A gene module responsible for morphological diversification of leaf form within a species may have been co-opted during plant evolution to regulate morphological variation among species.
Abstract
Plants show leaf form alteration in response to changes in the surrounding environment, and this phenomenon is called heterophylly. Although heterophylly is seen across plant species, the regulatory mechanisms involved are largely unknown. Here, we investigated the mechanism underlying heterophylly in Rorippa aquatica (Brassicaceae), also known as North American lake cress. R. aquatica develops pinnately dissected leaves in submerged conditions, whereas it forms simple leaves with serrated margins in terrestrial conditions. We found that the expression levels of KNOTTED1-LIKE HOMEOBOX (KNOX1) orthologs changed in response to changes in the surrounding environment (e.g., change of ambient temperature; below or above water) and that the accumulation of gibberellin (GA), which is thought to be regulated by KNOX1 genes, also changed in the leaf primordia. We further demonstrated that exogenous GA affects the complexity of leaf form in this species. Moreover, RNA-seq revealed a relationship between light intensity and leaf form. These results suggest that regulation of GA level via KNOX1 genes is involved in regulating heterophylly in R. aquatica. The mechanism responsible for morphological diversification of leaf form among species may also govern the variation of leaf form within a species in response to environmental changes.
INTRODUCTION
Plants display amazing morphological diversity and leaves are among the most diverse organs in plants (Bell, 2008; Nicotra et al., 2011; Tsukaya, 2014). Recent studies have revealed the molecular mechanisms underlying leaf form diversification between species (Kimura et al., 2008; Piazza et al., 2010; Yamaguchi et al., 2010; Vlad et al., 2014). However, morphological variation within a single plant is also seen in nature. For instance, many plants can alter their leaf form in response to surrounding environmental conditions (Arber, 1920). In the amphibious angiosperm Hippuris vulgaris (Plantaginaceae), small and linear leaves with a branched leaf venation develop on aerial shoots, while narrower leaves with an unbranched midvein develop on submerged shoots (Kane and Albert, 1987). In Sium latifolium (Apiaceae), compound leaves with toothed leaflets develop on aerial shoots, while compound leaves with weaker expansion of lamina are seen on submerged shoots (Arber, 1920).
Such phenotypic alteration within individuals in a species in response to their surrounding environment is known as phenotypic plasticity (Pigliucci, 2010; Donohue, 2013), and phenotypic plasticity in leaf form within a single plant in response to environmental conditions is called heterophylly (Zotz et al., 2011). Some leaf form alterations are thought to be an adaptive response to the environment (Cook and Johnson, 1968). In addition, heterophylly is seen across diverse plant species (Wells and Pigliucci, 2000; Wanke, 2011; Nakayama et al., 2012a). Hence, heterophylly provides a good model to understand gene-environment interactions (Pigliucci, 2010).
Morphological changes in heterophyllous plants have been described (Arber, 1920; Fassett, 1930), and more recent studies have revealed a hormonal influence on heterophylly (Kane and Albert, 1987; Goliber and Feldman, 1990). For example, studies with Ludwigia arcuata (Onagraceae) suggested that ethylene induces cell elongation in leaves (Sato et al., 2008) and that ethylene and abscisic acid (ABA) affect leaf form (Kuwabara et al., 2003). However, the exact molecular mechanisms regulating these changes remain largely unclear. Revealing the relationship among environmental fluctuation, plant hormones, and genes is critical for understanding the morphological switch induced in heterophylly.
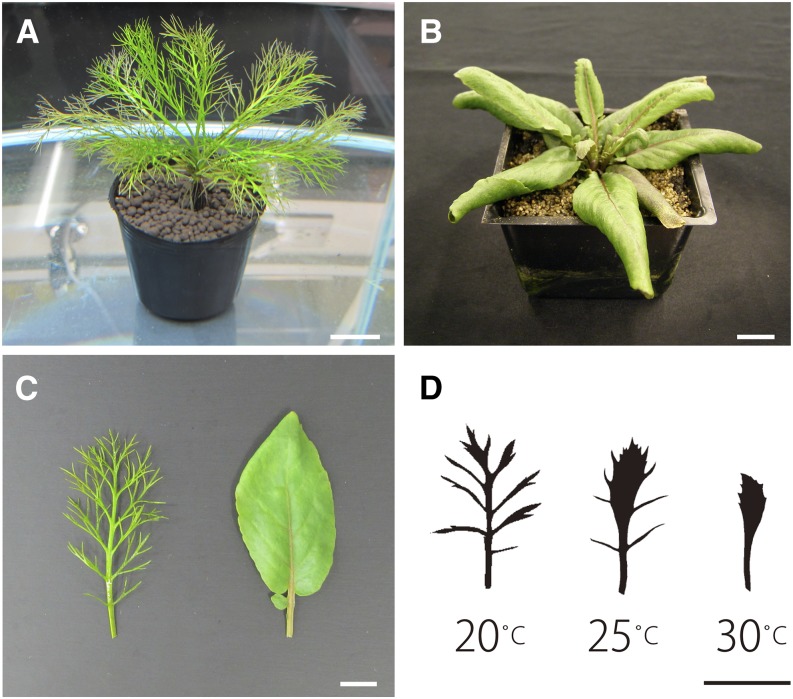
Rorippa aquatica (Brassicaceae) is a perennial herbaceous and semiaquatic plant whose habitat includes bays of lakes, ponds, and streams in North America (La Rue, 1943). Of particular interest is the distinct heterophylly between submerged and terrestrial conditions that R. aquatica shows in nature (Figures 1A and 1B). In submerged conditions, pinnately deeply dissected leaves develop. On the other hand, in terrestrial conditions, simple leaves with serrated or lobed margins develop (Fassett, 1930; Figure 1C). Molecular phylogenetic analysis revealed that R. aquatica is a member of the Cardamineae and closely related to Cardamine hirsuta (Brassicaceae) (Les, 1994; Couvreur et al., 2010), a well-characterized model plant for studies of compound leaf development (Hay and Tsiantis, 2006; Canales et al., 2010; Hay et al., 2014). In addition, R. aquatica also belongs to the same family as Arabidopsis thaliana, the most researched model in molecular plant biology, which displays simple leaf morphology. Therefore, the extensive knowledge on leaf development accumulated from these compound- and simple-leafed model species may help to reveal the molecular mechanism regulating heterophylly in R. aquatica. Indeed, studies on submergence tolerance in Rorippa species have been successfully performed based on comparisons with Arabidopsis (Akman et al., 2012; Sasidharan et al., 2013), and phylogenetic and modeling-based analyses have also been reported (Nakamasu et al., 2014; Nakayama et al., 2014). These features make R. aquatica an excellent model species to investigate the mechanism regulating heterophylly.
Figure 1.
Gross Morphology of R. aquatica.
(A) Side view of a shoot under submerged condition.
(B) Side view of a shoot in terrestrial condition.
(C) Mature leaf morphology. Left, submerged condition; right, terrestrial condition.
(D) Effects of change in water temperature on leaf morphology. Left, 20°C; center, 25°C; right, 30°C. All silhouettes of leaves are based on photographic images of the 7th leaves.
Bars = 2 cm in (A) and (B) and 1 cm in (C) and (D).
The molecular framework of the mechanisms behind simple and compound leaf development in Brassicaceae has so far been investigated with Arabidopsis and C. hirsuta (Hay and Tsiantis, 2006; Barkoulas et al., 2008; Moon and Hake, 2011; Yamaguchi et al., 2012; Tsukaya, 2013; Hepworth and Lenhard, 2014). Leaf primordia initiate as bulges at the flanks of the shoot apical meristem (SAM). The SAM has pluripotent cells marked by the expression of class I KNOTTED1-LIKE HOMEOBOX (KNOX1) genes, among which SHOOT MERISTEMLESS (STM) is involved in the initiation and maintenance of the SAM (Shani et al., 2006). The expression of KNOX1 genes is repressed to suppress the undifferentiated state of the SAM in regions where leaf primordia will initiate (Jackson et al., 1994; Long et al., 1996). This repression of KNOX1 genes at the initiation site of leaf primordia is maintained throughout leaf development in the simple-leafed Arabidopsis. However, the expression pattern is different in compound leaf primordia. In C. hirsuta, a compound-leafed species, KNOX1 genes are re-expressed during leaf development (Hay and Tsiantis, 2006). Since the determinacy of leaves may be associated with the repression of KNOX1 genes (Kim et al., 2003; Scofield and Murray, 2006; Veit, 2009; Jun et al., 2010), their expression in compound-leafed plants is thought to promote a transient state of indeterminacy in the marginal meristem of leaf primordia, during which leaflets form. These studies indicate that KNOX1 genes are among the key factors regulating leaf morphological differences among species. Interestingly, such morphological variations are also seen in the leaves of R. aquatica, raising the question of whether the KNOX1 framework could explain morphological differences in leaves within an individual species.
To elucidate the mechanism underlying heterophylly, we first investigated the anatomy and development of leaves of R. aquatica. We then measured the levels of plant hormones with ultraperformance liquid chromatography coupled with tandem quadruple mass spectrometry and determined how leaf primordia respond to phytohormone applications. Moreover, we examined the expression patterns of genes known to be involved in leaf development. We finally obtained an overview of the transcriptional changes associated with heterophylly by RNA-seq. These studies provide insights into the developmental and molecular mechanisms underlying heterophylly in R. aquatica.
RESULTS
Leaf Form Alteration Is Induced by Change in Ambient Temperature
Previous studies showed that pinnately dissected leaves developed in submerged conditions in R. aquatica, whereas serrated or lobed simple leaves developed in terrestrial conditions (Fassett, 1930; Nakayama et al., 2012a; Figures 1A to 1C). We also found that a change in water temperature could affect leaf form under submergence (Figure 1D). Submerged plants at the higher water temperature of 30°C developed simpler leaves than did those grown at lower water temperatures (Figure 1D), indicating that R. aquatica leaf form can be affected by multiple environmental factors including temperature.
