Abstract
PKDREJ is a testis-specific protein thought to be located on the sperm surface. Functional studies in the mouse revealed that loss of PKDREJ has effects on sperm transport and the ability to undergo an induced acrosome reaction. Thus, PKDREJ has been considered a potential target of post-copulatory sexual selection in the form of sperm competition. Proteins involved in reproductive processes often show accelerated evolution. In many cases, this rapid divergence is promoted by positive selection which may be driven, at least in part, by post-copulatory sexual selection. We analysed the evolution of the PKDREJ protein in primates and rodents and assessed whether PKDREJ divergence is associated with testes mass relative to body mass, which is a reliable proxy of sperm competition levels. Evidence of an association between the evolutionary rate of the PKDREJ gene and testes mass relative to body mass was not found in primates. Among rodents, evidence of positive selection was detected in the Pkdrej gene in the family Cricetidae but not in Muridae. We then assessed whether Pkdrej divergence is associated with episodes of sperm competition in these families. We detected a positive significant correlation between the evolutionary rates of Pkdrej and testes mass relative to body mass in cricetids. These findings constitute the first evidence of post-copulatory sexual selection influencing the evolution of a protein that participates in the mechanisms regulating sperm transport and the acrosome reaction, strongly suggesting that positive selection may act on these fertilization steps, leading to advantages in situations of sperm competition.
Keywords: positive selection, PKDREJ, sperm competition, primates, rodents
Introduction
Proteins involved in reproductive processes (the so-called ‘reproductive proteins’) were once regarded as being highly conserved, meaning that new mutations were removed by negative selection. Nonetheless, some reproductive proteins evolve rapidly, in most cases driven by positive selection, indicating that sequence divergence has key effects for reproduction (Swanson and Vacquier, 2002; Clark et al., 2006). The underlying selective forces driving the divergence of reproductive proteins are poorly understood. Several lines of evidence suggest that the adaptive evolution of reproductive proteins is the result of post-copulatory sexual selection. A powerful mechanism of post-copulatory sexual selection is sperm competition, which arises when sperm from multiple males compete to be the first to fertilize the egg(s) in a polyandrous system (Parker, 1970; Birkhead and Møller, 1998). Sperm competition generates selective pressures that shape reproductive traits increasing the fertilization success of an ejaculate under competitive conditions (Birkhead and Møller, 1998; Pizzari and Parker, 2008). Several traits important for fertilization have been identified as targets of sperm competition. For example, sperm competition favours improvements in both sperm quantity and sperm quality (Gómez Montoto et al., 2011) and selects for increases in sperm swimming velocity (Gomendio and Roldan, 2008; Fitzpatrick et al., 2009; Lüpold et al., 2009; Tourmente et al., 2011). It is also assumed that sperm competition selects for sperm with the best ability to bind to the components of the egg's extracellular coat, penetrate the egg envelope and fuse with the egg (Swanson and Vacquier, 2002).
A study of murine rodents revealed that sperm competition selects for a larger proportion of sperm undergoing changes required for fertilization (i.e. capacitation) and sperm more sensitive to the signals emitted by the egg (i.e. acrosomal exocytosis) (Gomendio et al., 2006). It has been thus suggested that adaptive changes in proteins controlling the processes that prepare sperm for fertilization in the oviduct could provide paternity advantages under situations of sperm competition (Gomendio et al., 1998; Birkhead and Pizzari, 2002).
A sperm protein which has attracted much interest both in functional and in evolutionary studies is PKDREJ. This is a plasma membrane protein (Zigo et al., 2013) belonging to the polycystin-1 gene family, presumably localized in the extracellular portion of the acrosomal region of the sperm head (Butscheid et al., 2006). Pkdrej is expressed exclusively in the testis, and Pkdrej transcript expression has only been detected in the spermatogenic lineage (Butscheid et al., 2006). Several arguments have led to the suggestion that PKDREJ could be involved in the response of mammalian sperm to the egg's zona pellucida (Gallagher et al., 2002; Neill et al., 2004; Butscheid et al., 2006; Hamm et al., 2007). A more recent study revealed that PKDREJ controls sperm transport and the timing of zona pellucida-evoked exocytosis of the sperm acrosome, a proxy for sperm fertility (Sutton et al., 2008). In this study, male mice homozygous for a targeted mutation of the Pkdrej gene were fertile, but when they were examined under conditions of sperm competition with sperm from wild-type mice, the mutation in Pkdrej resulted in sperm with lower fertilization efficiency due to delays in the transport to the oviduct. In vitro studies indicated that sperm from these animals showed a delayed acquisition of the ability to undergo the zona pellucida-induced acrosome reaction. Given that a functional PKDREJ provides a fertility advantage when sperm from two or more males are present in the female reproductive tract, it was suggested that PKDREJ could be an important factor in sperm competition (Sutton et al., 2008).
Evolutionary analyses of PKDREJ have been carried out previously in primates (Hamm et al., 2007) and cetaceans (Amaral et al., 2011). In primates, evidence of adaptive evolution was detected across PKDREJ sequences, but a direct association between the evolutionary rate of PKDREJ and differences in mating systems was not observed (Hamm et al., 2007). In cetaceans, PKDREJ showed very low levels of amino acid divergence and no signal of positive selection (Amaral et al., 2011). The different evolutionary patterns of PKDREJ observed between these groups suggest that PKDREJ could be subjected to different selective constraints across mammals. In this regard, analyses in other taxa, showing a faster rate of genome evolution as well as a wide variety of mating strategies, could clarify if sexual selection influences the evolution of PKDREJ in mammals.
In this study we reassessed whether post-copulatory sexual selection may drive the evolution of PKDREJ in primates. We used testes mass relative to body mass (i.e. relative testes mass), a good proxy of sperm competition levels, as opposed to earlier work which employed mating system as an indicator of post-copulatory sexual selection (Hamm et al. 2007), and which may have limitations to estimate levels of sperm competition. In addition, we analysed the evolution of PKDREJ in species belonging to two muroid families, Muridae and Cricetidae. These species show a great variety of mating systems, with differences in the degree of promiscuity and, as a result, a wide range of sperm competition levels (Gómez Montoto et al., 2011). Thus, they constitute an ideal model to study the effects of sexual selection on the evolution of reproductive proteins. We tested whether PKDREJ is undergoing adaptive molecular evolution in rodents and whether such divergence is linked to episodes of post-copulatory sexual selection.
In both primates and in rodents we focused on the REJ domain (receptor for egg jelly) in the extracellular N-terminal region, where a high concentration of sites under positive selection has been detected for sea urchins and primates (Mah et al., 2005; Hamm et al., 2007). In mammals, the REJ domain is thought to interact with the extracellular matrix and with adjacent cells, coupling these signalling events to intracellular responses through multimeric complexes formed by polycystin family members (Delmas, 2005). On the other hand, the REJ modules of sea urchin homologues (suREJ1, suREJ3) of polycystin kidney disease (PKD1) are receptors for the fucose sulphate polymer of egg jelly, participating in the initiation of the acrosome reaction (Moy et al., 1996; Mengerink et al., 2002).
