Subcellular localization of abscisic acid receptors as peripheral proteins in the plasma membrane is mediated in a calcium-dependent manner by C2-domain abscisic acid-related proteins.
Abstract
Membrane-delimited abscisic acid (ABA) signal transduction plays a critical role in early ABA signaling, but the molecular mechanisms linking core signaling components to the plasma membrane are unclear. We show that transient calcium-dependent interactions of PYR/PYL ABA receptors with membranes are mediated through a 10-member family of C2-domain ABA-related (CAR) proteins in Arabidopsis thaliana. Specifically, we found that PYL4 interacted in an ABA-independent manner with CAR1 in both the plasma membrane and nucleus of plant cells. CAR1 belongs to a plant-specific gene family encoding CAR1 to CAR10 proteins, and bimolecular fluorescence complementation and coimmunoprecipitation assays showed that PYL4-CAR1 as well as other PYR/PYL-CAR pairs interacted in plant cells. The crystal structure of CAR4 was solved, which revealed that, in addition to a classical calcium-dependent lipid binding C2 domain, a specific CAR signature is likely responsible for the interaction with PYR/PYL receptors and their recruitment to phospholipid vesicles. This interaction is relevant for PYR/PYL function and ABA signaling, since different car triple mutants affected in CAR1, CAR4, CAR5, and CAR9 genes showed reduced sensitivity to ABA in seedling establishment and root growth assays. In summary, we identified PYR/PYL-interacting partners that mediate a transient Ca2+-dependent interaction with phospholipid vesicles, which affects PYR/PYL subcellular localization and positively regulates ABA signaling.
INTRODUCTION
Abscisic acid (ABA) elicits plant responses through binding to soluble PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) receptors, which constitute a 14-member family. PYR/PYL/RCAR receptors perceive ABA intracellularly and, as a result, form ternary complexes with clade A PP2C phosphatases, thereby inactivating them (Ma et al., 2009; Park et al., 2009; Santiago et al., 2009; Nishimura et al., 2010). This allows the activation of downstream targets of the PP2Cs, such as the sucrose nonfermenting 1-related protein kinase (SnRK) subfamily 2 (i.e., SnRK2.2/D, SnRK2.3/I, and SnRK2.6/OPEN STOMATA1 [OST1]/E), which are key players in the regulation of transcriptional response to ABA and stomatal aperture (Cutler et al., 2010). Additional targets of clade A PP2Cs have been described, such as SnRK1, SnRK3s/calcineurin B-like (CBL)-interacting protein kinases (CIPKs), calcium-dependent protein kinases (CDPKs/CPKs), ion transporters such as the K+ channel AKT1 and AKT2 or the slow anion channel 1 (SLAC1) and SLAC1 homolog 3 (SLAH3), and transcriptional regulators, such as bZIP transcription factors and chromatin-remodeling complexes (Chérel et al., 2002; Guo et al., 2002; Lee et al., 2007, 2009; Saez et al., 2008; Geiger et al., 2009; Antoni et al., 2012; Brandt et al., 2012; Lynch et al., 2012; Pizzio et al., 2013; Rodrigues et al., 2013). Some of these interactions have been shown to be modulated by PYR/PYL/RCAR receptors (Geiger et al., 2010, 2011; Brandt et al., 2012; Pizzio et al., 2013; Rodrigues et al., 2013). Therefore, clade A PP2Cs act as key negative regulators of ABA signaling and as a hub for the regulation of different environmental responses.
Genetic evidence about the function of PYR/PYLs indicates that they play a major role in the quantitative regulation of ABA responses, affecting both seed and vegetative responses to ABA (Park et al., 2009; Nishimura et al., 2010; Gonzalez-Guzman et al., 2012; Antoni et al., 2013). Analysis of their gene expression patterns together with their biochemical and genetic characterization have served to establish common and divergent properties of PYR/PYL ABA receptors (Dupeux et al., 2011b; Hao et al., 2011; Gonzalez-Guzman et al., 2012). Analyses of combined pyr/pyl mutants indicate that PYR/PYL function is partially redundant; however, PYL8 plays a nonredundant role to regulate root sensitivity to ABA (Antoni et al., 2013). Structural and biochemical studies also reveal several divergences among PYR/PYLs, particularly with respect to oligomeric structure and the perception of chemical agonists (Dupeux et al., 2011b; Hao et al., 2011; Okamoto et al., 2013). Activation of dimeric receptors by the ABA agonist quinabactin, which also activates to some extent PYL5 and PYL7, is enough to elicit both seed and vegetative responses to ABA (Cao et al., 2013; Okamoto et al., 2013).
The regulation of cellular processes involves intermolecular interactions that alter the location and/or activity of signaling proteins, and cellular membranes are a platform for intracellular communication involving lipid-protein and protein-protein complexes (Cho and Stahelin, 2005; Scott and Pawson, 2009). PYR/PYL proteins are intracellular ABA receptors localized both at the cytosol and the nucleus; however, detailed knowledge of their subcellular localization or putative transient interactions with membrane systems of the cell is currently lacking. Cytosolic proteins can reside partially in vesicles as peripheral proteins or interact transiently with membranes for trafficking or signaling purposes driven by protein modules that recognize specific features of proteins or membranes (Cho and Stahelin, 2005; Seet et al., 2006; Lemmon, 2008). In addition to the plasma membrane, eukaryotic cells possess an elaborate membrane system with multiple intracellular membranes (e.g., at the nucleus, organelles, and endocytic and secretory pathways) (Mellman and Emr, 2013; Voeltz and Barr, 2013). Thus, lipid bilayers take part in myriad processes in the plant cell, and cytosolic/nuclear proteins can interact transiently with membranes for signaling, transport, or other purposes (Cho and Stahelin, 2005; Voeltz and Barr, 2013). PYR/PYL ABA receptors, together with clade A PP2Cs and ABA-activated SnRK2s, play a key role in the regulation of ion transporters and membrane-associated enzymes that generate second messengers involved in ABA signaling (Geiger et al., 2009, 2011; Lee et al., 2009; Sato et al., 2009; Sirichandra et al., 2009; Cutler et al., 2010); however, it is not understood how PYR/PYL proteins (or PP2Cs/SnRK2s) can reach the proximity of cellular membranes beyond random diffusion. It is possible that auxiliary proteins might be involved in transiently approaching receptor, phosphatase, or kinase complexes next to cellular membranes, where early ABA signaling events take place. Recently, it was shown that ABA signaling modulates through ABSCISIC ACID INSENSITIVE1 (ABI1) and PYL9 the association of the signaling and transport complex CPK21/SLAH3 within plasma membrane domains, which is reminiscent of animal lipid rafts (Demir et al., 2013). These results imply that PYL9 must be able to inhibit ABI1 in the proximity of lipid nanodomains to allow the activation of SLAH3 by CPK21 (Demir et al., 2013).
In order to identify putative regulatory proteins of PYR/PYL receptors in Arabidopsis thaliana (e.g., auxiliary proteins that might regulate their subcellular localization or activity), we performed yeast-two hybrid (Y2H) screening using PYL4 as bait. PYL4, a representative member of the PYR/PYL family, shows high expression levels in different tissues, and its inactivation is required to strongly generate ABA-insensitive combined pyr/pyl mutants (Park et al., 2009; Gonzalez-Guzman et al., 2012). The search for new interacting partners of PYL4 resulted in the discovery of a family of small proteins containing a lipid binding C2 domain, named CAR proteins for C2-domain ABA-related protein, which interact with PYR/PYLs and positively regulate ABA sensitivity. The C2 domain comprises ∼130 residues and was first identified in protein kinase C (PKC), located between the C1 domain and the PKC catalytic domain (Nishizuka, 1988). The C2 domains of classical PKCs bind to phospholipid membranes in a calcium-dependent manner and are involved in targeting PKC activity to cell membranes in response to extracellular signals (Guerrero-Valero et al., 2007). C2 domains share functional characteristics with annexins, which also bind phospholipids in a calcium-dependent manner, but they are structurally unrelated (Lemmon, 2008). C2 domains are usually found in a large variety of eukaryotic proteins, where the C2 module is combined with a wide range of other modules encoding different enzymatic activities involved in intracellular signal transduction and membrane trafficking (Zhang and Aravind, 2010). The C2 domain acts in these proteins as a Ca2+-activated module that promotes targeting to membranes of the catalytic activity encoded in another region of the polypeptide. However, small C2-domain proteins, such as CAR proteins, that lack additional catalytic domains also have been identified in plants (Kim et al., 2003; Wang et al., 2009; Yokotani et al., 2009). Finally, the C2 domain and the EF-hand motif are the two most frequently occurring calcium sensors, and at least 123 proteins contain C2 domains in Arabidopsis (http://smart.embl-heidelberg.de/). However, not all C2 domains are able to bind calcium, and some of them have diverged evolutionarily into Ca2+-independent lipid binding variants (Cho and Stahelin, 2006).
RESULTS
Identification of CAR Proteins as Interacting Partners of PYR/PYLs
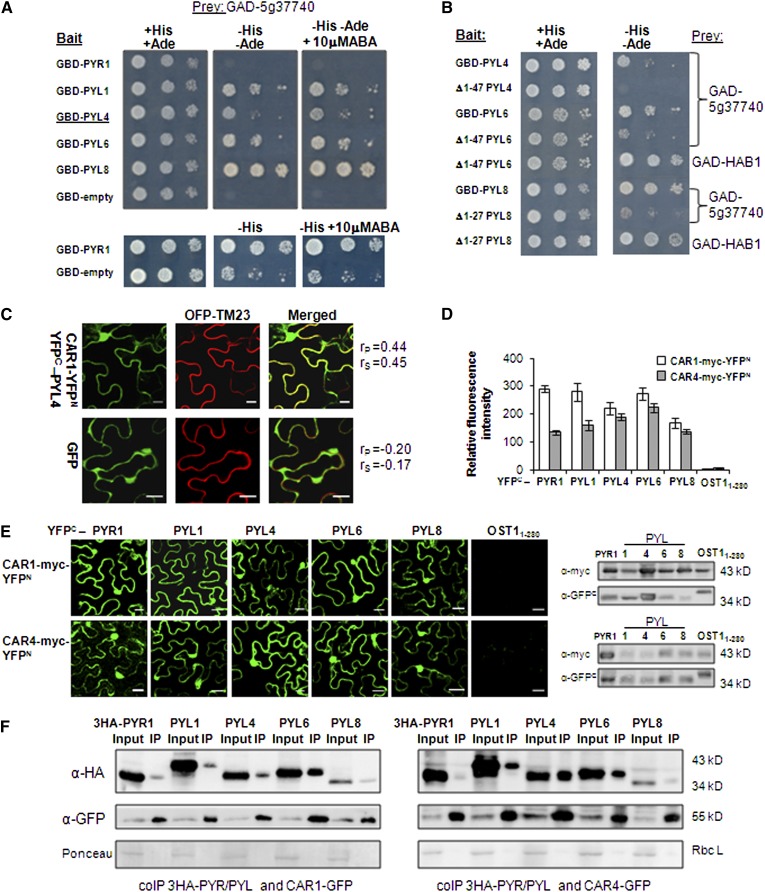
We performed a Y2H screen in the absence of ABA using PYL4 as a bait. As a result, we found a PYL4-interacting protein, At5g37740, whose binding to PYL4 was not ABA-dependent (Figure 1A). Analysis using the Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/) revealed that At5g37740 belongs to a branch of the C2-domain superfamily represented by single-C2 domain proteins that lack additional catalytic domains (Supplemental Figure 1). Other PYL receptors, such as PYL1, PYL6, and PYL8, also were able to interact in Y2H assays (minus histidine and adenine) with this small C2-domain protein (Figure 1A). Interaction with PYR1 in a Y2H assay was only detected under less demanding conditions (minus histidine). Deletion of the N-terminal region of PYL4, PYL6, and PYL8 severely impaired the interaction with At5g37740 (Figure 1B). This N-terminal deletion did not affect the binding of Δ1-47 PYL6 or Δ1-27 PYL8 to HAB1, which reflects that this region is not involved in the formation of the receptor-ABA-phosphatase complex (Melcher et al., 2009; Miyazono et al., 2009; Dupeux et al., 2011a). We named At5g37740 as CAR1, and BLAST search at TAIR revealed a CAR gene family composed of 10 members (CAR1 to CAR10) in Arabidopsis (Supplemental Figure 2). The CAR family was also found in other plant species such as tomato (Solanum lycopersicum) and rice (Oryza sativa) (Supplemental Figure 3). Arabidopsis CAR proteins range between 165 and 185 amino acid residues with estimated molecular masses of 18 to 20 kD (Supplemental Figure 2).
