Induction of miR172c in response to rhizobial inoculation activates ENOD40 expression and nodule formation by repressing an AP2/ERF transcription factor, called NNC1, that acts to inhibit ENOD40 expression.
Abstract
MicroRNAs are noncoding RNAs that act as master regulators to modulate various biological processes by posttranscriptionally repressing their target genes. Repression of their target mRNA(s) can modulate signaling cascades and subsequent cellular events. Recently, a role for miR172 in soybean (Glycine max) nodulation has been described; however, the molecular mechanism through which miR172 acts to regulate nodulation has yet to be explored. Here, we demonstrate that soybean miR172c modulates both rhizobium infection and nodule organogenesis. miR172c was induced in soybean roots inoculated with either compatible Bradyrhizobium japonicum or lipooligosaccharide Nod factor and was highly upregulated during nodule development. Reduced activity and overexpression of miR172c caused dramatic changes in nodule initiation and nodule number. We show that soybean miR172c regulates nodule formation by repressing its target gene, Nodule Number Control1, which encodes a protein that directly targets the promoter of the early nodulin gene, ENOD40. Interestingly, transcriptional levels of miR172c were regulated by both Nod Factor Receptor1α/5α-mediated activation and by autoregulation of nodulation-mediated inhibition. Thus, we established a direct link between miR172c and the Nod factor signaling pathway in addition to adding a new layer to the precise nodulation regulation mechanism of soybean.
INTRODUCTION
Through a symbiotic relationship with nitrogen-fixing rhizobial bacteria, most legume plants can use atmospheric dinitrogen gas to help satisfy their nitrogen needs. The process of symbiotic nitrogen fixation takes place in specialized lateral organs called nodules (Ferguson et al., 2010). Nodulation is a complex developmental process involving a direct interaction between rhizobium and legume signals (Desbrosses and Stougaard, 2011; Oldroyd, 2013; Ferguson and Mathesius, 2014). Understanding the mechanisms underlying nodulation and nitrogen fixation could aid in improving nitrogen use efficiency of crops, with the aim of reducing nitrogen fertilizer inputs and improving agricultural and environmental sustainability (Salvagiotti et al., 2008; Peoples et al., 2009; Jensen et al., 2012; Gresshoff et al., 2014).
The process of nodulation is initiated by legume roots secreting flavonoid molecules into the surrounding rhizosphere, which attracts compatible rhizobium strains and stimulates them to synthesize lipochitin oligosaccharide signals, called Nod factors (NFs; e.g., in soybean [Glycine max]) (Sanjuan et al., 1992). NFs are perceived by root LysM Nod Factor Receptors (NFRs), which activate signaling cascades that promote root hair deformation and microsymbiont infection as well as cortical and pericycle cell division (nodule primordium formation) (Broghammer et al., 2012; Moling et al., 2014). The NFRs of soybean are encoded by NFR1α and NFR5α (Indrasumunar et al., 2010, 2011).
Downstream components of NF perception have been thoroughly reviewed (Ferguson et al., 2010; Desbrosses and Stougaard, 2011; Oldroyd, 2013). Among them is ENOD40, which is upregulated at the onset of nodulation (Yang et al., 1993; Crespi et al., 1994; Mathesius et al., 2000; Compaan et al., 2001) and is expressed in pericycle cells of root vascular bundles, dividing cortical cells, the nodule primordium, and developing nodules (reviewed in Ferguson and Mathesius, 2014). In soybean roots, ENOD40 expression is upregulated following either Bradyrhizobium japonicum inoculation or NF treatment (Kouchi and Hata, 1993; Yang et al., 1993; Minami et al., 1996; Hayashi et al., 2012). Alterations in ENOD40 expression can significantly influence legume nodule numbers, suggesting that it plays a pivotal role in nodule organogenesis (Charon et al., 1999; Kumagai et al., 2006; Wan et al., 2007). ENOD40 may function in nodulation as a cell-cell signaling molecule; it lacks an open reading frame but does encode two small peptides (Sousa et al., 2001; Röhrig et al., 2002). Notably, ENOD40 peptides can bind to, and enhance the stability of, sucrose synthase (Hardin et al., 2003; Röhrig et al., 2004). Thus, ENOD40 may promote nodule organogenesis by increasing the carbon sink strength of the dividing cells. Despite ENOD40 playing a key role in nodulation, and for many years being used as a marker for nodule primordium initiation, it remains unknown how ENOD40 transcription is modulated during nodule initiation and development.
The formation and maintenance of legume nodules is energy-consuming to the host plant. Thus, to avoid excessive nodulation, the process is tightly regulated by a negative feedback system, termed autoregulation of nodulation (AON; reviewed in Caetano-Anollés and Gresshoff, 1991; Reid et al., 2011a). The regulatory process involves a critical leucine-rich repeat Ser/Thr receptor-like kinase (Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005; Ferguson et al., 2014) with strong homology to Arabidopsis thaliana CLAVATA1 (Clark et al., 1997). In soybean, this leucine-rich repeat receptor is encoded by NODULE AUTOREGULATION RECEPTOR KINASE (NARK; Searle et al., 2003). Mutations in NARK (e.g., nts382 and nts1007) cause abundant nodulation (termed hypernodulation or supernodulation; Carroll et al., 1985). Reciprocal grafting demonstrated that NARK functions in the shoot in the AON pathway to regulate nodulation in the root (Delves et al., 1986; Caetano-Anollés and Gresshoff, 1991; Nishimura et al., 2002).
Recent work has indicated that Gm-NARK (or its homolog in Lotus japonicus, Lj-HAR1; Okamoto et al., 2013) acts in the leaf vascular tissue to perceive root-derived peptides, called RIC1 and RIC2 in soybean (for rhizobia-induced CLV3/ESR-related peptides) (Reid et al., 2011b), that originate during nodule primordium formation. This perception results in the production of a shoot-derived inhibitor (SDI), which travels to the roots to inhibit further nodule development (Delves et al., 1986; Lin et al., 2010; Sasaki et al., 2014). Excitingly, recent evidence has revealed that cytokinins, whose production is activated by Lj-HAR1 perception of AON CLE peptides in the shoot, may function as an SDI to systemically suppress nodulation in L. japonicus (Sasaki et al., 2014). Despite this great progress, many questions remain unanswered, including the identity of downstream signaling components of AON and how legumes integrate the NF and AON signaling pathways to optimize nodule numbers.
MicroRNAs (miRNAs) are short regulatory RNAs that destabilize or translationally repress target mRNAs (Hutvágner and Zamore, 2002; Llave et al., 2002). They play key roles in plant growth and development (Mallory and Vaucheret, 2006). Deep sequencing studies have identified a number of miRNAs that are upregulated during nodulation, and ecotopic expression has demonstrated that some are involved in early nodule organogenesis. For example, a specific isoform of miR171 represses critical NODULATION SIGNALING PATHWAY2 (NSP2) expression in M. truncatula and L. japonicus (Devers et al., 2011; Bazin et al., 2012; De Luis et al., 2012). In soybean, numerous miRNAs are differentially expressed during nodule organogenesis (Subramanian et al., 2008; Wang et al., 2009; Dong et al., 2013). Studies have shown that overexpression of several of these soybean miRNAs, including isoforms of miR160, miR156, miR1515, and miR172, can affect nodule numbers (Li et al., 2010; Turner et al., 2013; Yan et al., 2013). However, to date, there are no reports on the consequences of reducing endogenous miRNA activity on soybean nodule development. Moreover, no functional miRNA target modules have been identified in soybean nodulation; thus, their effect on the regulation of nodule development has not been investigated. Molecular mechanisms underlying the regulation of nodulation by miRNAs also remain unaddressed, and evidence for a direct link between miRNAs and the NF or AON signaling pathway is lacking.
Previously, we used cloning and sequencing methods to detect high levels of miR172c expression in mature soybean nodules (Wang et al., 2009). This prompted us to investigate the role of miR172c in soybean nodulation. Attempts were also made to discover the functional target of miR172c. Here, we report that miR172c is required for B. japonicum infection and nodule primordium initiation in soybean. Expression of miR172c is induced following inoculation with B. japonicum and continues to rise during nodule development. Overexpression of miR172c increased soybean nodule numbers, whereas diminished endogenous activity of miR172c resulted in reduced nodulation. We also show that soybean NODULE NUMBER CONTROL1 (NNC1) is a functionally relevant target that is directly cleaved by miR172c, both in vitro and in vivo. Additionally, we show that miR172c regulates nodule formation downstream of NF signaling in an NFR-dependent manner. Intriguingly, we demonstrate that NNC1 directly binds to AP2 cis-elements of ENOD40 promoters, which consequently represses ENOD40 expression and negatively regulates nodulation. This repression is alleviated by miR172c, which cleaves NNC1 transcripts, leading to the promotion of nodule development. Finally, we provide evidence showing that NARK diminishes the upregulation of miR172c during nodule primordium initiation, thereby governing both nodule formation and nodule number regulation.
RESULTS
miRNA172 Family Members Are Differentially Expressed, with miR172c Upregulated during Nodulation
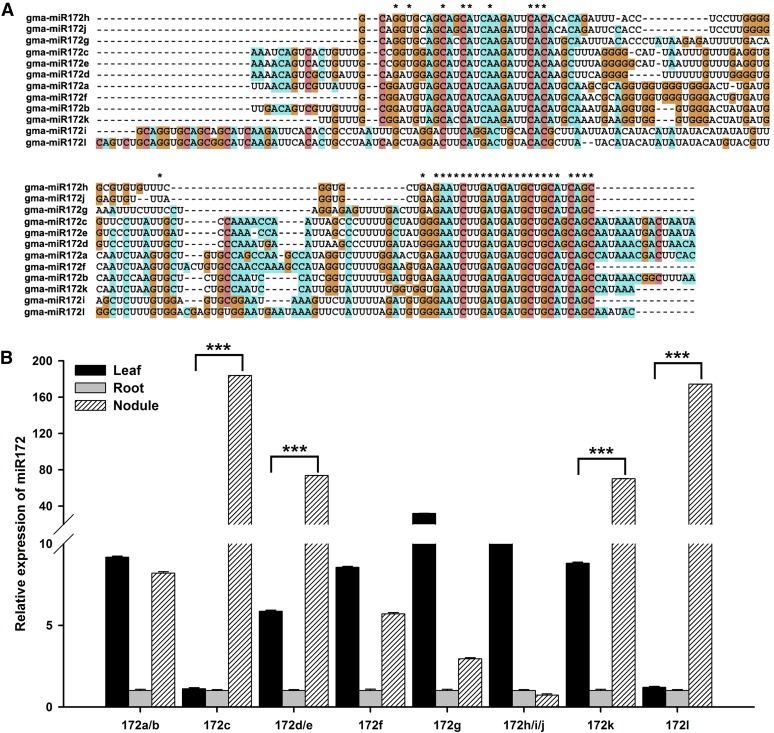
The miR172 family of miRNAs is highly conserved, with a variable number of members present in different plant species (from 1 to 15; Supplemental Table 1). There are 12 soybean miR172 members (Supplemental Table 2), which is the second largest miR172 family among plants analyzed thus far, suggesting functional diversity among the members. The sequence similarity between mature miR172 family members is very high (Supplemental Figure 1), but it varies among the pre-miRNAs (Figure 1A).
Figure 1.
Sequence Alignments and Tissue-Specific Expression Analysis of miR172 Family Members.
(A) Pre-miRNA sequence alignment of miR172 family members. All sequences of the miR172 family members were aligned using the software MEGA5. Asterisks represent conserved nucleotides in all pre-miRNAs.
(B) Tissue-specific expression analysis of miR172 family members. Seven-day-old seedlings were inoculated with B. japonicum strain USDA110. Leaves, roots, and nodules were harvested at 28 DAI (n = 5). Transcript abundance in the different samples was normalized to that of miR1520d. Expression levels are shown as means ± se from three replicates. Asterisks indicate statistically significant differences (***P < 0.001, Student’s t test).
[See online article for color version of this figure.]
