Phospholipase D1 and its product, phosphatidic acid, facilitate muscle fiber regeneration in vivo and are required by mononuclear myocytes to fuse with nascent myotubes during second-phase myoblast fusion in vitro.
Abstract
Myoblast differentiation and fusion is a well-orchestrated multistep process that is essential for skeletal muscle development and regeneration. Phospholipase D1 (PLD1) has been implicated in the initiation of myoblast differentiation in vitro. However, whether PLD1 plays additional roles in myoblast fusion and exerts a function in myogenesis in vivo remains unknown. Here we show that PLD1 expression is up-regulated in myogenic cells during muscle regeneration after cardiotoxin injury and that genetic ablation of PLD1 results in delayed myofiber regeneration. Myoblasts derived from PLD1-null mice or treated with PLD1-specific inhibitor are unable to form mature myotubes, indicating defects in second-phase myoblast fusion. Concomitantly, the PLD1 product phosphatidic acid is transiently detected on the plasma membrane of differentiating myocytes, and its production is inhibited by PLD1 knockdown. Exogenous lysophosphatidylcholine, a key membrane lipid for fusion pore formation, partially rescues fusion defect resulting from PLD1 inhibition. Thus these studies demonstrate a role for PLD1 in myoblast fusion during myogenesis in which PLD1 facilitates the fusion of mononuclear myocytes with nascent myotubes.
INTRODUCTION
Myoblast differentiation and fusion is key for skeletal muscle development and for muscle repair in aging or diseased states. The process is dynamic and tightly orchestrated, involving cell withdrawal from the cell cycle, cell–cell recognition, adhesion, migration, and subsequent membrane fusion (reviewed in Horsley and Pavlath, 2004; Rochlin et al., 2010; Abmayr and Pavlath, 2012). Myoblast fusion proceeds in two steps. On initiation of differentiation, mononucleated myoblasts first fuse together to form small nascent myotubes, which is termed first-phase fusion. During second-phase fusion, the nascent myotubes recruit and fuse with additional myocytes or other myotubes. With accretion of nuclei and increase in myotube size, mature myotubes form eventually. The molecular mechanisms underlying both phases of fusion have been studied intensively over the last few decades yet remain relatively poorly understood.
Myoblast fusion, a cell–cell fusion event, is not as extensively studied as intracellular vesicle fusion or virus–cell fusion (Chen and Olson, 2005; Chen et al., 2007). For membrane fusion to occur, first two lipid bilayers must juxtapose and protrude to form a contact site where the outer membrane leaflets break down to form a hemifusion stalk with the outer/proximal leaflets fused and inner/distal leaflets unfused. This step is followed by fusion pore formation and expansion, which leads to fusion between opposed inner leaflets and mixing of the aqueous contents of the fused cells (reviewed in Chernomordik and Kozlov, 2008). Evidence suggests that both protein and lipid molecules play essential roles in the membrane fusion event (Lang et al., 2008), yet little is known about their specific roles during cell–cell fusion.
Mammalian phospholipase D (PLD) is a membrane-associated enzyme that catalyzes the hydrolysis of phosphatidylcholine (PC) to generate the signaling lipid phosphatidic acid (PA). There are two mammalian isoforms of canonical PLD, denoted PLD1 and PLD2, which share conserved regulatory and catalytic domains yet have distinct regulatory mechanisms and functional roles (reviewed in Liscovitch et al., 2000; Cockcroft, 2001; Jenkins and Frohman, 2005; Roth, 2008). Recent investigations by us and others have shown that PLD1 and PA play pivotal roles in membrane fusion between intracellular compartments and the plasma membrane (PM), such as in the translocation and fusion of glucose transporter Glut4-containing vesicles to the PM in adipocytes (Huang et al., 2005b) and the release of insulin and catecholamines by pancreatic β-cells and adrenal chromaffin cells, respectively (Vitale et al., 2001; Hughes et al., 2004). However, whether PLD1 exerts any function on cell–cell fusion processes such as myoblast fusion has not yet been elucidated.
A role for PLD1 in myoblast differentiation has been reported in vasopressin-stimulated rat L6 myoblasts through actin cytoskeleton remodeling (Komati et al., 2005) and in mouse C2C12 myoblasts through sequential activation of the mammalian target of rapamycin (mTOR) and insulin-like growth factor 2 (IGF2) signaling (Yoon and Chen, 2008). Thus PLD1 is a multifunctional regulator of myoblast differentiation. However, whether PLD1 has a physiological function in myogenesis in vivo has not been explored. Here we use in vivo and in vitro approaches to investigate how PLD1 regulates myoblast differentiation and fusion. Our data suggest that PLD1 expression is transiently up-regulated during myoblast fusion, and its genetic ablation results in delayed myofiber regeneration after chemical injury. Blocking PLD1 activity with a PLD1-specific inhibitor or ablation of PLD1 expression either by RNA interference or genetic knockout revealed a novel role for PLD1 in regulating fusion of myocytes to existing myotubes, that is, during second-phase myoblast fusion.
RESULTS
PLD1 is down-regulated in diseased muscle but becomes increased during muscle regeneration in vivo and myogenesis in vitro
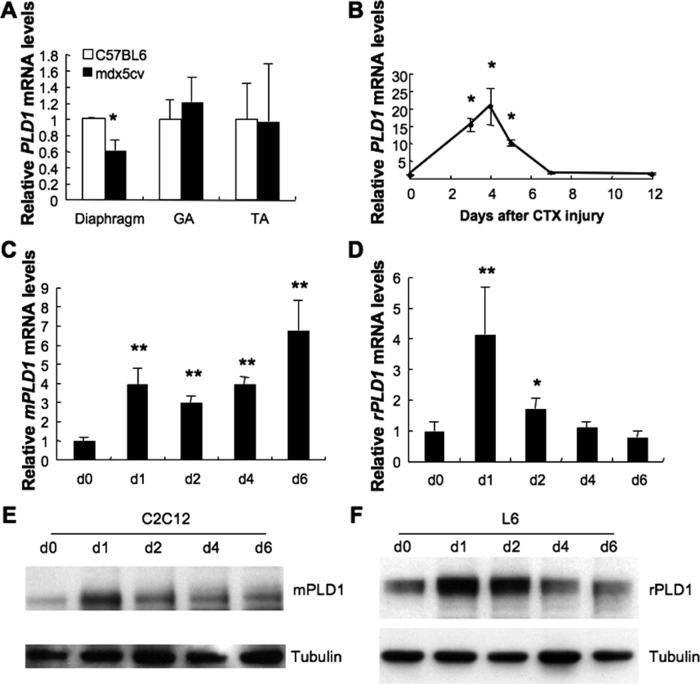
The mdx mouse models Duchenne muscular dystrophy (DMD), caused by dystrophin deficiency. In mdx mice, different muscle groups exhibit substantial divergence in dystrophy severity, with the diaphragm being the most severely affected and phenotypically the closest to DMD patients (Stedman et al., 1991). In a microarray analysis of gene expression profiles in skeletal muscles isolated from 8-wk-old wild-type, mdx, and mdx5cv (an mdx variant with a more severe phenotype) mice (Chapman et al., 1989), a fivefold down-regulation of PLD1 was detected in the diaphragms of mdx5cv mice (Haslett et al., 2005) but not in limb muscles. To validate this finding, we analyzed PLD1 transcript levels in the diaphragm, gastrocnemius (GA), and tibialis anterior (TA) muscles of 8-wk-old wild-type and mdx5cv mice (Figure 1A). In accordance with the previous report, quantitative reverse transcription PCR (RT-PCR) revealed a significant decrease of PLD1 expression in the diaphragm but not in the GA and TA muscles of mdx5cv mice. Given that different skeletal muscles respond to dystrophin deficiency heterogeneously, it has been suggested that other factors have critical effects on the phenotype severity (Porter et al., 2004). Accordingly, PLD1 could be one of the regulatory molecules dictating this selectivity.
FIGURE 1:
PLD1 expression is down-regulated in diseased muscle and up-regulated during myoblast differentiation and fusion. (A) Reduction of PLD1 mRNA expression in the diaphragm of 8-wk-old mdx5cv mice (n = 5–6, *p < 0.05) but not in gastrocnemius (GA) and tibialis anterior (TA) muscles. (B) PLD1 gene expression was temporarily up-regulated in mouse CTX-injured TA muscle (n = 3, *p < 0.05). (C, E) Mouse C2C12 and (D, F) rat L6 myoblasts were grown to confluence, induced to differentiate for up to 6 d, and analyzed by quantitative RT-PCR (C, D) and Western blotting (E, F) for PLD1 expression. Results are shown as mean ± SD (n = 3). Asterisks indicate statistically significant differences between cells collected at day 0 and the subsequent time points (*p < 0.05; **p < 0.01).
