The arrestin Art1 and the Rho1 guanine nucleotide exchange factor Rgf3 are interdependent for their localizations to the division site during fission yeast cytokinesis. Art1 physically interacts with Rgf3 to modulate active Rho1 GTPase levels for successful septal formation.
Abstract
Rho GTPases, activated by guanine nucleotide exchange factors (GEFs), are essential regulators of polarized cell growth, cytokinesis, and many other cellular processes. However, the regulation of Rho-GEFs themselves is not well understood. Rgf3 is an essential GEF for Rho1 GTPase in fission yeast. We show that Rgf3 protein levels and localization are regulated by arrestin-related protein Art1. art1∆ cells lyse during cell separation with a thinner and defective septum. As does Rgf3, Art1 concentrates to the contractile ring starting at early anaphase and spreads to the septum during and after ring constriction. Art1 localization depends on its C-terminus, and Art1 is important for maintaining Rgf3 protein levels. Biochemical experiments reveal that the Rgf3 C-terminus binds to Art1. Using an Rgf3 conditional mutant and mislocalization experiments, we found that Art1 and Rgf3 are interdependent for localization to the division site. As expected, active Rho1 levels at the division site are reduced in art1∆ and rgf3 mutant cells. Taken together, these data reveal that the arrestin family protein Art1 regulates the protein levels and localization of the Rho-GEF Rgf3, which in turn modulates active Rho1 levels during fission yeast cytokinesis.
INTRODUCTION
Cytokinesis is the last step of the cell cycle that partitions a mother cell into two daughter cells. Most proteins involved in cytokinesis are evolutionarily conserved (Barr and Gruneberg, 2007; Pollard and Wu, 2010). From yeast to humans, cytokinesis requires the coordination of six key events: division-plane specification, actomyosin contractile-ring assembly, constriction and disassembly of the contractile ring, targeted plasma membrane deposition/fusion, extracellular matrix remodeling/formation, and abscission or cell separation (Balasubramanian et al., 2004; Green et al., 2012). The last two steps involve extracellular matrix remodeling and midbody abscission in animal cells and septal formation and daughter cell separation in yeasts (Gould and Simanis, 1997; Cabib et al., 2001; Sipiczki, 2007; Xu and Vogel, 2011; Wloka and Bi, 2012; Elia et al., 2013)
The septum in yeasts is a trilaminar structure. The primary septum is synthesized in a centripetal manner as the actomyosin contractile ring constricts and is sandwiched on both sides by the secondary septa. The primary septum and the surrounding cell wall on the cell sides are digested during cell separation, and the secondary septum becomes the cell wall at the new cell end in the resulting daughter cells (Cabib et al., 2001; Humbel et al., 2001; Sipiczki, 2007). Proper synthesis and maintenance of the septum is essential for cell survival. The septum in the fission yeast Schizosaccharomyces pombe is composed mainly of the polysaccharides α- and β-glucans and galactomannans (Humbel et al., 2001; Sipiczki, 2007). The enzymes involved in glucan synthesis are α- and β-glucan synthases, which consist of a catalytic subunit and a regulatory subunit (Kang and Cabib, 1986; Ribas et al., 1991).
The Rho GTPases Rho1 and Rho2 act as regulatory subunits for glucan synthases in yeasts (Drgonova et al., 1996; Nakano et al., 1997; Cabib et al., 1998; Arellano et al., 1999). Rho GTPases are switched from the inactive GDP-bound form to the active GTP-bound form by Rho guanine nucleotide exchange factors (GEFs). While the GEF for Rho2 is unknown, Rho1 is activated by at least three Rho-GEFs (Rgf1–Rgf3) for its roles in cell polarity and cytokinesis (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005). The Rho-GEF Rgf3 is essential for cytokinesis (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005). Cells with mutated Rgf3 or Rho1 lyse mainly during cell separation. Rgf3-depleted cells have abnormal contractile-ring formation, aberrant septal material deposition, and defective cell separation occurring unevenly from one side of the septum (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005). The findings that Rgf3 interacts with GDP-Rho1 and cells overexpressing Rgf3 have increased glucan synthase activity support Rgf3 being a positive regulator of Rho1 function (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005). However, not much is known about Rgf3 binding partners and regulation.
Septins are conserved GTPases that function as scaffolds or diffusion barriers during cytokinesis, cell polarization, and many other processes in fungal and animal cells (Longtine et al., 1996; Gladfelter et al., 2001; Caudron and Barral, 2009). Unlike in budding yeast, septins are not essential in S. pombe (Longtine et al., 1996; Berlin et al., 2003; Tasto et al., 2003; Wu et al., 2003; An et al., 2004). The synthetic lethal screen with septin-depleted mutants identified Rgf3 and the arrestin-related protein Art1 to be components of parallel pathways that are required to recognize and repair cell wall damage (Wu et al., 2010). The art1-s34 mutant can be rescued by overexpression of Rho1 and Rho-GEFs Rgf1–Rgf3 (Wu et al., 2010). However, the relationship between the arrestin Art1 and Rho-GEFs was unknown. Arrestins are known for abrogating G protein–coupled receptor (GPCR)-mediated signaling in eukaryotes (Kang et al., 2014). They typically have arrestin-N and arrestin-C domains separated by a linker. Both the N and C domains fold into a characteristic immunoglobulin-like β-sandwich fold connected by the linker (Kang et al., 2014). Arrestins are involved in receptor recognition and binding. In addition, they have clathrin-binding sites involved in binding to the endocytotic machinery (Premont and Gainetdinov, 2007; Aubry et al., 2009; Kendall and Luttrell, 2009). In budding yeast, structurally related arrestin proteins (ARTs) are known to act as endocytic adaptors in recycling of cell surface receptors (Lin et al., 2008; Nikko and Pelham, 2009; Herrador et al., 2010). There are at least 10 arrestin-related proteins in S. pombe (Supplemental Figure S1). They are not well studied, except for Ste7 having a known role in meiosis and Any1/Arn1 being involved in regulating amino acid transporters by interacting with the ubiquitin ligase Pub1 (Matsuyama et al., 2000; Nakase et al., 2013; Nakashima et al., 2014).
In this study, we found that the arrestin-related protein Art1 and the Rho1 GEF Rgf3 are interdependent for their localizations to the division site. Art1 physically interacts with Rgf3 and is important for regulating Rgf3 protein levels. Art1 and Rgf3 cooperate to modulate active Rho1 levels for septal formation. Collectively our results reveal a novel role for the arrestin Art1 in cytokinesis by regulating Rho-GEF and Rho GTPase activities.
RESULTS
art1 mutants lyse during cell separation with defective septa
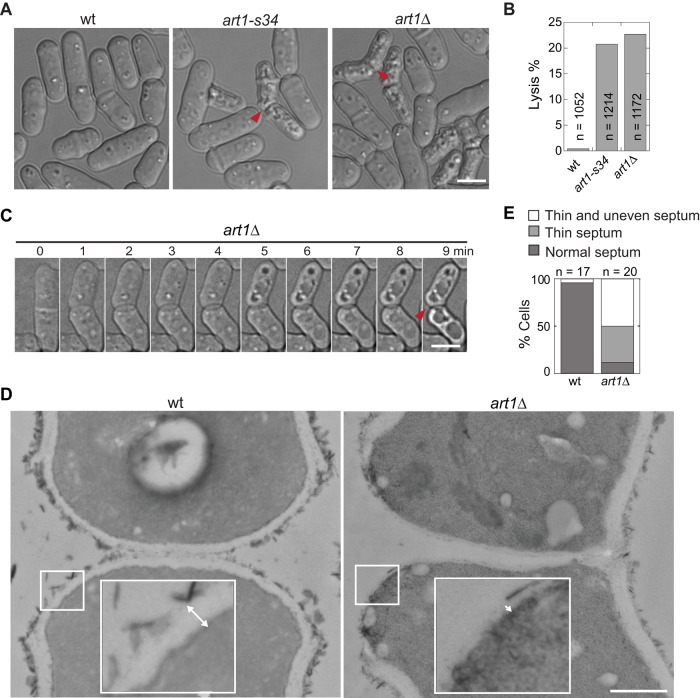
The art1-s34 mutant was isolated in a synthetic lethal/sick screen carried out in a septin-depletion strain along with several genes that are known or predicted components of the cell-integrity pathway (Wu et al., 2010). art1-s34 cells have a mild cell lysis defect (∼5% of asynchronous cells) when grown in minimal medium EMM5S at 30°C (Wu et al., 2010). However, when grown in rich YE5S medium at 25°C, ∼20% of art1-s34 cells lysed (Figure 1, A and B). To further study the role of Art1, we deleted its whole open reading frame and found that cell lysis was similar (∼22%) in art1∆ cells (Figure 1, A and B), which is consistent with the prediction that art1-s34 is a null allele (Wu et al., 2010). The lysis phenotype of the art1 mutants resembled that of the Rho-GEF rgf3(lad1-1) mutant (Morrell-Falvey et al., 2005), in which the lysed daughter cells often attach to each other in the shape of “boomerangs” (Figure 1A, arrowheads). Time-lapse imaging of art1∆ cells revealed that lysis occurred during cell separation (Figure 1C and Supplemental Video S1). The lysis phenotype suggests that the plasma membrane and/or the septum at the division site are defective in the art1 mutants.
FIGURE 1:
art1 mutants have defective division septa. (A–C) Cell lysis phenotype of art1 mutant cells. Arrowheads mark boomerang-shaped lysed cells. (A) Differential interference contrast images and (B) quantification of cell lysis in wt (strain JW81), art1-s34 (JW404), and art1∆ (JW3563) cells. (C) Time-lapse imaging of an art1∆ (JW3563) cell lysed during cell separation. (D and E) Micrographs (D) and quantification (E) of septa under EM in wt (JW81) and art1∆ (JW2543) cells. Insets in D show magnification of the cell wall region in the white box. Double-headed arrows mark cell wall thickness. (E) A thin septum means the cell wall at the division site is continuous but ≤ 50% the thickness of that in wt cells. Thin and uneven septum is the same as thin septum, except that the septum is discontinuous. Scale bars: A and C, 5 μm; D, 0.5 μm.
Plasma membrane deposition and septal formation at the division site are closely coordinated with the actomyosin contractile-ring constriction (Gould and Simanis, 1997; Liu et al., 1999; Laporte et al., 2010; Wloka and Bi, 2012). So, using myosin II heavy chain Myo2 as a marker, we first tested whether the contractile ring was defective in art1∆ cells. While ring formation and ring morphology were normal, rates of ring maturation and constriction were significantly delayed in art1∆ cells (Supplemental Figure S2). We next tested plasma membrane closure/integrity during cytokinesis, using fluorescence loss in photobleaching (FLIP) assays. Fluorescence exchange of diffusible green fluorescent protein (GFP) between the two halves of a dividing cell stopped at approximately the same time following the completion of contractile-ring constriction in wild-type (wt) and art1∆ cells (Supplemental Figure S3, A and B). Together these data indicate that Art1 has no obvious role in the plasma membrane integrity during cytokinesis, although ring maturation and constriction take longer times without Art1. Therefore cell lysis in art1∆ cells could be due to a defect in the newly formed cell wall.
