Abstract
Glioblastoma (GBM) with oligodendroglioma component (GBMO) is a newly described GBM subtype in the 2007 World Health Organization classification. However, its biological and genetic characteristics are largely unknown. We investigated the clinicopathological and molecular features of 34 GBMOs and compared the survival rate of these patients with those of patients with astrocytoma, oligodendroglioma, anaplastic oligoastrocytoma (AOA), and conventional GBMs in our hospital. GBMO could be divided into two groups based on the presence of an IDH1 mutation. The IDH1 mutation was more frequently found in secondary GBMO, which had lower frequencies of EGFR amplification but higher MGMT methylation than the wild type IDH1 group, and patients with mutant IDH1 GBMO were on average younger than those with wild-type IDH1. Therefore, GBMO is a clinically and molecularly heterogeneous subtype, largely belonging to a proneural and classical subtype of GBM. The survival rate of GBMO patients itself was worse than that of AOA patients but not significantly better than that of conventional GBM patients. GBMO survival was independent of the dominant histopathological subtype i.e., astrocyte-dominant or oligodendroglioma -dominant, but it was significantly associated with the IDH1 mutation and MGMT methylation status. Therefore, GBMO should be regarded as a separate entity from AOA and must be classified as a subtype of GBM. However, further study is needed to determine whether it is a pathologic variant or a pattern of GBM because GBMO has a similar prognosis to conventional GBMs.
Introduction
Glioblastoma (GBM) is the most common primary brain tumor and is associated with a short survival time of approximately 14.6 to 15.1 months following current multimodal treatment [1]. GBM is known to have heterogeneous histological features, which allow further subdivision into variants. GBM with oligodendroglioma component (GBMO) is a new variant that has been added to the updated 2007 World Health Organization (WHO) classification of tumors of the central nervous system (CNS) [2]. In the WHO blue book of brain tumors, 7th edition, a variant was defined as “a significant subtype with sufficiently different biological behavior than the main entity” [3]. Histological pattern was defined as “a particular differentiation pattern that does not correspond to a unique clinical/biological behavior” [3].
Two features distinguish this entity as a new variant. First, it contains foci that resemble oligodendroglioma on histological examination. Second, this subgroup shows a biological difference from preexisting tumor types in large studies [4], [5], because anaplastic oligoastrocytoma (AOA) with necrosis is associated with a significantly worse prognosis than AOA without necrosis, but is associated with a better overall survival rate than that of conventional GBM patients (median overall survival: AOA with necrosis, 22.8 months; AOA without necrosis, 86.9 months; GBM, 9.8 months). However, in 1996, Nelson et al. reported a slightly better median survival in GBMO patients than in conventional GBM patients (14.3 months vs. 10.4 months), which was quoted by Vordermark et al. [6]. Therefore, AOA with necrosis has been renamed GBMO, although at the consensus meeting there was debate as to whether this should be sanctioned, as it is still not clearly defined. Hence, collection of clinicopathological and biological data has been necessary in order to accurately classify GBMO [7], [8].
The reported biological behavior of GBMO has varied, with some studies reporting a better survival rate for GBMO compared to conventional GBM [6], [9], [10], whilst others have found no difference [11], [12], [13]. There have been many studies to assess the difference between GBMO and conventional GBM based on their clinicopathological and molecular genetic characteristics [8], [9], [11], [13], [14], [15], [16]. Despite this, there have been neither definitive diagnostic criteria nor a sufficient description of the clinical and genetic features of GBMO until now. Here, we report the genetic abnormalities of GBMO cases in our hospital and compare our results with previously reported data to delineate the genetic characteristics and biological behavior of this malignancy.
Materials and Methods
Tissue Samples
We selected cases that showed distinct morphological features of both GBM and oligodendroglioma in the same tumor from the pathology archives of the Department of Pathology, Seoul National University Hospital, collected between July 2007 and January 2013. Histopathological slides were reviewed independently by at least two neuropathologists (S. H. Park and J. K. Myung), and representative paraffin blocks were selected for the immunohistochemical and genetic studies. All of the specimens contained a classic GBM area with vascular hyperplasia and necrosis, together with oligodendroglial morphology (Figure 1). The classic GBM area was designated as astrocytic differentiation with glial fibrillary acidic protein-positive cells as well as micro vascular hyperplasia and necrosis. The oligodendroglioma component was characterized by oligodendroglioma morphology, including uniform round cells with rounded nuclei, a perinuclear halo, and delicate capillaries forming a chicken-wire pattern. Thirty-four brain tumors fulfilled the criteria of GBMO. We used a 10% cutoff for the minimum oligodendroglioma or astrocytoma component, as used in the studies by Ha et al. and Donahue et al. [11], [17], but sufficient counter-component was usually present in our series of GBMOs. We strictly excluded the small cell variant of GBM. Additionally, we sub-classified GBMO into two histopathological subtypes according to the dominant component comprising > 50% of tumor. Astrocyte-dominant type (GBMO-A) and oligodendroglioma dominant type (GBMO-O) accounted for 18 and 16 cases, respectively. We then compared overall survival according to the dominant histological subtype using Kaplan-Meier survival analysis. Secondary GBMO was defined as those cases that had previous pathology-proven lower grade tumors. We tested for the presence of the isocitrate dehydrogenase 1/2 (IDH1/2) and B-Raf protooncogene (BRAF) V600E mutations, amplification or high polysomy of epidermal growth factor receptor (EGFR), homozygous deletion of 9p21.3, allelic loss of 1p/19q, and O-6-methylguanine DNA methyltransferase (MGMT) gene methylation in our series of GBMOs. Immunohistochemical staining for IDH1 (H09) and other markers were performed in 34 cases.
