Abstract
Common fragile sites (CFSs) are large regions with profound genomic instability that often span extremely large genes a number of which have been found to be important tumor suppressors. RNA sequencing previously revealed that there was a group of six large CFS genes which frequently had decreased expression in oropharyngeal squamous cell carcinomas (OPSCCs) and real-time reverse transcriptase polymerase chain reaction experiments validated that these six large CFS genes (PARK2, DLG2, NBEA, CTNNA3, DMD, and FHIT) had decreased expression in most of the tumor samples. In this study, we investigated whether the decreased expression of these genes has any clinical significance in OPSCCs. We analyzed the six CFS large genes in 45 OPSCC patients and found that 27 (60%) of the OPSCC tumors had decreased expression of these six genes. When we correlated the expression of these six genes to each patient’s clinical records, for 11 patients who had tumor recurrence, 10 of them had decreased expression of almost all 6 genes. When we divided the patients into two groups, one group with decreased expression of the six genes and the other group with either slight changes or increased expression of the six genes, we found that there is significant difference in the incidence of tumor recurrence between these two groups by Kaplan-Meier plot analysis (P < .05). Our results demonstrated that those OPSCC tumors with decreased expression of this select group of six large CFS genes were much more likely to be associated with tumor recurrence and these genes are potential prognostic markers for predicting tumor recurrence in OPSCC.
Introduction
Head and neck cancer is the sixth most common malignancy worldwide, but its overall incidence in the United States has declined due to the decreased incidence of smoking [1]. However, oropharyngeal squamous cell carcinoma (OPSCC), one subtype of head and neck cancer with tumors derived from the tonsil or the base of the tongue, has been dramatically increasing in recent decades. This is most probably a result of the dramatic increase in the proportion of OPSCCs that have human papillomavirus (HPV) infection due to changing sexual practices [2], [3]. The presence of HPV in OPSCC has important clinical significance as many reports have shown that HPV-positive OPSCC patients are associated with significantly improved overall survival as compared to HPV-negative OPSCC patients [4], [5]. The evaluation of the presence of HPV has been incorporated into the clinical treatment of the OPSCC, and there is considerable discussion about de-escalation of the therapies for the patients with HPV-positive OPSCC [6], [7]. Currently, prognostic evaluation of OPSCC patients is based on pathological staging on tumor, nodal status, and distant metastasis (DM) and histopathological parameters. What is lacking, however, are good molecular markers to help determine which patients are more likely to have tumor recurrence either with local recurrence or DM, as this clinical outcome is highly predictive of overall patient survival.
Common fragile sites (CFSs) are large regions of profound genomic instability that are observed cytogenetically when cells are cultured in the presence of inhibitors of replication such as the DNA polymerase α inhibitor aphidicolin [8]. These sensitive regions are also found to be hot spots for deletions, translocations, and other alterations in different cancers. CFSs are hot spots for viral integrations as over 50% of human papillomavirus 16 and 18 integration sites in the human genome in cervical cancers occur within one of the CFS regions [9], [10]. There is a group of genes which span extremely large genomic regions which were found to be localized within CFSs. The three most unstable CFS regions in lymphoctyes are FRA3B (3p14.2), FRA16D (16q23.2), and FRA6E (6q26) [11], [12], [13]. Each of these CFS regions extends for 2 or more megabases, and each spans at least one extremely large gene [14]. These genes are FHIT, WWOX, and PARK2, respectively, and each has been found to function as tumor suppressors involved in the development of many different cancers and is also related to disease progression in different cancers [12], [13], [15]. Many of the other large CFS genes are also targets for alteration in cancers, but they have not yet been functionally tested to determine if they too are tumor suppressors.