Using electron microscopy (EM), we found that wt daughter cells separated symmetrically, with the primary septum and surrounding cell wall being digested evenly from the cell equator (Figure 1D, left). In art1∆, however, daughter cells separated primarily from one side (Figure 1D, right). Moreover, in art1∆ cells, the septum was severely defective, being wavy and thinner (Figure 1, D and E, and Supplemental Figure S3C). The cell wall at the new cell end, which derives from the secondary septum, had uniform thickness (∼70% of the thickness of the cell wall at other locations) in wt cells (Supplemental Figure S3C). In contrast, in the majority of art1∆ cells, it was thinner, uneven, or discontinuous, with less than half of the thickness of wt cell wall (Figure 1, D and E, and Supplemental Figure S3C). This presumably leaves the plasma membrane partially exposed at the new cell end (Figure 1D, right, inset). The septal defect is similar to that seen in the Rho-GEF rgf3(lad1-1) mutant (Morrell-Falvey et al., 2005). Genetic evidence also supports Art1 playing a role in septal formation. art1∆ is synthetic lethal with sid2-250 and mob1-M17, two mutations in the septation initiation network (SIN) pathway that regulate septal formation (Salimova et al., 2000; Hou et al., 2004; Jin et al., 2006). No double mutants were recovered, even at the permissive temperature of 25°C. Therefore Art1 may have a role, most likely involving septal integrity, in later stages of cytokinesis.
Art1 localizes to the contractile ring and the septation site
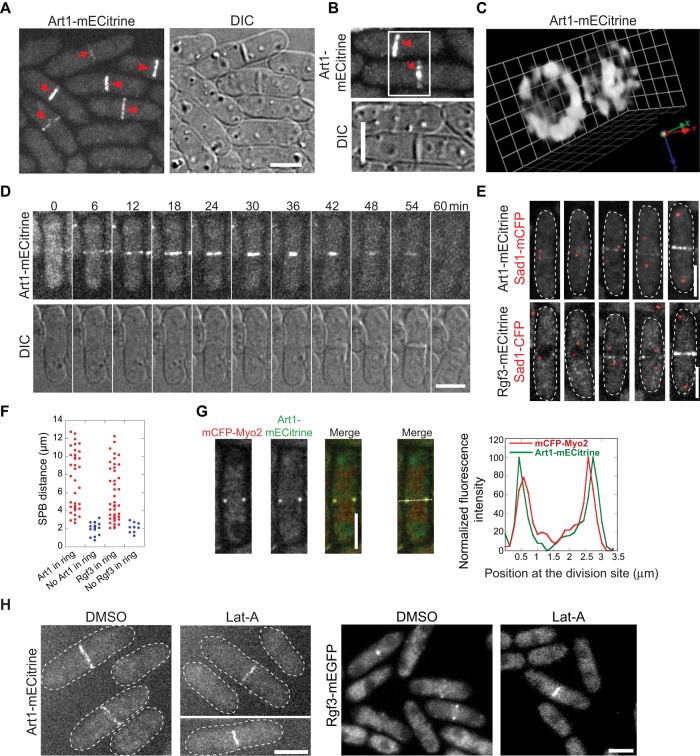
To further understand Art1 function during later stages of cytokinesis, we examined its localization using a monomeric enhanced Citrine (mECitrine; yellow fluorescent protein [YFP] variant)-tagged strain. Art1 concentrated to the contractile ring during cytokinesis (Figure 2, A–C, arrowheads). After ring constriction, the Art1 signal spread unevenly onto the septal disk (Figure 2, A–C, arrows). In addition, Art1 also diffused in the cytoplasm at all stages of the cell cycle. A compact contractile ring forms from cytokinetic nodes at the end of anaphase A in wt cells, when the spindle is ∼2.5 μm long (Wu et al., 2003; Lee et al., 2012). Using time-lapse microscopy, we found that Art1 appeared directly in the contractile ring (not in the precursor nodes), increased in fluorescence intensity, constricted, and then spread to the septum (Figure 2D). Art1 appeared in the contractile ring when spindle pole bodies (SPBs) were ∼3 μm apart (Figure 2, E and F), corresponding to the maturing contractile ring in wt cells at early anaphase B (Nabeshima et al., 1998; Mallavarapu et al., 1999; Wu et al., 2003). It remained at the division site until cell separation. Thus the pattern of Art1 localization is consistent with its role in contractile-ring maturation, ring constriction, and septal integrity.
FIGURE 2:
Localization of Art1 to the contractile ring and the septation site during cytokinesis. (A–C) Micrographs of Art1-mECitrine (JW2674) localization at the contractile ring (arrowheads) and septal disk (arrows). (C) Three-dimensional projection of the cells in B imaged with 0.2-μm z-spacing. 1 grid = 0.95 μm. (D) Time course of Art1-mECitrine (JW2674) shows its appearance, constriction, and septal localization. (E and F) Micrographs (E) and quantification (F) of the arrival of Art1 (JW2694) and Rgf3 (JW2748) at the division site using SPB protein Sad1-mCFP as a cell cycle marker. (G) Art1 locates outside of Myo2 in the contractile ring. Micrographs (left) and line scans of fluorescence intensity at the contractile ring (right) in cells expressing both Art1-mECitrine and mCFP-Myo2 (JW4427). The central focal plane was used. (H) Actin-filament independence of Art1 (JW2674) and Rgf3 (JW1105) localization. Scale bars: 5 μm.
To further characterize Art1 localization, we tested the domain required for its localization (Supplemental Figure S4). Art1 is annotated as an arrestin family protein with a conserved Arrestin_C terminal-like domain (www.pombase.org/spombe/result/SPBC19G7.08c; Supplemental Figure S4A). We constructed C- and N-terminal truncations of Art1 with an mECitrine tag. Full-length (FL) Art1 and the C-terminal truncations Art1(∆429–483) and Art1(∆287–483) were expressed at expected sizes as detected by Western blotting (Supplemental Figure S4B). The two truncations were diffused in the cytoplasm and abolished from the contractile ring and septum (Supplemental Figure S4C). However, the expression of Art1 N-terminal or internal truncations Art1(∆1–286) and Art1(∆287–428) was not detectable by Western blotting or observable via microscopy (Supplemental Figure S4, B and C). Consistent with the loss of localization or expression, all the truncations had a cell lysis phenotype similar to art1∆ (Supplemental Figure S4, C and D). These results indicate that the Art1 C-terminal region aa 429–483 is important for its localization, and Art1 localization is critical for its function. In addition, the Art1 N-terminus and the Arrestin_C like domain are important for its stability.
Of interest, the localization of Art1 to the division site and its timing resemble those of Rho-GEF Rgf3 (Tajadura et al., 2004; Mutoh et al., 2005; Wu et al., 2010). First, Rgf3 appeared in the contractile ring when SPBs were ∼3 μm apart (Figure 2, E and F). Second, the pattern of Art1 localization was identical with that of Rgf3. As reported for Rgf3 (Mutoh et al., 2005), the Art1 ring was at the trailing edge of the myosin II heavy-chain Myo2 ring (Figure 2G) and myosin II regulatory light-chain Rlc1-mCFP ring (unpublished data), which indicates that Art1 is closer to the plasma membrane than myosin II. Third, like Rgf3 localization, Art1 localization was independent of actin filaments. When cells were treated with latrunculin A (Lat-A), a sequesterer of actin monomer, that disrupts actin filaments, both Rgf3 and Art1 still localized to the contractile ring and septum (Figure 2H). Together the data presented so far on the mutant phenotype and localization suggest that Art1 may work together with the Rho-GEF Rgf3 during septal formation.
Art1 physically interacts with the Rho-GEF Rgf3
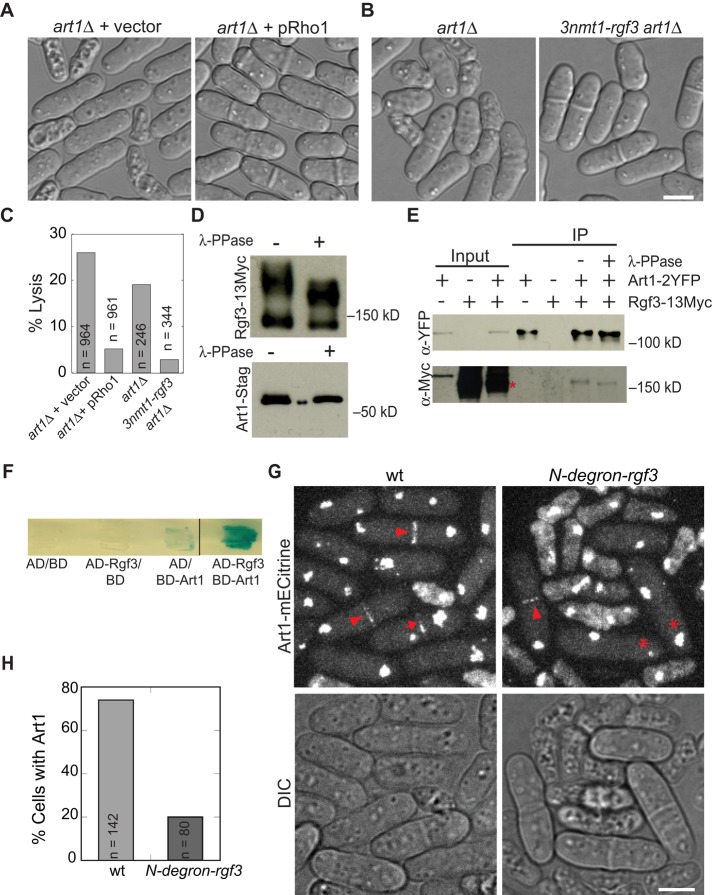
Our previous study showed that the art1-s34 mutant can be rescued by overexpression of Rho-GEFs Rgf1, Rgf2, and Rgf3 or Rho1 GTPase (Wu et al., 2010), but the mechanism is unknown. Rgf1 and Rgf2 have localizations and/or deletion phenotypes different from those of Art1 (Figures 1 and 2; Mutoh et al., 2005; Garcia et al., 2006; Wu et al., 2010). Thus we focused on Rgf3. Rgf3 is a known GEF for Rho1 that activates the cell wall–synthesizing enzymes β-glucan synthases and other effectors (Arellano et al., 1997, 1999; Nakano et al., 1997; Tajadura et al., 2004; Mutoh et al., 2005). We hypothesized that Art1 functions in the same pathway as Rgf3 and Rho1 for cell wall synthesis and cell integrity. To test this hypothesis, we investigated whether art1∆ cells can be rescued by Rgf3 or Rho1 overexpression. Cell lysis of art1∆ cells decreased to ≤ 5% when Rho1 or Rgf3 was overexpressed (Figure 3, A–C). In addition, we found that art1∆ was synthetic lethal at 25°C with rgf3-s44, a point mutation in the GEF domain of Rgf3 (Wu et al., 2010). Thus Art1 indeed functions in the same or a related pathway as Rgf3 and Rho1. Rho1 localizes to both the division site and cell tips (Arellano et al., 1997; Mutoh et al., 2005), whereas Art1 and Rgf3 only concentrate at the division site (Figure 2); thus it is more likely that Art1 interacts with Rgf3.
