An extensive analysis of small RNAs in soybean identified many miRNAs and phased, secondary siRNA (phasiRNA) loci; some of these miRNAs were the triggers of the phasiRNA loci.
Abstract
Small RNAs are ubiquitous, versatile repressors and include (1) microRNAs (miRNAs), processed from mRNA forming stem-loops; and (2) small interfering RNAs (siRNAs), the latter derived in plants by a process typically requiring an RNA-dependent RNA polymerase. We constructed and analyzed an expression atlas of soybean (Glycine max) small RNAs, identifying over 500 loci generating 21-nucleotide phased siRNAs (phasiRNAs; from PHAS loci), of which 483 overlapped annotated protein-coding genes. Via the integration of miRNAs with parallel analysis of RNA end (PARE) data, 20 miRNA triggers of 127 PHAS loci were detected. The primary class of PHAS loci (208 or 41% of the total) corresponded to NB-LRR genes; some of these small RNAs preferentially accumulate in nodules. Among the PHAS loci, novel representatives of TAS3 and noncanonical phasing patterns were also observed. A noncoding PHAS locus, triggered by miR4392, accumulated preferentially in anthers; the phasiRNAs are predicted to target transposable elements, with their peak abundance during soybean reproductive development. Thus, phasiRNAs show tremendous diversity in dicots. We identified novel miRNAs and assessed the veracity of soybean miRNAs registered in miRBase, substantially improving the soybean miRNA annotation, facilitating an improvement of miRBase annotations and identifying at high stringency novel miRNAs and their targets.
INTRODUCTION
Small noncoding RNAs have important roles in development, cell differentiation, adaptation to biotic and abiotic stresses, and genome stability. The main activity of small RNAs is the negative regulation of specific mRNAs or gene expression via target degradation, translational repression, or by directing chromatin modifications (Chen, 2009). Several different classes of small RNAs have been identified to date. In plants, the best-characterized small RNAs are microRNAs (miRNAs) and small interfering RNAs (siRNAs); these are generated from different precursors and via distinct pathways (Carthew and Sontheimer, 2009). miRNAs, typically 21 to 22 nucleotides in length, are derived from long noncoding RNA precursors transcribed from MIRNA genes by RNA polymerase II. The miRNA precursor forms a stem-loop structure processed by DICER-LIKE1 (DCL1), or other DCLs in rare cases, yielding a single small RNA duplex (miRNA/miRNA*) with two-nucleotide 3′ overhangs (Kim, 2005). One strand of the small RNA duplex is the mature miRNA, the guide strand that is loaded onto an Argonaute (AGO) protein to form an effector complex (the so-called RISC for RNA-induced silencing complex) that directs cleavage or translational repression of target miRNAs (Mallory and Bouché, 2008). The other strand of the duplex, the miRNA* or passenger strand, is rapidly degraded and normally does not accumulate. siRNAs are canonically processed from perfectly complementary, long double-stranded RNA (dsRNA) precursors that are typically formed by RNA-dependent RNA polymerase (RDRs), or perhaps formed from annealed sense and antisense transcripts (Carthew and Sontheimer, 2009). Several classes of siRNAs have been defined in plants (reviewed in Axtell, 2013), with the major class of heterochromatic siRNAs playing a key role in both establishment and maintenance of cytosine methylation and repressive histone modifications (Kanno and Habu, 2011; Saze et al., 2012). siRNAs are also able to act as a mobile signal with silencing effects spreading from cell to cell, or longer distances, through the movement of siRNAs (Mlotshwa et al., 2002; Dunoyer et al., 2010).
