The ER quality control factor UDP-glucose:glycoprotein glucosyltransferase 1 modifies folding intermediates but most efficiently terminally misfolded targets. Whereas on-pathway targets are retained in the ER when trapped in monoglucosylated states, off-pathway proteins are degraded rapidly, exposing a dominant ERAD selection process.
Abstract
UDP-glucose:glycoprotein glucosyltransferase 1 (UGT1) is a central quality control gatekeeper in the mammalian endoplasmic reticulum (ER). The reglucosylation of glycoproteins supports their rebinding to the carbohydrate-binding ER molecular chaperones calnexin and calreticulin. A cell-based reglucosylation assay was used to investigate the role of UGT1 in ER protein surveillance or the quality control process. UGT1 was found to modify wild-type proteins or proteins that are expected to eventually traffic out of the ER through the secretory pathway. Trapping of reglucosylated wild-type substrates in their monoglucosylated state delayed their secretion. Whereas terminally misfolded substrates or off-pathway proteins were most efficiently reglucosylated by UGT1, the trapping of these mutant substrates in their reglucosylated or monoglucosylated state did not delay their degradation by the ER-associated degradation pathway. This indicated that monoglucosylated mutant proteins were actively extracted from the calnexin/calreticulin binding-reglucosylation cycle for degradation. Therefore trapping proteins in their monoglucosylated state was sufficient to delay their exit to the Golgi but had no effect on their rate of degradation, suggesting that the degradation selection process progressed in a dominant manner that was independent of reglucosylation and the glucose-containing A-branch on the substrate glycans.
INTRODUCTION
Proteins that traverse the eukaryotic secretory pathway fold in the oxidizing environment of the endoplasmic reticulum (ER) (Braakman and Hebert, 2013). Because protein folding and maturation are error-prone steps, a quality control process is in place in the ER to evaluate the structural integrity of maturing proteins (Hebert and Molinari, 2007; Vembar and Brodsky, 2008; Lederkremer, 2009). Properly folded proteins are permitted to traffic to various locations within the cell to perform their activities, whereas misfolded proteins are targeted for retention and subsequent degradation. The triaging decision that evaluates proteins in the ER as native or aberrant is based on the exposure of general nonnative structural hallmarks such as exposed hydrophobic residues or free thiols (Ellgaard et al., 1999; Anelli and Sitia, 2008). Because of these different fundamental properties, it is relatively straightforward to conceptualize how a quality control process could differentiate between native and aberrant substrates for a diverse array of proteins. The more difficult decision to envision is how the cellular quality control machinery discriminates between immature folding intermediates that transiently display nonnative characteristics and terminally misfolded structures. In other words, how does the quality control process distinguish an on-pathway foldable intermediate that should be provided further time to mature compared with an irreparable off-pathway misfolded product that should be targeted for turnover? Understanding how these fundamental quality control sorting decisions are made and implemented is important for understanding protein homeostasis and the basis for a large number of disease states that are caused by inefficient maturation, subsequent degradation, or toxic accumulation of secretory pathway cargo or an overzealous quality control process that inappropriately retains and degrades active client proteins (Hebert and Molinari, 2007; Balch et al., 2008; Hartl et al., 2011).
Mammalian secretory pathway cargoes are commonly glycosylated, and the process of glycoprotein quality control uses glycans as protein maturation or quality control tags (Cabral et al., 2001; Helenius and Aebi, 2004; Caramelo and Parodi, 2008; Lederkremer, 2009; Hebert and Molinari, 2012; Hebert et al., 2014). The composition of a substrate's N-linked glycans dictates downstream carbohydrate-based interactions that support ER retention, exit, or degradation. Carbohydrates are generally added to the nascent chains cotranslationally as they enter the ER lumen (Aebi et al., 2010). A preassembled Glc3Man9GlcNAc2 (Glc, glucose; Man, mannose; GlcNAc2, N-acetylglucosamine) glycan is transferred to the consensus site en bloc by the oligosaccharyltransferase complex. The glycan is then rapidly trimmed to the monoglucosylated form by the sequential actions of glucosidases I and II. The monoglucosylated glycoform is recognized by the carbohydrate-binding chaperone calnexin and its soluble paralogue calreticulin, which assist in the maturation of the nascent chain (Helenius and Aebi, 2004; Hebert et al., 2005; Pearse and Hebert, 2010). Trimming of the last glucose releases the glycoproteins from these lectin chaperones or ablates their rebinding. Uridine diphosphate (UDP)-glucose:glycoprotein glucosyltransferase 1 (UGT1) is an ER-resident protein that helps in the maturation of glycoproteins by reglucosylating maturing proteins, therefore supporting additional rounds of lectin chaperone binding (Caramelo and Parodi, 2008). Although UGT1 appears to be a central protein sensor in the ER, its role in quality control is not completely understood.
An ER reglucosylation activity was discovered by Parodi and colleagues, and in these early studies, they found that microsomal extracts glucosylated isolated thyroglobulin, a large and heavily glycosylated secretory protein (Trombetta et al., 1989). The reglucosylation activity from ER extracts preferentially modified denatured thyroglobulin compared with the native protein. These results were supported by subsequent studies using UGT purified from rat livers, Schizosaccharomyces pombe, and Drosophila, and smaller, more-defined substrates such as ribonuclease B, phytohemagglutinin, and soybean agglutinin (Sousa et al., 1992; Trombetta and Parodi, 1992; Fernandez et al., 1994; Parker et al., 1995). Although these studies were in agreement with the conclusion that UGT1 efficiently modified nonnative substrates, more recent studies have investigated the properties of the nonnative proteins that most effectively supported reglucosylation to characterize the specificity of UGT1. Isolated UGT1 was found to favor the modification of near-native folding intermediates or orphan subunits over native or extensively misfolded substrates (Trombetta and Helenius, 2000; Caramelo et al., 2003, 2004; Taylor et al., 2004; Keith et al., 2005; Ritter et al., 2005). The reglucosylation activity of UGT1 even serves to retain assembled major histocompatibility class I antigen presentation complex (MHC I) in the ER until it is loaded with a high-affinity peptide (Zhang et al., 2011; Blum et al., 2013b). These results suggested that UGT1 has the ability to monitor slight structural aberrations, which has led to the postulation that UGT1 can differentiate between on-pathway foldable substrates and off-pathway terminal misfolded secretory cargo, thereby directing the rebinding of the lectin chaperones toward on-pathway salvageable targets that might eventually fold and oligomerize properly.
UGT1 is an essential gene for mouse viability, but an UGT1-deficient mouse embryonic fibroblast cell line has been used to explore the role of UGT1 in the ER (Molinari et al., 2005; Solda et al., 2007; Pearse et al., 2010; Zhang et al., 2011). The absence of UGT1 has diverse consequences for glycosylated secretory cargo, including the following: 1) premature release from the lectin chaperones, resulting in a decrease in trafficking or secretion efficiency; 2) delay in the release from the lectin chaperones and ER exit; or 3) no effect on lectin chaperone binding and secretion. Determining whether UGT1 is providing a direct or indirect effect using these knockout cells is complicated by the fact that UGT1 modifies a wide range of substrates that may be influenced by its absence and that UGT1 is found in a protein complex (Korotkov et al., 2001; Meunier et al., 2002; Pearse et al., 2010).
Although the substrate specificity of UGT1 has been studied using purified components, its activity in mammalian cells has been poorly defined due to a number of experimental obstacles hindering the ability to monitor the reglucosylation process (Pearse et al., 2008, 2010). Here a cell-based reglucosylation assay was used to characterize the activity of UGT1 in its native environment. The hypothesis that UGT1 favors the reglucosylation of on-pathway over off-pathway targets was tested. In contrast to what has previously been proposed (Caramelo et al., 2004; D’Alessio et al., 2010), UGT1 was found to reglucosylate off-pathway maturation substrates more efficiently and persistently than wild-type maturing secretory pathway clients. Trapping wild-type UGT1 substrates in the monoglucosylated state delayed their secretion, whereas trapping off-pathway substrates in the monoglucosylated state did not affect their rate of degradation. This suggested that terminally misfolded substrates were efficiently extracted from the lectin chaperone binding cycle and targeted for degradation through a dominant process that was unaffected by the presence of monoglucosylated side chains on the substrate.
RESULTS
The reglucosylation of ER-associated degradation substrates
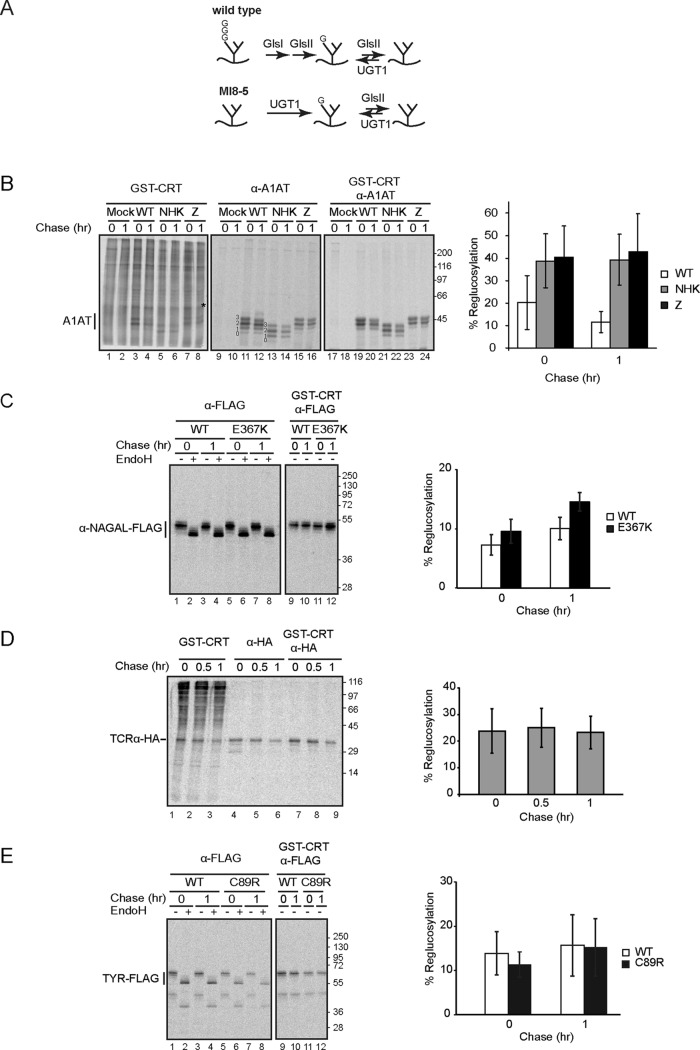
To explore the substrate specificity of UGT1 in live cells, the reglucosylation of model maturation and quality control substrates was analyzed using a cellular reglucosylation assay. MI8-5 Chinese hamster ovary cells are deficient in the dolichol-P glucose–dependent glycosyltransferase termed Alg6 (Quellhorst et al., 1999). Truncated unglucosylated N-linked glycans lacking glucoses (Man9GlcNAc2) are transferred instead of the normal triglucosylated species (Glc3Man9GlcNAc2). Reglucosylation is the sole mechanism through which glycoproteins can reach the monoglucosylated state in these cells (Figure 1A, bottom scheme). The reglucosylated or monoglucosylated proteins can be isolated based on their affinity for glutathione S-transferase (GST)–calreticulin (Pearse et al., 2008, 2010). Glucosidase inhibition traps reglucosylated glycans in their monoglucosylated state in MI8-5 Chinese hamster ovary cells since these Chinese hamster ovary–derived cells lack an endomannosidase activity (Karaivanova et al., 1998), further facilitating the ability to monitor the reglucosylation reaction.