Figure 1.
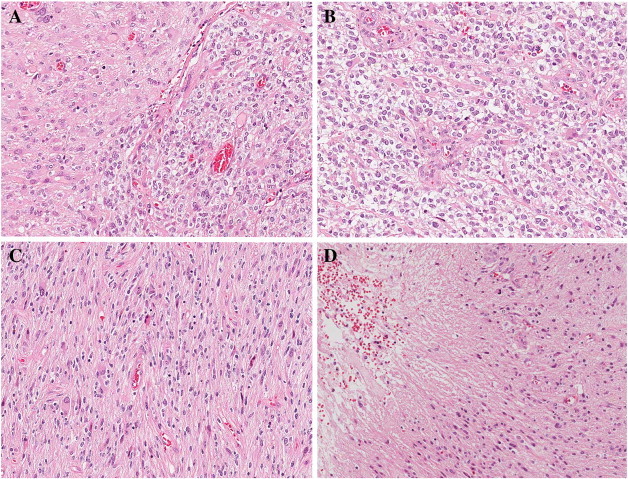
Histopathological features of glioblastoma (GBM) with oligodendroglioma component (GBMO). A) This picture shows transitional area of astrocytic and oligodendroglioma-like tumor. B) The oligodendroglioma component was composed of monomorphic cells with uniform round nuclei and perinuclear halos (giving a honeycomb appearance). C) Concomitantly, the same tumor had sheets of irregular or elongated cells with pleomorphic nuclei, and D) necrosis, consistent with glioblastoma area. (A–D: H&E, A: × 100, B–D: × 200).
This study was approved by the institutional review board of Seoul National University Hospital (IRB No. 1310-008-521).
DNA Extraction
Tumor areas were manually micro-dissected using 6-μm unstained tissue sections made from formalin-fixed and paraffin-embedded (FFPE) tissue. DNA was isolated from the micro-dissected tissue using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions.
Polymerase Chain Reaction (PCR) Amplification and Sequencing of IDH1, IDH2, and BRAF V600E
Template DNA (1 μl) was added to 100 μl of PCR solution (10 μl of 10 × MagnesiumTaq-High Fidelity [HF] buffer, 10 μl deoxynucleotide triphosphate [dNTP] mixture with 2 mM magnesium, 5 μl of 10 pmol primer [× 2], 1 μl of magnesium Taq-HF polymerase, and distilled water). The IDH1-Forward(F)/IDH1-Rerverse(R), IDH2-F/IDH2-R, and BRAF-F/BRAF-R primers (Supplementary Table 1) were used with the following program: 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 60 seconds for IDH1/IDH2 sequencing; and 35 cycles of 94°C for 45 seconds, 55°C for 45 seconds, and 72°C for 45 seconds for BRAF sequencing. The product sizes were 130-base pair (bp) (IDH1), 293-bp (IDH2), and 200-bp (BRAF). Unincorporated PCR primers and dNTPs were removed from the PCR products using a Montage PCR Clean-up Kit (Millipore, Billerica, MA).
The purified products were sequenced using the same primers. Sequencing was performed using a BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Foster City, CA). The sequencing products were resolved on an Applied Biosystems model 3730XL Automated DNA Sequencing System.
Fluorescence In Situ Hybridization (FISH)
FISH with Vysis probes was used to assess 1p/19q, 9p21.3 (CDKN2A), and EGFR gene status. Sections (3 μm thick) were deparaffinized in xylene, incubated with 0.3% pepsin in 10 mM HCl at 37°C for 10 minutes, boiled with citrate buffer (pH 6.0) in a microwave, incubated in 1 M NaSCN for 35 minutes at 80°C, immersed in the pepsin solution, and fixed in 10% neutral buffered formalin.
Labeled locus-specific (LSI) EGFR/CEP7 dual-color probes (Abbott Molecular), LSI CDKN2A/CEP9 dual-color probes (Abbott Molecular), and 1p36/1q25 and 19q13/19p13 LSI dual-color probes (Abbott Molecular) were used according to the manufacturer’s protocol. We applied the probe mixture to the slides and incubated them in a humidified atmosphere with HYBrite (Abbott Molecular, Des Plaines, IL) at 73°C for 5 minutes for simultaneous denaturation of the probe and target DNA. Then, we cooled the samples to 37°C and incubated for 19 hours to hybridize the probes and target DNA. The slides were submerged in 0.4 × SSC buffer/0.3% NP-40 for 2 minutes at room temperature, followed by 2 × SSC/0.1% NP-40 for 5 minutes at 73°C.