To evaluate whether PLD1 regulates adult myogenesis, we analyzed cardiotoxin (CTX)-injured TA muscle. New regenerating myofibers with centrally localized nuclei would suggest muscle regeneration. PLD1 mRNA expression was strongly up-regulated from days 3–5 postinjury (Figure 1B; [15.2 ± 1.8]-, [20.5 ± 5.2]-, and [10.2 ± 0.9]-fold increase at days 3–5, respectively, as compared with the day 0 control), a period when major myoblast fusion and formation of regenerating fibers occurs. To determine whether PLD1 is up-regulated in the myogenic cells that actively regenerate myofibers, we obtained myogenic cells (CD45− Sca-1− PDGFRα−; Joe et al., 2010; Lawlor et al., 2012) using fluorescence-activated cell sorting (see Supplemental Methods) from control and regenerating muscle. The myogenicity of these cells was confirmed by up-regulation of MyoD expression after injury and the formation of myotubes derived from these cells. As expected, PLD1 expression increased by 18-fold in myogenic cells at day 4 postinjury (Supplemental Figure S1, A and B).
Next we examined PLD1 expression during myoblast differentiation and fusion in vitro in myogenic cell lines C2C12 and L6, which originated from mouse and rat, respectively. PLD1 transcript level in C2C12 cells increased threefold 1 d after differentiation and was sustained during the 6-d differentiation process (Figure 1C). In contrast, PLD1 up-regulation in L6 cells was more transient, peaking at day 1 during differentiation with a fourfold increase and gradually decreasing to basal levels by day 4 (Figure 1D). PLD1 protein level was dramatically elevated at day 1 after differentiation in both C2C12 and L6 cells and gradually decreased to basal level afterward (Figure 1, E and F). Together the results show that PLD1 expression was up-regulated during myoblast differentiation and fusion in vivo and in vitro, suggesting that PLD1 may function in these processes.
Skeletal muscle regeneration is compromised in PLD1−/− mice
To investigate further the functional role of PLD1 in myogenesis, we turned to a PLD1-knockout (KO) mouse model (PLD1−/−). Mice lacking PLD1 display impaired platelet aggregation (Elvers et al., 2010) and reduced tumor angiogenesis (Chen et al., 2012), but the effect of PLD1 ablation on myogenesis remains unknown.
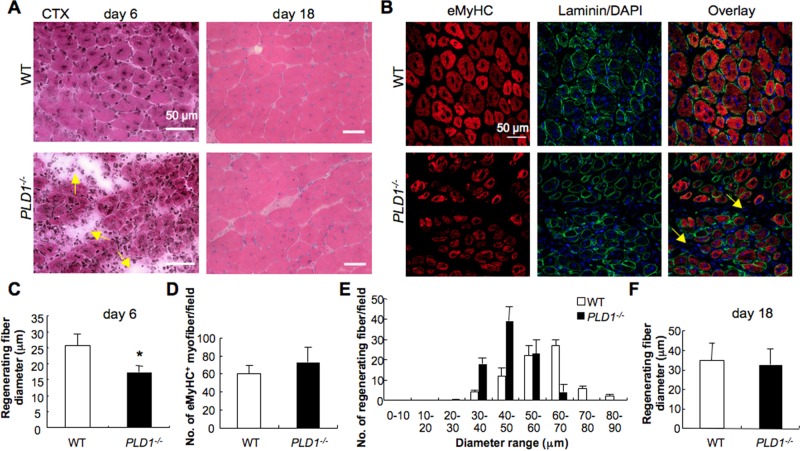
PLD1−/− mouse muscle developed normally, and adult limb muscles and the diaphragm did not show any significant histological abnormalities (Supplemental Figure S2, A and C). The average diameter of postnatal and adult myofibers in PLD1−/− mice was comparable to that of the wild-type (WT) control (Supplemental Figure S2, B and D). PLD2 mRNA expression was also not altered in PLD1−/− muscles (Supplemental Figure S2E). To investigate whether muscle regeneration is affected in KO mice, we injured TA muscle by CTX and compared muscle regeneration in age- and sex-matched WT and KO adult mice. Consistent with previous reports (Harris and Johnson, 1978; Maltin et al., 1983), by day 6 after injury, WT muscle formed small, regenerating myofibers characterized by the presence of centrally located nuclei (Figure 2A) and by the expression of embryonic myosin heavy chain (eMyHC; Figure 2B). In contrast, KO muscle displayed signs of delayed regeneration, including smaller regenerating myofibers, increased numbers of interstitial cells, and increased interstitial space between myofibers (Figure 2, A and B, arrows). Quantitative analysis revealed that the average diameter of regenerating myofibers was significantly reduced in PLD1−/− mice (Figure 2C; 16.82 ± 2.94 μm for PLD1−/− vs. 22.26 ± 10 μm for WT, n = 3 per group, p < 0.01). Moreover, although the number of eMyHC-expressing regenerating myofibers was comparable to that of controls (Figure 2D), PLD1−/− mice contained more small myofibers of 30–50 μm in diameter, whereas WT mice contained more myofibers in the range of 60–70 μm in diameter (Figure 2E). These results suggest that PLD1 is essential for normal myofiber formation during adult myogenesis. Eighteen days after injury, muscle morphology was similar in control and PLD1−/− muscles, and the diameters of regenerating myofibers were comparable (Figure 2, A and F). To assess whether the defect in early muscle regeneration is due to impaired myogenic activity of myoblasts, we analyzed expression of myogenic transcription factors Pax7, Myf5, MyoD, and myogenin in TA muscles of both genotypes (Supplemental Figure S2F). No significant differences were observed. Thus ablation of PLD1 impairs early muscle regeneration, but ultimately it can be compensated for by alternate mechanisms in vivo.
FIGURE 2:
Lack of PLD1 expression impairs skeletal muscle regeneration in vivo. (A) Representative images of H&E staining of TA muscles from WT and PLD1−/− mice at 6 and 18 d after CTX injury (n ≥ 3/genotype). (B) TA muscle was immunostained for eMyHC and laminin at 6 d after CTX injury. Nuclei were stained with DAPI. Arrows in A and B indicate the increased interstitial space between myofibers in PLD1−/− muscle. (C, F) The mean diameters of regenerating myofibers at 6 (C) and 18 (F) d after CTX injury were measured to assess myofiber size (n ≥ 3/genotype, *p < 0.05). (D) The total number of eMyHC-positive regenerating myofibers was not different in WT and PLD1−/− muscle at 6 d after CTX injury. (E) The numbers of regenerating myofibers (6 d postinjury) were measured and expressed as a histogram plot (n ≥ 3/genotype). PLD1−/− mice contained more regenerating myofiber of smaller size than did WT. (F) By day 18, regenerating myofiber size was similar in WT and PLD1−/− mice.
Lack of PLD1 impairs mature myotube formation in vitro
To explore the mechanisms underlying impaired muscle regeneration in PLD1−/− mice, we examined whether myogenic differentiation and fusion was affected in primary myoblasts in vitro. The fusion index (number of nuclei in myotubes divided by total number of nuclei in myoblasts and myotubes) was monitored during the differentiation process to assess the ability of PLD1−/− myoblasts to form myotubes. At day 1, when nascent myotubes (two to four nuclei) were forming, the fusion index was similar in WT and KO cultures (Figure 3, A, top, and B), indicating that PLD1 ablation did not affect differentiation and nascent myotube formation. However, by day 4, WT myoblasts had formed elongated multinucleated mature myotubes, whereas myoblasts derived from PLD1−/− muscle exhibited smaller and fewer myotubes (Figure 3A, bottom). The total fusion index was significantly decreased for the PLD1−/− cultures (33% ± 7%) compared with WT ones (54 ± 3%; Figure 3C; p < 0.05). Further analysis revealed that the decrease of total fusion index was caused by the decrease of mature myotube formation (Figure 3C; 28 ± 3% in WT vs. to 8 ± 5% in KO, p < 0.01), whereas nascent myotube formation was unchanged. In accordance with this result, more unfused mononuclear myoblasts were found in the PLD1−/− cultures (Figure 3D).