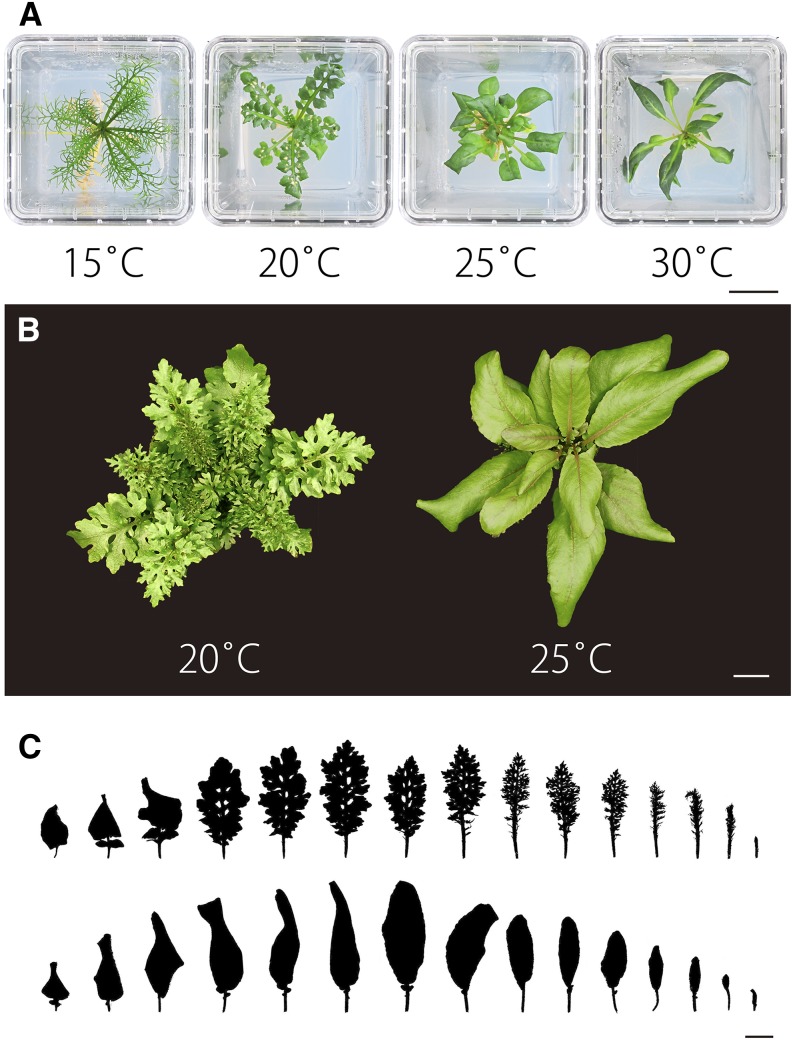
Next, we tested the effect of ambient temperature on leaf morphology in terrestrial conditions. Higher ambient temperatures induced simpler leaves in terrestrial conditions compared with lower temperatures (Figure 2A). At 15°C, pinnately dissected leaves developed and their morphology closely resembled the leaf form seen under submerged conditions (Figures 1C and 2A; Supplemental Figure 1). Thus, pinnately dissected leaves developed at lower temperatures under both submerged and terrestrial conditions and probably utilize a common molecular framework for their morphogenesis under both of these conditions. On the other hand, simple leaves with smooth margins developed at 30°C, while intermediate leaf forms were observed at 20°C and 25°C (Figure 2A). Thus, leaf form alteration in R. aquatica appears to be regulated by a progressive process or morphogenic gradient, rather than a dimorphism switch.
Figure 2.
Comparison of Leaf Morphology at Various Ambient Temperatures.
(A) Effects of change in ambient temperature on leaf morphology using in vitro cultured plants.
(B) Top view of shoots. Left, 20°C; right, 25°C.
(C) Comparison of leaf morphology at 20°C (upper) and 25°C (lower). All silhouettes of leaves are based on photographic images. The youngest leaf is at the right and the oldest is at the left.
Bars = 2 cm in (A) to (C).
Because the growth of the plants at 15°C was slower than at higher temperatures, and some experiments were not easily performed in submerged conditions, we used terrestrial plants grown at 20°C and 25°C to investigate the mechanisms underlying heterophylly. Under terrestrial conditions, dissected leaves developed at 20°C, whereas serrated simple leaves developed at 25°C (Figure 2B). The morphology of leaves under both temperatures changed and development progressed to become more complex. This change in leaf morphology during the course of development is known as heteroblasty, and it was more noticeable in the case of dissected leaves (20°C) than of simple leaves (25°C; Figure 2C).
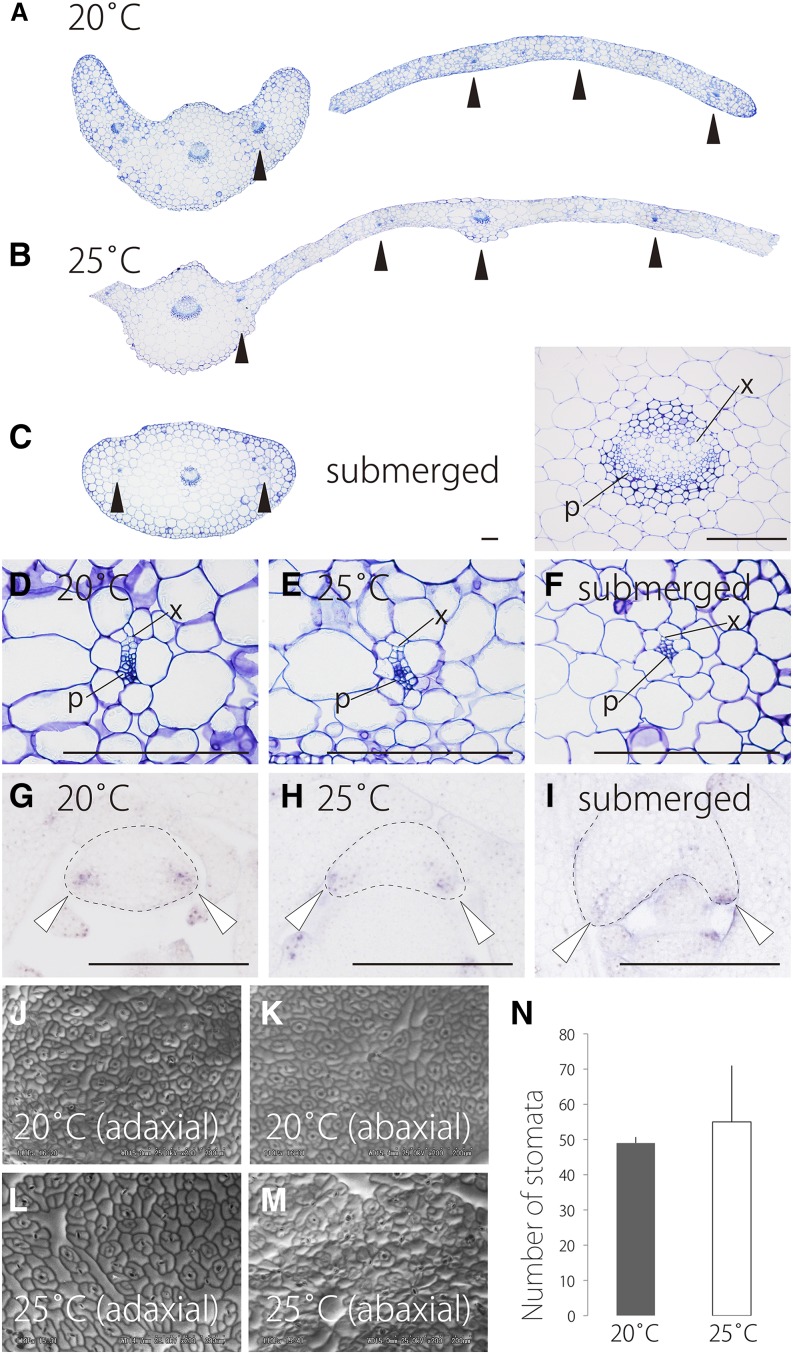
Pinnately Deeply Dissected Leaves under Submerged Conditions Have Adaxial-Abaxial Polarity
In addition to morphology, we determined whether various growth conditions affect leaf anatomy. We focused particularly on the establishment of adaxial-abaxial (ad-ab) polarity, since pinnately dissected leaves at lower temperature and under submergence resembled radialized unifacial leaves with an abaxialized leaf lamina. In leaves of R. aquatica at 20°C and 25°C, the midrib was well developed, and vascular bundles were arranged in line along the medio-lateral (central-marginal) axis in the lamina as is typical of bifacial leaves (Figures 3A and 3B). In submerged leaves, the central vascular bundle had ad-ab polarity as was seen in terrestrial leaves, and the vascular bundles were arranged along the medio-lateral axis. However, an extended leaf lamina was lacking compared with terrestrial leaves (Figures 3C to 3F). Vascular bundle polarity and arrangement are hallmarks of ad-ab polarity (Waites and Hudson, 1995). Therefore, pinnately dissected leaves that develop under submerged conditions have ad-ab polarity even though their needle-like morphology implies otherwise. Gene expression analysis confirms this observation. The PRESSED FLOWER (PRS) gene in Arabidopsis is known to be expressed at the margins of leaf primordia and act in leaf blade outgrowth downstream of establishment of ad-ab polarity (Matsumoto and Okada, 2001; Nakata et al., 2012). In leaf primordia of R. aquatica, the ortholog of PRS (Ra PRS) was expressed at the margin of leaf primordia as seen in that of Arabidopsis at different ambient temperatures and under water (Figures 3G to 3I; Supplemental Figure 2 and Supplemental Data Set 1). These results suggest that all examined leaves had ad-ab polarity.
Figure 3.
Comparison of Leaf Anatomy.
(A) to (C) Transverse sections of leaves at 20°C (A) and 25°C (B) and under submerged conditions (C). The inset shows an enlarged view of a central vascular bundle of a submerged leaf. Black arrowheads indicate small vascular bundles.
(D) to (F) Enlarged views of small vascular bundles at 20°C (D) and 25°C (E) and under submerged conditions (F). p, phloem; x, xylem. Top of the image is the adaxial side.
(G) to (I) In situ localization of Ra PRS transcripts at 20°C (G) and 25°C (H) and under submerged conditions (I). White arrowheads indicate expression region in a leaf primordium.
(J) to (M) Scanning electron micrographs of the adaxial side of a leaf at 20°C (J), the abaxial side of a leaf at 20°C (K), the adaxial side of a leaf at 25°C (L), and the abaxial side of a leaf at 25°C (M).
(N) Comparison of stomatal density: the number of stomata in the adaxial side per unit area. Data are means ± sd (n = 6).
Bars = 100 μm in (A) to (I).
