Abstract
Genome-wide association studies of the related chronic inflammatory bowel diseases (IBD) known as Crohn’s disease and ulcerative colitis have shown strong evidence of association to the major histocompatibility complex (MHC). This region encodes a large number of immunological candidates, including the antigen-presenting classical HLA molecules1. Studies in IBD have indicated that multiple independent associations exist at HLA and non-HLA genes, but lacked the statistical power to define the architecture of association and causal alleles2,3. To address this, we performed high-density SNP typing of the MHC in >32,000 patients with IBD, implicating multiple HLA alleles, with a primary role for HLA-DRB1*01:03 in both Crohn’s disease and ulcerative colitis. Significant differences were observed between these diseases, including a predominant role of class II HLA variants and heterozygous advantage observed in ulcerative colitis, suggesting an important role of the adaptive immune response to the colonic environment in the pathogenesis of IBD.
Meta-analyses of genome-wide association studies (GWAS) have recently shown that Crohn’s disease (CD) (MIM266600) and ulcerative colitis (UC) (MIM191390) share the majority of the 163 known genetic risk factors for IBD, with the MHC being one of the notable exceptions4. Data from these GWAS, however, have had insufficient variant density to define the association signals within the MHC. Targeted studies of IBD with higher variant density within the MHC region but with modest sample sizes have indicated that multiple independent associations are likely to exist at human leukocyte antigen (HLA) genes and non-HLA genes, with the most consistent associations being to HLA class II loci, mainly HLA-DRB1 and HLA-DQB1, with some reports of association at the HLA-C class I locus and potentially also at non-HLA genes2,3,5–8. In the current study we generated high quality genotypes for 7,406 SNPs within the MHC region on a total of 18,405 patients with CD, 14,308 patients with UC and 34,241 controls subjects. Using this SNP data, we imputed and benchmarked the genetic variation within the class I (HLA-B, HLA-C, and HLA-A) and class II (HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5, HLA-DQA1, HLA-DQB1, HLA-DPA1 and HLA-DPB1) HLA genes at the level of classical HLA alleles and amino acid positions (please refer to the Online Methods).
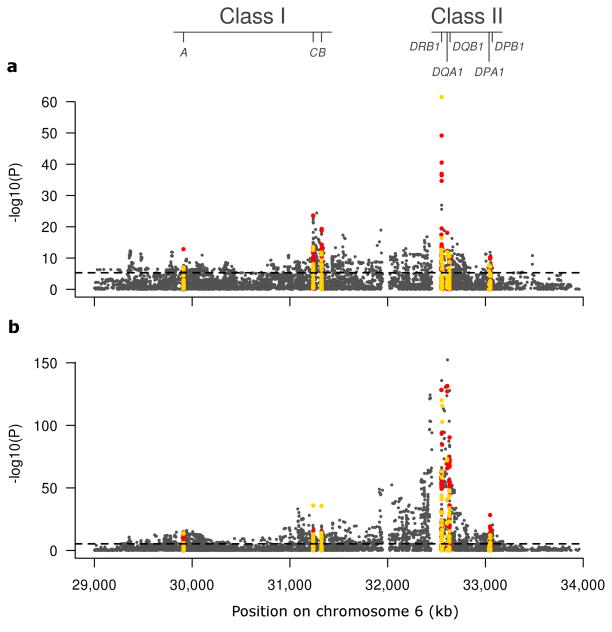
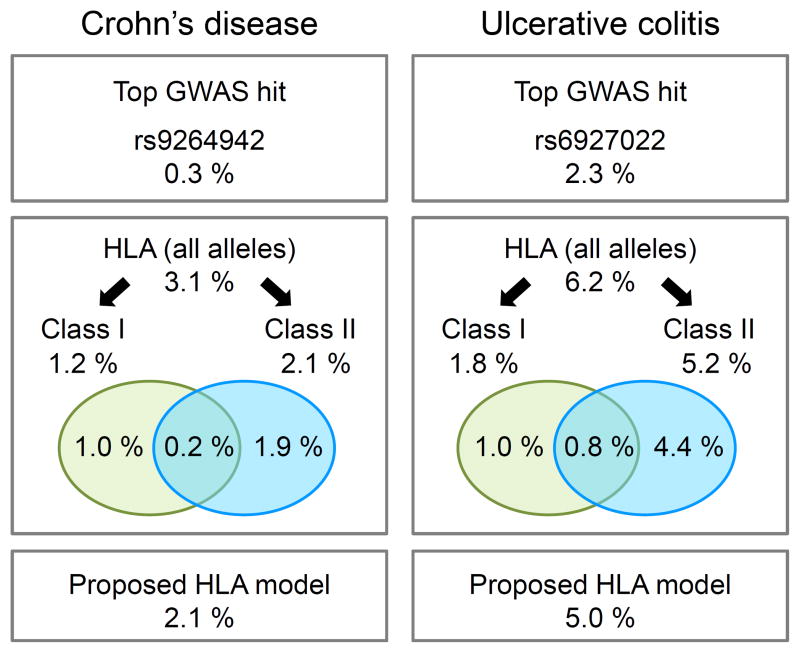
As a first step to defining the nature of the association to CD and UC within the MHC, we performed univariate analyses of the SNPs, classical HLA alleles, and HLA amino acids. These analyses revealed a very large number of variants across the MHC region with significant association to these phenotypes (Fig. 1), with major peaks of association centered in and around the classical HLA genes, suggesting a role for classical HLA alleles in CD and UC risk. This observation is consistent with gene-based analyses, which show strong association at the HLA genes for both UC and CD (e.g. P<1×10−300 for HLA-DRB1 in UC) (Supplementary Table 1). In particular, these analyses demonstrated a role of HLA-DRB1 that cannot be attributed to other HLA genes, with evidence of residual association in class I and class II regions (Supplementary Table 1). In order to be more quantitative, we calculated the variance explained by the class I and class II alleles. Whereas the contribution of class I and class II alleles are relatively equivalent in CD, not only is the overall impact of HLA on disease risk greater in UC, but the alleles in the class II region have nearly three-fold greater impact than class I alleles (Fig. 2). Moreover, these analyses have revealed that classical HLA alleles explain three- to ten-fold more of the disease variance than that explained by the index SNPs that were previously identified (~3% vs ~0.3% in CD; ~6% vs ~2% in UC) (Fig. 2).
Figure 1. Primary univariate association analyses of CD and UC.
Univariate association analysis results for 8,939 SNPs (dark grey) (Supplementary Table 12), 90 2-digit and 138 4-digit resolution HLA alleles (yellow) (Supplementary Table 13), as well as 741 single amino acid variants (red) (Supplementary Table 4) in the MHC region are shown for 18,405 CD cases and 14,308 UC cases (with 34,241 common control subjects). Given that previous genetic analyses have identified distinct effects in the MHC for CD and UC, with different non-correlated alleles identified in each disease, we opted to perform the finemapping analyses for CD and UC separately. (a) The primary univariate association analysis in CD reveals over 1,789 markers showing study-wide significant association (P<5×10−6) across the MHC, including 32 4-digit resolution classical HLA alleles (Fig. 3 and Supplementary Table 2). The single most significant variant for CD is HLA-DRB1*01:03 (P=3×10−62, OR= 2.51). (b) The primary univariate association analysis in UC reveals over 2,762 markers showing study-wide significant association across the MHC, including 50 4-digit resolution classical HLA alleles (Fig. 3 and Supplementary Table 3). The single most significant variant for UC is rs6927022 (P=8×10−154, OR= 1.49) while the best HLA allele is HLA-DRB1*01:03 (P=3×10−119, OR=3.59); each acting independently. Twenty-nine SNPs and 9 amino acid variants surpass HLA-DRB1*01:03 as the next most significant variants in the primary analysis however all of these are correlated to rs6927022 and their significance is dramatically reduced by conditional logistic regression.
Figure 2. Variance explained by 4-digit HLA alleles in CD and UC.