FIGURE 3:
Genetic ablation of PLD1 inhibits myoblast fusion in vitro. (A) Immunofluorescence analysis of desmin and myogenin expression in primary myoblasts derived from WT and PLD1−/− skeletal muscles and subjected to differentiation for up to 4 d. Nuclei were counterstained with DAPI. (B, C) Fusion index was determined by dividing the number of nuclei within myotubes by the total number of nuclei in a microscopic field (three to four microscopic fields/sample). The data were collected from three independent experiments and are shown as mean ± SD (*p < 0.05; **p < 0.01). (D) Percentage of desmin-positive mononuclear cells was determined in WT and PLD1−/− cultures after 4 d in differentiation (n = 3, *p < 0.05). (E) Percentages of desmin- and myogenin-positive cells were determined in WT and PLD1−/− cultures after 4 d in differentiation (n = 3).
The fusion defects in PLD1−/− culture were neither due to impurity of myogenic cells, as the level of desmin (a myogenic marker) expression was similar for both WT and KO myoblasts (Figure 3E), nor due to impaired myogenin expression, which is prerequisite for myoblast differentiation (Figure 3, A and E). Overall these data imply that myoblast fusion in vitro, especially second-phase fusion (i.e., addition of nuclei to nascent myotubes), is impaired in the absence of PLD1.
Knocking down PLD1 in rat myoblasts delays the onset of differentiation and inhibits mature myotube formation
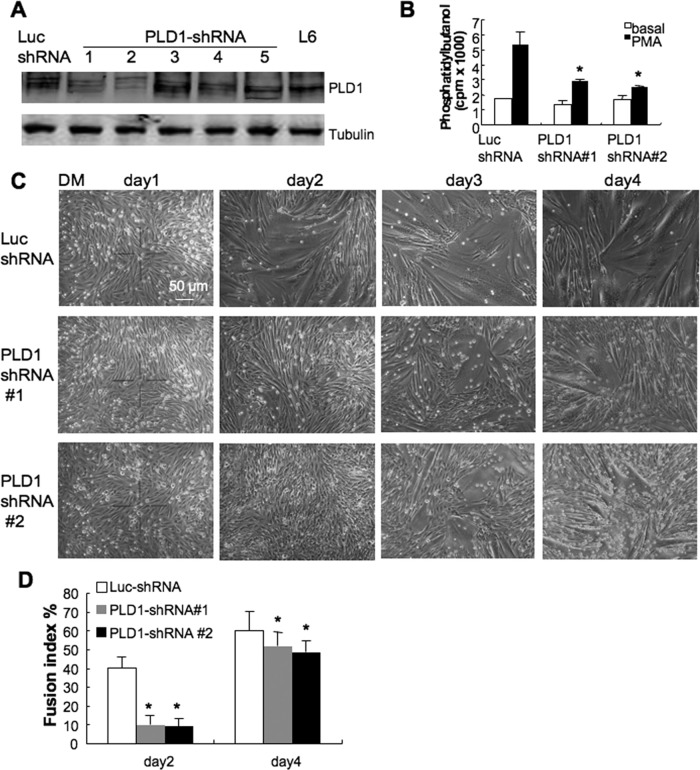
To confirm the requirement for PLD1 in myoblast fusion in a different model, we knocked down PLD1 expression in rat L6 myoblasts. As shown in Figure 4A, PLD1 protein levels were dramatically reduced in two of the five clones (clones 1 and 2) treated with short hairpin RNA (shRNA) against PLD1 (PLD1-shRNA; Huang et al., 2005b), whereas luciferase shRNA (Luc-shRNA; control) had no effect on PLD1 expression. Down-regulation of PLD1 significantly inhibited PLD activity in cells stimulated with phorbol myristic acid (PMA) but not the basal PLD activity, which was generated by the PLD2 isoform (Figure 4B; Colley et al., 1997; Singer et al., 1997).
FIGURE 4:
Inhibition of myogenesis by PLD1 silencing in L6 muscle cells. (A) L6 myoblasts were transfected with either pSuper-PLD1 shRNA (PLD1-shRNA) or luciferase shRNA (Luc-shRNA). Western blot analysis showed that PLD1 expression was significantly reduced by RNA interference in clones 1 and 2 compared with Luc-shRNA–treated cells and L6 myoblasts. (B) PLD1 knockdown inhibited PLD activity stimulated by PMA, whereas the basal activity was not affected (n = 3, *p < 0.05). (C, D) PLD1 knockdown resulted in delayed onset of myoblast fusion. Luc- and PLD1-shRNA myoblasts were cultured until nearly confluent and switched to low-serum differentiation medium. Myoblast fusion was monitored daily for 4 d using phase-contrast microscopy, and fusion index was analyzed at days 2 and 4. Data are shown as mean ± SD (n = 3, *p < 0.05).
To verify whether similar fusion defects exist in PLD1-shRNA cells as displayed in PLD1−/− myoblasts, we conducted fusion assay in L6 myoblasts treated with PLD1-shRNA. Luc-shRNA cells were used as control. In control culture, myoblasts started to form nascent myotubes by day 2 in low serum differentiation medium (DM: DMEM plus 2% horse serum), and large, elongated, mature myotubes were observed by days 3 and 4 (Figure 4C, top). In contrast, the onset of myoblast differentiation was delayed in PLD1-shRNA sublines 1 and 2, the fusion index of which was just 30 and 23% that of the control, respectively, at day 2 of differentiation (Figure 4D). By day 4, the overall fusion indexes of PLD1-shRNA sublines were still significantly less than that of control (Figure 4D), and this difference persisted even after prolonged differentiation for another 3 d (unpublished data).
The myogenic transcriptional factors myogenin and MRF4 function to execute the differentiation program for committed myoblasts and are assisted by the myocyte enhancer-binding factor 2 (MEF2) to mediate expression of muscle-specific genes (Braun et al., 1989; Molkentin et al., 1995; Black and Olson, 1998). In PLD1-shRNA cells, myogenin and MEF2 protein levels were much lower than those of Luc-shRNA cells at day 1 of differentiation but increased to levels similar to controls by day 2 (Supplemental Figure S3, A–C), suggesting that delayed onset of myoblast differentiation could be due to the delayed myogenin and MEF2 expression. However, by day 2, myogenin and MEF2 protein expression had increased to normal levels, allowing the knockdown cells to enter the differentiation program. Therefore reduced myotube formation in PLD1-shRNA cells was unlikely to have been caused by the delayed onset of differentiation.
The protein expression of the late myogenic differentiation marker myosin was greatly decreased at all time points examined in PLD1-shRNA cells (Supplemental Figure S3D), consistent with less myotube formation, as shown in Figure 4C. On day 4 of differentiation, the myosin-positive myotubes were much smaller and contained fewer nuclei in PLD1-shRNA cultures compared with control cultures (Supplemental Figure S3, E and F), a phenotype similar to PLD1−/− cultures, suggesting that lack of PLD1 inhibits myoblast fusion, especially mature myotube formation. As a measure of the specificity of RNA interference targeting, an shRNA-resistant PLD1 cDNA (wobble mutation; Huang et al., 2005b) was expressed in PLD1-shRNA cells via adenoviral infection, which rescued the secondary fusion defect and restored mature myotube formation (Supplemental Figure S3, E and F). Thus these data are in line with previous findings suggesting that PLD1 can assist second-phase myoblast fusion.
PLD1 facilitates mononucleated myoblasts fusion with nascent myotubes during second-phase fusion
To investigate the roles of PLD1 in myoblasts and nascent myotubes in second-phase myoblast fusion, we performed a nascent myotube-myoblast fusion assay in which PLD1-shRNA knockdown myoblasts or nascent myotubes were pulse labeled with CellTracker Green and CellTracker Red, respectively, to trace their contributions in mature myotubes. By calculating the ratio of green to red nuclei in mature myotubes, we could determine whether PLD1 acts on myoblasts, nascent myotubes, or both in this setting.