Palisade and spongy parenchyma did not appear to be well differentiated in all conditions. The absence of differentiation between the palisade and the spongy parenchyma is a relatively common feature of leaves on aquatic plants (Arber, 1920). These results indicated that dissected leaves at 20°C and under submerged conditions are largely the same except for a difference in the degree of lamina expansion. Indeed, the terrestrial leaves from plants grown at 15°C were needle-like and pinnately dissected with reduced lamina expansion (Supplemental Figure 1). Additionally, in the leaves at 20°C and 25°C, we could not see any significant differences in epidermal cell shape between the adaxial and abaxial sides or in the distribution pattern of stomata between 20°C and 25°C: Cell shapes were simple jigsaw type, and stomata were observed on both sides (Figures 3J to 3M). The stomatal density did not change between 20°C and 25°C (Figure 3N). Trichomes were not seen on all observed leaf primordia or mature leaves.
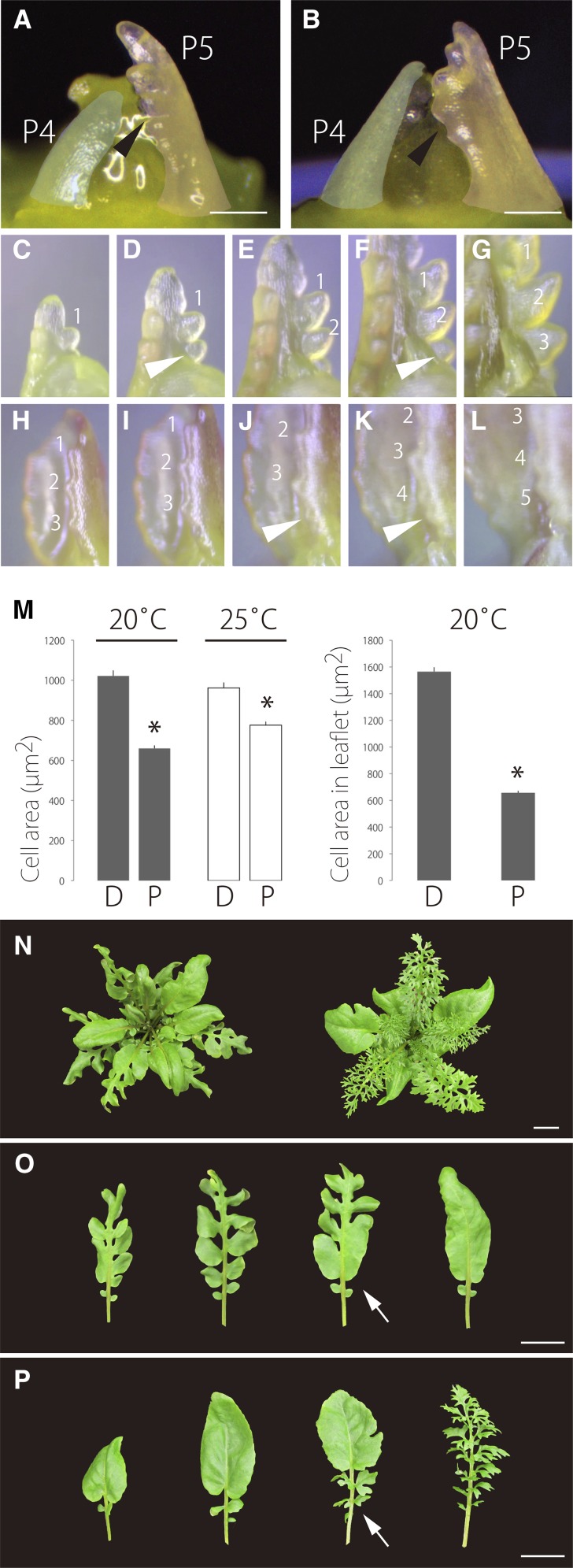
Proximal Leaflet Initiation Is Important for Final Leaf Form
To investigate the key developmental processes of leaf form alteration in heterophylly, we first examined the early stages of leaf morphogenesis and development. Observation of the SAM subtending leaf primordia showed that the morphology of the leaf blade at the P4 stage was not strikingly different between plants grown at 20°C and 25°C. On the other hand, differences in leaf blade morphology were clearly visible at the P5 stage (Figures 4A and 4B). Thus, the morphological differences between leaves grown at 20°C and 25°C appear to be determined between stages P4 and P5. Because a major morphological difference is the appearance of leaflets on leaf primordia, we examined leaflet initiation in the dissected leaf primordia by time-lapse image analysis. In dissected leaf primordia, a new leaflet primordium initiated below existing leaflet primordia, and leaflet initiation progressed basipetally (Figures 4C to 4G). In simple leaf primordia, which do not have leaflets, serrations were initiated basipetally instead of leaflet primordia (Figures 4H to 4L). Additionally, we noted that the cell areas in the proximal region of the leaves and the leaflets were smaller than those in the distal region (Figure 4M). These observations suggested that cell differentiation occurs basipetally and that morphogenesis in the proximal region of leaf primordia and leaves is particularly important for the final leaf form.
Figure 4.
Comparison of Leaf Development Dynamics.
(A) and (B) The shoot apex at 20°C (A) and at 25°C (B). P4 and P5 primordium are false-colored in blue and red, respectively. Black arrowheads indicate initiation of a leaflet or serration on a leaf primordium.
(C) to (G) Time-lapse analysis of leaflet initiation.
(H) to (L) Time-lapse analysis of serration initiation in simple leaves. White arrowheads indicate a newly emerged leaflet or serration.
(M) Comparison of cell area. Leaflets and leaves were counted at 20°C and 25°C, respectively. D, distal part of a leaf or a leaflet; P, proximal part of a leaf or a leaflet. The 7th leaves were used. Data are means ± sd (n = 6). *P < 0.01 (Student’s t test).
(N) to (P) Effect of temperature shift on leaf morphology.
(N) Top view of shoots. Left, shifted from 20°C to 25°C; right, shifted from 25°C to 20°C.
(O) Leaves from a plant shifted from 20°C to 25°C.
(P) Leaves from a plant shifted from 25°C to 20°C. Leaves are indicated in phyllotaxy order from left to right. White arrows indicate a leaf that displays intermediate morphology in response to temperature shifts.
Bars = 100 μm in (A) and (B) and 2 cm in (N) to (P).
To test this hypothesis, we performed a temperature shift analysis. When the temperature was shifted from 20°C to 25°C, newly emerged leaves became simpler leaves, and the proximal part of leaves in the transition phase was altered from dissected to smooth, simple margin form (Figures 4N, left, and 4O). Shifting from 25°C to 20°C resulted in newly emerged leaves developing as dissected leaves, with the proximal part of the leaves in the transition phase changing from smooth to dissected form (Figure 4N, right, and 4P). Next, we monitored cell proliferation in the primordia by 5′-ethynyl-2′-deoxyuridine (EdU) labeling. EdU signal was uniformly distributed in early simple leaf primordia (∼500 μm in length) (Figure 5A). By contrast, the signal in the distal region of the lamina was sparser than that in the proximal region at a later stage (Figures 5B to 5E). A similar pattern was observed in dissected leaves, with a dense, uniform signal in early simple leaf primordia (∼500 μm in length) (Figure 5F) and a sparser signal in the distal region of later stage leaflets than in the proximal part (Figures 5G to 5J). We obtained similar results from developing leaflets (Figures 5K to 5M). In summary, cell proliferation is concentrated in the proximal part of leaves and leaflets as they develop, indicating that morphogenesis in the proximal region of leaf primordia is important for determination of the final leaf form.
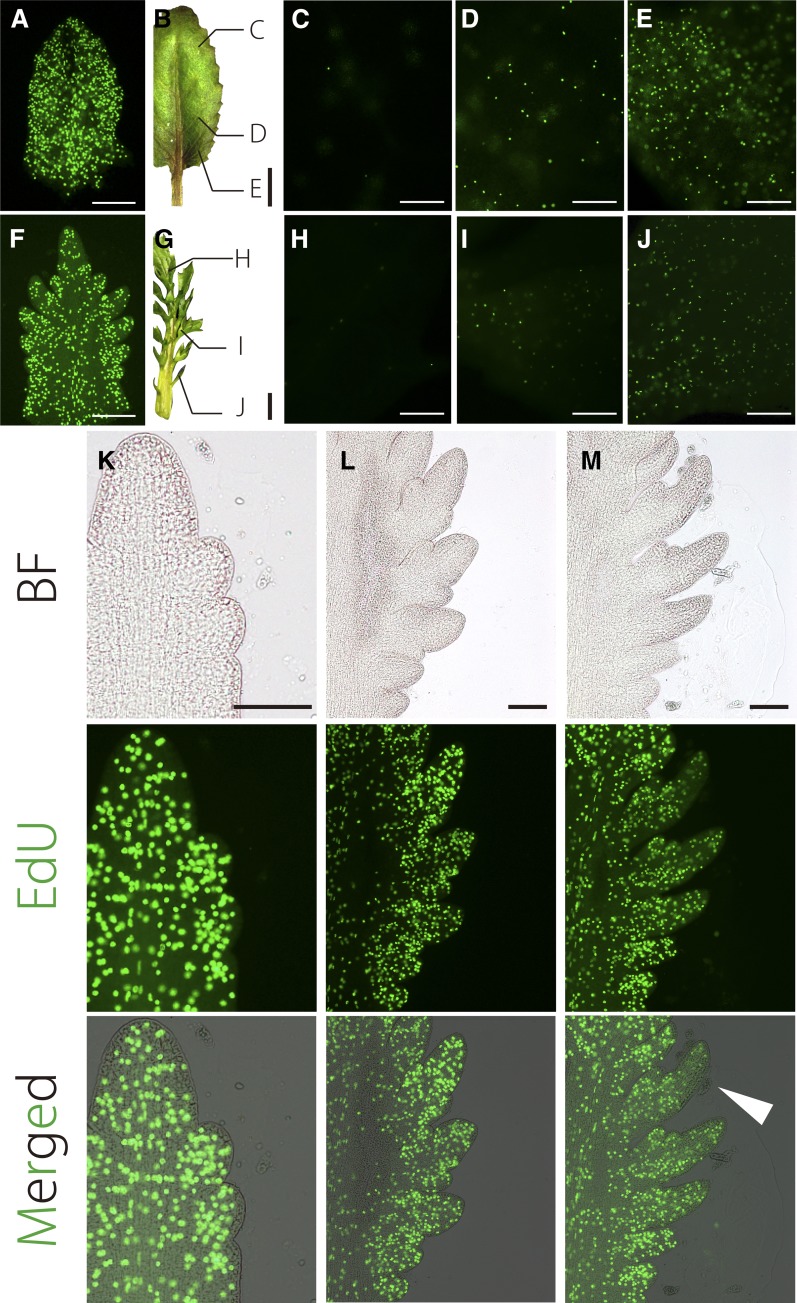
Figure 5.
EdU Visualization of Cell Proliferation in Leaves.
