Abstract
The mammalian gastrointestinal tract is home to trillions of commensal microorganisms that collectively make up the intestinal microbiota. These microbes are important environmental factors that regulate homeostasis and alterations in the composition of the microbiota have been associated with several diseases, including inflammatory bowel disease (IBD), diabetes, and cancer. New research is beginning to uncover epigenomic pathways that may regulate this relationship with the microbiota. Epigenomic modifications alter the structure of the chromatin and therefore alter the transcriptional program of a cell. These modifications are maintained by the dynamic activity of various modifying and demodifying enzymes, the activities of which can be influenced by metabolites and other environmental cues. Histone deacetylases (HDACs) are a class of epigenomic-modifying enzymes that are regulated by both endogenous and exogenous factors and recent studies have suggested that host HDAC expression is important for regulating communication between the intestinal microbiota and mammalian host cells.
Keywords: epigenomics, microbiota, HDAC, butyrate
Introduction
An individual’s genetic makeup is highly influential with regards to the development of disease, but often the genome is not the sole factor. In addition to genetic susceptibility, there is significant evidence that the environment influences the pathogenesis of a variety of chronic diseases, such as inflammatory bowel disease (IBD), diabetes, cancer, heart disease, and allergy (Kaser et al, 2010; Kau et al, 2011; Mukherjee & Zhang, 2011; Renz et al, 2011). Therefore, understanding the fundamental mechanisms that influence how environmental factors regulate gene expression could aid in the development of novel therapeutics to prevent or limit chronic human diseases. There is increasing evidence that the commensal microbes that normally colonize our bodies, referred to as our microbiota, can act as environmental factors that influence both health and disease (Cadwell et al, 2010; Ivanov & Honda, 2012; Kamada et al, 2013; Kaser et al, 2010; Kau et al, 2011).
Commensal bacteria colonize all body surfaces that are exposed to the external environment, such as the gastrointestinal and respiratory tracts as well as the skin. These bacteria have co-evolved with their hosts and have formed complex ecosystems in specific anatomical locations of the mammalian body (Kamada et al, 2013; Ley et al, 2008). The number of bacteria that normally colonize the intestinal and skin surfaces greatly exceeds the number of human cells in the body. It is estimated that there are approximately ten times more resident microbes than human cells. Further, the microbiome, which encompasses the collective genomes of these commensal microbes, makes up more than 99 percent of the genetic information in the human body (Backhed et al, 2005).
Recent molecular advances in high-throughput metagenomic deep sequencing analyses have greatly expanded our ability to identify bacterial communities throughout the body in human health and disease, as well as in animal models. Specifically, bacterial DNA can be isolated from distinct anatomical locations, sequenced, and compared computationally to determine microbial diversity and phylogenetic composition. These approaches have resulted in a plethora of collaborations and publications characterizing core bacterial communities in specific regions of the body under healthy and disease conditions (Caporaso et al, 2010; Cho & Blaser, 2012; Huttenhower C, 2012).
The host-microbiota relationship and intestinal homeostasis
The majority of the commensal bacteria reside within the intestine where they directly interact with a single layer of intestinal epithelial cells (IECs) that line the lumen. It has become evident that the mammalian host has formed a symbiotic relationship with these intestinal commensal bacteria. In particular, the intestinal microbiota plays a critical role in the normal digestion of food, resulting in the release of nutrients and metabolites that are essential for the maintenance of mammalian health (Tremaroli & Backhed, 2012). As the vast majority of the microbiota resides in the mammalian intestine, one strategy to decipher how the host and microbiota interact has been to focus on evaluating commensal bacteria-dependent homeostatic regulation of mammalian cells in the intestinal microenvironment.