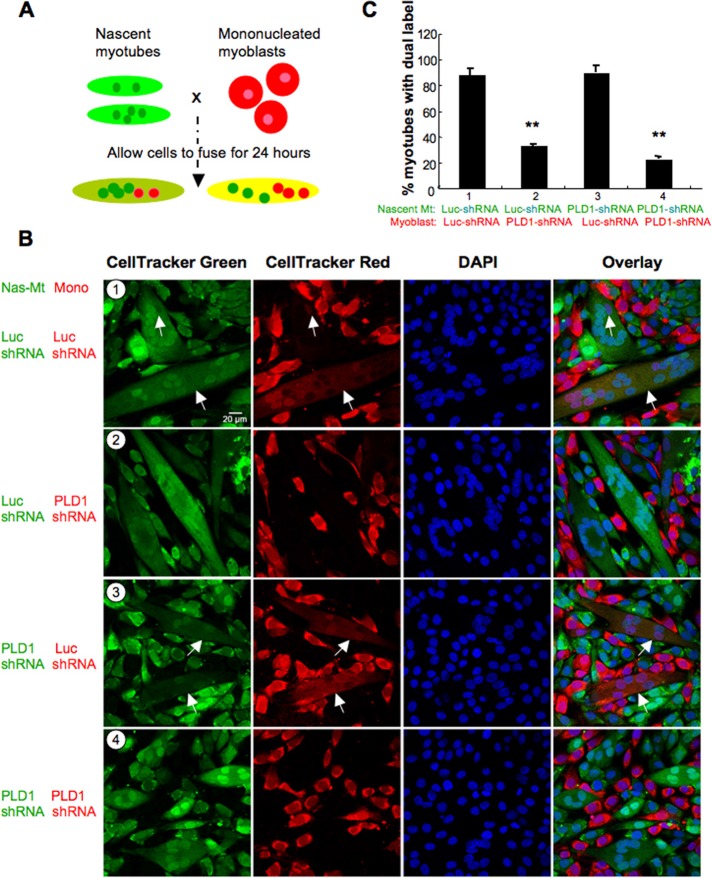
As depicted in the schematic in Figure 5A, the labeled cells were trypsinized, mixed, and cocultured in DM for 24 h before fixation and scoring for the dual-labeled myotubes, with ≥5 nuclei indicating mature myotube formation. Owing to the delayed onset of differentiation in PLD1-shRNA cultures (compared with Luc-shRNA control), nascent myotubes formed by day 3 in DM were used in the assay. The coculture strategies are as follows (Figure 5B): 1) Luc-shRNA myotubes (day 2) plus Luc-shRNA myoblasts; 2) Luc-shRNA myotubes (day 2) plus PLD1-shRNA myoblasts; 3) PLD1-shRNA myotubes (day 3) plus Luc-shRNA myoblasts; and 4) PLD1-shRNA myotubes (day 3) plus PLD1-shRNA myoblasts. In PLD1-shRNA coculture (Figure 5B4), only 22.61 ± 2.2% of myotubes (Figure 5C) showed dual label, in contrast to Luc-shRNA coculture (Figure 5B1), for which 87.76 ± 5.4% of myotubes were double stained (Figure 5C). These results reconfirmed that lack of PLD1 impaired second-phase myoblast fusion. More important, mixing of PLD1-shRNA myoblasts with Luc-shRNA myotubes (Figure 5B2) significantly hindered maturation of nascent myotubes and resulted in only 32.95 ± 1.32% of myotubes with double staining (Figure 5C), suggesting that myoblasts lacking PLD1 expression were unable to fuse properly with existing myotubes. Conversely, heterotypic mixing of Luc-shRNA myoblasts with PLD1-shRNA myotubes (Figure 5B3) led to adequate development of nascent myotubes (89.9 4± 5.33% of myotubes with dual labeling; Figure 5C), indicating that nascent myotubes from PLD1-shRNA cultures had no defect in fusion. Hence these data show that mononucleated myoblasts are the primary sites of PLD1 function during their fusion with nascent myotubes.
FIGURE 5:
PLD1 is required by mononucleated myoblasts to fuse with nascent myotubes during second-phase myoblast fusion. (A) Schematic diagram showing coculture experiments. Nascent myotubes and mononucleated myoblasts were labeled with CellTracker green and CellTracker red, respectively, and cocultured for 24 h in DM before assessing the fused myotubes with dual labeling. (B, C) Confocal imaging analysis of myoblast and myotube coculture derived from Luc-shRNA or PLD1-shRNA cells. See text for details. Myoblasts from PLD1-shRNA culture exhibited impaired fusion capacity with control (Luc-shRNA) myotubes, whereas nascent myotubes from PLD1-shRNA culture fused normally with control myoblasts. Arrows indicate myotubes with dual labeling. Data are shown as mean ± SD from three independent analyses (**p < 0.01).
PA production on plasma membrane is required for myocytes to fuse with nascent myotubes
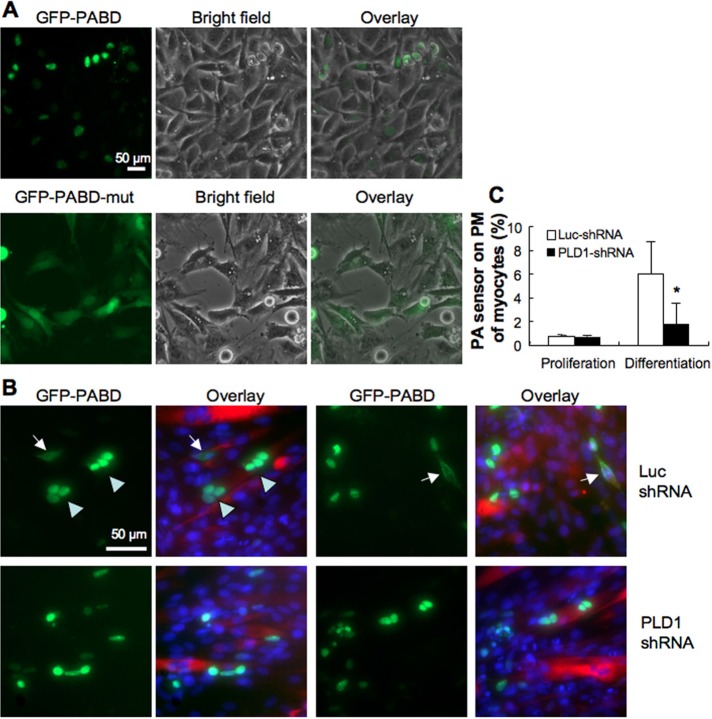
Because PLD1 is required for myoblasts to fuse with nascent myotubes, we wondered whether PLD1 activity could be detected on the PM of myoblasts. The PLD product PA can be monitored using a fluorescent sensor, which consists of enhanced GFP fused with the PA-binding domain (PABD) of the yeast Spo20 protein (GFP-Spo2051-91; Nakanishi et al., 2004). This sensor, called GFP-PABD hereafter, translocates to the PM to bind acidic phospholipid PA, and monitoring the GFP fluorescence has been successfully used to track PA production in mammalian cells (Zeniou-Meyer et al., 2007; Su et al., 2009), whereas the mutant sensor GFP-Spo20L67P, which does not bind PA, localizes uniformly in the cell. In proliferating L6 myoblasts, the majority of the sensor localized to the nucleus (Figure 6A). On differentiation, GFP-PABD localized to the PM, as the percentage of myocytes expressing GFP-PABD on the cell surface increased dramatically (Figure 6, B, arrow, and C). In addition, PM localization of GFP-PABD was observed only in mononuclear myocytes and not in the fused myotubes (Figure 6B, arrowhead), and knocking down PLD1 significantly decreased PA production on the PM of differentiated myocytes (Figure 6C). Together these data imply that PLD1 activity on the PM of myocytes is essential for their fusion into nascent myotubes.
FIGURE 6:
PA production on the plasma membrane is required for myocytes to fuse with nascent myotubes. (A) Localization of GFP-PABD and mutant GFP-PABD in proliferating L6 myoblasts. (B) At day 2 of differentiation, GFP-PABD was detected on the PM of mononuclear myocytes (arrows) but inside myotubes (arrowhead). In PLD1-shRNA culture, fewer myocytes with cell surface localization of GFP-PABD were detected. (C) Quantification of PA sensor (GFP-PABD) on the PM of Luc-shRNA and PLD1-shRNA cells (n = 3, *p < 0.05).