We previously did RNA seq analysis in a small group of head and neck tumors and matched normal tissue from the same patients [16]. When we specifically focused on the expression of the largest human genes, we observed that there was a select group of six large CFS genes which consistently had decreased expression in the tumors. These genes were DLG2, NBEA, CTNNA3, DMD, and the two tumor suppressors, FHIT and PARK2 [17]. We validated these observations using real-time reverse transcriptase polymerase chain reaction (RT-PCR) examining a large number of OPSCC specimens. Almost 60% of OPSCC tumors that we tested had decreased expression of all six of these large CFS genes. In this report, we examined the clinical data which were available for the OPSCC tumors analyzed and correlated them with CFS gene expression to evaluate if the decreased expression of these genes has any clinical significance.
Materials and Methods
Patient Information
OPSCC samples were collected from patients undergoing surgical treatment from the Department of Otorhinolaryngology at Mayo Clinic, Rochester, MN, from 2008 to 2010. The study was approved by the Institutional Review Board of the Mayo Clinic. After achieving negative surgical margins, the normal tissue was collected from the immediately adjacent site. All tissue samples were snap frozen in liquid nitrogen for storage, and a hematoxylin and eosin slide for each sample was also prepared for evaluating the presence or absence of tumor by the pathologist. For all the tumor sections, over 80% of cells being neoplastic were considered optimal. Normal tissue was also evaluated to make sure that it did not have too many other cell types (such as infiltrating lymphocytes or necrotic cells).
RNA Extraction and Real-Time RT-PCR
RNA from each tumor-normal pair was isolated using Qiagen RNeasy mini kits (Qiagen, Valencia, CA) according to manufacturer’s protocol. Extracted RNA was quantified by NanoDrop ND1000 (Thermo Fisher Scientific, Waltham, MA). RNA isolated from each tumor-normal pairs was reverse transcribed using oligo dT for cDNA synthesis (Life Technologies, Grand Island, NY). The cDNA produced were then utilized for real-time PCR to determine the large genes’ expression. Each sample for real-time PCR was run in duplicate, and the difference in Ct between any two duplicated samples was always less than 0.3. The average Ct value for each sample was calculated for analysis. β-Actin and GAPDH were used as an internal control. Each primer used for analysis is as previously described [17].
Statistical Analysis
The statistical analysis for the recurrence Kaplan-Meier curve was performed using SPSS software (Version 21.0).
Results
Patients’ Characteristics
Forty-five OPSCC patients who had surgical treatment from the Department of Otorhinolaryngology at Mayo Clinic, Rochester, MN, from 2008 to 2010 were used in this study. The patients’ clinical characteristics are listed in Table 1. Among these 45 patients, 80% of the patients were male, consistent with what is observed for OPSCC in the United States. There are 24 patients (53%) whose tumors were from the base of the tongue and 21 patients (47%) whose tumors were from the tonsil. Among these tumors, 80% were HPV 16 positive. Among these 45 patients analyzed, there are 11 patients who developed disease recurrence, with 7 of them having distant pulmonary metastasis and 4 of them having just local recurrence. We had previously analyzed 47 OPSCCs to validate the decreased expression of six large CFS genes. Unfortunately, the clinical data for tumor recurrence were not available for over half of these patients. Thus, in this report, we have found a total of 45 patients (including 23 from the previous paper) that did have this information, and these were the patients analyzed to make our conclusions.
Table 1.
