Kinesin-13, a microtubule-end depolymerase, has been shown to affect the length of cilia, but its ciliary function is unclear. In Tetrahymena thermophila, kinesin-13 positively regulates the axoneme length, influences the properties of ciliary tubulin, and affects the ciliary dynein-dependent motility.
Abstract
Kinesin-13, an end depolymerizer of cytoplasmic and spindle microtubules, also affects the length of cilia. However, in different models, depletion of kinesin-13 either lengthens or shortens cilia, and therefore the exact function of kinesin-13 in cilia remains unclear. We generated null mutations of all kinesin-13 paralogues in the ciliate Tetrahymena. One of the paralogues, Kin13Ap, localizes to the nuclei and is essential for nuclear divisions. The remaining two paralogues, Kin13Bp and Kin13Cp, localize to the cell body and inside assembling cilia. Loss of both Kin13Bp and Kin13Cp resulted in slow cell multiplication and motility, overgrowth of cell body microtubules, shortening of cilia, and synthetic lethality with either paclitaxel or a deletion of MEC-17/ATAT1, the α-tubulin acetyltransferase. The mutant cilia assembled slowly and contained abnormal tubulin, characterized by altered posttranslational modifications and hypersensitivity to paclitaxel. The mutant cilia beat slowly and axonemes showed reduced velocity of microtubule sliding. Thus kinesin-13 positively regulates the axoneme length, influences the properties of ciliary tubulin, and likely indirectly, through its effects on the axonemal microtubules, affects the ciliary dynein-dependent motility.
INTRODUCTION
Members of the kinesin motor superfamily form 14 conserved subfamilies (Lawrence et al., 2004). Kinesin-8 and kinesin-13 are atypical members that act as microtubule end depolymerizers (reviewed in Su et al., 2012). Kinesin-13 has a centrally located catalytic domain (Aizawa et al., 1992) and depolymerizes the ends of microtubules in the presence of ATP in vitro (Desai et al., 1999). Kinesin-13 moves to the ends of microtubules either by diffusion (Cooper et al., 2010) or by riding on microtubule plus end–tracking proteins (Honnappa et al., 2009) or on a motor (Piao et al., 2009). At the microtubule end, kinesin-13 imposes shear between the tubulin subunits, which leads to lattice depolymerization (Asenjo et al., 2013). Kinesin-13 promotes depolymerization of interphase microtubules before mitosis (Kline-Smith and Walczak, 2002; Mennella et al., 2005), shortens the kinetochore microtubules during mitosis (Walczak et al., 1996; Maney et al., 1998; Kline-Smith et al., 2004; Rogers et al., 2004; Wickstead et al., 2010), and promotes reorganization of microtubules during neuronal differentiation (Ghosh-Roy et al., 2012).
Kinesin-13 homologues are uniformly present in the genomes of ciliated eukaryotes and are absent in some nonciliated lineages, such as the nonciliated species of fungi (Wickstead and Gull, 2006), suggesting that kinesin-13 coevolved with cilia. A dominant-negative mutation of kinesin-13 in Giardia intestinalis results in longer cilia, suggesting that in cilia, kinesin-13 acts in a canonical manner by depolymerizing the ends of axonemal microtubules (Dawson et al., 2007). Surprisingly, in Chlamydomonas reinhardtii, an RNA interference (RNAi) knockdown of kinesin-13 leads to short cilia that assemble slowly (Piao et al., 2009; Wang et al., 2013). The Chlamydomonas observations opened a possibility that kinesin-13 contributes to cilia indirectly, by depolymerizing the cell body microtubules to produce soluble tubulin for transport into cilia (Piao et al., 2009; Wang et al., 2013). On the other hand, in Chlamydomonas, kinesin-13 moves into cilia both during assembly and disassembly (Piao et al., 2009). Thus, whereas kinesin-13 appears important for cilia length, its sites of activity in the cell and its function in the context of cilia remain unclear.
Here we used the efficient homologous DNA recombination activity in the ciliate Tetrahymena thermophila to create knockouts for all three kinesin-13 homologues. We find that one of the three paralogues is required for nuclear divisions, whereas the remaining two act in the cell body and cilia. In the cell body, kinesin-13 activity shortens the cortical microtubules. In addition, in the absence of the nonnuclear kinesin-13, cilia become shorter and beat more slowly. A pharmacological approach suggests that the soluble ciliary tubulin is more concentrated at the tips of assembling mutant cilia, likely as a result of slow addition of the incoming tubulin dimers to the ends of growing axonemal microtubules. We suggest that the ciliary function of kinesin-13 extends beyond what the earlier studies suggested, namely, the canonical activity of a microtubule-end depolymerizer. Our observations can be reconciled by proposing that inside cilia, kinesin-13 functions as an axoneme assembly–promoting factor.
RESULTS
T. thermophila has three kinesin-13 homologues that differ in subcellular localization
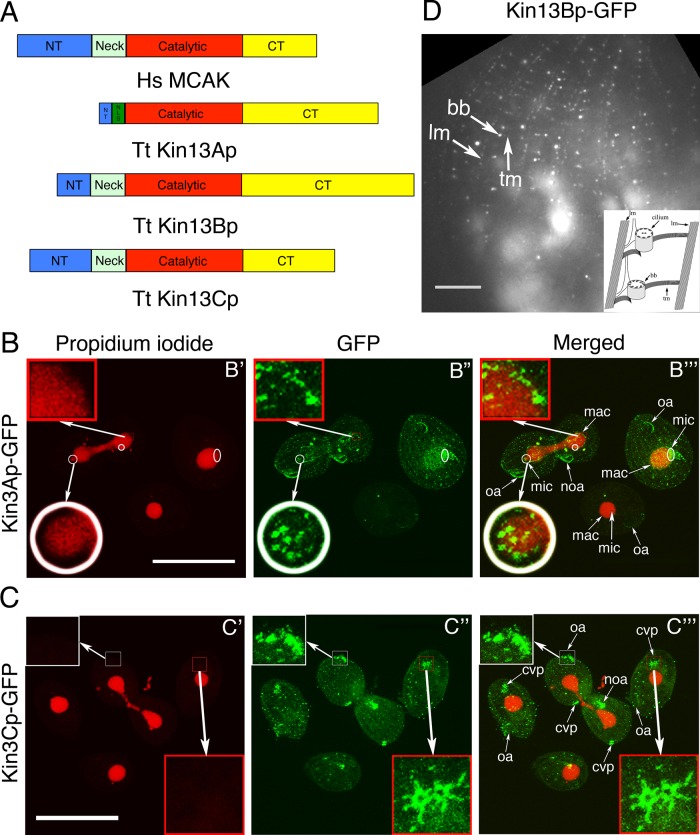
The genome of T. thermophila contains three genes encoding kinesin-13 homologues, KIN13A (TTHERM_00790940), KIN13B (TTHERM_00429870), and KIN13C (THERM_00648540) (Wickstead et al., 2010). Each of the predicted gene products encodes a protein with an organization typical of kinesin-13 (Figure 1A and Supplemental Figure S1), including a catalytic domain with KEC/KVD motifs in the loop 2 that are important for the microtubule- depolymerization activity (Ogawa et al., 2004; Shipley et al., 2004). Kin13Bp and Kin13Cp (but not Kin13Ap) have a positively charged neck, an ∼70-residue extension N-terminal to the catalytic domain, that contributes to the recruitment of kinesin-13 to microtubules (Moores et al., 2006; Cooper et al., 2010; Wang et al., 2012).
FIGURE 1:
Tetrahymena expresses three homologues of kinesin-13, each with a distinct pattern of localization. (A) A comparison of predicted domain organizations of the well-studied human kinesin-13 (MCAK) and homologues of T. thermophila. CT, C-terminal domain; NT, N-terminal domain; NLS, nuclear localization signal (predicted using cNLS mapper). (B, C) Confocal immunofluorescence images of cells in which either Kin13Ap or Kin13Cp is tagged with a C-terminal GFP expressed in the native locus. The cells show a direct kinesin-13–GFP signal (green) and nuclear DNA stained with propidium iodide (red). (B) Kin13Ap localizes to the nuclei when they divide. The cells on the left and right are in an advanced (left) or early (right) stage of cell division, respectively, whereas the middle bottom cell is in interphase. In the cell on the left, the macronucleus undergoes amitosis, whereas the micronucleus is in the telophase of mitosis. The insets show a higher magnification of the micronucleus (white circle) and the macronucleus (red box) in the boxed area. In the cell on the right, the micronucleus is in early anaphase. The white circles and oval in B′ mark the micronuclei in mitosis. The two dividing cells have weak green dots in the cell cortex, which are likely the somatic and oral basal bodies. Bar, 50 μm. (C) Kin13Cp associates with cortical microtubules and cilia. The images show a dividing cell that is surrounded by three interphase cells. All cells show weak dots of cortical labeling consistent with basal bodies. Both dividing and two of the three nondividing cells show a strong CVP signal (red box). The dividing cell shows a very strong signal in the growing cilia of oral apparatuses (the anterior one is magnified in the white box) in both the anterior and posterior daughter cells. Bar, 50 μm. (D) TIRF image of a cell with a natively tagged Kin13Bp-GFP that is detected near the basal bodies and cortical microtubules (transverse and longitudinal). The structures are identified based on their shape and relative locations. The schematic organization of the cell cortex microtubules viewed from the ventral side is shown in the right bottom corner (modified from Sharma et al., 2007). Bar, 20 μm. bb, basal body; cvp, contractile vacuole pore; lm, longitudinal microtubule; mac, macronucleus; mic, micronucleus; noa, new oral apparatus; oa, oral apparatus; tm, transverse microtubule.
We tagged each paralogue with green fluorescent protein (GFP) at the C-terminus by modifying its gene at the native locus. Tetrahymena has two functionally distinct nuclei in a single cytoplasm: the micronucleus (containing a transcriptionally silent, diploid, germline genome) and the macronucleus (containing a transcriptionally active, polyploid, somatic genome). Kin13Ap-GFP was detected inside the micronucleus at the time of mitosis and inside the dividing macronucleus during amitosis (a nuclear division that does not involve a bipolar spindle formation or chromosome condensation; Figure 1B). Kin13Cp-GFP was enriched at the microtubules of the contractile vacuole pore (CVP) and weakly present near the basal bodies. A strong signal of Kin13Cp-GFP was seen uniformly along the length of oral cilia of dividing cells (when these cilia assemble; Figure 1C). Although we could not detect Kin13Bp-GFP in fixed cells using confocal microscopy, total internal reflection fluorescence microscopy (TIRFM) of live cells detected dots arranged in a pattern consistent with the basal bodies and cortical microtubule bundles (transverse and longitudinal; Figure 1D). To conclude, one of the kinesin-13 paralogues (Kin13Ap) is mainly confined to the dividing nuclei, whereas the remaining two paralogues (Kin13Bp and Kin13Cp) are extranuclear and localize to the cortical microtubules and cilia. In agreement with these observations, a putative nuclear localization signal is present near the N-terminus of Kin13Ap but not in Kin13Bp and Kin13Cp (Figure 1A).
Kin13Ap is required for divisions of micronuclei and macronuclei
We used homologous DNA recombination to construct strains lacking one or more of the kinesin-13 genes. Homozygotes expressing a knockout phenotype were obtained by mating heterokaryons (Hai et al., 1999). The knockout cells lacking the nuclear paralogue, Kin13Ap (13A-KO cells), were not viable (Table 1) but managed to divide a few times. Most of the 13A-KO cells examined before their death lacked either a micronucleus or a macronucleus (Supplemental Figure S2). This phenotype is consistent with defects in the nuclear divisions, and fits with the localization of KIN13A-GFP to the dividing micronuclei and macronuclei. Thus Kin13Ap is required for both mitosis and amitosis, likely by regulating the spindle microtubules within the micronucleus (LaFountain and Davidson, 1979, 1980) and microtubules that form inside the macronucleus during amitosis (Kushida et al., 2011).
