Only a small fraction of the maize genome undergoes RNA-directed DNA methylation and, for several characteristics, the chromatin in these areas resembles euchromatin more than heterochromatin.
Abstract
RNA-directed DNA methylation (RdDM) in plants is a well-characterized example of RNA interference-related transcriptional gene silencing. To determine the relationships between RdDM and heterochromatin in the repeat-rich maize (Zea mays) genome, we performed whole-genome analyses of several heterochromatic features: dimethylation of lysine 9 and lysine 27 (H3K9me2 and H3K27me2), chromatin accessibility, DNA methylation, and small RNAs; we also analyzed two mutants that affect these processes, mediator of paramutation1 and zea methyltransferase2. The data revealed that the majority of the genome exists in a heterochromatic state defined by inaccessible chromatin that is marked by H3K9me2 and H3K27me2 but that lacks RdDM. The minority of the genome marked by RdDM was predominantly near genes, and its overall chromatin structure appeared more similar to euchromatin than to heterochromatin. These and other data indicate that the densely staining chromatin defined as heterochromatin differs fundamentally from RdDM-targeted chromatin. We propose that small interfering RNAs perform a specialized role in repressing transposons in accessible chromatin environments and that the bulk of heterochromatin is incompatible with small RNA production.
INTRODUCTION
Eukaryotic genomes typically contain a large fraction of repetitive DNA that appears to be dispensable, at least in terms of supporting the production of proteins. The abundance of the transposons and tandem repeats that make up this repetitive DNA is highly variable between species and even between populations within a single species, as illustrated by studies in maize (Zea mays) (Tenaillon et al., 2011; Kanizay et al., 2013). Repetitive regions of the genomes were identified early on by their dense staining, both by light and electron microscopy. Importantly, these regions were not just condensed during cell division but remained so in interphase cells. The term “heterochromatin” was first used by Heitz (1928), who was cytologically characterizing moss. At about the same time, McClintock (1929) described an extreme example of heterochromatin, the knobs of maize, which are now known to be made up of megabase-scale expansions of tandem repeats on chromosome arms that are still poorly understood and difficult to accurately assemble into genome reference sequences (Ghaffari et al., 2013). Similar to knobs in maize, the repeat-rich centromeric regions of many other organisms, including Drosophila melanogaster, mouse, and Arabidopsis thaliana, form visible heterochromatic structures called chromocenters (Peacock et al., 1974; Fransz et al., 2002; Guenatri et al., 2004). Another early observation was that entire inactive mouse X chromosomes adopt such densely staining structures (Barr and Bertram, 1949; Lyon, 1961), as does the entire set of paternal chromosomes of male mealybugs (Brown and Nur, 1964). These cytological observations have led to much research into the genetic features, behavior, and biochemical makeup of heterochromatin (reviewed in Slotkin and Martienssen, 2007). While the composition of heterochromatin varies within and among species, a unifying theme is the inhibition of gene expression; in fact, the term heterochromatin has become synonymous with the presence of specific chromatin modifications associated with gene silencing rather than with a condensed appearance in interphase nuclei. This broad concept of heterochromatin also includes facultative heterochromatin, which, among others, contains developmental genes silenced by Polycomb group complexes and associated histone modifications (reviewed in Schwartz and Pirrotta, 2013).
One of the biggest surprises in the last decade is that, although heterochromatin is associated with transcriptional gene silencing, transcription appears compatible with heterochromatin and actually contributes toward its establishment and maintenance (reviewed in Sabin et al., 2013). Forms of RNA-mediated silencing have been observed in plants, fungi, and animals. A well-studied example is RNA-directed DNA methylation (RdDM), which was initially discovered as a response to viral infection in tobacco (Nicotiana tabacum) (Wassenegger et al., 1994) and involves a process related to RNA interference that directs de novo DNA methyltransferase to specific loci (reviewed in Matzke and Mosher, 2014). The link between RdDM and gene silencing has led some authors to refer to the short RNA molecules involved in RdDM as heterochromatic short interfering RNAs (siRNAs) (Axtell, 2013; Pontes et al., 2013; Shin et al., 2013; Bologna and Voinnet, 2014; Creasey et al., 2014). RdDM is primarily associated with the repression of transposons, but it can indirectly affect gene expression through genomic imprinting (Vu et al., 2013) and paramutation (reviewed in Hollick, 2012). Multiple studies have suggested that RdDM is not a uniform feature of heterochromatin in plant genomes but is enriched in particular heterochromatic regions, including 5S ribosomal loci, long terminal repeats of retrotransposons, DNA transposons, and repetitive elements in general when close to genes (Onodera et al., 2005; Zheng et al., 2012; Zhong et al., 2012; Gent et al., 2013; Regulski et al., 2013; Stroud et al., 2013; Sun et al., 2013; Zemach et al., 2013; Diez et al., 2014; Wei et al., 2014). In Arabidopsis, methylation in long transposons near centromeres can be mediated by the DNA methyltransferase CMT2 without the involvement of RdDM (Zemach et al., 2013). Little is known about what triggers or sustains RdDM, but expression of transposons (Nuthikattu et al., 2013) and proximity to genes (Gent et al., 2013) have been implicated as well as specific chromatin modifications (Law et al., 2013; Johnson et al., 2014).
Maize presents a valuable model system for elucidating the roles and mechanisms underlying heterochromatin formation and maintenance, not only because of its historical significance in discoveries related to paramutation, genomic imprinting, and transposons (McClintock, 1950; Brink, 1956; Kermicle, 1970) but also because its heterochromatin is implicated in its phenotypic diversity (Eichten et al., 2011, 2012, 2013; Barber et al., 2012; Freeling et al., 2012; Regulski et al., 2013). Maize has an abundance of transposons and tandem repeats, including a diverse collection within and close to genes (Baucom et al., 2009; Schnable et al., 2009; Han et al., 2013), making it an attractive model for studying interactions between gene expression and transposon-related heterochromatin (Eichten et al., 2012; Gent et al., 2013). At a molecular level, the most thorough descriptions of maize heterochromatin have been related to DNA methylation and siRNAs (Eichten et al., 2011; Gent et al., 2013; Regulski et al., 2013). Multiple other studies have provided valuable insights into the distribution of DNA methylation, nucleosomes, and histone modifications (Eichten et al., 2012; Fincher et al., 2013; Labonne et al., 2013; Makarevitch et al., 2013; He et al., 2014; Madzima et al., 2014). These studies have been limited in their application to heterochromatin, however, because of poor resolution over repetitive regions, shortage of experimental data, or a focus on genic chromatin.
To characterize heterochromatin landscapes in the maize genome and their dependence on small RNA regulation, we performed chromatin immunoprecipitation and Illumina sequencing (ChIP-seq) for two histone H3 modifications, dimethylation of lysine 9 and lysine 27 (H3K9me2 and H3K27me2). In Arabidopsis, both of these modifications are associated with heterochromatin, while H3K27me2 is also located in silenced genes, as is trimethylation of lysine 27 (Roudier et al., 2011; Park et al., 2012). We observed H3K9me2 and H3K27me2 to be enriched in heterochromatin in maize as well. In regions undergoing RdDM, however, H3K27me2 remained similarly enriched, but H3K9me2 tended to be depleted relative to regions not undergoing RdDM. We also assayed chromatin accessibility by micrococcal nuclease digestion followed by sequencing (MNase-seq). We compared chromatin accessibility and histone modifications with existing siRNA and DNA methylation data (Gent et al., 2012, 2013; Regulski et al., 2013) as well as with new data we generated from mutants known to affect DNA methylation: two mutants lacking the chromomethylase ZEA METHYLTRANSFERASE2 (ZMET2 [or DMT102]) (Papa et al., 2001), which is predicted to function independently of RdDM, and one mutant lacking the RNA-dependent RNA polymerase (RdRP) MEDIATOR OF PARAMUTATION1 (MOP1) (Alleman et al., 2006), which is expected to be a key component of RdDM in maize. These data reveal that the maize genome is dominated by inaccessible heterochromatin that is depleted of small RNAs. Loci undergoing RdDM represent a distinct and scarce form of chromatin that, at least in maize, is not representative of heterochromatin as originally defined.
RESULTS
siRNA Profiling in mop1 Mutants and Control Samples as a Means of Defining Loci Undergoing RdDM
To identify candidate loci undergoing RdDM, we made use of a mop1 mutant (mop1-1). MOP1 is an RdRP homologous to Arabidopsis RDR2, which is a key factor in Arabidopsis RdDM because of its role in the synthesis of 24-nucleotide siRNAs (reviewed in Matzke and Mosher, 2014). The maize genome encodes nine putative RdRPs, including one (GRMZMG481730) with 35% amino acid identify to MOP1, but MOP1 alone is required for accumulation of the vast majority of 24-nucleotide siRNAs and for DNA methylation at specific loci in maize, as evidenced by small RNA sequencing and RNA and DNA gel blots in wild-type and mop1 mutant plants (Lisch et al., 2002; Woodhouse et al., 2006; Nobuta et al., 2008). The primary function of the 24-nucleotide class of siRNAs is to mediate RdDM (reviewed in Matzke and Mosher, 2014). Therefore, we sequenced siRNAs from developing ears from mop1-1 and wild-type siblings and compared the two samples to identify candidate RdDM loci. This MOP1-based definition of RdDM is used with the understanding that many other proteins besides MOP1 are known to contribute to RdDM, and we cannot exclude that the full range of sequences under the control of RdDM may be larger.
