ABSTRACT
WD40-repeat protein 62 (WDR62) is a spindle pole protein required for normal cell division and neuroprogenitor differentiation during brain development. Microcephaly-associated mutations in WDR62 lead to mitotic mislocalization, highlighting a crucial requirement for precise WDR62 spatiotemporal distribution, although the regulatory mechanisms are unknown. Here, we demonstrate that the WD40-repeat region of WDR62 is required for microtubule association, whereas the disordered C-terminal region regulates cell-cycle-dependent compartmentalization. In agreement with a functional requirement for the WDR62–JNK1 complex during neurogenesis, WDR62 specifically recruits JNK1 (also known as MAPK8), but not JNK2 (also known as MAPK9), to the spindle pole. However, JNK-mediated phosphorylation of WDR62 T1053 negatively regulated microtubule association, and loss of JNK signaling resulted in constitutive WDR62 localization to microtubules irrespective of cell cycle stage. In contrast, we identified that Aurora A kinase (AURKA) and WDR62 were in complex and that AURKA-mediated phosphorylation was required for the spindle localization of WDR62 during mitosis. Our studies highlight complex regulation of WDR62 localization, with opposing roles for JNK and AURKA in determining its spindle association.
KEY WORDS: WDR62, JNK, Aurora kinase A, Spindle, Mitosis
INTRODUCTION
Mitotic spindle assembly and positioning during mammalian cell division can regulate the segregation of cell fate determinants to control the balance between proliferation and differentiation (Morin and Bellaïche, 2011). Defects in spindle regulatory mechanisms might result in tissue overgrowth (as observed in cancer; Knoblich, 2010) or, conversely, growth insufficiency [such as that observed in the hereditary condition known as autosomal recessive primary microcephaly (MCPH); Thornton and Woods, 2009]. In defining molecular determinants of the severely reduced head and brain size at birth observed in MCPH, genetic studies have identified pathogenic mutations in centrosome- and spindle-pole-associated proteins (Mahmood et al., 2011; Thornton and Woods, 2009). Importantly, the most commonly mutated MCPH-associated genes encode spindle pole proteins such as ASPM (abnormal spindle-like microcephaly associated; also known as MCPH5) and WDR62 (WD40-repeat protein 62; also known as MCPH2) (Bond et al., 2002; Wollnik, 2010). Although non-mitotic functions of spindle pole proteins might contribute to MCPH disease progression, these observations suggest that mitotic spindle architecture and/or regulation are crucial factors in neural stem cell survival and/or fate decisions.
WDR62 is the second most commonly mutated MCPH-associated gene, with >30 nonsense, missense and truncating frame-shift mutations identified (Nicholas et al., 2010). WDR62 mutations are also linked to a broader spectrum of gross cortical malformations (Bilgüvar et al., 2010; Yu et al., 2010), which indicates a requirement for WDR62 in multiple, albeit largely undefined, functions during brain formation. The requirement for WDR62 expression during neural stem cell expansion is reinforced by the observations that most WDR62 mutations linked to MPCH are predicted to be null mutations. In support of this, in utero depletion of WDR62 alters the symmetry of neuroprogenitor division events in the neurogenic ventricular and subventricular regions, resulting in reduced proliferation and premature differentiation into immature neurons (Xu et al., 2014). These studies highlight key contributions by WDR62 towards neuroprogenitor fate determination.
WDR62, a large (∼175 kDa) multi-domain protein product of the WDR62 gene, contains multiple WD40 protein interaction repeats within its N-terminus that are predicted to form β-propeller structures. In contrast, the largely disordered C-terminal region of WDR62 is predicted to contain serine/threonine phosphorylation motifs targeted by proline-directed kinases such as mitogen-activated protein kinases (MAPKs) and cyclin-dependent kinases (CDKs) (Wasserman et al., 2010). Indeed, WDR62 was originally identified as a scaffold protein that binds to c-Jun N-terminal kinase (JNK) through a defined motif on the C-terminal region to coordinate stress-activated signaling pathways (Cohen-Katsenelson et al., 2011; Wasserman et al., 2010). The intracellular distribution of WDR62 is dynamic and cell-cycle-dependent, being predominantly cytoplasmic in interphase but spindle-pole-localized during mitosis (Bogoyevitch et al., 2012; Nicholas et al., 2010). WDR62 expression is required for normal mitotic progression (Bogoyevitch et al., 2012, Chen et al., 2014). In addition, the mitotic contributions of WDR62 require JNK activity (Bogoyevitch et al., 2012, Xu et al., 2014), which suggests a signaling role, although the precise function of WDR62 on the mitotic spindle is not fully defined. Previous studies on the intracellular distribution of ectopically expressed WDR62 with missense MCPH mutations that altered evolutionarily conserved residues revealed lost or substantially reduced spindle pole accumulation (Nicholas et al., 2010). Similar defects in spindle localization were also reported in primary cells isolated from an MCPH patient (Farag et al., 2013). MCPH-associated WDR62 mutants failed to rescue altered neuroprogenitor cell divisions induced by embryonic WDR62 depletion (Xu et al., 2014). Thus, the precise control of WDR62 localization is required for its mitotic functions that likely contribute to binary cell fate decisions in vivo. However, the mechanisms that regulate the spatiotemporal distribution of WDR62 remain undefined.
In this study, we revealed how the domains of WDR62 and their regulation by signaling events impact on WDR62 cell-cycle-dependent association with the microtubule cytoskeleton. Importantly, we demonstrated for the first time that WDR62 specifically recruits JNK1 (also known as MAPK8) to spindle microtubules, consistent with the essential requirement for the WDR62–JNK interaction in spindle regulation, mitotic progression and neural development (Bogoyevitch et al., 2012; Xu et al., 2014). Furthermore, although JNK association is required for mitotic regulation, we demonstrated that WDR62 microtubule association was negatively regulated by JNK-mediated phosphorylation. In contrast, WDR62 association with, and phosphorylation by, mitotic Aurora A kinase (AURKA) increased WDR62 spindle localization during mitotic entry. These studies are the first to define signaling mechanisms regulating WDR62 spatiotemporal distribution, highlighting the crucial and opposing roles played by JNK- and AURKA-mediated phosphorylation.
RESULTS
Increased WDR62 association with microtubules is coordinated with mitotic entry and metaphase-anaphase transition
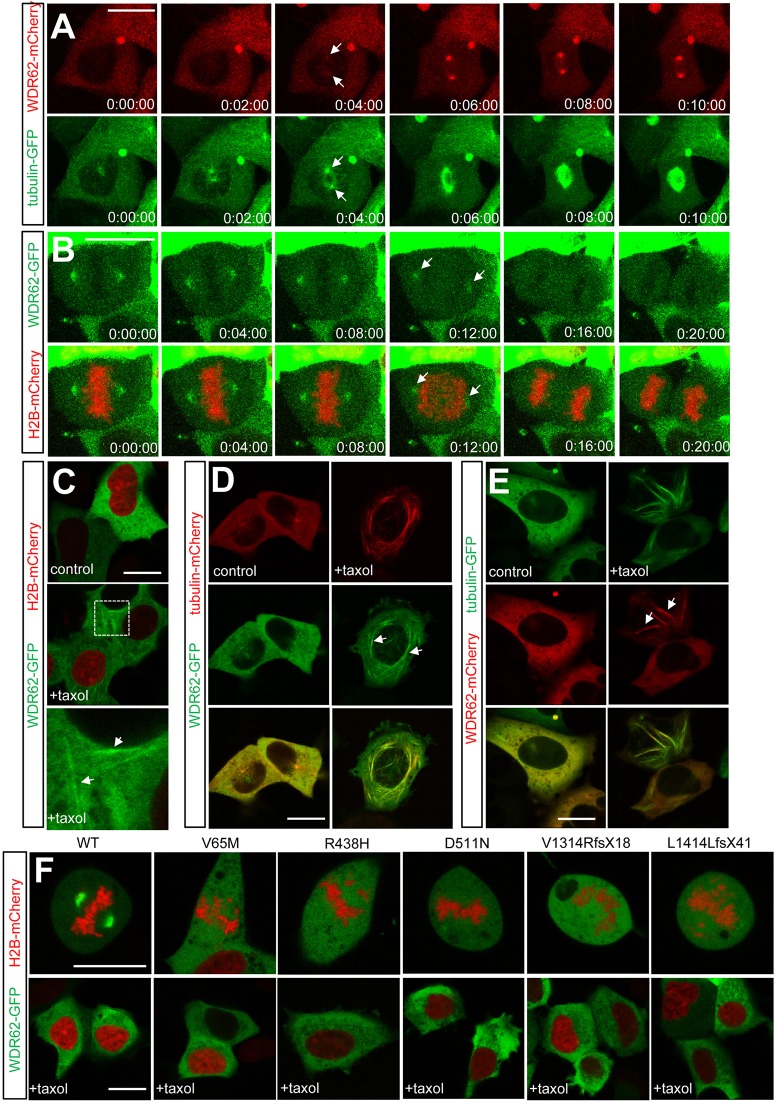
During cell division, WDR62 localization shifts between cytoplasmic and cytoskeletal compartments (Bogoyevitch et al., 2012; Nicholas et al., 2010) but precisely when and how rapidly these transitions occur are not known. We have addressed this through live imaging of single cells expressing GFP or mCherry-tagged WDR62 with improved temporal resolution of WDR62 distribution during mitotic stages to reveal how WDR62 intracellular movements are coordinated with cell cycle progression and mitotic signaling mechanisms. We observed that WDR62 association with centrosome-nucleated (astral) microtubules was first evident after separation of duplicated centrosomes and this increased markedly following nuclear envelope breakdown and mitotic entry (Fig. 1A; supplementary material Fig. S1A). WDR62 localization at spindle poles persisted throughout prometaphase and metaphase, and decreased following chromosome segregation and transition into anaphase I. WDR62 returned to the cytoplasm by anaphase II/telophase (Fig. 1B). Thus, WDR62 association with spindle microtubules during mitosis is stage specific, correlating precisely with mitotic entry and metaphase-to-anaphase transition.
Fig. 1.