TABLE 1:
Viability of knockout progeny of mating heterokaryons.
| Strain name | Macronuclear genotype | Viable progeny of heterokaryons (%) | n |
|---|---|---|---|
| 13A-KO | kin13a::neo4 | 0 | 100 |
| 13B-KO | kin13b::neo4 | 79 | 48 |
| 13C-KO | kin13c::neo4 | 85 | 48 |
| 13BC-KO | kin13b::neo4; kin13C::neo4 | 47 | 101 |
| 13BC-MEC-KO | kin13b::neo4; kin13C::neo4; mec17:neo4 | 0 | 101 |
Two nonnuclear paralogues, Kin13Bp and Kin13Cp, act synergistically to support cell multiplication and ciliary functions
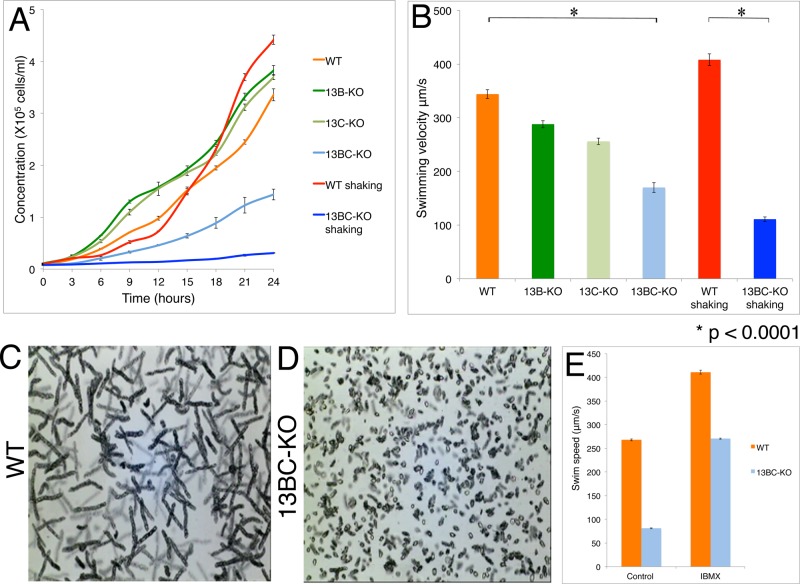
Deletions of either KIN13B or KIN13C did not affect the rate of cell multiplication (Figure 2A) or the gross phenotype, except for a mild decrease in the motility rate in the absence of KIN13C (Figure 2B). Kin13Bp and Kin13Cp have a similar domain organization (Figure 1A), and the sequences of their motor domains indicate that they originated from a recent gene duplication (Wickstead et al., 2010). We hypothesized that Kin13Bp and Kin13Cp have partially redundant functions. About half of the homozygotes lacking both Kin13Bp and Kin13Cp (13BC-KO) were viable (Table 1) but grew and swam more slowly than the single knockouts (Figure 2, A–D, Supplemental Figure S3, A–D, and Supplemental Movie S1). A phosphodiesterase inhibitor, IBMX, increases the swim speed of Tetrahymena by increasing the ciliary beat frequency (Hennessey and Lampert, 2012). IBMX (1 mM) increased the swimming rate of the 13BC-KO mutants, but they remained slower than the similarly treated wild-type cells (Figure 2E and Supplemental Movies S2 and S3). The slow cell motility indicates an abnormal function of the locomotory cilia. The 13BC-KO cells also had a reduced rate of phagocytosis, a function that depends on the motility of oral cilia (Supplemental Figure S3E). Shaking of the 13BC-KO flask cultures caused further reduction in the multiplication and motility rates, whereas this treatment had little effect on the wild-type cells (Figure 2, A and B). The phenotypes of some ciliary mutants are enhanced by increased aeration (Brown et al., 2003; Hou et al., 2007), again indicating that Kin13Bp and Kin13Cp affect cilia. Whereas Kin13Cp localized strongly to the CVP, a microtubule-rich organelle involved in osmoregulation (Allen and Naitoh, 2002), we did not detect a change in the sensitivity of 13BC-KO cells to either hypoosmotic or hyperosmotic conditions (unpublished data).
FIGURE 2:
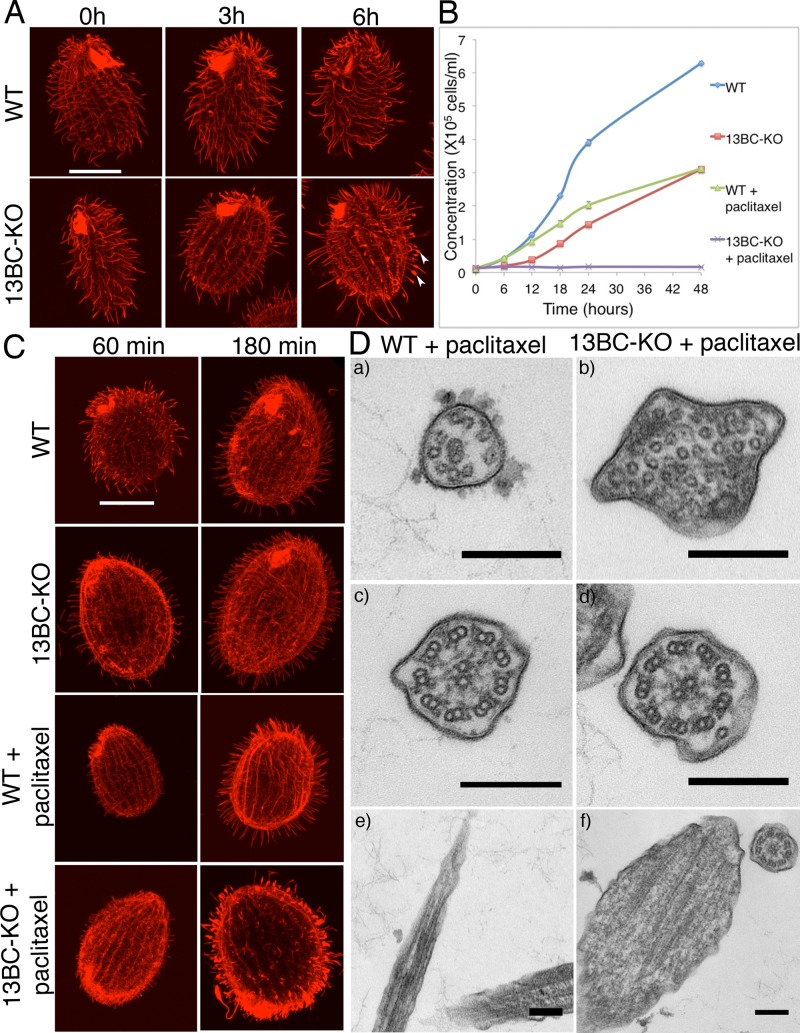
Cells lacking both Kin13Bp and Kin13Cp are deficient in multiplication and ciliary functions. (A) Average growth curves of strains grown with or without shaking (three experiments). (B) Average swimming velocity of cells grown with or without shaking (25 measured cells for each strain/condition). Bars represent standard errors. *p < 0.0001. (C, D) Images of swimming paths of live wild-type and 13BC-KO cells recorded for 10 s. (E) Histogram showing average swim speeds of wild-type and 13BC-KO cells that are either untreated or treated with 1 mM IBMX.
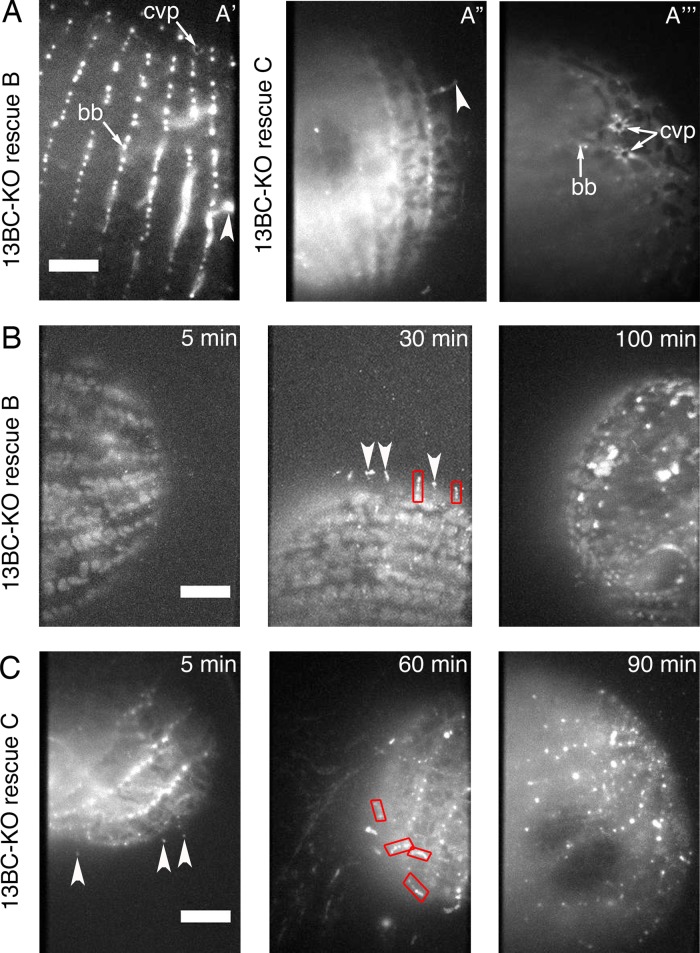
The 13BC-KO cells died in the presence of a microtubule-stabilizing drug, paclitaxel, at the concentration in which wild-type cells multiply (see later discussion), indicating that the loss of kinesin-13 leads to abnormal microtubules. We took advantage of the paclitaxel sensitivity to confirm that the mutant phenotype of 13BC-KO cells is caused by the absence of Kin13Bp and Kin13Cp. Introduction of either GFP-Kin13Bp or GFP-Kin13Cp transgenes into the 13BC-KO cells resulted in paclitaxel-resistant rescue transformants that displayed nearly normal cell multiplication and motility rates, whereas no rescues were observed in a mock transformation experiment (107 cells). Based on TIRFM, the rescuing proteins had localization patterns that were similar but not identical to their natively tagged versions. Namely, the rescuing GFP-KIN13B was found at the CVP (Figure 3A and Supplemental Movies S6 and S7), whereas the natively tagged protein was not (Figure 1D). Likely, when both proteins are present, Kin13Cp has higher affinity for the CVP sites, but Kin13Bp can occupy the same sites in the absence of Kin13Cp. This can explain how Kin13Bp and Kin13Cp can be functionally redundant despite having nonoverlapping localizations when tagged in their native loci (Figure 1). Strikingly, in the live rescue cells observed in TIRFM, both GFP-Kin13Bp and GFP-Kin13Cp localized to a subset of cilia, which appeared short and likely were assembling (Figure 3A, arrowheads, and Supplemental Movies S6 and S7). To test further whether GFP-Kin13Bp and GFP-Kin13Cp localize preferentially to the assembling cilia, we observed the rescue cells as they regenerate cilia after deciliation. GFP-positive structures consistent with short assembling cilia were frequently seen on the surface of regenerating cells between 30 and 60 min after deciliation, when the majority of the regenerating cilia grow, and rarely at earlier and later time points (Figure 3, B and C, and Supplemental Movies S8 and S9). We conclude that Kin13Bp and Kin13Cp are partially functionally redundant and localize to the CVP, cortical microtubule bundles, basal bodies, and assembling cilia.