FIGURE 3:
Rgf3 physically interacts with Art1 and is important for Art1 localization to the division site. (A–C) Overexpression of Rho1 or Rgf3 rescues the lysis phenotype of art1∆. (A) art1∆ cells with empty vector (JW3632) or pRho1 plasmid (JW3633). (B) art1∆ (JW2543) and 3nmt1-rgf3 art1∆ (JW3868) cells were grown in YE5S liquid medium for 40 h. (C) Quantification of lysis in cells as shown in A and B. (D and E) Art1 and Rgf3 co‑IP independent of the phosphorylation status of proteins. (D) Rgf3 (top; strain JW2421) and Art1 (bottom; strain JW3381) are phosphoproteins. (E) Rgf3 co‑IPs with Art1. IPs using anti-YFP antibody and cell lysates of strains expressing Art1-2YFP (JW3711), Rgf3-13Myc (JW2421), and Art1‑2YFP Rgf3-13Myc (JW2696) with or without λ-phosphatase. (F) Art1 and Rgf3 interact in the yeast two-hybrid assays. X-gal overlay assay between VP16-activation domain (AD)-Rgf3 and GAL4-DNA-binding domain (BD)-Art1 along with empty vector controls. The dark blue color indicates a positive reaction. (G and H) Rgf3 is important for Art1 localization. art1-mECitrine sad1-mCFP (JW2694) and art1-mECitrine sad1-mCFP N-degron-rgf3 strains (JW3351) were grown at 25°C and shifted to 36°C for 4 h before imaging at 36°C. (G) Art1 localizes to the contractile ring (arrowheads) and septal region (arrows) in wt cells. Asterisks indicate cells with a complete septum that lack Art1 signal at the division site. (H) Quantification of dividing cells (cells with two separated SPBs) with Art1-mECitrine signal at the division site. Scale bars: 5 μm.
To test whether Art1 and Rgf3 physically interact, we examined their interaction using coimmunoprecipitation (co-IP) and yeast two-hybrid assays. Both Rgf3 and Art1 were seen to be phosphoproteins, because they migrated faster after λ-phosphatase treatment (Figure 3D). Art1-2YFP pulled down Rgf3-13Myc from extracts of cells expressing both proteins at their native levels (Figure 3E). This interaction was not disrupted by phosphatase treatment of the bound complex (Figure 3E). In addition, the interaction between FL Art1 and Rgf3 was confirmed by a yeast two-hybrid assay (Figure 3F). We then used this assay to map out the domains involved in the interaction (Supplemental Figure S5). We first tested the interaction of Art1 C-terminal truncations with FL Rgf3. Rgf3 did not interact with Art1 C-terminal truncations Art1(1–286) or Art1(1–428) (Supplemental Figure S5B), suggesting that Art1(429–483) is important for the Art1–Rgf3 interaction or the interaction only occurs when Art1 is localized to the division site. We then tested the interactions of Rgf3 truncations with FL Art1 and found that truncating the Rgf3 N-terminus greatly enhanced Art1–Rgf3 binding. FL Art1 interacted much more strongly with the truncations Rgf3(657–1275) and Rgf3(856–1275) than with FL Rgf3 (Supplemental Figure S5B). This suggested that the N-terminus of Rgf3 could inhibit Art1 binding to Rgf3. Thus we tested whether the Rgf3 C-terminus binds to Rgf3 N-terminus for the autoinhibition. However, we detected no interaction between Rgf3(1–656) and Rgf3(657–1275) (Supplemental Figure S5B). In addition, we found that the integrity of the Rgf3(856–1275) fragment was important for the strong interaction between Rgf3 and Art1 (Supplemental Figure S5B).
The interaction between Art1 and Rgf3 plays an important role in Art1 localization. When the essential protein Rgf3 was degraded using a temperature-inducible rgf3 (N-degron-rgf3) mutant strain, Art1 localization to the division site was severely affected (Figure 3, G and H). After 4 h at 36°C, only 20% of live N-degron-rgf3 cells with two separated SPBs had an Art1 signal at the division site, compared with 70% in wt cells (Figure 3, G and H). Art1 localized only to the contractile ring (arrowhead), and the localization to the septum (arrows) was not detected in the mutant cells (asterisks). Furthermore, no noticeable difference in Art1 global fluorescence intensity was seen in the two strains. Thus Art1 partially depends on Rgf3 for its ring localization and completely depends on Rgf3 for its septal localization.
Art1 is important for maintaining Rho-GEF Rgf3 protein levels
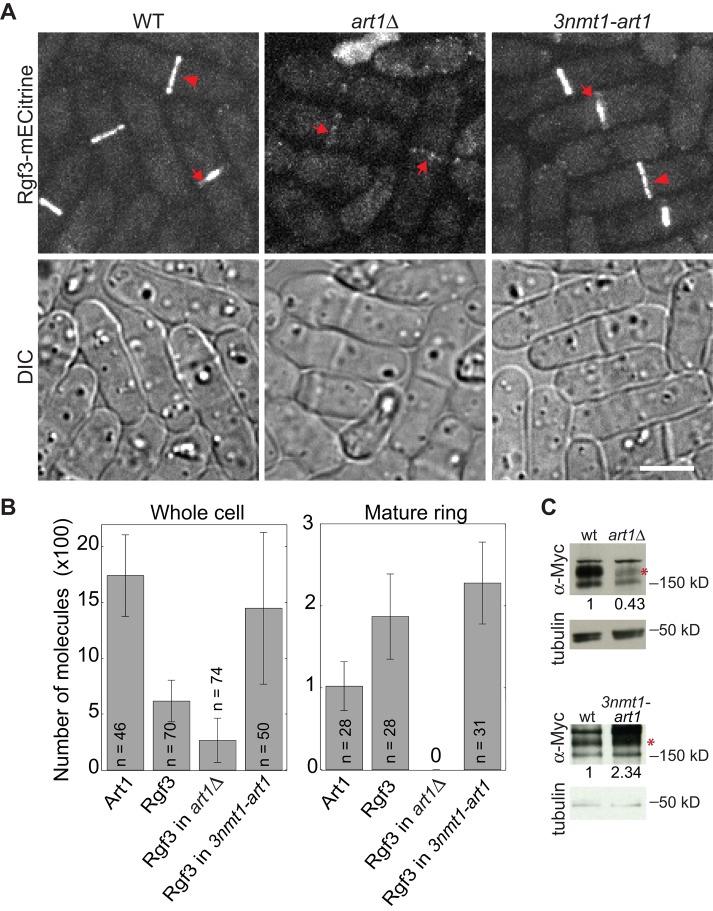
Because Art1 and Rgf3 interact with each other and Rgf3 plays a role in Art1 localization, it is interesting to know whether Art1 is involved in Rgf3 localization. In art1∆ cells, Rgf3 localization to the contractile ring was essentially abolished, and the intensity of Rgf3 in the cytoplasm was also reduced compared with wt (Figure 4A). However, a weak Rgf3 signal at the division site was detected in cells with a complete septum (Figure 4A, arrows). In contrast, Rgf3 intensities in the cytoplasm and the division site obviously increased when Art1 was overexpressed (Figure 4A, right).
FIGURE 4:
Art1 affects Rgf3 localization and protein levels. (A and B) Micrographs (A) and quantifications (B) of Rgf3-mECitrine in wt (JW2748), art1∆ (JW5232), 3nmt1-art1 (JW4889), and Art1-mECitrine (JW2694). (A) Arrows mark septa and arrowheads mark the contractile ring. (B) The numbers of Art1 and Rgf3 molecules globally in the whole cell and locally in mature but full-sized contractile rings. (C) Art1 affects Rgf3 protein levels. Anti-Myc antibody was used to detect Rgf3-13Myc (asterisks) in cell extracts from rgf3-13Myc (wt, JW2421) and rgf3-13Myc art1∆ (JW4816) strains grown in YE5S (top), or from rgf3-13Myc (wt, JW2421) and rgf3-13Myc 3nmt1-art1 (JW5129) strains induced in EMM5S for 24 h (bottom). The relative ratios of Rgf3 protein levels are given (n = 2). Scale bar: 5 μm.
We counted Rgf3 molecules in wt, art1∆, and art1 overexpression cells using quantitative fluorescence microscopy (Wu and Pollard, 2005; Coffman and Wu, 2012) and compared them with the number of Art1 molecules in wt cells. On average, each wt cell had 1740 ± 370 Art1 molecules (Figure 4B, left). However, only a small fraction of them, 100 ± 30 molecules, were in a mature, unconstricting contractile ring (Figure 4B, right). Compared with Art1, each wt cell had less Rgf3 globally (620 ± 190 molecules) but more in the mature ring (190 ± 50) (Figure 4B). Consistent with the microscopy images (Figure 4A), however, the total number of Rgf3 molecules decreased to 270 ± 200 per cell, and Rgf3 was essentially absent in the mature, unconstricting contractile ring in art1∆ cells (Figure 4B). When Art1 was moderately overexpressed, each cell had 1450 ± 680 Rgf3 molecules with 230 ± 50 Rgf3 molecules in the mature ring (Figure 4B). Thus Art1 protein levels influence Rgf3 levels in cells, which was confirmed by measuring Rgf3 levels in art1∆ and art1 overexpression cells using Western blotting (Figure 4C). Together these data imply that Art1 plays a role in maintaining Rgf3 protein levels.
