Abstract
Detecting the cancer cells in the peripheral blood, i.e. circulating tumor cell (CTC), have been considered as the “liquid biopsy” and become a particular area of focus. A deep insight into CTC provides a potential alternative method for early diagnosis of solid tumor. Previous studies showed that CTC counts could be regarded as an indicator in tumor diagnosis, predicting clinical outcomes and monitoring treatment responses. In this report, we utilize our facile and efficient CTC detection device made of hydroxyapatite/chitosan (HA/CTS) for rare cancer cells isolation and enumeration in clinical use. A biocompatible and surface roughness controllable nanofilm was deposited onto a glass slide to achieve enhanced topographic interactions with nanoscale cellular surface components, anti-EpCAM (epithelial cell adhesion molecule, EpCAM) were then coated onto the surface of nanosubstrate for specific capture of CTCs. This device performed a considerable and stable capture yields. We evaluated the relationship performance between serial CTC changes and the changes of tumor volume/serum tumor marker in gastrointestinal cancer patients undergoing anti-cancer treatments. The present study results showed that changes in the number of CTC were associated with tumor burden and progression. Enumeration of CTCs in cancer patients may predict clinical response. Longitudinal monitoring of individual patients during the therapeutic process showed a close correlation between CTC quantity and clinical response to anti-cancer therapy. Effectively capture of this device is capable of CTCs isolation and quantification for monitoring of cancer and predicting treatment response.
Introduction
Rare circulating tumor cells (CTCs) are some special cancer cells which detach from the primary solid tumor and/or metastasis then circulate in peripheral blood [1]. CTCs have been considered as the probable cause of fatal metastatic disease [1], [2], [3]. A better understanding of the biology of CTCs may provide a potential alternative access to solid tumor and further the study of tumor metastasis and disease progression (regarded as a “liquid biopsy” for epithelial cancers) [4]. However, the acquisition and analysis of CTCs are still one of the technically challenges because these cells are extremely rare (a few to hundreds cells per milliliter) and mixed with a large number of hematologic cells (109 cells per milliliter) [5], [6]. Over the past decade, some technologies have been developed to detect CTCs, including flow cytometry [7] and size-based separation [8], [9]. The immunomagnetic separation-based CellSearch™ Assay is the only FDA-approved CTC diagnostic technology for metastatic breast, prostate and colorectal cancers. Consequently, metastatic breast cancer patients with 5 or more CTCs counts (enumerated by CellSearch-Veridex) in 7.5 mL blood before treatments may suffer poor progression-free survival and overall survival [5]. However, due to its low capture yield and high cost of isolating CTCs, monitoring CTCs from gastrointestinal cancer patients to guide therapeutic choices has not been established [10], [11].
Previously, we have reported a high efficient CTCs detection assay using a horizontally packed TiO2 nanofibers [12]. The capture of CTCs by this method mainly depends on the enhanced local topographic interactions [13], [14] between nano-components on cancer cell membrane (e.g., microvilli and filopodia) and the antibody (anti-EpCAM [15]) coated nanofibers, which improves the CTCs capture efficiency. Due to the complex process control and special equipment for preparing these special nanostructures, this method have been limited for high throughput fabrication. It's urgent to develop a new device that is worth popularizing in clinical practice for its simple manufacture, low cost and convenient use.
Herein, based on our previous publications, we utilize our recent CTC detection device made of hydroxyapatite/chitosan (HA/CTS), optimized from our previous works (TiO2 nanoparticles [16] and HA-CTS nanofilms [17]) for rare cancer cells isolation and enumeration in clinical use. Results showed that capture yields of more than 80% can be achieved through the use of our substrate (new version of HA-CTSNF). Given the high sensitivity and specificity of the new version of HA-CTSNF substrate, we validated the ability of isolation and enumeration of CTCs in totally 32 localized and metastatic gastrointestinal cancer patients and tested its potential utility in monitoring response to anti-cancer therapy.