Clinical Characteristics of the 45 OPSCC patients
| All Tumors | Recur | W/ Decreased Expression of Six Large Genes | |
|---|---|---|---|
| Age | |||
| Median | 59 | 60 | 60 |
| Range | 37-79 | 42-79 | 37-79 |
| Sex, No. (%) | |||
| Male | 36 (80%) | 9 (25%) | 22(61%) |
| Female | 9 (20%) | 2 (22%) | 5(56%) |
| Site, No. (%) | |||
| Base of tongue | 24 (53%) | 6 (25%) | 14(51%) |
| Tonsil | 21 (47%) | 5 (24%) | 13(49%) |
| HPV status | |||
| Positive | 36 (80%) | 9 (25%) | 22(61%) |
| Negative | 9 (20%) | 2 (22%) | 5(56%) |
| AJCC stage, No. (%) | |||
| I | 1 (2%) | 1 (n/a) | 1(n/a) |
| II | 2 (4%) | 0 (n/a) | 0(n/a) |
| III | 5 (11%) | 0 (n/a) | 3(n/a) |
| IVA | 35 (79%) | 9 (n/a) | 22(n/a) |
| IVB | 2 (4%) | 1 (n/a) | 1(n/a) |
Decreased Expression of the Selected Six CFS Large Genes Was Associated with Disease Recurrence
We previously reported that there were six CFS large genes that frequently had decreased expression in the OPSCC tumor samples. These six genes are PARK2, DLG2, NBEA, CTNNA3, DMD, and FHIT. Each gene’s full name and its potential function are listed in Table 2. In this group of genes, each of them has been shown to play some important role in cancer, and all have been shown to have decreased expression in a number of different cancers. PARK2 and FHIT have been demonstrated as tumor suppressors, while the other four genes are very attractive potential tumor suppressors. In this study, we analyzed expression of these six large CFS genes by quantitative real-time PCR in each individual tumor and matched normal tissue samples from 45 patients. Each individual gene’s expression difference in tumor was calculated as a ∆∆Ct by comparing the ∆Ct in the tumor to the ∆Ct in its matched normal tissue using GAPDH as an internal normalization control. Since the Ct value is in log2 format, if the ∆∆Ct value is larger than 1, it means that the mRNA expression difference between the tumor and normal is over greater than two times. In this study, the expression of all six large CFS genes appeared to be coordinated in most samples. Thus, the expression of these six genes was analyzed as a group in this study. Of the 45 OPSCC tumors analyzed, there were 27 (60%) that had decreased expression of all 6 large CFS genes, 9 (20%) that had had modest or no changes in the expression of the 6 genes, and 9 (20%) with slightly increased expression of all 6 genes (Figure 1A). The criteria for determining whether an individual tumor had increased or decreased expression of the six genes were based on the ∆∆Ct value of the six genes: in this group of six genes, if the ∆∆Ct value of at least four genes was above or lower than 1, we classified this group of genes as having decreased or increased expression; if most genes’ ∆∆Ct value of difference was less than 1 or some were upregulated and some were downregulated, it was defined as unchanged in this study. When we correlated gene expression to each patient’s clinical characteristics, what was most striking was that of the 11 patients that had tumor recurrence, 10 of them had decreased expression of almost all 6 genes. There was only one patient (patient 642) who had recurrence and yet no decrease in the expression of these genes (actually a slightly increased expression of almost all six genes in his tumor) (Figure 1A and B).
Table 2.
The Six CFS Large Genes’ Chromosome Regions, Size and Their Implications in Different Cancers
| Chromosome Locations | CFS Region | Size (bp) | Gene Description | Implications in Different Cancers | |
|---|---|---|---|---|---|
| PARK2 | 6q26 | FRA6E | 1379130 | parkin, RBR E3 ubiquitin protein ligase | Reduced or absent PARK2 transcripts were found in ovarian cancer, breast cancer, renal cancer, lung cancer, and sporadic colorectal cancer. |
| DLG2 | 11q14.1 | FRA11F | 1463760 | discs, large homolog 2(Drosophila) | It is involved in epithelial polarity during cell division and has been implicated in cancer cell invasion. Recurrent somatic alteration was observed in pediatric osteosarcoma. |
| NBEA | 13q13 | FRA13A | 730451 | neurobeachin | It is a target of recurrent interstitial deletions in patients with monoclonal gammopathy of undetermined significance and multiple myeloma. It is also a translocation partner of PVT1 in multiple myeloma. |
| CTNNA3 | 10q21.3 | FRA10D | 1775996 | catenin (cadherin-associated protein), alpha 3 | Belongs to catenines family and necessary for the formation of a stable complex with the other catenines and cadherins contributing to solid cell–cell adhesion. Decreased expression of CTNNA3 was found in bladder urothelial carcinoma. |
| DMD | Xp21.1 | FRAXC | 2092287 | dystrophin | The reduced expression was found in brain tumors and frequent inactivation was found in malignant melanoma. Its expression is associated with genetic risk and survival in chronic lymphocyte leukemia. |
| FHIT | 3p14.2 | FRA3B | 1499181 | fragile histidine triad | Loss of expression was observed in breast cancer, lung cancer, cervical cancer, and B-cell lymphoma. Loss of FHIT expression was found to be associated with lymph node metastasis, cancer progression, and poor outcome. |
Figure 1.
