Abstract
The superfamily of phytochrome (Phy) photoreceptors regulates a wide array of light responses in plants and microorganisms through their unique ability to reversibly switch between stable dark-adapted and photoactivated end states. Whereas the downstream signaling cascades and biological consequences have been described, the initial events that underpin photochemistry of the coupled bilin chromophore and the ensuing conformational changes needed to propagate the light signal are only now being understood. Especially informative has been the rapidly expanding collection of 3D models developed by x-ray crystallographic, NMR, and single-particle electron microscopic methods from a remarkably diverse array of bacterial Phys. These structures have revealed how the modular architecture of these dimeric photoreceptors engages the buried chromophore through distinctive knot, hairpin, and helical spine features. When collectively viewed, these 3D structures reveal complex structural alterations whereby photoisomerization of the bilin drives nanometer-scale movements within the Phy dimer through bilin sliding, hairpin reconfiguration, and spine deformation that ultimately impinge upon the paired signal output domains. When integrated with the recently described structure of the photosensory module from Arabidopsis thaliana PhyB, new opportunities emerge for the rational redesign of plant Phys with novel photochemistries and signaling properties potentially beneficial to agriculture and their exploitation as optogenetic reagents.
INTRODUCTION
The perception of light is vital to the survival and fitness of most cellular organisms and is mediated by a collection of specially adapted photoreceptors bearing intrinsic or prosthetic chromophores. In plants, these include the UVR8 photoreceptor that exploits a cluster of tryptophans to detect UV-B light (Jenkins, 2014), the phototropins, cryptochromes, and Zeitlupe-type F-box proteins that harbor one or more flavin moieties for blue light detection (Kim et al., 2007; Chaves et al., 2011; Liscum et al., 2014), and the phytochrome (Phy) family that absorbs red and far-red light through a bilin (or open-chain tetrapyrrole) chromophore (Franklin and Quail, 2010; Chen and Chory, 2011). Collectively, these photoreceptors enable continuous sensation of the ambient light environment with respect to presence/absence, fluence rate, direction, duration, and color, which in concert allow plants to adjust appropriately their growth and development to optimize photosynthetic light capture and entrain their life cycles to diurnal and seasonal light fluctuations.
The influence of Phys was first realized almost 100 years ago in seed plants through the study of various photomorphogenic processes, such as seed germination, deetiolation, stem growth, pigmentation, and flowering in response to the long-wavelength region of the visible light spectrum (Schafer and Nagy, 2006). Biochemical studies on the purified chromoproteins in the 1960s later revealed their unique ability to exist in two stable, interconvertible forms, a red-light-absorbing Pr state that represents the dark-adapted, biologically inactive conformer and a far-red-light-absorbing Pfr state that is generated upon red-light excitation and is biologically active (Butler et al., 1959). By photointerconverting between Pr and Pfr and reverting thermally from Pfr back to Pr, Phys act as light-regulated master switches in various signaling cascades. Studies over the past two decades have revealed an intricate network of Phy isoforms (PhyA-E in Arabidopsis thaliana; Sharrock and Quail, 1989) and associated signaling components, including a set of PHY-INTERACTING FACTOR (PIF) transcriptional repressors, that ultimately control much of plant gene expression in response to light (Franklin and Quail, 2010; Chen and Chory, 2011; Leivar and Monte, 2014). But how Phys translate this light information into conformational and/or chemical signals interpretable by these cascades have remained largely unknown.
Here, we describe how the more recent discoveries of Phy relatives throughout the microbial world followed by the 3D structural analyses of representatives in their dark-adapted and photoactivated states have provided tantalizing clues. The emerging “toggle” mechanism shows that Phys are exquisitely designed photosensors that amplify light-driven, angstrom-scale alterations in bilin conformation into nanometer-scale domain movements within the dimeric chromoprotein, which ultimately trigger photomorphogenesis.
MODULAR ARCHITECTURE OF PHYs
Long before any structural appreciation, it was well known that plant Phys are homodimeric complexes with each subunit containing an N-terminal photosensing module (PSM) that binds bilin followed by a C-terminal output module (OPM) that promotes dimerization and might relay the light signal (Rockwell et al., 2006). The PSM in turn sequentially harbors a PAS (Period/Arnt/Single-Minded) domain of unknown function, a signature GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domain that binds the bilin, and a PHY (Phy-specific) domain that is related to GAF domains and stabilizes the photoactivated Pfr state (Figure 1). Upstream of the PAS domain is a long N-terminal extension (NTE) that differs among plant Phy isoforms. The NTE is important for Pfr stability and signaling and appears to be negatively impacted by phosphorylation (Cherry et al., 1992; Nito et al., 2013).
Figure 1.
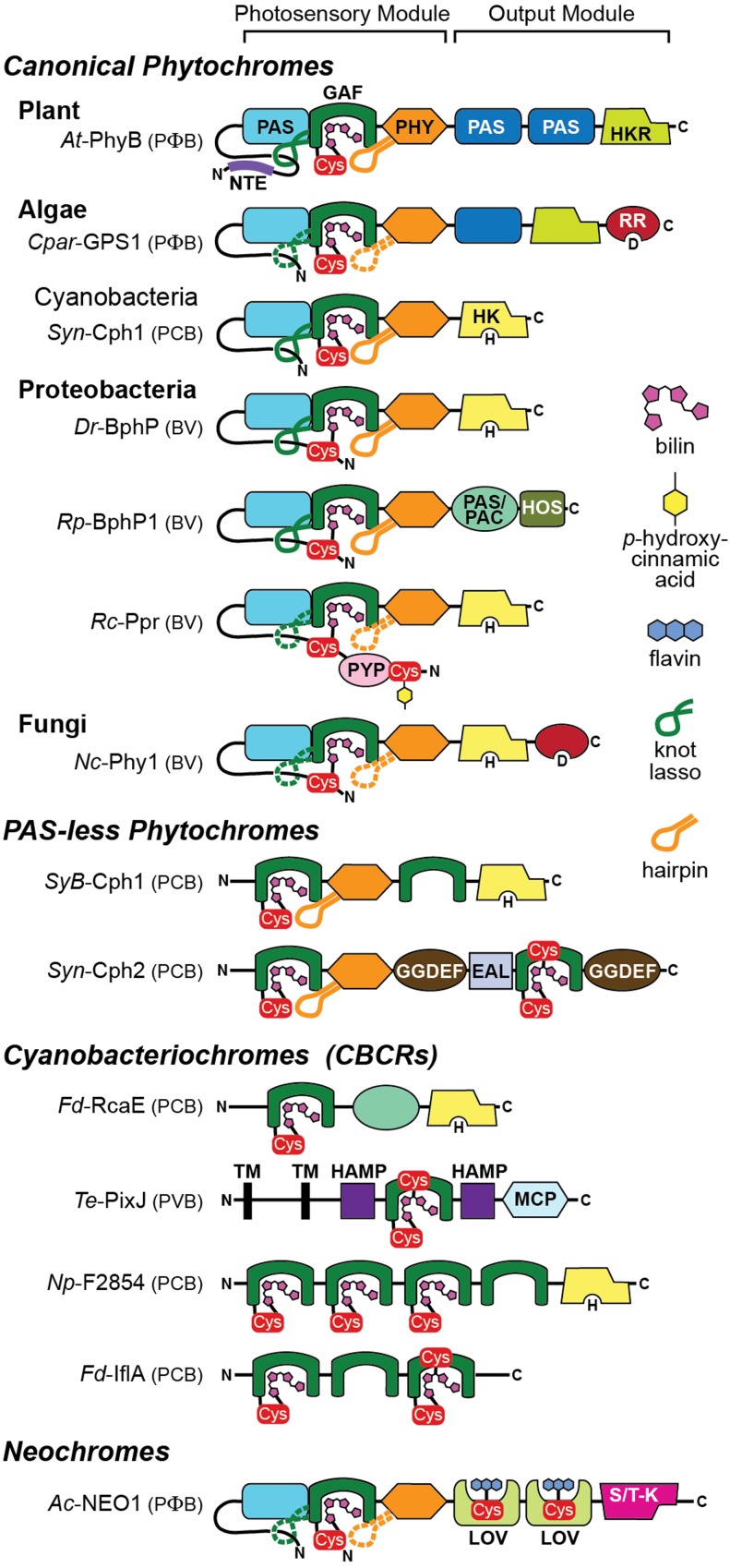
Architecture of Representative Members of the Phy Superfamily within Seed and Seedless Plants (Neochromes), Algae (Glaucophyte), Proteobacteria, and Cyanobacteria (PAS-Less and CBCRs).
Shown are Arabidopsis (At) PhyB, Cyanophora paradoxa CCMP329 (glaucophyte, Cpar) GPS1, Synechocystis PCC6803 (Syn) Cph1 and Cph2, D. radiodurans (Dr) BphP, R. palustris (Rp) BphP1, R. centenum (Rc) Ppr, Neurospora crassa (Nc) Phy1, Synechococcus OSB’ (SyB) Cph1, T. elongatus (Te) PixJ, N. punctiforme (Np) F2854, F. diplosiphon (Fd) RcaE and IflA, and A. capillus-veneris (Ac) neochrome NEO1. The known or likely bilin type for each Phy is indicated in parenthesis. Domains include conserved residues in diguanylate phosphodiesterase (EAL), cGMP phosphodiesterase/adenylyl cyclase/FhlA (GAF), conserved residues in diguanylate cyclase (GGDEF), histidine kinase/adenylyl cyclase/methyl binding protein/phosphatase (HAMP), histidine kinase (HK), histidine-kinase-related (HKR), 2-helix output sensor (HOS), light/oxygen/voltage (LOV), methyl-accepting chemotaxis protein (MCP), Phy N-terminal extension (NTE), Period/Arnt/Single-Minded (PAS), PAS domain followed by C-terminal motif similar to PAS domain (PAS/PAC), Phy-specific (PHY), photoactive yellow protein (PYP), response regulator (RR), serine/threonine-kinase (S/T-K), and predicted transmembrane (TM). The knot lasso and hairpin motifs are indicated by the green and orange loops, respectively. Confirmed and predicted lassos/hairpins are in solid and dashed lines, respectively. Cys, cysteine residue that covalently binds the chromophores: bilin, flavin, or p-hydroxycinnamic acid. H, phosphoacceptor histidine in HK domains; D, phosphoacceptor aspartate in RR domains; C, C terminus; N, N terminus.
Using a lyase activity intrinsic to the GAF domain (Wu and Lagarias, 2000), the plant Phy apoproteins covalently attach the bilin, phytochromobilin (PΦB), through a thioether linkage between a conserved cysteine (Cys-357 in At-PhyB) and the ethylidene side chain of PΦB (Figure 2A). PΦB is synthesized in chloroplasts via the oxidative cleavage of heme by a heme oxygenase to generate biliverdin IXα (BV) followed by reduction of the C3 vinyl side chain and neighboring C2 carbon of BV using a PΦB synthase (HY1 and HY2 in Arabidopsis, respectively; Davis et al., 1999a; Frankenberg et al., 2001). Various NMR analyses and spectroscopic studies proposed that photoconversion involves a Z to E isomerization within the methine bridge between the C and D pyrrole rings that flips the D ring (e.g., Rüdiger et al., 1983; Kneip et al., 1999; Rockwell et al., 2006), but how this subtle change ultimately drives a conformational/enzymatic switch was unclear. That plant Phys have weak phosphotransferase activity in vitro and that the OPM includes a sequence related to histidine kinase domains common among bacterial two-component sensors have led to speculation that they instigate a phosphorelay (Yeh and Lagarias, 1998), but direct proof is currently lacking. More recent interaction studies proposed that light-driven conformational remodeling of Phys promotes the phosphorylation-induced turnover of the PIF family, thus alleviating the inhibitory activity of these repressors on photomorphogenesis (Al-Sady et al., 2006; Ni et al., 2014). Irrespective of the signaling pathways, a key step in the action of plant Phys is their light-induced transport from the cytoplasm into the nucleus (Kevei et al., 2007).
Figure 2.