A class of siRNAs of growing interest has been identified as the product of processive cleavage in increments of 21 nucleotides from a long dsRNA precursor, generating a phased or perfectly spaced arrangement of small RNAs. These siRNAs, so-called phased siRNAs (phasiRNAs), are triggered by particular miRNA-guided cleavage following either the one-hit (122) or two-hit (221) models capacitated by target sites of one 22-nucleotide or two 21-nucleotide miRNAs, respectively (Fei et al., 2013). The cleaved, uncapped mRNA product serves as a substrate for RDR6, generating a dsRNA precursor, which is then cleaved by DCL4 to produce the 21-nucleotide phased siRNAs. Some of the resulting phased siRNAs have been shown to function in trans regulation of target genes; hence, this class of siRNAs was originally called tasiRNAs, yet many more loci generate siRNA of the same phased pattern (PHAS loci) with unknown trans-activity and hence are labeled with the general title of “phasiRNAs.” tasiRNAs regulate mRNA via cleavage at complementary target sites, like many plant miRNAs (Allen et al., 2005). The most well known tasiRNAs are the set of trans-acting short interfering RNA-auxin response factors (tasiARFs) generated from TRANS-ACTING SIRNA GENE3 (TAS3) (Allen et al., 2005; Yoshikawa et al., 2005; Axtell et al., 2006). tasiARFs function in the suppression of auxin-responsive factor genes (ARF2, ARF3/ETTIN, and ARF4) (Adenot et al., 2006; Fahlgren et al., 2006; García et al., 2006; Hunter et al., 2006). Many phasiRNAs have been identified in a number of plant species, including Arabidopsis thaliana (Axtell et al., 2006), rice (Oryza sativa) (Johnson et al., 2009), and Vitis vinifera (grapevine) (Zhang et al., 2012). The number of known PHAS loci varies substantially between species, from over 800 in wild rice (Oryza rufipogon; Liu et al., 2013) to fewer than 30 in Arabidopsis (Axtell et al., 2006; Fei et al., 2013). In legumes, 114 and 41 PHAS loci were identified in Medicago truncatula and soybean (Glycine max), respectively (Zhai et al., 2011).
Soybean is economically the most important legume in the world, and it is one of the major sources of protein and edible oil. A genome sequence of soybean is now publicly available (Schmutz et al., 2010; phytozome.org). Genome sequences, together with data generated by next-generation sequencing technologies, have enabled the identification and quantification of small RNAs on a genome-wide scale. To date, hundreds of miRNAs have been identified in soybean (Song et al., 2011; Li et al., 2012). However, many newly annotated miRNAs and their targets have not been well verified, and even annotated miRNAs are often later corrected based on more robust experimental data (Jeong et al., 2011). PHAS loci are even more poorly annotated than miRNAs. Compared with M. truncatula, far fewer PHAS loci have been identified in soybean (Zhai et al., 2011). With extensive small RNA data and higher sequencing depth, many more PHAS could be discovered. In this study, we analyzed a large set of small RNA libraries created from diverse tissues to build an expression atlas of small RNAs and comprehensively identify PHAS loci in soybean. We demonstrated that many protein-coding genes in soybean are PHAS loci. In addition to NB-LRRs, previously identified as a primary class of PHAS loci in legumes (Zhai et al., 2011), we discovered hundreds of other protein-coding genes that generate phasiRNAs. We integrated parallel analysis of RNA end (PARE) data to identify the miRNA triggers of these PHAS loci. From these data, we verified the soybean miRNAs registered in miRBase (version 20) and identified novel miRNAs as well, demonstrating that many of previously reported miRNAs have characteristics of siRNAs. Based on the expression analysis, we demonstrated the tissue- or treatment-specific expression of phasiRNAs, as well as of both known and novel miRNAs.
RESULTS
Construction and Sequencing of Small RNA and PARE Libraries from Soybean
We constructed and analyzed a total of 69 small RNA libraries from vegetative and reproductive parts of soybean, including flowers, leaves, and developing nodules; in addition, we integrated public data from seed and seed coat tissues (Supplemental Data Set 1A) (Tuteja et al., 2009; Song et al., 2011). The leaf tissues were from plants under well-watered or drought stress, or using treatments to mimic biotic stresses (i.e., flagellin and chitin treatments). The small RNA libraries for flower tissues were prepared from unopened flowers, open flowers, ovaries, and anthers. The small RNA libraries for nodules were prepared from the developing nodules at 10, 15, 20, 25, and 30 d after inoculation. The libraries we constructed (i.e., all except the seed-related data from published sources) included two to three biological replicates for each sample.
