Applying a combination of algorithms on an Arabidopsis abiotic stress gene expression compendium generated system-wide predictions of transcription regulatory interactions and functional relationships. This joined computational and experimental approach identified a set of transcription factors that highly regulate one another and function in detoxification during oxidative stress.
Abstract
The abiotic stress response in plants is complex and tightly controlled by gene regulation. We present an abiotic stress gene regulatory network of 200,014 interactions for 11,938 target genes by integrating four complementary reverse-engineering solutions through average rank aggregation on an Arabidopsis thaliana microarray expression compendium. This ensemble performed the most robustly in benchmarking and greatly expands upon the availability of interactions currently reported. Besides recovering 1182 known regulatory interactions, cis-regulatory motifs and coherent functionalities of target genes corresponded with the predicted transcription factors. We provide a valuable resource of 572 abiotic stress modules of coregulated genes with functional and regulatory information, from which we deduced functional relationships for 1966 uncharacterized genes and many regulators. Using gain- and loss-of-function mutants of seven transcription factors grown under control and salt stress conditions, we experimentally validated 141 out of 271 predictions (52% precision) for 102 selected genes and mapped 148 additional transcription factor-gene regulatory interactions (49% recall). We identified an intricate core oxidative stress regulatory network where NAC13, NAC053, ERF6, WRKY6, and NAC032 transcription factors interconnect and function in detoxification. Our work shows that ensemble reverse-engineering can generate robust biological hypotheses of gene regulation in a multicellular eukaryote that can be tested by medium-throughput experimental validation.
INTRODUCTION
Plants are continuously exposed to changing environmental conditions, such as low and high temperatures, shortage of water, high salinity, radiation, and nutrient deficiencies. Responses to this fluctuating environment are complex and involve a multitude of signaling molecules triggering rapid changes in gene expression to reprogram the plant’s metabolism and achieve a new state of homeostasis (Kilian et al., 2007; Cramer et al., 2011). Functional genomics studies have identified a portfolio of transcription factors (TFs) and cis-regulatory motifs that are involved in stress-inducible regulation (Chen et al., 2002; Zou et al., 2011; Petrov et al., 2012; Naika et al., 2013).
However, we still lack a systems-level understanding of the gene regulatory networks (GRNs) that orchestrate the complex abiotic stress response through fine-tuned regulation (Cramer et al., 2011). As genes that share a biological function tend to be coregulated, coexpression analysis through statistical correlation and clustering can infer functional associations based on the guilt-by-association principle. Besides the availability of expression database resources like Genevestigator (Zimmermann et al., 2004), ATTED-II (Obayashi et al., 2007), CORNET (De Bodt et al., 2010), and GeneMANIA (Warde-Farley et al., 2010), coexpression-based gene association networks were generated for Arabidopsis thaliana with different computational sophistication (Ma et al., 2007, 2014; Horan et al., 2008; Bassel et al., 2011; Kourmpetis et al., 2011; Less et al., 2011; Heyndrickx and Vandepoele, 2012; Bhosale et al., 2013). Although these studies provide “functional modules” of genes operating in abiotic stress, information on their specific regulators is lacking.
Due to experimental challenges, only a limited number of gene regulatory interactions between TFs and their target genes have been experimentally mapped for Arabidopsis by yeast one-hybrid (Y1H) (Brady et al., 2011; Gaudinier et al., 2011), chromatin immunoprecipitation (ChIP) (Kaufmann et al., 2010; Xie et al., 2010), or TF perturbation studies (Bassel et al., 2012). Whereas the first two monitor a direct physical association of a TF with genomic DNA, the latter report on both direct and indirect regulatory effects of a TF on downstream gene expression. AtRegNet, the Arabidopsis thaliana Regulatory Network database, currently documents 11,355 binding and regulatory interactions between 67 TFs and their target genes (Palaniswamy et al., 2006; Yilmaz et al., 2011).
Potential interactions between TFs and their targets can be predicted through reverse-engineering of transcriptomics data. Since TFs are themselves often regulated at the transcriptional level, the activity of TFs is embedded in their expression profiles. Therefore, the causal relationship between TFs and their target genes is implicitly present in the correspondence between their expression profiles. There are several reverse-engineering methods available ranging from correlation, mutual information, regression, Bayesian networks, Gaussian mixture models, Hidden Markov models, and Boolean networks to ordinary differential equations (De Smet and Marchal, 2010; Friedel et al., 2012; Marbach et al., 2012). Reverse-engineering is a high-dimensional, underdetermined problem: The number of possible interactions between TFs and target genes by far exceeds the number of different experimental conditions for which expression profiles are available. To reduce the search space, additional measures can be taken. For instance, in contrast to direct network inference, module-based network inference assigns the same regulatory program to all genes with a similarly coordinated expression behavior (De Smet and Marchal, 2010). Each reverse-engineering method generates a confidence score for a link between a TF and target gene.
However, benchmark studies have shown that no single best reverse-engineering method exists: Different methods show different biases in detecting regulatory relationships and act complementary in revealing the true underlying GRNs (Michoel et al., 2009; Marbach et al., 2012). Therefore, the combination of the results of different network inference algorithms into one ensemble solution has recently been explored in bacteria and yeast. Ensemble solutions such as average rank aggregation and union are consistently as good as or better than the top-performing individual methods and are more robust across different data sets (Marbach et al., 2012; Qi et al., 2012; Hase et al., 2013). Moreover, the more diverse the individual inference solutions are, the better the performance of the integrated solution.
Prediction of GRNs from abiotic stress gene expression profiles could not only advance the holistic understanding of the abiotic stress response and its key regulators, but also offer high potential as hypothesis generators for time- and cost-efficient design of experiments. Until now, reverse-engineering has only limitedly been applied to plant transcriptomics data (Street et al., 2011; Yu et al., 2011; Friedel et al., 2012; Hickman et al., 2013; Misra and Sriram, 2013; Chávez Montes et al., 2014). Despite generating useful biological hypotheses in plants, these studies have applied one network inference algorithm accompanied by no or very limited experimental validation.
Here, we applied different methods for network inference to an abiotic stress-specific microarray compendium of Arabidopsis: two different parameter settings of the stochastic Bayesian module network algorithm LeMoNe (LearningModuleNetworks) (Joshi et al., 2009), the mutual information direct algorithm CLR (Context Likelihood of Relatedness) (Faith et al., 2007), and the double two-way t test direct algorithm TwixTrix (Qi and Michoel, 2012). Previously, these methods have been shown to act complementary for the reverse-engineering of GRNs in bacteria, yeast, and worm (Michoel et al., 2009; Vermeirssen et al., 2009). We constructed an abiotic stress GRN of the top 200,014 regulatory interactions from the ensemble solution obtained by average rank aggregation of these four predictions. Through extensive validation with experimental data from literature, we captured many reported regulatory interactions and known biological information. In addition, we performed an unbiased, medium-throughput experimental validation, where we identified an intricate abiotic stress GRN, where NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUP-SHAPED COTYLEDON (NAC) TFs NAC13, NAC053, NAC032, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR6 (ERF6), and WRKY DNA BINDING PROTEIN6 (WRKY6) function in detoxification processes during oxidative stress. Not only does our study provide insights into the gene regulation of the abiotic stress response in plants at a systems level, it also exemplifies the potential of ensemble reverse-engineering in combination with medium-throughput experimental validation for the mapping of GRNs in multicellular eukaryotes.
RESULTS
Integrating Different Reverse-Engineering Algorithms into an Ensemble Abiotic Stress GRN
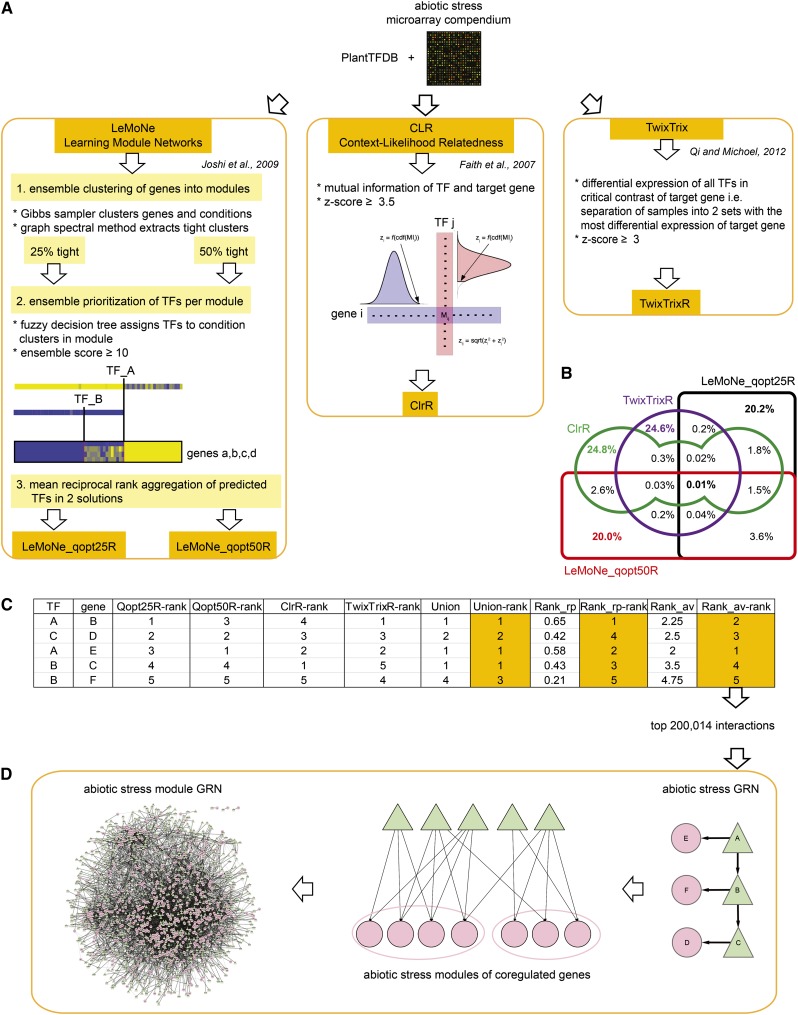
Three different network inference algorithms were applied to an Arabidopsis abiotic stress-dedicated microarray expression ratio compendium (199 conditions; Supplemental Data Set 1), using 1340 TFs listed in the Plant TF Database (PlantTFDB) (Zhang et al., 2011) as putative regulators: LeMoNe (Joshi et al., 2009), CLR (ClrR) (Faith et al., 2007), and TwixTrix (TwixTrixR) (Qi and Michoel, 2012). For LeMoNe, we assigned regulators to 25% tightly clustered modules (LeMoNe_qopt25R) and 50% tightly clustered modules (LeMoNe_qopt50R), respectively (Figure 1A; see Methods). Approximately 200,000 ranked regulatory interactions were obtained for each reverse-engineering solution leading to a total of 785,913 uniquely predicted regulatory interactions. As expected, we observed only 10% overlap between the different regulatory predictions and less than 0.01% of all interactions were found by all four methods (Figure 1B).
Figure 1.
Construction of the Abiotic Stress GRN by Ensemble Reverse-Engineering.
(A) An abiotic stress microarray compendium and TFs from PlantTFDB were subjected to reverse-engineering, resulting in four network inference solutions: LeMoNe_qopt25R, LeMoNe_qopt50R, ClrR, and TwixTrixR.
(B) The Venn diagram illustrates the percentage of 785,913 unique regulatory interactions predicted by each of the four network inference solutions and their overlap.
(C) The regulatory predictions were combined by rank aggregation into three ensembles: union, mean reciprocal rank, and average rank.
(D) The top 200,014 predictions from the average rank ensemble made the abiotic stress GRN. Target genes were subsequently clustered into modules of coregulated genes and only the most important regulating TFs per module (≤10) were retained, generating the abiotic stress module GRN.
To assess the biological relevance of these complementary predictions, we evaluated them against a set of 52,328 known regulatory interactions that we created from the databases AtRegNet and CORNET on the one hand and from Y1H and ChIP protein-DNA interactions and TF perturbed expression profiles found in literature on the other hand (Supplemental Table 1; see Methods). This reference set contains interactions for only 15% of TFs for which we had predicted regulatory interactions, and it consists of both direct and indirect regulatory interactions. We designate a regulatory interaction as “direct” if a TF binds nearby a target gene and thereby controls the target gene’s expression. In addition, we extended this reference set by calculating, based on transitivity, all possible indirect paths between TFs and target genes, obtaining 789,068 interactions: We derived a regulatory interaction between protein A and target gene B if there is a path through a combination of reported protein-DNA interactions and regulatory interactions present from A to B, e.g., A binds C, C regulates D, D binds E, and E regulates B (see Methods). In this way, we allow for a maximal overlap between benchmark data and predicted regulatory interactions. On the other hand, we restricted the literature reference set to contain only 1307 direct regulatory interactions, for which there was evidence of both binding between a TF and its target gene and regulatory effect of a TF on transcription of its target gene. Hence, we removed all interactions that might be due to indirect regulatory effects or nonfunctional binding (Gitter et al., 2009). From the precision, recall, F-measure, and the area under the precision-recall (AUPR) curve, evaluating the top 200,000 predictions, we observed that ClrR ranked first for the initial reference set, as well as for the direct regulatory interaction set (Table 1). For the extended reference set, LeMoNe_qopt25R was the first in rank. F-measures and AUPR values corresponded to what has previously been reported for yeast (Marbach et al., 2012; Qi and Michoel, 2012).