Proportion of variance explained on a logit scale (McKelvey and Zavoina’s Pseudo R2, see Online Methods) for different models in CD (left) and UC (right). The top boxes show the variance explained by previously identified GWAS index SNPs within the MHC4. The middle boxes illustrate the variance explained by HLA models including all 4-digit alleles of frequency > 0.5% (126 alleles in CD and UC) and models restricted to 4-digit alleles within either class I (63 alleles) or class II regions (63 alleles), respectively. The Venn diagram illustrates the proportion of variance explained that is unique to class I, class II or shared. The bottom boxes indicate the variance explained by the proposed HLA models (15 and 16 alleles in CD and UC, respectively). To be noted, these estimations of variance explained were performed on the logit scale for practical reasons, and should not be directly compared to heritability estimates computed on the (Gaussian) liability scale.
Specifically, in our univariate analyses, the most significant association in CD is to HLA-DRB1*01:03 (P<4×10−62, OR= 2.53), with a p-value over 10 orders of magnitude more significant than the next best associated variants in the region. Importantly, HLA-DRB1*01:03 has an effect in CD which is statistically independent from the other most associated variants in the MHC, as shown by reciprocal conditional logistic regression (Supplementary Fig. 1). In the UC univariate analysis the single most significant variant is a non-coding SNP (rs6927022, P<5×10−153, OR=1.49) near HLA-DQA1, previously identified in the recent meta-analysis of GWAS 4; while multiple additional variants show highly significant association, most are correlated to this top signal (Fig. 1 and Supplementary Fig. 2). Strikingly the next strongest independent association is to HLA-DRB1*01:03, having a much greater OR (P<1×10−120, OR=3.63; Pcond<2×10−89, OR=3.06) (Supplementary Fig. 2). Reciprocal conditioning on HLA-DRB1*01:03 did not abolish the effect seen at rs6927022 (P<9×10−123, OR=1.43), indicating that these have mostly statistically independent effects in UC. Taken together, our analyses point to HLA-DRB1*01:03 as likely being causal in both diseases, with additional causal alleles in the class II and class I regions. Given this observation, it is probable that additional alleles within HLA-DRB1 contribute to IBD risk.
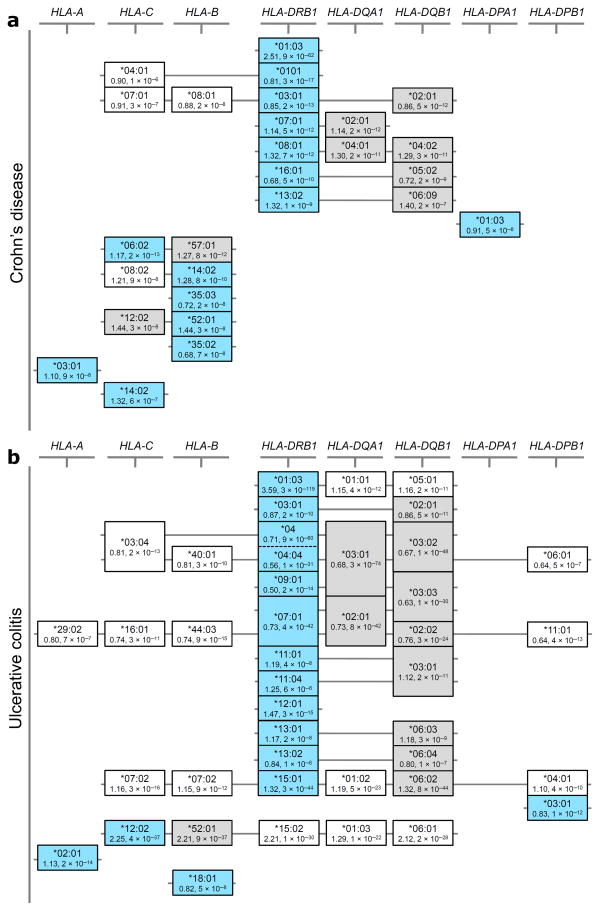
We thus examined an HLA-DRB1-centric model and identified seven HLA-DRB1 alleles with independent effects on CD risk (study-wide significance threshold of 5×10−6) (Supplementary Table 2). Moreover, when controlling for these seven HLA-DRB1 alleles, we identified only a single additional class II allele (HLA-DPA1*01:03) independently associated with CD. Using the same conditional logistic regression framework for the analysis of the class I locus, we identified seven class I HLA alleles that are significantly associated with CD, after conditioning on the eight class II alleles (Fig. 3 and Supplementary Table 2). This HLA-DRB1-centric model explains about 2% of disease variance (Fig. 2). In UC, we identified a total of 12 HLA-DRB1 alleles, 1 HLA-DPB1 allele and 3 class I alleles (Supplementary Table 3) that can explain the association to the MHC and which account for about 5% of disease variance (Fig. 2).
Figure 3. Correlated association signals at HLA alleles support potential alternate association models for both CD and UC.
Equivalence of effect at the different study-wide significant associated 4-digit HLA alleles is shown for (a) CD and (b) UC. The structures illustrated in the figure are not classically defined haplotype structures, but were identified entirely based on the correlation of signal defined through pairwise reciprocal conditional logistic regression analyses (see Supplementary tables 2 and 3); although such correlations are clearly dependent on the underlying haplotypic structure of the region. Alleles identified as primary tags for independent association signals in our HLA-DRB1 focused models are shown in light blue boxes, while alternate alleles with equivalent effects are shown in grey boxes. Alleles in white boxes show study-wide significant secondary effects that can be explained entirely by the selected HLA alleles. Alleles at the HLA-DRB3, -DRB4 and -DRB5 genes were omitted in order to simplify the display; many of the alleles at these genes show high frequency and as such are correlated to many different alleles (both risk and protective) at the other class II genes. Of note, the HLA-DRB4*null allele is the second strongest associated allele in UC (see Supplementary table 3).
As can be seen in Figure 3, for many of the alleles identified in the HLA-DRB1-centric model, a few other candidate alleles in class I or class II can be considered. In particular, multiple HLA-DRB1 alleles have equivalent associations at HLA-DQA1 and HLA-DQB1 (e.g. HLA-DQA1*03:01 is equivalent to HLA-DRB1*04 and HLA-DRB1*09 alleles in UC) equally supporting a role for genetic variation within HLA-DQA1 and/or HLA-DQB1 in disease susceptibility, particularly for UC (Fig. 3). However, several of the alleles in these models, including HLA-DRB1*01:03, do not have any such proxies and thus are strong candidates for being causal (Fig. 3). Further dissection of these class II correlated signals for identifying potential causal alleles may only be feasible in admixed or ethnically diverse populations 9. Further refinement may also be possible by examining the impact of clinical sub-phenotype and associated autoimmune co-morbidities on observed associations, although functional studies will be needed to infer causality. For the present analysis we were able to assess the impact of colonic vs. non-colonic inflammation, and found that HLA-DRB1*01:03 is associated with colonic CD and that HLA-DRB1*07:01 is associated with the absence of colon involvement (Supplementary Fig. 3), in line with previous suggestions10. This explains the shared associations for CD and UC at HLA-DRB1*01:03 and strongly suggests that this allele is critically involved in determining the colonic immune response to local flora.
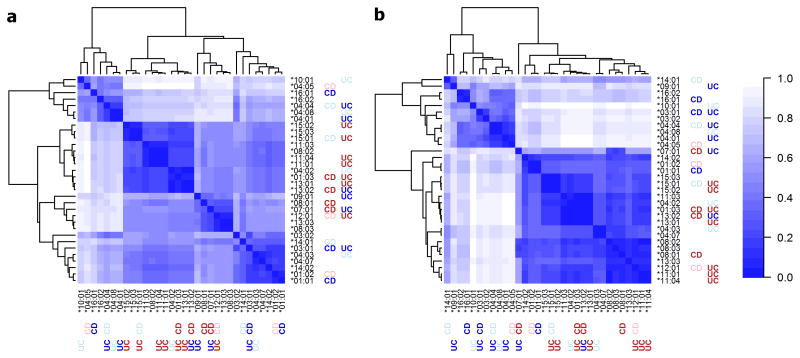
Given that classical HLA alleles consist of combinations of specific amino acids at multiple positions, we tested whether the association to disease could be better explained by single amino acid positions. Indeed we observed very strong association signals at many single amino acid variants in CD (e.g. five amino acids of HLA-DRβ at positions 67, 70 and 71) and in UC (e.g. 4 amino acid variants of HLA-DQα at positions 50 and 53 and 215 and 4 amino acid variants in HLA-DRβ at positions 98 and 104) and also performed per position omnibus analyses that confirm the predominant association to HLA-DRβ position 11 in UC, as previously reported 5, and to HLA-DRβ position 70 in CD (Supplementary Tables 4–5 and Supplementary Fig. 4). While the hypothesis of a positional effect is appealing, the interpretation of these position-based tests is not straightforward in the context of likely multiple causal alleles (Supplementary Note on amino acids, Supplementary Table 6 and Supplementary Fig. 5). Furthermore in this study the amino acid-based models did not capture the association at HLA-DRB1 in a more parsimonious way than the HLA allele-based models (Supplementary Note on amino acids). To further explore the basis for the observed HLA associations, we performed three-dimensional protein structure modeling followed by analysis of the electrostatic properties of the binding groove of associated (P<10−4) and common (frequency>1%) HLA-DRB1 alleles. These analyses suggest that HLA-DR alleles associated with increased risk of UC and CD, share common structural and electrostatic properties within or near their peptide binding groove that are largely distinct from those of HLA-DR alleles associated with decreased risk of UC and CD (Fig. 4).