To help distinguish the role of specific miR172 family members, we analyzed their expression in leaves, roots, and mature nodules of soybean plants 28 d after inoculation (DAI) with compatible B. japonicum strain USDA110. Because several mature miR172 members have identical sequences (Supplemental Figure 1), the expression of some could not be separated. As shown in Figure 1B, miR172 family members were differentially expressed in different tissues, with miR172c, miR172d/e, miR172k, and miR172l showing higher levels of expression in nitrogen-fixing nodules than in leaves and roots. Among these, miR172c exhibited the highest expression in nodules (Figure 1B), highlighting it as a strong candidate for having a role in nodulation.
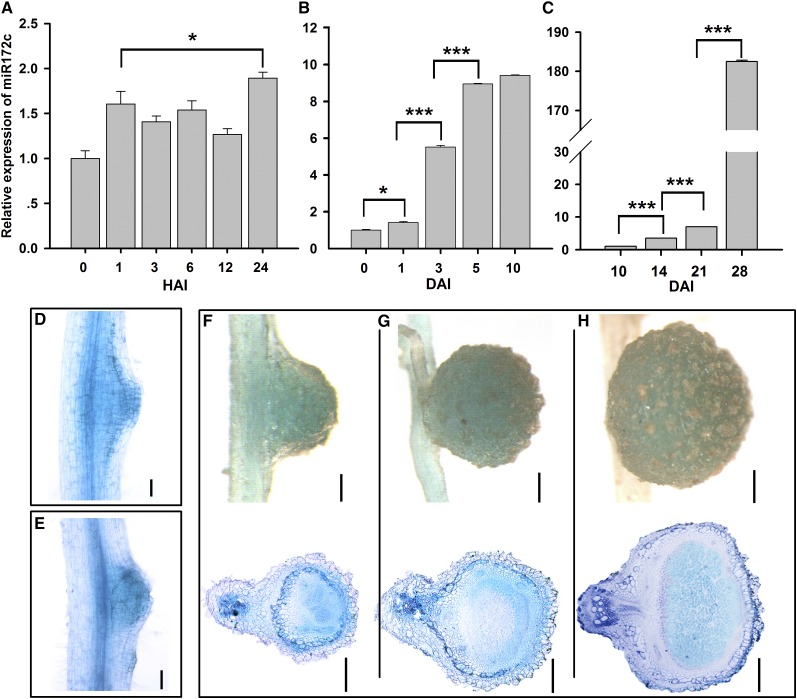
To establish whether miR172c functions during nodulation, we examined its expression in roots inoculated with B. japonicum at time points corresponding to key nodulation events (Yang et al., 1993; Patriarca et al., 2004). The expression of miR172c was slightly upregulated at 24 h after inoculation (Figure 2A) and gradually increased throughout nodule development, reaching its highest level at the maturation stage (Figures 2B and 2C). Notably, its expression increased sharply at 3 and 5 DAI, during extensive nodule primordium formation (Figure 2B). These results indicate that miR172c may be involved in rhizobial infection and the initiation of nodule organogenesis.
Figure 2.
The Expression Pattern of miR172c.
(A) and (B) qRT-PCR relative expression of miR172c in roots. Seven-day-old seedlings were inoculated with B. japonicum strain USDA110, and roots were harvested at 0, 1, 3, 6, 12, and 24 h after inoculation (HAI) (A) or 0, 1, 3, 5, and 10 DAI (B) (n = 5). miR1520d was used as an endogenous control for gene expression. Expression levels are shown as means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001, *P < 0.05).
(C) qRT-PCR of miR172c in nodules. Seven-day-old seedlings were inoculated with B. japonicum strain USDA110, and nodules were harvested at 10, 14, 21, and 28 DAI. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001).
(D) and (E) miR172c histochemical analysis of promiR172c:GUS during nodule initiation. Bars = 200 μm.
(F) to (H) GUS staining of promiR172c:GUS in 10-d-old (F), 14-d-old (G), and 28-d-old (H) nodules. Bars in (F) = 100 μm; bars in (G) = 250 μm; bars in (H) = 400 μm.
Analysis of the miR172c promoter revealed 13 nodule-specific cis-elements (NODCON1GM/NODCON2GM; Stougaard et al., 1990; Jørgensen et al., 1991) and multiple other cis-regulatory elements associated with cytokinin, salicylic acid, and pathogen responses (Supplemental Figure 2). The tissue-specific expression pattern of miR172c was determined by expressing miR172cpro:GUS (for β‑glucuronidase) using an Agrobacterium rhizogenes-mediated transformation system (Kereszt et al., 2007; Jian et al., 2009). In the absence of B. japonicum, GUS activity was almost completely lacking in the majority of the transgenic roots, with only ∼20% exhibiting GUS staining in some epidermal, cortical, and vascular tissues (Supplemental Figure 3). In the presence of B. japonicum, nearly all transformed roots exhibited GUS expression, with over 50% showing strong GUS activity in these same root tissues (Supplemental Figure 3). These results confirm the transcriptional upregulation of miR172c by B. japonicum.
Strong GUS expression was observed throughout nodulation, including in the dividing cortical cells and nodule primordium and in both young and mature nodule structures (Figures 2D to 2H). In young nodules, GUS expression occurred in all tissues and root cells (Figures 2D to 2G), whereas in mature nodules, it was predominantly localized to the infection zone (Figure 2H). These results imply an important role for miR172c throughout nodule development.
miR172c Expression Regulates Early Nodulation Events and Nodule Numbers
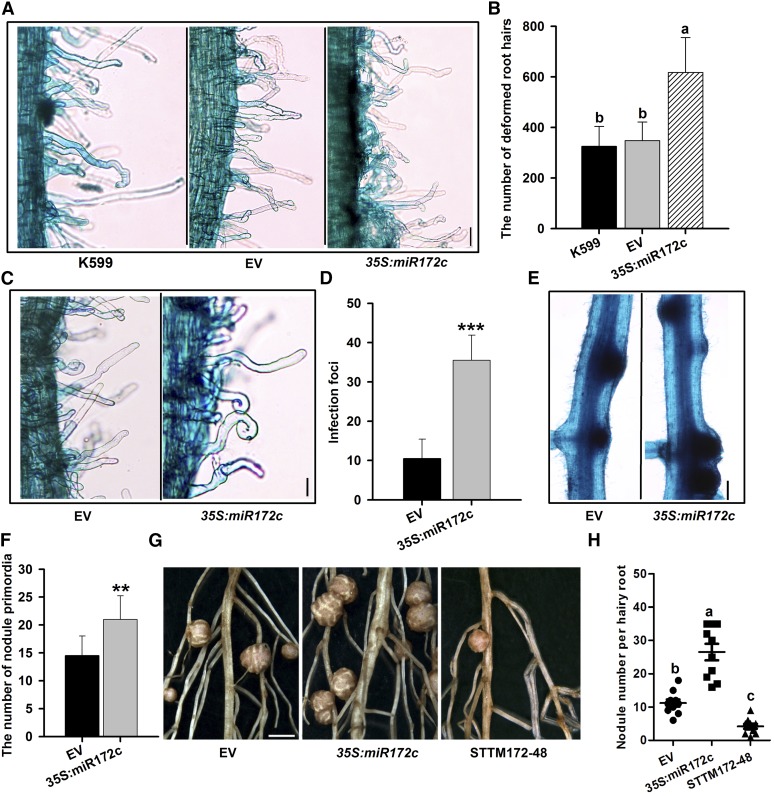
Overexpression of 35S:miR172c was used to help determine the role of miR172c in early nodulation. Six days after inoculation with B. japonicum, overexpression of miR172c markedly increased root hair deformation (Figures 3A and 3B), the number of infection foci, and the number of nodule primordia (Figures 3C to 3F) compared with control plants. These findings indicate that miR172c may be a key regulator of NF perception, bacterial infection, and nodule formation. By 28 DAI, miR172c-overexpressing roots developed significantly more nodules than empty vector control roots (Figure 3G and 3H; Supplemental Figure 4A), consistent with recently reported data (Yan et al., 2013).
Figure 3.
Effect of the Overexpression or Knockdown of miR172c on Nodulation.
(A) to (C) Overexpression of miR172c increased the number of deformed root hairs. At 6 DAI, 2-cm root segments of hairy roots overexpressing miR172c below the root-hypocotyl junction were cut and stained with 1% (w/v) methylene blue. Considerably deformed root hairs were counted (n = 10 to 12).
(A) Root hair deformation in transgenic roots harboring empty vector (EV) and 35S:miR172c vector. Bar = 40 μm.
(B) Quantification of deformed root hairs in the transgenic lines (n = 10 to 12). Values are averages ± sd from three independent experiments. Different letters indicate a significant difference (Student-Newman-Kuels test, P < 0.05).
(C) Highly magnified view of deformed root hairs in the roots transformed with empty and 35S:miR172c vectors. Bar = 40 μm.
(D) The number of infection foci observed in transgenic roots overexpressing miR172c (n = 10 to 12). Asterisks indicate statistically significant differences (Student’s t test, ***P < 0.001). Values are averages ± sd from three independent experiments.
(E) Nodule primordia of individual hairy roots expressing the empty vector and 35S:miR172c at 6 DAI. Bar = 400 μm.
(F) The number of nodule primordia in roots transformed with empty and 35S:miR172c vectors (n = 10 to 12). Values are averages ± sd from three independent experiments. Asterisks represent statistically significant differences (Student’s t test, **P < 0.01).
(G) Nodules of individual hairy roots expressing the empty vector, 35S:miR172c, and STTM172-48 at 28 DAI. Bar = 3 mm.
(H) Quantitative analysis of the nodule number per hairy root expressing empty vector, 35S:miR172c, and STTM172-48 (n = 10 to 12). Values are averages ± sd from three independent experiments. Different letters indicate a significant difference (Student-Newman-Kuels test, P < 0.05).
Overexpression of a short tandem target mimic (STTM) can effectively reduce the activity of target miRNA (Yan et al., 2012). To determine whether miR172c regulates nodule development, a construct was designed to contain an STTM composed of two short sequences mimicking the miR172c target site separated by a linker (STTM172-48; Supplemental Figure 5). Quantitative real-time PCR (qRT-PCR) confirmed that STTM172-48 was highly overexpressed in the resultant transgenic roots (Supplemental Figure 4B), and, as expected, this overexpression correlated with a reduction in the transcript abundance of endogenous miR172c compared with control roots (Supplemental Figure 4A). The number of nodules formed on STTM172-48-overexpressing roots 28 DAI with B. japonicum was reduced significantly compared with the control roots (Figure 3G and 3H). Thus, activation of miR172c during nodule initiation is required for soybean nodulation.
miR172c Acts in the NF Signaling Pathway in an NFR1α-Dependent Manner
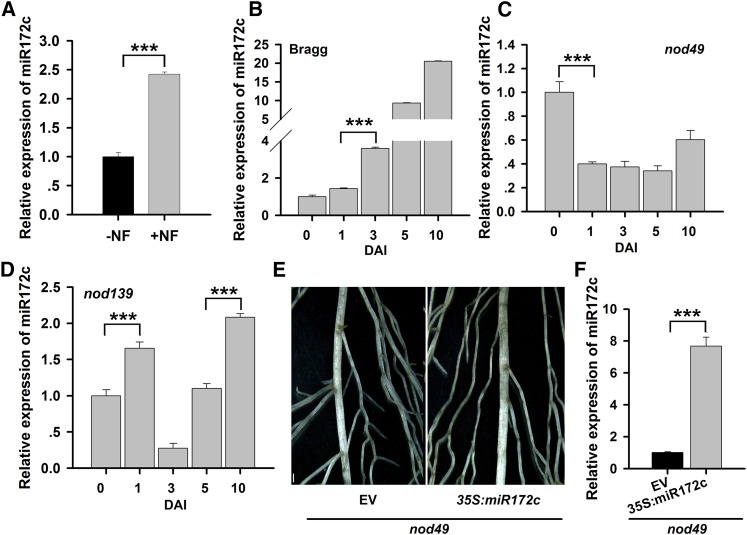
To establish whether miR172c is induced by NF, miR172c expression was determined following NF application. NFs of B. japonicum were induced and extracted as described previously (Banfalvi et al., 1988; Sanjuan et al., 1992). qRT-PCR analysis showed that miR172c expression was markedly upregulated in roots 3 DAI with 10−8 M NF compared with untreated control roots (Figure 4A). This suggests that miR172c is induced by NF and may function in the NF signaling pathway.
