This study is a step toward understanding the minimal requirements for MVB biogenesis and for ESCRT machinery in this process. Early ESCRT complexes (ESCRT-0 and -I) seem to have redundant functions in cargo sorting. Furthermore, vesicle formation seems to require cargo sorting in addition to late ESCRT machinery.
Abstract
The endosomal sorting complex required for transport (ESCRT) protein complexes function at the endosome in the formation of intraluminal vesicles (ILVs) containing cargo proteins destined for the vacuolar/lysosomal lumen. The early ESCRTs (ESCRT-0 and -I) are likely involved in cargo sorting, whereas ESCRT-III and Vps4 function to sever the neck of the forming ILVs. ESCRT-II links these functions by initiating ESCRT-III formation in an ESCRT-I–regulated manner. We identify a constitutively active mutant of ESCRT-II that partially suppresses the phenotype of an ESCRT-I or ESCRT-0 deletion strain, suggesting that these early ESCRTs are not essential and have redundant functions. However, the ESCRT-III/Vps4 system alone is not sufficient for ILV formation but requires cargo sorting mediated by one of the early ESCRTs.
INTRODUCTION
The multivesicular body (MVB) pathway is the major route for proteins destined for the lumen of the vacuole/lysosome. This pathway is conserved from yeast to human cells, where it functions in transmembrane protein degradation and trafficking of resident hydrolases to the vacuolar lumen (reviewed in Katzmann et al., 2002). MVBs are endosomes that internalize part of their limiting membrane as intralumenal vesicles (ILVs) along with transmembrane proteins destined for the vacuolar lumen. MVBs fuse with the vacuole and release their contents into the hydrolytic lumen of the vacuole. The formation of ILVs is catalyzed by the endosomal sorting complexes required for transport (ESCRTs). For most of the known ILV cargo proteins, ubiquitination serves as a sorting signal that is recognized by the ESCRT complexes.
The ESCRT machinery is composed of five multimeric complexes: ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and the Vps4 complex (reviewed in Henne et al., 2011). These soluble complexes are recruited from the cytosol to the MVB, where they function in cargo sorting and ILV formation. “Early-acting” ESCRT complexes (ESCRT-0 and ESCRT-I) possess numerous ubiquitin-binding domains and are therefore proposed to concentrate ubiquitinated cargo on the endosomal membrane (Bilodeau et al., 2002, 2003; Pornillos et al., 2002; Shih et al., 2002; Mizuno et al., 2003; Shields et al., 2009; Ren and Hurley, 2010). The later-acting ESCRT-III proteins form helical and circular filaments that can deform membrane both in vivo and in vitro (Hanson et al., 2008; Bodon et al., 2011; Henne et al., 2012). The Vps4 complex then disassembles ESCRT-III, an ATP-dependent activity that seems to drive ILV abscission (Babst et al., 1998, 2002a, b; Adell et al., 2014). Therefore these “late-acting” ESCRTs are implicated in the formation of ILVs. ESCRT-II links the cargo sorting activity of the early-acting ESCRTs to ILV formation mediated by the late-acting ESCRTs. However, mechanistic insights are lacking as to how ESCRT-II executes this process.
ESCRT-II is a heterotetrameric protein complex consisting of one copy of Vps36 and Vps22 and two copies of Vps25 (Hierro et al., 2004; Teo et al., 2004). The N-terminal GRAM-like ubiquitin-binding in Eap45 (GLUE) domain (Slagsvold et al., 2005) of Vps36 interacts with ubiquitin (either on cargo proteins or other ubiquitinated ESCRT proteins; Meyer et al., 2002; Alam et al., 2004), with the endosomal lipid phosphatidylinositol 3-phosphate (PI3P; Slagsvold et al., 2005; Teo et al., 2006), and with the Vps28 C-terminus of ESCRT-I (Teo et al., 2006; Gill et al., 2007). These interactions are predicted to play an important role in the recruitment of ESCRT-II to the MVB. Furthermore, an ESCRT-I–ESCRT-II supercomplex has been described that is proposed to function in the membrane deformation process leading to the formation of ILVs (Wollert and Hurley, 2010; Boura et al., 2012). Finally, the Vps25 subunits of ESCRT-II bind to Vps20, a myristoylated subunit of ESCRT-III (Ashrafi et al., 1998; Teo et al., 2004). This interaction between ESCRT-II and Vps20 is believed to initiate the assembly of the ESCRT-III filament (Teis et al., 2008).
In this study, we report that ESCRT-II is regulated not only by recruitment to the MVB but also by an ESCRT-I–mediated activation step. However, this regulatory function of ESCRT-I is not essential for the MVB pathway and can be partially bypassed by a constitutively active ESCRT-II mutant. Furthermore, we show that assembly of ESCRT-III, which is considered the core ESCRT machinery, is not sufficient for efficient ILV formation and that cargo concentration may be essential for this process.
RESULTS
Regulation of ESCRT-II by membrane recruitment
The early ESCRTs (ESCRT-0 and -I) contain numerous ubiquitin-binding domains and are thus likely to be involved in cargo sorting (reviewed in Henne et al., 2011). ESCRT-III and Vps4 have been proposed to function in membrane fission, severing the neck of the forming MVB vesicles. ESCRT-II links these early and late functions of the ESCRT machinery by initiating ESCRT-III formation in an ESCRT-I–dependent manner. This interaction between ESCRT-I and ESCRT-II seems to be regulatory, since the MVB sorting defect observed in an ESCRT-I–deletion strain can be partially bypassed by overexpressing ESCRT-II (Babst et al., 2002b). ESCRT-I binds to ESCRT-II via the Vps36 GLUE domain, a domain that also interacts with PI3P and ubiquitin (Figure 1A; Meyer et al., 2002; Alam et al., 2004; Slagsvold et al., 2005; Teo et al., 2006; Gill et al., 2007). These three GLUE domain interactions might regulate ESCRT-II function by aiding the recruitment of ESCRT-II from the cytoplasm to the MVB. Alternatively, the GLUE domain interactions might trigger a conformational change in ESCRT-II that switches ESCRT-II from an inactive to an active state. To differentiate between these two models, we first determined which of the GLUE domain interactions are important for endosomal recruitment of ESCRT-II by mutating the different interaction sites.
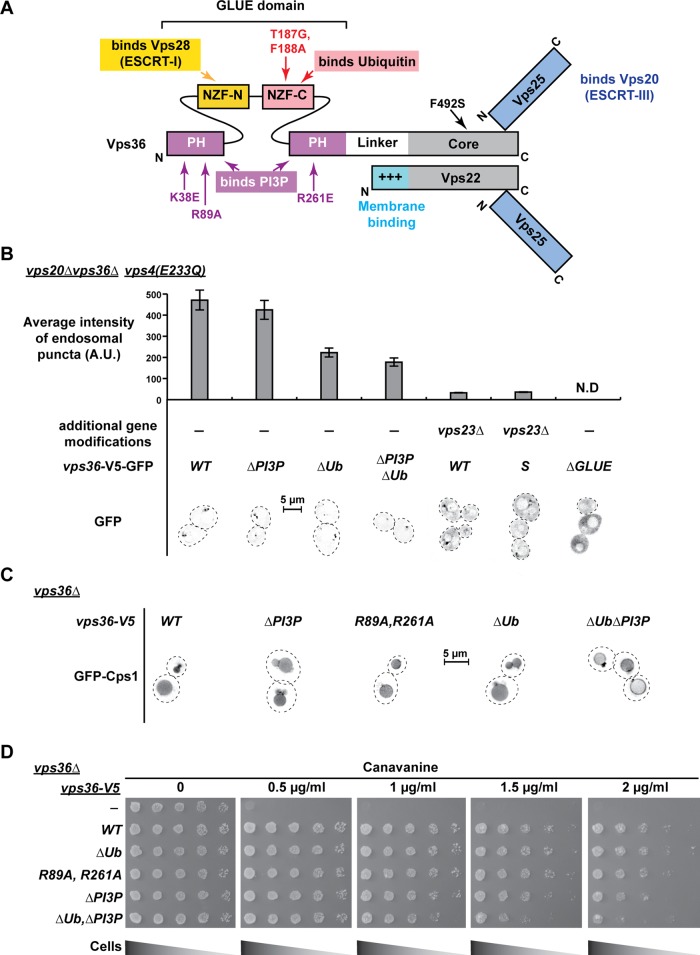
FIGURE 1:
ESCRT-II binding partners and their role in endosomal localization and function of ESCRT-II. All strains in this figure lack the genomic copy of VPS36. Tagged versions of wild-type (WT) or mutant copies of VPS36 are expressed from a low-copy CEN plasmid under native promoter. (A) A schematic of ESCRT-II domain architecture, interacting partners, and point mutations used in this study to disrupt these interactions. (B) Endosomal localization of ESCRT-II by fluorescence microscopy and its quantification. Microscopic images are presented in inverted gray scale, where black denotes GFP signal. The GFP puncta represent endosomes since they colocalized with FM4-64 as shown in Supplemental Figure S1C. n = 552, 544, 545, 558, 521, and 461 (in the order of strain presentation). Error bars denote the 95% confidence intervals over mean. N.D., not determined. (C) Localization of GFP-Cps1 in different vps36-mutant strains. (D) Growth assays on plates containing different concentrations of canavanine.