WDR62 associates with microtubules during interphase and mitosis. (A) WDR62–mCherry and tubulin–GFP were coexpressed in AD293 cells and their association with astral microtubules during mitotic entry was revealed by live-cell fluorescence imaging. Arrows indicate spindle association. (B) Decreased spindle pole localization of GFP-tagged WDR62 in cells undergoing metaphase-anaphase transition. Chromosome separation and anaphase transition were determined by H2B–mCherry coexpression. Arrows highlight reduced levels of centrosome-associated WDR62. (C) Taxol treatment (10 µM, 30 min) revealed that WDR62–GFP was partially localized to cytoplasmic filaments (arrows) during interphase. The highlighted area of interest indicated in the middle panel is shown at higher magnification in the lower panel. (D,E) Filamentous WDR62 is colocalized with microtubule bundles (arrows) in taxol-treated non-mitotic AD293 cells. (F) Defective microtubule localization by GFP-tagged MCPH-associated WDR62 missense (V65M, R438H, D511N) and truncated frame-shift (V1314RfsX18, L1414LfsX41) mutants in mitotic or taxol-treated (10 µM, 30 min) interphase cells. Scale bars: 20 µm.
In non-dividing cells, WDR62–GFP was predominantly cytoplasmic and excluded from the nucleus (Fig. 1C). However, taxol-induced stabilization of interphase microtubules revealed a proportion of WDR62–GFP as cytoplasmic filaments (Fig. 1C). These colocalized with microtubules marked by tubulin–GFP or tubulin–mCherry (Fig. 1D,E). Moreover, WDR62 association with interphase microtubules was only observed in live cells and was lost following chemical fixation (not shown), indicating low-affinity association of WDR62 with microtubules in non-dividing cells. To assess the direct association of WDR62 with microtubules, we reconstituted recombinant WDR62 with purified tubulin in vitro (supplementary material Fig. S1B,C). Although the inclusion of WDR62 did not alter tubulin polymerization (supplementary material Fig. S1B), a proportion of WDR62 pelleted with the polymerized tubulin fraction in our sedimentation analysis (supplementary material Fig. S1C), consistent with a direct association of WDR62 with microtubules. Our studies indicate the capacity of WDR62 to bind to microtubules, even during interphase. However, the extent to which endogenous WDR62 binds to interphase microtubules remains undetermined. We had not previously observed endogenous protein colocalization with tubulin in fixed samples (Bogoyevitch et al., 2012). It is also likely that the small proportion of microtubule-associated protein in interphase would not be preserved following chemical fixation.
Previous studies have shown that MCPH-associated WDR62 mutants fail to rescue neural proliferation defects resulting from WDR62 depletion in vivo (Xu et al., 2014). In this study, we also observed that WDR62 MCPH mutants failed to localize to the spindle pole and remained cytoplasmic throughout cell division (Fig. 1F). In addition, these pathogenic WDR62 mutations showed defects in microtubule binding, as revealed by taxol treatment, which did not enhance microtubule colocalization of MCPH-associated WDR62 mutants as observed for the wild-type WDR62 (Fig. 1F; supplementary material Fig. S1D). Our findings thus demonstrate the specific enhancement of the microtubule association of WDR62 during mitotic entry, reinforcing the notion that the stage-specific intracellular localization of WDR62 is tightly regulated and is likely important for WDR62 functions in neural development.
Domain mapping analysis of WDR62 association with microtubules
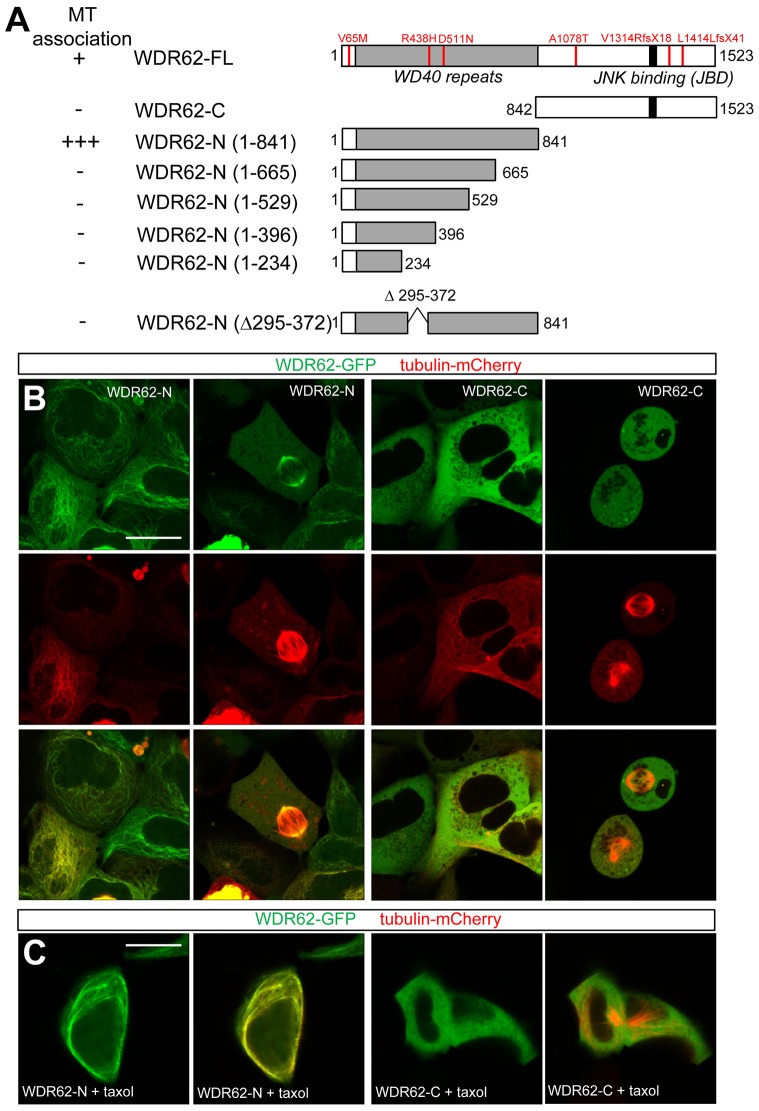
To identify WDR62 domains involved in microtubule association, we generated GFP-labeled WDR62 truncation mutants (illustrated in Fig. 2A) lacking the C-terminal disordered region (WDR62-N, residues 1–841) or the N-terminal WD40-repeat rich region (WDR62-C, residues 842–1523). Firstly, we showed that GFP-tagged WDR62-N decorated the mitotic spindle (Fig. 2A,B), indicating that the C-terminal half of WDR62, which includes JNK-binding and putative Ser/Thr-Pro phosphorylation sites (Wasserman et al., 2010), was not required for spindle localization in accord with our previous immunofluorescence analysis (Bogoyevitch et al., 2012). Interestingly, GFP-tagged WDR62-N prominently decorated microtubule filaments during interphase in live cells (Fig. 2B). Taxol treatment increased the proportion of WDR62-N localized to microtubules during interphase (Fig. 2C). Further truncation of WDR62-N, or WD40 domain deletion from the WDR62–GFP construct, resulted in cytoplasmic localization and a complete loss of microtubule association in mitotic and non-dividing cells (Fig. 2A; representative images in supplementary material Fig. S1E). Thus, WDR62 association with microtubules required an intact WD40 repeat region (Fig. 2). In contrast, GFP-tagged WDR62-C, lacking the WD40 repeat region, was localized in the cytoplasm regardless of cell cycle stage (Fig. 2B,C). Thus, the N-terminal WD40 repeat region of WDR62 was necessary and sufficient for microtubule association, whereas cell-cycle-dependent spatiotemporal regulation could be attributed to the C-terminus of WDR62.
Fig. 2.
The WD40-repeat rich N-terminal region of WDR62 is required and sufficient for microtubule localization. (A) The microtubule localization of GFP-tagged WDR62-N with progressive truncations and deletions was determined in interphase and mitotic cells. Schematic depicts truncation or deletion, the MCPH mutations tested and the extent of microtubule (MT) binding. +, MT association; +++, strong MT association; −, no association. Representative images are shown in supplementary material Fig. S1E. (B) GFP-tagged WDR62-N (amino acids 1–841) or WDR62-C (amino acids 842–1523) truncation mutants were coexpressed with tubulin–mCherry, and microtubule association was determined in AD293 cells during mitosis and interphase. (C) WDR62-N–GFP and WDR62-C–GFP are localized to microtubule and cytoplasmic compartments, respectively, in taxol-treated (10 µM, 30 min) non-mitotic AD293 cells. Scale bars: 20 µm.