Art1 plays a role in Rgf3 localization
In art1∆ cells, Rgf3 localized to the septal region, but its localization to the mature contractile ring was severely affected (Figure 4A), which suggested that Art1 was important for Rgf3 localization to the contractile ring. However, the Rgf3 levels were also significantly reduced in art1∆ cells (Figure 4, A–C). Thus it was hard to know whether the lack of Rgf3 localization was due to the reduction in overall protein levels. To test whether Art1 plays a direct role in localizing Rgf3, we mislocalized Art1 to various cellular structures using the GFP-binding protein (GBP; Figure 5 and Supplemental Figure S6; Rothbauer et al., 2008). GBP binds to the YFP variant mECitrine but not to cyan fluorescent protein (CFP; Rothbauer et al., 2008; Grallert et al., 2013); we therefore tested whether mislocalized Art1-mECitrine recruited Rgf3-mCFP. We mislocalized Art1 to cortical nodes using anillin Mid1-GBP (Figure 5A) and to SPBs using SIN kinase Cdc7-GBP (Figure 5, B and C). Mislocalized Art1 successfully recruited Rgf3-mCFP to cortical nodes and SPBs (Figure 5, A–C). Only a fraction of Art1 was mislocalized; we observed that Art1 still localized to its native localizations, the constricting ring and the septum (Figure 5, B and C, and Supplemental Figure S6), where Mid1 and Cdc7 are normally absent (Sohrmann et al., 1998; Paoletti and Chang, 2000; Mehta and Gould, 2006). Two lines of evidence indicate the mislocalization is specific: 1) No bleed-through was seen from the YFP to the CFP (440 nm) channel (Supplemental Figure S6A, middle panel); and 2) although Rlc1 is more abundant in cells than Rgf3 (Wu and Pollard, 2005), Rlc1 was not recruited by mislocalized Art1 to SPBs (Supplemental Figure S6B). Together these data indicate that Art1 is important for both the localization and protein levels of Rgf3.
FIGURE 5:
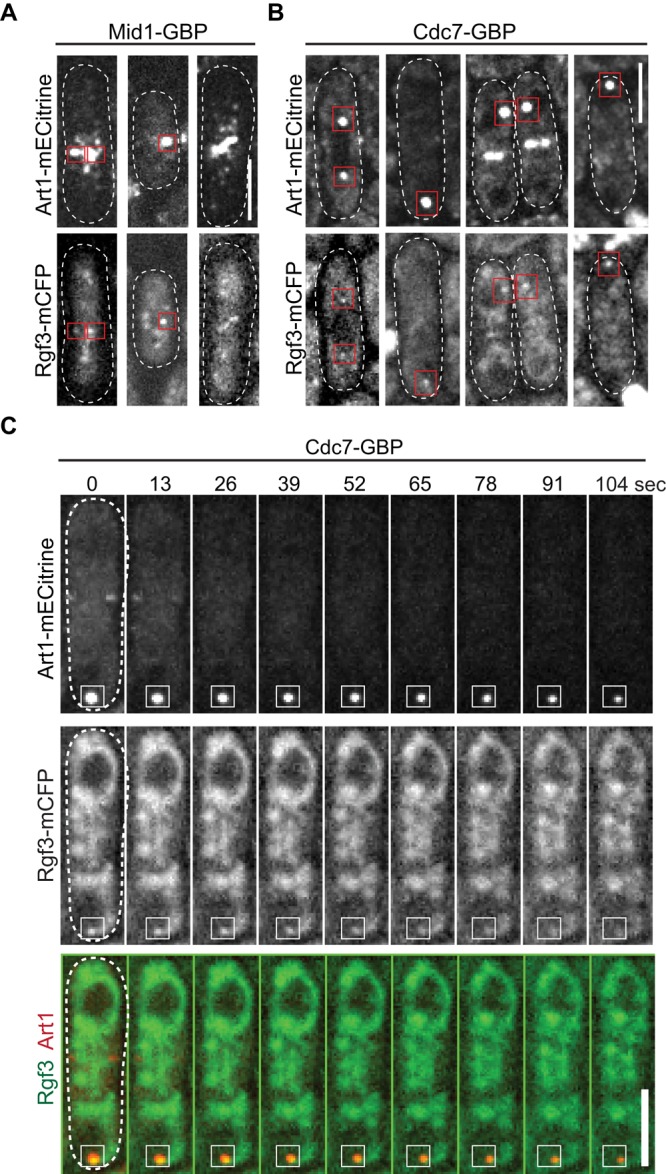
Art1 plays a role in Rgf3 localization. (A) Art1 mislocalizes Rgf3 to Mid1 cortical nodes in single-plane (near cell surface) images of cells expressing Art1-mECitrine Rgf3-mCFP Mid1-GBP (JW5798). (B and C) Art1 mislocalizes Rgf3 to SPBs with Cdc7 in cells expressing Art1-mECitrine Rgf3-mCFP Cdc7-GBP (JW5799). (B) Single focal-plane images. Red boxes show examples of Rgf3 colocalization with mislocalized Art1. (C) Each image shown in the time course is a rolling average of the sum projection (of three z-slices spaced at 0.4 μm) from three consecutive time points at ∼13 s apart. The cell boundary of all cells (A and B) or the cell at 0 s (C) was marked with dashed lines. Scale bars: 5 μm.
Art1 helps to maintain active Rho1 activity at the division site
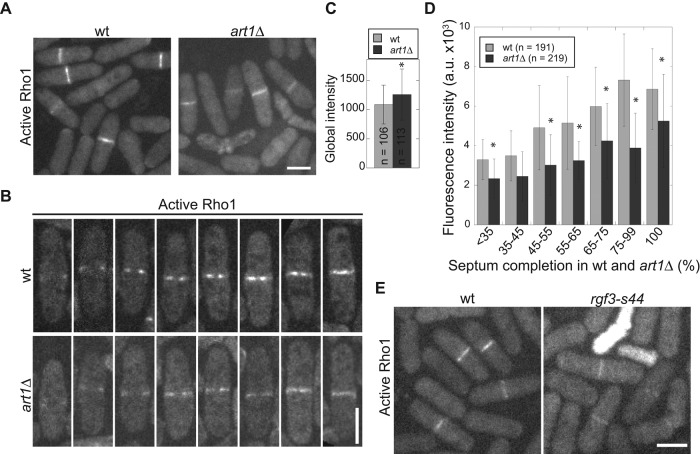
Our data have shown that Art1 interacts with Rgf3 and is important for its localization and protein levels. Rgf3 is a GEF known to activate Rho1 (Tajadura et al., 2004). Thus we hypothesized that art1∆ negatively affected active Rho1 levels at the division site. We used the Rho1 biosensor Pkc1(HR-C2) (Kono et al., 2012) tagged with mECitrine to measure active GTP-Rho1 in wt and art1∆ cells (Figure 6, A–D). Although the global level of active Rho1 was slightly higher in art1∆ cells than in wt cells (Figure 6C), the amount of active Rho1 at the division sites in cells with forming and complete septa was significantly lower in art1∆ cells than in wt cells (Figure 6, A, B, and D). This result was confirmed using an rgf3-s44 mutant (Figure 6E) in which Rho1 activity is expected to be affected, because rgf3-s44 has a mutation in the GEF domain (Wu et al., 2010).
FIGURE 6:
Active Rho1 GTPase at the division site is reduced in art1∆ cells. (A–D) Micrographs (A and B) and quantifications (C and D) of active Rho1 using biosensor Pkc1(HR-C2)-mECitrine in wt (JW5593) and art1∆ (JW5877) cells. Cells were grown in YE5S liquid medium for 48 h before imaging. (B) Representative cells quantified in D. (C) The global intensity of the Rho1 biosensor in wt and art1∆ cells. *, p < 0.05 in t test. (D) Rho1 biosensor intensity at the division site during septal formation in wt and art1∆ cells. Septal growth was measured as percentage completed septa over cell width. *, p ≤ 0.05. (E) Localization of the Rho1 biosensor in wt (JW5593) and rgf3-s44 mutant (JW6165). Cells were grown in YE5S + 1.2 M sorbitol medium due to the severe lysis of rgf3-s44. Scale bars: 5 μm.
Rho1 regulates β-glucan synthases in yeasts (Arellano et al., 1996; Drgonova et al., 1996; Qadota et al., 1996). Thus we measured the levels of β-glucan synthase Bgs1, which is essential for primary septal synthesis (Liu et al., 1999), in art1∆ cells. We detected no significant difference in the Bgs1 levels globally in whole cells or locally at the division site in art1∆ cells compared with wt cells (Supplemental Figure S7, A–C). This suggests that Art1 may indirectly affect Bgs1 enzyme activity through Rho1 but not Bgs1 localization. We also tested the specificity of the Rho1 biosensor in other Rho-GEF mutants (Supplemental Figure S7D). Except for rgf1∆ cells showing slightly higher Rho1 levels at the division site, no obvious difference in active Rho1 levels was observed in other GEF mutants tested (Supplemental Figure S7D). Taking these results together, we conclude that Art1 and Rgf3 work together to activate the Rho1 GTPase during cytokinesis in fission yeast.
DISCUSSION
In this study, we found that Art1, a previously uncharacterized arrestin family protein, plays an important role in fission yeast cytokinesis. Art1 binds to Rgf3, a GEF for Rho1 GTPase, and recruits it to the contractile ring during cytokinesis. Moreover, we found that Art1 also affects Rgf3 protein levels. Thus we have discovered that the arrestin Art1 is a positive regulator of Rho-GEF Rgf3 function in cytokinesis.
Roles of the arrestin Art1 in septal formation during cytokinesis
Our results provide several lines of evidence to support that Art1 is important for septal formation during cytokinesis. First, the septum is defective, and cell lysis occurs during cell separation in art1∆ cells (Figure 1). This defective septal formation may lead to the delay in ring constriction in art1∆ cells (Supplemental Figure S2). Second, art1∆ is synthetic lethal with sid2-250 and mob1‑M17 mutants in the SIN pathway, which is essential for regulating septal formation. Third, Art1 localizes to the contractile ring and septal region, which is consistent with its function (Figure 2). Fourth, Art1 physically interacts with Rgf3, which is essential for septal formation by activating Rho1 GTPase (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005), to regulate its protein levels and localization (Figures 3–5). Fifth, we found that cell lysis in art1∆ cells can be rescued by overexpression of Rgf3 and Rho1 GTPase (Figure 3, A–C), and Art1 modulates the levels of active Rho1 at the division site (Figure 6), although the specificity of the Rho1 biosensor in fission yeast needs to be fully tested in the future. Taken together, our data indicate that Art1 plays a role in septal formation.
Both Art1 and Rgf3 localize to the contractile ring and the septal region (during and after the contractile-ring disassembly). They are partially interdependent for localization. Interestingly, Rgf3 is important for Art1 localization to the septation site but not to the contractile ring (Figure 3G). In contrast, Art1 is crucial for Rgf3 localization to the contractile ring but less important for its localization to the septum. Thus both proteins have other means to localize to the division site. This partial interdependence is consistent with the fact that Rgf3 is essential and Art1 is not. We found that the Art1 C-terminus is important for its localization, and the Rgf3 C-terminus is important for its interaction with Art1 (Supplemental Figures S4 and S5). The DBL homology and pleckstrin homology domain (DH–PH domain; aa 469–855) seems to hinder the interaction of the Rgf3 C-terminal region with Art1. Rgf3 can be recruited to the division site via its N-terminus (Mutoh et al., 2005). The importance of the domain downstream of the DH–PH domain has been previously documented (Morrell-Falvey et al., 2005). A point mutation in this region, F867S, leads to the loss of Rgf3 localization to the contractile ring and causes a reduction in cellular Rgf3 levels (Morrell-Falvey et al., 2005). This indicates that Art1 binding of the Rgf3 C-terminus is another mechanism that recruits Rgf3 to the division site during cytokinesis.