Previous studies suggested that endosomal localization of ESCRT-II is dependent on the GLUE–PI3P interaction (Teo et al., 2006) or the combination of both PI3P binding and the interaction of a positive-charged helix of Vps22 with negative-charged phospholipids (Im and Hurley, 2008). Furthermore, a study showed that deletion of ESCRT-I did not impair MVB recruitment of ESCRT-II (Babst et al., 2002b). These localization studies were performed either with just the GLUE domain or in absence of functional Vps4, an ATPase that recycles ESCRTs back to the cytoplasm. Lack of Vps4 results in accumulation of ESCRT-II–ESCRT-III complexes that are thoroughly anchored on the endosomal membrane (Babst et al., 2002b). Because of this trapping effect, even strong defects in ESCRT-II recruitment to the endosome would not be detectable. To circumvent these problems, we performed the localization studies using a green fluorescent protein (GFP)–tagged version of full-length Vps36 in a strain that lacked both the ESCRT-III subunit Vps20 (to prevent anchoring by ESCRT-III) and a functional Vps4 (expressing dominant-negative vps4(E233Q) to accumulate endosomal membranes) in addition to lacking genomic VPS36. Supplemental Figure S1A shows that the GFP-tagged version of Vps36 used in localization studies (VPS36-V5-GFP, where V5 acts as a linker) is functional and is able to complement the lack of genomic VPS36. ESCRT-II localization to endosomes was quantified by measuring the fluorescence intensity of the accumulated endosomes (class E compartments) found in ∼500 cells. Western blot analysis indicated that the different mutant forms of vps36-V5-GFP were expressed at levels similar to that of the wild-type fusion protein (Supplemental Figure S1B).
We first tested the contribution of the PI3P-binding domain to the recruitment of ESCRT-II by constructing the double mutant vps36(R89A,R261A)-V5-GFP, which, based on data from a previous report, was predicted to abolish PI3P binding (Teo et al., 2006). Surprisingly, this mutant form of ESCRT-II was as efficiently recruited to endosomes as the wild-type complex (unpublished data). Therefore a triple mutant was constructed (K38E, R89A, R261E) that replaced two key positively charged amino acids of the PI3P-binding pocket with glutamate (referred to as ∆PI3P). As observed for the double mutant, cells expressing vps36(∆PI3P)-V5-GFP showed wild-type like localization of ESCRT-II to endosomes (Figure 1B). Together these data suggested that the PI3P interaction does not significantly contribute to the endosomal recruitment of ESCRT-II.
Consistent with the localization data, we found that both vps36(R89A, R261A)-V5 and vps36(∆PI3P)-V5 complemented the MVB-sorting defect of a VPS36-deletion strain. Both Vps36 mutants were able to function in the efficient delivery of GFP-Cps1, a cargo of the MVB pathway (Odorizzi et al., 1998), to the vacuolar lumen (Figure 1C). Furthermore, both PI3P- binding mutants complemented the growth defect of a VPS36-deletion strain on canavanine-containing plates (Figure 1D). Canavanine, a toxic arginine analogue, is imported by yeast via the transporter Can1. Because Can1 is regulated in part by degradation via the MVB pathway, ESCRT mutants stabilize Can1 and thus show increased canavanine sensitivity compared with wild-type cells (Teis et al., 2010). Together our data indicate that interaction of Vps36 and PI3P plays a minor role in the localization and activity of ESCRT-II. At this point, we are not able to reconcile why our results contradict previously published data (Teo et al., 2006).
Next we analyzed the importance of the ubiquitin-binding site of Vps36 by introducing the mutations T187G and F188A (a double mutant we refer to as ∆Ub; Figure 1A). These mutations were previously shown to block binding of the Vps36 GLUE domain to ubiquitin (Shields et al., 2009). Localization studies of strains expressing vps36(∆Ub)-V5-GFP indicated that loss of ubiquitin binding caused a ∼50% drop of ESCRT-II association with the endosomal membrane (in vps20∆vps36∆ in the presence of vps4(E233Q); Figure 1B). However, this recruitment defect did not result in a MVB-sorting phenotype. Both the vacuolar delivery of GFP-Cps1 and growth on canavanine were found to be wild type–like in cells expressing vps36(∆Ub)-V5 (Figure 1, C and D). This result was consistent with previously published data (Shields et al., 2009).
The combination of the ubiquitin-binding mutations along with the PI3P-binding mutations (vps36(∆PI3P∆Ub)-V5-GFP) resulted in no additional ESCRT-II recruiting defect compared with the ubiquitin-binding mutation alone (Figure 1B). However, cells expressing vps36(∆PI3P∆Ub)-V5 exhibited partial missorting of GFP-Cps1 to the vacuolar membrane and marginally increased canavanine sensitivity (Figure 1, C and D), suggesting a possible regulatory role for ubiquitin and PI3P binding independent of their function in ESCRT-II recruitment. In addition, the lack of any observable GFP-Cps1 sorting defect with either ∆Ub or ∆PI3P alone suggests redundant functions for ubiquitin and PI3P binding.
To analyze the importance of ESCRT-I, we deleted VPS23, the gene encoding the major subunit of the ESCRT-I protein complex. In the strain vps23∆vps20∆vps36∆vps4(E233Q), GFP-tagged WT copy of ESCRT-II localized mainly to the cytoplasm, and endosomal recruitment was <10% compared to the recruitment in cells with functional ESCRT-I (Figure 1B). This result suggested that ESCRT-I provides a significant recruitment signal for ESCRT-II, most likely through direct interaction of the Vps28 C-terminus (ESCRT-I) with the Vps36 GLUE domain (ESCRT-II).
In summary, the analysis of the Vps36 GLUE-domain mutants indicated that the interaction between ESCRT-I and ESCRT-II plays a key role in the endosomal recruitment of ESCRT-II. Binding of the GLUE domain to ubiquitin further contributes to recruitment efficiency. In contrast, the interaction of the GLUE domain with PI3P seems to be less relevant for ESCRT-II localization or function.
ESCRT-I is not essential for ILV formation
The data presented so far suggested that the ESCRT-I interaction is important for localization of ESCRT-II. However, the observed suppression of the ESCRT-I phenotype by overexpression of ESCRT-II suggested that ESCRT-I is not an essential component of the ESCRT machinery (Babst et al., 2002b). To further test this idea, we carefully analyzed the functionality of the MVB pathway in strains deleted for VPS23 (loss of ESCRT-I) or VPS36 (loss of ESCRT-II). Using fluorescence microscopy, we confirmed the previously published results that both deletions cause severe missorting of GFP-Cps1 to the vacuolar membrane (Babst et al., 2002b; Alam et al., 2004; Figure 2A). To better quantify the sorting defects, we analyzed GFP-Cps1 sorting by Western blot using anti-GFP antibodies. When GFP-Cps1 enters the vacuolar lumen, the resident hydrolases clip the protein and release free GFP, resulting in a 25-kDa band (band b in Figure 2B). In contrast, the proteolytic clipping of GFP-Cps1, which mislocalizes to the vacuolar membrane and class E endosomes, causes the appearance of bands in the 30-kDa range (GFP fused to the transmembrane domain of Cps1; bands labeled a in Figure 2B). The ratio of the free GFP signal to the total GFP signal from proteolytic processing indicates the extent of GFP-Cps1 sorting into the MVB pathway (quantification in Figure 2B). This Western blot analysis indicated that the MVB-sorting defect in vps36∆ is more severe than that observed in the strain lacking VPS23 (Figure 2, A, #2 and 3, and B, #2 and 3). An additional analysis of the trafficking phenotype was performed using the canavanine-sensitivity assay. Consistent with the data of the GFP-Cps1 Western blot analysis, we observed slightly higher canavanine sensitivity in vps36∆ than with cells lacking VPS23 (Figure 3, #2 and 3). Together both assays indicated that the loss of VPS23 is less detrimental for the MVB pathway than the deletion of VPS36, suggesting that some ILV formation might still occur in vps23∆.
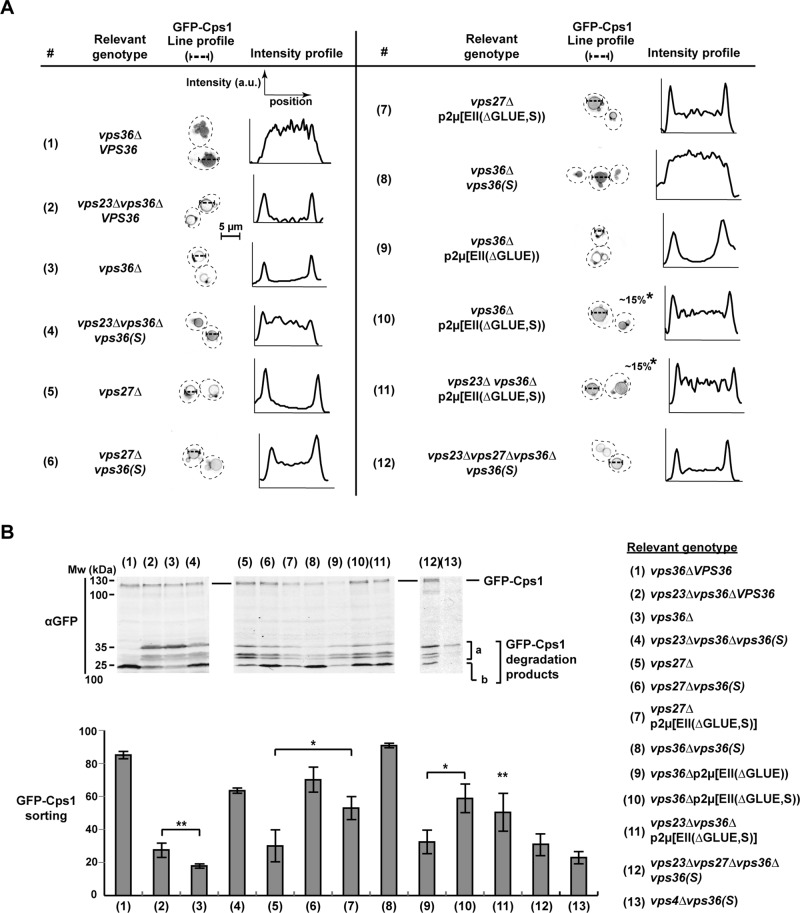
FIGURE 2:
Trafficking of GFP-Cps1 in different ESCRT mutants. All strains except #5–7 and 13 lack the genomic copy of VPS36. Strains #1, 2, and 5 express WT copy of VPS36 under its native promoter from a CEN plasmid, and strains #4, 6, 8, and 12 similarly express the mutant vps36(S). (A) Fluorescence microscopy showing the localization of GFP-Cps1. The images are presented in inverted gray scale. The intensities along the line profiles plotted as a graph give an estimate of the efficiency of GFP-Cps1 sorting into vacuolar lumen. Peaks at either ends of the graphs represents membrane intensity, whereas the intensities in between represent the lumen. Asterisk, only a subpopulation of cells show the particular phenotype. (B) Western blotting with total cell extracts of yeast strains expressing GFP-Cps1. Bottom, quantification of GFP-Cps1 sorting into vacuolar lumen from Western blots (example shown in B). GFP-Cps1 sorting = b/(a + b) percent. n = 4, 4, 4, 7, 3, 3, 3, 6, 3, 3, 7, 5, and 3 (in the order of strain presentation). Error bars represent 95% confidence intervals.