Figure 4.
The Function of miR172c in Regulating Nodule Development Is Dependent on NFRs.
(A) qRT-PCR analysis of miR172c in response to NF treatment. Four-day-old seedlings were treated with distilled, deionized water (−NF) or 10−8 M B. japonicum NF (+NF), and the roots were collected at 3 DAI (n = 5).
(B) to (D) qRT-PCR analysis of miR172c expression in wild-type cv Bragg (B) and its isogenic nonnodulation mutants nod49 (C) and nod139 (D), which harbor mutations in NFR1α and NFR5α, respectively (n = 5). Seven-day-old seedlings were inoculated with B. japonicum strain USDA110. The roots were harvested at 0, 1, 3, 5, and 10 DAI.
(E) Image of hairy roots from the NFR1α mutant nod49 expressing empty vector (EV) or 35S:miR172c at 28 DAI. Bar = 700 μm.
(F) qRT-PCR analysis of miR172c expression in transgenic roots of the NFR1α mutant nod49 expressing 35S:miR172c (n = 5).
For all gene expression results shown, miR1520d was used as an internal control. Expression levels are shown as means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001).
[See online article for color version of this figure.]
To test whether the induction of miR172c is dependent on NF receptors, its expression was evaluated in the nonnodulating soybean mutants nod49 and nod139, which carry defects in NFR1α and NFR5α, respectively (Indrasumunar et al., 2010, 2011). As shown in Figure 4B, miR172c expression was upregulated following B. japonicum inoculation in wild-type cv Bragg roots. However, this upregulation was completely blocked in nod49 (Figure 4C) and reduced in nod139 (Figure 4D) roots. Moreover, overexpression of miR172c in nod49 roots was not able to overcome the nonnodulating phenotype of the mutant (Figures 4E and 4F). These data suggest that miR172c expression is dependent on the NFRs, mainly NFR1α.
Identification and Validation of NNC1 as a Candidate Target of miR172c
To determine whether miR172c suppresses one or more target genes to modulate nodulation, psRNATarget (Dai and Zhao, 2011) and Phytozome (Goodstein et al., 2012) were used to identify genes containing putative miR172c target sites. Eleven candidate gene targets were found (Supplemental Table 3), all of which encode AP2/ERF family proteins, with an AP2 domain(s) and a single miR172c target site located in either the coding sequence or the 3′ untranslated region. A phylogenetic analysis based on protein sequence showed that the predicted target genes are highly conserved and could be divided into two clades (Supplemental Figure 6A). To determine which are targeted by miR172c, a 5′ rapid amplification of cDNA ends (RACE) PCR assay was performed. Six of the predicted 11 targets were cleaved by miR172c, with cleavage frequently occurring between bases 10 and 11 (Supplemental Figure 6B). These results suggest that miR172 family members may regulate distinct aspects of plant growth and development by targeting different genes.
The most thoroughly studied target gene of miR172 in Arabidopsis is the AP2/ERF transcription factor encoding TOE1 (Mathieu et al., 2009). Bioinformatic analysis revealed that soybean has six putative TOE1 orthologs, all belonging to the same subclade. Among them, only three genes (glyma13g40470, glyma12g07800, and glyma11g15650) have been experimentally validated as targets of miR172c (Supplemental Figure 6B). All three exhibit reduced expression in nodules of transgenic roots overexpressing miR172c, with glyma12g07800 having the greatest reduction in expression, followed by glyma13g40470 and glyma11g15650 (Supplemental Figure 7A). Expression of glyma12g07800 was also increased significantly in the STTM lines (Supplemental Figure 7B). This pattern of expression was opposite to that of miR172c (Supplemental Figure 7C). Collectively, these data imply that glyma12g07800 is the main target of soybean miR172c during nodulation. The glyma12g07800 coding sequence contains nine exons and eight introns, with a miR172c binding site in exon 9 (Supplemental Figure 7D); it was named NNC1 based on its influence on nodule number.
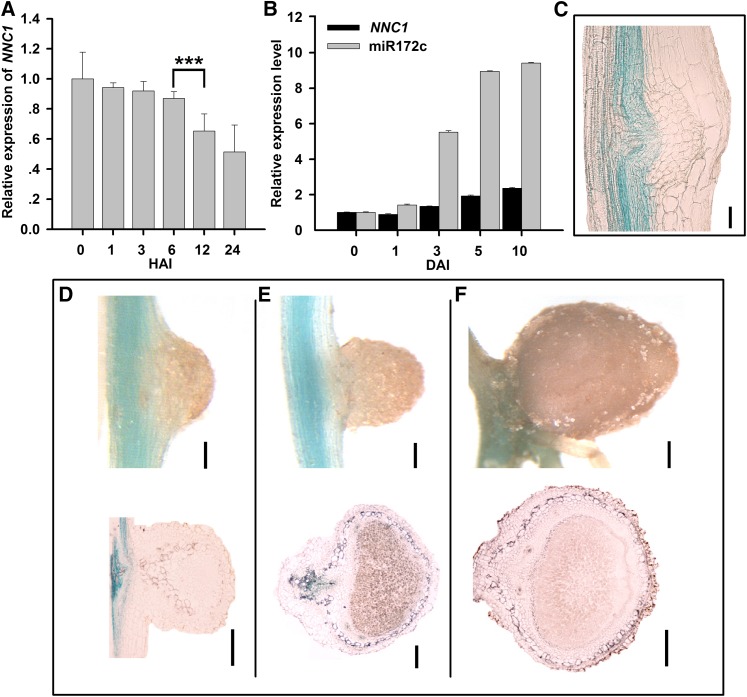
The promoter of NNC1 contains multiple cis-elements, which are similar to those identified in miR172c (Supplemental Figure 8), suggesting that NNC1 and miR172c are regulated similarly during nodulation. NNC1 expression was downregulated 24 h after inoculation with B. japonicum (Figure 5A), in contrast with that of miR172c at the same time point (Figure 2A). During nodule formation, NNC1 expression was slightly upregulated (Figure 5B), as opposed to the strong upregulation of miR172c (Figure 2B). Histochemical analysis of proGmNNC1:GUS showed that NNC1 was predominantly expressed in the vascular tissue of infected roots but was not detected in nodule primordia or in developing and mature nodules (Figures 5C to 5F), in contrast with that of miR172c (Figures 2D to 2H). This opposite pattern in expression suggests that NNC1 may be a direct target of miR172c during nodulation.
Figure 5.
The Expression Pattern of the miR172c Target Gene NNC1.
(A) and (B) qRT-PCR of NNC1 in roots at various times after B. japonicum inoculation during early infection (A) and nodule development (B) (n = 5). Transcript levels were normalized to the expression of GmELF1b in each sample. Expression levels are shown as means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001). HAI, h after inoculation.
(C) Expression of proGmNNC1:GUS at the nodule initiation stage at 10 DAI. Bar = 50 μm.
(D) to (F) Expression of proGmNNC1:GUS in developing nodules at 10 (D), 14 (E), or 28 (F) DAI. Bars in (D) = 100 μm; bars in (E) = 300 μm; bars in (F) = and 400 μm.
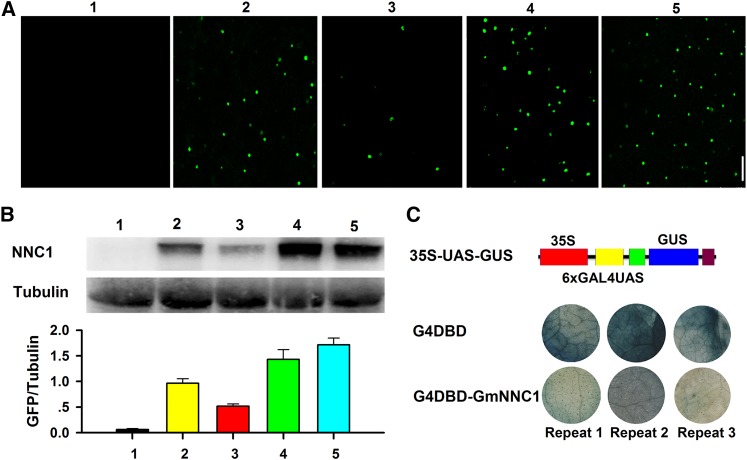
To determine whether NNC1 mRNA is directly cleaved by miR172c, green fluorescent protein (GFP) reporter-based cellular assays were used to assess NNC1 protein levels. Constructs harboring wild-type NNC1 or NNC1m6, a mutant NNC1 cDNA with six mismatches (Chen, 2004) to miR172c, were fused with GFP under the control of the cauliflower mosaic virus 35S promoter. The constructs were transformed (either individually or cotransformed with 35S:miR172c) into Nicotiana benthamiana cells, and GmNNC1-GFP expression was evaluated by confocal microscopy and immunoblotting. NNC1 contains a classical nuclear localization sequence (NLS) in its C-terminal region and consequently is localized in the nucleus (Supplemental Figure 9A). Strong GFP activity was detected in nuclei of transformed cells expressing wild-type NNC1 or NNC1m6 alone (Figure 6A), consistent with it having a putative role in the control of transcription (Supplemental Figures 9A and 9B). Notably, the GFP signal was substantially reduced in cells coexpressing wild-type NNC1 and 35S:miR172c (Figure 6A). The protein levels in cells cotransformed with NNC1 and 35S:miR172c were also markedly reduced (Figure 6B). By contrast, the levels of GFP and NNC1 were not greatly affected in cells expressing NNC1m6 and 35S:miR172c (Figure 6B). These results demonstrate that miR172c directly targets and represses NNC1 mRNA.
Figure 6.
Experimental Validation of NNC1 as a Target Gene of miR172c and Transcriptional Activity Analysis.
(A) and (B) Analysis of the cleavage of NNC1 by miR172c. The indicated constructs were transformed or cotransformed into N. benthamiana leaves, and the expression of NNC1 was imaged (A). Experiments were performed three times. Immunoblot analysis using antibody against GFP and relative GmNNC1-GFP accumulation in the different agroinfiltration assays are indicated in bar graphs below each panel (B). Experiments were performed three times. Different numbers indicate (as follows): 1, 35S:miR172c; 2, 35S:GmNNC1-GFP; 3, 35S:GmNNC1-GFP + 35S:miR172c; 4, 35S:GmNNC1m6-GFP; 5, 35S:GmNNC1m6-GFP + 35S:miR172c.
(C) Transcriptional activity of Gm-NNC1 was tested in N. benthamiana leaves using a GAL4/UAS-based system. 35S, the 35S promoter without the TATA box; 6×GAL4 UAS, six copies of the GAL4 binding site (UAS); G4DBD, the GAL4 DNA binding domain; G4DBD-GmNNC1, G4DBD fused with Gm-NNC1. Experiments were performed three times, and each experiment contained at least three replicates.
NNC1 Encodes a Transcription Factor That Negatively Regulates Nodule Number
In Arabidopsis, TOE1 is a transcription factor with DNA binding activity that represses the expression of downstream genes (Mathieu et al., 2009). Bioinformatic analysis of soybean NNC1 revealed that it contains a bipartial NLS and two typical EAR motifs (LDLNLN and LDLNLG; Supplemental Figure 9A), indicating that it also functions as a transcriptional repressor.
To verify whether NNC1 is a transcriptional repressor, we performed a classical GAL4/UAS-based system assay to study the transcriptional repression of a protein (Tao et al., 2013). As shown in Figure 6C, the GAL4 DNA binding domain (G4DBD) that binds to the six copies of GAL4 UAS can activate the GUS gene expression. Then we generated a fusion protein of G4DBD-NNC1 and cotransformed 35S-UAS-GUS with G4DBD-NNC1 in N. benthamiana leaves to determine Gm-NNC1 transcriptional activity. We found that GUS expression was markedly repressed (Figure 6C), confirming that it is repressed by NNC1.