The aim of this work was to test for positive selection in PKDREJ of primates and rodents and examine whether, within these groups, lineages experiencing high levels of sperm competition show a higher evolutionary rate of PKDREJ.
Materials and Methods
Primate and rodent species
For primates, we analysed species used in a previous study (Hamm et al., 2007) and PKDREJ sequences were obtained from NCBI GenBank. The species, with accession numbers in parentheses, were: Homo sapiens (EF517278), Pan troglodytes (EF517279), Pan paniscus (EF517280), Gorilla gorilla (EF517281), Pongo pygmaeus (EF517282), Macaca nemestrina (EF517283), Macaca mulatta (EF517284), Erythrocebus patas (EF517286), Ateles geoffroyi (EF517287), Saguinus labiatus (EF517289), Callithrix jacchus (EF517290) and Lemur catta (EF517291). Species for which PKDREJ sequences are known, but for which data of relative testes mass are not available (Macaca nigra, Lagothrix lagothrica and Otolemur garnetti), were not included in this study.
We analysed 24 rodent species belonging to the superfamily Muroidea. A total of 11 species belonged to the family Muridae, namely Mus musculus musculus, Mus m. domesticus, Mus m. castaneus, Mus m. bactrianus, Mus spretus, Mus spicilegus, Mus macedonicus, Mus famulus, Mus cookii, Mus pahari and Apodemus sylvaticus. The remaining 13 species belonged to the family Cricetidae, with 8 of the subfamily Arvicolinae, Arvicola sapidus, Arvicola terrestris, Clethrionomys (=Myodes) glareolus, Chionomys nivalis, Microtus arvalis, Microtus cabrerae, Pitymys duodecimostatus and Pitymys lusitanicus, and 5 of the subfamily Cricetinae, Cricetulus griseus, Mesocricetus auratus, Phodopus campbelli, Phodopus roborovskii and Phodopus sungorus. These groups of species have experienced rapid evolutionary radiations and diversification (Steppan et al., 2004; Suzuki et al., 2004), and cover a wide range of levels of sperm competition (Gómez Montoto et al., 2011). Moreover, the set of lineages chosen within each family have split in different stages of evolutionary time. In this way, we evaluated whether the degree of species relatedness has effects in detecting positive selection (Anisimova et al., 2001).
Males of Mus species were purchased from the Institut des Sciences de l'Evolution, CNRS-Université de Montpellier 2, France. Males of Apodemus sylvaticus and the Arvicolinae subfamily were trapped in the field, with the required permits, during the breeding season at different locations in Spain. Males of the Cricetinae subfamily were obtained from commercial suppliers and they were unrelated. PKDREJ sequences from at least three individuals per species were obtained for the analyses. Animal handling and housing followed the standards of the Spanish Animal Protection Regulation RD1201/2005, which conforms to European Union Regulation 2003/65. This study was approved by the Bioethics Committee of the Consejo Superior de Investigaciones Científicas (CSIC, Spain).
Rodent PKDREJ sequence data
Pkdrej gene sequences of rodent species were obtained by amplification using PCR. Genomic DNA was extracted from frozen tissues using E.Z.N.A.® Tissue DNA kit (Omega, Madrid, Spain) following the manufacturer's recommendations. PCR mixtures were prepared in a 50 μl-volume containing PCR Gold buffer 1× (Roche, Barcelona, Spain), 2.5 mM MgCl2 (Roche), 0.8 mM dNTPs mix supplying 0.2 mM of each deoxynucleotide triphosphate (Applied-Biosystems, Barcelona, Spain), 0.25 mM of forward and reverse primers (Life Technologies, Madrid), 2 U of DNA polymerase (Biotools, Madrid), and 20–200 ng/μl of genomic DNA template. PCRs were performed in a Veriti thermocycler (Applied-Biosystems). The thermocycler protocol consisted of an initial denaturation of 94°C for 5 min, followed by 35 cycles with a denaturation step of 94°C for 30–40 s, an annealing step at 58–62°C (depending on annealing temperature of primers) for 60 s and an elongation step at 72°C for 90 s, followed by a final extension of 7 min. PCR primers for murid species were designed based on the Pkdrej sequences of Mus musculus and Rattus norvegicus available from GenBank (accession numbers NM_011105 and NM_001134866, respectively). For cricetid species, primers were designed using as reference the genomic sequences of Mesocricetus auratus and Cricetulus griseus available from The National Center for Biotechnology Information (NCBI) Genome database. In both cases, when Pkdrej sequences of several closely related species were obtained, more specific primers were designed for those divergent species in order to improve the PCR efficiency. Primer sequences and combinations used for each species are given in Supplementary Table SI. PCR products were purified by using the E.Z.N.A.® Cycle Pure kit (Omega). Purified products were sequenced directly in an automatic sequencer (Secugen S.L., Madrid, Spain), in both directions and using internal primers (Supplementary Table SI).
Sequence and phylogenetic analyses
For primates, two alignments were built. A first alignment included the complete sequence of PKDREJ in order to reanalyse the evolution of the whole gene as done previously (Hamm et al., 2007). In a second analysis, we aligned the REJ domain to compare the evolution of this region with its homologue in rodents.
In the case of rodents, sequences obtained in the laboratory were manually curated based on the chromatogram and heterozygous sites were coded based on IUPAC nucleotide code. Sequence quality was checked by translating codon sequences using BioEdit software (Hall, 1999), confirming the absence of any frame shift mutation able to generate false positives in the evolutionary analyses. The identification of the sequenced fragments as Pkdrej homologues in rodents was confirmed using BLASTn algorithm (NCBI) against the genome of model species (Mus musculus in Muridae and Mesocricetus auratus in Cricetidae). Codon-based alignments were performed using the Muscle algorithm (Edgar, 2004). Alignments were quality-checked manually and edited in problematic codon positions when required. Consensus sequences for each species were obtained aligning sequences of at least three individuals.
PKDREJ phylogenetic trees covering the analysed species were constructed using Maximum-Likelihood (PhyML 3.0) and Bayesian (MrBayes 3.1) methods (Ronquist and Huelsenbeck, 2003; Guindon et al., 2010). In the case of Maximum-likelihood analyses, statistical support for each internal branch was estimated using non-parametric bootstrap of 100 replicates. In all cases, we applied the generalized time-reversible (GTR) model of nucleotide substitution estimating the gamma distribution parameter and the proportion of invariable sites based on the hierarchical likelihood-ratio tests implemented in JmodelTest 0.1.1 (Posada, 2008).
Site models of positive selection
The nonsynonymous/synonymous substitutions ratio (dN/dS = ω) is a robust indicator of the selective pressure on a gene, with ω = 1 indicating neutral evolution, ω < 1 purifying selection and ω > 1 diversifying positive selection (Goldman and Yang, 1994).