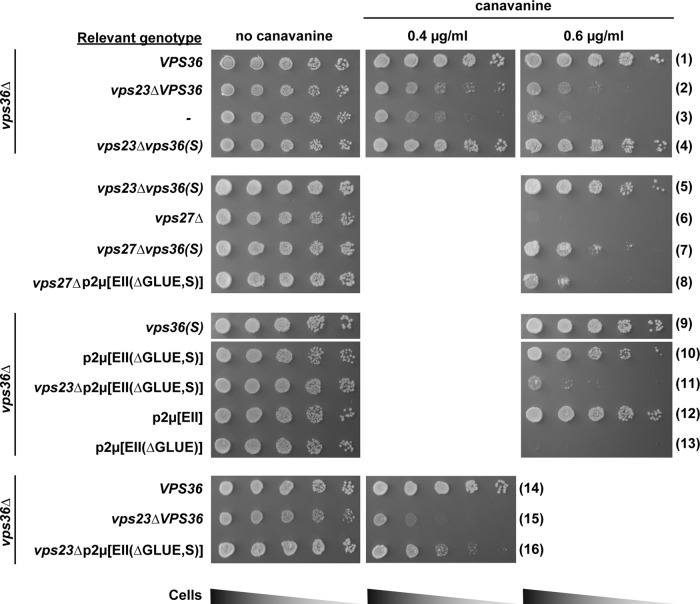
FIGURE 3:
Growth assay in the presence of canavanine. Serial dilutions of cells from each strain are plated on agar plates containing different concentrations of canavanine. All strains except #5–8 lack the genomic copy of VPS36. Strains #1, 2, 6, 14, and 15 express WT copy of VPS36 under its native promoter from a CEN plasmid, and strains #4, 5, 7, and 9 similarly express the mutant vps36(S).
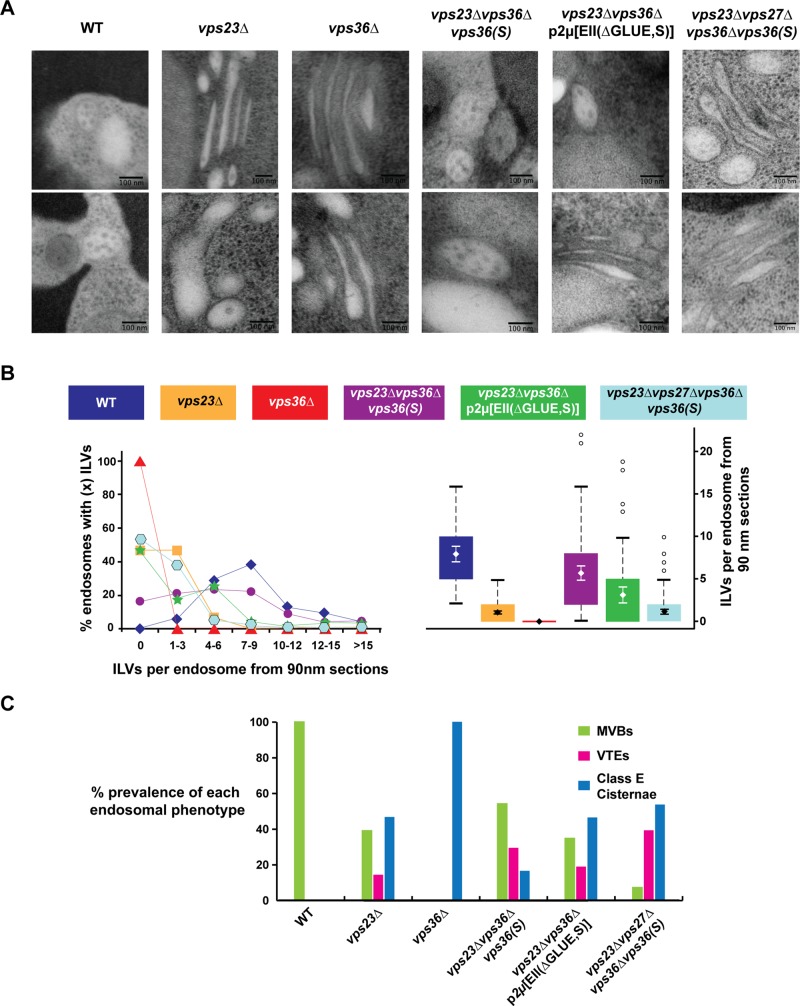
Transmission electron microscopy (TEM) was used to visualize the late endosomal compartments of the ESCRT-I– and ESCRT-II–deletion strains. The cross-sections of vps36∆ cells showed no MVBs but instead the stacked and flattened endosomal structures referred to as class E compartments (Figure 4, A and C; Rieder et al., 1996). In contrast, in cells deleted for VPS23, we observed a mixture of class E compartments and endosomes containing ILVs (Figure 4, A– C). However, compared with wild-type MVBs, the number of ILVs in these endosomal cross-sections was dramatically reduced (from approximately eight in wild type approximately one in vps23∆; Figure 4B). Furthermore, some of these ILV-containing endosomes displayed the flattened morphology of class E compartments (termed vesicular tubular endosomes [VTEs]; Nickerson et al., 2006). Together the results from the TEM analysis were consistent with the trafficking data and supported the notion that in contrast to ESCRT-II, ESCRT-I is not essential for ILV formation.
FIGURE 4:
TEM analysis of MVB formation. The strains vps23∆vps36∆vps36(S), vps23∆vps36∆p2μ[EII(∆GLUE,S)], and vps23∆vps27∆vps36∆vps36(S) lack the genomic VPS36 but instead express WT or mutant copy of VPS36/ESCRT-II under its native promoter from CEN/2μ plasmids. (A) Representative images of endosomes for each strain. (B) ILVs per endosome from 90-nm thin sections shown as a distribution and box plot. The error bars show the 95% confidence interval over the mean. All differences, except between the strains vps23∆ and vps23∆vps27∆vps36∆vps36(S), are significant (with p < 0.001). (C) Endosomal morphology distribution. Number of endosomal profiles counted for each strain: 52, 105, 34, 109, 69, and 97 (in the order of strain presentation).
Identification of a bypass suppressor of vps23∆
To study further the potential regulatory role of ESCRT-I on ESCRT-II function, we performed a genetic screen to identify an ESCRT-II mutant that suppresses the MVB-sorting defect caused by loss of ESCRT-I. We randomly mutagenized plasmid-encoded VPS36 and expressed these mutants in a strain deleted for VPS36 and VPS23. The resulting strains were grown in the presence of canavanine at a concentration that is lethal for vps23∆. This screen identified vps36(F492S) (Phe at 492 to Ser, referred to as vps36(S); Figure 1A) as a mutant that suppresses the canavanine sensitivity of vps23∆ (Figure 3, #4). Using a functional V5-tagged version of Vps36, Western blot analysis showed that the mutation did not affect the expression levels of the Vps36 protein (Supplemental Figure S2A).
The vps36(S) mutant gene was able to complement the phenotype of a VPS36 deletion strain (Figures 2, A, #8, and B, #8, and 3, #9, and Supplemental Figure S2B). Moreover, vps36(S) was found to be dominant. Even in the presence of wild-type VPS36, the suppressor mutant was able to partially rescue canavanine sensitivity of a vps23∆ strain (Supplemental Figure S3, #2). Furthermore, compared with vps23∆, TEM analysis of vps23∆vps36∆vps36(S) cells showed improvement of MVB morphology both in ILV number and endosomal morphology (fewer class E compartments and more globular endosomes; Figure 4, A– C). Canavanine sensitivity tests also indicated that vps36(S) suppressed not only the phenotype of vps23∆ but also the loss of any of the other ESCRT-I subunits (Supplemental Figure S3, #3–8). Furthermore, vps36(S) suppressed the MVB-sorting phenotype of a strain deleted for the ESCRT-0 subunit Vps27 (Figures 2, A, #5 and 6, and B, #5 and 6, and 3, #6 and 7). However, the suppression of vps27∆ by vps36(S) was greatly impaired in the absence of the ESCRT-I subunit VPS23 (Figures 2, A, #12, and B, #12, and 4, A– C), suggesting that at least one of the early ESCRTs has to be present for a functioning MVB pathway and that the functions of ESCRT-0 and ESCRT-I seemed to be redundant. Of importance, canavanine sensitivity caused by mutations in ESCRT-II or ESCRT-III subunits were not suppressed by vps36(S) (except vps36∆; Supplemental Figure S3, #9–16). Furthermore, GFP-Cps1 sorting with vps36(S) was severely defective in the absence of VPS4 (Figure 2B, #13).