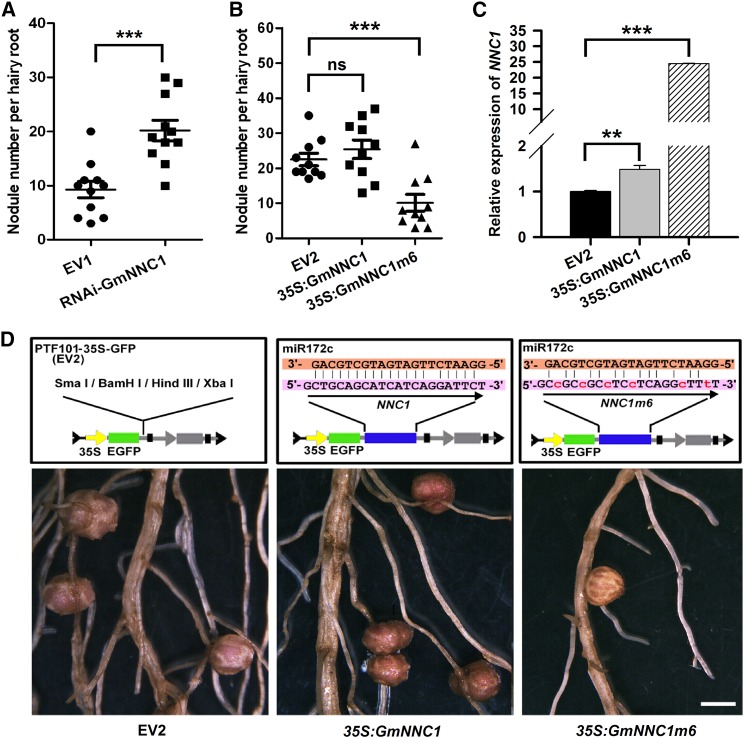
Given that miR172c overexpression results in increased nodule numbers (Figures 3G and 3H), RNA interference (RNAi) of NNC1 was performed to assess whether NNC1 knockdown would produce the same phenotype. As shown in Supplemental Figures 10A and 10B, the abundance of NNC1 transcripts was confirmed to be greatly reduced in transgenic roots, and this knockdown was accompanied by a substantial promotion in nodule formation, similar to that observed in miR172c-overexpressing roots (Figures 3G, 3H, and 7A). These results demonstrate a central role for NNC1 as a negative regulator of nodulation.
Figure 7.
miR172c Regulates Soybean Nodule Numbers through Direct Inhibition of Its Target Gene NNC1.
(A) and (B) Nodule numbers per hairy root transformed with empty vector (EV1, pTCK303) or RNAi-GmNNC1 (A) or 35S:GmNNC1 and 35S:GmNNC1m6 (B) at 28 DAI (n = 10 to 12). Experiments were performed three times. Values are averages ± sd. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001; ns, not significant at P > 0.05).
(C) qRT-PCR analysis of GmNNC1 in roots transformed with empty vector and constructs harboring 35S:GmNNC1 and 35S:GmNNC1m6 (n = 10 to 12). ELF1b was used as an endogenous control for gene expression. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001, **P < 0.05).
(D) Nodules of representative roots overexpressing NNC1, NNC1m6, or GFP (EV2; control) at 28 DAI. Bar = 3 mm.
[See online article for color version of this figure.]
To further establish whether miR172c promotes nodulation by negatively regulating NNC1 expression, NNC1 or its mutant sequence NNC1m6 with six mismatches to miR172c were overexpressed. Overexpression of NNC1 did not significantly affect nodulation; however, NNC1m6 overexpression substantially reduced the number of nodules that formed (Figures 7B to 7D). NNC1 mRNA levels increased less than 2-fold in 35S:GmNNC1 roots but were 25 times higher in 35S:GmNNC1m6 roots compared with empty vector controls (Figure 7C). This confirms that NNC1 is a key target of miR172c and that it is intimately involved in the regulation of soybean nodulation.
NNC1 Directly Targets the Promoters of ENOD40-1/2 and Negatively Regulates Their Expression
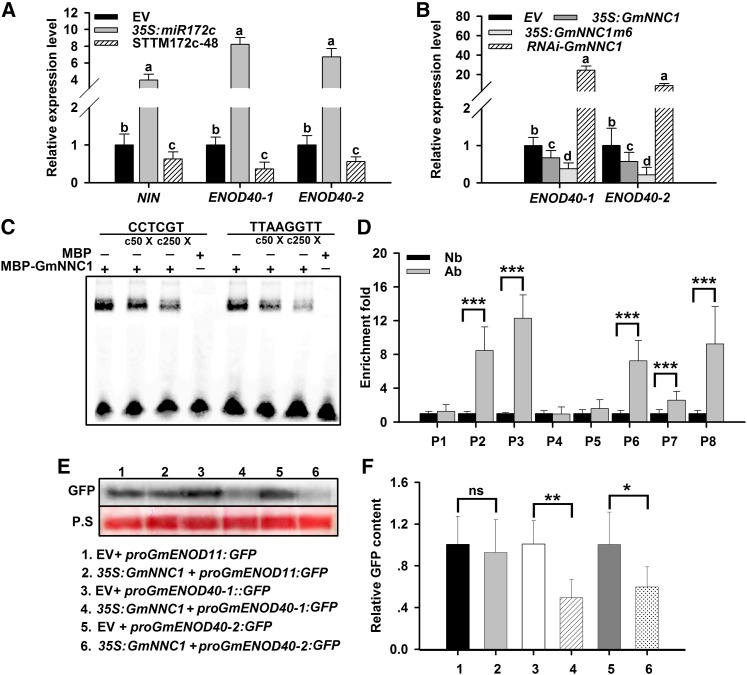
To discover the molecular basis of miR172c-GmNNC1 activity in the NF signaling pathway, the expression profiles of the symbiosis marker genes NIN (Schauser et al., 1999) and ENOD40 (Minami et al., 1996) were examined in B. japonicum-inoculated soybean roots with altered expression of miR172c. Overexpression of miR172c increased the relative expression of NIN and the ENOD40 genes; notably, both ENOD40-1 and ENOD40-2 were highly upregulated (Figure 8A). By contrast, the expression of these genes was downregulated in STTM-miR172c transgenic roots having reduced activity of miR172c. These results indicate that the transcriptional activities of NIN, ENOD40-1, and ENOD40-2 were positively regulated by miR172c and that the miR172c-NNC1 module may function upstream of NIN and the ENOD40 genes in the NF signaling pathway.
Figure 8.
NNC1 Directly Targets the Promoters of ENOD40 Genes.
(A) qRT-PCR analysis of NIN and ENOD40-1 and -2 in roots transformed with empty vector (EV), 35S:miR172c, or STTM172-48 at 28 DAI (n = 10 to 12). Transcript amounts in each sample were normalized to those of ELF1b. Expression levels shown are means ± se from three replicates. Different letters indicate a significant difference (Student-Newman-Kuels test, P < 0.05).
(B) qRT-PCR analysis of ENOD40 genes in roots transformed with empty vector, 35S:GmNNC1, 35S:GmNNC1m6, or RNAi-GmNNC1 at 28 DAI (n = 10 to 12). Transcript amounts in each sample were normalized to those of ELF1b. Expression levels are means ± se from three replicates. Different letters indicate a significant difference (Student-Newman-Kuels test, P < 0.05).
(C) EMSA showing that MBP-GmNNC1 binds to the CCTCGT and TTAAGGTT motifs of the ENOD40 promoters in vitro following incubation. Competition for binding was performed using 50× (c50×) and 250× (c250×) competitive ENOD40 probes; MBP was used as a negative control. Three biological replications were performed.
(D) ChIP assay for binding NNC1 to the ENOD40 promoters. The sequence regions marked by P1 to P8 indicate regions examined in the ChIP assays. ELF1b was employed as an internal control for expression. Three biological replications were performed. Each value is the average ± sd from three independent experiments. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001). Ab, antibody for fragment; Nb, no antibody for fragment.
(E) and (F) Repression of ENOD40 genes by NNC1. Constructs harboring proGmENOD40:GFP were transformed with 35S:GmNNC1 into N. benthamiana leaves. ENOD40 expression was analyzed by immunoblot (E), and GFP intensity was measured by fluorospectrophotometer (F). P.S, Ponceau S-stained gel representing equal loading. Nine independent plants were assessed. The experiment was repeated three times and always exhibited a similar trend. ENOD11 was used as a negative control. Each value is the average ± sd from three independent experiments. Asterisks represent statistically significant differences (Student’s t test, **P < 0.01, *P < 0.05; ns, not significant at P > 0.05).
[See online article for color version of this figure.]
Previous studies have demonstrated that Arabidopsis TOE1 functions as a transcriptional repressor of its target genes through binding to TOE1 cis-elements (CCTCGT and TTAAGGTT) located in the promoter regions of these genes (Franco-Zorrilla et al., 2014). In soybean, bioinformatic analysis identified CCTCGT and TTAAGGTT TOE1 binding cis-elements in the promoters of ENOD40-1 and ENOD40-2 but not in the promoter of NIN, suggesting that the ENOD40 genes may be potential targets of NNC1. Both ENOD40-1 and ENOD40-2 were downregulated in expression in roots overexpressing NNC1m6, whereas they were greatly upregulated in NNC1-RNAi roots (Figure 8B), further supporting that the ENOD40 genes may be functional targets of NNC1.
To test for in vitro binding of NNC1 to the ENOD40 promoter regions, electrophoretic mobility-shift assays (EMSAs) were conducted with the NNC1-MBP protein. Shifted bands were clearly detected when the probes containing CCTCGT and TTAAGGTT in the ENOD40-1 and ENOD40-2 promoters were incubated with the NNC1 protein (Figure 8C). By contrast, shifted bands were not observed when the probes were incubated with the MBP protein. This demonstrates conclusively that NNC1 interacts directly with the promoter regions of ENOD40-1 and ENOD40-2.
A chromatin immunoprecipitation (ChIP) analysis was performed to further verify whether NNC1 binds to the ENOD40 promoters in vivo. Chromatin suspensions were prepared from roots transformed with either 35S:GmNNC1-GFP or an empty vector. Eight primer pairs, four for each ENOD40 gene (Supplemental Table 4), were designed to detect the promoter fragments of both ENOD40 copies. Among them, two primer sets for each ENOD40 promoter contained the TOE1 binding cis-elements (TTAAGGTT and CCTCGT; Supplemental Figure 11). Anti-GFP immunoblot detected high levels of enrichment of ENOD40 promoter regions containing the TOE1 binding cis-elements in immunoprecipitates of chromatin suspensions derived from roots expressing NNC1-GFP (Figure 8D). These results demonstrate that NNC1 directly targets the promoter regions of the ENOD40 genes via the specific nucleotide sequences in their promoters.