Small RNA reads in the range of 18 to 34 nucleotides were retained, yielding from all libraries a total of 1,967,153,698 reads. After removal of sequences that matched to structural RNAs (predominantly rRNA- or tRNA-derived), 1,158,661,201 genome-matched reads (58.9% of the total) were retained, corresponding to 138,436,684 unique or distinct sequences (11.9% of genome-matched reads and 7.0% of total reads) (Supplemental Data Set 1A). The abundance of sequences in each library was normalized to transcripts per five million (TP5M). The highest proportion of unique sequences was found in nodule libraries (27.5%), while the lowest was found in leaf libraries (6.6%), possibly reflecting saturation of the sRNA complexity in leaves, which had the highest read abundance (Supplemental Data Set 1A). An analysis of the size distributions revealed that the proportion of small RNAs in different size classes varied among different tissues (Supplemental Figure 1). In almost all tissues, the proportion of total read abundance in the sizes of 21 and 24 nucleotides was higher than those of other size classes and consistent across replicates and tissues; the one exception was in leaf tissues in which the proportion of total abundance of reads in the 24-nucleotide class was highly reduced (Supplemental Figures 1A, 1C, 1E, and 1G). This latter case was different from Arabidopsis leaves in which the abundance of the 24-nucleotide class is high (Supplemental Figure 2). The proportion of distinct reads in the 24-nucleotide class was greater than the 21-nucleotide class in all tissues, likely reflecting that these are typically heterochromatic siRNAs from a wide set of genomic repeats (Supplemental Figures 1B, 1E, 1F, and 1H). As mentioned above, the leaf libraries had relatively few unique reads among which the most prominent type (68% of all) was miRNAs (Supplemental Figure 2). In the leaf libraries, the miRNAs largely comprised just three species: miR398c, a miR3522 variant, and miR166a, and among these sequences, miR398c accounted for ∼22.5% of the 21-nucleotide small RNAs. A rich source of 21-nucleotide small RNAs in leaves was from intergenic regions (19%). These intergenic region-associated sequences were the most diverse, accounting for 69% of the distinct reads. In the reproductive tissues, the proportion of 22-nucleotide distinct reads was high and comparable to that of 21-nucleotide small RNAs (Supplemental Figure 1B), whereas in nodule and seed tissues, a pattern of higher levels of 22- than 21-nucleotide distinct reads was apparent (Supplemental Figure 1F). All of the genome-matched reads were utilized for miRNA evaluation and phasing locus identification (see below).
Reevaluation of Annotated miRNAs
miRBase version 20 (http://www.mirbase.org) dating from November, 2013, contains over 6000 MIRNA genes from more than 70 plant species. Among these, ∼554 mature miRNAs derived from 505 precursors have been registered for soybean. Many miRNAs registered in miRBase were computationally identified based on similarity to conserved miRNAs in other species, some were verified by deep-sequencing small RNA libraries, and an even smaller set have their functions verified by PARE data (aka degradome data). In the absence of experimental target validation, like PARE data or 5′-rapid amplification of cDNA ends, predictions of miRNA function may provide ambiguous results. Analysis of rice miRNAs has shown that many predicted miRNAs are marginal, lacking conventional miRNA features, or they are siRNA-like miRNAs rather than typical miRNAs (Jeong et al., 2011). Properties of siRNA-like miRNAs include that the small RNAs at the locus are diverse, distributed, of low abundance and are found on both strands of their generating loci. This analysis of annotated rice miRNAs in miRBase using deep-sequencing small RNA data in conjunction with PARE libraries greatly improved the characterization of typical miRNAs (Jeong et al., 2011). In our study, the use of the largest small RNA data set for soybean generated to date, as well as PARE data, allowed us to evaluate miRBase-annotated soybean miRNAs (from version 20) and to discover novel miRNAs. The criteria for characterizing typical plant miRNAs were based on Meyers et al. (2008), and the process for evaluating miRNAs was essentially as described by Jeong et al. (2011). After removal of annotated miRNAs unmatched to the soybean version 1.1 genome, 530 previously reported miRNAs were subjected to a process of reevaluation to characterize each one as either (A) a weakly expressed miRNA that is difficult to assess, but resembles a heterochromatic siRNA; (B) highly similar to and likely an siRNA; (C) a miRNA that marginally meets the strict definition (and could include newly evolved miRNAs); and (D) a typical miRNA that meets all standards of well-defined miRNAs (see Methods; examples of each class are shown in Supplemental Figure 3). We also evaluated the conservation of the soybean miRNAs by comparison to Arabidopsis, based on the criteria for miRNA families of Meyers et al. (2008), yielding 231 miRNAs conserved between soybean and Arabidopsis, with names assigned accordingly in the miRNA list (Supplemental Data Set 1B); these miRNAs clearly fit into category D, as well-defined miRNAs.