The processing and analysis of the FISH studies were conducted as described previously [18], as was the analysis of the chromosome 1p/19q deletion and CDKN2A (9p21.3) [18], [19]. For analysis of EGFR gene status, at least 100 tumor cell nuclei were counted per case. High polysomy (≥ 4 copies in ≥ 40% of cells) and gene amplification (the presence of tight EGFR gene clusters and ≥ 2 EGFR copies per chromosome or ≥ 15 EGFR copies per genome in ≥ 10% of analyzed cells) was regarded as an EGFR-positive FISH result.
MGMT Methylation-Specific PCR (MSP) Analysis
Tumor DNA was extracted after manual micro-dissection from FFPE, and MSP was conducted using an EZ DNA Methylation Kit (Zymo Research, Irvine, CA) to determine the methylation status of the MGMT promoter, as described previously [20].
Hematoxylin and Eosin (H&E) Staining and Immunohistochemistry
FFPE (10% neutral buffered formalin, routinely processed, and paraffin embedded) tissue sections (2–4 μm thick) were cut for H&E staining and immunohistochemistry. Tissue sections were stained with anti-IDH1 R132H (H09) monoclonal antibody (Dianova, Hamburg, Germany) using a 1:20 dilution. Immunohistochemical staining was carried out using a standard avidin–biotin peroxidase method.
Survival Analysis
We compared overall survival of GBMO to other gliomas, including WHO grade II tumors (low grade oligodendroglioma, LO, n = 39), WHO grade III tumors (anaplastic oligodendroglioma, AO, n = 44; anaplastic oligoastrocytoma, n = 37), and conventional GBM (n = 44). The clinical features of these patients are summarized in Supplementary Table 2.
Statistical Analysis
The chi-square test and Fisher’s exact test were used to measure the strength of interaction between two independent variables. An independent t test was used to compare the mean age in the two groups. Overall survival was estimated using the Kaplan–Meier survival analysis, and was compared using the log-rank test. All statistical analyses were performed with SPSS version 18 (Chicago, IL); P < .05 was considered significant.
Results
Clinicopathological Results
We identified GBMO tumors from 34 subjects whose ages ranged from 19 to 77 years (mean age, 53.3 years). The ratio of men to women was 3.25:1. The follow-up periods ranged from 1 month to 37 months (median 16.0 months). For post-operative treatment, 24 patients (70.6%) received concurrent chemoradiation therapy with temozolomide treatment, six received either chemotherapy or radiation therapy alone, and 5 received neither adjuvant treatment. GBMO was the primary tumor in 25 (73.5%) patients, and secondary in 9 patients (26.5%). The primary tumors for the secondary GBMOs were astrocytic tumors (grade 2 or 3) in 7 patients and oligodendroglioma in 2 patients. The clinical features of all patients are summarized in Supplementary Table 3.
Molecular Studies
We found 13 (13/34; 38%) IDH1 (H09) positive cases, all of which were found to carry a mutation in codon 132 of IDH1 on direct sequencing. A 1p/19q co-deletion was found in three of 33 patient tumors (9.1%), and two of them (2/3, 66.7%) had concomitant IDH1 mutations, but none of them showed positive results on EGFR FISH or homozygous deletion of CDKN2A. EGFR gene amplification or high polysomy were present in 13 cases (38.2%, amplification in seven cases and high polysomy in six cases), and two of them carried the IDH1 mutation. Nine (33.3%) of 27 cases revealed homozygous deletion in CDKN2A (9p21.3), five of which (5/9, 55.6%) had positive results on EGFR FISH and two (2/9, 22.2%) had the IDH1 mutation. Eighteen of 33 patient tumors (54.5%) revealed methylation of the MGMT promoter. Of these, ten of 18 patients (10/18, 55.6%) had the IDH1 mutation and five patients (5/18, 27.8%) had positive results on EGFR FISH. The BRAF V600E mutation was found in only one case. These results are summarized in Supplementary Table 4.
We divided the GBMOs into two subgroups according to IDH1 mutation status. Patients with the IDH1 mutation were on average younger than those without the mutation (46.5 years [range, 34 to 62 years] vs. 57.5 years [range, 19 to 77 years]; P < .05) (Table 1). Both groups contained more men than women. Only one of 21 cases (4.8%) of the wild-type IDH1 group involved secondary GBMO, compared with eight cases (8/13, 61.5%) of the IDH1 mutant group.
Table 1.
Molecular Features of GBMO According to IDH1 Mutation
| Variable | Result | IDH1⁎ |
Total | P Value | |
|---|---|---|---|---|---|
| Mutation# (n = 13) | Wild Type (n = 21) | ||||
| Age | Mean (range) | 46.5 (39-62) | 57.5 (19-77) | 53.3 (19-77) | .007 |
| > 45 y | 6 | 17 | 23 | .036 | |
| > 60 y | 1 | 10 | 11 | .007 | |
| Gender | Male | 9 | 17 | 26 | .449 |
| Female | 4 | 4 | 8 | ||
| GBM type | Primary GBMO | 5/13 (38.5) | 20/21 (95.2%) | 25 (73.5%) | .001 |
| Secondary GBMO | 8/13 (61.5%) | 1/21 (4.8%) | 9 (26.5%) | ||
| EGFR FISH | Amplification (7)/high polysomy (6) | 2/13 (15.4%) | 11/21 (52.4%) | 13 (38.2%) | .031 |
| CDKN2A FISH | HD | 3/10 (30.0%) | 6/17 (35.3%) | 9 (33.3%) | .788 |
| MGMT MSP | Methylation | 10/12 (83.3%) | 8/21 (38.1%) | 18 (54.5%) | .011 |
| 1p 19q FISH | Co-deletion | 2/13 (15.4%) | 1/21 (4.8%) | 3 (8.8%) | .303 |
| BRAF V600E⁎ | Mutation | 1/10 (10.0%) | 0/12 (0%) | 1 (4.5%) | .333 |
| PTEN IHC | Loss | 3/13 (23.1%) | 3/21(14.3%) | 6 (17.6%) | .528 |
| P53 IHC | Positive | 11/13 (84.6%) | 15/21 (71.4%) | 26 (76.5%) | .569 |
HP: high polysomy, FISH: fluorescence in situ hybridization, HD: homozygous deletion, MGMT-MSP: O-6-methylguanine DNA methyltransferase-methylation specific PCR, IHC: immunohistochemistry.