A small-molecule PLD1 inhibitor blocks mature myotube formation, whereas lysophosphatidylcholine addition partially rescues the fusion defect
Chronic depletion of PLD1 protein throughout myoblast differentiation and fusion revealed a second-phase fusion defect. To verify this result, we used the PLD1-specific inhibitor VU0359595 to block PLD1 activity at a distinct phase of myoblast fusion and to test whether the effect of PLD1 inhibition is reversible (Lewis et al., 2009; Scott et al., 2009; Supplemental Figure S4A). To inhibit PLD1 activity in the first-phase fusion, we differentiated L6 myoblasts in DM with 4 μM VU0359595 for 48 h (Bach et al., 2010; Scott et al., 2009). We then removed the inhibitor from the culture and differentiated the cells in inhibitor-free DM for another 3 d. Under such conditions, myoblasts differentiated and fused similarly to the dimethylsulfoxide (DMSO)-treated controls (Supplemental Figure S4, B and C), suggesting that acute PLD1 inhibition in early differentiation did not hinder myoblast fusion. To block PLD1 activity specifically during second-phase myoblast fusion, we differentiated L6 myoblasts in normal DM for 48 h, followed by addition of PLD1 inhibitor for another 3 d. This led to remarkable reduction of mature myotube formation and accumulation of nascent myotubes (Supplemental Figure S4, D and E). Of note, selective blockade of PLD1 activity at the onset of second-phase fusion is sufficient to recapitulate the myogenic fusion defect shown in PLD1−/− myoblasts and PLD1-knockdown cells, again demonstrating that PLD1 plays an essential role in second-phase myoblast fusion.
PLD1 and its product, PA, have been shown to facilitate vesicle–PM fusion during exocytosis, possibly through regulation of hemifusion and/or fusion pore dynamics (Huang et al., 2005b; Zeniou-Meyer et al., 2007; Xu et al., 2011). The opening and expansion of a fusion pore—a rate-limiting step of fusion (Cohen and Melikyan, 2004)—is facilitated by lysophosphatidylcholine (LPC). To test whether addition of LPC might reverse the PLD1 inhibition phenotype and restore mature myotube formation, we differentiated L6 myoblasts in DM containing PLD1 inhibitor VU0359595 for 4 d and then replaced the medium with DM with 50 μM LPC for another 15 h. LPC at this concentration does not change cell viability significantly (Huang et al., 2005a) and enhances fusion of WT myocytes (Supplemental Figure S5). As shown in Figure 7A, acute perturbation of PLD1 abrogated mature myotube formation and led to the accumulation of nascent myotubes, whereas LPC could partially reverse this second-phase fusion defect. Moreover, LPC specifically increased myotube maturation instead of nascent myotube formation (Figure 7B). Similarly, LPC partially rescued the fusion arrest of the nascent PLD1−/− myotubes and promoted more mature myotube formation (Figure 7, C and D). Taken together, these data suggest that PLD1 and its product, PA, may be required for a late step of fusion of adjacent membranes between myoblasts and myotubes.
FIGURE 7:
PLD1 inhibitor blocks mature myotube formation in L6 myoblasts, which can be partially rescued by addition of LPC. (A) Near-confluent L6 myoblasts were treated with either DM plus DMSO or DM plus PLD1 inhibitor (4 μM) for 4 consecutive days. Then the medium was replaced with DM with or without 50 μM LPC for another 15 h. Severe retardation of mature myotube formation was observed in PLD1 inhibitor-treated cells, whereas LPC treatment partially reversed the PLD1 inhibition. L6 cells were immunostained with myosin and myogenin antibodies, and the nuclei were counterstained with DAPI. (B) The fusion indexes of mature and nascent myotubes were calculated in the treated cells (n = 3, *p < 0.05, **p < 0.01). (C, D) Primary myoblasts isolated from WT and PLD1−/− muscle were differentiated for 4 d, and PLD1−/− culture was then treated with or without 50 μM LPC for another 15 h before immunofluorescence staining for myosin and myogenin expression. The fusion indexes of mature and nascent myotubes were calculated (n = 3, *p < 0.05, **p < 0.01). Again, addition of LPC could partially reverse the PLD1−/− fusion defect.
DISCUSSION
In this study, we demonstrated that PLD1 facilitates muscle regeneration in vivo and validated that its expression is decreased in diseased muscle. In an acute muscle injury model, cardiotoxin is known to cause acute muscle damage on existing myofibers but spares satellite cells, which are reactivated from quiescent stage upon injury and reenter the cell cycle for proliferation and differentiation to make new myofibers (Harris and Johnson, 1978; Dixon and Harris, 1996). We detected a significant increase in PLD1 transcript levels in myogenic cells (satellite cells) at a time when myoblast fusion is actively ongoing and nascent myofibers have formed but not yet matured. Concordant with PLD1 up-regulation during muscle regeneration, ablation of PLD1 in vivo attenuates myofiber regeneration. In the mdx mouse, diaphragm is the most severely affected muscle, which exhibits a chronic degeneration phenotype starting at 6–8 wk of age, comparable to that of DMD patient limb muscle (Stedman et al., 1991). Given that PLD1 expression was greatly down-regulated in the diaphragm of adult mdx5cv mice, it is likely that PLD1 expression level is associated with the severity of muscle degeneration. Although this is intriguing, further research is required to corroborate the role of PLD1 in the development of DMD.
In vitro analysis of primary myoblasts from PLD1−/− mice and L6 myoblasts treated with either PLD1-shRNA or PLD1 inhibitor further defined a role for PLD1 in secondary myoblast fusion—that is, PLD1 facilitates mononuclear myocyte fusion into nascent myotubes for generation of mature myotubes. This role of PLD1 in myogenesis is distinct from what is found in previous reports. For example, knocking down PLD1 in C2C12 cells was shown to inhibit myoblast differentiation through the mTOR-IGF2 pathway (Yoon and Chen, 2008). In a different model of myogenesis, Nemoz's group demonstrated that vasopressin-induced L6 differentiation is PLD1 dependent and occurs through activation of mTORC2-PKCα pathways (Jaafar et al., 2011). In both cases, PLD1 is implied to be involved in the early phase of differentiation and regulates myogenin expression, which is prerequisite for myoblast differentiation and fusion (Hasty et al., 1993; Nabeshima et al., 1993). In the present study, L6 cells treated with PLD1-shRNA displayed delayed expression of myogenin and delayed onset of differentiation. However, after 1 d of differentiation, myogenin expression rose to a level similar to WT, whereas primary myoblasts derived from WT and PLD1−/− mice exhibit comparable level of myogenin expression upon differentiation. Thus our data indicate that ultimately myoblasts lacking PLD1 were able to express myogenin to enter the differentiation program; however, these cells could not complete secondary fusion to form mature myotubes, implying a novel role of PLD1 in secondary myoblast fusion.
Our results further indicate that PA transiently accumulated on the PM of myocytes and appeared to act on myocytes but not nascent myotubes during second-phase fusion. Secondary myoblast fusion is asymmetric, with many proteins required exclusively by unfused myoblasts (reviewed in Chen et al., 2007). Reports, including ours, also suggest that PLD1 and its product, PA, promote intracellular vesicle fusion to the PM (reviewed in Roth, 2008) and do so most probably asymmetrically because this enhancement occurs only when PLD/PA is present on the acceptor membrane (equivalent to PM) in a cell-free system (Vicogne et al., 2006).
PA generation on the PM predisposes these cells for subsequent fusion events. In an earlier study using chick embryo myoblasts, Santini et al. (1990) showed that right before fusion occurs, PC and cholesterol on the PM break down, whereas PA and PE phospholipids increase inversely (which is in agreement with our finding). These kinds of lipid composition changes result in a highly destabilized fusion-competent myoblast membrane. In addition to PLD hydrolysis of PC, PA can be generated from lysophosphatidic acid (LPA) through LPA acyltransferases (LPAAT) and from 1,2-diacyl-sn-glycerols (DAG) by the action of diacylglycerol kinases (DGKs). Several LPAAT isoforms have been found to be highly expressed in skeletal muscle, although their function in myogenesis has not been fully understood (Leung, 2001; Li et al., 2003). DGK-catalyzed PA production increases in fusion-competent myoblasts with Ca2+ stimulation (Wakelam, 1983). More recently, DGKζ has been shown to mediate mechanical activation of PA-mTORC1 signaling in skeletal muscle (You et al., 2014). Here our data indicate that in vitro PLD1 and its product, PA, facilitate mononuclear myocyte fusion with nascent myotubes.