Together the phenotypic analyses indicated that the vps36(S)-mutant form of ESCRT-II partially bypassed the need for the early ESCRTs (ESCRT-0 or ESCRT-I) but remained dependent on the function of the late ESCRTs. However, this suppression of the ESCRT-I–mutant phenotype was not due to improved endosomal recruitment of ESCRT-II in this strain. Localization studies in vps23∆vps20∆vps36∆vps4(E233Q) cells expressing vps36(S)-V5-GFP as the only copy of VPS36 indicated that the bypass mutation did not improve the amount of endosome-associated ESCRT-II over the wild-type copy (Figure 1B). This result suggested that ESCRT-II function was regulated not only by endosomal recruitment but also by a mechanism that switched membrane-associated ESCRT-II from an inactive to an active form. In the case of wild-type ESCRT-II, this activation might be triggered by the interactions of the Vps36 GLUE domain with ESCRT-I, PI3P, and ubiquitin. In contrast, the vps36(S)-mutant form of ESCRT-II seemed to be constitutively active and is therefore independent of the upstream factors for activation.
vps36(S) causes ESCRT-I–independent ESCRT-III assembly
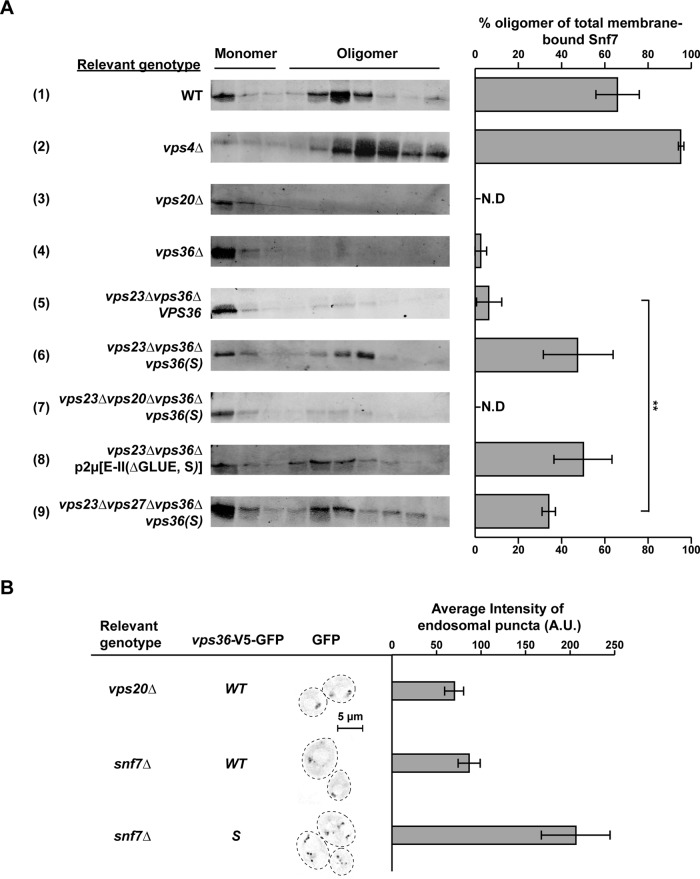
The observation that vps36(S) suppressed the trafficking and morphology defects of vps23∆ suggests that this mutant form of ESCRT-II is able to initiate ESCRT-III formation without the ESCRT-I interaction. We tested this prediction by analyzing the oligomeric state of the most common ESCRT-III subunit, Snf7, using a glycerol density gradient centrifugation assay (Figure 5A). In these experiments, the monomeric Snf7 protein was found in the top three fractions of the gradient, whereas the oligomers were identified in fractions 4–10 (consistent with previously published data; Teis et al., 2008). The ratio of Snf7 oligomers relative to total endosomally bound Snf7 was determined from three independent experiments. Similar to the previously published results, we found that wild-type cells contained 65% of the total Snf7 as large oligomers, whereas in cells lacking the ESCRT-III disassembly factor Vps4, close to 100% of Snf7 accumulated in the oligomeric form (Figure 5A, #1 and 2). The vps20∆ strain contained no ESCRT-III oligomers, consistent with the model in which Vps20 initiates Snf7 polymerization (Figure 5A, #3). In cells deleted for VPS36, only ∼3% of Snf7 was present in oligomers (Figure 5A, #4). Similarly, in cells lacking VPS23, Snf7 was predominantly monomeric (only 6% oligomers), indicating a defect in ESCRT-III assembly in this mutant strain (Figure 5A, #5). However, the presence of the bypass mutant vps36(S) in vps23∆ restored ESCRT-III oligomer assembly to almost wild-type levels (∼50% of Snf7 was present as oligomers; Figure 5A, #6). Vps20 was required for this suppression of ESCRT-III formation, as a vps23∆vps20∆vps36∆ strain expressing vps36(S) showed greatly impaired Snf7 oligomerization (Figure 5A, #7). Of interest, the presence of vps36(S) triggered ESCRT-III oligomerization even in the absence of both VPS27 and VPS23 (Figure 5A, #9), although the same strain was severely defective in MVB vesicle formation (Figure 4, A– C). This result suggested that ESCRT-III assembly is not sufficient to drive ILV formation.
FIGURE 5:
ESCRT-II–ESCRT-III interaction. (A) Snf7 oligomerization assay using glycerol density gradients. Bar graphs represent mean for each strain, where n = 3 for all strains. The error bars denote the 95% confidence over the mean. N.D., not determined. Strains #4–9 lack the genomic copy of VPS36. Strain #5 expresses the WT copy of VPS36 under its native promoter from a CEN plasmid, and strains #6, 7, and 9 similarly express the mutant vps36(S). Strain #8 overexpresses ESCRT-II(∆GLUE,S) from a 2μ plasmid. (B) Fluorescence microscopy showing endosomal localization of ESCRT-II and ESCRT-II(S) in the absence of VPS23 and VPS20/SNF7. n = 142, 210, and 159 (in the order of strain presentation). The strains additionally lack the genomic copy of VPS36, and the WT/mutant copy of VPS36-V5-GFP is expressed under the native promoter of VPS36 from a CEN plasmid. There was no significant difference in the overall expression of WT/mutant copy of VPS36-V5-GFP (as quantified by microscopy).
So far the data support the model in which the GLUE domain interactions drive both recruitment and activation of ESCRT-II. One possible mechanism for this regulation would be that the GLUE domain inhibits ESCRT-II functions unless the GLUE domain is bound to the upstream effectors ESCRT-I, PI3P, and ubiquitinated cargo. This type of autoinhibitory system is found in several ESCRT components, including ESCRT-III subunits and Vps4, which ensures that the ESCRTs are only assembling at the proper place and time (reviewed in Babst et al., 2011; McCullough et al., 2013).
ESCRT-II can function without the GLUE domain
To test whether the GLUE domain indeed functions as an autoinhibitory system, we deleted this domain of Vps36 (amino acids 1–289; Vps36(∆GLUE); Teo et al., 2006) and analyzed the consequences for localization and function of ESCRT-II. We expected that the GLUE-domain deletion would impair recruitment of ESCRT-II to MVBs. Localization studies of cells expressing Vps36(∆GLUE)-V5-GFP confirmed this prediction; GLUE-deleted ESCRT-II was almost exclusively found in the cytoplasm of the cell (Figure 1B). To overcome the recruitment defect, we overexpressed ESCRT-II(∆GLUE) by introducing a high-copy plasmid containing the genes of all three ESCRT-II subunits (vps36(∆GLUE), VPS22, and VPS25; referred to as p2μ[EII(∆GLUE)]). In contrast to overexpression of wild-type ESCRT-II (p2μ[EII]), p2μ[EII(∆GLUE)] showed minimal complementation for the impaired GFP-Cps1 trafficking of vps36∆ (Figure 2, A, #9, and B, #9) and no complementation for canavanine growth defect in the same strain (Figure 3, #12 and 13). However, introducing the suppressor mutation F492S into the ESCRT-II(∆GLUE) overexpression construct resulted in a clear rescue of the vps36∆ phenotype (p2μ[EII(∆GLUE,S)] in Figures 2, A, #10, and B, #10, and 3, #10). Furthermore, high levels of ESCRT-II(∆GLUE,S) were able to partially suppress the trafficking phenotypes caused by deletion of VPS23 (Figures 2, A, #11, and B, #11, and 3, #15 and 16), as well as the phenotypes of vps27∆ (Figures 2, A, #7, and B, #7, and 3, #8). Note that the effectiveness of suppression by p2μ[EII(∆GLUE,S)] varied dramatically from cell to cell, which is likely caused by the cell-to-cell variation of copy numbers of 2μ-type plasmids (Futcher and Cox, 1984). Consistent with the trafficking phenotypes seen in the foregoing, the presence of p2μ[EII(∆GLUE,S)] in a strain lacking VPS23 increased both the oligomerization of ESCRT-III (Figure 5, #8) and the formation of ILVs (approximately three per endosome; Figure 4, A and B). However, the endosomal morphology defect of vps23∆ did not improve by overexpressing ESCRT-II(∆GLUE,S) (Figure 4C).