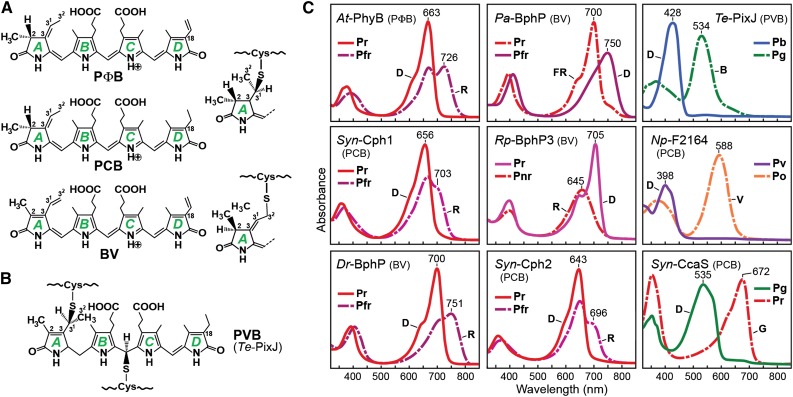
Bilin Chromophores and Absorption Spectra of Representative Phys within the Superfamily.
(A) Chemical structure and apoprotein thioether linkage position for various bilins used in Phys. The four pyrrole rings (A-D) and important carbon positions in the bilin are indicated.
(B) Chemical structure and dual thioether linkage specific to CBCRs with blue/green photocycles.
(C) Absorption spectra of the dark-adapted (D) and photoactivated states after blue (B), far-red (FR), green (G), red (R), and violet (V) excitation for canonical Phys with Pr dark-adapted states (At-PhyB, Syn-Cph1, and Dr-BphP), a bathy-Phy (Pa-BphP), a Pnr Phy (Rp-BphP3), a PAS-less Phy (Syn-Cph2), and three CBCRs (Te-PixJ, Np-F2164, and Syn-CcaS). Dashed lines denote the photoactivated state. The bilin type is indicated in parenthesis for each Phy. Absorption maxima are indicated.
PHY DIVERSITY IN MICROORGANISMS
While originally thought to be restricted to plants and some algae, we now know that Phy-type photoreceptors pervade the microbial world (Karniol and Vierstra, 2006; Auldridge and Forest, 2011) (Figure 1). The first prokaryotic representative discovered was RcaE from the cyanobacterium Fremyella diplosiphon that harbors a Phy-like GAF sequence capable of binding bilins followed by a prototypic histidine kinase OPM (Kehoe and Grossman, 1996). Genome sequence gazing subsequently identified numerous relatives in various cyanobacteria, proteobacteria (both photosynthetic and nonphotosynthetic), actinobacteria, and filamentous fungi and found Phys to be widely distributed but not ubiquitous in green, brown, and heterokont algae, thus greatly expanding the known realm of these photoreceptors (e.g., Hughes et al., 1997; Yeh et al., 1997; Davis et al., 1999b; Bhoo et al., 2001; Giraud et al., 2002; Karniol and Vierstra, 2003; Karniol et al., 2005; Ikeuchi and Ishizuka, 2008; Ulijasz et al., 2009; Rockwell et al., 2011, 2014). In many cases, confirmation was provided by demonstration that the respective GAF domain(s) would covalently bind bilins when expressed recombinantly. For simplicity, we defined members of the Phy superfamily here as proteins containing one or more GAF domains capable of housing a covalently bound bilin but acknowledge that close sequence relatives that do not bind bilins exist (e.g., Vuillet et al., 2007; Rockwell et al., 2012a). In most cases, these Phy chromoproteins have photoreversible absorption.
Collectively, cyanobacterial Phys conjugate phycocyanobilin (PCB), which is abundant in this phylum given its use as an antennae pigment in photosynthesis. Like PΦB in plant Phys, PCB is synthesized by reduction of BV and then becomes covalently linked through a positionally conserved cysteine in the GAF domain. PCB differs from PΦB by containing an ethyl side chain at the C18 position (instead of a vinyl for PΦB), which hypsochromically shifts absorption (Figures 2A and 2C). In contrast, proteobacterial and fungal Phys use BV directly, which is tethered via a thioether linkage to a cysteine upstream of the PAS domain; the additional double bond at the C3-C31 position of BV bathochromically shifts absorption (Figures 2A and 2C). For some proteobacteria, the gene encoding the heme oxygenase that synthesizes BV is within the same operon as the Phy apoprotein, thus genetically connecting synthesis of the two moieties (Bhoo et al., 2001; Karniol and Vierstra, 2003). Regardless of the linkage used, the bilins in proteobacterial and cyanobacterial Phys are positioned similarly in the GAF domain pocket (Figure 3D).
Figure 3.
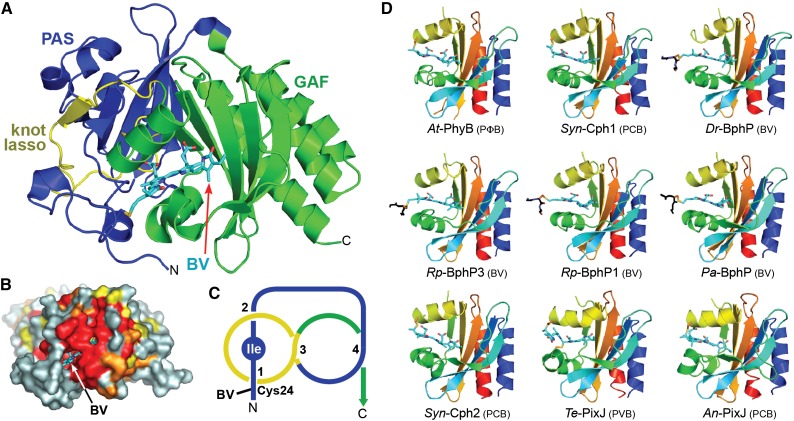
3D Structures of the Bilin Binding Region in Phys.
(A) Ribbon diagram of the PAS-GAF region of Dr-BphP (see Protein Databank [PDB] codes 1ZTU, 2O9C, and 4Q0H). Shown are the PAS (blue) and GAF (green) domains and the knot lasso (yellow). BV (arrow) is displayed in cyan with the nitrogens and oxygens colored in blue and red, respectively. N, N terminus; C, C terminus. The sulfur moiety in Cys-24 that forms the thioether linkage with BV is in yellow.
(B) Surface view of the PAS-GAF region of Dr-BphP showing the buried chromophore. Residues with 90, 75, and 60% identity within the Phy superfamily are colored in red, orange, and yellow, respectively. BV (arrow) is displayed in cyan.
(C) Diagram of the figure-of-eight knot connecting the PAS (blue) and GAF domains (yellow) in Dr-BphP. The isoleucine at the nexus of the knot is labeled.
(D) Ribbon diagrams of GAF domains in their dark-adapted states from representative canonical Phys (At-PhyB, PDB code 4OUR; Syn-Cph1, 2VEA; and Dr-BphP, 2O9C), bathy-Phys (Pa-BphP, 3C2W; Rp-BphP1, 4GW9), a Pnr Phy (Rp-BphP3, 2OOL), a PAS-less Phy (Syn-Cph2, 4BWI), and CBCRs (Te-PixJ, 4GLQ; An-PixJ, 3W2Z). Polypeptide chains are displayed in rainbow with the N-terminal ends in blue and the C-terminal ends in red. Bilins are colored in cyan, and the cysteine(s) forming the thioether linkage(s) are in orange with the sulfur atoms in yellow. A portion of the polypeptide for proteobacterial Phys that adjoins the site of thioether linkage is included and shown in black.
([A] and [B] are adapted from Wagner et al. [2005], Figures 1B and 4A.)
What has become clear through domain alignments and photochemical studies is that organisms from multiple kingdoms of life have evolved a dizzying panoply of structural and spectral Phy variants in addition to those first seen in seed plants. Some species retained the canonical PAS-GAF-PHY architecture of the PSM but employ photocycles substantially different from the plant paradigm (Figures 1 and 2C). These include algal forms that have adapted to sense different color ranges (Rockwell et al., 2014); bathy-Phys in proteobacteria that use the canonical architecture but prefer Pfr as the dark-adapted state and need far-red light to generate a photoactivated Pr state (Pseudomonas aeruginosa BphP, Rhodopseudomonas palustris BphP1, Bradyrhizobium ORS278 BphP, and Agrobacterium tumefaciens BphP2; Giraud et al., 2002; Karniol and Vierstra, 2003; Tasler et al., 2005; Yang et al., 2008; Bellini and Papiz, 2012); proteobacterial forms that photoconvert from Pr to a shorter wavelength-absorbing Pnr species (e.g., Rp-BphP3; Giraud et al., 2005; Yang et al., 2007); neochromes from mosses, hornworts, and ferns (e.g., Adiantum capillus-veneris NEO1) that retain the canonical PSM in series with light/oxygen/voltage sensor domains that bind flavins (Nozue et al., 1998; Suetsugu et al., 2005; Li et al., 2014); and a proteobacterial Phy (Rhodospirillum centenum Ppr) fused with a photoactive yellow protein domain that incorporates p-hydroxycinnamic acid for blue light sensing (Jiang et al., 1999). Cyanobacterial Phys with divergent PAS-less PSMs are also present that retain full photochromicity with just the GAF and PHY domains in the absence of the PAS domain (Synechocystis [Syn]-Cph2 and Synechococcus OSB’ [SyB]-Cph; Ulijasz et al., 2010; Anders et al., 2013).
The most distinctive members of the Phy superfamily with respect to domain composition and spectral variety are the cyanobacteriochromes (CBCRs), which as the name implies are common among cyanobacteria (Hirose et al., 2008; Ikeuchi and Ishizuka, 2008; Ulijasz et al., 2009; Rockwell et al., 2011, 2014). CBCRs typically require only their GAF domains for full photoconversion and by modification of the bilin binding pocket have spectrally tuned PCB absorption to accommodate a rich array of end-state colors, ranging from violet/blue, violet/orange, blue/green, green/red, to red/green (Figures 1 and 2C). For example, by autocatalytically isomerizing the π-conjugation system of the bound PCB to generate phycoviolobilin combined with a second, light-labile thioether linkage to the C10 carbon, CBCRs like Thermosynechococcus elongatus PixJ interconvert between blue and green-light-absorbing end states (Ishizuka et al., 2011; Rockwell et al., 2012b; Burgie et al., 2013) (Figures 2B and 2C). Some CBCRs harbor multiple GAF domains linked in tandem; each domain has the potential to bind their own bilin with different spectral end states, thus enabling integration of multiple light signals simultaneously (Nostoc punctiforme F2854 and Fd-Ifla; Chen et al., 2012; Rockwell et al., 2012a; Bussell and Kehoe, 2013; Figure 1).
Microbial Phys typically terminate in prototypic histidine kinase domains that would presumably initiate signaling by phosphotransfer from an active-site histidine to the aspartate in a cognate response regulator (Yeh et al., 1997; Karniol and Vierstra, 2006; Auldridge and Forest, 2011). Some Phys, especially those in filamentous fungi (Blumenstein et al., 2005; Froehlich et al., 2005), even contain the response regulator sequence translationally appended downstream of the histidine kinase domain, thus encouraging intramolecular transfer (Figure 1). However, other signaling motifs are also employed, including diguanyl cyclase, diguanylate phosphodiesterase, methyl-accepting, phosphatase 2C, two-helix output sensor, and histidine kinase/methyl-accepting/phosphatase (HAMP) domains, demonstrating that the Phy-type PSM is amenable to actuating a wide variety of other signaling pathways (Figure 1). In fact, it is possible to attach the PSM of Phys to non-native signal output motifs to invent novel light-responsive pathways (Gasser et al., 2014; Ryu et al., 2014). Phenotypic outputs from these microbial Phys include the regulation of pigmentation, photoprotection, redox sensing, phototaxis, sporulation, entrainment of circadian rhythms, and the chromatic acclimation of the photosynthetic machinery to the available light spectrum (Auldridge and Forest, 2011; Bussell and Kehoe, 2014). In one extreme case of adaptation, several R. palustris strains have evolved a photochemically inert variant of canonical BphPs that no longer binds BV and functions instead in redox sensing (Vuillet et al., 2007).
