Abstract
Context
Glutaredoxins (GRX) are involved in the regulation of thiol redox state. GRX-1 is a cytosolic enzyme responsible for the catalysis of deglutathionylation of proteins. To date, very few inhibitors of GRX-1 have been reported.
Objective
The objective of this paper is to report 2-acetylamino-3-[4-(2-acetylamino-2-carboxyethyl-sulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid (2-AAPA) as an inhibitor of human GRX-1.
Materials and methods
The mechanism of inhibition of GRX-1 was investigated using dialysis, substrate protection, and mass spectrometry.
Results
2-AAPA inhibits GRX-1 in a time and concentration dependent manner. The activity did not return following dialysis indicating that inhibition is irreversible. Results of substrate protection and mass spectrometry indicate that the inhibition is occurring at the active site. The compound also produced GRX inhibition in human ovarian cancer cells.
Discussion
2-AAPA is an irreversible GRX-1 inhibitor with similar or greater potency compared to previously reported inhibitors.
Conclusion
The inhibition of GRX-1 by 2-AAPA could be used as a tool to study thiol redox state.
Keywords: glutathione, glutathionylation, thiol, glutaredoxin
Introduction
Glutaredoxins (GRX), also known as thioltransferases, are enzymes that are important in the maintenance of thiol redox state. Specifically, GRX is the enzyme that catalyzes the removal of glutathione (GSH) from a protein mixed disulfide (PS-SG) formed between a protein thiol (PSH) and glutathione, a process known as deglutathionylation. GRXs can be classified as monothiol or dithiol enzymes depending on the number of active site cysteine residues.(1) GRX-1 is a 12 kDa cytosolic isoform and contains two cysteine residues in the active site (Cys-22 and Cys-25 in human GRX-1). However, only one of the thiols is required for the deglutathionylation reaction. The N-terminal active site cysteine (Cys-22) has a very low pKa (~3.5), which means that it will be in the thiolate (−S−) form at physiologic pH and capable of reacting with the mixed disulfide PS-SG. During deglutathionylation, the GSH unit (−SG) of PS-SG is transferred to this cysteine to form a mixed disulfide bond (GRX-S-SG). Subsequent removal of the GSH unit from the GRX-S-SG is achieved by another molecule of GSH to regenerate GRX and produce a molecule of glutathione disulfide (GSSG). The GSSG is reduced to GSH by glutathione reductase (GR) (Figure 1).(2–6) The other GRX isoforms found in mammalian cells include the mitochondrial and nuclear dithiol GRX-2, the cytosolic monothiol GRX-3, and the mitochondrial monothiol GRX-5.(7)
Figure 1.
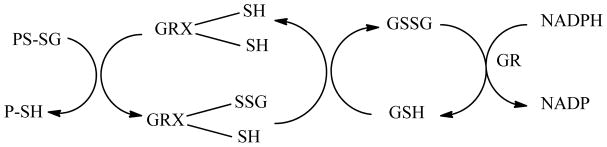
Mechanism of catalysis of deglutathionylation by GRX. A glutathionylated protein (PS-SG) is restored by the action of GRX. GRX operates by a monothiol mechanism in the process of deglutathionylation. The glutathione (GSH) is transferred from the protein to GRX. A second GSH molecule restores the normal GRX active site structure with the production of GSSG. The GSSG produced is reduced to GSH by the action of GR which uses NADPH as a cofactor.
To date, only a few GRX inhibitors have been reported.(8–15) Cadmium is one of the most commonly utilized inhibitors of GRX. Cadmium chloride 100 μM was reported to inhibit GRX activity in lung cancer cells by 32%.(8) An earlier examination of the effect of cadmium on GRX activity reported almost complete inhibition at 100 μM in H9 and Jurkat cells.(9) A few nonmetal inhibitors have also been reported. 100 μM L-DOPA treatment resulted in around 60% inhibition of GRX activity in a dopaminergic neuron model; analysis revealed that a quinone metabolite of L-DOPA was responsible for the enzyme inhibition.(10) Sporidesmin, a fungal toxin, inhibited GRX-1 activity to around 15% of control activity at a concentration of 1 mM; however the inhibition only occurred in the absence of GSH.(11) A GSH-platinum complex, a major metabolite of cisplatin, inhibited human GRX with an IC50 of 350 μM.(12) Peroxynitrite produced good inhibition of GRX activity at concentrations above 200 μM.(13)
Because of the lack of potent GRX inhibitors, the development of agents that can inhibit the activity of this enzyme is needed. Previously, this group reported 2-acetylamino-3-[4-(2-acetylamino-2-carboxyethylsulfanylthiocarbonylamino)phenylthiocarbamoylsulfanyl]propionic acid (2-AAPA) (Figure 2) as an irreversible inhibitor of GR with a Ki of 56 μM and a kinact of 0.143 min−1 against yeast GR.(16) 2-AAPA was also shown to inhibit GR, increase GSSG, and produce increased glutathionylation in CV-1 (monkey kidney) cells.(16, 17) In this study, the potential for human GRX-1 inhibition by 2-AAPA was evaluated.
