Abstract
Definitive diagnosis of vesicular or vesicular-like lesions in livestock animals presents challenges both for veterinary clinicians and diagnostic laboratories. It is often impossible to diagnose the causative disease agent on a clinical basis alone and difficult to collect ample vesicular epithelium samples. Due to restrictions of time and sample size, once laboratory tests have ruled out foot-and-mouth disease, vesicular stomatitis and swine vesicular disease a definitive diagnosis may remain elusive. With the ability to test a small quantity of sample for a large number of pathogens simultaneously, DNA microarrays represent a potential solution to this problem. This study describes the application of a long oligonucleotide microarray assay to the identification of viruses known to cause vesicular or vesicular-like lesions in livestock animals. Eighteen virus isolates from cell culture were successfully identified to genus level, including representatives of each foot-and-mouth disease virus serotype, two species of vesicular stomatitis virus, swine vesicular disease virus, vesicular exanthema of swine virus, bovine herpesvirus 1, orf virus, pseudocowpox virus, bluetongue virus serotype 1 and bovine viral diarrhoea virus 1. Vesicular stomatitis virus and vesicular exanthema of swine virus were also identified in vesicular epithelium samples, with varying levels of sensitivity. The results indicate that with further development this microarray assay could be a valuable tool for the diagnosis of vesicular and vesicular-like diseases.
1. Introduction
To date, the application of microarray technology to the detection of pathogens causing disease in animals has remained somewhat limited. Baxi et al. (2006) have described a microarray assay developed to detect and type Foot-and-mouth disease virus (FMDV). Using a universal PCR primer set to amplify RNA extracted from infected cell cultures prior to hybridization with 155 virus-common and serotype-specific oligonucleotide probes, this assay correctly differentiated the seven serotypes of FMDV. A microarray-based assay using 36 padlock probes designed to detect FMDV, vesicular stomatitis virus (VSV) and swine vesicular disease virus (SVDV) has been shown to identify successfully 39 of 41 isolates derived from infected cell culture, including discrimination of the New Jersey and Indiana species of VSV (Baner et al., 2007). An assay coupling multiplex PCR with a small microarray based on short oligonucleotide probes accurately detected five important marine fish pathogens (Gonzalez et al., 2004); a low density oligonucleotide suspension array for the detection and differentiation of animal pestiviruses has also been described (Deregt et al., 2006).
Foot-and-mouth disease, vesicular stomatitis, swine vesicular disease and vesicular exanthema are characterised by vesicular lesions in the mouth and on the snout and feet of infected animals. Vesicular-like lesions are also a feature of several other viral diseases. Infectious bovine rhinotracheitis, bovine papular stomatitis, orf, pseudocowpox, bovine viral diarrhoea, malignant catarrhal fever, sheeppox, rinderpest, peste des petits ruminants and bluetongue can all cause lesions indistinguishable from those associated with viral vesicular diseases, particularly in their advanced clinical stages (Geering et al., 1995). Detection of virus in epithelium samples collected from lesions is a critical step in the definitive diagnosis of viral vesicular or vesicular-like diseases. Currently, routine methods include isolation in cell culture, ELISA and PCR. While in combination these tests represent a rapid, accurate strategy for virus detection, they cannot easily detect a wide range of viruses. Although at present a less sensitive approach, microarray technology offers the ability to screen a small amount of clinical material for a large number of pathogens simultaneously and as such could provide a valuable adjunct to frontline laboratory tests. This study aimed to determine the ability of a prototype DNA microarray assay to identify viruses known to cause vesicular or vesicular-like lesions in livestock and to evaluate its potential as a diagnostic tool.