A dynamic and complex relationship exists between the intestinal microbiota, the immune system, and the intestinal epithelium (Artis, 2008; Hooper et al, 2012). IECs function as a crucial cell lineage that resides at the interface between the mammalian host and commensal bacteria. These cells form a physical barrier, sense bacterial-derived signals, and secrete antimicrobial peptides and cytokines/chemokines that, in turn, regulate the microbiota and immune cell homeostasis (Gallo & Hooper, 2012; Roda et al, 2010). Further, epithelial permeability, proliferation, and expression of antimicrobial proteins can be regulated by cytokines produced by immune cells (Dahan et al, 2007). The microbiota, itself, influences immune cell homeostasis and is critical in the development/maturation of both the innate and adaptive system (Khosravi et al, 2014; Littman & Pamer, 2011). In fact, recent studies have identified specific species of commensal bacteria in the intestine that drive differentiation of intestinal CD4+ T cells to distinct lineages, suggesting that there may be core species of microbes that are required for development of select immune cell populations (Atarashi et al, 2011; Littman & Pamer, 2011).
In addition to intestinal homeostasis, a wide range of disease entities, including inflammatory bowel disease (IBD) obesity, cancer, and allergy have been associated with dysregulation of the host-microbiota relationship and alterations in the diversity of intestinal commensal bacteria (dysbiosis) (Arthur et al, 2012; Kamada et al, 2013; Kau et al, 2011; McLoughlin & Mills, 2011). The pathogenesis of IBD, in particular, appears to be driven both by altered immune responses to commensal bacteria in addition to alterations in the composition of these commensal bacteria (Kamada et al, 2013; Kaser et al, 2010; Maloy & Powrie, 2011). Further, commensal bacteria help to prevent pathogen infection in part by stimulating the mammalian host’s immune defenses in a manner that preserves their own survival and simultaneously limits infection (Abt & Pamer, 2014). A large field of research is now dedicated towards better understanding the effects of manipulating commensal bacterial communities. Many of these studies investigate how altering the diversity of the microbiota, possibly through the use of antibiotics, could negatively impact health and disease susceptibility. Further, investigation into probiotics has significantly expanded in order to identify potential cocktails of commensal bacteria or bacterial-derived products that can beneficially regulate the immune system, confer protection against infections, or aid in digestion (Sanders et al, 2013).
Chromatin and the epigenome
Dynamic transcriptional regulation is essential in mediating the symbiotic host-microbiota relationship. Eukaryotic cells package their DNA around histone proteins to form a higher order structure termed chromatin. The repetitive element within chromatin, called the nucleosome, is composed of 147bp of DNA tightly wound around a histone octamer. Histone H1 functions as a linker between nucleosomes that permits further condensation of the chromatin structure. Chromatin, itself, is generally repressive by limiting access of transcriptional machinery to the genome. However, covalent nucleosomal modifications as well as ATP dependent chromatin remodeling ATPases enable chromatin flexibility in response to specific cellular signals. The chromatin structure can, therefore, undergo local condensation or decondensation which facilitates various processes such as DNA replication, repair, or transcription (Richmond & Davey, 2003). The most well characterized mechanisms that regulate the chromatin structure are DNA methylation and histone modifications, each of which can modify gene expression without altering the associated DNA sequence. Histone N-terminus tails extend from the nucleosomal core and provide a template for various covalent modifications such as acetylation, phosphorylation, methylation, SUMOylation, ubiquitination, and more. The pattern of modifications on these histone tails establish a “histone code” which guides changes in chromatin structure and/or directs recruitment of specific cofactors (Jenuwein & Allis, 2001; Strahl & Allis, 2000).
While covalent DNA and histone modifications are most commonly discussed as epigenetic phenomena, the term epigenetics often suggests heritability of the associated changes in gene expression. A broader concept of the epigenome has more recently been adopted to refer to the combination of histone and DNA modifications and associated proteins that package the genome and define the transcriptional program of a cell (Arrowsmith et al, 2012). Epigenomic modifications are maintained by the balanced activity of various enzymes. Alterations in epigenomic modifications provide a mechanism by which the environment can alter gene expression and influence disease pathogenesis. For this reason, epigenomic mechanisms have been implicated in the development of most chronic conditions with complex gene-environment etiologies, including cancer, diabetes, allergy, atherosclerosis, and IBD (Begin & Nadeau, 2014; Feil & Fraga, 2011; Handel et al, 2010; Renz et al, 2011).