In summary, the analysis of the GLUE domain–deleted ESCRT-II indicated that, although not essential for function, this domain plays an important role in the localization and activation of the ESCRT-II complex. However, the GLUE domain does not function as a simple inhibitory domain that blocks ESCRT-II activity when not engaged with ESCRT-I. The deletion of the GLUE domain did not result in a constitutively active ESCRT-II. The switch to the constitutively active state still required the suppressor mutation F492S. Therefore the GLUE domain seems to have activation rather than inhibitory functions.
Interaction of ESCRT-II with Vps20 might be regulated
ESCRT-II interacts with the downstream ESCRT machinery via Vps20. Like all other ESCRT-III subunits, monomeric Vps20 is believed to exist in an autoinhibited or closed state (Muzioł et al., 2006; Shim et al., 2007; Lata et al., 2008; Bajorek et al., 2009). To start the assembly of the ESCRT-III polymer, Vps20 has to be switched into an open conformation, a conformational change believed to be caused by binding to ESCRT-II. In one possible scenario, ESCRT-II activation by ESCRT-I triggers a conformational change that allows ESCRT-II to bind to Vps20, causing Vps20 to switch into the open state. To test this model, we quantified the endosomal localization of wild-type and F492S mutant ESCRT-II in strains lacking the ESCRT-I subunit Vps23 and the ESCRT-III subunit Snf7. In this double-mutant strain, the endosomal localization of ESCRT-II is independent of the upstream ESCRTs (because of VPS23 deletion) and the downstream ESCRT-III polymer (because of SNF7 deletion). Therefore the localization of ESCRT-II to the endosome in vps23∆snf7∆ should be sensitive to the interaction strength between ESCRT-II and Vps20. The analysis showed that in the presence of the activating mutation F492S, ESCRT-II localization to endosomes is increased (Figure 5B). This increase in ESCRT-II(S) localization was not observed in the absence of Vps20 (Figure 1B), supporting the idea that an increased interaction between ESCRT-II(S) and Vps20 might be responsible for the enhanced endosomal recruitment of activated ESCRT-II. This observation is consistent with the model that activation of ESCRT-II increases binding to Vps20, which then initiates the polymerization of ESCRT-III. However, we cannot exclude the possibility that the increased endosomal recruitment of ESCRT-II(S) is due to an unknown factor bridging Vps20 and ESCRT-II.
The linker region is not essential for ESCRT-II function
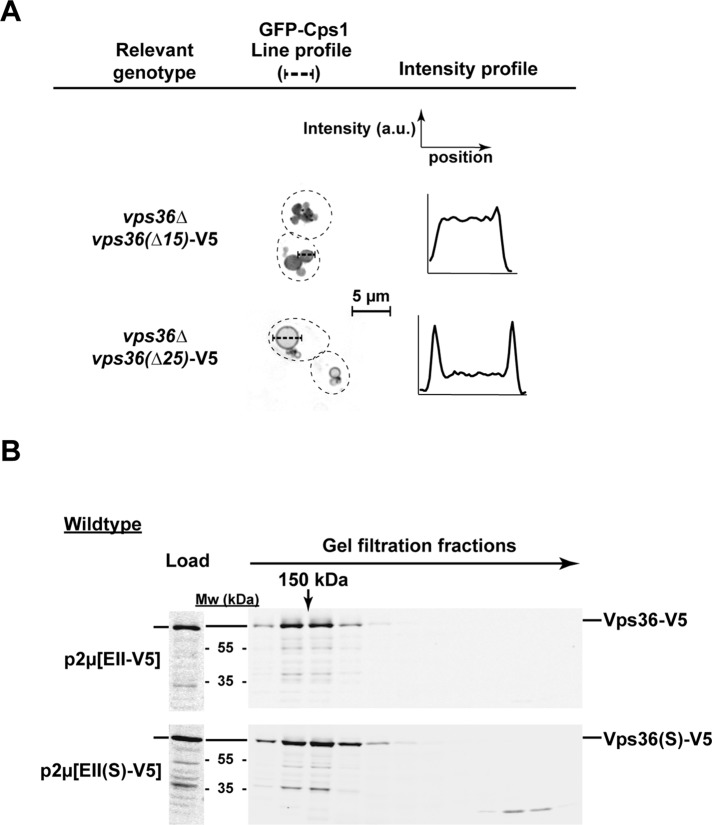
Based on comparisons with the human ESCRT-II structure, the yeast Vps36 core is connected to the GLUE domain via an ∼30–amino acid flexible linker region (Figure 1A; Im and Hurley, 2008). Our model predicts that the interaction of the GLUE domain with ESCRT-I is able to affect the binding of the ESCRT-II subunit Vps25 to Vps20. Therefore the linker region might not be as unstructured as predicted and thus might be able to relay the information regarding the GLUE domain interactions to the rest of the ESCRT-II complex. To test this idea, we deleted either 15 or 25 amino acids from the central section of the linker region of Vps36 and analyzed these mutant proteins in a complementation assay (in the absence of genomic VPS36). With ∆15 linker deletion, GFP-Cps1 localized mainly to the vacuolar lumen, indicating that this mutation did not impair ESCRT-II function. However, the larger deletion of the linker region (∆25) caused a GFP-Cps1 sorting defect (Figure 6A). These results suggested that the linker is not essential to relay GLUE domain interactions to the rest of the ESCRT-II complex but possibly provides a flexible connection between the GLUE domain and Vps36 core.
FIGURE 6:
Function of Vps36 linker domain. (A) Fluorescence microscopy of strains expressing GFP-Cps1. Images are presented in inverted gray scale. The intensities along the line profiles plotted as a graph give an estimate of the efficiency of GFP-Cps1 sorting into vacuolar lumen. The strains lack genomic VPS36 and the mutant forms of vps36 (having C-terminal V5 tag) are present on CEN plasmids under the native promoter. (B) Gel filtration analysis of a wild-type strain overexpressing V5-tagged ESCRT-II or ESCRT-II(S) from a 2μ plasmid.
The results from the linker deletions suggested that the GLUE domain might directly interact with other domains or subunits of ESCRT-II in order to regulate the interaction with Vps20. This model is supported by previous studies that indicated a compact structure of the ESCRT-II complex (in contrast to a longer, flexible connection between GLUE and the ESCRT-II core; Im and Hurley, 2008). To test whether ESCRT-II activation changes the overall structure of the complex, we compared by gel filtration the size of wild-type and F492S mutant forms of ESCRT-II (using antibodies against the V5 tag at the C-terminus of Vps36). The results indicated that both protein complexes have the same apparent molecular weight of ∼150 kDa, suggesting that activation does not cause a dramatic change in the Stokes radius of ESCRT-II (Figure 6B). Therefore it is likely that the GLUE domain is “tucked in” by binding to the ESCRT-II core in both the inactive and active conformations. However, ESCRT-II(S) was found to be more prone to degradation during the sample preparation, causing the formation of truncated, monomeric Vps36 fragments (Figure 6B).
DISCUSSION
ESCRT-0 and ESCRT-I contain numerous ubiquitin-binding sites. In addition, ESCRT-0 seems to be able to form higher-ordered oligomers on the endosomal membrane (Mayers et al., 2011). These properties make these early ESCRTs prime candidates for the role of a cargo-capturing system. ESCRT-III polymers together with Vps4 seem to execute the membrane abscission reaction during both cytokinesis and HIV viral budding. Therefore it is likely that ESCRT-III/Vps4 performs the same function at the MVB, constricting and severing the neck of the forming ILV. Besides its role in initiating ESCRT-III assembly, little is known about the function of ESCRT-II in ILV formation. Based on structural studies, a model has been proposed in which the ESCRT-I–ESCRT-II supercomplex is involved in deforming the membrane to drive the invagination of the forming ILV (Wollert and Hurley, 2010; Boura et al., 2012). However, direct evidence for this role of ESCRT-II in membrane bending is lacking.
A surprising conclusion from our study is the observation that ESCRT-0 and ESCRT-I are not essential for the MVB pathway but have redundant roles, most likely in cargo sorting and concentration. Even the GLUE domain of ESCRT-II, which binds to ESCRT-I, ubiquitinated cargo, and the lipid PI3P, is not essential for the MVB pathway. In contrast, the ESCRT-II–mediated initiation of ESCRT-III assembly is critical for this pathway. These observations suggested that the interactions of ESCRT-II with the upstream ESCRT-I complex are not essential for vesicle formation (e.g., driving membrane invagination) but play a regulatory role in ensuring ESCRT-III formation at the proper time and place. Our data suggest that the regulation of ESCRT-II function is mediated by both the recruitment of the complex from the cytoplasm to the MVB and an activation mechanism. The key for both of these regulation mechanisms is the interaction between the GLUE domain and ESCRT-I. The binding of the GLUE domain to ubiquitin and PI3P plays only a minor role in ESCRT-II regulation (Table 1).
TABLE 1:
Summary of phenotypic analyses.
| ESCRT-II or strain | ESCRT-II localization | Cargo sorting | ILV formation | ESCRT-III formation |
|---|---|---|---|---|
| WT | + | + | + | + |
| vps36∆ | − | − | − | − |
| vps23∆ | − − + | − − + | − − + | − |
| ∆PI3P | + | + | nd | nd |
| ∆Ub | + | + | nd | nd |
| S | + | + | + | nd |
| vps23∆, S | − − + | + − | + − | + |
| vps27∆, S | nd | + − | nd | nd |
| vps23∆, vps27∆, S | nd | − − + | − − + | + − |
| vps23∆, 2 μ[∆GLUE,S] | nd | + − | + − | + |
nd, not determined.