To detect variable selective pressures among amino acid sites and infer positive selection across the PKDREJ sequence we used Maximum Likelihood-based methods implemented in Codeml as part of the package PAML v 4.4 (Yang, 2007). A likelihood-ratio test (LRT) was performed to compare the likelihood of a null model, which does not allow sites under selection, with the likelihood of a selection model including an additional class allowing values of ω > 1. Three classes of tests were applied. The first compared a nearly neutral model M1a, which assumes values for ω between 0 and 1, with a selection model M2a which includes an additional class of sites with ω estimated from the data. The second test was more robust and compared two models assuming a beta distribution for ω values. In this case, the null model M7 that limits ω between 0 and 1 is compared with the alternative model M8, which adds an extra class of sites with the ω ratio allowed to be greater than 1 (Nielsen and Yang, 1998; Yang et al., 2000). A third test compared the likelihood of model M8 to the likelihood of a null model M8a in which ω in the extra site class was fixed to one in order to avoid detecting false signatures of positive selection as a result of functional relaxation (Wong et al., 2004). The LRT compared twice the log-likelihoods of selection and neutral models to critical values from a chi-square distribution with the degrees of freedom equal to the difference in the number of parameters between the two models. If the LRT is significant, this implies that selection models show a better fit and thus positive selection at, at least, some sites can be inferred. A Bayes empirical Bayes (BEB) approach (Yang et al., 2005) was used to calculate the probability that each site belongs to a particular class in terms of the posterior ω value, and those residues with posterior probabilities higher than 0.95 of having ω > 1 were inferred to be under significant positive selection.
Codeml analyses were performed without setting a molecular clock and arbitrary parameters, such as codon frequency and the number of gamma categories, were optimized for the best fit of data.
Identification of lineages under positive selection
To detect episodes of adaptive evolution on specific lineages, branch-site analyses that allow ω to vary both among sites and among lineages were applied (Yang and Nielsen, 2002; Zhang et al., 2005). In these tests, the phylogenetic tree is divided into foreground and background branches, where only foreground lineages are allowed to exhibit positive selection. A LRT is used to compare the likelihood of a null model with that of an alternative model. In the null model, sites are partitioned into four site classes: class 0, where sites are conserved in all lineages with 0 < ω0 < 1; class 1, including neutral sites in which ω1 = 1 for all branches; class 2a, with purifying selection for background branches and neutral evolution (ω2 = 1) for foreground branches; and class 2b, assuming neutral evolution (ω2 = 1) along both background and foreground branches. This model is compared with the alternative model, where the foreground lineages are allowed to have ω2 > 1 for classes 2a and 2b. As the alternative model has ω2 > 1 constrained and the test is one-sided, the asymptotic null distribution is a 50:50 mixture of point mass 0 and chi-square with the critical values to be 2.71 and 5.41, at the 5 and 1% levels, respectively (Zhang et al., 2005).
Measures of post-copulatory sexual selection
An almost universal response to increased levels of sperm competition is an increase in testes mass relative to body mass (Birkhead and Moller, 1998; Gomendio et al., 1998). Thus, relative testes mass has been used as a suitable proxy to measure the influence of post-copulatory sexual selection on reproductive traits. Data of body mass and testes mass for primates were collected from the literature (see Results section). For rodents, body mass and testes mass data were empirically obtained in our laboratory. A total of five males for each species were sacrificed by cervical dislocation and weighed. Testes were then removed and weighed. Values of relative testes mass were calculated as the residual testis mass obtained from a linear regression using body mass as predictor variable and testes mass as dependent variable (see Results section). We used relative testes mass as an index for levels of sperm competition (Supplementary Tables SII and SIII).
Evolutionary rates and levels of sperm competition
To examine the association between PKDREJ divergence and sperm competition, we carried out multiple regression analyses including both testes mass and body mass as predictors and individual ω values for each species as the dependent variable. Since predictor variables are related to each other, we applied a sequential Type I sum of squares by which body mass and testes mass were added in this order. Lineage-specific ω ratios were calculated using a free-ratio model implemented in Codeml allowing variation of ω among terminal branches of a tree (Yang and Nielsen, 1998). The evolutionary rate of Pkdrej for each species was then calculated by comparing dN and dS values from the root of the phylogeny to the terminal branch giving an ω ratio from the root. In this way, the total accumulated selective pressures during the evolution of the Pkdrej gene are accounted for, which is more suitable for testing associations with phenotypic data as it reflects the entire phenotypic evolution from the common ancestor (Lüke et al., 2011; Montgomery et al., 2011). In addition, estimating the rate of evolution since the last common ancestor forces that all branches have the same length and therefore the analysis is not subject to temporal effects on ω (Wolf et al., 2009). Rates of evolution through the phylogeny were calculated using the ETE2 toolkit (Huerta-Cepas et al., 2010).
Phenotypic data are not free of phylogenetic association, since species may share character values as a result of a common ancestry rather than independent evolution (Harvey and Pagel, 1991), and thus may not be truly independent. To control for this phylogenetic inertia, regression analyses were performed using a generalized least-squares approach in a phylogenetic framework (PGLS) (Freckleton et al., 2002). This model allows for the control of phylogenetic effects on the associations of variables to be correlated. The PGLS analyses were performed using a code written by R. Freckleton for the statistical package R v.2.10.1 (R Foundation for Statistical Computing 2010). Statistical significance was set at P < 0.05.
Results
Primate sequences and phylogenetic analyses
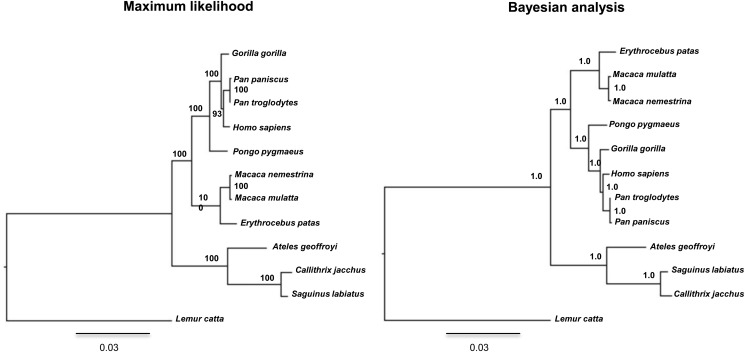
We used primate PKDREJ sequences obtained in previous interspecific studies (Hamm et al., 2007) and built phylogenies using Maximum Likelihood and Bayesian methods; they yielded the same topology as the neighbour-joining tree constructed by Hamm et al. (2007) (Fig. 1). Such topology is in concordance with the supported phylogeny of primates (Goodman et al., 1998) and, therefore, we used this topology for Codeml analyses.
Figure 1.
Primate phylogenetic trees constructed using PKDREJ sequences. Phylogenetic analyses were performed using Maximum likelihood and Bayesian methods. Branching support from a non-parametric bootstrap of 100 replicates is shown for each internal node in the Maximum likelihood tree. Posterior probabilities are indicated in the internal nodes of the Bayesian phylogeny. Scale bars represent the genetic distances.
Sperm competition and evolution of PKDREJ in primates
A previous study in primates did not find any correlation between the evolutionary rate of PKDREJ and differences in mating systems (Hamm et al., 2007). Because this lack of relationship could be due to limitations in the estimates of the level of sperm competition, we reanalysed the evolution of PKDREJ in primates to test whether the evolution of this sperm protein is influenced by post-copulatory sexual selection using, instead, testes mass relative to body mass as a proxy of sperm competition (Supplementary Table SII).