JNK phosphorylation of WDR62 negatively regulates microtubule association
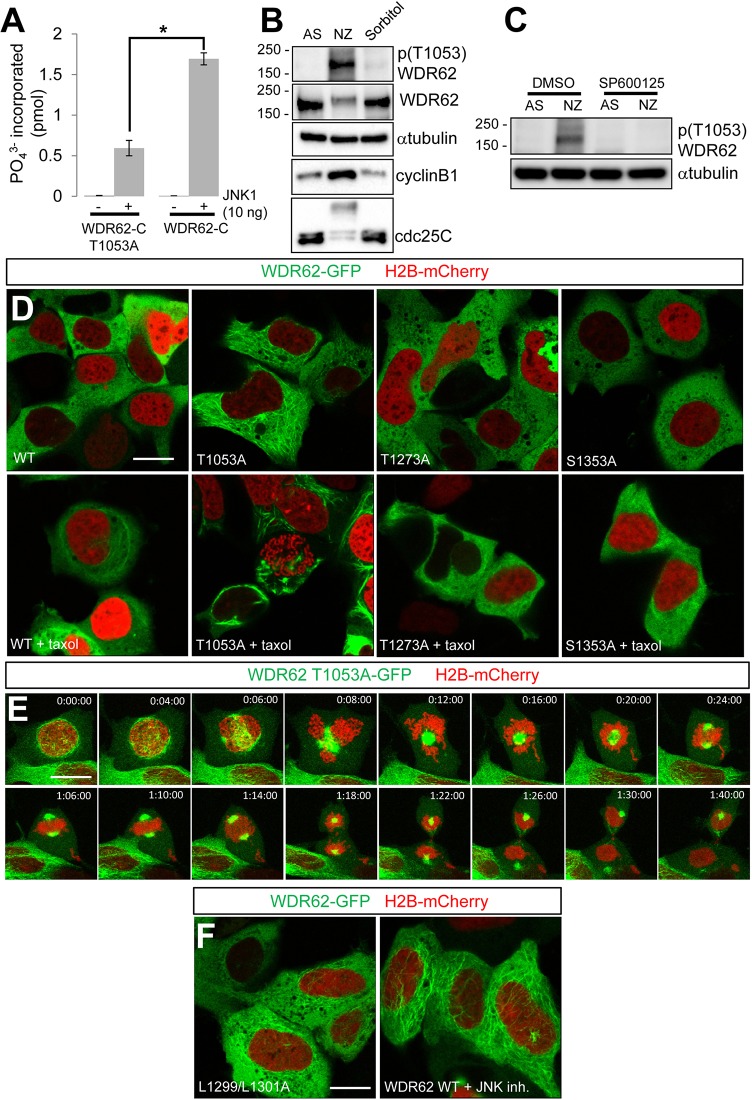
In considering the mechanisms regulating the association of WDR62 with microtubules, we investigated the potential contributions by JNK signaling. WDR62 is phosphorylated by JNK, although the specific sites targeted and their contributions to mitotic localization and function have not been characterized (Bogoyevitch et al., 2012; Wasserman et al., 2010). Our previous in vitro phosphorylation studies have indicated that JNK phosphorylates residues within the C-terminal half of WDR62 (WDR62-C) (Bogoyevitch et al., 2012). In cells, the phosphorylation-dependent mobility shift of WDR62-C was induced by mitotic arrest or hyperosmotic stress stimulation (supplementary material Fig. S2A). Moreover, the phosphorylation-dependent mobility shift of WDR62-C correlated with the shift observed for endogenous WDR62 (supplementary material Fig. S2A). In contrast, the mobility of a truncated WDR62 mutant containing the N-terminal WD40 repeats (WDR62-N) was not altered by mitotic or stress stimulation (supplementary material Fig. S2A). Phosphopeptide mapping by mass spectrometry of recombinant WDR62-C phosphorylated in vitro by active JNK1 identified three putative JNK target sites (T1053, T1273, S1353), including the highly conserved T1053 residue (supplementary material Fig. S2B,C). Thus, we focused our investigation on this specific phosphorylation site. JNK1-mediated phosphorylation of the recombinant WDR62-C T1053A mutant was substantially reduced in our in vitro kinase assay (Fig. 3A). We generated a phospho-T1053 WDR62 antibody, validated its specificity with ectopically expressed WDR62 T1053A (supplementary material Fig. S2D) and demonstrated that endogenous WDR62 T1053 was phosphorylated in response to nocodazole-induced mitotic synchronization but was also modestly phosphorylated following stress stimulation (Fig. 3B). Furthermore, WDR62 T1053 phosphorylation in mitotically arrested cells was attenuated by JNK inhibition (Fig. 3C). Thus, WDR62 T1053 was identified as a novel phosphorylation site targeted by JNK in vitro and in cells.
Fig. 3.
JNK-mediated phosphorylation of WDR62 maintains cytoplasmic localization during interphase. (A) In vitro phosphorylation of recombinantly expressed WDR62-C T1053A or WDR62-C by JNK1. Data show phosphate incorporation (pmol) and are expressed as the mean±s.e.m. (n = 3); *P<0.05 (Student's t-test). (B) AD293 cells were synchronized in mitosis (NZ, 350 nM, 16 h), stimulated with hyperosmotic stress (Sorbitol, 0.5 M, 30 min) or left untreated in asynchrony (AS), and the lysates were blotted as indicated. (C) JNK inhibition (SP600125, 20 µM, 60 min) decreased WDR62 T1053 phosphorylation in mitotic cells (NZ, 350 nM, 16 h). (D) The localization of GFP-tagged alanine-substituted WDR62 mutants (T1053A, T1273A, S1353A) or wild-type (WT) WDR62, coexpressed with mCherry-tagged H2B, was evaluated in non-dividing AD293 cells. The increased association of the WDR62 T1053A mutant with microtubules (compared to that of WDR62 T1273A or S1353A) was more evident following taxol-treatment (10 µM, 30 min). (E) Mitotic progression and division of a single AD293 cell expressing the GFP-tagged WDR62 T1053A mutant and H2B–mCherry. (F) Cytoplasmic microtubule association of the GFP-tagged JNK-binding-domain mutant of WDR62 (L1299/1301A) or wild-type WDR62 treated with JNK inhibitor [JNK inhibitor VIII (JNK inh.), 20 µM, 60 min]. Scale bars: 20 µm.
To determine the contribution of novel JNK target sites to the mitotic localization of WDR62, we next generated alanine substitution mutants of full-length WDR62–GFP. We found a striking association of GFP-tagged WDR62 T1053A with interphase microtubules in non-dividing cells, whereas WDR62 T1273A or S1353A mutants did not show altered cytoplasmic localization (Fig. 3D). The enhanced filamentous distribution of WDR62 T1053A was exacerbated by taxol treatment, and this mutant protein colocalized with microtubules (Fig. 3D; supplementary material Fig. S3A). In contrast, although WDR62 T1273A and S1353A mutants also localized to microtubules in taxol-treated cells, the extent of association was more comparable to that of wild-type WDR62 (supplementary material Fig. S3A). Thus, alanine substitution of T1053, but not T1273 or S1353, increased WDR62 association with microtubules.
Furthermore, the microtubule association of WDR62 T1053A persisted throughout mitosis, including in early prophase and late anaphase/telophase when WDR62 localization was predominantly cytoplasmic (Fig. 3E). Moreover, the intracellular distribution of the WDR62 JNK-binding mutant (L1299/L1301A) as defined under live-cell conditions showed clear microtubule association in the cytoplasm and in taxol-treated cells (Fig. 3F; supplementary material Fig. S3A). Similarly, chemical inhibition of JNK activity was sufficient to increase the microtubule association of wild-type WDR62–GFP during interphase (Fig. 3F; supplementary material Fig. S3A). Thus, JNK phosphorylation of the novel WDR62 target site, WDR62 T1053, antagonized the association of WDR62 with microtubules.
Dynamic WDR62 association with microtubules is regulated by T1053 phosphorylation
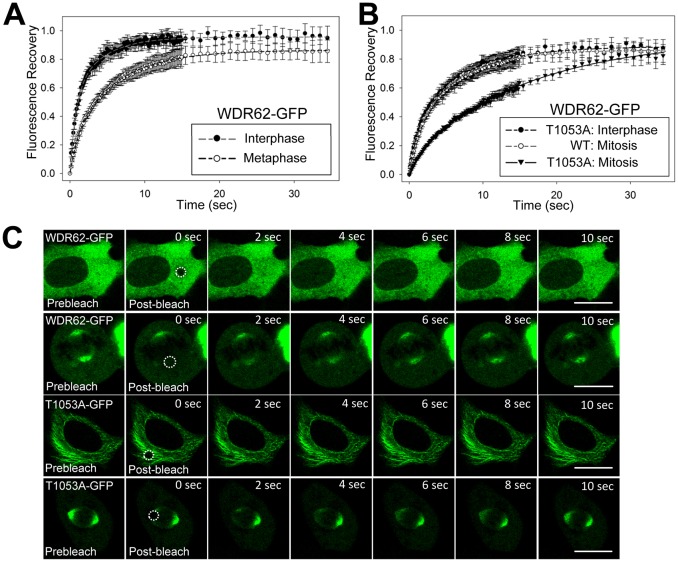
To probe the dynamics and mobility of WDR62 association with microtubules, we conducted fluorescence recovery measurements after photobleaching (FRAP) in mitotic and non-dividing cells. We observed rapid and near complete fluorescence recovery of cytoplasmic WDR62–GFP during interphase, indicating that the majority of protein was highly mobile and readily diffusible (Fig. 4A). In comparison, the recovery of WDR62–GFP during mitosis was substantially slower, which we attributed to a greater proportion of WDR62–GFP participating in microtubule binding (Fig. 4A). We next compared the fluorescence recoveries of WDR62 T1053A and found that our measurements in interphase superimposed with the recovery curve of spindle-pole-localized wild-type WDR62 (WDR62-WT), consistent with a role for T1053 in regulating the kinetics of WDR62 microtubule association (Fig. 4B). Interestingly, the mobility of spindle-pole-localized WDR62 T1053A during metaphase was further reduced in comparison to that of spindle-localized WDR62-WT (Fig. 4B). We calculated the half-time to maximum recovery (t½) of metaphase-spindle-localized WDR62-WT as 4.7±0.2 s. In comparison, the t½ of spindle-localized WDR62 T1053A was substantially extended, at 11.2±0.4 s. Representative images depict prebleach and postbleach fluorescence recovery in regions measured (Fig. 4C). The mobile fraction (∼95%) was not significantly different for WDR62–GFP in interphase and mitosis or for the WDR62 T1053A mutant (Fig. 4A,B). Thus, WDR62 is mobile with rapid exchange between cytoplasmic and microtubule-bound populations. Our results indicate that increased microtubule retention during mitosis might contribute to WDR62 spindle-microtubule localization during cell division. We also conclude that the WDR62 T1053A mutant is less mobile and therefore more stably associated with microtubules, which reinforces the negative regulation of WDR62 retention at microtubules by T1053 phosphorylation. Furthermore, our results highlight mitotic factors that determine the kinetics of WDR62 association with microtubules in addition to JNK signaling.
Fig. 4.
WDR62 T1053 regulates dynamic association with microtubules. GFP-tagged WDR62-WT or T1053A mutant were expressed in AD293 cells, and single cells were subjected to FRAP analysis. (A) Fluorescence intensity of GFP-tagged WDR62 in interphase and metaphase is plotted as a function of time (s) post-bleach. (B) Fluorescence intensity of the GFP-tagged dephosphomimetic mutant WDR62 T1053A in interphase or metaphase cells is plotted as a function of time (s) post-bleach. Fluorescence recovery of wild-type (WT) WDR62 during metaphase is also included for comparison. Values show the percentage fluorescence recovery as the mean±s.d. (n = 15). Dashed lines indicated fitted curves. (C) Representative pre-bleach and post-bleach images of GFP-tagged WDR62 or WDR62 T1053A in non-dividing and mitotic cells. Circles indicate the bleached region. Scale bars: 20 µm.