Do epigenomic pathways regulate the host-microbiota relationship?
As discussed earlier, the immune system is a key component that shapes the microbiota and the epigenome clearly mediates development and differentiation of immune cells (Cedar & Bergman, 2011; Cuddapah et al, 2010; Natoli, 2010; O’Shea et al, 2011). Recent work demonstrates more direct links between commensal bacterial-derived signals and host epigenomic pathways, particularly though histone acetylation. Acetylation of histone tails is believed to disrupt the DNA-histone interaction, causing local relaxation of the chromatin and permitting access for transcription machinery (Chen et al, 2001). Furthermore, histone acetylated-lysines are the preferred substrate for proteins that propagate increased acetylation and promote transcriptional activation (Eberharter & Becker, 2002).
Conversely, removal of the acetyl groups by histone deaceytlases (HDACs) generally promotes tighter DNA-histone associations and inhibits transcriptional activity. There are 18 known HDACs that are classified into four groups based on their homology to yeast HDACs and subcellular location. The class I, II, and IV HDACs require zinc for their enzymatic deacetylase activities, whereas class III HDACs (sirtuins) depend on nicotine adenine dinucleotide as a cofactor (Sauve et al, 2006). These enzymes are often found in large complexes that are recruited to the chromatin through interactions with transcription factors. Further, their activity and recruitment to the genome can be altered by hormones, dietary compounds, and bacterial-derived products (Chen et al, 2012; Dashwood & Ho, 2007; Donohoe & Bultman, 2012; Haberland et al, 2009; Kim et al, 2010; Perissi & Rosenfeld, 2005). In vivo studies suggest that the specificity of HDACs in regulating distinct gene programs differs with cell identity, available associating proteins, and the cell signaling environment (Haberland et al, 2009).
Importantly, the zinc-dependent HDACs can be targeted by a large class of inhibitors that are currently being utilized or examined for the treatment of various types of cancer, as well as inflammatory and degenerative conditions (Haberland et al, 2009; Huang, 2006). The therapeutic potential of HDAC inhibitors (HDACi) is promising, however the mechanisms underlying their clinical effects are not fully understood. Studies directed towards better understanding their specificity and mode of action are ongoing (Ververis et al, 2013).
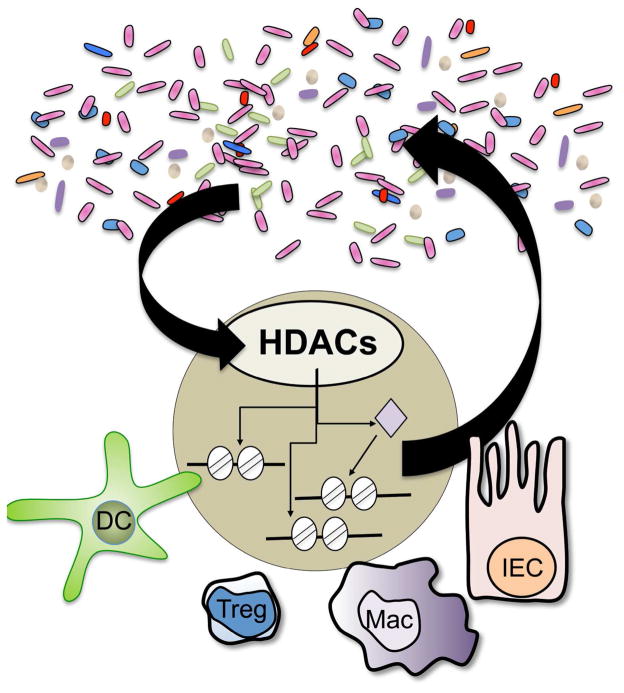
While studies have indicated that the microbiota regulates DNA and histone methylation dependent pathways in the host (Ganal et al, 2012; Kellermayer et al, 2011; Obata et al, 2014; Olszak et al, 2012; Takahashi et al, 2011), recent work over the last year has brought histone acetylation and HDACs to the forefront as a critical level of epigenomic regulation that mediates the interplay between mammalian host cells and the intestinal microbiota (Figure 1). The class I HDAC, HDAC3 regulates transcription through histone deacetylation, but has also been suggested to deacetylate non-histone targets and possess enzyme-independent effects (Choudhary et al, 2009; Sun et al, 2013; You et al, 2010). As discussed earlier, IECs are central to integrating signals from the intestinal microenvironment to regulate intestinal homeostasis. Recent work revealed that loss of IEC-specific HDAC3 expression led to extensive alterations in gene expression, changes in histone acetylation, impaired Paneth cell survival, and decreased intestinal barrier function. Lack of