FIGURE 1:
Reglucosylation of ERAD and disease-associated mutants is efficient. (A) Schematic of glycan processing in MI8-5 and wild-type cells. Whereas wild-type cells allow the transfer of triglucosylated glycans onto the nascent chain, unglucosylated glycans are transferred in MI8-5 cells. Thus, monoglucosylated glycans can only be generated through reglucosylation by UGT1 in MI8-5 cells. G, glucose; GlsI, glucosidase I; GlsII, glucosidase II. (B) Right, MI8-5 Chinese hamster ovary cells were transiently transfected with an empty vector (mock) or with wild-type α-1-antitryspin (WT A1AT), α-1-antitrypsin null Hong Kong (NHK), and α-1-antitrypsin Z (A1ATZ). Cells were radiolabeled for 1 h and chased for the indicated times. DNJ, 0.5 mM, was added to the pulse and chase media. Monoglucosylated proteins were isolated by GST-calreticulin (CRT) pull down from 10% of the cell lysate each, and WT A1AT, NHK, and A1ATZ were isolated with A1AT antisera. Monoglucosylated WT A1AT, NHK, and A1ATZ were isolated by GST-CRT pull down, followed by immunoprecipitation with A1AT antisera from 80% of the cell lysate. All samples were resolved on 9% SDS–PAGE reducing gels. The numbers next to the bands indicate the number of glycans on A1AT. The asterisk indicates the previously identified endogenous UGT1 substrate prosaposin. Left, quantification of the percentage of reglucosylation of WT A1AT, NHK, and A1ATZ. The quantifications of the fully glycosylated A1AT (top band) from lanes 19–24 were divided by the quantifications from lanes 11–16, which were multiplied by eight to account for lower percentage of sample used. The error bars are representative of the SD of five independent experiments. (C) Right, MI8-5 Chinese hamster ovary cells were transiently transfected with FLAG tagged α-NAGAL, wild type, and E367K mutant and treated as in B. α-NAGAL was isolated with antisera recognizing the FLAG epitope from 10% of the cell lysate, and an equal fraction was followed by EndoH treatment where indicated. Monoglucosylated α-NAGAL was isolated by GST-CRT pull down, followed by immunoprecipitation with FLAG antisera from 70% of the cell lysate. All samples were resolved on a 9% SDS–PAGE reducing gel. Left, quantification of the percentage of reglucosylation of α-NAGAL was performed as in B. The error bars are representative of the SD of three independent experiments. (D) Right, MI8-5 Chinese hamster ovary cells were transiently transfected with HA-tagged TCRα and treated as in B. TCRα was isolated with antisera recognizing the HA epitope and total radiolabeled monoglucosylated proteins with GST-CRT from 10% of the cell lysate. Monoglucosylated TCRα was isolated by GST-CRT pull down, followed by immunoprecipitation with HA antisera from 80% of the cell lysate. All samples were resolved on a 12% SDS–PAGE reducing gel. Left, quantification of the percentage of reglucosylation of TCRα was performed as in B. The error bars are representative of the SD of three independent experiments. (E) Right, MI8-5 Chinese hamster ovary cells were transiently transfected with FLAG-tagged tyrosinase, wild type, and C89R mutant and treated as in B. Total and monoglucosylated tyrosinase were isolated as in C. Left, quantification of the percentage of reglucosylation of tyrosinase was performed as in B. Statistical analysis using single-factor analysis of variance test gave p of 0.04<0.05 for WT A1AT compared with NHK and Z mutants at 0 h, p of 0.001 and 0.004<0.01 for WT A1AT compared with NHK and Z mutant, respectively, at 1 h, and p of 0.03<0.05 for WT α-NAGAL compared with the E367K at 1 h, indicating that the increase in reglucosylation of the mutants relative to WT at the indicated time points is statistically significant.
α-1-Antitrypsin (A1AT) was chosen as the initial substrate to evaluate for reglucosylation since it has a number of mutations with varying levels of structural perturbations and is associated with disease. A1AT is a soluble secretory glycoprotein synthesized in hepatocytes that belongs to the serine protease inhibitor or serpin family of proteins. Serpins are monomeric proteins comprised of three sheets and nine helices. Three different variants of A1AT were expressed in MI8-5 cells, including wild type and two mutant variants termed null Hong Kong (NHK) and A1ATZ. NHK is a frameshift and premature truncation mutation that is associated with gross misfolding, ER retention, and rapid subsequent turnover by ER-associated degradation (ERAD) (Sifers et al., 1988; Liu et al., 1999). It was named null because it was found to be absent from serum. A common pathological variant of A1AT is A1ATZ, which is caused by a single site mutation (E342K) (Blanco et al., 2006). A1ATZ contains structural perturbations that slow its folding and favor polymerization (Bottomley, 2011; Kass et al., 2012). Although ∼15% of A1ATZ is secreted, the remainder is ER retained and degraded by both ERAD and autophagy (Lomas et al., 1992; Hidvegi et al., 2010). The retention of polymerized A1ATZ in the ER of hepatocytes is associated with cirrhosis.
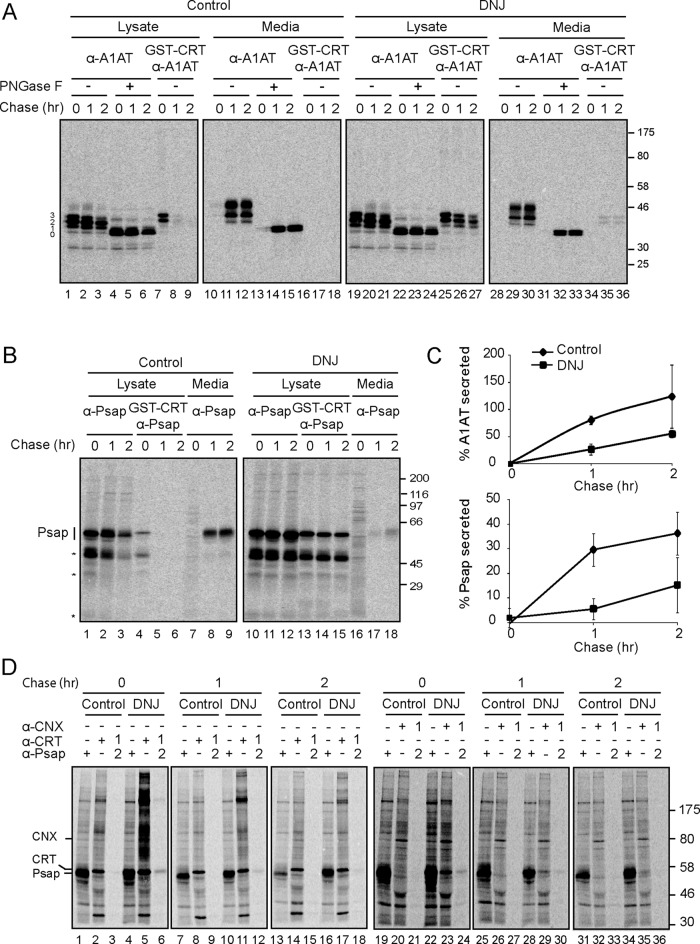
Cells transfected with A1AT were pulsed for 1 h with [35S]Met/Cys and analyzed either directly after the pulse or after a 1-h chase (Figure 1B). The pulse and chase were performed in the presence of the glucosidase inhibitor N-butyl deoxynojirimycin (DNJ) to trap reglucosylated glycans. Cell lines defective in assembling the complete dolichol precursor, such as MI8-5, are commonly associated with the hypoglycosylation of nascent proteins since the oligosaccharyltransferase frequently transfers the incompletely assembled glycan inefficiently (Huffaker and Robbins, 1983). The hypoglycosylation of A1AT was observed for all three forms after immunoprecipitation of the samples with anti-A1AT sera, resulting in the formation of a ladder of glycosylation states ranging from the faster-migrating unglycosylated protein to the slower-migrating, fully glycosylated protein containing all three glycans (Figure 1B, lanes 11–16). To confirm that the multiple bands of A1AT were caused by hypoglycosylation, we treated samples with peptide N-glycosidase F (PNGase F) to remove any heterogeneity attributed to N-linked carbohydrates. All protein bands observed collapsed into a single band migrating with the same mobility as unglycosylated A1AT, demonstrating that the different protein bands resolved were due to different levels of glycosylation (Figure 2A, lanes 1–6).
FIGURE 2:
Trapping WT A1AT and the endogenous substrate prosaposin (Psap) in the monoglucosylated state reduced their secretion efficiency and prolonged chaperone binding. (A) MI8-5 Chinese hamster ovary cells were transiently transfected with WT A1AT, radiolabeled for 30 min, and chased for the indicated times. DNJ, 0.5 mM, was added to the pulse and chase media where indicated. WT A1AT was isolated with A1AT antisera from 12% of the cell lysate or the media, and an equal fraction was treated with PNGase F. Monoglucosylated WT A1AT was isolated by GST-CRT pull down, followed by immunoprecipitation with A1AT antisera from 50% of the cell lysate or the media. All samples were resolved on a 9% SDS–PAGE reducing gel. (B) MI8-5 Chinese hamster ovary cells were radiolabeled for 30 min and chased for the indicated times. DNJ, 0.5 mM, was added to the pulse and chase media where indicated. Psap was isolated with Psap antisera from 30% of the lysate or the media. Monoglucosylated Psap was isolated by GST-CRT pull down, followed by immunoprecipitation with Psap antisera from 60% of the cell lysate. All samples were resolved on a 9% SDS–PAGE reducing gel. The asterisk indicates cleaved Psap products. (C) Quantification of the percentage of the fully glycosylated (top band) WT A1AT (top) and endogenous prosaposin (bottom) secreted into the media as a percentage of synthesis. The error bars are representative of the SD from three independent experiments. (D) MI8-5 Chinese hamster ovary cells were radiolabeled for 1 h and chased for the indicated times. Psap was isolated from 5% of the lysate with Psap antisera and calreticulin (CRT)- and calnexin (CNX)-bound species from 15% of the lysate with their respective antisera. For sequential immunoprecipitations, 65% of the lysate was sequentially immunoprecipitated as indicated. All samples were resolved via 7.5% reducing SDS–PAGE.
Reglucosylation or the presence of monoglucosylated glycans was probed by monitoring binding to GST-calreticulin, a bacterially expressed and purified lectin fusion protein (Pearse et al., 2008, 2010). A large number of proteins of a variety of sizes were found to be monoglucosylated (Figure 1B, lanes 1–8). To characterize A1AT reglucosylation, the GST-calreticulin pull down was followed by the immunoprecipitation with anti-A1AT sera (Figure 1B, lanes 17–24). Twenty percent of wild-type A1AT was reglucosylated immediately after the pulse, and this level diminished to 11% after a 1-h chase. Of interest, the level of reglucosylation doubled for A1AT mutants NHK and A1ATZ, and this level of ∼40% reglucosylation persisted after 1 h of chase. These values were calculated using A1AT possessing all three of its glycans; however similar reglucosylation efficiencies were obtained when all A1AT glycoforms were grouped. The fastest-migrating protein bands (unglycosylated A1AT) were not recognized by GST-calreticulin. Therefore, although all glycoforms of A1AT were reglucosylated, the terminally misfolded mutant forms were more efficiently reglucosylated than the wild-type protein, and the level of reglucosylation of the two mutant variants was similar even though they possessed different types of abnormalities.