Materials and Methods
Materials
Hydroxyapatite (HA) in powder form was purchased from Sigma Aldrich (St Louis, MO), chitosan (CTS) was obtained from Haisheng Co., Ltd (Qingdao, China), 4-Maleimidobutyric acid N-hydrosuccinimide (≥ 98% GMBS), 3-mercaptopropyl trimethoxysilane (95% MTPMS), paraformaldehyde (PFA), streptavidin (1 mg mL− 1 SA), biotinylated anti-human EpCAM/TROP1 antibody (Goat IgG) were supplied by R&D systems (Minneapolis, MN). Anti-human CD45-FITC (Ms IgG1, clone H130) and anti-cytokeratin-PE (CAM5.2, conjugated with phycoerythrin) were purchased from BD Biosciences (San Jose, CA). Triton X-100 and 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) were purchased from Sigma-Aldrich (St Louis, MO). Phosphate-Buffered Saline (PBS 1X, 0.0067 M PO43 −), RPMI-1640 growth medium, dulbecco's modified eagle medium (DMEM), Tween-20, fetal bovine serum (FBS) and 0.25% Trypsin-EDTA (Gibco, 1X) were supplied by Invitrogen (Carlsbad, CA). Colorectal cancer cell line (HCT116), gastric cancer cell line (MGC803), cervical cancer cell line (HeLa) and chronic myelogenous leukaemia cell line (K562) were harvested from Hubei Key Laboratory of Tumor Biological Behaviors.
Blood samples
Whole blood samples from healthy donors were obtained from Department of Clinical Laboratory, Zhongnan Hospital of Wuhan University according to a protocol by the Institutional Review Board (IRB) and cancer patients' blood samples were obtained from Department of Oncology, Zhongnan Hospital of Wuhan University under a separate IRB-approved protocol. All blood samples were collected into Anticoagulant tubes (EDTAK2, 2 ml, violet cap) and processed within 12 h.
Fabrication of HA-CTS nanofilm substrate
According to our previously established method[17], HA-CTS nanofilm (HA-CTSNF) was prepared as below. Firstly, the glass substrate was boiled in Piranha solution (3:1 (v/v) H2SO4/H2O2) at 100 °C for 1 h, washed several times with deionized water, then treated by oxygen plasma treatment (PDC-32G, Harrick Plasma, USA) for 2 min, and dried with nitrogen gas. A chitosan aqueous solution of 2 wt% was prepared by dissolving chitosan powder into distilled water containing 2% acetic acid and then 5 wt% nano-hydroxyapatite (HA) powder was added slowly. The mixed solution was blended by magnetic stirrer for 12 h and then uniformly coated on the clean glass by using a photoresist spinner (10000 r/h, 60 seconds). In the baking process the coated solution was heated on bake table for 1 h. Following the baking process the HA-CTS nanofilm substrate was soaked in 10 wt% NaOH solution for 10 h. Finally, the substrate was washed with DI water, dried and ready for chemical modification. The morphology of the HA-CTS nanofilm substrate was observed by Field Emission Scanning Electron Microscopy (SEM, 6700 F, JEOL, Japan). Compared to our previous works (Table 1), the new version of HA-CTSNF has a larger surface area, higher transmittance, and equally high capture efficiency. It may increase the “loading capacity” and enable the anti-EpCAM modified HA-CTSNF to directly monitor the captured cancer cells.
Table 1.