Figure 2.
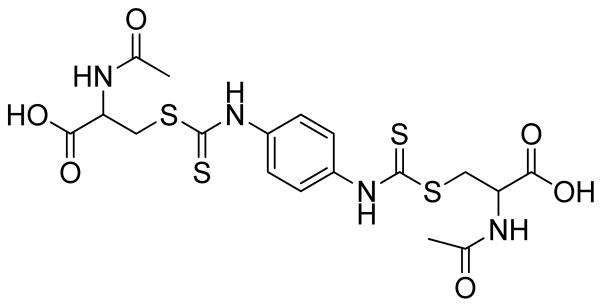
Structure of 2-AAPA.
Methods
Materials
All reagents for enzyme assays, including human recombinant GRX-1 and yeast GR, were purchased from Sigma-Aldrich Chemical Co (Milwaukee, WI). RPMI 1640 growth medium, penicillin/streptomycin, phosphate buffered saline (PBS), and trypsin were purchased from Mediatech (Herndon, VA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals (Lawrenceville, GA). OVCAR-3 cells were obtained from the National Institutes of Health National Cancer Institute. 2-AAPA was synthesized in this laboratory according to a previously published method.(16) The 2-AAPA was prepared as a 6.67 mM stock solution in a 3:1 solution of water and tetrahydrofuran (THF) for all enzyme assays except the cell based assay. For the incubation of 2-AAPA with OVCAR-3 cells, a 2 mM stock solution was prepared in RPMI 1640 growth medium; the stock solution was prepared fresh and used immediately for each treatment.
GRX Assay
GRX activity was determined from a coupled reaction with GR. In this assay, a mixed disulfide between GSH and the mercaptoethanol moiety derived from 2-hydroxyethyl disulfide (HED) served as the substrate for GRX; briefly GSH (10 mM) and HED (7 mM) were premixed in water for 5 minutes before transferring onto ice. The final GRX assay solution contained GSH (1 mM), HED (0.7mM), GR (0.02 units/mL), NADPH (0.2 mM), and bovine serum albumin (BSA, 1 mg/mL) in Tris buffer (pH 8, 0.1 M). The activity was determined by monitoring the disappearance of NADPH spectrophotometrically at λ=340 nm.(18)
Kinetics of GRX-1 Inhibition
The time and concentration dependence of GRX-1 inhibition by 2-AAPA was evaluated and used to determine parameters of enzyme inhibition kinetics. Human GRX-1 (0.25 unit/mL) was incubated at 25°C with increasing concentrations of 2-AAPA (25, 50, 100, and 200 μM) and BSA (1 mg/mL). Aliquots were withdrawn for determination of GRX activity at 3, 10, and 20 minutes. Control incubations without 2-AAPA were conducted in parallel. The aliquots were added to the GRX assay solution, and enzyme activity was determined as described above. Inhibitory parameters, Ki and kinact, were determined by the method of Kitz and Wilson.(19)
Irreversibility of GRX inhibition
Human GRX-1 (0.9 units/mL) was incubated with 2-AAPA (1 mM) for 1 hour at 25°C to obtain complete inactivation. The incubation solution was then transferred to a 5 mL DisPoDialyzer (Spectrum) with a molecular weight cut-off of 3.5–5 kDa and extensively dialyzed in Tris buffer (2 x 800 mL, pH 8, 0.1 M). Aliquots were withdrawn after 30 min, 1 hr, 2 hr, and 4 hr for analysis of GRX-1 activity. A control incubation without 2-AAPA was conducted simultaneously.