To determine whether UGT1 efficiently modified other ERAD or disease-associated substrates, we studied the reglucosylation of several additional proteins. These included proteins with varying topologies, such as the soluble α-N-acetylgalactosaminidase (α-NAGAL), as well as two membrane proteins, T-cell receptor α-subunit (TCRα) and tyrosinase.
α−NAGAL is a lysosomal protein that possesses five N-linked glycans (Clark and Garman, 2009). There are a number of loss-of-function mutations in α-NAGAL that are associated with Schindler/Kanzaki disease (Clark et al., 2012). The reglucosylation of C-terminally tagged FLAG constructs of wild-type α−NAGAL and the missense E367K mutant was studied as described earlier. Samples were treated with endoglycosidase H (EndoH) to differentiate EndoH-sensitive ER glycoforms of α-NAGAL from protein that had trafficked through the Golgi and received complex sugars rendering them EndoH resistant. During the time frame studied, all forms of α-NAGAL appeared to be EndoH sensitive, consistent with their ER residency. The level of reglucosylation for wild-type protein (7%) was lower than that observed for the E367K mutant (10%) immediately after the pulse (Figure 1C). This trend continued after the 1-h chase. The α-NAGAL E367K mutant was more efficiently modified than the wild-type protein.
TCRα is a membrane glycoprotein that forms a larger heterocomplex in T lymphocytes; however, when it is unable to assemble, it is rapidly degraded through the ERAD pathway (Bonifacino et al., 1990; Huppa and Ploegh, 1997; Call et al., 2002). MI8-5 cells were transfected with a TCRα construct containing a C-terminal hemagglutinin (HA) tag and pulsed for 1 h in [35S]Met/Cys, followed by a chase for the indicated times in the presence of a glucosidase inhibitor. TCRα was found to be efficiently (23–25%) and persistently reglucosylated regardless of the duration of the chase period (Figure 1D).
Tyrosinase is a type I membrane glycoprotein with seven N-glycans that is involved in melanin biosynthesis (Újvári et al., 2001). A C89R missense mutation for tyrosinase is an ERAD substrate, and this loss-of-function mutation is associated with albinism (Halaban et al., 1997). MI8-5 cells were transfected with C-terminal FLAG constructs of either human wild-type tyrosinase or a C89R mutant, and a pulse-chase procedure was performed as described. Both wild-type and the C89R mutant of tyrosinase were efficiently reglucosylated, with only a very slight increase in the level for reglucosylation observed for C89R over wild type immediately after 1 h of chase (Figure 1E).
MI8-5 cells supported the efficient glycosylation and reglucosylation of a variety of glycoproteins. Three of four glycoproteins studied were efficiently glycosylated, whereas a fourth protein (A1AT) was found to be hypoglycosylated, indicating that hypoglycosylation of glycoproteins in the MI8-5 cells was protein dependent. All four proteins were efficiently reglucosylated. The level for reglucosylation for the two mutants of A1AT was double that observed for wild-type A1AT. Wild-type α-NAGAL and tyrosinase were efficiently reglucosylated, but the reglucosylation of their mutants was associated with only a modest increase in modification, and in the case if tyrosinase, it was seen only after a 1-h chase.
Trapping proteins in their monoglucosylated state delays their secretion
Reglucosylation of proteins by UGT1 supports their rebinding to the lectin chaperones calnexin and calreticulin (Caramelo and Parodi, 2008). To determine whether trapping glycoproteins in their monoglucosylated state would affect their secretion, we performed a pulse-chase experiment using MI8-5 cells transfected with wild-type A1AT in the absence and presence of DNJ. Cell lysates and the culture medium were collected and analyzed.
The ability of glucosidase inhibition to trap wild-type A1AT in a monoglucosylated state was verified, as the addition of DNJ greatly increased the level of reglucosylation observed with the lysate samples after sequential precipitation with GST-calreticulin and anti-A1AT sera (Figure 2A, compare lanes 7–9 to lanes 25–27). Trapping A1AT in the monoglucosylated state was also associated with a significant decrease in secretion, as less than half the amount of A1AT was found in the culture medium after a 2-h chase when DNJ was present compared with in its absence (Figure 2, A, compare lanes 10–12 to lanes 28–30, and C). The secreted protein found in the medium was largely not glucosylated as probed by GST-calreticulin binding (Figure 2A, lanes 16–18 and 34–36). These results demonstrated that the inhibition of deglucosylation of reglucosylated protein delayed the secretion of ectopically expressed wild-type A1AT.
Because quality control pathways can be saturated by protein overexpression, the effect of trapping a reglucosylated substrate on the secretion of an endogenous UGT1 substrate was analyzed. Previously we found that prosaposin was an obligate substrate for UGT1, as it was efficiently reglucosylated in MI8-5 cells, and its maturation was inefficient in ugt1−/− mouse embryonic fibroblast cells (Pearse et al., 2010). As previously observed, glucosidase inhibition trapped a significant fraction of the prosaposin in the monoglucosylated state (Figure 2B, lanes 13–15). Furthermore, glucosidase inhibition and monoglucosylation trapping were associated with less than half the amount of prosaposin being secreted into the culture medium after 2 h of chase (Figure 2B, lanes 9 and 18, and C). The delay in secretion of prosaposin in the presence of DNJ appeared to be due to calreticulin and calnexin binding to prosaposin, as chaperone binding levels increased in the presence of the glucosidase inhibitor (Figure 2D, compare lanes 3–6 and lanes 21–24). Taken together, these results demonstrated that trapping either ectopically expressed wild-type A1AT or endogenously expressed prosaposin in the monoglucosylated state significantly inhibited their secretion.
Trapping ERAD substrates in their monoglucosylated state does not delay their turnover
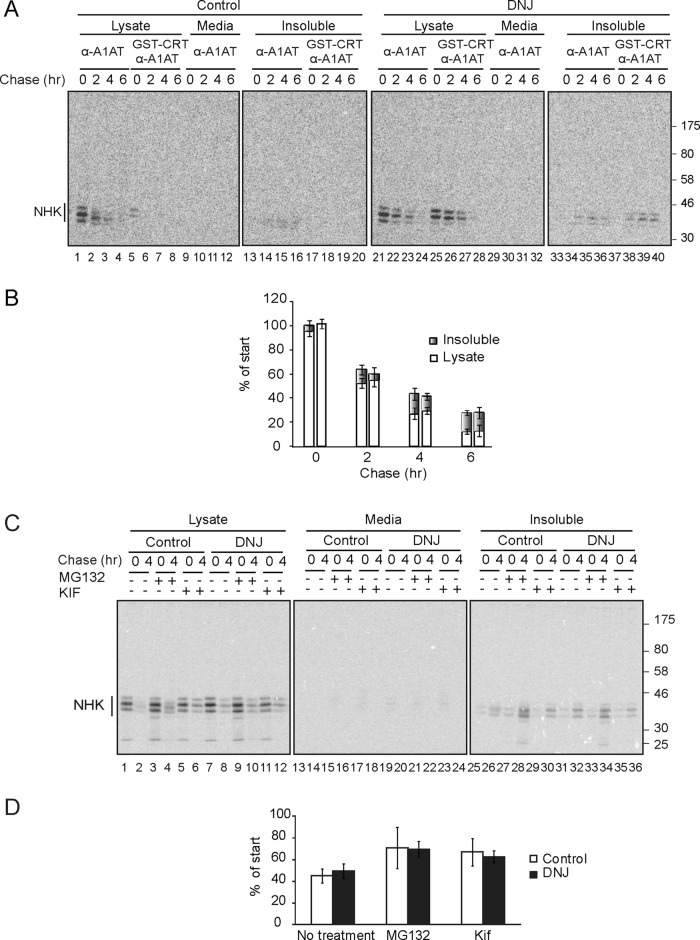
Next we determined whether trapping an efficiently reglucosylated ERAD substrate in the monoglucosylated state would influence its turnover. First, the reglucosylation and secretion of NHK was followed in MI8-5 cells in the absence and presence of DNJ by pulse-chase analysis. NHK was not secreted regardless of whether DNJ was excluded or included in the pulse and chase conditions, demonstrating the effectiveness of the quality control process in retaining and degrading this terminally misfolded protein and recapitulating the null phenotype (Figure 3A, lanes 9–12 and 29–32). A fraction of NHK was also observed in the Triton X-100–insoluble material (Figure 3A, lanes 13–16 and 33–36). This fraction was included in the quantification of the amount of NHK remaining before and after trapping in the monoglucosylated state. Glucosidase inhibition efficiently captured NHK in the monoglucosylated state as observed by sequential pull downs with GST-calreticulin and anti-A1AT sera (Figure 3A, compare lanes 25–28 to lanes 5–8). Of interest, trapping NHK in the monoglucosylated glycoforms did not influence the half-life of NHK regardless of the conditions used (Figure 3B). Monoglucosylated NHK was detected in the detergent insoluble fraction even after 6 h of chase, indicating that insoluble NHK was also reglucosylated (Figure 3A, lanes 37–40). The increase in NHK levels observed after the addition of proteasome (MG132) or the mannosidase (kifunensine) inhibitors demonstrated that NHK continued to be degraded by ERAD in the absence or presence of glucosidase inhibition (Figure 3, C and D).
FIGURE 3:
Trapping NHK in the monoglucosylated state did not change its degradation rate. (A) MI8-5 Chinese hamster ovary cells were transiently transfected with NHK, radiolabeled for 30 min, and chased for the indicated times. DNJ, 0.5 mM, was added to the pulse and chase media where indicated. NHK was isolated with A1AT antisera from 10% of the lysate, the medium, or the Triton X-100–insoluble fraction. Monoglucosylated NHK was isolated by GST-CRT pull down, followed by immunoprecipitation with A1AT antisera from 80% of the cell lysate or the Triton X-100–insoluble fraction. All samples were resolved on a 9% SDS–PAGE reducing gel. (B) Quantification of the percentage of NHK from the lysate and the Triton X-100–insoluble fraction as a percentage of synthesis. The first bar for each time point corresponds to control conditions, and the second corresponds to DNJ treatment. All bands were quantified because the fully glycosylated (NHK top band) was not always well resolved in the later time points or the insoluble fraction. The error bars are representative of the SD of three independent experiments. (C) Cells were treated as in A. MG132 (20 μM) or kifunensine (100 μM) was added 4 and 2 h, respectively, before the pulse and included in the pulse and the chase where indicated. All samples were resolved on a 9% reducing SDS–PAGE gel. (D) Quantification of the percentage of the sum of NHK from the cell lysate and the Triton X-100–insoluble fraction as a percentage of synthesis (4-h chase). The error bars are representative of the SD of three independent experiments.