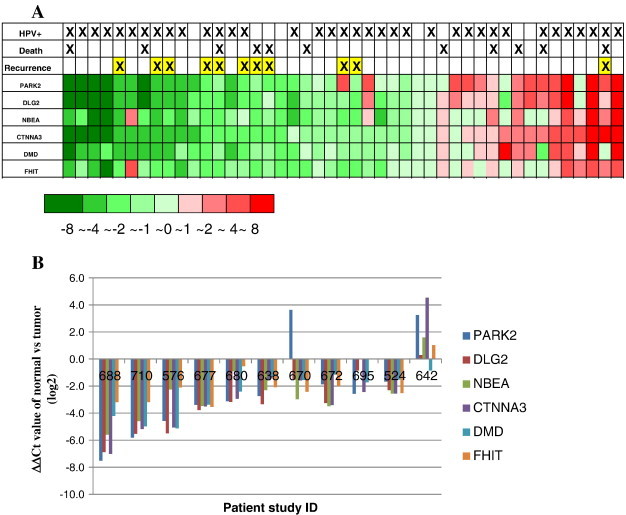
(A) Heat map of the six CFS large genes’ expression and prognostic parameters in 45 OPSCC patients examined.
Each column is a sample, and each row is a gene showing the expression of the six genes (ΔΔCt) in each patient tumor sample as compared to each individual matched normal. Prognostic parameters such as recurrence, live/dead, and HPV status are indicated above for each patient.
(B) The expression of the six large CFS genes in 11 OPSCC tumors which had tumor recurrence.
The expression of the six large CFS genes was tested by real-time PCR in OPSCC in both normal and tumor tissues. The difference of each gene’s expression (ΔΔCt) in these 11 patients was listed in the Y-axis.
We then divided the patients into two groups, one group which had decreased expression of all six genes (n = 27) and the other group which had either no changes in the expression of all six genes or increased expression of all six (n = 18), and we found that there is a significant difference in the incidence of tumor recurrence in these two groups: 37.0% (10/27) in the first group and 5.6% (1/18) in the other group. Kaplan-Meier plot analysis and a log-rank test analyzing the time to recurrence on these two groups showed a significant difference in recurrence (P = .037) (Figure 2).
Figure 2.
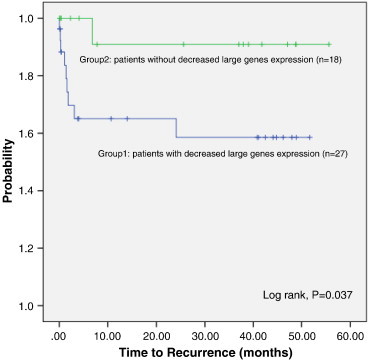
Kaplan-Meier analysis of recurrence curve for the 45 OPSCC patients.
The patients were divided into two groups based upon whether they had decreased expression of the six large CFS genes or not. One group of patients has decreased expression in most of the six large genes, and the other group of patients has no change or increased expression of the six large genes. The Kaplan-Meier curve (log-rank) analyzed the time to recurrence in these two groups.