Table 1. Performance Evaluation of the Top 200,000 Predictions of the Four Individual Reverse-Engineering Methods and Their Ensemble Solutions on Correctly Predicting Known Regulatory Interactions.
| Npred | TP | Precision | Recall | F | AUPR | |
|---|---|---|---|---|---|---|
| Set of 52,328 known protein-DNA and/or regulatory interactions | ||||||
| LeMoNe_qopt25R | 31886 | 972 | 0.030 | 0.019 | 0.023 | 0.00118 |
| LemoNe_qopt50R | 30056 | 862 | 0.029 | 0.016 | 0.021 | 0.00104 |
| ClrR | 31290 | 1301 | 0.042 | 0.025 | 0.031 | 0.00190 |
| TwixTrixR | 26353 | 955 | 0.036 | 0.018 | 0.024 | 0.00072 |
| Union | 31847 | 1092 | 0.034 | 0.021 | 0.026 | 0.00130 |
| Rank_rp | 31824 | 1091 | 0.034 | 0.021 | 0.026 | 0.00137 |
| Rank_av | 31546 | 1182 | 0.037 | 0.023 | 0.028 | 0.00158 |
| Extended set of 789,068 known and “hidden” interactions | ||||||
| LeMoNe_qopt25R | 31886 | 10696 | 0.335 | 0.014 | 0.026 | 0.00516 |
| LemoNe_qopt50R | 30056 | 8686 | 0.289 | 0.011 | 0.021 | 0.00352 |
| ClrR | 31290 | 10056 | 0.321 | 0.013 | 0.025 | 0.00466 |
| TwixTrixR | 26353 | 7647 | 0.290 | 0.010 | 0.019 | 0.00292 |
| Union | 31847 | 10798 | 0.339 | 0.014 | 0.026 | 0.00517 |
| Rank_rp | 31824 | 10677 | 0.336 | 0.014 | 0.026 | 0.00512 |
| Rank_av | 31546 | 10280 | 0.326 | 0.013 | 0.025 | 0.00461 |
| Set of 1307 direct regulatory interactions | ||||||
| LeMoNe_qopt25R | 561 | 26 | 0.046 | 0.020 | 0.028 | 0.00128 |
| LemoNe_qopt50R | 452 | 12 | 0.027 | 0.009 | 0.014 | 0.00032 |
| ClrR | 727 | 51 | 0.070 | 0.039 | 0.050 | 0.00537 |
| TwixTrixR | 311 | 16 | 0.051 | 0.012 | 0.020 | 0.00065 |
| Union | 569 | 40 | 0.070 | 0.031 | 0.043 | 0.00259 |
| Rank_rp | 574 | 40 | 0.070 | 0.031 | 0.043 | 0.00248 |
| Rank_av | 575 | 45 | 0.078 | 0.034 | 0.048 | 0.00272 |
The table shows the individual reverse-engineering methods LeMoNe_qopt25R, LemoNe_qopt50R, ClrR, and TwixTrixR, as well as their ensemble solutions by union, mean reciprocal rank (Rank_rp), and average rank (Rank_av) aggregation (i.e., the abiotic stress GRN; underlined), against three reference sets: (1) an assembled interaction set of 52,328 experimental protein-DNA and regulatory interactions, (2) an extended set of (1) containing all indirect hidden paths of 789,068 interactions (paths of length greater than one), and (3) a confined set of (1) containing only 1307 direct regulatory interactions. Npred = number of predictions made for TFs and target genes belonging to the reference set; TP = number of true positives; Precision = TP/Npred; Recall = TP/number of interactions in the reference set; F = 2 × precision × recall/(precision + recall) = 2 × TP/(2 × TP + FP + FN); AUPR = estimated AUPR curve. Due to the size of the reference sets, the AUPR calculation of the confined set is based on only a few hundreds of points, while for the other two reference sets, this is thousands to tens of thousands of points.
Over a total of 785,913 unique predictions from all four methods, 35,034 true positives were uncovered in the extended reference set. All four methods largely inferred different parts of the true gene regulatory networks: Only 12.5% of true positives were found by at least two methods, but this was a significant enrichment compared with all predictions (hypergeometric P value = 0), indicating that interactions inferred by multiple algorithms were more likely to be biologically relevant.
The benchmarking results of the different methods suggest that we could obtain a better predictive power by integrating all four solutions. Therefore, we computed ensemble solutions by union, mean reciprocal rank aggregation, and average rank aggregation (Figure 1C; see Methods). When we compared the top 200,000 interactions to the reference sets (Table 1), we observed that all ensemble methods generated a similar number of true positives, F-measure, and AUPR and showed an equal or higher performance compared with the individual solutions, in accordance with what has been observed for ensembles in bacteria and yeast (Marbach et al., 2012). We continued with the average rank aggregation ensemble, since this ensemble was first in rank over the other ensembles for the initial and the direct reference sets. Moreover, this ensemble performed at least as well as the best individually inferred solution in predicting direct regulatory interactions. We constructed an abiotic stress GRN by taking the top 200,014 predictions of the average rank aggregation ensemble, containing 1290 TFs and 11,938 target genes (Supplemental Data Set 2). In this abiotic stress GRN, 55% of the predictions were made by LeMoNe_qopt25R, 52% by LeMoNe_qopt50R, 36% by ClrR, and 3% by TwixTrixR, with 40% of the interactions inferred by at least two algorithms and covering 99% of the overlap between LeMoNe_qopt25R, LeMoNe_qopt50R, ClrR, and TwixTrixR (Supplemental Figure 1). Predictions inferred solely by TwixTrixR were not included in the network because the more local nature of this algorithm differs the most from the more global inference behavior of the others: Since TwixTrix is based on differential expression testing, interactions predicted by TwixTrix have significantly lower Pearson correlations between the expression profiles of predicted TFs and target genes than interactions predicted by LeMoNe and CLR (Qi and Michoel, 2012).
Functionally Coherent Modules of Coregulated Genes and Stress-Related TF Hubs
To identify regulators of coregulated genes, we clustered the abiotic stress GRN into modules based on the Jaccard similarity index of shared predicted TFs (see Methods). We retained at most 10 TFs per module, regulating the highest number of genes (≥50%) and displaying the highest average rank per module. In this abiotic stress module GRN, we obtained 572 modules of between 3 and 92 coregulated genes, regulated by 853 TFs. Since each gene ended up in only one module, we recovered the most important regulators and functional environment for each gene in the abiotic stress response. Modules of coregulated genes and their predicted regulating TFs were visualized as heat maps of the expression profile ratios in function of annotated abiotic stress conditions, together with information on functional coherence of the module and biological relevance of the predicted regulators (Figure 2; ModuleViewer; see Methods). We also generated a network view of the module and its predicted regulators. All modules and regulating TFs can be queried at http://bioinformatics.psb.ugent.be/supplementary_data/vamei/module_display/. Many regulating TFs appeared to target multiple modules, leading to a big well-connected component (Figure 1D). Several TFs were also present in the modules as targets and, in turn, regulated other modules, generating connections between different modules of coregulated genes. The hubs of the abiotic stress GRN were largely overlapping with the hubs of the module network, with half of them having a known Gene Ontology (GO) annotation for “response to stress” and/or “response to abiotic stimulus” (Supplemental Table 2; see Methods).
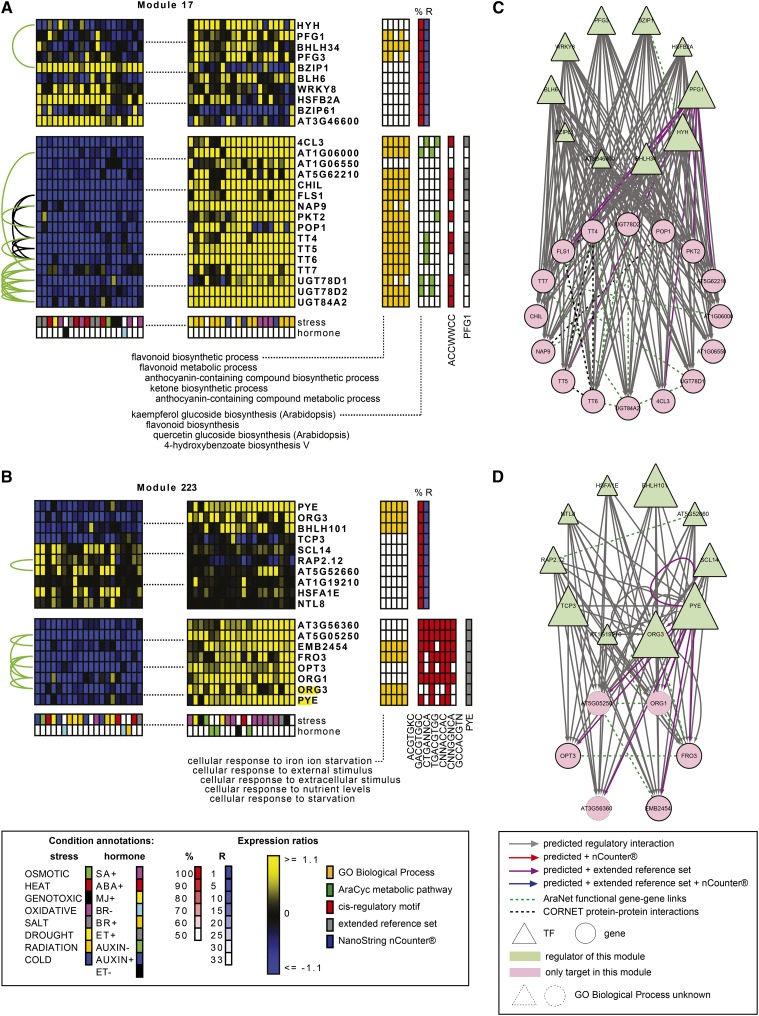
Figure 2.
Literature-Based Evidence for the Biological Relevance of the Abiotic Stress Module GRN.
(A) and (C) Module 17 was implicated in flavonoid biosynthesis.
(B) and (D) Module 223 functioned in the iron deficiency response.
(A) and (B) Illustrations by ModuleViewer (see text) were modified to display only the 20 most up- and downregulated conditions. Upper block = log2 expression ratios of regulatory TFs ordered by rank; lower block = log2 expression ratios of coregulated target genes. Blocks on the right indicate functional coherence: top 5 BINGO GO Biological Process enrichment, top 5 AraCyc metabolic pathway enrichment, cis-regulatory motif enrichment, true positives in extended reference set, and true positives from the nCounter experiments. The regulating TFs are ranked by the percentage of the genes in the module they are predicted to regulate (%) and their average rank over the module interactions (R). It is also indicated if the TFs have the GO Biological Process annotation that is enriched in the module. Abiotic stress conditions are annotated according to abiotic stress or/and hormone treatment (SA, salicylic acid; MJ, methyl jasmonate; BR, brassinosteroids; ET, ethylene). Arrows on the left: green = AraNet links; black = CORNET experimental protein-protein interaction links.
(C) and (D) Network illustrations by Cytoscape. The size of the regulating TF node is proportional to the average rank of this regulator in the module, i.e., the importance of the TF for the module. Edges are colored if the predictions were validated by the extended reference set (purple), nCounter experiments (red), or both (blue). Dotted node borders indicate module genes (including TFs) that lack a known GO Biological Process annotation.
Since coregulated genes are more likely to be coexpressed and to function in a similar biological process, we analyzed the functional coherence of the modules as a measure for the true biological nature of the abiotic stress GRN (Supplemental Data Set 3). First, 70% of the modules displayed a significant Biological Process GO enrichment, 55% of which had one that was directly related to “response to stress” or “response to abiotic stimulus.” Secondly, 14% of all modules had a significant plant metabolic pathway enrichment from AraCyc, the Plant Metabolic Network. Third, 60% of all modules contained genes that are also connected with one another in AraNet, with 90 modules having 50% or more of the genes being linked. AraNet is a probabilistic functional gene network of Arabidopsis that integrates species-wide and diverse omics data (Lee et al., 2010). Since genes encoding physically interacting proteins tend to be coregulated, we found experimentally validated protein-protein interactions from the CORNET database to be present in 9% of all modules (De Bodt et al., 2012). Besides, 37% of the modules were significantly enriched for genes related to “oxidative stress,” which is a stress that accompanies other abiotic stresses (Gadjev et al., 2006). Finally, since coregulated genes, certainly when sharing a biological function, tend to be coexpressed, we found a higher average Pearson Correlation Coefficient (PCC) within modules of the abiotic stress GRN (average PCC = 0.45) than within “random” modules where we randomly distributed the 10,350 genes 1000 times in modules of the same size as the abiotic stress module GRN (average PCC = 0.01) (Wilcoxon P value < 2.2e-16) (Supplemental Figure 2).
As a result of this functional coherence in the abiotic stress module GRN, we could predict functional relationships for uncharacterized genes based on the guilt by association principle. Of the 10,350 module genes, 3016 lacked a known GO Biological Process annotation, 1966 of which we attributed with one based on the enriched GO annotation(s) of the module in which they resided (Supplemental Data Set 3).
cis-Regulatory Motifs Link Modules and Predicted TFs
We investigated if cis-regulatory motifs listed in the PLACE (Higo et al., 1999) and AGRIS (Palaniswamy et al., 2006) databases, together with a complementary set of motifs identified using the network-level conservation principle from ATCOECIS (Vandepoele et al., 2009), and several binding sites from literature (Supplemental Table 3) were uncovered significantly more frequently in the promoters of genes belonging to the same module compared with all promoters in the Arabidopsis genome (see Methods). We detected 141 different motifs in 124 modules (Supplemental Data Set 3). Motifs AAACCCTA (UP2) (Tatematsu et al., 2005), ACGTGKC (ABRE), VCGCGB (CGCG BOX) (Yang and Poovaiah, 2002), and CACGTG (ABRE) were all found in 10 or more modules.
The abscisic acid (ABA)-responsive element (ABRE), with the core sequence ACGTG, is a major cis-acting regulatory element in ABA-dependent gene expression in adaptation to abiotic stresses such as drought and high salinity, as well as in seed maturation and dormancy. ABA promotes stomatal closure in guard cells and regulates the expression of many genes that may function in dehydration tolerance (Yamaguchi-Shinozaki and Shinozaki, 2006; Lumba et al., 2014).
Only one module with an overrepresented ABRE motif was predicted to be regulated by an ABRE binding factor (module 78; ABSCISIC ACID RESPONSIVE ELEMENT BINDING FACTOR1 [ABF1]). Activation of the ABRE binding AREB/ABF TFs requires ABA-dependent posttranscriptional phosphorylation (Furihata et al., 2006). Therefore, the transcriptional expression of these TFs is less a proxy for their activity, which explains why they are more difficult to be predicted as regulators by network inference algorithms. Nevertheless, 12 modules shared ABA-responsive TFs functioning in the dehydration stress response as predicted regulators, such as homeodomains ARABIDOPSIS THALIANA HOMEOBOX7 and 12 (Valdés et al., 2012), NAC TFs RESPONSIVE TO DESICCATION26 (RD26) (Fujita et al., 2004), ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN2 (ATAF1), NAC019, and NAC WITH TRANSMEMBRANE MOTIF1-LIKE6 (NTL6) (Kim et al., 2012; Nakashima et al., 2012), basic helix-loop-helix (bHLH) TF JASMONATE INSENSITIVE1 (ZBF1/MYC2) (Yadav et al., 2005), and basic leucine zipper (bZIP) TF G-BOX BINDING FACTOR3 (GBF3) (Lu et al., 1996).
Promoters of genes in module 440 were significantly enriched for the dehydration-responsive element (DRE), with core sequence G/ACCGAC (Stockinger et al., 1997). The module genes are mostly upregulated upon cold and osmotic stress (Yamaguchi-Shinozaki and Shinozaki, 2006). Accordingly, we detected APETALA2 TFs, RELATED TO AP2 1 (RAP2.1) and FLORAL MUTANT2 (FLO2), as prominent regulators of this module, for which predictions were confirmed by the extended reference set (Dong and Liu, 2010; Akhtar et al., 2012).
A MYB binding site was found in six modules, five of which displayed a function and predicted regulators that clearly corresponded to this motif. Module 503 and 580, as well as their top-ranked regulators the GRAS TF AT1G63100 and MYB DOMAIN PROTEIN 3R-4 (MYB3R-4), function in cytokinesis by cell plate formation. MYB3R-4 regulates multiple cell cycle G2/M phase-specific genes, several of which were present in the modules, and binds AACGG, the MYB cis-regulatory motif that we found to be overrepresented in these modules (Haga et al., 2011).