WDR62 recruits JNK1 to the spindle pole
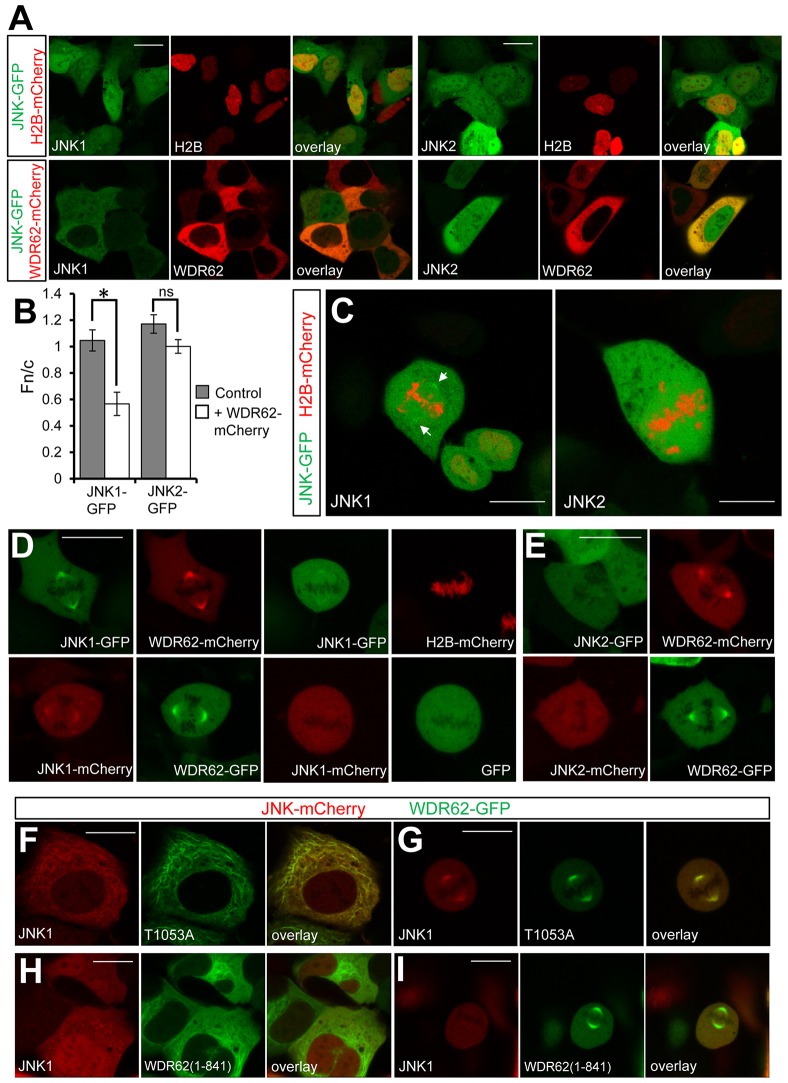
The intracellular distribution of JNK family members is diverse and regulated by complex mechanisms, including direct interaction with protein binding partners. Centrosome localization and spindle regulatory functions for JNK have been described previously (Huang et al., 2011; MacCorkle-Chosnek et al., 2001) and we have shown that mitotic functions of WDR62 required JNK interaction (Bogoyevitch et al., 2012). Therefore, we evaluated the involvement of WDR62 in recruiting JNK to the mitotic spindle. Firstly, we investigated the effect of ectopically expressed WDR62 on the intracellular distribution of GFP-tagged JNK. During interphase, JNK1–GFP or JNK2–GFP were present in nuclear and cytoplasmic compartments (Fig. 5A). However, WDR62 was predominantly cytoplasmic, and coexpression with JNK1, but not JNK2 (also known as MAPK9), was sufficient to alter the localization of the kinase from the nucleus to a predominantly cytoplasmic distribution (Fig. 5A,B). Quantitative measurements of the ratio of nuclear to cytoplasmic JNK1–GFP fluorescence (Fn/c) revealed that coexpression of WDR62 substantially decreased both Fn/c and the cytoplasmic retention of JNK1 (Fig. 5B). In contrast, the Fn/c of JNK2–GFP was not significantly altered by WDR62 coexpression (Fig. 5B). Our findings indicate a capacity for WDR62 to determine the subcellular localization of the JNK1 isoform specifically. Therefore, we postulated that WDR62 might also recruit JNK1 to the spindle pole.
Fig. 5.
WDR62 recruits JNK1 to the spindle pole. (A) GFP-tagged JNK1 or JNK2 were coexpressed with mCherry-tagged WDR62 or histone 2B (H2B), as a negative control. The cytoplasmic or nuclear localization of JNK isoforms was evaluated in interphase. (B) Nuclear and cytoplasmic fluorescence of GFP-tagged JNK1 or JNK2, in the presence or absence of WDR62, was quantified and expressed as the nuclear to cytoplasmic ratio (Fn/c). Values ∼1 indicate nuclear and cytoplasmic localization, whereas values <1 indicate predominantly cytoplasmic distribution. Data show the mean±s.d. (25 cells evaluated in three independent analyses); *P<0.05; ns, non-significant (Student's t-test). (C) Intracellular localization of GFP-tagged JNK1 or JNK2, with H2B–mCherry, in mitotic cells. Arrows indicate JNK1–GFP localized to spindle poles. (D) The localization of fluorescent-protein-tagged JNK1 coexpressed with WDR62 was determined in mitotic cells. Fluorescent-protein-tagged JNK1 coexpressed with GFP or H2B–mCherry served as negative controls. (E) The localization of fluorescent-protein-tagged JNK2 coexpressed with WDR62 was determined in mitotic cells. (F) Colocalization of JNK1–mCherry and WDR62 T1053A–GFP on interphase microtubules and (G) mitotic spindle poles. (G,H) JNK1–mCherry does not associate with interphase (H) or spindle (I) microtubules when coexpressed with GFP-tagged WDR62-N (amino acids 1–841). Scale bars: 20 µm.
Live imaging of dividing cells expressing JNK1–GFP indicated that a small subset of JNK1–GFP was localized to the spindle pole during mitosis (Fig. 5C). Interestingly, this was not observed with cells expressing JNK2–GFP (Fig. 5C). Moreover, the spindle pole localization of fluorescent-protein-tagged JNK1 increased when it was coexpressed with WDR62 and did not increase with the coexpression of negative controls such as H2B or GFP (Fig. 5D). This was not dependent on JNK activity, as identical results were obtained with the inactive JNK1 K55A mutant (supplementary material Fig. S3B,C). In addition, whereas JNK1 and WDR62 were colocalized at the spindle pole, JNK2 remained cytoplasmic and did not colocalize with WDR62 when coexpressed (Fig. 5E). In extending our findings on the capacity of WDR62 to direct the intracellular distribution of JNK1, we showed that coexpression of WDR62 T1053A and JNK1 resulted in their colocalization to microtubule filaments during interphase and mitosis (Fig. 5F,G). In contrast, the WDR62-N (amino acids 1–841) mutant, lacking the JNK-binding motif, localized constitutively to microtubules but failed to recruit coexpressed JNK1 to the cytoskeletal compartment (Fig. 5H,I). Taken together, our findings reinforce a role for WDR62 in directing the localization of the JNK1 isoform specifically to spindle microtubules. WDR62-directed JNK1 localization at the spindle pole is likely required for mitosis or spindle regulation and is consistent with the recently described role for the WDR62–JNK1 complex in neuroprogenitors (Xu et al., 2014).
AURKA activity promotes WDR62 spindle localization
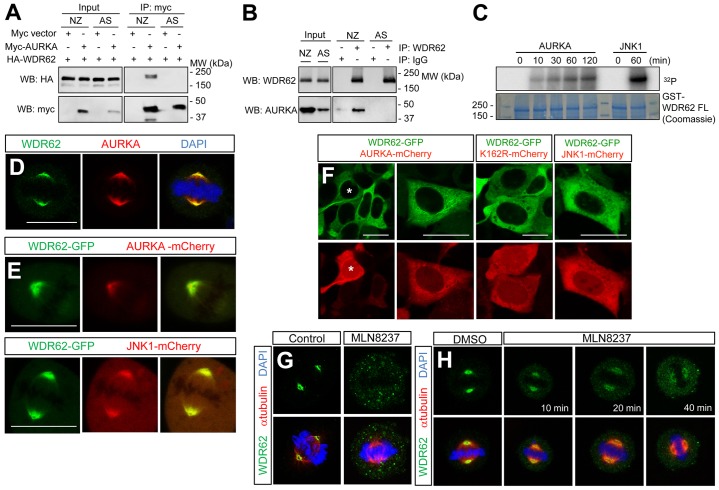
Our findings show that although JNK-mediated WDR62 phosphorylation was increased during mitosis, this was not required for spindle localization. To gain insights into the mechanism that triggers rapid spindle accumulation of WDR62 upon mitotic entry, we performed affinity pulldown of Myc-tagged WDR62 from mitotically synchronized cells and identified AURKA as a highly significant mitosis-specific binding partner using mass spectrometry in repeated analyses (supplementary material Fig. S3D,E). We demonstrated the interaction between coexpressed HA–WDR62 and Myc–AURKA in co-immunoprecipitation studies (Fig. 6A) and verified the interaction between endogenous WDR62 and AURKA proteins in mitotically arrested cells (Fig. 6B). We next purified recombinant full-length WDR62 (GST–WDR62-FL) for in vitro kinase assays with active AURKA and demonstrated that WDR62 was a direct phosphorylation target of AURKA (Fig. 6C). We found that WDR62 and AURKA were partially colocalized at the spindle pole (Fig. 6D,E). Although AURKA is localized to centrosomes and the mitotic spindle, WDR62 specifically colocalized with AURKA on spindle microtubules (Fig. 6E). In contrast, spindle-associated WDR62 and JNK1 were closely colocalized (Fig. 6E). In investigating the requirement of WDR62 in regulating AURKA, depletion of WDR62 did not substantially alter AURKA expression or mitotic localization of AURKA (supplementary material Fig. S4A,B). In contrast, we found that AURKA activity regulated WDR62 localization. The ectopic expression of AURKA–mCherry augmented the microtubule association of WDR62–GFP during interphase (Fig. 6F). As negative controls, inactive AURKA (K162R) or JNK1 overexpression did not alter WDR62–GFP distribution in non-dividing cells (Fig. 6F). Moreover, AURKA-enhanced WDR62–GFP association with microtubules was reversed by treatment with an AURKA-selective inhibitor (Manfredi et al., 2011) (MLN8237, supplementary material Fig. S4C). Spindle pole accumulation of endogenous WDR62 was also disrupted by MLN8237 administered during mitosis (Fig. 6G). Furthermore, inhibition of AURKA in mitotically arrested cells rapidly decreased WDR62 spindle localization (Fig. 6H). These results indicate that the microtubule localization of WDR62 during mitosis is enhanced and sustained by AURKA activity.