Substrate Protection
Human GRX-1 (1 unit/mL) was incubated with 2-AAPA 100 μM at 25°C in the presence or absence of increasing concentrations of GSH-HED, the GRX substrate. Aliquots were withdrawn for determination of GRX activity. Controls were also conducted in the presence or absence of GSH-HED.
Determination of Covalent Binding
The potential for covalent bond formation between 2-AAPA and GRX was determined by liquid chromatography/mass spectrometry (LC/MS) analysis. GRX-1 (200 ng/μL) was incubated with 2-AAPA 100 μM for 20 minutes. A nano Acquity UPLC connected to a Waters Micromass QTOF MS was used for the sample analysis. The separation was conducted on a C18 column (flow rate 0.4 μL/min) using a gradient program where solvent B was increased from 3% to 80% in 17 minutes (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile). Analytes were detected in positive ion mode over an m/z range of 600–2000, and the deconvoluted spectra were analyzed to determine the protein molecular weight.
Inhibition of GRX in OVCAR-3 Cells
OVCAR-3 (human ovarian cancer) cells (2.5 million) were placed in a 185 cm2 flask in RPMI 1640 growth medium containing 10% FBS and 1% penicillin/streptomycin. The cells were placed in a humidified 5% CO2 incubator for 24 hours prior to treatment. A solution of 2-AAPA was prepared in growth medium, and the compound was added to the cells at a final concentration of 100 μM. After a 20 minute incubation, the cells were detached by trypsinization and collected by centrifugation. The cell pellet was washed with ice cold PBS containing 1 mM EDTA and resuspended in 1 mL of hypotonic (1 mM) phosphate buffer containing 1 mM EDTA. The cells were homogenized over ice using an OMNI 5000 homogenizer. The homogenate was centrifuged at 120,000 x g at 4°C for 30 minutes. The supernatant was collected and used for determination of GRX activity. The activity was standardized by protein content as determined by the BCA method.
Results
Kinetics of GRX Inhibition
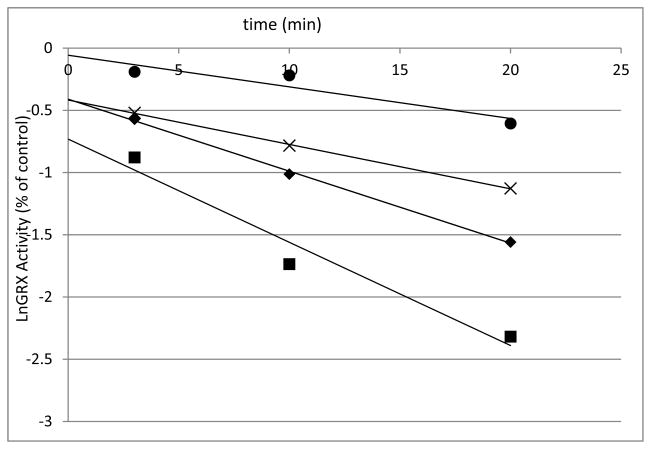
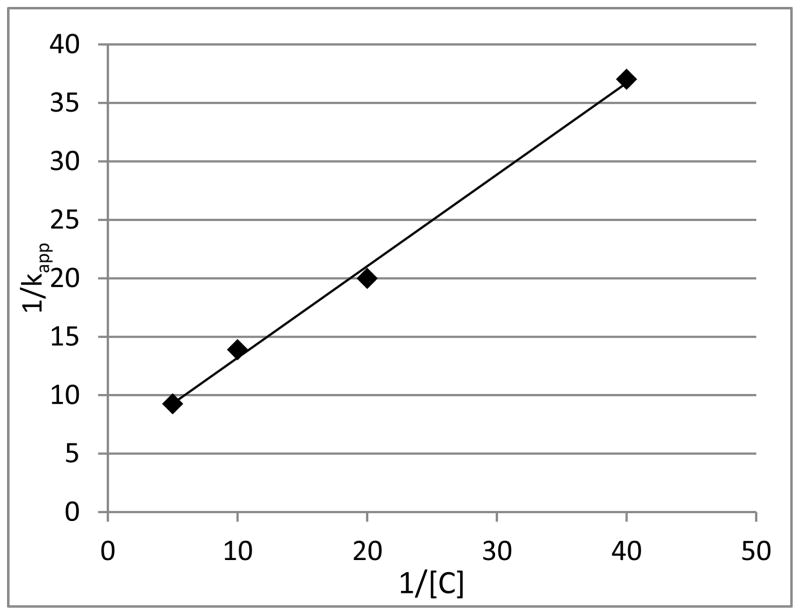
Human GRX-1 was shown to be inhibited by 2-AAPA in a concentration and time dependent manner. Figure 3 shows the natural logarithm of GRX activity versus time at increasing concentrations of 2-AAPA. The loss of enzyme activity over time shown in the plot is characteristic of irreversible enzyme inhibitors. The inhibitory parameters were determined by plotting the reciprocal of the apparent rate constants of inhibition (kapp, slopes from Figure 3) versus the reciprocal of the inhibitor concentration (Figure 4).(19) The Ki and kinact of 2-AAPA against human GRX-1 were determined to be 91.3 μM and 0.127 min−1, respectively.