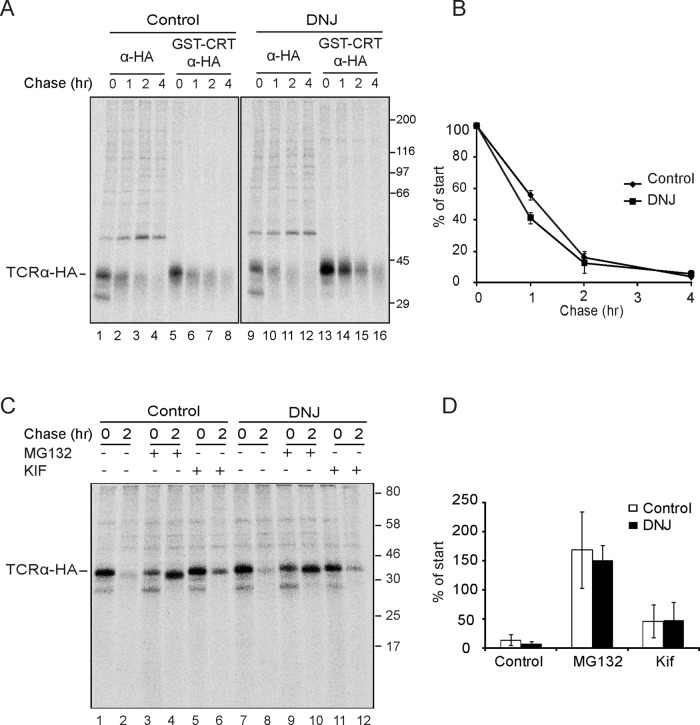
To test the generality of this result, we investigated whether trapping another ERAD substrate, TCRα, in the monoglucosylated state would affect its disposal by ERAD. Persistent monoglucosylation of TCRα was observed in the presence of the glucosidase inhibitor as observed by pull down with GST-calreticulin followed by immunoprecipitation with anti-HA sera (Figure 4A, compare lanes 13–16 to lanes 5–8). The half-life of TCRα was not significantly affected by the presence of the glucosidase inhibitor, as the half-life of TCRα was ∼1 h in the absence or presence of DNJ (Figure 4B). TCRα was not detected in the Triton X-100–insoluble material (unpublished data), indicating that TCRα analyzed using the detergent-soluble fraction was representative of the total amount of the protein remaining. TCRα was degraded by ERAD, as MG132 inhibited its turnover; however, mannosidase inhibition stabilization was less efficient (Figure 4, C and D). Taken together, these results demonstrated that degradation of the ERAD substrates NHK and TCRα was unaffected by being trapped in the monoglucosylated state.
FIGURE 4:
Trapping TCRα in the monoglucosylated state did not change its degradation rate. (A) MI8-5 Chinese hamster ovary cells were transiently transfected with TCRα, radiolabeled for 30 min, and chased for the indicated times. DNJ, 0.5 mM, was added to the pulse and chase media where indicated. TCRα was isolated with HA antisera from 40% of the lysate. Monoglucosylated TCRα was isolated by GST-CRT pull down, followed by immunoprecipitation with HA antisera from 50% of the cell lysate. All samples were resolved on a 9% SDS–PAGE reducing gel. (B) Quantification of the percentage of TCRα from the lysate as a percentage of synthesis. The error bars are representative of the SD of three independent experiments. (C) Cells were treated as in A. MG132 (20 μM) or kifunensine (100 μM) was added 4 and 2 h, respectively, before the pulse and included in the pulse and the chase where indicated. (D) Quantification of the percentage of TCRα from the cell lysate as a percentage of synthesis (2-h chase). The error bars are representative of the SD of four independent experiments for no treatment and three independent experiments for MG132 and kifunensine treatments.
The timing and persistence of reglucosylation
The previous experimental strategy trapped reglucosylated substrates in their monoglucosylated state to query the influence on secretion or degradation. Because the persistent trap did not allow the determination of the timing for reglucosylation, a transient 15-min window of glucosidase inhibition was applied at various time points with respect to the radioactive pulse and the cold chase to trap reglucosylated substrates over a range of times. Reglucosylated substrates were isolated using GST-calreticulin, followed by the immunoprecipitation of the specific substrate.
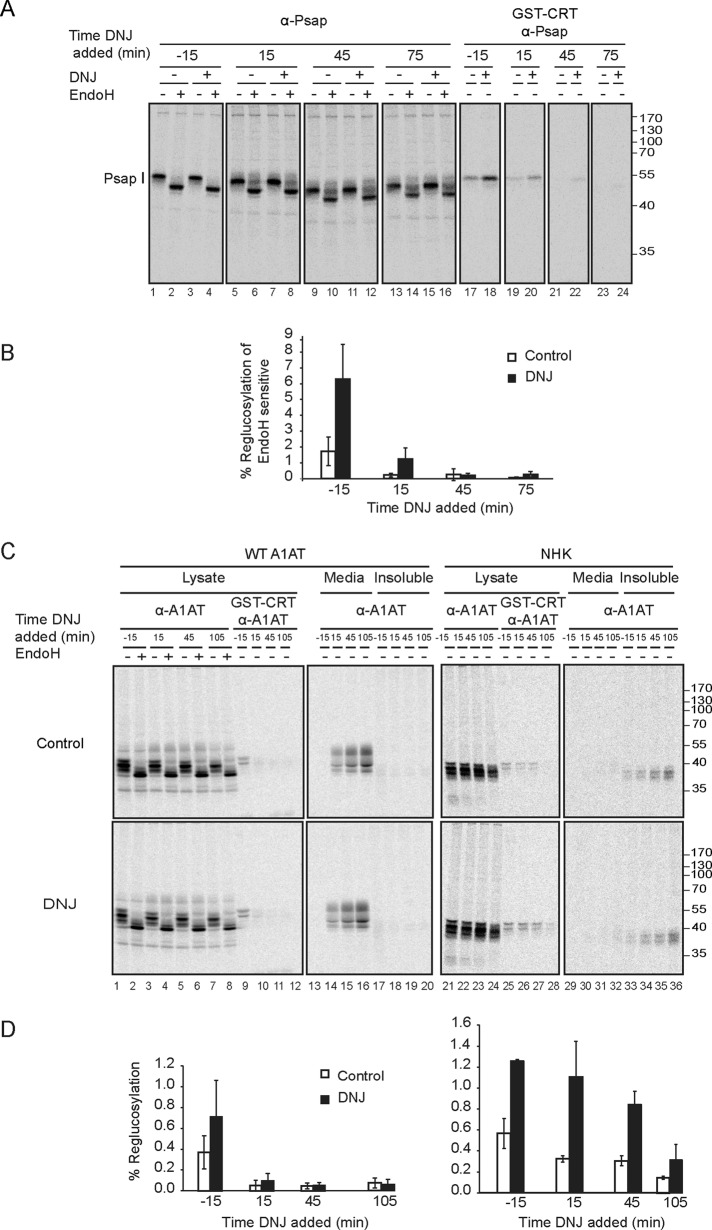
After maturing properly in the ER, prosaposin has two separate fates: it can traffic to lysosomes, where it is processed by cathepsin D into four separate proteins that act as cofactors for different lipid hydrolases, or it can be secreted as a full-length protein, with only its signal sequence removed (O’Brien et al., 1994; Lefrancois et al., 2003). To differentiate ER-resident prosaposin in cell lysates from lysosomal prosaposin, we treated prosaposin immunoprecipitated from cell lysates with EndoH. High-mannose glycoforms consistent with ER residence remain EndoH sensitive, whereas complex sugar glycoforms that have trafficked through the Golgi become EndoH resistant (Halaban et al., 1997). Most of the prosaposin from cell lysates displayed an EndoH-sensitive profile throughout the experiment; however, after 15 min of chase, a fraction of prosaposin that increased slightly with time was found to be EndoH resistant (Figure 5A, lanes 6 and 8).
FIGURE 5:
WT A1AT and prosaposin are transiently reglucosylated by UGT1, whereas reglucosylation of NHK is more persistent. (A) MI8-5 Chinese hamster ovary cells were radiolabeled for 30 min. Where indicated, 0.5 mM DNJ was added 15 min before the end of the pulse. DNJ was also added at the indicated times of the chase, followed by 15-min incubation. Prosaposin was isolated with prosaposin antisera, followed by treatment with EndoH where indicated. Monoglucosylated prosaposin was isolated by GST-CRT pull down, followed by immunoprecipitation with prosaposin antisera. All samples were resolved on a 9% SDS–PAGE reducing gel. (B) The quantification of the percentage of reglucosylation of prosaposin was performed as in Figure 1, accounting for differences in amounts of sample used for each treatment as a percentage of EndoH-sensitive prosaposin, since this represents ER-localized protein compared with EndoH-resistant protein localized to the lysosome. The error bars are representative of the SD of three or more independent experiments. (C) MI8-5 Chinese hamster ovary cells where transfected with either WT or NHK A1AT and treated as in A. (D) The quantification of the percentage of reglucosylation of all bands was performed as in Figure 1 accounting for differences in amounts of sample used for each treatment. The error bars are representative of the SD of three or more independent experiments.
The reglucosylation of prosaposin was transient, as monoglucosylated protein was isolated by GST-calreticulin most effectively when DNJ was added during the 15-min pulse (Figure 5, A, lanes 17 and 18, and B). The amount of prosaposin trapped in the monoglucosylated state with the 15 min of DNJ treatment greatly decreased during the chase even though a significant fraction of prosaposin continued to be EndoH sensitive, signifying ER residence. A similar profile was observed for transiently expressed wild-type A1AT (Figure 5, C and D). Monoglucosylated A1AT was generated only during the 15-min pulse period and did not accumulate after a chase period of 15 min or more.
In contrast, reglucosylation of the ERAD substrate NHK A1AT was more efficient during the pulse period, and reglucosylation persisted for the next hour (Figure 5, C and D). After 105 min of chase, the reglucosylation level dropped to roughly one-third of its maximum. Whereas the Triton-insoluble fraction with wild-type A1AT remained empty throughout the chase period, the Triton-insoluble fraction for NHK increased in a time-dependent manner. The persistent reglucosylation of NHK appeared to contribute to its ER retention and the lack of appearance in the media to create the null phenotype. Overall these results indicated that for both prosaposin and wild-type A1AT, reglucosylation occurred early in the maturation process. Although protein remained in the ER during the extended chase period, it did not appear to be monoglucosylated. For the A1AT NHK mutant variant, reglucosylation occurred at a higher level and persisted, consistent with the conclusion that UGT1 reglucosylates off-pathway aberrant structures most efficiently.