Figure 4. HLA-DR peptide binding groove electrostatic properties and risk of IBD.
The electrostatic potential of all HLA-DR alleles associated with UC or CD, and of all common HLA-DR alleles (frequency >1%), was calculated. HLA-DR alleles associated with increased or decreased risk of IBD at study wide-significance level (P< 5×10−6) are shown in dark red or dark blue, respectively. Respective risk associations at suggestive level (1×10−4<P<5×10−6) are shown in pale red and pale blue. Electrostatic potential comparisons among HLA-DR molecules were performed in a pairwise, all-versus-all, fashion (see Online Methods) to produce distance matrices that are displayed as symmetrical heatmaps (scale ranges from 0 [identical] to 1 [maximum difference]). (a) The electrostatic potential in seven regions within the peptide binding groove (see Online Methods and Supplementary Fig. 10), which interact with the presented peptide, were compared among the HLA-DR alleles and pooled onto a single Euclidian distance matrix. The distance-based clustering identifies four clusters, with an enrichment of risk alleles in two of these. Comparison of the electrostatic potential at individual peptide binding groove regions is shown in Supplementary Fig. 13. (b) Heatmap representing electrostatic potential differences among the HLA-DR alleles at a spherical region that encompasses amino acid residues 67, 70 and 71 of the HLA-DRβ chain (associated with risk for UC and CD; Supplementary Table 13). The distance-based clustering identifies two clusters that correlate with directionality of effect in IBD.
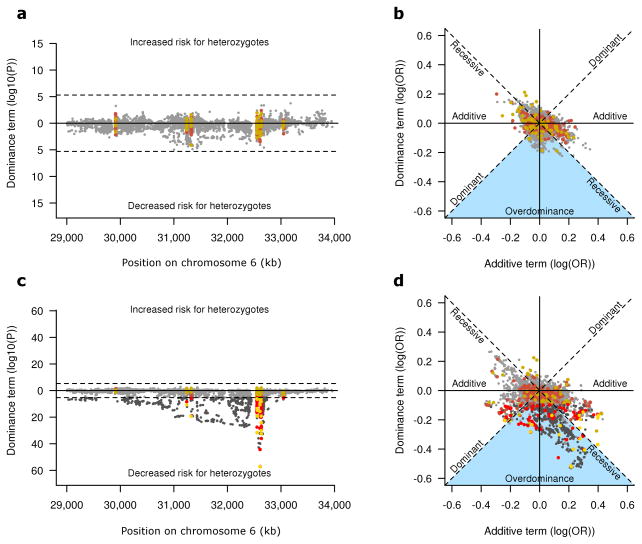
While we performed the primary analyses based on a dose effect model, our sample size allowed us to investigate the effects further, by testing for non-additive effects. In fact we found significant departure from additive effects in UC, but not for CD (Fig 5a–c). Specifically, we found evidence of decreased heterozygosity in UC patients for genotyped and imputed variants across the MHC and at HLA genes, mostly in class II (Supplementary Tables 7–8). This heterozygote advantage could be explained by an enrichment of dominant protective and recessive risk alleles11, that is absent or much less important in CD (Fig. 5 and Supplementary Fig. 6). Notably, we also detected multiple overdominant effects in UC, the strongest of which is captured by HLA-DRB1*03:01 (Fig. 5, Supplementary Fig. 6–7, and Supplementary Table 9). This allele is mostly found on the ancestral haplotype 8.1, a relatively common (~5–10%) haplotype that is conserved in European populations and that is implicated in other immune diseases 12–14. The overdominance effect of this haplotype in UC is possibly due to the presence of both dominant protective and recessive risk alleles, which would be consistent with the reported recessive risk of this haplotype in the UC-related biliary disease primary sclerosing cholangitis (Supplementary Fig. 8–9) 15,16. Analogous with an infectious paradigm 11, these data may suggest that decreased HLA class II heterozygosity may impair the ability to appropriately control colonic microbiota in UC.
Figure 5. Non-additive effect models in CD and UC.
Evidence for non-additive effect of common variants (frequency >5%) across the MHC tested under a general model of additive and dominance effects (Online Methods) in CD (a,b) and UC (c,d). The p-values and directionality for departure from additive effect (dominance term) are represented on the y-axis (a,c). HLA alleles and amino acids variants are in yellow and red respectively, while SNPs are represented in dark grey. Variants with non-significant (P>5×10−6) dominance term are plotted in less pronounced colors. A clear enrichment for lower risk in heterozygotes is observed in UC (c) as suggested by the large number of significant negative dominance term (lower part of the plot). This effect is absent in CD (a), or much less important. The dominance term OR is illustrated (y-axis) versus the additive term (x-axis) (b,d). Protective and risk minor alleles are shown on the left and right sides of the plot respectively. Strictly recessive or dominant variants are expected to fall on the diagonals, while strictly additive variants lay on or close to the x-axis. The y-axis is the expected position for pure over/under dominance. In UC (d), many alleles fall into the region of the plot for protective dominant, risk recessive or overdominance (blue triangle) (see Supplementary Table 9 for pairwise comparison of HLA-DRB1 alleles). These non-additive effects are observed for many variants in UC (c,d) (e.g HLA-DRB1*03:01 and HLA-DQB1*02:01) but are mostly absent in CD (a,b); notable exception being the HLA-B*08 allele (Supplementary Fig. 6).
Although there is a significant challenge in defining the causal alleles for CD and UC in the MHC given the LD structure in the region, a number of conclusions can be drawn regardless of the models tested. First, the high density mapping of this region in a large cohort revealed the significant contribution of the MHC to disease risk, a contribution that is not apparent in the previous GWAS. Second, for both CD and UC it would appear that variation within HLA genes as opposed to variation in other genes within the MHC plays a predominant role in disease susceptibility. Third, while the contribution of class I and class II HLA variants to disease risk is relatively equivalent in CD, HLA class II variation plays a more important role in UC. Fourth, in contrast to the majority of non-MHC susceptibility loci being shared between CD and UC, most associated HLA alleles have a predominant role in either CD or UC, with very few having shared IBD risk Fig. 6). Finally, the decreased heterozygosity in UC suggests that the ability to recognize a broader set of antigens, potentially of colonic microbial origin, is important to mount protective immunity.