Figure 3.
Time and concentration dependence of GRX-1 inhibition by 2-AAPA. The natural logarithm of GRX-1 remaining activity is plotted against time. The enzyme was incubated with increasing concentrations of 2-AAPA, and aliquots were withdrawn at various time points for determination of remaining GRX-1 activity. The graph shows a representative plot from one of triplicate experiments. ■, 200 μM; ◆ 100 μM; x, 50 μM; ●, 25 μM.
Figure 4.
Double reciprocal plot. The reciprocals of the apparent rate constants of inhibition (slopes from Figure 3) are plotted against the reciprocals of 2-AAPA concentration. The graph shows a representative plot from one of triplicate experiments.
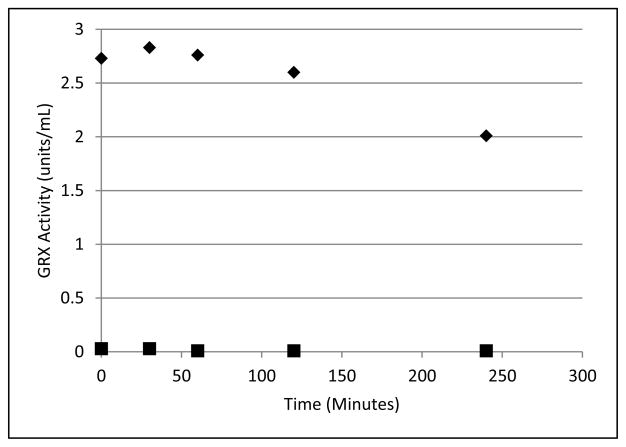
Irreversibility of Inhibition
Dialysis of the inhibited enzyme was conducted to confirm the irreversibility of the inhibition by 2-AAPA. No recovery of enzyme activity was observed after four hours of dialysis, thus confirming irreversible inactivation of GRX-1 by 2-AAPA (Figure 5).
Figure 5.
Irreversibility of GRX-1 inhibition by 2-AAPA. GRX-1 was incubated with 2-AAPA 1 mM for 1 hour to achieve complete inhibition. Then the solution was transferred to a DisPoDialyzer and dialyzed in Tris buffer for 4 hours. No enzyme activity returned in the 2-AAPA sample over a 4 hour period. The graph shows a representative plot from one of triplicate experiments. ◆, control; ■, 2-AAPA treated.
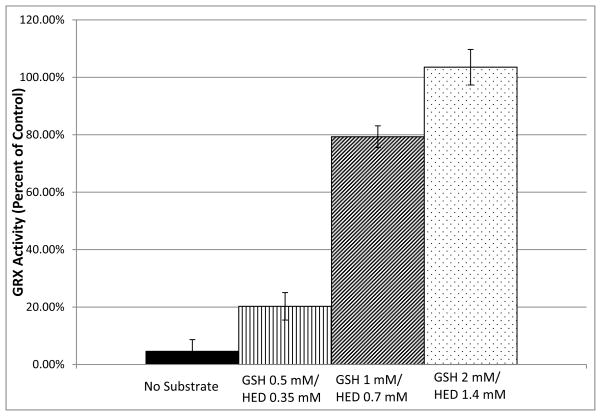
Substrate Protection
Inhibition of GRX-1 by 2-AAPA was conducted in the presence and absence of GSH-HED in order to evaluate whether 2-AAPA is a competitive inhibitor of GRX and acting at the active site of the enzyme. The presence of GSH-HED in the incubation solution did protect the enzyme from inhibition by 2-AAPA. The impact of the substrate on GRX-1 inhibition occurred in a concentration dependent manner; higher concentrations of the substrate were able to completely protect the enzyme from inhibition (Figure 6). This indicates that 2-AAPA and GSH-HED are competing for the same site on GRX-1 and shows that the inhibitor is binding at the enzyme active site.