The evening element, a marker of circadian control of gene expression that induces peak expression in the evening, was significantly enriched in module 159. This module contained genes that function in the circadian rhythmic starch metabolic process and that are highly upregulated by cold stress. The circadian clock controls starch degradation to ensure continued optimal growth during the night (Graf and Smith, 2011). Soluble sugars protect plant cells from cellular damage caused by cold stress by acting as osmoprotectants and nutrients as well as interacting with the lipid bilayer. The circadian clock and cold acclimation are intimately linked in plants (Bieniawska et al., 2008), and the evening element was previously suggested to integrate cold- and clock-regulated transcription (Mikkelsen and Thomashow, 2009). Similarly, the module was predicted to be regulated by circadian rhythmic TFs such as B-box zinc finger TFs, which have been implicated in the regulation of light- and cold-influenced processes (Soitamo et al., 2008; Gangappa and Botto, 2014); GATA TRANSCRIPTION FACTOR28 (ZML2), which is a regulator of the cryptochrome-mediated response to excess light (Shaikhali et al., 2012); SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7, which controls the induction of sucrose-responsive microRNAs (Ren and Tang, 2012); and DRE binding protein TFs with a role in cold acclimation (Maruyama et al., 2009).
Module 531 was significantly GO enriched for “response to endoplasmic reticulum (ER) stress,” which refers to a condition where stress leads to the accumulation of unfolded or misfolded proteins in the ER. This triggers the protective cellular unfolded protein response, which aims to restore normal cell function by halting protein translation and activating the production of molecular chaperones involved in protein folding. Correspondingly, seven module genes out of 17 contained the ER stress response element CCAAT-N9-CCACG and 11 module genes had the unfolded protein response element core TGACGT in their promoters (Yamamoto et al., 2004).
In addition, several other abiotic stress-related cis-regulatory motifs were detected, such as the I-box (GATAAG), a conserved sequence in light-regulated promoters (Donald and Cashmore, 1990); the W-box (TTGAC(C/T), which is recognized by WRKY TFs and is often involved in the defense response (Yamasaki et al., 2012); and the heat shock element (GAANNTTC) that is targeted by HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2) (Barros et al., 1992) (Supplemental Data Set 3). Moreover, several of the motifs were previously identified to be singlet oxygen-responsive motifs: ACGTGTC (ABRE), TTCACY (W-box), GCCGCC (GCC-box), and ACCWWCC (MYB) (Petrov et al., 2012).
Taken together, many modules were enriched for a cis-regulatory motif that often corresponded to the specific function of the module genes in the abiotic stress response and to the binding site of the predicted regulators. The latter points to the likely direct nature of the predicted regulatory interactions.
Literature-Based Evidence for the Biological Relevance of Predicted TF-Target Gene Interactions
Matching the “known” TF-target gene interactions from the extended reference set to the module network predictions, we retrieved 4172 interactions in 228 modules (Supplemental Data Set 3), compared with 10,280 interactions in the abiotic stress GRN (Table 1). Despite the fact that modules facilitate the interpretation of the abiotic stress GRN, this emphasizes the importance of also studying the whole GRN: More than 1500 genes did not fit our definition of modules of coregulated genes and TFs that regulate many genes in a module were favored over TFs with a high ranking for a specific gene.
Although the reverse-engineering methods do not directly predict cooperativity between TFs, we investigated whether we could detect known heterodimers within the predicted TFs of a coregulated module through the experimental protein-protein interactions from the CORNET database. Among the predicted regulators of 22 modules, we observed heterodimers between members of various TF families, with most heterodimer formations occurring within a TF family (Supplemental Data Set 3). It is very likely that there are many more, but the data set of experimental heterodimer TF-TF interactions in CORNET is limited to 1100 interactions between 462 TFs. For instance, the regulators BASIC LEUCINE-ZIPPER1 (BZIP1) and GBF6/BZIP11 of module 154, which functioned in glycolysis and the tricarboxylic acid cycle, form a heterodimer. These TFs belong to the C/S1 bZIP family, and heterodimers of this family involving BZIP1 and GBF6 are able to reprogram sugar and amino acid metabolism during low energy stress (Hanson et al., 2008; Dietrich et al., 2011; Ma et al., 2011). To further illustrate the biological relevance of our predictions, we will discuss below three modules and one transcription factor.
Module 17 was enriched for flavonoid biosynthesis genes and flavonoid transporters (Yonekura-Sakakibara et al., 2008). The bZIP TF HY5-HOMOLOG (HYH) and the MYB TF PRODUCTION OF FLAVONOL GLYCOSIDES1 (PFG1/MYB12) were top-ranked regulators, and the module was enriched for a MYB binding site (Figure 2A). From the extended reference set, it was known that PFG1 activates 9 out of 16 module genes (Supplemental Data Set 3). PFG1 and the related, third-ranked predicted TF, PFG3 (MYB111), have very similar target specificity in flavonoid biosynthesis but function in different parts of the plant (Stracke et al., 2007). HYH is a functionally redundant homolog of ELONGATED HYPOCOTYL5, which is known to regulate PFG1 and PFG3 in flavonoid biosynthesis (Stracke et al., 2010). The second-ranked predicted TF, BHLH34, is part of a functional plant module with a role in flavonoid biosynthesis and response to sucrose (Heyndrickx and Vandepoele, 2012).
Module 223 was enriched for the GO Biological Process “cellular response to iron starvation” containing five out of eight genes with a reported function in iron homeostasis (Figure 2B). The top-ranked regulators bHLH TFs OBP3-RESPONSIVE GENE3 (ORG3/BHLH039) and POPEYE (PYE/BHLH047) are important regulators of the iron deficiency response (Yuan et al., 2008; Long et al., 2010). The second-ranked regulator BHLH101 also governs iron homeostasis (Yuan et al., 2008; Sivitz et al., 2012). In addition to the TF FE-DEFICIENCY INDUCED TRANSCRIPTION FACTOR1 (FIT1/FRU) regulatory network in the epidermis, PYE, PYE homologs, and BRUTUS (BTS/EMB2454) form a regulatory network for maintaining iron homeostasis in low Fe conditions in the vasculature (Hindt and Guerinot, 2012), the latter being clearly represented in module 223. In pye-1 mutants, significant expression changes and a strong coexpression of all module genes, including BHLH101, were reported and ChIP-on-chip analysis detected the module gene FERRIC REDUCTION OXIDASE3 as a direct target of PYE (Long et al., 2010). Accordingly, the module was enriched for the presence of GCCACGTN (Supplemental Data Set 3), which resembles the E-box CANNTG known to be bound by bHLH TFs.
Module 402 functioned in jasmonic acid biosynthesis and was highly upregulated by salt stress and methyl jasmonate (Supplemental Figure 3). In correspondence with the module function and the presence of ABRE motifs, the top-ranked regulator JA-ASSOCIATED MYC2-LIKE1 (ATAIB/JAM1) acts as an ABA-inducible transcriptional repressor of jasmonic acid signaling (Nakata et al., 2013). The third-ranked predicted regulator ZBF1 (MYC2) transcriptionally modulates the jasmonic acid signaling pathway (Dombrecht et al., 2007) and was known from the extended reference set to regulate all module genes. ZBF1, together with ATAIB, binds to the target sequence of ZBF1 (Nakata et al., 2013). The regulators HIGH INDOLIC GLUCOSINOLATE1 (MYB51) and WRKY6 have also been implicated in jasmonic acid signaling (Skibbe et al., 2008; Laluk et al., 2012). Jasmonic acid signaling is not only involved in plant development, but also in response to biotic and abiotic stress, especially salt stress (Santino et al., 2013). This module also illustrates the success of the ensemble approach, since half of the interactions with the four relevant predicted TFs was found by only one inference method. Whereas most interactions with ATAIB were predicted by LeMoNe_qopt25R, LeMoNe_qopt50R, and ClrR, ZBF1 was identified as a regulator of the module by LeMoNe_qopt50R and ClrR, most interactions with MYB51 were found only by LeMoNe_qopt25R, and most interactions with WRKY6 were inferred only by LeMoNe_qopt50R (Supplemental Data Set 2).
Modules 61, 193, 381, and 491 had HSFA2 as the top-ranked regulator (Supplemental Table 5). HSFA2 is highly induced in response to oxidative stress caused by high light intensity and/or heat, conditions where the module genes were mostly upregulated (Nishizawa et al., 2006). Moreover, HSFA2 steers a heat shock factor signaling network and the response to misfolded protein accumulation in the cytosol in a later response to environmental stress (Sugio et al., 2009). In the extended reference set, HSFA2 was a known or hidden regulator of most module genes, including several heat shock proteins. Whereas the other HSFA2-regulated modules were significantly enriched for GO Biological Process “response to heat,” 8 out of 14 genes in module 61 had an unknown GO Biological Process annotation.
Experimental Confirmation of Predicted Interactions Reveals an Intricate Oxidative Stress GRN
To experimentally assess the performance of the ensemble reverse engineering approach, we analyzed gain- and loss-of-function mutants of seven TFs: the NAC TFs NAC13, NAC032, and NAC053; the DRE binding protein TF RAP2.1; the ERF TFs RELATED TO AP2 6L (RAP2.6L) and ERF6; and the WRKY TF WRKY6. These TFs were predicted to jointly regulate multiple target genes and modules (e.g., module 10 and module 293; Supplemental Data Set 3), which were induced upon salt, osmotic, and/or oxidative stresses. Six of these TFs function in plant stress responses: NAC13, and likely NAC053, as positive regulators in mitochondrial retrograde regulation (MRR) (De Clercq et al., 2013); NAC053 as a positive regulator in drought-induced leaf senescence (Lee et al., 2012); RAP2.1 as a negative regulator in cold and drought stress responses (Dong and Liu, 2010); RAP2.6L as a positive regulator in salt and drought stress responses (Krishnaswamy et al., 2011), in waterlogging stress (Liu et al., 2012), and in tissue regeneration upon wounding (Asahina et al., 2011); ERF6 as a positive regulator in jasmonic acid or ethylene-mediated pathogen defense (Moffat et al., 2012; Meng et al., 2013) and the osmotic stress response in leaves (Dubois et al., 2013); and WRKY6 as a positive and negative regulator of senescence and biotic defense responses (Robatzek and Somssich, 2002; Skibbe et al., 2008; Chai et al., 2014) and nutrient deficiency stress (Chen et al., 2009; Kasajima et al., 2010; Castrillo et al., 2013). Two-week-old transgenic and wild-type plants were mock-treated or salt-stressed for 12 h, and transcript levels of 92 predicted target genes, the 7 TFs, and 10 housekeeping genes were quantified by the NanoString nCounter Analysis System in a total of 15 times three biological replicate experiments (Geiss et al., 2008) (see Methods; Supplemental Figure 4). Differential expression analysis by a combination of DESeq and edgeR resulted in 289 unique TF-target gene interactions (Supplemental Data Set 4). Apparently, most TFs could act as activator and repressor, very likely through indirect regulatory effects.
We compared the performance of the experimentally derived regulatory interactions to those of the reference sets from literature and the reverse-engineering predictions (Table 2). Only seven interactions between the selected TFs and target genes were reported in literature, i.e., for ERF6 and WRKY6, of which six interactions were recovered by the nCounter experiments (Supplemental Data Set 4). For the extended reference set, where indirect regulatory effects of TFs were explicitly modeled through transitivity, 94 out of 192 relevant interactions were confirmed experimentally by nCounter. This significant increase in recall justified the construction of the extended reference set for benchmarking. Upon evaluation of the reverse-engineering predictions by nCounter, the integrated abiotic stress GRN (rank_av) ranked first in performance over the other ensembles and the individual inference methods as indicated by the F-measure. We experimentally validated 100 (50% precision) out of 199 predictions from the abiotic stress GRN and found an additional 189 (35% recall) regulatory interactions that were not predicted. For these additional regulatory interactions, we identified 25 extra true positives in the set of 785,913 total unique predictions, which were predicted only by LeMoNe_qopt25R, LeMoNe_qopt50R, or ClrR, and did not end up in the abiotic stress GRN. We retrieved 80 true positives in 14 modules, and found four additional true positive interactions that were not originally predicted, but whose prediction was inferred by the construction of the module abiotic stress GRN of coregulated genes and their regulators. Many of the differentially expressed genes might be the result of indirect regulatory effects or secondary effects in stable mutants. To account for the indirect influence of gain- and loss-of-function TF mutants and to increase the number of predictions, we also considered the relevant predicted targets of CYTOKININ RESPONSE FACTOR6, NAC032, NAC13, SALT-INDUCIBLE ZINC FINGER1 (SZF1), and WRKY6, which are TF targets of the perturbed TFs in nCounter, hence paths of length two in the GRN (see Methods; Supplemental Data Set 4). If target genes are considered the “children” of TF nodes, then we refer here to the indirect links between the TF nodes and their “grandchildren” nodes. In this way, 141 (52% precision) out of 271 predictions were experimentally validated and 148 (49% recall) additional regulatory interactions were recovered, leading to an F-score increase of 23% (Table 2). When evaluating the predictions for each TF separately, the performance of prediction was good for all TFs and increased upon considering paths of length two.
Table 2. Performance of the Literature Reference Set, the Extended Reference Set, the Individual Reverse-Engineering Methods, and Their Ensemble Solutions on Correctly Predicting the NanoString nCounter Experimental Data.
| Npred | Nknown | TP | Precision | Recall | F | |
|---|---|---|---|---|---|---|
| Literature reference set | 7 | 289 | 6 | 0.857 | 0.021 | 0.041 |
| Extended reference set | 192 | 289 | 94 | 0.490 | 0.325 | 0.391 |
| LeMoNe_qopt25R | 155 | 289 | 78 | 0.503 | 0.270 | 0.351 |
| LemoNe_qopt50R | 136 | 289 | 60 | 0.441 | 0.208 | 0.282 |
| ClrR | 195 | 289 | 96 | 0.492 | 0.332 | 0.397 |
| Union | 172 | 289 | 84 | 0.488 | 0.291 | 0.364 |
| Rank_rp | 177 | 289 | 86 | 0.486 | 0.298 | 0.369 |
| Rank_av | 199 | 289 | 100 | 0.503 | 0.346 | 0.410 |
| All_pred | 249 | 289 | 125 | 0.502 | 0.433 | 0.465 |
| Rank_av_2 | 271 | 289 | 141 | 0.520 | 0.488 | 0.504 |
| ERF6 | 11 | 49 | 10 | 0.909 | 0.204 | 0.333 |
| NAC13 | 30 | 85 | 27 | 0.900 | 0.318 | 0.470 |
| NAC032 | 41 | 42 | 22 | 0.537 | 0.524 | 0.530 |
| NAC053 | 32 | 57 | 13 | 0.406 | 0.228 | 0.292 |
| RAP2.1 | 18 | 6 | 2 | 0.111 | 0.333 | 0.167 |
| RAP2.6L | 26 | 4 | 3 | 0.115 | 0.750 | 0.200 |
| WRKY6 | 41 | 46 | 23 | 0.561 | 0.500 | 0.529 |
| ERF6_2 | 13 | 49 | 11 | 0.846 | 0.224 | 0.355 |
| NAC13_2 | 32 | 85 | 29 | 0.906 | 0.341 | 0.496 |
| NAC032_2 | 61 | 42 | 35 | 0.574 | 0.833 | 0.680 |
| NAC053_2 | 52 | 57 | 23 | 0.442 | 0.404 | 0.422 |
| WRKY6_2 | 69 | 46 | 38 | 0.551 | 0.826 | 0.661 |
The table shows the literature reference set of 52,328 experimental protein-DNA and regulatory interactions, the extended reference set, the top 200,000 predictions of the individual reverse-engineering methods LeMoNe_qopt25R, LeMoNe_qopt50R, and ClrR, and their ensemble solutions by union (Union), mean reciprocal rank (Rank_rp), and average rank (Rank_av) aggregation, against the 289 NanoString nCounter experimental data. The “_2” also takes the predictions of the TF targets of the perturbed TFs into account (paths of length two). For the final abiotic stress gene regulatory network (Rank_av and Rank_av_2; underlined), each TF was also evaluated individually. All_pred points to the 785,913 predictions of all four individual inference methods. Npred = number of predictions made for TFs and target genes belonging to the nCounter experimental data; Nknown = number of experimental nCounter interactions; TP = number of true positives; Precision = TP/Npred; Recall = TP/Nknown; F = 2 × precision × recall/(precision + recall) = 2 × TP/(2 × TP + FP + FN).