We identified the amino acid exchange F492S in Vps36 as a mutation that seemed to result in a constitutively active form of ESCRT-II (referred to as ESCRT-II(S)). The localization of ESCRT-II(S) to endosomes was not increased compared with the wild-type complex, indicating that recruitment and activation of ESCRT-II are independent processes. ESCRT-II(S) was able to suppress the MVB-sorting phenotypes of an ESCRT-0 or ESCRT-I deletion by triggering ESCRT-III polymer formation in the absence of these upstream factors. ESCRT-III polymerization is initiated by the interaction between ESCRT-II and Vps20. Our ESCRT-II localization data indicated increased Vps20-dependent endosomal recruitment of ESCRT-II(S) compared with the wild-type ESCRT-II complex (Figure 7A). One interpretation of this observation is that activation of ESCRT-II increases the interaction strength between the ESCRT-II subunit Vps25 and Vps20. This increased interaction with Vps25 might cause Vps20 to switch from the inactive, closed conformation to the polymer-forming, open conformation (Muzioł et al., 2006; Shim et al., 2007).
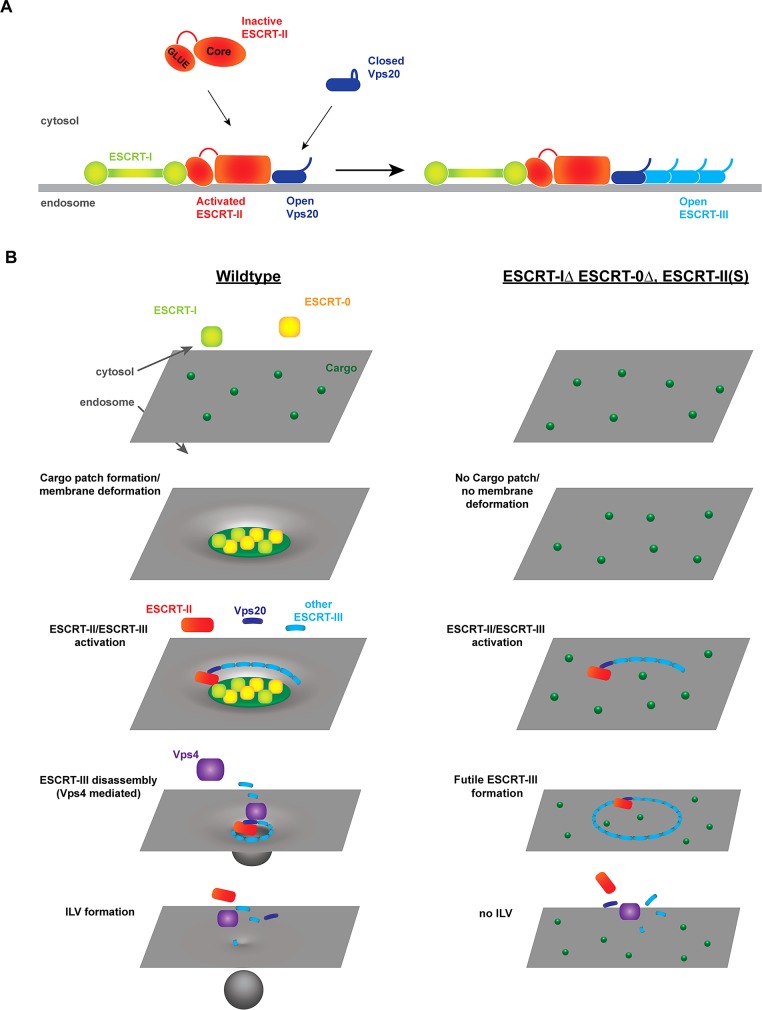
FIGURE 7:
A model for ESCRT-II–mediated coupling of ILV formation with cargo sorting. The early ESCRT complexes (ESCRT-0 and ESCRT-I) concentrate transmembrane cargo along with lipids interacting with the cargoes. The resulting cargo–ESCRT–lipid patch deforms the endosomal membrane. Subsequent recruitment and activation of ESCRT-II causes ESCRT-III polymerization. Vps4-mediated constriction of ESCRT-III results in deepening of the membrane invagination and ultimately fission of the ILV neck. In the absence of ESCRT-0 and ESCRT-I, ESCRT-II(S) randomly initiates ESCRT-III polymerization. However, in the absence of a cargo patch, Vps4-mediated remodeling of ESCRT-III does not result in ILV formation.
Another interesting observation from this study is that the constitutively active ∆GLUE version of ESCRT-II (ESCRT-II(∆GLUE,S)) efficiently rescued ESCRT-III polymerization in vps23∆ strain but showed only a marginal improvement in ILV formation in the same strain (Table 1). This difference in phenotypic suppression suggested that some of the ESCRT-III polymers formed by ESCRT-II(∆GLUE,S) were unproductive, most likely because ESCRT-III polymerization was triggered at the wrong place and/or time. In addition, this result suggested that ESCRT-III alone is not sufficient to deform the membrane and cause vesicle budding. The concentration of cargo into a cargo–ESCRT patch at the endosome might play an important role in initiating vesicle formation. Consistent with this idea, ESCRT-II(S) exhibited very poor ILV formation and GFP-Cps1 sorting in a strain lacking both ESCRT-0 and ESCRT-I despite showing substantial levels of ESCRT-III polymerization (Table 1). Furthermore, a previous study indicated that the presence of ubiquitinated cargo at the endosome is essential for ILV formation (MacDonald et al., 2012). Together our data suggest that ESCRT-0 and ESCRT-I have redundant functions and that at least one of these cargo-sorting complexes has to be present to drive ILV formation (Table 1).
In summary, we propose a model in which the assembly of a cargo–ESCRT patch is an essential first step toward ILV formation (Figure 7B). A cargo patch is formed by the interaction of ESCRT-0 and ESCRT-I with transmembrane proteins (ubiquitin dependent or independent). Because transmembrane proteins surround themselves with specific lipids, the cargo patch is expected to have a unique lipid composition compared with the rest of the endosomal membrane. We propose that this assembly of ESCRTs, cargo, and lipids causes an inward deformation of the cargo patch (discussed in Babst, 2011). The ESCRT-I component of the cargo patch, together with ubiquitinated cargo and PI3P, recruits ESCRT-II by interacting with the GLUE domain. This interaction also changes the ESCRT-II conformation into the active state, which in turn increases the affinity of ESCRT-II to Vps20 (Figure 7A). ESCRT-II–bound Vps20 changes its conformation from the closed to the open state, which recruits Snf7 and initiates the polymerization of the ESCRT-III complex. Based on previous in vitro studies, the ESCRT-III polymer might form a corral that defines the boundaries of the forming vesicle (Henne et al., 2012). Finally, ESCRT-III, together with the Vps4 complex, drives the constriction of the membrane neck, which ultimately causes the abscission of the forming ILV (Figure 7B).
In this model, constitutively active ESCRT-II(S) can partially bypass the loss of ESCRT-I by interacting with the ubiquitinated cargo, which ensures ESCRT-III formation at the proper place. However, deletion of the GLUE domain of ESCRT-II(S) results in an ESCRT-II complex that lacks interaction with the cargo patch. As a consequence, ESCRT-III polymerization occurs randomly on the endosomal membrane, and only by chance will ESCRT-III form close to a cargo patch, explaining the inefficiency in ILV formation in this mutant strain. Similar decoupling of ESCRT-III polymerization from ILV formation occurs with ESCRT-II(S) in a strain lacking both ESCRT-0 and ESCRT-I complexes (Figure 7B).
The data presented here add to the growing evidence that the ESCRT machinery does not cause ILV formation by physically invaginating the membrane but instead by stabilizing and constricting the neck of an existing invagination. Membrane constriction is mediated by ESCRT-III and Vps4, the essential part of the ESCRT machinery that is common to all ESCRT-mediated processes, including cytokinesis, viral budding, and plasma membrane repair. Therefore the essential ESCRTs are, first and foremost, a membrane-abscission machinery.
MATERIALS AND METHODS
Strains, media, and plasmids
The strains and plasmids used in this study are described in Table 2. Yeast gene knockouts were constructed as previously described (Longtine et al., 1998). Yeast strain KFY7 was constructed in 6210.1 WT yeast by replacing VPS36 with URA3 gene and VPS20 with HIS3 gene. SKY1 strain was constructed by introducing URA3 gene into the VPS36 locus of EEY6-2. SKY8, SKY14, and SKY15 strains were constructed by further knocking out the genomic copy of VPS20, VPS27, or SNF7 with G418 cassette (KanMX6) in SKY1. Plasmids were constructed using PCR/conventional restriction enzymes, the sequence and ligation-independent cloning (SLIC; Li and Elledge, 2007) method, or by QuikChange protocol. pSD2 and pSK211 were constructed by inserting the previously described GFP-Cps1 construct (Odorizzi et al., 1998) into pRS415 and pRS414 vectors, respectively. Similar vector swap was performed to construct pMB187 from the previously described pMB66 (Babst et al., 1998). F492S mutation in VPS36 was isolated from the canavanine screen by using homologous recombination of error-prone PCR product of VPS36 and a gapped plasmid of pMB131 that lacked most of the coding sequence of VPS36 but retained the 5′ and 3′ untranslated regions. pSK163 was constructed by replacing the promoter region of the mutant VPS36 isolated in the screen with wild-type promoter region. F492S mutation was introduced into pMB175 and its derivatives by QuikChange protocol using Phusion polymerase enzyme. Similar QuikChange protocol was used to make ubiquitin and PI3P mutants; linker deletions and V5- and GFP-tagged VPS36 in centromere (CEN) plasmids were derived from pMB131. V5 tagging of VPS36 in pMB175 was performed by SLIC technique (using two PCR products for the vector and ESCRT-II sequences, whereas the V5 sequence was reconstituted by annealing two oligonucleotides). The GLUE domain deletions in pMB131, pMB175 and their derivatives were similarly made using SLIC technique using two PCR products for the vector and ESCRT-II sequences (without GLUE domain). Yeast strains were grown in rich yeast extract/peptone/dextrose (YPD) medium or in the appropriate synthetic drop-out media in yeast nitrogen base (YNB; Becton, Dickinson and Company, Sparks, MD) as published (Sherman et al., 1979). In every set of experiments, strains were grown in the same growth medium.