Figure 1.
PYL4 and Other PYR/PYL Receptors Interact with CAR1 in Y2H Assays.
BiFC and coIP assays reveal interactions between CAR1/CAR4 and PYR/PYLs in N. benthamiana epidermal cells.
(A) ABA-independent interaction between CAR1 and different PYR/PYLs. Interaction was determined by growth assay on media lacking His and adenine (−His and −Ade), which were supplemented or not with 10 mM ABA. Dilutions (10−1, 10−2, and 10−3) of saturated cultures were spotted onto the plates. GAD, GAL4 activation domain; GBD, GAL4 DNA binding domain.
(B) Deletion of the N-terminal region of PYL4, PYL6, and PYL8 impairs the interaction with CAR1 but not with HAB1 (assayed for Δ1-47 PYL6 and Δ1-27 PYL8).
(C) CAR1 and PYL4 interact in the plasma membrane and nucleus of N. benthamiana cells. Confocal images show transiently transformed N. benthamiana epidermal cells coexpressing CAR1-YFPN/YFPC-PYL4-interacting proteins and the plasma membrane marker OFP-TM23 (top row) or GFP and OFP-TM23 (bottom row). BiFC interaction of CAR1-YFPN and YFPC-PYL4 was observed, and this interaction colocalizes with the plasma membrane marker OFP-TM23 (Merged). Epifluorescence confocal images of epidermal N. benthamiana leaves infiltrated with the indicated constructs were merged to quantitatively estimate the colocalization of YFP/GFP and OFP fluorescence. Statistical analysis of the colocalization of CAR1-YFPN/YFPC-PYL4-interacting proteins and OFP-TM23 was done using Pearson’s (rP) and Spearman’s (rS) correlation factors (French et al., 2008). The degree of colocalization between the two fluorescent signals was analyzed using Zeiss software. Bars = 20 μm.
(D) Relative fluorescence intensity of the BIFC interactions detected for either CAR1-YFPN or CAR4-YFPN and the indicated constructs. Each BiFC experiment was scanned and measured in 25 randomly chosen microscopic fields (n = 25) and repeated three times.
(E) BiFC assays show both nuclear and nonnuclear interactions of CAR1/CAR4 and PYR/PYLs in N. benthamiana epidermal cells coinfiltrated with Agrobacterium suspensions containing the indicated constructs and the silencing suppressor p19. Immunoblot analyses (at right) confirm the expression of myc-tagged CAR1 (top)/CAR4 (bottom) and YFPC-tagged PYR/PYL proteins in N. benthamiana epidermal cells.
(F) coIP assays demonstrate the interaction of CAR1-GFP or CAR4-GFP and PYR/PYLs. Nuclear protein extracts obtained from N. benthamiana leaves infiltrated with Agrobacterium suspensions harboring the indicated constructs were analyzed using anti-HA or anti-GFP antibody. Input levels of epitope-tagged proteins in crude protein extracts (20 μg of total protein) were analyzed by immunoblotting. Immunoprecipitated (IP) CAR1-GFP and CAR4-GFP proteins were probed with anti-HA antibodies to detect coIP of HA-tagged PYR/PYLs. Ponceau staining from the large subunit of Rubisco (Rbc L) is shown as a loading control.
C2 domains are usually found in mammals as regulatory modules of different polypeptides that also include a catalytic domain, such as the typical PKC-C2, phosphatidylinositol 3-kinase-C2, and phospholipase A2-C2 combinations (Zhang and Aravind, 2010). Thus, C2 domains are able to translocate to membrane compartments the associated catalytic activity in response to Ca2+ peaks. Analysis of the Arabidopsis genome reveals combinations of the C2 domain with different catalytic domains, such as phospholipase D, lysine decarboxylase, phosphoribosylanthranilate transferase, endonucleases, inositol 1,4,5-trisphosphate phosphatases, and phospholipase C (Supplemental Figure 1). However, the Arabidopsis CAR family hereby identified represents a plant-specific C2-domain family of small proteins not associated with catalytic domains. Therefore, we suggest that CAR proteins as well as other short C2 proteins lacking additional domains might function through interaction with lipids or other proteins and display a dual function as a calcium-dependent phospholipid binding protein and a protein-protein interaction module.
Bimolecular fluorescence complementation (BiFC) assays were used to test the CAR1-PYL4 interaction in plant cells. To this end, 35S:CAR1-YFPN (also harboring the 35S cauliflower mosaic virus promoter and the N-terminal yellow fluorescent protein gene) and 35S:YFPC-PYL4 constructs were delivered into leaf cells of Nicotiana benthamiana by Agrobacterium tumefaciens infiltration (agroinfiltration); as a result, fluorescence was observed in the nucleus or visualized as a thin layer that could reflect plasma membrane or cytosolic localization (Figure 1C). As a marker of plasma membrane localization, we used the red fluorescence emitted by the orange/red fluorescent protein OFP-TM23, a modified version of OFP (orange fluorescent protein) containing a transmembrane domain that results in plasma membrane targeting (Batistic et al., 2012). Therefore, we coexpressed CAR1-YFPN, YFPC-PYL4, and OFP-TM23 into leaf cells of N. benthamiana by agroinfiltration. Next, we followed the protocol described by French et al. (2008) in order to perform statistical analysis of the putative colocalization of the fluorescent markers. We found that Pearson-Spearman correlation coefficients indicated colocalization of OFP-TM23 and reconstituted YFP proteins; therefore, a significant amount of the CAR1-PYL4 interaction was localized to the plasma membrane (Figure 1C). By contrast, green fluorescent protein (GFP) did not show colocalization with OFP-TM23 when both proteins were coexpressed in N. benthamiana cells (Figure 1C). We also found that other PYR/PYLs, such as PYR1, PYL1, PYL6, and PYL8, also interacted with CAR1 in BiFC assays (Figures 1D and 1E). In contrast with the interaction observed with YFPC-PYR/PYLs, the YFPC-OST11-280 protein was not able to interact with CAR1 (Figures 1D and 1E). We wondered whether other members of the CAR family were able to interact with PYR/PYLs. As a result, we found that CAR4 was able to interact with PYR1, PYL1, PYL4, PYL6, and PYL8 using BiFC assays, whereas it did not interact with YFPC-OST11-280 (Figures 1D and 1E). Although the expression levels of the YFP-tagged proteins varied in the different agroinfiltrations of N. benthamiana plants, we could confirm by immunoblot analyses the expression of the receptors and CAR proteins in all the BiFC experiments (Figure 1E).
Finally, we coexpressed in N. benthamiana epidermal cells HA-tagged PYR/PYLs and either CAR1-GFP or CAR4-GFP proteins through agroinfiltration in order to conduct coimmunoprecipitation (coIP) experiments (Figure 1F). To avoid precipitation of the membrane fractions (not mediated by antibody) where CAR1-PYL4 interaction occurred, we used the soluble nuclear fractions where this interaction also occurs (Figures 1C and 1E). To this end, we precipitated nuclear CAR-GFP proteins using α-GFP and tested the simultaneous presence of PYR/PYLs using α-HA. As a result, we could detect coIP of either CAR1-GFP or CAR4-GFP and the five receptors assayed to various extents (Figure 1F). The recovery of coimmunoprecipitated CAR1-GFP and HA-PYL6 was apparently higher than that of the other interactions; however, we also recovered more CAR1-GFP in the α-GFP precipitated fraction. We also found a consistently higher recovery of coimmunoprecipitated CAR4-GFP and HA-PYL1/PYL4/PYL6 compared with HA-PYR1. Expression of HA-PYL8 was lower than that of the other HA-tagged PYLs; therefore, the initial input was not comparable to that of the other PYLs.
Subcellular Localization of CAR and PYL4 Proteins
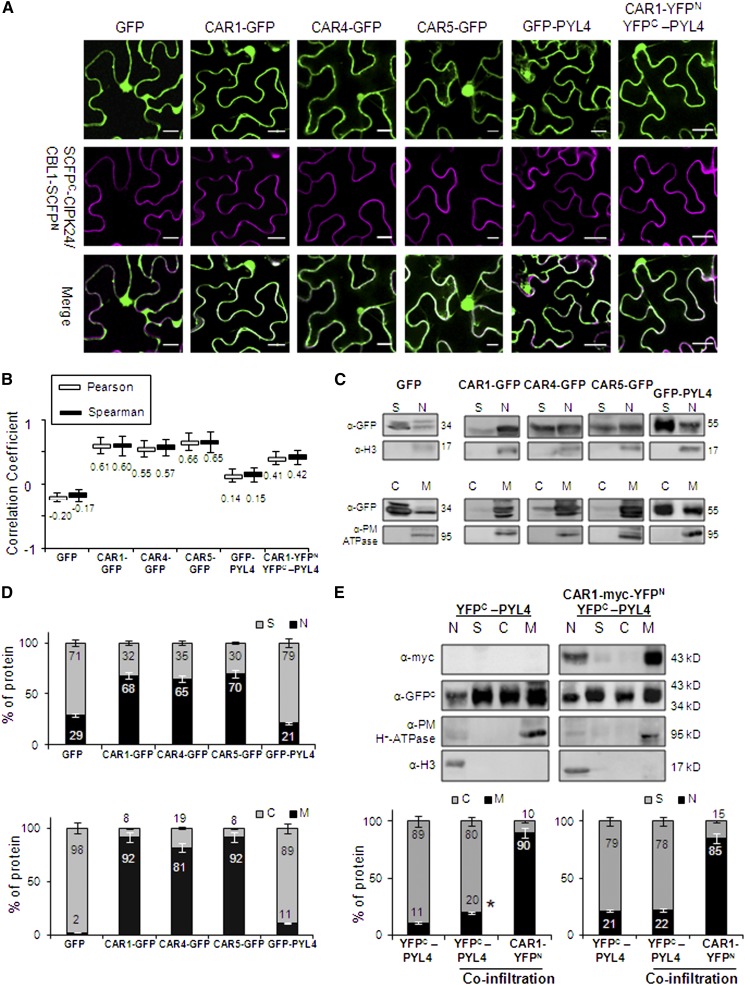
In order to further explore the subcellular localization of individual CAR and PYL4 proteins, we expressed CAR1-GFP, CAR4-GFP, CAR5-GFP, and GFP-PYL4 fusion proteins in N. benthamiana epidermal cells by agroinfiltration. In addition to their nuclear localization, CAR-GFP proteins decorated the perimeter of the cell, which could reflect plasma membrane or cytosolic localization (Figure 2A). As a marker of plasma membrane localization, we used the cyan fluorescence emitted by the reconstituted super cyan fluorescent protein (SCFP) of the SCFPC-CIPK24/CBL1-SCFPN interaction, which has been reported previously to be localized at the plasma membrane as a peripheral protein (Waadt et al., 2008). We coexpressed individual CAR-GFP proteins with this marker and performed statistical analysis of the putative colocalization of the fluorescent markers (fluorescence emission spectra in the green range for GFP and the cyan range for SCFP). As a result, we found colocalization of both fluorescent markers (Pearson-Spearman correlation coefficients in the range 0.55 to 0.66; Figure 2B). Therefore, a significant fraction of CAR-GFP proteins was localized to the plasma membrane. By contrast, GFP alone did not show colocalization with reconstituted SCFP (Pearson-Spearman correlation coefficients below zero). GFP-PYL4 alone did not show a substantial localization in the plasma membrane; however, when YFPC-PYL4 was coexpressed with CAR1-YFPN, it showed partial colocalization with the plasma membrane marker used in this experiment (reconstituted SCFP) (Figures 2A and 2B). Independent evidence for localization in the plasma membrane of CAR proteins came from data mining in the results published by Demir et al. (2013). In that work, proteins associated with detergent-resistant membranes from leaf plasma membrane preparations were identified using mass spectrometry. Two CAR proteins, Arabidopsis CAR6 and N. benthamiana CAR2 homolog, were identified in data sets S2 and S3, respectively, further confirming the presence of CAR proteins in the plasma membrane.