(A) Cell proliferation in ∼500-μm simple leaf primordia and leaves at 25°C.
(B) to (E) Cell proliferation in later stage leaves. The image in (B) illustrates leaf regions shown in (C) to (E). Cell proliferation in the distal region (C), middle region (D), and proximal region (E).
(F) Cell proliferation in ∼500-μm dissected leaf primordia and leaflets (at 20°C).
(G) to (J) Cell proliferation in dissected leaf primordia at a later stage. The image in (G) illustrates leaf regions shown in (H) to (J).
(K) to (M) Cell proliferation in leaflet primordia of increasing age. White arrowhead indicates a region showing decreased cell cycle activity. Top, bright field (BF); middle, EdU; bottom: merged. All leaves are 7th leaves.
Bars = 100 μm in (A), (C) to (F), and (H) to (M) and 1 mm in (B) and (G).
Gibberellin Is Sufficient to Alter Leaf Form in R. aquatica
Plant hormones are involved in the regulation of heterophylly (Goliber and Feldman, 1990; Kuwabara et al., 2003). Previous studies also demonstrated that changes in the endogenous gibberellin (GA) or cytokinin (CK) levels in leaf primordia affect leaf complexity in compound leaves of tomato (Solanum lycopersicum; Shani et al., 2010; Yanai et al., 2011). We therefore reasoned that plant hormones are likely to be involved in leaf form alteration in R. aquatica. To test this hypothesis, we measured the endogenous hormone concentrations of ABA, CK, GA, and jasmonic acid in leaf primordia by ultraperformance liquid chromatography coupled with tandem quadruple mass spectrometry (Figure 6; Supplemental Figure 3).
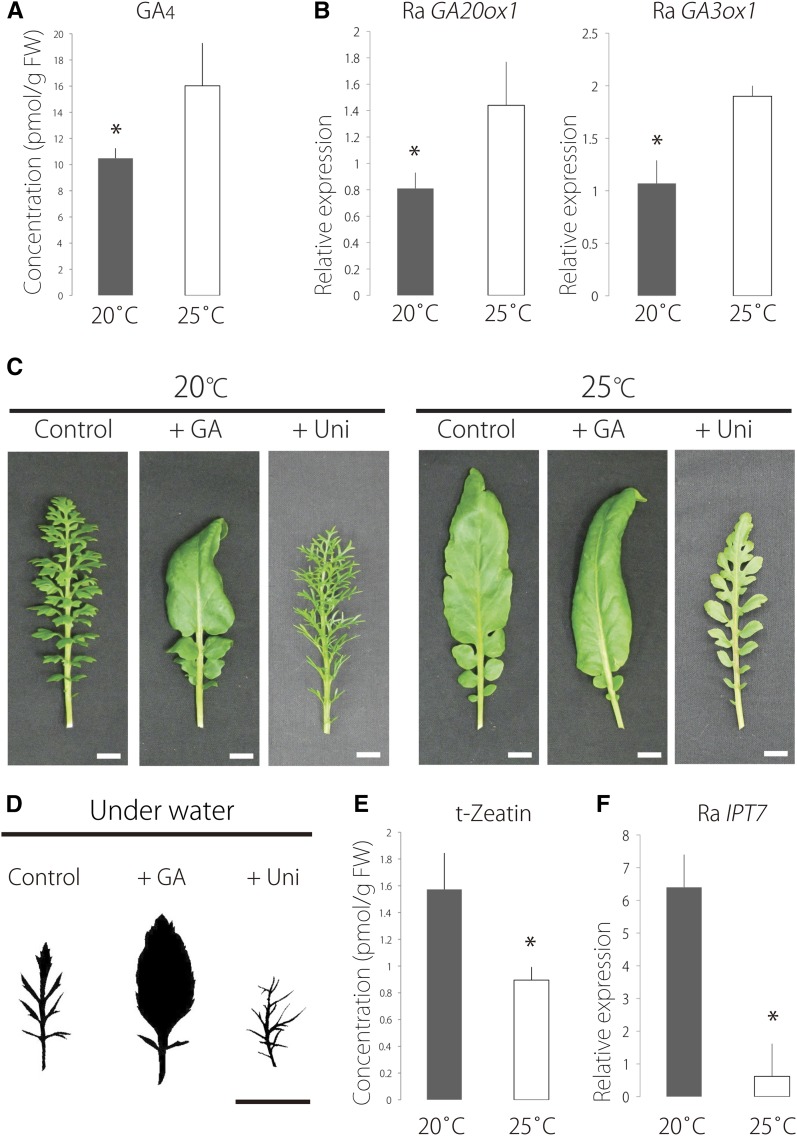
Figure 6.
Plant Hormone Profiling and Application of GA and Uniconazole.
(A) Accumulation level of GA4 in leaf primordia at 20°C and 25°C. Data are means ± sd (n = 3). FW, fresh weight.
(B) Expression levels of Ra GA20ox1 and Ra GA3ox1 in leaf primordia at 20°C and 25°C. Data are means ± sd (n = 3).
(C) Effects of GA or uniconazole application to the SAM. +GA and +Uniconazole (+Uni) plants were treated with 50 μM GA and 10 μM uniconazole, respectively.
(D) Effects of GA or uniconazole. All plants were grown underwater. +GA and +Uni plants were treated with 50 μM GA and 0.1 μM uniconazole, respectively. All silhouettes of leaves are based on photographic images of 7th leaves.
(E) Accumulation level of t-Zeatin in leaf primordia at 20°C and 25°C. Data are means ± sd (n = 3).
(F) Expression level of Ra IPT7 in leaf primordia at 20°C and 25°C. Data are means ± sd (n = 3). *P < 0.05 (Student’s t test). Bars = 1 cm.
Of particular note were GA and CK because their concentrations were significantly altered between 20°C and 25°C. The concentration of endogenous GA4, the bioactive form of GA, in leaf primordia was higher at 25°C than at 20°C (Figure 6A).
Subsequently, we investigated the expression levels of genes involved in GA biosynthesis. Specifically, we focused on genes involved in rate-limiting or final processes of hormone biosynthesis. The orthologs in R. aquatica were identified by BLAST searches, which revealed that putative amino acid sequences encoded by the isolated fragments were similar to those of each target gene in Arabidopsis: GIBBERELLIN 20-OXIDASE1 (GA20ox1) and GA3ox1, which are involved in GA biosynthesis. Additionally, multiple sequence alignments showed that each putative protein had characteristic functional domain(s), conserved among the homologs in a diverse array of species (Supplemental Figures 4 and 5 and Supplemental Data Sets 2 and 3). Phylogenetic analyses also supported the identity of the orthologs of the targeted genes: Ra GA20ox1 was located in a GA20ox1 clade (Supplemental Figure 4 and Supplemental Data Set 2), and Ra GA3ox1 was in a GA3ox1 clade (Supplemental Figure 5 and Supplemental Data Set 3).
Quantitative RT-PCR showed that the expression levels of Ra GA20ox1 and Ra GA3ox1 were higher at 25°C than at 20°C (Figure 6B). In light of these findings, we applied GA and the GA biosynthesis inhibitor uniconazole to the SAM to investigate the effect of GA on leaf morphology. GA treatment simplified leaf forms at 20°C and 25°C compared with the control, whereas uniconazole treatment increased leaf complexity (Figure 6C). Moreover, we investigated the effect of GA on leaf morphology in submerged plants. Exogenous GA treatment simplified the leaf form under submergence (Figure 6D).
On the other hand, the concentration of endogenous t-Zeatin, the bioactive form of CK, in leaf primordia was lower at 25°C than at 20°C (Figure 6E). To investigate the expression level of the gene involved in CK biosynthesis, the ortholog of ISOPENTENYL TRANSFERASE7 (IPT7), which is one of the well-characterized genes involved in CK biosynthesis, was identified by BLAST searches (Supplemental Figure 6 and Supplemental Data Set 4). The expression level of Ra IPT7 was lower at 25°C than at 20°C (Figure 6F). Additionally, we examined the effect of exogenous t-Zeatin (1 μM) on leaf morphology; however, we did not detect any change in the leaf morphology upon cytokinin treatment.
These results suggest that GA biosynthesis is promoted at 25°C, leading to the higher concentration of GA4 in leaf primordia, whereas CK biosynthesis is repressed at 25°C, leading to the lower concentration of t-Zeatin in leaf primordia. The endogenous concentrations of GA and CK seem to respond to the changes in ambient temperature.
Expression of KNOX1 Genes Is Altered in Response to Environmental Conditions
GA biosynthesis is regulated by KNOX1 genes (Hay et al., 2002; Bolduc and Hake, 2009), and the KNOX1 protein directly regulates GA20ox expression in some species (Sakamoto et al., 2001). CK biosynthesis is also regulated by KNOX1 genes (Jasinski et al., 2005). Previous studies with C. hirsuta and relatives of Arabidopsis have demonstrated that KNOX1 genes are involved in the development of compound and dissected leaves (Hay and Tsiantis, 2006; Blein et al., 2008; Piazza et al., 2010). Because the accumulation levels of GA and CK and the expression levels of genes involved in their biosynthesis were changed in response to ambient temperature in R. aquatica, it is likely that KNOX1 genes are involved in leaf form alteration in this species.
To elucidate the expression level and pattern of these genes, we isolated orthologs of KNOX1 and CUC from R. aquatica and confirmed their identity using BLAST, multiple sequence alignments, and phylogenetic analyses. Ra STM and Ra BP were located in an At (Arabidopsis) STM and KNAT1 clades, respectively, in the KNOX1 phylogeny (Supplemental Figure 7 and Supplemental Data Set 5). Among the CUC genes, we focused our analysis on the CUC3, since CUC3 is thought to have a conserved role in leaf development (Blein et al., 2008). Ra CUC3 was placed in an At CUC3 clade in the NAC phylogeny (Supplemental Figure 8 and Supplemental Data Set 6). Next, we determined the expression level of these orthologs by quantitative RT-PCR and found that the levels of Ra STM, Ra BP, and Ra CUC3 expression were higher at 20°C than at 25°C (Figure 7A). We also examined their expression patterns by in situ hybridization. At 25°C, Ra STM was mainly expressed in the SAM, as seen in Arabidopsis, whereas at 20°C it was expressed at the base of developing leaf primordia in addition to the SAM (Figures 7B and 7C). Similarly, Ra STM was expressed at the base of developing leaf primordia and in the SAM in submerged conditions (Figure 7D). Meanwhile, the expression pattern of Ra BP appeared not to change either at different ambient temperatures or under submerged conditions (Figures 7E to 7G). Both at 20°C and under submerged conditions, Ra CUC3 was expressed in the boundary domain between the leaflet primordia (Figures 7H to 7L). In summary, the expression patterns and levels of R. aquatica STM and CUC3 orthologs were sensitive to changes in the surrounding environment such as ambient temperature or inside or outside water.