TABLE 2:
Strains and plasmids used in this study.
| Strain or plasmid | Descriptive name | Genotype or description | Reference or source |
|---|---|---|---|
| Strains | |||
| SEY6210 | WT | MATα leu2-3112 ura3-52 his3-∆200 trp1-∆901 lys2-801 suc2-∆9 GAL | Robinson et al. (1988) |
| KFY7 | vps36∆vps20∆ | SEY6210.1, VPS36::URA3, VPS20::HIS3 | This study |
| SKY8 | vps23∆vps36∆vps20∆ | SEY6210, VPS23::HIS3, VPS36::URA3, VPS20::G418 | This study |
| MBY30 | vps36∆ | SEY6210, VPS30::HIS3 | Babst et al. (2002b) |
| SKY1 | vps23∆vps36∆ | SEY6210, VPS23::HIS3, VPS36::URA3 | This study |
| MBY21 | vps27∆ | SEY6210, VPS27::HIS3 | Odorizzi et al. (1998) |
| EEY6-2 | vps23∆ | SEY6210, VPS23::HIS3 | Babst et al. (2000) |
| MBY3 | vps4∆ | SEY6210, VPS4::TRP1 | Babst et al. (1997) |
| EEY2-1 | vps20∆ | SEY6210, VPS20::HIS3 | Babst et al. (2002a) |
| SKY15 | vps23∆vps36∆snf7∆ | SEY6210, VPS23::HIS3, VPS36::URA3, SNF7::G418 | This study |
| SKY14 | vps23∆vps36∆vps27∆ | SEY6210, VPS23::HIS3, VPS36::URA3, VPS27::G418 | This study |
| MCY23 | vps28∆ | SEY6210, VPS28::HIS3 | Curtiss et al. (2007) |
| DKY48 | vps37∆ | SEY6210, VPS37::HIS3 | Katzmann et al. (2001) |
| MBY65 | mvb12∆ | SEY6210, MVB12::HIS3 | Curtiss et al. (2007) |
| MBY31 | vps22∆ | SEY6210.1, VPS22::HIS3 | Babst et al. (2002b) |
| BWY101 | vps25∆ | SEY6210, VPS25::HIS3 | Babst et al. (2002b) |
| EEY2-1 | vps20∆ | SEY6210, VPS20::HIS3 | Babst et al. (2002a) |
| EEY9 | snf7∆ | SEY6210, SNF7::HIS3 | Babst et al. (2002a) |
| BWY102 | vps24∆ | SEY6210, VPS24::HIS3 | Babst et al. (1998) |
| MBY28 | vps2∆ | SEY6210, VPS2::HIS3 | Babst et al. (2002a) |
| Plasmids | |||
| pSK212 | VPS36-V5-EGFP | VPS36-V5-EGFP (pRS414) | This study |
| pSK243 | vps36(∆PI3P)-V5-GFP | vps36(K38E,R89A,R261E)-V5-EGFP (pRS414) | This study |
| pSK253 | vps36(R89A,R261A)-V5-GFP | vps36(R89A,R261A)-V5-EGFP (pRS414) | This study |
| pSK239 | vps36(∆Ub)-V5-GFP | vps36(T187G,F188A)-V5-EGFP (pRS414) | This study |
| pSK240 | vps36(∆Ub∆PI3P)-V5-GFP | vps36(K38E,R89A,R261E,T187G,F188A)-V5-EGFP (pRS414) | This study |
| pSK241 | vps36(S)-V5-GFP | vps36(F492S)-V5-EGFP (pRS414) | This study |
| pSK215 | vps36(∆GLUE)-V5-GFP | vps36(∆1-289)-V5-EGFP (pRS414) | This study |
| pMB187 | vps4(E233Q) | vps4(E233Q) (pRS415) | Babst et al. (1998) |
| pSK179 | VPS36-V5 | VPS36-V5 (pRS414) | This study |
| pSK213 | vps36(∆PI3P) | vps36(K38E,R89A,R261E)-V5 (pRS414) | This study |
| pSK250 | vps36(R89A,R261A) | vps36(R89A,R261A)-V5 (pRS414) | This study |
| pSK183 | vps36(∆Ub) | vps36(T187G,F188A)-V5 (pRS414) | This study |
| pSK222 | vps36(∆Ub∆PI3P) | vps36(K38E,R89A,R261E,T187G,F188A)-V5 (pRS414) | This study |
| pSD2 | GFP-Cps1 | GFP-Cps1 (pRS415) | Odorizzi et al. (1998) |
| pMB131 | VPS36 | VPS36 (pRS414) | Alam et al. (2004) |
| pSK163 | vps36(S) | vps36(F492S) (pRS414) | This study |
| pSK180 | vps36(S)-V5 | vps36(F492S)-V5 (pRS414) | This study |
| pMB196 | CFP-Cps1 | CFP-Cps1 (pRS425) | This study |
| pSK143 | p2μ[EII-V5] | VPS36-V5, VPS22-HA, VPS25 (pRS425) | This study |
| pMB175 | p2μ[EII] | VPS36, VPS22-HA, VPS25 (pRS425) | Babst et al. (2002b) |
| pSK165 | p2μ[EII(∆GLUE)] | vps36(∆1-289), VPS22-HA, VPS25 (pRS425) | This study |
| pSK223 | p2μ[EII(∆GLUE,S)] | vps36(∆1-289,F492S)-V5, VPS22-HA, VPS25 (pRS425) | This study |
| pSK197 | vps36(∆15) | vps36(∆301-315)-V5 (pRS414) | This study |
| pSK200 | vps36(∆25) | vps36(∆301-325) (pRS414) | This study |
| pSK147 | p2μ[EII(S)] | vps36(F492S)-V5, VPS22-HA, VPS25 (pRS425) | This study |
Fluorescence microscopy and quantification
Yeast cells were grown in YNB selective medium (Anachem Bio101 Systems, Luton, UK, and USBiological, Swampscott, MA) to logarithmic growth phase (0.7 OD600). Fluorescence microscopy was performed at 100× magnification on a deconvolution microscope (DeltaVision; Applied Precision, Issaquah, WA) using the accompanying software. Images were taken as Z-stacks and deconvolved.
For quantification of endosomal localization of Vps36-V5-GFP, a projection of the Z-stacks (25 stacks, 5 μm total) of 500 cells was analyzed. The brightest intensity of puncta for each cell was recorded after background subtraction. For cells showing no discernible puncta, the value was recorded as zero. The error bars denote the 95% confidence interval over the mean for each strain.
For staining with FM4-64, cells in logarithmic growth phase were incubated with the dye (at a final concentration of 1 μg/ml) at 30°C for 15 min. The cells were washed with fresh growth medium and incubated at 30°C for 30 min. Manders’ colocalization coefficient (M1) was used to quantify colocalization (Dunn et al., 2011). Deconvolved images from Z-stacks containing multiple cells were converted into a two-dimensional array of pixel intensities before background subtraction. For background subtraction, local background (median of a 40 × 40 region around the particular pixel) for each pixel was calculated. The local background value for each pixel was then subtracted from the respective pixel value. An additional small value was subtracted from each pixel to remove signal from nonspecific regions. M1 was calculated over several images for each strain and presented as mean ± range for 95% confidence interval.
Canavanine plate assay
Yeast cells were grown in YNB (selective media wherever necessary) to logarithmic growth phase. After removal of the growth medium, cells were resuspended in 1 M sorbitol to final OD600 of 0.5. Serial dilutions were made for each strain in 96-well plates, so that the first column had 0.5 OD600 cells and every successive column was five times diluted from the preceding column. For each strain, 3 μl of cells were plated from the foregoing dilutions. Agar plates containing canavanine (Sigma-Aldrich, St. Louis, MO) were made with 2% glucose and 6.7 g/l YNB without arginine. In addition, uracil (20 μg/ml), leucine (30 μg/ml), histidine (20 μg/ml), adenine (20 μg/ml), lysine (30 μg/ml), and tryptophan (20 μg/ml) were provided wherever necessary to support the growth of auxotrophic strains.