Figure 2.
CAR-GFP Fusion Proteins Localize to Nucleus, Plasma Membrane, and Cytosol upon Transient Expression in N. benthamiana.
(A) Confocal images of transiently transformed N. benthamiana epidermal cells coexpressing GFP, GFP-tagged CAR/PYL4 proteins or the CAR1-YFPN/YFPC-PYL4-interacting proteins, and the plasma membrane marker resulting from the SCFPN-CIPK24/CBL1-SCFPC interaction. The degree of colocalization between the two fluorescent signals was analyzed using merged images and Zeiss software (ZEN Lite 2012). The magenta color of the reconstituted SCFP is a pseudocolor generated from the original cyan fluorescence. Bars = 20 μm.
(B) Pearson-Spearman correlation coefficients indicate the colocalization of CAR-GFP proteins or the CAR1-YFPN/YFPC-PYL4 interaction and the plasma membrane marker. Epifluorescence confocal images of epidermal leaves coinfiltrated with the indicated constructs were merged to quantitatively estimate the colocalization of GFP/YFP and SCFP fluorescence (French et al., 2008). At least 10 single-scanned cell images per experiment were collected and analyzed using the same conditions of laser intensity, pinhole size, and gain levels.
(C) Biochemical fractionation and immunoblot analyses of protein extracts prepared from N. benthamiana leaves infiltrated with Agrobacterium harboring the indicated constructs. Protein extracts from the different fractions were analyzed by immunoblotting using anti-GFP, anti-Histone3 (H3), and anti-plasma membrane (PM) H+-ATPase antibodies. The positions of the molecular mass standards (kD) are indicated. C, cytosolic fraction; M, microsomal fraction; N, nuclear fraction; S, nonnuclear soluble fraction.
(D) Quantification of the subcellular localization of GFP and GFP-tagged proteins transiently expressed in N. benthamiana epidermal cells. Immunoblot signals obtained in (C) were captured using the image analyzer LAS3000, and quantification of the protein signal was done using Image Guache version 4.0 software.
(E) Overexpression of CAR1 together with PYL4 in coinfiltration experiments increases the PYL4 presence in membranes. Biochemical fractionation, immunoblot analyses, and quantification of protein extracts prepared from N. benthamiana leaves infiltrated with Agrobacterium harboring the indicated constructs were performed.
Finally, we examined the subcellular localization of CAR1-GFP, CAR4-GFP, CAR5-GFP, and GFP-PYL4 proteins by standard biochemical techniques (Figure 2C). First, we followed a fractionation technique to separate nuclei (pelleted at 1000g) from the soluble nonnuclear fraction (Saez et al., 2008; Antoni et al., 2012). Compared with GFP, the CAR-GFP proteins were enriched in the nuclear fraction. Additionally, we submitted the soluble fraction to a centrifugation of 100,000g to separate the soluble cytosolic fraction from the pelleted microsomal fraction (Antoni et al., 2012). We found that, in contrast with GFP, which was localized mostly to the cytosolic soluble fraction, the CAR proteins were localized mostly to the microsomal fraction (Figure 2D). GFP-PYL4 also showed a dual nuclear and nonnuclear localization, although a lower percentage of nuclear protein was found compared with CAR-GFP proteins. In contrast with CAR-GFP proteins, most of the soluble nonnuclear fraction of GFP-PYL4 was localized to cytosol, although a significant amount was localized to the microsomal fraction (Figure 2D). Overexpression of CAR1 together with PYL4 in coinfiltration experiments increased the presence of PYL4 in membranes relative to the cytosolic location, although to a modest extent (Figure 2E). This result suggests that membrane recruitment of PYL4 by CAR1 affects only a fraction of the total receptor pool.
CAR Proteins Show an α1βAβB CAR-Signature Extra-Domain Inserted into a Canonical C2 Fold
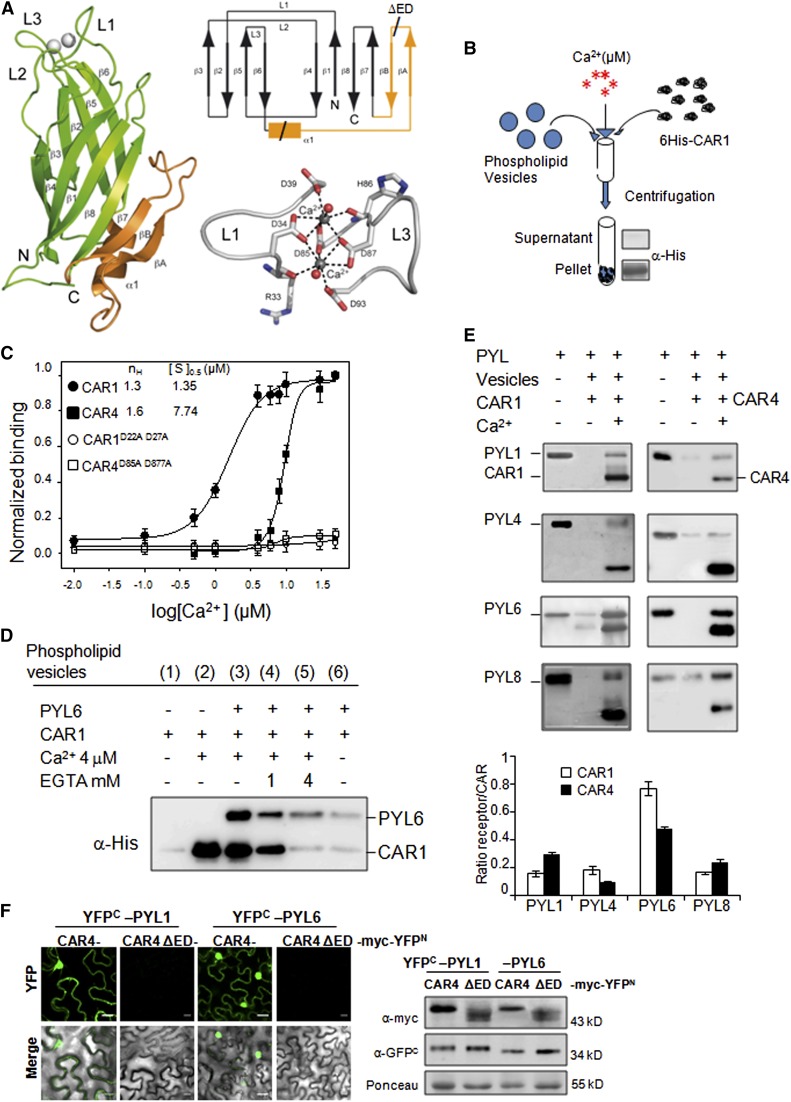
In order to obtain molecular insights into CAR proteins, the x-ray structure of CAR4 in complex with Ca2+ was solved by molecular replacement at 1.6 Å resolution (Figure 3A; Supplemental Table 1 and Supplemental Methods). The overall structure of CAR4 is almost identical to that found for other C2 domains. CAR4 folds as a compact β sandwich that is composed by two four-stranded β sheets with type II domain topology (Rizo and Südhof, 1998). In addition, it contains an extra-domain insertion consisting of 43 amino acids that connect the two four-stranded β sheets (β3β2β5β6 with β4β1β8β7) and folds as an α helix followed by a β hairpin (α1βAβB) (Figure 3A, see CAR4 topology). This insertion is conserved among the members of the CAR family (Supplemental Figures 2 and 3) and represents a unique CAR signature when the fold is compared with other known families of C2 domains using the PDBeFold structure similarity service (http://www.ebi.ac.uk/msd-srv/ssm/; Krissinel and Henrick, 2004). The CAR-signature extra-domain is situated in the protein face opposite to loops L1 and L3, which bind Ca2+ ions that bridge C2 proteins to membranes (Figure 3A; Verdaguer et al., 1999). The crystallographic analysis revealed two CAR4 molecules in the asymmetric unit. The structures of these independent molecules are nearly identical (Cα backbone root-mean square deviation of 0.14 Å).
Figure 3.
Calcium-Dependent Phospholipid Binding of CAR Proteins.
Ca2+ and CAR-dependent recruitment of PYR/PYLs to phospholipid vesicles was performed.
(A) Crystal structure, Ca2+ coordination, and topology of CAR4. A ribbon representation of the CAR4 crystal structure (left) showing overall fold together with a scheme of the topology (top right) and a detailed representation of the calcium binding sites (bottom right) are shown. The α1βAβB extra-domain is highlighted in orange.
(B) Scheme of the biochemical assay to detect Ca2+-dependent protein-phospholipid interaction through pelleting of phospholipid vesicles and immunoblot analysis. Phospholipid vesicles were composed of 25% phosphatidylserine and 75% phosphatidylcholine. The vesicles were precipitated by centrifugation, and bound proteins were revealed by SDS-PAGE and immunoblot analysis using α-His antibody.
(C) CAR1, CAR4, CAR1D22A D27A, and CAR4D85A D87A proteins were incubated with phospholipid vesicles in the presence of increasing concentrations of Ca2+ to determine the half-maximal binding for the ion. Introduction of two Asp-to-Ala mutations into amino acid residues 22 and 27 of CAR1 or residues 85 and 87 of CAR4 abolished phospholipid binding. The Hill coefficient (nH) and calcium concentration leading to half-maximal binding ([S]0.5) are indicated at the top of the graph.
(D) Ca2+- and CAR1-dependent recruitment of PYL6 to phospholipid vesicles. Ca2+- and CAR1-dependent vesicle pelleting of PYL6 can be reversed by EGTA treatment. Pelleted vesicles bound to PYL6 and CAR1 were EGTA treated, precipitated again by centrifugation, and analyzed by SDS-PAGE and immunoblot.
(E) Ca2+-dependent vesicle pelleting assay of CAR1 and CAR4 using different PYR/PYLs. The Ca2+ concentration was either 4 or 20 μM for CAR1 or CAR4, respectively. The quantification of the assays shows the relative ratio of receptor bound (minus background in the absence of calcium) per molecule of CAR1 or CAR4.
(F) A CAR4ΔED mutant is unable to interact with PYL1/PYL6 ABA receptors. A BiFC assay was performed in N. benthamiana epidermal cells coinfiltrated with Agrobacterium suspensions containing the indicated constructs and the silencing suppressor p19. Immunoblot analyses (at right) confirm the expression of myc-tagged CAR4/CAR4 ΔED and YFPC-tagged PYL1/PYL6 proteins in N. benthamiana epidermal cells. Bars = 20 μm.