We visualized predictions for the 102 selected target genes in the abiotic stress GRN and whether they were previously known from literature, inferred in the extended reference set, experimentally validated by nCounter, or any combination of these in Supplemental Figure 5. This illustrates the limited availability of known regulatory interactions with few TFs and the potential of reverse-engineering, generating regulatory hypotheses on a systems-wide scale.
We analyzed the presence of the GCC-box (ERF6 and RAP2.6L) (Hao et al., 1998), ERF6 GCC-box (ERF6) (Wang et al., 2013), DRE (RAP2.1) (Dong and Liu, 2010), NAC (NAC13, NAC032, and NAC053) (Duval et al., 2002; Tran et al., 2004; Olsen et al., 2005), MDM (NAC13 and NAC053) (De Clercq et al., 2013), W-box (WRKY6) (Yamasaki et al., 2012), and ARE (WRKY6) (Castrillo et al., 2013) cis-regulatory elements at most 1000 bp upstream of the translation start site of the nCounter target genes (Supplemental Table 4). For 170 target genes, multiple copies of a relevant binding site were present in their promoters (Supplemental Data Set 4). Since the response to perturbation of a TF is strongest for its direct targets and dissipates rapidly as it propagates through the network (Haynes et al., 2013), we found that in 38 interactions, the target genes were at least one log2-fold induced or repressed by the TF mutants. Finally, eight NAC13 target gene promoters were previously shown to be bound by NAC13 in ChIP experiments (De Clercq et al., 2013). These observations suggest that 65% of the nCounter interactions and 76% of the true positives that were directly predicted are direct transcriptional regulatory interactions.
As NAC032 had not yet been functionally characterized, we analyzed whether altered NAC032 levels affect tolerance to osmotic stress, a condition under which its target module genes were most prominently upregulated. Interestingly, NAC032 overexpression lines displayed increased plant biomass and rosette area under osmotic stress conditions (Supplemental Figure 6).
From the visualization of the nCounter data in a network, we observed many regulatory interactions between the selected TFs and detected multiple copies of relevant cis-regulatory motifs in their promoters (Figure 3A; Supplemental Table 4). The most influential TF in the experimental network appeared to be NAC13, since it targeted the largest number of genes (83%), including six out of the seven TFs, and gave rise to the highest transcriptional responses (Supplemental Data Set 4). NAC053, ERF6, WRKY6, and NAC032 targeted between 56 and 41% of the genes, while RAP2.1 and RAP2.6L regulated only 5% of the genes. The topological overlap, i.e., the number of overlapping interacting nodes/modules between the TFs, normalized over their out-degrees, was highest for NAC13 and NAC053 (Figure 3B; see Methods). The next best topological overlap was observed between WRKY6, ERF6, and NAC032. Overall, these five TFs showed a high overlap in transcription regulatory targets, both at the module and the gene level.
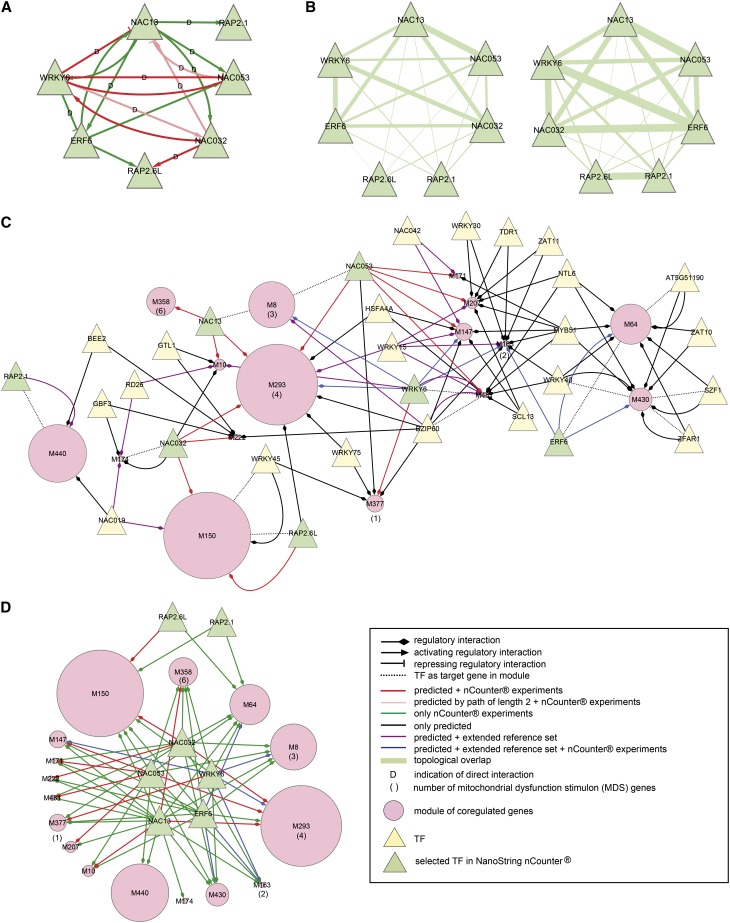
Figure 3.
Experimental Confirmation of Predicted Interactions Reveals a Core Oxidative Stress GRN with Intertwined Regulation by NAC13, NAC053, ERF6, WRKY6, and NAC032.
(A) In gain- and loss-of-function mutants of seven TFs upon 12 h salt stress treatment, transcript changes of a hundred predicted target genes, including the seven TFs, were quantified by nCounter and statistical analysis by a combination of DESeq and edgeR. These experiments revealed even more interconnected regulatory interactions between the perturbed TFs than was predicted by ensemble reverse-engineering. NAC13 activated all TFs except RAP2.6L. WRKY6 was found to repress NAC13, NAC053, and ERF6. In turn, NAC053 had an inhibitory effect on WRKY6, creating a negative feedback loop between these two TFs. NAC13 and NAC032 activated WRKY6, and there was a mixed feedback loop between NAC13 and WRKY6. Similarly, ERF6 activated NAC13, which resulted in a positive feedback loop between NAC13 and ERF6. D = indication of direct interaction by multiple copies of a relevant binding site in the promoter, more than one log2-fold transcriptional change upon TF perturbation, and/or reported TF binding by ChIP (see text).
(B) Topological overlap between the perturbed TFs in the nCounter experiments on a gene basis (left) or module basis (right). This is the number of overlapping target genes or modules between two TFs normalized over their out-degrees. The width of the edge is proportional to the topological overlap coefficient. NAC13, NAC053, WRKY6, ERF6, and NAC032 showed a high overlap in transcription regulatory targets, both at the gene and the module level.
(C) Predicted regulatory network for modules containing nCounter target genes that were predicted to be regulated by the seven selected TFs or contained these TFs themselves as target gene. The size of the circular node, i.e., the module, is proportional to the number of nCounter target genes present in the module. In addition to the selected TFs, only TFs predicted to regulate multiple modules were displayed. Modules that contained or were predicted to be regulated by any of the seven selected TFs shared predicted regulators and GO Biological Process annotations in detoxification and oxidative stress responses.
(D) Experimental nCounter regulatory network for modules containing nCounter target genes that were predicted to be regulated by the seven selected TFs or contained these TFs themselves as target gene. Interpretation is as in (C). We found many modules to be regulated by combinations of NAC13, NAC053, NAC032, WRKY6, and ERF6 or all these TFs together.
To gain insight into the functional role of the selected TFs within the abiotic stress GRN, we visualized the modules that contained or were predicted to be regulated by any of the seven selected TFs and were tested in the nCounter experiments. Figure 3C represents the predicted regulatory interactions of the abiotic stress GRN, some of which were confirmed by the extended reference set, while Figure 3D illustrates the experimental regulatory interactions. These modules shared the following GO Biological Process annotations: response to chitin, respiratory burst in the defense response, regulation of the plant-type hypersensitive response, regulation of programmed cell death, ER unfolded protein response, toxin catabolic process, response to cyclopentenone, and response to ethylene. They contained 16 mitochondrial dysfunction stimulon (MDS) genes, which are implicated in MRR and contain the cis-regulatory motif MDM in their promoter (De Clercq et al., 2013) (Figures 3C and 3D). Many of these modules shared other predicted TFs: We counted the highest number of outgoing edges for MYB51 and BZIP60, followed by WRKY15, WRKY40, and NTL6, with MYB51 and WRKY15 being first rank regulators. Other predicted first rank regulators of multiple modules included JUNGBRUNNEN1 (NAC042), RD26, WRKY30, BR ENHANCED EXPRESSION2, and SZF1. Several predicted interactions with BZIP60, WRKY15, NAC042, and RD26 were confirmed in the extended reference set. Even more so in the nCounter regulatory network, we found many modules to be regulated by combinations of NAC13, NAC053, NAC032, WRKY6, and ERF6 or all these TFs together (modules 8, 64, 150, 163, 293, 358, and 430). We assessed modules 150 and 293 in more detail, since they contained the highest number of target genes tested experimentally (Figure 4).
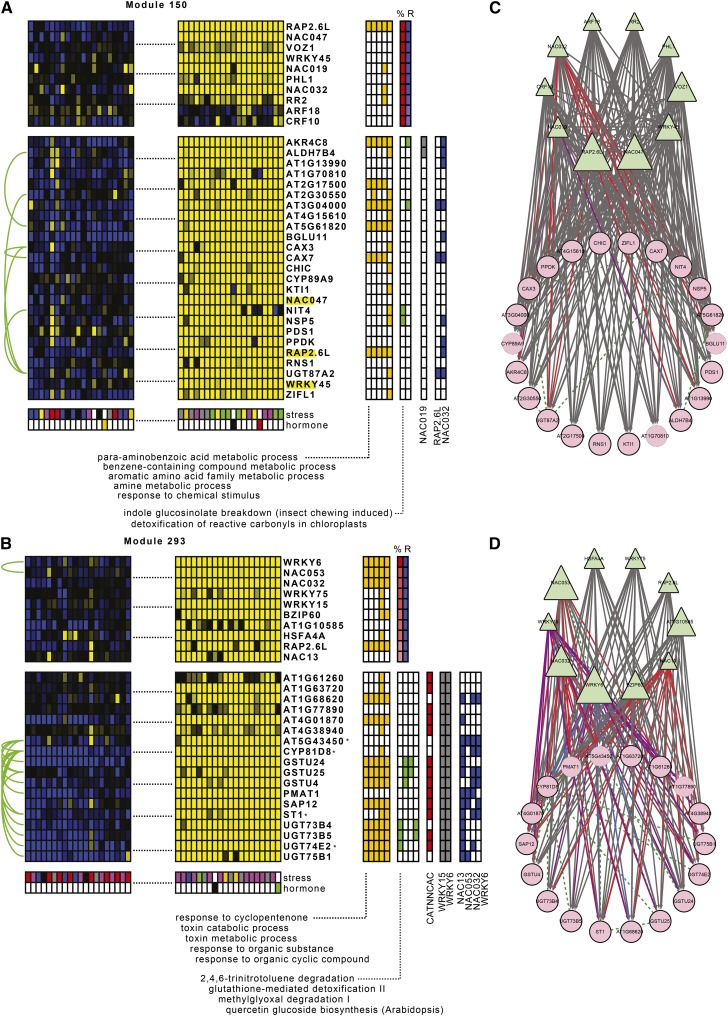
Figure 4.
Experimental and Literature-Based Evidence for Modules Involved in Detoxification Processes of the Oxidative Stress Response.
(A) and (C) Module 150 had as top significant GO Biological Process term “PABA metabolism.” Conditions where the module genes were most highly upregulated consisted of osmotic, salt, and oxidative stress. Two regulatory interactions with NAC019 were confirmed by the extended reference set. Out of 15 module genes tested by nCounter, we experimentally validated 10 targets for NAC032, all of which contained multiple NAC binding motifs in their promoters, and three targets for RAP2.6L. Additionally, we detected one interaction with RAP2.1, four interactions with WRKY6, seven with ERF6, eight with NAC053, and 14 with NAC13.
(B) and (D) Module 293 was highly significantly GO enriched for “response to cyclopentenone” and “toxin catabolic process.” Conditions where the module genes were most highly induced included oxidative stress and auxin inhibition. All module genes were found to be regulated by WRKY6 and WRKY15 in the extended reference set. Through nCounter experiments on 14 module genes, we confirmed four targets for NAC053, seven for WRKY6, eight for NAC032, and 13 for NAC13. Additionally, we found 12 targets for ERF6. Several of these targets contain multiple NAC or W-box binding sites in their promoter, as well as the NAC13/NAC053 MDM motif and the WRKY6 ARE motif. This module is likely involved in MRR.
Interpretation is as in Figure 2. Asterisk indicates MDS gene.
The highly significant GO Biological Process terms for module 150 were “para-aminobenzoic acid (PABA) metabolism,” “toxin catabolic process,” “response to cyclopentenone,” and “response to water deprivation.” According to AraCyc, it was involved in “indole glucosinolate breakdown” and “detoxification of reactive carbonyls.” It was mostly upregulated by osmotic, salt, and oxidative stress as well as ABA. Two regulatory interactions with NAC019 were validated by the extended reference set. Out of 15 module genes tested by nCounter, we experimentally validated 10 targets for NAC032, all of which contained multiple NAC binding motifs in their promoters, and three targets for RAP2.6L. Additionally, we detected one interaction with RAP2.1, four interactions with WRKY6, seven with ERF6, eight with NAC053, and 14 with NAC13 (Supplemental Data Set 4).