A transient expression system was used to analyze the effect of NNC1 on the expression of the ENOD40 genes in vivo. Constructs harboring proENOD40-1/-2:GFP were transformed into N. benthamiana leaf cells cotransformed with either an empty vector control or 35S:GmNNC1. In the absence of 35S:GmNNC1, a strong GFP band was detected in the transformed cells by immunoblotting against the GFP antibody. By contrast, the intensity of the GFP band was substantially reduced in the presence of 35S:GmNNC1 (Figure 8E). We also coexpressed 35S:GmNNC1 with proENOD11:GFP, which acted as a negative control, since the ENOD11 promoter does not contain NNC1 binding sites. As expected, the intensity of the GFP band was not affected by coexpression with NNC1 (Figure 8E). Thus, the repression of ENOD40-1 and ENOD40-2 expression by NNC1 was further confirmed using another quantitative technique (Figure 8F). Taken together, these results demonstrate that NNC1 directly targets the ENOD40 promoters and represses their transcription activity.
miR172c Is Negatively Regulated by NARK in the AON Pathway
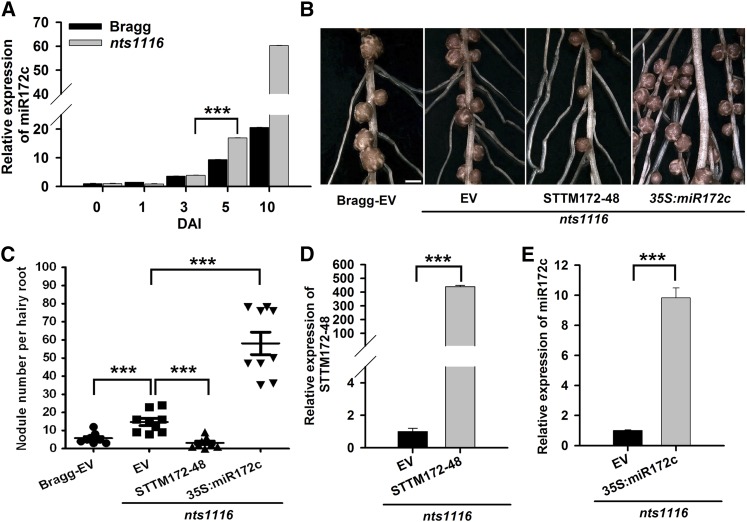
The increase in nodule numbers attributed to miR172c overexpression prompted us to test whether its expression is controlled by the AON pathway. To this end, we examined miR172c expression in the soybean mutant nts1116, which carries a missense mutation in NARK (Searle et al., 2003). qRT-PCR showed that miR172c expression was not significantly influenced during infection and infection thread formation (i.e., 1 and 3 DAI) in nts1116 mutant roots compared with wild-type control roots (Figure 9A). However, it was elevated in nts1116 roots during nodule primordium formation (5 DAI) and remained high at later stages of nodule development compared with that of control roots (Figure 9A). This suggests that hypernodulation of nts1116 is closely linked to elevated miR172c expression.
Figure 9.
The Function of miR172c in Regulating Nodule Development Is Negatively Regulated by NARK.
(A) qRT-PCR analysis of miR172c in wild-type cv Bragg and its isogenic nodulation mutant nts1116, which carries a mutation in NARK (n = 10 to 12). miR1520d was used as an internal control for gene expression. Expression levels shown are means ± se from three replicates.
(B) Nodules from hairy roots of nts1116 mutant plants expressing empty vector (EV), STTM172-48, or 35S:miR172c at 28 DAI. Bar = 700 μm.
(C) Quantitative analysis of the nodule number per hairy root of nts1116 mutant plants expressing empty vector, STTM172-48, and 35S:miR172c (n = 10 to 12). Nodule number per hairy root of wild-type cv Bragg plants expressing the empty vector was used as a control. Each value is the average ± sd from three independent experiments. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001).
(D) qRT-PCR analysis of STTM172-48 in transgenic hairy roots of nts1116 mutant plants (n = 10 to 12). The y axis indicates the expression levels of the gene relative to the expression of ELF1b. Expression levels are means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001)
(E) qRT-PCR analysis of miR172c in transgenic hairy roots of nts1116 mutant plants. The expression levels were normalized against the geometric mean of miR1520d. Expression levels are means ± se from three replicates. Asterisks represent statistically significant differences (Student’s t test, ***P < 0.001).
[See online article for color version of this figure.]
To confirm that NARK negatively regulates miR172c during nodulation, we overexpressed STTM172c-48 to reduce the activity of miR172c and evaluate the resultant number of nodules that formed on transgenic nts1116 mutant roots. At 28 DAI with B. japonicum, nts1116 mutant roots transformed with the empty vector had approximately twice the number of nodules seen in wild-type roots (Figures 9B and 9C), which is consistent with previous findings investigating untransformed roots (Carroll et al., 1985; Searle et al., 2003). By contrast, the numbers of primordia and nodules of transgenic roots expressing STTM172-48, including nts1116 roots, were significantly reduced compared with those of the wild-type control (Figures 9B to 9D). This confirms that hypernodulation in the nts1116 mutant may be due to increased expression of miR172c, and it suggests that miR172c expression is negatively regulated by NARK.
To further validate the genetic relationship between miR172c and NARK, nts1116 plants overexpressing miR172c were generated (Figure 9E). A quantitative analysis of nodule numbers at 28 DAI showed that miR172c overexpression markedly exacerbated the hypernodulation phenotype of nts1116 (Figures 9B and 9C). The average number of nodules per transgenic root exceeded 50, which was much higher than that of the control roots. This demonstrates that the transcript level of miR172c is critical for determining soybean nodule numbers. Taken together, our data suggest that NARK negatively regulates miR172c transcription during nodule primordium formation to prevent excess nodulation.
DISCUSSION
The most fundamental challenges in understanding the symbiosis between nitrogen-fixing bacteria and their respective legume partners are to determine how the association arises, how nodule organogenesis and thus nodule number are regulated, and how nitrogen fixation is controlled. Recently, there has been a great deal of interest in understanding the roles of miRNAs during nodulation in legumes (Wang et al., 2009; Bazin et al., 2012; Bustos-Sanmamed et al., 2013; Turner et al., 2013). Our analysis here identified miR172c-NNC1 as a critical regulatory module of nodulation that targets early nodulin ENOD40 genes and is essential for nodule initiation and nodule organogenesis in soybean.
The miR172 gene family is conserved in plants, although the number of family members varies by species (Supplemental Table 1), indicating the essential roles of miR172 in plant growth and development. Soybean has 12 miR172 family members (Figure 1A) and is one of the few species analyzed to have more than 10, suggesting that the soybean miR172 family may have more diverse functions. Indeed, soybean miR172 family members were differentially expressed in leaves, roots, and nodules (Figure 1B), implying that miR172 participates in regulating the growth and development of a range of soybean tissues. Interestingly, miR172c, miR172d/e, miR172k, and miR172l exhibited high levels of expression in mature nodules (Figure 1B), suggesting that these evolutionarily close family members may differentially regulate soybean nodulation. The fact that ectopic expression of miR172c (Figure 3) and miR172l (Yan et al., 2013) dramatically increased soybean nodule numbers supports the notion that multiple members of the family modulate nodulation.
In Arabidopsis, the upregulation of miR172 during vegetative growth plays a key role in the phase transition from vegetative to reproductive growth (Wu et al., 2009). In soybean, such upregulation of miR172c was in response to B. japonicum infection and NFs (Figures 2A, 2B, and 4A) and was increasingly evident during nodule development (Figures 2B to 2H). Most intriguingly, high levels of miR172c are actually required for rhizobial infection and nodule organogenesis (Figure 3). Therefore, miR172c is a positive regulator of nodulation in soybean, and upregulation of miR172c may be involved in the onset of infection, nodule initiation, and organogenesis and/or the developmental phase transition during nodule development.
In soybean, NFR1α and NFR5α are key receptors of B. japonicum NFs (Indrasumunar et al., 2010, 2011; Nakagawa et al., 2011). Thus far, it is unclear what other key signaling components are transmitted to activate downstream targets of NF perception. Our results reveal that soybean miR172c is a signal component downstream of NF recognition but upstream of the NIN and ENOD40 genes. We provide three pieces of evidence that favor the above hypothesis. First, miR172c expression was induced by both B. japonicum (Figures 2A and 2B) and NFs (Figure 4A). Second, the upregulation of miR172c in response to B. japonicum infection and during nodule organogenesis is dependent upon NFR1α and NFR5α, especially NFR1α (Figures 4C, 4E, and 4F). Third, we showed that overexpression of miR172c results in substantial increases in the expression of NIN and the early nodulin ENOD40 genes (Figure 8A). Therefore, miR172c represents a key regulator of NFR-mediated microsymbiont infection and nodule formation.
In Arabidopsis, multiple AP2 transcription factors are downregulated by miR172 through a translational mechanism (Aukerman and Sakai, 2003; Chen, 2004; Lauter et al., 2005; Wu et al., 2009; Sarkar, 2010). Here, we demonstrated that NNC1, a putative soybean ortholog of Arabidopsis TOE1, is the direct target of miR172c in nodulation (Figures 6A, 6B, and 7; Supplemental Figures 7A to 7C). Since multiple genes are cleaved by miR172c (Supplemental Figure 6B) and overexpression of glyma11g15650, the closest homolog to NNC1, can also reduce the number of nodules in soybean (Yan et al., 2013), we do not exclude the possibility that other targets are also involved in miR172-mediated regulation of soybean nodulation. Our RACE results demonstrated direct cleavage of target mRNAs by miR172c (Supplemental Figure 6B), but we cannot exclude the possibility that miR172c may also regulate its targets at the posttranslational level.
Several miRNAs have been shown to participate in nodulation regulation (Li et al., 2010; Turner et al., 2013). However, there is no evidence for a direct link between these miRNAs and the NF signaling pathway. In this study, we provide solid evidence that the miR172c-NNC1 module regulates nodule initiation and development through modulation of the expression of ENOD40 genes. We showed that NNC1, the transcription factor targeted by miR172c, directly binds to the promoters of ENOD40-1 and ENOD40-2 (Figures 8C and 8D) to repress their transcriptional activity (Figures 8E and 8F). Based on this evidence, we conclude that the miR172c-NNC1 module is a direct upstream regulator of the ENOD40 genes. Since overexpression or knockdown of the ENOD40 genes mainly affects nodule primordium formation and nodule development, but not rhizobial infection (Wan et al., 2007), it is likely that miR172c-NNC1 modulates nodule organogenesis, such as primordium formation and nodule development via the ENOD40 gene products (either as RNA or encoded small peptides). The similar expression pattern of miR172c and ENOD40 in dividing cortical cells, nodule primordia, and nodules (Figures 2B to 2G; Wan et al., 2007) favors their common regulatory role in nodule organogenesis.
In addition to the role of miR172c in nodule initiation and development, our results also showed that miR172c is involved in early infection steps (Figures 3A to 3D). It is conceivable that the miR172c-NNC1 module may regulate nodulation through targeting multiple promoters of genes functioning at different stages of nodulation. In addition, although we established a link between miR172c with soybean NIN expression (Figure 8), the lack of NNC1 binding sites in the promoter of NIN suggests that the miR172c-NNC1 module does not directly regulate NIN gene expression.
Optimal nodulation is triggered by NF signaling but inhibited by the AON pathway (Kinkema et al., 2006). However, the molecular basis of how the NF symbiotic pathway and the AON signaling pathway coordinately modulate nodule formation and development remains completely unknown. Here, we provided evidence to support the idea that miR172c may be a gene targeted by AON signaling. Our results showed that knockdown of NARK resulted in extremely high levels of miR172c expression during nodule initiation and primordium formation (Figure 9A) and that the reduction of miR172c activity in the mutant nts1116, which carries a mutation in NARK, completely restored the supernodulation phenotype of the mutant (Figures 9B to 9E). These results suggest that NARK negatively regulates the miR172c expression level to influence nodulation. Further upregulation of miR172c in nts1116 enhanced the hypernodulation phenotype of the mutant (Figures 9B, 9C, and 9E). This strongly supports the notion that activation of NARK at the primordium formation stage may maintain miR172c expression below the threshold level required for nodule formation as a means of preventing excessive nodulation. Thus, our studies highlight miR172c as a key factor involved in crosstalk between the NF and AON signaling pathways that ultimately helps to determine nodule number in soybean. Recent results have shown that shoot-derived cytokinin is a strong candidate for the SDI signal that inhibits nodule formation (Sasaki et al., 2014). Interestingly, there are cytokinin-activated ARR1 binding cis-regulatory elements in the miR172c promoter that are responsible for the cytokinin-mediated regulation of genes (Sakai et al., 2000; Oka et al., 2002). Thus, it is possible that the AON signaling pathway negatively regulates miR172c through shoot-derived cytokinin-activated signaling. Further elucidation of the mechanism by which miR172c is repressed will give a better understanding of the regulation of nodulation inhibition by cytokinins. Furthermore, it has been hypothesized that AON may inhibit nodule formation through repressing NSP2 expression downstream of the cytokinin receptor LOTUS HISTIDINE KINASE1 (Sasaki et al., 2014). Characterization of the relationship between NSP2 and miR172c will help us to decipher the molecular mechanism through which AON inhibits nodulation.