Figure 6.
Substrate protection of GRX from 2-AAPA inhibition. GRX substrate GSH-HED protected the enzyme from inhibition by 2-AAPA in a concentration dependent manner indicating that 2-AAPA is a competitive inhibitor of GRX-1 (n=3).
Determination of Covalent Binding
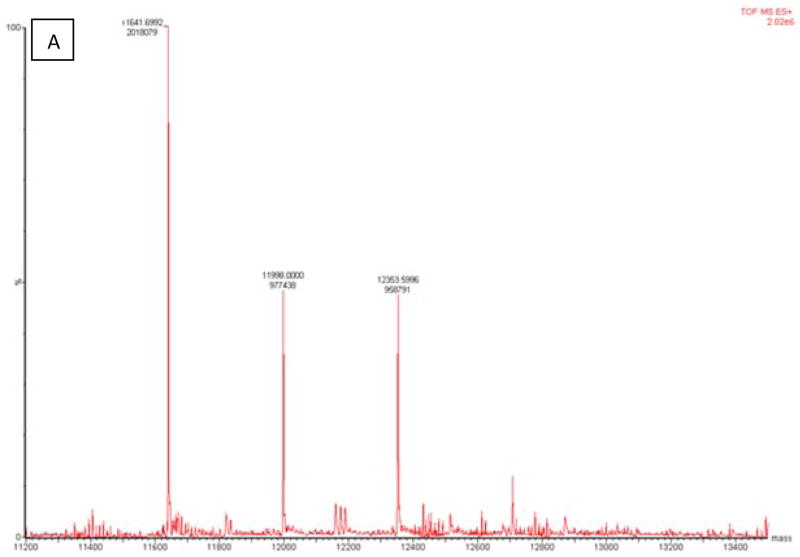
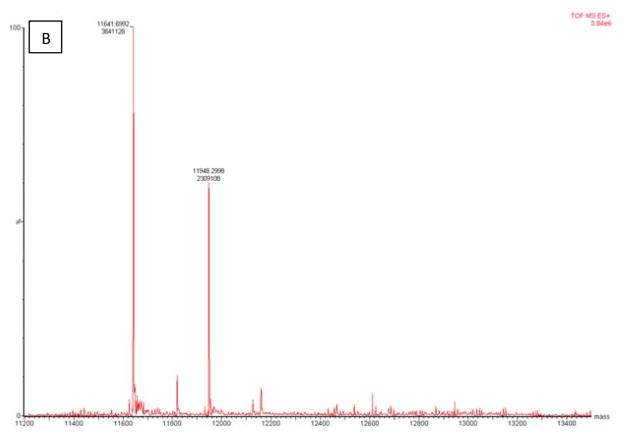
LC/MS analysis of the inhibited GRX-1 was conducted in the absence and presence of the substrate GSH-HED. The unmodified enzyme was found to have an m/z of 11641, which is consistent with the reported molecular weight of human GRX-1. LC/MS analysis of the 2-AAPA inhibited enzyme revealed additional peaks at m/z 11997 and 12353, corresponding to mass increases of 356 and 712, respectively. The increase of 356 results from monothiocarbamoylation of GRX-1 by 2-AAPA while the increase of 712 indicates monothiocarbamoylation at two different sites on the enzyme. The presence of the substrate prevented the formation of these two additional peaks indicating that the binding is occurring with the cysteine residues at the active site (Figure 7).
Figure 7.
LC/MS analysis of covalent binding of 2-AAPA to GRX. Panel A: GRX-1 inhibited by 2-AAPA 0.1 mM for 20 minutes; Panel B: GRX-1 with 2-AAPA 0.1 mM and substrate (GSH 2 mM/HED 1.4 mM). The native enzyme has an m/z of 11641. In the inhibited sample, additional peaks are observed at m/z 11997 and 12353, corresponding to monothiocarbamoylation at one or two cysteines, respectively. These signals are not observed in the sample incubated with inhibitor and substrate; the signal at m/z 11948 corresponds to the addition of glutathione.