IDH1 and BRAF mutations were studied by direct sequencing.
All IDH1 mutation was Arg132His.
The frequency of EGFR gene abnormality was significantly higher in the wild-type IDH1 group than in the IDH1 mutant group (52.4% vs. 15.4%). In contrast, MGMT promoter methylation occurred significantly more frequently in IDH1-mutant GBMOs than in IDH1-wild type GBMOs (83.3% vs. 38.1%). However, 1p/19q co-deletion, BRAF mutation, and PTEN loss rarely occurred, and were not significantly different between IDH1 mutant- and IDH1 wild-type GBMOs. The key findings were therefore that EGFR gene abnormalities were negatively associated with IDH1 mutation (P = .031) and MGMT methylation was positively associated with IDH1 mutation (P = .011). In addition, as described above, IDH1 mutant GBMO had a higher MGMT methylation rate (83.3%); however, about half (55.6%) of MGMT methylated tumors carried the IDH1 mutation. These results are summarized in Table 1.
Survival Analysis
The cumulative survival rate of patients with GBMO was worse than that of AOA patients, but it was not significantly different from that of patients with conventional GBM (Figures 2A, 3A, B).
Figure 2.
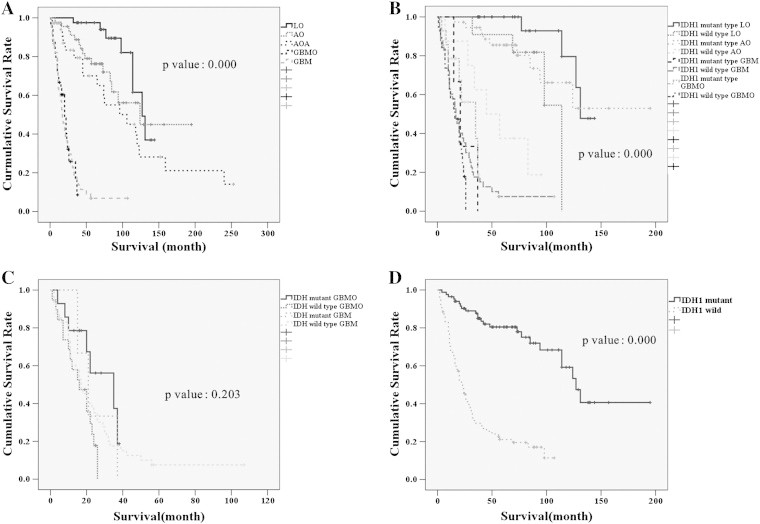
Kaplan-Meier survival analysis of patients with low grade oligodendroglioma (LO), anaplastic oligodendroglioma (AO), anaplastic oligoastrocytoma (AOA), gliobloastoma with oligodendroglial component (GBMO) and conventional glioblastoma (GBM). A) GBMO patients had significantly shorter survival than those with AOA without necrosis, LO and AO, but had not better survival than conventional GBM patients. B) As expected, patients with IDH1 mutant tumors survived for longer than those with IDH1 wild-type tumors. The patients with IDH1 mutant AO survived longer than patients with wild-type AO and had similar survival to patients with IDH1 wild-type LO, and patients with IDH1 wild-type AO had better survival than patients with GBMO or conventional GBM. C) A survival analysis of GBM and GBMO patients according to IDH1 mutation status revealed no statistically significant difference (P = .203). D) If all tumors are subdivided according to IDH1 mutation status regardless of tumor type, patients with IDH1 mutant tumors survived for considerably longer than those with IDH1 wild-type tumors. We did not include AOAs in B and D because we did not study IDH1 mutations in all of the AOA cases.
Figure 3.