FIGURE 3:
GFP-Kin13Bp and GFP-Kin13Cp rescue the mutant phenotype of 13BC-KO cells and localize to assembling cilia. (A) TIRF images of 13BC-KO cells rescued with either GFP-Kin13Bp (A′) or GFP-Kin13C (A′′, A′′′). GFP-Kin13Bp and GFP-Kin13Cp are detected near the basal bodies, CVP, and in a subset of cilia, possibly enriched at their tips. (B) TIRF images of 13BC-KO cells rescued with GFP-Kin13Bp that were deciliated and allowed to regenerate their cilia for 5, 30, and 100 min. The background of faintly autofluorescent mitochondria is visible. Note the GFP-positive cilia on the cell surface at 30 min. (C) TIRF imaging of 13BC-KO cells rescued with GFP-Kin13Cp that were deciliated and allowed to regenerate cilia for 5, 60, and 90 min. Note short cilia at 60 min. Red boxes outline growing cilia. Arrowheads mark growing cilia where the pattern of fluorescence is consistent with the strong presence of kinesin-13 limited to the distal tips. bb, basal body; cvp, contractile vacuole pore. Bar, 20 μm.
The loss of Kin13B and Kin13Cp has opposite effects on the length of microtubules in the cell body and cilia
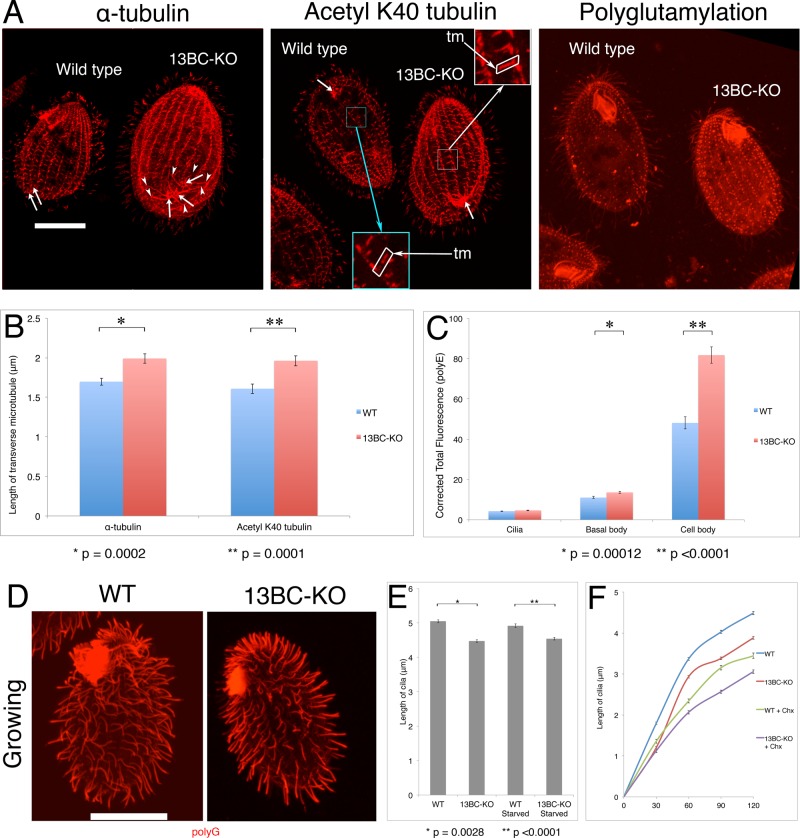
When the wild-type and 13BC-KO cells were analyzed side by side by immunofluorescence using anti–α-tubulin antibodies, the 13BC-KO cells showed an increased signal of tubulin in the cell bodies (Figure 4A, left). The organization of cortical microtubules seemed normal, except that some of the microtubule types were excessively long. Whereas in the wild-type cells, the CVP rootlets are limited to the vicinity of the CVP rings within two ciliary rows, in the 13BC-KO cells, the same microtubules were exceptionally long, covering almost the entire posterior region of the cell (Figure 4A, left and middle). The mutant transverse microtubule bundles were slightly but significantly longer (Figure 4, A, middle, and B). We conclude that Kin13Bp and Kin13Cp function in the shortening of subtypes of cortical microtubules. In the cell body, the consequences of the absence of kinesin-13 can be explained by its canonical activity as a microtubule-end depolymerizer.
FIGURE 4:
The 13BC-KO cells have overgrown cortical microtubules and shorter axonemes. (A) Confocal immunofluorescence images of mixed wild-type (labeled by loading the food vacuoles with India ink) and 13BC-KO cells labeled with anti–α-tubulin monoclonal antibody (mAb; 12G10), anti–acetyl K40 mAb (6-11B-1), or anti-polyglutamylation antibodies (polyE). Insets represent a threefold magnification of the selected areas. Bar, 20 μm. Arrows point at the central rings of CVPs. Arrowheads identify the ends of the CVP rootlet microtubules that are overgrown in the 13BC-KO cells. (B) Average length of the transverse microtubule bundles based on immunofluorescence using either an anti–α-tubulin 12G10 or anti–acetyl-K40 α-tubulin 6-11B-1 antibody. (C) Corrected total fluorescence signal of cilia, basal body, and cell body areas of wild-type and 13BC-KO cells labeled with the anti-polyglutamylation antibodies (polyE). The intensity of twenty-five 200-pixel circles (in five cells) was measured for each strain. Bars represent SE. *p = 0.00012 for basal body, and **p < 0.0001 for cell body. (D) Confocal immunofluorescence images of growing wild-type and 13BC-KO cells labeled by immunofluorescence with a mixture of anti–α-tubulin mAb (12G10) and anti-polyglycylation antibodies (polyG), which strongly label cilia. Bar, 20 μm. (E) Average length of cilia of growing or starved cells (20 cells for each strain). The values are 5.05 ± 0.04 μm for WT and 4.47 ± 0.04 μm for 13BC-KO growing and 4.92 ± 0.06 μm for WT and 4.54 ± 0.04 μm for 13BC-KO starved for 24 h. *p = 0.0028 for growing cells, and **p < 0.0001 for starved cells. (F) Average length of cilia in regenerating cells (taken from growing conditions and deciliated with a pH shock) in the presence or absence of 20 μg/ml cycloheximide (Chx; 10 cells for each sample). For each time point, there is a significant difference between the lengths of the wild-type and 13BC-KO cilia (p < 0.0001). tm, transverse microtubules.
The 13BC-KO cells were covered by a normal number of cilia (Table 2) whose axonemes were slightly but significantly shorter than in the wild type (4.52 ± 0.07 vs. 4.83 ± 0.08 μm; Figure 4, D and E). In standard transmission electron microscopy (TEM), the mutant axonemes and basal bodies of 13BC-KO cells appeared structurally normal (Supplemental Figure S4) and the basal bodies had a normal length (wild type, 0.53 ± 0.02 μm; 13BC-KO, 0.54 ± 0.01 μm; n = 7 for each genotype). Thus kinesin-13 selectively affects the length of axonemes. When Tetrahymena cells starve, they arrest the cell cycle in the (macronuclear) G1 phase and do not assemble new cilia (Mowat et al., 1974; Ross et al., 2013). Thus starved cells can be used to measure the length of mature cilia reliably. Cilia in both growing and starved 13BC-KO cells were shorter than in the wild type under the same conditions (Figure 4E), indicating that the loss of kinesin-13 causes a defect in ciliary length regulation. Next we tested whether the rate of elongation of cilia is also affected. After deciliation, the 13BC-KO cilia elongated at a reduced rate and failed to reach the proper maximal length (Figure 4F). We conclude that the loss of kinesin-13 reduces the rate of axoneme elongation and its maximal length.
TABLE 2:
Number of cilia per micrometer of cell circumference.
| No shaking | Shaking | |
|---|---|---|
| WT | 0.271 ± 0.063 | 0.271 ± 0.052 |
| 13BC-KO | 0.238 ± 0.047 | 0.300 ± 0.074 |
n = 20 for each condition.
Chlamydomonas cells with a knockdown of kinesin-13 fail to regenerate cilia in the presence of the protein translation inhibitor cycloheximide, which led to a hypothesis that kinesin-13 produces cilia-destined tubulin by depolymerizing the cell body microtubules (Wang et al., 2013). In the presence of 20 μM cycloheximide at a concentration that shuts down protein synthesis in Tetrahymena (Hallberg and Hallberg, 1983), both wild-type (as reported; Hadley and Williams, 1981) and 13BC-KO cells regenerated cilia, and the drug decreased the elongation rate and the maximal length in both genetic backgrounds to a similar extent; at 120 min postdeciliation, the wild-type and 13BC-KO cilia were shorter by 23 and 21%, respectively (Figure 4F). Thus the loss of kinesin-13 does not make ciliary assembly more dependent on the synthesis of new proteins. Hence it appears unlikely that in Tetrahymena, the depolymerization of cell body microtubules by kinesin-13 is a major source of cilia-destined tubulin.
Paclitaxel reveals abnormal tubulin inside cilia of kinesin-13–null cells
Although the strong effect of the loss of kinesin-13 on cortical microtubules, some of which are in proximity of the ciliated basal bodies (transverse bundles), can explain the entire contribution of kinesin-13 to ciliogenesis (see Discussion), we were intrigued by the enrichment of Kin13Bp and Kin13Cp in assembling cilia (Figures 1 and 3). The multiplication of 13BC-KO cells is inhibited by paclitaxel (Figure 5B). Paclitaxel could act synergistically with the loss of kinesin-13, making certain microtubules excessively long or too stable. Paclitaxel (40 μM) increased the abundance of cell body microtubules in both the wild-type and 13BC-KO cells (unpublished data). In addition, the paclitaxel-treated wild-type cells developed hyperelongated cilia (as described; Wloga et al., 2009); in 6 h, the average length of cilia in the wild type increased by 40% (from 5.0 ± 1.2, n = 177, at t = 0 to 7.0 ± 1.7, n = 220, at t = 6 h). In the similarly treated 13BC-KO cells, the cilia lengthen only by 9% (from 4.5 ± 0.9 at 0 h, n = 206, to 4.9 ± 1.8, n = 427 at t = 6 h). Unexpectedly, a subset of the drug-treated mutant cilia were excessively short and had tubulin-positive swollen tips (10.5%, n = 427 cilia, at 6 h), whereas such cilia were not seen in the similarly treated wild type (Figure 5A). The swollen mutant cilia were located mostly in the midposterior region, where new cilia assemble from the newly duplicated basal bodies (Frankel, 2000), suggesting that paclitaxel interferes with the elongation of mutant cilia. To test this hypothesis, we applied paclitaxel to cilia-regenerating cells. Whereas wild-type cells regenerated hyperelongated cilia, the 13BC-KO cells regenerated predominantly short cilia with tubulin-filled tips (Figure 5C). At 3 h postdeciliation, the wild-type cilia treated with paclitaxel were 9% longer than the wild-type untreated cilia, whereas the 13BC-KO paclitaxel-treated cilia were 40% shorter than untreated mutant cilia, and 42% of the mutant cilia had tubulin-filled swollen tips (wild type, 4.55 ± 0.08, n = 122; wild type/paclitaxel, 4.92 ± 0.13, n = 158; 13BC-KO, 4.34 ± 0.08, n = 157; 13BC-KO/paclitaxel, 2.71 ± 0.09, n = 195). TEM showed a normal ultrastructure of cilia in the wild-type paclitaxel-treated cells (Figure 5D), except that the cross sections of distal segments (which contain singlet peripheral microtubules; Figure 5D) were more frequent than with the nontreated control (Table 3). Thus, in the wild-type cilia, paclitaxel hyperelongates the distal axonemal segment. Remarkably, the 13BC-KO paclitaxel-treated cells had cilia with tips filled by numerous singlet microtubules (Figure 5D). The large number of these microtubules in cross sections indicates that most if not all are ectopic and not extensions of the axoneme. Such abnormal microtubules were also occasionally seen on cross sections of the middle axoneme segment in the drug-treated 13BC-KO cells. The ectopic singlet microtubules were not present in the untreated 13BC-KO or in treated or untreated wild-type cells (Figure 5D, Supplemental Figure S4, and Table 3). The formation of ectopic microtubules at the tips of assembling cilia of 13BC-KO cells indicates that the delivery of precursor tubulin into kinesin-13–deficient cilia is not inhibited by paclitaxel. To the contrary, the 13BC-KO mutants may have excessive levels of soluble tubulin around the tips of growing axonemes, which could drive the formation of ectopic microtubules in the presence of paclitaxel. To conclude, paclitaxel reveals an abnormal tubulin inside assembling cilia of cells lacking nonnuclear kinesin-13.