CAR Proteins Show Ca2+-Dependent Phospholipid Binding Activity and Recruit PYR/PYLs to Membranes
C2 domains display a well-defined calcium-dependent lipid binding site that relies on the unspecific interaction of the phosphate moiety of phospholipids with the calcium ions coordinated in the conserved L1 and L3 loops and the specific interaction of the phospholipid headgroup with amino acid residues from the loops that conform the cup-shaped calcium binding site (Verdaguer et al., 1999). The crystallographic analysis shows that CAR4 binds two calcium atoms at this site (Figure 3A). They are coordinated with conserved Asp residues in loops L1 (D34 and D39) and L3 (D85 and D87), which would make it possible that Ca2+ bridges the C2 domain to phospholipids (Perisic et al., 1998; Verdaguer et al., 1999; Guerrero-Valero et al., 2009). Since the Ca2+-dependent phospholipid binding is a hallmark of many C2 domains, we tested whether CAR proteins were able to bind negatively charged phospholipid vesicles (25% phosphatidylserine/75% phosphatidylcholine) in a Ca2+-dependent manner. The vesicle-pelleting assay is a standard method to detect the lipid binding of peripheral proteins and is summarized in Figure 3B (Cho et al., 2001). This assay was performed in the absence or presence of different Ca2+ concentrations using CAR1 and CAR4 as well as the CAR1D22A D27A and CAR4D85A D87A mutants, which contain double Asp-to-Ala mutations in the loops L1 and L3, respectively. As a result, we could observe that Ca2+ promoted the binding of CAR1 and CAR4 to phospholipid vesicles (Figure 3C). We calculated half-maximal calcium concentrations of 1.3 and 7.7 μM for in vitro phospholipid binding of CAR1 and CAR4, respectively, and a certain cooperative effect for Ca2+ binding according to Hill coefficient value (Figure 3C, top of the graph). Transient increases in Ca2+ to micromolar levels have been described in different signaling pathways, so the values found for CAR proteins are into the physiological range of Ca2+ signaling (Swanson et al., 2011). Additionally, the limited analysis of two members of the CAR family suggests that different CAR proteins might sense and respond differentially to an increase of intracytosolic Ca2+ levels, as has been described in different C2 domains of classical PKCs type α, β, and γ (Guerrero-Valero et al., 2007). Finally, we confirmed that the coordination of Ca2+ ions by L1 and L3 loops is crucial for phospholipid binding, since the CAR1D22A D27A and CAR4D85A D87A mutations abolished phospholipid binding (Figure 3C).
One of the receptors that interacted well with CAR proteins in BiFC and coIP assays was PYL6 (Figure 1). Only a residual presence of PYL6 in the pellet was detected upon coincubation with CAR1 and phospholipid vesicles lacking calcium (Figure 3D, lane 6). By contrast, coincubation of PYL6 with CAR1 and phospholipid vesicles in the presence of Ca2+ promoted the recruitment of PYL6 to membranes (Figure 3D, lane 3). Such an effect could be reversed by treating the pelleted vesicles with increasing concentrations of EGTA, a chemical acting as a Ca2+ chelating agent (Figure 3D, lanes 4 and 5). These results indicate that binding of PYL6 to phospholipid vesicles was dependent both on Ca2+ and CAR1, and the Ca2+-dependent recruitment of PYL6 to membranes by CAR1 was reversible, excluding unspecific effects, such as protein aggregation or protein insolubility induced by the CAR1-PYL6 interaction.
We also tested whether other PYR/PYL receptors could be recruited to phospholipid vesicles by either CAR1 or CAR4 in a Ca2+-dependent manner. PYL1, PYL4, PYL6, and PYL8 were recruited to phospholipid vesicles by CAR1, whereas PYL1, PYL6, and PYL8 were recruited by CAR4 (Figure 3E). The starting receptor:CAR ratio was 1:1 in the in vitro assay, and after performing the vesicle pelleting assay, we measured the ratio of receptor to CAR protein in the pelleted vesicles to estimate the affinity of each receptor to either CAR1 or CAR4. Clearly, PYL6 was efficiently recruited by both CAR1 and CAR4. The other receptors were less efficiently recruited, ranging between 0.2 and 0.3 molecules of receptor bound per molecule of CAR protein. Taken together, these assays reveal that CAR1 and CAR4 proteins were able to selectively bridge the interaction of PYR/PYL proteins with phospholipid vesicles in a Ca2+-dependent manner.
Finally, we generated a CAR4 internal deletion (named Δextra-domain [ΔED]) affecting amino acid residues 107 to 140 to partially eliminate the extra-domain insertion of CAR4 (Figure 3A), and we tested its effect on the interaction with PYL1 and PYL6 (Figure 3F). As a result, we found that CAR4ΔED did not interact with either PYL1 or PYL6 in BiFC assays, in spite of being synthesized at roughly similar levels to the corresponding wild-type control (Figure 3F, right panel). This result suggests that the extra-domain present in CAR proteins is required for interaction with PYL ABA receptors.
Triple Mutants Impaired in CAR Genes Show Reduced Sensitivity to Both ABA-Mediated Inhibition of Seedling Establishment and Root Growth
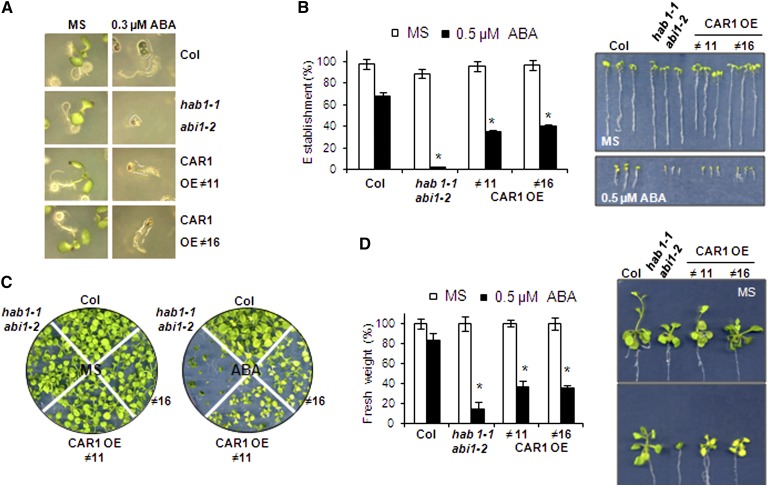
Both gain-of-function and loss-of-function approaches were followed in order to investigate whether CAR genes affect ABA signaling. First, we analyzed the ABA sensitivity of 35S:CAR1 lines with respect to the ABA-mediated inhibition of seedling establishment and shoot growth (Figure 4). 35S:CAR1 lines showed enhanced sensitivity to ABA-mediated inhibition of seedling establishment and shoot growth compared with the wild type (Figure 4). Thus, establishment at 0.3 to 0.5 μM ABA was impaired in 35S:CAR1 lines compared with the wild type (Figures 4A and 4B). Those seedlings from 35S:CAR1 lines that were able to establish in 0.5 μM ABA showed a clear impairment of shoot growth compared with the wild type after 20 d of growth in medium supplemented with ABA (Figure 4C). Likewise, seedlings from 35S:CAR1 lines transferred from Murashige and Skoog (MS) plates to plates supplemented with 0.5 μM ABA showed reduced fresh weight after 20 d of growth compared with the Columbia (Col) wild type (Figure 4D).
Figure 4.
Overexpression of CAR1 Leads to Enhanced ABA-Mediated Inhibition of Seedling Establishment and Shoot Growth.
(A) Photographs of the Col wild type, the hab1-1abi1-2 ABA-hypersensitive mutant (Saez et al., 2006), and two CAR1-overexpressing (OE) lines (#11 and #16) grown for 4 d on MS medium either lacking or supplemented with 0.3 μM ABA.
(B) Quantification of ABA-mediated inhibition of seedling establishment of the Col wild type compared with the hab1-1abi1-2 double mutant and two CAR1-overexpressing lines. Approximately 100 seeds of each genotype were sown on MS plates lacking or supplemented with 0.5 μM ABA and scored for the presence of green expanded cotyledons 7 d later. The photographs show representative seedlings removed at 7 d from MS plates lacking or supplemented with 0.5 μM ABA and rearranged on agar plates.
(C) 35S:CAR1 lines show enhanced sensitivity to ABA-mediated inhibition of vegetative growth. Photographs were taken of the Col wild type, the hab1-1abi1-2 double mutant, and two CAR1-overexpressing lines grown for 12 d on MS medium lacking ABA or 21 d in medium supplemented with 0.5 μM ABA.
(D) Quantification of fresh weight after 21 d of growth in medium lacking or supplemented with 0.5 μM ABA.
Asterisks in (B) and (D) indicate P < 0.05 (Student’s t test) when comparing data of 35S:CAR1 lines and the hab1-1abi1-2 mutant with Col wild-type plants in the same assay conditions.
[See online article for color version of this figure.]
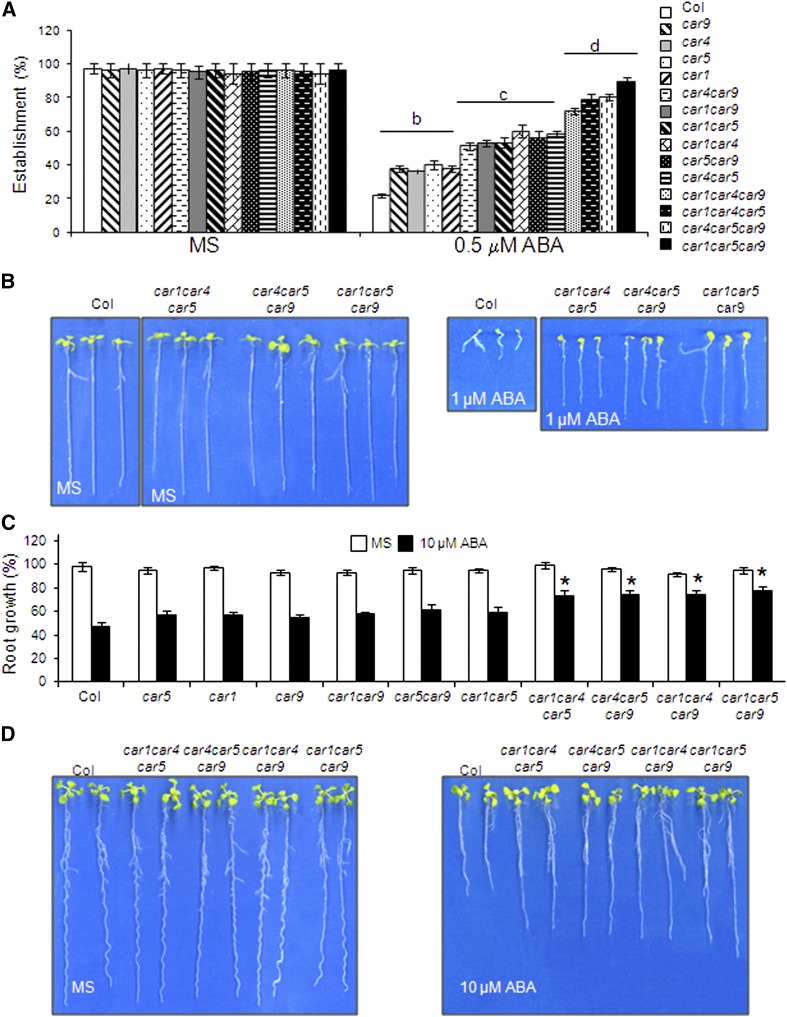
Next, we identified loss-of-function knockout mutants for different members of the CAR family that were available from T-DNA mutant collections. At the beginning of this work, we identified T-DNA homozygous mutants for the loci At5g37740 (car1), At3g17980 (car4), At1g48590 (car5), and At1g70790 (car9) (Supplemental Figure 4). Since some functional redundancy might be expected among CAR proteins as was observed previously for PYR/PYL proteins, we generated different double and triple mutants impaired in CAR genes and analyzed their sensitivity to ABA (Figure 5). ABA-mediated inhibition of seedling establishment assays revealed both a reduced sensitivity to ABA in the combined mutants compared with the wild type and functional redundancy among CAR genes, since the generation of triple mutants was required to obtain robust phenotypes (Figures 5A and 5B). Inhibition of root growth in the single and double mutants was slightly lower than in the wild type, whereas triple mutants showed a significant ABA insensitivity compared with the wild type (Figures 5C and 5D).
Figure 5.
Triple car Mutants Show Reduced Sensitivity to ABA-Mediated Inhibition of Seedling Establishment and Root Growth.
(A) Quantification of ABA-mediated inhibition of seedling establishment of the Col wild type compared with single, double, and triple car mutants. Approximately 100 seeds of each genotype were sown on each plate and scored for the presence of green expanded cotyledons 5 d later. The letters denote significant differences among the different genetic backgrounds (P < 0.05, Fisher’s least significant difference test).
(B) Photographs of the Col wild type and triple car mutants grown for 7 d on MS medium either lacking or supplemented with 1 μM ABA.