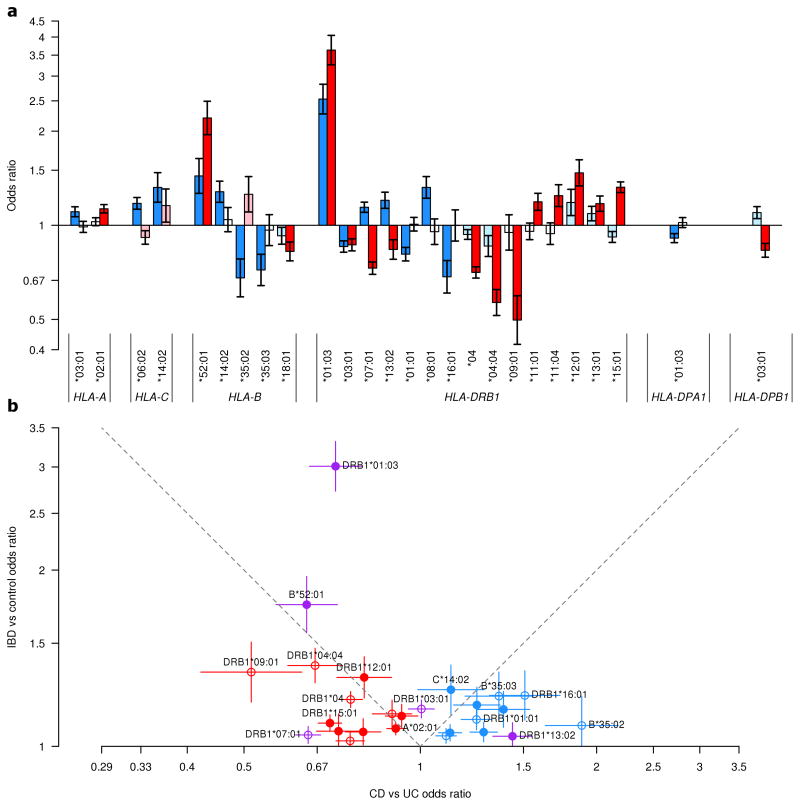
Figure 6. Comparison of odds ratio in CD and UC for HLA alleles identified from HLA-focused models.
Odds ratio (OR) from the primary univariate association analyses in CD and UC for all alleles identified in the HLA-focused models of CD and/or UC are presented with 95% confidence intervals (a). Odds ratio for CD and UC are in blue and red respectively; darker colors indicate study-wide significant effect (P<5×10−6), lighter colors indicate nominal significance level (0.05>P 5×10−6) and white indicates non-significance (P 0.05) (for specific effect and significance values refer to Fig. 3 and Supplementary Tables 2 and 3). Allele HLA-B*52:01 is indicated for UC in place of the equivalent HLA-C*12:02 to simplify the display of this shared signal. For the same HLA alleles, odds ratio (with 95% confidence intervals) for an IBD analysis are plotted against the odds ratio for the CD versus UC analysis with the IBD risk allele as the reference (b). Empty circles represent variants where the absence of the allele is the reference. Alleles identified as significant in CD or UC only are plotted in blue and red, respectively. Variants identified as significant in both are shown in purple. To be noted, HLA-DRB1*07:01 and HLA-DRB1*13:02 have opposite direction of effect between CD and UC. Shared association signals are expected to fall in the upper triangle of the plot. Most variants fall outside of this region, highlighting the difference between CD and UC in the MHC.
Online Methods
Genotype dataset
The cohorts used in the current study were collected from 15 countries across Europe, North America, and Australia, and have previously been described4. In total 19,802 CD cases, 14,864 UC cases and 34,872 controls of European ancestries, from the International IBD genetics consortium (IIBDGC; www.ibdgenetics.org), were included in the study.
Genotyping of the IIBDGC cohorts was performed in 34 different batches across 11 different genotyping centers, and additional genotyping data for 5,815 controls was obtained from the International MS Genetics Consortium (IMSGC)17. All participating centers received approval from their local and national institutional review boards, and informed consent was obtained from all participants in the study. All DNA samples included in the study were genotyped using the Immunochip custom genotyping array (Illumina, San Diego, California, USA)4,18.
Genotype calling and quality control
After an initial genotype calling using Illumina’s Bead Studio and a first stage of quality control, all data was centrally recalled using optiCall v.0.6.219. OptiCall clustering was performed for each batch separately, with a Hardy-Weinberg Equilibrium (HWE)-threshold of 1×10−15, HWE blanking disabled and a genotype call threshold of 0.7. HWE was calculated conditional on predicted ethnicity, and related individuals were removed from this calculation.
After recalling, a single unified QC procedure was performed across all genotyping batches, including the IMSGC controls. Variants that either failed the Hardy-Weinberg Equilibrium test in unaffected individuals, had different missing genotype rates in affected and unaffected individuals, or had significantly different allele frequencies across the batches based on false-discovery rate (FDR) threshold of 10−5 for each test were removed. We also removed variants that had missing genotype rate >2% across the entire collection, or > 10% in any single batch. Variants that only failed one QC criteria in a single batch were set to missing in the failed batch. Individuals were removed if they showed a missing genotype rate >2%, had a significantly higher or lower inbred coefficient (F) (plink)20 at FDR <0.01, and showed a high level of relation (PI_HAT ≥ 0.4) calculated on IBS distance between all individuals. The coefficient of inbreeding and inter-sample relationships were calculated on a LD-pruned dataset of independent variants. In order to control for population stratification while avoiding the possible bias introduced by the enrichment of associated alleles in the dataset, principal components were computed on control samples, based on a set of 18,123 independent (LD-pruned) SNPs across the Immunochip, and then applied to the affected samples. To generate the LD-pruned SNPs, we removed variants in long range LD and pruned the common variants (MAF>0.05) three times (plink20). Genomic inflation factor (λ) was estimated from a set of 3120 “null” SNPs (chosen based on GWAS of schizophrenia, psychosis and reading/mathematics ability), using different subset of principal components. Based on these and investigation of contribution of individual SNPs to the components (loadings), we selected to use the first five principal components to control for population stratification (Supplementary Fig. 11).
For the purpose of this study, the chr6:25Mb-34Mb region, encompassing MHC region, was extracted from the post-QC Immunochip dataset. In total, after QC, 18,405 CD cases, 14,308 UC and 34,241 healthy controls subjects were successfully genotyped for 8,001 SNPs within the MHC region.
Imputation of missing genotype data
Missing genotype data, from failed genotype calls or failed QC in single batches, were imputed using the Beagle SNP imputation package(v.3.0.4); imputation was performed using only the high density information contained within the dataset, no external reference dataset was used21.
Imputation of HLA alleles
In order to avoid cohort specific asymmetry in the dataset, variants that failed QC in only one genotyping batch were removed before imputation of HLA alleles. Imputation of HLA alleles was done using two independent HLA imputation pipelines HLA*imp222 and SNP2HLA (v2)23; imputation of polymorphic amino acid positions and SNP variants was performed using SNP2HLA (v2). A set of additional SNP variants were also included using version 1 of SNP2HLA, which used a different reference dataset and imputed SNP variants not found in the SNP2HLA (v2) reference dataset.
Imputation accuracy
To benchmark HLA imputation results generated by HLA*IMP2 and SNP2HLA, we used two cohorts from the current study (Italian and Norwegian) for which classical HLA typing was available. The Italian dataset consisted of 450 ulcerative colitis cases and 280 controls for which 4-digit HLA types at HLA-A, HLA-B, HLA-C, HLA-DQB1 and HLA-DRB1 generated by Sequence-based typing (SBT)24. The Norwegian dataset contained 244 ulcerative colitis cases and 254 controls with 2-digit HLA types and a subset of 92 cases and 250 controls with 4-digit HLA types at HLA-DRB1 generated at the Oslo University Hospital. We only considered individuals of whom both HLA alleles were successfully typed at 2- and 4-digit resolution at the locus under validation. For validation of HLA allele imputations, we compared imputation results of HLA*IMP2 and SNP2HLA to the lab-derived types in a locus-specific and allele-specific manner. We calculated per locus concordance (Supplementary Table 10) and sensitivity, specificity, positive predictive value, negative predictive value and accuracy for each allele (Supplementary Table 11).
Let’s denote true positives, true negatives, false positives and false negatives by TP, TN, FP and FN, respectively. Given our analyses were performed using expected allele doses from posterior probabilities, we used the expected doses for computation:
where xi and yi are respectively the typed and imputed dose for individual, and the sum over all individuals gives the TP, FP, TN and FN (Supplementary Table 11). For a given HLA allele, we calculated sensitivity, specificity, positive predictive value, negative predictive value and accuracy using the usual definitions:
For a given locus, we calculated the concordance as TPall/(total number of chromosomes) (Supplementary Table 10).
Final dataset
As an additional QC step, we performed manual cluster inspection for any genotyped SNPs in the region tagging (r2>0.8) imputed SNPs, amino acid variants and HLA alleles reported in our proposed models for CD and UC.