HDAC3-dependent regulation in IECs also resulted in significant alterations in the composition of the intestinal microbiota as well as increased susceptibility to intestinal damage and inflammation. Interestingly, elimination of the microbiota by generating a germ-free HDAC3-deficient mouse strain restored Paneth cell homeostasis and the functionality of the intestinal barrier, suggesting that HDAC3 in IECs mediates functional crosstalk between commensal bacteria and host cells (Alenghat et al, 2013). However, the underlying microbiota-dependent mechanisms that orchestrate HDAC3 function in this pathway remain to be defined.
Figure 1.
Recent studies indicate that HDACs, a family of epigenomic-modifying enzymes, mediate dynamic regulation between the microbiota and multiple cell lineages in the mammalian intestine. Manipulation of these pathways may result in altered intestinal homeostasis and susceptibility to inflammation, as well as changes in the diversity of the microbiota. HDAC, Histone deacetylase; IEC, intestinal epithelial cell; Treg, regulatory T cell; DC, dendritic cell; Mac, Macrophage.
The microbiota can produce byproducts that modify the epigenome of host cells and in turn alter the cell’s function and the intestinal environment. HDACs in immune cells have been highlighted as targets of microbiota-derived short chain fatty acids (SCFAs). SCFAs that are produced by commensal bacteria through fermentation of dietary carbohydrates in the colon can function as HDAC inhibitors (Macfarlane & Macfarlane, 2003) and have been found to regulate the development and function of several immune cell lineages (Brestoff & Artis, 2013) A series of recent high profile publications demonstrated that SCFAs derived from commensal bacteria in the large intestine exert anti-inflammatory effects in the colon by stimulating histone acetylation of the FoxP3 locus and driving differentiation of regulatory T cells (Tregs) (Arpaia et al, 2013; Furusawa et al, 2013; Smith et al, 2013). Smith et al. suggested a mechanism by which SCFAs decreased expression of HDAC6 and/or HDAC9 in Tregs in a G-protein-coupled receptor-dependent manner. Consistent with these SCFA findings, synthetic pan-HDACi were previously found to limit colitis through expansion of Foxp3+ Tregs and associated differences in HDAC9 expression (de Zoeten et al, 2010; Glauben et al, 2008; Tao et al, 2007).
The effects of butyrate, a microbiota-derived SCFA, on histone acetylation in myeloid cell lineages have also been examined. Arpaia et al. found that butyrate increased histone H3 acetylation in dendritic cells, enhancing their ability to facilitate Treg differentiation and Chang et al. recently demonstrated that treatment of macrophages with butyrate increased histone acetylation and decreased expression of pro-inflammatory cytokines (Arpaia et al, 2013; Chang et al, 2014). While butyrate clearly increases levels of histone acetylation in immune cells, it remains unclear how this relates to physiologic concentrations of SCFAs in the colon and whether commensal-derived SCFAs increase histone acetylation primarily through direct or indirect regulation of HDAC expression versus inhibition of HDAC enzymatic activity. Further, multiple outcomes and mechanisms have been suggested for butyrate treatment during colitis, which warrants a more thorough examination of the differential effects of SCFAs on specific HDACs in different host cells, in the context of protective and pathologic immunity (Berndt et al, 2012; Chang et al, 2014; Hamer et al, 2010; Kim et al, 2013; Tarrerias et al, 2002).