Phylogenetically-corrected regression analyses were carried out to examine a link between PKDREJ divergence and levels of sperm competition in primates. No significant associations between lineage-specific ω and testes mass corrected for body mass were found for the complete PKDREJ sequence (P = 0.443) or for the region of the extracellular REJ domain (P = 0.117).
We further used branch-site models to identify specific primate lineages in which PKDREJ is subjected to positive selection. Using this method we did not detect a signal of positive selection in any lineage for the entire PKDREJ sequence or for the REJ domain (Table I).
Table I.
Branch-site analyses of PKDREJ in primate species.
| Foreground branch | Complete PKDREJ |
REJ domain |
||||||
|---|---|---|---|---|---|---|---|---|
| M2 | M2 fix | LRT | P | M2 | M2 fix | LRT | P | |
| Gorilla gorilla | −14 225.85 | −14 225.85 | 0.000 | 1.000 | −14 225.85 | −14 225.85 | 0.000 | 1.000 |
| Homo sapiens | −14 225.87 | −14 225.87 | 0.000 | 1.000 | −14 225.87 | −14 225.87 | 0.000 | 1.000 |
| Pan troglodytes | −14 225.94 | −14 225.94 | 0.000 | 0.987 | −14 225.94 | −14 225.94 | 0.000 | 0.987 |
| Pan panyscus | −14 225.94 | −14 225.97 | 0.060 | 0.807 | −14 225.94 | −14 225.97 | 0.060 | 0.807 |
| Pongo pygmaeus | −14 226.06 | −14 226.06 | 0.003 | 0.955 | −14 226.06 | −14 226.06 | 0.003 | 0.955 |
| Erythrocebus patas | −14 223.52 | −14 223.52 | 0.000 | 1.000 | −14 223.52 | −14 223.52 | 0.000 | 1.000 |
| Macaca mulatta | −14 226.06 | −14 226.06 | 0.000 | 1.000 | −14 226.06 | −14 226.06 | 0.000 | 1.000 |
| Macaca nemestrina | −14 226.06 | −14 226.06 | 0.000 | 0.983 | −14 226.06 | −14 226.06 | 0.000 | 0.983 |
| Ateles geoffroyi | −14 223.52 | −14 223.52 | 0.000 | 0.986 | −14 223.52 | −14 223.52 | 0.000 | 0.986 |
| Saguinus labiatus | −14 224.30 | −14 224.40 | 0.190 | 0.663 | −14 224.30 | −14 224.40 | 0.190 | 0.663 |
| Callithrix jacchus | −14 223.72 | −14 224.40 | 1.358 | 0.244 | −14 223.72 | −14 224.40 | 1.358 | 0.244 |
| Lemur catta | −14 226.06 | −14 226.06 | 0.000 | 1.000 | −14 226.06 | −14 226.06 | 0.000 | 1.000 |
LRT (likelihood-ratio test): 2 times log likelihood difference of alternative (M2) versus null (M2 fix) models (see Materials and Methods).
Rodent sequences and phylogenetic analyses
We examined PKDREJ sequences of 24 rodent species of the superfamily Muroidea (11 of the family Muridae and 13 of the family Cricetidae). Sequences were obtained by amplification using PCR. Sequencing of PCR products gave results for Pkdrej fragments with lengths ranging from 1248 to 1257 nucleotides, and translation resulted in amino acid fragments homologous to positions 213–629 of Mus musculus PKDREJ (Supplementary Fig. S1). This fragment falls within the extracellular REJ domain.
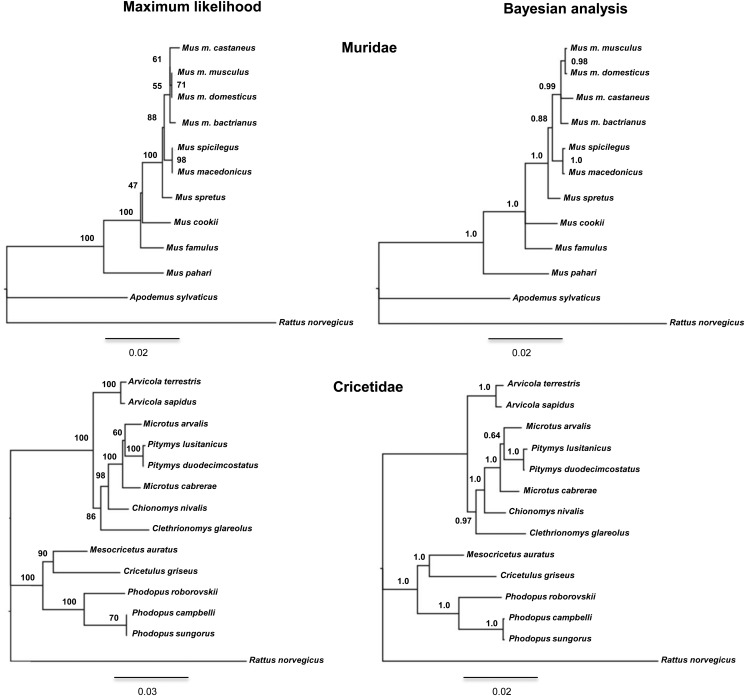
The construction of PKDREJ phylogenies by Maximum Likelihood and Bayesian methods produced the same topologies both for Muridae and for Cricetidae (Fig. 2). Furthermore, these topologies were in agreement with resolved trees presented in previous phylogenetic studies (Steppan et al., 2004; Suzuki et al., 2004; Galewski et al., 2006; Neumann et al., 2006; Martín-Coello et al., 2009). In Muridae, the clade grouping the Mus musculus subspecies was resolved with low support, where we found a polytomy. The branch leading to Mus cookii was resolved as adjacent to the Mus clade with low probability, as Mus famulus is assumed to be closer to these species. In Cricetidae, only the clade grouping Microtus arvalis with the Pitymys species was resolved with low probability. Therefore, we used the phylogenetic trees of Supplementary Fig. S2 for evolutionary analyses. Cricetidae showed a higher average evolutionary divergence of PKDREJ (d = 0.117) than Muridae (d = 0.039).
Figure 2.
Phylogenetic trees constructed with rodent Pkdrej sequences. Branching support and posterior probabilities are indicated in each internal node of the Maximum likelihood and Bayesian phylogenies, respectively. Scale bars represent the genetic distances. Rattus norvegicus was used as outgroup in both analyses.
Evidence of positive selection in rodent PKDREJ
Likelihood models accounting for heterogeneous ω values among sites were applied to search for evidence of positive selection in rodent Pkdrej gene. A preliminary site-analysis was performed on a phylogeny including all muroid lineages. Both selection models (M2a and M8) fitted significantly better the data than the respective neutral models (M1a, M7 and M8a) (Table II). Model M8 detected an amino acid under significant positive selection falling in position 47 (PBEB = 0.978), whereas no significant positively selected site with a PBEB > 0.95 was identified by model M2a. BEB analyses are able to identify amino acids conserved by purifying selection (with ω ratios between 0 and 1) and those undergoing neutral evolution (with ω around 1) under M2a model. Most sites in Pkdrej were found to be under purifying selection (287 amino acids out of 405; 71%), whereas 114 codon sites (28%) are evolving neutrally by relaxation of functional constraints. Finally, 1% of sites were inferred to belong to a site class with ω > 1 (ω = 6.475), though none of these sites could be identified with high confidence (all PBEB < 0.95).