Characterization of HPV Status and Its Relationship to Other Clinical Parameters
In each of these 45 patients studied, the HPV status was evaluated by real-time RT-PCR for E6 and E7 expression. There are 36 patients (80%) that were HPV positive and 9 patients (20%) that were HPV negative. The high HPV-positive infection rate is similar to other recent reports from other groups. Among the HPV-positive OPSCCs, we observed a spectrum of expression of the E6 and E7 transcripts, with some tumors having robust expression of these transcripts, while other HPV-positive tumors had quite low expression of these transcripts. This could be representative of what is now referred to as “active” as compared to latent HPV-infected OPSCCs. We were also curious if the HPV was associated with the patients’ disease outcome especially the recurrence in this study. For the limited patient number of this group of patients, we did not observe any significant difference in disease recurrence between the HPV-positive and HPV-negative patients. There are 9 out of 36 (25%) HPV-positive patients, and 2 out of the 9 (22%) HPV-negative patients had disease recurrence. The tumor recurrence was also not associated with higher or lower E6 and E7 expression in the HPV-positive group. Then, we also examined if the decreased expression of the six large genes was associated with the patients’ HPV status. There were 22 out of 36 (61%) HPV-positive patients and 5 out of 9 (56%) HPV-negative patients who showed decreased expression of these six CFS large genes. Thus, there are no significant differences observed between these 6 large genes’ expression and the HPV status either.
We then examined if other clinical characteristics have any association with disease recurrence or the six large genes’ expression in this group of patients. As shown in Table 1, there were no significant differences observed between the groups who developed recurrence vs those who did not in terms of age, gender, or tumor sites. For the 45 patients analyzed in this study, the vast majority (37 patients, 82%) were in stage IV. While we found that 10 out of 11 patients who developed recurrence were in stage IV, we could not do a valuable estimation whether there is association between the clinical stages and tumor recurrence, as there were insufficient tumors of other stages for the analysis. Thus, from this study, the single determination that did have important clinical significance was which tumors had decreased expression of these specific six large CFS genes.
Discussion
Our results demonstrate that those OPSCC tumors with decreased expression of this select group of six large CFS genes were much more likely to be associated with tumor recurrence. It is already well known that the decreased expression of specific large CFS genes, most notably FHIT, is usually associated with an overall worse prognosis in a number of different tumor types. In addition, our previous RNA seq analysis from 11 head and neck cancer patients revealed that the 3 tumors which had the most dramatically decreased expression for 2 of these genes, PARK2 and DMD, were derived from patients whose tumors recurred (data not shown). In this group of large CFS genes, FHIT and PARK2 have been demonstrated to function as important tumor suppressors involved in the development of many different cancers [18], [19]. Decreased FHIT expression has been shown to be associated with tumor progression in sporadic colon adenocarcinoma and poor prognosis in gastric cancer and oral squamous carcinoma [20], [21], [22]. PARK2 is commonly downregulated in clear-cell renal carcinoma and is associated with aggressive disease and a poor clinical outcome [23].
The other four large CFS genes, DLG2, NBEA, CTNNA3, and DMD, are each very attractive tumor suppressor candidates (Table 2). DLG2 belongs to the membrane-associated guanylate kinase family. A previous report indicated that HPV E6 could efficiently degrade members of the membrane-associated guanylate kinase family [24], and DLG2 has been implicated in cancer cell invasion [25]. DMD is the second largest known human gene and is localized within the chromosomal band Xp21.2. It is abundantly expressed in normal brain but was dramatically decreased in glioblastomas, and homozygous deletion of DMD has also been reported to be observed in esophageal adenocarcinoma [26]. DMD has been shown to be associated with genetic risk and prognosis in chronic lymphocyte leukemia [25], [27]). A recent report also showed that DMD acted as a tumor suppressor and an antimetastatic factor in cancers with a myogenic program [28]. NBEA located in the FRA13A CFS is known as a target of recurrent interstitial deletions in multiple myeloma [29]. CTNNA3 is located in FRA10D and was found to be frequently mutated in laryngeal carcinomas, non–small-cell lung carcinoma, and breast cancer [30]. Hence, each of these genes is thus an extremely attractive tumor suppressor candidate.