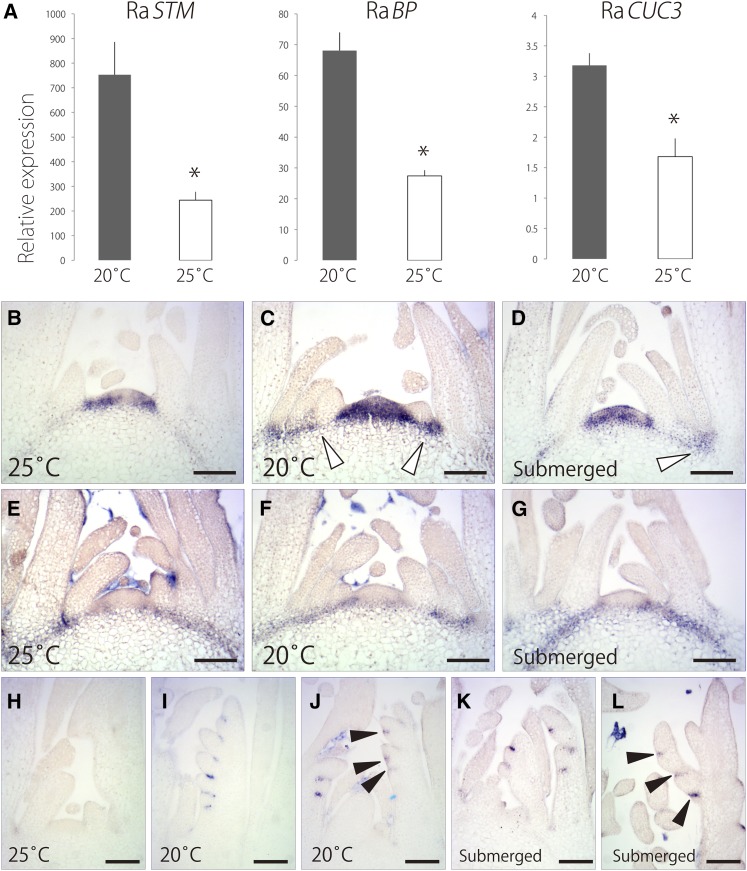
Figure 7.
Temperature-Dependent Expression of Ra STM, Ra BP, and Ra CUC3.
(A) Expression levels of Ra STM, Ra BP, and Ra CUC3 in shoot at 20°C and 25°C. Data are means ± sd (n = 3). *P < 0.01 (Student’s t test).
(B) to (D) Expression pattern of Ra STM at 25°C (B) and 20°C (C) and under submerged conditions (D).
(E) to (G) Expression pattern of Ra BP at 25°C (E) and 20°C (F) and under submerged conditions (G).
(H) to (L) Expression pattern of Ra CUC3. A shoot at 25°C (H) and 20°C (I). A shoot with leaf primordia subtending secondary leaflet at 20°C (J). A shoot under submerged conditions (K). A shoot with leaf primordia subtending secondary leaflet under submerged conditions (L).
All images show longitudinal sections. White arrowheads indicate expression of Ra STM at the basal part of leaf primordia. Black arrowheads indicate expression of Ra CUC3 in the boundary domain between the leaflet primordia. Bars = 100 μm.
Heterophylly in R. aquatica Is Also Regulated by Light Intensity
To understand global transcriptional alterations associated with heterophylly in R. aquatica, we performed RNA-seq analysis. RNA was extracted from the shoots including the leaf primordia of plants grown at 20°C (the dissected leaf condition) and 25°C (the simple leaf condition). The total numbers of reads obtained by single-end sequencing of cDNA derived from the dissected and simple leaf conditions were 12,107,279 and 11,948,965, respectively. Because R. aquatica is closely related to Arabidopsis, we used TAIR10 cDNA as a reference for mapping. Such transcriptome analysis, exploiting the close relationship between Rorippa species and Arabidopsis, has been successfully performed using Arabidopsis GeneChip microarrays (Sasidharan et al., 2013). Of the respective total reads, 3,657,900 and 3,417,848 were mapped to the reference Arabidopsis TAIR10 cDNA. Finally, 630 and 471 differentially expressed genes (false discovery rate [FDR] < 0.05) were defined in the dissected leaf and simple leaf conditions, respectively (Figure 8A; Supplemental Data Set 7).
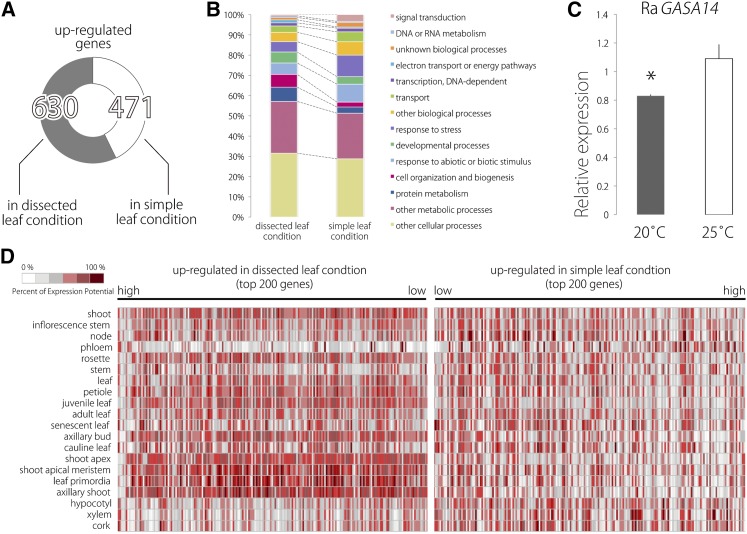
Figure 8.
RNA-seq Profiling of Differential Transcriptome in Heterophylly.
(A) Numbers of genes upregulated in dissected and simple leaf conditions (FDR < 0.05).
(B) Distribution of differentially expressed genes in GO biological process categories.
(C) Expression level of Ra GASA14 in leaf primordia at 20°C and 25°C. Data are means ± sd (n = 3). *P < 0.03 (Student’s t test).
(D) Expression profiles of differentially expressed genes in various tissues and organs in Arabidopsis. Two hundred of the most strongly upregulated genes in the dissected or simple leaf conditions were compared with publicly available Arabidopsis microarray data sets using the Genevestigator platform.
To gain an overview of changes in the transcriptome that are associated with heterophylly, we performed Gene Ontology (GO) category overrepresentation analysis using the GO database (http://www.arabidopsis.org/tools/bulk/go/index.jsp). This revealed that genes involved in protein metabolism and cell organization and biogenesis were overrepresented in the dissected leaf condition, suggesting that dissected leaf primordia undergo active morphogenesis to attain their more complex leaf form (Figure 8B). On the other hand, in the simple leaf condition, genes annotated in the response to abiotic or biotic stimuli and response to stress categories were overrepresented (Figure 8B; Supplemental Table 1). Additionally, in the simple leaf condition, we found that an ortholog of GA-stimulated in Arabidopsis 14 (GASA14) was upregulated. This is not surprising since this ortholog regulates leaf lamina expansion and is also induced by GA (Sun et al., 2013). Quantitative RT-PCR showed that the expression level of Ra GASA14 is higher at 25°C than at 20°C (Figure 8C; Supplemental Figure 9 and Supplemental Data Set 8).
Moreover, we compared these results with publicly available Arabidopsis microarray data sets using the Genevestigator platform (https://www.genevestigator.com/gv/plant.jsp) to gain better insight into the differences in gene expression. We found that the 200 most strongly upregulated genes in the dissected leaf condition overlap predominantly with genes expressed in the shoot apex, the SAM, and leaf primordia, indicating the undifferentiated state of the dissected leaf primordia. On the other hand, the top 200 genes upregulated in the simple leaf condition did not show this overlap (Figure 8D). Moreover, we found that the top 200 genes upregulated in the dissected leaf condition largely overlapped with genes that are upregulated in response to cold treatment and metabolism of glucose, whereas those upregulated in the simple leaf condition overlapped with genes that are downregulated in response to cold treatment and metabolism of glucose (Figure 9A). These results reveal striking differences between dissected and simple leaf transcriptomes.
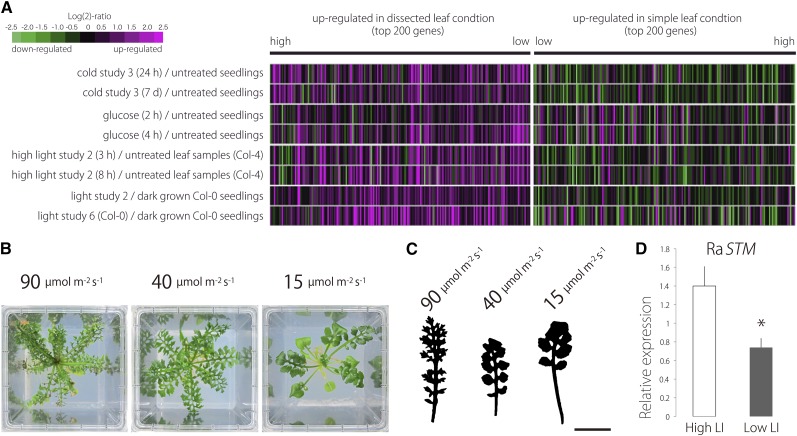
Figure 9.
Effects of Light Intensity on Leaf Form.
(A) Response of differentially expressed genes to various perturbations in Arabidopsis. Two hundred of the most strongly upregulated genes in the dissected or simple leaf conditions were compared with publicly available Arabidopsis microarray data sets using the Gene Search Perturbations tool (Genevestigator) to identify genes that are specifically upregulated in individual experimental conditions.
(B) Effects of change in light intensity on leaf morphology using plants grown at 20°C. Left, 90 μmol photons m−2 s−1; middle, 40 μmol photons m−2 s−1; right, 15 μmol photons m−2 s−1.
(C) Comparison of leaf morphology. All silhouettes of leaves are based on photographic images of 6th leaves. Bar = 2 cm.
(D) Expression levels of Ra STM in shoot at high and low light intensities at 20°C. High LI, high light intensity (70 μmol photons m−2 s−1); Low LI, low light intensity (15 μmol photons m−2 s−1). Data are means ± sd (n = 3). *P < 0.01 (Student’s t test).