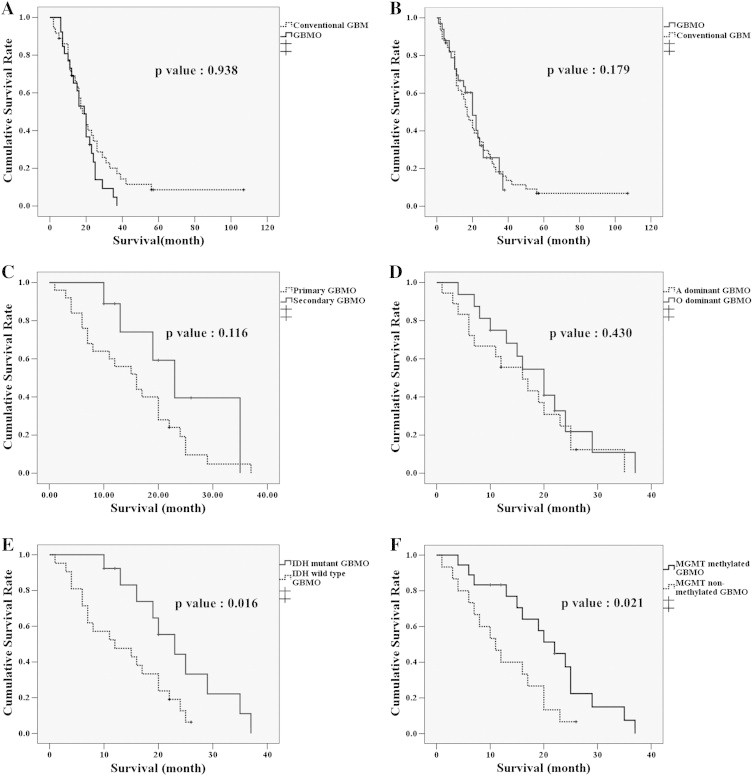
A, B) The survival rate of GBMO patients was similar to that of conventional GBM patients, even with age- and treatment matching. C, D) The survival rate of GBMO patients is not statistically different regardless of whether it is primary or secondary, or whether it has a dominant oligodendroglioma or astrocytic component. E, F) However, patients with IDH1 mutant and MGMT methylated GBMO had significantly better survival than patients with GBMO without IDH1 mutation or MGMT methylation.
Interestingly, patients with IDH1 wild-type AO showed intermediate survival between IDH1 wild-type LO and AOA patients (Figure 2B). There was no prognostic difference between GBM and GBMO regardless of whether the IDH1 mutation was present (P = .203, Figure 2C). However, there was a distinct survival difference according IDH1 mutation status, regardless of tumor type (Figure 2D). We did not include AOAs in the Kaplan–Meier survival analysis according to IDH1 mutation status, because we did not study IDH1 mutation status in all of the AOAs. Furthermore, there was no survival difference with respect to treatment between age matched patients with conventional GBM or GBMO (Figure 3B), primary or secondary GBMO (Figure 3C), or dominant histological subtype (Figure 3D). However, the cumulative survival rates of GBMO patients with tumors carrying the IDH1 mutation and the methylated MGMT promoter were significantly better than the rates of patients with IDH1 wild-type and MGMT unmethylated GBMOs (Figure 3E, F). On multiple regression analysis, none of the above mentioned clinicopathological parameters were associated with the overall survival of GBMO patients except primary and secondary nature of GBMO (P = .009).
Discussion
Clinical Aspects of GBMO
The reported incidence of GBMO varies between 11.9% and 17.6% [10], [21], and in our hospital it was 14.0% (34 GBMO/243 GBM). Our cases of GBMO showed characteristics similar to those described in previous reports. The overall mean age of GBMO patients and of primary GBMO patients was similar to that of patients with conventional GBM, although patients with secondary GBMO were younger (45.3 years) than patients with primary GBMO (56.1 years) (Supplementary Table 5). In our series, secondary GBMO accounted for 26.5% of all GBMO cases, which is a similar proportion to that previously reported (24% to 32%, Table 2) but more frequent than secondary conventional GBM (about 5%). Also consistent with previous reports was the predominance of male patients with primary and secondary GBMO, and conventional GBM (Supplementary Table 5) [9], [21].
Table 2.
Summary of Clinical and Molecular Features of GBMO from the Literatures and our Cases
| Variable | Appin et al. | Ha et al. | Wang et al. | Our Series | |
|---|---|---|---|---|---|
| Case number | 28 | 42 | 40 | 34 | |
| Primary GBM | 19 (67.9%) | 32 (76.2%) | Not record | 25 (73.5%) | |
| Secondary GBM | 9 (32.1%) | 10 (23.8%) | Not record | 9 (26.5%) | |
| Mean age (range) | 50.7 | 49.2 (26-73) | 43.2 (19-62) | 53.3 (19-77) | |
| Sex ratio(M:F) | 3:1 | 2:1 (28:14) | 23:17 | 3.25:1 | |
| IDH1 mutation | Positive | 7/20 (35%) | 11/42 (26.2%) | 9/29 (31.0%)⁎ | 13/34 (38.2%) |
| EGFR FISH | amp | 6/26(23%) | Not record | 19 (47.5%)-IHC | 13/34 (38.2%) |
| 1p 19q FISH | Co-deletion | 8/27 (29.6%) | 7/39 (17.9%) | 1/28 (3.6%) | 3/33(9.1%) |
| MGMT MSP | Methylation | 9/20 (45.6%) | 13/42 (31.0%) | 8/18 (44.4%) | 18/33 (54.5%) |
| CDKN2A FISH | HD | No record | No record | No record | 9/27 (33.3%) |
GBMO: glioblastoma with oligodendroglioma component, GBM:glioblastoma, IDH1: isocitrate dehydrogenase 1, amp: amplification, FISH: fluorescence in situ hybridization, IHC: immunohistochemistry, MGMT-MSP: O-6-methylguanine DNA methyltransferase-methylation specific PCR.