In order to not duplicate HLA alleles in our dataset, and given the mostly equivalent imputation quality of the 2 pipelines, we opted to keep HLA imputations for HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, HLA-DQB1, HLA-DPA1, and HLA-DPB1 derived from the SNP2HLA pipeline since amino acid and additional SNPs were also obtained from this pipeline, whereas the HLA allele information for the HLA-DRB3, HLA-DRB4, and HLA-DRB5 genes were obtained from the HLA*imp2 pipeline.
Genotyped SNPs variants, as well as imputed SNPs, HLA alleles and amino acid variants at polymorphic amino acid positions were combined into a single dataset for analysis. Variants with a minor allele frequency of less than 0.05% in controls and variants showing imputation quality (INFO) score <0.5 were removed from the analyses. The final dataset contained 8,939 SNP variants, 138 4-digit resolution HLA alleles, 90 2-digit resolution HLA alleles and 741 single amino acid variants.
Heterogeneity
In order to evaluate heterogeneity of effects and allele frequencies between subgroups of different European ancestry, we clustered the individuals into relatively large population subgroups. These clusters were determined using k-means on the principal components. Based on the decline within cluster sum of squares, we determined that the optimal choice for the number of clusters (k) was in a range of 6–10. For each value of k in this range, we compared the clustering obtained to self-reported country of origin, when available. The objective was to be able to identify known population structures, while keeping homogeneous group in a single cluster. Based on these criteria, we stratified the dataset into 9 clusters (Supplementary Fig. 12). For every reported variant, we evaluated and illustrated heterogeneity of effect sizes between these 9 clusters using forest plots (Data on heterogeneity of effects is available on http://www.medgeni.org/goyette_nature_gen_2015).
Association testing and conditional analyses
Unless otherwise stated, all analyses were corrected for five principal components and performed in R (v 2.15.2) on expected allele counts (additive dose from posterior probability)(see Supplementary Tables 4, 12 and 13 for primary univariate association results for single amino acid variants, SNP and HLA alleles respectively). Also, 4-digit HLA alleles were prioritized over 2-digit alleles in the final selection of association signals included in our models. We calculated the threshold for statistical significance in our study at p<5×10−6 for a study-wide type 1 error rate of 5% with 9,852 independent tests.
Given the imputed nature of the HLA allele data within our dataset, complexity of signals and burden of dimensionality, a standard forward conditional logistic regression approach was avoided. We opted instead to identify all HLA alleles showing study-wide significant association across the MHC region, in the primary univariate association dataset, and evaluate their independent effects through single pair-wise reciprocal conditional logistic regression (Supplementary Tables 2–3). This approach allowed the identification of independent association signals, composed either of single HLA alleles or groups of equivalent alleles (Fig. 3).
Gene based analyses (omnibus)
We tested the association to phenotypes at each HLA gene using logistic regression under an additive model of effect, including all 4-digit alleles with a frequency greater than 0.5%. Evidence of association at the gene is given as the p-value of the likelihood test for this regression model versus the null model (including only principal components). Evidence of association at a given gene, conditional on other HLA genes, is given as the p-value of the likelihood ratio test for the full model (including all HLA alleles) versus the partial model (without including the alleles at the given gene). In a similar fashion, we also tested the remaining association at each HLA gene, conditional on our final set of associated alleles (final model). To be noted, the different HLA genes have different number of distinct alleles, thus variable level of complexity (degrees of freedom) for the model and the interpretation of these gene-based tests is not straightforward in the context of likely multiple causal alleles (see the Supplementary Note on amino acid).
Variance explained (pseudo R2)
In order to represent the importance of genetic variation in the MHC for CD and UC, we computed an estimate of variance explained for different models. Variance explained is not well defined for binary outcomes and many different metrics exist to represent it25. Given the correlation between the variants in the MHC, we computed McKelvey Zavoina’s pseudo R2 on the logit scale26. This metric can be computed using correlated variables and is independent of disease prevalence. Let β be the vector of fitted coefficients from logistic regression and S the estimated covariance matrix of the predictor, the McKelvey Zavoina’s pseudo R2 is then given by:
To be noted, this estimation of variance explained shouldn’t be directly compared to values given by other metrics or to heritability estimates based on Gaussian liability. We computed the variance explained by a regression model including all the HLA alleles with a frequency greater than 0.5%. We did the same separately for HLA alleles within class I and class II. Variance explained by class I, after inclusion of class II, was computed as the improvement in R2 for the full model (all HLA alleles), compared to class II only. This difference in R2 estimates the specific contribution of class I to the total variance explained that cannot be attributed to class II alleles. We did the same for class II. In order to be able to compare our results to the SNPs identified by the GWAS4, we computed R2 for the published GWAS index SNP in the MHC for CD and UC.
Subphenotype analyses
The IIBDGC has collected detailed subphenotype information for a subset of samples included in this study. These phenotypes include: demographics, disease location and behavior in CD, disease extent in UC, surgery and primary sclerosing cholangitis (PSC). Quality control of this subphenotype information and genotype-phenotype association testing has been performed in the context of another project from this IIBDGC (personal communication with Charlie Lee). In the context of this fine-mapping project, we considered disease location in CD and PSC in UC to evaluate the impact of disease heterogeneity. Association tests were performed within subset of samples with known subphenotype.
Non-additive effects, heterozygote advantage and overdominance
We tested evidence of non-additive effects at each variant using a logistic regression including terms for additive and non-additive effect, as described below. Rare variants with minor allele frequencies below 5% were excluded from this analysis, given that the very low number of homozygote precludes the evaluation of the model. For this particular analysis, we used best guess genotype data, unless otherwise stated. We computed evidence of non-additive effect as the p-value of the Wald statistic for the dominance term. We also computed the evidence of association for an allele under the general model using the likelihood ratio test.
Suppose a genetic variant with alleles G and g. The genotypes (GG, Gg, gg) can be coded as u = (1, 0, −1) and v = (0, 1, 0), respectively. In this context, u = dose - 1 and v = 1 − |u|. The effect of a specific genotype is then given as au + dv, where a and d are respectively the additive and dominance effect, as estimated by logistic regression. This parameterization can be generalized to expected allele counts (additive dose from posterior probability).
Under this parameterization, the effects of the genotypes GG, Gg and gg are given by a, d and −a, respectively. A strictly additive model would be d = 0 and a ≠ 0, while a dominant or recessive model would be d = a or d = −a. If the dominance term has a higher protective effect than the additive term, that is d < −|a|, the model is one of overdominance, where being a heterozygote provides protection, compared to both homozygotes.
For each HLA gene, we also tested for evidence of heterozygote advantage. We coded each individual as homozygote or heterozygote at the gene, as determined from the imputed 2-digit and 4-digit alleles. Association of phenotype and zygosity at each gene was tested using logistic regression. Evidence of association is given as the p-value of the likelihood test.
Pairwise comparisons of common 2-digit alleles at HLA-DRB1 were conducted to better understand the non-additive effects (Supplementary Table 9). For each pair or alleles, analysis was constrained to the subset of individual carrying only these two alleles as homozygotes or heterozygotes. Genotype effects were evaluated within each subset. Overdominance was tested as heterozygote vs the lowest risk homozygote. Heterozygote advantage was tested as heterozygote vs the pooled homozygote. Non-additive effect was tested based on the dominance term, as described previously.
Comparative structural modeling of HLA-DR alleles
Comparative structure models for all HLA-DR alleles associated with UC or CD, and of all common HLA-DR alleles (population frequency >1%), were generated using the program MODELLER27. Templates for comparative structure modeling were identified by querying the RCSB Protein Database28 using the sequence of HLA-DRB1*01:01 and the DELTA-BLAST algorithm, for humans (Taxonomy ID: 9606, E-value threshold of 0.05). Of 36 templates identified, 10 crystallographically resolved HLA-DRB1 structures (PDB codes: 4MD5, 1FV1, 1KLU, 3PDO, 1D5M, 1JH8, 4MDI, 3L6F, 4I5B, 2IPK) were retained, which had high resolution (<2.5 Å) and favorable markers of structural quality (Ramachandran plot, DOPE, Verfiy3D and WHAT_CHECK scores)29–32. The sequences of the extracellular domain of target HLA molecules were retrieved from the EBI sequence database (ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/) and aligned using Clustal W2 using the BLOSUM matrix and the Neighbor Joining clustering algorithm and manually adjusted as indicated33. The modeled peptide was standardized to an alanine 12-mer, which was removed prior to calculating protein electrostatics to allow comparison of the electrostatic potential within the peptide-binding groove.