Fig. 6.
AURKA interacts with WDR62 and regulates its accumulation on spindle microtubules. (A) Myc–AURKA and HA-tagged WDR62 were transiently coexpressed, and their interaction in asynchronous (AS) or mitotically arrested (NZ, 350 nM, 16 h) cells was evaluated by co-immunoprecipitation (IP) analysis. Coexpression with a Myc vector served as a negative control. WB, western blot. (B) Endogenous AURKA co-immunoprecipitation with WDR62 in mitotically arrested cells (NZ, 350 nM, 16 h). Immunoprecipitation with IgG antibodies served as a negative control. (C) In vitro kinase assays to determine the phosphorylation of full-length WDR62 (GST–WDR62 FL) by AURKA or JNK1. (D) Mitotic AD293 cells were immunostained to determine the localization of endogenous WDR62 and AURKA. (E) WDR62–GFP and AURKA–mCherry or JNK1–mCherry were coexpressed in AD293 cells, and their localization assessed in mitotic cells by live-cell imaging. (F) mCherry-tagged AURKA, kinase-dead AURKA mutant (K162R) or JNK1 were coexpressed with GFP-tagged WDR62, and the microtubule association of WDR62 was determined using live-cell microscopy. The asterisk (*) indicates a cell with enhanced microtubule association of WDR62 with AURKA coexpression. Scale bars: 20 µm. (G) Following MLN8237 (0.5 µM) treatment, spindle pole association of endogenous WDR62 was determined by immunofluorescence staining. (H) The spindle-pole localization of endogenous WDR62 in M-phase-arrested (taxol, 10 nM, 16 h) cells was evaluated at the indicated time intervals following MLN8237 (0.5 µM) treatment.
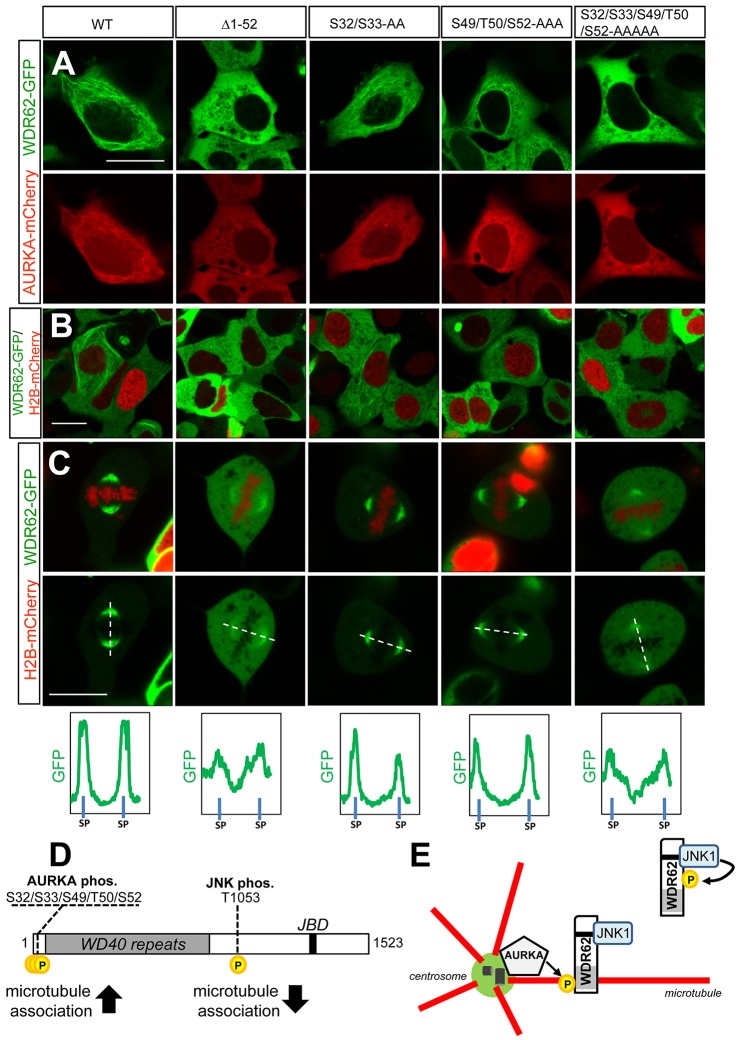
In considering the AURKA target residues on WDR62 potentially involved in regulating microtubule association, phosphorylated Ser/Thr residues (S32, S33, S49, T50 and S52) within the N-terminus of WDR62 had previously been identified in global phosphoproteomic profiling of mitotic AURKA substrates (Kettenbach et al., 2011). In addition, our quantitative phosphoproteomic analysis of in-vitro-phosphorylated WDR62 identified S32 and S33 as significantly phosphorylated in the presence of active AURKA (supplementary material Fig. S4D). In determining the contribution of these residues to the localization of WDR62, we showed that AURKA overexpression did not enhance microtubule association of N-terminally truncated WDR62 (Δ1–52) (Fig. 7A). This is consistent with a requirement for AURKA-mediated phosphorylation of the N-terminal region of WDR62 in promoting microtubule association. To identify the specific residues involved, we generated alanine-substituted mutants in the N-terminal region of WDR62. Alanine replacement of the five putative phosphorylation sites (S32/S33/S49/T50/S52-AAAAA) of WDR62 attenuated interphase microtubule association induced by AURKA coexpression (Fig. 7A), recapitulating our observations with N-terminally truncated WDR62 (Δ1–52). In contrast, WDR62 S32/S33-AA or WDR62 S49/T50/S52-AAA retained some capacity to associate with interphase microtubules when coexpressed with AURKA (Fig. 7A). Similarly, taxol treatment of non-dividing cells, in the absence of ectopically expressed AURKA, revealed reduced microtubule association of WDR62 mutants defective in AURKA phosphorylation (Fig. 7B; supplementary material Fig. S4E). This indicates redundancy in AURKA-targeted residues or, alternatively, that multiple residues might be involved in phosphorylation-mediated localization of WDR62 to microtubules.
Fig. 7.
AURKA-mediated phosphorylation of residues within the WDR62 N-terminal region regulates microtubule association. (A) GFP-tagged N-terminally truncated (Δ1–52) WDR62, full-length wild-type (WT) WDR62, WDR62 S32/S33-AA, WDR62 S49/T50/S52-AAA or WDR62 S32/S33/S49/T50/S52-AAAAA mutants were coexpressed with mCherry–AURKA and localization was evaluated. (B) GFP-tagged N-terminally truncated or alanine-substituted WDR62 mutants were coexpressed with H2B–mCherry and taxol treated (10 µM, 30 min), and protein localization was evaluated. (C) GFP-tagged N-terminally truncated or alanine-substituted WDR62 mutants were coexpressed with H2B–mCherry and the spindle pole association of WDR62 proteins was determined in mitotic cells. Relative GFP signal intensities were quantified along line scans bisecting spindle poles (SP). Scale bars: 20 µm. (D) Schematic of WDR62 depicting the position of AURKA and JNK phosphorylation sites and their respective effects on protein association with microtubules. (E) Proposed model of WDR62 spatiotemporal regulation by JNK and AURKA. Centrosome-associated AURKA phosphorylates WDR62 to trigger association with astral microtubules following mitotic entry. This is required for the accumulation of WDR62–JNK signaling complexes at the spindle pole. JNK-mediated phosphorylation of WDR62 promotes dissociation from microtubules for cytoplasmic localization.
During mitosis, we observed perturbed spindle pole accumulation of WDR62 Δ1–52 and WDR62 S32/S33/S49/T50/S52-AAAAA (Fig. 7C). The spindle pole association of WDR62 Δ1–52 and WDR62 S32/S33/S49/T50/S52-AAAAA was reduced compared to that of WDR62-WT, although a proportion of mutant WDR62 was observed on the spindle (Fig. 7C). This contrasts the complete loss of spindle-pole localization observed with MCPH-associated WDR62 mutants (Fig. 1F). Thus, whereas WDR62 N-terminal phosphorylation can promote microtubule association, MCPH mutations in the N-terminal region likely impact on an intrinsic capacity of WDR62 to bind to microtubule filaments, resulting in lost spindle pole localization. Taken together, our results indicate that AURKA-mediated phosphorylation of the WDR62 N-terminal region promotes WDR62 microtubule association (Fig. 7D,E). In contrast, JNK phosphorylation negatively regulates microtubule association (Fig. 7D,E). Our findings indicate an AURKA-directed mechanism crucial for spatiotemporal regulation of WDR62–JNK1 during mitosis (Fig. 7E).
DISCUSSION
The mitotic accumulation of MCPH-associated spindle pole proteins such as ASPM and WDR62 is required for spindle organization and mitotic regulation that is necessary for neural stem cell expansion or maintenance (Bogoyevitch et al., 2012; Fish et al., 2006; Higgins et al., 2010; Xu et al., 2014). Our study has revealed the mechanisms regulating the intracellular distribution of WDR62, a MCPH protein. Specifically, we have demonstrated opposing roles of JNK and AURKA in mediating WDR62 microtubule association for spatiotemporal control of mitotic signaling events (Fig. 7D,E).