FIGURE 5:
13BC-KO cells are hypersensitive to paclitaxel. (A) Confocal immunofluorescence images of wild-type and 13BC-KO cells grown in the presence of 40 μM paclitaxel for 0–6 h. The cilia were labeled with a mixture of anti–α-tubulin mAb (12G10) and anti-polyglycylation antibodies (polyG). Bar, 20 μm. Note that whereas the cilia in the wild-type cells grown in paclitaxel get longer, many cilia of mutant cells grown with paclitaxel are short and have tubulin-filled swellings at the tips (arrowheads). (B) Average growth curves of wild-type and 13BC-KO cells in the presence and absence of 20 μM paclitaxel. Bars represent SE (three experiments). (C) Confocal immunofluorescence images of wild-type and 13BC-KO cells that were deciliated and allowed to regenerate cilia for 60 and 180 min in the presence or absence of 20 μM paclitaxel and labeled with a mixture of anti–α-tubulin mAb (12G10) and anti-polyglycylation antibodies (polyG). Bar, 20 μm. Note that the majority of paclitaxel-treated 13BC-KO cilia are short and have tubulin- filled swollen tips. (D) TEM images of wild-type and 13BC-KO cells treated with 40 μM paclitaxel for 6 h, showing cross section of distal cilia segments with peripheral singlet microtubules (a and b), cross section of the middle segments with doublet microtubules in cilia (c and d), and longitudinal sections of axonemes distal segments (e and f). Bar, 0.2 μm.
TABLE 3:
Analysis of cross sections in TEM.
| Wild type | Wild type plus paclitaxel | 13BC-KO | 13BC-KO plus paclitaxel | |
|---|---|---|---|---|
| Ratio of distal segments (singlets) to total axoneme cross sections | 25/110 (23%) | 57/111 (51%) | 23/105 (22%) | 60/112 (54%) |
| Distal segment cross sections with ectopic microtubules | 0/25 (0%) | 0/57 (0%) | 0/23 (0%) | 15/60 (25%) |
| Middle segment cross sections with ectopic microtubules | 1/110 (1%) | 3/111 (3%) | 2/105 (2%) | 22/112 (20%) |
| Longitudinal sections with visible ectopic microtubules | 0/85 (0%) | 0/85 (0%) | 2/86 (2%) | 18/80 (23%) |
Overexpressed kinesin-13 has an axoneme distal end–depolymerizing activity
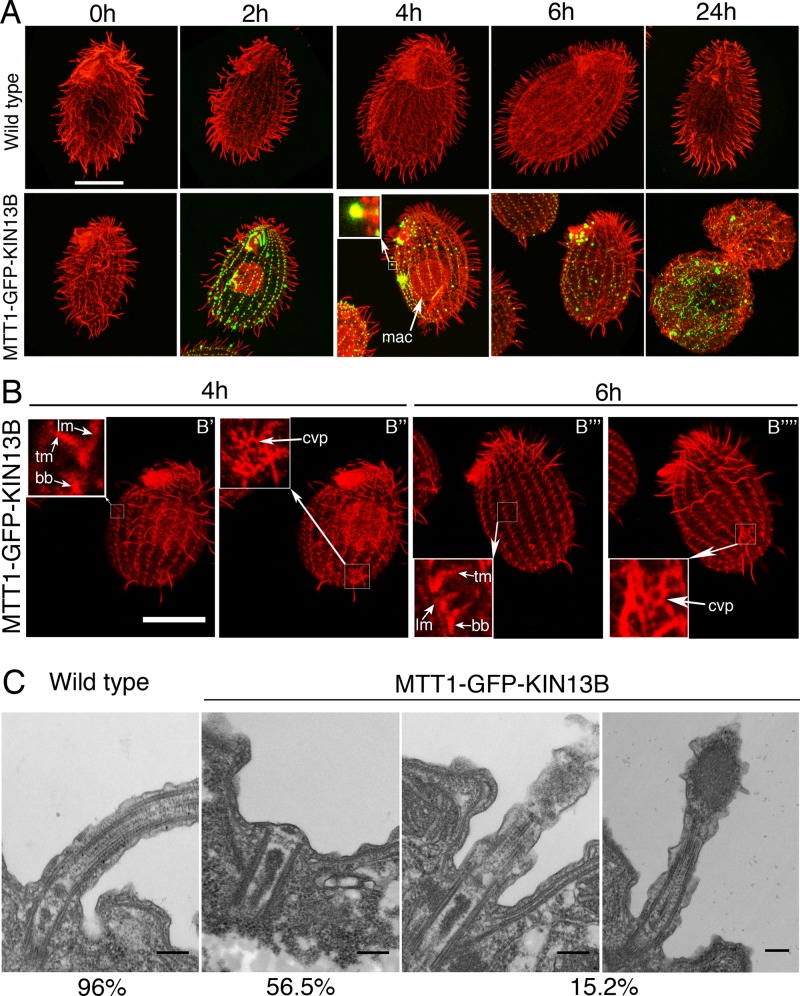
Our results so far opened a possibility that Kin13Bp and Kin13Cp act inside cilia as an axoneme assembly–promoting factor. These observations seem inconsistent with the only known biochemical activity of kinesin-13, a microtubule-end depolymerase. Thus we tested whether Kin13Bp and Kin13Cp promote microtubule depolymerization in vivo by overproduction of either GFP-Kin13Bp or GFP-Kin13Cp under the cadmium-inducible MTT1 promoter. No transgenic strains were obtained for GFP-Kin13Cp, possibly because the transgene's uninduced expression was lethal. Overexpressed GFP-Kin13Bp localized to the basal bodies and formed large clusters in the middle or at the tips of cilia (Figure 6A, bottom). Overproduction of GFP-Kin13Bp led to shortening and complete loss of cilia within 24 h (Figure 6A). Some Kin13Bp-overproducing cells also showed accumulation of microtubules inside the macronucleus (Figure 6A, 2 and 4 h), suggesting that the soluble tubulin released from depolymerization of the extranuclear microtubules enters the macronucleus. Strikingly, the GFP-Kin13Bp–overproducing cells that lacked most axonemes had mostly intact cortical microtubules (Figure 6B), indicating a degree of selectivity for axonemes in the GFP-Kin13Bp–mediated depolymerization activity. TEM of GFP-Kin13Bp cells induced for 4 h showed that most of the basal bodies lacked an axoneme entirely (56.5%). An additional 15.2% of cilia had partly depolymerized distal portions with electron-dense aggregates that likely contained overproduced GFP-Kin13Bp (compare the inset in Figure 6, A, 4 h, and C, right), indicating that the depolymerization occurs mainly at the axoneme plus ends. The ciliated and nonciliated basal bodies did not show signs of depolymerization at the proximal side corresponding to the minus ends of microtubules (Figure 6C). Thus the overproduced Kin13Bp has a strong axoneme plus end–depolymerizing activity in vivo.
FIGURE 6:
Overexpressed GFP-Kin13Bp induces shortening and complete loss of axonemes. (A) Confocal immunofluorescence images of wild-type and MTT1-GFP-Kin13Bp expressing cells induced with 2.5 μg/ml CdCl2 for 0–24 h. The cells show a direct GFP signal (green) and an immunofluorescence signal (red) obtained with a mixture of anti–α-tubulin mAb (12G10) and anti-polyglycylation antibodies (polyG). In A, the inset shows a higher magnification of a shortening cilium. Bar, 20 μm. (B) Confocal immunofluorescence images of MTT1-GFP-KIN13B cells (red) induced with 2.5 μg/ml CdCl2 for 4 and 6 h and labeled with a mixture of anti–α-tubulin mAb (12G10) and anti-polyglycylation antibodies (polyG). Subsets of confocal sections were combined to show the cell cortex. Note that the overexpressing cells, while having lost most of their cilia, have rows with apparently normal basal bodies, transverse microtubules, and longitudinal microtubules (B′ and B′′′), as well as contractile vacuole pores (B′′ and B′′′′). The inset shows a higher magnification of basal bodies, transverse microtubules, and longitudinal microtubules (B′ and B′′′) and contractile vacuole pores (B′′ and B′′′′). Bar, 20 μm. (C) TEM images of wild-type and MTT1-GFP-Kin13Bp cells induced for 4 h with 2.5 μg/ml CdCl2. bb, basal body; cvp, contractile vacuole pore; lm, longitudinal microtubule; oa, oral apparatus; tm, transverse microtubule. Bar, 0.2 μm.
Defective kinesin-13 affects ciliary beating and the microtubule-sliding activity of dynein arms
The motility defect of 13BC-KO cells seems too severe to be explained by a modest reduction in the length of cilia. Using high-speed video recording of live cells, we found that the 13BC-KO cilia beat 35% more slowly (20.8 ± 2.9 beats/s) than wild-type cilia (32.0 ± 0.4 beats/s). The mutant cilia showed an apparently normal waveform, but the neighboring cilia were less coordinated (Supplemental Movies S4 and S5). To test whether the reduced beat frequency is due to a defect in the activity of the dynein arms, we isolated 13BC-KO and wild-type axonemes and measured the rate of microtubule sliding in vitro in the presence of ATP. The velocity of sliding of microtubules was significantly decreased in the 13BC-KO axonemes (5.44 ± 0.14 μm/s, n = 79, compared with wild type, 6.99 ± 0.1 μm/s, n = 75). TEM of cross sections of 13BC-KO axonemes showed apparently normal inner and outer dynein arms (Supplemental Figure S4), and doublets extruded from the reactivated axonemes and stained with uranyl acetate had a normal density of outer dynein arms (Supplemental Figure S3). Thus the simplest interpretation is that the reduced beat frequency and swimming speed are consequences of defective dynein motor activity.