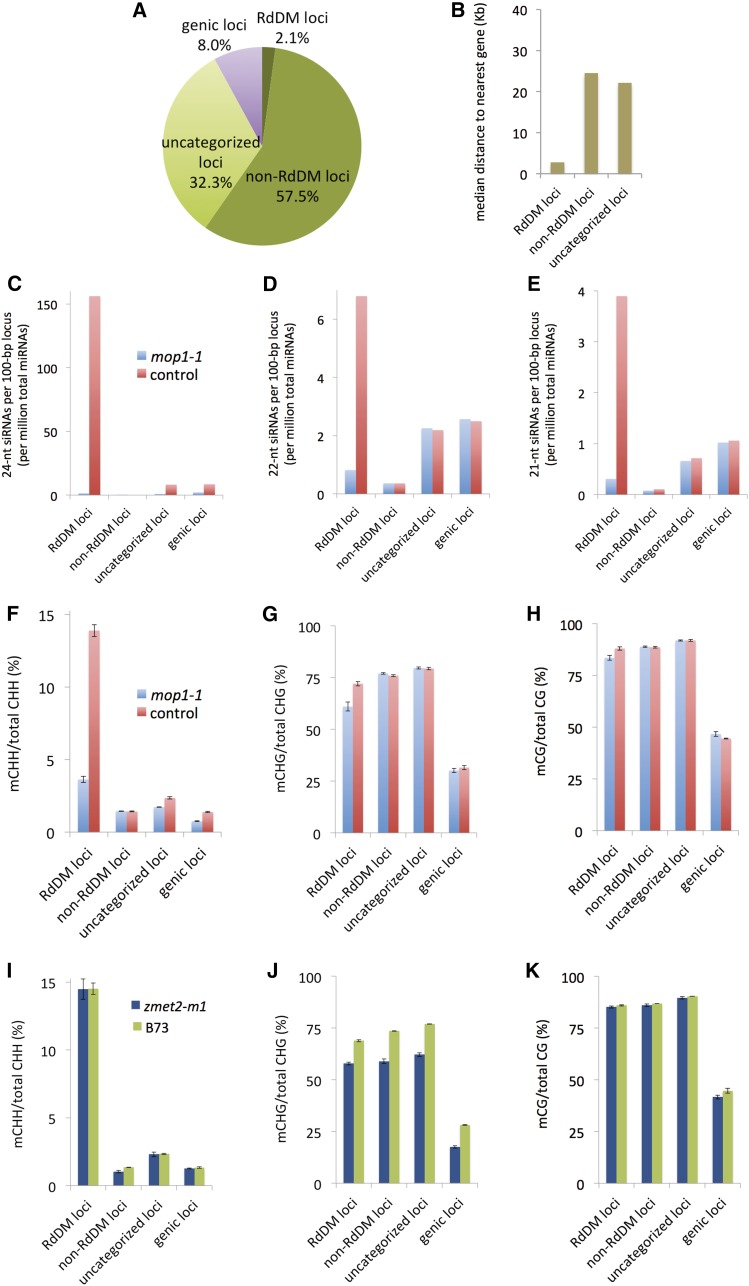
As a first step in the data analysis, we split the genome into nonoverlapping 100-bp loci and separated them by whether they were intergenic or genic (within exons or introns). From the intergenic loci, we then identified “RdDM loci” based on whether 24-nucleotide siRNA abundance was reduced by at least 3-fold in the mop1 mutant (Figure 1A). For quantification of siRNA abundance, we counted all uniquely mapping siRNAs with at least a 50% overlap in the 100-bp windows and normalized the values over total microRNA (miRNA) counts. Using miRNAs as a standard is expected to be more accurate than using total small RNAs because of the dramatic loss of siRNAs in mop1 mutants. We defined “non-RdDM loci” by the absence of 24-nucleotide siRNAs (unique or nonunique) in both of two different wild-type samples (B73 and wild-type siblings of mop1 individuals). Loci that did not fit into either the RdDM or the non-RdDM category were assigned to “uncategorized loci.” Additional details of the categorization scheme can be found in Methods. By these definitions, RdDM loci constituted 2% of the genome, non-RdDM loci constituted 58% of the genome, and uncategorized loci constituted 32% of the genome. Consistent with recent evidence suggesting that RdDM can be promoted by unidentified cis-acting effects of genes (Gent et al., 2013), the median distance between the edges of RdDM loci and the edges of the nearest gene was 2.7 kb, while the median distance between the edges of non-RdDM loci and the edges of the nearest gene was 24.5 kb (Figure 1B). The median distance for uncategorized loci was 22.2 kb, similar to non-RdDM loci.
Figure 1.
An siRNA- and MOP1-Based Subdivision of the Genome, and Corresponding Patterns of siRNA and DNA Methylation Levels.
(A) Pie chart of the genome according to genic and RdDM status in B73 developing ears. The genome was divided into 100-bp loci, and each locus was categorized as either genic or one of three categories of intergenic: RdDM, non-RdDM, and uncategorized.
(B) Median distance to genes for each locus category. The height of each bar indicates the median distance from the edge of a locus to the edge of the closest gene.
(C) to (E) The 21-, 22-, and 24-nucleotide siRNA abundance in each locus category. Raw counts were normalized both by the number of loci in each category and by the number of miRNAs in each sequencing library. Both uniquely and nonuniquely mapping siRNA reads were included in the counts. For nonunique reads, one of multiple, equally likely positions was selected at random. Note the difference in scales in (C), (D), and (E).
(F) to (K) DNA methylation in each locus category, with CHH, CHG, and CG sequence contexts separated, in the mop1-1 mutant ([F] to [H]), the zmet2-m1 mutant ([I] to [K]), and wild-type controls. The values are percentages of methylated 5-methylcytosines (mC) per total cytosines in each context. Error bars represent se derived from two or three BS-seq biological replicates.
RdDM loci on average exhibited a 112-fold decrease in 24-nucleotide siRNAs in the mop1-1 mutant when we counted both unique and nonunique siRNAs (Figure 1C). Uncategorized loci, which we defined to include loci putatively associated with MOP1-independent siRNAs, exhibited a 9.2-fold decrease in 24-nucleotide siRNAs in the mop1 mutant. We note, however, that in wild-type individuals, uncategorized loci already had 20-fold lower 24-nucleotide siRNA levels than RdDM loci. These data suggest that the majority of uncategorized loci were either not undergoing RdDM or were undergoing RdDM at much lower levels than RdDM loci.
The next most-abundant sizes of siRNAs in the maize genome are 21- and 22-nucleotide siRNAs (Nobuta et al., 2008), which represent diverse classes of small RNAs produced from different sources, including large hairpins, overlapping transcripts, and RdRP templates (reviewed in Axtell, 2013). Certain transposon-derived 21- and 22-nucleotide siRNAs have been shown to function in RdDM in Arabidopsis (Nuthikattu et al., 2013). We found that the accumulation of 21- and 22-nucleotide siRNAs was also dependent upon MOP1 at RdDM loci (Figures 1D and 1E). In the mop1-1 mutant, 21-nucleotide siRNAs exhibited a 12.6-fold decrease and 22-nucleotide siRNAs exhibited an 8.3-fold decrease. Note that these smaller siRNAs were of lower abundance to begin with (see the different scales in Figures 1C to 1E). These results suggest a role for MOP1 in the production of 21- and 22-nucleotide siRNAs, which would be surprising because 21- and 22-nucleotide siRNAs in Arabidopsis are produced by RDR6 rather than by its MOP1 homolog RDR2 (Nuthikattu et al., 2013). We also compared our new siRNA data from developing ears with siRNA levels from published studies in root tips (Gent et al., 2012) and coleoptile-stage shoots (Regulski et al., 2013). While siRNAs were less abundant in these other tissues (relative to miRNAs), similar trends were evident: highest levels of all three classes in RdDM loci and a lack of 21-, 22-, and 24-nucleotide siRNAs in the non-RdDM loci (Supplemental Figures 1A to 1C).
DNA Methylation Confirms siRNA-Based Identification of RdDM Loci
Although the immediate output of MOP1 activity is predicted to be double-stranded RNA, a downstream consequence is DNA methylation. To determine the effect of MOP1 on DNA methylation genome-wide, we performed bisulfite sequencing (BS-seq) in the same developing ear samples we used for siRNA sequencing and measured DNA methylation in each of the three major sequence contexts (CG, CHG, and CHH, where H = A, C, or T). While RdDM methylates cytosines in all three sequence contexts, CHH methylation is the best indicator of RdDM, as CG and CHG methylation can be maintained by other DNA methylation pathways (reviewed in Matzke and Mosher, 2014). In maize, for this reason, loci undergoing RdDM were called CHH islands (Gent et al., 2013). We measured CHH methylation as being nearly 10-fold higher in RdDM loci than in non-RdDM loci (Figure 1F). In the mop1 mutant, compared with the wild type, CHH methylation exhibited a nearly 4-fold reduction in RdDM loci but no reduction in other loci. Nevertheless, mop1-1 did not remove all CHH methylation at RdDM loci, and the remaining levels (3.6%) were substantially higher than in non-RdDM loci (1.4%). This residual CHH methylation may be triggered by MOP1-independent siRNAs or by siRNA-independent DNA methylase activity at these loci. The low levels of CHH methylation observed in uncategorized and genic loci showed a modest decrease in the mop1 mutant (a 1.4-fold decrease, from 2.3% down to 1.7%), also indicating that the majority of the uncategorized loci were not undergoing RdDM or were undergoing RdDM at very low levels. CHG and CG methylation was significantly higher than CHH methylation overall and did not differ appreciably between RdDM, non-RdDM, and uncategorized loci (Figures 1F to 1H). CHG and CG methylation was significantly lower in genic loci, as expected. In the mop1 mutant, the only substantial reduction we observed for CG and CHG methylation was in RdDM loci (Figures 1G and 1H).