Comparison of the Fabrication Specifications Between the Previous Reported Methods and the New Version of the HA-CTSNF
| TiO2 Nanoparticles | HA-CTSNF | New Version of HA-CTSNF | |
|---|---|---|---|
| Photoresist spinner parameter | - | 6000 r/h, 60 s | 10000 r/h, 60 s |
| Nanosubstrate roughness | 100-140 nm | 170-250 nm | 120-170 nm |
| Nanosubstrate area | 1 cm × 1 cm | 1 cm × 1 cm | 2 cm × 2 cm |
Surface modification with antibody
Surface modification procedure for the HA-CTSNF was reported earlier [18]. The HA-CTSNF substrate was modified with 4% (v/v) 3-mercaptopropyl trimethoxysilane (MPTMS) in ethanol at room temperature for 1 h [19]. Then, the substrate was treated with the coupling agent N-y-maleimidobutyryloxy succinimide ester (GMBS, 0.25 mM) for 1 h make for GBMS attachment to the substrate. Next, the substrate was treated with streptavidin (SA) at room temperature for 45 min, resulting in immobilization onto GMBS, and washed with 1 x PBS to remove excess SA before immersed in biontinylated anti-EpCAM solution (10 μg mL− 1 in PBS) for 60 min (RT).
Cell capture and identification
1 mL cell suspension (i.e., HCT116, MGC803, HeLa and K562, 105 cells mL− 1) was introduced onto the new version of HA-CTSNF substrate and incubated for 1 h (37°C and 5% CO2). Then, the chip was rinsed with PBS for 5 times. The captured cells on the chip were fixed with 4% paraformaldehyde (PFA) in PBS for 10 min. Then, the chip was loaded with 0.2% Triton X-100 in PBS and incubated for 10 min in order to improve the permeability of the cell and to allow for intracellular staining, 40 ul blocking solution (2% donkey serum in PBS/0.2% triton X-100) was added onto the chip and incubated for 30 min at room temperature, followed by staining with PE-labeled anti-CK and FITC-labeled anti-CD45 overnight. 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI, 0.33 μg mL− 1 in DI water) was used for staining nuclear for 10 min. Finally, detecting and counting of targeted cells using the fluorescence microscope (IX81, Olympus, Tokyo, Japan). Captured CTCs on chip were photographed by using IPP software (Media Cybernetics Inc., Silver Spring, MD).
Cell capture from cancer patient blood samples
The circulating tumor cells from cancer patient were captured according to the procedure described below. About 4.0 mL patient peripheral blood sample was introduced onto our chip and captured for 1 h. After rinsing, followed by staining of anti-CK, anti-CD45 and DAPI, the specifically captured cancer cells were identified and counted on the substrates directly.
Results and Discussion
The cancer cells capture principle of the HA-CTSNF substrate was illustrated in Figure 1. The improved HA-CTSNF substrate (New version of HA-CTSNF) was prepared by HA/CTS composites (Figure 2A, see details inMaterials and Methods). A layer of nanostructured film was fabricated onto a slide glass (Figure 2B). The roughness of the nanostructured substrates were controlled by changing the HA:CTS ratio. First, in order to demonstrate the surface roughness of nanostructured film impacts the capture yields, the biotin-HA-CTSNF substrate were prepared before cell capture (Figure 2C), and two EpCAM-positive cancer cell lines (HCT116, MGC803) were chosen and spiked into the Dulbecco's Modified Eagle's Medium (DMEM) at a concentration of 105 mL− 1, then, these spiked cells were captured by anti-EpCAM-grafted substrates which has been prepared with different surface roughness (ranging from 20 to 280 nm). For comparison, two EpCAM-negative cancer cell lines (K562 and HeLa) were chosen for specific cell capture comparison experiments. As shown in Figure 2D, when the roughness of the substrates was around 150 nm, maximal cell capture yields were achieved for HCT116 cell line (approximately 81%) and MGC803 cell line (approximately 64%) while minor changes were observed for EpCAM-negative cells. These results revealed that the effect of roughness on the cell capture yields can be attributed to the local topographic interactions between nano-substrate and cancer cells.
Figure 1.
