12.3 M prewarmed formaldehyde, transferred overnight to a Hybond N+ nylon membrane (Amersham) in 20×saline-sodium citrate (SSC) using the TurboBlotter (Whatman) capillary transfer system, and crosslinked by ultraviolet irradiation. To produce the probe, pcDNA3 (10 µg) vector containing PR3 (nucleotides 336–627) was linearized. In vitro transcription was carried out with Sp6 RNA Polymerase using the MAXIscript Kit (Ambion) generating a 291-bp probe. PR-3 probe (500 ng) was biotinylated using the BrightStar Psoralen-Biotin Nonisotopic Labeling Kit (Ambion) and stored at −80°C. Membrane was blocked with preheated ULTRAhyb (Ambion) buffer at 68°C for 30 minutes. After addition of 10 ng psoralen-biotin–labeled probe/ml ULTRAhyb buffer, blot was incubated for 16 hours at 68°C. Washes were (two at 5 minutes each) at 68°C in 2× SSC and 0.1% SDS solution followed by high-stringency washes (two at 15 minutes each) with 0.1 SSC and 0.1% SDS. A Brighstar Biodetect Kit (Ambion) with manufacturer’s guidelines was used for nonisotopic detection of hybridized RNA probe. Digital images were captured on a Fluorochem Q imaging system (Cell Biosciences).
RACE
RACE was performed to determine the 3′ end of PR-3 mRNA comparing patient leukocyte RNA with that of healthy donors as previously described35 (detailed in Supplemental Figure 1).
RNA Amplification and Detection
TaqMan PCR was performed as previously described.9 RT-PCR protocol is detailed in Supplemental Figure 2, and primer sequences are listed in Supplemental Table 1.
ELISA for GM-CSF
GM-CSF was measured by ELISA using the sera of 31 samples from 27 separate patients. The GM-CSF concentrations for patients’ samples used are listed in Supplemental Table 1. The ELISA was performed with the Human GM-CSF ELISA MAX Standard (432001; BioLegend).
Metabolic Labeling by Click-It Reaction
A detailed protocol is in Supplemental Figure 3. Briefly, neutrophils were purified and cultured in methionine free RPMI1640 supplemented with 10% dialyzed FBS for 1 hour. Methionine analog l-aziodohomoalanine (40 μM; Invitrogen) was added to medium, and neutrophils were incubated for 20 hours at 37°C and 5% CO2 under gentle agitation. Incorporated methionine analog was subsequently biotinylated, and proteins were detected by streptavidin conjugated to alkaline phosphatase (Pierce, Rockford, IL) and/or antibody-specific immunnoprecipitations and immunoblotting. Anti-PR3 antibodies included Epitope A (PR3.4A3), Epitope B (PR3.4A5), and Epitope C (PR3.6A6; gift from Jörgen Weislander), Sc-28818 (Santa Cruz Biotechnology, Santa Cruz, CA), HM2172 (Hy Clone), AF6134 (R&D Systems), and MS-1333-P1 (Neomark). Anti-MPO antibody was from Dako (Carpinteria, CA), and anti-COX5B antibody WAS from Abcam, Inc. (Cambridge, MA).
Electrospray MS
Gel bands were excised, destained, dehydrated, and washed one time with 50 mM ammonium bicarbonate (pH 7.8), two times with 50 mM ammonium bicarbonate and 50% (vol/vol) acetonitrile, and one time with 100% acetonitrile. Gel pieces were dehydrated under vacuum before rehydration with 50 mM ammonium bicarbonate (pH 7.8), reduction with 5 mM dithiothreitol at 50°C for 30 minutes, and carbamidomethylation using 15 mM iodoacetamide for 30 minutes at room temperature in the dark. After reduction/alkylation, gel pieces were dehydrated and rehydrated with sequencing grade trypsin (125 ng µl−1 in 50 mM ammonium bicarbonate [pH 7.8]) and incubated overnight at 37°C. Electrospray MS analyses were performed on an Agilent 1100 Series HPLC-Chip/MS (Santa Clara, CA) interfaced to an Agilent 6340 Ion Trap and/or a Waters Q-TOF Global Mass Spectrometer equipped with a NanoAcquity UPLC System (Waters, Milford, MA). An automated database searching of data acquired on the ion trap was performed using the Data Extractor feature of the SpectrumMill software from Agilent. Data were searched against the National Center for Biotechnology Information (NCBI) nonredundant database using the MS/MS Search function in the SpectrumMill software. Quadrupole time-of-flight mass spectrometer data were processed using ProteinLynx Global Server software (Waters) and searched against the NCBI nonredundant database.
Western Blots for p24PR3/MBN
Whole-cell extracts from MCF-7 cells were separated under nonreducing conditions using precast Novex 4%–12% Bis-Tris Plus polyacrylamide gels (BG04127BOX; Life Technologies). The proteins were transferred to nitrocellulose membranes using standard conditions. Individual segments of the nitrocellulose membrane with MCF-7 extracts were probed separately with sera from 10 patients and four healthy donors. The sera were diluted 1:50 and incubated with the separate membranes overnight at 4°C. A goat anti-human IgG, IgM, and IgA secondary antibody conjugated to alkaline phosphatase (31316; Pierce) was diluted 1:5000 and incubated with membranes for 1 hour at room temperature. The immunoblots were developed using the Western Blue Stabilized Substrate for Alkaline Phosphatase (S3841; Promega).