Kinesin-13 affects the posttranslational modifications on ciliary tubulin and genetically interacts with MEC-17/ATAT1 tubulin acetyltransferase
Multiple observations so far indicate that the loss of kinesin-13 affects the dynamics of microtubules (changes in length and sensitivity to paclitaxel). Posttranslational modifications (PTMs) on tubulin are reporters of the microtubule dynamics; namely, most PTMs accumulate on long-lived microtubules. As in other ciliated models, in Tetrahymena, axonemal tubulin is modified by acetylation at K40 on α-tubulin and polyglutamylation and polyglycylation of multiple glutamates in the C-terminal tail domains of α- and β-tubulin (reviewed in Gaertig and Wloga, 2008). Based on immunofluorescence using PTM-specific antibodies, the 13BC-KO cells showed increased signal of acetyl-K-40 α-tubulin in the subtypes of microtubules in the cell cortex, including the basal body–associated postciliary and transverse bundles, as well as the CVP rootlets (Figure 4A, middle). The CVP rootlets (Figure 4A left) and transverse microtubules (Figure 4, A and B) are also excessively long when labeled with an antibody that recognizes unmodified α-tubulin (the postciliary microtubules are too small to be measured reliably). Therefore, the increased signal of acetyl-K40 α-tubulin on cortical microtubule bundles may simply reflect the increased mass of these microtubules and not their excessive acetylation. However, a stronger signal of polyglutamylated tubulin was detected in the basal bodies of 13BC-KO cells (Figure 4, A, right, and C). Because the basal bodies were normal in structure and length in TEM (Supplemental Figure S4), it appears that their microtubules are more polyglutamylated in the 13BC-KO cells. Of interest, the mutants also had a greatly increased diffused signal of polyglutamylated tubulin in the cell body, which could represent hypermodified soluble tubulin (Figure 4, A, right, and C; such a tubulin was also detected inside cilia; see later discussion).
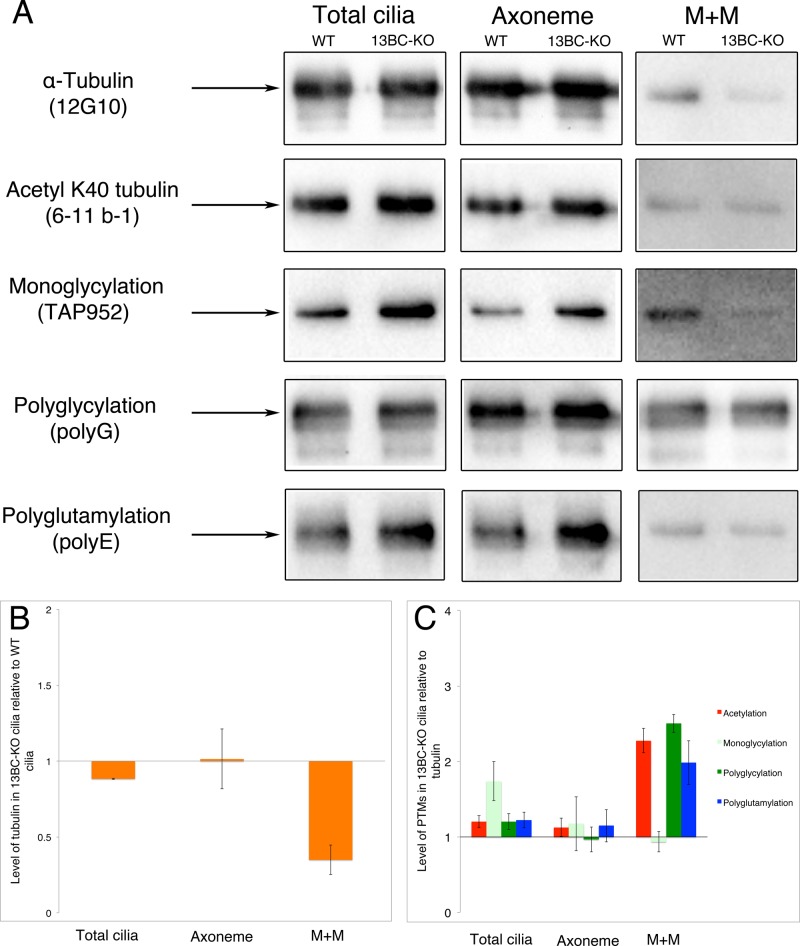
We used Western blotting to quantify the levels of modified and total tubulin in the wild-type and 13BC-KO cilia. The 13BC-KO and wild-type cilia showed a nearly normal level of total α-tubulin in the axoneme but had reduced levels of soluble (membrane plus matrix [M+M]) α-tubulin (Figure 7, A and B). For quantification of the PTM levels, we averaged data obtained from three independent preparations of cilia and normalized the signals to the signal of total tubulin. Although there is some variability in the patterns of PTM signals between the experiments, on average, 13BC-KO cilia had increased levels of tubulin PTMs, especially monoglycylation. The average levels of PTMs on axonemes were also mildly elevated except for polyglycylation (Figure 7C). Surprisingly, the tubulin in the soluble fraction of 13BC-KO cilia, while less abundant, contained 2- to 2.5-fold increased levels of K-40 acetylated, polyglycylated, and polyglutamylated isoforms (Figures 7, A, right, and C). Thus kinesin-13 affects the levels of tubulin PTMs, particularly in the soluble compartment of cilia.
FIGURE 7:
Double-knockout cells have altered patterns and levels of PTMs on tubulin. (A) Western blots of cilia, M+M, and axoneme fractions of wild-type and 13BC-KO cells. The lanes were loaded using the amount of material obtained from the same number of processed cells. Arrows represents a size marker of 50 kDa. The antibodies recognize either α-tubulin (12G10) or various modified tubulin isoforms. The Coomassie blue–stained gels loaded with the same samples are shown in Supplemental Figure S5. (B, C) Charts documenting the levels of α-tubulin (B) and posttranslational modifications on tubulin (C) in the 13BC-KO cilia relative to wild-type levels. The data represent an average of band signals on three blots (including blots shown in A). In B, the signal values of tubulin were corrected to account for slightly unequal loading on each gel by measuring the signal intensity of five non-tubulin bands on the image of a corresponding Coomassie blue–stained gel. The level of wild type equals 1. In C, the levels of modified tubulins are normalized to the amount of α-tubulin, and the normalized signal is expressed as a ratio to the normalized wild-type signal that equals 1.
Because Kin13Bp and Kin13Cp affect the levels of tubulin modifications, there is a possibility that excessive levels of tubulin PTMs mediate some if not all of the consequences of loss of kinesin-13. Thus we tested whether the phenotype of KIN13BC-KO can be rescued by a deletion of Mec17p, a homologue of conserved MEC-17/ATAT1 α-tubulin (K40) acetyltransferase. Mec17p was chosen among the known tubulin modifying enzymes because it produces most if not all acetylated α-tubulin acetylation in Tetrahymena, and its loss does not affect the cell phenotype except for a mild change in the resistance to anti-tubulin drugs (Akella et al., 2010). Surprisingly, the triple-knockout cells lacking Kin13Bp, Kin13Cp, and Mec17p were not viable (Supplemental Figure S6C). Before their death, the triple-knockout homozygotes had an excessively round shape and disorganized cell body microtubules and had lost most cilia (Supplemental Figure S6A, bottom). We rescued the progeny of the triple-knockout heterokaryons with transgenes encoding either Kin13Bp or Kin13Cp (Supplemental Figure S6B). For unclear reasons, we could not rescue the triple knockouts with a transgene encoding GFP-Mec17p. To test further whether the triple deletion of Kin13Bp, Kin13Cp, and Mec17p is lethal, we outcrossed a triple-knockout heterokaryon to a wild type, produced and genotyped the F2 progeny, and, again, could not recover viable triple-knockout progeny cells (unpublished data). We conclude that the loss of kinesin-13 is synthetically lethal with a loss of the α-tubulin acetyltransferase MEC-17/ATAT1. Whereas in Tetrahymena, Mec17p is not important (Akella et al., 2010), it becomes essential in the absence of kinesin-13. This indicates that kinesin-13 not only affects the levels of tubulin modifications but also functionally synergizes with a tubulin-modifying enzyme, MEC-17/ATAT1.
DISCUSSION
Kinesin-13 shortens microtubules in the cell body, but its activity is not required for generation of ciliary precursor tubulin
We show that Tetrahymena has three kinesin-13 proteins, one of which, Kin13Ap, functions inside the nuclei, and two, Kin13Bp and Kin13Cp, function in the cell body and cilia. A similar division of labor among the kinesin-13 homologues exists in Trypanosoma brucei (Chan et al., 2010).
How kinesin-13 homologues contribute to cilia remains unclear. Some studies indicate that kinesin-13 shortens the axoneme, acting as a microtubule-end depolymerase. A rigor mutation of kinesin-13 in Giardia induced excessively long cilia (Dawson et al., 2007). Although an RNAi knockdown of a kinesin-13 homologue in Trypanosoma (Blaineau et al., 2007) led to the lengthening of cilia, no such effect was observed when the same gene was deleted (Chan and Ersfeld, 2010). However, in mammalian cells, depletion of Kif24 kinesin-13 induced premature ciliogenesis (Kobayashi et al., 2011). Furthermore, overexpression of kinesin-13 in Leishmania (Blaineau et al., 2007) and Tetrahymena (this study) shortened cilia. Kinesin-13 is enriched in the resorbing cilia of Chlamydomonas (Piao et al., 2009). Thus kinesin-13 may act inside cilia in a manner similar to kinesin-8, which depolymerizes the distal end of the axoneme (Niwa et al., 2012).
Unexpectedly, in Chlamydomonas (Piao et al., 2009; Wang et al., 2013) and Tetrahymena (this study), a loss of function of kinesin-13 leads to shorter cilia that assemble slowly. Wang et al. (2013) proposed that kinesin-13 produces cilia-destined tubulin by depolymerizing the cell body microtubules. In agreement, we found that in Tetrahymena lacking nonnuclear kinesin-13, the cell body microtubules are overgrown, and cilia have less of soluble tubulin. However, unlike the Chlamydomonas kinesin-13–knockdown cells (Wang et al., 2013), the Tetrahymena kinesin-13 knockouts regenerate cilia in the absence of protein synthesis. Thus, in Tetrahymena, kinesin-13 is not required for the generation of precursor tubulin from the preexisting sources in the cell body. However, by maintaining the length of cortical microtubules, kinesin-13 could contribute to cilia indirectly (e.g., by supporting transport along the cortical microtubules toward the basal body). Later we argue that in addition to its cell body function, kinesin-13 also acts inside cilia. A similarly complex picture of kinesin-13 activities emerged for Drosophila, for which a loss of kinesin-13 led to hyperelongation of centrioles and the formation of short and abnormal axonemes in the spermatocytes (Delgehyr et al., 2012).
Our observations indicate that in Tetrahymena, the nonnuclear kinesin-13 also acts inside cilia. We detected kinesin-13 inside the assembling cilia, as shown in Chlamydomonas (Wang et al., 2013). When the Tetrahymena kinesin-13–knockout cells were treated with paclitaxel, the assembling axonemes failed to elongate, and ectopic singlet microtubules formed at the distal tips of cilia. Paclitaxel lowers the critical concentration of tubulin (Schiff et al., 1979). Without kinesin-13, the addition of new tubulin to the growing axoneme end is slower, and this could build up excessive levels of soluble tubulin arriving by intraflagellar transport (IFT; Hao et al., 2011; Bhogaraju et al., 2013) to reach the critical concentration with paclitaxel. Although the Tetrahymena kinesin-13–mutant cilia have lower total soluble tubulin in cilia, this does not exclude a possibility that an excessively high concentration of tubulin builds up around the tips of assembling mutant cilia.