Interestingly, the genes upregulated in the dissected leaf condition also overlapped with those that respond to changes in light intensity (Figure 9A), suggesting that light intensity affects the leaf morphology of R. aquatica in addition to temperature. We therefore grew plants at various light intensities at 20°C and found that changing light intensity affects leaf complexity in R. aquatica (Figure 9B). Under high light intensity (90 μmol photons m−2 s−1), dissected leaves with deep biserrated leaflets developed, whereas in low light intensity (15 μmol photons m−2 s−1), dissected leaves with a relatively smooth margin developed (Figure 9C). Indeed, the expression level of the STM ortholog was sensitive to the changes in light intensity (Figure 9D). Therefore, light intensity also affects leaf form in R. aquatica, leading to the hypothesis that temperature and light intensity affect leaf form via a common developmental mechanism.
DISCUSSION
The molecular mechanism underlying heterophylly is largely unknown, although the phenomenon is widely seen in nature. We investigated the molecular developmental mechanisms of heterophylly in R. aquatica. During the formation of dissected leaves, Ra STM was expressed in the leaf primordia, whereas its expression was reduced in simple leaf primordia. Additionally, the expression of Ra GA20ox1, which is thought to be negatively regulated by KNOX1 genes, was lower in the dissected leaf primordia, as was the accumulation level of bioactive GA4. Application of GA was sufficient to alter leaf form in R. aquatica. Furthermore, RNA-seq analysis revealed a relationship between light intensity and leaf form. Based on these results, we propose that regulation of GA level via KNOX1 genes is the key developmental switch for heterophylly in this species, which may be regulated by the combination of multiple environmental cues such as temperature and light.
Regulation of GA Level via KNOX1 Genes Induces Heterophylly
Low temperature induced higher expression of the KNOX1 ortholog at the basal part of emerging leaf primordia in R. aquatica. Additionally, expression of the CUC ortholog was also observed in the boundary region between the leaflets on the dissected leaf primordia (Figure 7). In C. hirsuta, RNA interference of the STM ortholog produced plants with fewer leaflets. Conversely, 35S:KN1-GR plants produce leaves with more leaflets. These results confirm that KNOX1 genes are involved in maintaining the undifferentiated state and in regulating cell division during leaflet development (Hay and Tsiantis, 2006).
In R. aquatica, cells in the proximal region in leaves were smaller at 20°C than at 25°C and leaflet initiation mainly progressed basipetally (Figures 4 and 5), suggesting a more prolonged undifferentiated state at the lower temperature. RNA-seq analysis showed that genes associated with the SAM, shoot apex, and leaf primordia are highly expressed in dissected leaves (Figure 8D). Collectively, these results suggest that the expression of KNOX1 orthologs at the basal part of leaf primordia is involved in maintaining the undifferentiated state to form leaflets in dissected leaves in R. aquatica. On the other hand, CUC expression leads to suppression of marginal outgrowth in leaf primordia, and it has been demonstrated that this CUC function is conserved within eudicots (Hay and Tsiantis, 2006; Blein et al., 2008). Therefore, in the dissected leaf primordia of R. aquatica, the expression of CUC ortholog is likely to suppress cell division activity in the boundary regions between the leaflet primordia in dissected leaves.
In tobacco (Nicotiana tabacum), the KNOX1 protein NTH15 directly binds to an intron sequence in the GA20ox to represses its expression (Sakamoto et al., 2001). Expression of the Ra GA20ox1, which is thought to be regulated by KNOX1 in R. aquatica, was reduced in dissected leaf primordia at 20°C (Figure 6). Moreover, in tomato, the KNOX1 overexpression mutant Tkn2 showed repressed SlGA20ox1 expression, and the highly complex compound leaf phenotype of Tkn2 was suppressed by GA application (Hay et al., 2002). These studies demonstrated that KNOX1 affects leaf complexity by suppressing GA biosynthesis (hereafter referred to as the KNOX-GA gene module). In addition to reduced Ra GA20ox1 and Ra GA3ox1 expression, the level of bioactive GA4 in leaf primordia at 20°C was lower than at 25°C in R. aquatica (Figures 6A and 6B).
Moreover, GA or uniconazole treatment demonstrated that manipulation of GA levels affects leaf complexity and the degree of lamina expansion in leaves and leaflets in R. aquatica (Figures 6C and 6D). The study with tomato demonstrated that GA promotes differentiation, which disables transient organogenetic activity in the leaf margins, from which marginal serrations and leaflets arise (Yanai et al., 2011). Hence, it is likely that low levels of GA delay the differentiation of leaves and facilitate morphogenic activity in the basal part of leaves in dissected leaf primordia at 20°C in R. aquatica. On the other hand, high levels of GA promote differentiation and cell expansion, limiting leaflet initiation due to a reduction in morphogenic activity. The expression of the ortholog of GASA14, which is upregulated by GA and regulates leaf lamina expansion in Arabidopsis, increased at 25°C (Figure 8C; Sun et al., 2013). Moreover, the expression levels appeared to correlate with the changes in the concentration of endogenous GA4 (Figures 6A and 8C). These results suggested that this gene is involved in lamina expansion in response to environmental change.
This hypothesis that alteration of morphogenic activity by the KNOX-GA gene module affects leaf morphology is also supported by the behavior of another plant hormone, CK. The accumulation level of t-Zeatin was higher in dissected leaf primordia than in simple leaf primordia (Figure 6E). CK biosynthesis is positively regulated by KNOX1 expression, and CK is involved in prolonged morphogenetic activity in the leaf margin (Jasinski et al., 2005; Shani et al., 2006, 2010). Therefore, the observed CK accumulation also implies higher morphogenetic activity in the dissected leaf primordia.
We showed that, in addition to ambient temperature, light intensity also affects leaf form (Figure 9). Studies in Arabidopsis have demonstrated that such environmental signals feed into a pathway of PHYTOCHROME INTERACTING FACTORs (PIFs), which in turn regulate auxin biosynthesis (Franklin, et al., 2011; Leivar and Monte, 2014). Additionally, auxin is known to repress the expression of KNOX1 genes (Heisler et al., 2005). Therefore, one possibility is that KNOX1 expression is regulated by PIFs via auxin in R. aquatica, resulting in alteration in the concentrations of hormones such as GA and CK. If so, the use of PIFs as network hubs may allow integration of various environmental and developmental cues to optimize final leaf form. This existing gene regulatory network may have been further modified to regulate heterophylly in the genus Rorippa during evolution. The RNA-seq analysis did not detect significant expression change of PIFs. Since PIF proteins are regulated via the ubiquitin-proteasome system (Leivar and Monte, 2014), it may be necessary to consider the protein level of PIFs. In addition, methylation of DNA plays a key role in the regulation of gene expression (Zhang et al., 2006), and nucleosomes containing the histone H2A.Z are essential for perceiving ambient temperature correctly and thereby regulating gene expression (Kumar and Wigge, 2010). Therefore, DNA methylation and/or nucleosome assembly may also be important for the regulation of the key developmental modules in response to the surrounding environment.
The KNOX-GA Gene Module Diversifies Morphology both among Species and within a Species
Compound or dissected leaves arose multiple independent times during the evolution of plants (Bharathan et al., 2002). Nevertheless, the responsible mechanisms seem to be relatively converged. For instance, differences in KNOX1 expression have led to morphological differences of leaves between Arabidopsis and C. hirsuta (Hay and Tsiantis, 2006). The cis-regulatory divergence in KNOX1 has led to the morphological differences seen in leaves in the genus Arabidopsis (Piazza et al., 2010). The reduction in KNOX1 activity by competitive inhibition has led to differences in leaf complexity between tomato species (Kimura et al., 2008). Gene network analysis confirmed the role of KNOX1 activity in regulating interspecific diversity in leaf shape in the tomato clade (Ichihashi et al., 2014). Additionally, the KNOX protein NTH15 directly regulates GA biosynthesis (Sakamoto et al., 2001), and GA level affects leaf complexity (Yanai et al., 2011). Therefore, the KNOX-GA gene module is an important factor in dissected and compound leaf development and may be responsible for variation in leaf form between species. Interestingly, our results indicate that the activity of the KNOX-GA gene module alters in response to the surrounding environment within individuals in R. aquatica (Figures 6 and 7). As a result, leaf form is altered from dissected to simple and vice versa. These data suggest that the module participates not only in the generation of morphological variation between species but also in morphological variation within a species in response to environmental changes. Hence, the KNOX-GA gene module may be considered a hotspot contributing to leaf form variation in plants during evolution in response to selective pressure or environmental conditions.
However, all heterophyllous plants do not utilize the same mechanism. Previous studies have shown that ethylene and/or ABA have a role in the regulation of heterophylly (Anderson, 1978; Kuwabara et al., 2003; Sato et al., 2008). For instance, in L. arcuata (Onagraceae), ethylene and ABA induce submerged-type and terrestrial-type leaves, respectively (Kuwabara et al., 2003), and in Potamogeton nodosus (Potamogetonaceae), ABA induces floating leaves (Anderson, 1978). In contrast with these reports, our results shed light on the relation between GA and leaf form alteration in heterophylly. Therefore, it appears that the core mechanism regulating leaf form in R. aquatica differs from that of L. arcuata and P. nodosus. Parallels are seen between this situation and that of leaf complexity, where other mechanisms besides the KNOX-GA gene module have been shown to operate in a derived clade of Fabaceae (Hofer et al., 1997; Champagne et al., 2007; Zhou et al., 2014). Since the acquisition of heterophylly has also occurred independently multiple times (Wells and Pigliucci, 2000; Wanke, 2011; Nakayama et al., 2012a), the core regulatory mechanisms for acquisition of heterophylly may not be conserved during evolution.
Taken together, our findings suggest that the regulation of GA and CK levels via KNOX1 is involved in leaf form alteration in response to the environment in R. aquatica, and a regulatory module that can be responsible for variation of leaf form may be used in the case of morphological variation both between species and within a species. Understanding the molecular mechanism by which the key network is regulated to allow for heterophylly should yield novel insights into the relationship between plant form and environment.
METHODS
Plant Materials
All plants were grown in a growth chamber under continuous illumination with a light intensity of ∼50 μmol photons m−2 s−1. Seedlings were planted in pots containing soil and watered every 2 d. Most of the plants used in experiments (morphological observation, time-lapse analysis, EdU incorporation assay, quantification of hormones, real-time PCR, in situ hybridization, and gene expression profiling) were grown in each condition for a month. Agarose medium (8 g/L) containing half-strength Murashige and Skoog (MS) salts (WAKO) and 2% (w/v) sucrose was used for in vitro culture. The shoots or leaf primordia were frozen in liquid nitrogen just after sampling and then stored at –80°C until use for RNA extraction and plant hormone profiling.