Statistical Analyses
P values were calculated by Wilcoxon two-sample tests for two-sample comparisons.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Jason Williams, Katina Johnson, and Andrea Adams from the protein microcharacterization core facility of the National Institute of Environmental Health Sciences (NIEHS) and Leesa Deterding from the Collaborative Mass Spectrometry Support Group at NIEHS for helpful suggestions and assistance. We also thank Dr. Jorgen Wieslander for the gift of anti-PR3 antibodies. We thank Dr. Julie Anne McGregor for reading the manuscript and providing helpful comments. We acknowledge Candace Henderson, our liaison with the patient community, who receives and processes blood samples. Our deepest gratitude goes to the patients who so generously donate blood in support of the research.
This work was supported by National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases Grant P01-DK058335-06.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013101092/-/DCSupplemental.
References
- 1.Rönnblom L, Eloranta ML: The interferon signature in autoimmune diseases. Curr Opin Rheumatol 25: 248–253, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Alcorta DA, Barnes DA, Dooley MA, Sullivan P, Jonas B, Liu Y, Lionaki S, Reddy CB, Chin H, Dempsey AA, Jennette JC, Falk RJ: Leukocyte gene expression signatures in antineutrophil cytoplasmic autoantibody and lupus glomerulonephritis. Kidney Int 72: 853–864, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Ohlsson S, Hellmark T, Pieters K, Sturfelt G, Wieslander J, Segelmark M: Increased monocyte transcription of the proteinase 3 gene in small vessel vasculitis. Clin Exp Immunol 141: 174–182, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang JJ, Pendergraft WF, Alcorta DA, Nachman PH, Hogan SL, Thomas RP, Sullivan P, Jennette JC, Falk RJ, Preston GA: Circumvention of normal constraints on granule protein gene expression in peripheral blood neutrophils and monocytes of patients with antineutrophil cytoplasmic autoantibody-associated glomerulonephritis. J Am Soc Nephrol 15: 2103–2114, 2004 [DOI] [PubMed] [Google Scholar]
- 5.De'Oliviera J, Gaskin G, Dash A, Rees AJ, Pusey CD: Relationship between disease activity and anti-neutrophil cytoplasmic antibody concentration in long-term management of systemic vasculitis. Am J Kidney Dis 25: 380–389, 1995 [DOI] [PubMed] [Google Scholar]
- 6.Falk RJ, Jennette JC: Anti-neutrophil cytoplasmic autoantibodies with specificity for myeloperoxidase in patients with systemic vasculitis and idiopathic necrotizing and crescentic glomerulonephritis. N Engl J Med 318: 1651–1657, 1988 [DOI] [PubMed] [Google Scholar]
- 7.Falk RJ, Terrell RS, Charles LA, Jennette JC: Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci U S A 87: 4115–4119, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross WL, Schmitt WH, Csernok E: ANCA and associated diseases: Immunodiagnostic and pathogenetic aspects. Clin Exp Immunol 91: 1–12, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciavatta DJ, Yang J, Preston GA, Badhwar AK, Xiao H, Hewins P, Nester CM, Pendergraft WF, 3rd, Magnuson TR, Jennette JC, Falk RJ: Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J Clin Invest 120: 3209–3219, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris KV, Vogt PK: Long antisense non-coding RNAs and their role in transcription and oncogenesis. Cell Cycle 9: 2544–2547, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nordang GB, Viken MK, Amundsen SS, Sanchez ES, Flatø B, Førre OT, Martin J, Kvien TK, Lie BA: Interferon regulatory factor 5 gene polymorphism confers risk to several rheumatic diseases and correlates with expression of alternative thymic transcripts. Rheumatology (Oxford) 51: 619–626, 2012 [DOI] [PubMed] [Google Scholar]
- 12.Stone RC, Du P, Feng D, Dhawan K, Rönnblom L, Eloranta ML, Donnelly R, Barnes BJ: RNA-Seq for enrichment and analysis of IRF5 transcript expression in SLE. PLoS ONE 8: e54487, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labbaye C, Musette P, Cayre YE: Wegener autoantigen and myeloblastin are encoded by a single mRNA. Proc Natl Acad Sci U S A 88: 9253–9256, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabay JE, Almeida RP: Antibiotic peptides and serine protease homologs in human polymorphonuclear leukocytes: Defensins and azurocidin. Curr Opin Immunol 5: 97–102, 1993 [DOI] [PubMed] [Google Scholar]
- 15.Bories D, Raynal MC, Solomon DH, Darzynkiewicz Z, Cayre YE: Down-regulation of a serine protease, myeloblastin, causes growth arrest and differentiation of promyelocytic leukemia cells. Cell 59: 959–968, 1989 [DOI] [PubMed] [Google Scholar]