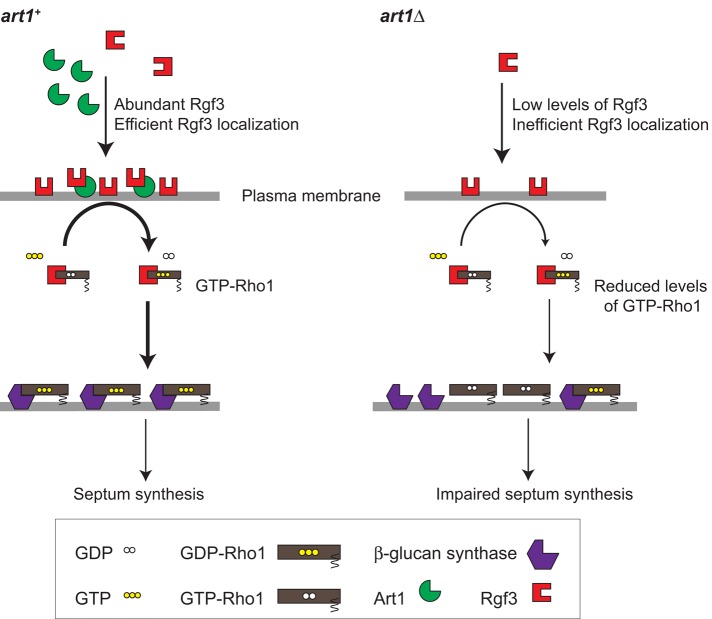
Our data suggest a model in which Art1 recruits Rgf3 to the division site, where it catalyzes the exchange of GDP for GTP on Rho1 GTPase (Figure 7). GEF activity of Rgf3 leads to increased levels of active Rho1, which in turn activates the β-glucan synthases Bgs1 and Bgs4 for septal and cell wall synthesis (Figure 7, left). In art1∆ cells, low levels of Rgf3 at the division site lead to inefficient Rho1 activation, which in turn leads to poor activation of β-glucan synthases, resulting in an impaired septum and cell lysis (Figure 7, right). The fact that Art1 does not make a significant contribution toward Bgs1 localization suggests that Rho1 plays no prominent role in Bgs1 localization. Bgs1 localization may be dependent on the exocyst complex, Cdc42, and the F-BAR protein Cdc15 (Bendezu and Martin, 2011; Arasada and Pollard, 2014).
FIGURE 7:
A model for the role of Art1 in regulating Rgf3 and Rho1 functions. Rgf3 localization, Rho1 activation, and septal formation are compared in art1+ (left) versus art1∆ (right) cells. See the Discussion for details.
Art1 in cytokinesis: a novel role for arrestins
Our finding that the arrestin Art1 regulates the localization and protein levels of Rgf3 has highlighted a novel role for the arrestin family of proteins in cytokinesis. Arrestins have diverse functions in eukaryotes. They were originally identified as proteins that bind to GPCRs (Kendall and Luttrell, 2009). GPCRs are transmembrane receptors activated by ligand binding and can act like GEFs to activate heterotrimeric G proteins. Arrestins bind to active GPCRs and prevent their association with the G proteins, which disrupts G protein–mediated signaling from the GPCRs (Kendall and Luttrell, 2009; Kang et al., 2014). Arrestins then act as adaptors connecting the receptors to the endocytic machinery (Moore et al., 2007; Kendall and Luttrell, 2009; Kang et al., 2014). Besides GPCRs, arrestins also bind to endocytic receptors involved in other signaling pathways to regulate a diverse array of cellular functions. They regulate trafficking of receptors such as Frizzled, Notch, and IGF-1 in developmental pathways such as Hedgehog and Wnt (Kovacs et al., 2009). Arrestins have also been implicated in mitosis due to their constitutive association with centrosomes in human cells (Shankar et al., 2010). The budding yeast Saccharomyces cerevisiae has 10 arrestin-related proteins (ARTs) that have an arrestin fold and also contain the PPXY motif, which aids the endocytosis of associated membrane proteins (Lin et al., 2008). In S. pombe, the arrestin-like protein Any1/Arn1 regulates endocytosis of amino acid transporters, and its PPXY motif interacts with the ubiquitin ligase Pub1 (Nakase et al., 2013; Nakashima et al., 2014). However, S. pombe Art1 has no PPXY motif and does not localize to actin patches, the sites of clathrin‑mediated endocytosis.
Arrestins were previously implicated in the regulation of GEFs and GTPases. The presence of arrestins along with Gα proteins was found to be essential for the activation of Rho A GTPase, which is important for stress fiber formation and cell migration in response to activation of angiotensin II type 1A receptor (Barnes et al., 2005). In another example, the arrestin acts as a scaffold and binds the GEF ARNO, which then binds and activates the GTPase ARF6 upon receptor activation. The activated ARF6 is necessary for the internalization of the β(2)-adrenergic receptor via endocytosis (Claing et al., 2001). It will be interesting to explore whether Art1 acts as a scaffold for Rgf3–Rho1 interaction. In the third example, β-arrestin binds to the GEF RalGDS for the Ras family GTPase RalA that controls cytoskeletal reorganization (Bhattacharya et al., 2002). The arrestin forms an inactive complex with RalGDS in the cytosol and recruits it to the plasma membrane upon chemoattractant stimulation. After recruitment, RalGDS is released for interaction with RalA GTPase. In this case, β-arrestin–mediated recruitment of RalGDS to the plasma membrane is crucial for RalA activity (Bhattacharya et al., 2002). However, to our knowledge, a direct role for arrestins in cytokinesis has not been reported before.
Regulation of Rho-GEFs by adaptor proteins
Rho-GEF activities in cells, like those of Rho GTPases, are also regulated. Disruption of Rho-GEF activities is associated with developmental defects, cancer, and neurological disorders in mice and humans (Rossman et al., 2005; Fields and Justilien, 2010; Cook et al., 2011). Rho-GEFs are known to be regulated by intramolecular binding, which prevents their association with GTPases, because many of them become constitutively active GEFs when truncated N-terminally to the GEF domain (Fritz et al., 2002; Yohe et al., 2007). Deletion of the N-terminus of RhoA GEF Ect2 leads to increased GEF activity, but the mechanism of release from the autoinhibition is not clearly understood (Fields and Justilien, 2010). Rho-GEFs are also regulated by localized activation, sometimes by GTPases themselves, and by different adaptor proteins (Rossman et al., 2005; Buchsbaum, 2007).
The regulation mechanism of the Rho1 GEF Rgf3 in S. pombe was unknown. In this study, we found that Rgf3 protein levels and localization to the division site are regulated by Art1, which has only ∼38% of the mass of Rgf3. Thus Art1 may be an adaptor protein for Rgf3. Further studies are needed to clarify whether Art1 regulates Rgf3 protein levels by affecting Rgf3 stability. Previously we found that the putative Rho-GEF Gef2 interacts with the adaptor protein Nod1 to regulate division site selection and contractile-ring maintenance in fission yeast (Zhu et al., 2013). Gef2 and Nod1 are completely interdependent for localization to the cortical nodes but partially dependent on each other for the contractile-ring localization (Jourdain et al., 2013; Zhu et al., 2013). Similarly, the activity of Scd1, a GEF of S. pombe Cdc42, is enhanced by its adaptor, Scd2. Scd2 has been proposed to act as scaffold for the formation of the Scd1/Cdc42 complex (Endo et al., 2003).
In budding yeast, the Cdc42 GEF Cdc24 is regulated by the adaptor Bem1 for cell polarization. However, the molecular mechanism of intra- and intermolecular interactions among Cdc24, Bem1, and Cdc42 is not fully understood and remains controversial (Bose et al., 2001; Shimada et al., 2004; Wiget et al., 2004; Smith et al., 2013). The localizations of S. cerevisiae Rho-GEFs Tus1 and Rom2 to the bud neck/division site depend on adaptor proteins Rgl1 and Ack1, respectively (Krause et al., 2012). In the absence of Rgl1, the intensity of Tus1 at the division site is reduced fivefold. Similarly, in the absence of Ack1, Rom2 localization to the bud neck is abolished and that to the bud cortex is greatly reduced (Krause et al., 2012). Considering that the divergence between budding yeast and fission yeast is similar to their evolutionary distances to humans, the regulations of Rho-GEFs by small adaptor proteins may be widely conserved during evolution.
In conclusion, we found that the arrestin family protein Art1 binds to the Rho-GEF Rgf3 and recruits it to the division site for septal formation during cytokinesis. To our knowledge, no arrestin has been reported to play a direct role in cytokinesis. It will be interesting to learn whether arrestins also regulate Rho-GEFs during cytokinesis in other systems.
MATERIALS AND METHODS
Strains and molecular, cellular, and genetic methods
Table 1 lists the strains used in this study. We used PCR-based gene targeting and classical yeast genetics to construct strains (Moreno et al., 1991; Bähler et al., 1998). All tagged and truncation strains are regulated under native promoters and integrated into endogenous chromosomal loci. The exceptions include strains for overexpression that are under the control of the 3nmt1 promoter, which is repressed by thiamine (Maundrell, 1990). For rescue of the art1∆ phenotype by Rho1 overexpression, art1∆ cells were transformed with pUR19-Rho1 or the control pUR19 plasmid. Cells with the plasmids were grown exponentially in EMM5S – uracil medium for 36 h and subsequently transferred to YE5S medium for 3 h to observe and quantify the phenotype.