3D STRUCTURES OF BACTERIAL PHYs
Important to our understandings of how Phys work at the atomic level, microbial members often express well in Escherichia coli-based systems and provided for the first time samples with sufficient integrity, purity, and quantity for many biophysical and structure-based analyses (Bhoo et al., 2001; Gambetta and Lagarias, 2001). Either native or chemical variants of the bilin can be added in pure form to the recombinant Phy apoprotein (Karniol et al., 2005; Inomata et al., 2009), or photochemically active chromoproteins can be assembled in vivo by expression of the PHY gene with those encoding the appropriate bilin biosynthetic enzyme(s) for BV or PCB (Bhoo et al., 2001; Gambetta and Lagarias, 2001).
Using such recombinant preparations, Wagner et al. (2005, 2007) generated by x-ray crystallography the first 3D structures of the bilin pocket using the PAS-GAF region of a proteobacterial Phy from Deinococcus radiodurans (Dr-BphP) assembled with BV (Figure 3A). The models described the PAS and GAF domain folds and confirmed prior biochemical studies showing that the bilin becomes bound by a thioether linkage between Cys-24 upstream of the PAS domain and the C32 carbon of the A-ring vinyl group (Lamparter et al., 2003; Figures 2A and 4C). Unexpectedly, the structures revealed that the chromophore is almost completely buried within the GAF domain cleft in a contorted 5Zsyn, 10Zsyn, 15Zanti (ZZZssa) configuration with the D ring tilted substantially out of the plane created by the A-C rings (Figures 3A and 3B). The bilin is firmly anchored by electrostatic contacts between surrounding amino acids and the B and C ring propionates that project deep into the GAF cleft. Space can be observed around the D ring, which presumably would facilitate its photoisomerization-induced flip. The structures also identified a surprising figure-of-eight knot that strengthens the interactions between the PAS and GAF domains. The knot is formed by a loop extending between strand β4 and helix α4 of the GAF domain that lassos the polypeptide chain upstream of the PAS domain with a conserved isoleucine (Ile35) at its nexus (Figure 3C). Such knots are rare and imply that complicated gymnastics are required to properly fold the Phy apoprotein prior to bilin attachment (Wagner et al., 2005). The Dr-BphP PAS-GAF structures also revealed an unanticipated dimerization interface involving the GAF domain helices α1, α2, and α6, indicating that the PSM also contributes to assembly of the Phy dimer (Wagner et al., 2007).
Figure 4.
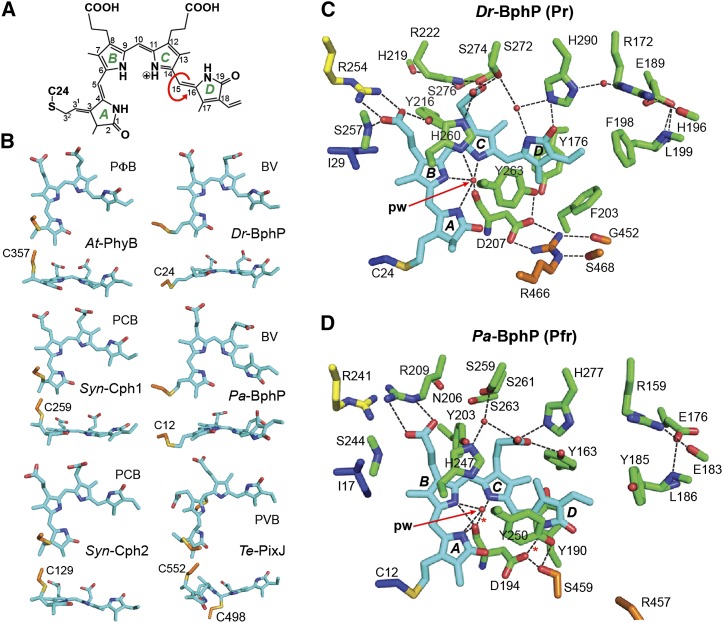
Structure of the Bilin and Bilin Binding Pocket in Representative Phys.
(A) Chemical diagram of BV bound to Dr-BphP. The pyrrole rings (A-D), carbon positions, and location of the Cys-24 linkage are labeled. Arrow shows the location of the prototypic Z to E isomerization of the bilin during photoconversion.
(B) Conformation in side and top views of the bilin bound to representative Phys in their dark-adapted states. The bilin type is indicated for each Phy. The cysteine(s) that links the bilin via a thioether bond is shown.
(C) and (D) Bilin and surrounding amino acids in the canonical Phy Dr-BphP (C) and the bathy-Phy Pa-BphP (D) in their dark-adapted Pr and Pfr end states, respectively (PDB codes 4Q0J and 3C2W). Amino acids in the PAS domain and upstream region, GAF domain, PHY domain, and the knot lasso are indicated in blue, green, orange, and yellow, respectively. The sulfur moiety in the cysteine forming the thioether linkage with the bilin is in yellow. Fixed waters are in red spheres. pw, pyrrole water. Dashed lines indicate hydrogen bonds.
Following the Dr-BphP PAS-GAF model, there has been a bevy of additional 3D structures from an assortment of recombinant Phy fragments, which have provided further clues about signaling. Included are x-ray crystallographic and/or solution 2D-NMR structures of the GAF domain from PAS-less Phys (Cornilescu et al., 2008; Anders et al., 2013), a Phy that photoconverts from Pr to the Pnr state (Yang et al., 2007), and CBCRs (Burgie et al., 2013; Narikawa et al., 2013; Cornilescu et al., 2014), crystal structures of the entire PSM region (PAS-GAF-PHY) from canonical Phys (Essen et al., 2008; Burgie et al., 2014b; Takala et al., 2014) and bathy-Phys (Yang et al., 2008; Bellini and Papiz, 2012), and even images of entire Phy dimers by small-angle x-ray scattering (Evans et al., 2006) and single-particle electron microscopy (SPEM) (Li et al., 2010). Particularly informative have been paired structures of the dark-adapted and photoactivated states (Ulijasz et al., 2010; Cornilescu et al., 2014; Takala et al., 2014), comparisons of canonical and bathy-Phys (Yang et al., 2009), and solid-state NMR analyses of the bilin and temperature-scanning cryocrystallography following sample irradiation (Song et al., 2011; Yang et al., 2011) that have collectively illuminated the structural changes associated with photointerconversion. As will be described below, these models provided unifying principles related to Phy architecture, photochemistry, and signaling mechanisms, as well as demonstrated how subtle modifications of key chromophore/protein contacts have been exploited by the superfamily to generate the rich diversity of spectral end states from a common scaffold.
GAF Domain and Bilin Architecture
All available Phy 3D structures show that the signature GAF domain provides the primary interactions with the chromophore. Its C-shaped configuration forms a deep crevice, framed by a six-stranded β-sheet (3-2-1-6-5-4 β-strand ordering) and helices α2-α4, which intimately cradles the bilin (Figure 3A). On the opposite face of the β-sheet is a three-helix bundle (helices α1, α5, and α6) that often creates the core surface for PSM dimerization (Wagner et al., 2007; Yang et al., 2007, 2008). As evidenced by the montage of GAF domains shown in Figure 3D, the main-chain conformation of this domain is remarkably conserved within the superfamily with strong sequence identity near the chromophore. The net effect is to restrain the bilin in remarkably similar configurations (typically ZZZssa) even among Phys with substantially different spectral end states and linkage sites (e.g., the blue/green CBCR Te-PixJ versus the canonical red/far-red Phy Dr-BphP; Figure 4B). For Phys with canonical PSMs, structures of the PAS-GAF region revealed that the knot lasso generates a tight water-excluded interface with the PAS domain and directs the sequence upstream of the PAS domain toward the bilin regardless of whether the bilin is attached by a cysteine within this sequence or by a cysteine within the GAF domain (Wagner et al., 2005; Essen et al., 2008; Yang et al., 2008; Burgie et al., 2014b). Other than the absence of the knot lasso, the GAF domain folds from the two representative CBCRs diverge only subtly from those of canonical Phys (Burgie et al., 2013; Narikawa et al., 2013); the largest alteration is in the region surrounding helix α4, which results in a more fully enveloped chromophore (Figure 3D). This increased coverage might help CBCRs maintain full photointerconversion between their end states in the absence of flanking PAS and PHY domains.
As illustrated by the high-resolution structures of the PAS-GAF region from Dr-BphP in its dark-adapted Pr state (Wagner et al., 2007; Burgie et al., 2014b), the chemical features of the contorted bilin dovetail extensively with complementary surfaces in the GAF pocket (Figure 4C). For example, the negatively charged propionates extending from the B and C pyrrole rings are well anchored as Pr via electrostatic interactions with a set of conserved arginine/histidine/serine residues (e.g., using Dr-BphP numbering: Arg-222, Arg-254, Ser-257, His-260, Ser-272, and Ser-274). In most of the Phys studied (the known exceptions being CBCRs with green/red photocycles; Hirose et al., 2013), all four pyrrole nitrogens of the bilin are protonated in the dark and photoactivated states (Strauss et al., 2005; von Stetten et al., 2007; Ulijasz et al., 2008; Burgie et al., 2014b). For canonical Phys, the crescent configuration of the A-C pyrrole moieties as Pr are in turn stabilized by an extensive hydrogen bond lattice involving the protonated nitrogens, an adjacent imidazole nitrogen (His-260), and the main-chain oxygen of an aspartate (Asp-207) within a nearly invariant Asp-Ile-Pro (DIP) motif, with a uniquely fixed pyrrole water as its focal point (Figure 4C). The pyrrole water presumably aids the protonation/deprotonation cycle envisioned for the bilin during Pr-to-Pfr photoconversion (von Stetten et al., 2007). The D-ring itself is chemically asymmetric with aliphatic methyl and vinyl groups on one side of the methine bridge and hydrophilic pyrrole nitrogen and carbonyl moieties on the other. The aliphatic face is flanked by a compatible environment provided by conserved hydrophobic residues (e.g., Tyr-176, Phe-198, Phe-203, and Tyr-263), whereas the hydrophilic face hydrogen bonds with an ordered water and the imidazole of an invariant histidine (His-290). Not surprisingly, mutational studies showed that most, if not all, of the conserved amino acids surrounding the bilin are critical to proper photochemistry (Fischer and Lagarias, 2004; Hahn et al., 2006; Yang et al., 2007, 2008; Wagner et al., 2008; Ulijasz et al., 2009; Auldridge et al., 2012; Burgie et al., 2013, 2014b). In fact, a number of variants impacting the chromophore binding pocket fail to photoconvert to Pfr in red light and instead are highly fluorescent. Of special importance is the central DIP aspartate (Asp-207) where even the subtle glutamic acid substitution in Dr-BphP and other Phys abolishes photoconversion.
A proposed key step in photoconversion for most Phys involves a light-driven Z to E isomerization of the C15=C16 double bond in the bilin that flips the D pyrrole ring (Rüdiger et al., 1983; Song et al., 2011; Yang et al., 2011). The immediate consequences of this flip can now be seen from the paired solution 2D NMR and crystal structures of the CBCR Te-PixJ (Burgie et al., 2013; Narikawa et al., 2013; Cornilescu et al., 2014) and by comparing the PSM models for canonical and bathy-Phys in their dark-adapted Pr and Pfr end states, respectively (Yang et al., 2009, 2011; Anders et al., 2013; Burgie et al., 2014b). Using Dr-BphP for illustration, rotation of the D ring would expose its functionalities to a non-ideal environment, which recovers by sliding of the bilin within the GAF pocket. Whereas the pyrrole nitrogen of the flipped D-ring assumes a large displacement to hydrogen bond with the carboxylate of Asp-207, the aliphatic functional groups remain nestled in their hydrophobic environment, which shifts commensurately by rotamerization of the Tyr-176 and Phe-203 side chains (Yang et al., 2009). As revealed by spectroscopic and crystallographic analyses of the Asp-207 mutants (Hahn et al., 2006; von Stetten et al., 2007; Wagner et al., 2008; Auldridge et al., 2012; Burgie et al., 2014b), this D-ring/Asp207 connection is critical for acquisition of the Pfr state as well as for preventing incorporation of more cyclic tetrapyrroles into the GAF domain pocket.