2. Materials and methods
2.1. Microarray design and printing
The microarray used in this study contained 60-mer oligonucleotide probes designed to detect Foot-and-mouth disease virus (FMDV), Vesicular stomatitis New Jersey virus (VSNJV), Vesicular stomatitis Indiana virus (VSIV), Vesicular exanthema of swine virus (VESV), Swine vesicular disease virus (SVDV), Orf virus (ORFV), Bovine papular stomatitis virus (BPSV), Bovine viral diarrhoea virus 1 (BVDV-1), Bovine viral diarrhoea virus 2 (BVDV-2), Classical swine fever virus (CSFV), Bovine herpes virus 1 (BoHV-1), Ovine herpes virus 2 (OvHV-2), Malignant catarrhal fever virus in white-tailed deer (MCFDV), Alcelaphine herpesvirus 1 (AlHV-1), Alcelaphine herpesvirus 2 (AlHV-2), Rinderpest virus (RPV), Peste-des-petits-ruminants virus (PPRV), Sheeppox virus (SPPV), Lumpy skin disease virus (LSDV), and Bluetongue virus (BTV) (Table 1). Where possible, 2 distinct genomic target regions were represented for each virus, including a highly conserved region within an enzyme such as the polymerase, and a more variable region often corresponding to a structural protein. Probes were selected with the aim of detecting as wide a range of variants of a virus species or strain as possible given the available sequence information in the National Center for Biotechnology Information (NCBI) database. A viral sequence was considered to be covered on the array if it aligned with at least 1 probe with no more than 5 nt mismatches. In addition to the 412 probes representing viruses causing vesicular or vesicular-like diseases, the subarray also contained 177 probes representing arboviruses commonly isolated in Australia’s Northern Territory and 95 probes representing selected avian viruses, for use in parallel studies (data not shown). Oligonucleotide probes were synthesised commercially (lllumina Inc., San Diego, California) and printed in replicates of 6 on Corning® UltraGAPS™ slides (Corning Inc. Life Sciences, Lowell, Massachusetts), 2 arrays per slide. Refer to Supplementary Materials and Methods for array printing details.
Table 1.
Microarray representation of viruses causing vesicular or vesicular-like lesions in livestock animals
| Genus: | Probes designed to detect: | Number of probes: |
|---|---|---|
| Aphtdovirus | FMDV | 23 |
| Vesiculovirus | VSNJV, VSIV | 42 |
| Vesivirus | VESV | 67 |
| Enterovirus | Human enterovirus B, strain SVDV | 24 |
| Parapoxvirus | ORFV, BPSV | 45 |
| Pestivirus | BVDV-1, BVDV-2, CSFV | 123 |
| Varicellovirus | BoHV-1 | 32 |
| Rhadinovirus | OHV-2, MCFDV, AlHV-1, AlHV-2 | 20 |
| Morbillivirus | RPV, PPRV | 17 |
| Capripoxvirus | SPPV, LSDV | 9 |
| Orbivirus | BTV | 10 |
2.2. Viruses and cell culture
GenBank accession numbers (GenBank) for virus isolates have been provided where possible, otherwise NCBI taxonomy identification numbers (tax.ID) are listed. BVDV-1/Z937, BVDV-1/A531, BVDV-1/P92-4151 (tax.ID 11099); VSIV/Chimayo (tax.ID 11277); VSNJV/Atlanta/bull (tax.ID 11280); VESV/A48 (GenBank AJ131384); SVDV/UKG/72 (GenBank X54521); BoHV-1/V155 (tax.ID 79890); ORFV/Websters vaccine (tax.ID 10258); pseudocowpox virus PCPV/Australia/05 (tax.ID 129726); BTV/Serotype 1/Australia (tax.ID 35327); and BTV/Serotype 3/South Africa (tax.ID 36423) were grown in cell monolayers as detailed in Supplementary Table 1. Upon reaching an adequate level of cytopathic effect (CPE), infected cell culture supernatants were decanted and clarified by centrifugation prior to storage at − 70 °C. ORFV and PCPV-infected cells were additionally harvested into phosphate-buffered saline. All viruses were titrated on 96-well plates (Nalge Nunc International, Rochester, New York) using serial 10-fold dilutions with 12 replicates at each dilution. CPE was assessed at 5–10 days post-infection and the tissue culture infectious dose 50 (TCID50) calculated using the method of Spearman-Kärber (Villegas and Purchase, 1980). For the non-cytopathic BVDV-1 isolate A531, infection of cells was determined by immunoperoxidase staining using pestivirus-specific anti-NS3 monoclonal antibodies. FMDV serotypes A/A24/Cruzeiro/55 (GenBank J02183), C/Phillipine/84 (tax.ID 12116), O/Manisa/Turkey/78 (tax.ID 12118), Asia 1/ISR/89 (tax.ID 110195), SAT1/Rho/78 (tax.ID 12122) and SAT2/Zimbabwe/83 (GenBank AF540910) were purchased as binary ethyleneimine-inactivated vaccine antigens from Merial Animal Health Ltd (Pirbright, United Kingdom). Binary ethyleneimine-inactivated SAT3/KNP/90/3 (GenBank DQ009739) was obtained from the Onderstepoort Veterinary Institute (Onderstepoort, South Africa). All strains had been grown in BHK 21 cells which upon reaching adequate CPE were clarified by centrifugation or filtration prior to inactivation. The antigens purchased from Merial Animal Health Ltd had been purified by industrial scale chromatography; SAT3/KNP/90/3 had been concentrated by ultracentrifugation.