It is currently unclear whether overall genomic instability is responsible for the decreased expression of all six large CFS genes in 60% of the OPSCCs. Also unclear is why these genes appear to have concordant expression in an individual OPSCC. However, those OPSCCs that had decreased expression of the two important tumor suppressors, FHIT and PARK2, and the four additional candidate tumor suppressors appear more likely to have local recurrence and DM, which indicates that the loss of expression of all six of these genes could have a profound impact upon the resulting phenotype of the cells.
Although it has been suggested that HPV is associated with better overall prognosis in OPSCC, different reports have indicated that both HPV-positive and -negative patients were found to have similar rates of DM, and DM seems to be the leading cause of death in HPV-positive patients. Thus, finding biomarkers that are able to predict different recurrence risk would help to stratify patients for better treatment options. In this group of patients studied, for the 11 patients who developed recurrence, there are 7 patients who developed DM, and 6 of them showed decreased expression of the six large genes. However, due to limited cases in this study and tertiary care that Mayo Clinic provides to many patients, we did not have full clinical follow-up on each of the patients that provided tumor and matched normal tissue. Incomplete data on the cause of death of some of the 11 OPSCC patients who died made it difficult to do survival analysis.
To determine if our observations on the expression of the large CFS genes in OPSCC is relevant to other cancers, we analyzed available data provided by the Cancer Genome Atlas on breast cancers. We found that there is indeed a group of large CFS genes which have lower expression in many breast tumors. The specific large CFS genes that have decreased expression in breast cancers are different from those observed in the OPSCCs. Thus, the expression of selected specific large CFS genes might prove useful as diagnostic and prognostic markers for different cancer types.
Competing Interests
All authors declare that they have no competing interests.
Acknowledgment
We want to thank Ann Oberg for her kind suggestions in statistical analysis. This work is supported by the Experimental Pathology Development Fund from the Department of Laboratory Medicine and Pathology at Mayo Clinic.
Contributor Information
Ge Gao, Email: gao.ge@mayo.edu.
Jan L. Kasperbauer, Email: Kasperbauer.jan@mayo.edu.
Nicole M. Tombers, Email: tombers.nicole@mayo.edu.
Melissa D. Cornell, Email: cornell.melissa@mayo.edu.
David I. Smith, Email: smith.david@mayo.edu.
References
- 1.Murar S., Forastiere A.A. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clin Proc. 2008;83:489–501. doi: 10.4065/83.4.489. [DOI] [PubMed] [Google Scholar]
- 2.Chaturvedi A.K., Engels E.A., Pfeiffer R.M., Hernandez B.Y., Xiao W., Kim E., Jiang B., Goodman M.T., Sibug-Saber M., Cozen W. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panwar A., Batra R., Lydiatt W.M., Ganti A.K. Human papilloma virus positive oropharyngeal squamous cell carcinoma: a growing epidemic. Cancer Treat Rev. 2014;40:215–219. doi: 10.1016/j.ctrv.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Mroz Edmund A., Forastiere Arlene A., Rocco James W. Implications of the oropharyngeal cancer epidemic. J Clin Oncol. 