Our studies have addressed the outstanding question of the role of WDR62 in mitosis. We have shown previously that the mitotic functions of WDR62 require JNK binding (Bogoyevitch et al., 2012). JNK signaling has been implicated in the regulation of spindle assembly and metaphase progression (Gutierrez et al., 2010; Huang et al., 2011; MacCorkle and Tan, 2004), although the underlying mechanisms have not been completely defined. Thus, we postulated that WDR62 might coordinate spindle-localized signaling events by directing mitotic-stage-specific recruitment of JNK to the spindle pole. In agreement with this, a subset of JNK1–GFP was observed at spindle poles and this was increased by ectopic expression and colocalization of WDR62. Interestingly, despite previously described biochemical association with JNK1 and JNK2 isoforms (Cohen-Katsenelson et al., 2011; Wasserman et al., 2010), we found that WDR62 recruited the JNK1 isoform specifically to the mitotic spindle. Thus, our findings suggest a scaffolding function for WDR62 in spatiotemporal coordination of isoform-specific JNK signaling at the mitotic spindle. Additionally, it was recently reported that interaction with the JNK1 isoform was specifically required for the neural functions of WDR62 in the mouse embryo (Xu et al., 2014). Although the molecular basis behind WDR62 discrimination of JNK isoforms is unclear, our studies support the increased appreciation of JNK isoform-selective functions in biological and disease processes and are consistent with a role for the WDR62–JNK1 complex in neuroprogenitor cell regulation.
WDR62 joins a list of JNK-binding partners (for example, JIP and DCX) that are associated, either directly or indirectly, with the microtubule cytoskeleton (Gdalyahu et al., 2004; Verhey et al., 2001). We demonstrated that the association of WDR62 with microtubules, mediated through its WD repeat region, was highly dynamic and tightly regulated. In addition, the steady-state cytoplasmic distribution of WDR62 during interphase was regulated by JNK signaling. In the absence of JNK signaling, as achieved through mutation of target residues (T1053A), truncation (WDR62-N) or kinase inhibition, WDR62 decorated the microtubules constitutively irrespective of cell cycle stage. Furthermore, our photobleaching studies indicated the rapid turnover and recovery kinetics for microtubule-associated WDR62, indicative of its low-affinity microtubule binding. In the absence of JNK activity, we observed increased WDR62 retention times on microtubules, leading to its steady-state decoration of interphase microtubules. Thus, while WDR62 directed JNK1 localization in the cytoplasm and on spindle microtubules, JNK activity reciprocally regulated the intracellular compartmentalization of WDR62, and WDR62 T1053 phosphorylation negatively regulated microtubule binding to promote cytoplasmic localization (Fig. 7D,E). We further demonstrated that the WD40-repeat region of WDR62 (WDR62-N) was sufficient for microtubule and spindle pole association. This raises the question of why the MCPH-associated WDR62 truncation mutants V1314RfsX18 and L1414fsX41, which lack C-terminal regions but contain the requisite WD40 repeats, are defective in their localization to spindle microtubules. The C-terminus of WDR62 is required for protein dimerization, although our findings would also suggest that this is dispensable for microtubule association (Cohen-Katsenelson et al., 2011). Thus, the mechanistic basis underlying the loss of microtubule association by the C-terminally truncated MCPH mutants remains undefined but might involve negative regulation by aberrant JNK signaling.
Our current study indicates that WDR62 association with spindle microtubules is triggered during mitotic entry. In living single cells, the intracellular distribution of WDR62 rapidly shifts from predominantly cytoplasmic to microtubule-associated immediately following nuclear envelope breakdown, and this persists until chromosome segregation, highlighting the fact that mitotic entry and anaphase-promoting signaling events are likely to be involved in regulating WDR62 localization. Moreover, our photobleaching analysis showed that mitosis further impacted on the fluorescence recovery of spindle-associated WDR62 T1053A, and this suggests that distinct mechanisms promote enhanced microtubule association upon mitotic entry. Indeed, we revealed that the mitotic localization of WDR62 was coordinated with, and dependent on, AURKA signaling. AURKA is a centrosome- and spindle-associated mitotic kinase with well-defined functions during early mitotic phases, including regulation of centrosome maturation and separation, mitotic entry and bipolar spindle assembly (Barr and Gergely, 2007; Nikonova et al., 2013). Recent studies have also revealed important roles in central spindle assembly in late mitotic stages and during interphase (Lioutas and Vernos, 2013; Nikonova et al., 2013; Reboutier et al., 2013). The expression of AURKA is upregulated in G2 and its activity is increased through interactions with protein activators and by kinase autophosphorylation primarily to facilitate mitotic entry (Bayliss et al., 2003; Littlepage et al., 2002). AURKA is inactivated by proteasomal degradation initiated during anaphase by the APC/C complex (Castro et al., 2002; Littlepage and Ruderman, 2002). Therefore, WDR62 association with spindle microtubules from mitotic entry to anaphase transition correlates closely with the kinetics of AURKA activation during mitosis.
A quantitative phosphoproteomic screen of AURKA substrates has previously identified WDR62 as a phosphorylation target (Kettenbach et al., 2011). Moreover, during the preparation of this manuscript, the biochemical and genetic interactions between AURKA and WDR62 involved in murine neural stem cell mitosis were revealed (Chen et al., 2014). Our study confirms AURKA interaction with and phosphorylation of WDR62 and provides an expanded mechanistic understanding of AURKA and WDR62 signaling interactions. The mitotic accumulation of AURKA is mediated through an interaction with centrosomal proteins such as CEP192 (Joukov et al., 2010). However, unlike Chen et al. (Chen et al., 2014), we excluded a requirement for WDR62 in regulating AURKA expression or mitotic localization. The reasons for these discrepancies are unclear but they are likely to be accounted for by differences in our experimental approaches. Previously, reduced AURKA expression and spindle accumulation were observed in mutant embryonic fibroblasts and neural progenitors with hypomorphic alleles. In contrast, we did not observe altered AURKA expression or spindle localization with specific depletion of WDR62. Rather, the specific inhibition of AURKA activity prevented WDR62 spindle accumulation, placing WDR62 downstream of AURKA in a mitotic signaling pathway. AURKA signaling regulates multiple centrosome and spindle components for centrosome maturation and bipolar spindle formation (Barr and Gergely, 2007; Nikonova et al., 2013). Abrogated AURKA signaling has been found to lead to profound mitotic and spindle defects, including attenuated astral microtubule formation, centrosome expansion, misaligned chromosomes and monopolar spindles (Barr and Gergely, 2007). In comparison, mitotic defects as a result of WDR62 loss were less severe and were restricted predominantly to metaphase spindle organization and progression (Bogoyevitch et al., 2012). Therefore, the spindle recruitment of the WDR62–JNK complex could represent another aspect of the AURKA signaling network involved in spindle regulation and provides additional insights into the diverse downstream mechanisms regulated by mitotic AURKA activity. Nevertheless, our study supports the contribution of AURKA–WDR62 to neural development (Chen et al., 2014). Further studies will clarify the precise mechanistic links between AURKA and MCPH-associated WDR62.
In summary, our study has identified opposing roles for JNK and AURKA in regulating the spindle-microtubule association of WDR62 and a role for WDR62 in directing spindle-associated JNK1 signaling (Fig. 7D,E). The further characterization of WDR62-directed signaling events on the mitotic spindle will provide novel insights into how mitotic phosphorylation networks contribute to neuroprogenitor cell division and brain size determination.
MATERIALS AND METHODS
Antibodies, enzymes, inhibitors and reagents
Anti-WDR62 was from Bethyl Laboratories (A301-560A). Anti-cdc25C was from Cell Signaling Technology. Myc and HA antibodies were from Santa Cruz Biotechnology. WDR62 phospho-T1053 (pT1053 WDR62) antibody was custom generated by Abmart through inoculation of rabbits with SSLPQ(pT)PEQEK phosphopeptides (>90% purity) conjugated to keyhole limpet hemocyanin and supplied as affinity-purified phospho-specific antibodies. Active JNK1 was purified as described previously (Ngoei et al., 2011). Active AURKA and horseradish peroxidase (HRP)-conjugated secondary antibodies were obtained from Millipore. JNK inhibitor VIII was from Calbiochem and MLN8237 was from SelleckChem. Cell culture reagents, including DMEM and fetal bovine serum, were from Invitrogen-GibcoBrl. α-tubulin, γ-tubulin and standard laboratory chemicals including nocodazole and taxol were obtained from Sigma-Aldrich.
Plasmids
WDR62 truncation mutants (Fig. 2A) were generated by PCR using siRNA-resistant codon-optimized full-length WDR62 (WDR62-FL) as template. Alanine substitutions of Ser/Thr phosphorylation sites were made by site-directed mutagenesis and DpnI digestion (Stratagene). JNK-binding domain and MCPH-associated missense and frame-shift mutants of WDR62 were generated as described previously (Bogoyevitch et al., 2012; Xu et al., 2014). WDR62 mutants were labeled with fluorescent protein (GFP, mCherry) or small affinity (Myc) epitope tags by cloning into pEGFP-N3/C3, pmCherry-N1/C1 or pXJ40-Myc vectors as appropriate. All constructs were validated by restriction digestion and full sequencing analysis. AURKA cDNA was obtained from Origene. H2B–mCherry and tubulin–GFP were obtained from the Addgene plasmid repository.
In vitro kinase assays
Purified recombinant GST–WDR62 (10 µg) was incubated with active AURKA or JNK1 (10 ng) and 32P-radiolabeled ATP ([γ32P]-ATP, 1 µCi/reaction) in a kinase reaction buffer (20 mM HEPES pH 7.6, 20 mM MgCl2•6H2O, 75 µM ATP, 20 mM β-glycerophosphate, supplemented with 25 µM Na3VO4 and 100 µM DTT) over a 120-min timecourse at 30°C. Reactions were stopped with the addition of Laemmli sample buffer. Samples were then resolved by SDS-PAGE, stained with Gelcode Blue Stain reagent (Pierce) and analyzed by autoradiography and Cerenkov counting.