The process for evaluating miRNAs and sorting the loci into the categories mentioned above primarily involved three criteria, including their abundance, abundance ratio, and strand ratio (Jeong et al., 2011). The abundance was calculated by examining the count of reads from the two most abundant small RNAs (“top1+top2”) matching each miRNA locus which, for a real miRNA, would typically represent the two strands of the miRNA duplex. The overall summed abundance of 530 miRNAs ranged from as low as 1 TP5M to the highest abundances of 44.1 million TP5M (the two most abundant sequence variants of miR166) and 36.9 million TP5M (miR1507). We designated 191 miRNA precursors as “weakly expressed” loci; these had matched read abundances <924 TP5M, lower than 95% of the conserved miRNA loci (Supplemental Data Set 1B). For the second criterion, the abundance ratio, we examined the ratio of abundances between two most abundant small RNAs (top1+top2) and all small RNAs matching each miRNA locus, while for the third criterion, strand bias, for each stem-loop we divided the summed abundance of small RNA sequences from the sense strand by those from both strands. Among the conserved miRNAs, 95% had an abundance ratio of 0.565 or higher, whereas only 17.5% of nonconserved miRNAs had abundance ratios of 0.565 or higher (Supplemental Data Set 1B). Following Jeong et al. (2011), we designated miRNA loci with abundance ratios of less than 0.4 as “siRNA-like” miRNA loci and those with ratios between 0.4 and 0.5 as “marginal” miRNA loci, consistent with the examples shown in Supplemental Figure 3. In addition, 95% of conserved miRNA precursors had strand ratios of 0.978 or higher, while only 23% (71/299) of nonconserved miRNAs fit this value. We considered miRNA precursors with strand ratios of less than 0.8 as “siRNA-like” miRNAs and those with a ratio between 0.8 and 0.9 as “marginal miRNAs.” Integrating the second and third criteria, we were able to classify 312 miRNAs as typical miRNAs, 203 miRNAs as siRNA-like miRNAs, and 15 miRNAs as marginal miRNAs; this set of 312 miRNAs included the 191 weakly expressed miRNAs defined from the first criterion (Supplemental Data Set 1B). The majority of miRNAs in the “typical miRNA” class were 21 and 22-nucleotides in length, whereas the “siRNA-like” class was rich in annotated miRNAs with a size of 24 nucleotides (Supplemental Data Set 1B). This latter group of siRNA-like, 24-nucleotide miRBase miRNAs is likely incorrectly annotated.
Identification of Novel miRNAs and miRNA Variants in Soybean
In addition to the reevaluation of previously reported miRNAs, we also used the small RNA data to identify novel miRNAs and to annotate miRNA variants. The pipeline used for the identification of novel miRNAs was adapted from Jeong et al. (2011) (Supplemental Figure 4). Using 124,526,477 distinct reads after excluding t/r/sn/snoRNAs, all genome-matched reads between 18 and 26 nucleotides were filtered for read abundance, to include only those ≥50 TP5M in at least one library. The reads that mapped to more than 20 locations in the soybean chromosomes were also discarded as too repetitive to be miRNAs. Of the 124,526,447 reads, there were 29,133 sequences that passed the first filter set, including 198 sequences that matched to the known miRNAs. The candidate precursors that passed the first filter set were analyzed by miREAP (https://sourceforge.net/projects/mireap) as described by Jeong et al. (2011). In all, 2523 sequences corresponding to 4047 precursors were obtained. Of the 198 reported miRNAs, only 120 passed this second filter. The third filter set then was used to evaluate the single-strand bias (sense/total ≥0.9) and an abundance bias ([top1 + top2]/total ≥ 0.7) to yield only one or two most prominent miRNAs for each given precursor. In total, 180 small RNA sequences corresponding to 361 precursors passed this filter, including 71 known miRNAs. The fourth filter was applied in order to identify only high quality stem-loop structures, analyzed using CentroidFold (Sato et al., 2009). A total of 151 candidates from 332 precursors passed this filter; all of the 71 known miRNAs from the previous step also passed. Among the 71 known miRNAs, we found 44 variants compared with the miRNAs registered in miRBase (Supplemental Figure 4). After excluding known miRNAs, 22 high-confidence candidates were designated as novel miRNAs (Supplemental Data Set 1C). We also identified miRNA variants by comparing the small RNA reads with those registered in the miRBase (Supplemental Data Set 1D). We found ∼20 sequences that varied in length and/or contained nucleotide substitutions relative to the miRNAs registered in miRBase. The length of these miRNA variants varied from 19 to 24 nucleotides, including one- to four-nucleotide substitutions. There were also 10 novel miRNAs identified from different locations on the same precursors of the previously reported miRNAs (Supplemental Data Set 1D). Thus, we were able to identify a large number of new and known soybean miRNAs from our data set.