Sequencing.
Molecular Aspects of GBMO
IDH1 Mutation
The frequency of IDH1 was slightly higher (38.2%) in our series than previous reports (Supplementary Table 4 and Table 2). IDH1 mutation is the most important molecular event in low-grade glioma and secondary GBMs [19], [22], [23]. Joseph et al. also suggested that mutation of the IDH1 gene could be a key step in the tumorigenesis of GBMO [24]. However, as expected, fewer primary GBMOs in this study carried an IDH1 mutation than secondary GBMOs (20% vs. 88.9%) (Supplementary Table 5). While this figure is a simplistic depiction of the difference in IDH1 mutation rates between GBMO and conventional GBM, and between primary and secondary GBMO, it provides further evidence for a similar heterogeneity in IDH1 mutation status in these tumor types. This might also explain the heterogeneous clinical and molecular features of GBMO.
Other Molecular Profiles of GBMO
Kraus et al. and He et al. found that GBMOs more frequently showed a 1p and 19q deletion, but less commonly exhibited loss of heterozygosity (LOH) of 10q, loss of PTEN, and homozygous deletion of CDKN2A compared to conventional GBM [8], [21]. According to the He et al. report, there was LOH 1p in 40%, LOH 19q in 60%, EGFR amplification in 44%, p16 deletion in 48%, LOH 10q in 64%, PTEN loss in 20%, and TP53 mutation in 24% of GBMOs [21]. We found far less co-deletion of 1p/19q (9.1%) in our study of 34 GBMOs, but a similar rate of EGFR amplification or high polysomy (38.2%) and homozygous deletion of 9p21.3 (CDKN2A) (33.3%). Kraus et al. and Pinto et al. found that the 1p/19q co-deletion was present in as few as 10% of GBMOs [8], [25], which is similar to the observed 1p/19q co-deletion rate (8.8%) in our study. Co-deletion of 1p/19q is an important diagnostic and prognostic factor in oligodendroglioma [26]. Based on our findings, GBMO has a different genetic background from that of pure oligodendroglial tumors.
The previously reported frequency of MGMT MSP in GBMO ranged from 44.4% to 47% [9], [16], which is similar to that of conventional GBM; however, we found a higher rate of MGMT methylation (54.5%). When we divided the GBMO cases into IDH1 wild type and mutant subgroups, the latter showed an even higher frequency (83.3%) of MGMT methylation, but a lower frequency (15.4%) of EGFR gene amplification. The mean age of the IDH1 mutant GBMO group was younger than that of the wild type IDH1 GBMO group (46.5 years vs. 57.5 years) and the former had a higher proportion of secondary GBMO (61.5% vs. 4.8%).
In 2010, four molecular classifications of GBM based on the Cancer Genome Atlas Network data were widely accepted [27]. According to that classification, classical, neural, proneural, and mesenchymal subtypes were defined by mutations and differing expression levels of a number of genes including EGFR, neurofibromin1 (NF1), platelet-derived growth factor receptor alpha polypeptide (PDGFRA), and IDH1. GBMO carrying an IDH1 mutation in our study shared similar molecular profiles to the proneural type of GBM, whilst wild-type IDH1 GBMO belonged to the classical type of GBM. Similar results were also reported by Hegi et al. [16]. More meaningful sub-classification is possible when IDH1 mutation status is taken into consideration (Table 1). Therefore, GBMO should be classified according to its molecular features when investigating differences in clinical findings and prognosis.
Sequencing for the BRAF V600E mutation was performed in only 22 cases of GBMO, and only a single mutation was found (1/22, 4.5%), which is concordant with previously published data [9], [11], [13].
The BRAF V600E mutation rate in GBMO has not been established. Our previous study of BRAF V600E in diverse CNS tumors revealed a low mutation rate (3.7–8.6%) in GBMs and oligodendroglial tumors [18]. Therefore, we believe that the BRAF V600E mutation does not have particular importance in the pathogenesis of GBM and GBMO. In Table 2, we have summarized the diverse molecular genetic profiles of GBMO that have been previously published [9], [10], [11].
Survival of Patients with GBMO
GBMO was initially considered to be a separate entity because it was associated with a worse prognosis than WHO grade III mixed oligoastrocytoma, but a better survival rate than conventional GBM [6], [9], [10]. However, some studies stressed that better survival might be associated with chemosensitive molecular genetic alterations of oligodendroglioma component [6], [25]. Wang et al. found that aggressive treatment did not generally improve the survival of GBMO patients, although it significantly reduced the mortality of patients with conventional GBM [9]. In our series, the cumulative survival of GBMO patients was clearly worse than that of patients with AOA, but it was not significantly different from the survival of patients with conventional GBM (Figure 3, A and B). Furthermore, there was no survival difference between primary and secondary GBMO patients and between those with GBMO-A and GBMO-O subtypes, although secondary GBMO had tendency of better prognosis than primary GBMO (Figure 3, C and D). However, IDH1 mutation status or MGMT methylation status were important predictive factors for survival. The overall survival of patients with IDH1 mutant or MGMT methylated GBMO was significantly better than that of the patients with wild type IDH1 and MGMT unmethylated GBMO (Figure 3E and F). Our findings concur with those of Ha et al.’s results in that the outcome of GBMO in general was not different from conventional GBMO; however, their finding that GBMO-O patients had a significantly better outcome than patients with GBMO-A (cutoff: > 50%) was not in agreement with ours [11]. Elmahdi et al. also observed that GBMO had a similar clinical profile to conventional GBM, even with respect to age distribution and survival [12]. Conversely, Appin et al. found that GBMO patients had a longer median survival than conventional GBM patients (16.2 vs. 8.1 months), although this might have reflected a younger age at presentation and a 1p deletion [10].