Bilin sliding induces a striking reorientation of the B and C ring propionates (Figures 4C and 4D). Whereas the B-ring propionate transfers its hydrogen bonding from one arginine (Arg-254) to another (Arg-222), the C-ring propionate repositions substantially to directly engage His-290 and the newly arranged Tyr-176. As vividly illustrated by the photoconversion of the CBCR Te-PixJ from its blue to green light-absorbing end states (Cornilescu et al., 2014), the net effect is to substantially displace the bilin across the main-chain ribs in the GAF domain β-sheet (Figure 7A). Notably, toggling between the end-state positions appears to be partially delineated by the knobby features of the bilin articulating between these ribs. The importance of this sliding is supported by spectroscopic and crystallographic analyses of GAF-domain mutants, especially those surrounding the D-ring or providing electrostatic contacts to the propionates moieties. Examples include amino acid substitutions predicted to stabilize and destabilize the propionate positions as Pr, which in turn generated variants with short-lived or locked Pfr states, respectively (Burgie et al., 2014b). It is even possible to generate a Phy with more typical spectral behavior (i.e., Pr dark-adapted state) from a bathy-Phy by a single amino acid substitution near these contact points (Yang et al., 2008, 2009).
Figure 7.
Conformational Changes Associated with Phy Photoconversion.
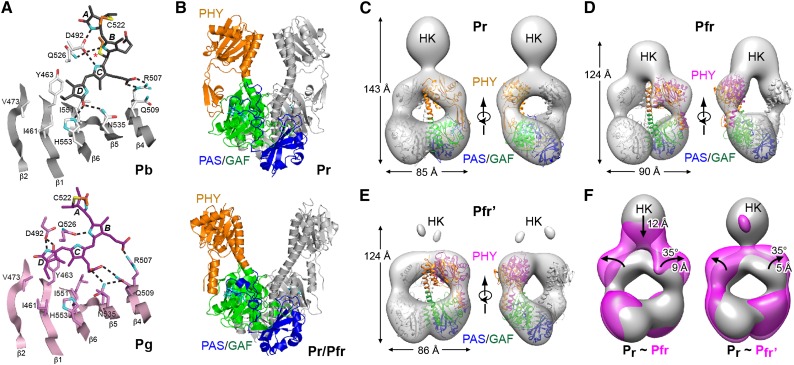
(A) Photoconversion of Te-PixJ from Pb to Pg drives bilin sliding within the GAF pocket. Shown are portions of the GAF domain β-sheet that impinges upon the bilin. Selected residues that contact the bilin directly are labeled. Hydrogen bonds are indicated with dashed lines. Pb and Pg carbons are colored white and pink, respectively, with the exception that cysteine carbons of the thioether linkages are colored orange. Carbons of Pb and Pg state bilins are colored gray and magenta, respectively. Oxygens, red; nitrogens, cyan; sulfurs, yellow. Asterisk identifies the light-labile thioether linkage.
(B) Paired Pr (4Q0J) and Pr/Pfr mixed structures (4O01) of the PSM from Dr-BphP showing the PAS (blue), GAF (green), and PHY (orange) domains.
(C) to (E) Negative-staining SPEM images of full-length Dr-BphP in its dark-adapted Pr and photoactivated Pfr end states. The crystal structures of the corresponding Pr (PDB ID code 4Q0J) and red-light-treated states (4O01) were superposed on the Pr (C) and Pfr/Pfr’ models ([D] and [E]). Each micrograph is shown in two orientations, and height and width of the respective species are indicated. Domain coloring of the crystal structures is the same as in (B). The positions of the PHY domains in (D) and (E) were adjusted to better fit the SPEM density (magenta).
(F) Superimposed SPEM density of the Pr versus the Pfr (left) and Pfr’ conformers (right). The position of the histidine kinase domain is indicated (HK). Arrows highlight the direction of displacement of the PHY or HK domains from the Pr state to the Pfr or Pfr’ states.
([A] is adapted from Cornilescu et al. [2014], Figure 4; [B] from Takala et al. [2014], Figure 2A, and Burgie et al. [2014b], Figure 5A; and [C] to [F] from Burgie et al. [2014b], Figures 7C to 7F.)
Although such movements might represent the prototypic photoconversion route, it appears that species-specific nuances and alternative pathways are possible. For example, to help anchor the B-ring propionate, some proteobacterial Phys employ a tyrosine that is not conserved in their plant or cyanobacterial relatives (e.g., Tyr-216 of Figure 4C; Wagner et al., 2007). More strikingly, photoconversion of CBCRs such as Te-PixJ with blue/green-light-absorbing end states also involves rupture of the second cysteine linkage, which would untether the bilin in addition to extending its π-conjugation system (Ishizuka et al., 2011; Rockwell et al., 2011; Burgie et al., 2013; Narikawa et al., 2013; Cornilescu et al., 2014). CBCRs with green/red photocycles include a deprotonation step during photoconversion in addition to a proposed D-ring rotation (Hirose et al., 2013). Additionally, detailed solution 2D-NMR analyses of the GAF domain of the PAS-less Phy SyB-Cph1 in its Pr and Pfr end states implied that the A pyrrole ring rotates during photoconversion of this Phy with only minor movements at the D-ring (Ulijasz et al., 2010). More recent solid-state NMR studies of the same fragment also showed that the most robust chemical shift changes during photoconversion occur around the A-ring as opposed to the D-ring (Song et al., 2014). We believe that these observations bolster the possibility of an A-ring rotation even though the authors concluded otherwise. Clearly, additional studies, including the paired crystal structures of the SyB-Cph1, are needed to resolve this issue.
Structure of the PHY Domain and Associated Hairpin
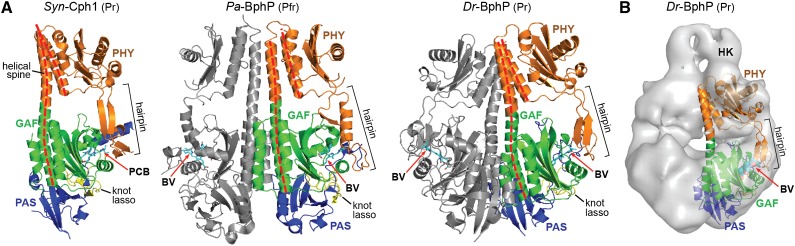
In canonical and PAS-less Phys, the GAF domain is immediately followed by a single PHY domain. The PHY domain is configured much like the GAF domain (and in some cases too similar to be considered distinct; Anders et al., 2013), including a central five-stranded β-sheet with an similar core β-strand ordering (2-1-5-4-3; Figure 5A). α-Helices flank both sides of the β-sheet. To one side are the main segments of a striking helical spine, which comprises the exiting helix α6 from the GAF domain that transitions into the PHY domain, and a second α-helix that connects the PHY domain to the OPM. For the Dr-BphP PSM, this spine includes 18 helical turns spanning at least 72 Å and likely extends further into the OPM (Figure 5A). Notably, the longer α-helix connecting the GAF and PHY domains is substantially bowed in the PSM crystal structures of Dr-BphP (Burgie et al., 2014b; Takala et al., 2014), which may have importance to signaling at least for this Phy (see below). Although the core PHY fold generates a C-shaped architecture similar to that seen in the GAF domain, to date no associated prosthetic group has been detected.
Figure 5.
Structure of the PSM from Representative Phys in Their Dark-Adapted States.
(A) Ribbon diagrams of PSMs from Syn-Cph1 (PDB code 2VEA), Pa-BphP (3C2W), and Dr-BphP (4Q0J) in their dark-adapted states. Helical spine is highlighted by the dashed red lines. Hairpin is located by the bracket. The paired subunits of the Pa-BphP and Dr-BphP dimers are in gray. Other coloring is as in Figure 3. The spectral state of each structure is indicated in parenthesis.
(B) SPEM model of the full-length Dr-BphP dimer. Only a portion of the histidine kinase domain was imaged due to conformational flexibility. The crystal structure of the PSM is included for one subunit with the space-filling model of the bilin shown.
([A] is adapted from data presented in Essen et al. [2008], Figure 1B; Yang et al. [2008], Figure 1A; and Burgie et al. [2014b], Figure 5A.)
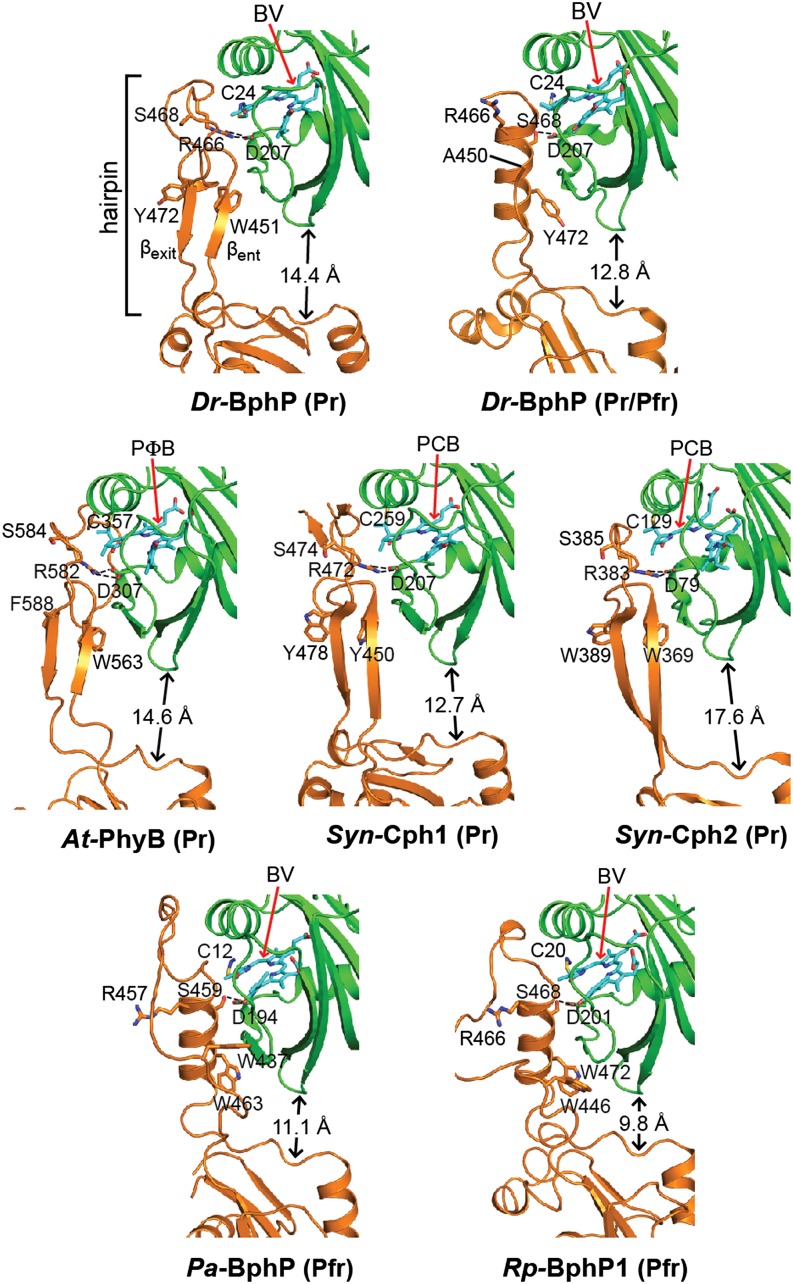
The PHY domain is also connected noncovalently to the GAF domain through a remarkable hairpin (or tongue/arm; Essen et al., 2008; Yang et al., 2008) feature that emanates from between strand β5 and helix α6 and projects toward the GAF domain surface near the chromophore binding site (Figure 5A). For canonical Phys with dark-adapted Pr states, the stem of the hairpin is created by two antiparallel β-strands designated βent and βexit (Figure 6). They loosely associate with the GAF domain as a continuation of its β-sheet and make several conserved contacts near the bilin, including a salt bridge between the arginine (Arg-466 in Dr-BphP) in a conserved Pro-Arg-X-Ser-Phe (PRXSF) motif and the DIP aspartate (Asp-207), and a hydrophobic interaction between the tryptophan in a conserved Trp-Gly-Gly (WGG) motif and a hydrophobic pocket formed at the junction of strand β3 and helix α2 of the GAF domain (Figure 6). Whereas the hairpin stems for currently available Pr models show strong similarity in conformation and sequence, the intervening loops differ substantially in length, sequence, and GAF-domain contact points, suggesting that this loop functions in a species-specific manner (Essen et al., 2008; Anders et al., 2013; Burgie et al., 2014a, 2014b).