TABLE 1:
S. pombe strains used in this study. Continued
| Strain | Genotype | Source/reference |
|---|---|---|
| JW81 | h− ade6-M210 leu1-32 ura4-D18 | Wu et al., 2003 |
| JW404 | h+ art1-s34-F3 leu1-32 ura4-D18 | Wu et al., 2010 |
| JW739 | h− ade6-M216 leu1-32 ura4-D18 | Lab stock |
| JW797 | h− sad1-CFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | Wu et al., 2003 |
| JW1105 | h+ rgf3-mEGFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | Wu et al., 2010 |
| JW2245 | h+ rgf3-mECitrine-kanMX6 ade6-M210 leu1-32 ura4-D18 | Lab stock |
| JW2421 | h+ rgf3-13Myc-hphMX6 ade6-M210 leu1-32 ura4-D18 | Lab stock |
| JW2543 | h− art1Δ::ura4+ ade6-M216 leu1-32 ura4-D18 | This study |
| JW2674 | h− art1-mECitrine-kanMX6 ade6-210 ura4-D18 leu1-32 | This study |
| JW2694 | art1-mECitrine-kanMX6 sad1-mCFP-kanMX6 ade6-M210 ura4-D18 leu1-32 | This study |
| JW2696 | h− art1-2YFP-kanMX6 rgf3-13Myc-hphMX6 ade6 leu1-32 ura4-D18 | This study |
| JW2748 | h+ rgf3-mECitrine-kanMX6 sad1-CFP-kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW3055 | h− bgs1Δ::ura4+ Pbgs1-GFP-bgs1-leu1+ leu1-32 ura4-D18 | Cortes et al., 2012 |
| JW3313 | h− leu1-32-kanMX6-3nmt1-mEGFP rlc1-tdTomato-natMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW3344 | h− art1(Δ287-483)-mECitrine-kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| JW3345 | h− art1(Δ429-483)-mECitrine-kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| JW3351 | art1-mECitrine-kanMX6 kanMX6-Prgf3-N-degron-HA-rgf3 sad1-mCFP-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW3381 | h− art1-Stag-kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| JW3395 | art1Δ::ura4+ leu1-32-kanMX6-3nmt1-mEGFP rlc1-tdTomato-natMX6 ade6 leu1-32 ura4-D18 | This study |
| JW3563 | h− art1Δ::kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| JW3632 | h− art1Δ::kanMX6 ade6-210 leu1-32 ura4-D18 + pUR19 | This study |
| JW3633 | h− art1Δ::kanMX6 ade6-210 leu1-32 ura4-D18 + pUR19-Rho1 | This study |
| JW3675 | art1(Δ287-428)-mECitrine-kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| JW3711 | art1-2YFP-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW3795 | art1(Δ1-286)-mECitrine-kanMX6 ade6-210 leu1-32 ura4-D18 | This study |
| JW3868 | art1Δ::ura4+ kanMX6-3nmt1-rgf3 ade6 leu1-32 ura4-D18 | This study |
| JW4427 | art1-mECitrine-kanMX6 kanMX6-Pmyo2-mCFP-myo2 ade6-M210 leu1-32 ura4-D18 | This study |
| JW4816 | art1Δ::ura4+ rgf3-13Myc-hphMX6 ade6 leu1-32 ura4-D18 | This study |
| JW4889 | rgf3-mECitrine-kanMX6 kanMX6-P3nmt1-art1 ade6 leu1-32 ura4-D18 | This study |
| JW5129 | rgf3-13Myc-hphMX6 kanMX6-P3nmt1-mECitrine-art1 ade6 leu1-32 ura4-D18 | This study |
| JW5232 | rgf3-mECitrine-kanMX6 art1Δ::ura4+ sad1-mCFP-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5593 | h− leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine ade6-M210 leu1-32 ura4-D18 | This study |
| JW5712 | art1-mECitrine-kanMX6 mid1-GBP-RFP-hphMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5796 | cdc7-GBP-hphMX6 art1-mECitrine-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5798 | art1-mECitrine-kanMX6 rgf3-mCFP-hphMX6 mid1-GBP-hphMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5799 | cdc7-GBP-hphMX6 art1-mECitrine-kanMX6 rgf3-mCFP-hphMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5857 | leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine ade6 leu1-32 | This study |
| JW5858 | cdc7-GBP-hphMX6 art1-mECitrine-kanMX6 rlc1-mCFP-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5859 | art1-mECitrine-kanMX6 rlc1-mCFP-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW5877 | art1Δ::kanMX6 leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine ade6-M210 leu1-32 ura4-D18 | This study |
| JW6165 | rgf3-s44-F3 leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine ade6-M210 leu1-32 | This study |
| JW6664 | leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine gef2Δ::kanMX4 ade6 leu1-32 ura4-D18 | This study |
| JW6685 | kanMX6-Pmyo2-mEGFP-myo2 art1Δ::kanMX6 sad1-mCherry-natMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW6686 | bgs1Δ::ura4+ Pbgs1-GFP-bgs1-leu1+ art1∆::kanMX6 leu1-32 ura4-D18 | This study |
| JW6696 | leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine gef3Δ::kanMX6 ade6-M210 leu1-32 ura4-D18 | This study |
| JW6697 | rgf1-∆1::kanMX6 leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine ade6 leu1-32 ura4-D18 | This study |
| JW6698 | leu1::kanMX6-P3nmt1-pkc1(HR1-C2)-mECitrine rgf2-Δ1::kanMX6 ade6 leu1-32 ura4-D18 | This study |
| JW6716 | kanMX6-Pmyo2-mEGFP-myo2 sad1-mCherry-natMX6 ade6-M210 leu1-32 ura4-D18 | This study |
For creation of the temperature-sensitive mutant of Rgf3, the N-terminus of Rgf3 was fused with N-degron-HA, an engineered temperature-sensitive variant of mouse dihydrofolate reductase (DHFRts; Rajagopalan et al., 2004). The N-degron-HA fragment was amplified using PCR from plasmid pPW58 (a gift from Jurgen Dohmen, University of Cologne, Cologne, Germany). The PCR fragment was then cloned downstream of the 1-kb promoter region of rgf3. The resulting plasmid pFA6a-Prgf3-N-degron-HA (JQW248) was used as a template to tag native Rgf3 at its N-terminus. For construction of the diffusible monomeric enhanced green fluorescent protein (mEGFP) strain (JW3313), the 3nmt1-mEGFP fragment under the 3nmt1 promoter was amplified from JQW72 and integrated at the C-terminus of the leu1-32 locus.
Art1 N-terminal truncations and N-terminal–tagged strains were under the control of the art1 promoter that was constructed by replacing the 3nmt1 promoter in plasmid pFA6a-P3nmt1-mECitrine with art1 5′UTR + 200 base pairs (−338 to +6) at BglII and PacI sites. The resulting plasmid pFA6a-kanMX6-Part1-mECitrine (JQW728) was then used as the template for PCR amplification and gene targeting. Primers were designed according to desired truncation sites, and the PCR products were transformed into wt cells. The resulting truncations were sequenced.
For mislocalization of Art1, the art1-mECitrine strain was crossed to strains expressing SIN kinase Cdc7 and anillin Mid1 tagged at their C-termini with GBP (Rothbauer et al., 2008). A Rho1 biosensor Pkc1(HR-C2) was constructed using the plasmid PB2729 (PKC1 [1–1173 base pairs]-GFP [ Denis and Cyert, 2005; Kono et al., 2012], a gift from the David Pellman lab [Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA]). The Pkc1(HR-C2) fragment was amplified from PB2729 and fused with mECitrine that was amplified separately from pFA6a-mECitrine-kanMX6 (JQW228). Both fragments were amplified with overlapping overhangs that allowed them to fuse with a BamHI restriction site in between in a fusion PCR. The fused product was then cloned downstream of the 3nmt1 promoter and resulted in plasmid JQW740 (pFA6a-kanMX6-P3nmt1-pkc1-mECitrine). The entire fragment was then amplified and used to transform wt S. pombe cells at the leu1 locus.
To depolymerize actin filaments, we used 100 μM Lat-A as described previously (Wu et al., 2001). We treated cells with dimethyl sulfoxide (DMSO) or Lat-A for 15 min. Cells were then imaged on bare slides.
Microscopy and data analysis
Cells were restreaked from −80°C stocks and grown for 1–2 d on YE5S plates at 25°C and then inoculated into YE5S liquid media except cells with plasmids. Cells were grown at exponential phase for ∼48 h at 25°C before microscopy, except where noted. For microscopy, cells were collected by centrifugation, washed twice with EMM5S medium to reduce autofluorescence, and resuspended in EMM5S plus 5 μM n-propyl-gallate. The cells were imaged on EMM5S + 20% gelatin pads with 5 μM n-propyl-gallate as previously described (Laporte et al., 2012; Wang et al., 2014). For imaging of Rho1 biosensor strains, cells were washed with YE5S with n-propyl-gallate, then spotted on YE5S agar pads with 5 μM n-propyl-gallate. For experiments in Figures 1C, 2D, and 3G and Supplemental Figures S2 and S7, A–C, cells were washed with fresh YE5S medium once, then with YE5S + 5 μM n-propyl-gallate once, and ∼5 μl of concentrated cells were spotted on a 35-mm dish with a glass coverslip bottom (0420041500C; Bioptechs, Butler, PA). The cells were then covered with a piece of YE5S agar before imaging. Microscopy was carried out at 23.5–25°C, except where noted. For imaging at restrictive temperatures, a preheated climate chamber (stage-top incubator INUB-PPZI2-F1 equipped with a UNIV2-D35 dish holder; Tokai Hit, Shizuoka-ken, Japan) was used.
For visualizing cell morphology and septum only, cells were imaged with a 100 ×/1.4 numerical aperture (NA) Plan-Apo objective lens on a Nikon Eclipse Ti inverted microscope (Nikon, Melville, NY) equipped with a Nikon cooled digital camera DS-Ql1. Other experiments were performed using 100 ×/1.4 NA Plan-Apo objective lens (Nikon) on a spinning-disk confocal system (UltraVIEW ERS; Perkin Elmer-Cetus Life and Analytical Sciences Waltham, MA) with 440- and 514-nm lasers and an ORCA-AG camera (Hamamatsu, Bridgewater, NJ) with 2 × 2 binning, or on a spinning-disk confocal system (UltraVIEW Vox CSUX1 system; Perkin Elmer-Cetus Life and Analytical Sciences) with 440-, 488-, 515-, and 561-nm solid-state lasers and a back-thinned, electron-multiplying charge-coupled device camera (C9100-13; Hamamatsu) without binning.
Microscopy data were analyzed using ImageJ (National Institutes of Health [NIH], Bethesda, MD), UltraVIEW, or Volocity (Perkin Elmer-Cetus) software. Fluorescence images shown in the figures are maximum intensity projections of image stacks at 0.4- to 0.6-μm spacing, except where noted. Art1 and Rgf3 molecules in cells and in the ring were counted globally or locally by measuring fluorescence intensity as previously described (Coffman et al., 2011; Laporte et al., 2011). Briefly, 14 z-sections with 0.45 spacing were taken using the exact same imaging conditions for all strains. The images were subtracted by the offset. Mean intensities in whole cells were measured using sum-intensity projections. The wt strain (JW739) was used for background subtraction. Mean intensity at the division site was measured using the polygon region of interest (ROI) tool in ImageJ on the sum-intensity projection of the 14-slice stack. A region ≥3× the ROI, including the contractile ring, was used to calculate the background intensity. For estimating numbers of Art1 and Rgf3 molecules, the measured global and local intensities of Art1 and Rgf3 were normalized to Mid1 intensities measured in the same way and then converted to known Mid1 molecule numbers (Wu and Pollard, 2005). p Values in statistical analyses are from comparison with wt in a two-tailed t test.