Inhibition of GRX in OVCAR-3 Cells
In order to evaluate the effect of 2-AAPA on intracellular GRX activity, the inhibitor was incubated with OVCAR-3 cells, and the GRX activity was determined. After a 20 minute treatment, 67% ±5% of the GRX activity was inhibited by 2-AAPA at 0.1 mM concentration, a concentration close to its Ki value, indicating that 2-AAPA can also inhibit intracellular GRX.
Discussion
Thiols and enzymes involved in regulation of cellular thiol status are important for the cell’s protection against oxidative stress. GRX is involved in the regulation of glutathionylation and maintenance of thiol redox state. Therefore, inhibitors of GRX would be valuable in order to further study the role of GRX in the cell and the impact of glutathionylation on cellular function.
In this study, we have shown that a previously reported inhibitor of GR is also an inhibitor of GRX-1. The inhibition of GRX-1 by 2-AAPA occurs at similar or lower concentrations than previously reported GRX inhibitors indicating favorable potency. The time dependent inhibition and the lack of return of enzyme activity following extensive dialysis confirms that the inhibition of GRX-1 by 2-AAPA is irreversible. The results of the substrate protection experiment indicate that this inhibition is occurring at the enzyme active site. Previous studies with 2-AAPA report that the compound preferentially reacts with thiols and not with other nucleophiles.(16) This would indicate that the compound is likely binding to a cysteine in the active site. The LC/MS analysis of covalent bonding between 2-AAPA and GRX-1 demonstrate that monocarbamoylation was occurring at one or two cysteine residues. Human GRX-1 has two cysteine residues in the active site and three non-active site cysteine residues. The non-active site cysteines are not believed to be required for GRX activity.(3, 20) The catalysis of deglutathionylation only requires one of the active site cysteines, Cys-22.(2–6) It is likely that the carbamoylation leading to loss of GRX activity then is occurring at Cys-22. This would be consistent with previous reports of irreversible GRX inhibitors that exhibited covalent bonding between the inhibitor and Cys-22.(10, 11, 14) The proposed mechanism of 2-AAPA binding to the active site of human GRX-1 is shown in figure 8.
Figure 8.
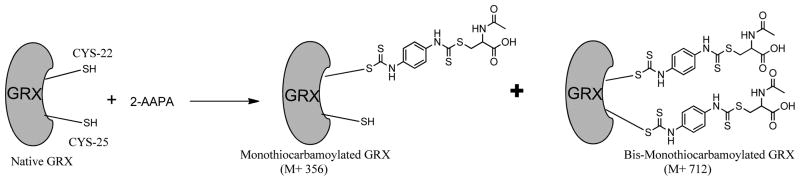
Proposed mechanism of 2-AAPA binding to GRX-1. Data from mass spectrometry analysis of the inhibited enzyme show that 2-AAPA binds covalently to GRX-1. Monothiocarbamoylation of the enzyme can occur at one or two cysteines in GRX. Based on the pKa, it is likely that the monothiocarbamoylation leading to GRX-1 inhibition is occurring at CYS-22.
In this investigation, the enzyme inhibition by 2-AAPA was evaluated using recombinant human GRX-1. Inhibition of the other GRX isoforms by 2-AAPA has not been specifically investigated, although the cell homogenate would contain other isoforms. The 67% inhibition observed with OVCAR-3 cell homogenate at 0.1 mM 2-AAPA does fit with the observed Ki of 91 μM with the GRX-1 isoform. The other dithiol GRX isoform, GRX-2, has a similar active site as GRX-1, but the two isoforms share less than 40% sequence identity.(21, 22) Also, GRX-2 has been reported to have significantly lower specific activity in the HED assay compared to GRX-1.(22) In addition, there is conflicting evidence about whether monothiol GRXs from various species have activity with the HED-based assay.(23, 24) Based on this information, it is likely that the inhibition observed in cell homogenate is reflective of GRX-1 inhibition.