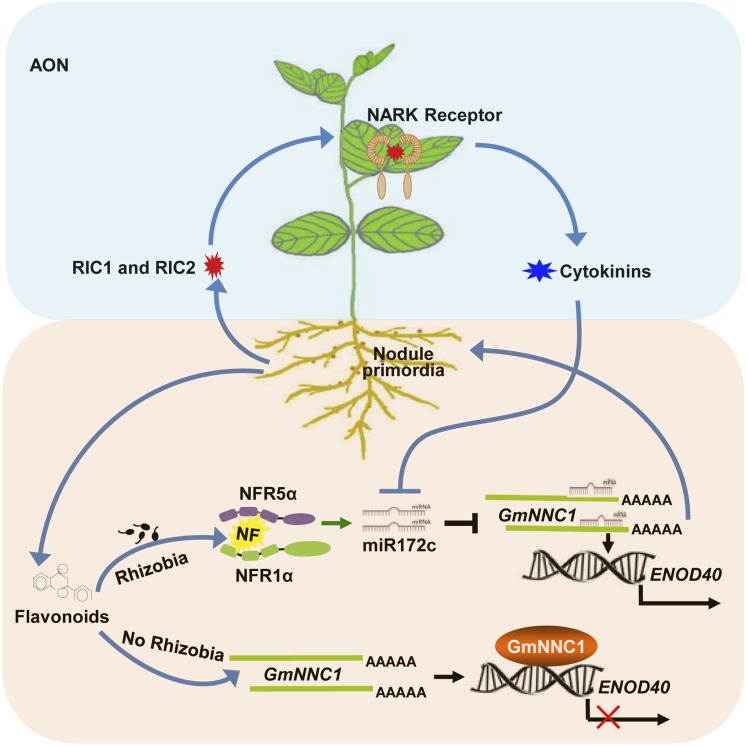
Based on our data, we propose that, in the absence of rhizobia or NFs, NNC1 binds to the promoter of the ENOD40 genes and represses their transcription. In the presence of rhizobia, the perception of NF by NFRs activates a signaling cascade that triggers the upregulation of miR172c to target and cleave NNC1 mRNAs. This reduction in NNC1 transcript reduces its inhibitory effect on the transcription of the ENOD40 genes, resulting in their activation and subsequent nodule organogenesis (Figure 10). To avoid overproducing nodules, AON signaling occurs, commencing with short CLE peptides (RIC1 and RIC2) activating NARK located on the plasma membrane of leaf phloem parenchyma cells. This activation of NARK stimulates SDI production (e.g., shoot-derived cytokinins) that, in turn, represses the transcriptional activity of miR172c through an unknown mechanism to prevent excessive nodulation. A model of this network is illustrated in Figure 10. Our results provide insight into miRNA-mediated regulatory gene expression and the molecular mechanism of nodulation in soybean. Based on the profound influence of miR172c on nodulation, and the differential expression of other miR172 family members during nodulation, we anticipate that miR172 family members may function in combination to fine-tune rhizobium-legume symbiosis, nodule number, nitrogen fixation, and plant growth and development through different target genes.
Figure 10.
A Proposed Model of the miR172c-NNC1-Mediated Regulation of Nodule Formation and Nodule Number Control in Soybean.
When rhizobia are absent, NNC1 represses ENOD40 gene transcription via promoter binding. In the presence of rhizobia, NFRs recognize NFs and induce signaling that upregulates miR172c, which targets and cleaves NNC1 mRNAs. The resulting decrease in NNC1 transcript releases the inhibition of ENOD40 expression, leading to ENOD40 activation and ultimately to nodule organogenesis. Nodule overproduction is prevented by AON signaling, in which short CLE peptides (RIC1 and RIC2) activate NARK. NARK, which is found on the plasma membrane of leaf phloem parenchyma cells, induces shoot-derived cytokinins that, in turn, repress the transcriptional activity of miR172c and thereby promote nodulation.
METHODS
Plant and Rhizobium Growth Conditions
Soybean (Glycine max cv Williams 82) was used to clone the miRNAs and target genes, for 5′ RACE, and in the functional analysis of miR172c and its target. Soybean cv Bragg and its isogenic mutant lines nod49, nod139, and nts1116 containing mutations in NFR1α, NFR15α, and NARK, respectively, were also used. The plant growth conditions and inoculation procedures using Bradyrhizobium japonicum USDA110 were modified from Wang et al. (2009). For RNA extraction, seedling roots were rinsed briefly in PBS buffer, pH 7.5, to remove vermiculite and perlite particles. Harvested tissues were frozen immediately in liquid nitrogen and stored at −80°C until used for RNA extraction.
Extraction and Application of NF
The lipooligosaccharide NF signals were purified from B. japonicum strain USDA110 as described previously (Sanjuan et al., 1992; Carlson et al., 1993). B. japonicum cultures were induced for Nod gene expression by the addition of soybean seed extract (Banfalvi et al., 1988; Smit et al., 1992). Ten milliliters of sterilized, deionized water containing 10−8 M NF was used to irrigate 4-d-old seedlings. After 3 d, the root samples were collected and used to analyze the expression of miR172c. Root samples irrigated with 10 mL of distilled, deionized water were used as control.
RNA Extraction and Quantitative PCR Analysis
Total RNA and small RNAs were extracted from leaves, roots, and nodules using Trizol reagent (Tiangen Biotech). Total RNA samples were treated with DNase I (Invitrogen) to remove contaminating genomic DNA. First-strand cDNA was synthesized from the total RNA using the FastQuant RT Kit (Tiangen Biotech). qRT-PCR was performed using SuperReal PreMix Plus (SYBR Green; Tiangen Biotech) and gene-specific primers for the genes analyzed (listed in Supplemental Table 4). ELF1b and CYP2 were used as housekeeping genes (Jian et al., 2008).
Stem-Loop qRT-PCR
Stem-loop-specific reverse transcription for miRNAs was performed as described previously (Chen et al., 2005; Kulcheski et al., 2010). Soybean miR1520d was used as a reference miRNA gene to normalize the samples as outlined previously (Kulcheski et al., 2010). The expression pattern of all soybean miR172 family members was assessed in leaves, roots, and nodules 28 DAI with B. japonicum. Mature sequences of miR172 members were used to design primers according to Chen et al. (2005). All primers used for stem-loop qRT-PCR are listed in Supplemental Table 4. To distinguish between individual members of the miR172 family, the stem-loop primers used for cDNA synthesis included 44 conserved and 6 reverse 3′ end nucleotides of the miR172 family members. The forward primers used for qRT-PCR were designed to be specific to their miR172 family member sequence, with the exception of the last six nucleotides at the 3′ end of the miRNA. To enable the melting temperatures of the primers to be raised, six GC-rich nucleotides were added at the 5′ end (Guleria and Yadav, 2011). Due to the high similarity between the mature miR172 sequences, only the same stem-loop reverse transcription primers were transcribed in one reaction. Likewise, the same miR172-specific quantitative real-time forward primer was used for qRT-PCR in one reaction. To further confirm the specificity of the amplified miR172 family members, the PCR products were sequenced.
Prediction of miRNA Targets and 5′ RACE Mapping of miRNA Target Cleavage Sites
Putative miRNA targets were predicted using psRNATarget (Dai and Zhao, 2011). Total RNA was isolated from a mixture of roots, leaves, and nodules collected from 5-week-old cv Williams 82 plants using Plant RNA Reagent (Invitrogen) according to the manufacturer’s recommendations. A GeneRacer Kit (Invitrogen) was used to process the total RNA and map the 5′ terminus of the primary transcript. The cDNA samples were amplified by nested PCR according to the manufacturer’s protocols. Gene-specific primers (Supplemental Table 4) were designed by Invitrogen.
Vector Construction
For the promoter:GUS reporter fusion construct, putative promoter regions of miR172c (788 bp) and NNC1 (1113 bp) were amplified from cv Williams 82 genomic DNA and inserted upstream of the GUS gene in pTF102 or pCAMBIA3301 vector using BamHI or EcoRI/BglII, respectively. For the miR172c overexpression construct, the pre-miRNA fragment of miR172c (220 bp) was amplified and inserted into the plant expression vector pEGAD under the control of the cauliflower mosaic virus 35S promoter using SmaI and BamHI. For the RNAi construct for the target gene, RNAi-NNC1 containing 510 bp of the coding sequence was amplified by PCR from cv Williams 82 cDNA and inserted into the vector pTCK303 in the sense and antisense orientations using KpnI/PstI and BamHI/SacI. To reduce the activity of miR172c, the STTM172-48 construct was made according to Yan et al. (2012). The STTM module was inserted between the 35S promoter and the 35S terminator in pEGAD vector using AgeI and BamHI. For the overexpression construct, the coding sequence of NNC1 was amplified and inserted into the vector pTF101-GFP using HindIII and BamHI. For site-directed mutagenesis, six point mutations in the miRNA binding site of NNC1 were designed according to the procedure of Chen (2004) and inserted into the vector pTF101-GFP using HindIII and BamHI. All primers used for plasmid construction are listed in Supplemental Table 4.
Soybean Hairy Root Transformation and B. japonicum Inoculation Assay
Soybean transformation to generate hairy root composite plants was done using Agrobacterium rhizogenes K599 according to previously described methods (Kereszt et al., 2007; Jian et al., 2009). For nodulation assays, transgenic composite plants were transplanted to pots (10 × 10 cm) containing a 3:1 mixture of vermiculite and perlite and grown for 1 week (16 h of light, 25°C, and 50% RH) to allow recovery. The plants were then inoculated with a suspension of B. japonicum strain USDA110 (OD600 = 0.08). At 10 or 28 DAI, roots and nodules were assayed and harvested. To examine the expression of NIN, ENOD40-1, ENOD40-2, NFR1α, and NFR5α, miR172c-overexpressing roots identified by qRT-PCR were used.
Root Hair Deformation and Infection Assays
To assay for infection events, 2-cm root segments of hairy roots overexpressing miR172c below the root-hypocotyl junction were cut and harvested at 6 DAI and then rinsed briefly in sterile PBS buffer to remove vermiculite/perlite particles. The roots were later fixed with ethanol:glacial acetic acid (3:1) for 2 h and then washed three times with deionized water. The roots were then stained with 0.01% methylene blue for 15 min and washed three times with deionized water as described previously (Subramanian et al., 2004). The stained transgenic roots overexpressing miR172c were observed with an Olympus CX31 biological microscope for infection events. Root hairs with swelling tips and distinct growth direction (wavy) in the specified root segments were considered as deformed root hairs (Heidstra et al., 1994; Geurts et al., 2005) and counted blindly by two persons under light microscopy (Olympus CX31) (n = 10 to 12); the number of root hairs forming tight curls that were referred to as “infection foci” was also estimated (n = 10 to 12).
Histochemical Localization of GUS Expression
For the histochemical analysis of GUS expression in roots and nodules, multiple roots or nodules at different developmental stages from at least 16 independent lines were stained for each construct. GUS activity was analyzed histochemically as described previously (Jefferson et al., 1987).
Microscopy and Image Analysis
All microscopic observations were performed on an Olympus dissecting or Olympus compound light microscope. Both microscopes were integrated with a Canon digital camera. Longitudinal and cross sections of nodule segments were generated by embedding specimens in paraffin wax and sectioning to 25 μm thickness using a VT 1000S vibratome (Leica Microsystems).
Subcellular Localization and Validation of the Direct Inhibition of NNC1 by miR172c
To assess the subcellular localization of NNC1, pTF101-GFP-NNC1 and the empty vector were introduced into Agrobacterium tumefaciens EHA101. The infiltration of Nicotiana benthamiana was performed as described by Hu et al. (2013). Transformed A. tumefaciens cells were then grown at 28°C in Luria-Bertani liquid medium. The cells were incubated at room temperature for at least 3 h and then infiltrated into the abaxial air spaces of N. benthamiana plants. Epidermal cell layers of N. benthamiana leaves were assayed for fluorescence 3 d after infiltration. GFP and 4′,6-diamidino-2-phenylindole fluorescence were observed with a confocal laser scanning microscope (Leica TCS SP8).