Differentially Abundant miRNAs Across Soybean Tissues and Treatments
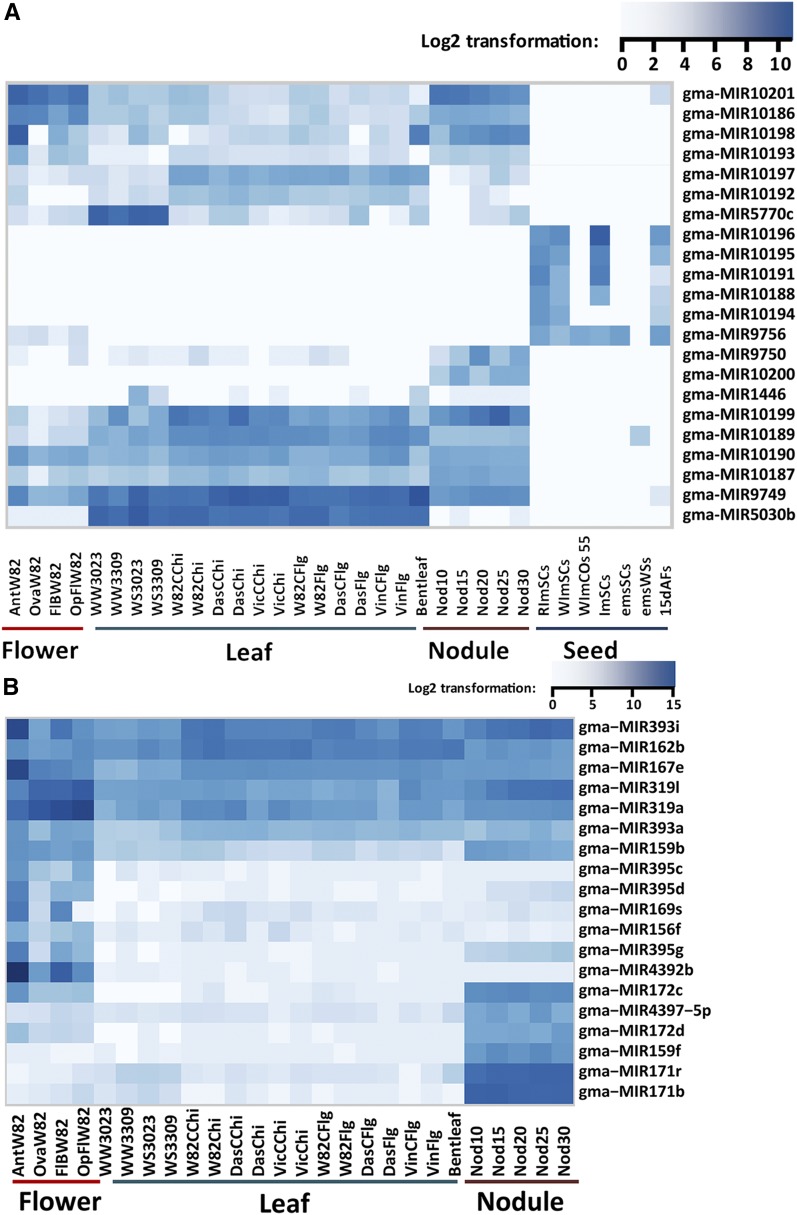
An assessment of differential abundance counts was performed for the novel and known miRNAs and variants across all 69 small RNA libraries. Hierarchical clustering (Eisen et al., 1998) of our data revealed that many miRNAs demonstrate tissue-preferential accumulation. We selected three sets of miRNAs for more detailed analysis. The first set was all novel miRNAs showing tissue-preferential levels (Figure 1A). Of 22 novel miRNAs, six were exclusively observed in seed tissues, including gma-miR10196, gma-miR10195, gma-miR10191, gma-miR10188, gma-miR10194, and gma-miR9756 (Figure 1A). Similarly, gma-miR10200 was enriched in nodules and gma-miR5030b was enriched in leaves. Several of these novel miRNAs were enriched in more than one tissue; i.e., gma-miR10201, gma-miR10186, gma-miR10198, gma-miR10193, and gma-miR9749 were enriched in both reproductive tissues and nodules (Figure 1A). The second set was miRNAs highly enriched in the reproductive tissues. This set included gma-miR395c, gma-miR395d, gma-miR395g, gma-miR169s, gma-miR156f, and gma-miR4392 (Figure 1B). Among the miRNAs that were preferentially observed in the flower tissues, a few of them showed high enrichment in anthers, i.e., gma-miR4392, gma-miR393l, and gma-miR167e. Intriguingly, gma-miR4392 was highly abundant in reproductive tissues, especially in anthers, but nearly absent in other tissues (Figure 1B, and analyzed in more detail below). There were also miRNAs preferentially present in reproductive tissues as well as in nodules, i.e., miR172c, miR159b, and miR395g (Figure 1B). The last set of miRNAs that were observed in a tissue-preferential manner comprised those strongly present in developing nodules but sparingly in other tissues. These included miR171b, miR171r, miR159f, miR172d, and miR4394-5p (Figure 1B). Not fitting into any of our three sets was a number of miRNAs enriched in seed tissues, i.e., gma-miR176e/f and gma-miR1512c. These seed-specific miRNAs were well described in the original study from which they were derived (Zabala et al., 2012).