(C) Quantification of ABA-mediated root growth inhibition of the Col wild type compared with single, double, and triple car mutants. Asterisks indicate P < 0.05 (Student’s t test) when comparing data of car mutants with Col wild-type plants in the same assay conditions.
(D) Photographs of representative seedlings 10 d after the transfer of 4-d-old triple car mutants seedlings from MS medium to plates lacking or supplemented with 10 μM ABA.
[See online article for color version of this figure.]
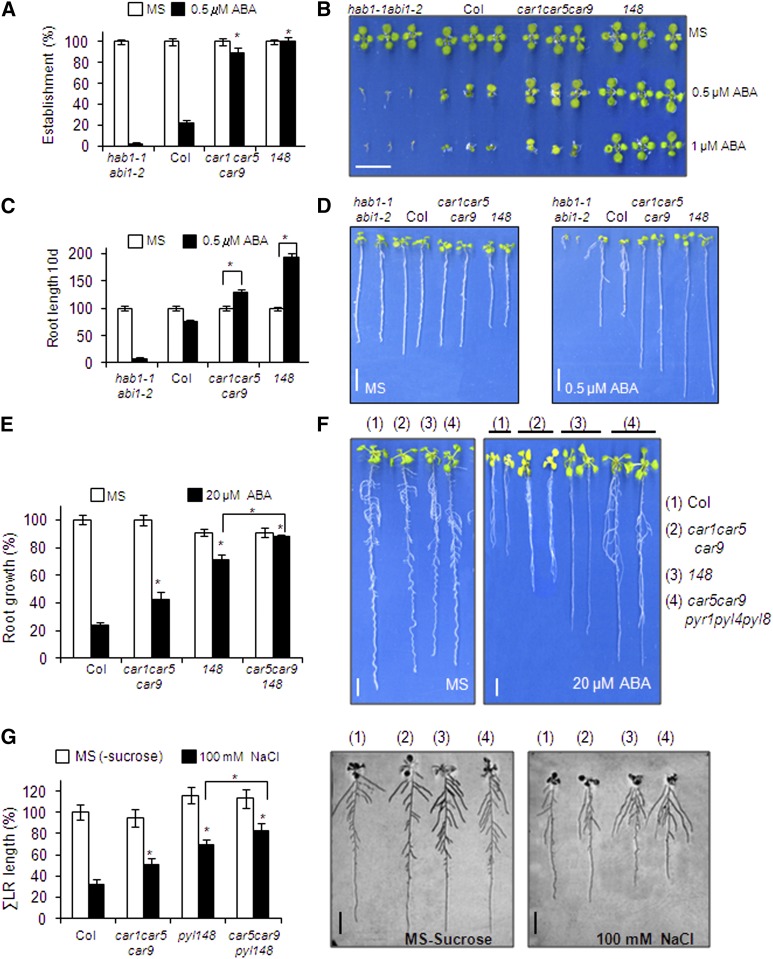
Finally, we selected one of the car triple mutants that showed reduced sensitivity to ABA and compared it with a triple mutant impaired in three ABA receptors, pyr1, pyl4, and pyl8, abbreviated as 148 (Figure 6). Seedling establishment of the car1 car5 car9 triple mutant was less sensitive to ABA-mediated inhibition than the wild type (Figures 6A and 6B). The percentage of seedlings that established at 0.5 μM ABA was similar in car1 car5 car9 and 148; however, further development of the seedlings was less inhibited in 148 compared with car1 car5 car9 (Figure 6B). The root length of the seedlings that were established in 0.5 μM ABA was larger in car1 car5 car9 compared with the wild type (Figure 6C). Interestingly, 0.5 μM ABA supplementation enhanced the root length of car1 car5 car9 seedlings compared with medium lacking ABA (Figures 6C and 6D), an effect that was described previously in some combined pyr/pyl ABA-insensitive mutants (Gonzalez-Guzman et al., 2012; Antoni et al., 2013). To further study the genetic interaction between C2 and PYR/PYL proteins, we performed a cross between the car1 car5 car9 and 148 triple mutants. We were able to recover a car5 car9 pyr1 pyl4 pyl8 pentuple mutant (CAR1 and PYL8 show linkage in the lower arm of chromosome 5), and root growth assays in medium supplemented with 20 μM ABA showed that the pentuple mutant was less sensitive to ABA-mediated inhibition of primary root growth than the 148 triple mutant (Figures 6E and 6F). Therefore, both CAR and PYR/PYL genes additively regulate root sensitivity to ABA. Lateral root growth is also dependent on ABA, since endodermal ABA signaling promotes lateral root quiescence during salt stress and, accordingly, ABA-insensitive mutants show reduced inhibition of lateral root growth induced by NaCl (Duan et al., 2013). According to this notion, the 148 triple mutant was more resistant to salt-induced inhibition of lateral root growth than the wild type. Lateral roots of car1 car5 car9 also showed a lower sensitivity to NaCl compared with the wild type, which was additive with the 148 phenotype when the car5 car9 pyr1 pyl4 pyl8 pentuple mutant was assayed (Figure 6G). Therefore, taken together, these results indicate that CAR proteins regulate ABA sensitivity in both primary and lateral roots. Reporter gene analysis of the CAR1 promoter showed predominant expression of CAR1 in the vascular bundle of the primary root as well as in the cortex of the upper part of the root (Supplemental Figure 5). In lateral roots, CAR1 expression was also detected in epidermis and root tips.
Figure 6.
ABA Sensitivity of the car1car5car9 Triple Mutant Compared with the pyr1 pyl4 pyl8 Triple Mutant.
The additive effect of the car5 car9 pyr1 pyl4 pyl8 pentuple mutant is shown.
(A) Quantification of the ABA-mediated inhibition of seedling establishment of the Col wild type compared with car and pyr/pyl triple mutants. Approximately 100 seeds of each genotype were sown on MS plates lacking or supplemented with 0.5 μM ABA and scored for the presence of green expanded cotyledons 5 d later. Asterisks indicate P < 0.05 (Student’s t test) when comparing data of car and pyr/pyl triple mutants with Col wild-type plants in the same assay conditions.
(B) Photographs of the Col wild type and car1 car5 car9 and pyr1 pyl4 pyl8 triple mutants grown for 7 d on MS medium either lacking or supplemented with 0.5 or 1 μM ABA.
(C) ABA supplementation improves root growth of the car1 car5 car9 triple mutant. Quantification of root length was done in the 10-d-old seedlings described in (A). Asterisks indicate P < 0.05 (Student’s t test) when comparing data obtained in medium lacking or supplemented with 0.5 μM ABA.
(D) Photographs show representative seedlings removed at 10 d from MS plates lacking or supplemented with 0.5 μM ABA and rearranged on agar plates.
(E) Reduced sensitivity to ABA-mediated inhibition of root growth in the car5 car9 pyr1 pyl4 pyl8 pentuple mutant compared with other genetic backgrounds. Seedlings were grown on vertically oriented MS plates for 4 d. Next, 20 plants were transferred to new MS plates lacking or supplemented with 20 mM ABA. Quantification of ABA-mediated root growth inhibition was performed after 20 d. Asterisks indicate P < 0.05 (Student’s t test) when comparing data of mutants with Col wild-type plants in the same assay conditions.
(F) Photographs show representative seedlings removed at 20 d from MS plates lacking or supplemented with 20 μM ABA and rearranged on agar plates.
(G) Reduced inhibition of lateral root (LR) growth by NaCl in the car5 car9 pyr1 pyl4 pyl8 pentuple mutant compared with other genetic backgrounds. Inhibition of lateral root growth by NaCl was assayed on MS plates lacking sucrose and supplemented or not with 100 mM NaCl. Asterisks indicate P < 0.05 (Student’s t test) when comparing data of mutants with Col wild-type plants in the same assay conditions or when comparing the car1 car5 pyr1 pyl4 pyl8 pentuple mutant with the pyr1 pyl4 pyl8 triple mutant. Representative seedlings were removed at 10 d and rearranged on agar plates.
[See online article for color version of this figure.]
DISCUSSION
In this work, we describe a family of C2-domain calcium binding proteins that interact with PYR/PYL ABA receptors and mediate their approach to the plasma membrane in a Ca2+-dependent manner. We suggest that the high local calcium concentration found at cellular membranes in response to different stimuli (ABA, abiotic stress, and pathogen attack) might allow CAR proteins to translocate to cell membranes in response to calcium oscillations, as we have demonstrated using in vitro assays. Therefore, CAR-interacting proteins, such as the PYR/PYL ABA receptors, are membrane-recruited in a Ca2+-dependent manner (Figure 3). We have demonstrated that PYL4 and CAR1 interact in the plasma membrane of plant cells; however, we cannot exclude interaction in other membrane compartments for the full set of 10 CAR and 14 PYR/PYL proteins. Depending on the membrane system targeted by CAR proteins, we suggest that it might affect the activity, half-life, trafficking, or targeting of the interacting PYR/PYLs by changing their subcellular localization, which would in turn affect the receptor-mediated regulation of clade A PP2Cs. Genetic evidence obtained with combined car mutants supports the notion that CAR proteins regulate at least a subset of ABA responses, so CAR-dependent transient interactions of ABA receptors with the plasma membrane affect ABA signaling.
Different abiotic stresses induce Ca2+ fluctuations, which serve as a second messenger to elicit plant responses to a changing environment (McAinsh and Pittman, 2009; Dodd et al., 2010). For instance, both osmotic stress and cold require Ca2+ signaling in order to regulate gene expression and to cope with cellular damage, such as repair of the plasma membrane (Schapire et al., 2008; Yamazaki et al., 2008; Dodd et al., 2010). Members of the CAR family are transcriptionally regulated by different abiotic stresses (Kilian et al., 2007; Supplemental Figure 6), and a homologous rice gene of the CAR family, Os-SMCP1, was previously found to confer tolerance to both abiotic and biotic stresses in transgenic Arabidopsis (Yokotani et al., 2009). Therefore, it is possible that CAR proteins, either through the regulation of ABA signaling or additional downstream targets regulated by PP2Cs, are also involved in responses to abiotic stress involving calcium fluctuations. ABA signaling involves increases in intracellular [Ca2+] levels, which have primarily been studied in guard cells (Kim et al., 2010). However, it is likely that other plant tissues responsive to ABA, such as root, also utilize Ca2+ as a second messenger of ABA signaling or that some interplay occurs between Ca2+ signaling induced by abiotic stress and ABA. For instance, it is well known that the root response to abiotic stress involves increases in cytoplasmic free calcium (Kiegle et al., 2000). Osmotic and salt stress cause increases in Ca2+ levels in the endodermis, and this tissue is the target cell layer for ABA-dependent regulation of lateral root growth in response to osmotic stress (Kiegle et al., 2000; Duan et al., 2013). Therefore, a Ca2+-mediated connection between osmotic/salt stress and ABA signaling is envisaged in the root response to environmental stress. CAR proteins might mediate such crosstalk, since car mutants showed reduced sensitivity to ABA-mediated inhibition of primary root growth, and they were also less sensitive to salt-mediated inhibition of lateral root growth (Figure 6).
It also has been reported that ABA can prime the sensitivity of Ca2+-dependent processes (Young et al., 2006). Although a molecular explanation for this priming mechanism has not been reported yet, it is possible that ABA affects the function of several Ca2+-associated proteins, such as CDPKs/CPKs, calmodulins (CaMs), CBLs, and CIPKs, which mediate plant responses to environmental stress (Hubbard et al., 2010). Indeed, some of these components are involved in the interplay between abiotic stress and ABA signaling (reviewed in Dodd et al., 2010). In this work, we provide an additional point of crosstalk for Ca2+ and ABA signaling, since we describe a family of Ca2+ binding proteins that are able to modify the subcellular localization of ABA receptors, which presumably affects their ability to regulate downstream targets (i.e., PP2Cs and SnRK2s). A lipid nanodomain plasma membrane localization of core ABA signaling components, PYL9 and ABI1, has been reported to be required to regulate the activity of CPK21 and SLAH3 and ABA signaling (Demir et al., 2013), and other PYL-PP2C targets are localized to the plasma membrane (Chérel et al., 2002; Lee et al., 2007, 2009; Geiger et al., 2009; Brandt et al., 2012; Pizzio et al., 2013).