Western blot analysis
For quantification of GFP-Cps1 sorting into vacuole and for visualizing expression of the various tagged-Vps36, yeast cell extracts were obtained from strains grown to logarithmic phase. Cells were pelleted, resuspended in SDS–PAGE sample buffer (2% SDS, 0.1 M Tris, pH 6.8, 10% glycerol, 0.01% bromophenol blue, 5% β-mercaptoethanol), lysed using glass beads, and boiled for 5 min at 95°C. For Western blotting, monoclonal antibodies against V5 and GFP (anti-V5 antibody from AbD Serotec [Kidlington, UK] and anti-GFP antibody from Roche Diagnostics [Basel, Switzerland]) were used at 1:2500 dilutions. The anti-Snf7 antibody used was previously described (Babst et al., 1998). IRDye-conjugated secondary antibody were purchased from LI-COR Biosciences (Lincoln, NE), and Western blots were imaged using the accompanying Odyssey Imaging System.
Transmission electron microscopy
Yeast were grown at 30°C to logarithmic phase, transferred to aluminum hats, high-pressure frozen, and placed in vials with a freeze substitution solution of 0.1% uranyl acetate and 2% glutaraldehyde in anhydrous acetone. Vials were placed in an automatic freeze substitution device (EM AFS2; Leica, Buffalo Grove, IL) at –140°C and warmed to –80°C over 24 h. Cells were extracted from the hats at −80°C, placed in chilled tubes with fresh freeze substitution solution for 48 h, and then warmed to −60°C over 20 h. Fixative was replaced with Lowicryl HM20 over the next 96 h with three acetone washes, three washes of increasing HM20:acetone (1:3, 1:1, and 3:1), and six washes with 100% HM20. Polymerization with ultraviolet light began at −60°C for 12 h and continued up to 20°C over 36 h. The 90-nm sample thin sections were placed on rhodium-plated Formvar-coated copper grids and viewed with a Philips (Eindhoven, The Netherlands) CM10 transmission electron microscope, and images were processed using ImageJ64.
Glycerol density gradient centrifugation assay
This assay was adapted from Teis et al. (2008). Before harvesting cells and preparing cell lysates for the assay, we prepared 10–40% linear glycerol gradients in Beckman Ultra-Clear SW-41 tubes (14 × 89 mm; 344059; Beckman Coulter, Brea, CA). We prepared 10, 20, 30, and 40% stocks of glycerol in phosphate-buffered saline (PBS) containing 0.5% Tween 20 and protease inhibitors. Gradients were made in SW-41 tubes by carefully layering the different glycerol stock solutions in decreasing order of glycerol concentration (2.5 ml of each of 40, 30, 20, and 10% glycerol solutions), so that 40% glycerol solution was at the bottom of the tube. The 10% glycerol solution was made in lysis buffer containing 1% Triton X-100. The resulting 10–40% step gradients were converted into 10–40% linear gradients by spinning at 30,000 rpm for 1 h or until the step interfaces were no longer visible. To prepare cell extracts for the gradient assay, 30 OD600 equivalents of logarithmic growth phase cells were harvested, washed, spheroplasted, and Dounced for each experiment. Unbroken cells (P5 fraction) were removed by spinning them out for 3 min at 3000 rpm. Supernatants were collected and spun for 10 min at 13,000 rpm to separate the endosomal fraction (P13). Proteins in the P13 fraction, which are mostly membrane associated, were solubilized in 1 ml PBS containing 0.5% Tween 20. Subsequently, the solubilized P13 fraction was passed five times through a 25G5/8 needle and spun again for 10 min at 13,000 rpm. The resulting supernatants were carefully isolated for gradient analysis. Simultaneously, two different sizing control standards were prepared, one containing aldolase (159 kDa) and ferritin (440 kDa), and the other containing catalase (247 kDa) and thyroglobulin (667 kDa), with each protein at 1.67 mg/ml final concentration in 1.2 ml of lysis buffer (also containing 1% Triton X-100). The stock solutions of sizing standard proteins were prespun at 13,000 rpm before use to remove aggregates. The two sizing standards and the experimental samples (solubilized P13 fractions) were added to the top of different linear glycerol gradients and subjected to centrifugation at 100,000 × g (30,000 rpm) for 4 h at 4°C. After the spin, 10 fractions of 1 ml each were manually collected from top to bottom of each resulting gradient. Fractions collected from the P13-gradients were trichloroacetic acid precipitated and analyzed by Western blotting, whereas those collected from sizing standard gradients were analyzed by absorbance at different wavelengths (OD280 for aldolase, catalase, and thyroglobulin and OD420 for ferritin).
Gel filtration analysis
Yeast cells were spheroplasted and lysed in PBS containing protease inhibitor cocktail (Complete; Roche Molecular Biochemicals, Indianapolis, IN) and 200 μg/ml phenylmethylsulfonyl fluoride (Sigma-Aldrich Life Science). The lysates were centrifuged at 100,000 × g to remove cellular debris and aggregates. The resulting supernatants were loaded on a Sephacryl S300 column (16/60; GE Healthcare) and separated in the presence of PBS.
Statistical analyses
A Kolmogorov–Smirnov test was performed for endosomal localization of ESCRT-II in Figures 1B and 5B and for ILV counts in Figure 4B using the application R (www.r-project.org). Student's t test (unequal variance, two-tailed) was performed for GFP-Cps1 sorting in Figure 2B and Snf7 oligomerization assay in Figure 5A. All differences are significant with p < 0.05. p values for the less obvious differences are denoted in the figures as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Material
Acknowledgments
We thank Kelsey Fulkerson and Stacey Drosner for help with yeast strain/plasmid construction and Matt Curtiss for helping with the preparation of reagents used in this study. We also thank Srivishnu Satyavolu for help with microscopy quantification and Matthew West for help with TEM.
Abbreviations used:
- CEN
centromere
- ESCRT
endosomal sorting complex required for transport
- GLUE
GRAM-like ubiquitin-binding in Eap45
- ILV
intralumenal vesicle
- MVB
multivesicular body
- PI3P
phosphatidylinositol 3-phosphate
- TEM
transmission electron microscopy
- YNB
yeast nitrogen base.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1469) on December 10, 2014.
REFERENCES
- Adell MAY, Vogel GF, Pakdel M, Muller M, Lindner H, Hess MW, Teis D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI. Ubiquitin interactions of NZF zinc fingers. EMBO J. 2004;23:1411–1421. doi: 10.1038/sj.emboj.7600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Farazi TA, Gordon JI. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem. 1998;273:25864–25874. doi: 10.1074/jbc.273.40.25864. [DOI] [PubMed] [Google Scholar]
- Babst M. MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr Opin Cell Biol. 2011;23:452–457. doi: 10.1016/j.ceb.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Davies BA, Katzmann DJ. Regulation of Vps4 during MVB sorting and cytokinesis. Traffic. 2011;12:1298–1305. doi: 10.1111/j.1600-0854.2011.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002a;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002b;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- Babst M, Odorizzi G, Estepa EJ, Emr SD. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic. 2000;1:248–258. doi: 10.1034/j.1600-0854.2000.010307.x. [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield W-MB, Uter NT, Myszka DG, Hill CP, Sundquist WI. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau PS, Urbanowski JL, Winistorfer SC, Piper RC. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat Cell Biol. 2002;4:534–539. doi: 10.1038/ncb815. [DOI] [PubMed] [Google Scholar]
- Bilodeau PS, Winistorfer SC, Kearney WR, Robertson AD, Piper RC. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J Cell Biol. 2003;163:237–243. doi: 10.1083/jcb.200305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Hulsik DL, Belly A, Goldberg Y, Chatellard-Causse C, Blot B, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Róz˙ycki B, Chung HS, Herrick DZ, Canagarajah B, Cafiso DS, Eaton WA, Hummer G, Hurley JH. Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission. Structure. 2012;20:874–886. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss M, Jones C, Babst M. Efficient cargo sorting by ESCRT-I and the subsequent release of ESCRT-I from multivesicular bodies requires the subunit Mvb12. Mol Biol Cell. 2007;18:636–645. doi: 10.1091/mbc.E06-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher AB, Cox BS. Copy number and the stability of 2-micron circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol. 1984;157:283–290. doi: 10.1128/jb.157.1.283-290.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Emr SD, Williams RL. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 2007;26:600–612. doi: 10.1038/sj.emboj.7601501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- Hierro A, Sun J, Rusnak AS, Kim J, Prag G, Emr SD, Hurley JH. Structure of the ESCRT-II endosomal trafficking complex. Nature. 2004;431:221–225. doi: 10.1038/nature02914. [DOI] [PubMed] [Google Scholar]
- Im YJ, Hurley JH. Integrated structural model and membrane targeting mechanism of the human ESCRT-II complex. Dev Cell. 2008;14:902–913. doi: 10.1016/j.devcel.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann DJ, Babst M, Emr SD. ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- MacDonald C, Buchkovich NJ, Stringer DK, Emr SD, Piper RC. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 2012;13:331–338. doi: 10.1038/embor.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Fyfe I, Schuh AL, Chapman ER, Edwardson JM, Audhya A. ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J Biol Chem. 2011;286:9636–9645. doi: 10.1074/jbc.M110.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E, Kawahata K, Kato M, Kitamura N, Komada M. STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol Biol Cell. 2003;14:3675–3689. doi: 10.1091/mbc.E02-12-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzioł T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175:715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Rich RL, Myszka DG, Davis DR, Sundquist WI. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 2002;21:2397–2406. doi: 10.1093/emboj/21.10.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29:1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Köhrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics: A Laboratory Manual, Cold Spring Harbor. NY: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Katzmann DJ, Schnell JD, Sutanto M, Emr SD, Hicke L. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat Cell Biol. 2002;4:389–393. doi: 10.1038/ncb790. [DOI] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, Wakatsuki S, Stenmark H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- Teo H, Perisic O, González B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.