To test the generality of our observations in other tissues, we compared our new DNA methylation results with those from a previous study in coleoptile shoots (Regulski et al., 2013) and from mature but unfertilized ear tissue (Gent et al., 2013) (Supplemental Figures 1D to 1F). Regardless of differences in total methylation levels between tissues, all three tissues showed a similar trend: relatively high CHH methylation at RdDM loci, low CHH methylation at other loci, and high CG and CHG methylation at all loci except genes.
Limited Effect of Chromomethylase Activity on CHH Methylation
In Arabidopsis, the majority of CHH methylation occurs independent of RdDM and is mediated by two chromomethylases: CMT2, which is absent in maize, and CMT3, which contributes primarily to CHG methylation and to a lesser extent also to CHH methylation (Stroud et al., 2013, 2014; Zemach et al., 2013). Maize has two CMT3 homologs, one of which is ZMET2 (alias DMT102). ZMET2 has been characterized at multiple levels (Papa et al., 2001; Makarevitch et al., 2007; Garcia-Aguilar et al., 2010; Du et al., 2012), but its genome-wide contributions to DNA methylation remain unclear. Therefore, we performed BS-seq in developing ears of two different zmet2 mutants (zmet2-m1 and zmet2-mu1013094). Both zmet2 mutants exhibited a clear effect only on CHG methylation, with a drop in methylation levels across both genic and intergenic regions regardless of the involvement of RdDM (Figures 1I to 1K; Supplemental Figures 2G to 2L). The only statistically significant change in CHH methylation was an ∼11% decrease in non-RdDM loci (one-tailed Student’s t test, P = 0.015 for zmet2-m1 versus its control and P = 0.001 for zmet2-mu1013094 versus its control). Although zmet2 has a close homolog and its overall phenotype is fairly weak (reducing CHG methylation by 20% on average), these data support the assertions that CHH methylation at RdDM loci is primarily directed by MOP1 and that the majority of its activity is localized to a small portion of intergenic regions (Figures 1A and 1F).
Accessibility of RdDM Loci to Micrococcal Nuclease
A theoretical explanation for the relatively small median distance between RdDM and genes (Figure 1B) is that the chromatin within and flanking genes is in a relatively open state to be accessible to RNA polymerases, which are required for genic transcription as well as RdDM. To determine the chromatin accessibility at RdDM loci, we measured the sensitivity of DNA to nuclease digestion by micrococcal nuclease (MNase). This enzyme is well known for its ability to selectively digest DNA that is not bound by nucleosomes but can also be used to distinguish accessible from inaccessible chromatin (reviewed in Telford and Stewart, 1989). To measure genome-wide chromatin accessibility, we performed MNase-seq using developing ear tissue. Briefly, we isolated chromatin and treated it with MNase for varying lengths of time to increase the extent of digestion (Figure 2A). With extensive digestion, the majority of the chromatin was digested to 150-bp fragments, the amount of DNA protected by single nucleosomes. With less extensive digestion, longer fragments were also produced, corresponding to arrays of nucleosomes and their intervening linker DNA. The most accessible nucleosomes in the genome should be rapidly released from the bulk chromatin, while the inaccessible nucleosomes should resist digestion for longer. Hence, sequencing the 150-bp DNA fragments from the shorter versus longer MNase digestions reveals which portions of the genome are accessible and inaccessible. We chose a series of three digestion times for our analysis, sequenced the 150-bp fragments of each sample, and called them “accessible chromatin,” “mid-accessible chromatin,” and “inaccessible chromatin.”
Figure 2.
Chromatin Accessibility Revealed by MNase-seq.
(A) Ethidium bromide-stained agarose gel showing the extent of MNase digestion. Bands of DNA of ∼150-bp were excised from the indicated lanes and sequenced with Illumina HiSeq 2500.
(B) Enrichment for each set of DNA fragments near genes in B73 developing ears. All genes were defined by their annotated transcription start and stop sites and split into nonoverlapping 100-bp intervals. The number of uniquely mapping reads in each interval was summed from 4 kb upstream to 1 kb downstream of the transcription start site and from 1 kb upstream to 4 kb downstream of the stop site. To calculate enrichment relative to the genome average, the read counts for the 100-nucleotide intervals in the set of all genes were normalized by read counts derived from randomly sheared DNA.
(C) Chromatin accessibility ratio near genes. All genes were defined by their annotated transcription start and stop sites and the number of uniquely mapping reads summed for each 100 bp. The normalized read counts from the accessible chromatin library were divided by the normalized read counts from the inaccessible chromatin library to determine accessibility ratios. Each line represents a different set of genes, defined by their gene expression levels (RNA sequencing) in developing ears. Genes with zero matching reads were assigned to the zero category, and the remaining genes were split into four quartiles, low to high expression.
(D) Chromatin accessibility ratio for each locus category described in Figure 1A.
(E) Histogram of AT content in reads from samples representing the different accessibility categories. The average percentage of A + T was calculated for the reads in each set. Randomly fragmented reads were the same ones used for normalization in (B).
(F) AT content in each locus category. The average percentage of A + T was calculated for the 100-bp loci in each category. Error bars represent sd.
(G) Chromatin accessibility ratio in each locus category, as in (D), but only including 100-bp loci with moderate AT content: at least 50% but less than 60% AT.
To determine whether the digestion method efficiently separated accessible from inaccessible chromatin, we examined the enrichment of each sample relative to the genome average (Figure 2B). Accessible chromatin was enriched for genes and their immediate flanking regions, and distance from genes correlated with an increased enrichment of inaccessible chromatin, which is expected since genes are generally flanked by heterochromatin in maize. Mid-accessible chromatin exhibited an intermediate distribution. Using 100-bp windows, we divided the normalized read count for accessible chromatin by the normalized read count for inaccessible chromatin to produce a “chromatin accessibility ratio.” This is a measure of chromatin accessibility where the genome average is 1 and more accessible chromatin is represented by numbers higher than 1. There was a clear correlation between steady state gene expression levels in developing ears and the accessibility ratio, with the transcription starts and ends of highly expressed genes being most accessible (Figure 2C). The average accessibility ratio across all genic loci was 1.52 (Figure 2D). RdDM loci had an average accessibility ratio of 1.33, lower than genic loci but substantially higher than non-RdDM loci (accessibility ratio of 0.96) or uncategorized loci (accessibility ratio of 0.89).
A complicating factor in the accessibility assays is that AT content within nucleosome-bound DNA is known to positively influence its susceptibility to MNase digestion. As a result, AT-rich DNA may artificially appear to have reduced nucleosome occupancy upon extensive MNase digestion (McGhee and Felsenfeld, 1983; Tillo and Hughes, 2009). To determine if such an effect was introducing biases in coverage in our MNase-seq assay, we measured the abundance of A and T in each of our samples. We found that reads with <50% AT were enriched while reads with ≥60% AT were depleted from the most extensive MNase digestion (“inaccessible chromatin”), confirming that AT content could affect the outcome of this experiment (Figure 2E). Thus, we measured the AT content of each category of loci to determine whether what we attributed to chromatin accessibility might actually be attributed simply to differing AT content. However, we found no substantial differences in AT content between the different categories of loci (Figure 2F). We also removed all loci with <50% AT or ≥60% AT and measured the chromatin accessibility ratio. The results were nearly identical to the analysis of all loci, except that the accessibility ratio for uncategorized loci increased from 0.89 to 0.96 (Figure 2G). The fact that we obtained consistent results whether we included all loci or just those with moderate AT content provides strong evidence that chromatin accessibility indeed differs between the four categories of loci, with genic and RdDM loci being the most accessible.
Enrichment of Both H3K9me2 and H3K27me2 in Heterochromatin but Relative Depletion of H3K9me2 at RdDM Loci
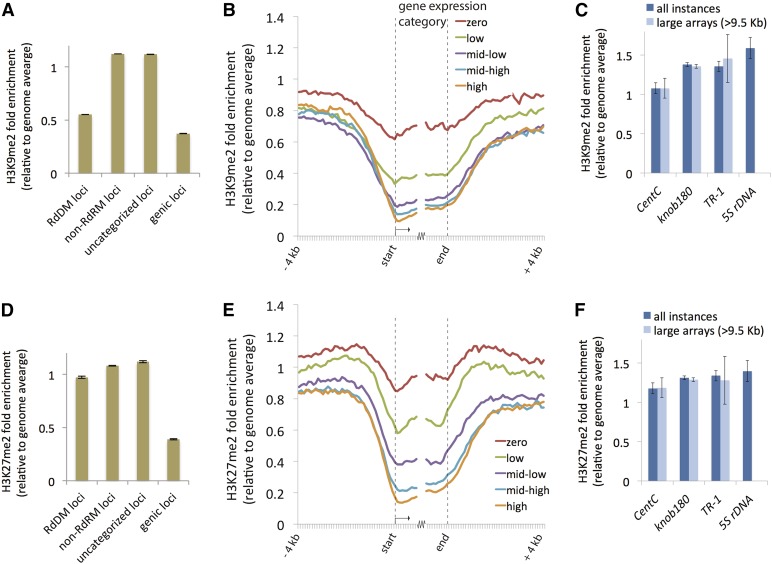
Our finding that the chromatin at RdDM loci is relatively accessible led us to ask what other chromatin features might associate with these loci. We performed ChIP-seq using chromatin isolated from young stalk tissue. H3K9me2 is a general marker of inactive chromatin (Law and Jacobsen, 2010) and marks inaccessible chromatin in Arabidopsis (Shu et al., 2012), while H3K27me2 is involved in the repression of repetitive elements as well as genes (Roudier et al., 2011; Ferrari et al., 2014). H3K9me2 levels showed an inverse relationship to chromatin accessibility (compare Figure 2D with Figure 3A) and were reduced by approximately 2-fold in RdDM loci relative to non-RdDM and uncategorized loci. However, H3K9me2 levels at RdDM loci were more similar to genic loci than to non-RdDM and uncategorized loci (Figure 3A). By contrast, H3K27me2 was nearly as abundant in RdDM as in non-RdDM and uncategorized loci (Figure 3D). H3K9me2 and H3K27me2 exhibited a strong tendency for genome-wide enrichment in the same regions, although H3K27me2 appeared to be enriched at a larger set of loci (Supplemental Figure 3). Both modifications were depleted within genes relative to intergenic DNA and exhibited an inverse correlation with gene expression levels (Figures 3B and 3E). H3K9me2 was primarily enriched in poorly expressed genes, while H3K27me2 was enriched in a broader set of moderately expressed genes. The two marks may directly regulate gene expression within gene bodies, but it is also possible that transposon insertions within genes attract H3K9me2 and H3K27me2, resulting in decreased gene expression.