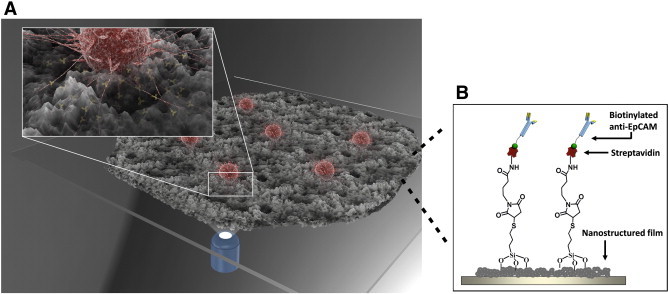
Schematic of cancer cells capture principle of the HA-CTSNF substrate. Capture efficiency was improved by combining cell-capture-agent and cancer cell preferred nano-scaled topography of substrate.
Figure 2.
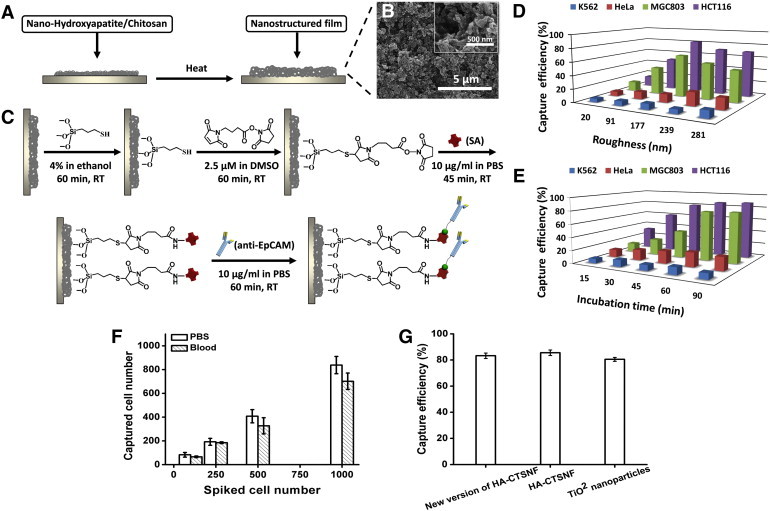
(A) Schematic procedure for the fabrication of the nanostructured film. (B) Scanning electron microscopy (SEM) image of HA-CTSNF substrate. (C) Grafting of biotinylated epithelial-cell adhesion-molecule antibody (anti-EpCAM) onto HA-CTSNF substrate. The cell capture yields of the HA-CTSNF substrate with (D) different surface roughness of nanostructured film ranging from 20 to 280 nm and (E) at different incubation time (with incubation time of 15, 30, 45, 60 and 90 min). (F) Capture efficiencies of four different cell lines at the optimal cell capture conditions. (G) Comparison of capture efficiency of new version of HA-CTSNF with two different controls: (i) HA-CTSNF, (ii) TiO2 nanoparticles. Error bars show standard deviations (n = 3).
To determine the optimum incubation time for efficient cell capture, we performed experiment about incubation times by using the optimal substrate modified with anti-EpCAM as studied above. Two EpCAM-positive cancer cell lines (HCT116 and BGC803) and two EpCAM-negative cancer cell lines (K562 and Hela) were examined. The cell capture yields with different incubation time were shown in Figure 2E. For the EpCAM-positive cell line, the recovery yields of cell capture increased with increasing incubation time and significant changes had been observed, and the maximal recovery yields achieved saturation at an incubation of 60 min or longer. There was several times difference between the maximal recovery yields and the minimal recovery yields in our test, leading us to ascertain 60 min to be the minimum time for optimal capture.
Finally, we tested these four cell lines on the new version of HA-CTSNF under the optimal cell capture conditions, showed a high capture efficiency of 72% to 84% with EpCAM-positive cell lines (Figure 2F). New version of HA-CTSNF exhibited capture efficiency equal to that of our previous works when we compare the capture performance of new version of HA-CTSNF with HA-CTSNF and TiO2 nanoparticles (Figure 2G).