2.3. Clinical samples
Six-week-old Landrace × Large White pigs were infected with VESVA48: 2 pigs (pig 1, pig 13) were inoculated with a total of approximately 2 × 107 TCID50 spread over multiple sites within the dermis of the coronary band and the mucosal surface of the oral cavity; 4 additional pigs, which shared the same pen, became infected after close contact with the inoculated pigs. Epithelium samples were collected from vesicular lesions at days 2, 3, 6 and 7 post-inoculation. Day 7 samples were collected post134 mortem. Six-month-old Holstein-Friesian steers were inoculated with approximately 4 × 107 TCID50 VSNJV spread over multiple sites within the dermis of the coronary band and interdigital space and the lingual and mucosal surfaces of the oral cavity. Epithelium samples were collected from vesicular lesions at days 3, 4, 6 and 8 post-inoculation. Day 8 samples were collected post-mortem. Samples for RNA extraction were stored in RNAlater®(Ambion, Austin, Texas) reagent immediately following collection; parallel samples for virus isolation were stored directly at −70 °C. Animal husbandrymethods and experimental design were endorsed by the CSIRO Australian Animal Health Laboratory's Animal Ethics Committee.
2.4. Nucleic acid extraction and amplification
For RNA viruses, total RNA was extracted from 100 µl of infected cell culture supernatant or inactivated antigen using the RNeasy Mini Kit (QIAGEN, Venlo, The Netherlands), including on-column DNase treatment. Prior to RNA extraction using the same method, VESVA48 and VSNJV infected epithelial tissues (10 mg) were homogenised in RLT buffer (QIAGEN) using a MagNA Lyser and MagNA Lyser Green Beads (Roche Diagnostics Australia Pty Ltd, Castle Hill, New South Wales). A modified version of a random PCR protocol (Wang et al., 2002; Palacios et al., 2007) was used to amplify extracted RNA (refer to Supplementary Materials and Methods for details). For DNA viruses, DNA was extracted from 100 µl of infected cell culture supernatant or cell suspension with 100 µl of extraction buffer (10 mM Tris pH 7.6–7.8, 50 mM β-mercaptoethanol, 100 mM NaCl, 10 mM EDTA, 1 % Sarcosyl NL-97, 20 % sucrose). Following 2 rounds of phenol/chloroform/iso-amyl alcohol purification, DNA was ethanol precipitated and resuspended in Tris-EDTA buffer (pH 8.0) containing 20 µl/ml RNase. DNA was randomly amplified using a protocol adapted from Bohlander et al. (1992) (refer to Supplementary Materials and Methods for details).
2.5. Microarray hybridization and data analysis
Two virus samples were hybridized on each microarray slide as detailed in the Supplementary Materials and Methods section. Hybridized microarrays were scanned in the 595 nm and 685 nm channels using an ArrayWoRx® scanner (Applied Precision, Issaquah, Washington). Images were analysed using softWoRx® Tracker software, version 2 (Applied Precision) and numerical data exported to Microsoft Excel. The signal-to-noise ratio (SNR), computed from the background corrected spot median intensity divided by the standard deviation of the background, was calculated for each microarray spot in the 595 nm channel. The SNR value for each probe was obtained by calculating the mean SNR value of the probe’s 6 replicate spots. To display the results of each hybridization graphically, probe SNR values were plotted on a continuous colour spectrum using the PermutMatrix program, Version 1.8.7 (Caraux and Pinloche, 2005). The upper limit (brightest green colour) of the spectrum was independently set for each hybridization to the probe with the highest SNR value. For the display of results in tabular form, average SNR values were calculated across probes grouped according to virus genus (refer to Table 1).
3. Results
3.1. Detection of viruses from cell culture
The microarray was initially tested using a range of viruses grown in cell culture that had been selected to represent different genome types. RNA or DNA was extracted directly from infected culture supernatants or cell suspensions, ranging in titre from 1 × 104 to 4 × 107 TCID50 per 100 µl (Supplementary Table 1), with the exception of the FMDV preparations, which had undergone prior binary ethyleneimine inactivation and purification. Randomly amplified target DNA was hybridized on the array. Raw signal data were used to calculate a signal-to-noise ratio (SNR) for each microarray spot. Measuring signal strength relative to background noise, the SNR is a gauge of signal quality. In comparison to measuring signal strength by standard deviation units after within array normalization of background corrected log-transformed raw signals, the SNR appeared to be a more robust measure of target-probe hybridization. For several hybridizations, the SNR value for each microarray spot was plotted against the corresponding signal strength measurement: spots with a high SNR always had high signal strength, whereas a significant proportion of spots of high signal strength had a low SNR (Supplementary Fig. 1).