Table II.
Tests of positive selection in rodent Pkdrej.
| Species | N | L | Site model | LRT | Parameter estimates | Positively selected sites |
|---|---|---|---|---|---|---|
| Muroidea (superfamily) | 25 | 417 | M1a-M2a | 8.052* | p0 = 0.646, p1 = 0.350, p2 = 0.003, ω0 = 0.184, ω1 = 1, ω2 = 6.471 | None |
| M7-M8 | 12.195** | p0 = 0.982, p = 0.680, q = 0.919, p1 = 0.018, | 47* (0.978) | |||
| M8a-M8 | 52.8845*** | |||||
| Muridae (family) | 12 | 437 | M1a-M2a | 0.179 | p0 = 0.552, p1 = 0.448, ω0 = 0.117, ω1 = 1 | Not allowed |
| M7-M8 | 0.381 | p = 0.178, q = 0.168 | Not allowed | |||
| M8a-M8 | 0.292 | p0 = 0.569, p = 14.512, q = 98.963, p1 = 0.430, ω = 1 | Not allowed | |||
| Cricetidae (family) | 13 | 426 | M1a-M2a | 9.376** | p0 = 0.598, p1 = 0.393, p2 = 0.009, ω0 = 0.123, ω1 = 1, ω2 = 6.551 | 47* (0.984) |
| M7-M8 | 11.167** | p0 = 0.989, p = 0.265, q = 0.312, p1 = 0.011, ω = 5.679 | 47* (0.981), 206* (0.957) | |||
| M8a-M8 | 8.242* |
LRT 2 times log likelihood difference of free selection versus neutral models. Significant tests with a threshold of *P < 0.05, **P < 0.01 and ***P < 0.001 in a chi-square distribution with 2 d.f. are indicated.
Parameter estimates are shown for the best fit model.
Predicted sites under positive selection with Bayes empirical Bayes (BEB) posterior probability >95% are indicated with an asterisk.
N: number of sequences. L: sequence length (number of codons).
To assess whether the signal of adaptive evolution detected in muroid Pkdrej is a result of positive selection acting at different stages of evolution, we conducted separate site analyses for the two muroid families (Muridae and Cricetidae). Selection models (M2a and M8) were rejected by LRTs in favour of neutral models (M1a, M7 and M8a) in the case of Muridae, which suggests that Pkdrej is not undergoing adaptive evolution in this family (Table II). The same results were obtained using each of the two possible topologies to resolve the Mus musculus polytomy. In contrast, selection models fitted significantly better the evolution of Pkdrej in the family Cricetidae (Table II). The proportion of sites estimated to belong to a site class with ω > 1 was 0.9%, and both models calculated a PBEB > 0.95 for the residue in position 47, whereas the amino acid in position 220 showed significant positive selection under the M8 model (Table II). These two sites were also detected in the analysis of Muroidea, with nine non-significant additional sites detected in the superfamily.
Sperm competition and evolution of PKDREJ in rodents
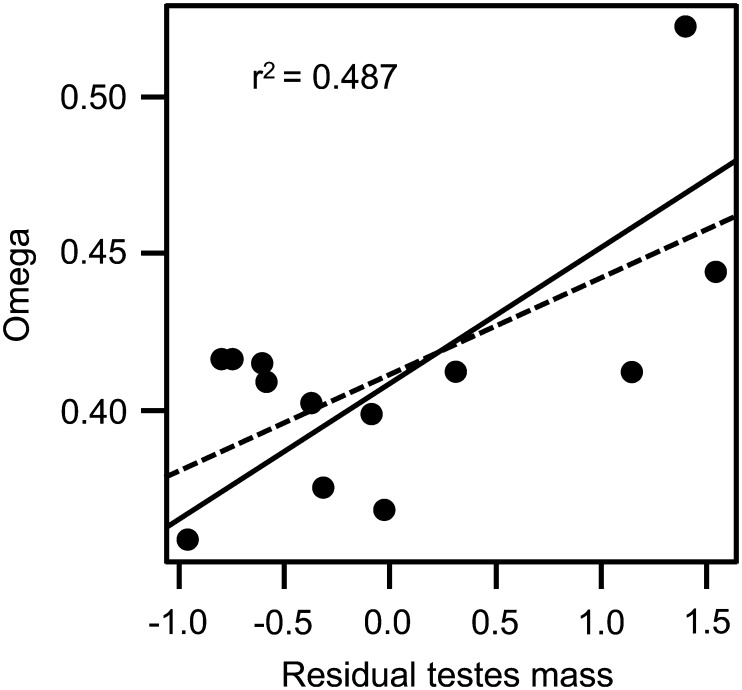
As done for primates, we tested for a relationship between Pkdrej divergence and the intensity of sperm competition in rodents. We calculated dN and dS substitution rates in each branch under a free-ratio model in PAML (Nielsen and Yang, 1998) and ω ratios were averaged from the ancestral node to the terminal branches corresponding to each lineage (Montgomery et al., 2011). Estimates of ω over the entire (root-to-tip) branches were <1 for all lineages of Muridae and Cricetidae (Supplementary Table SIV). After controlling for phylogenetic effects, a multiple regression analysis revealed a significant positive correlation between lineage-specific ω ratios and testes mass corrected for body mass in Cricetidae (Table III). We plotted ω values of cricetid species against their residual testes mass and obtained a significant correlation between these two variables (P = 0.003, Fig. 3). In order to examine if these results were driven just by the inclusion of Cricetulus griseus in the dataset, we redid the analyses without this lineage. We found that, when this lineage was not included, significant positive correlation of ω ratios with testes mass corrected for body mass was still obtained (P = 0.036). No significant correlation was found for Muridae in these regression analyses (Table III).
Table III.
PKDREJ divergence and sperm competition in rodents.
| Group | Predictor | Slope | F | P | d.f. | λ | r | CL− | CL+ |
|---|---|---|---|---|---|---|---|---|---|
| Muridae | Body mass | 0.0002 | 0.256 | 0.628 | 7 | 0.584 (n.s., n.s.) | 0.188 | −0.551 | 0.931 |
| Testes mass | 0.0737 | 0.917 | 0.37 | 0.34 | −0.386 | 1.095 | |||
| Cricetidae | Body mass | −0.0005 | 1.693 | 0.222 | 10 | 0.999 (*, n.s.) | 0.381 | −0.219 | 1.02 |
| Testes mass | 0.0343 | 10.131 | 0.0097** | 0.709 | 0.266 | 1.506 |
Lineage-specific omegas were used as dependent variables.
Statistical significance of correlation analysis is shown as **P < 0.01.
λ: phylogenetic scaling parameter that indicates the transformation that makes the data fit the Brownian motion evolutionary model. Between parentheses, the significance against models assuming λ = 0 and λ = 1 respectively, where *P > 0.05.
CLs: 95% confidence limits for the regression slope. Statistical significance is obtained as confidence interval does not include zero.
Figure 3.