During the clinical utility of our platform for rare cancer cells capture from cancer patient peripheral blood samples, we evaluated the relationship performance between serial CTCs changes and the changes of tumor volume/serum tumor marker (i.e. CEA, CA19-9) in gastrointestinal cancer patients who have received anti-cancer treatments. Patients were followed up after discharge by physical examination, routine blood tests, serum tumor marker tests, and computed tomograms (CT scans) according to standard clinical practice. In the patient information table (Table 2), CTC numbers were calculated before and 10–18 weeks after the treatment initiation. Medians and ranges were calculated on patients' age and their serum CA19-9 and CEA concentrations. The frequencies and percentages were calculated for the patients' clinical status. About 4.0 mL peripheral blood samples were introduced onto our substrate for cell capture, at the same time, 5 healthy donors' epithelia blood samples were tested as control. Specifically captured CTCs (CK +/CD45 −/DAPI +, 10 μm < cell sizes < 30 μm) were identified from WBCs (CK −/CD45 +/DAPI +, sizes < 15 μm) using fluorescence microscopy [20], [21] (Figure 3A). CTC enumeration results obtained from 5 healthy donors and 32 cancer patients before treatment (12 gastric cancer patients and 20 colorectal cancer patients) were showed in Figure 3B. Cell counts of cancer patients ranging from 3 to 162 per 4.0 mL that were significantly higher than cells counts of healthy donors (0-5 CTCs/4 mL). The CTCs changes corresponded with tumor size (Figure 3, C–D), while the changes in CTCs did not always keep consistent in serum tumor marker (Figure 3, E–F), as a common phenomenon reported in early study (serum tumor marker flare after starting chemo). The results showed that enumeration of CTCs in cancer patients may predict clinical response to anti-cancer treatment. In general, these data demonstrated that our platform performed considerable capture efficiency and allowed for monitoring CTCs to evaluate the clinical treatment response and guide therapeutic choices.
Table 2.
Clinicopathologic Characteristics and CTC Numbers of Patients
| Cancer Species | Age Median (Range) |
CTC/4 ml Median (Range) |
Serum CA19-9 [U/ml] Median (Range) |
Serum CEA [ng/ml] Median (Range) |
Clinical Status |
|---|---|---|---|---|---|
| Healthy control (n = 5) | 25 (22-29) | 2.2 (0-5) | - | - | - |
| Gastric cancer (n = 12) | 58.2 (39-75) | 35.75 (3-96) | 121.8 (2.69->1000) | 4.55 (0.74-35.5) | I: 0 (0%) II: 2 (16.67%) III: 6 (50%) IV: 4 (33.33%) |
| Colon cancer (n = 20) | 59.1 (38-80) | 47.55 (4-162) | 30.3 (2-304.7) | 8.24 (0.49-39.72) | I: (10%) II: 7 (35%) III: 7 (35%) IV: 4 (20%) |
Figure 3.
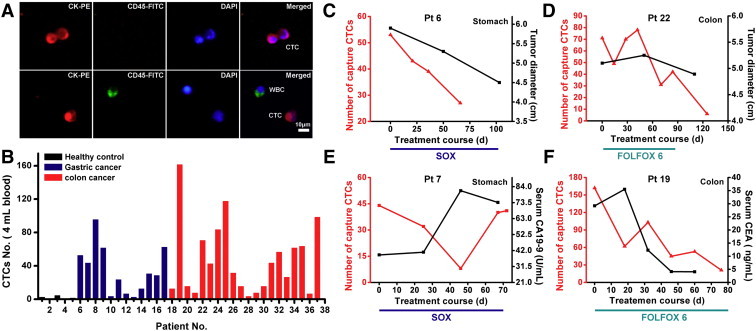
(A) CTCs isolated from a colorectal cancer patient. Three-color immunocytochemistry method based on FITC-labeled anti-CD45, PE-labeled anti-CK, and DAPI nuclear staining was applied to identify and enumerate CTCs from non-specifically trapped WBCs. (B) CTC enumeration results obtained from healthy donors and cancer patients. Numerical analysis of 4 patients showing the number of CTCs (red), concentration of serum CA19-9, CEA and tumor size (black) measured as the unidimensional sum of all significant tumor sites on a CT scan. Imaging studies were assessed by a single reference radiologist (Haibin Song), who graded responses according to Response Evaluation Criteria in Solid Tumors (RECIST). Type of treatment and duration are noted for each case. Patients with diagnoses and specific therapies shown are (C) patient 6, gastric, SOX: S-1 plus Oxaliplatin, (D) patient 22, colorectal, FOLFOX 6: Oxaliplatin, 5-FU and leucovorin, (E) Patient 7: gastric, SOX, f) Patient 19, colorectal, FOLFOX 6.