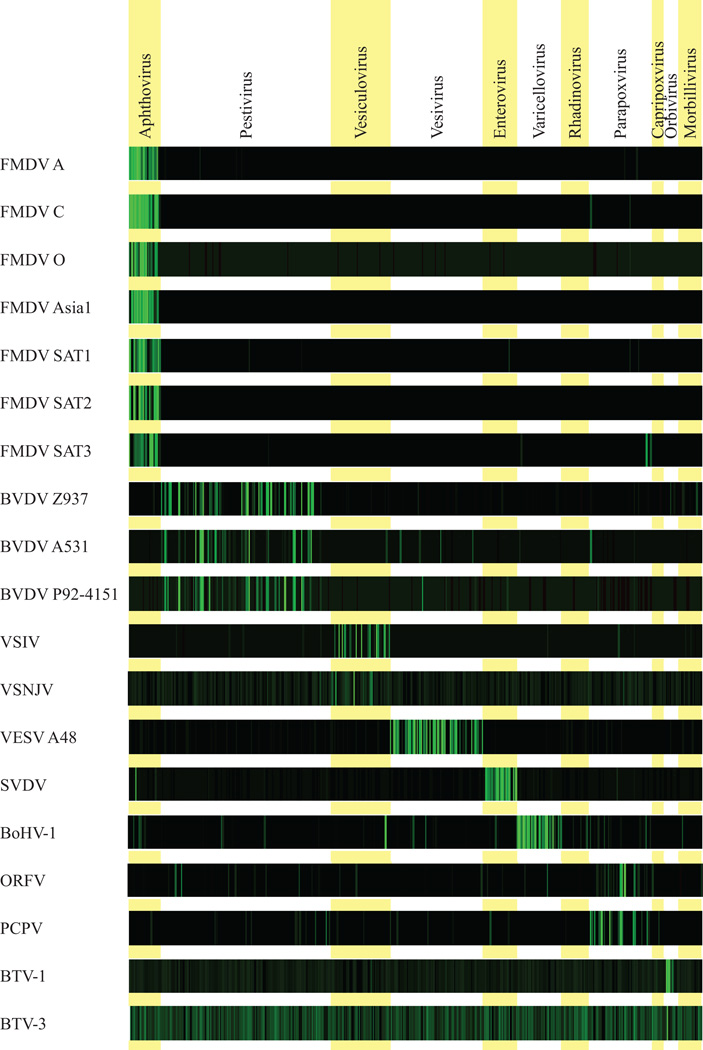
For each hybridization, the mean SNR for each of the virus probes on the array was plotted on a continuous green colour scale (Fig. 1). To assess the outcome of hybridizations in a more quantitative manner, mean probe SNR values were averaged across each genus represented on the array (Table 2). It should be noted that in the course of this study SNR values can be compared within hybridizations but not between them. Both methods of data presentation allowed identification of 18 of the 19 virus isolates hybridized on the array. The array failed to identify the BTV-3 isolate, returning similar average SNR values for all genera represented. Although the probe with the highest mean SNR was a BTV probe, it was not sufficiently elevated to be specific for the orbivirus genus. For 16 of the 18 positive hybridizations, the average SNR was raised above background levels for the relevant virus genus alone. In the FMDV SAT3 hybridization, although the aphthovirus genus average SNR (282) was the clear outlier, the parapoxvirus genus average SNR (9.68) was also significantly higher than those of the remaining genera (<1). This elevation was due to 2 probes, neither of which showed significant sequence similarity to the FMDV SAT3 reference genome (GenBank NC011452). In the SVDV hybridization, the enterovirus genus average SNR (494.15) was approximately 50-fold higher than the other genera with the exception of the aphthovirus genus (46.78). The raised aphthovirus SNR was due to cross-hybridization of the SVDV isolate with an FMDV probe that was identical to the SVDV strain UKG 72 genome (GenBank X54521) at 49 of 60 nt.
Figure 1.
Microarray hybridization results of samples derived from virus-infected cell cultures. Within the bar graphics representing each hybridization, oligonucleotide probes are represented by vertical stripes and grouped by virus genera. The mean signal-to-noise ratio (SNR) value for each probe within the array is indicated by the colour of the stripe on a continuous green colour scale. To accommodate array to array variation in SNR range, the upper limit (brightest green colour) of the scale was independently set for each hybridization to the probe with the highest SNR value.
Table 2.