Figure 6.
Conformation of the PHY-Domain Hairpin from Representative Phys and Its Interaction with the GAF Domain.
Ribbon diagrams of crystal structures were drawn from Dr-BphP as Pr (PDB ID code 4Q0J) and a mixed Pr/Pfr state (4O01), Syn-Cph1 as Pr (2VEA), Syn-Cph2 as Pr (4BWI), At-PhyB as Pr (4OUR), Pa-BphP as Pfr (3C2W), and Rp-BphP1 (4GW9) as Pfr. The bilin type (arrow) and the spectral state of each parent model are indicated. The coloring is as in Figure 3. Side chains are shown for relevant amino acids. Dashed lines highlight hydrogen bond contacts between the DIP (Asp-Ile-Pro) motif aspartate in the GAF domain and either the conserved arginine or serine residues in the PRXSF motif from the hairpin stem. The distance separating the GAF and PHY domain globular regions is shown; it was measured from the loop separating the β1 and β2 strands of the GAF domain and the α carbon of a conserved tryptophan (Trp-483 in Dr-BphP) just proximal to the exiting α-helix of the PHY domain. βent and βexit label the entrance (N-terminal) and exit (C-terminal) β-strands in the hairpin. (Adapted from Burgie et al. [2014b], Figure 6.)
Given the novelty of the hairpin, it was immediately expected to be pivotal to signaling by PHY-domain bearing Phys (see Anders et al., 2013). First clues came from the structural solution of two bathy-Phys in their dark-adapted Pfr states (Yang et al., 2008; Bellini and Papiz, 2012). Here, a striking difference in stem conformation was evident compared with canonical Phys in their dark-adapted Pr states. Instead of forming a two-stranded β-sheet, the entrance and exit hairpin stem sequences were rotated 180° and assumed disordered and α-helical conformations, respectively (Figure 6). This swivel in turn modified the interactions between the hairpin stem and GAF domain, the most substantial of which was an alternative nest of hydrogen bonds as the serine within the PRXSF motif bound both the DIP aspartate and a conserved helix α5 tyrosine (Tyr-263 in Dr-BphP) to effectively replace the arginine/aspartate salt bridge. A new hydrophobic contact was also created that replaced the WGG contact with the GAF domain. Here, a hydrophobic residue, “Hb” (Trp451 in Dr-BphP) within the conserved Hb-X-Glu (HbXE) motif of the hairpin specifically bound the Pfr GAF domain surface after displacement of the βent strand (Figure 6).
Based on these structural comparisons and additional studies with the PAS-less Phy Syn-Cph2, Anders et al. (2013) proposed a “tryptophan switch” mechanism for hairpin-containing Phys whereby photoactivation melts the β-strand interaction between the hairpin stem and the GAF domain, and then the stem swivels and reforms a new set of interactions with the GAF domain and the now helical hairpin, one of which involves a switch in hairpin tryptophan/GAF domain contacts. The presumed end result is to alter the orientation of the PHY domain relative to the GAF, which would reverberate into the OPM. In support of this hairpin conformational switch, mutational studies on several Phys showed that the hairpin/GAF domain interactions are critical to Phy chemistry despite limited contact with the chromophore. For example, single amino acid substitutions within the PRXSF, WGG, and HbXE motifs either preclude photoconversion of Pr to Pfr, or enhance or stall thermal reversion if the Pfr state was generated, consistent with their predicted ability to impact the hairpin/GAF domain interface as Pr or Pfr (Essen et al., 2008; Anders et al., 2013, 2014; Burgie et al., 2014a, 2014b).
More convincingly, Takala et al. (2014) succeeded in solving low-resolution but informative crystal structures of the Dr-BphP PSM in both its dark-adapted Pr state and following red light irradiation. Whereas the Pr hairpin stem had β-strand character as seen in the more resolved Dr-BphP structure (Burgie et al., 2014b), the stem of the mixed Pr/Pfr species studied was obviously helical. Although the core folds of the internally rigid PAS, GAF, and PHY domains appeared unaffected, the gap between the GAF and PHY domains became significantly shorter, presumably fostered by the hairpin swivel and contraction. The net effect would be to pull the GAF and PHY domains together at this noncovalent tether.
Signal Transmission within the Phy Dimer
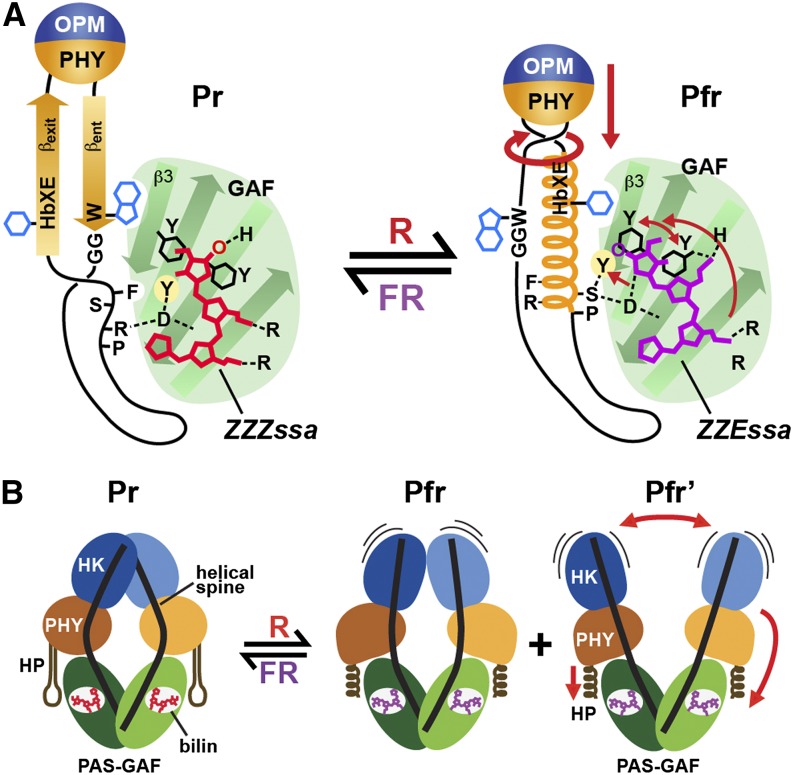
Clearly, the next question is: How would bilin sliding and hairpin reconfiguration affect the OPM and subsequent signaling? Canonical PSMs are generally thought to dimerize in a head-to-head fashion such that the helical spines of sister subunits are roughly parallel (Wagner et al., 2007; Yang et al., 2007, 2008; Bellini and Papiz, 2012; Burgie et al., 2014b; Takala et al., 2014) (Figure 5A). This arrangement is clearly advantageous for Phys in which the OPMs need to work in concert, such as two-component sensors that often require phosphorylation in trans across the paired histidine kinase domains for signal output. Both the crystal structures of the PSM and cryo-SPEM models of the full-length Dr-BphP dimer as Pr support this head-to-head arrangement and, interestingly, revealed a substantial bow in the opposing helical spines coupled with a slight left-handed twist (Li et al., 2010; Burgie et al., 2014b; Takala et al., 2014). The outward splay of each spine begins after exiting the GAF domain with the return generated by a prominent kink involving Ala-326 at the helical junction between the GAF and PHY domains (Figure 5). The end result is to create a sizable cavity (∼20 Å) at the center of the dimer with only the PAS-GAF and histidine kinase domains providing contact points (Figure 5B).
By contrast, the crystal structure of the Dr-BphP PSM after photoactivation revealed a substantial straightening of the helical spine at the Ala-326 kink, presumably caused by light-induced transformation of the hairpin/GAF domain interaction pulling the PHY domain and its helical spine into a more open configuration (Takala et al., 2014) (Figure 7B). Negative-staining SPEM images of full-length Dr-BphP supported this deformation and revealed that the open configuration for the PHY domain broadens the dimer by increasing the diameter of the cavity and downwardly displacing the OPMs (Figures 7C to 7E). In fact, two Pfr conformations could be detected, one in which the paired OPMs remained in contact, albeit with a modified interface, and another in which the OPMs appeared to separate from each other and become more dynamic. If we assume that Dr-BphP participates in a two-component kinase cascade with Pfr being more active, then this light-driven separation of the OPMs should encourage phosphotransfer in trans (or possibly promote a switch to phosphotransfer in cis; Casino et al., 2009), simply by physically altering the relative positions of the paired histidine kinase domains to a more productive catalytic state.
Whereas such movements might be widely employed by canonical, hairpin-containing Phys, alternative mechanisms are possible for Phys without this feature (CBCRs) or that use OPMs where pairing is not essential for signal output. For example, it appears that the head-to-head arrangement seen in most Phys is not universal. The bathy-Phy Rp-BphP1 likely forms a head-to-tail dimer interface using the opposing PHY and PAS/PAC domains within the OPM to the exclusion of the PSM PAS and GAF domains (Bellini and Papiz, 2012). Similarly, the 3D structure of the GAF-PHY region from the PAS-less Phy Syn-Cph2 supports a head-to-tail dimer arrangement (Anders et al., 2013), which might be functionally significant as this quaternary architecture mimics the head-to-tail arrangement found in other diguanylate cyclase signaling proteins that exploit GGDEF-type OPMs. CBCRs do not have an obvious PHY domain and likely not a loop resembling the hairpin and thus should couple conformational changes in the bilin to their downstream OPMs by alternative mechanisms. For Te-PixJ, a light-driven swivel of the GAF domain helical bundle involved in dimerization could trigger rotation of the connected helical bundles found in its flanking HAMP domains (Cornilescu et al., 2014). Consequently, this CBCR could activate signaling through the gear box-type mechanism predicted for other HAMP domain-containing sensory receptors (Ferris et al., 2012).
STRUCTURAL RESOLUTION OF A PLANT PHY
With respect to the mode of action of plant Phys, it was unclear how relevant the bacterial Phy 3D structures would be given the substantial differences in sequence and domain architecture. Examples of such distinctions include a longer NTE in plant Phys beyond the PAS domain, pervasive sequence variation within the predicted PAS, GAF, and PHY domains, and a substantially reorganized OPM that includes tandem PAS domains followed by a histidine kinase-related domain. While similar to archetypical histidine kinase domains, including the presence of a consensus ATP binding site (Yeh and Lagarias, 1998), these histidine kinase-related sequences are notably missing the positionally conserved histidine essential for phosphotransfer (Figure 1). Analysis of plant Phy mutants and chimeras also suggested that unlike bacterial Phys, the OPM is not essential for signaling but serves only to enhance dimerization and possibly nuclear import (Krall and Reed, 2000; Matsushita et al., 2003; Kevei et al., 2007).