Longitudinal monitoring of individual patients during the therapeutic process also showed a close correlation between CTC quantity and clinical response. We serially monitored CTCs in patient 9 with gastric cancer receiving SOX regimens (S-1 plus Oxaliplatin) and total gastrectomy. As result shown in Figure 4 that declines in CTC number paralleled effective therapeutic interventions. With clinical benefit, CTC number of this patient rapidly decreased while serum CA19-9 concentrations also declined gradually, at the same time, the tumor size and lymph node sizes reduced obviously; irregularly thickening of gastric body near by the pancreas was disappeared and could be separated from mass tissue as shown in computed tomography image (Figure 4A). Another phenomenon could be observed that a temporally increment in CTC number during and after total gastrectomy and followed by a drop. This result may indicate that surgical operation increase the chance of cancer cells shed from solid tumor into the circulation. Above all our study showed that the capture efficiency of HA-CTSNF substrate is reliable and capable of longitudinal monitoring of cancer and this approach may have early diagnostic potential. A prospective trial would be needed to explore its predictive value.
Figure 4.
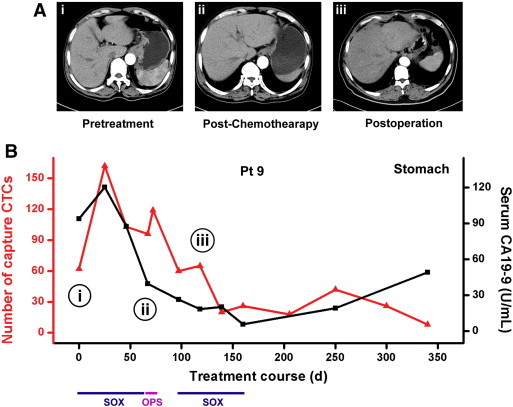
Serial Changes of CTCs in cancer patient. (A) Computed tomography (CT) images in three different stages, i) before treatment, ii) after chemo and iii) after dissection. (B) Serial CTCs and serum CA19-9 changes of patient 9 are plotted. Type of treatment and duration are noted: Sox: S-1 plus Oxaliplatin, OPS: total gastrectomy.
In conclusion, CTCs isolation based on our platform allowed the longitudinal enumeration of cancer cells from cancer patients with high capture yield. Regarding the ability of monitoring CTCs from patient who are receiving clinical therapy, we expect this method is of application potential for treatment responses monitor, patient prognosis indication and cancer-related biological studies.
Acknowledgments
The work was supported by National High Technology Research and Development Program of China (Grant No. 2012AA02A502, 2012AA02A506).
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest.