Analysis of the Transcriptional Activity of NNC1
For transcriptional activity analysis, a 35S-UAS-GUS reporter construct was obtained from Genji Qin. Coding sequence for a fusion protein, G4DBD-GmNNC1, was produced in pYF503 using the primers NNC1-503-F and GmNNC1-503-R with SalI and NotI. Next, the G4DBD and G4DBD-GmNNC1 fragments were amplified from pYF503 and pYF503-G4DBD-GmNNC1 using the primers G4DBD-F/G4DBD-R and G4DBD-F/GmNNC1DBD-R, respectively. The products were cloned into pDORNER207 (Invitrogen) to generate pDORNER207-G4DBD and pDORNER207-G4DBD-GmNNC1, respectively. The two entry plasmids were used in an LR reaction with pGWB6 to generate the effector constructs. The plasmids of the reporter 35S-UAS-GUS and effector constructs were transformed into A. tumefaciens strain GV3101 cells, and the two effectors were coinfiltrated with the reporter 35S-UAS-GUS into N. benthamiana leaves as described previously (Voinnet et al., 2003). After incubation in the dark for 24 h and subsequent incubation in the light for 72 h, the leaves were used for histochemical GUS staining. All primers used are listed in Supplemental Table 4.
Transcription Repression of Gm-ENOD40 Gene Promoters by Gm-NNC1 in N. benthamiana Leaves and Immunoblot Analysis
Transient expression assays were performed in N. benthamiana leaves as described previously. The promoters of Gm-ENOD40-1/2 were cloned into pGWB4 to generate the reporter constructs pGmENOD40-1:GFP and pGmENOD40-2:GFP. For the negative control ENOD11, primers 5′-AGTTGTTAGATACCTAAAAGTCAG-3′ and 5′-AGGTTTTTAGTTACTGGTGAAT-3′ were used for cloning its promoter. The Gm-NNC1 effector construct used was the 35S:GmNNC1. For the 35S:GmNNC1 construct, the coding sequence of NNC1 was amplified and inserted into pEGAD using AgeI and BamHI. GFP and tubulin antibody (Clontech) were used for immunoblot analysis. Immunoreactive proteins were detected using an ECL Plus Chemiluminescence Kit (GE Healthcare). GFP quantification was performed using a BioVision GFP Quantification Kit (BioVision; K815-100) according to the manufacturer’s protocol with nine independent plants assessed. Immunoreactive proteins were detected using the ECL Plus Chemiluminescence Kit (GE Healthcare).
EMSA
EMSAs were performed using the LightShift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s protocol and as described by Yoo et al. (2010). For the MBP-GmNNC1 construct, the coding sequence of NNC1 was amplified and inserted into pMAL-2CX using BamHI and XbaI. Recombinant MBP-GmNNC1 was expressed and purified using amylose resin (New England Biolabs) according to the manufacturer’s instructions. The binding activity of the protein was analyzed using an oligonucleotide containing CCTCGT and TTAAGGTT motifs present in the ENOD40-1 and ENOD40-2 promoters, labeled with biotin at the 5′ end (Invitrogen) (Kagaya et al., 1999). After incubation for 30 min at room temperature, the protein-probe mixture was separated on a 6% polyacrylamide gel and transplanted to a Biodyne B nylon membrane (Pall). Migration of the biotin-labeled probes was detected using streptavidin-horseradish peroxidase conjugates that bind to biotin and the chemiluminescent substrate according to the manufacturer’s protocol. This experiment was performed three times.
ChIP-Quantitative PCR Assay
One gram of 10-DAI transgenic roots containing 35S:GmNNC1-GFP and 35S:GFP were used for the ChIP assay. The roots were cross-linked with 1% formaldehyde for 30 min under a vacuum; cross-linking was stopped with 0.125 M glycine. The seedlings were ground in liquid nitrogen, and their nuclei were isolated. Immunoprecipitations were performed with the anti-GFP antibody and protein G beads. Immunoprecipitation in the absence of anti-GFP served as the control. qRT-PCR analysis was performed using specific primers corresponding to different promoter regions of ENOD40-1 and ENOD40-2. ELF1b was used as an internal control (primers used are shown in Supplemental Table 1).
Phylogenetic Analysis
At-TOE1 and its 11 homologous protein sequences from soybean were obtained from TAIR (www.Arabidopsis.org) and Phytozome (www.phytozome.net) and imported into MEGA5 (Tamura et al., 2011) for complete alignment using Explorer/Clustal (Thompson et al., 1997). The phylogenetic tree was built using MEGA5 (Tamura et al., 2011). Phylogenies were built using the neighbor-joining method with the bootstrapping value set at 1000 replications. The sequence alignments used for the analysis are available as Supplemental Data Set 1.
Bioinformatics Analysis
Pre-miRNA sequence alignment of miR172 family members (http://www.mirbase.org/) was performed using the software MEGA5 (Tamura et al., 2011). Promoter analyses of miR172c and NNC1 were performed using PLACE (http://www.dna.affrc.go.jp/PLACE). The regions located 2000 bp upstream of pre-miR172c or the start codon (ATG) of NNC1 (http://www.phytozome.net) were used as promoter sequences for analyzing cis-elements using PLACE (http://www.dna.affrc.go.jp/PLACE). The amino acid sequences of Gm-NNC1 and At-TOE1 were aligned using MEGA5. The bipartial NLS motif was analyzed using PredictNLS (https://rostlab.org/owiki/index.php/Predict_NLS).
Statistical Analysis
All data were analyzed using SigmaPlot 10.0 (Systat Software) and GraphPad Prism 5 (GraphPad Software) software. The averages and sd of all results were calculated, and ANOVA and Student’s t test were performed to generate P values. When there were statistically significant differences, Student-Newman-Kuels tests were conducted.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: gma-miR172a (MI0001780), gma-miR172b (MI0001781), gma-miR172c (MI0010727), gma-miR172d (MI0010728), gma-miR172e (MI0010729), gma-miR172f (MI0016574), gma-miR172g (MI0018668), gma-miR172 h (MI0018669), gma-miR172i (MI0018670), gma-miR172j (MI0018671), gma-miR172k (MI0019746), gma-miR172l (MI0019750), ENOD40-1 (glyma01g03470), ENOD40-2 (glyma02g04180), ENOD11 (glyma09g12198), NFR1α (glyma02g43860), NFR5α (glyma11g06740), NIN (glyma04g00210), NARK (glyma12g04390), NNC1 (glyma12g07800), TOE1 (At2g28550), ELF1B (glyma02g44460), CYP2 (glyma12g02790), glyma11g15650, glyma13g40470, glyma15g04930, glyma19g36200, glyma03g33470, glyma17g18640, glyma05g18041, glyma01g39520, glyma11g05720, and glyma02g09600.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Analysis of Mature miRNA Sequences of the miR172 Family Members.
Supplemental Figure 2. Analysis of the miR172c Promoter.
Supplemental Figure 3. Histochemical Analysis of miR172c.
Supplemental Figure 4. qRT-PCR Analysis of the miR172c Transcript Level.
Supplemental Figure 5. Diagram of STTM172c-48 Structure Showing the Design Strategy.
Supplemental Figure 6. Prediction and Experimental Validation of the Target Genes of miR172c.
Supplemental Figure 7. Gene Expression Analysis of Target Genes and Structures of NNC1.
Supplemental Figure 8. Promoter Analysis of NNC1.
Supplemental Figure 9. NNC1 Protein Analysis.
Supplemental Figure 10. Identification of NNC1 RNAi Knockdown Roots.
Supplemental Figure 11. Analysis of ENOD40 Promoters.
Supplemental Table 1. The Number of miR172 Family Members in Plants.
Supplemental Table 2. Sequence of gma-miR172 Family Members.
Supplemental Table 3. Results of miR172c Target Prediction in psRNATarget.
Supplemental Table 4. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree of miR172c Target Genes Shown in Supplemental Figure 6.
Supplementary Material
Acknowledgments
We thank members of X.L.’s laboratory for their helpful comments and discussions. We thank Kan Wang (Iowa State University) and Guiliang Tang (University of Kentucky) for providing the pTF101.1 vector and for technical support. We thank Genji Qin (Peking University) for kindly providing the 35S-UAS-GUS reporter construct. We also thank Wenxin Chen (China Agricultural University) for kindly providing B. japonicum strain USDA110 and members of the Centre for Integrative Legume Research for support. This study was funded by the National Natural Science Foundation of China (Grants 31230050 and 30971797), the Ministry of Agriculture of the People’s Republic of China (Grants 2014ZX0800929B and 2009ZX08009-132B), the Youth Innovation Promotion Association of the Chinese Academy of Sciences, and the ARC for AON funding (Grants DP130103084 and DP130102266).
AUTHOR CONTRIBUTIONS
Y.W., Y.Z., and X.L. conceived the study. Y.W., Y.Z., P.M.G., and X.L. designed the experiments. Y.Z., L.W., and L.C. performed the miRNA and GmNNC1 analyses and the miR172 cleavage of target gene experiments. Y.W., Y.Z., L.W., Z.C., and S.Z. performed phenotypic analyses of transgenic roots. F.Z. and Y.T. conducted hairy root transformation. Y.W., Y.Z., L.C., and Q.J. analyzed the experimental data. Y.W., Y.Z., B.J.F., P.M.G., and X.L. wrote the article.