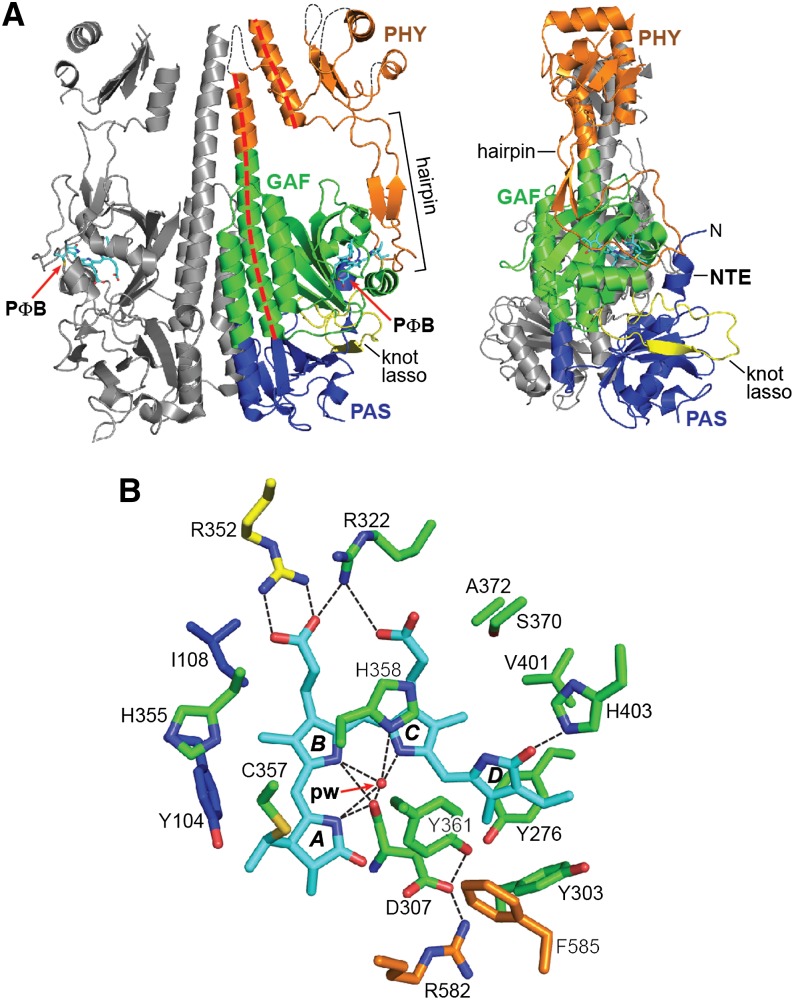
Burgie et al. (2014a) recently addressed this question through the first structural resolution of the PSM (minus the first 89 residues) from a plant Phy (Arabidopsis PhyB) assembled with PΦB and in its dark-adapted Pr state. As shown in Figure 8A, the overall 3D structure of the At-PhyB PSM is largely congruent with the PSMs of canonical bacterial Phys with PAS-GAF-PHY architectures and likewise includes a head-to-head dimer arrangement and obvious knot, hairpin, and helical spine features (Figures 3, 5C, and 8A). The bilin is similarly arranged in a ZZZssa configuration with the A-C rings mostly coplanar and the D-ring rotated out of this plane by 58° (Figure 4B). An analogous web of electrostatic and hydrophobic interactions that cradle PΦB in the binding pocket was also found, including the hydrogen bonding lattice involving the A-C ring nitrogens, Asp-307, His-358, and a similarly fixed pyrrole water (Figure 8B).
Figure 8.
3D Crystal Structure of the PSM from Arabidopsis PhyB.
(A) Ribbon diagram of the PSM with the PAS (blue), GAF (green), and PHY (orange) domains indicated. The knot-lasso is colored yellow, the helical spine is indicated with a dashed red line, and the hairpin is delineated with a bracket. Connectivity of the PHY domain in the disordered regions is illustrated with dashed black lines. The second subunit of the dimer is colored gray. PΦB is colored cyan. N, N terminus.
(B) Bilin and surrounding amino acids. NTE, GAF domain, knot lasso, hairpin, and PΦB carbons are colored blue, green, yellow, orange, and cyan, respectively. Amino acid side chains are labeled. Hydrogen bonds are indicated with black dashed lines. Oxygens, red; nitrogens, blue; and sulfurs, gold. pw, pyrrole water. (Adapted from Burgie et al. [2014a], Figure 2C.)
However, unique features were also seen in the At-PhyB PSM that are potentially relevant to signaling by the plant photoreceptors (Burgie et al., 2014a), some of which have been confirmed as critical by a plethora of engineered and natural variants, and mutations identified by forward genetic screens. The helical region of the PAS domain varies substantially from its bacterial counterparts and includes a large flexible loop at residues 145 to 155 (150s loop). Phenotypic analysis of Arabidopsis plants expressing the PhyB(Pro149-Leu) mutant implied that this loop is significant to plant Phy signaling without altering photochemistry, possibly by binding PIFs (Oka et al., 2008). Adjacent to the 150s loop is a conserved methionine (Met-159 in At-PhyB) that stabilizes residues 335 to 337 of the uniquely structured knot lasso via its hydrophobic interaction with the PAS domain surface. This distinctive plant-Phy knot is of significant interest as it has also been implicated in PIF binding (Kikis et al., 2009). The sequence connecting the PAS and GAF domains is also distinctive, as it forms a continuous α-helix that extends the helical spine to span the entire PSM (Figure 8A). In addition, the GAF domain contains a large hydrophilic loop (residues 379 to 393) of unknown function that we speculate might bind accessory or effector proteins.
Unlike bacterial Phys, plant Phys include a long glycine/serine-rich NTE beyond the PAS domain that is especially important for the thermal stability of the Pfr state (Burgie et al., 2014a) and proper biological activity, possibly via its light-dependent phosphorylation (Cherry et al., 1992; Medzihradszky et al., 2013; Nito et al., 2013). The 3D model of At-PhyB containing much of the NTE revealed an unexpected contact between this extension and PΦB that helps explain some of these effects. In particular, an α-helix encompassing residues 104 to 110 forms a steric barrier for the A and B pyrrole rings with residues Tyr-104 and Ile-108 directly abutting the bilin (Figure 8A). The direct interaction between the Tyr-104 side chain and the bilin is enlightening as this residue was previously shown to be key to At-PhyB nuclear localization and photoactivity in planta through its Pfr-dependent phosphorylation (Nito et al., 2013). Interestingly, Tyr-104 is buried by the hairpin in the At-PhyB Pr structure, suggesting that light-induced reorganization of the NTE/hairpin region facilitates its modification (Burgie et al., 2014a).
The PSM of At-PhyB is lined by a helical spine with a similar construction as those found in canonical bacterial Phys (Essen et al., 2008; Yang et al., 2008; Burgie et al., 2014a). Although the main α-helix of the spine is relatively linear, there is little contact between opposing PHY domains in the dimer via this feature (Figure 8A). The resulting diffuse density at the core PHY fold in the At-PhyB model suggests that the GAF/PHY domain connections, including the helical spine and hairpin, are relatively pliable within the dimer as one might expect if their interdomain movements are critical to photoconversion.
By contrast, the signature PHY hairpin was well defined and consistent between sister subunits with a central feature being the two antiparallel β-strand stem as seen in bacterial Pr-state Phys (Figure 6). Extensive site-directed mutagenesis within the conserved PRXSF, WGG, and HbXE motifs revealed that the hairpin significantly impacts At-PhyB photochemistry in a similar manner as in bacterial Phys. Strikingly, a nearly million-fold difference in Pfr to Pr thermal reversion rate could be generated by single point mutations in this region (Burgie et al., 2014a). For example, the Gly564-Glu substitution affecting the middle position in the hairpin WGG motif greatly slows Pfr thermal reversion. This mutation appears to provide a steric barrier against reformation of the Pr-type hairpin, which in turn could extend the lifetime of Pfr by favoring its helical hairpin conformation and Pfr binding pocket geometry. The biochemical effect of the Gly564-Glu mutation is consequential to PhyB signaling as it was previously shown that this allele creates a substantially hyperactive PhyB variant in planta (Kretsch et al., 2000; Ádám et al., 2011). Conversely, the Met579-Thr natural variant impacting the hairpin loop attenuates phenotypic signaling (Maloof et al., 2001). This threonine replacement likely stabilizes the hairpin/GAF domain contact for Pr by an adventitious interaction with His-355 and thus might enhance avidity of the Pr-type hairpin connection with the GAF domain to speed Pfr to Pr thermal reversion or potentially dampen Pr to Pfr photoconversion.
Of interest to plant Phy signaling is the ability of At-PhyB to bind PIFs via just the NTE-PAS-GAF region and that their Pfr-specific interactions appear independent of the histidine kinase-related domain (Shimizu-Sato et al., 2002; Kikis et al., 2009; Levskaya et al., 2009). We speculate that the increased affinity of PIFs for the Pfr state are instigated by rearrangement of the hairpin to expose a complementary PIF binding surface in the hairpin or elsewhere (e.g., exposing residue Tyr-104 of At-PhyB; Nito et al., 2013). Although the plant Phy OPMs might also contribute directly to signaling through their currently enigmatic kinase activity (Yeh and Lagarias, 1998), it appears that many of the phenotypic functions of plant Phys can be performed in the absence of this activity (Krall and Reed, 2000; Matsushita et al., 2003). These observations suggest that plant Phys have diverged significantly from bacterial Phys with bona fide histidine kinase activities and now exploit other features within the PSM for signaling despite appearing to share a common PSM architecture. Clearly, full appreciation of the necessary features within plant Phys and how and where on the Phy surface the downstream signaling partners such as PIFs interact now require 3D structures of an entire plant Phy dimer both alone and in association with these partners.
OVERVIEW OF PHY SIGNALING: THE TOGGLE MODEL
Although the details of Phy signaling at an atomic level are still in their infancy, we propose a toggle model for canonical Phy photoconversion that integrates the ensemble of chromoprotein structures and the “rotate and flip” and “tryptophan switch” mechanisms presented previously (Yang et al., 2009; Anders et al., 2013) (Figure 9). Using Dr-BphP as the blueprint, absorption of red light by the bilin induces a Z to E isomerization of the bilin that flips the D pyrrole ring. Release of its hydrogen bond with a proximal histidine (His-290) combined with its chemical asymmetry (with aliphatic and hydrophilic faces) places the D-ring in a nonideal environment. The bilin recovers by sliding within the GAF domain pocket, which is not followed by substantial changes in the GAF domain fold but by more subtle displacements of surrounding amino acids. Included are a new contact between the D-ring nitrogen and the carboxylate of Asp-207, substantially altered interactions between the B- and C-ring propionates and the bilin binding pocket, and rotation of the Tyr-176 and Phe-203 side chains.
Figure 9.
Toggle Model for Photoconversion of Canonical Phys That Translates Light into a Conformational Signal.
(A) Conformational changes within the GAF domain and hairpin associated with bilin photoisomerization and sliding and hairpin deformation using At-PhyB(Pr) and Pa-BphP(Pfr) as the examples. Upon light-induced rotation of the D pyrrole ring, the bilin breaks its D-ring/His-403 connection and slides (see arrow) within the GAF domain crevice to form a new contact between the D-ring and Asp-307 and the C-ring propionate and His-403. The tryptophan pair, Tyr-276 and Tyr-303 adjacent to the D-ring, rotate in directions opposite to the D-ring rotation. Together, the effects initiate a collision of Tyr-361 with Phe-585, breakage of the Asp-307/Arg-582 contact, and other changes that collectively release the hairpin stem from the GAF domain. The freed hairpin stem becomes helical, swivels, reforms a new contact between Asp-307 and Ser-584 in the PRXSF motif, and swaps the βent for a βexit/Phe-588 connection with the GAF-domain surface. The rotation and helical conversion of strand βexit presumably reorients the PHY domain relative to the GAF domain and/or tugs on the helical spine connecting the PHY domain to OPM (not shown) to eventually actuate signaling changes in the OPM. The conformations of the bilin in the two end states are indicated.
(B) Global conformational changes within the Dr-BphP Phy dimer. The PHY domains adjust their positions relative to their GAF domains through the light-induced conformational changes in the PHY domain hairpin stem from β-strand to helical followed by straightening of the bowed helical spines. Ultimately, the position and/or mobility of the paired OPMs are impacted to alter their signaling potential, in this case phosphotransferase activity by a histidine kinase domain (HK). HP, hairpin. Pfr and Pfr' represent the two photoactivated states observed by SPEM.