Efficient reglucosylation of reduced protein
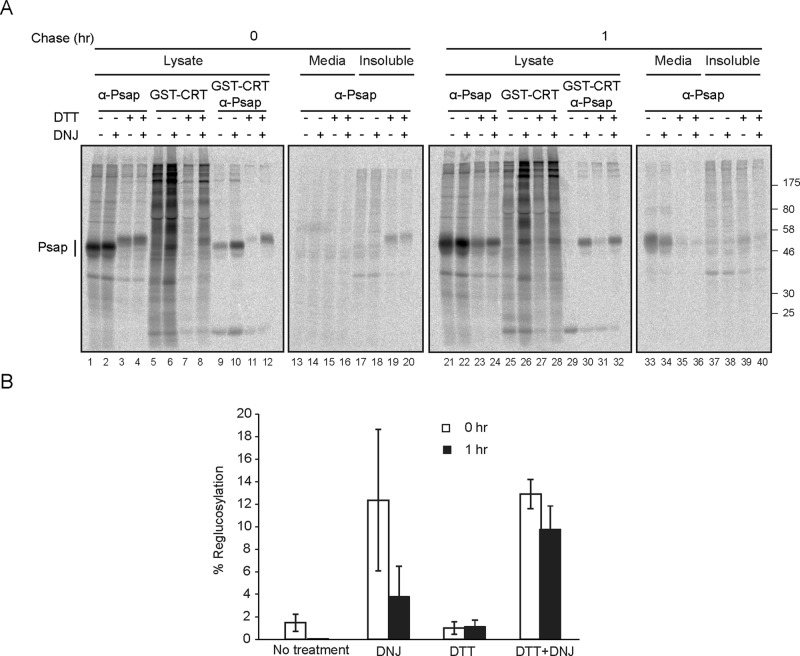
Previous results suggested that UGT1 does not modify grossly misfolded proteins. This case was in part supported by the observation that in some cases, ER glucosyltransferases did not recognize reduced substrates (Fernandez et al., 1998; Ritter and Helenius, 2000; Pearse et al., 2008). The timing of dithiothreitol (DTT) addition, its activation of the unfolded protein response (UPR), and the use of different species complicate the interpretation of these results (Travers et al., 2000). We determined whether DTT affected the reglucosylation of the obligate UGT1 substrate prosaposin using our cell-based reglucosylation assay. Cells were pulsed with [35S]Met/Cys for 30 min in the presence or absence of DTT and processed either immediately after the pulse or after an oxidative chase for 1 h. DNJ was added to trap reglucosylated substrates, and the samples were processed by immunoprecipitation and using GST-calreticulin pull downs.
Reduced prosaposin migrated more slowly than prosaposin that accumulated under oxidizing conditions, as free thiols were modified by the alkylating agent N-ethyl maleimide included in the lysis buffer (Figure 6A, lanes 1–4). The amount of prosaposin was also significantly diminished by DTT treatment, likely due to the activation of UPR and translation attenuation. Maximal reglucosylation of prosaposin was observed immediately after the pulse when glucosidases were inhibited by DNJ (Figure 6, A, lane 10, and B), and this level dropped significantly after a 1-h oxidative chase (Figure 6, A, lane 30, and B). Optimal reglucosylation also occurred in the presence of DTT immediately after the pulse (Figure 6, A, lane 12, and B), but this level of reglucosylation remained elevated even after the chase. When oxidation was initiated posttranslationally, prosaposin was more efficiently reglucosylated compared with commencing oxidation during translation. This suggested that the posttranslation oxidation of prosaposin was not as efficient and created aberrant conformations that continued to be recognized by UGT1 even after 1 h of oxidation. Therefore optimal reglucosylation occurred immediately after the pulse regardless of whether DTT was present; however, DTT supported efficient and persistent reglucosylation, consistent with UGT1 preference for nonnative substrates.
FIGURE 6:
Reduced prosaposin is efficiently reglucosylated by UGT1. (A) Cells were radiolabeled for 30 min and chased for 1 h. Where indicated, 0.5 mM DNJ was added in the pulse and chase media. DTT, 5 mM, was added in the pulse medium where indicated and excluded from the chase medium. Prosaposin was isolated with prosaposin antisera. Monoglucosylated prosaposin was isolated by GST-CRT pull down, followed by immunoprecipitation with prosaposin antisera. All samples were resolved on a 9% SDS–PAGE reducing gel. (B) The quantification of the percentage of reglucosylation of prosaposin was performed as in Figure 1, accounting for differences in amounts of sample used for each treatment. The error bars are representative of the SD of three or more independent experiments.
DISCUSSION
A main question concerning the biological role of UGT1 as a central decision maker in the fate of glycoproteins in the ER is whether UGT1 is able to recognize productively folding immature and/or terminally misfolded proteins and how it affects the subsequent cellular fates of the proteins it modifies. Using a cell-based reglucosylation assay, we found that UGT1 generally glucosylated mutant off-pathway substrates more efficiently than wild-type substrates. Secretion of wild-type substrates was slowed and less efficient when they were trapped in the monoglucosylated state. Surprisingly, the turnover of off-pathway substrates was not affected by locking them in monoglucosylated states. These results suggested that UGT1 was capable of differentiating between native and nonnative proteins, but downstream factors appeared to play a dominant role in determining the fate of terminally misfolded proteins.
The initial studies using cell-free or purified protein assays showed that UGT1 favored the modification of nonnative compared with native conformations (Trombetta et al., 1989; Trombetta and Parodi, 1992; Sousa et al., 1992; Fernandez et al., 1994; Parker et al., 1995). More recent and detailed studies using biophysically characterized and engineered substrates further examined the specificity of UGT1 and found preferential reglucosylation of proteins that are partially structured or possess minor local folding defects (Ritter and Helenius, 2000; Trombetta and Helenius, 2000; Caramelo et al., 2003, 2004; Taylor et al., 2004; Ritter et al., 2005). In some of these reports, grossly misfolded structures were poorly modified. The glucosylation of biophysically characterized neoglycoproteins derived from C-terminal truncation fragments of chymotrypsin inhibitor 2 showed that UGT1 had a preference for fragments with molten globule–like conformations when compared with random coil or native conformations (Caramelo et al., 2003, 2004). Consistent with the preference of ER glucosyltransferases for near-native targets, in some cases they did not efficiently modify reduced proteins (Fernandez et al., 1996; Ritter and Helenius, 2000; Pearse et al., 2008). The reglucosylation of cruzipain in Trypanosoma cruzi was directed toward later oxidative intermediates that enabled binding by its lone lectin chaperone, calreticulin (Labriola et al., 1999). An earlier study using MI8-5 cells found that late but not early oxidative intermediates of hemagglutinin were reglucosylated on membrane-proximal glycans, which supported persistent calnexin binding (Pearse et al., 2008). From these results, it was concluded that UGT1 recognizes near-native substrates more efficiently than native or severely misfolded substrates. This suggested that UGT1 might possess the ability to redirect the binding of lectin chaperones to maturing substrates that are expected to be salvageable for eventual proper on-pathway folding and trafficking (D’Alessio et al., 2010).
Our cellular results indicate a less discriminatory role for UGT1 in live cells, as UGT1 generally reglucosylated terminally misfolded mutant proteins more efficiently than immature wild-type proteins. UGT1 was capable of modifying both on-pathway and off-pathway targets. The Z-variant involves a missense mutation and self-associates to form ER-retained polymers (Lomas et al., 1992). The less common NHK mutation consists of a frameshift at position Leu-318 that creates a premature termination codon at position 334, producing a more severe disruption to A1AT (Sifers et al., 1988). NHK is ER retained and subsequently degraded by ERAD. Because A1AT comprises three sheets and nine helices, NHK is missing or has mutations in one or two of the strands from each of the three sheets. Although Z and NHK likely have very different levels of structural perturbations, they are both reglucosylated at approximately twice what is observed for wild-type A1AT, indicative of the ability of UGT1 to recognize terminally misfolded substrates. Ferris et al. (2013) found that the level of mutant A1AT in Triton-insoluble fractions was increased in MEF cells lacking UGT1, suggestive of a role for UGT1 in maintaining the solubility for glycoproteins. However, in our study, reglucosylated mutant A1AT was also found in the Triton-insoluble fractions, suggesting that reglucosylation did not ablate aggregation. These differences might be due to the hypoglycosylation or the constitutive activation of UPR found in MI8-5 cells (Pearse et al., 2008).
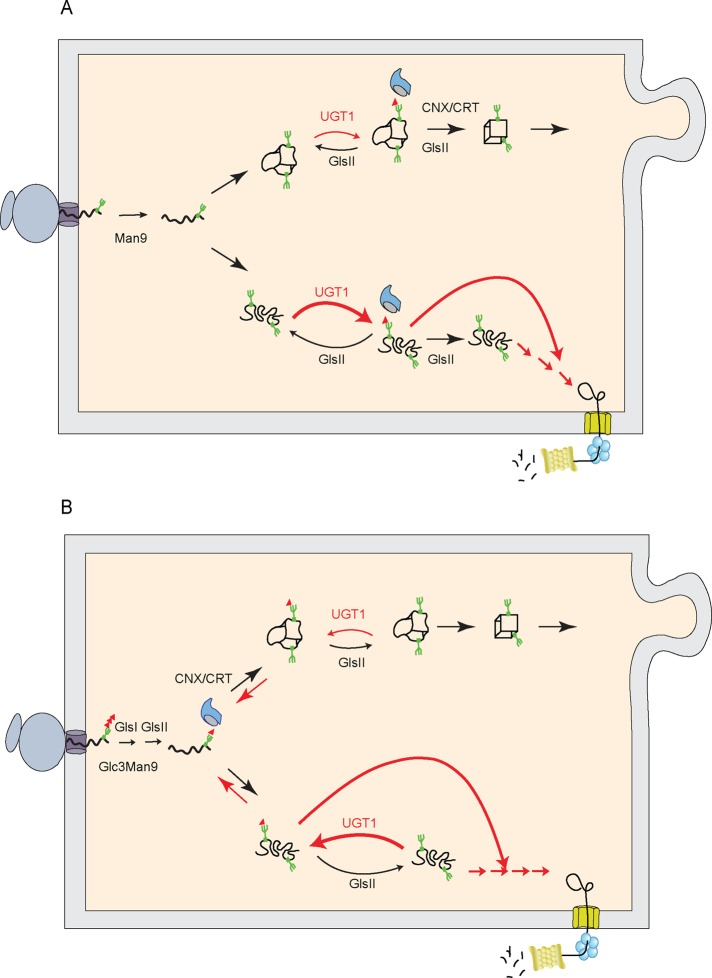
The ERAD substrate TCRα was also efficiently reglucosylated, as were the disease-associated mutants for α-NAGAL and tyrosinase. However, the increase in the reglucosylation of the mutant for α-NAGAL compared with its wild-type counterpart was not as significant as was observed for A1AT, and no increase in reglucosylation was observed for the mutant of tyrosinase compared with wild type. The reglucosylation assay traps reglucosylated glycans after a single reglucosylation event. Therefore it cannot differentiate between the reglucosylation of folding intermediates that are transiently glucosylated and a terminally misfolded protein that might be persistently reglucosylated unless additional glycosylations sites on a protein are modified, supporting the more efficient affinity isolation with the GST-calreticulin pull down. Wild-type tyrosinase appears to be a slow and inefficient folder (Halaban et al., 1997; Francis et al., 2003). α-NAGAL also appears to fold inefficiently in Chinese hamster ovary cells. This likely also helps to favor the efficient modification of their wild-type proteins, dampening the difference in reglucosylation between wild type and mutants. Maturation of these difficult folders may be further derailed in MI8-5 cells, since the transfer of Man9GlcNAc2 glycans bypasses the initial binding to calnexin and calreticulin initiated by glucosidase I and II trimming of the triglucosylated glycan. Lectin chaperone binding is solely directed by UGT1 reglucosylation in MI8-5 cells (Figures 1A and 7A).