Evolutionary rate of rodent Pkdrej and sperm competition. Relationship between residual testes mass and omega (ω) calculated for each species of Cricetidae. Regression lines for standard linear regression (dashed line) and phylogenetically-corrected linear regression (solid line) are shown.
Site analyses described above identified residues under positive selection across all cricetid lineages, but they were not able to detect episodes of adaptive evolution in specific lineages. We thus used the branch-site method, as employed above for primates. Analyses were carried out separately for Muridae and Cricetidae after observing that Pkdrej exhibits different evolutionary patterns between these families. In Cricetidae, we detected evidence of positive selection on the branches leading to Cricetulus griseus and Mesocricetus auratus (Table IV) and a residue (206A) was inferred with a PBEB > 0.95 for C. griseus. Positive selection was detected when the branches leading to species with the highest levels of sperm competition (C. griseus and M. auratus) were assigned to the foreground (LRT = 5.96, P = 0.012), but no significant positively selected site was found in this case. Branch-site tests on the murid phylogeny did not detect evidence of positive selection in any lineage of this family.
Table IV.
Branch-site analyses of Pkdrej in rodent species.
| Foreground branch | M2 | M2 fix | LRT | P |
|---|---|---|---|---|
| Arvicola sapidus | −4711.24 | −4711.24 | 0.000 | 1.000 |
| Arvicola terrestris | −4711.24 | −4711.24 | 0.000 | 1.000 |
| Clethrionomys glareolus | −4711.14 | −4711.14 | 0.000 | 1.000 |
| Chionomys nivalis | −4710.82 | −4710.82 | 0.000 | 1.000 |
| Microtus arvalis | −4711.24 | −4711.24 | 0.000 | 1.000 |
| Microtus cabrerae | −4711.24 | −4711.24 | 0.000 | 1.000 |
| Pitymys duodecimostatus | −4711.24 | −4711.24 | 0.000 | 1.000 |
| Pitymys lusitanicus | −4711.24 | −4711.24 | 0.000 | 1.000 |
| Mesocricetus auratus | −4711.21 | −4708.86 | 4.789 | 0.029* |
| Cricetulus griseus | −4711.22 | −4706.81 | 8.810 | 0.003** |
| Phodopus sungorus | −4711.23 | −4710.91 | 0.644 | 0.422 |
| Phodopus campbelli | −4711.23 | −4710.91 | 0.644 | 0.422 |
| Phodopus roborovskii | −4711.24 | −4711.24 | 0.000 | 1.000 |
LRT: comparing the likelihood values of alternative (M2) and null (M2 fix) models (see Materials and Methods).
Statistical significance of LRTs is indicated as *P < 0.05 and **P < 0.01.
Discussion
We have analysed the evolution of PKDREJ, a protein involved in the regulation of sperm function, in two orders of mammals: primates and rodents. In particular, we focused on the extracellular REJ domain of this protein, a region that presumably acts as a receptor for other cells or extracellular signals (Moy et al., 1996; Delmas, 2005), and where a strong signal of positive selection has previously been identified in sea urchin and primate PKDREJ homologues (Mah et al., 2005; Hamm et al., 2007).
In primates, evidence of a link between divergence of this region and sperm competition was not found. In rodents, analyses on species of the superfamily Muroidea revealed a higher divergence of Pkdrej in lineages with high levels of sperm competition within the family Cricetidae. This relationship was revealed by a significant correlation between relative testes mass (a reliable index of sperm competition) and lineage-specific evolutionary rates of Pkdrej.
Evolution of primate PKDREJ and sexual selection
Evidence of post-copulatory sexual selection driving adaptive evolution of reproductive proteins has been documented more frequently for primates (Herlyn and Zischler, 2007; Ramm et al., 2008; Finn and Civetta, 2010). In the case of primate PKDREJ, evidence of associations between bouts of adaptive evolution and differences in mating systems were not found (Hamm et al., 2007). Because mating system may not be an entirely reliable proxy of post-copulatory sexual selection, we reanalysed the evolution of PKDREJ in primates employing testes mass relative to body mass as an index of sperm competition. Divergence of primate PKDREJ was not associated with residual testes mass, giving further evidence that sperm competition is not a predominant force driving the evolution of PKDREJ in primates. Nonetheless, this does not necessarily imply that primate PKDREJ is evolving free of the influence of sexual selection. The detection of positive selection on this protein with testis-specific expression and with a role in sperm function indicates that PKDREJ is likely subjected to sexual selection forces. A possible explanation for a lack of association between PKDREJ evolution and sexual selection is that multiple selection forces may participate simultaneously in the evolution of primate PKDREJ. In this scenario, the adaptive changes driven by sexual selection would likely be masked by those arising from other underlying forces. This is suggested by the fact that the REJ domain of primates showed, overall, a stronger signal of positive selection than that of rodents (Hamm et al., 2007). Another possible interpretation is that different sites are under selection in different primate lineages as, in the context of sexual selection, species may undergo different mutational routes to increase the reproductive fitness.
Adaptive evolution of PKDREJ in rodents
Our analyses revealed evidence of positive selection when all rodent species were compared. However, when the two families of muroid rodents included in this study (Muridae and Cricetidae) were analysed separately, significant evidence of positive selection in the Pkdrej gene was detected only for Cricetidae. Thus, the signal of positive selection exhibited by the superfamily Muroidea is likely reflecting the adaptive divergence of Pkdrej in the family Cricetidae.
It is possible that the adaptive evolution of PKDREJ is a generalized phenomenon across rodents, but the high degree of conservation of a large portion of the gene makes it difficult to detect positive selection when very closely related species are compared (Anismova et al., 2001; Zhang et al., 2005). A similar pattern was observed in previous work analysing the evolution of protamine sequences in murids (Martín-Coello et al., 2009) and cricetids (Lüke et al., 2011). An alternative explanation could be that positive selection is limited to the cricetid clade. Comparative studies analysing other rodent families could clarify the evolutionary trends of PKDREJ.
PKDREJ as target of sexual selection in rodents
It is generally assumed that the adaptive evolution of proteins involved in reproductive processes is driven by sexual selection and coevolution between the sexes, being particularly strong in promiscuous species (Swanson and Vacquier, 2002; Clark et al., 2006; Turner and Hoekstra, 2008). Analyses by Sutton et al. (2008) revealed that sperm with mutant Pkdrej showed lower reproductive success than wild-type sperm in the mouse, which suggested that PKDREJ could be a protein evolving under the influence of post-copulatory sexual selection. We thus tested whether post-copulatory sexual selection influences the adaptive evolution of PKDREJ in rodents using relative testes mass as proxy of sperm competition (Birkhead and Moller, 1998; Gomendio et al., 1998). A positive correlation was found between lineage-specific evolutionary rates of Pkdrej and residual testis mass in cricetids, supporting that rate of changes in PKDREJ increases with intensity of post-copulatory sexual selection. Branch-site models testing for adaptive evolution lineage by lineage revealed that the signal of positive selection in Cricetidae is mainly concentrated on Cricetulus griseus and Mesocricetus auratus, which are indeed the species with the highest inferred levels of sperm competition. Our results thus suggest that adaptive changes in PKDREJ can be driven by post-copulatory sexual selection.