([A] is adapted from Burgie et al. [2014a], Figure 5; and [B] from Burgie et al. [2014b], Figure 11.)
While currently speculative, these combined motions impinge on Asp-207 and Tyr-263 (and potentially other residues), causing a conflict at the interface between the GAF domain surface and the hairpin (Figure 9). The weakened connection now enables the release of the hairpin (and its Arg-466/Asp-207 salt bridge) from the GAF domain. The freed hairpin rotates, its βent strand refolds to become helical, and subsequently the hairpin restores contact with the GAF domain surface through the Ser-468/Asp-207 hydrogen bond and additional hydrophobic contacts with its βexit strand and the GAF domain strand β3. Much like a twisting rope, the new hairpin connection would pull the outward side of the PHY domain toward the GAF domain through both translational and rotational motions that ultimately distorts the helical spine as seen for Dr-BphP at the Ala-326 hinge (Takala et al., 2014). The role(s) of the PAS domain is currently unclear, but one possibility is that its tight connection to the GAF domain via the knot provides a backstop to direct motion from the GAF domain toward the PHY domain.
As seen by the SPEM images of the full-length Dr-BphP dimer (Burgie et al., 2014b), the resulting splay of the sister PHY domains alters the topology of the paired OPMs in which the adjacent histidine kinase domains are either teased apart or completely released from each other. Through this exquisite cascade of events, the light signal is translated from subtle angstrom-scale conformational changes in the bilin into nanometer-scale movements of the paired PHY domains and ultimately into substantial destabilization/reorientation of the downstream OPMs. While the OPMs are the likely targets of this toggle motion in microbial Phys, for plant Phys, altered binding of partners like PIFs to the more upstream region might be the final outcome of this toggle to the Pfr end state.
TRANSLATION OF PHY STRUCTURES INTO PRACTICAL USES
Beyond revealing the basic principles of Phy signaling, the structural appreciation of these photoreceptors offers opportunities for engineering plant photomorphogenesis to benefit agriculture and to better exploit Phys as cell biological and optogenetic reagents. The findings that GAF-domain fragments bearing mutations in key bilin/protein contacts are highly fluorescent (e.g., Asp-207 mutants in Dr-BphP; Wagner et al., 2008) and that these fragments remain functional as tags have led to the current interest in Phys, particularly BV-containing bacterial versions, as cell biological reporters (Fischer and Lagarias, 2004; Wagner et al., 2008; Ulijasz et al., 2009). Their advantages over other commonly used reporters, such as the suite of green fluorescent protein derivatives, are their smaller monomeric sizes (220 amino acids), the ability to regulate chromophore availability and spectrally tune light absorption, and, importantly, the fact that the excitation and emission spectra of some are in the red/far-red regions of the visible spectrum, which extends their utility to otherwise opaque tissues or even whole organisms (Shu et al., 2009). Improvements have been made recently in fluorescence brightness and extension to near-infrared wavelengths through directed evolution, thus adding Phy mutant-based tags to the arsenal of fluorescence imaging tools (Filonov et al., 2011; Lecoq and Schnitzer, 2011; Auldridge et al., 2012; Yu et al., 2014).
In the rapidly expanding field of optogenetics, a variety of semisynthetic photoreceptors have been engineered to allow precise light control of various internal cellular processes both spatially and temporally. Here, plant Phys have been particularly useful by exploiting their ability to bind PIFs in a light-dependent fashion (Shimizu-Sato et al., 2002; Levskaya et al., 2005). Recent applications have included the fusion of the PAS-GAF-PHY-PAS-PAS region of At-PhyB to membrane tethers followed by coexpression with PIFs fused to various signaling factors, thus enabling the reversible subcellular recruitment of membrane-associated signaling events by light (Levskaya et al., 2009; Toettcher et al., 2013; Yang et al., 2013). Such optogenetic controls appear amenable to any organism simply by feeding the At-PhyB-expressing cells/organism either PΦB or PCB. Using the At-PhyB PSM structure as a blueprint (Burgie et al., 2014a), it should now be possible to redesign these photoreceptors with more desirable affinities for PIFs and longer or shorter retentions of the Pfr end state for more exacting control.
And last but not least, the At-PhyB PSM model now provides the structural backdrop for the rational redesign of plant Phys with new photochemical and signaling properties beneficial to plant science. At an experimental level, these modified forms could facilitate testing various models of plant Phy signaling (e.g., Rausenberger et al., 2011), while at the applied level, these new variants might have agricultural utility. This potential was first shown by the expression of the highly fluorescent, photochemical compromised Tyr176-His variant of At-PhyB (Su and Lagarias, 2007). When used to replace the wild-type version of PhyB in Arabidopsis, the PhyBY176H mutant induced photomorphogenesis even in dark-grown seedlings, indicative of constitutive PhyB signaling. While the extreme phenotype of this mutant limits its commercial value, Zhang et al. (2013) recently showed that more subtle variants could have better utility. For example, the structure-guided mutation Tyr361-Phe of At-PhyB, which is at the center of the D-ring flip and bilin sliding, generated Arabidopsis plants with relatively normal etiolated and green seedling phenotypes but now the plants were 50 times more sensitive to red light and with greatly suppressed shade avoidance in low light.
Clearly, using the available array of At-PhyB photochemical mutants (e.g., Maloof et al., 2001; Oka et al., 2008; Kikis et al., 2009; Ádám et al., 2011; Nito et al., 2013; Zhang et al., 2013; Burgie et al., 2014a) along with new mutational opportunities that should arise as more complete plant Phy models emerge, it should be possible to alter aspects of Phy signaling important to various agronomic processes such as seed germination, shade avoidance, and flowering time. For example, the suite of mutants dramatically impacting Pfr thermal reversion (Burgie et al., 2014a) will enable testing of the photoperiod measurement model that evokes thermal reversion of Phys as part of an hourglass mechanism. And by modification of plant Phy GAF domains to mimic the spectral properties of their CBCR or algal brethren, it might even be possible to color tune plant photomorphogenesis to wavelengths other than red and far-red light.
Acknowledgments
Work in our lab was supported by grants from the U.S. National Science Foundation (Grants MCB 1022010 and 1329956) and the UW College of Agricultural and Life Sciences (Hatch WIS01440) to R.D.V. We thank Adam Bussell, Erin Weber, and Robert Augustine for reviewing the article. We apologize in advance for the omission of other publications in this review due to space constraints.
AUTHOR CONTRIBUTIONS
E.S.B. and R.D.V. wrote the article and prepared the figures.
References
- Ádám É., Hussong A., Bindics J., Wüst F., Viczián A., Essing M., Medzihradszky M., Kircher S., Schäfer E., Nagy F. (2011). Altered dark- and photoconversion of phytochrome B mediate extreme light sensitivity and loss of photoreversibility of the phyB-401 mutant. PLoS ONE 6: e27250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Anders K., Gutt A., Gärtner W., Essen L.O. (2014). Phototransformation of the red light sensor cyanobacterial phytochrome 2 from Synechocystis species depends on its tongue motifs. J. Biol. Chem. 289: 25590–25600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders K., Daminelli-Widany G., Mroginski M.A., von Stetten D., Essen L.O. (2013). Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288: 35714–35725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auldridge M.E., Forest K.T. (2011). Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46: 67–88. [DOI] [PubMed] [Google Scholar]
- Auldridge M.E., Satyshur K.A., Anstrom D.M., Forest K.T. (2012). Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 287: 7000–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini D., Papiz M.Z. (2012). Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20: 1436–1446. [DOI] [PubMed] [Google Scholar]
- Bhoo S.H., Davis S.J., Walker J., Karniol B., Vierstra R.D. (2001). Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414: 776–779. [DOI] [PubMed] [Google Scholar]
- Blumenstein A., Vienken K., Tasler R., Purschwitz J., Veith D., Frankenberg-Dinkel N., Fischer R. (2005). The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15: 1833–1838. [DOI] [PubMed] [Google Scholar]
- Burgie E.S., Bussell A.N., Walker J.M., Dubiel K., Vierstra R.D. (2014a). Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. USA 111: 10179–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie E.S., Walker J.M., Phillips G.N.J. Jr., Vierstra R.D. (2013). A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21: 88–97. [DOI] [PubMed] [Google Scholar]
- Burgie E.S., Wang T., Bussell A.N., Walker J.M., Li H., Vierstra R.D. (2014b). Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J. Biol. Chem. 289: 24573–24587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell A.N., Kehoe D.M. (2013). Control of a four-color sensing photoreceptor by a two-color sensing photoreceptor reveals complex light regulation in cyanobacteria. Proc. Natl. Acad. Sci. USA 110: 12834–12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell, A.N., and Kehoe, D.M. (2014). Chromatic acclimation: a many-coloured mechanism for maximizing photosynthetic light harvesting efficiency. In The Cell Biology of Cyanobacteria, E. Flores and A. Herrero, eds (Poole, UK: Caister Academic Press), pp. 149–170. [Google Scholar]
- Butler W.L., Norris K.H., Siegelman H.W., Hendricks S.B. (1959). Detection, assay, and preliminary purification of the pigment controlling photoresponsive development in plants. Proc. Natl. Acad. Sci. USA 45: 1703–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casino P., Rubio V., Marina A. (2009). Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction. Cell 139: 325–336. [DOI] [PubMed] [Google Scholar]
- Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.O., van der Horst G.T., Batschauer A., Ahmad M. (2011). The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62: 335–364. [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhang J., Luo J., Tu J.M., Zeng X.L., Xie J., Zhou M., Zhao J.Q., Scheer H., Zhao K.H. (2012). Photophysical diversity of two novel cyanobacteriochromes with phycocyanobilin chromophores: photochemistry and dark reversion kinetics. FEBS J. 279: 40–54. [DOI] [PubMed] [Google Scholar]
- Cherry J.R., Hondred D., Walker J.M., Vierstra R.D. (1992). Phytochrome requires the 6-kDa N-terminal domain for full biological activity. Proc. Natl. Acad. Sci. USA 89: 5039–5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu C.C., Cornilescu G., Burgie E.S., Markley J.L., Ulijasz A.T., Vierstra R.D. (2014). Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 289: 3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G., Ulijasz A.T., Cornilescu C.C., Markley J.L., Vierstra R.D. (2008). Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. J. Mol. Biol. 383: 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Kurepa J., Vierstra R.D. (1999a). The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc. Natl. Acad. Sci. USA 96: 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S.J., Vener A.V., Vierstra R.D. (1999b). Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286: 2517–2520. [DOI] [PubMed] [Google Scholar]
- Essen L.O., Mailliet J., Hughes J. (2008). The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. USA 105: 14709–14714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K., Grossmann J.G., Fordham-Skelton A.P., Papiz M.Z. (2006). Small-angle X-ray scattering reveals the solution structure of a bacteriophytochrome in the catalytically active Pr state. J. Mol. Biol. 364: 655–666. [DOI] [PubMed] [Google Scholar]
- Ferris H.U., Dunin-Horkawicz S., Hornig N., Hulko M., Martin J., Schultz J.E., Zeth K., Lupas A.N., Coles M. (2012). Mechanism of regulation of receptor histidine kinases. Structure 20: 56–66. [DOI] [PubMed] [Google Scholar]
- Filonov G.S., Piatkevich K.D., Ting L.M., Zhang J., Kim K., Verkhusha V.V. (2011). Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 29: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A.J., Lagarias J.C. (2004). Harnessing phytochrome’s glowing potential. Proc. Natl. Acad. Sci. USA 101: 17334–17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg N., Mukougawa K., Kohchi T., Lagarias J.C. (2001). Functional genomic analysis of the HY2 family of ferredoxin-dependent bilin reductases from oxygenic photosynthetic organisms. Plant Cell 13: 965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Quail P.H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich A.C., Noh B., Vierstra R.D., Loros J., Dunlap J.C. (2005). Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell 4: 2140–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta G.A., Lagarias J.C. (2001). Genetic engineering of phytochrome biosynthesis in bacteria. Proc. Natl. Acad. Sci. USA 98: 10566–10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser C., Taiber S., Yeh C.M., Wittig C.H., Hegemann P., Ryu S., Wunder F., Möglich A. (2014). Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc. Natl. Acad. Sci. USA 111: 8803–8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Fardoux J., Fourrier N., Hannibal L., Genty B., Bouyer P., Dreyfus B., Verméglio A. (2002). Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417: 202–205. [DOI] [PubMed] [Google Scholar]