FIGURE 7:
Model for UGT1 reglucosylation of on- and off-pathway substrates. (A) In MI8-5 Chinese hamster ovary cells, unglucosylated glycans are transferred onto the protein. On-pathway substrates in the near-native conformations are transiently reglucosylated by UGT1 until folding is completed (top route). DNJ addition favors accumulation of these substrates in the UGT1 cycle, supporting prolonged chaperone binding and secretion delay (represented by the red arrows). In contrast, aberrant off-pathway substrates are more efficiently and persistently reglucosylated by UGT1 (as indicated by the more prominent arrows on the bottom route). The addition of the glucosidase inhibitor DNJ, despite trapping misfolded proteins in their monoglucosylated state, does not protect substrates from ERAD, as they are efficiently extracted for degradation by a dominant ERAD selection process (exhibited by the long red arrows). (B) In WT Chinese hamster ovary cells, monoglucosylated substrates generated by GlsI and GlsII trimming bind to the lectin chaperones. After trimming of the last glucose by GlsII, substrates are released from the lectin chaperones and continue to be recognized by UGT1 and proceed in the top or bottom routes as described in A.
Earlier studies using a Chinese hamster ovary–derived cell line that transfers Man5GlcNAc2 carbohydrates (MadIA214 cells) found that maturing HA was reglucosylated, as was as a mutant of ribophorin that is an ERAD substrate (Ermonval et al., 2000, 2001). The fate of trapped monoglucosylated proteins was not followed, and the efficiency for the reglucosylation of the mutant of ribophorin was not compared with that for the native ER-resident protein. These studies showed the scope of proteins modified by the glucosyltransferase in live cells and that proteins possessing Man5 side chains are suitable substrates for modification. Given that mannosidase inhibitors stabilized mutant ribophorin expressed in MadIA214 cells that lacked B and C branch mannose residues, these results also suggest that mannose trimming of the A branch might be required for ERAD. Alternatively, these mannose derivatives might also inhibit the binding of ERAD factors to the downstream ERAD adapter SEL1L, as found for EDEM1, EDEM3, and OS-9 (Christianson et al., 2008; Cormier et al., 2009; Saeed et al., 2011).
Proteomic and morphological studies indicated that UGT1 is largely situated in the smooth ER near ER-exit sites (Zuber et al., 2001; Gilchrist et al., 2006). This positioning is consistent with the observation that reglucosylation occurs posttranslationally after the nascent chain has been released from the ribosome-associated translocon, as ribosome-arrested nascent chains were unable to be reglucosylated (Pearse et al., 2008). This provides an explanation for the observation that UGT1 displayed a preference for late oxidative intermediates or folding domains in MI8-5 Chinese hamster ovary cells and T. cruzi for influenza hemagglutinin and cruzipain, respectively (Labriola et al., 1999; Pearse et al., 2008). Therefore the localization and accessibility of the enzyme rather than an inherent specificity toward more mature substrates likely explain these findings.
Conflicting results have been obtained for the ability of UGT to modify reduced proteins. Several studies found that UGT did not modify reduced proteins, favoring the conclusion that UGT does not recognize grossly misfolded substrates (Fernandez et al., 1998; Ritter and Helenius, 2000; Trombetta and Helenius, 2000; Pearse et al., 2008). In some instances, reglucosylation was dependent on the alkylating agent used (Trombetta and Helenius, 2000), whereas in other cases, UGT1 was found to modify reduced proteins (Zapun et al., 1997; Taylor et al., 2004). The absence of reglucosylation in microsomes or cells treated with DTT could be explained by either the timing being soon after synthesis, as reglucosylation appears to occur posttranslationally, or by problems with the protein bookkeeping in cells, as DTT induces the UPR that leads to protein translation attenuation. We found that DTT supported the efficient and persistent reglucosylation of endogenous prosaposin in cells, favoring the conclusion that UGT1 can modify severely misfolded nonnative substrates.
UGT1 can also recognize slight structural perturbations, as it efficiently modified the orphan subunit of the heteromeric T-cell receptor, TCRα. A recent study found that unassembled TCRα caused the release of its single transmembrane region into the ER lumen, where it was then targeted for ERAD (Feige and Hendershot, 2013). The ability of UGT1 to recognize slight alterations is also exploited for the regulation of cellular processes such as antigen presentation and calcium homeostasis by supporting the recruitment of lectin chaperones and their associated oxidoreductase, ERp57 (Camacho and Lechleiter, 1995; Li and Camacho, 2004; John et al., 1998; Blum et al., 2013; Kunte et al., 2013). The reglucosylation of fully folded MHC class I heavy chain or SERCA2b modulates their function and localization. It is clear that UGT1 can recognize subtle aberrations in protein structure, but it also efficiently recognizes terminally misfolded substrates. UGT1 does not appear to have the ability to preferentially select on-pathway or repairable substrates for further lectin chaperone intervention but instead simply modified nonnative substrates.
The secretion of glucosylated trapped wild-type proteins was significantly delayed. A similar finding was previously observed for endogenous cruzipain in T. cruzi, which naturally transfers high-mannose glycans (Labriola et al., 1995). In sharp contrast, the more efficient reglucosylation of ERAD substrates did not appear to influence their fate or the turnover of terminally misfolded proteins when glucosylated glycans were trapped on the substrate. This was suggestive of a dominant downstream ERAD sorting receptor playing an important role that was unaffected by the presence of glucosylated glycans or lectin chaperone binding. As mannoses were trimmed from the B and C branches in fission yeast S. pombe, glucosidase II activity toward the demannosylated substrate decreased, whereas reglucosylation was unaffected (Aebi et al., 2010; Stigliano et al., 2011). This favored the accumulation of monoglucosylated side chains on slow-folding substrates, which would be ER retained, assisted, and protected by calnexin and calreticulin binding. However, cell-free mammalian assays have shown that mannose trimming diminishes the activity of UGT1 toward substrates (Sousa et al., 1992), as well as the binding affinity of calreticulin and possibly calnexin (Spiro et al., 1996). Mannose trimming also exposes α1,6-linked mannoses side chains on C chains, which act as degradation tags for recognition by the luminal ERAD receptors (Quan et al., 2008; Clerc et al., 2009). Alternatively, A branch demannosylation would remove a glycoprotein from being a substrate for reglucosylation and subsequent lectin chaperone binding to possibly favor recognition by ERAD machinery (Olivari et al., 2006). Therefore mannose trimming likely plays a key role in rapidly sorting the trapped glucosylated proteins for destruction. Understanding the precise mechanism by which mannose trimming contributes to the ERAD process is complicated by the dual role that N-linked glycans play in the secretory pathway, as they act as sorting and quality control tags, as well as docking tags, for ER machinery complex formation and targeting (Hebert and Molinari, 2012).
The recognition and modification of ERAD substrates by UGT1 appear to be more a consequence of the ability of UGT1 to recognize structural imperfections akin to molecular chaperones than of functional significance that directly affects the fate for defective proteins. Downstream ERAD receptors evidently efficiently sort defective cargo for destruction regardless of their glucosylation status. Our results favor the model that UGT1 modifies nonnative proteins regardless of whether they are on- or off-pathway (Figure 7, A and B). This query likely occurs after an initial period of maturation that is dictated by the localization of UGT1 and its access to the maturing nascent chain. Of importance, reglucosylation inhibits secretion of on-pathway substrates either by supporting persistent chaperone binding (bulk flow hypothesis) or inhibiting binding to anterograde targeting receptors (receptor-mediated selective transport hypothesis; Wieland et al., 1987; Balch et al., 1994; Aridor and Balch, 1996). The significance of the more efficient reglucosylation of off-pathway terminally misfolded substrates is uncertain, as they evidently display a signal that is efficiently recognized by a downstream ERAD receptor. Perhaps persistent lectin chaperone binding of the monoglucosylated substrate localizes the ERAD substrate to a location in the ER where ERAD receptors or mannosidases are concentrated, such as the proposed ER quality control compartment (ERQC), where calnexin, calreticulin, ER ManI, and EDEM1 have been localized but not UGT1 and ERp57 (Avezov et al., 2008). Likely candidates to shepherd glycosylated proteins through this retrograde trafficking route include ER ManI/Man1B1, EDEM1-3, Os-9, and XTP3-B (Hosokawa et al., 2003, 2009; Molinari et al., 2003; Oda et al., 2003; Groisman et al., 2011; Hebert and Molinari, 2012; Pan et al., 2013; Aikawa et al., 2014; Ninagawa et al., 2014). There is vigorous debate over the location, roles, and substrate selectivity for some of these factors.
Some diseases are caused by the ER retention of an otherwise active protein by an apparently overzealous quality control process (Guerriero and Brodsky, 2012) that is capable of recognizing slight imperfections found even associated with active proteins. If the quality control process could be relaxed for these substrates, proper trafficking could alleviate the loss-of-function disease. UGT1 is a prime candidate for modulation for diseases of this category, as the reglucosylation of wild-type on-pathway targets supported ER retention and delayed the secretion of potentially active substrates. The fact that the terminally misfolded substrates were still cleared efficiently and rapidly when proteins were trapped with monoglucosylated glycans suggests that the modulation of the activity of UGT1 might not disrupt the ERAD process and might provide an effective therapy for the treatment of some of these loss-of-function diseases. A recent study showed that recombinant ER ManI/Man1B1 was capable of trimming glucosylated glycans and favored the trimming of denatured proteins (Aikawa et al., 2014); however, there is debate on whether this exomannosidase is localized to the ER or Golgi and the necessity of its activity for ERAD (Avezov et al., 2008; Pan et al., 2013; Ninagawa et al., 2014). Further studies will be required to identify the mechanism by which reglucosylated ERAD substrates are efficiently recognized and targeted for degradation. Sorting out these concerns and the mechanism by which reglucosylated ERAD substrates are efficiently recognized and targeted for degradation will require further cellular experiments, as well as approaches using purified components.
MATERIALS AND METHODS
Reagents
MI8-5 Chinese hamster ovary cells were a gift from S. Krag (Johns Hopkins University, Baltimore, MD; Quellhorst et al., 1999). Plasmids for pGEX-3X GST-calreticulin, T-cell receptor α (TCRα) with HA tag at its C-terminus, human α1-antitrypsin, and α-NAGAL were from M. Michalak (University of Alberta, Edmonton, Canada), S. Fang (University of Maryland, Baltimore, MD), M. Ziak and J. Roth (University of Zurich, Zurich, Switzerland), and S. Garman (University of Massachusetts-Amherst, Amherst, MA), respectively. Polyclonal rabbit α1-antitrypsin (Dako, Glostrup, Denmark) and calreticulin antisera (Thermo Fisher Scientific, Waltham, MA), monoclonal mouse HA antibodies (Roche, Mannheim, Germany), and polyclonal goat antisera against murine prosaposin (G. Grabowski and Y. Sun, University of Cincinnati College of Medicine) were obtained as indicated. Cell culture material and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA) and polyethyleneimine from Polysciences (Warrington, PA). [35S]Met/Cys was acquired from PerkinElmer (Waltham, MA). Reduced glutathione Sepharose 4B and protein A Sepharose 4B were from GE Healthcare (Uppsala, Sweden) and protein G-plus agarose beads from Santa Cruz Biotechnology (Santa Cruz, CA). DNJ and kifunensine (KIF) were obtained from Toronto Research Chemicals (Toronto, Canada). PNGase F and EndoH were acquired from New England Biolabs (Ipswich, MA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO).