Cell culture and transient transfection
HeLa and AD293 cells were maintained in DMEM supplemented with 10% fetal calf serum and 100 U/ml penicillin-streptomycin, and cells were cultured in a humidified 5% CO2 environment. Liposome-mediated transfection was performed with LipofectamineTM 2000 and antibiotic-free Opti-MEM medium according to the manufacturer's instructions.
Cell lysates, immunoblots and immunoprecipitation
Total cell lysates were prepared in RIPA buffer [50 mM Tris-HCl pH 7.3, 150 mM NaCl, 0.1 mM EDTA, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100, 0.2% (w/v) NaF and 100 µM Na3VO4] supplemented with protease inhibitors. Cell lysates were incubated on ice (10 min) and cleared by centrifugation (16,000 g, 10 min). Protein concentrations were determined by Bradford assay and Laemmeli buffer was added prior to SDS-PAGE. Proteins were transferred onto polyvinylidene fluoride (PVDF) membrane, which was blocked with 5% (w/v) non-fat milk powder in Tris-buffered saline with 0.01% (v/v) Tween 20 and immunoblotted with the appropriate primary and HRP-conjugated secondary antibodies. Immunoprecipitation was performed by incubating protein lysates (2 mg) with Myc or HA antibodies conjugated to Protein-A–agarose beads as appropriate for 2 h at 4°C with continuous end-to-end mixing on a rotating wheel. Protein-A–agarose beads were washed with lysis buffer before precipitated proteins were eluted by the addition of Laemmeli buffer. Immunoprecipitated proteins and total cell lysates (input) were resolved by SDS-PAGE and blotted as above.
Immunofluorescence and live-cell imaging
For immunostaining, cells were cultured and treatments were performed on uncoated glass coverslips. Cells were washed in PBS before fixation with either 4% (w/v) paraformaldehyde (20 min, room temperature) or cold methanol (5 min, −20°C) as appropriate. Sample preparation and confocal microscopy was then performed as described previously (Ng et al., 2011). Cells were permeabilized [0.2% (v/v) Triton X-100 in PBS] and pre-blocked [10% (v/v) fetal calf serum in PBS] before incubation with primary antibodies and Cy2/Cy3-conjugated secondary antibodies diluted in 1% (w/v) BSA in PBS. The cells were then mounted (GelMount, Biomeda Corp) and images captured on a Leica SP5 confocal microscope using 100× 1.35 NA objectives. In live-imaging studies, cells were seeded on eight-well μ-slides (Ibidi) and transiently transfected with plasmids encoding GFP- or mCherry-tagged proteins. At 24 h post-transfection, the culture medium was replaced with Phenol-Red-free DMEM supplemented with 10% fetal calf serum and 100 U/ml penicillin-streptomycin. Images were captured at 37°C on a Leica TCS SP5 confocal microscope equipped with an environmental chamber using a 40× 0.9 NA objective. We excluded highly expressing cells in our analysis to avoid non-specific aggregation of WDR62 as we have reported previously (Bogoyevitch et al., 2012). Importantly, our live fluorescence analysis circumvents the need for specific antibody detection and cell fixation, which we have found to alter the association of WDR62 with microtubules.
FRAP
Photobleach analyses were performed with cells maintained in Phenol-Red-free growth medium at 37°C with 5% CO2 supplied to the microscope chamber. Images were captured at 37°C on a Leica TCS SP5 confocal microscope. Five prebleach images were collected with excitation at 488 nm prior to photobleaching. A region of interest in the cytosol or spindle pole was then bleached for 1 s with the laser power at 100%. Fluorescence recovery was recorded at 150-ms intervals for 25 s and then 1-s intervals for a further 40 s. FRAP measurements (n = 20) from three independent experiments were normalized as described previously (Phair et al., 2003):
where Ffrap(t) is the fluorescence recovery in bleached region of interest (ROI) at time t, Fwhole(t) is whole-cell fluorescence and Fbg(t) is the fluorescence intensity in a background region outside the cell. Ffrap-prebleach and Fwhole-prebleach represent mean prebleach fluorescence intensity of bleached ROI and whole cell, respectively. Normalized FRAP measurements were plotted against postbleach recovery time and the resulting data fitted with the double exponential equation [F(t) = A1(1−exp−τ1.t) + A2(1−exp−τ2.t)].
Affinity pulldown of WDR62 and binding partner identification by mass spectrometry
Myc–WDR62 or Myc-vector control was transiently expressed in AD293 cells. Cells were synchronized in mitosis (nocodazole, 350 nM, 16 h) or in interphase prior to protein lysate preparation and immunoprecipitation with Myc-conjugated beads. Proteins were eluted from the bead fraction with 100% trifluoroethanol (TFE)/0.1% formic acid (1∶3 ratio, pH 2.0), before the addition of 1 M triethylammonium bicarbonate buffer pH 8.0. This was followed by reduction and alkylation with TCEP (final concentration 1 M) and 100 mM iodoacetamide, respectively. In-solution tryptic digestion was carried out with 12.5 ng/ml sequencing-grade modified trypsin solution (Thermo Pierce) in 25 mM triethylammonium bicarbonate buffer pH 8.0 (TEAB) at 37°C overnight. The volume was reduced in vacuo, and tryptic peptides were analyzed on an ESI-qTOF® 5600 LC-MS/MS system (AB SCIEX) coupled to an Eskigent ULTRA nanoflow LC equipped with a nanoflex chip cube. The peptides were loaded onto a microfluidic cHiPLC trap column (ChromXP C18-CL, 120 Å nominal pore size, Eskigent) with a microfluidic cHiPLC separating column (15 cm×75 µM ChromXP, C18-CL, 3 µm particles, 120 Å nominal pore size, Eskigent) running on an 5–50% CH3CN-containing 0.1% formic acid gradient over 25 min.
Resultant MS/MS data was analyzed using the Mascot search engine (Matrix Science version 2.4) against the Uniprot database. A false discovery rate threshold of 1% was applied to exclude false positives. In addition, only proteins repeatedly identified in three separate experiments were considered.
Multiple-reaction monitoring
In vitro JNK phosphorylation sites on WDR62 were identified by mass spectrometry and confirmed using a multiple-reaction monitoring protocol as described previously (Ciccimaro et al., 2006). In brief, following in vitro phosphorylation of WDR62 (10 µg) by JNK, samples were prepared and analyzed by the same setup used for the binding-partner identification studies. Following the determination of sequence coverage and initial identification of JNK-specific phosphorylation sites, a list of transitions was generated for selective targeting.
Subsequently, peptide samples were analyzed on a 5500 QTRAP (AB SCIEX) coupled to a Tempo nanoLC-1Dplus-linked cHiPLC-nanoflex system (Eksigent). Peptides were separated by a step gradient with 0.1% (v/v) formic acid (mobile phase A) and 0.1% formic acid/95% acetonitrile (v/v) (mobile phase B) (300 nl/min). Eluted peptides were ionized, and analyzed in the positive-ion mode. Upon detection, a full-scan MS/MS spectrum of the target peptide was produced and manually inspected using Peakview software (AB SCIEX).
Stable isotope dimethyl labeling quantitative phosphopeptide mapping studies
AURKA–WDR62 quantitative phosphopeptide mapping studies were carried out through dimethyl methylation and phosphopeptide enrichment. Post in vitro kinase reactions, equal volumes of denaturation solution containing 8 M urea and 25 mM triethylammonium bicarbonate were added to the protein mix followed by reduction and alkylation with 10 mM TCEP and 55 mM iodoacetamide, respectively. The samples were diluted to 1 M urea with 25 mM TEAB, followed by overnight digestion with sequencing-grade modified trypsin at 37°C. The digestion mix was quenched by the addition of formic acid to 1% (v/v) followed by solid-phase extraction on an Oasis HLB cartridge (Waters). Eluted peptides were freeze dried overnight prior to dimethyl labeling. Samples were resuspended in 100 mM TEAB, and GST–WDR62 was labeled with the light version of formaldehyde (CH2O, +28.0313) and GST–WDR62/AURKA was labeled with the heavy version of formaldehyde (CD2O, +32.0564). The light- and heavy-labeled samples were mixed at a ratio of 1∶1 and a 2-µl aliquot of this mix was analyzed by LC-MS/MS. The remaining samples were then used to enrich for phosphopeptides using titanium dioxide.
Dimethyl-labeled samples (both phosphopeptide-enriched and non-enriched samples) were analyzed on a LTQ Orbitrap Elite (Thermo Scientific) coupled to an Ultimate 3000 RSLC nanosystem (Dionex). The nanoLC system was equipped with an Acclaim Pepmap nano-trap column (Dionex) and an Acclaim Pepmap analytical column (Dionex) running on a 3–80% CH3CN-containing 0.1% formic acid gradient over 25 min. The LTQ Orbitrap Elite mass spectrometer was operated in the data-dependent mode, whereby spectra were acquired first in positive mode followed by either collision-induced activation (CID) or high-energy collisional dissociation (HCD). Ten of the most intense peptide ions with charge states ≧2 were isolated and fragmented using normalized collision energy of 35 and activation Q of 0.25 (CID) or activation time of 0.1 ms (HCD).
The Orbitrap MS data was analyzed using Proteome Discoverer (Thermo Scientific version 1.4) with the Mascot search engine against the Uniprot database. A false discovery rate threshold of 1% was applied, and phosphopeptide identification was validated with PhosphoRS, requiring at least 90% confidence (Taus et al., 2011).
In vitro tubulin turbidity and sedimentation assay
Purified porcine tubulin (300 µg, cytoskeleton) was incubated with WDR62 (20 µg) to a final volume of 100 µl in tubulin assembly buffer (80 mM PIPES pH 6.8, 0.5 mM EGTA, 2 mM MgCl2, 1 mM GTP and 10% v/v glycerol) on clear 96-well plates. Tubulin polymerization was initiated with incubation at 37°C, and absorbance (at 340 nm) was measured every minute for 100 min on a temperature-controlled plate reader (POLARstar Optima, BMG Labtech). At the completion of turbidity analysis, samples were fractionated to separate soluble and polymerized tubulin pelleted by ultracentrifugation (100,000 g, 30 min, 37°C). Soluble fractions were removed, pellets were washed and depolymerized with 2 mM CaCl2 (4°C). Soluble and polymerized tubulin were resolved by SDS-PAGE, detected and quantified by blotting for WDR62 and tubulin.