CAR1 and CAR4 proteins also localize into the nucleus and interact there with PYR/PYL receptors, although the role of nuclear CAR-PYR/PYL interactions as well as the putative role of Ca2+ in this interaction remain to be investigated. Interestingly, other small C2-domain proteins described in plants also show a dual localization at the plasma membrane and nucleus (Wang et al., 2009), and several CBLs show both cytosolic and nuclear localization, further confirming that Ca2+ fluctuations induced by abiotic stress can be sensed in the nucleus (Batistic et al., 2010). Ca2+ signals are not exclusively from the cytosol, since they also exist in noncytosolic locations, such as mitochondria, chloroplast, and the nucleus (McAinsh and Pittman, 2009; Dodd et al., 2010). In the latter case, Ca2+ may permeate from the cytosol into the nucleus, or it can be released by different transporters from Ca2+ stores in the lumen of the nuclear envelope contiguous with the endoplasmic reticulum. Many mechanisms account for nuclear calcium signaling and calcium-regulated transcription in plants (Galon et al., 2010; Charpentier and Oldroyd, 2013). Recently, a nuclear calcium-sensing pathway required for the salt stress response has been reported in Arabidopsis, and it is well known that calcium/CaM binding transcription activators (CAMTAs) mediate abiotic stress responses (Galon et al., 2010; Guan et al., 2013). CAMTAs include a C-terminal CaM binding domain and an N-terminal domain that mediates binding to DNA cis-elements, such as ABA-responsive elements, and ABA-responsive elements confer transcriptional regulation to stress-dependent calcium-responsive genes, linking nuclear calcium signaling to transcription (Kaplan et al., 2006; Dodd et al., 2010). Calcium-mediated transcriptional regulation can be achieved through phosphorylation-dephosphorylation events. For instance, nuclear CDPKs can regulate ABA-responsive element binding factors by phosphorylation (Galon et al., 2010). Nuclear CBLs and their interacting CIPKs also regulate nuclear targets and interact with the ABA signaling components ABI1, ABI2, and ABI5 (Lyzenga et al., 2013). It is tempting to speculate that PYR/PYLs, either through the regulation of PP2C activity or downstream kinase targets, might link calcium nuclear signals perceived through CAR proteins to transcriptional regulation.
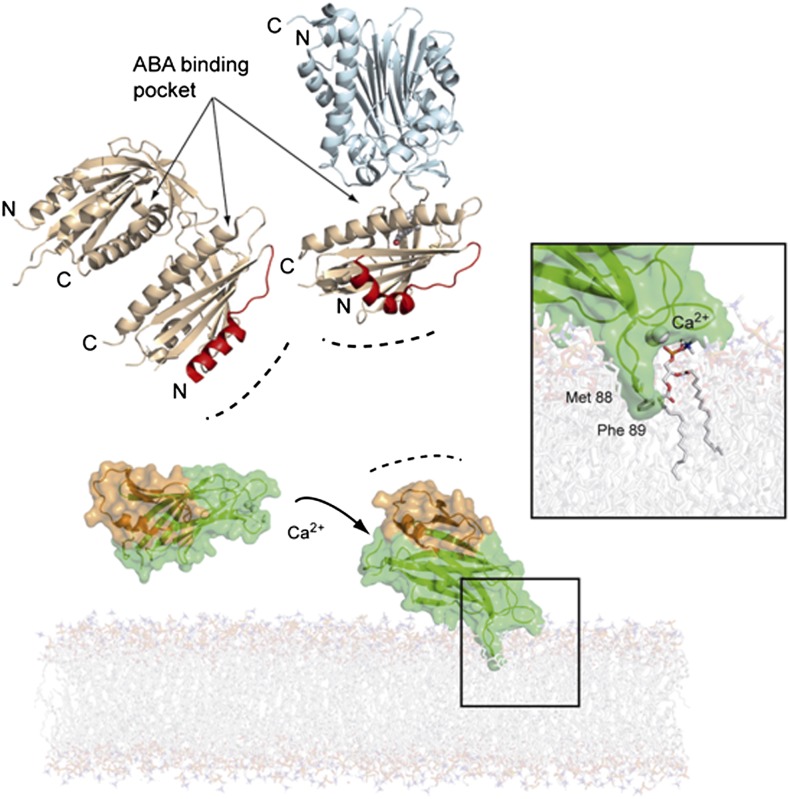
The analyses of our structural data and the available structural information on the interaction of various C2 domains with membranes (Davletov et al., 1998; Medkova and Cho, 1998; Verdaguer et al., 1999; Frazier et al., 2003; Kohout et al., 2003) allowed us to simulate the CAR4 interaction with a phospholipid bilayer (Figure 7). C2 domains use a combined mechanism for membrane binding based on phospholipid headgroup binding, electrostatic interaction, and membrane insertion of hydrophobic residues (Lemmon, 2008). In this model, the calcium atoms bridge CAR4 with the phosphate moiety of phospholipids, and the hydrophobic tip of loop L3 (Met88-Phe89) is inserted into the membrane, as has been described for other C2 domains (Cho and Stahelin, 2006; Ausili et al., 2011). In this situation, the characteristic α1βAβB CAR-signature domain is fully solvent-accessible, as it is placed opposite to the calcium binding site. This suggests a role for the α1βAβB CAR-signature domain in the recruitment of the PYR/PYL receptor to an area near the membrane. Interestingly, the protein face opposite to Ca2+ binding loops has been reported to be involved in C2-mediated protein-protein interactions (Law et al., 2010). For instance, membrane binding by lymphocyte perforin relies on the Ca2+ binding loops of a C2 domain, which in the opposite face is linked to a membrane attack complex/perforin-like fold (Law et al., 2010). We have also shown that the N-terminal helix of ABA receptors is involved in CAR binding using limited deletion analysis (Figure 1B). This receptor area is opposite to the ABA and PP2C phosphatase binding sites and is not involved in the receptor-phosphatase interaction or receptor dimer formation (Figure 7). This suggests that CAR proteins might bind a functional ABA receptor or facilitate the formation of receptor-ABA-PP2C ternary complexes at cell membranes. Since early events of ABA signaling are linked to membrane proteins regulated by PP2Cs and SnRK2s, the reported interaction might facilitate the connection of ABA perception to downstream regulatory events.
Figure 7.
A Working Model for Calcium-Dependent CAR4 Membrane Binding and Its Interaction with the PYR/PYL ABA Receptors.
CAR4, represented as a semitransparent surface, has been docked into a phosphatidylcholine model membrane. The dimeric structures of the apo-PYL1 receptor (Protein Data Bank code 3KAY) and its complex with ABA and ABI1 phosphatase (Protein Data Bank code 3JRQ) are displayed as wheat and pale cyan ribbons, respectively. The CAR4 α1β1β2 extra-domain and the N-terminal receptor-interacting areas are highlighted in orange and red, respectively. The inset represents a closeup of the modeled CAR4 calcium-phospholipid complex. Membrane insertion of CAR4 exposes the CAR-signature extra-domain to the cytosol.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana plants were grown as described by Pizzio et al. (2013). CAR knockout insertion lines car1 (SALK_080173.54.40.X), car4 (SM 3-1727), car5 (SAIL_802_B08), and car9 (SALK_088115.56.00.X) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk). To confirm and identify homozygous T-DNA individuals, seedlings of each insertion line were grown individually, and DNA from each plant was extracted and submitted to PCR-mediated genotyping using the primers described in Supplemental Table 2. In order to generate 35S:3HA-CAR1-overexpressing lines, the CAR1 coding sequence was cloned into pCR8/GW/TOPO entry vector (Invitrogen) and recombined by LR reaction into the Gateway-compatible pALLIGATOR2 vector (Bensmihen et al., 2004). The pALLIGATOR2-CAR1 construct was transferred to Agrobacterium tumefaciens C58C1 (pGV2260) (Deblaere et al., 1985) by electroporation and used to transform Col wild-type plants by the floral dip method (Clough and Bent, 1998). T1 transgenic seeds were selected based on GFP visualization and sown in soil to obtain the T2 generation. Homozygous T3 progeny were used for further studies, and the expression of HA-tagged protein was verified by immunoblot analysis using anti-HA peroxidase (Roche).
Y2H Assays
Protocols were similar to those described previously (Saez et al., 2008). Briefly, an oligo(dT)-primed cDNA library prepared in plasmid pACT2 using mRNA from an Arabidopsis cell suspension was kindly provided by K. Salchert (Saez et al., 2008). The library was shuttled to yeast AH109 by cotransformation with pGBKT7-PYL4. Yeast transformants were pooled, and clones able to grow in the absence of exogenous ABA in medium lacking histidine and adenine were selected. Yeast plasmids were extracted, sequenced, and retransformed in yeast cells to recapitulate the phenotype. Arabidopsis ABA receptors were fused by Gateway recombination to the GAL4 DNA binding domain in pGBKT7GW. N-terminal deletions of PYL4, PYL6, and PYL8 were generated using the primers described in Supplemental Table 2. The CAR1 prey was fused to the GAL4 activation domain in pACT2 vector.
Transient Protein Expression in Nicotiana benthamiana
Agrobacterium infiltration of N. benthamiana leaves was performed basically as described by Voinnet et al. (2003). Constructs to investigate the subcellular localization of CAR and PYL4 proteins were used in pMDC83 and pMDC43 vectors, respectively. The constructs encoding the plasma membrane markers OFP-TM23 and SCFPC-CIPK24/CBL1-SCFPN were reported by Batistic et al. (2012) and Waadt et al. (2008), respectively. To investigate the interaction of CAR and PYR/PYL proteins in planta, we used the pSPYNE-35S and pYFPC43 vectors (Walter et al., 2004; Belda-Palazón et al., 2012). The coding sequences of CAR1, CAR4, and CAR5 were doubly digested with BamHI-EcoRV and cloned into BamHI-SmaI pSPYNE-35S. The coding sequences of PYR1, PYL1, PYL4, PYL6, and PYL8 were recombined by LR reaction from pCR8 entry vector to pYFPC43 destination vector. The different binary vectors described above were introduced into Agrobacterium strain C58C1 (pGV2260) (Deblaere et al., 1985) by electroporation, and transformed cells were selected on Luria-Bertani plates supplemented with kanamycin (50 mg/L). Then, they were grown in liquid Luria-Bertani medium to late exponential phase, and cells were harvested by centrifugation and resuspended in 10 mM MES-KOH, pH 5.6, containing 10 mM MgCl2 and 150 mM acetosyringone to an OD600 of 1. These cells were mixed with an equal volume of Agrobacterium strain C58C1 (pCH32 35S:p19) expressing the silencing suppressor p19 of Tomato bushy stunt virus (Voinnet et al., 2003) so that the final density of Agrobacterium solution was ∼1. Bacteria were incubated for 3 h at room temperature and then injected into young fully expanded leaves of 4-week-old N. benthamiana plants. Leaves were examined 48 to 72 h after infiltration using confocal laser scanning microscopy.
Confocal Laser Scanning Microscopy
Confocal imaging was performed using a Zeiss LSM 780 AxioObserver.Z1 laser scanning microscope with a C-Apochromat 40×/1.20 W corrective water immersion objective. The following fluorophores, which were excited and fluorescence emission detected by frame switching in the single or multiple tracking mode at the indicated wavelengths, were used in N. benthamiana leaf infiltration experiments: SCFP (405 nm/464 to 486 nm), GFP (488 nm/500 to 530 nm), YFP (488 nm/529 to 550 nm), and OFP (561 nm/575 to 600 nm). Pinholes were adjusted to 1 air unit for each wavelength. Postacquisition image processing was performed using ZEN (ZEISS Efficient Navigation) Lite 2012 imaging software and ImageJ (http://rsb.info.gov/ij/).
Epifluorescence confocal images of epidermal N. benthamiana leaves coinfiltrated with the constructs described were merged to quantitatively estimate the colocalization of fluorescent markers (French et al., 2008). Statistical analyses for fluorescence colocalization were performed through the determination of linear Pearson’s and nonlinear Spearman’s correlation coefficients between fluorescent signals. Nuclear fluorescent signals of GFP, reconstituted YFP, CAR-GFP, and GFP-PYL4 proteins were not taken into account for the colocalization analysis.