2-AAPA was previously reported as a GR inhibitor. Inhibition of both GR and GRX may raise a concern about the selectivity of enzyme inhibition by 2-AAPA. There are three enzyme systems that are closely related in the regulation of thiol redox state: GR, GRX and the thioredoxin system (consisting of thioredoxin reductase and thioredoxin). All of these enzymes achieve their function through reduction of a disulfide bond. GR is responsible for the reduction of GSSG back to GSH. GRX is responsible for the reduction of a disulfide bond between a protein thiol and glutathione (deglutathionylation). The thioredoxin system is responsible for the reduction of a disulfide bond between two protein thiols. An inhibitor which can inhibit more than one of these enzymes or all of the enzymes will more effectively increase intracellular thiol oxidative stress. In fact, we have obtained preliminary data which demonstrates that 2-AAPA also inhibits thioredoxin reductase. Therefore, inhibition of both GR and GRX, and potentially the thioredoxin system, is considered to be an advantage of using 2-AAPA as a tool to modulate intracellular thiol oxidative stress. Actually, inhibition of all of the enzymes involved in regulation of thiol redox state may not be a surprise since there is structural similarity in the active site of the enzymes; structurally, all of these enzymes contain cysteine residues at the active site facilitating the reduction of the disulfide bond of the substrate. It needs to be noted that the selectivity of 2-AAPA against other enzymes involved in the glutathione pathway has been reported earlier. 2-AAPA exhibited no or minimal inhibition of other enzymes including glutathione S-transferase, glutathione peroxidase, γ-glutamylcysteine synthetase, and glutathione synthetase.(16)
Conclusions
In summary, 2-AAPA is an irreversible inhibitor of human GRX-1 with favorable potency compared to previously reported GRX inhibitors. Inhibition of intracellular GRX was also observed with 2-AAPA. The enzyme inhibition can be useful as a tool to study thiol redox state and the role of the enzymes involved in regulation of thiol redox state.
Footnotes
Declaration of Interest
Funding for this project was provided by the National Institutes of Health (CA120062-01).
References
- 1.Kim MJ, Lee BC, Jeong J, Lee KJ, Hwang KY, Gladyshev VN, et al. Tandem use of selenocysteine: adaptation of a selenoprotein glutaredoxin for reduction of selenoprotein methionine sulfoxide reductase. Mol Microbiol. 2011;79(5):1194–203. doi: 10.1111/j.1365-2958.2010.07500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanschmann EM, Lonn ME, Schutte LD, Funke M, Godoy JR, Eitner S, et al. Both thioredoxin 2 and glutaredoxin 2 contribute to the reduction of the mitochondrial 2-Cys peroxiredoxin Prx3. J Biol Chem. 2010;285(52):40699–705. doi: 10.1074/jbc.M110.185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jensen KS, Winther JR, Teilum K. Millisecond dynamics in glutaredoxin during catalytic turnover is dependent on substrate binding and absent in the resting states. J Am Chem Soc. 2011;133(9):3034–42. doi: 10.1021/ja1096539. [DOI] [PubMed] [Google Scholar]
- 4.Iversen R, Andersen PA, Jensen KS, Winther JR, Sigurskjold BW. Thiol-disulfide exchange between glutaredoxin and glutathione. Biochemistry. 2010;49(4):810–20. doi: 10.1021/bi9015956. [DOI] [PubMed] [Google Scholar]
- 5.Gallogly MM, Starke DW, Mieyal JJ. Mechanistic and kinetic details of catalysis of thiol-disulfide exchange by glutaredoxins and potential mechanisms of regulation. Antioxid Redox Signal. 2009;11(5):1059–81. doi: 10.1089/ars.2008.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100(2):213–9. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 7.Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochim Biophys Acta. 2008;1780(11):1304–17. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y, Rahmanto YS, Hawkins CL, Richardson DR. The potent and novel thiosemicarbazone chelators di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone and 2-benzoylpyridine-4,4-dimethyl-3-thiosemicarbazone affect crucial thiol systems required for ribonucleotide reductase activity. Mol Pharmacol. 2011;79(6):921–31. doi: 10.1124/mol.111.071324. [DOI] [PubMed] [Google Scholar]