Morphological Observations
Shoots and leaves were fixed overnight in 45% ethanol, 2.5% acetic acid, and 2.5% formaldehyde at 4°C. The fixed samples were dehydrated in an ethanol series (50, 60, 70, 80, 90, 95, 99.5, and 100% [v/v]; 30 min per step) and stored overnight in 100% (v/v) ethanol at 4°C. Fixed samples embedded in Technovit resin (Kulzer and Co.) were sectioned as described previously (Nakayama et al., 2012b). Photographs were taken using an ECLIPSE 80i (Nikon). To determine the cell area of leaves, the 7th leaves were fixed in a formalin-acetic acid-alcohol solution and cleared using a chloral hydrate solution. The samples were then photographed under the microscopes, and cell area was determined using ImageJ software (http://rsb.info.nih.gov/ij/). For each leaf, more than 10 palisade cells in the subepidermal layer in the center of the leaf blade between the midvein and the leaf margin were analyzed.
Time-Lapse Analysis
To perform time-lapse analysis of developing leaf primordia, we used a Dino-Lite Pro digital microscope (AnMo Electronics). Plants were grown on MS plus 1% sucrose agar plates. Before the analysis, relatively mature leaves and leaf primordia were dissected out, leaving the two or three youngest primordia. Dissected shoots were grown on MS plus 1% sucrose agar plates again and were used for the analysis.
Detecting the Cell Proliferation Zone in Leaves with EdU
To identify dividing cells, we performed an EdU incorporation assay. EdU is incorporated into DNA during active DNA synthesis (i.e., during S-phase) (Kotogány et al., 2010). Dissected shoot apices were incubated in water containing 10 μM EdU (Click-iT EdU Alexa Fluor 488 imaging kit; Invitrogen). After 3 h, the samples were fixed in 3.7% formaldehyde in PBS (pH 7.4) for 30 min and then washed three times in PBS with shaking. Coupling of EdU to the Alexa Fluor substrate was performed in the dark in the Click-iT reaction mixture, according to the manufacturer’s instructions. Photographs were taken using an ECLIPSE 80i.
Quantification of Plant Hormones and GA Application
For quantification of hormones, plants were grown in a chamber for a month for each condition. Leaf primordia were dissected and frozen in liquid nitrogen just after sampling. Extraction and determination of ABA, cytokinin (t-Zeatin), gibberellin (GA4), and jasmonic acid contents were performed as described previously (Kojima et al., 2009; Kojima and Sakakibara, 2012). Experiments were conducted in triplicate from three independent tissues. GA solutions used in the experiment reported in Figure 6 contained 1% (w/v) ethanol; the control solution comprised 1% (w/v) ethanol alone. +GA and +Uniconazole plants were treated with 50 μM GA and 10 μM uniconazole, respectively. A 100-μL drop of each solution was applied to the SAM every day for a month. These plants were grown in each condition for 2 months until the leaves became mature.
RNA Extraction, cDNA Synthesis, and Gene Isolation
Total RNA was extracted from shoots including leaf primordia of plants grown for a month using an RNeasy Plant Mini Kit (Qiagen) with DNase I treatment, and cDNA was synthesized from 1 μg total RNA using Transcriptor Universal cDNA Master (Roche). Diluted reverse-transcribed cDNA (0.5 μL) was subjected to 30 to 40 cycles of PCR amplification (94°C for 30 s, 55°C for 30 s, and 72°C for 30 s) using gene-specific primers (Supplemental Table 2). The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced using an automated sequencer (model 3100; Applied Biosystems) for at least eight clones per fragment, according to the manufacturer’s protocol. Primer sequences are listed in Supplemental Table 2.
5′ and 3′ Rapid Amplification of cDNA Ends
To obtain 5′ and 3′ sequences of the isolated genes, we performed 5′ and 3′ rapid amplification of cDNA ends using the SMARTer cDNA amplification kit (Takara) according to the manufacturer’s instructions. The primers were designed to amplify the 5′ and 3′ regions of the isolated fragment (Supplemental Table 2), and PCR was performed as follows: 94°C for 2 min; five cycles at 94°C for 30 s and 72°C for 3 min; five cycles at 94°C for 30 s and 70°C for 3 min; 20 cycles at 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min; and a final extension at 72°C for 10 min. As necessary, nested PCR using specific primers was then performed as follows: 94°C for 2 min, followed by 25 cycles at 94°C for 30 s, 68°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 10 min. Gene-specific primers for Ra STM and Ra CUC3 were designed based on the sequences of Arabidopsis thaliana STM and CUC3. The PCR products were cloned into pGEM-T Easy and sequenced using a model 3100 automated sequencer for at least eight clones per fragment, according to the manufacturer’s protocol. Primer sequences are listed in Supplemental Table 2.
Phylogenetic Analyses of Isolated Genes
The predicted amino acid sequences of isolated genes were aligned using ClustalW and readjusted manually. Phylogenetic trees were reconstructed using MEGA5 (Tamura et al., 2011) using the neighbor-joining method (Perrière and Gouy, 1996). Bootstrap values were derived from 1000 replicate runs.
Real-Time PCR
Total RNA was extracted from shoots including leaf primordia or only leaf primordia of plants grown for a month and used to synthesize cDNA, as described above. Quantitative RT-PCR analysis was conducted using the following gene-specific primer pairs: NaSTM_RT_F and NaSTM_RT_R; NaBP_RT_F and NaBP_RT_R; NaCUC3_RT_F and NaCUC3_RT_R; NaGA20ox_RT_F and NaGA20ox_RT_R; NaGA3ox_RT_F and NaGA3ox_RT_R; and NaGASA14_RT_F and NaGASA14_RT_R (Supplemental Table 3). Real-time PCR amplification was performed using the KAPA SYBR Fast qPCR kit (KAPA Biosystems) in a 7500 Real Time PCR system (Applied Biosystems). Experiments were performed in triplicate from three independent tissue RNA extractions. Expression was normalized to the Ra TUB4 beta control.
In Situ Hybridization
Portions of genes isolated in pGEM-T Easy were amplified by PCR using the universal primers M13-Fw (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) and M13-Rv (5′-TCACACAGGAAACAGCTATGAC-3′). The amplified fragments were then used to produce digoxigenin (DIG)-labeled sense and antisense RNA probes using a DIG RNA labeling kit (Roche). Shoots were fixed in 100 mM sodium phosphate (pH 7.5) containing 3% (w/v) paraformaldehyde, 0.25% (w/v) glutaraldehyde, and 1 M NaOH. Fixed samples were dehydrated in an ethanol series and embedded in Paraplast Plus (Sigma-Aldrich). The embedded samples were sectioned to a thickness of 8 μm, and the sections were affixed to glass slides by an overnight incubation at 42°C. DIG-labeled sense and antisense RNA probes were synthesized with SP6 RNA polymerase or T7 RNA polymerase (Roche). Hybridizations were performed according to Nakayama et al. (2012b). For immunological detection, the slides were incubated in detection buffer containing nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3′-indolylphosphatase p-Toluidine salt (Roche) at room temperature overnight. Photographs were taken using an ECLIPSE 80i. The experiments were performed at least three times.
Gene Expression Profiling with RNA-seq
For RNA-seq analysis, plants were grown in each condition for a month. Total RNAs were extracted from the shoot apex subtending the leaf primordia and treated with DNaseI using an RNeasy Plant Mini Kit. RNA-seq libraries were prepared using an mRNA-Seq Sample Prep Kit (Illumina) with barcodes for each library (Kumar et al., 2012). Multiplex sequencing was performed on an Illumina GA II, and 36-bp reads were obtained. After separating reads by barcode, we used BWA (http://bio-bwa.sourceforge.net) to map raw reads. Because Rorippa aquatica is closely related to Arabidopsis, we used TAIR10 cDNA as a reference for the mapping. Transcript expression and differentially expressed (DE) genes were defined with EdgeR (Robinson et al., 2010), and genes with FDRs of < 0.05 were regarded as DE genes. GO analysis was performed with GOHyperGAll (Horan et al., 2008). To obtain an overview of DE genes, Genevestigator (https://www.genevestigator.com/gv/) was used for comparison with the publicly available microarray data sets (Hruz et al., 2008).
Accession Numbers
Nucleotide sequence data reported here are available in the DDBJ under the following accession numbers: Ra GA20ox1 (AB971251), Ra GA3ox1 (AB971252), Ra STM (AB971253), Ra BP (AB971254), Ra CUC3 (AB971255), Ra GASA14 (AB972534), Ra IPT7 (AB972535), and Ra PRS (AB972536). RNA-seq data reported here are available in the DDBJ Sequenced Read Archive under accession numbers DRX 019546 to DRX019553.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gross Morphology of Leaves at 15°C.
Supplemental Figure 2. A Phylogenetic Tree and Alignments of PRS Orthologs.
Supplemental Figure 3. Plant Hormone Profiles.
Supplemental Figure 4. A Phylogenetic Tree and Alignments of GA20ox1 Orthologs.
Supplemental Figure 5. A Phylogenetic Tree and Alignments of GA3ox1 Orthologs.
Supplemental Figure 6. A Phylogenetic Tree and Alignments of IPT7 Orthologs.
Supplemental Figure 7. A Phylogenetic Tree and Alignments of KNOX1 Orthologs.
Supplemental Figure 8. A Phylogenetic Tree and Alignments of CUC Orthologs.
Supplemental Figure 9. A Phylogenetic Tree and Alignments of GASA14 Orthologs.
Supplemental Table 1. Functional Categorization by GO Annotation.
Supplemental Table 2. List of Oligonucleotide Primers Used in This Study.
Supplemental Data Set 1. The Sequences Used to Generate the Phylogenies of the PRS Family.
Supplemental Data Set 2. The Sequences Used to Generate the Phylogenies of the GA20ox1 Family.
Supplemental Data Set 3. The Sequences Used to Generate the Phylogenies of the GA3ox1 Family.
Supplemental Data Set 4. The Sequences Used to Generate the Phylogenies of the IPT7 Family.
Supplemental Data Set 5. The Sequences Used to Generate the Phylogenies of the KNOX Family.
Supplemental Data Set 6. The Sequences Used to Generate the Phylogenies of the CUC Family.
Supplemental Data Set 7. List of Differentially Expressed Genes.
Supplemental Data Set 8. The Sequences Used to Generate the Phylogenies of the GASA14 Family.