Highly significant GO enrichments for module 293 were “response to cyclopentenone,” “toxin catabolic process,” “response to ethylene stimulus,” “response to chitin,” and “hyperosmotic salinity response.” The module was enriched for the AraCyc metabolic pathways “2,4,6-trinitrotoluene degradation,” “glutathione-mediated detoxification II,” and “quercetin glucoside biosynthesis.” An additional connection with module 150 was that UDP-GLUCOSE TRANSFERASE1 (UGT75B1) in this module glucosylates the folate precursor PABA into the storage form of PABA as a glucose ester (Eudes et al., 2008). Module genes were highly induced by oxidative stress and auxin inhibitors and were known to be regulated by WRKY6 and WRKY15 from the extended reference set. Through experimental validation on 14 module genes, we confirmed four targets for NAC053, seven for WRKY6, eight for NAC032, and 13 for NAC13, and we found in addition 12 targets for ERF6 (Supplemental Data Set 4). Several of these targets contained multiple NAC or W-box binding sites in their promoter, as well as the NAC13/NAC053 MDM motif and the WRKY6 ARE motif (Supplemental Table 4). Four module genes were MDS genes and three of them were previously shown to be direct NAC13 targets in a ChIP experiment (De Clercq et al., 2013).
Both modules 150 and 293 contained many catabolic detoxification enzymes such as cytochrome P450, oxidoreductase, hydrolase, dehydrogenase, UDP-glucose transferase (UGT), and glutathione S-transferase (GST). Like module 293, module 191 contained multiple UGTs and GSTs, was highly upregulated by oxidative stress and auxin inhibitors, and was involved in the AraCyc metabolic pathway “glutathione-mediated detoxification II” and “ABA glucose ester biosynthesis” (Supplemental Figure 7). Similarly to modules 150 and 293, it was highly enriched for GO Biological Processes “PABA metabolic process” and “response to cyclopentenone,” and predicted to be regulated by WRKY6, which was confirmed by the extended reference set, NAC032, NAC053, and WRKY45 (Supplemental Table 3). Moreover, this module contained an overrepresented TGACGT motif, which is found in as-1-like elements in promoters of early salicylic acid-induced genes and is bound by class II TGACG SEQUENCE-SPECIFIC BINDING PROTEIN (TGA) TFs that drive xenobiotic detoxification (Fode et al., 2008; Blanco et al., 2009). This motif was identified in 14 out of 20 module genes, many having two copies in their promoter, the ideal as-1-like element (Qin et al., 1994). This motif was also found, although not significantly enriched and in single copy, in 10 genes of module 150 and five genes of module 293. Of the selected TFs, the NAC032 promoter encodes three TGA motifs and its expression is highly dependent on TGA TFs and its transcriptional coactivator SCARECROW-like 14 (SCL14) (Fode et al., 2008; Zander et al., 2014). The WRKY6 promoter had one TGA motif. Treatment with salicylic acid induced genes of module 191 more than module 293 and even more than module 150. Approximately 60% of all cyclopentenone-inducible genes, many of them encoding detoxification-related genes such as GSTs, cytochrome P450s, UDPs, and transporters, are dependent on the TFs TGA2, TGA5, and TGA6 for their expression and 40% of these genes contain a TGA motif in their promoter (Mueller et al., 2008). We found large similarities, but also differences between the detoxification modules. Detoxification genes have been shown to differ in their level of induction by cyclopentenone but also in the specificity of induction by different TGA factors (Stotz et al., 2013), which could be regulated by the copy number of TGA binding sites in their promoter (Zander et al., 2014). Several of these cyclopentenone-inducible genes and TGA2/TGA5/TGA6-dependent genes were present in modules 150, 191, and 293, but none of the class II TGA TFs were predicted as regulators of these modules (Mueller et al., 2008; Zander et al., 2014).
In a recent study on the role of the module 293 gene UGT73B5 in the redox balance during the hypersensitive stress response to pathogen treatment, multiple genes of modules 191 and 293, and to a lesser extent module 150, as well as NAC032 and NAC053, were coexpressed with this UGT, which has a TGA motif in its promoter (Simon et al., 2014).
Hence, these modules are likely implicated in the detoxification of cyclopentenone oxylipins and by-products of secondary metabolites, such as camalexin (Mueller et al., 2008; Simon et al., 2014), hormone metabolism and transport (e.g., ZINC INDUCED FACILITATOR-LIKE1 [Remy et al., 2013], UGT73B1 and UGT75B1 [Lim et al., 2005], INDOLE-3-ACETATE β-d-GLUCOSYLTRANSFERASE, UGT74E2 [Tognetti et al., 2010], SULFURTRANSFERASE1 [Marsolais et al., 2007], UGT74F2 [Dean and Delaney, 2008]), and protection against oxidative stress (e.g., PHYTOENE DESATURATION1 [Norris et al., 1995]; UGT73B1, UGT73B4, and UGT73B5 [Lim et al., 2004]). The detoxification process starts with the introduction of functional groups by enzymes like cytochrome P450 (phase I), which are subsequently conjugated to glucose or glutathione by enzymes such as UGTs and GSTs (phase II).
In conclusion, we validated the ensemble reverse-engineering approach and hence revealed an intricate oxidative stress network regulated by NAC13, NAC053, ERF6, WRKY6, and NAC032 that is implicated in detoxification processes aimed at removing reactive compounds created by cellular stress.
DISCUSSION
Reverse-engineering of gene expression profiles or network inference offers great potential for elucidating GRNs. Here, we applied three different, highly complementary reverse-engineering algorithms to a microarray expression compendium of abiotic stress conditions resulting in four network inference solutions. Next, an abiotic stress GRN was created by taking the top 200,014 predictions of an ensemble solution through average rank aggregation of the different individual predictions. Through benchmark analysis against a data set of known protein-DNA and regulatory interactions, as well as unbiased, medium-throughput expression profiling of TF gain- and loss-of-function mutants, we demonstrated that this ensemble solution was the most robust in predicting regulatory interactions and attained precision and recall as high as 50%. Evaluation of the biological relevance of the predicted regulators also indicated that the ensemble provided a more complete picture of the regulatory landscape than individual inference methods could. Ensemble reverse-engineering by average rank aggregation significantly increases the coverage of biologically meaningful regulatory interactions and compensates for the peculiar biases of the different reverse-engineering algorithms (Michoel et al., 2009; Marbach et al., 2012).
Gene regulatory network validation is difficult, since each benchmark set or experimental validation setup has its own limitations and the best practice is to combine multiple methods (Walhout, 2011). Despite the fact that the in silico benchmark set contained regulatory interactions for only 15% of TFs with predicted regulatory interactions, that GO, AraCyc, CORNET protein-protein interactions and known cis-regulatory elements are incomplete and might contain false positives, and that TF perturbation experiments and nCounter analysis might create false positives and negatives, we still recovered several interactions of which the biological relevance was supported by the extended reference set, the functional and regulatory coherence analysis, and the nCounter experiments together. Nevertheless, due to the limited coverage of the benchmarking in general (Supplemental Figure 5), it is justified to consider all abiotic stress GRN predictions as useful hypotheses, with the ones being validated obtaining a higher confidence (Walhout, 2011).
Furthermore, we showed that the abiotic stress GRN can be clustered into functionally coherent coregulated gene modules, which facilitated the biological interpretation of the GRN network. These gene modules made it also possible to attribute GO Biological Process annotations to 1966 uncharacterized genes using the “guilt by association” principle. Interestingly, we detected stress-related modules that operate in response to environmental conditions that were not included in the microarray compendium, such as biotic stress (module 443) and iron deficiency (module 223). This could be explained by the fact that the modules are part of a “general plant core environmental stress response” (Kilian et al., 2007) but could also indicate specific crosstalk (Suzuki et al., 2014).
Reverse-engineering based only on expression profiles has limitations in recovering true direct regulatory interactions and will generate false positives. First, the predicted interactions are not necessarily direct interactions but might constitute longer paths consisting of hidden direct regulators between the detected regulator and target gene (Vermeirssen et al., 2009). This can partially be overcome by identifying cis-regulatory motifs in the promoters of target genes, either within the network inference algorithm or postprocessing, as is done in this study. We demonstrated coregulation of module genes through the significant overrepresentation of abiotic stress cis-regulatory motifs, which for several modules corresponded to the predicted TFs, indicative of direct regulatory interactions. The availability of known cis-regulatory motifs for specific TFs is limited, even for a well-studied model species such as Arabidopsis (Higo et al., 1999; Palaniswamy et al., 2006; Vandepoele et al., 2009). Furthermore, there is not a one-to-one relation between a specific TF and a cis-regulatory motif (Badis et al., 2009). Hence, reverse-engineering methods that do incorporate cis-regulatory motifs might be too strict. Additionally, we had binding evidence from Y1H and ChIP experiments in the reference sets indicating that several of the predicted interactions were direct (Supplemental Table 1). Moreover, we were able to correctly predict the direct regulation by NAC13 of eight genes, promoters of which were bound by NAC13 in a ChIP experiment in our previous study (Supplemental Data Set 4) (De Clercq et al., 2013). Second, many network inference methods, including LeMoNe and CLR, have difficulty in distinguishing between coexpression and regulation based on expression profiles (Michoel et al., 2009). Nevertheless, average rank aggregation will result in the top predictions having a higher chance of being predicted by multiple methods, although the ranks of a specific interaction in the individual methods also play a role. Our benchmarking points out that regulatory interactions predicted by multiple reverse-engineering algorithms were more likely to be true positives.
Experimental validation indicated that many regulatory interactions were also missed by network inference. Only regulators with a clear correspondence between their transcriptional readout and their regulatory activity can be readily retrieved. Interactions with TFs that are tightly regulated through posttranslational modifications including redox modifications, protein-protein interactions, phosphorylation, ubiquitination, sumoylation, proteolytic activation, or a combination thereof (Qin et al., 2011; Vaahtera and Brosché, 2011), or that are controlled by microRNAs, might be missed. However, we were able to recover functional heterodimers among the predicted regulators, despite the limited availability of experimental protein-protein interactions. Additionally, TF perturbation combined with differential expression analysis also identified direct and indirect interactions, complicating the recovery of the true underlying direct GRN. We partially circumvented this by considering paths of length two in the network, hence increasing our predictions. False negatives could possibly be further reduced by adding more diverse algorithms to the ensemble. Overall, experimental protein-DNA interaction mapping combined with gene expression data could provide an even more biologically relevant picture of the GRN.
TF perturbation experiments revealed an interconnected GRN, with many genes targeted by multiple TFs from the selected set, as predicted. Moreover, the selected TFs regulated one another, with NAC13 as a potential major regulator. Several of the selected TFs have previously been reported to act in oxidative stress responses. WRKY6 is a positive regulator of a thioredoxin (Laloi et al., 2004). RAP2.6L induces antioxidant defense under waterlogging stress by an ABA-dependent pathway (Liu et al., 2012). ERF6 is a positive regulator of reactive oxygen species (ROS)-responsive genes in the oxidative stress response (Wang et al., 2013). NAC053 promotes ROS production by binding directly to the promoters of genes encoding ROS biosynthetic enzymes during drought-induced leaf senescence (Lee et al., 2012). NAC13 and likely also NAC053 are direct regulators of the mitochondrial retrograde regulation of the oxidative stress response (De Clercq et al., 2013). These membrane-bound NAC proteins (NTL TFs) are part of a phylogenetic subgroup (NAC2) of the NTL TFs (Kim et al., 2010) and share the MDM cis-regulatory element present in mitochondrial retrograde regulated genes (De Clercq et al., 2013). Therefore, it is not surprising that their topological overlap was the largest. NAC032 interacts with NAC019 upon salt and osmotic stress in an ABA hormone interactome (Lumba et al., 2014), in agreement with both TFs targeting modules 150 and 174, which are highly induced upon these stresses and ABA (Figure 3C). Therefore, together with our observed phenotype of improved growth performance during osmotic stress, it is likely that the ABA-responsive NAC032 mediates the oxidative stress aspect of the osmotic stress response. While RAP2.1 and RAP2.6L targeted only a minority of genes and modules, NAC032 and ERF6 mostly activated, NAC053 and WRKY6 mostly repressed, and NAC13 both activated and repressed over 40% of the target genes experimentally tested. These five TFs regulated modules that function in detoxification processes during oxidative stress in plants, such as modules 150, 191, and 293. Abiotic stress is associated with the perturbation of ROS homeostasis. Increased ROS levels are able to damage cellular components and might also act as secondary messengers (Apel and Hirt, 2004; Baxter et al., 2014). Nonenzymatically formed compounds derived from the oxidative metabolism of polyunsaturated fatty acids, cyclopentenone oxylipins, are triggered by increasing ROS levels and serve important roles in diverse processes such as detoxification, defense response by the production of secondary metabolites, and programmed cell death (Thoma et al., 2003; Mueller et al., 2008; Stotz et al., 2013). These oxylipins induce detoxification enzymes, which in turn metabolize them. NAC032, WRKY6, NAC053, and RAP2.6L have been associated with response to cyclopentenone and detoxification processes (Mueller et al., 2008; Heyndrickx and Vandepoele, 2012; Simon et al., 2014; Zander et al., 2014). TFs that govern detoxification of xenobiotics and possibly endogenous harmful metabolites, such as cyclopentenone, are TGA2, TGA5, and TGA6, which bind a motif with consensus sequence TGACGT in promoters of salicylic acid-inducible genes, as a heterodimer with SCL14 (Fode et al., 2008; Blanco et al., 2009). We did not detect these TFs as predicted regulators of our modules. However, NAC032 is most likely a direct target of the TGA-SCL14 complex (Fode et al., 2008; Zander et al., 2014), and WRKY6 as well, since both TFs contain TGA motifs in their promoters and were found to regulate detoxification-related genes, several of which are reported to be TGA regulated (Mueller et al., 2008; Zander et al., 2014; this study). Therefore, detoxification-related genes in the oxidative stress network could be regulated by TGA TFs indirectly, through NAC032 or WRKY6, or other TFs in the oxidative stress GRN. In addition, the expression of 40% of all cyclopentenone-inducible genes was independent of TGA2, TGA5, and TGA6 (Mueller et al., 2008), and TGA’s TFs can recruit different interacting proteins for heterodimerization (Fode et al., 2008), suggesting that other regulatory factors influence cyclopentenone oxylipin detoxification. In this respect, treatment with an ethylene precursor of the triple mutant tga2 tga5 tga6 identified 136 induced genes whose expression is TGA dependent, including genes encoding the TFs NAC032, WRKY45, and ZAT10 of our oxidative stress GRN, while 227 TGA-independent upregulated genes were detected, including those encoding NAC019, RAP2.6L, WRKY40, and WRKY75 (Zander et al., 2014).
Several of the modules of the oxidative stress GRN were also predicted to be regulated by other TFs with a reported function in the oxidative stress response, such as MYB51 (Gigolashvili et al., 2007), BZIP60 (Iwata et al., 2008), WRKY15 (Vanderauwera et al., 2012), WRKY40 (Van Aken et al., 2013), WRKY30 (Scarpeci et al., 2008), NTL6 (Yang et al., 2014), and NAC042 (Wu et al., 2012); with a reported function in osmotic stress, such as WRKY15 (Vanderauwera et al., 2012), and salt stress, such as SZF1 (Sun et al., 2007); or with a reported function in detoxification, such as RD26 (Fujita et al., 2004). Regulatory interactions with BZIP60, WRKY15, NAC042, RD26, and NAC019 were even confirmed by the reference sets from literature (Figure 3C). Whereas previous studies elaborate on the individual roles of these TFs in the oxidative stress response, we demonstrated here the coordinated and intertwined regulation by NAC13, NAC053, ERF6, WRKY6, and NAC032, and very likely MYB51, BZIP60, WRKY15, and the other TFs, of the oxidative stress and detoxification response in Arabidopsis.