We propose that two models on kinesin-13 can be reconciled with its axoneme end–depolymerizing activity. First, the ciliary kinesin-13 could function in a complex with another protein—a microtubule polymerase—and the loss of kinesin-13 could destabilize the entire complex. In mammalian cells, Kif24 kinesin-13 is required for loading of its binding partner, CP110, onto the ends of the mother centriole microtubules (Kobayashi et al., 2011). Alternatively, kinesin-13 could act directly as an assembly-promoting factor, in analogy to kinesin-8, a depolymerizer that has a stabilizing activity at a low concentration (Stumpff et al., 2008; Du et al., 2010; Su et al., 2011). Alternatively, the axoneme assembly–promoting function of kinesin-13 could be a direct result of its lattice-destabilizing activity. The ends of axonemal microtubules are blocked by caps (Dentler, 1980) that prevent the addition of new tubulin subunits in vitro (Dentler and Rosenbaum, 1977). Kinesin-13 could use its protofilament-curving activity to bend the microtubule ends away from the caps and make them more accessible for addition of new tubulin.
Kinesin 13 alters posttranslational modifications of tubulin and synergizes with tubulin-modifying enzyme MEC-17/ATAT1
The loss of kinesin-13 alters the levels of posttranslationally modified tubulin in cilia. We detected moderate increases in the levels of modified tubulin isoforms in the axoneme. Intriguingly, large increases in PTMs were observed on the soluble ciliary tubulin. The tubulin PTMs studied here accumulate on microtubules that are relatively stable, such as those forming the axonemes and basal bodies. The acetylation of K40 on α-tubulin has evolved as a marker of long-lived microtubules. The responsible enzyme, MEC17/ATAT1, acetylates K40 inside the microtubule at a slow rate caused by its naturally “suboptimal” active site, providing a time-stamping function for long-lived microtubules (Szyk et al., 2014). Thus the elevation in the levels of PTMs could be a result of an improper dynamics of axonemal tubulin, which could increase the residence time of tubulin subunits in the mutant axoneme.
We speculated that the excessive levels of tubulin PTMs contribute to the mutant phenotype observed in the kinesin-13–null cells, in particular the slow ciliary motility. We explored this hypothesis by testing whether the mutant phenotype can be rescued by deletion of a tubulin-modifying enzyme, the K40 α-tubulin acetyltransferase, MEC-17/ATAT1 (Akella et al., 2010; Shida et al., 2010). To our surprise, the loss of both nonnuclear kinesin-13 and MEC-17/ATAT1 is synthetically lethal. It is likely that the synthetic lethality of kinesin-13 and MEC17/ATA1 deletions is caused by a malfunction of microtubules in the cell body, since we could not recover the triple-knockout cells on a specialized medium that supports growth of cells lacking cilia (unpublished data). MEC-17/ATAT1 also affects microtubules without acetylating them (Topalidou et al., 2012), likely by depolymerization (Kalebic et al., 2013a, b). Thus the synthetic lethality of kinesin-13 and MEC17/ATAT1 could be caused by an insufficient depolymerization of the cell body microtubules. At present, we cannot exclude a possibility that accumulation of K40 α-tubulin acetylation specifically in cilia (or excessive levels of other PTMs) contributes to the ciliary phenotypes of kinesin-13–deficient cilia.
Alternatively, the changes in the levels of tubulin PTMs in cilia may not be a consequence of an altered dynamics of ciliary microtubules but could be caused by the reduced length of the mutant axoneme. Cilia that are excessively short have hyperglutamylated axonemes (Sharma et al., 2007; Dave et al., 2009b). None of the foregoing models explains the hypermodified state of soluble ciliary tubulin in the absence of kinesin-13. More work is needed to dissect the apparently complex relationships between kinesin-13, the axoneme length, microtubule dynamics, and the PTMs of tubulin.
Kinesin-13 affects ciliary beating
The most unexpected observation is that kinesin-13 affects ciliary motility: cells lacking Kin13Bp and Kin13Cp swim slowly, and the mutant cilia beat at a reduced frequency. In Drosophila, a loss of kinesin-13, Klp10, also impaired sperm motility, but this phenotype was associated with hyperelongation of centrioles and structural defects in the axoneme (Delgehyr et al., 2012). We show that in Tetrahymena, the kinesin-13–deficient axonemes are normal (by standard TEM) but display a reduced rate of sliding of doublet microtubules when reactivated with ATP in vitro. The velocity of sliding microtubules is primarily determined by the activity of outer dynein arms (reviewed in Kamiya, 2002). Thus, in the absence of kinesin-13, outer dynein arms could be less active. This hypothesis also fits with the observed reduction in the beat frequency caused by the loss of kinesin-13, as the outer dynein arms are required for proper control of beat frequency (reviewed in Kamiya, 2002). Dynein arms are stably attached to the A-tubule while they exert force on the B-tubule of an adjacent doublet. The B-tubule is highly enriched in modified tubulins (reviewed in Wloga and Gaertig, 2010). Whereas tubulin glycylation and acetylation play a minor role in cilia motility (Wloga et al., 2009; Akella et al., 2010), tubulin polyglutamylation on the B-tubule is a key regulator of ciliary motility. In Tetrahymena, Chlamydomonas, and mouse, losses of enzymes that glutamylate tubulin compromise ciliary motility (Ikegami et al., 2010; Kubo et al., 2010; Suryavanshi et al., 2010). Thus kinesin-13, by affecting tubulin modifications on the axonemal microtubules, could indirectly influence the dynein arm activity.
MATERIALS AND METHODS
Multiple sequence alignment
The amino acid sequences of kinesin-13 homologues were aligned using Clustal W (Larkin et al., 2007) and T-Coffee (Notredame et al., 2000). The sequences used were T. thermophila KIN13A XP_001026192.1, KIN13B XP_001011311.1, KIN13C XP_001032255.1; Homo sapiens KIF2A ABQ59038.1, KIF2B NP_115948.4, MCAK AAC27660.1; Mus musculus KIF2A AAH06803.2, KIF2B AAI00485.1, MCAK NP_608301.3; G. intestinalis KIN13 ABD60079.1; Leishmania major KIN13-2 XP_001681757.1; T. brucei KIN13-2 XP_828542.1, and C. reinhardtii CrKIN13, Chlre4|149713.
Strains and cultures
T. thermophila strains were grown at 30°C in either SPP (Gorovsky, 1973) or MEPP (Orias and Rasmussen, 1976) medium with 2 μg/ml dextrose (MEPPD) and antibiotics (Gaertig et al., 2013).
GFP tagging
To overexpress kinesin-13 homologues tagged at the N-terminus with GFP, fragments of the genomic coding regions were amplified with addition of MluI and BamHI sites and cloned into the pMTT1-GFP plasmid (Wloga et al., 2006). KIN13B was amplified with primers carrying MluI (5′-GAAAACGCGTCCTGAGAAAGCAAATTAACA-3′) and BglII (5′-CCCAGATCTTCAATTTTCTATTTTTTTCTTC-3′). KIN13C was amplified with primers carrying MluI (5′-TTAACGCGTCATGAAGGGCACAGC-3′) and BglII (5′-TATAGATCTTCAAAAGTAGAAGGTATC-3′). The plasmids were digested with ApaI and SacII and introduced into the CU522 strain by biolistic bombardment and selection with paclitaxel (Gaertig et al., 2013). For overexpression, transformants were grown in SPP medium with antibiotics (SPPA) to 2 × 105 cells/ml and induced with 2.5 μg/ml CdCl2. For GFP tagging of the kinesin-13 genes in their native loci, plasmids were made carrying DNA fragments in the following order: 1) a terminal fragment of a targeted coding region without a stop codon, 2) an in-frame GFP coding region with a stop codon TGA, 3) the transcription terminator region of BTU1, 4) the neo3 gene for selection with paromomycin, and 5) a 3′-untranslated region (UTR) fragment of a targeted gene. For KIN13A, 2.5 kb of the coding region was amplified with addition of SacII and MluI restriction sites (primers 5′-ATTACCGCGGAGCCTTAGTTTTCTCACTTAT-3′ and 5′-TATTACGCGTGCATTATCATATAGATCAGG-3′ ), and 1.3 kb of 3′-UTR was amplified with the addition of ClaI and SacI restriction sites (primers 5′-TTAAATCGATAAACTTACAACAATCAATCAATA-3′ and 5′-ATAAGAGCTCATTTAAGGGATTGGAATATCAT-3′). For KIN13B, 2.8 kb of 5′-UTR was amplified with addition of SacII and MluI restriction sites (primers 5′-AAATCCGCGGAAGACTTCTAGTTTGATAACG-3′ and 5′-TATTACGCGTGCCTTCAAAAGAAATGATTACCTC-3′), and 1.3 kb of 3′-UTR was amplified with the addition of ClaI and SacI restriction sites (primers 5′-TTAAATCGATTGCATAAAAAGAAATATCTATCT-3′ and 5′-ATAAGAGCTCAAATCAGTAACCACAGACGA-3′). For KIN13C, 2.5 kb of coding region was amplified with addition of SacII and MluI restriction sites (primers 5′-TTTTCCGCGGCATACCTAATCCAGTCCAG-3′ and 5′-AATACGCGTAAGTAGAAGGTATCTTCAACT-3′), and 0.9 kb of 3′-UTR was amplified with the addition of ClaI and SacI restriction sites (primers 5′-TTATATCGATTTTATCTTCCTAATATTAGTTATA-3′ and 5′-ATAAGAGCTCAATAACTACTGTAATTTAAACCA-3′). These plasmids were digested with SacI and SacII and introduced into the starved CU428 cells by biolistic bombardment and selection with paromomycin and cadmium chloride (Gaertig et al., 2013).
Germline gene knockouts and rescues
To construct single germline-knockout strains with deletions of KIN13A, KIN13B, and KIN13C, plasmids were made carrying DNA fragments in the following order: 1) a 5′ fragment of the targeted gene, 2) the neo4 cassette for selection with paromomycin, and 3) a 3′ fragment of a targeted gene. For KIN13A, 1.4 kb of 5′-UTR was amplified with addition of ApaI and SmaI restriction sites (primers 5′-TATTGGGCCCTCTTATGATAATTCTTCTATTTCA-3′ and 5′-TTATCCCGGGTCTTTTCCTTTTCCTTTTTCGT-3′), and 1.4 kb of 3′-UTR was amplified with the addition of PstI and SacII restriction sites (primers 5′-TATTCTGCAGATAAAAATGGATTGGGACAGTT-3′ and 5′-ATATCCGCGGGTTTTAATCGATAACATCAGCA-3′ ). For KIN13B, 1.4 kb of 5′ fragment was amplified with addition of ApaI and SmaI restriction sites (primers 5′-ATAAGGGCCCTGACAAGATAATTTACTTACTTA-3′ and 5′-AATTCCCGGGTCACACTTTTAGATGCTTAAC-3′), and 1.4 kb of the 3′ fragment was amplified with the addition of PstI and SacII restriction sites (primers 5′-ATAACTGCAGAGATGTTTACAAGTATTCCTTA-3′ and 5′-TTATCCGCGGTTCATTAAATATTATGTGTGATC-3′). For KIN13C, 1.4 kb of the 5′-UTR was amplified with addition of ApaI and SmaI restriction sites (primers 5′-ATATGGGCCCCCTATTTTTAATTTTCTCAAGTTT-3′ and 5′-TTATCCCGGGAAGTTAACTATTTATATAGCTTGA-3′), and 1.4 kb of 3′-UTR was amplified with the addition of PstI and SacII restriction sites (primers 5′-AATACTGCAGAAGAAGAGCTAAAATGGAAGAA-3′ and 5′-TTATCCGCGGTTCTTTCAAACGATCTGCATAT-3′). The resulting plasmids were digested with ApaI and SacII and biolistically transformed into mating cells of CU428 and B2086. Heterokaryon strains homozygous for the deleted gene in the micronucleus and having wild-type copies in the macronucleus were made, and homozygotes were generated by a heterokaryon-to-heterokaryon cross (Dave et al., 2009a). The absence of the targeted genomic region in the homozygote was confirmed by PCR of genomic DNA with primers designed to amplify the deleted region. To confirm the deletion of KIN13A, 2.1 kb was amplified with primers 5′-TATTGGGCCCTCTTATGATAATTCTTCTATTTCA-3′ and 5′-TATGTAGCTTTTATTATTCTGAC-3′. To confirm the deletion of KIN13B, 2 kb was amplified with primers 5′-ATAAGGGCCCTGACAAGATAATTTACTTACTTA-3′ and 5′-CACATGCTATAAACTGAAGTT-3′. To confirm the deletion of KIN13C, 2 kb was amplified with primers 5′-ATATGGGCCCCCTATTTTTAATTTTCTCAAGTTT-3′ and 5′-TCTATTTCTTCTGCTAGTCATT-3′. Double-knockout heterokaryon strains lacking both KIN13B and KIN13C (13BC-KO) and triple-knockout heterokaryons also lacking MEC17 were obtained by crosses. For rescue of 13BC-KO cells, overexpression-type plasmids with either GFP-KIN13B or GFP-KIN13C transgene were biolistically introduced into the BTU1 locus, and transformants were selected with 20 μM paclitaxel. The rescues of the triple-knockout cells were done using the foregoing BTU1-based fragments, including a similar GFP-MEC17 transgene, using a mating of heterokaryons and selection of progeny based on the knockout gene cassettes with 100 μg/ml paromomycin and 2.5 μg/ml cadmium.