Figure 3.
H3K9me2 and H3K27me2 Distribution within the Maize Genome.
(A) H3K9me2 ChIP enrichment in each locus category in B73 young stalk tissue. Error bars represent se derived from two biological replicates.
(B) H3K9me2 ChIP enrichment profile near genes in B73 young stalk tissue. All genes were defined by their annotated transcription start and stop sites and split into nonoverlapping 100-bp intervals. The numbers of uniquely mapping reads in each interval were summed from 4 kb upstream to 1 kb downstream of the transcription start site and from 1 kb upstream to 4 kb downstream of the stop site. To produce enrichment relative to the genome average, the counts for the 100-nucleotide intervals in each set of genes were normalized by read counts derived from randomly sheared DNA. Each line represents a different set of genes, defined by their gene expression levels as in Figure 2C except that gene expression data were obtained from stem and shoot apical meristem tissue of V3-stage plants (Sekhon et al., 2013).
(C) H3K9me2 ChIP enrichment in tandem repeats in B73 young stalk tissue. All instances includes all homologous loci in the reference genome; large arrays (>9.5 kb) is the subset of loci long enough to consist of at least 50 nucleosomes. As in (A) and (B), only uniquely mapping reads were included in both the all-instances and large-arrays analyses. For nonunique reads, see Supplemental Figure 4.
(D) H3K27me2 ChIP enrichment in each locus category, as in (A).
Heterochromatic Structure and RdDM Depletion of Knobs and Centromeres
The maize genome is notable not only for its numerous transposons but also for its tandem repeats. The heterochromatic knobs discovered by McClintock (1929) are composed primarily of repeated units of 180 and 350 bp (called knob180 and TR-1, respectively) that can occur in arrays of diverse sizes and positions throughout the genome (Ghaffari et al., 2013; Kanizay et al., 2013). Maize centromeres contain a 156-bp repeating unit called CentC (Wolfgruber et al., 2009). knob180 alone is estimated to make up between 1 and 6% of the genome (Peacock et al., 1981; Estep et al., 2013) and CentC another 0.2% (Bilinski et al., 2014).
Although most of the tandem repeats in maize are not present in the genome reference assembly, there is sufficient polymorphism among repeat monomers that a few long arrays have been assembled, although they are not currently annotated as such (as illustrated by a recent analysis of CentC [Bilinski et al., 2014]). To identify locations of CentC, knob180, and TR-1 as well as the repetitive 5S ribosomal subunit plus spacer sequence, we aligned a reference sequence of each repeat to the assembled genome. We determined H3K9me2 and H3K27me2 enrichment for each tandem repeat locus, and all four exhibited high levels of both modifications, even higher than the genome average (Figures 3C and 3F). This result held true whether we included all loci homologous to the repeats (including single monomers and short arrays) or restricted the analysis to arrays longer than 9.5 kb (see Methods).
We also looked at chromatin accessibility, siRNAs, and DNA methylation of the four tandem repeat loci. Unfortunately, tandem repeats cannot be reliably assayed by our chromatin accessibility assay because each sequence has a different AT content and may have a different propensity toward MNase digestion (in particular, the TR-1 repeat is 64% AT and shows an extreme, highly suspicious accessibility ratio of 2.4). We also found that 24-nucleotide siRNA reads and 100-bp bisulfite-treated reads (where C’s are converted to T’s) rarely mapped uniquely within tandem repeat arrays. Therefore, we aligned siRNA and bisulfite reads directly to CentC, knob180, TR-1, and 5S reference sequences to determine their enrichment in each data set. All but the 5S repeat behaved as non-RdDM loci; they displayed small RNA depletion, low CHH methylation, and H3K9me2 and H3K27me2 enrichment (Supplemental Figure 4). Consistent with work from Arabidopsis (Onodera et al., 2005), 5S repeats showed evidence of targeting by RdDM.
CentC is of special interest because of its enrichment in centromeres. CentC displayed lower levels of H3K9me2, H3K27me2, and CHG methylation than knob180 and TR-1 (Figures 3C and 3F; Supplemental Figures 4C, 4E, and 4F). This makes sense because, in centromeres, a subset of histone H3 is replaced by the histone H3 variant cenH3/CENP-A, which lacks the Lys-9 and Lys-27 residues (Bailey et al., 2013). Besides CentC, maize centromeres (defined as regions of cenH3 enrichment) contain a complex mixture of other DNA elements, including abundant and polymorphic retrotransposons that allow for the mapping of unique reads (excluding most CentC and other sequences with near-perfect copies). Previous work indicated that maize centromeres are depleted of siRNAs and CHH methylation but enriched in CHG and CG methylation (Gent et al., 2012; Regulski et al., 2013). In addition to confirming these observations (Supplemental Figures 2A to 2L), we found that centromeric loci were relatively inaccessible to MNase (Supplemental Figure 2M) and had intermediate enrichment for H3K9me2 and H3K27me2 (Supplemental Figure 2N). Taken together, these results indicate that regions traditionally viewed as heterochromatic are enriched in H3K9me2 and H3K27me2 and in CG and CHG methylation but are depleted of both CHH methylation and small RNAs.
DISCUSSION
Similar to the pioneering studies linking small RNAs to histone modifications in the fission yeast Schizosaccharomyces pombe, a rich and extensive set of observations in plants has outlined a molecular mechanism linking 24-nucleotide siRNAs to DNA methylation and heterochromatin (reviewed in Sabin et al., 2013). Recent studies from Arabidopsis further suggest that RdDM is enriched in specific genomic contexts such as recently inserted transposons and promoters (Zheng et al., 2012; Zhong et al., 2012). In maize, both 24-nucleotide siRNAs and CHH methylation are more abundant near genes than in the expanses of repetitive intergenic DNA, giving rise to the term CHH islands (Gent et al., 2013). In addition, 24-nucleotide siRNAs in maize are much more highly associated with DNA transposons, which tend to be close to genes, than with LTR retrotransposons, which make up most of the intergenic spaces (Gent et al., 2013; Diez et al., 2014). The results of our genome-level studies using BS-seq in maize mutants, chromatin immunoprecipitation (ChIP), and MNase digestion extend and quantify these observations and indicate that heterochromatin in the traditional sense is structurally distinct from chromatin engaged in RdDM.
Using the effect of the mop1-1 mutant on 24-nucleotide siRNA levels to identify RdDM regions, we conservatively estimated RdDM and non-RdDM loci to make up 2 and 58% of the genome, respectively (Figure 1A). The real extent of non-RdDM loci is larger, however, because the 32% of the genome we left uncategorized consistently looked more like non-RdDM loci than RdDM loci. We also note, however, that lack of RdDM at particular loci in ears, root tips, and coleoptile-stage seedlings does not mean that such loci can never undergo RdDM. In Arabidopsis, conditions that disrupt heterochromatin can increase siRNA production and RdDM in genomic regions where siRNAs are normally not produced (Tanurdzic et al., 2008; Schoft et al., 2009; Slotkin et al., 2009; Creasey et al., 2014). We found that MOP1 activity was also associated with 21- and 22-nucleotide siRNAs derived from the same loci as 24-nucleotide siRNAs (Figures 1C to 1E).
DNA methylation analysis confirmed that in maize ears and coleoptile-stage seedlings, RdDM is a minor feature of chromatin regulation genome-wide, even though the vast majority of the genome is heterochromatic. In the intergenic genome, there is very high DNA methylation in the CG and CHG contexts (∼90% CG methylation and 75% CHG methylation) and very little CHH methylation (∼2%) (Figures 1A and 1F to 1H). The scarcity of CHH methylation in the maize genome can theoretically be explained by the fact that maize lacks a homolog of Arabidopsis CMT2, the major chromomethylase that functions independently of RdDM in CHH methylation (Zemach et al., 2013; Stroud et al., 2014). In Arabidopsis, CMT3 also contributes in a minor but significant way to CHH methylation (Stroud et al., 2013). We tested mutants in a maize homolog of CMT3 (ZMET2) and found that, in maize, the contribution to CHH methylation by ZMET2 was also small but significant and limited to non-RdDM loci (Figure 1I; Supplemental Figures 2G and 2J). The fact that ZMET2 has a close homolog in maize (ZMET5) might explain some of the residual CHH methylation in the zmet2 mutant. In contrast with ZMET2, MOP1 had no detectable affect on CHH methylation in non-RdDM loci.