Transfection and metabolic labeling
MI8-5 Chinese hamster ovary cells were cultured in MEMα supplemented with 5 or 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin at 34°C in 5% CO2. Nearly confluent cells were transfected with the indicated plasmid using Lipofectamine 2000 or polyethyleneimine according to manufacturer's instructions.
Cells were starved for 30 min or 1 h in Cys/Met-free medium with 0.5 mM DNJ where indicated and then pulse labeled with 60 μCi of [35S]Cys/Met in 3.5-cm plates for 30 min or 1 h as indicated. When analyzing degradation of TCRα or NHK, no prior amino acid starvation was performed. Instead, cells were preincubated with 0.5 mM DNJ for 30 min before pulse labeling. Immediately after the pulse, cells were washed with phosphate buffer solution (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4) and chased with regular growth medium for the indicated times. DNJ was present throughout the chase where indicated except for Figure 5, where DNJ was added at the indicated times relative to the start of chase. Radiolabeled cells were washed twice with PBS on ice, followed by lysis with MNT buffer (20 mM 2-(N-morpholino) ethanesulfonic acid, 100 mM NaCl, 0.5% Triton X-100, 30 mM Tris-HCl, pH 7.5) containing 50 μM calpain inhibitor I, 1 μM pepstatin, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 0.4 mM phenylmethylsulfonyl fluoride, and 20 mM N-ethyl maleimide. Media and the Triton X-100–insoluble pellet were collected where indicated. Cells were incubated as designated with 20 μM MG132 or 100 μM KIF for 4 or 2 h before the pulse, respectively.
Immunoprecipitations and SDS–PAGE
The postnuclear fraction was precleared with 10% zysorbin for 1 h at 4°C and then incubated with antibody and protein A–Sepharose beads and rotated end over end for 14 h at 4°C, except with the prosaposin antibody, for which protein G–agarose was used. The Triton X-100–insoluble pellet was solubilized in 1% SDS in 100 mM Tris-HCl, pH 8, by high-speed vortexing at 22°C for 5 min, followed by heating for 10 min at 100°C and then sonication. The SDS was quenched by dilution with excess MNT buffer. The media and Triton X-100–insoluble fractions were also precleared with 10% zysorbin, and immunoprecipitations were conducted as described for the postnuclear supernatant. Immunopellets were washed with 100 mM Tris, pH 8.6, 300 mM NaCl, 0.1% SDS, and 0.05% Triton X-100. Proteins were eluted from beads with reducing sample buffer, and then SDS–PAGE was performed. Radiolabeled samples were visualized by phosphorimaging (FLA-500; Fujifilm, Tokyo, Japan) and quantified using MultiGauge software or ImageQuant (Fujifilm).
GST-calreticulin pull down
Recombinant GST-calreticulin was expressed in Escherichia coli and purified as previously described (Baksh and Michalak, 1991; Pearse et al., 2008). A fraction of the cell lysate was incubated with 8 μg of purified GST-calreticulin prebound to reduced glutathione beads at 4°C and rotated end over end for 14 h. Samples were washed with 100 mM Tris, pH 8.6, 300 mM NaCl, 0.1% SDS, and 0.05% Triton X-100 and eluted with reducing sample buffer. If followed by immunoprecipitation, samples were eluted with 1% SDS in 10 mM Tris, pH 7.5, and 150 mM NaCl at 100°C and quenched with excess MNT buffer, followed by incubation with the corresponding antisera and Sepharose beads overnight and washing as described.
Coimmunoprecipitation studies with calnexin and calreticulin
Cells were grown in 10-cm plates, lysed with 2% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid and 200 mM NaCl, pH 7.5 (HBS), and incubated with the respective antibody for 3 h. Immunopellets were washed with 0.5% CHAPS in HBS and eluted from beads with reducing sample buffer. When followed by a second immunoprecipitation, samples were eluted with 1% SDS in 10 mM Tris, pH 7.5, and 150 mM NaCl at 100°C and quenched with excess 2% CHAPS in HBS, followed by incubation with the corresponding antisera and Sepharose beads overnight and washing with 0.5% CHAPS in HBS.
Calculation of percentages of reglucosylation
The percentage of reglucosylation was calculated by dividing the number obtained from quantifying the bands in the lanes with the GST-calreticulin pull down followed by immunoprecipitation, by the number obtained from quantifying the bands in the lanes with the corresponding immunoprecipitation directed against the given substrate. Because different percentages of lysate were used to perform the GST-calreticulin pull down followed by immunoprecipitation and the immunoprecipitation alone, this number was accounted for in the division. The fully glycosylated bands were used for the quantification when they were well resolved. Otherwise, all bands were quantified.
Acknowledgments
We are grateful to S. Krag for the generous gift of the MI8-5 Chinese hamster ovary cell line, S. Garman for the α-NAGAL constructs, and G. Grabowski and Y. Sun for providing the prosaposin antibody. We acknowledge the following current or former members of the Hebert laboratory for helpful discussions: B. Pearse, J. Sunryd, K. Giorda, L. Lamriben, and J. Graham. This work was supported by National Institutes of Health Grant GM08674 (U.S. Public Health; to D.N.H.), the Chemistry Biology Interface Training Program (National Research Service Award T32 GM08515 to A.T.), and the Uehara Memorial Foundation (to T. T.).
Abbreviations used:
- A1AT
α-1-antitrypsin
- α-NAGAL
α-N-acetylgalactosaminidase
- DNJ
N-butyl deoxynojirimycin
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum–associated degradation
- Glc
glucose
- GlcNAc
N-acetylglucosamine
- Man
mannose
- NHK
null Hong Kong
- TCRα
T-cell receptor α subunit
- UGT1
uridine diphosphate (UDP)-glucose:glycoprotein glucosyltransferase 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1254) on November 26, 2014.
*Present address: Department of Life Sciences, Faculty of Engineering and Resource, Akita University, Akita 010-8502, Japan.
REFERENCES
- Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Aikawa J, Takeda Y, Matsuo I, Ito Y. Trimming of glucosylated N-glycans by human ER alpha1,2-mannosidase I. J Biochem. 2014;155:375–384. doi: 10.1093/jb/mvu008. [DOI] [PubMed] [Google Scholar]
- Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–327. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Balch WE. Principles of selective transport: coat complexes hold the key. Trends Cell Biol. 1996;6:315–320. doi: 10.1016/0962-8924(96)10027-1. [DOI] [PubMed] [Google Scholar]
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5 6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 2008;19:216–225. doi: 10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baksh S, Michalak M. Expression of calreticulin in Escherichia coli and identification of its Ca2+ binding domains. J Biol Chem. 1991;266:21458–21465. [PubMed] [Google Scholar]
- Balch WE, McCaffery JM, Plutner H, Farquhar MG. Vesicular stomatitis virus is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Blanco I, de Serres FJ, Fernandez-Bustillo E, Lara B, Miravitlles M. Estimated numbers and prevalence of PI*S and PI*Z alleles of alpha1-antitrypsin deficiency in European countries. Eur Respir J. 2006;27:77–84. doi: 10.1183/09031936.06.00062305. [DOI] [PubMed] [Google Scholar]
- Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Bottomley SP. The structural diversity in alpha1-antitrypsin misfolding. EMBO Rep. 2011;12:983–984. doi: 10.1038/embor.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral CM, Liu Y, Sifers RN. Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem Sci. 2001;26:619–624. doi: 10.1016/s0968-0004(01)01942-9. [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho P, Lechleiter JD. Calreticulin inhibits repetitive intracellular Ca2+ waves. Cell. 1995;82:765–771. doi: 10.1016/0092-8674(95)90473-5. [DOI] [PubMed] [Google Scholar]
- Caramelo JJ, Castro OA, Alonso LG, de Prat-Gay G, Parodi AJ. UDP-Glc:glycoprotein glucosyltransferase recognizes structured and solvent accessible hydrophobic patches in molten globule-like folding intermediates. Proc Natl Acad Acad Sci USA. 2003;100:86–91. doi: 10.1073/pnas.262661199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo JJ, Castro OA, de Prat-Gay G, Parodi AJ. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J Biol Chem. 2004;279:46280–46285. doi: 10.1074/jbc.M408404200. [DOI] [PubMed] [Google Scholar]
- Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1?SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NE, Garman SC. The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases. J Mol Biol. 2009;393:435–447. doi: 10.1016/j.jmb.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NE, Metcalf MC, Best D, Fleet GW, Garman SC. Pharmacological chaperones for human alpha-N-acetylgalactosaminidase. Proc Natl Acad Sci USA. 2012;109:17400–17405. doi: 10.1073/pnas.1203924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc S, Hirsch C, Oggier DM, Deprez P, Jakob C, Sommer T, Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier JH, Tamura T, Sunryd JC, Hebert DN. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio C, Caramelo JJ, Parodi AJ. UDP-GlC:glycoprotein glucosyltransferase-glucosidase II, the ying-yang of the ER quality control. Semin Cell Dev Biol. 2010;21:491–499. doi: 10.1016/j.semcdb.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Ermonval M, Duvet S, Zonneveld D, Cacan R, Buttin G, Braakman I. Truncated N-glycans affect protein folding in the ER of CHO-derived mutant cell lines without preventing calnexin binding. Glycobiology. 2000;10:77–87. doi: 10.1093/glycob/10.1.77. [DOI] [PubMed] [Google Scholar]
- Ermonval M, Kitzmuller C, Mir AM, Cacan R, Ivessa NE. N-glycan structure of a short-lived variant of ribophorin I expressed in the MadIA214 glycosylation-defective cell line reveals the role of a mannosidase that is not ER mannosidase I in the process of glycoprotein degradation. Glycobiology. 2001;11:565–576. doi: 10.1093/glycob/11.7.565. [DOI] [PubMed] [Google Scholar]
- Feige MJ, Hendershot LM. Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol Cell. 2013;51:297–309. doi: 10.1016/j.molcel.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F, D’Alessio C, Fanchiotti S, Parodi AJ. A misfolded protein conformation is not a sufficient condition for in vivo glucosylation by the UDP-Glc:glycoprotein glucosyltransferase. EMBO J. 1998;17:5877–5886. doi: 10.1093/emboj/17.20.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F, Jannatipour M, Hellman U, Rokeach LA, Parodi A. A new stress protein: synthesis of Schizosaccharomyces pombe UDP-glc:glycoprotein glycosyltransferase mRNA is induced by stress conditions but the enzyme is not essential for viability. EMBO J. 1996;15:705–713. [PMC free article] [PubMed] [Google Scholar]
- Fernandez FS, Trombetta SE, Hellman U, Parodi AJ. Purification to homogeneity of UDP-glucose:glycoprotein glucosyltransferase from Schizosaccharomyces pombe and apparent absence of the enzyme from Saccharomyces cerevisiae. J Biol Chem. 1994;269:30701–30706. [PubMed] [Google Scholar]