PLD1 and its product, PA, could regulate myoblast fusion by changing the composition of lipid bilayers to favor fusion. Topologically, membrane lipids with distinct curvature will favor or disfavor certain steps of fusion. According to their geometric features, PA, a cone-shaped lipid, produces negative curvature; by contrast, the inverted-cone-shaped LPC induces positive curvature and exhibits opposite effects in fusion. The effects of different cone-shaped lipids on membrane fusion also depend on their location on membrane bilayers. For instance, during cell–cell fusion, LPC in the contacting membrane monolayers (outer leaflets) is expected to inhibit hemifusion, whereas the same lipid in the inner membrane leaflet should promote fusion pore development (Kozlovsky et al., 2002). During myoblast fusion, the hemifusion of myoblasts could be blocked by LPC at a concentration of 300 μM (Leikina et al., 2013).
In the present study, addition of 50 μM LPC to the differentiation medium enhanced the membrane fusion of PLD1 inhibitor-treated myoblasts, supporting its role in inducing fusion pore formation in the inner membrane bilayer. Transbilayer flip-flop of LPC from the outer leaflet to the inner leaflet of the PM is an energy-independent process with a translocation rate of 1.87%/h in human erythrocytes (Mohandas et al., 1982). The amount of LPC that translocates to the inner monolayer increases linearly with total LPC concentration ranging from 8 to 80 μM. Thus it is feasible to speculate that in our study, LPC translocates to the inner membrane leaflets to promote the opening of fusion pore and subsequently removes the block that keeps membranes from merging in PLD1 inhibitor-arrested cells. In addition to its role as a lipid component of the cell membrane, LPC has been shown to stimulate various signaling molecules through the activation of the membrane G-protein–coupled receptors (Wang et al., 2005). Of particular relevance to our study, L6 myotubes treated with 20 μM exogenous LPC for 3 h resulted in increased phosphorylation of Jun N-terminal kinase (JNK; Han et al., 2011). However, JNK activation impairs myogenesis (Meriane et al., 2000), which is contradictory to LPC's effect in this setting. Therefore it is reasonable to deduce that LPC-mediated fusion rescue is unlikely through signal transduction, even though it cannot be ruled out that LPC may modulate myoblast fusion via an unidentified signaling network.
Apart from influencing lipid composition during fusion, PLD1 and its product, PA, could also regulate myoblast fusion by 1) serving as an anchor for recruiting fusogenic proteins to the PM or 2) playing a role in signal transduction to promote membrane fusion. For example, PA binds to and activates PI4P5 kinase on PM to generate phosphatidylinositol 4,5-bisphosphate (PIP2), which is known to promote myoblast fusion (Bach et al., 2010). More recently, PA-induced PIP2 generation was shown to be specifically required for membrane inner leaflet fusion, which ensures the mixing of cellular contents from fused cells (Leikina et al., 2013).
In conclusion, our data extend previous studies on the role of PLD1 in skeletal myogenesis and reveal the specific requirement of PLD1 for myocytes to fuse into existing myotubes through regulation of a late step of membrane fusion possibly involving fusion pore opening and expansion. Increased knowledge of additional protein players in myoblast fusion would have significant advantages for developing new strategies for accelerating muscle regeneration after injuries and treating neuromuscular disorders.
MATERIALS AND METHODS
Mouse strains
PLD1−/− mice were generated and characterized as described (Elvers et al., 2010; Chen et al., 2012). C57BL/6Ros-5cv (mdx5cv) mice were obtained from Jackson Laboratories (Bar Harbor, ME). All animal procedures were approved by the Institutional Animal Care and Use Committee at Boston Children's Hospital.
Cardiotoxin injury and analysis of muscle regeneration
Muscle injury was made by injection of cardiotoxin (15 μl of 0.5 μg/μl stock) from Naja mossambica (Sigma-Aldrich, St. Louis, MO) into the TA muscle of 8- to 12-wk-old C57BL/6 mice or wild-type and PLD1-knockout mice. Muscles were harvested at various times (3, 4, 5, 7, and 12 d) after injection to extract RNA. Contralateral, uninjected TA muscle was used as a control. For regeneration studies, muscles were embedded and rapidly frozen in isopentane cooled with liquid nitrogen for cryostat sectioning at days 6 and 18 after injury. Muscle tissue sections (10 μm) were stained for hematoxylin and eosin (H&E), and regeneration was analyzed by measuring the diameter of regenerating myofibers with centrally localized nuclei. Measurements were performed using ImageJ software (National Institutes of Health, Bethesda, MD).
RNA isolation and quantitative RT-PCR analysis
Total RNA was isolated from tissues or cultured cells using Trireagent (Molecular Research Center, Cincinnati, OH). Reverse transcription was performed using the Superscript III First Strand Synthesis kit for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's instruction. Quantitative RT-PCR was performed with the SYBR Green PCR master mix kit (Applied Biosystems, Carlsbad, CA) using an ABI7900HT PCR machine under ‘‘default’’ conditions: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of amplification at 94°C for 15 s and 60°C for 1 min. All transcripts levels were normalized to glyceraldehyde-3-phosphate dehydrogenase transcript levels. The sequences of primers used in this study are listed in Supplemental Table S1.
Immunoblot analysis
Protein lysates were collected from cultured C2C12 and L6 cells before differentiation and at the specified days after differentiation. Briefly, cells were rinsed in cold phosphate-buffered saline (PBS) and lysed on ice for 30 min in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% NP-40, 0.1% SDS, and Halt Protease Inhibitor Cocktail [Pierce Biotechnology, Rockford, IL]). Protein concentration was measured using a bicinchoninic acid protein assay (Pierce Biotechnology). Twenty to fifty micrograms of protein extracts was separated on 10% SDS–PAGE gels and transferred onto nitrocellulose membranes. Western blot was performed using the antibodies against PLD1 (1:1000), tubulin (1:1000; Sigma-Aldrich), myogenin (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA), MEF2 (1:1000; Santa Cruz Biotechnology), and myosin heavy chain (1:10,000; Developmental Studies Hybridoma Bank, Iowa City, IA), as described (Huang et al., 2005b).
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, blocked with PBS containing 2% bovine serum albumin (BSA) plus 5% normal goat serum for 1 h, and incubated overnight at 4°C with mouse anti-desmin antibody (1:50; DakoCytomation, Carpinteria, CA), rabbit anti-myogenin (1:50; Santa Cruz Biotechnology), or mouse anti–myosin heavy chain (1:20; Developmental Studies Hybridoma Bank). After three washes with PBS, cells were incubated with Alexa Fluor 546–conjugated or Alexa Fluor 488–conjugated secondary antibodies (Molecular Probes, Eugene, OR) for 1 h at room temperature and washed with PBS. Nuclei were counterstained with either 4′,6-diamidino-2-phenylindole (DAPI; 100 ng/ml) or TOTO-3 (2.4 nM; Molecular Probes). Slides were then mounted in Vectashield (Vector Laboratories, Burlingame, CA). Cells were visualized using an Eclipse E-1000 microscope (Nikon, Melville, NY) and photographed using a Hamamatsu digital camera (Hamamatsu, Bridgewater, NJ), and images were acquired using Openlab software, version 3.1.5 (Improvision, PerkinElmer, Waltham, MA). For confocal imaging, all images were acquired with a 40×/1.25 numerical aperture oil immersion objective using a TCS2 confocal microscope (Leica, Wetzlar, Germany).
Cell culture
Primary muscle cultures were derived from the pooled muscles of 6-wk-old wild-type and PLD1-knockout mice and cultured as described (Rando and Blau, 1994; Sohn et al., 2009). Briefly, muscles were minced under sterile conditions and incubated for 45 min at 37ºC in 1× Hanks’ balanced salt solution (HBSS) containing 35 mg of collagenase D (Roche, Indianapolis, IN) and 8.4 U of Dispase II (Roche) per gram of tissue with gentle trituration every 15 min. Digestion was terminated by addition of growth medium (1:1 mixture of DMEM-high glucose and Ham's F-10 medium; 20% fetal bovine serum, 5 ng/ml basic fibroblast growth factor, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamate), and the cell suspension was filtered through 100- and 40- μm filters. After centrifugation at 1100 rpm for 5 min, cells were resuspended in 1:7 mixture of growth medium and RBC lysis buffer (Invitrogen). Cells were then preplated on noncoated tissue culture (TC) dishes for 2 h; afterward, the cells in suspension were transferred to collagen type I (BD Biosciences, Bedford, MA)–coated TC dishes. C2C12 and L6 myoblasts were grown in DMEM supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamate. All cells were incubated in a humidified incubator with 5% CO2 at 37°C. The PLD1 inhibitor (VU0359595; Avanti Polar Lipids, Alabaster, AL) was used at 4 μM (Scott et al., 2009), and the inhibitor-containing medium was replenished every day. Aliquots of 16:0 Lyso PC (Avanti Polar Lipids) were evaporated under nitrogen in glass tubes, resuspended in HBSS, and added to the cultures to give a final concentration of 50 μM for 15 h.