Active genes lie in open, accessible chromatin, while heterochromatin shields DNA from exposure to enzymes and other proteins and is differentiated from euchromatin by specific histone modifications such as H3K9me2 (Shu et al., 2012). Given that RdDM in maize targets repetitive DNA close to genes by a mechanism that depends on transcription, it was unclear whether RdDM loci would resemble genes or repetitive DNA in MNase accessibility and ChIP assays. Our data revealed that the accessibility of RdDM loci more closely resembles genes than classical heterochromatin, as RdDM regions, like genic regions, are relatively rapidly released upon short digestion (Figure 2). H3K9me2 is a hallmark of heterochromatin, and in Arabidopsis it correlates closely with CHG methylation (Du et al., 2012). Chromomethylases of the Arabidopsis CMT3 type contain chromodomains that recognize and bind to H3K9me2 and methylate DNA primarily in the CHG context. The enzymes that mediate H3K9me2 methylation reinforce this association by binding to methylated cytosines in the CHG context, creating a feedback loop between DNA methylation and histone methylation (reviewed in Law and Jacobsen, 2010). We saw the expected genome-wide correlation between H3K9me2 and CHG in maize; however, H3K9me2 methylation was relatively low in RdDM loci even though CHG methylation was relatively high (Figures 1G and 3A). Some of the CHG methylation in RdDM loci is contributed by the RdDM pathway itself, which methylates DNA in all sequence contexts (Figure 1G). We also performed ChIP for H3K27me2, which is involved in the repression of repetitive elements as well as genes in Arabidopsis (Roudier et al., 2011), and found that H3K27me2 did not differentiate RdDM loci from other repetitive regions (Figure 3D). Rather, it was enriched in all categories of intergenic loci. It was also inversely correlated with gene expression levels in and near genes (Figure 3E).
Heterochromatin was originally defined by its dense staining at the cytological level, and maize knobs are classic examples, being huge ball-shaped structures that remain condensed throughout the cell cycle (McClintock, 1929). Although in prior work we did not find a clear H3K9me2 knob staining pattern in meiotic chromosome spreads or interphase mitotic root cells (Shi and Dawe, 2006), more recent cytogenetic data from root tip chromosomes (He et al., 2014) and ChIP-seq data from developing ear tissue reported herein clearly show that knobs are marked by H3K9me2 (Figure 3C). Neither type of knob repeat showed any evidence of RdDM as assayed by 24-nucleotide siRNAs (Supplemental Figure 4A). Together, these data indicate that both the vast intergenic spaces and the visible heterochromatic knob domains in maize are not subject to RdDM.
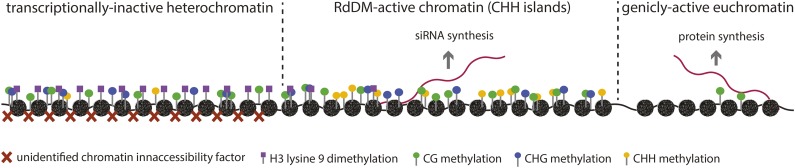
Our results are qualitatively summarized in Figure 4. The key results are that RdDM loci are primarily limited to regions close to genes, where they are characterized by accessible chromatin and relatively low H3K9me2. By contrast, the large expanses of intergenic DNA, primarily consisting of retrotransposons and tandem repeats, are enriched for H3K9me2 and inaccessible chromatin. Interestingly, for Arabidopsis, a subdivision of intergenic chromatin has been suggested (Veiseth et al., 2011) that may be analogous to RdDM and non-RdDM loci in maize. What we refer to as RdDM may be equivalent to the “intermediate heterochromatin” of Veiseth et al. (2011), and what we refer to as non-RdDM may be equivalent to their “centromeric and pericentromeric heterochromatin.” This Arabidopsis classification in part reflects a difference in H3K9me2 abundance similar to what we report. Similarly, we propose that heterochromatin marked by the activity of the chromatin remodeler DDM1 in Arabidopsis (Zemach et al., 2013) is equivalent to maize non-RdDM loci, and we expect that the maize homologs of DDM1 will function primarily in inaccessible, transcriptionally inactive regions.
Figure 4.
Conceptual Illustration of Three Major Chromatin Types.
In a simplified model, DNA regions we identified as non-RdDM loci correspond to transcriptionally inactive heterochromatin, while RdDM loci correspond to a distinct form of chromatin that is transcriptionally active. Transcription in RdDM loci, however, templates siRNA formation, which in turn leads to RdDM and reinforces silencing. Genically active euchromatin is accessible to nucleases and carries relatively low levels of DNA methylation, allowing for mRNA production.
The RdDM and non-RdDM pathways repress repetitive DNA through different mechanisms, raising the question of why there are two means to the same end. Hypothetically, forms of heterochromatin that completely seal off the DNA would be detrimental to nearby genes. By contrast, RdDM inactivates transposons close to genes in a transcriptionally compatible chromatin environment similar to what genes require. The key difference between genes and RdDM loci is that the transcription of one leads to translatable mRNAs while the transcription of the other leads to siRNAs and the inhibition of mRNA production (Figure 4). RdDM may be more compatible with gene expression, but it presumably comes at higher metabolic costs and incurs some risk, as transposable elements and other repeats are transcribed in the process. Conversely, it is possible that non-RdDM intergenic heterochromatin may have its own benefits in addition to transposon repression, such as in regulating centromere activity (Folco et al., 2008), telomere function (Gonzalo et al., 2006), recombination frequency (Fedoroff, 2012), and hybrid vigor (Freeling et al., 2012). We anticipate that further research into heterochromatin regulation will lead to a better understanding of how genomes are structured to accommodate the conflicting demands of repressing transposable elements while allowing genes to be precisely controlled and will also help us understand how to best utilize this information for agricultural purposes.
METHODS
Plant Materials and Genetic Stocks
For all experiments, developing maize (Zea mays) ears of 4 to 6 cm in length were used as the source tissue, with the exception of ChIP experiments, for which young stalk tissue was used (1-month-old plants approximately in the V4 stage with roots and exposed leaf blades removed). The inbred stock B73 (Schnable et al., 2009) was used for chromatin accessibility experiments, ChIP, and the randomly fragmented DNA control. A stock that was segregating mop1-1 in a B73 genetic background (Madzima et al., 2014) was used for small RNA and DNA methylation analyses. Siblings that were homozygous wild type for mop1 were used as controls. These wild-type, B73-derived siblings were also used as the source material for mRNA sequencing. Two mutant alleles of zmet2 were included in the DNA methylation analysis. zmet2-m1 had been introgressed previously into a B73 background (Makarevitch et al., 2007), and B73 was used as its wild-type control. The second allele, zmet2-mu1013094, was produced by the UniformMu resource (McCarty et al., 2005), and a stock segregating the allele, UFMu-00744, was obtained from the Maize Genetics Cooperation Stock Center. Heterozygous siblings were used as wild-type controls.
Preparation of Small RNA Sequencing Libraries
Small RNA-enriched RNA was extracted from developing ear tissue of four mop1-1 homozygotes and four wild-type homozygotes using the mirVana miRNA Isolation Kit (Life Technologies; AM1560) with Plant RNA Isolation Aid (Life Technologies; AM9690). Individual small RNA-enriched samples from each set of four were combined in equal mass into two pairs; 600 ng of each sample was ligated to an adenylated 3′ adapter (1.25 μM) with 5% polyethylene glycol 8000 and 10 units/μL T4 RNA Ligase 2 (truncated K227Q; New England Biolabs; M0351S) in a 10-μL reaction volume. The entire product was then directly ligated to an RNA/DNA hybrid 5′ adapter (1 μM) with 0.4 units/μL T4 RNA Ligase 1 (New England Biolabs; M0204S) in a 12-μL reaction volume. Six microliters of the adapter-ligated product was directly reverse transcribed with SuperScript II Reverse Transcriptase (Life Technologies; 18064-014) in a 10-μL volume with 1 μM primer (same indexed reverse primer as used for PCR; see below). Six microliters of cDNA was directly PCR-amplified using FailSafe PCR 2X premix buffer (Epicentre; FSP995E), 200 nM forward primer, 100 nM reverse primer (in addition to the reverse primer remaining after reverse transcription), and 1 μL of FailSafe PCR Enzyme Mix (Epicentre; FS99100) in a 50-μL total volume. Thermocycler conditions were as follows: an initial denaturation at 95°C for 1 min; no more than 15 cycles of 95°C for 30 s, 50°C for 30 s, and 68°C for 1 min; and a final extension at 68°C for 7 min. Amplicons of ∼140 to 150 bp in length were selected by gel electrophoresis using a 4% NuSieve GTG agarose gel (Lonza; 50080). The DNA was extracted from the gel with a QIAquick gel extraction kit (Qiagen; 28704) without heating. Adapter and primer sequences are listed in Supplemental Table 1.
Preparation of mRNA Sequencing Libraries
Total RNA was extracted from the same six B73 near-isogenic developing ear samples that were used as wild-type controls for mop1-1 BS-seq experiments, using the mirVana miRNA Isolation Kit (Life Technologies; AM1560) with Plant RNA Isolation Aid (Life Technologies; AM9690). The six individual isolated RNA samples were combined in equal mass into three pairs, and 5 μg of each pair was treated with DNase I using the DNA-Free RNA Kit (Zymo Research; R1013) using the small RNA elimination procedure. Sequencing libraries were prepared using a dUTP-stranded procedure according to the Illumina guidelines (TruSeq Stranded Total RNA Sample Preparation Guide, Part 15031048, Rev. C)
Preparation of BS-seq Libraries
For each sample, DNA was extracted from two developing ears (∼5 cm in length) using the DNeasy Plant Mini Kit (Qiagen; 69106), and DNA from each of the two ears was combined in equal mass. Illumina guidelines (Whole-genome Bisulfite Sequencing for Methylation Analysis, Part 15,021,861, Rev. B) were followed, with the following changes: 6 μg of DNA was fragmented by sonication, purified using the Agencourt AMPure XP protocol (1.8:1 ratio), and ligated to adapters in a reaction with 1.85 μM Y adapter consisting of the premethylated DNA oligonucleotides listed in Supplemental Table 1, which had previously been annealed to form a partial duplex. Approximately 40 ng of adapter-ligated DNA was treated with sodium bisulfite and amplified with no more than 13 cycles of PCR following the Illumina guidelines and using the same primers as used for small RNA libraries. PCR products were purified using the Agencourt AMPure XP protocol (0.9:1 ratio).