More specifically, we found several connections of the core oxidative stress GRN with conditions in which mitochondrial function is impaired due to adverse environmental conditions and mitochondria signal to the nucleus to trigger feedback responses (De Clercq et al., 2013). Several TFs in the network bind the promoters of nuclear genes encoding mitochondrial proteins, such as GBF3, NAC13, NAC053, WRKY15, WRKY30, WRKY40, WRKY45, and WRKY75 (Ng et al., 2014). Several MDS genes were part of the selected modules (Figure 3C). In addition to the MRR regulatory role of NAC13 and NAC053, WRKY TFs have also been reported to regulate MRR (Van Aken et al., 2013). There is also a connection between detoxification and MRR. Genes of modules involved in detoxification were highly upregulated by auxin transcriptional inhibitors, which have been reported to induce MRR (Kerchev et al., 2014). In mammalian cells, cyclopentenone prostaglandins are potent inhibitors of nuclear factor-kappaB activation (Rossi et al., 1997), which also mediates MRR upon mitochondrial dysfunction (Butow and Avadhani, 2004). This points to MRR steering detoxification processes during oxidative stress, likely through the TFs in the oxidative stress GRN.
Taken together, through extensive in silico and experimental validation of an abiotic stress regulatory network constructed by ensemble reverse-engineering, we demonstrated its value in unraveling system biological insights into the plant abiotic stress response.
METHODS
Expression Profile Compendium
We gathered Raw Affymetrix ATH1 expression profile data (CEL files) from GEO, TAIR, ArrayExpress, NASCArrays, and in house for 45 series of experiments, representing 642 arrays and 283 different control and experimental conditions (Supplemental Data Set 1) (Petrov et al., 2012). Experimental conditions were annotated according to the type of abiotic stress and hormone treatment. The microarray data were preprocessed in Bioconductor R. Through the robust multiarray average method, a background-adjusted, quantile normalized, and summarized log-transformed expression value was obtained for each Arabidopsis thaliana probe set. Different replicate conditions were summarized and 199 ratio expression values were derived for each probe set by dividing experiment over control. To limit off-target hybridization, we used a custom Arabidopsis cdf file that consists of 19,937 probe sets of at least eight probes, each targeting with perfect sequence identity to its transcript and not aligning to any other gene’s transcript with zero or one mismatches (Casneuf et al., 2007). Gene symbols and TAIR functional descriptions throughout the article are based on gene_aliases.20130130.txt and TAIR10_functional_descriptions from TAIR (www.arabidopsis.org). We removed genes for which the ratio hardly changed over all conditions (sd lower than 0.25) to identify regulation programs for different types of abiotic stresses as opposed to the overall regulation of the general abiotic stress response. Hence, we obtained an Arabidopsis gene expression profile compendium consisting of average expression ratios for 13,805 genes in 199 conditions.
Regulator List
We selected 1340 TFs based on the presence on the ATH1 array and described in the database Plant TFDB v2.0 Peking University, which contains 1709 TFs in 58 families (only ATG identifiers, PlantGDB-generated Unique Transcripts, and UniGene identifiers) (Zhang et al., 2011).
LeMoNe Analysis
We ran 20 independent Gibbs sampler LeMoNe runs (software available at http://bioinformatics.psb.ugent.be/software/details/LeMoNe) (Joshi et al., 2008, 2009), generating 20 local optima module clusters solutions, from which an ensemble-averaged solution of coexpression modules was created. We varied the ensemble clustering parameter qopt (at least cluster together in 5 out of the 20 runs _ qopt25 and at least cluster together in 10 out of the 20 runs _ qopt50, with a higher stringency leading to smaller size modules). LeMoNe assigns each gene to only one cluster. For qopt25 and qopt50, we obtained 380 and 998 module clusters, respectively, containing three or more genes, with a total of 6683 and 7698 genes. Using the regulator list described above and now also including regulators with low variation in gene expression for the regulator assignment, LeMoNe predicted a ranked list of weighted regulators for each module (Vermeirssen et al., 2009). We considered only the top 2% of all regulators assigned, having a weight of 10 or higher. For both cluster solutions qopt25 and qopt50, the regulator assignment was performed twice and the solutions were combined by mean reciprocal rank aggregation, i.e., the final rank of a predicted interaction is the reciprocal mean of the ranks of the underlying LeMoNe_qopt25R and LeMoNe_qopt50R solutions. Finally, we obtained 215,656 and 220,841 predicted regulatory interactions for LeMoNe_qopt25R and LeMoNe_qopt50R, respectively.
CLR Analysis
We applied CLR (Faith et al., 2007) to the microarray compendium described above, to which the expression profiles of remaining TFs from Plant TFDB present on ATH1 with low expression ratio variability were added and now consisted of average expression ratios for 14,219 genes in 199 conditions. We retrieved mutual information z-scores for target gene interactions with the 1340 regulators defined above. With a cutoff for the z-score at 3.5 (P value < 2.15 × 10−5), we obtained a total of 243,530 predicted regulatory interactions.
TwixTrix Analysis
We also applied TwixTrix (software available at https://code.google.com/p/twixtrix/) to the same microarray compendium of 14,219 genes (Qi and Michoel, 2012). We obtained a total of 199,985 predicted regulatory interactions with a cutoff for the z-score at 3 (P value < 0.0027).
Combining Predictions Made by LeMoNe, CLR, and TwixTrix
The four predictions made by different, individual reverse-engineering methods were combined by unweighted rank aggregation. Every predicted regulatory interaction received a final rank that was calculated by averaging the ranks of the four individual inference solutions (average rank aggregation), by taking the highest ranking given by the individual solutions (union), or by averaging the reciprocal ranks of the individual solutions (mean reciprocal rank aggregation) for that specific interaction. When an interaction was not predicted by a certain method, it received for that method a rank that was equal to the number of predictions (around 200,000) plus one (Marbach et al., 2012).
Reference Sets
From different resources we compiled a set of “known” experimental regulatory and protein-DNA interactions to benchmark our predictions (we incorporated only interactions that contain genes and TFs for which predictions were made): (1) AtRegNet (last updated September 15, 2010; Yilmaz et al., 2011): 7221 interactions between 57 TFs and 4903 genes (of which 648 interactions between 37 TFs and 581 genes were direct confirmed interactions); (2) TF perturbation expression interactions from literature (mostly microarray): 8772 interactions between 43 TFs and 5456 genes (Supplemental Table 5); (3) Brady Y1H (Brady et al., 2011; Gaudinier et al., 2011): 157 interactions between 82 TFs and 20 genes; (4) TF tool from CORNET: 36,550 interactions between 48 TFs and 10,336 genes (De Bodt et al., 2012); (5) additional ChIP experiments from literature, where high-confidence targets were identified (combination of binding and expression profiling): 659 interactions between 7 TFs and 624 genes (Supplemental Table 5). In total, this known reference set has 52,328 interactions between 201 TFs and 12,072 genes (Supplemental Table 1). From this reference set, we assembled two additional reference sets. We constructed an extended reference set of 789,068 interactions in an iterative way as follows: We derived a regulatory interaction between protein A and target gene B if there is a path through a combination of reported protein-DNA interactions and regulatory interactions present from A to B, e.g., A binds C, C regulates D, D binds E, and E regulates B (Supplemental Table 1). The derived regulatory interactions are the “hidden” interactions present in the known protein-DNA interactions and regulatory interactions data sets, with paths of length greater than one in the known reference set. We also built a direct reference set that consisted of 1307 “known” regulatory interactions between 43 TFs and 1115 genes that had evidence of both binding between TF and target gene and regulatory effect of TF on transcription of target gene (Supplemental Table 1). The latter reference set likely consists of “true” direct transcriptional regulatory interactions.
Measurement of Algorithm Performance
We constrained the predicted interactions to include only the TFs and target genes of the reference sets, and we also restricted the reference sets to the 14,219 genes and 1340 TFs for which predictions were made. For every algorithm and every reference set, we evaluated the algorithm performance as follows. We calculated precision, i.e., number of true positives over number of predictions, and recall, i.e., number of true positives over number of known interactions, for every rank for which predictions for the specific reference set were made to make precision-recall curves and to estimate AUPR, using trapezoid rule numerical integration. We computed the F-measure, which is the harmonic mean of precision and recall with a best value of 1, for the top 200,000 total predictions.
Clustering Genes into Coregulation Modules
We calculated the overlap in predicted regulators for all 11,938 genes in the abiotic stress GRN using the Jaccard similarity index and used this to group the genes into modules of coregulated genes through k-means clustering. The optimal number of clusters (k = 600) was chosen based on coregulation correctness and functional coherence of the obtained coregulated modules. We retained at most 10 regulating TFs per module: TFs were ordered by the number of genes in the module they regulate, where we required that they regulate at least half of the genes in the module and by their average rank calculated over all the genes they regulate in the module. We removed few clusters containing less than three genes, one cluster containing over 1000 genes, and few clusters that had no single TF regulating 50% or more of the cluster genes.
We calculated the out-degree of the TF nodes in the abiotic stress GRN and the module network to identify the 5% most connected TFs and denoted them “hubs.”
Visualization of Gene Modules
To generate figures that visualize coregulated modules as heat maps, both for the article and for the online supplemental data, we developed a standalone Java tool called ModuleViewer. ModuleViewer can read an expression matrix in combination with data that cluster genes into coregulated modules and assigns regulators to each module. On top of this, biological data can be loaded that link information on the functional coherence of the module and the biological relevance of the predicted regulators, such as GO and AraCyc annotations, cis-regulatory motifs, condition categories, and protein-protein interactions to each module. These data can be represented in several visual styles, like color matrices, interaction arrows, or gene highlights. After loading the data, the tool will automatically compose the figures and return a GUI allowing the user to browse through the generated figures and export a selection as vector graphics. The network figures for the article were drawn in Cytoscape. To create the interactive networks in the online supplemental data, we used the cytoscape.js JavaScript libraries.
Functional Analysis on Gene Modules
Each module was analyzed for GO Biological Process enrichment with BiNGO using a gene-based custom annotation file for Arabidopsis (created from annotation and ontology files downloaded from www.geneontology.org on May 3, 2013; Berardini et al., 2004), the whole annotation as a reference set (22,813 Arabidopsis genes with Arabidopsis Gene ID have a Biological Process annotation), and Benjamini and Hochberg false discovery rate multiple hypothesis testing correction for multiple testing with a confidence level of 95% (Maere et al., 2005). In a similar manner, each module was assessed for AraCyc 10.0 metabolic pathway enrichment (2785 Arabidopsis genes are annotated with a specific metabolic pathway). We kept the significant GO or AraCyc enrichment only if more than one gene in the module had the GO Biological Process or AraCyc metabolic pathway annotation. The functional coherence of genes in the modules was independently assessed by identifying the percentage of genes in the module that shared functional gene-gene links in AraNet (1,062,222 gene-gene links in total between 19,647 genes) and the percentage of AraNet gene-gene links compared with all possible gene-gene interactions within the module (Lee et al., 2010). Through 33,202 experimental protein-protein interactions between 7936 different proteins present in CORNET (De Bodt et al., 2012), we also calculated for each module the percentage of genes in the module that shared protein-protein interactions in CORNET and the percentage of CORNET protein-protein interactions compared with all possible gene-gene interactions within the module. Finally, the modules were analyzed for the enrichment in 831 known oxidative stress response marker genes (Supplemental Tables II to IV from Gadjev et al. [2006]).
cis-Regulatory Motif Detection
We searched Arabidopsis promoters for known cis-regulatory motifs from the PLACE (Higo et al., 1999) and AGRIS databases (Palaniswamy et al., 2006), motifs identified using the network-level conservation principle present in ATCOECIS (Vandepoele et al., 2009), and additional binding sites reported in the literature (Supplemental Table 3) through dna-pattern and RSA-tools (van Helden et al., 2000). Promoters are defined as 1 kb upstream of translation start sites or in the intergenic region if the adjacent upstream gene is located within a smaller distance, of all Arabidopsis genes, which were obtained from PLAZA2.5 and correspond to TAIR10 (Van Bel et al., 2012). In total, 33,104 genes are associated with a cis-regulatory motif. cis-regulatory motif enrichment was calculated for each module using hypergeometric enrichment and Bonferroni multiple hypothesis testing correction with a confidence level of 95%. To further reduce the inclusion of false positives, we considered only motifs that were present in at least 50% of the module gene promoters and that were more than 1.5 times enriched in the module compared with the genome.
Generation of Transgenic Arabidopsis Plants
Several transgenic Arabidopsis lines were previously described: NAC13_OE (overexpression) and NAC053_OE (De Clercq et al., 2013), ERF6_KO (knockout) (Dubois et al., 2013), RAP2.6L_OE (Krishnaswamy et al., 2011), RAP2.6L_KO (Che et al., 2006), and WRKY6_OE lines (Robatzek and Somssich, 2002). For ERF6_OE and NAC032_OE, the full-length open reading frames of ERF6 and NAC032 were amplified by PCR from first-strand cDNA of Arabidopsis ecotype Columbia-0 (wild type) with gene-specific primers extended with the attB sites for Gateway cloning (Invitrogen; Supplemental Table 6). PCR reactions were run with high-fidelity Phusion DNA polymerase (Finnzymes), and fragments were cloned into the Gateway entry vectors according to the manufacturer’s instructions. Next, a constitutive promoter-driven expression construct was generated for NAC032 and ERF6 in the binary destination vector pK7WG2D (Karimi et al., 2007). Constructs were transformed into Arabidopsis by Agrobacterium tumefaciens-mediated floral dipping (Clough and Bent, 1998). Homozygous lines with a single T-DNA locus and high transgene expression were selected via segregation analysis, followed by RNA gel blot and quantitative RT-PCR analysis. For NAC13_ami, NAC13-specific sequences were identified with the Web MicroRNA Designer (www.weigelworld.org). The miR precursors were constructed according to Schwab et al. (2006) and cloned into pK7WG2D as described by De Clercq et al. (2013). For the loss-of-function plants, we obtained T-DNA insertion mutants from the ABRC at Ohio State University (Supplemental Table 7). Homozygous plants were selected by genomic PCR with gene-specific and T-DNA-specific primers. Residual expression levels of the genes were determined by quantitative RT-PCR.