Supplementary Material
Acknowledgments
We thank Kaoru O. Yoshiyama, Akiko Nakamasu, and Yasunori Ichihashi for helpful discussion throughout our study. The hormone analysis reported here was supported by Japan Advanced Plant Science Network. This work was partially supported by Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) (KAKENHI Grant Numbers 22870031 and 24770047) and The Science Research Promotion Fund from the Promotion and Mutual Aid Corporation for Private Schools of Japan to S.K. and by a Research Fellowship from JSPS to H.N. N.S. was supported by a UC Davis RISE grant.
AUTHOR CONTRIBUTIONS
H.N., N.N., N.S., and S.K. conceived and designed the project. H.N., N.N., M.K., H.S., S.S., and S.K. performed the experiments. H.N., N.N., N.S., and S.K. wrote the article.
Glossary
- ABA
abscisic acid
- SAM
shoot apical meristem
- ad-ab
adaxial-abaxial
- EdU
5′-ethynyl-2′-deoxyuridine
- GA
gibberellin
- CK
cytokinin
- FDR
false discovery rate
- GO
Gene Ontology
- MS
Murashige and Skoog
- DIG
digoxigenin
- DE
differentially expressed
Footnotes
Online version contains Web-only data.
References
- Akman M., Bhikharie A.V., McLean E.H., Boonman A., Visser E.J.W., Schranz M.E., van Tienderen P.H. (2012). Wait or escape? Contrasting submergence tolerance strategies of Rorippa amphibia, Rorippa sylvestris and their hybrid. Ann. Bot. (Lond.) 109: 1263–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber A. (1920). Water Plants. (London: Cambridge University Press; ). [Google Scholar]
- Anderson L.W.J. (1978). Abscisic acid induces formation of floating leaves in the heterophyllous aquatic angiosperm Potamogeton nodosus. Science 201: 1135–1138. [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Hay A., Kougioumoutzi E., Tsiantis M. (2008). A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 40: 1136–1141. [DOI] [PubMed] [Google Scholar]
- Bell A.D. (2008). Plant Form: An Illustrated Guide to Flowering Plant Morphology. (Portland, OR: Timber Press; ). [Google Scholar]
- Bharathan G., Goliber T.E., Moore C., Kessler S., Pham T., Sinha N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296: 1858–1860. [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., Johansen I.E., Tsiantis M., Laufs P. (2008). A conserved molecular framework for compound leaf development. Science 322: 1835–1839. [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C., Barkoulas M., Galinha C., Tsiantis M. (2010). Weeds of change: Cardamine hirsuta as a new model system for studying dissected leaf development. J. Plant Res. 123: 25–33. [DOI] [PubMed] [Google Scholar]
- Champagne C.E., Goliber T.E., Wojciechowski M.F., Mei R.W., Townsley B.T., Wang K., Paz M.M., Geeta R., Sinha N.R. (2007). Compound leaf development and evolution in the legumes. Plant Cell 19: 3369–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S.A., Johnson M.P. (1968). Adaptation to heterogenous environments I. Variation in heterophylly in Ranunculus flammula L. Evolution 22: 496–516. [DOI] [PubMed] [Google Scholar]
- Couvreur T.L., Franzke A., Al-Shehbaz I.A., Bakker F.T., Koch M.A., Mummenhoff K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27: 55–71. [DOI] [PubMed] [Google Scholar]
- Donohue K. (2013). Development in the wild: Phenotypic plasticity. Ann. Plant Rev. 45: 321–356. [Google Scholar]
- Fassett N.C. (1930). A Manual of Aquatic Plants. (Madison, WI: The University of Wisconsin Press; ). [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goliber T.E., Feldman L.J. (1990). Developmental analysis of leaf plasticity in the heterophyllous aquatic plant Hippuris vulgaris. Am. J. Bot. 77: 399–412. [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay A.S., et al. (2014). Cardamine hirsuta: a versatile genetic system for comparative studies. Plant J. 78: 1–15. [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hepworth J., Lenhard M. (2014). Regulation of plant lateral-organ growth by modulating cell number and size. Curr. Opin. Plant Biol. 17: 36–42. [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., Ellis N. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7: 581–587. [DOI] [PubMed] [Google Scholar]
- Horan K., Jang C., Bailey-Serres J., Mittler R., Shelton C., Harper J.F., Zhu J.K., Cushman J.C., Gollery M., Girke T. (2008). Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147: 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y., Aguilar-Martínez J.A, Farhi M., Chitwood D.H., Kumar R., Millon L.V., Peng J., Maloof J.N., Sinha N.R. (2014). Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. USA 111: E2616–E2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413. [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565. [DOI] [PubMed] [Google Scholar]
- Jun J.H., Ha C.M., Fletcher J.C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22: 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M.E., Albert L.S. (1987). Abscisic acid induces aerial leaf morphology and vasculature in submerged Hippuris vulgaris L. Aquat. Bot. 28: 81–88. [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.-S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F.Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18: 672–677. [DOI] [PubMed] [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., Ashikari M., Ueguchi-Tanaka M., Matsuoka M., Suzuki K., Sakakibara H. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M., Sakakibara H. (2012). Highly sensitive high-throughput profiling of six phytohormones using MS-probe modification and liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 918: 151–164. [DOI] [PubMed] [Google Scholar]
- Kotogány E., Dudits D., Horváth G.V., Ayaydin F. (2010). A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Ichihashi Y., Kimura S., Chitwood D.H., Headland L.R., Peng J., Maloof J.N., Sinha N.R. (2012). A high-throughput method for Illumina RNA-Seq library preparation. Front. Plant Sci. 3: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Kuwabara A., Ikegami K., Koshiba T., Nagata T. (2003). Effects of ethylene and abscisic acid upon heterophylly in Ludwigia arcuata (Onagraceae). Planta 217: 880–887. [DOI] [PubMed] [Google Scholar]
- La Rue C.D. (1943). Regeneration in Radicula aquatica. Mich. Acad. 28: 51–61. [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Les D.H. (1994). Molecular systematics and taxonomy of lake cress (Neobeckia aquatica; Brassicaceae), an imperiled aquatic mustard. Aquat. Bot. 49: 149–165. [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Matsumoto N., Okada K. (2001). A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 15: 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J., Hake S. (2011). How a leaf gets its shape. Curr. Opin. Plant Biol. 14: 24–30. [DOI] [PubMed] [Google Scholar]
- Nakamasu A., Nakayama H., Nakayama N., Suematsu N.J., Kimura S. (2014). A developmental model for branching morphogenesis of lake cress compound leaf. PLoS ONE 9: e111615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M., Matsumoto N., Tsugeki R., Rikirsch E., Laux T., Okada K. (2012). Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell 24: 519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H., Fukushima K., Fukuda T., Yokoyama J., Kimura S. (2014). Molecular phylogeny determined using chloroplast DNA inferred a new phylogenetic relationship of Rorippa aquatica (Eaton) EJ Palmer & Steyermark (Brassicaceae) — lake cress. Am. J. Plant Sci. 5: 48–54. [Google Scholar]
- Nakayama H., Nakayama N., Nakamasu A., Sinha N., Kimura S. (2012a). Toward elucidating the mechanisms that regulate heterophylly. Plant Morph. 24: 57–63. [Google Scholar]
- Nakayama H., Yamaguchi T., Tsukaya H. (2012b). Acquisition and diversification of cladodes: leaf-like organs in the genus Asparagus. Plant Cell 24: 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra A.B., Leigh A., Boyce C.K., Jones C.S., Niklas K.J., Royer D.L., Tsukaya H. (2011). The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 38: 535–552. [DOI] [PubMed] [Google Scholar]
- Perrière G., Gouy M. (1996). WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78: 364–369. [DOI] [PubMed] [Google Scholar]
- Piazza P., et al. (2010). Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr. Biol. 20: 2223–2228. [DOI] [PubMed] [Google Scholar]
- Pigliucci, M. (2010). Phenotypic plasticity. In Evolution: The Extended Synthesis, M. Pigliucci and G.M. Muller, eds (London: MIT Press), pp. 355–378. [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan R., Mustroph A., Boonman A., Akman M., Ammerlaan A.M., Breit T., Schranz M.E., Voesenek L.A., van Tienderen P.H. (2013). Root transcript profiling of two Rorippa species reveals gene clusters associated with extreme submergence tolerance. Plant Physiol. 163: 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Tsutsumi M., Ohtsubo A., Nishii K., Kuwabara A., Nagata T. (2008). Temperature-dependent changes of cell shape during heterophyllous leaf formation in Ludwigia arcuata (Onagraceae). Planta 228: 27–36. [DOI] [PubMed] [Google Scholar]
- Scofield S., Murray J.A. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60: 929–946. [DOI] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9: 484–489. [DOI] [PubMed] [Google Scholar]
- Shani E., Ben-Gera H., Shleizer-Burko S., Burko Y., Weiss D., Ori N. (2010). Cytokinin regulates compound leaf development in tomato. Plant Cell 22: 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wang H., Yu H., Zhong C., Zhang X., Peng J., Wang X. (2013). GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. J. Exp. Bot. 64: 1637–1647. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya, H. (2013). Leaf development. The Arabidopsis Book 11: e0163, doi/10.1199/tab.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2014). Comparative leaf development in angiosperms. Curr. Opin. Plant Biol. 17: 103–109. [DOI] [PubMed] [Google Scholar]
- Veit B. (2009). Hormone mediated regulation of the shoot apical meristem. Plant Mol. Biol. 69: 397–408. [DOI] [PubMed] [Google Scholar]
- Vlad D., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154. [Google Scholar]
- Wanke D. (2011). The ABA-mediated switch between submersed and emersed life-styles in aquatic macrophytes. J. Plant Res. 124: 467–475. [DOI] [PubMed] [Google Scholar]
- Wells C.K., Pigliucci M. (2000). Adaptive phenotypic plasticity: the case of heterophylly in aquatic plants. Perspect. Plant Ecol. Evol. Syst. 3: 1–18. [Google Scholar]
- Yamaguchi T., Nukazuka A., Tsukaya H. (2012). Leaf adaxial-abaxial polarity specification and lamina outgrowth: evolution and development. Plant Cell Physiol. 53: 1180–1194. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Yano S., Tsukaya H. (2010). Genetic framework for flattened leaf blade formation in unifacial leaves of Juncus prismatocarpus. Plant Cell 22: 2141–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68: 571–582. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W.L., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhou C., Han L., Li G., Chai M., Fu C., Cheng X., Wen J., Tang Y., Wang Z.Y. (2014). STM/BP-like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula. Plant Cell 26: 1464–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotz G., Wilhelm K., Becker A. (2011). Heteroblasty-A review. Bot. Rev. 77: 109–151. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.