Lipofectamine transfection and generation of PLD1-shRNA stable cell lines
GFP-PABD, GFP-PABD-mutant constructs, and shRNA construct for PLD1 (target nucleotides 547–565 of the open reading frame; Huang et al., 2005b) were transfected in L6 cells with Lipofectamine reagents (Invitrogen), following manufacturer's instructions. Stable PLD1-shRNA cell lines were selected 48 h later in growth medium containing G418 (1 mg/ml), and single colonies were picked for Western blot analysis and PLD enzyme activity assays. As a control, luciferase-shRNA was transfected into L6 cells as well. One control and two PLD1-shRNA sublines were chosen for subsequent experiments and maintained in medium supplemented with G418 (400 μg/ml). For rescue assays, adenoviral vectors containing PLD1 cDNA with wobble mutations was generated using the AdEasy system (Huang et al., 2005b). PLD1-shRNA myoblasts were infected at a multiplicity of infection of 100 plaque-forming units/cell in DMEM with 0.5% BSA overnight. The medium was replaced with DMEM containing 10% serum the next morning, and the cells were induced for myoblast differentiation after 48 h of infection. This protocol resulted in an infection efficiency of at least 80%.
Myoblast differentiation and fusion index determination
Myoblasts were plated in a 12-well plate at a density of 1 × 105 cells/well in growth medium. Differentiation was induced when the cells reach 90% confluency by switching to differentiation medium (DMEM with low glucose, 1 g/l, 2% horse serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamate). The fusion index was determined from three to five randomly chosen microscopic fields as the ratio of the number of nuclei within myotubes over the total number of nuclei.
Cocultures of myoblasts with nascent myotubes
Coculture of labeled myoblasts and myotubes has been described (Horsley et al., 2003; Sohn et al., 2009). In brief, L6 myoblasts and nascent myotubes were labeled with 5 μM CellTracker red and green CMFDA (Molecular Probes), respectively, for 30 min at 37°C, washed with PBS twice, and incubated with fresh growth medium for another 30 min at 37°C. Labeled cells were trypsinized, plated in 12-well plates at equal cell numbers, and cocultured for another 24 h. Cells were then fixed with 3.7% formaldehyde, stained with DAPI, and visualized with a Leica confocal microscope. Fusion between myoblasts with nascent myotubes was analyzed in double-stained myotubes with ≥3 nuclei. More than 300 nuclei per sample were counted for analysis.
PLD activity assay
PLD activities were determined by an in vivo transphosphatidylation reaction to measure the accumulation of phosphatidylbutanol in intact cells (Morris et al., 1997). A detailed protocol for the PLD activity assay in L6 cells has been described (Cazzolli et al., 2009). For PMA-stimulated PLD activity, myoblasts were incubated with 100 nM PMA and 0.3% butan-1-ol for 30 min.
Statistical analysis
All values are presented as the mean ± SD from at least three independent experiments unless otherwise stated. Two-tailed Student's t test and one-way ANOVA were used for statistical analyses with two groups and more than two groups, respectively. p < 0.05 was considered to represent statistically significant differences.
Supplementary Material
Acknowledgments
We thank Emanuela Gussoni for helpful discussion and Melissa Wu for critical reading of the manuscript. Funding for this work was generously provided by the American Heart Association (Grant SDG 0730285N to P.H.), The Key Technology Project of Jilin Province (20076023), and the Deutsche Forschungsgemeinschaft (Grant Ni556/8-1 to B.N.). The myosin heavy chain antibody (MF20, developed by Donald Fischman) was obtained from the Development Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biology, University of Iowa, Iowa City, IA.
Abbreviations used:
- CTX
cardiotoxin
- GA
gastrocnemius
- LPC
lysophosphatidylcholine
- PA
phosphatidic acid
- PABD
PA-binding domain
- PC
phosphatidylcholine
- PLD
phospholipase D
- TA
tibialis anterior.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0802) on November 26, 2014.
REFERENCES
- Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach AS, Enjalbert S, Comunale F, Bodin S, Vitale N, Charrasse S, Gauthier-Rouviere C. ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways. Mol Biol Cell. 2010;21:2412–2424. doi: 10.1091/mbc.E09-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Braun T, Bober E, Buschhausen-Denker G, Kohtz S, Grzeschik KH, Arnold HH. Differential expression of myogenic determination genes in muscle cells: possible autoactivation by the Myf gene products. EMBO J. 1989;8:3617–3625. doi: 10.1002/j.1460-2075.1989.tb08535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzolli R, Huang P, Teng S, Hughes WE. Measuring phospholipase D activity in insulin-secreting pancreatic beta-cells and insulin-responsive muscle cells and adipocytes. Methods Mol Biol. 2009;462:241–251. doi: 10.1007/978-1-60327-115-8_16. [DOI] [PubMed] [Google Scholar]
- Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci USA. 1989;86:1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Unveiling the mechanisms of cell-cell fusion. Science. 2005;308:369–373. doi: 10.1126/science.1104799. [DOI] [PubMed] [Google Scholar]
- Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo JA, van der Velden A, Tian H, Di Paolo G, Nieswandt B, et al. Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci Signal. 2012;5:ra79. doi: 10.1126/scisignal.2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nat Struct Mol Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft S. Signalling roles of mammalian phospholipase D1 and D2. Cell Mol Life Sci. 2001;58:1674–1687. doi: 10.1007/PL00000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen FS, Melikyan GB. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J Membrane Biol. 2004;199:1–14. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol. 1997;7:191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- Dixon RW, Harris JB. Myotoxic activity of the toxic phospholipase, notexin, from the venom of the Australian tiger snake. J Neuropathol Exp Neurol. 1996;55:1230–1237. doi: 10.1097/00005072-199612000-00006. [DOI] [PubMed] [Google Scholar]
- Elvers M, Stegner D, Hagedorn I, Kleinschnitz C, Braun A, Kuijpers ME, Boesl M, Chen Q, Heemskerk JW, Stoll G, et al. Impaired alpha(IIb)beta(3) integrin activation and shear-dependent thrombus formation in mice lacking phospholipase D1. Sci Signal. 2010;3:ra1. doi: 10.1126/scisignal.2000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MS, Lim YM, Quan W, Kim JR, Chung KW, Kang M, Kim S, Park SY, Han JS, Park SY, et al. Lysophosphatidylcholine as an effector of fatty acid-induced insulin resistance. J Lipid Res. 2011;52:1234–1246. doi: 10.1194/jlr.M014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JB, Johnson MA. Further observations on the pathological responses of rat skeletal muscle to toxins isolated from the venom of the Australian tiger snake, Notechis scutatus scutatus. Clin Exp Pharmacol Physiol. 1978;5:587–600. doi: 10.1111/j.1440-1681.1978.tb00714.x. [DOI] [PubMed] [Google Scholar]
- Haslett JN, Kang PB, Han M, Kho AT, Sanoudou D, Volinski JM, Beggs AH, Kohane IS, Kunkel LM. The influence of muscle type and dystrophin deficiency on murine expression profiles. Mamm Genome. 2005;16:739–748. doi: 10.1007/s00335-005-0053-8. [DOI] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–494. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- Horsley V, Pavlath GK. Forming a multinucleated cell: molecules that regulate myoblast fusion. Cells Tissues Organs. 2004;176:67–78. doi: 10.1159/000075028. [DOI] [PubMed] [Google Scholar]
- Huang F, Subbaiah PV, Holian O, Zhang J, Johnson A, Gertzberg N, Lum H. Lysophosphatidylcholine increases endothelial permeability: role of PKCalpha and RhoA cross talk. Am J Physiol Lung Cell Mol Physiol. 2005a;289:L176–185. doi: 10.1152/ajplung.00003.2005. [DOI] [PubMed] [Google Scholar]