Plant Growth Conditions and Stress Treatments
Arabidopsis ecotype Columbia-0 plants were grown until stage 1.04 (Boyes et al., 2001) on half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie) at 21°C with a 16-h/8-h light/dark photoperiod and 100 μmol m−2 s−1 light intensity. For the nCounter experiments, a combination of wild-type and three transgenic lines were grown for 2 weeks on a nylon mesh (20-µm pore size; PROSEP) on half-strength MS agar medium and subsequently transferred to half-strength MS supplemented with 150 mM NaCl. Twelve hours after transfer, three biological samples of 5 to 10 seedlings were harvested. Total RNA was prepared with TRIzol reagent (Invitrogen). The nCounter analysis of 100 ng RNA from each sample was performed according to the gene expression assay protocol of the manufacturer NanoString Technologies (Geiss et al., 2008). For the mannitol stress assays, the wild type, two independent transgenic NAC32_OE plants (NAC032_OE1, which was also used for nCounter analysis, and NAC032_OE2 as a second independent transgenic line for the mannitol stress assays), and NAC032_KO were grown together on a nylon mesh on half-strength MS supplemented with 75 mM mannitol for 17 d. Rosette area was measured using ImageJ software (http://imagej.nih.gov/ij/). Green biomass was separately harvested from the root tissue to determine the shoot biomass.
nCounter Data Analysis
We identified the differential expression analysis between wild-type and perturbed TF plants using both the DESeq and edgeR packages for count data in Bioconductor R (Anders and Huber, 2010; Robinson et al., 2010; McCarthy et al., 2012). DESeq and edgeR are compatible statistical methods that are based on a negative binomial distribution model of read counts (Nookaew et al., 2012; Robles et al., 2012). The raw nCounter data needed to be rescaled by dividing (per experiment) by two factors: (1) the sum of the positive controls per experiment divided by the median of the sums of the positive controls over all experiments, to correct for technical errors, and (2) the geometric mean of the three most stable household genes, selected by geNorm (Vandesompele et al., 2002), per experiment divided by the total geometric mean of these three household genes over all experiments, to correct for differences in mRNA content of the samples. The raw nCounter data were fed into the R packages and the above rescaling factors were written in the sizeFactors slot of the CountDataSet object for the package DESeq and in the norm.factors slot of the samples slot of the DGElist object for the package edgeR. For DESeq, we applied a local fit for the estimation of the dispersions using the method “per-condition.” In a nonpaired design, we compared three replicate perturbed TF plants against three replicate wild-type plants that were together with the former on the same experimental plates. For edgeR, we estimated the dispersions per group of samples that were together on the same plates. Plotting the sample relations by multidimensional scaling indicated a batch effect for the salt-treated NAC13_ami, NAC032_KO, and NAC053_KO samples and the Wassilewskija background samples (WS-RAP2.6L_OE) (data not shown). Hence, these samples were analyzed in a paired design, using the GLM functionality and the Cox-Reid profile-adjusted likelihood for the estimation of the dispersions (McCarthy et al., 2012). The other samples were analyzed in a nonpaired design, using the classical quantile-adjusted condition maximal likelihood for the estimation of the dispersions (Robinson et al., 2010). Raw P values were adjusted for multiple comparisons by the Benjamini-Hochberg procedure, which controls the false discovery rate. Genes with an adjusted P value < 0.1 were considered to be differentially expressed. Upon evaluation of the reverse-engineering predictions by nCounter, EdgeR detected 56% of the true positives, 24% by DESequation, and 20% by both methods. We found that true positive interactions were more likely to be detected by both DESeq and edgeR (two-tailed Fisher exact test, P value = 0.0022).
Topological Overlap Coefficient
For the nCounter experimental network, a topological overlap matrix was created for promoters of target genes and interacting TFs, respectively, by calculating the (directed) topological overlap coefficient for every node that had more than one link (Vermeirssen et al., 2007). The topological overlap coefficient or mutual clustering coefficient is a relative measure of the number of neighboring nodes that are shared between two nodes. The geometric formula was used (Goldberg and Roth, 2003).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the accession numbers provided in Supplemental Table 8.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Venn Diagram Illustrating the Percentage of the Top 200,014 Predictions of the Rank Aggregation Ensemble Predicted by Each of the Four Network Inference Solutions and Their Overlap.
Supplemental Figure 2. Histograms of the Pearson Correlation Coefficient on the Expression Profile Ratios within Modules of the Abiotic Stress GRN and within Random Modules.
Supplemental Figure 3. Literature-Based Evidence for the Biological Relevance of the Abiotic Stress Module GRN: Module 402 Was Implicated in Jasmonic Acid Biosynthesis.
Supplemental Figure 4. Number of Differentially Expressed Genes in the Different TF Perturbation Experiments Analyzed by nCounter and Considered Statistically Significant by edgeR, DESeq, or Both.
Supplemental Figure 5. Visualization of all the Predictions in the Abiotic Stress GRN for the 102 Selected Genes in the nCounter Experiments.
Supplemental Figure 6. Osmotic Stress Resistance Phenotype of NAC032 Overexpression Plants.
Supplemental Figure 7. Module 191 Was Involved in Detoxification Processes of the Oxidative Stress Response.
Supplemental Table 1. Number of Interactions by Source and Type of Experiment of the Reference Sets of Known Protein-DNA and Regulatory Interactions.
Supplemental Table 2. Properties of the 5% Most Connected TFs in the Abiotic Stress Gene Regulatory Network and the 5% TFs That Regulate Most Modules, Which Are Defined as “Hubs” in the Network.
Supplemental Table 3. Known cis-Regulatory Motifs from the PLACE, AGRIS, and ATCOECIS Databases and Some Additional cis-Regulatory Motifs from Literature, Which Are Listed Below, Were Searched in 1-kb Arabidopsis Promoters.
Supplemental Table 4. Known cis-Regulatory Motifs for the Tested TFs Present in 1-kb Promoters of nCounter Target Genes.
Supplemental Table 5. ChIP and Perturbed TF Expression Profiling Experiments from Literature Used in the Reference Sets.
Supplemental Table 6. Sequence of Primers Used to Clone the Coding Sequence of NAC032 and ERF6.
Supplemental Table 7. SALK T-DNA Insertion Lines Used in This Study as Knockout Lines.
Supplemental Table 8. Accession Numbers of the Genes Mentioned in This Study.
Supplemental Data Set 1. Microarray Compendium from Which Expression Profile Ratios (Perturbation over Control) in 199 Different Abiotic Stress Conditions Were Calculated.
Supplemental Data Set 2. The Abiotic Stress Gene Regulatory Network: Predictions, Rank, Symbolic Names, Individual Methods, and Module Number.
Supplemental Data Set 3. Overview of the 572 Modules of Coregulated Genes, Their Functional Coherence, and the Biological Relevance of Their Predicted TFs.
Supplemental Data Set 4. Overview of the 289 nCounter Experimental Interactions.
Supplementary Material
Acknowledgments
We thank the following people for fruitful discussions: Jens Hollunder on integration of reverse-engineering solutions, Bram Slabbinck on performance assessment of network inference and ModuleViewer, Frederik Coppens on the NanoString nCounter experimental setup, and Eric Bonnet on the combination of network inference with experimental validation by nCounter. WRKY6_OE lines were kindly provided by Wei-Hua Wu; NAC13_ami, NAC32_KO, NAC53_KO, and NAC overexpression lines by Frank Hoeberichts and Sandy Vanderauwera; and the ERF6_OE line by Marieke Dubois. Cezary Waszczak helped sort out transgenic lines. Debbie Rombout and Brigitte Van De Cotte delivered excellent technical assistance. This work was supported by grants from Ghent University Multidisciplinary Research Partnership (“Bioinformatics: from nucleotides to networks” [Project 01MR0310W] and “Ghent BioEconomy” [Project 01MRB510W]) and VIB (Technology Watch Fund for nCounter). I.D.C. is indebted to the Research Foundation-Flanders for a postdoctoral fellowship.
AUTHOR CONTRIBUTIONS
V.V. designed the study, analyzed the computational and experimental data, interpreted the results, and wrote the article with the help of I.D.C., F.V.B., and Y.V.D.P. I.D.C. compiled the abiotic stress microarray compendium and designed the nCounter experiments with the help of V.V. I.D.C. performed the wet-lab experiments. T.V.P. developed ModuleViewer to illustrate the coregulated gene modules and made the Web application with figures, interactive networks, and search options available.
Glossary
- TF
transcription factor
- GRN
gene regulatory network
- Y1H
yeast one-hybrid
- ChIP
chromatin immunoprecipitation
- AUPR
area under the precision-recall
- GO
Gene Ontology
- PCC
Pearson Correlation Coefficient
- ABA
abscisic acid
- ABRE
ABA-responsive element
- bHLH
basic helix-loop-helix
- bZIP
basic leucine zipper
- DRE
dehydration-responsive element
- ER
endoplasmic reticulum
- MRR
mitochondrial retrograde regulation
- MDS
mitochondrial dysfunction stimulon
- PABA
para-aminobenzoic acid
- UGT
UDP-glucose transferase
- GST
glutathione S-transferase
- ROS
reactive oxygen species
- MS
Murashige and Skoog
Footnotes
Online version contains Web-only data.
References
- Akhtar M., Jaiswal A., Taj G., Jaiswal J.P., Qureshi M.I., Singh N.K. (2012). DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91: 385–395. [DOI] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399. [DOI] [PubMed] [Google Scholar]
- Asahina M., et al. (2011). Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 16128–16132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G., et al. (2009). Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros M.D., Czarnecka E., Gurley W.B. (1992). Mutational analysis of a plant heat shock element. Plant Mol. Biol. 19: 665–675. [DOI] [PubMed] [Google Scholar]
- Bassel G.W., Gaudinier A., Brady S.M., Hennig L., Rhee S.Y., De Smet I. (2012). Systems analysis of plant functional, transcriptional, physical interaction, and metabolic networks. Plant Cell 24: 3859–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel G.W., Glaab E., Marquez J., Holdsworth M.J., Bacardit J. (2011). Functional network construction in Arabidopsis using rule-based machine learning on large-scale data sets. Plant Cell 23: 3101–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A., Mittler R., Suzuki N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65: 1229–1240. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhosale R., Jewell J.B., Hollunder J., Koo A.J., Vuylsteke M., Michoel T., Hilson P., Goossens A., Howe G.A., Browse J., Maere S. (2013). Predicting gene function from uncontrolled expression variation among individual wild-type Arabidopsis plants. Plant Cell 25: 2865–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F., Salinas P., Cecchini N.M., Jordana X., Van Hummelen P., Alvarez M.E., Holuigue L. (2009). Early genomic responses to salicylic acid in Arabidopsis. Plant Mol. Biol. 70: 79–102. [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., et al. (2011). A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 7: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butow R.A., Avadhani N.G. (2004). Mitochondrial signaling: the retrograde response. Mol. Cell 14: 1–15. [DOI] [PubMed] [Google Scholar]
- Casneuf T., Van de Peer Y., Huber W. (2007). In situ analysis of cross-hybridisation on microarrays and the inference of expression correlation. BMC Bioinformatics 8: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G., et al. (2013). WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 25: 2944–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai J., Liu J., Zhou J., Xing D. (2014). Mitogen-activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. J. Exp. Bot. 65: 6513–6528. [DOI] [PubMed] [Google Scholar]
- Chávez Montes R.A., Coello G., González-Aguilera K.L., Marsch-Martínez N., de Folter S., Alvarez-Buylla E.R. (2014). ARACNe-based inference, using curated microarray data, of Arabidopsis thaliana root transcriptional regulatory networks. BMC Plant Biol. 14: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P., Lall S., Nettleton D., Howell S.H. (2006). Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 141: 620–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14: 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.F., Li L.Q., Xu Q., Kong Y.H., Wang H., Wu W.H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cramer G.R., Urano K., Delrot S., Pezzotti M., Shinozaki K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J.V., Delaney S.P. (2008). Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant. 132: 417–425. [DOI] [PubMed] [Google Scholar]
- De Bodt S., Carvajal D., Hollunder J., Van den Cruyce J., Movahedi S., Inzé D. (2010). CORNET: a user-friendly tool for data mining and integration. Plant Physiol. 152: 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S., Hollunder J., Nelissen H., Meulemeester N., Inzé D. (2012). CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol. 195: 707–720. [DOI] [PubMed] [Google Scholar]
- De Clercq I., et al. (2013). The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R., Marchal K. (2010). Advantages and limitations of current network inference methods. Nat. Rev. Microbiol. 8: 717–729. [DOI] [PubMed] [Google Scholar]
- Dietrich K., Weltmeier F., Ehlert A., Weiste C., Stahl M., Harter K., Dröge-Laser W. (2011). Heterodimers of the Arabidopsis transcription factors bZIP1 and bZIP53 reprogram amino acid metabolism during low energy stress. Plant Cell 23: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R.G., Cashmore A.R. (1990). Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 9: 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.J., Liu J.Y. (2010). The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M., Skirycz A., Claeys H., Maleux K., Dhondt S., De Bodt S., Vanden Bossche R., De Milde L., Yoshizumi T., Matsui M., Inzé D. (2013). Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162: 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M., Hsieh T.F., Kim S.Y., Thomas T.L. (2002). Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50: 237–248. [DOI] [PubMed] [Google Scholar]
- Eudes A., Bozzo G.G., Waller J.C., Naponelli V., Lim E.K., Bowles D.J., Gregory J.F. III, Hanson A.D. (2008). Metabolism of the folate precursor p-aminobenzoate in plants: glucose ester formation and vacuolar storage. J. Biol. Chem. 283: 15451–15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J.J., Hayete B., Thaden J.T., Mogno I., Wierzbowski J., Cottarel G., Kasif S., Collins J.J., Gardner T.S. (2007). Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 5: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode B., Siemsen T., Thurow C., Weigel R., Gatz C. (2008). The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 20: 3122–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel S., Usadel B., von Wirén N., Sreenivasulu N. (2012). Reverse engineering: a key component of systems biology to unravel global abiotic stress cross-talk. Front. Plant Sci. 3: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39: 863–876. [DOI] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I., Vanderauwera S., Gechev T.S., Laloi C., Minkov I.N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa S.N., Botto J.F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19: 460–470. [DOI] [PubMed] [Google Scholar]
- Gaudinier A., et al. (2011). Enhanced Y1H assays for Arabidopsis. Nat. Methods 8: 1053–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss G.K., et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26: 317–325. [DOI] [PubMed] [Google Scholar]
- Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. (2007). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50: 886–901. [DOI] [PubMed] [Google Scholar]
- Gitter A., Siegfried Z., Klutstein M., Fornes O., Oliva B., Simon I., Bar-Joseph Z. (2009). Backup in gene regulatory networks explains differences between binding and knockout results. Mol. Syst. Biol. 5: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg D.S., Roth F.P. (2003). Assessing experimentally derived interactions in a small world. Proc. Natl. Acad. Sci. USA 100: 4372–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Smith A.M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16: 169–175. [DOI] [PubMed] [Google Scholar]
- Haga N., Kobayashi K., Suzuki T., Maeo K., Kubo M., Ohtani M., Mitsuda N., Demura T., Nakamura K., Jürgens G., Ito M. (2011). Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157: 706–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J., Hanssen M., Wiese A., Hendriks M.M., Smeekens S. (2008). The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 53: 935–949. [DOI] [PubMed] [Google Scholar]