Evidence for post-copulatory sexual selection influencing the molecular evolution of reproductive genes is limited and often controversial in rodents (Ramm et al., 2008; Lüke et al., 2011). A rodent gene coding for an ejaculate protein involved in the formation of the copulatory plug (Svs2) was shown to exhibit signs of positive selection in species with high levels of sperm competition (Ramm et al., 2008). Evolutionary analyses of protamines, a family of nuclear proteins involved in condensing sperm chromatin during spermiogenesis, revealed a negative correlation between rate of divergence in Prm2 and sperm competition intensity, suggesting that sexual selection may have a role in halting the degeneration of this protein (Lüke et al., 2011). The findings in the present study would thus represent the first evidence in rodents of adaptive evolution of an integral sperm protein associated with sexual selection.
A functional study on PKDREJ suggests that this protein controls important aspects of male fertility, as sperm lacking Pkdrej show a reduction in the efficiency of ascent in the oviduct and acquire the competence to undergo the zona pellucida-induced acrosome reaction more slowly (Sutton et al., 2008). The time of insemination relative to ovulation is crucial for the success of fertilization (Krzanowska, 1986; Gomendio et al., 1998). Thus, it is expected that spermatozoa that become ready to interact with the ovum faster (i.e. capacitation), or which are more efficient at responding to the signals of the ovum (i.e. sperm chemotaxis, acrosome reaction), may be able to fertilize more rapidly. In fact, sperm competition favours an increase in the proportion of spermatozoa that undergo capacitation and acrosome reaction at the time of fertilization in rodents (Gomendio et al., 2006). Thus, proteins regulating these processes may be targets of sexual selection if, under positive selection, they provide an advantage in the timing and success of fertilization (Gomendio et al., 1998, 2006; Birkhead and Pizzari, 2002). Our present findings support this hypothesis since rodent species subjected to more intense sperm competition undergo adaptive changes in PKDREJ. On the other hand, we cannot completely discard the possibility that our results are driven by specific processes occurring in the species with adaptive divergence of PKDREJ. For instance, it is possible that Cricetulus griseus and Mesocricetus auratus may have evolved some mechanism regulating sperm function in which PKDREJ is involved, and that this protein therefore, may be subjected to different functional constraints.
Functional significance of the adaptive evolution of PKDREJ
Because the precise role of PKDREJ in mammals is still unknown, we cannot establish the functional effects of adaptive changes in this protein. Given that Pkdrej mutant sperm require longer to reach the oocyte-cumulus complex (Sutton et al., 2008), it is possible that PKDREJ has a role in sperm-oviduct interactions. The oviduct is likely to be the site of the selection of spermatozoa prior to fertilization, acting as the environment for post-copulatory sperm selection by females (cryptic female choice) or acting as an arena for sperm competition (Holt and Fazeli, 2010). In this scenario, it would be predicted that proteins on the sperm surface are potential mediators of sperm-oviduct interactions which would impact either the rate of fertility acquisition and/or migration to the oocyte (Talevi and Gualtieri, 2010). Such receptor-ligand pairs would likely be the targets of selective forces. Nevertheless, it should be borne in mind that PKDREJ could have other roles in the sperm cell. As previously mentioned, PKDREJ has been suggested to participate in the signalling pathway that drives the mammalian sperm acrosome reaction (Sutton et al., 2006; Hamm et al., 2007). In this way, mutations in PKDREJ could affect to the efficacy of the acrosome reaction induction pathway.
New approaches to characterizing the molecular function of PKDREJ in mammals will be necessary to assess the effects of its genetic divergence on sperm phenotype. For example, experimental studies analysing the phenotypic effect of point mutations in residues under positive selection would be worth pursuing to evaluate the adaptive significance of such changes. Since PKDREJ regions different from the extracellular REJ domain are likely to regulate sperm function, it would also be desirable to analyse the evolution of the entire rodent PKDREJ. This may reveal, for instance, if the transmembrane segments of PKDREJ, which could have an analogous function to the polycystin-2 protein as a permeable Ca2+ channel, show signals of adaptive evolution.
Conclusions
Our results revealed that PKDREJ, a sperm protein that appears to modulate the timing of fertilization through a role in sperm transport and the acrosome reaction, shows bouts of adaptive evolution associated with sperm competition in rodents. These findings thus provide the first support for the idea that post-copulatory sexual selection can act on proteins involved in the processes that lead to the acquisition of the ability of mammalian spermatozoa to bind and penetrate the egg.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/ online.
Authors’ roles
E.R.S.R., K.A.S. and A.V. designed the study, A.V., L.G.M., F.C.-F and K.A.S. carried out laboratory work, all authors analysed data, A.V. K.A.S. and E.R.S.R. wrote the paper; all authors approved the final manuscript.
Funding
This work was funded by the Spanish Ministry of Economy and Competitiveness (grant number CGL2011-26341 to E.R.S.R; postgraduate studentship BES-2009-029239 to A.V.; postgraduate studentship BES-2005-11028 to L.G.M.) and the National Institute of Child Health and Human Development (grant number HD060034) to K.A.S.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We are grateful to Pedro Castañera and to Grupo de Interacción Planta-Insecto of Centro de Investigaciones Biológicas (CSIC) for allowing us to use their research facilities in the initial phase of this work. We thank Lena Lüke for help and advice with laboratory work and comments on the manuscript.