- Huang P, Altshuller YM, Hou JC, Pessin JE, Frohman MA. Insulin-stimulated plasma membrane fusion of Glut4 glucose transporter-containing vesicles is regulated by phospholipase D1. Mol Biol Cell. 2005b;16:2614–2623. doi: 10.1091/mbc.E04-12-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WE, Elgundi Z, Huang P, Frohman MA, Biden TJ. Phospholipase D1 regulates secretagogue-stimulated insulin release in pancreatic beta-cells. J Biol Chem. 2004;279:27534–27541. doi: 10.1074/jbc.M403012200. [DOI] [PubMed] [Google Scholar]
- Jaafar R, Zeiller C, Pirola L, Di Grazia A, Naro F, Vidal H, Lefai E, Nemoz G. Phospholipase D regulates myogenic differentiation through the activation of both mTORC1 and mTORC2 complexes. J Biol Chem. 2011;286:22609–22621. doi: 10.1074/jbc.M110.203885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AW, Yi L, Natarajan A, Le Grand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komati H, Naro F, Mebarek S, De Arcangelis V, Adamo S, Lagarde M, Prigent AF, Nemoz G. Phospholipase D is involved in myogenic differentiation through remodeling of actin cytoskeleton. Mol Biol Cell. 2005;16:1232–1244. doi: 10.1091/mbc.E04-06-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky Y, Chernomordik LV, Kozlov MM. Lipid intermediates in membrane fusion: formation, structure, and decay of hemifusion diaphragm. Biophys J. 2002;83:2634–2651. doi: 10.1016/S0006-3495(02)75274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Halemani ND, Rammner B. Interplay between lipids and the proteinaceous membrane fusion machinery. Prog Lipid Res. 2008;47:461–469. doi: 10.1016/j.plipres.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Lawlor MW, Alexander MS, Viola MG, Meng H, Joubert R, Gupta V, Motohashi N, Manfready RA, Hsu CP, Huang P, et al. Myotubularin-deficient myoblasts display increased apoptosis, delayed proliferation, and poor cell engraftment. Am J Pathol. 2012;181:961–968. doi: 10.1016/j.ajpath.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leikina E, Melikov K, Sanyal S, Verma SK, Eun B, Gebert C, Pfeifer K, Lizunov VA, Kozlov MM, Chernomordik LV. Extracellular annexins and dynamin are important for sequential steps in myoblast fusion. J Cell Biol. 2013;200:109–123. doi: 10.1083/jcb.201207012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW. The structure and functions of human lysophosphatidic acid acyltransferases. Front Biosci. 2001;6:D944–953. doi: 10.2741/leung. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Scott SA, Lavieri R, Buck JR, Selvy PE, Stoops SL, Armstrong MD, Brown HA, Lindsley CW. Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I: Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg Med Chem Lett. 2009;19:1916–1920. doi: 10.1016/j.bmcl.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Yu L, Wu H, Shan Y, Guo J, Dang Y, Wei Y, Zhao S. Cloning and identification of the human LPAAT-zeta gene, a novel member of the lysophosphatidic acid acyltransferase family. J Hum Genet. 2003;48:438–442. doi: 10.1007/s10038-003-0045-z. [DOI] [PubMed] [Google Scholar]
- Liscovitch M, Czarny M, Fiucci G, Tang X. Phospholipase D: molecular and cell biology of a novel gene family. Biochem J. 2000;345:401–415. [PMC free article] [PubMed] [Google Scholar]
- Maltin CA, Harris JB, Cullen MJ. Regeneration of mammalian skeletal muscle following the injection of the snake-venom toxin, taipoxin. Cell Tissue Res. 1983;232:565–577. doi: 10.1007/BF00216429. [DOI] [PubMed] [Google Scholar]
- Meriane M, Roux P, Primig M, Fort P, Gauthier-Rouviere C. Critical activities of Rac1 and Cdc42Hs in skeletal myogenesis: antagonistic effects of JNK and p38 pathways. Mol Biol Cell. 2000;11:2513–2528. doi: 10.1091/mbc.11.8.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohandas N, Wyatt J, Mel SF, Rossi ME, Shohet SB. Lipid translocation across the human erythrocyte membrane. Regulatory factors. J Biol Chem. 1982;257:6537–6543. [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Morris AJ, Frohman MA, Engebrecht J. Measurement of phospholipase D activity. Anal Biochem. 1997;252:1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, de los Santos P, Neiman AM. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol Biol Cell. 2004;15:1802–1815. doi: 10.1091/mbc.E03-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter JD, Merriam AP, Leahy P, Gong B, Feuerman J, Cheng G, Khanna S. Temporal gene expression profiling of dystrophin-deficient (mdx) mouse diaphragm identifies conserved and muscle group-specific mechanisms in the pathogenesis of muscular dystrophy. Hum Mol Genet. 2004;13:257–269. doi: 10.1093/hmg/ddh033. [DOI] [PubMed] [Google Scholar]
- Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin K, Yu S, Roy S, Baylies MK. Myoblast fusion: when it takes more to make one. Dev Biol. 2010;341:66–83. doi: 10.1016/j.ydbio.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MG. Molecular mechanisms of PLD function in membrane traffic. Traffic. 2008;9:1233–1239. doi: 10.1111/j.1600-0854.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- Santini MT, Indovina PL, Cantafora A, Blotta I. The cesium-induced delay in myoblast membrane fusion is accompanied by changes in isolated membrane lipids. Biochim Biophys Acta. 1990;1023:298–304. doi: 10.1016/0005-2736(90)90426-o. [DOI] [PubMed] [Google Scholar]
- Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, Thomas AL, Armstrong MD, Arteaga CL, Lindsley CW, Brown HA. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer WD, Brown HA, Sternweis PC. Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D. Annu Rev Biochem. 1997;66:475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci USA. 2009;106:9274–9279. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman HH, Sweeney HL, Shrager JB, Maguire HC, Panettieri RA, Petrof B, Narusawa M, Leferovich JM, Sladky JT, Kelly AM. The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature. 1991;352:536–539. doi: 10.1038/352536a0. [DOI] [PubMed] [Google Scholar]
- Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2009;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicogne J, Vollenweider D, Smith JR, Huang P, Frohman MA, Pessin JE. Asymmetric phospholipid distribution drives in vitro reconstituted SNARE-dependent membrane fusion. Proc Natl Acad Sci USA. 2006;103:14761–14766. doi: 10.1073/pnas.0606881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N, Caumont AS, Chasserot-Golaz S, Du G, Wu S, Sciorra VA, Morris AJ, Frohman MA, Bader MF. Phospholipase D1: a key factor for the exocytotic machinery in neuroendocrine cells. EMBO J. 2001;20:2424–2434. doi: 10.1093/emboj/20.10.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelam MJ. Inositol phospholipid metabolism and myoblast fusion. Biochem J. 1983;214:77–82. doi: 10.1042/bj2140077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Radu CG, Yang LV, Bentolila LA, Riedinger M, Witte ON. Lysophosphatidylcholine-induced surface redistribution regulates signaling of the murine G protein-coupled receptor G2A. Mol Biol Cell. 2005;16:2234–2247. doi: 10.1091/mbc.E04-12-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Rubin BR, Orme CM, Karpikov A, Yu C, Bogan JS, Toomre DK. Dual-mode of insulin action controls GLUT4 vesicle exocytosis. J Cell Biol. 2011;193:643–653. doi: 10.1083/jcb.201008135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon MS, Chen J. PLD regulates myoblast differentiation through the mTOR-IGF2 pathway. J Cell Sci. 2008;121:282–289. doi: 10.1242/jcs.022566. [DOI] [PubMed] [Google Scholar]
- You JS, Lincoln HC, Kim CR, Frey JW, Goodman CA, Zhong XP, Hornberger TA. The role of diacylglycerol kinase zeta and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy. J Biol Chem. 2014;289:1551–1563. doi: 10.1074/jbc.M113.531392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeniou-Meyer M, Zabari N, Ashery U, Chasserot-Golaz S, Haeberle AM, Demais V, Bailly Y, Gottfried I, Nakanishi H, Neiman AM, et al. Phospholipase D1 production of phosphatidic acid at the plasma membrane promotes exocytosis of large dense-core granules at a late stage. J Biol Chem. 2007;282:21746–21757. doi: 10.1074/jbc.M702968200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.