- Hao D., Ohme-Takagi M., Sarai A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J. Biol. Chem. 273: 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hase T., Ghosh S., Yamanaka R., Kitano H. (2013). Harnessing diversity towards the reconstructing of large scale gene regulatory networks. PLOS Comput. Biol. 9: e1003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.C., Maier E.J., Kramer M.H., Wang P.I., Brown H., Brent M.R. (2013). Mapping functional transcription factor networks from gene expression data. Genome Res. 23: 1319–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx K.S., Vandepoele K. (2012). Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 159: 884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman R., et al. (2013). A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75: 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt M.N., Guerinot M.L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823: 1521–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K., Jang C., Bailey-Serres J., Mittler R., Shelton C., Harper J.F., Zhu J.K., Cushman J.C., Gollery M., Girke T. (2008). Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 147: 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Fedoroff N.V., Koizumi N. (2008). Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20: 3107–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A., De Smet R., Marchal K., Van de Peer Y., Michoel T. (2009). Module networks revisited: computational assessment and prioritization of model predictions. Bioinformatics 25: 490–496. [DOI] [PubMed] [Google Scholar]
- Joshi A., Van de Peer Y., Michoel T. (2008). Analysis of a Gibbs sampler method for model-based clustering of gene expression data. Bioinformatics 24: 176–183. [DOI] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasajima I., Ide Y., Yokota Hirai M., Fujiwara T. (2010). WRKY6 is involved in the response to boron deficiency in Arabidopsis thaliana. Physiol. Plant. 139: 80–92. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89. [DOI] [PubMed] [Google Scholar]
- Kerchev P.I., De Clercq I., Denecker J., Mühlenbock P., Kumpf R., Nguyen L., Audenaert D., Dejonghe W., Van Breusegem F. (2014). Mitochondrial perturbation negatively affects auxin signaling. Mol. Plant 7: 1138–1150. [DOI] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Park M.J., Seo P.J., Song J.S., Kim H.J., Park C.M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448: 353–363. [DOI] [PubMed] [Google Scholar]
- Kim S.G., Lee S., Seo P.J., Kim S.K., Kim J.K., Park C.M. (2010). Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95: 56–65. [DOI] [PubMed] [Google Scholar]
- Kourmpetis Y.A., van Dijk A.D., van Ham R.C., ter Braak C.J. (2011). Genome-wide computational function prediction of Arabidopsis proteins by integration of multiple data sources. Plant Physiol. 155: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S., Verma S., Rahman M.H., Kav N.N. (2011). Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 75: 107–127. [DOI] [PubMed] [Google Scholar]
- Laloi C., Mestres-Ortega D., Marco Y., Meyer Y., Reichheld J.P. (2004). The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 134: 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K., Prasad K.V., Savchenko T., Celesnik H., Dehesh K., Levy M., Mitchell-Olds T., Reddy A.S. (2012). The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 53: 2008–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E.M., Rhee S.Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Seo P.J., Lee H.J., Park C.M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70: 831–844. [DOI] [PubMed] [Google Scholar]
- Less H., Angelovici R., Tzin V., Galili G. (2011). Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23: 1264–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E.K., Ashford D.A., Hou B., Jackson R.G., Bowles D.J. (2004). Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87: 623–631. [DOI] [PubMed] [Google Scholar]
- Lim E.-K., Doucet C.J., Hou B., Jackson R.G., Abrams S.R., Bowles D.J. (2005). Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron Asymmetry 16: 143–147. [Google Scholar]
- Liu P., Sun F., Gao R., Dong H. (2012). RAP2.6L overexpression delays waterlogging induced premature senescence by increasing stomatal closure more than antioxidant enzyme activity. Plant Mol. Biol. 79: 609–622. [DOI] [PubMed] [Google Scholar]
- Long T.A., Tsukagoshi H., Busch W., Lahner B., Salt D.E., Benfey P.N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Paul A.L., McCarty D.R., Ferl R.J. (1996). Transcription factor veracity: is GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 8: 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumba S., Toh S., Handfield L.F., Swan M., Liu R., Youn J.Y., Cutler S.R., Subramaniam R., Provart N., Moses A., Desveaux D., McCourt P. (2014). A mesoscale abscisic acid hormone interactome reveals a dynamic signaling landscape in Arabidopsis. Dev. Cell 29: 360–372. [DOI] [PubMed] [Google Scholar]
- Ma C., Xin M., Feldmann K.A., Wang X. (2014). Machine learning-based differential network analysis: a study of stress-responsive transcriptomes in Arabidopsis. Plant Cell 26: 520–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Hanssen M., Lundgren K., Hernández L., Delatte T., Ehlert A., Liu C.M., Schluepmann H., Dröge-Laser W., Moritz T., Smeekens S., Hanson J. (2011). The sucrose-regulated Arabidopsis transcription factor bZIP11 reprograms metabolism and regulates trehalose metabolism. New Phytol. 191: 733–745. [DOI] [PubMed] [Google Scholar]
- Ma S., Gong Q., Bohnert H.J. (2007). An Arabidopsis gene network based on the graphical Gaussian model. Genome Res. 17: 1614–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449. [DOI] [PubMed] [Google Scholar]
- Marbach D., Costello J.C., Küffner R., Vega N.M., Prill R.J., Camacho D.M., Allison K.R., Kellis M., Collins J.J., Stolovitzky G.; DREAM5 Consortium (2012). Wisdom of crowds for robust gene network inference. Nat. Methods 9: 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolais F., Boyd J., Paredes Y., Schinas A.M., Garcia M., Elzein S., Varin L. (2007). Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 225: 1233–1244. [DOI] [PubMed] [Google Scholar]
- Maruyama K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150: 1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40: 4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Xu J., He Y., Yang K.Y., Mordorski B., Liu Y., Zhang S. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25: 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michoel T., De Smet R., Joshi A., Van de Peer Y., Marchal K. (2009). Comparative analysis of module-based versus direct methods for reverse-engineering transcriptional regulatory networks. BMC Syst. Biol. 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M.D., Thomashow M.F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60: 328–339. [DOI] [PubMed] [Google Scholar]
- Misra A., Sriram G. (2013). Network component analysis provides quantitative insights on an Arabidopsis transcription factor-gene regulatory network. BMC Syst. Biol. 7: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat C.S., Ingle R.A., Wathugala D.L., Saunders N.J., Knight H., Knight M.R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S., Hilbert B., Dueckershoff K., Roitsch T., Krischke M., Mueller M.J., Berger S. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20: 768–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naika M., Shameer K., Mathew O.K., Gowda R., Sowdhamini R. (2013). STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 54: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 97–103. [DOI] [PubMed] [Google Scholar]
- Nakata M., Mitsuda N., Herde M., Koo A.J., Moreno J.E., Suzuki K., Howe G.A., Ohme-Takagi M. (2013). A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., De Clercq I., Van Aken O., Law S.R., Ivanova A., Willems P., Giraud E., Van Breusegem F., Whelan J. (2014). Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 7: 1075–1093. [DOI] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Yoshida E., Maruta T., Yoshimura K., Shigeoka S. (2006). Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 48: 535–547. [DOI] [PubMed] [Google Scholar]
- Nookaew I., Papini M., Pornputtapong N., Scalcinati G., Fagerberg L., Uhlen M., Nielsen J. (2012). A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res. 40: 10084–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Barrette T.R., DellaPenna D. (1995). Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Kinoshita K., Nakai K., Shibaoka M., Hayashi S., Saeki M., Shibata D., Saito K., Ohta H. (2007). ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35: D863–D869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Lo Leggio L., Skriver K. (2005). DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169: 785–797. [Google Scholar]
- Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. (2006). AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 140: 818–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V., Vermeirssen V., De Clercq I., Van Breusegem F., Minkov I., Vandepoele K., Gechev T.S. (2012). Identification of cis-regulatory elements specific for different types of reactive oxygen species in Arabidopsis thaliana. Gene 499: 52–60. [DOI] [PubMed] [Google Scholar]
- Qi J., Michoel T. (2012). Context-specific transcriptional regulatory network inference from global gene expression maps using double two-way t-tests. Bioinformatics 28: 2325–2332. [DOI] [PubMed] [Google Scholar]
- Qi J., Michoel T., Butler G. (2012). An integrative approach to infer regulation programs in a transcription regulatory module network. J. Biomed. Biotechnol. 2012: 245968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Shinozaki K., Yamaguchi-Shinozaki K. (2011). Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 52: 1569–1582. [DOI] [PubMed] [Google Scholar]
- Qin X.F., Holuigue L., Horvath D.M., Chua N.H. (1994). Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E., Cabrito T.R., Baster P., Batista R.A., Teixeira M.C., Friml J., Sá-Correia I., Duque P. (2013). A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25: 901–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Tang G. (2012). Identification of sucrose-responsive microRNAs reveals sucrose-regulated copper accumulations in an SPL7-dependent and independent manner in Arabidopsis thaliana. Plant Sci. 187: 59–68. [DOI] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles J.A., Qureshi S.E., Stephen S.J., Wilson S.R., Burden C.J., Taylor J.M. (2012). Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing. BMC Genomics 13: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Elia G., Santoro M.G. (1997). Inhibition of nuclear factor kappa B by prostaglandin A1: an effect associated with heat shock transcription factor activation. Proc. Natl. Acad. Sci. USA 94: 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santino A., Taurino M., De Domenico S., Bonsegna S., Poltronieri P., Pastor V., Flors V. (2013). Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 32: 1085–1098. [DOI] [PubMed] [Google Scholar]
- Scarpeci T.E., Zanor M.I., Carrillo N., Mueller-Roeber B., Valle E.M. (2008). Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol. Biol. 66: 361–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikhali J., de Dios Barajas-Lopéz J., Ötvös K., Kremnev D., Garcia A.S., Srivastava V., Wingsle G., Bako L., Strand Å. (2012). The CRYPTOCHROME1-dependent response to excess light is mediated through the transcriptional activators ZINC FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM LIKE1 and ZML2 in Arabidopsis. Plant Cell 24: 3009–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Langlois-Meurinne M., Didierlaurent L., Chaouch S., Bellvert F., Massoud K., Garmier M., Thareau V., Comte G., Noctor G., Saindrenan P. (2014). The secondary metabolism glycosyltransferases UGT73B3 and UGT73B5 are components of redox status in resistance of Arabidopsis to Pseudomonas syringae pv. tomato. Plant Cell Environ. 37: 1114–1129. [DOI] [PubMed] [Google Scholar]
- Sivitz A.B., Hermand V., Curie C., Vert G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbe M., Qu N., Galis I., Baldwin I.T. (2008). Induced plant defenses in the natural environment: Nicotiana attenuata WRKY3 and WRKY6 coordinate responses to herbivory. Plant Cell 20: 1984–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soitamo A.J., Piippo M., Allahverdiyeva Y., Battchikova N., Aro E.M. (2008). Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol. 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H.U., Mueller S., Zoeller M., Mueller M.J., Berger S. (2013). TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 64: 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50: 660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Favory J.J., Gruber H., Bartelniewoehner L., Bartels S., Binkert M., Funk M., Weisshaar B., Ulm R. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33: 88–103. [DOI] [PubMed] [Google Scholar]
- Street N.R., Jansson S., Hvidsten T.R. (2011). A systems biology model of the regulatory network in Populus leaves reveals interacting regulators and conserved regulation. BMC Plant Biol. 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Dreos R., Aparicio F., Maule A.J. (2009). The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21: 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Jiang H., Xu Y., Li H., Wu X., Xie Q., Li C. (2007). The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 48: 1148–1158. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Rivero R.M., Shulaev V., Blumwald E., Mittler R. (2014). Abiotic and biotic stress combinations. New Phytol. 203: 32–43. [DOI] [PubMed] [Google Scholar]
- Tatematsu K., Ward S., Leyser O., Kamiya Y., Nambara E. (2005). Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol. 138: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma I., Loeffler C., Sinha A.K., Gupta M., Krischke M., Steffan B., Roitsch T., Mueller M.J. (2003). Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 34: 363–375. [DOI] [PubMed] [Google Scholar]
- Tognetti V.B., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtera L., Brosché M. (2011). More than the sum of its parts—how to achieve a specific transcriptional response to abiotic stress. Plant Sci. 180: 421–430. [DOI] [PubMed] [Google Scholar]
- Valdés A.E., Overnäs E., Johansson H., Rada-Iglesias A., Engström P. (2012). The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 80: 405–418. [DOI] [PubMed] [Google Scholar]
- Van Aken O., Zhang B., Law S., Narsai R., Whelan J. (2013). AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162: 254–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., Proost S., Wischnitzki E., Movahedi S., Scheerlinck C., Van de Peer Y., Vandepoele K. (2012). Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 158: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Quimbaya M., Casneuf T., De Veylder L., Van de Peer Y. (2009). Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 150: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S., Vandenbroucke K., Inzé A., van de Cotte B., Mühlenbock P., De Rycke R., Naouar N., Van Gaever T., Van Montagu M.C., Van Breusegem F. (2012). AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 20113–20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., André B., Collado-Vides J. (2000). A web site for the computational analysis of yeast regulatory sequences. Yeast 16: 177–187. [DOI] [PubMed] [Google Scholar]
- Vermeirssen V., Barrasa M.I., Hidalgo C.A., Babon J.A., Sequerra R., Doucette-Stamm L., Barabási A.L., Walhout A.J. (2007). Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 17: 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V., Joshi A., Michoel T., Bonnet E., Casneuf T., Van de Peer Y. (2009). Transcription regulatory networks in Caenorhabditis elegans inferred through reverse-engineering of gene expression profiles constitute biological hypotheses for metazoan development. Mol. Biosyst. 5: 1817–1830. [DOI] [PubMed] [Google Scholar]
- Walhout A.J. (2011). What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 12: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Zhao X., Miao Y., Song C.P. (2013). The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 161: 1392–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D., et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38: W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24: 482–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Lee E., Lucas J.R., Morohashi K., Li D., Murray J.A., Sack F.D., Grotewold E. (2010). Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. Plant Cell 22: 2306–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V., Mallappa C., Gangappa S.N., Bhatia S., Chattopadhyay S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Yoshida H., Kokame K., Kaufman R.J., Mori K. (2004). Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 136: 343–350. [DOI] [PubMed] [Google Scholar]
- Yamasaki K., Kigawa T., Watanabe S., Inoue M., Yamasaki T., Seki M., Shinozaki K., Yokoyama S. (2012). Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. J. Biol. Chem. 287: 7683–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277: 45049–45058. [DOI] [PubMed] [Google Scholar]
- Yang Z.T., Lu S.J., Wang M.J., Bi D.L., Sun L., Zhou S.F., Song Z.T., Liu J.X. (2014). A plasma membrane-tethered transcription factor, NAC062/ANAC062/NTL6, mediates the unfolded protein response in Arabidopsis. Plant J. 79: 1033–1043. [DOI] [PubMed] [Google Scholar]
- Yilmaz A., Mejia-Guerra M.K., Kurz K., Liang X., Welch L., Grotewold E. (2011). AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 39: D1118–D1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., Saito K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65: 634–646. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., Wang D., Ling H.Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18: 385–397. [DOI] [PubMed] [Google Scholar]
- Zander M., Thurow C., Gatz C. (2014). TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 165: 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jin J., Tang L., Zhao Y., Gu X., Gao G., Luo J. (2011). PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 39: D1114–D1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C., Sun K., Mackaluso J.D., Seddon A.E., Jin R., Thomashow M.F., Shiu S.H. (2011). Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 14992–14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.