Electrostatic potential calculations
Atom charges and radii were assigned and side-chains protonated for pH 7.4 using the PARSE force field in PDB2PQR34. Protein electrostatic potential was calculated by solving the linearized Poisson-Boltzmann equation in APBS for a cubic grid of 353 points at a spacing of 0.33 Å35. Other parameters were set as follows: ionic solution of 0.15 M of univalent positive and negative ions; protein dielectric of 2; solvent dielectric of 78; temperature of 310 K; and a probe radius of 1.4 Å. HLA class II molecules typically make contact with nine amino acid residues of the presented peptide; of these, 7 peptide residues (at peptide positions 1, 2, 3, 4, 6, 7 and 9) make contact within the peptide binding groove36. Peptide residues at positions 5 and 8 are elevated away from the peptide-binding groove. For comparison of the electrostatic potential of the peptide-binding groove, radii were chosen so that the space around each coordinate encompassed all side chain atoms of the relevant peptide amino acid residue; the electrostatic potential within 1Å from the molecular surface of the HLA molecule was not examined.
The peptides of the template HLA structures were used to define the geometric average coordinate of the positions of the side chain atoms of peptide amino acid residues 1, 2, 3, 4, 6, 7 and 9. The electrostatic potential within a 3.5–5 Å radius from each coordinate was considered for comparison of the electrostatic properties of the peptide-binding groove among HLA-DR alleles (Supplementary Fig. 10). For comparison of the electrostatic potential around amino acid positions 67, 70 and 71, (associated to both UC and CD in different analyses; Supplementary Tables 4–6), the geometric average coordinate of the positions of these amino acids’ side chain atoms was calculated and the electrostatic potential within a 5 Å radius from this coordinate was considered for comparisons among different alleles. Electrostatic potential comparisons were performed using the Hodgkin’s index as described previously27,28 in a pairwise, all-versus-all, fashion to produce a distance matrix37,38. Distance matrices were displayed as a symmetrical heatmap with re-ordering such that electrostatically similar alleles are clustered together, according to the dendrogram (Supplementary Fig. 13). A pooled heatmap was created using the Euclidian distance between the individual distance matrices from the 7 peptide-binding groove regions (Fig. 4a). Heatmaps were created for electrostatic potential comparisons at individual regions of the HLA-DR molecule (7 regions in the peptide-binding groove and one region defined by amino acid residues 67, 70 and 71, as detailed above) (Fig. 4b).
Acknowledgments
J.D.R. holds a Canada Research Chair and his current work is supported by grants from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (DK064869; DK062432). A.F.’s lab is supported by the German Ministry of Education and Research (BMBF) grant program e:Med (sysINFLAME). A.F. receives infrastructure support from the Deutsche Forschungsgemeinschaft (DFG) Cluster of Excellence “Inflammation at Interfaces” and holds an endowment professorship (Peter Hans Hofschneider Professorship) of the Foundation for Experimental Biomedicine (Zurich, Switzerland). Grant support for T.H.K. and A.F. was received from EU 7th Framework Programme (FP7/2007-2013, grant number 262055, ESGI). This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research (M.N.C). J.B. was supported by a Wellcome Trust grant (#WT098051). D.H.M. and V.K. are supported by the NIHR Cambridge Biomedical Research Centre. P.S. is supported by the NIDDK IBD Genetics Consortium Data Coordinating Center grant (7 U01 DK062429-14). J.T. is supported by the Medical Research Council. D.P.B.M. is supported by The Leona M. and Harry B. Helmsley Charitable Trust, The European Union (305479), and grants DK062413, DK046763-19, AI067068, HS021747 and U54DE023789-01. R.H.D holds the Inflammatory Bowel Disease Genetic Research endowed chair at the University of Pittsburgh and was supported by an NIDDK IBD Genetics Consortium Genetic Research Center grant (DK062420) and a U.S. National Cancer Institute grant (CA141743). S.L.H. and J.R.O. would like to also acknowledge the support of National Institutes of Health (R01 NS049477; 1U19A1067152), and the National Multiple Sclerosis Society (RG 2899-D11). S.L. wishes to acknowledge the support from the Australian National Health and Medical Research Council (RD Wright Career Development Fellowship, APP1053756). We would like to thank the International PSC study group (www.ipscsg.org) for sharing data. We are grateful to Benedicte A. Lie and Kristian Holm for helpful discussions.
Members of the International Inflammatory Bowel Disease Genetics Consortium
Clara Abraham30, Jean-Paul Achkar31,32, Tariq Ahmad33, Leila Amininejad34,35, Ashwin N Ananthakrishnan36,37, Vibeke Andersen38,39, Carl A Anderson19, Jane M Andrews40, Vito Annese11,12, Guy Aumais29,41, Leonard Baidoo17, Robert N Baldassano42, Tobias Balschun4, Peter A Bampton43, Murray Barclay44, Jeffrey C Barrett19, Theodore M Bayless45, Johannes Bethge46, Joshua C Bis47, Alain Bitton48, Gabrielle Boucher1, Stephan Brand49, Steven R Brant45, Carsten Büning50, Angela Chew51,52, Judy H Cho53, Isabelle Cleynen54, Ariella Cohain55, Anthony Croft56, Mark J Daly7,8, Mauro D’Amato57, Silvio Danese58, Dirk De Jong59, Martine De Vos60, Goda Denapiene61, Lee A Denson62, Kathy L Devaney36, Olivier Dewit63, Renata D’Inca64, Marla Dubinsky65, Richard H Duerr17,18, Cathryn Edwards66, David Ellinghaus4, Jonah Essers67,68, Lynnette R Ferguson69, Eleonora A Festen70, Philip Fleshner20, Tim Florin71, Denis Franchimont34,35, Andre Franke4, Karin Fransen72, Richard Gearry44,73, Michel Georges74,75, Christian Gieger76, Jürgen Glas48, Philippe Goyette1, Todd Green8,67, Anne M Griffiths77, Stephen L Guthery78, Hakon Hakonarson42, Jonas Halfvarson79,80, Katherine Hanigan56, Talin Haritunians20, Ailsa Hart81, Chris Hawkey82, Nicholas K Hayward83, Matija Hedl30, Paul Henderson84,85, Xinli Hu86, Hailiang Huang7,8, Ken Y Hui53, Marcin Imielinski42, Andrew Ippoliti20, Laimas Jonaitis87, Luke Jostins5,6, Tom H Karlsen26,27,28, Nicholas A Kennedy88, Mohammed Azam Khan89,90, Gediminas Kiudelis87, Subra Kugathasan91, Limas Kupcinskas92, Anna Latiano11, Debby Laukens60, Ian C Lawrance52, James C Lee93, Charlie W Lees88, Marcis Leja94, Johan Van Limbergen95, Paolo Lionetti96, Jimmy Z Liu19, Edouard Louis97, Gillian Mahy98, John Mansfield99, Dunecan Massey93, Christopher G Mathew100,101, Dermot PB McGovern20, Raquel Milgrom102, Mitja Mitrovic72,103, Grant W Montgomery83, Craig Mowat104, William Newman89,90, Aylwin Ng36,105, Siew C Ng106, Sok Meng Evelyn Ng30, Susanna Nikolaus46, Kaida Ning30, Markus Nöthen107, Ioannis Oikonomou30, Orazio Palmieri11, Miles Parkes93, Anne Phillips104, Cyriel Y Ponsioen108, Urõs Potocnik103,109, Natalie J Prescott100,101, Deborah D Proctor110, Graham Radford-Smith111,112, Jean-Francois Rahier113, Soumya Raychaudhuri86, Miguel Regueiro17, Florian Rieder31, John D Rioux1,29, Stephan Ripke7,8, Rebecca Roberts44, Richard K Russell84, Jeremy D Sanderson114, Miquel Sans115, Jack Satsangi88, Eric E Schadt55, Stefan Schreiber4,46, L Philip Schumm21, Regan Scott17, Mark Seielstad116,117, Yashoda Sharma30, Mark S Silverberg118, Lisa A Simms111, Jurgita Skieceviciene87, Sarah L Spain101, A. Hillary Steinhart102, Joanne M Stempak102, Laura Stronati119, Jurgita Sventoraityte92, Stephan R Targan20, Kirstin M Taylor114, Anje ter Velde108, Emilie Theatre74,75, Leif Torkvist120, Mark Tremelling121, Andrea van der Meulen122, Suzanne van Sommeren70, Eric Vasiliauskas20, Severine Vermeire54,123, Hein W Verspaget122, Thomas Walters77,124, Kai Wang42, Ming-Hsi Wang31,45, Rinse K Weersma70, Zhi Wei125, David Whiteman126, Cisca Wijmenga72, David C Wilson84,85, Juliane Winkelmann127,128, Ramnik J Xavier8,36, Sebastian Zeissig46, Bin Zhang55, Clarence K Zhang129, Hu Zhang130,131, Wei Zhang30, Hongyu Zhao129, Zhen Z Zhao83, Australia and New Zealand IBDGC, Belgium IBD Genetics Consortium, Italian Group for IBD Genetic Consortium, NIDDK Inflammatory Bowel Disease Genetics Consortium, Quebec IBD Genetics Consortium, United Kingdom IBDGC, Wellcome Trust Case Control Consortium
Footnotes
Section of Digestive Diseases, Department of Internal Medicine, Yale School of Medicine, NewHaven, Connecticut, USA.