MNase Digestion of Chromatin for the Chromatin Accessibility Experiment
Chromatin was extracted from developing B73 ears by grinding to a fine powder in liquid N2, lysing in a 35-mL volume of cold nuclei isolation buffer (2 mM MgCl2, 3 mM CaCl2, and 10 mM Tris-HCl, pH 7.5) containing 0.5% Tween 40 with vigorous shaking for 1 min, and filtering twice through 35-μm-pore Nitex bolting cloth. The nuclei were then pelleted by centrifuging at 6000g for 10 min at 4°C and, after pouring off the supernatant, washed by resuspending in nuclei isolation buffer (without Tween 40) and then pelleted by centrifuging at 6000g for 5 min at 4°C. Approximately 200 μL of purified nuclei was recovered.
The nuclei were treated with MNase (New England Biolabs; M0247S) by adding 2 μL of MNase and 800 μL of MNase buffer (1×) to a final MNase concentration of ∼4 gel units/μL. This mixture was divided into six 150-μL aliquots while still on ice. All aliquots were transferred simultaneously to a 37°C water bath and removed one at a time at the following time points: 30 s and 1, 2, 4, 6, 8, and 16 min (at which time nearly all the chromatin had been reduced to mononucleosomes; Figure 2A). To stop the digestions, the samples were immediately placed on ice, and 150 μL of histone removal buffer (1% SDS, 100 mM NaCl, 50 mM EDTA, and 100 mM Tris-HCl, pH 8) and 15 μL of 20 mg/mL Proteinase K (New England Biolabs; P8102S) were added to each tube. Samples were incubated for 45 min at 65°C to digest histones, then the DNA was purified by adding an equal volume (315 μL) of phenol:chloroform:isoamyl alcohol (25:24:1) followed by centrifugation. An equal volume (300 μL) of chloroform:isoamyl alcohol (24:1) was added to the supernatant, and the samples were centrifuged again. The supernatant was collected (240 μL), and the DNA was precipitated at −20°C after adding 1 μL of 20 mg/mL glycogen, 10 μL of 5 M NaCl, and 625 μL of ethanol. The DNA was pelleted by centrifugation, washed with 75% ethanol, air-dried, and resuspended in 100 μL of 10 mM Tris, pH 7.5. Finally, the fragments were separated by gel electrophoresis (3.5% agarose) of up to 24 μL of each DNA sample, and the ∼150-bp DNA fragments were extracted with a QIAquick gel extraction kit (Qiagen; 28704) without heating. A total of 150 to 300 ng of DNA was recovered from each sample in a 30-μL volume. The 1-, 4-, and 16-min digestions were selected for Illumina library preparation (corresponding to the accessible, mid-accessible, and inaccessible samples, respectively, in Figure 2).
Fragmentation of DNA for ChIP-seq and MNase-seq Controls
Five micrograms of genomic maize DNA was fragmented using the NEBNext dsDNA Fragmentase (New England Biolabs; M0348S) according to the standard genomic fragmentation protocol with a 45-min incubation time at 37°C. The DNA was purified using the Agencourt AMPure XP protocol (1.8:1 ratio). As with the MNase fragments, fragments around 150 bp in size were selected by gel electrophoresis with a 3.5% agarose gel.
ChIP
ChIP was performed essentially as described previously (Haring et al., 2007) with minor modifications. Cross-linked, isolated nuclei were sonicated using the Branson Double Step Microtip (Digital Sonifier; 101-148-063) at an amplitude of 10% in 1.5-mL microfuge tubes. The samples were sonicated for 5 s, followed by 20 s on ice, and this was repeated 45 times. Instead of Protein A-Agarose beads, DiaMag Protein A-coated magnetic beads were used (Diagenode; kch-802-150). Immediately before use, the required amounts of beads were washed two times in ChIP antibody incubation buffer (100 μL/30 μL beads) and resuspended in the original volume. Instead of spinning, a magnetic separation rack (DYNAL; MPC-S) was used. No salmon sperm DNA was added to the immunoprecipitation reaction. Instead, the same volume of ChIP antibody incubation buffer was used. A 1:100 dilution of antibodies recognizing H3K9me2 (Cell Signaling; 4658) and H3K27me2 (Cell Signaling; 9728) was used in the ChIP reactions. The quality of the precipitated material was checked by quantitative PCR as described previously (Haring et al., 2010). For each ChIP-seq sample, the precipitated DNA of three independent ChIP reactions was pooled. For each antibody, two ChIP-seq samples were sequenced.
Preparation of ChIP-seq, MNase-seq, and Fragmentase Sequencing Libraries
For all samples, 100 to 300 ng of double-stranded, fragmented DNA was end-repaired and 5′-phosphorylated using the End Repair Module (New England Biolabs; E6050L), A-tailed with Klenow exo- (New England Biolabs; M0212S), ligated to adapters (8.3 nM) with the Quick Ligase Kit (New England Biolabs; M2200S), and PCR-amplified using Phusion 2X HF master mix (New England Biolabs; M0531S) in a 50-μL total volume containing 200 nM of each primer. Thermocycler conditions were as follows: an initial denaturation at 98°C for 30 s; no more than 12 cycles of 98°C for 30 s, 65°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. The primers and adapters were the same ones used in the BS-seq libraries, except that additional ScriptSeq Index PCR primers were included (Epicentre; SSIP1202). DNA was purified between each enzymatic treatment using Agencourt AMPure XP at a ratio of 1.8:1, except after PCR, where the ratio was 0.9:1. Prior to sequencing, amplicons of ∼150 bp in length were selected by gel electrophoresis on a 2.5% agarose gel and purified using the QIAquick gel extraction kit (Qiagen; 28704).
Processing and Genomic Alignments of Small RNA Reads
Mutant and wild-type small RNA libraries prepared for this study were sequenced with an Illumina HiSeq 2500, producing single-end, 101-nucleotide reads. Low-quality reads and portions of reads were removed using fastq_quality_trimmer (parameters “-t 20 -l 30”) and fastq_quality_filter (parameters “-q 20 -p 80”) from the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The 3′ adapters were trimmed from the reads using FAR, The Flexible Adapter Remover (parameters “-trim-end right-adaptive-overlap yes-min-readlength 19”) with the adapter sequence appropriate for each small RNA library. To remove miRNAs from the data set, the reads were aligned to a list of maize miRNAs from miRBase, release 20 (Kozomara and Griffiths-Jones, 2011). Blastall software was used for the alignment (parameters “-e 1e-5”). After removal of miRNA matches, the resulting siRNA-enriched set of small RNA reads was split into individual lengths, 21, 22, and 24 nucleotides, and aligned to the RefGen_v3 genome assembly using BWA-ALN (Li and Durbin, 2009) (parameters “-t 8 -l 10”). Uniquely mapping reads were defined by MAPQ values of at least 20. The two mutant and two wild-type data sets were combined into single data sets for each of the three siRNA lengths.
Processing and Genomic Alignments of mRNA Reads
mRNA libraries from developing ears were sequenced with an Illumina HiSeq 2500, producing single-end, 101-nucleotide reads. Low-quality reads and portions of reads were removed using fastq_quality_trimmer (parameters “-t 20 -l 75”) and fastq_quality_filter (parameters “-q 20 -p 80”) from the FASTX-Toolkit. Reads were aligned to the genome using TopHat2 and Cufflinks (Trapnell et al., 2012; Kim et al., 2013) with a transcriptome index built from the complete set of protein-coding gene annotations in Zea_mays.AGPv3.21.gff3 (default parameters). Genes were then grouped into expression categories based on fragments per kilobase of exon model per million mapped fragments (FPKM) values. The 12,088 genes with FPKM values of zero were put into the zero expression category, and the remaining 27,045 genes were split into quartiles based on FPKM values: low, mid-low, mid-high, and high. The mRNA reads from V3 stem and shoot apical meristem tissue (Sekhon et al., 2013) were processed similarly except that fastq_quality_trimmer parameters were adjusted to “-t 20 -l 50.” The 8978 genes with FPKM values of zero were put into the zero expression category, and the remaining 30,155 genes were split into quartiles based on FPKM values: low, mid-low, mid-high, and high.
Processing and Genomic Alignments of BS-seq Reads
All libraries prepared for this study were sequenced with an Illumina HiSeq 2500, producing single-end, 101-nucleotide reads. Low-quality reads and portions of reads were removed using fastq_quality_trimmer (parameters “-t 20 -l 90”) and fastq_quality_filter (parameters “-q 20 -p 80”) from the FASTX-Toolkit. The 3′ adapters were trimmed using FAR, The Flexible Adapter Remover (parameters “-trim-end right-adaptive-overlap yes-min-readlength 90”). Reads were aligned to the genome and used to measure methylation using BS-Seeker2 (Guo et al., 2013) (parameters “-m 2-aligner=bowtie2”). For the BS-seq data set from coleoptile-stage shoots (Regulski et al., 2013), which was produced by paired-end sequencing, quality filtration and adapter removal were omitted and BS-Seeker2 parameters were adjusted (“-maxins=500 -m 1-aligner=bowtie2”).