Figure 1.
Expression Profiling of Novel and Tissue-Preferential miRNAs.
(A) Novel miRNAs identified in this study included many with enriched abundances in specific tissues or organs.
(B) Analysis of previously described soybean miRNAs also reveals a range of tissue preferences in flower, leaf, and nodule.
miRNAs within a family accumulated differentially across tissues; for example, the large miR171 family, which contains 22 members, shows a rich diversity of abundance patterns (Supplemental Figure 5). Some were nodule-enriched, i.e., gma-miR171s, gma-miR171r, and gma-miR171b-3p, while others were both flower- and leaf-enriched. Processing variants of miRNAs from a single precursor also accumulated differently; the variant gma-miR156c.2 was highly abundant in cotyledon, whereas gma-miR156c.1 was absent (Supplemental Data Set 1D). gma-miR156c is in most or all tissues, but preferentially expressed in seed coat tissues. Similarly, gma-miR3522.1 was identified preferentially in seed tissues and leaf tissues, while gma-miR3522 was measurable only in seed tissues and at low levels (Supplemental Data Set 1D).
We next identified miRNAs differentially expressed in stress treatments. This was done using the R package baySeq (Hardcastle and Kelly, 2010), with a likelihood of ≥0.95 and false discovery rate at < 0.01. With this cutoff, no miRNAs were differentiated in water-stressed leaves of two genotypes (IA3023 and LD003309); however, the closest was gma-miR1446, enriched in drought-stressed leaves (Supplemental Data Set 1E; Figure 1A). We found nine miRNAs upregulated in the flagellin-treated Dassel genotype, perhaps mimicking biotic stress (Supplemental Data Set 1E), while we were unable to identify any differentially expressed miRNAs resulting from chitin treatments. Across our libraries, differential miRNAs were much more enriched comparing tissues than treatments.