To quantify the relative fluorescence intensities of BiFC experiments, all images were captured using the same laser, pinhole, and gain settings of the confocal microscope to maintain high reproducibility of the data (laser, 2.0%; pinhole diameter, 34 μm; master gain, 740; digital gain, 1.00; digital offset, 0.00) as well as zoom factor (1.2) covering two to three living cells per image. As negative controls in the interaction assays, Agrobacterium expressing either CAR1-myc-YFPN or CAR4-myc-YFPN was coinfiltrated with YFPC-OST11-280 (Vlad et al., 2009). Image quantification of relative fluorescence intensities was performed using ImageJ software by measuring the fluorescence intensity corrected for mean background fluorescence subtracted from corresponding areas showing no green fluorescence. Each BiFC experiment was scanned and measured in 25 randomly chosen microscopic fields (n = 25) and repeated tree times.
Biochemical Fractionation, Protein Extraction, Analysis, and Immunoprecipitation
Constructs to express GFP- or HA-tagged proteins were generated in pMDC43/83 or pALLIGATOR2 vector, respectively. The generation of PYL4 and PYL8 constructs in pALLIGATOR2 has been described previously (Antoni et al., 2013; Pizzio et al., 2013), and similar constructs were used for PYR1, PYL1, and PYL6. The different binary vectors described above were introduced into Agrobacterium strain C58C1 (pGV2260) and used for infiltration of N. benthamiana leaves. Protein extracts for immunodetection experiments were prepared from N. benthamiana leaves 48 to 72 h after infiltration. Plant material (∼100 mg) for direct protein gel blot analysis was extracted in 2× Laemmli buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 2% mercaptoethanol, and 0.001% bromophenol blue), and proteins were run on a 10% SDS-PAGE gel and analyzed by immunoblotting. Cytosolic and microsomal fractionation of GFP- or HA-tagged proteins was performed as described previously (Antoni et al., 2012). The microsomal fractionation procedure used a lysis buffer supplemented with 25 mM CaCl2 (Antoni et al., 2012). Nuclear fractionation was performed as described previously (Saez et al., 2008; Antoni et al., 2012), and the soluble nuclear fraction was used for immunoprecipitation experiments. Soluble proteins from the nuclear fraction were immunoprecipitated using superparamagnetic micro MACS beads coupled to monoclonal anti-GFP antibody according to the manufacturer’s instructions (Miltenyi Biotec). Purified immunocomplexes were eluted in Laemmli buffer, boiled, and run on a 10% SDS-PAGE gel. Proteins immunoprecipitated with anti-GFP antibody were transferred onto Immobilon-P membranes (Millipore) and probed with anti-HA peroxidase to detect the coIP of HA-tagged receptors. Immunodetection of GFP fusion proteins was performed with an anti-GFP monoclonal antibody (clone JL-8; Clontech) as primary antibody and ECL anti-mouse peroxidase (GE Healthcare) as secondary antibody. Antibodies were used at a 1:10,000 dilution. Detection was performed using the ECL Advance protein gel blotting chemiluminiscent detection kit (GE Healthcare). Image capture was done using the image analyzer LAS3000, and quantification of the protein signal was done using Image Guache version 4.0 software.
Phospholipid Binding Assays
Calcium-dependent protein binding to phospholipid vesicles was assessed as described by Schapire et al. (2008). A 25:75 (w/w) mixture of phosphatidylserine:phosphatidylcholine (Sigma-Aldrich) was prepared in chloroform and dried under a stream of nitrogen to obtain a thin layer. The dried lipids were resuspended in buffer A (100 mM NaCl, 50 mM HEPES, pH 6.8, and 4 mM EGTA) and mixed by vortexing for 20 min. We pelleted the large multilamellar vesicles by 20 min of centrifugation at 16,000g, then they were resuspended in 1 mL of buffer A supplemented or not with the indicated calcium concentration. Free calcium concentrations were calculated using the WEBMAXC program (http://www.stanford.edu/~cpatton/maxc.html). The vesicles (∼100 μg of phospholipids) were used immediately after preparation and mixed with the indicated His-tagged recombinant proteins (5 μg). Next, they were incubated with gentle shaking (250 rpm) on a platform shaker. The vesicles and the bound proteins were pelleted by centrifugation for 10 min at 16,000g at 4°C, and pellets were washed twice with 0.5 mL of buffer A. Proteins that were bound to the vesicles were revealed by immunoblot analysis using anti-His antibody and ECL anti-mouse peroxidase (GE Healthcare) as secondary antibody. Detection was performed using the ECL Advance protein gel blotting detection kit (GE Healthcare). Quantification of the binding was determined using ImageJ software, and mathematical analysis of calcium binding was performed based on nonlinear least-squares fitting to the three-parameter Hill’s equation using Sigma Plot 12 software.
Protein Preparation and Crystallization
CAR4 was obtained from cultures of Escherichia coli, purified to homogeneity, and crystallized as described (Diaz et al. 2011). Briefly, CAR4 coding sequence was cloned into the pETM11 vector, and the overexpressed His-tagged protein was purified to homogeneity in a single chromatographic step. Prior to crystallization, CAR4 protein was dialyzed to a buffer containing 20 mM Tris-HCl, pH 8.5, 200 mM NaCl, and 0.1 mM CaCl2. The stock protein was concentrated to 8.0 mg/mL. CAR4 prismatic crystals were grown in 0.01 M LiCl2, 0.1 M MES, pH 6, and 20% (w/v) polyethylene glycol 6K. The crystals were mounted in a fiber loop and soaked in cryoprotectant consisting of mother liquor containing 20% (w/v) polyethylene glycol 400 and flash-cooled in liquid nitrogen. CAR1 coding sequence was cloned into pCOLADuet-1 (Novagen) through BamHI digestion, and the overexpressed His-tagged protein was purified to homogeneity in a single chromatographic step. CAR1D22A D27A, CAR4D85A D87A, and CAR4ΔED mutants were generated using the PCR-overlap extension procedure using the oligonucleotides described in Supplemental Table 2. CAR1D22A D27A and CAR4D85A D87A were cloned into pETM11, whereas CAR4ΔED was cloned into pSPYNE-35S. PYR/PYL proteins were prepared as described previously (Santiago et al., 2009).
Data Collection and Structure Determination and Refinement
CAR4 x-ray diffraction data were collected in an ADSC detector using the European Synchrotron Radiation Facility radiation source at 0.94 Å wavelengths at the ID14.4 beamline. The refinement statistics and data analyses are summarized in Supplemental Table 1 and Supplemental Methods, respectively.
Seed Germination and Seedling Establishment Assays
After surface sterilization of the seeds, stratification was conducted in the dark at 4°C for 3 d. Approximately 100 seeds of each genotype were sown on MS plates supplemented with different ABA concentrations per experiment. To score seed germination, radicle emergence was analyzed at 72 h after sowing. Seedling establishment was scored as the percentage of seeds that developed green expanded cotyledons and the first pair of true leaves at 5 or 7 d.
Root and Shoot Growth Assays
Seedlings were grown on vertically oriented MS plates for 4 to 5 d. Afterward, 20 plants were transferred to new MS plates lacking or supplemented with the indicated concentrations of ABA. The plates were scanned on a flatbed scanner after 10 or 20 d to produce image files suitable for quantitative analysis of root growth using ImageJ version 1.37. As an indicator of shoot growth, fresh weight was measured after 21 d. Inhibition of lateral root growth by NaCl was assayed on MS plates lacking sucrose and supplemented or not with 100 mM NaCl. After 10 d, plates were scanned as described above, and total lateral root growth per plant (n = 30) was measured.
Accession Numbers
The atomic coordinates and structure factor amplitudes of CAR4 in complex with calcium have been deposited in the Protein Data Bank (www.pdb.org) with identifier 4v29. Arabidopsis Genome Initiative locus identifiers for CAR1, CAR2, CAR3, CAR4, CAR5, CAR6, CAR7, CAR8, CAR9, and CAR10 are At5g37740, At1g66360, At1g73580, At3g17980, At1g48590, At1g70800, At1g70810, At1g23140, At1g70790, and At2g01540, respectively. Arabidopsis Genome Initiative identifiers for PYR1, PYL1, PYL4, PYL6, and PYL8, are At4g17870, At5g46790, At2g38310, At2g40330, and At5g53160, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Scheme of Some Representative Proteins Harboring C2 Domains in Arabidopsis.
Supplemental Figure 2. Amino Acid Sequence and Secondary Structure Alignment of Arabidopsis CAR Proteins.
Supplemental Figure 3. Amino Acid Sequence Alignment of Representative Members of the CAR Family in Arabidopsis, Tomato (Solanum lycopersicum), and Rice (Oryza sativa).
Supplemental Figure 4. Scheme of CAR1, CAR4, CAR5, and CAR9 Genes and Location of the Corresponding T-DNA Insertion in car Mutants.
Supplemental Figure 5. Photographs Showing GUS Expression Driven by ProCAR1:GUS Gene in Different Tissues and Developmental Stages.
Supplemental Figure 6. Induction of CAR Genes by Cold, Osmotic, and Salt Stress in Shoot or Root Tissues.
Supplemental Table 1. Data Collection and Refinement Statistics for CAR4 Structure.
Supplemental Table 2. List of Oligonucleotides Used in This Work.
Supplementary Material
Acknowledgments
We thank Joerg Kudla (University of Münster) for kindly providing plasma membrane markers. This work was supported by the Ministerio de Ciencia e Innovacion, Fondo Europeo de Desarrollo Regional, and Consejo Superior de Investigaciones Cientificas (Grants BIO2011-23446 to P.L.R and BFU2011-25384 to A.A.; fellowships to L.R., R.A., and A.C.I.-G.; BES-2009-016569; JAE-DOC contract to M.G.-G.) as well as the Senacyt-Ifarhu (Panama) (fellowship to M.D.).
AUTHOR CONTRIBUTIONS
P.L.R. conceived the project. L.R., M.G.-G., M.D., A.R., J.M.M., A.A., and P.L.R. designed research. L.R., M.G.-G., M.D., A.R., A.C.I.-G., M.P.-L., M.A.F., R.A., D.F., J.A.M., J.M.M., A.A., and P.L.R. performed research. L.R., M.G.G., M.D., A.R., A.C.I.-G., M.P.L., M.A.F., R.A., D.F., J.A.M., J.M.M., A.A., and P.L.R. analyzed data. A.A. and P.L.R. wrote the article.