Detection of viruses from infected cell cultures
| Hybridization: | Average signal-to-noise ratios of probes representing: | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Aphtho.* | Pesti. | Vesiculo. | Vesi. | Entero. | Varicello. | Rhadino. | Parapox. | Capripox. | Orbi. | Morbilli. | |
| FMDV A | 148.70 | 0.21 | 0.05 | 0.06 | 0.04 | 0.09 | 0.05 | 0.30 | 0.32 | 0.07 | 0.05 |
| FMDV C | 429.48 | 0.40 | 0.62 | 0.30 | 0.43 | 0.29 | 0.50 | 2.16 | 0.49 | 0.31 | 0.28 |
| FMDV O | 343.54 | 0.33 | 0.23 | 0.29 | 0.42 | 0.18 | 0.19 | 0.53 | 0.24 | 0.19 | 0.22 |
| FMDV Asia1 | 223.47 | 0.13 | 0.10 | 0.01 | 0.18 | −0.02 | 0.01 | 0.06 | 0.07 | 0.36 | 0.01 |
| FMDV SAT1 | 690.84 | 0.66 | 0.17 | 0.25 | 3.52 | 0.16 | 0.21 | 2.06 | 0.32 | 1.00 | 0.23 |
| FMDV SAT2 | 463.08 | 0.32 | 0.29 | 0.33 | 0.23 | 0.20 | 0.32 | 0.26 | 0.35 | 0.30 | 0.27 |
| FMDV SAT3 | 281.94 | 0.46 | 0.56 | 0.61 | 0.50 | 0.91 | 0.47 | 9.68 | 0.63 | 0.43 | 0.34 |
| BVDV Z937 | 0.03 | 4.31 | 0.02 | −0.06 | −0.01 | −0.04 | −0.10 | 0.02 | −0.04 | 0.21 | 0.16 |
| BVDV A531 | 0.20 | 39.44 | 0.71 | 2.46 | 0.17 | 0.59 | 0.23 | 2.19 | 0.47 | 0.31 | 0.18 |
| BVDV P92-4151 | 0.15 | 33.52 | 0.30 | 1.77 | 0.42 | 0.17 | 0.16 | 0.37 | 0.30 | 0.14 | 0.10 |
| VSIV | 0.43 | 0.42 | 36.33 | 0.26 | 0.33 | 0.37 | 0.31 | 0.63 | 0.45 | 0.41 | 0.51 |
| VSNJV | 4.45 | 4.88 | 16.18 | 4.47 | 4.86 | 4.83 | 4.81 | 5.53 | 4.91 | 4.58 | 4.56 |
| VESV A48 | 5.55 | 3.71 | 3.58 | 128.40 | 3.12 | 2.94 | 5.88 | 2.94 | 3.11 | 3.41 | 3.23 |
| SVDV | 46.78 | 8.60 | 5.89 | 4.06 | 494.15 | 9.95 | 6.82 | 10.55 | 7.88 | 6.74 | 8.16 |
| BoHV-1 | 3.37 | 0.91 | 3.52 | 1.76 | 1.44 | 58.18 | 4.88 | 3.08 | 0.57 | 0.64 | 1.71 |
| ORFV | 0.02 | 0.02 | 0.01 | −0.04 | −0.01 | 0.04 | −0.05 | 0.62 | 0.05 | −0.07 | −0.03 |
| PCPV | 0.03 | 0.07 | 0.02 | 0.03 | 0.07 | 0.01 | 0.00 | 1.23 | 0.10 | 0.01 | 0.02 |
| BTV-1 | 6.20 | 6.09 | 5.41 | 5.64 | 5.07 | 6.19 | 6.76 | 6.02 | 6.30 | 68.11 | 5.89 |
| BTV-3 | 13.33 | 12.60 | 10.86 | 12.39 | 12.58 | 12.57 | 13.97 | 12.23 | 14.23 | 12.21 | 12.97 |
| Cell-only control | 11.21 | 11.49 | 12.42 | 17.68 | 11.67 | 11.35 | 13.17 | 11.13 | 13.59 | 11.76 | 11.01 |
For ease of presentation, genus names have been abbreviated by omitting ‘virus’.