Phenotypic studies
The growth rates were determined by inoculating cells in 25 ml SPPA at 2 × 104 cells/ml and incubating at 30°C with or without shaking at 160 rpm. To measure the swimming velocity, 1 ml of cells at 1 × 104 cells/ml was placed in a well of a 12-well polystyrene plate, and paths of the moving cells were recorded for 10 s at a maximal speed by AxioCam HS camera mounted on a Nikon Observer.A1 microscope with 10× lens. The path lengths were measured using the AxioVision particle tracking software. To measure the beat frequency, cells (at 2 × 105 cells/ml) were recorded at 500 frames/s by a Photronics 1280 PCI FastCam on a Nikon Eclipse E600 microscope. To measure the swim speeds of cells treated with IBMX (1 mM, 20 min), digital videos were recorded on the Nikon Eclipse E600 microscope with 40× magnification for 10 s. These videos were converted into 1-s still images of swimming paths, and swimming distances were determined using ImageJ 1.46 (National Institutes of Health, Bethesda, MD). To measure the length and number of cilia, cells were labeled by immunofluorescence with a mix of the monoclonal anti–α-tubulin antibody, 12G10 (Jerka-Dziadosz et al., 2001), and the polyclonal anti–polyglycylated tubulin antibody, polyG (Duan and Gorovsky, 2002), as described later. For consistency, we determined the number and average length of cilia on 20 cells using confocal optical sections that include the widest diameter of the macronucleus. The length measurements were done using ImageJ 1.46. To study cilia regeneration, 1 × 106 cells were washed with 10 mM Tris-HCl, pH 7.4 and suspended in 0.5 ml of deciliation medium (10% Ficoll 400, 10 mM sodium acetate, 10 mM CaCl2, 10 mM EDTA, pH 4.2). The cells were passed thrice through an 18 ½-gauge needle, and 27.5 ml of the regeneration medium (15 mM Tris-HCl, 2 mM CaCl2, pH 7.95) was added (Calzone and Gorovsky, 1982). The cells were incubated at room temperature and allowed to regenerate their cilia either in the presence or absence of 20 μM cycloheximide (Sigma-Aldrich, St. Louis, MO) or 20 μM paclitaxel (LC Laboratories, Woburn, MA).
For immunofluorescence, the cells were stained as described (Gaertig et al., 2013) with primary antibody 12G10 monoclonal anti–α-tubulin (1:30; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA; Jerka-Dziadosz et al., 2001) or 6-11B-1 anti–acetyl-K40 α-tubulin (1:200; Sigma-Aldrich; Piperno and Fuller, 1985) and viewed with a Zeiss (Jena, Germany) LSM 510 or LSM 710 confocal microscope (using 63× oil immersion with 1.2 numerical aperture [NA]). Images were assembled using either LSM (Zeiss), Zen black (Zeiss), or ImageJ software.
For measuring the corrected total fluorescence signal, the cells were labeled with the anti-polyglutamylation antibodies (polyE). The intensity of the signal in cilia, basal body, and cell body areas of the cells were measured using ImageJ. A total of twenty-five 200-pixel circles (in five cells) were measured for each strain, and the corrected total fluorescence signal was calculated as described in Burgess et al. (2010). For TIRF, 10 μl of cells (2 × 105 cells/ml) in SPPA or MEPPD medium with 2–3 mM NiCl2 was placed on a slide with a 22 × 22–mm #1.5 coverglass and viewed on a TIRF system home built around a Nikon Eclipse Ti-U inverted microscope equipped with a 60 × NA 1.49 TIRF objective (Lechtreck, 2013).
To measure the rate of phagocytosis, cells were grown to a concentration of 2 × 104 cells/ml and fed with 0.2% India ink for 20 min. The fraction of cells that took up the ink into food vacuoles was determined.
For negative staining of axonemes, 1 × 106 cells were washed with 10 mM Tris-HCl, pH 7.4, and suspended in 0.5 ml of deciliation medium (10% Ficoll 400, 10 mM sodium acetate, 10 mM CaCl2, 10 mM EDTA, pH 4.2). The cells were passed thrice through a 18 ½-gauge needle, and 27.5 ml of the regeneration medium (15 mM Tris-HCl, 2 mM CaCl2, pH 7.95) was added (Calzone and Gorovsky, 1982). Cells were then spun down twice at 3000 rpm for 5 min to separate the cilia (supernatant) from the cell bodies (pellet). The supernatant was then spun at 14,000 rpm for 20 min at 4°C to obtain cilia. The pelleted cilia were suspended in 220 μl of the motility buffer (1 mM dithiothreitol [DTT], 50 mM potassium acetate, 5 mM MgSO4, 1 mM ethylene glycol tetraacetic acid [EGTA], 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], poly(ethylene glycol) [PEG] 1%, pH 7.6) with protease inhibitors (Complete; Roche, Indianapolis, IN). To demembranate, we added 30 μl of 1% NP-40 in the motility buffer to 150 μl of purified cilia and mixed gently. The samples were spun at 13,000 rpm for 20 min at 4°C to separate the axonemes. Reactivation of the axonemes was done by adding 0.5 μl of 100 mM ATP to 50 μl of the axoneme fraction and leaving it at room temperature for 5 min. Ten microliters of the reactivated axonemes was placed on a carbon-coated Formvar-covered grid and negatively stained with fresh 2% uranyl acetate. The negatively stained samples were then analyzed on a JEOL 1200 EX transmission electron microscope.
For TEM, 2 × 106 cells were concentrated by centrifugation to 100 μl and fixed with 1 ml of 2% glutaraldehyde (in 0.1 M sodium cacodylate buffer, pH 7.2) on ice for 1 h. Ten microliters of 1% tannic acid was added, and the cells was kept on ice for 1 h. Cells were washed five times in the cold sodium cacodylate buffer (10 min on ice) and postfixed with 1 ml of 1% osmium tetroxide for 1 h on ice. The pellet was washed five times in water, followed by dehydration in an ethanol/water concentration series and embedding in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate and analyzed on a JEOL 1200 EX transmission electron microscope.
Western blotting
For Western blotting, 2 × 107 cells of wild-type and 13BC-KO strains were deciliated using a pH shock (Gaertig et al., 2013). Cell bodies and cilia were collected and suspended in 3 ml and 300 μl of ice-cold axoneme buffer (20 mM potassium acetate, 5 mM MgSO4, 0.5 mM EDTA, 20 mM HEPES, pH 7.6) with protease inhibitors (Complete), respectively. One hundred microliters of the ciliary extract were demembranated by addition of 100 μl of 1% NP-40 with protease inhibitors in the axoneme buffer on ice for 2 min. The demembranated cilia were spun at 13,000 rpm for 15 min at 4°C to obtain the supernatant fraction (M+M) and the pellet fraction (axoneme). Equivalent amounts (based on the cell numbers) of cell bodies, cilia, M+M, and axoneme fractions were separated on a 10% SDS–PAGE and transferred to a polyvinylidene fluoride membrane. Western blots were done using the primary antibodies at the following dilutions: 12G10, 1:1000; 6-11B-1, 1:10,000; TAP952 monoclonal anti–monoglycylated tubulin antibody (Callen et al., 1994; Bré et al., 1998), 1:20,000; polyG, 1:20,000; and polyE, 1:4000. The blots were visualized using Amersham ECL Prime reagents (GE Healthcare Life Sciences, Pittsburgh, PA). Images were taken using BioRad (Hercules, CA) ChemiDoc MP imaging system, and quantification of the band intensity was done using Imagelab (Bio-Rad) software.
Microtubule sliding in isolated axonemes
Cilia were purified from 1 l of cells grown to a concentration of 2 × 105 cells/ml and suspended at 0.05 mg protein/ml in 500 μl of the axoneme buffer (20 mM potassium acetate, 5 mM MgSO4, 0.5 mM EDTA, 20 mM HEPES, pH 7.6) without protease inhibitors. To demembranate, we added 10 μl of 1% NP-40 in the motility buffer (1 mM DTT, 50 mM potassium acetate, 5 mM MgSO4, 1 mM EGTA, 30 mM HEPES, PEG 1%, pH 7.6) to 50 μl of diluted cilia. The axoneme suspension was pipetted into a perfusion chamber constructed with a glass slide and coverslip separated by double-sided stick tape. The perfusion chamber was washed with 50 μl of motility buffer, followed by perfusion with 50 μl of 1 mM of ATP in the motility buffer. The sliding of microtubules was recorded on a Zeiss Axiovert 35 microscope equipped with dark-field optics (40× PlanApo) on a silicon-intensified camera (VE-1000; Dage-MTI, Michigan City, IN). The sliding velocity was determined by measuring microtubule end displacement as a function of time.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Science Foundation (MBC-033965) and the National Institutes of Health (RO1GM089912 to J.G., RO1GM0571173 to W.S.S., and RO1GM110413 to K.L.). We thank the following researchers for providing reagents: Martin A. Gorovsky (University of Rochester, Rochester, NY) for polyE and polyG antibodies, Joseph Frankel (University of Iowa, City, IA) for 12G10 antibodies (available from the Developmental Studies Hybridoma Bank, University of Iowa), and Marie-Helene Bré and Nicolette Levilliers (University Paris-Sud, Orsay, France) for TAP952 antibodies. We are grateful to Claire Walczak (Indiana University, Bloomington, IN) for suggestions about the domain organization of Tetrahymena kinesin-13 homologues. We acknowledge the excellent technical assistance with TEM by Mary Ard at the University of Georgia College of Veterinary Medicine (Athens, GA).
Abbreviations used:
- CVP
contractile vacuole pore
- PTM
posttranslational modification
- TEM
transmission electron microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-09-1354) on December 10, 2014.