References
- Amaral AR, Möller LM, Beheregaray LB, Coelho MM. Evolution of 2 reproductive proteins, ZP3 and PKDREJ, in cetaceans. J Hered. 2011;102:275–282. doi: 10.1093/jhered/esq131. [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- Birkhead TR, Møller AP. Sperm Competition and Sexual Selection. San Diego, CA: Academic Press; 1998. [Google Scholar]
- Birkhead TR, Pizzari T. Postcopulatory sexual selection. Genetics. 2002;3:262–273. doi: 10.1038/nrg774. [DOI] [PubMed] [Google Scholar]
- Butscheid Y, Chubanov V, Steger K, Meyer D, Dietrich A, Gudermann T. Polycystic kidney disease and receptor for egg jelly is a plasma membrane protein of mouse sperm head. Mol Reprod Dev. 2006;73:350–360. doi: 10.1002/mrd.20410. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Delmas P. Polycystins: polymodal receptor/ion-channel cellular sensors. Pflugers Arch. 2005;451:264–276. doi: 10.1007/s00424-005-1431-5. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn S, Civetta A. Sexual selection and the molecular evolution of ADAM proteins. J Mol Evol. 2010;71:231–240. doi: 10.1007/s00239-010-9382-7. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JL, Montgomerie R, Desjardins JK, Stiver KA, Kolm N, Balshine S. Female promiscuity promotes the evolution of faster sperm in cichlid fishes. Proc Natl Acad Sci USA. 2009;106:1128–1132. doi: 10.1073/pnas.0809990106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am Nat. 2002;160:712–726. doi: 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Galewski T, Tilak M, Sanchez S, Chevret P, Paradis E, Douzery EJP. The evolutionary radiation of Arvicolinae rodents (voles and lemmings): relative contribution of nuclear and mitochondrial DNA phylogenies. BMC Evol Biol. 2006;17:1–17. doi: 10.1186/1471-2148-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher AR, Hidaka S, Gretz N, Witzgall R. Molecular basis of autosomal-dominant polycystic kidney disease. Cell Mol Life Sci. 2002;59:682–693. doi: 10.1007/s00018-002-8457-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Gomendio M, Roldan ERS. Implications of diversity in sperm size and function for sperm competition and fertility. Int J Dev Biol. 2008;52:439–447. doi: 10.1387/ijdb.082595mg. [DOI] [PubMed] [Google Scholar]
- Gomendio M, Harcourt AH, Roldan ERS. Sperm competition in mammals. In: Birkhead TR, Moller AP, editors. Sperm Competition and Sexual Selection. London: Academic press; 1998. pp. 667–751. [Google Scholar]
- Gomendio M, Martín-Coello J, Crespo C, Magaña C, Roldan ERS. Sperm competition enhances functional capacity of mammalian spermatozoa. Proc Natl Acad Sci USA. 2006;103:15113–15117. doi: 10.1073/pnas.0605795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Montoto L, Magaña C, Tourmente M, Martín-Coello J, Crespo C, Luque-Larena JJ, Gomendio M, Roldan ERS. Sperm competition, sperm numbers and sperm quality in muroid rodents. PLoS One. 2011;6:e18173. doi: 10.1371/journal.pone.0018173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hamm D, Mautz BS, Wolfner MF, Aquadro CF, Swanson WJ. Evidence of amino acid diversity-enhancing selection within humans and among primates at the candidate sperm-receptor gene PKDREJ. Am. J Hum Genet. 2007;81:44–52. doi: 10.1086/518695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PH, Pagel MD. The comparative method in evolutionary biology. In: May RM, Harvey PH, editors. Trends in Ecology and Evolution. Vol. 239. Oxford: Oxford University Press; 1991. [Google Scholar]
- Herlyn H, Zischler H. Sequence evolution of the sperm ligand zonadhesin correlates negatively with body weight dimorphism in primates. Evolution. 2007;61:289–298. doi: 10.1111/j.1558-5646.2007.00035.x. [DOI] [PubMed] [Google Scholar]
- Holt WV, Fazeli A. The oviduct as a complex mediator of mammalian sperm function and selection. Mol Reprod Dev. 2010;77:934–943. doi: 10.1002/mrd.21234. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzanowska H. Interstrain competition amongst mouse spermatozoa inseminated in various proportions, as affected by the genotype of the Y chromosome. J Reprod Fertil. 1986;77:265–270. doi: 10.1530/jrf.0.0770265. [DOI] [PubMed] [Google Scholar]
- Lüke L, Vicens A, Serra F, Luque-Larena JJ, Dopazo H, Roldan ERS, Gomendio M. Sexual selection halts the relaxation of protamine 2 among rodents. PLoS One. 2011;6:e29247. doi: 10.1371/journal.pone.0029247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüpold S, Calhim S, Immler S, Birkhead TR. Sperm morphology and sperm velocity in passerine birds. Proc R Soc B. 2009;276:1175–1181. doi: 10.1098/rspb.2008.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah SA, Swanson WJ, Vacquier VD. Positive selection in the carbohydrate recognition domains of sea urchin sperm receptor for egg jelly (suREJ) proteins. Mol Biol Evol. 2005;22:533–541. doi: 10.1093/molbev/msi037. [DOI] [PubMed] [Google Scholar]
- Martín-Coello J, Dopazo H, Arbiza L, Ausió J, Roldan ERS, Gomendio M. Sexual selection drives weak positive selection in protamine genes and high promoter divergence, enhancing sperm competitiveness. Proc R Soc B. 2009;276:2427–2436. doi: 10.1098/rspb.2009.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengerink KJ, Moy GW, Vacquier VD. suREJ3, a polycystin-1 protein, is cleaved at the GPS domain and localizes to the acrosomal region of sea urchin sperm. Biochemistry. 2002;277:943–948. doi: 10.1074/jbc.M109673200. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 2011;28:625–638. doi: 10.1093/molbev/msq237. [DOI] [PubMed] [Google Scholar]
- Moy GW, Mendoza LM, Schulz JR, Swanson WJ, Glabe CG, Vacquier VD. The sea urchin sperm receptor for egg jelly is a modular protein with extensive homology to the human polycystic kidney disease protein, PKD1. J Cell Biol. 1996;133:809–817. doi: 10.1083/jcb.133.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill AT, Moy GW, Vacquier VD. Polycystin-2 associates with the polycystin-1 homolog, suREJ3, and localizes to the acrosomal region of sea urchin spermatozoa. Mol Reprod Dev. 2004;67:472–477. doi: 10.1002/mrd.20033. [DOI] [PubMed] [Google Scholar]
- Neumann K, Michaux J, Lebedev V, Yigit N, Colak E, Ivanova N, Poltoraus A, Surov A, Markov G, Maak S, et al. Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene. Mol Phylogenet Evol. 2006;39:135–148. doi: 10.1016/j.ympev.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GA. Sperm competition and its evolutionary consequences in insects. Biol Rev. 1970;45:525–567. [Google Scholar]
- Pizzari T, Parker GA. Sperm competition and sperm phenotype. In: Birkhead TR, Hosken DJ, Pitnick SS, editors. Sperm Biology: An Evolutionary Perspective. San Diego, CA: Academic Press; 2008. pp. 207–246. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol. 2008;25:207–219. doi: 10.1093/molbev/msm242. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Steppan SJ, Adkins RM, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Sutton KA, Jungnickel MK, Ward CJ, Harris PC, Florman HM. Functional characterization of PKDREJ, a male germ cell-restricted polycystin. J Cell Physiol. 2006;209:493–500. doi: 10.1002/jcp.20755. [DOI] [PubMed] [Google Scholar]
- Sutton KA, Jungnickel MK, Florman HM. A polycystin-1 controls post-copulatory reproductive selection in mice. Proc Natl Acad Sci USA. 2008;105:8661–8666. doi: 10.1073/pnas.0800603105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol Phylogenet Evol. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Genetics. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Talevi R, Gualtieri R. Molecules involved in sperm-oviduct adhesion and release. Theriogenology. 2010;73:796–801. doi: 10.1016/j.theriogenology.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Tourmente M, Gomendio M, Roldan ERS. Sperm competition and the evolution of sperm design in mammals. BMC Evol Biol. 2011;11:12. doi: 10.1186/1471-2148-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Hoekstra HE. Causes and consequences of the evolution of reproductive proteins. Int J Dev Biol. 2008;52:769–780. doi: 10.1387/ijdb.082577lt. [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Künstner A, Nam K, Jakobsson M, Ellegren H. Nonlinear dynamics of nonsynonymous (dN) and synonymous (dS) substitution rates affects inference of selection. Genome Biol Evol. 2009;1:308–319. doi: 10.1093/gbe/evp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong WSW, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen A. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zigo M, Jonáková V, Sulc M, Manásková-Postlerová P. Characterization of sperm surface protein patterns of ejaculates and capacitated boar sperm, with the detection of ZP binding candidates. Int J Biol Macromol. 2013;61:322–328. doi: 10.1016/j.ijbiomac.2013.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.