3.2. Detection of viruses in vesicular epithelium samples
The ability of the microarray assay to identify viruses in infected vesicular epithelium was tested using samples collected from pigs experimentally infected with VESVA48, and calves experimentally infected with VSNJV. RNA extracted from epithelium was randomly amplified in an identical manner to that employed for the cultured viruses. VESVA48 was identified in 7 of the 8 samples obtained on days 2 through 7 of infection (Table 3a). The titre of VESVA48 in the positive samples tested ranged between 60 and 107 TCID50/10 mg of tissue. The negative sample had a titre of 3000 TCID50/10 mg of tissue (a full list of sample virus titres is contained in Supplementary Table 2a). For 3 of the samples the average SNR for the varicellovirus genus was moderately elevated above background levels. This was due to raised SNR values for 5 BoHV-1 probes, differing in sequence to one another at only 1 or 2 nt sites. This result does not suggest presence of the related Suid herpesvirus 1 (SuHV-1), as none of the other probes representing genome regions conserved between BoHV-1 and SuHV-1 were elevated. Results were less reliable with VSNJV-infected vesicular epithelium. VSNJV was identified in 3 of the 6 samples hybridized (Table 3b) with a sensitivity threshold of approximately 105 TCID50/10 mg of tissue (refer to Supplementary Table 2b for sample virus titres). In addition to a clear elevation of the vesiculovirus average SNR, the Calf 3 Day 3 sample hybridization also showed elevation above background of the parapoxvirus average SNR, a result of a high SNR reading for a single ORFV probe. Similarly, the Calf 4 Day 3 sample hybridization showed an elevation of the aphthovirus average SNR that was due to a high SNR for a single FMDV probe.
Table 3.
| a) Detection of VESV A48 in infected pig vesicular epithelium samples | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample: | Average signal-to-noise ratio of probes representing: | ||||||||||
| Aphtho.* | Pesti. | Vesiculo. | Vesi. | Entero. | Varicello. | Rhadino. | Parapox. | Capripox. | Orbi. | Morbilli. | |
| Pig 1 Day 3 | 10.84 | 9.22 | 8.62 | 180.75 | 8.22 | 11.29 | 9.54 | 11.06 | 8.01 | 7.98 | 8.00 |
| Pig 1 Day 7 | 8.36 | 8.19 | 8.26 | 181.82 | 8.13 | 8.26 | 8.39 | 8.50 | 8.80 | 8.28 | 8.28 |
| Pig 3 Day 3 | 9.22 | 8.88 | 9.75 | 185.49 | 8.41 | 10.10 | 8.20 | 10.24 | 8.97 | 11.89 | 8.14 |
| Pig 3 Day 6 | 7.72 | 7.65 | 7.82 | 487.12 | 10.50 | 27.59 | 8.01 | 7.42 | 7.38 | 7.58 | 7.53 |
| Pig 6 Day 7 | 0.19 | 0.09 | −0.04 | 28.42 | −0.02 | 0.04 | 0.00 | 0.05 | −0.01 | 0.10 | 0.37 |
| Pig 7 Day 2 | 9.23 | 9.14 | 8.70 | 335.54 | 8.86 | 12.26 | 9.09 | 9.01 | 9.63 | 9.07 | 9.51 |
| Pig 7 Day 7 | 12.36 | 12.04 | 11.49 | 13.90 | 11.84 | 27.21 | 11.91 | 12.00 | 14.29 | 12.91 | 13.93 |
| Pig 13 Day 3 | 10.63 | 9.95 | 10.02 | 71.10 | 9.82 | 23.90 | 10.42 | 10.49 | 11.14 | 10.21 | 9.29 |
| b) Detection of VSNJV in infected calf vesicular epithelium samples | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample: | Average signal-to-noise ratio of probes representing: | ||||||||||
| Aphtho.* | Pesti. | Vesiculo. | Vesi. | Entero. | Varicello. | Rhadino. | Parapox. | Capripox. | Orbi. | Morbilli. | |
| Calf 1 Day 6 | 7.26 | 8.37 | 8.03 | 7.86 | 8.86 | 8.26 | 7.67 | 9.18 | 8.01 | 7.92 | 8.70 |
| Calf 3 Day 3 | 10.60 | 7.65 | 67.70 | 8.07 | 7.50 | 8.06 | 7.55 | 24.82 | 7.53 | 7.88 | 7.42 |
| Calf 3 Day 6 | 10.09 | 8.03 | 12.07 | 7.62 | 7.75 | 8.00 | 7.56 | 8.70 | 8.46 | 8.15 | 7.94 |
| Calf 4 Day 3 | 20.42 | 7.26 | 29.63 | 7.43 | 7.27 | 7.32 | 7.01 | 7.65 | 7.22 | 7.26 | 7.08 |
| Calf 5 Day 8 | 8.20 | 7.31 | 26.84 | 7.26 | 7.27 | 7.57 | 8.40 | 7.63 | 7.42 | 7.23 | 7.54 |
| Calf 6 Day 4 | 9.63 | 6.87 | 7.35 | 7.01 | 6.89 | 7.98 | 7.46 | 7.13 | 6.99 | 6.81 | 7.10 |
For ease of presentation, genus names have been abbreviated by omitting ‘virus’.