FLIP analysis
FLIP analysis was performed using the photokinesis unit on the UltraVIEW ERS confocal system. The best focal plane for bleaching was chosen from a z-stack. Cells with constricting rings (marked with Rlc1–tandem dimer Tomato [Rlc1-tdTomato]) at their final stage of constriction were chosen from cells with an almost complete septum. ROIs (∼1 μm2) were drawn in half of the cells and were bleached to <50% of the original intensity after two prebleach images were collected. Then two postbleach images were collected, and the ROI was bleached again after a 30-s delay. The bleach-imaging cycle was repeated 40 times. For data analysis, the images were corrected for uneven illumination, background, and photobleaching during image acquisition using nonbleached neighboring cells as previously described (Coffman et al., 2013). The mean intensities of the bleached and unbleached half of the cell were manually collected using the measuring tool in ImageJ. We set the intensity before the first bleach to 100%. The intensity values from the bleached and unbleached halves of cells were compared to detect the diffusion of mEGFP molecules between two daughter cells connected by the septum. Time zero was defined as being when the cell had a fully constricted ring, at which point the length of the ring equals its height in fluorescence images. The time that fluorescence exchange stopped between the two daughter cells after time zero was compared between wt and art1∆ cells.
EM
WT and art1∆ cells were grown in YE5S medium at exponential phase at 25°C for ∼48 h and fixed using phosphate buffer (2.5% glutaraldehyde and 0.1 M sucrose in a 0.1 M sodium phosphate buffer, pH 7.4) for 1 h. The samples were then rinsed 3× (with 0.1 M sucrose in the 0.1 M sodium phosphate buffer, pH 7.4), and the pellet was submitted to the Campus Microscopy and Imaging facility at the Ohio State University for further preparations. The samples were further fixed with 1% osmium tetroxide and embedded in agarose. The sample, in the agarose block, was dehydrated in a graded series of alcohol and embedded in Epon8 epoxy resin. Thin sections (70–90 nm) were cut using a Leica EM UC6 ultramicrotome. Sections were stained with uranyl acetate and lead citrate and imaged on a FEI Tecnai G2 Spirit transmission electron microscope at 80 kV (Hayat, 1986, 2000; Watanabe et al., 1988).
IP and Western blotting
IP assays and Western blotting were carried out as previously described (Lee and Wu, 2012; Ye et al., 2012). For the phosphatase treatment and IPs, proteins were pulled down with beads. Briefly, Myc- (Figure 3D) or YFP-tagged (Figure 3E) protein expressed at its native level was pulled down from fission yeast cell extracts by protein G covalently coupled Dynabeads (100.04D; Invitrogen, Carlsbad, CA) with anti-Myc antibody (sc-40; Santa Cruz Biotechnology, Dallas, Texas) or polyclonal anti-GFP antibodies (NB600-308; Novus Biologicals, Littleton, CO), respectively. S-tagged protein (Figure 3D) was pulled down from fission yeast cell extract using S-protein agarose slurry (69704; Novagen, San Diego, CA). To test whether Art1 and Rgf3 are phosphoproteins, we incubated the beads with 400 U of lambda protein phosphatase (P0753; 400,000 U/ml; New England Biolabs, Ipswich, MA) for 40 min at 30°C. Sample buffer was added to the beads, and the mobility shift in the treated samples was examined using an SDS–PAGE gel. For testing whether Art1–Rgf3 interaction depends on the phosphorylation status of the two proteins, the beads coupled to polyclonal anti-GFP antibodies were used to pull down Art1 from a cell extract of strain art1-2YFP rgf3-13Myc. After pull down, the beads were washed 3× with 1 ml 1% NP-40 buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% NP-40, 50 mM NaF, 20 mM glycerophosphate, and 0.1 mM Na3VO4) and 2× with 1 ml 1% NP-40 buffer without phosphatase inhibitors (50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1% NP-40). Then the beads were treated with 400 U of lambda protein phosphatase for 40 min at 30°C and then washed 3× with 1 ml 1% NP-40 buffer without phosphatase inhibitors before being boiled in sample buffer.
The protein samples were separated on SDS–PAGE, and Western blotting was performed using monoclonal antibodies: anti-GFP antibody (11814460001, 1:5000 dilution; Roche, Mannheim, Germany), anti-Myc antibody (9E10, 1:1000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA), anti–S-tag antibody (1:5000 dilution; ICL Lab, Portland, OR). The anti-tubulin monoclonal TAT1 antibody was used at 1:20,000 dilution as the loading control (Woods et al., 1989). Anti-mouse secondary antibody was used at 1:5000 dilution.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed as described previously using the X-gal overlay and β-galactosidase activity assays (Laporte et al., 2011; Zhu et al., 2013). Art1 and Rgf3 DNAs were cloned into vectors with either viral VP16 transcription activation domain (AD) or GAL4 transcription factor DNA-binding domain (BD). The pairs of plasmids were then cotransformed into S. cerevisiae strain MAV203 (11281-011; Invitrogen), and cells were plated on solid medium lacking leucine and tryptophan (SD-L-W). Yeast strains to be tested were streaked onto fresh yeast–peptone–dextrose plates and grown overnight. The cells on the plates were then permeabilized using 10-12 ml of chloroform per plate. After 10 min, the chloroform was poured off, and the colonies were dried. Then 1% agarose in 25 ml of 100 mM potassium phosphate buffer (pH 7.0) was prepared and melted. After cooling, 500 μl of 20 mg/ml X-gal in DMSO was added and mixed thoroughly. The X-gal–containing agarose was overlaid onto the colonies. The plates were incubated at 30°C, and the coloration was checked every 30 min. The positive interactions in the X-gal overlay assays were then quantified by measuring β-d-galactosidase activity in the o-nitrophenyl β-d-galactopyranoside assay (Sigma-Aldrich, St. Louis, MO) as previously described (Laporte et al., 2011; Zhu et al., 2013).
Supplementary Material
Acknowledgments
We thank John Pringle, Thomas Pollard, David Pellman, Dannel McCollum, Jurgen Dohmen, Sophie Martin, Valerie Coffman, I-Ju Lee, Ning Wang, Yanfang Ye, and Yihua Zhu for strains and plasmids; the James Hopper, Anita Hopper, and Steve Osmani laboratories for sharing their equipment; Richard Montione and the campus microscopy and imaging facility at the Ohio State University for technical support with EM; and current and former members of the Wu lab for helpful discussions. This work was supported by the National Institute of General Medical Sciences/NIH grant GM086546 and American Cancer Society grant RSG-13-005-01-CCG to J.‑Q.W.
Abbreviations used:
- AD
activation domain
- BD
DNA-binding domain
- CFP
cyan fluorescent protein
- DH
DBL homology
- EM
electron microscopy
- FL
full length
- FLIP
fluorescence loss in photobleaching
- GBP
GFP-binding protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GPCR
G protein–coupled receptor
- IP
immunoprecipitation
- Lat-A
latrunculin A
- mCFP
monomeric cyan fluorescent protein
- mECitrine
monomeric enhanced Citrine
- PH
pleckstrin homology
- ROI
region of interest
- SIN
septation initiation network
- SPB
spindle pole body
- tdTomato
tandem dimer Tomato
- wt
wild type
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1252) on December 3, 2014.
*Present address: Institut de Biochimie et Génétique Cellulaires, Université de Bordeaux, F‑33077 Bordeaux Cedex, France.