2011;29:4222–4223. doi: 10.1200/JCO.2011.37.8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao G., Chernock R.D., Gay H.A., Thorstad W.L., Zhang T.R., Wang H., Ma X.J., Luo Y., Lewis J.S., Jr., Wang X. A novel RT-PCR method for quantification of human papillomavirus transcripts in archived tissues and its application in oropharyngeal cancer prognosis. Int J Cancer. 2013;132:882–890. doi: 10.1002/ijc.27739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quon H., Forastiere A.A. Controversies in treatment deintensification of human papillomavirus–associated oropharyngeal carcinomas: should we, how should we, and for whom? J Clin Oncol. 2013;29:520–522. doi: 10.1200/JCO.2012.46.7746. [DOI] [PubMed] [Google Scholar]
- 7.Mirghani H., Amen F., Blanchard P., Moreau F., Guigay J., Hartl D.M., Lacau St Guily J. Treatment de-escalation in HPV-positive oropharyngeal carcinoma: Ongoing trials, critical issues and perspectives. Int J Cancer. 2014 doi: 10.1002/ijc.28847. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 8.Glover T.W., Berger C., Coyle J., Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet. 1984;67:136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 9.Thorland E.C., Myers S.L., Gostout B.S., Smith D.I. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22:1225–1237. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- 10.Ferber M.J., Thorland E.C., Brink A.A., Rapp A.K., Phillips L.A., McGovern R., Gostout B.S., Cheung T.H., Chung T.K., Fu W.Y. Preferential integration of human papillomavirus type 18 near the c-myc locus in cervical carcinoma. Oncogene. 2003;22:7233–7242. doi: 10.1038/sj.onc.1207006. [DOI] [PubMed] [Google Scholar]
- 11.Ohta M., Inouse H., Cotticelli M.G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 12.Krummel K.A., Roberts L.R., Kawakami M., Glover T.W., Smith D.I. The characterization of the common fragile site FRA16D and its involvement in multiple myeloma translocations. Genomics. 2000;69:37–46. doi: 10.1006/geno.2000.6321. [DOI] [PubMed] [Google Scholar]
- 13.Denison S.R., Callahan G., Becker N.A., Phillips L.A., Smith D.I. Characterization of FRA6E and its potential role in autosomal recessive juvenlile parkinsonism and ovarian cancer. Genes Chromosomes Cancer. 2003;38:40–52. doi: 10.1002/gcc.10236. [DOI] [PubMed] [Google Scholar]
- 14.Becker N.A., Thorland E.C., Denison S.R., Philips L.A., Smith D.I. Evidence that instability within the FRA3B region extends four metgabases. Oncogene. 2002;21:8713–8722. doi: 10.1038/sj.onc.1205950. [DOI] [PubMed] [Google Scholar]
- 15.Bednarek A.K., Keck-Waggoner C.L., Daniel R.L., Laflin K.J., Bergsagel P.L., Kiguchi K., Brenner A.J., Aldaz C.M. WWOW, the FRA16D gene, behaves as a suppressor of tumor growth. Cancer Res. 2001;61:8068–8073. [PubMed] [Google Scholar]
- 16.Laborde R.R., Wang V.W., Smith T.M., Olson N.E., Olsen S.M., García J.J., Olsen K.D., Moore E.J., Kasperbauer J.L., Tombers N.M. Transcriptional profiling by sequencing of oropharyngeal cancer. Mayo Clin Proc. 2012;87:226–232. doi: 10.1016/j.mayocp.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao G., Kasperbauer J.L., Tombers N.M., Wang V., Mayer K., Smith D.I. A selected group of large common fragile site genes have decreased expression in oropharyngeal squamous cell carcinomas. Genes Chromosomes Cancer. 2014;53:392–401. doi: 10.1002/gcc.22150. [DOI] [PubMed] [Google Scholar]