REFERENCES
- Aizawa H, Sekine Y, Takemura R, Zhang Z, Nangaku M, Hirokawa N. Kinesin family in murine central nervous system. J Cell Biol. 1992;119:1287–1296. doi: 10.1083/jcb.119.5.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–222. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD, Naitoh Y. Osmoregulation and contractile vacuoles of protozoa. Int Rev Cytol. 2002;215:351–394. doi: 10.1016/s0074-7696(02)15015-7. [DOI] [PubMed] [Google Scholar]
- Asenjo AB, Chatterjee C, Tan D, DePaoli V, Rice WJ, Diaz-Avalos R, Silvestry M, Sosa H. Structural model for tubulin recognition and deformation by kinesin-13 microtubule depolymerases. Cell Rep. 2013;3:759–768. doi: 10.1016/j.celrep.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA, Lorentzen E. Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science. 2013;341:1009–1012. doi: 10.1126/science.1240985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pages M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Bré MH, Redeker V, Vinh J, Rossier J, Levilliers N. Tubulin polyglycylation: differential posttranslational modification of dynamic cytoplasmic and stable axonemal microtubules in Paramecium. Mol Biol Cell. 1998;9:2655–2665. doi: 10.1091/mbc.9.9.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Fine NA, Pandiyan G, Thazhath R, Gaertig J. Hypoxia regulates assembly of cilia in suppressors of Tetrahymena lacking an intraflagellar transport subunit gene. Mol Biol Cell. 2003;14:3192–3207. doi: 10.1091/mbc.E03-03-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA. 2010;107:12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen A-M, Adoutte A, Andrew JM, Baroin-Tourancheau A, Bré M-H, Ruiz PC, Clérot J-C, Delgado P, Fleury A, Jeanmaire-Wolf R, et al. Isolation and characterization of libraries of monoclonal antibodies directed against various forms of tubulin in Paramecium. Biol Cell. 1994;81:95–119. doi: 10.1016/s0248-4900(94)80002-2. [DOI] [PubMed] [Google Scholar]
- Calzone FJ, Gorovsky MA. Cilia regeneration in Tetrahymena. Exp Cell Res. 1982;140:474–476. doi: 10.1016/0014-4827(82)90144-6. [DOI] [PubMed] [Google Scholar]
- Chan KY, Ersfeld K. The role of the Kinesin-13 family protein TbKif13-2 in flagellar length control of Trypanosoma brucei. Mol Biochem Parasitol. 2010;174:137–140. doi: 10.1016/j.molbiopara.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Matthews KR, Ersfeld K. Functional characterisation and drug target validation of a mitotic kinesin-13 in Trypanosoma brucei. PLoS Pathog. 2010;6:e1001050. doi: 10.1371/journal.ppat.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D, Wloga D, Gaertig J. Manipulating ciliary protein-encoding genes in Tetrahymena thermophila. Methods Cell Biol. 2009a;93:1–20. doi: 10.1016/S0091-679X(08)93001-6. [DOI] [PubMed] [Google Scholar]
- Dave D, Wloga D, Sharma N, Gaertig J. DYF-1 Is required for assembly of the axoneme in Tetrahymena thermophila. Eukaryot Cell. 2009b;8:1397–1406. doi: 10.1128/EC.00378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson SC, Sagolla MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ. Kinesin-13 regulates flagellar, interphase, and mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell. 2007;6:2354–2364. doi: 10.1128/EC.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Rangone H, Fu J, Mao G, Tom B, Riparbelli MG, Callaini G, Glover DM. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr Biol. 2012;22:502–509. doi: 10.1016/j.cub.2012.01.046. [DOI] [PubMed] [Google Scholar]
- Dentler WL. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- Dentler WL, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. III. structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J Cell Biol. 1977;74:747–759. doi: 10.1083/jcb.74.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Duan J, Gorovsky MA. Both carboxy terminal tails of alpha and beta tubulin are essential, but either one will suffice. Curr Biol. 2002;12:313–316. doi: 10.1016/s0960-9822(02)00651-6. [DOI] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Frankel J. Cell Biology of Tetrahymena. Methods Cell Biol. 2000;62:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Wloga D. Ciliary tubulin and its post-translational modifications. Curr Top Dev Biol. 2008;85:83–113. doi: 10.1016/S0070-2153(08)00804-1. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Wloga D, Vasudevan KK, Guha M, Dentler WL. Discovery and functional evaluation of ciliary proteins in Tetrahymena thermophila. Methods Enzymol. 2013;525:265–284. doi: 10.1016/B978-0-12-397944-5.00013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Goncharov A, Jin Y, Chisholm AD. Kinesin-13 and tubulin posttranslational modifications regulate microtubule growth in axon regeneration. Dev Cell. 2012;23:716–728. doi: 10.1016/j.devcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky MA. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973;20:19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Hadley GA, Williams NE. Control of initiation and elongation of cilia during ciliary regeneration in Tetrahymena. Mol Cell Biol. 1981;1:865–870. doi: 10.1128/mcb.1.9.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Gaertig J, Gorovsky MA. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 1999;62:513–531. doi: 10.1016/s0091-679x(08)61554-x. [DOI] [PubMed] [Google Scholar]
- Hallberg RL, Hallberg EM. Characterization of a cycloheximide-resistant Tetrahymena thermophila mutant which also displays altered growth properties. Mol Cell Biol. 1983;3:503–510. doi: 10.1128/mcb.3.4.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L, Thein M, Brust-Mascher I, Civelekoglu-Scholey G, Lu Y, Acar S, Prevo B, Shaham S, Scholey JM. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol. 2011;13:790–798. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessey TM, Lampert TJ. Behavioral bioassays and their uses in Tetrahymena. Methods Cell Biol. 2012;109:393–410. doi: 10.1016/B978-0-12-385967-9.00015-3. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K, Sato S, Nakamura K, Ostrowski LE, Setou M. Tubulin polyglutamylation is essential for airway ciliary function through the regulation of beating asymmetry. Proc Natl Acad Sci USA. 2010;107:10490–10495. doi: 10.1073/pnas.1002128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Strzyewska-Jowko I, Wojsa-Lugowska U, Krawczynska W, Krzywicka A. The dynamics of filamentous structures in the apical band, oral crescent, fission line and the postoral meridional filament in Tetrahymena thermophila revealed by the monoclonal antibody 12G9. Protist. 2001;152:53–67. doi: 10.1078/1434-4610-00043. [DOI] [PubMed] [Google Scholar]
- Kalebic N, Martinez C, Perlas E, Hublitz P, Bilbao-Cortes D, Fiedorczuk K, Andolfo A, Heppenstall PA. Tubulin acetyltransferase alphaTAT1 destabilizes microtubules independently of its acetylation activity. Mol Cell Biol. 2013a;33:1114–1123. doi: 10.1128/MCB.01044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebic N, Sorrentino S, Perlas E, Bolasco G, Martinez C, Heppenstall PA. alphaTAT1 is the major alpha-tubulin acetyltransferase in mice. Nat Commun. 2013b;4:1962. doi: 10.1038/ncomms2962. [DOI] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Walczak CE. The microtubule-destabilizing kinesin XKCM1 regulates microtubule dynamic instability in cells. Mol Biol Cell. 2002;13:2718–2731. doi: 10.1091/mbc.E01-12-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145:914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa HA, Yagi T, Hirono M, Kamiya R. Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol. 2010;20:441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- Kushida Y, Nakano K, Numata O. Amitosis requires gamma-tubulin-mediated microtubule assembly in Tetrahymena thermophila. Cytoskeleton (Hoboken) 2011;68:89–96. doi: 10.1002/cm.20496. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Jr, Davidson LA. An analysis of spindle ultrastructure during prometaphase and metaphase of micronuclear division in Tetrahymena. Chromosoma. 1979;75:293–308. doi: 10.1007/BF00293473. [DOI] [PubMed] [Google Scholar]
- LaFountain JR, Jr, Davidson LA. An analysis of spindle ultrastructure during anaphase of micronuclear division in Tetrahymena. Cell Motil. 1980;1:41–61. doi: 10.1002/cm.970010105. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LSB, Goodson HV, Hirokawa N, Howard J, et al. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF. In vivo imaging of IFT in Chlamydomonas flagella. Methods Enzymol. 2013;524:265–284. doi: 10.1016/B978-0-12-397945-2.00015-9. [DOI] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Moores CA, Cooper J, Wagenbach M, Ovechkina Y, Wordeman L, Milligan RA. The role of the kinesin-13 neck in microtubule depolymerization. Cell Cycle. 2006;5:1812–1815. doi: 10.4161/cc.5.16.3134. [DOI] [PubMed] [Google Scholar]
- Mowat D, Pearlman RE, Engberg J. DNA synthesis following refeeding of starved Tetrahymena pyriformis GL: starved cells are arrested in G 1. Exp Cell Res. 1974;84:282–286. doi: 10.1016/0014-4827(74)90407-8. [DOI] [PubMed] [Google Scholar]
- Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev Cell. 2012;23:1167–1175. doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- Orias E, Rasmussen L. Dual capacity for nutrient uptake in tetrahymena. IV. growth without food vacuoles. Exp Cell Res. 1976;102:127–137. doi: 10.1016/0014-4827(76)90307-4. [DOI] [PubMed] [Google Scholar]
- Piao T, Luo M, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106:4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of a-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Ross I, Clarissa C, Giddings TH, Jr, Winey M. epsilon-tubulin is essential in Tetrahymena thermophila for the assembly and stability of basal bodies. J Cell Sci. 2013;126:3441–3451. doi: 10.1242/jcs.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178:1065–1079. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107:21517–21522. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley K, Hekmat-Nejad M, Turner J, Moores C, Anderson R, Milligan R, Sakowicz R, Fletterick R. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ohi R, Pellman D. Move in for the kill: motile microtubule regulators. Trends Cell Biol. 2012;22:567–575. doi: 10.1016/j.tcb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS, et al. Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity. Curr Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Spector J, Goodman B, Valenstein ML, Ziolkowska NE, Kormendi V, Grigorieff N, Roll-Mecak A. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell. 2014;157:1405–1415. doi: 10.1016/j.cell.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Keller C, Kaleboc N, Nguyen KC, Somhegyi H, Politi KA, Heppenstall P, Hall DH, Chalfie M. Enzymatic and non-enzymatic activities of the tubulin acetyltransferase MEC-17 are required for microtubule organization and mechanosensation in C. elegans. Curr Biol. 2012;22:1057–1065. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wang W, Jiang Q, Argentini M, Cornu D, Gigant B, Knossow M, Wang C. Kif2C minimal functional domain has unusual nucleotide binding properties that are adapted to microtubule depolymerization. J Biol Chem. 2012;287:15143–15153. doi: 10.1074/jbc.M111.317859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Piao T, Cao M, Qin T, Huang L, Deng H, Mao T, Pan J. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J Cell Sci. 2013;126:1531–1540. doi: 10.1242/jcs.124255. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Carrington JT, Gluenz E, Gull K. The expanded Kinesin-13 repertoire of trypanosomes contains only one mitotic kinesin indicating multiple extra-nuclear roles. PLoS One. 2010;5:e15020. doi: 10.1371/journal.pone.0015020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. A “holistic” kinesin phylogeny reveals new kinesin families and predicts protein functions. Mol Biol Cell. 2006;17:1734–1743. doi: 10.1091/mbc.E05-11-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol Biol Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Gaertig J. Post-translational modifications of microtubules. J Cell Sci. 2010;123:3447–3455. doi: 10.1242/jcs.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Webster DM, Rogowski K, Bre MH, Levilliers N, Jerka-Dziadosz M, Janke C, Dougan ST, Gaertig J. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell. 2009;16:867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.