REFERENCES
- An H, Morrell JL, Jennings JL, Link AJ, Gould KL. Requirements of fission yeast septins for complex formation, localization, and function. Mol Biol Cell. 2004;15:5551–5564. doi: 10.1091/mbc.E04-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep. 2014;8:1533–1544. doi: 10.1016/j.celrep.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Duran A, Perez P. Rho 1 GTPase activates the (1–3)β-d-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15:4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Duran A, Perez P. Localisation of the Schizosaccharomyces pombe rho1p GTPase and its involvement in the organisation of the actin cytoskeleton. J Cell Sci. 1997;110:2547–2555. doi: 10.1242/jcs.110.20.2547. [DOI] [PubMed] [Google Scholar]
- Arellano M, Valdivieso MH, Calonge TM, Coll PM, Duran A, Perez P. Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J Cell Sci. 1999;112:3569–3578. doi: 10.1242/jcs.112.20.3569. [DOI] [PubMed] [Google Scholar]
- Aubry L, Guetta D, Klein G. The arrestin fold: variations on a theme. Curr Genomics. 2009;10:133–142. doi: 10.2174/138920209787847014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Barnes WG, Reiter E, Violin JD, Ren XR, Milligan G, Lefkowitz RJ. β-Arrestin 1 and Gαq/11 coordinately activate RhoA and stress fiber formation following receptor stimulation. J Biol Chem. 2005;280:8041–8050. doi: 10.1074/jbc.M412924200. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bendezu FO, Martin SG. Actin cables and the exocyst form two independent morphogenesis pathways in the fission yeast. Mol Biol Cell. 2011;22:44–53. doi: 10.1091/mbc.E10-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya M, Anborgh PH, Babwah AV, Dale LB, Dobransky T, Benovic JL, Feldman RD, Verdi JM, Rylett RJ, Ferguson SS. β-Arrestins regulate a Ral-GDS Ral effector pathway that mediates cytoskeletal reorganization. Nat Cell Biol. 2002;4:547–555. doi: 10.1038/ncb821. [DOI] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Buchsbaum RJ. Rho activation at a glance. J Cell Sci. 2007;120:1149–1152. doi: 10.1242/jcs.03428. [DOI] [PubMed] [Google Scholar]
- Cabib E, Drgonova J, Drgon T. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu Rev Biochem. 1998;67:307–333. doi: 10.1146/annurev.biochem.67.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Roh DH, Schmidt M, Crotti LB, Varma A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J Biol Chem. 2001;276:19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Claing A, Chen W, Miller WE, Vitale N, Moss J, Premont RT, Lefkowitz RJ. β-Arrestin-mediated ADP-ribosylation factor 6 activation and β2-adrenergic receptor endocytosis. J Biol Chem. 2001;276:42509–42513. doi: 10.1074/jbc.M108399200. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Sees JA, Kovar DR, Wu J-Q. The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J Cell Biol. 2013;203:101–114. doi: 10.1083/jcb.201305022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu J-Q. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, Solski PA, Bultman SJ, Kauselmann G, Schoor M, Kuehn R, Friedman LS, Cowley DO, Van Dyke T, Yeh JJ, et al. The Ect2 Rho guanine nucleotide exchange factor is essential for early mouse development and normal cell cytokinesis and migration. Genes Cancer. 2011;2:932–942. doi: 10.1177/1947601912437035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes JC, Sato M, Munoz J, Moreno MB, Clemente-Ramos JA, Ramos M, Okada H, Osumi M, Duran A, Ribas JC. Fission yeast Ags1 confers the essential septum strength needed for safe gradual cell abscission. J Cell Biol. 2012;198:637–656. doi: 10.1083/jcb.201202015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis V, Cyert MS. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot Cell. 2005;4:36–45. doi: 10.1128/EC.4.1.36-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonova J, Drgon T, Tanaka K, Kollar R, Chen GC, Ford RA, Chan CS, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- Elia N, Ott C, Lippincott-Schwartz J. Incisive imaging and computation for cellular mysteries: lessons from abscission. Cell. 2013;155:1220–1231. doi: 10.1016/j.cell.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shirouzu M, Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J Biol Chem. 2003;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- Fields AP, Justilien V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 2010;50:190–200. doi: 10.1016/j.advenzreg.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Tajadura V, Garcia I, Sanchez Y. Rgf1p is a specific Rho1-GEF that coordinates cell polarization with cell wall biogenesis in fission yeast. Mol Biol Cell. 2006;17:1620–1631. doi: 10.1091/mbc.E05-10-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Grallert A, Patel A, Tallada VA, Chan KY, Bagley S, Krapp A, Simanis V, Hagan IM. Centrosomal MPF triggers the mitotic and morphogenetic switches of fission yeast. Nat Cell Biol. 2013;15:88–95. doi: 10.1038/ncb2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Hayat MA. Basic Techniques for Transmission Electron Microscopy. San Diego, CA: Academic; 1986. [Google Scholar]
- Hayat MA. Principles and Techniques of Electron Microscopy: Biological Applications. Cambridge, UK: Cambridge University Press; 2000. [Google Scholar]
- Herrador A, Herranz S, Lara D, Vincent O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Guertin DA, McCollum D. Initiation of cytokinesis is controlled through multiple modes of regulation of the Sid2p-Mob1p kinase complex. Mol Cell Biol. 2004;24:3262–3276. doi: 10.1128/MCB.24.8.3262-3276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbel BM, Konomi M, Takagi T, Kamasawa N, Ishijima SA, Osumi M. In situ localization of β-glucans in the cell wall of Schizosaccharomyces pombe. Yeast. 2001;18:433–444. doi: 10.1002/yea.694. [DOI] [PubMed] [Google Scholar]
- Jin Q-W, Zhou M, Bimbo A, Balasubramanian MK, McCollum D. A role for the septation initiation network in septum assembly revealed by genetic analysis of sid2–250 suppressors. Genetics. 2006;172:2101–2112. doi: 10.1534/genetics.105.050955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain I, Brzezinska EA, Toda T. Fission yeast Nod1 is a component of cortical nodes involved in cell size control and division site placement. PLoS One. 2013;8:e54142. doi: 10.1371/journal.pone.0054142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DS, Tian X, Benovic JL. Role of β-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr Opin Cell Biol. 2014;27:63–71. doi: 10.1016/j.ceb.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Cabib E. Regulation of fungal cell wall growth: a guanine nucleotide-binding, proteinaceous component required for activity of (1→3)-β-d-glucan synthase. Proc Natl Acad Sci USA. 1986;83:5808–5812. doi: 10.1073/pnas.83.16.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall RT, Luttrell LM. Diversity in arrestin function. Cell Mol Life Sci. 2009;66:2953–2973. doi: 10.1007/s00018-009-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 2012;150:151–164. doi: 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: emerging roles for β-arrestins in developmental signaling pathways. Dev Cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause SA, Cundell MJ, Poon PP, McGhie J, Johnston GC, Price C, Gray JV. Functional specialization of the yeast Rho1 GTP exchange factors. J Cell Sci. 2012;25:2721–2731. doi: 10.1242/jcs.100685. [DOI] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee I-J, Wu J-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Ojkic N, Vavylonis D, Wu J-Q. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol Biol Cell. 2012;23:3094–3110. doi: 10.1091/mbc.E12-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Zhao R, Wu J-Q. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol. 2010;21:892–898. doi: 10.1016/j.semcdb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Coffman VC, Wu J-Q. Contractile-ring assembly in fission yeast cytokinesis: recent advances and new perspectives. Cytoskeleton (Hoboken) 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wu J-Q. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J Cell Sci. 2012;125:2973–2985. doi: 10.1242/jcs.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- Liu J, Wang H, McCollum D, Balasubramanian MK. Drc1p/Cps1p, a 1,3-β-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 1999;153:1193–1203. doi: 10.1093/genetics/153.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Matsuyama A, Yabana N, Watanabe Y, Yamamoto M. Schizosaccharomyces pombe Ste7p is required for both promotion and withholding of the entry to meiosis. Genetics. 2000;155:539–549. doi: 10.1093/genetics/155.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- Mehta S, Gould KL. Identification of functional domains within the septation initiation network kinase, Cdc7. J Biol Chem. 2006;281:9935–9941. doi: 10.1074/jbc.M600160200. [DOI] [PubMed] [Google Scholar]
- Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu Rev Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, Haese GD, Gould KL. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J Cell Sci. 2005;118:5563–5573. doi: 10.1242/jcs.02664. [DOI] [PubMed] [Google Scholar]
- Mutoh T, Nakano K, Mabuchi I. Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells. 2005;10:1189–1202. doi: 10.1111/j.1365-2443.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Arai R, Mabuchi I. The small GTP-binding protein Rho1 is a multifunctional protein that regulates actin localization, cell polarity, and septum formation in the fission yeast Schizosaccharomyces pombe. Genes Cells. 1997;2:679–694. doi: 10.1046/j.1365-2443.1997.1540352.x. [DOI] [PubMed] [Google Scholar]
- Nakase Y, Nakase M, Kashiwazaki J, Murai T, Otsubo Y, Mabuchi I, Yamamoto M, Takegawa K, Matsumoto T. The fission yeast β-arrestin-like protein Any1 is involved in TSC-Rheb signaling and the regulation of amino acid transporters. J Cell Sci. 2013;126:3972–3981. doi: 10.1242/jcs.128355. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Kamada S, Tamanoi F, Kikkawa U. Fission yeast arrestin-related trafficking adaptor, Arn1/Any1, is ubiquitinated by Pub1 E3 ligase and regulates endocytosis of Cat1 amino acid transporter. Biol Open. 2014;3:542–552. doi: 10.1242/bio.20148367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont RT, Gainetdinov RR. Physiological roles of G protein-coupled receptor kinases and arrestins. Annu Rev Physiol. 2007;69:511–534. doi: 10.1146/annurev.physiol.69.022405.154731. [DOI] [PubMed] [Google Scholar]
- Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin DE, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S, Liling Z, Liu J, Balasubramanian M. The N-degron approach to create temperature-sensitive mutants in Schizosaccharomyces pombe. Methods. 2004;33:206–212. doi: 10.1016/j.ymeth.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Ribas JC, Diaz M, Duran A, Perez P. Isolation and characterization of Schizosaccharomyces pombe mutants defective in cell wall (1–3)β-d-glucan. J Bacteriol. 1991;173:3456–3462. doi: 10.1128/jb.173.11.3456-3462.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Shankar H, Michal A, Kern RC, Kang DS, Gurevich VV, Benovic JL. Non-visual arrestins are constitutively associated with the centrosome and regulate centrosome function. J Biol Chem. 2010;285:8316–8329. doi: 10.1074/jbc.M109.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Wiget P, Gulli MP, Bi E, Peter M. The nucleotide exchange factor Cdc24p may be regulated by auto-inhibition. EMBO J. 2004;23:1051–1062. doi: 10.1038/sj.emboj.7600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Splitting of the fission yeast septum. FEMS Yeast Res. 2007;7:761–770. doi: 10.1111/j.1567-1364.2007.00266.x. [DOI] [PubMed] [Google Scholar]
- Smith SE, Rubinstein B, Mendes Pinto I, Slaughter BD, Unruh JR, Li R. Independence of symmetry breaking on Bem1-mediated autocatalytic activation of Cdc42. J Cell Biol. 2013;202:1091–1106. doi: 10.1083/jcb.201304180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura V, Garcia B, Garcia I, Garcia P, Sanchez Y. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall β-glucan biosynthesis through the GTPase Rho1p. J Cell Sci. 2004;117:6163–6174. doi: 10.1242/jcs.01530. [DOI] [PubMed] [Google Scholar]
- Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lo Presti L, Zhu Y-H, Kang M, Wu Z, Martin SG, Wu J-Q. The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J Cell Biol. 2014;205:357–375. doi: 10.1083/jcb.201308146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Sasaki J, Wada T, Tanaka Y, Otsuka N. Low gel temperature agarose encapsulation of small specimens for electron microscopy. J Electron Microsc (Tokyo) 1988;37:89–91. [PubMed] [Google Scholar]
- Wiget P, Shimada Y, Butty AC, Bi E, Peter M. Site-specific regulation of the GEF Cdc24p by the scaffold protein Far1p during yeast mating. EMBO J. 2004;23:1063–1074. doi: 10.1038/sj.emboj.7600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Bähler J, Pringle JR. Roles of a fimbrin and an α-actinin-like protein in fission yeast cell polarization and cytokinesis. Mol Biol Cell. 2001;12:1061–1077. doi: 10.1091/mbc.12.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Ye Y, Wang N, Pollard TD, Pringle JR. Cooperation between the septins and the actomyosin ring and role of a cell-integrity pathway during cell division in fission yeast. Genetics. 2010;186:897–915. doi: 10.1534/genetics.110.119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Vogel BE. A new job for ancient extracellular matrix proteins: hemicentins stabilize cleavage furrows. Commun Integr Biol. 2011;4:433–435. doi: 10.4161/cib.4.4.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Lee I-J, Runge KW, Wu J-Q. Roles of putative Rho-GEF Gef2 in division-site positioning and contractile-ring function in fission yeast cytokinesis. Mol Biol Cell. 2012;23:1181–1195. doi: 10.1091/mbc.E11-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe ME, Rossman KL, Gardner OS, Karnoub AE, Snyder JT, Gershburg S, Graves LM, Der CJ, Sondek J. Auto-inhibition of the Dbl family protein Tim by an N-terminal helical motif. J Biol Chem. 2007;282:13813–13823. doi: 10.1074/jbc.M700185200. [DOI] [PubMed] [Google Scholar]
- Zhu Y-H, Ye Y, Wu Z, Wu J-Q. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol Biol Cell. 2013;24:3187–3204. doi: 10.1091/mbc.E13-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.