- 18.Siprashvili Z., Sozzi G., Barnes L.D., McCue P., Robinson A.K., Eryomin V., Sard L., Tagliabue E., Greco A., Fusetti L. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci U S A. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cesari R., Martin E.S., Calin G.A., Pentimalli F., Bichi R., McAdams H., Trapasso F., Drusco A., Shimizu M., Masciullo V. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapitanović S., Čačev T., Lončar B., Catela Ivković T., Križanac Š., Pavelić K. Reduced FHIT expression is associated with tumor progression in sporadic colon adenocarcinoma. Exp Mol Pathol. 2014;96:92–97. doi: 10.1016/j.yexmp.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Wang H.L., Zhou P.Y., Liu P., Zhang Y. Abnormal FHIT protein expression may be correlated with poor prognosis in gastric cancer: a meta-analysis. Tumour Biol. 2014;35:6815–6821. doi: 10.1007/s13277-014-1936-7. [DOI] [PubMed] [Google Scholar]
- 22.Joo Y.H., Park S.W., Jung S.H., Lee Y.S., Nam I.C., Cho K.J., Park J.O., Chung Y.J., Kim M.S. Recurrent loss of the FHIT gene and its impact on lymphatic metastasis in early oral squamous cell carcinoma. Acta Otolaryngol. 2013;133:992–999. doi: 10.3109/00016489.2013.795289. [DOI] [PubMed] [Google Scholar]
- 23.Toma M.I., Wuttig D., Kaiser S., Herr A., Weber T., Zastrow S., Koch R., Meinhardt M., Baretton G.B., Wirth M.P. PARK2 and PACRG are commonly downregulated in clear-cell renal cell carcinoma and are associated with aggressive disease and poor clinical outcome. Genes Chromosomes Cancer. 2013;52:265–273. doi: 10.1002/gcc.22026. [DOI] [PubMed] [Google Scholar]
- 24.Accardi R., Rubino R., Scalise M., Gheit T., Shahzad N., Thomas M., Banks L., Indiveri C., Sylla B.S., Cardone R.A. E6 and E7 from human papillomavirus type 16 cooperate to target the PDZ protein Na/H exchange regulatory factor 1. J Virol. 2011;85:8208–8216. doi: 10.1128/JVI.00114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts S., Delury C., Marsh E. The PDZ protein discs-large (DLG): the 'Jekyll and Hyde' of the epithelial polarity proteins. FEBS J. 2012;279:3549–3558. doi: 10.1111/j.1742-4658.2012.08729.x. [DOI] [PubMed] [Google Scholar]
- 26.Gu J., Ajani J.A., Hawk E.T., Ye Y., Lee J.H., Bhutani M.S., Hofstetter W.L., Swisher S.G., Wang K.K., Wu X. Genome-wide catalogue of chromosomal aberrations in barrett's esophagus and esophageal adenocarcinoma: a high-density single nucleotide polymorphism array analysis. Cancer Prev Res (Phila) 2010;3:1176–1186. doi: 10.1158/1940-6207.CAPR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikitin E.A., Malakho S.G., Biderman B.V., Baranova A.V., Lorie Y.Y., Shevelev A.Y., Peklo M.M., Vlasik T.N., Moskalev E.A., Zingerman B.V. Expression level of lipoprotein lipase and dystrophin genes predict survival in B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 2007;48:912–922. doi: 10.1080/10428190701245112. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Marino-Enriquez A., Bennett R.R., Zhu M., Shen Y., Eilers G., Lee J.C., Henze J., Fletcher B.S., Gu Z. Dystrophin is a tumor suppressor in human cancers with myogenic programs. Nat Genet. 2012;46:601–606. doi: 10.1038/ng.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Neal J., Gao F., Hassan A., Monahan R., Barrios S., Kilimann M.W., Lee I., Chng W.J., Vij R., Tomasson M.H. Neurobeachin (NBEA) is a target of recurrent interstitial deletions at 13q13 in patients with MGUS and multiple myeloma. Exp Hematol. 2009;37:234–244. doi: 10.1016/j.exphem.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fanjul-Fernández M., Quesada V., Cabanillas R., Cadiñanos J., Fontanil T., Obaya A., Ramsay A.J., Llorente J.L., Astudillo A., Cal S. Cell–cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat Commun. 2013;4:2531–2539. doi: 10.1038/ncomms3531. [DOI] [PubMed] [Google Scholar]