To summarize, our findings show that GBMO is genetically heterogeneous in a similar manner to conventional GBM, which largely belongs to the proneural and classical subtype. The survival rate of GBMO patients in general is significantly worse than that of AOA patients, but not different from that of conventional GBM patients. However, as expected, GBMO patients with IDH1 mutant or MGMT methylated tumors survived longer than those with wild-type IDH1 or MGMT unmethylated tumors. Therefore, GBMO should be regarded as a separate entity from AOA without necrosis, and should be considered as a GBM subtype. Moreover, GBMO with the IDH1 mutation and/or MGMT methylation should be considered as a biologically significant variant since its prognosis and clinical features differ from that of GBMO without the IDH1 mutation and/or MGMT methylation. For these reasons, IDH1 mutation and MGMT methylation status should be considered when we make a diagnosis of GBMO. Despite this, the histopathological dominant subtype did not show prognostic value. In addition, on multivariate analysis, none of the clinical, genetic, or molecular parameters were associated with the survival of GBMO patients except primary or secondary status. A review of more cases is needed to determine whether GBMO is a pathologic variant or a pattern of GBM because our study failed to show any significant differences between GBMO and conventional GBM.
The previous reports showed that the prognosis of GBMO was better than that of GBM might be affected by high proportion of IDH1 mutated or MGMT methylated or secondary tumors among GBMOs than those of conventional GBM.
The following are the supplementary data related to this article.
The IDH1-F/IDH1-R, IDH2-F/IDH2-R and BRAF-F/BRAF-R primers
Clinical information of patients analyzed for Kaplan-Meier analysis
Clinical features of GBMO in general
Molecular features of our series of GBMO
Clinical features of GBMO according to primary and secondary GBMO
Acknowledgment
This work was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HIA112005) and the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (No. 2010-0028631).
Footnotes
Conflict of Interest: We have no conflicts of interest to declare.
References
- 1.Karayan-Tapon L, Quillien V, Guilhot J, Wager M, Fromont G, Saikali S, Etcheverry A, Hamlat A, Loussouarn D, Campion L. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97:311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. IARC WHO classification of tumours. IARC press; 2007. WHO classification of tumours of the central nervous system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller CR, Dunham CP, Scheithauer BW, Perry A. Significance of necrosis in grading of oligodendroglial neoplasms: a clinicopathologic and genetic study of newly diagnosed high-grade gliomas. J Clin Oncol. 2006;24:5419–5426. doi: 10.1200/JCO.2006.08.1497. [DOI] [PubMed] [Google Scholar]
- 5.van den Bent MJ, Carpentier AF, Brandes AA, Sanson M, Taphoorn MJ, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006;24:2715–2722. doi: 10.1200/JCO.2005.04.6078. [DOI] [PubMed] [Google Scholar]
- 6.Vordermark D, Ruprecht K, Rieckmann P, Roggendorf W, Vince GH, Warmuth-Metz M, Kolbl O, Flentje M. Glioblastoma multiforme with oligodendroglial component (GBMO): favorable outcome after post-operative radiotherapy and chemotherapy with nimustine (ACNU) and teniposide (VM26) BMC Cancer. 2006;6:247–253. doi: 10.1186/1471-2407-6-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Homma T, Fukushima T, Vaccarella S, Yonekawa Y, Di Patre PL, Franceschi S, Ohgaki H. Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol. 2006;65:846–854. doi: 10.1097/01.jnen.0000235118.75182.94. [DOI] [PubMed] [Google Scholar]
- 8.Kraus JA, Lamszus K, Glesmann N, Beck M, Wolter M, Sabel M, Krex D, Klockgether T, Reifenberger G, Schlegel U. Molecular genetic alterations in glioblastomas with oligodendroglial component. Acta Neuropathol. 2001;101:311–320. doi: 10.1007/s004010000258. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Li S, Chen L, You G, Bao Z, Yan W, Shi Z, Chen Y, Yao K, Zhang W. Glioblastoma with an oligodendroglioma component: distinct clinical behavior, genetic alterations, and outcome. Neuro Oncol. 2012;14:518–525. doi: 10.1093/neuonc/nor232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appin CL, Gao J, Chisolm C, Torian M, Alexis D, Vincentelli C, Schniederjan MJ, Hadjipanayis C, Olson JJ, Hunter S. Glioblastoma with oligodendroglioma component (GBM-O): molecular genetic and clinical characteristics. Brain Pathol. 2013;23:454–461. doi: 10.1111/bpa.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha SY, Kang SY, Do IG, Suh YL. Glioblastoma with oligodendroglial component represents a subgroup of glioblastoma with high prevalence of IDH1 mutation and association with younger age. J Neurooncol. 2013;112:439–448. doi: 10.1007/s11060-013-1073-y. [DOI] [PubMed] [Google Scholar]