Department of Gastroenterology and Hepatology, Digestive Disease Institute, Cleveland Clinic, Cleveland, Ohio, USA.
Department of Pathobiology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio, USA.
Peninsula College of Medicine and Dentistry, Exeter, UK.
Department of Gastroenterology, Erasmus Hospital, Brussels, Belgium.
Department of Gastroenterology, Free University of Brussels, Brussels, Belgium.
Gastroenterology Unit, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Division of Medical Sciences, Harvard Medical School, Boston, Massachusetts, USA.
Medical Department, Viborg Regional Hospital, Viborg, Denmark.
Organ Center, Hospital of Southern Jutland Aabenraa, Aabenraa, Denmark.
Inflammatory Bowel Disease Service, Department of Gastroenterology and Hepatology, Royal Adelaide Hospital, Adelaide, Australia.
Department of Gastroenterology, Hôpital Maisonneuve-Rosemont, Montréal, Québec, Canada.
Center for Applied Genomics, The Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Department of Gastroenterology and Hepatology, Flinders Medical Centre and School of Medicine, Flinders University, Adelaide, Australia.
Department of Medicine, University of Otago, Christchurch, New Zealand.
Meyerhoff Inflammatory Bowel Disease Center, Department of medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Department for General Internal Medicine, Christian-Albrechts-University, Kiel, Germany.
Cardiovascular Health Research Unit, University of Washington, Seattle, Washington, USA.
Division of Gastroenterology, Royal Victoria Hospital, Montréal, Québec, Canada.
Department of Medicine II, Ludwig-Maximilians-University Hospital Munich-Grosshadern, Munich, Germany.
Department of Gastroenterology, Campus Charité Mitte, Universitatsmedizin Berlin, Berlin, Germany.
IBD unit, Fremantle Hospital, Fremantle, Australia.
School of Medicine and Pharmacology, University of Western Australia, Fremantle, Australia.
Department of Genetics, Yale School of Medicine, New Haven, Connecticut, USA.
Department of Clinical and experimental medicine, Translational Research in GastroIntestinal Disorders (TARGID), Katholieke Universiteit (KU) Leuven, Leuven, Belgium.
Department of Genetics and Genomic Sciences, Mount Sinai School of Medicine, New York, New York, USA.
Inflammatory Bowel Diseases, Genetics and Computational Biology, Queensland Institute of Medical Research, Brisbane, Australia.
Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden.
IBD Center, Department of Gastroenterology, Istituto Clinico Humanitas, Milan, Italy.
Department of Gastroenterology and Hepatology, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands.
Department of Hepatology and Gastroenterology, Ghent University Hospital, Ghent, Belgium.
Center of hepatology, Gastroenterology and Dietetics, Vilnius University, Vilnius, Lithuania.
Pediatric Gastroenterology, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA.
Department of Gastroenterology, Université Catholique de Louvain (UCL) Cliniques Universitaires Saint-Luc, Brussels, Belgium.
Division of Gastroenterology, University Hospital Padua, Padua, Italy.
Department of Pediatrics, Cedars Sinai Medical Center, Los Angeles, California, USA.
Department of Gastroenterology, Torbay Hospital, Torbay, Devon, UK.
Center for Human Genetic Research, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Pediatrics, Harvard Medical School, Boston, Massachusetts, USA.
Faculty of Medical & Health Sciences, School of Medical Sciences, The University of Auckland, Auckland, New Zealand.
Department of Gastroenterology and Hepatology, University Medical Center Groningen, Groningen, The Netherlands.
Department of Gastroenterology, Mater Health Services, Brisbane, Australia.
Department of Genetics, University Medical Center Groningen, Groningen, The Netherlands.
Department of Gastroenterology, Christchurch Hospital, Christchurch, New Zealand.
Unit of Animal Genomics, Groupe Interdisciplinaire de Génoprotéomique Appliquée (GIGA-R) Research Center, University of Liege, Liege, Belgium.
Faculty of Veterinary Medicine, University of Liege, Liege, Belgium.
Institute of Genetic Epidemiology, Helmholtz Zentrum München - German Research Center for Environmental Health, Neuherberg, Germany.
Gastroenterology, Hepatology and Nutrition, The Hospital for Sick Children, Toronto, Ontario, Canada.
Department of Pediatrics, University of Utah School of Medicine, Salt Lake City, Utah, USA.
Department of Medicine, Örebro University Hospital, Örebro, Sweden.
School of Health and Medical Sciences, Örebro University, Örebro, Sweden.
Department of Medicine, St Mark’s Hospital, Harrow, Middlesex, UK.
Nottingham Digestive Diseases Centre, Queens Medical Centre, Nottingham, UK.
Genetic Epidemiology, Genetics and Computational Biology, Queensland Institute of Medical Research, Brisbane, Australia.
Paediatric Gastroenterology and Nutrition, Royal Hospital for Sick Children, Edinburgh, UK.
Child Life and Health, University of Edinburgh, Edinburgh, Scotland, UK.
Division of Rheumatology Immunology and Allergy, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Academy of Medicine, Lithuanian University of Health Sciences, Kaunas, Lithuania.
Gastrointestinal Unit, Wester General Hospital University of Edinburgh, Edinburgh, UK.
Genetic Medicine, Manchester Academic Health Science Centre, Manchester, UK.
The Manchester Centre for Genomic Medicine, University of Manchester, Manchester, UK.
Department of Pediatrics, Emory University School of Medicine, Atlanta, Georgia, USA.
Department of Gastroenterology, Kaunas University of Medicine, Kaunas, Lithuania.
Inflammatory Bowel Disease Research Group, Addenbrooke’s Hospital, Cambridge, UK.
Faculty of medicine, University of Latvia, Riga, Latvia.
Division of Pediatric Gastroenterology, Hepatology and Nutrition, Hospital for Sick Children, Toronto, Ontario, Canada.
Dipartimento di Neuroscienze, Psicologia, Area del Farmaco e Salute del Bambino (NEUROFARBA), Università di Firenze SOD Gastroenterologia e Nutrizione Ospedale pediatrico Meyer, Firenze, Italy.
Division of Gastroenterology, University Hospital CHU of Liege, Liege, Belgium.
Department of Gastroenterology, The Townsville Hospital, Townsville, Australia.
Institute of Human Genetics, Newcastle University, Newcastle upon Tyne, UK.
Department of Medical and Molecular Genetics, Guy’s Hospital, London, UK.
Department of Medical and Molecular Genetics, King’s College London School of Medicine, Guy’s Hospital, London, UK.
Inflammatory Bowel Disease Centre, Mount Sinai Hospital, Toronto, Ontario, Canada.
Center for Human Molecular Genetics and Pharmacogenomics, Faculty of Medicine, University of Maribor, Maribor, Slovenia.