References
- 1.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 4.Pachmann K, Camara O, Kroll T, Gajda M, Gellner AK, Wotschadlo J, Runnebaum IB. Efficacy control of therapy using circulating epithelial tumor cells (CETC) as “liquid biopsy”: trastuzumab in HER2/neu-positive breast carcinoma. J Cancer Res Clin Oncol. 2011;137:1317–1327. doi: 10.1007/s00432-011-1000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LW, Uhr JW. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zieglschmid V, Hollmann C, Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42:155–196. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 7.Terstappen LW, Rao C, Gross S, Kotelnikov V, Racilla E, Uhr J, Weiss A. Flow cytometry—principles and feasibility in transfusion medicine. Enumeration of epithelial derived tumor cells in peripheral blood. Vox sanguinis. 1998;74(Suppl 2):269–274. doi: 10.1111/j.1423-0410.1998.tb05431.x. [DOI] [PubMed] [Google Scholar]
- 8.Davis JA, Inglis DW, Morton KJ, Lawrence DA, Huang LR, Chou SY, Sturm JC, Austin RH. Deterministic hydrodynamics: taking blood apart. Proc Natl Acad Sci U S A. 2006;103:14779–14784. doi: 10.1073/pnas.0605967103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang LR, Cox EC, Austin RH, Sturm JC. Continuous particle separation through deterministic lateral displacement. Science. 2004;304:987–990. doi: 10.1126/science.1094567. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, Jiang Z, Cheng J, Xiao BX. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl) 2010;88(7):709–717. doi: 10.1007/s00109-010-0617-2. [DOI] [PubMed] [Google Scholar]
- 11.Uen YH, Lin SR, Wu CH, Hsieh JS, Lu CY, Yu FJ, Huang TJ, Wang JY. Clinical significance of MUC1 and c-Met RT-PCR detection of circulating tumor cells in patients with gastric carcinoma. Clin Chim Acta. 2006;367:55–61. doi: 10.1016/j.cca.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S. Electrospun TiO2 nanofiber-based cell capture assay for detecting circulating tumor cells from colorectal and gastric cancer patients. Adv Mater. 2012;24:2756–2760. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 13.Fischer KE, Alemán BJ, Tao SL, Hugh Daniels R, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Biomimetic nanowire coatings for next generation adhesive drug delivery systems. Nano Lett. 2009;9(2):716–720. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis A, Varde M. Control of cell behavior: topological factors. Journal of the National Cancer Institute. 1964;33:15–26. [PubMed] [Google Scholar]
- 15.Went PTH, Lugli A, Meier S, Bundi M, Mirlacher M, Sauter G, Dirnhofer S. Frequent EpCam protein expression in human carcinomas. Human pathology. 2004;35:122–128. doi: 10.1016/j.humpath.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 16.He R, Zhao L, Liu Y, Zhang N, Cheng B, He Z, Cai B, Li S, Liu W, Guo S. Biocompatible TiO2 nanoparticle-based cell immunoassay for circulating tumor cells capture and identification from cancer patients. Biomed Microdevices. 2013;15:617–626. doi: 10.1007/s10544-013-9781-9. [DOI] [PubMed] [Google Scholar]
- 17.Cheng B, He Z, Zhao L, Fang Y, Chen Y, He R, Chen F, Song H, Deng Y, Zhao X. Transparent, biocompatible nanostructured surfaces for cancer cell capture and culture. Int J Nanomedicine. 2014;9:2569–2580. doi: 10.2147/IJN.S61233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupraz A, De Wijn J, De Groot K. Characterization of silane-treated hydroxyapatite powders for use as filler in biodegradable composites. Journal of biomedical materials research. 1996;30:231–238. doi: 10.1002/(SICI)1097-4636(199602)30:2<231::AID-JBM13>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S. Hydrophobic Interaction-Mediated Capture and Release of Cancer Cells on Thermoresponsive Nanostructured Surfaces. Advanced Materials. 2013;25:922–928. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Chen L, Xu T, Liu H, Liu X, Meng J, Yang G, Jiang L, Wang S. Programmable Fractal Nanostructured Interfaces for Specific Recognition and Electrochemical Release of Cancer Cells. Adv Mater. 2013;25:7603–7609. doi: 10.1002/adma.201300888. [DOI] [PubMed] [Google Scholar]