Collectively, it is becoming evident that proper innate and adaptive immune system development and function supports the maintenance of a healthy intestinal microbiota and that epigenomic pathways in host epithelial and immune cells mediate crosstalk between the microbiota and the mammalian host. Although deciphering the role of epigenomic regulation in mediating the mammalian relationship with commensal microbes is still in its infancy, these recent advances provoke several questions regarding the potential relevance of evaluating epigenomic endpoints and the microbiota in toxicologic pathology. Can we evaluate epigenomic modifications in the gut to assess intestinal health? How does clinical inhibition of epigenomic pathways affect the intestinal microbiota? What is the impact of drug-dependent alterations of the microbiota on epigenomic regulation? How does variability in the microbiota alter drug metabolism, efficacy, and toxicity? Continued basic mechanistic and translational studies will dictate how best to focus our efforts towards addressing these issues.
Acknowledgments
The author thanks T. Fung, J. Brestoff, and M. Abt for discussions and reading of the manuscript. This work is supported by the National Institutes of Health (DK093784) and the Burroughs Wellcome Fund Career Award for Medical Scientists.
Abbreviations
- HDAC
Histone deacetylase
- IBD
inflammatory bowel disease
- IEC
intestinal epithelial cell
- HDACi
HDAC inhibitors
- SCFA
short chain fatty acid
- Treg
regulatory T cell
References
- Abt MC, Pamer EG. Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol. 2014;29C:16–22. doi: 10.1016/j.coi.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat T, Osborne LC, Saenz SA, Kobuley D, Ziegler CG, Mullican SE, Choi I, Grunberg S, Sinha R, Wynosky-Dolfi M, Snyder A, Giacomin PR, Joyce KL, Hoang TB, Bewtra M, Brodsky IE, Sonnenberg GF, Bushman FD, Won KJ, Lazar MA, Artis D. Histone deacetylase 3 coordinates commensal-bacteria-dependent intestinal homeostasis. Nature. 2013;504:153–157. doi: 10.1038/nature12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. 2008;8:411–420. doi: 10.1038/nri2316. [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Begin P, Nadeau KC. Epigenetic regulation of asthma and allergic disease. Allergy Asthma Clin Immunol. 2014;10:27. doi: 10.1186/1710-1492-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt BE, Zhang M, Owyang SY, Cole TS, Wang TW, Luther J, Veniaminova NA, Merchant JL, Chen CC, Huffnagle GB, Kao JY. Butyrate increases IL-23 production by stimulated dendritic cells. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1384–1392. doi: 10.1152/ajpgi.00540.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11:478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tini M, Evans RM. HATs on and beyond chromatin. Curr Opin Cell Biol. 2001;13:218–224. doi: 10.1016/s0955-0674(00)00200-3. [DOI] [PubMed] [Google Scholar]
- Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Barski A, Zhao K. Epigenomics of T cell activation, differentiation, and memory. Curr Opin Immunol. 2010;22:341–347. doi: 10.1016/j.coi.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan S, Roth-Walter F, Arnaboldi P, Agarwal S, Mayer L. Epithelia: lymphocyte interactions in the gut. Immunol Rev. 2007;215:243–253. doi: 10.1111/j.1600-065X.2006.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–369. doi: 10.1016/j.semcancer.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoeten EF, Wang L, Sai H, Dillmann WH, Hancock WW. Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology. 2010;138:583–594. doi: 10.1053/j.gastro.2009.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe DR, Bultman SJ. Metaboloepigenetics: interrelationships between energy metabolism and epigenetic control of gene expression. J Cell Physiol. 2012;227:3169–3177. doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–229. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2011;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gallo RL, Hooper LV. Epithelial antimicrobial defence of the skin and intestine. Nat Rev Immunol. 