Processing and Genomic Alignments of MNase, ChIP, and Fragmentase Reads
All libraries were sequenced with an Illumina HiSeq 2500, producing single-end, 151-nucleotide reads. Low-quality reads and portions of reads were removed using fastq_quality_trimmer (parameters “-t 20 -l 125”) and fastq_quality_filter (parameters “-q 20 -p 80”) from the FASTX-Toolkit. The 3′ adapters were trimmed using FAR, The Flexible Adapter Remover (parameters “-trim-end right-adaptive-overlap yes-min-readlength 125”). Reads were aligned to the genome with the Burrows-Wheeler Aligner BWA-MEM (Li and Durbin, 2009) default parameters. Read counts were normalized by dividing the number of uniquely mapping reads at each locus by the number of uniquely mapping reads across the set of 10 chromosomes.
Determining Overlap between Reads and Genomic Loci
All reads were aligned to the most recent maize genome assembly, RefGen_v3, obtained from www.maizesequence.org. A combination of BEDTools (Quinlan and Hall, 2010), SAMtools (Li et al., 2009), and custom python3 scripts was used for converting between sequence read and alignment formats, calculating coverage, and calculating overlaps between genomic loci. For all reads except BS-seq reads, uniquely mapping reads were defined as those with MAPQ scores of at least 20. To identify and count reads that matched to specific loci, a >50% overlap was required between each read’s genomic position and the locus, except for small RNAs, where a perfect 50% overlap would allow a read to be mapped to both of two adjacent loci.
Categorization of Genomic Loci
The genome was divided into 100-bp loci, and loci that overlapped by even 1 bp with an annotated protein-coding gene were separated from those that did not. As a first step to identify putative RdDM loci and non-RdDM loci among the intergenic loci, 24-nucleotide siRNAs from a previous study with a high coverage of small RNAs (from mature, unfertilized earshoots minus the developing cob [Gent et al., 2013]) were used. First, putative RdDM loci were identified by having at least one matching read, either nonuniquely mapping or uniquely mapping (where matching means that at least 50% of the read overlaps). Second, putative non-RdDM loci were identified by having zero matching reads. Loci that met neither criterion for RdDM or non-RdDM were labeled “uncategorized.” To further refine this analysis, the small RNA sequencing libraries produced from mop1 mutant and wild-type developing ear tissue were added to the analysis. siRNA counts for each locus were normalized by the total number of miRNAs in each data set, as identified by comparison with miRBase miRNA annotations (described above). Putative RdDM loci that had at least three uniquely mapping siRNAs and whose normalized number of uniquely aligning 24-nucleotide siRNAs was reduced at least 3-fold in the mop1 mutant sample were kept as RdDM loci. Putative RdDM loci not meeting this criterion were moved into the uncategorized group. Putative non-RdDM loci that had no uniquely mapping 24-nucleotide siRNA reads from the control reads were kept as non-RdDM loci. Putative non-RdDM loci not meeting this criterion were moved into the uncategorized group.
Analysis of Tandem Repeats
The positions of tandem repeats in the genome were identified by aligning individual reference sequences for each repeat to the genome using Blastall BLASTn software (with a nonstringent E value so as to pick up variants of each repeat [“-e 1e-10”]). We defined large arrays as loci consisting of stretches of one particular repeat that were at least 9.5 kb in length, long enough to contain 50 nucleosomes with a spacing of 190 bp (Gent et al., 2011). In addition, we required that these loci had no interruptions of the repeat larger than five nucleosomes (950 bp). According to these definitions, large arrays make up 1161 kb of the total 1850 kb of CentC loci, 1083 kb of the total 1897 kb of knob180 loci, 50 kb of the total 271 kb of TR-1 loci, and none of the total 40 kb of 5S rDNA loci in the reference genome assembly.
For the genome reference independent analysis, filtered, trimmed, and adapter-free DNA fragment reads (ChIP, MNase, and Fragmentase) were aligned to known tandem repeat sequences using Blastall BLASTn software (“-e 1e-8”). As a control for 24-nucleotide siRNA reads, Fragmentase reads were also trimmed down to the first 24 nucleotides, and both Fragmentase and 24-nucleotide siRNA reads were processed identically, including removal of miRNAs after filtering and trimming. Both 24-nucleotide Fragmentase reads and siRNA reads were aligned to the repeat sequences using Blastall BLASTn software (parameters “-e 1e-4”). The filtered, trimmed, adapter-free bisulfite reads were aligned to the repeat sequences using BS Seeker (parameters “-e 100 -m 3”). Methylation calls from this analysis should be interpreted cautiously, as methylation-calling software cannot distinguish variants in tandem repeats from bisulfite-mediated C-to-T conversions. This effect is predicted to inflate methylation values mainly in the CHH context.
Accession Number
All sequence data are available at the National Center for Biotechnology Information Sequence Read Archive under accession number SRP047420.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of Small RNA Abundance and DNA Methylation in Different Tissues.
Supplemental Figure 2. Whole-Genome Summary: Small RNAs, DNA Methylation, Chromatin Accessibility, and Histone Methylation.
Supplemental Figure 3. Coenrichment Analysis of H3K9me2 and H3K27me2.
Supplemental Figure 4. Heterochromatin Features of Tandem Repeats.
Supplemental Table 1. Oligo Names, Descriptions, and Sequences.
Supplementary Material
Acknowledgments
We thank Nicole Riddle for testing the specificity of the antibodies; Savithri Nambeesan, Lin Guo, and Michael Regulski for help and advice on preparing BS-seq libraries; Kathryn Hodges for logistical support; Nathanael Ellis for insights on data analysis and interpretation; Phillip SanMiguel for Illumina sequencing services and advice on preparing sequencing libraries; the Maize Genetics Cooperation Stock Center, Nathan Springer, and Jack Gardiner for providing maize stocks; and Ludek Tikovsky for growing plants used for ChIP. This study was supported by resources and technical expertise from the Georgia Advanced Computing Resource Center, a partnership between the University of Georgia’s Office of the Vice President for Research and Office of the Vice President for Information Technology, by the National Science Foundation (Grants 0607123 and 0922703), and by the National Institutes of Health (Grant F32GM095223).
AUTHOR CONTRIBUTIONS
J.I.G., R.K.D., X.Z., M.S., and K.M.M. designed the research. J.I.G., T.F.M., K.M.M., R.B., and M.R.K. performed research. J.I.G. analyzed data. J.I.G., R.K.D., and M.S. wrote the article.
Glossary
- RdDM
RNA-directed DNA methylation
- siRNA
short interfering RNA
- ChIP-seq
chromatin immunoprecipitation and Illumina sequencing
- H3K9me2
dimethylation of lysine 9
- H3K27me2
dimethylation of lysine 27
- MNase-seq
micrococcal nuclease digestion followed by sequencing
- miRNA
microRNA
- BS-seq
bisulfite sequencing
- ChIP
chromatin immunoprecipitation
- FPKM
fragments per kilobase of exon model per million mapped fragments
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J.E., White J., Sikkink K., Chandler V.L. (2006). An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Bailey A.O., Panchenko T., Sathyan K.M., Petkowski J.J., Pai P.J., Bai D.L., Russell D.H., Macara I.G., Shabanowitz J., Hunt D.F., Black B.E., Foltz D.R. (2013). Posttranslational modification of CENP-A influences the conformation of centromeric chromatin. Proc. Natl. Acad. Sci. USA 110: 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber W.T., Zhang W., Win H., Varala K.K., Dorweiler J.E., Hudson M.E., Moose S.P. (2012). Repeat associated small RNAs vary among parents and following hybridization in maize. Proc. Natl. Acad. Sci. USA 109: 10444–10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr M.L., Bertram E.G. (1949). A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163: 676. [DOI] [PubMed] [Google Scholar]
- Baucom R.S., Estill J.C., Chaparro C., Upshaw N., Jogi A., Deragon J.M., Westerman R.P., Sanmiguel P.J., Bennetzen J.L. (2009). Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinski P., Distor K., Gutierrez-Lopez J., Mendoza G.M., Shi J., Dawe R.K., Ross-Ibarra J. (2014). Diversity and evolution of centromere repeats in the maize genome. Chromosoma, in press. [DOI] [PubMed] [Google Scholar]
- Bologna N.G., Voinnet O. (2014). The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65: 473–503. [DOI] [PubMed] [Google Scholar]
- Brink R.A. (1956). A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41: 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S.W., Nur U. (1964). Heterochromatic chromosomes in the coccids. Science 145: 130–136. [DOI] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez C.M., Meca E., Tenaillon M.I., Gaut B.S. (2014). Three groups of transposable elements with contrasting copy number dynamics and host responses in the maize (Zea mays ssp. mays) genome. PLoS Genet. 10: e1004298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2013). Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., Ellis N.A., Makarevitch I., Yeh C.T., Gent J.I., Guo L., McGinnis K.M., Zhang X., Schnable P.S., Vaughn M.W., Dawe R.K., Springer N.M. (2012). Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2011). Heritable epigenetic variation among maize inbreds. PLoS Genet. 7: e1002372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep M.C., DeBarry J.D., Bennetzen J.L. (2013). The dynamics of LTR retrotransposon accumulation across 25 million years of panicoid grass evolution. Heredity (Edinb.) 110: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N.V. (2012). Presidential address. Transposable elements, epigenetics, and genome evolution. Science 338: 758–767. [DOI] [PubMed] [Google Scholar]
- Ferrari K.J., Scelfo A., Jammula S., Cuomo A., Barozzi I., Stützer A., Fischle W., Bonaldi T., Pasini D. (2014). Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol. Cell 53: 49–62. [DOI] [PubMed] [Google Scholar]