4. Discussion
The microarray was initially tested using viruses grown to high titre in cell culture, correctly identifying all but one to genus level. Hybridization of the BTV-3 isolate, although returning a BTV probe in top place, failed to raise the average SNR of the 10 BTV probes above background levels. Sequence data for the isolate enabled alignments for 6 of the 10 probes (not including the top placed probe), which showed matches ≥ 54/60 nt with only 2 probes: 55/60 with a VP3 gene probe and 56/60 with a VP1 gene probe. Although only 4 probes (all VP3 gene) could be assessed for the BTV-1 isolate due to limited sequence data, all showed matches ≥ 54/60 nt: 54/60, 57/60, 57/60, and 58/60. We therefore speculate that the failure to identify the BTV-3 isolate may reflect limited probe coverage on the array. In the time elapsed since designing the BTV probes represented on this array the number of BTV nucleotide sequences available on GenBank has increased by 40 %, suggesting that a much more comprehensive probe design for BTV will now be possible using this updated sequence information. Indeed, in silica experiments indicate that BTV-3 would be captured on later generation arrays (Palacios et al. 2007). The PCPV isolate tested on the array was clearly identified as a member of the parapoxvirus genus, even though probes had not been specifically selected to detect it (Table 1). This result is due to the close genetic relationship between PCPV, BPSV and ORFV and the inclusion of probe sequences representing regions of conservation in related viruses.
The 8 VESVA48-infected samples tested on the array had been collected from the snout, oral mucosa and coronary band of infected pigs and varied in titre from 60 to 107 TCID50. The microarray detected VESVA48 in 7 samples, including 5 with virus titres less than 1000 TCID50. The only sample that the microarray did not identify as VESVA48-infected had a virus titre of 3000 TCID50. The observation that the sample yielded an unusually large quantity of RNA (8 µg/10 mg) compared with levels obtained from other samples (150–500 ng/10 mg) suggests that assay failure was due to difficulties with template preparation rather than probe representation. In comparison to the sensitivity of detection of the microarray assay for VESVA48 in vesicular epithelium samples, the sensitivity of detection for VSNJV was somewhat low. Of the 6 samples tested, VSNJV was clearly detected in only 3, all of which had virus titres of at least 105 TCID50. However, analysis of the cell culture-derived virus microarray hybridization results suggests that this disparity could be expected. Although both the VESVA48 and VSNJV cell culture supernatants had similar titres, VESVA48 returned an average SNR for the vesivirus genus 20 times higher than that of other genera represented on the array whereas VSNJV returned an average SNR for the vesiculovirus genus just 3 times higher than the other genera. As the cell culture-derived VSIV isolate tested returned an average SNR for the vesiculovirus genus 50-fold higher than the other genera, it can be surmised that the random PCR method was adequately amplifying the non-segmented negative strand rhabdovirus genome, and that the lower sensitivity of detection in vesicular epithelium samples observed for this VSNJV isolate is likely due to a suboptimal probe set. As with the BTV-3 hybridization described above, more recent arrays are predicted to identify the VSNJV isolate with greater sensitivity.
The aim of this study was to evaluate the potential of a DNA microarray as a diagnostic tool for vesicular or vesicular-like viral diseases of livestock animals. While the hybridization results of 19 different cell culture-derived virus isolates and 2 vesicular epithelium-derived isolates described here are encouraging, they also highlight challenges that need to be addressed before this microarray assay can achieve diagnostic standards. The foremost challenge is to determine the assay’s sensitivity and specificity of detection for each virus, a difficult task given the wide and ever-changing genetic variation of the viruses and the scarcity of infected epithelium samples. Probe sets will need to be modified as this validation proceeds, as has already been demonstrated for BTV and VSNJV. The microarray itself will need to adhere to high quality standards unlikely to be achieved by the in-house contact printing methods used here. In addition to ensuring a high-quality microarray, commercial on-slide probe synthesis would enable the probe sets to be easily updated and provide ample scope for additional probes, for example to facilitate serotyping of FMDV and differentiation of VSNJV and VSIV. Similar to all PCR-based diagnostic tests, strict laboratory procedures to prevent PCR contamination problems are essential to maintain the integrity of this assay. As described in the results section, some hybridization results have hinted that this may already be occurring at low levels. Time is always pertinent in a diagnostic setting and, with minor modification the assay could be completed comfortably within 2 days. A general approach to improve the sensitivity of detection of viruses in epithelium samples could be treatment of the total RNA sample to deplete large ribosomal RNA, thereby enriching for viral genome and/or transcripts, prior to the randomly primed cDNA synthesis step. Although a separate random DNA amplification protocol was used for DNA viruses in this study, it may be preferable both in terms of time and sensitivity to focus solely on viral transcripts in infected cells and tissues (Palacios et al., 2007) or alternatively to develop a combined DNA/RNA isolation and amplification strategy.