- 16.Gabay JE, Scott RW, Campanelli D, Griffith J, Wilde C, Marra MN, Seeger M, Nathan CF: Antibiotic proteins of human polymorphonuclear leukocytes. Proc Natl Acad Sci U S A 86: 5610–5614, 1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta SK, Niles JL, McCluskey RT, Arnaout MA: Identity of Wegener’s autoantigen (p29) with proteinase 3 and myeloblastin. Blood 76: 2162, 1990 [PubMed] [Google Scholar]
- 18.Horman S, Fokan D, Galand P: MCF-7 mammary tumour cells express the myeloid cell differentiation controlling factor, serine protease 3/myeloblastin. Cell Prolif 33: 331–340, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Z, Zhao MH, Segelmark M, Hellmark T: Natural autoantibodies to myeloperoxidase, proteinase 3, and the glomerular basement membrane are present in normal individuals. Kidney Int 78: 590–597, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Pendergraft WF, 3rd, Rudolph EH, Falk RJ, Jahn JE, Grimmler M, Hengst L, Jennette JC, Preston GA: Proteinase 3 sidesteps caspases and cleaves p21(Waf1/Cip1/Sdi1) to induce endothelial cell apoptosis. Kidney Int 65: 75–84, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Witko-Sarsat V, Canteloup S, Durant S, Desdouets C, Chabernaud R, Lemarchand P, Descamps-Latscha B: Cleavage of p21waf1 by proteinase-3, a myeloid-specific serine protease, potentiates cell proliferation. J Biol Chem 277: 47338–47347, 2002 [DOI] [PubMed] [Google Scholar]
- 22.Sköld S, Rosberg B, Gullberg U, Olofsson T: A secreted proform of neutrophil proteinase 3 regulates the proliferation of granulopoietic progenitor cells. Blood 93: 849–856, 1999 [PubMed] [Google Scholar]
- 23.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y: Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Landry JR, Mager DL, Wilhelm BT: Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet 19: 640–648, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Relle M, Becker M, Meyer RG, Stassen M, Schwarting A: Intronic promoters and their noncoding transcripts: A new source of cancer-associated genes. Mol Carcinog 53: 117–124, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Wiesner O, Litwiller RD, Hummel AM, Viss MA, McDonald CJ, Jenne DE, Fass DN, Specks U: Differences between human proteinase 3 and neutrophil elastase and their murine homologues are relevant for murine model experiments. FEBS Lett 579: 5305–5312, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Xiao H, Heeringa P, Hu P, Liu Z, Zhao M, Aratani Y, Maeda N, Falk RJ, Jennette JC: Antineutrophil cytoplasmic autoantibodies specific for myeloperoxidase cause glomerulonephritis and vasculitis in mice. J Clin Invest 110: 955–963, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borza DB, Bondar O, Colon S, Todd P, Sado Y, Neilson EG, Hudson BG: Goodpasture autoantibodies unmask cryptic epitopes by selectively dissociating autoantigen complexes lacking structural reinforcement: Novel mechanisms for immune privilege and autoimmune pathogenesis. J Biol Chem 280: 27147–27154, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Falk RJ, Hogan S, Carey TS, Jennette JC, The Glomerular Disease Collaborative Network : Clinical course of anti-neutrophil cytoplasmic autoantibody-associated glomerulonephritis and systemic vasculitis. Ann Intern Med 113: 656–663, 1990 [DOI] [PubMed] [Google Scholar]
- 30.Hogan SL, Cooper GS, Savitz DA, Nylander-French LA, Parks CG, Chin H, Jennette CE, Lionaki S, Jennette JC, Falk RJ: Association of silica exposure with anti-neutrophil cytoplasmic autoantibody small-vessel vasculitis: A population-based, case-control study. Clin J Am Soc Nephrol 2: 290–299, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogan SL, Falk RJ, Chin H, Cai J, Jennette CE, Jennette JC, Nachman PH: Predictors of relapse and treatment resistance in antineutrophil cytoplasmic antibody-associated small-vessel vasculitis. Ann Intern Med 143: 621–631, 2005 [DOI] [PubMed] [Google Scholar]
- 32.Hogan SL, Satterly KK, Dooley MA, Nachman PH, Jennette JC, Falk RJ, Glomerular Disease Collaborative Network : Silica exposure in anti-neutrophil cytoplasmic autoantibody-associated glomerulonephritis and lupus nephritis. J Am Soc Nephrol 12: 134–142, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Javierre BM, Hernando H, Ballestar E: Environmental triggers and epigenetic deregulation in autoimmune disease. Discov Med 12: 535–545, 2011 [PubMed] [Google Scholar]
- 34.Jennette JC, Falk RJ, Andrassy K, Bacon PA, Churg J, Gross WL, Hagen EC, Hoffman GS, Hunder GG, Kallenberg CG, et al. : Nomenclature of systemic vasculitides. Proposal of an international consensus conference. Arthritis Rheum 37: 187–192, 1994 [DOI] [PubMed] [Google Scholar]
- 35.Scotto-Lavino E, Du G, Frohman MA: 3′ end cDNA amplification using classic RACE. Nat Protoc 1: 2742–2745, 2006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.