- Fincher J.A., Vera D.L., Hughes D.D., McGinnis K.M., Dennis J.H., Bass H.W. (2013). Genome-wide prediction of nucleosome occupancy in maize reveals plant chromatin structural features at genes and other elements at multiple scales. Plant Physiol. 162: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco H.D., Pidoux A.L., Urano T., Allshire R.C. (2008). Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P., De Jong J.H., Lysak M., Castiglione M.R., Schubert I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99: 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M., Michaud C., Leblanc O., Grimanelli D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Dong Y., Jiang J., Dawe R.K. (2012). Strong epigenetic similarity between maize centromeric and pericentromeric regions at the level of small RNAs, DNA methylation and H3 chromatin modifications. Nucleic Acids Res. 40: 1550–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Ellis N.A., Guo L., Harkess A.E., Yao Y., Zhang X., Dawe R.K. (2013). CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Schneider K.L., Topp C.N., Rodriguez C., Presting G.G., Dawe R.K. (2011). Distinct influences of tandem repeats and retrotransposons on CENH3 nucleosome positioning. Epigenetics Chromatin 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaffari R., Cannon E.K., Kanizay L.B., Lawrence C.J., Dawe R.K. (2013). Maize chromosomal knobs are located in gene-dense areas and suppress local recombination. Chromosoma 122: 67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Jaco I., Fraga M.F., Chen T., Li E., Esteller M., Blasco M.A. (2006). DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8: 416–424. [DOI] [PubMed] [Google Scholar]
- Guenatri M., Bailly D., Maison C., Almouzni G. (2004). Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Fiziev P., Yan W., Cokus S., Sun X., Zhang M.Q., Chen P.Y., Pellegrini M. (2013). BS-Seeker2: A versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Qin S., Wessler S.R. (2013). Comparison of class 2 transposable elements at superfamily resolution reveals conserved and distinct features in cereal grass genomes. BMC Genomics 14: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M., Bader R., Louwers M., Schwabe A., van Driel R., Stam M. (2010). The role of DNA methylation, nucleosome occupancy and histone modifications in paramutation. Plant J. 63: 366–378. [DOI] [PubMed] [Google Scholar]
- Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. (2007). Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Yan S., Wang P., Zhu W., Wang X., Shen Y., Shao K., Xin H., Li S., Li L. (2014). Comparative analysis of genome-wide chromosomal histone modification patterns in maize cultivars and their wild relatives. PLoS ONE 9: e97364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. (1928). Das Heterochromatin der Moose. Jahrb. Wiss. Botanik 69: 762–818. [Google Scholar]
- Hollick J.B. (2012). Paramutation: A trans-homolog interaction affecting heritable gene regulation. Curr. Opin. Plant Biol. 15: 536–543. [DOI] [PubMed] [Google Scholar]
- Johnson L.M., Du J., Hale C.J., Bischof S., Feng S., Chodavarapu R.K., Zhong X., Marson G., Pellegrini M., Segal D.J., Patel D.J., Jacobsen S.E. (2014). SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanizay L.B., Pyhäjärvi T., Lowry E.G., Hufford M.B., Peterson D.G., Ross-Ibarra J., Dawe R.K. (2013). Diversity and abundance of the abnormal chromosome 10 meiotic drive complex in Zea mays. Heredity (Edinb.) 110: 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J.L. (1970). Dependence of the R-mottled aleurone phenotype in maize on mode of sexual transmission. Genetics 66: 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39: D152–D157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonne J.D., Dorweiler J.E., McGinnis K.M. (2013). Changes in nucleosome position at transcriptional start sites of specific genes in Zea mays mediator of paramutation1 mutants. Epigenetics 8: 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D., Carey C.C., Dorweiler J.E., Chandler V.L. (2002). A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc. Natl. Acad. Sci. USA 99: 6130–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon M.F. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Madzima T.F., Huang J., McGinnis K.M. (2014). Chromatin structure and gene expression changes associated with loss of MOP1 activity in Zea mays. Epigenetics 9: 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Eichten S.R., Briskine R., Waters A.J., Danilevskaya O.N., Meeley R.B., Myers C.L., Vaughn M.W., Springer N.M. (2013). Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Stupar R.M., Iniguez A.L., Haun W.J., Barbazuk W.B., Kaeppler S.M., Springer N.M. (2007). Natural variation for alleles under epigenetic control by the maize chromomethylase zmet2. Genetics 177: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- McCarty D.R., et al. (2005). Steady-state transposon mutagenesis in inbred maize. Plant J. 44: 52–61. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1929). Chromosome morphology in Zea mays. Science 69: 629. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1950). The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36: 344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.D., Felsenfeld G. (1983). Another potential artifact in the study of nucleosome phasing by chromatin digestion with micrococcal nuclease. Cell 32: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Nobuta K., et al. (2008). Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S., McCue A.D., Panda K., Fultz D., DeFraia C., Thomas E.N., Slotkin R.K. (2013). The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162: 116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Papa C.M., Springer N.M., Muszynski M.G., Meeley R., Kaeppler S.M. (2001). Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Oh S., van Nocker S. (2012). Genomic and gene-level distribution of histone H3 dimethyl lysine-27 (H3K27me2) in Arabidopsis. PLoS ONE 7: e52855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W.J., Brutlag D., Goldring E., Appels R., Hinton C.W., Lindsley D.L. (1974). The organization of highly repeated DNA sequences in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 38: 405–416. [DOI] [PubMed] [Google Scholar]
- Peacock W.J., Dennis E.S., Rhoades M.M., Pryor A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78: 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O., Vitins A., Ream T.S., Hong E., Pikaard C.S., Costa-Nunes P. (2013). Intersection of small RNA pathways in Arabidopsis thaliana sub-nuclear domains. PLoS ONE 8: e65652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M., et al. (2013). The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier F., et al. (2011). Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabin L.R., Delás M.J., Hannon G.J. (2013). Dogma derailed: The many influences of RNA on the genome. Mol. Cell 49: 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schoft V.K., Chumak N., Mosiolek M., Slusarz L., Komnenovic V., Brownfield L., Twell D., Kakutani T., Tamaru H. (2009). Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 10: 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. (2013). A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 14: 853–864. [DOI] [PubMed] [Google Scholar]
- Sekhon R.S., Briskine R., Hirsch C.N., Myers C.L., Springer N.M., Buell C.R., de Leon N., Kaeppler S.M. (2013). Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Dawe R.K. (2006). Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173: 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.H., Wang H.L., Lee J., Dinwiddie B.L., Belostotsky D.A., Chekanova J.A. (2013). The role of the Arabidopsis exosome in siRNA-independent silencing of heterochromatic loci. PLoS Genet. 9: e1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu H., Wildhaber T., Siretskiy A., Gruissem W., Hennig L. (2012). Distinct modes of DNA accessibility in plant chromatin. Nat. Commun. 3: 1281. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Guo W., Du J., Ni Z., Sun Q., Yao Y. (2013). Widespread, abundant, and diverse TE-associated siRNAs in developing wheat grain. Gene 522: 1–7. [DOI] [PubMed] [Google Scholar]
- Tanurdzic M., Vaughn M.W., Jiang H., Lee T.J., Slotkin R.K., Sosinski B., Thompson W.F., Doerge R.W., Martienssen R.A. (2008). Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 6: 2880–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford D.J., Stewart B.W. (1989). Micrococcal nuclease: Its specificity and use for chromatin analysis. Int. J. Biochem. 21: 127–137. [DOI] [PubMed] [Google Scholar]
- Tenaillon M.I., Hufford M.B., Gaut B.S., Ross-Ibarra J. (2011). Genome size and transposable element content as determined by high-throughput sequencing in maize and Zea luxurians. Genome Biol. Evol. 3: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D., Hughes T.R. (2009). G+C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiseth S.V., Rahman M.A., Yap K.L., Fischer A., Egge-Jacobsen W., Reuter G., Zhou M.M., Aalen R.B., Thorstensen T. (2011). The SUVR4 histone lysine methyltransferase binds ubiquitin and converts H3K9me1 to H3K9me3 on transposon chromatin in Arabidopsis. PLoS Genet. 7: e1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.M., Nakamura M., Calarco J.P., Susaki D., Lim P.Q., Kinoshita T., Higashiyama T., Martienssen R.A., Berger F. (2013). RNA-directed DNA methylation regulates parental genomic imprinting at several loci in Arabidopsis. Development 140: 2953–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Heimes S., Riedel L., Sänger H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. [DOI] [PubMed] [Google Scholar]
- Wei L., Gu L., Song X., Cui X., Lu Z., Zhou M., Wang L., Hu F., Zhai J., Meyers B.C., Cao X. (2014). Dicer-like 3 produces transposable element-associated 24-nucleotide siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. USA 111: 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgruber T.K., et al. (2009). Maize centromere structure and evolution: Sequence analysis of centromeres 2 and 5 reveals dynamic loci shaped primarily by retrotransposons. PLoS Genet. 5: e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse M.R., Freeling M., Lisch D. (2006). Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4: e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Rowley M.J., Böhmdorfer G., Sandhu D., Gregory B.D., Wierzbicki A.T. (2012). RNA polymerase V targets transcriptional silencing components to promoters of protein-coding genes. Plant J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Hale C.J., Law J.A., Johnson L.M., Feng S., Tu A., Jacobsen S.E. (2012). DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 19: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.