Department of Medicine, Ninewells Hospital and Medical School, Dundee, UK.
Center for Computational and Integrative Biology, Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts, USA.
Department of Medicine and Therapeutics, Institute of Digestive Disease, Chinese University of Hong Kong, Hong Kong.
Department of Genomics Life & Brain Center, University Hospital Bonn, Bonn, Germany.
Department of Gastroenterology, Academic Medical Center, Amsterdam, The Netherlands.
Faculty for Chemistry and Chemical Engineering, University of Maribor, Maribor, Slovenia.
Section of Digestive Diseases, Department of Medicine, Yale University, New Haven, Connecticut, USA.
Inflammatory Bowel Diseases, Genetics and Computational Biology, Queensland Institute of Medical Research, Brisbane, Australia.
Department of Gastroenterology, Royal Brisbane and Womens Hospital, Brisbane, Australia.
Department of Gastroenterology, Université Catholique de Louvain (UCL) Centre hospitalier (CHU) Mont-Godinne, Mont-Godinne, Belgium.
Department of Gastroenterology, Guy’s & St Thomas’ NHS Foundation Trust, St-Thomas Hospital, London, UK.
Department of Digestive Diseases, Hospital Quiron Teknon, Barcelona, Spain.
Human Genetics, Genome Institute of Singapore, Singapore.
Institute for Human Genetics, University of California San Francisco, San Francisco, California, USA.
Inflammatory Bowel Disease Centre, Mount Sinai Hospital, Toronto, Ontario, Canada.
Department of Biology of Radiations and Human Health, Agenzia nazionale per le nuove tecnologie l’energia e lo sviluppo economico sostenibile (ENEA), Rome, Italy.
Department of Clinical Science Intervention and Technology, Karolinska Institutet, Stockholm, Sweden.
Gastroenterology & General Medicine, Norfolk and Norwich University Hospital, Norwich, UK.
Department of Gastroenterology, Leiden University Medical Center, Leiden, The Netherlands.
Division of Gastroenterology, University Hospital Gasthuisberg, Leuven, Belgium.
Faculty of medicine, University of Toronto, Toronto, Ontario, Canada.
Department of Computer Science, New Jersey Institute of Technology, Newark, New Jersey, USA.
Molecular Epidemiology, Genetics and Computational Biology, Queensland Institute of Medical Research, Brisbane, Australia.
Institute of Human Genetics, Technische Universität München, Munich, Germany.
Department of Neurology, Technische Universität München, Munich, Germany.
Department of Biostatistics, School of Public Health, Yale University, NewHaven, Connecticut, USA.
Department of Gastroenterology, West China Hospital, Chengdu, Sichuan, China.
State Key Laboratory of Biotherapy, Sichuan University West China University of Medical Sciences (WCUMS), Chengdu, Sichuan, China
URLs
Additional data on heterogeneity of effects for associated alleles: http://www.medgeni.org/goyette_nature_gen_2015 EBI sequence database: ftp://ftp.ebi.ac.uk/pub/databases/ipd/imgt/hla/
Author Contributions
J.D.R., M.J.D., K.V.S., V.K., T.H.K., A.F. jointly supervised research. J.D.R., P.G., G.B., R.H.D., J.C.B., D.P.B.M., J.A.T., M.N.C., V.K., A.F. conceived and designed the experiments. P.G., G.B., D.M., L.J. performed statistical analysis. P.G., G.B., L.J., E.S.G. analyzed the data. V.A., S.L.H., J.R.O., I.T., S.L., L.P.S. contributed reagents/materials/analysis tools. P.G., G.B., E.E., H.H., S.R. performed data QC and imputation. J.D.R., P.G., G.B., D.M., D.P.B.M., V.K., T.H.K., A.F. wrote the paper. All authors read and approved the final manuscript before submission
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Horton R, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–99. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 2.Rioux JD, et al. Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A. 2009;106:18680–5. doi: 10.1073/pnas.0909307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395–401. doi: 10.1136/gut.45.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achkar JP, et al. Amino acid position 11 of HLA-DRbeta1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 2012;13:245–52. doi: 10.1038/gene.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones DC, et al. Killer Ig-like receptor (KIR) genotype and HLA ligand combinations in ulcerative colitis susceptibility. Genes Immun. 2006;7:576–82. doi: 10.1038/sj.gene.6364333. [DOI] [PubMed] [Google Scholar]
- 7.Kulkarni S, et al. Genetic interplay between HLA-C and MIR148A in HIV control and Crohn disease. Proc Natl Acad Sci U S A. 2013;110:20705–10. doi: 10.1073/pnas.1312237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satsangi J, et al. Contribution of genes of the major histocompatibility complex to susceptibility and disease phenotype in inflammatory bowel disease. Lancet. 1996;347:1212–7. doi: 10.1016/s0140-6736(96)90734-5. [DOI] [PubMed] [Google Scholar]
- 9.Oksenberg JR, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am J Hum Genet. 2004;74:160–7. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman B, et al. CARD15 and HLA DRB1 alleles influence susceptibility and disease localization in Crohn’s disease. Am J Gastroenterol. 2004;99:306–15. doi: 10.1111/j.1572-0241.2004.04038.x. [DOI] [PubMed] [Google Scholar]
- 11.Lipsitch M, Bergstrom CT, Antia R. Effect of human leukocyte antigen heterozygosity on infectious disease outcome: the need for allele-specific measures. BMC Med Genet. 2003;4:2. doi: 10.1186/1471-2350-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alper CA, Fleischnick E, Awdeh Z, Katz AJ, Yunis EJ. Extended major histocompatibility complex haplotypes in patients with gluten-sensitive enteropathy. J Clin Invest. 1987;79:251–6. doi: 10.1172/JCI112791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aly TA, et al. Multi-SNP analysis of MHC region: remarkable conservation of HLA-A1-B8-DR3 haplotype. Diabetes. 2006;55:1265–9. doi: 10.2337/db05-1276. [DOI] [PubMed] [Google Scholar]
- 14.Wiencke K, Spurkland A, Schrumpf E, Boberg KM. Primary sclerosing cholangitis is associated to an extended B8-DR3 haplotype including particular MICA and MICB alleles. Hepatology. 2001;34:625–30. doi: 10.1053/jhep.2001.27543. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson PT, Norris S. Evaluation of the role of MHC class II alleles, haplotypes and selected amino acid sequences in primary sclerosing cholangitis. Autoimmunity. 2002;35:555–64. doi: 10.1080/0891693021000054093. [DOI] [PubMed] [Google Scholar]
- 16.Liu JZ, et al. Dense genotyping of immune-related disease regions identifies nine new risk loci for primary sclerosing cholangitis. Nat Genet. 2013;45:670–5. doi: 10.1038/ng.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International Multiple Sclerosis Genetics C et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah TS, et al. optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics. 2012;28:1598–603. doi: 10.1093/bioinformatics/bts180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dilthey A, et al. Multi-population classical HLA type imputation. PLoS Comput Biol. 2013;9:e1002877. doi: 10.1371/journal.pcbi.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gourraud PAKP, Cereb N, Yang SY, Feolo M, Maiers M, Rioux JD, Hauser S, Oksenberg J. HLA diversity in the 1000 Genomes dataset. PloS One. 2014 doi: 10.1371/journal.pone.0097282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Veall MRZKF. Pseudo-R2 measures for some common limited dependent variable models. J econometric surveys. 10:241–259. [Google Scholar]
- 26.McKelvey RDZW. A statistical model for the analysis of ordinal level dependent variables. J Math Sociology. 1975;4:103–120. [Google Scholar]
- 27.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;Chapter 2(Unit 2):9. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 28.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 31.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–5. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 32.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–24. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 34.Dolinsky TJ, et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–5. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6:271–82. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 37.Richter S, Wenzel A, Stein M, Gabdoulline RR, Wade RC. webPIPSA: a web server for the comparison of protein interaction properties. Nucleic Acids Res. 2008;36:W276–80. doi: 10.1093/nar/gkn181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wade RC, Gabdoulline RR, De Rienzo F. Protein interaction property similarity analysis. Int J Quantum Chem. 2001;83:122–127. [Google Scholar]