Supplementary Material
Acknowledgments
We are grateful to Olga Plotnikova (Monash University, Clayton, VIC, Australia) for sharing the AURKA K162R construct, D. Lo and H.-C. Cheng for technical assistance in baculoviral expression of recombinant JNK1 and WDR62, and P. McMillan and the Biological Optical Microscopy Platform facility for assistance in cellular imaging.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
N.R.L. and Y.Y.C.Y. performed the majority of experimental work. T.T.T. identified JNK target sites. Y.Y.Y. contributed to FRAP analysis. D.X. and S.C.W. generated and characterized MCPH-associated WDR62 mutants. C.-S.A. contributed to the quantitative phosphoproteomic analyses. N.A.W., M.A.B. and Z.X. contributed towards experimental design and manuscript preparation. D.C.H.N. conceived of experiments and wrote the manuscript.
Funding
D.C.H.N. is a recipient of an Australian Research Council Future Fellowship [grant number FT120100193]; and this work was supported by a National Health and Medical Research Council Project Grant [grant number APP1046032].
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.157537/-/DC1
References
- Barr A. R., Gergely F. (2007). Aurora-A: the maker and breaker of spindle poles. J. Cell Sci. 120, 2987–2996 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- Bayliss R., Sardon T., Vernos I., Conti E. (2003). Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851–862 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bilgüvar K., Oztürk A. K., Louvi A., Kwan K. Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A. O., Gökben S. et al. (2010). Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 10.1038/nature09327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch M. A., Yeap Y. Y., Qu Z., Ngoei K. R., Yip Y. Y., Zhao T. T., Heng J. I., Ng D. C. (2012). WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J. Cell Sci. 125, 5096–5109 10.1242/jcs.107326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J., Roberts E., Mochida G. H., Hampshire D. J., Scott S., Askham J. M., Springell K., Mahadevan M., Crow Y. J., Markham A. F. et al. (2002). ASPM is a major determinant of cerebral cortical size. Nat. Genet. 32, 316–320 10.1038/ng995 [DOI] [PubMed] [Google Scholar]
- Castro A., Arlot-Bonnemains Y., Vigneron S., Labbé J. C., Prigent C., Lorca T. (2002). APC/Fizzy-Related targets Aurora-A kinase for proteolysis. EMBO Rep. 3, 457–462 10.1093/embo-reports/kvf095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-F., Zhang Y., Wilde J., Hansen K. C., Lai F., Niswander L. (2014). Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 5, 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccimaro E., Hevko J., Blair I. A. (2006). Analysis of phosphorylation sites on focal adhesion kinase using nanospray liquid chromatography/multiple reaction monitoring mass spectrometry. Rapid Commun. Mass Spectrom. 20, 3681–3692 10.1002/rcm.2783 [DOI] [PubMed] [Google Scholar]
- Cohen-Katsenelson K., Wasserman T., Khateb S., Whitmarsh A. J., Aronheim A. (2011). Docking interactions of the JNK scaffold protein WDR62. Biochem. J. 439, 381–390 10.1042/BJ20110284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag H. G., Froehler S., Oexle K., Ravindran E., Schindler D., Staab T., Huebner A., Kraemer N., Chen W., Kaindl A. M. (2013). Abnormal centrosome and spindle morphology in a patient with autosomal recessive primary microcephaly type 2 due to compound heterozygous WDR62 gene mutation. Orphanet J. Rare Dis. 8, 178 10.1186/1750-1172-8-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish J. L., Kosodo Y., Enard W., Pääbo S., Huttner W. B. (2006). Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc. Natl. Acad. Sci. USA 103, 10438–10443 10.1073/pnas.0604066103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdalyahu A., Ghosh I., Levy T., Sapir T., Sapoznik S., Fishler Y., Azoulai D., Reiner O. (2004). DCX, a new mediator of the JNK pathway. EMBO J. 23, 823–832 10.1038/sj.emboj.7600079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez G. J., Tsuji T., Chen M., Jiang W., Ronai Z. A. (2010). Interplay between Cdh1 and JNK activity during the cell cycle. Nat. Cell Biol. 12, 686–695 10.1038/ncb2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J., Midgley C., Bergh A. M., Bell S. M., Askham J. M., Roberts E., Binns R. K., Sharif S. M., Bennett C., Glover D. M. et al. (2010). Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 11, 85 10.1186/1471-2121-11-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Tong J. S., Wang Z. B., Yang C. R., Qi S. T., Guo L., Ouyang Y. C., Quan S., Sun Q. Y., Qi Z. Q. et al. (2011). JNK2 participates in spindle assembly during mouse oocyte meiotic maturation. Microsc. Microanal. 17, 197–205 10.1017/S1431927610094456 [DOI] [PubMed] [Google Scholar]
- Joukov V., De Nicolo A., Rodriguez A., Walter J. C., Livingston D. M. (2010). Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc. Natl. Acad. Sci. USA 107, 21022–21027 10.1073/pnas.1014664107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011). Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5 10.1126/scisignal.2001497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J. A. (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849–860 10.1038/nrm3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioutas A., Vernos I. (2013). Aurora A kinase and its substrate TACC3 are required for central spindle assembly. EMBO Rep. 14, 829–836 10.1038/embor.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage L. E., Ruderman J. V. (2002). Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16, 2274–2285 10.1101/gad.1007302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage L. E., Wu H., Andresson T., Deanehan J. K., Amundadottir L. T., Ruderman J. V. (2002). Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc. Natl. Acad. Sci. USA 99, 15440–15445 10.1073/pnas.202606599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCorkle R. A., Tan T. H. (2004). Inhibition of JNK2 disrupts anaphase and produces aneuploidy in mammalian cells. J. Biol. Chem. 279, 40112–40121 10.1074/jbc.M405481200 [DOI] [PubMed] [Google Scholar]
- MacCorkle-Chosnek R. A., VanHooser A., Goodrich D. W., Brinkley B. R., Tan T. H. (2001). Cell cycle regulation of c-Jun N-terminal kinase activity at the centrosomes. Biochem. Biophys. Res. Commun. 289, 173–180 10.1006/bbrc.2001.5948 [DOI] [PubMed] [Google Scholar]
- Mahmood S., Ahmad W., Hassan M. J. (2011). Autosomal Recessive Primary Microcephaly (MCPH): clinical manifestations, genetic heterogeneity and mutation continuum. Orphanet J. Rare Dis. 6, 39 10.1186/1750-1172-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi M. G., Ecsedy J. A., Chakravarty A., Silverman L., Zhang M., Hoar K. M., Stroud S. G., Chen W., Shinde V., Huck J. J. et al. (2011). Characterization of Alisertib (MLN8237), an investigational small-molecule inhibitor of aurora A kinase using novel in vivo pharmacodynamic assays. Clin. Cancer Res. 17, 7614–7624 10.1158/1078-0432.CCR-11-1536 [DOI] [PubMed] [Google Scholar]
- Morin X., Bellaïche Y. (2011). Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev. Cell 21, 102–119 10.1016/j.devcel.2011.06.012 [DOI] [PubMed] [Google Scholar]
- Ng D. C., Ng I. H., Yeap Y. Y., Badrian B., Tsoutsman T., McMullen J. R., Semsarian C., Bogoyevitch M. A. (2011). Opposing actions of extracellular signal-regulated kinase (ERK) and signal transducer and activator of transcription 3 (STAT3) in regulating microtubule stabilization during cardiac hypertrophy. J. Biol. Chem. 286, 1576–1587 10.1074/jbc.M110.128157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoei K. R., Catimel B., Church N., Lio D. S., Dogovski C., Perugini M. A., Watt P. M., Cheng H. C., Ng D. C., Bogoyevitch M. A. (2011). Characterization of a novel JNK (c-Jun N-terminal kinase) inhibitory peptide. Biochem. J. 434, 399–413 10.1042/BJ20101244 [DOI] [PubMed] [Google Scholar]
- Nicholas A. K., Khurshid M., Désir J., Carvalho O. P., Cox J. J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A. et al. (2010). WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 42, 1010–1014 10.1038/ng.682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonova A. S., Astsaturov I., Serebriiskii I. G., Dunbrack R. L., Jr, Golemis E. A. (2013). Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 70, 661–687 10.1007/s00018-012-1073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair R. D., Gorski S. A., Misteli T. (2003). Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375, 393–414 10.1016/S0076-6879(03)75025-3 [DOI] [PubMed] [Google Scholar]
- Reboutier D., Troadec M. B., Cremet J. Y., Chauvin L., Guen V., Salaun P., Prigent C. (2013). Aurora A is involved in central spindle assembly through phosphorylation of Ser 19 in P150Glued. J. Cell Biol. 201, 65–79 10.1083/jcb.201210060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., Mechtler K. (2011). Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 10.1021/pr200611n [DOI] [PubMed] [Google Scholar]
- Thornton G. K., Woods C. G. (2009). Primary microcephaly: do all roads lead to Rome? Trends Genet. 25, 501–510 10.1016/j.tig.2009.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K. J., Meyer D., Deehan R., Blenis J., Schnapp B. J., Rapoport T. A., Margolis B. (2001). Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152, 959–970 10.1083/jcb.152.5.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman T., Katsenelson K., Daniliuc S., Hasin T., Choder M., Aronheim A. (2010). A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell 21, 117–130 10.1091/mbc.E09-06-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollnik B. (2010). A common mechanism for microcephaly. Nat. Genet. 42, 923–924 10.1038/ng1110-923 [DOI] [PubMed] [Google Scholar]
- Xu D., Zhang F., Wang Y., Sun Y., Xu Z. (2014). Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Reports 6, 104–116 10.1016/j.celrep.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Yu T. W., Mochida G. H., Tischfield D. J., Sgaier S. K., Flores-Sarnat L., Sergi C. M., Topçu M., McDonald M. T., Barry B. J., Felie J. M. et al. (2010). Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 42, 1015–1020 10.1038/ng.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.