Glossary
- ABA
abscisic acid
- Y2H
yeast-two hybrid
- BiFC
bimolecular fluorescence complementation
- coIP
coimmunoprecipitation
- MS
Murashige and Skoog
- Col
Columbia
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Peirats-Llobet M., Pizzio G.A., Fernandez M.A., De Winne N., De Jaeger G., Dietrich D., Bennett M.J., Rodriguez P.L. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161: 931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni R., Gonzalez-Guzman M., Rodriguez L., Rodrigues A., Pizzio G.A., Rodriguez P.L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol. 158: 970–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausili A., Corbalán-García S., Gómez-Fernández J.C., Marsh D. (2011). Membrane docking of the C2 domain from protein kinase Cα as seen by polarized ATR-IR. The role of PIP2. Biochim. Biophys. Acta 1808: 684–695. [DOI] [PubMed] [Google Scholar]
- Batistic O., Rehers M., Akerman A., Schlücking K., Steinhorst L., Yalovsky S., Kudla J. (2012). S-Acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 22: 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic O., Waadt R., Steinhorst L., Held K., Kudla J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61: 211–222. [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B., Ruiz L., Martí E., Tárraga S., Tiburcio A.F., Culiáñez F., Farràs R., Carrasco P., Ferrando A. (2012). Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS ONE 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131. [DOI] [PubMed] [Google Scholar]
- Brandt B., Brodsky D.E., Xue S., Negi J., Iba K., Kangasjarvi J., Ghassemian M., Stephan A.B., Hu H., Schroeder J.I. (2012). Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc. Natl. Acad. Sci. USA 109: 10593–10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., et al. (2013). An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Res. 23: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M., Oldroyd G.E. (2013). Nuclear calcium signaling in plants. Plant Physiol. 163: 496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chérel I., Michard E., Platet N., Mouline K., Alcon C., Sentenac H., Thibaud J.B. (2002). Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W., Bittova L., Stahelin R.V. (2001). Membrane binding assays for peripheral proteins. Anal. Biochem. 296: 153–161. [DOI] [PubMed] [Google Scholar]
- Cho W., Stahelin R.V. (2005). Membrane-protein interactions in cell signaling and membrane trafficking. Annu. Rev. Biophys. Biomol. Struct. 34: 119–151. [DOI] [PubMed] [Google Scholar]
- Cho W., Stahelin R.V. (2006). Membrane binding and subcellular targeting of C2 domains. Biochim. Biophys. Acta 1761: 838–849. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Davletov B., Perisic O., Williams R.L. (1998). Calcium-dependent membrane penetration is a hallmark of the C2 domain of cytosolic phospholipase A2 whereas the C2A domain of synaptotagmin binds membranes electrostatically. J. Biol. Chem. 273: 19093–19096. [DOI] [PubMed] [Google Scholar]
- Deblaere R., Bytebier B., De Greve H., Deboeck F., Schell J., Van Montagu M., Leemans J. (1985). Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res. 13: 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F., Horntrich C., Blachutzik J.O., Scherzer S., Reinders Y., Kierszniowska S., Schulze W.X., Harms G.S., Hedrich R., Geiger D., Kreuzer I. (2013). Arabidopsis nanodomain-delimited ABA signaling pathway regulates the anion channel SLAH3. Proc. Natl. Acad. Sci. USA 110: 8296–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M., Rodriguez L., Gonzalez-Guzman M., Martínez-Ripoll M., Albert A. (2011). Crystallization and preliminary crystallographic analysis of a C2 protein from Arabidopsis thaliana. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67: 1575–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620. [DOI] [PubMed] [Google Scholar]
- Duan L., Dietrich D., Ng C.H., Chan P.M., Bhalerao R., Bennett M.J., Dinneny J.R. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F., Antoni R., Betz K., Santiago J., Gonzalez-Guzman M., Rodriguez L., Rubio S., Park S.Y., Cutler S.R., Rodriguez P.L., Márquez J.A. (2011a). Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol. 156: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupeux F., et al. (2011b). A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J. 30: 4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier A.A., Roller C.R., Havelka J.J., Hinderliter A., Cafiso D.S. (2003). Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry 42: 96–105. [DOI] [PubMed] [Google Scholar]
- French A.P., Mills S., Swarup R., Bennett M.J., Pridmore T.P. (2008). Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3: 619–628. [DOI] [PubMed] [Google Scholar]
- Galon Y., Finkler A., Fromm H. (2010). Calcium-regulated transcription in plants. Mol. Plant 3: 653–669. [DOI] [PubMed] [Google Scholar]
- Geiger D., Maierhofer T., Al-Rasheid K.A., Scherzer S., Mumm P., Liese A., Ache P., Wellmann C., Marten I., Grill E., Romeis T., Hedrich R. (2011). Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci. Signal. 4: ra32. [DOI] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Marten I., Ache P., Matschi S., Liese A., Wellmann C., Al-Rasheid K.A., Grill E., Romeis T., Hedrich R. (2010). Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc. Natl. Acad. Sci. USA 107: 8023–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D., Scherzer S., Mumm P., Stange A., Marten I., Bauer H., Ache P., Matschi S., Liese A., Al-Rasheid K.A., Romeis T., Hedrich R. (2009). Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 106: 21425–21430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernández M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., Rodriguez P.L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24: 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Wu J., Yue X., Zhang Y., Zhu J. (2013). A nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLoS Genet. 9: e1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Valero M., Ferrer-Orta C., Querol-Audí J., Marin-Vicente C., Fita I., Gómez-Fernández J.C., Verdaguer N., Corbalán-García S. (2009). Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc. Natl. Acad. Sci. USA 106: 6603–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Valero M., Marín-Vicente C., Gómez-Fernández J.C., Corbalán-García S. (2007). The C2 domains of classical PKCs are specific PtdIns(4,5)P2-sensing domains with different affinities for membrane binding. J. Mol. Biol. 371: 608–621. [DOI] [PubMed] [Google Scholar]
- Guo Y., Xiong L., Song C.P., Gong D., Halfter U., Zhu J.K. (2002). A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 3: 233–244. [DOI] [PubMed] [Google Scholar]
- Hao Q., Yin P., Li W., Wang L., Yan C., Lin Z., Wu J.Z., Wang J., Yan S.F., Yan N. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42: 662–672. [DOI] [PubMed] [Google Scholar]
- Hubbard K.E., Nishimura N., Hitomi K., Getzoff E.D., Schroeder J.I. (2010). Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 24: 1695–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan B., Davydov O., Knight H., Galon Y., Knight M.R., Fluhr R., Fromm H. (2006). Rapid transcriptome changes induced by cytosolic Ca2+ transients reveal ABRE-related sequences as Ca2+-responsive cis elements in Arabidopsis. Plant Cell 18: 2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegle E., Moore C.A., Haseloff J., Tester M.A., Knight M.R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23: 267–278. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kim C.Y., et al. (2003). Rice C2-domain proteins are induced and translocated to the plasma membrane in response to a fungal elicitor. Biochemistry 42: 11625–11633. [DOI] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout S.C., Corbalán-García S., Gómez-Fernández J.C., Falke J.J. (2003). C2 domain of protein kinase C alpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry 42: 1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60: 2256–2268. [DOI] [PubMed] [Google Scholar]
- Law R.H., et al. (2010). The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 468: 447–451. [DOI] [PubMed] [Google Scholar]
- Lee S.C., Lan W., Buchanan B.B., Luan S. (2009). A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 106: 21419–21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Lan W.Z., Kim B.G., Li L., Cheong Y.H., Pandey G.K., Lu G., Buchanan B.B., Luan S. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. USA 104: 15959–15964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A. (2008). Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9: 99–111. [DOI] [PubMed] [Google Scholar]
- Lynch T., Erickson B.J., Finkelstein R.R. (2012). Direct interactions of ABA-insensitive (ABI)-clade protein phosphatase (PP) 2Cs with calcium-dependent protein kinases and ABA response element-binding bZIPs may contribute to turning off ABA response. Plant Mol. Biol. 80: 647–658. [DOI] [PubMed] [Google Scholar]
- Lyzenga W.J., Liu H., Schofield A., Muise-Hennessey A., Stone S.L. (2013). Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 64: 2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- McAinsh M.R., Pittman J.K. (2009). Shaping the calcium signature. New Phytol. 181: 275–294. [DOI] [PubMed] [Google Scholar]
- Medkova M., Cho W. (1998). Differential membrane-binding and activation mechanisms of protein kinase C-alpha and -epsilon. Biochemistry 37: 4892–4900. [DOI] [PubMed] [Google Scholar]
- Melcher K., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I., Emr S.D. (2013). A Nobel Prize for membrane traffic: Vesicles find their journey’s end. J. Cell Biol. 203: 559–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., et al. (2009). Structural basis of abscisic acid signalling. Nature 462: 609–614. [DOI] [PubMed] [Google Scholar]
- Nishimura N., Sarkeshik A., Nito K., Park S.Y., Wang A., Carvalho P.C., Lee S., Caddell D.F., Cutler S.R., Chory J., Yates J.R., Schroeder J.I. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y. (1988). The molecular heterogeneity of protein kinase C and its implications for cellular regulation. Nature 334: 661–665. [DOI] [PubMed] [Google Scholar]
- Okamoto M., Peterson F.C., Defries A., Park S.Y., Endo A., Nambara E., Volkman B.F., Cutler S.R. (2013). Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc. Natl. Acad. Sci. USA 110: 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisic O., Fong S., Lynch D.E., Bycroft M., Williams R.L. (1998). Crystal structure of a calcium-phospholipid binding domain from cytosolic phospholipase A2. J. Biol. Chem. 273: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Pizzio G.A., Rodriguez L., Antoni R., Gonzalez-Guzman M., Yunta C., Merilo E., Kollist H., Albert A., Rodriguez P.L. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Südhof T.C. (1998). C2-domains, structure and function of a universal Ca2+-binding domain. J. Biol. Chem. 273: 15879–15882. [DOI] [PubMed] [Google Scholar]
- Rodrigues A., et al. (2013). ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25: 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Robert N., Maktabi M.H., Schroeder J.I., Serrano R., Rodriguez P.L. (2006). Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 141: 1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A., Rodrigues A., Santiago J., Rubio S., Rodriguez P.L. (2008). HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Rodrigues A., Saez A., Rubio S., Antoni R., Dupeux F., Park S.Y., Márquez J.A., Cutler S.R., Rodriguez P.L. (2009). Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 60: 575–588. [DOI] [PubMed] [Google Scholar]
- Sato A., Sato Y., Fukao Y., Fujiwara M., Umezawa T., Shinozaki K., Hibi T., Taniguchi M., Miyake H., Goto D.B., Uozumi N. (2009). Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 424: 439–448. [DOI] [PubMed] [Google Scholar]
- Schapire A.L., Voigt B., Jasik J., Rosado A., Lopez-Cobollo R., Menzel D., Salinas J., Mancuso S., Valpuesta V., Baluska F., Botella M.A. (2008). Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20: 3374–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.D., Pawson T. (2009). Cell signaling in space and time: Where proteins come together and when they’re apart. Science 326: 1220–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet B.T., Dikic I., Zhou M.M., Pawson T. (2006). Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7: 473–483. [DOI] [PubMed] [Google Scholar]
- Sirichandra C., Gu D., Hu H.C., Davanture M., Lee S., Djaoui M., Valot B., Zivy M., Leung J., Merlot S., Kwak J.M. (2009). Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 583: 2982–2986. [DOI] [PubMed] [Google Scholar]
- Swanson S.J., Choi W.G., Chanoca A., Gilroy S. (2011). In vivo imaging of Ca2+, pH, and reactive oxygen species using fluorescent probes in plants. Annu. Rev. Plant Biol. 62: 273–297. [DOI] [PubMed] [Google Scholar]
- Verdaguer N., Corbalan-Garcia S., Ochoa W.F., Fita I., Gómez-Fernández J.C. (1999). Ca2+ bridges the C2 membrane-binding domain of protein kinase Calpha directly to phosphatidylserine. EMBO J. 18: 6329–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Rubio S., Rodrigues A., Sirichandra C., Belin C., Robert N., Leung J., Rodriguez P.L., Laurière C., Merlot S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G.K., Barr F.A. (2013). Cell organelles. Curr. Opin. Cell Biol. 25: 403–405. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Waadt R., Schmidt L.K., Lohse M., Hashimoto K., Bock R., Kudla J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56: 505–516. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang X., Li Q., Niu X., Chen H., Xu L., Qi C. (2009). Characterization of a canola C2 domain gene that interacts with PG, an effector of the necrotrophic fungus Sclerotinia sclerotiorum. J. Exp. Bot. 60: 2613–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T., Kawamura Y., Minami A., Uemura M. (2008). Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20: 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N., Ichikawa T., Kondou Y., Maeda S., Iwabuchi M., Mori M., Hirochika H., Matsui M., Oda K. (2009). Overexpression of a rice gene encoding a small C2 domain protein OsSMCP1 increases tolerance to abiotic and biotic stresses in transgenic Arabidopsis. Plant Mol. Biol. 71: 391–402. [DOI] [PubMed] [Google Scholar]
- Young J.J., Mehta S., Israelsson M., Godoski J., Grill E., Schroeder J.I. (2006). CO2 signaling in guard cells: Calcium sensitivity response modulation, a Ca2+-independent phase, and CO2 insensitivity of the gca2 mutant. Proc. Natl. Acad. Sci. USA 103: 7506–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Aravind L. (2010). Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene 469: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.