- 9.Chrestensen CA, Starke DW, Mieyal JJ. Acute cadmium exposure inactivates thioltransferase (glutaredoxin), inhibits intracellular reduction of protein-glutathionyl-mixed disulfides, and initiates apoptosis. J Biol Chem. 2000;275(34):26556–65. doi: 10.1074/jbc.M004097200. [DOI] [PubMed] [Google Scholar]
- 10.Sabens EA, Distler AM, Mieyal JJ. Levodopa Deactivates Enzymes That Regulate Thiol-Disulfide Homeostasis and Promotes Neuronal Cell Death: Implications for Therapy of Parkinson’s Disease. Biochemistry. 2010;49(12):2715–24. doi: 10.1021/bi9018658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan U, Bala A, Jao SC, Starke DW, Jordan TW, Mieyal JJ. Selective inactivation of glutaredoxin by sporidesmin and other epidithiopiperazinediones. Biochemistry. 2006;45(29):8978–87. doi: 10.1021/bi060440o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arner ES, Nakamura H, Sasada T, Yodoi J, Holmgren A, Spyrou G. Analysis of the inhibition of mammalian thioredoxin, thioredoxin reductase, and glutaredoxin by cis-diamminedichloroplatinum (II) and its major metabolite, the glutathione-platinum complex. Free Radic Biol Med. 2001;31(10):1170–8. doi: 10.1016/s0891-5849(01)00698-0. [DOI] [PubMed] [Google Scholar]
- 13.Aykac-Toker G, Bulgurcuoglu S, Kocak-Toker N. Effect of peroxynitrite on glutaredoxin. Hum Exp Toxicol. 2001;20(7):373–6. doi: 10.1191/096032701680350578. [DOI] [PubMed] [Google Scholar]
- 14.Yang YF, Wells WW. Catalytic mechanism of thiol transferase. J Biol Chem. 1991;266(19):12766–71. [PubMed] [Google Scholar]
- 15.Hatakeyama M, Lee CP, Chon CG, Hayashi M, Mizoguchi T. Purification and some properties of rabbit liver cytosol thiol-transferase. J Biochem. 1985;97(3):893–7. doi: 10.1093/oxfordjournals.jbchem.a135131. [DOI] [PubMed] [Google Scholar]
- 16.Seefeldt T, Zhao Y, Chen W, Raza AS, Carlson L, Herman J, et al. Characterization of a Novel Dithiocarbamate Glutathione Reductase Inhibitor and Its Use as a Tool to Modulate Intracellular Glutathione. J Biol Chem. 2009;284(5):2729–37. doi: 10.1074/jbc.M802683200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, Seefeldt T, Chen W, Wang XQ, Matthees D, Hu YS, et al. Effects of glutathione reductase inhibition on cellular thiol redox state and related systems. Arch Biochem Biophys. 2009;485(1):56–62. doi: 10.1016/j.abb.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–92. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- 19.Kitz R, Wilson IB. Esters of methanesulfonic acid as irreversible inhibitors of acetylcholinesterase. J Biol Chem. 1962;237(10):3245. [PubMed] [Google Scholar]
- 20.Terada T, Okamoto K, Nishikawa J, Miura T, Nishinaka T, Nishihara T. Site-directed mutagenesis of rat thioltransferase: effects of essential cysteine residues for the protection against oxidative stress. J Biochem Mol Toxicol. 2010;24(1):60–5. doi: 10.1002/jbt.20312. [DOI] [PubMed] [Google Scholar]
- 21.Gladyshev VN, Liu A, Novoselov SV, Krysan K, Sun QA, Kryukov VM, et al. Identification and characterization of a new mammalian glutaredoxin (thioltransferase), Grx2. J Biol Chem. 2001;276(32):30374–80. doi: 10.1074/jbc.M100020200. [DOI] [PubMed] [Google Scholar]
- 22.Lundberg M, Johansson C, Chandra J, Enoksson M, Jacobsson G, Ljung J, et al. Cloning and expression of a novel human glutaredoxin (Grx2) with mitochondrial and nuclear isoforms. J Biol Chem. 2001;276(28):26269–75. doi: 10.1074/jbc.M011605200. [DOI] [PubMed] [Google Scholar]
- 23.Herrero E, de la Torre-Ruiz MA. Monothiol glutaredoxins: a common domain for multiple functions. Cell Mol Life Sci. 2007;64(12):1518–30. doi: 10.1007/s00018-007-6554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamarit J, Belli G, Cabiscol E, Herrero E, Ros J. Biochemical characterization of yeast mitochondrial Grx5 monothiol glutaredoxin. J Biol Chem. 2003;278(28):25745–51. doi: 10.1074/jbc.M303477200. [DOI] [PubMed] [Google Scholar]