- 12.Elmahdi A, Frary AJ, Scotton WJ, O'Donovan DG, Price SJ. Glioblastomas with oligodendroglial component have the same clinical phenotype as classical glioblastomas. Br J Neurosurg. 2013;27:419–424. doi: 10.3109/02688697.2013.767315. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura H, Makino K, Kuratsu J. Molecular and clinical analysis of glioblastoma with an oligodendroglial component (GBMO) Brain Tumor Pathol. 2011;28:185–190. doi: 10.1007/s10014-011-0039-z. [DOI] [PubMed] [Google Scholar]
- 14.Mizoguchi M, Hata N, Suzuki SO, Fujioka Y, Murata H, Amano T, Nakamizo A, Yoshimoto K, Iwaki T, Sasaki T. Pediatric glioblastoma with oligodendroglioma component: Aggressive clinical phenotype with distinct molecular characteristics. Neuropathology. 2013;33:652–657. doi: 10.1111/neup.12029. [DOI] [PubMed] [Google Scholar]
- 15.Klink B, Schlingelhof B, Klink M, Stout-Weider K, Patt S, Schrock E. Glioblastomas with oligodendroglial component-common origin of the different histological parts and genetic subclassification. Cell Oncol (Dordr) 2011;34:261–275. doi: 10.1007/s13402-011-0034-8. [DOI] [PubMed] [Google Scholar]
- 16.Hegi ME, Janzer RC, Lambiv WL, Gorlia T, Kouwenhoven MC, Hartmann C, von Deimling A, Martinet D, Besuchet Schmutz N, Diserens AC. Presence of an oligodendroglioma-like component in newly diagnosed glioblastoma identifies a pathogenetically heterogeneous subgroup and lacks prognostic value: central pathology review of the EORTC_26981/NCIC_CE.3 trial. Acta Neuropathol. 2012;123:841–852. doi: 10.1007/s00401-011-0938-4. [DOI] [PubMed] [Google Scholar]
- 17.Donahue B, Scott CB, Nelson JS, Rotman M, Murray KJ, Nelson DF, Banker FL, Earle JD, Fischbach JA, Asbell SO. Influence of an oligodendroglial component on the survival of patients with anaplastic astrocytomas: a report of Radiation Therapy Oncology Group 83-02. Int J Radiat Oncol Biol Phys. 1997;38:911–914. doi: 10.1016/s0360-3016(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 18.Myung JK, Cho H, Park CK, Kim SK, Lee SH, Park SH. Analysis of the BRAF(V600E) mutation in central nervous system tumors. Transl Oncol. 2012;5:430–436. doi: 10.1593/tlo.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myung JK, Cho HJ, Park CK, Kim SK, Phi JH, Park SH. IDH1 mutation of gliomas with long-term survival analysis. Oncol Rep. 2012;28:1639–1644. doi: 10.3892/or.2012.1994. [DOI] [PubMed] [Google Scholar]
- 20.Myung JK, Byeon SJ, Kim B, Suh J, Kim SK, Park CK, Chung CK, Chang KH, Park SH. Papillary glioneuronal tumors: a review of clinicopathologic and molecular genetic studies. Am J Surg Pathol. 2011;35:1794–1805. doi: 10.1097/PAS.0b013e31823456e6. [DOI] [PubMed] [Google Scholar]
- 21.He J, Mokhtari K, Sanson M, Marie Y, Kujas M, Huguet S, Leuraud P, Capelle L, Delattre JY, Poirier J. Glioblastomas with an oligodendroglial component: a pathological and molecular study. J Neuropathol Exp Neurol. 2001;60:863–871. doi: 10.1093/jnen/60.9.863. [DOI] [PubMed] [Google Scholar]
- 22.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 24.Joseph NM, Phillips J, Dahiya S, MF M, Tihan T, Brat DJ, Perry A. Diagnostic implications of IDH1-R132H and OLIG2 expression patterns in rare and challenging glioblastoma variants. Mod Pathol. 2013;26:315–326. doi: 10.1038/modpathol.2012.173. [DOI] [PubMed] [Google Scholar]
- 25.Pinto LW, Araujo MB, Vettore AL, Wernersbach L, Leite AC, Chimelli LM, Soares FA. Glioblastomas: correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Arch. 2008;452:481–490. doi: 10.1007/s00428-007-0562-9. [DOI] [PubMed] [Google Scholar]
- 26.Jeon YK, Park K, Park CK, Paek SH, Jung HW, Park SH. Chromosome 1p and 19q status and p53 and p16 expression patterns as prognostic indicators of oligodendroglial tumors: a clinicopathological study using fluorescence in situ hybridization. Neuropathology. 2007;27:10–20. doi: 10.1111/j.1440-1789.2006.00735.x. [DOI] [PubMed] [Google Scholar]
- 27.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The IDH1-F/IDH1-R, IDH2-F/IDH2-R and BRAF-F/BRAF-R primers
Clinical information of patients analyzed for Kaplan-Meier analysis
Clinical features of GBMO in general
Molecular features of our series of GBMO
Clinical features of GBMO according to primary and secondary GBMO