Glossary
- NF
Nod factor
- AON
autoregulation of nodulation
- SDI
shoot-derived inhibitor
- miRNA
microRNA
- DAI
days after inoculation
- STTM
short tandem target mimic
- qRT-PCR
quantitative real-time PCR
- RACE
rapid amplification of cDNA ends
- NLS
nuclear localization sequence
- EMSA
electrophoretic mobility-shift assay
- ChIP
chromatin immunoprecipitation
- RNAi
RNA interference
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfalvi Z., Nieuwkoop A., Schell M., Besl L., Stacey G. (1988). Regulation of nod gene expression in Bradyrhizobium japonicum. Mol. Gen. Genet. 214: 420–424. [DOI] [PubMed] [Google Scholar]
- Bazin J., Bustos-Sanmamed P., Hartmann C., Lelandais-Brière C., Crespi M. (2012). Complexity of miRNA-dependent regulation in root symbiosis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1570–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 109: 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos-Sanmamed P., Mao G., Deng Y., Elouet M., Khan G.A., Bazin J., Lelandais-Brière C. (2013). Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 40: 1208–1220. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G., Gresshoff P.M. (1991). Plant genetic control of nodulation. Annu. Rev. Microbiol. 45: 345–382. [DOI] [PubMed] [Google Scholar]
- Carlson R.W., Sanjuan J., Bhat U.R., Glushka J., Spaink H.P., Wijfjes A.H., van Brussel A.A.N., Stokkermans T.J.W., Peters N.K., Stacey G. (1993). The structures and biological activities of the lipo-oligosaccharide nodulation signals produced by type I and II strains of Bradyrhizobium japonicum. J. Biol. Chem. 268: 18372–18381. [PubMed] [Google Scholar]
- Carroll B.J., McNeil D.L., Gresshoff P.M. (1985). Isolation and properties of soybean [Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proc. Natl. Acad. Sci. USA 82: 4162–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon C., Sousa C., Crespi M., Kondorosi A. (1999). Alteration of enod40 expression modifies Medicago truncatula root nodule development induced by Sinorhizobium meliloti. Plant Cell 11: 1953–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., et al. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33: e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Williams R.W., Meyerowitz E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575–585. [DOI] [PubMed] [Google Scholar]
- Compaan B., Yang W.C., Bisseling T., Franssen H. (2001). ENOD40 expression in the pericycle precedes cortical cell division in Rhizobium-legume interaction and the highly conserved internal region of the gene does not encode a peptide. Plant Soil 230: 1–8. [Google Scholar]
- Crespi M.D., Jurkevitch E., Poiret M., d’Aubenton-Carafa Y., Petrovics G., Kondorosi E., Kondorosi A. (1994). enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 13: 5099–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Zhao P.X. (2011). psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 39: W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luis A., Markmann K., Cognat V., Holt D.B., Charpentier M., Parniske M., Stougaard J., Voinnet O. (2012). Two microRNAs linked to nodule infection and nitrogen-fixing ability in the legume Lotus japonicus. Plant Physiol. 160: 2137–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delves A.C., Mathews A., Day D.A., Carter A.S., Carroll B.J., Gresshoff P.M. (1986). Regulation of the soybean-rhizobium nodule symbiosis by shoot and root factors. Plant Physiol. 82: 588–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbrosses G.J., Stougaard J. (2011). Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348–358. [DOI] [PubMed] [Google Scholar]
- Devers E.A., Branscheid A., May P., Krajinski F. (2011). Stars and symbiosis: MicroRNA- and microRNA*-mediated transcript cleavage involved in arbuscular mycorrhizal symbiosis. Plant Physiol. 156: 1990–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Shi L., Wang Y., Chen L., Cai Z., Wang Y., Jin J., Li X. (2013). Identification and dynamic regulation of microRNAs involved in salt stress responses in functional soybean nodules by high-throughput sequencing. Int. J. Mol. Sci. 14: 2717–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Mathesius U. (2014). Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 40: 770–790. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Indrasumunar A., Hayashi S., Lin M.H., Lin Y.H., Reid D.E., Gresshoff P.M. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52: 61–76. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Li D., Hastwell A.H., Reid D.E., Li Y., Jackson S.A., Gresshoff P.M. (2014). The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnol. J. 12: 1085–1097. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352. [DOI] [PubMed] [Google Scholar]
- Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., Rokhsar D.S. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40: D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresshoff P.M., Hayashi S., Biswas B., Mirzaei S., Indrasumunar A., Reid D., Samuel S., Tollenaere A., van Hameren B., Hastwell A., Scott P., Ferguson B.J. (2014). The value of biodiversity in legume symbiotic nitrogen fixation and nodulation for biofuel and food production. J. Plant Physiol. 172C: 128–136. [DOI] [PubMed] [Google Scholar]
- Guleria P., Yadav S.K. (2011). Identification of miR414 and expression analysis of conserved miRNAs from Stevia rebaudiana. Genomics Proteomics Bioinformatics 9: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin S.C., Tang G.-Q., Scholz A., Holtgraewe D., Winter H., Huber S.C. (2003). Phosphorylation of sucrose synthase at serine 170: Occurrence and possible role as a signal for proteolysis. Plant J. 35: 588–603. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Reid D.E., Lorenc M.T., Stiller J., Edwards D., Gresshoff P.M., Ferguson B.J. (2012). Transient Nod factor-dependent gene expression in the nodulation-competent zone of soybean (Glycine max [L.] Merr.) roots. Plant Biotechnol. J. 10: 995–1010. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Geurts R., Franssen H., Spaink H.P., Van Kammen A., Bisseling T. (1994). Root hair deformation activity of nodulation factors and their fate on Vicia sativa. Plant Physiol. 105: 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Chen L., Wang H., Zhang L., Wang F., Yu D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74: 730–745. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., Zamore P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Indrasumunar A., Kereszt A., Searle I., Miyagi M., Li D., Nguyen C.D.T., Men A., Carroll B.J., Gresshoff P.M. (2010). Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol. 51: 201–214. [DOI] [PubMed] [Google Scholar]
- Indrasumunar A., Searle I., Lin M.H., Kereszt A., Men A., Carroll B.J., Gresshoff P.M. (2011). Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr). Plant J. 65: 39–50. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen E.S., Peoples M.B., Boddey R.M., Gresshoff P.M., Hauggaard-Nielsen H., Alves B., Morrison M.J. (2012). Legumes for mitigation of climate change and the provision of feedstock for biofuels and biorefineries. Agron. Sustain. Dev. 32: 329–364. [Google Scholar]
- Jian B., Hou W., Wu C., Liu B., Liu W., Song S., Bi Y., Han T. (2009). Agrobacterium rhizogenes-mediated transformation of Superroot-derived Lotus corniculatus plants: A valuable tool for functional genomics. BMC Plant Biol. 9: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian B., Liu B., Bi Y., Hou W., Wu C., Han T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen J.E., Stougaard J., Marcker K.A. (1991). A two-component nodule-specific enhancer in the soybean N23 gene promoter. Plant Cell 3: 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Ohmiya K., Hattori T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27: 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszt A., Li D., Indrasumunar A., Nguyen C.D.T., Nontachaiyapoom S., Kinkema M., Gresshoff P.M. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2: 948–952. [DOI] [PubMed] [Google Scholar]
- Kinkema M., Scott P.T., Gresshoff P.M. (2006). Legume nodulation: Successful symbiosis through short- and long-distance signaling. Funct. Plant Biol. 33: 707–721. [DOI] [PubMed] [Google Scholar]
- Kouchi H., Hata S. (1993). Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 238: 106–119. [DOI] [PubMed] [Google Scholar]
- Krusell L., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422–426. [DOI] [PubMed] [Google Scholar]
- Kulcheski F.R., Marcelino-Guimaraes F.C., Nepomuceno A.L., Abdelnoor R.V., Margis R. (2010). The use of microRNAs as reference genes for quantitative polymerase chain reaction in soybean. Anal. Biochem. 406: 185–192. [DOI] [PubMed] [Google Scholar]
- Kumagai H., Kinoshita E., Ridge R.W., Kouchi H. (2006). RNAi knock-down of ENOD40s leads to significant suppression of nodule formation in Lotus japonicus. Plant Cell Physiol. 47: 1102–1111. [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S.P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 102: 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Deng Y., Wu T., Subramanian S., Yu O. (2010). Misexpression of miR482, miR1512, and miR1515 increases soybean nodulation. Plant Physiol. 153: 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.H., Ferguson B.J., Kereszt A., Gresshoff P.M. (2010). Suppression of hypernodulation in soybean by a leaf-extracted, NARK- and Nod factor-dependent, low molecular mass fraction. New Phytol. 185: 1074–1086. [DOI] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Vaucheret H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38 (suppl.): S31–S36. [DOI] [PubMed] [Google Scholar]
- Mathesius U., Charon C., Rolfe B.G., Kondorosi A., Crespi M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13: 617–628. [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami E., Kouchi H., Cohn J.R., Ogawa T., Stacey G. (1996). Expression of the early nodulin, ENOD40, in soybean roots in response to various lipo-chitin signal molecules. Plant J. 10: 23–32. [DOI] [PubMed] [Google Scholar]
- Moling S., Pietraszewska-Bogiel A., Postma M., Fedorova E., Hink M.A., Limpens E., Gadella T.W.J., Bisseling T. (2014). Nod factor receptors form heteromeric complexes and are essential for intracellular infection in Medicago nodules. Plant Cell 26: 4188–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kaku H., Shimoda Y., Sugiyama A., Shimamura M., Takanashi K., Yazaki K., Aoki T., Shibuya N., Kouchi H. (2011). From defense to symbiosis: Limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J. 65: 169–180. [DOI] [PubMed] [Google Scholar]
- Nishimura R., Hayashi M., Wu G.J., Kouchi H., Imaizumi-Anraku H., Murakami Y., Kawasaki S., Akao S., Ohmori M., Nagasawa M., Harada K., Kawaguchi M. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426–429. [DOI] [PubMed] [Google Scholar]
- Oka A., Sakai H., Iwakoshi S. (2002). His-Asp phosphorelay signal transduction in higher plants: Receptors and response regulators for cytokinin signaling in Arabidopsis thaliana. Genes Genet. Syst. 77: 383–391. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4: 2191. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E. (2013). Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Patriarca E.J., Tatè R., Ferraioli S., Iaccarino M. (2004). Organogenesis of legume root nodules. Int. Rev. Cytol. 234: 201–262. [DOI] [PubMed] [Google Scholar]
- Peoples M.B., Unkovich M.J., Herridge D.F. (2009). Measuring symbiotic nitrogen fixation by legumes. In Nitrogen Fixation in Crop Production. Agronomy Monograph 52, Emerich D.W. and Krishnan H.B., eds (Madison, WI: American Society of Agronomy Publishing; ), pp. 125–170. [Google Scholar]
- Reid D.E., Ferguson B.J., Gresshoff P.M. (2011b). Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 24: 606–618. [DOI] [PubMed] [Google Scholar]
- Reid D.E., Ferguson B.J., Hayashi S., Lin Y.H., Gresshoff P.M. (2011a). Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. (Lond.) 108: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhrig H., John M., Schmidt J. (2004). Modification of soybean sucrose synthase by S-thiolation with ENOD40 peptide A. Biochem. Biophys. Res. Commun. 325: 864–870. [DOI] [PubMed] [Google Scholar]
- Röhrig H., Schmidt J., Miklashevichs E., Schell J., John M. (2002). Soybean ENOD40 encodes two peptides that bind to sucrose synthase. Proc. Natl. Acad. Sci. USA 99: 1915–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Aoyama T., Oka A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24: 703–711. [DOI] [PubMed] [Google Scholar]
- Salvagiotti F., Cassman K.G., Specht J.E., Walters D.T., Weiss A., Dobermann A. (2008). Nitrogen uptake, fixation and response to fertilizer N in soybeans: A review. Field Crops Res. 108: 1–13. [Google Scholar]
- Sanjuan J., Carlson R.W., Spaink H.P., Bhat U.R., Barbour W.M., Glushka J., Stacey G. (1992). A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 89: 8789–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D. (2010). Photoperiodic inhibition of potato tuberization: An update. Plant Growth Regul. 62: 117–125. [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5: 4983. [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Schnabel E., Journet E.P., de Carvalho-Niebel F., Duc G., Frugoli J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58: 809–822. [DOI] [PubMed] [Google Scholar]
- Searle I.R., Men A.E., Laniya T.S., Buzas D.M., Iturbe-Ormaetxe I., Carroll B.J., Gresshoff P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109–112. [DOI] [PubMed] [Google Scholar]
- Smit G., Puvanesarajah V., Carlson R.W., Barbour W.M., Stacey G. (1992). Bradyrhizobium japonicum nodD1 can be specifically induced by soybean flavonoids that do not induce the nodYABCSUIJ operon. J. Biol. Chem. 267: 310–318. [PubMed] [Google Scholar]
- Sousa C., Johansson C., Charon C., Manyani H., Sautter C., Kondorosi A., Crespi M. (2001). Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell. Biol. 21: 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard J., Jørgensen J.E., Christensen T., Kühle A., Marcker K.A. (1990). Interdependence and nodule specificity of cis-acting regulatory elements in the soybean leghemoglobin lbc3 and N23 gene promoters. Mol. Gen. Genet. 220: 353–360. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Fu Y., Sunkar R., Barbazuk W.B., Zhu J.K., Yu O. (2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Hu X., Lu G., Odelland J.T., Yu O. (2004). The promoters of two isoflavone synthase genes respond differentially to nodulation and defense signals in transgenic soybean roots. Plant Mol. Biol. 54: 623–639. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., Zhang J., Wei T., Gu H., Qu L.J., Qin G. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Nizampatnam N.R., Baron M., Coppin S., Damodaran S., Adhikari S., Arunachalam S.P., Yu O., Subramanian S. (2013). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 162: 2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of Tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wan X., Hontelez J., Lillo A., Guarnerio C., van de Peut D., Fedorova E., Bisseling T., Franssen H. (2007). Medicago truncatula ENOD40-1 and ENOD40-2 are both involved in nodule initiation and bacteroid development. J. Exp. Bot. 58: 2033–2041. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li P., Cao X., Wang X., Zhang A., Li X. (2009). Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochem. Biophys. Res. Commun. 378: 799–803. [DOI] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X., Chen X., Tang G. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Hossain M.S., Wang J., Valdés-López O., Liang Y., Libault M., Qiu L., Stacey G. (2013). miR172 regulates soybean nodulation. Mol. Plant Microbe Interact. 26: 1371–1377. [DOI] [PubMed] [Google Scholar]
- Yang W.C., Katinakis P., Hendriks P., Smolders A., de Vries F., Spee J., van Kammen A., Bisseling T., Franssen H. (1993). Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 3: 573–585. [DOI] [PubMed] [Google Scholar]
- Yoo C.Y., Pence H.E., Jin J.B., Miura K., Gosney M.J., Hasegawa P.M., Mickelbart M.V. (2010). The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 22: 4128–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.