We have described the design and use of a DNA microarray-based assay to detect viruses commonly known to cause vesicular or vesicular-like lesions in livestock. More extensive validation of probe sets and optimisation of sample preparation procedures are needed; nonetheless, the successful detection of a range of cell culture-derived viruses as well as of two viruses in infected vesicular epithelium samples demonstrates the potential of utility of this platform in veterinary diagnostic microbiology.
Supplementary Material
Analysis of microarray hybridization data. Each hybridization scatter plot presents the strength of the signal (on the vertical axis) against its quality (on the horizontal axis). The strength of the signal is measured by standard deviation units after within array normalization of the background corrected log-transformed raw signals and is bound to an upper limit of 6. The quality of the signal is measured by the signal369 to-noise ratio of the raw signal and is bound to an upper limit of 20. Each circle on the scatter represents a single spot on the array.
Acknowledgements
We thank Dr Deborah Middleton, Dr John Bingham and Maria Cardoso for their help in the collection of vesicular epithelium samples. The BVDV isolates were kindly provided by Dr Peter Kirkland of the Elizabeth Macarthur Agricultural Institute and Dr John Parkinson of the Department of Agriculture and Food Western Australia. This research was supported through funding from the Australian Biosecurity Cooperative Research Centre, AI57158 (Northeast Biodefense Center-Lipkin), AI070411, and HHSN266200400036C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baner J, Gyarmati P, Yacoub A, Hakhverdyan M, Stenberg J, Ericsson O, Nilsson M, Landegren U, Belak S. Microarray-based molecular detection of foot-and-mouth disease, vesicular stomatitis and swine vesicular disease viruses, using padlock probes. J. Virol. Methods. 2007;143:200–206. doi: 10.1016/j.jviromet.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Baxi MK, Baxi S, Clavijo A, Burton KM, Deregt D. Microarray-based detection and typing of foot-and-mouth disease virus. Vet. J. 2006;172:473–481. doi: 10.1016/j.tvjl.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Bohlander SK, Espinosa R, 3rd, Le Beau MM, Rowley JD, Diaz MO. A method for the rapid sequence-independent amplification of microdissected chromosomal material. Genomics. 1992;13:1322–1324. doi: 10.1016/0888-7543(92)90057-y. [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- Deregt D, Gilbert SA, Dudas S, Pasick J, Baxi S, Burton KM, Baxi MK. A multiplex DNA suspension microarray for simultaneous detection and differentiation of classical swine fever virus and other pestiviruses. J. Virol. Methods. 2006;136:17–23. doi: 10.1016/j.jviromet.2006.03.025. [DOI] [PubMed] [Google Scholar]
- Geering WA, Forman AJ, Nunn MJ. Canberra: Australian Government Publishing Service; 1995. Exotic Diseases of Animals: a field guide for Australian veterinarians; pp. 127–128. [Google Scholar]
- Gonzalez SF, Krug MJ, Nielsen ME, Santos Y, Call DR. Simultaneous detection of marine fish pathogens by using multiplex PCR and a DNA microarray. J. Clin. Microbiol. 2004;42:1414–1419. doi: 10.1128/JCM.42.4.1414-1419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G, Quan PL, Jabado OJ, Conlan S, Hirschberg DL, Liu Y, Zhai J, Renwick N, Hui J, Hegyi H, Grolla A, Strong JE, Towner JS, Geisbert TW, Jahrling PB, Buchen-Osmond C, Ellerbrok H, Sanchez-Seco MP, Lussier Y, Formenty P, Nichol MS, Feldmann H, Briese T, Lipkin WI. Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg. Infect. Dis. 2007;13:73–81. doi: 10.3201/eid1301.060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas P, Purchase HG. Titration of Biological Suspensions. In: Hitchner SB, Domermuth CH, Purchase HG, Williams JE, editors. Isolation and Identification of Avian Pathogens. Texas: American Association of Avian Pathologists; 1980. pp. 124–128. [Google Scholar]
- Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc. Natl. Acad. Sci. USA. 2002;99:15687–15692. doi: 10.1073/pnas.242579699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of microarray hybridization data. Each hybridization scatter plot presents the strength of the signal (on the vertical axis) against its quality (on the horizontal axis). The strength of the signal is measured by standard deviation units after within array normalization of the background corrected log-transformed raw signals and is bound to an upper limit of 6. The quality of the signal is measured by the signal369 to-noise ratio of the raw signal and is bound to an upper limit of 20. Each circle on the scatter represents a single spot on the array.