- Ferris SP, Jaber NS, Molinari M, Arvan P, Kaufman RJ. UDP-glucose:glycoprotein glucosyltransferase (UGGT1) promotes substrate solubility in the endoplasmic reticulum. Mol Biol Cell. 2013;24:2597–2608. doi: 10.1091/mbc.E13-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis E, Wang N, Parag H, Halaban R, Hebert DN. Tyrosinase maturation and oligomerization in the endoplasmic reticulum require a melanocyte-specific factor. J Biol Chem. 2003;278:25607–25617. doi: 10.1074/jbc.M303411200. [DOI] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Groisman B, Shenkman M, Ron E, Lederkremer GZ. Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J Biol Chem. 2011;286:1292–1300. doi: 10.1074/jbc.M110.154849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92:537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaban R, Chang E, Zhang Y, Moellmann G, Hanlon D, Michalak M, Setaluri V, Hebert DN. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc Natl Acad Sci USA. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Lamriben L, Powers ET, Kelly JW. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nat Chem Biol. 2014;10:902–910. doi: 10.1038/nchembio.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Molinari M. Flagging and docking: dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem Sci. 2012;37:404–410. doi: 10.1016/j.tibs.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Kamiya Y, Kamiya D, Kato K, Nagata K. Human OS-9, a lectin required for glycoprotein ERAD, recognizes mannose-trimmed N-glycans. J Biol Chem. 2009;284:17061–17068. doi: 10.1074/jbc.M809725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci USA. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa JB, Ploegh HL. The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity. 1997;7:113–122. doi: 10.1016/s1074-7613(00)80514-2. [DOI] [PubMed] [Google Scholar]
- John LM, Lechleiter JD, Camacho P. Differential modulation of SERCA2 isoforms by calreticulin. J Cell Biol. 1998;142:963–973. doi: 10.1083/jcb.142.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaivanova VK, Luan P, Spiro RG. Processing of viral envelope glycoprotein by the endomannosidase pathway: evaluation of host cell specificity. Glycobiology. 1998;8:725–730. doi: 10.1093/glycob/8.7.725. [DOI] [PubMed] [Google Scholar]
- Kass I, Knaupp AS, Bottomley SP, Buckle AM. Conformational properties of the disease-causing Z variant of alpha1-antitrypsin revealed by theory and experiment. Biophys J. 2012;102:2856–2865. doi: 10.1016/j.bpj.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith N, Parodi AJ, Caramelo JJ. Glycoprotein tertiary and quaternary structures are monitored by the same quality control mechanism. J Biol Chem. 2005;280:18138–18141. doi: 10.1074/jbc.M501710200. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Kumaraswamy E, Zhou Y, Hatfield DL, Gladyshev VN. Association between the 15-kDa selenoprotein and UDP-glucose:glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2001;276:15330–15336. doi: 10.1074/jbc.M009861200. [DOI] [PubMed] [Google Scholar]
- Kunte A, Zhang W, Paduraru C, Veerapen N, Cox LR, Besra GS, Cresswell P. Endoplasmic reticulum glycoprotein quality control regulates CD1d assembly and CD1d-mediated antigen presentation. J Biol Chem. 2013;288:16391–16402. doi: 10.1074/jbc.M113.474221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labriola C, Cazzulo JJ, Parodi AJ. Retention of glucose units added by the UDP-GLC:glycoprotein glucosyltransferase delays exit of glycoproteins from the endoplasmic reticulum. J Cell Biol. 1995;130:771–779. doi: 10.1083/jcb.130.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labriola C, Cazzulo JJ, Parodi AJ. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Mol Biol Cell. 1999;10:1381–1394. doi: 10.1091/mbc.10.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Lefrancois S, Zeng J, Hassan AJ, Canuel M, Morales CR. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 2003;22:6430–6437. doi: 10.1093/emboj/cdg629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Camacho P. Ca2+-dependent redox modulation of SERCA 2b by ERp57. J Cell Biol. 2004;164:35–46. doi: 10.1083/jcb.200307010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Choudhury P, Cabral CM, Sifers RN. Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 1999;274:5861–5867. doi: 10.1074/jbc.274.9.5861. [DOI] [PubMed] [Google Scholar]
- Lomas DA, Evans DL, Finch JT, Carrell RW. The mechanisms of Z a1-antitrypsin accumulation in the liver. Nature. 1992;357:605–607. doi: 10.1038/357605a0. [DOI] [PubMed] [Google Scholar]
- Meunier L, Usherwood Y-K, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes from multiprotein complexes in the endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari M, Calanca V, Galli C, Lucca P, Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299:1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- Molinari M, Galli C, Vanoni O, Arnold SM, Kaufman RJ. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol Cell. 2005;20:503–512. doi: 10.1016/j.molcel.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Ninagawa S, Okada T, Sumitomo Y, Kamiya Y, Kato K, Horimoto S, Ishikawa T, Takeda S, Sakuma T, Yamamoto T, Mori K. EDEM2 initiates mammalian glycoprotein ERAD by catalyzing the first mannose trimming step. J Cell Biol. 2014;206:347–356. doi: 10.1083/jcb.201404075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Hosokawa N, Wada I, Nagata K. EDEM as an acceptor of terminally misfolded glycoproteins released from calnexin. Science. 2003;299:1394–1397. doi: 10.1126/science.1079181. [DOI] [PubMed] [Google Scholar]
- Olivari S, Cali T, Salo KE, Paganetti P, Ruddock LW, Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- O’Brien JS, Carson GS, Seo HC, Hiraiwa M, Kishimoto Y. Identification of prosaposin as a neurotrophic factor. Proc Natl Acad Sci USA. 1994;91:9593–9596. doi: 10.1073/pnas.91.20.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Cheng X, Sifers RN. Golgi-situated endoplasmic reticulum alpha-1, 2-mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with gamma-COP. Mol Biol Cell. 2013;24:1111–1121. doi: 10.1091/mbc.E12-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CG, Fessler LI, Nelson RE, Fessler JH. Drosophila UDP-glucose:glycoprotein glucosyltransferase: sequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO J. 1995;14:1294–1303. doi: 10.1002/j.1460-2075.1995.tb07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BR, Gabriel L, Wang N, Hebert DN. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–320. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BR, Hebert DN. Lectin chaperones help direct the maturation of glycoproteins in the endoplasmic reticulum. Biochim Biophys Acta. 2010;1803:684–693. doi: 10.1016/j.bbamcr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse BR, Tamura T, Sunryd JC, Grabowski GA, Kaufman RJ, Hebert DN. The role of UDP-Glc:glycoprotein glucosyltransferase 1 in the maturation of an obligate substrate prosaposin. J Cell Biol. 2010;189:829–841. doi: 10.1083/jcb.200912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan EM, Kamiya Y, Kamiya D, Denic V, Weibezahn J, Kato K, Weissman JS. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quellhorst GJ, O’Rear JL, Cacan R, Verbert A, Krag SS. Nonglucosylated oligosaccharides are transferred to protein in MI8–5 Chinese hamster ovary cells. Glycobiology. 1999;9:65–72. doi: 10.1093/glycob/9.1.65. [DOI] [PubMed] [Google Scholar]
- Ritter C, Helenius A. Recognition of local glycoprotein misfolding by the ER folding sensor UDP-glucose:glycoprotein glucosyltransferase. Nat Struct Biol. 2000;7:278–280. doi: 10.1038/74035. [DOI] [PubMed] [Google Scholar]
- Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. EMBO J. 2005;24:1730–1738. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M, Suzuki R, Watanabe N, Masaki T, Tomonaga M, Muhammad A, Kato T, Matsuura Y, Watanabe H, Wakita T, Suzuki T. Role of the endoplasmic reticulum-associated degradation (ERAD) pathway in degradation of hepatitis C virus envelope proteins and production of virus particles. J Biol Chem. 2011;286:37264–37273. doi: 10.1074/jbc.M111.259085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifers RN, Brashears-Macatee S, Kidd VJ, Muensch H, Woo SL. A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem. 1988;263:7330–7335. [PubMed] [Google Scholar]
- Solda T, Galli C, Kaufman RJ, Molinari M. Substrate-specific requirements for UGT1-dependent release from calnexin. Mol Cell. 2007;27:238–249. doi: 10.1016/j.molcel.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Ferrero-Garcia MA, Parodi AJ. Recognition of the oligosaccharide and protein moieties of glycoproteins by the UDP-Glc:glycoprotein glucosyltransferase. Biochemistry. 1992;31:97–105. doi: 10.1021/bi00116a015. [DOI] [PubMed] [Google Scholar]
- Spiro RG, Zhu Q, Bhoyroo V, Soling HD. Definition of the lectin-like properties of the molecular chaperone, calreticulin, and demonstration of its copurification with endomannosidase from rat liver Golgi. J Biol Chem. 1996;271:11588–11594. doi: 10.1074/jbc.271.19.11588. [DOI] [PubMed] [Google Scholar]
- Stigliano ID, Alculumbre SG, Labriola CA, Parodi AJ, D’Alessio C. Glucosidase II and N-glycan mannose content regulate the half-lives of monoglucosylated species in vivo. Mol Biol Cell. 2011;22:1810–1823. doi: 10.1091/mbc.E11-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SC, Ferguson AD, Bergeron JJ, Thomas DY. The ER protein folding sensor UDP-glucose glycoprotein-glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat Struct Mol Biol. 2004;11:128–134. doi: 10.1038/nsmb715. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Helenius A. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J Cell Biol. 2000;148:1123–1129. doi: 10.1083/jcb.148.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta SE, Bosch M, Parodi AJ. Glucosylation of glycoproteins by mammalian, plant, fungal and trypanosomatid protozoa microsomal membranes. Biochemistry. 1989;28:8108–8116. doi: 10.1021/bi00446a022. [DOI] [PubMed] [Google Scholar]
- Trombetta SE, Parodi AJ. Purification to apparent homogeneity and partial characterization of rat liver UDP-glucose:glycoprotein glucosyltransferase. J Biol Chem. 1992;267:9236–9240. [PubMed] [Google Scholar]
- Újvári A, Aron R, Eisenhaure T, Cheng E, Parag HA, Smicun Y, Halaban R, Hebert DN. Translation rate of human tyrosinase determines its N-linked glycosylation level. J Biol Chem. 2001;276:5924–5931. doi: 10.1074/jbc.M009203200. [DOI] [PubMed] [Google Scholar]
- Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol 9, 944–957. 2008 doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland FT, Gleason ML, Serafini TA, Rothman JE. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell. 1987;50:289–300. doi: 10.1016/0092-8674(87)90224-8. [DOI] [PubMed] [Google Scholar]
- Zapun A, Petrescu SM, Rudd PM, Dwek RA, Thomas DY, Bergeron JJ. Conformation-independent binding of monoglucosylated ribonuclease B to calnexin. Cell. 1997;88:29–38. doi: 10.1016/s0092-8674(00)81855-3. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wearsch PA, Zhu Y, Leonhardt RM, Cresswell P. A role for UDP-glucose glycoprotein glucosyltransferase in expression and quality control of MHC class I molecules. Proc Natl Acad Sci USA. 2011;108:4956–4961. doi: 10.1073/pnas.1102527108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber C, Fan JY, Guhl B, Parodi A, Fessler JH, Parker C, Roth J. Immunolocalization of UDP-glucose:glycoprotein glucosyltransferase indicates involvement of pre-Golgi intermediates in protein quality control. Proc Natl Acad Sci USA. 2001;98:10710–10715. doi: 10.1073/pnas.191359198. [DOI] [PMC free article] [PubMed] [Google Scholar]