2012;12:503–516. doi: 10.1038/nri3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Glauben R, Batra A, Stroh T, Erben U, Fedke I, Lehr HA, Leoni F, Mascagni P, Dinarello CA, Zeitz M, Siegmund B. Histone deacetylases: novel targets for prevention of colitis-associated cancer in mice. Gut. 2008;57:613–622. doi: 10.1136/gut.2007.134650. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer HM, Jonkers DM, Vanhoutvin SA, Troost FJ, Rijkers G, de Bruine A, Bast A, Venema K, Brummer RJ. Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr. 2010;29:738–744. doi: 10.1016/j.clnu.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Handel AE, Ebers GC, Ramagopalan SV. Epigenetics: molecular mechanisms and implications for disease. Trends Mol Med. 2010;16:7–16. doi: 10.1016/j.molmed.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. Targeting histone deacetylases for the treatment of cancer and inflammatory diseases. J Cell Physiol. 2006;209:611–616. doi: 10.1002/jcp.20781. [DOI] [PubMed] [Google Scholar]
- Huttenhower CGD, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol. 2010;28:573–621. doi: 10.1146/annurev-immunol-030409-101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermayer R, Dowd SE, Harris RA, Balasa A, Schaible TD, Wolcott RD, Tatevian N, Szigeti R, Li Z, Versalovic J, Smith CW. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–1460. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, Mazmanian SK. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15:374–381. doi: 10.1016/j.chom.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GW, Gocevski G, Wu CJ, Yang XJ. Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery. Int J Cell Biol. 2010;2010:632739. doi: 10.1155/2010/632739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406. e391–310. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- McLoughlin RM, Mills KH. Influence of gastrointestinal commensal bacteria on the immune responses that mediate allergy and asthma. J Allergy Clin Immunol. 2011;127:1097–1107. doi: 10.1016/j.jaci.2011.02.012. quiz 1108–1099. [DOI] [PubMed] [Google Scholar]
- Mukherjee AB, Zhang Z. Allergic asthma: influence of genetic and environmental factors. J Biol Chem. 2011;286:32883–32889. doi: 10.1074/jbc.R110.197046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–250. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi M, Ikawa T, Otsubo T, Kawamura YI, Dohi T, Tajima S, Masumoto H, Ohara O, Honda K, Hori S, Ohno H, Koseki H, Hase K. The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol. 2014;15:571–579. doi: 10.1038/ni.2886. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Renz H, von Mutius E, Brandtzaeg P, Cookson WO, Autenrieth IB, Haller D. Gene-environment interactions in chronic inflammatory disease. Nat Immunol. 2011;12:273–277. doi: 10.1038/ni0411-273. [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Roda G, Sartini A, Zambon E, Calafiore A, Marocchi M, Caponi A, Belluzzi A, Roda E. Intestinal epithelial cells in inflammatory bowel diseases. World J Gastroenterol. 2010;16:4264–4271. doi: 10.3748/wjg.v16.i34.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sun Z, Feng D, Fang B, Mullican SE, You SH, Lim HW, Everett LJ, Nabel CS, Li Y, Selvakumaran V, Won KJ, Lazar MA. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Sugi Y, Nakano K, Tsuda M, Kurihara K, Hosono A, Kaminogawa S. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J Biol Chem. 2011;286:35755–35762. doi: 10.1074/jbc.M111.271007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- Tarrerias AL, Millecamps M, Alloui A, Beaughard C, Kemeny JL, Bourdu S, Bommelaer G, Eschalier A, Dapoigny M, Ardid D. Short-chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS-induced colonic inflammation in rats. Pain. 2002;100:91–97. doi: 10.1016/s0304-3959(02)00234-8. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Ververis K, Hiong A, Karagiannis TC, Licciardi PV. Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents. Biologics. 2013;7:47–60. doi: 10.2147/BTT.S29965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You SH, Liao X, Weiss RE, Lazar MA. The interaction between nuclear receptor corepressor and histone deacetylase 3 regulates both positive and negative thyroid hormone action in vivo. Mol Endocrinol. 2010;24:1359–1367. doi: 10.1210/me.2009-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]