- Giraud E., Zappa S., Vuillet L., Adriano J.M., Hannibal L., Fardoux J., Berthomieu C., Bouyer P., Pignol D., Verméglio A. (2005). A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 280: 32389–32397. [DOI] [PubMed] [Google Scholar]
- Hahn J., Strauss H.M., Landgraf F.T., Gimenèz H.F., Lochnit G., Schmieder P., Hughes J. (2006). Probing protein-chromophore interactions in Cph1 phytochrome by mutagenesis. FEBS J. 273: 1415–1429. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Rockwell N.C., Nishiyama K., Narikawa R., Ukaji Y., Inomata K., Lagarias J.C., Ikeuchi M. (2013). Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. USA 110: 4974–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Shimada T., Narikawa R., Katayama M., Ikeuchi M. (2008). Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc. Natl. Acad. Sci. USA 105: 9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J., Lamparter T., Mittmann F., Hartmann E., Gärtner W., Wilde A., Börner T. (1997). A prokaryotic phytochrome. Nature 386: 663. [DOI] [PubMed] [Google Scholar]
- Ikeuchi M., Ishizuka T. (2008). Cyanobacteriochromes: a new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 7: 1159–1167. [DOI] [PubMed] [Google Scholar]
- Inomata K., Khawn H., Chen L.Y., Kinoshita H., Zienicke B., Molina I., Lamparter T. (2009). Assembly of Agrobacterium phytochromes Agp1 and Agp2 with doubly locked bilin chromophores. Biochemistry 48: 2817–2827. [DOI] [PubMed] [Google Scholar]
- Ishizuka T., Kamiya A., Suzuki H., Narikawa R., Noguchi T., Kohchi T., Inomata K., Ikeuchi M. (2011). The cyanobacteriochrome, TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 50: 953–961. [DOI] [PubMed] [Google Scholar]
- Jenkins G.I. (2014). The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26: 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Swem L.R., Rushing B.G., Devanathan S., Tollin G., Bauer C.E. (1999). Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285: 406–409. [DOI] [PubMed] [Google Scholar]
- Karniol B., Vierstra R.D. (2003). The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA 100: 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol, B., and Vierstra, R.D. (2006). Structure, function, and evolution of microbial phytochromes. In Photomorphogenesis in Plant and Bacteria: Function and Signal Transduction Mechanisms, E. Schafer and F. Nagy, eds (Dordrecht, The Netherlands: Springer), pp. 65–98. [Google Scholar]
- Karniol B., Wagner J.R., Walker J.M., Vierstra R.D. (2005). Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392: 103–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe D.M., Grossman A.R. (1996). Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 273: 1409–1412. [DOI] [PubMed] [Google Scholar]
- Kevei E., Schafer E., Nagy F. (2007). Light-regulated nucleo-cytoplasmic partitioning of phytochromes. J. Exp. Bot. 58: 3113–3124. [DOI] [PubMed] [Google Scholar]
- Kikis E.A., Oka Y., Hudson M.E., Nagatani A., Quail P.H. (2009). Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet. 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Fujiwara S., Suh S.S., Kim J., Kim Y., Han L., David K., Putterill J., Nam H.G., Somers D.E. (2007). ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360. [DOI] [PubMed] [Google Scholar]
- Krall L., Reed J.W. (2000). The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl. Acad. Sci. USA 97: 8169–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneip C., Hildebrandt P., Schlamann W., Braslavsky S.E., Mark F., Schaffner K. (1999). Protonation state and structural changes of the tetrapyrrole chromophore during the Pr → Pfr phototransformation of phytochrome: a resonance Raman spectroscopic study. Biochemistry 38: 15185–15192. [DOI] [PubMed] [Google Scholar]
- Kretsch T., Poppe C., Schäfer E. (2000). A new type of mutation in the plant photoreceptor phytochrome B causes loss of photoreversibility and an extremely enhanced light sensitivity. Plant J. 22: 177–186. [DOI] [PubMed] [Google Scholar]
- Lamparter T., Michael N., Caspani O., Miyata T., Shirai K., Inomata K. (2003). Biliverdin binds covalently to Agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 278: 33786–33792. [DOI] [PubMed] [Google Scholar]
- Lecoq J., Schnitzer M.J. (2011). An infrared fluorescent protein for deeper imaging. Nat. Biotechnol. 29: 715–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A., Weiner O.D., Lim W.A., Voigt C.A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461: 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A., Chevalier A.A., Tabor J.J., Simpson Z.B., Lavery L.A., Levy M., Davidson E.A., Scouras A., Ellington A.D., Marcotte E.M., Voigt C.A. (2005). Synthetic biology: engineering Escherichia coli to see light. Nature 438: 441–442. [DOI] [PubMed] [Google Scholar]
- Li F.W., et al. (2014). Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc. Natl. Acad. Sci. USA 111: 6672–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhang J., Vierstra R.D., Li H. (2010). Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 107: 10872–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Askinosie S.K., Leuchtman D.L., Morrow J., Willenburg K.T., Coats D.R. (2014). Phototropism: growing towards an understanding of plant movement. Plant Cell 26: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof J.N., Borevitz J.O., Dabi T., Lutes J., Nehring R.B., Redfern J.L., Trainer G.T., Wilson J.M., Asami T., Berry C.C., Weigel D., Chory J. (2001). Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29: 441–446. [DOI] [PubMed] [Google Scholar]
- Matsushita T., Mochizuki N., Nagatani A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574. [DOI] [PubMed] [Google Scholar]
- Medzihradszky M., et al. (2013). Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell 25: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narikawa R., Ishizuka T., Muraki N., Shiba T., Kurisu G., Ikeuchi M. (2013). Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. USA 110: 918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.Y., Quail P.H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nito K., Wong C.C., Yates J.R. III, Chory J. (2013). Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Reports 3: 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K., Kanegae T., Imaizumi T., Fukuda S., Okamoto H., Yeh K.C., Lagarias J.C., Wada M. (1998). A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. USA 95: 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y., Matsushita T., Mochizuki N., Quail P.H., Nagatani A. (2008). Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet. 4: e1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Lagarias J.C. (2012a). Red/green cyanobacteriochromes: sensors of color and power. Biochemistry 51: 9667–9677. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Lagarias J.C. (2012b). Mechanistic insight into the photosensory versatility of DXCF cyanobacteriochromes. Biochemistry 51: 3576–3585. [DOI] [PubMed] [Google Scholar]
- Rockwell N.C., Martin S.S., Feoktistova K., Lagarias J.C. (2011). Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc. Natl. Acad. Sci. USA 108: 11854–11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Duanmu D., Martin S.S., Bachy C., Price D.C., Bhattacharya D., Worden A.Z., Lagarias J.C. (2014). Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. USA 111: 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Su Y.S., Lagarias J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüdiger W., Thümmler F., Cmiel E., Schneider S. (1983). Chromophore structure of the physiologically active form (P(fr)) of phytochrome. Proc. Natl. Acad. Sci. USA 80: 6244–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu M.H., Kang I.H., Nelson M.D., Jensen T.M., Lyuksyutova A.I., Siltberg-Liberles J., Raizen D.M., Gomelsky M. (2014). Engineering adenylate cyclases regulated by near-infrared window light. Proc. Natl. Acad. Sci. USA 111: 10167–10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock R.A., Quail P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3: 1745–1757. [DOI] [PubMed] [Google Scholar]
- Shimizu-Sato S., Huq E., Tepperman J.M., Quail P.H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Shu X., Royant A., Lin M.Z., Aguilera T.A., Lev-Ram V., Steinbach P.A., Tsien R.Y. (2009). Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324: 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Psakis G., Kopycki J., Lang C., Matysik J., Hughes J. (2014). The D-ring, not the A-ring, rotates in Synechococcus OS-B’ phytochrome. J. Biol. Chem. 289: 2552–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., Matysik J. (2011). Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. USA 108: 3842–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss H.M., Hughes J., Schmieder P. (2005). Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry 44: 8244–8250. [DOI] [PubMed] [Google Scholar]
- Su Y.S., Lagarias J.C. (2007). Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19: 2124–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu N., Mittmann F., Wagner G., Hughes J., Wada M. (2005). A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc. Natl. Acad. Sci. USA 102: 13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J.A., Westenhoff S. (2014). Signal amplification and transduction in phytochrome photosensors. Nature 509: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasler R., Moises T., Frankenberg-Dinkel N. (2005). Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 272: 1927–1936. [DOI] [PubMed] [Google Scholar]
- Toettcher J.E., Weiner O.D., Lim W.A. (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155: 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulijasz A.T., Cornilescu G., Cornilescu C.C., Zhang J., Rivera M., Markley J.L., Vierstra R.D. (2010). Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 463: 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulijasz A.T., Cornilescu G., von Stetten D., Kaminski S., Mroginski M.A., Zhang J., Bhaya D., Hildebrandt P., Vierstra R.D. (2008). Characterization of two thermostable cyanobacterial phytochromes reveals global movements in the chromophore-binding domain during photoconversion. J. Biol. Chem. 283: 21251–21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulijasz A.T., Cornilescu G., von Stetten D., Cornilescu C., Velazquez Escobar F., Zhang J., Stankey R.J., Rivera M., Hildebrandt P., Vierstra R.D. (2009). Cyanochromes are blue/green light photoreversible photoreceptors defined by a stable double cysteine linkage to a phycoviolobilin-type chromophore. J. Biol. Chem. 284: 29757–29772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stetten D., Seibeck S., Michael N., Scheerer P., Mroginski M.A., Murgida D.H., Krauss N., Heyn M.P., Hildebrandt P., Borucki B., Lamparter T. (2007). Highly conserved residues Asp-197 and His-250 in Agp1 phytochrome control the proton affinity of the chromophore and Pfr formation. J. Biol. Chem. 282: 2116–2123. [DOI] [PubMed] [Google Scholar]
- Vuillet L., Kojadinovic M., Zappa S., Jaubert M., Adriano J.M., Fardoux J., Hannibal L., Pignol D., Verméglio A., Giraud E. (2007). Evolution of a bacteriophytochrome from light to redox sensor. EMBO J. 26: 3322–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J.R., Brunzelle J.S., Forest K.T., Vierstra R.D. (2005). A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438: 325–331. [DOI] [PubMed] [Google Scholar]
- Wagner J.R., Zhang J., Brunzelle J.S., Vierstra R.D., Forest K.T. (2007). High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 282: 12298–12309. [DOI] [PubMed] [Google Scholar]
- Wagner J.R., Zhang J., von Stetten D., Günther M., Murgida D.H., Mroginski M.A., Walker J.M., Forest K.T., Hildebrandt P., Vierstra R.D. (2008). Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 283: 12212–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.H., Lagarias J.C. (2000). Defining the bilin lyase domain: lessons from the extended phytochrome superfamily. Biochemistry 39: 13487–13495. [DOI] [PubMed] [Google Scholar]
- Yang X., Kuk J., Moffat K. (2008). Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc. Natl. Acad. Sci. USA 105: 14715–14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Kuk J., Moffat K. (2009). Conformational differences between the Pfr and Pr states in Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. USA 106: 15639–15644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Stojkovic E.A., Kuk J., Moffat K. (2007). Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc. Natl. Acad. Sci. USA 104: 12571–12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Jost A.P., Weiner O.D., Tang C. (2013). A light-inducible organelle-targeting system for dynamically activating and inactivating signaling in budding yeast. Mol. Biol. Cell 24: 2419–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Ren Z., Kuk J., Moffat K. (2011). Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479: 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Lagarias J.C. (1998). Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95: 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K.C., Wu S.H., Murphy J.T., Lagarias J.C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277: 1505–1508. [DOI] [PubMed] [Google Scholar]
- Yu D., et al. (2014). An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 5: 3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Stankey R.J., Vierstra R.D. (2013). Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol. 161: 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]