miRNA Target Verification Using PARE Libraries
Experimental verification of miRNA-directed target cleavage is rapid and precise using PARE data (Addo-Quaye et al., 2008; Gregory et al., 2008; German et al., 2009; Zhai et al., 2011; Jeong et al., 2013). We constructed PARE libraries from flower, leaf, and nodule tissues and utilized public PARE data for seeds, comprising over 65 million distinct reads (Supplemental Data Set 1F). Among the PARE-validated targets of soybean miRBase-annotated miRNAs, we verified 392 targets for 263 miRNAs, most of which are typical miRNAs. Of these, 261 overlapped with annotated protein-coding genes, with the remainder in intergenic regions or unannotated genes (Supplemental Data Set 1G). The number of targets per miRNA ranged from one to 23. Among novel miRNAs and variants, nine targets were identified for eight novel miRNAs and 129 targets were identified for the 33 new miRNA variants. Of these, 8 and 86 targets for the novel miRNAs and new miRNA variants, respectively, overlapped with annotated genes and the remainder localized to intergenic regions (Supplemental Data Set 1H), which could be unannotated genes or noncoding transcripts like TAS loci.
Genome-Wide Identification of Loci Generating Phased siRNAs and Their Triggers
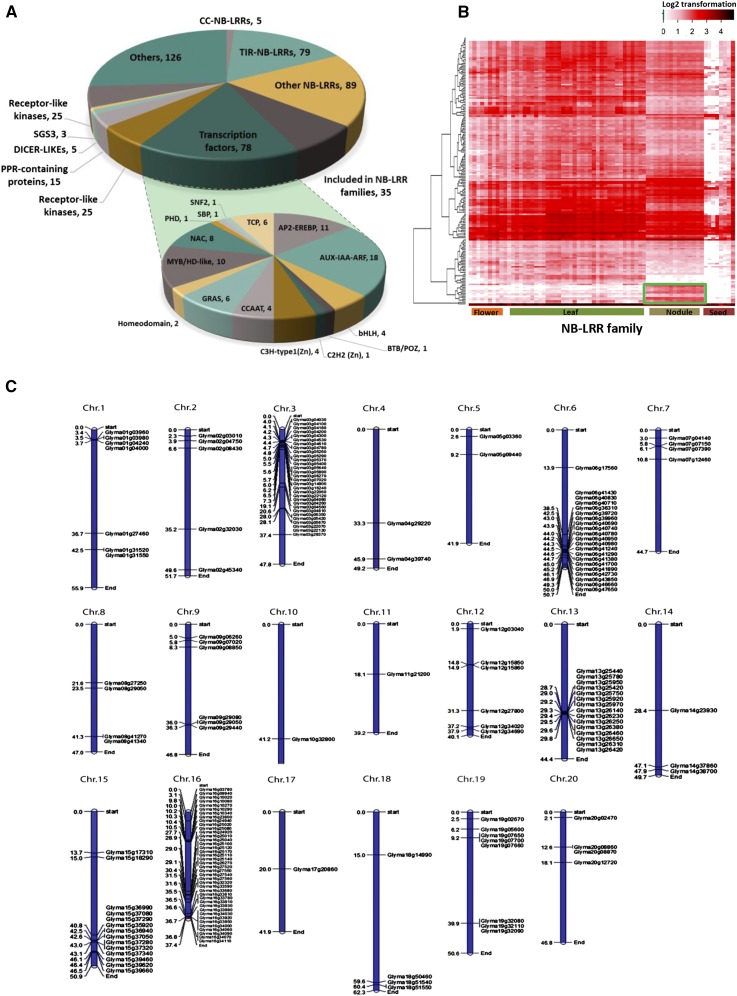
Plant loci producing phased, secondary siRNAs, so-called PHAS loci, include both protein-coding and noncoding transcripts; the legume M. truncatula is a rich source of such loci (Zhai et al., 2011), with variable numbers of loci in other plant species (Fei et al., 2013). We combined all 69 small RNA libraries to identify soybean PHAS loci and subsequently assess their miRNA triggers via a process of reverse computation (Xia et al., 2013). We identified 504 genomic PHAS loci at a phasing P value ≤ 0.001, a stringent threshold (Figure 2A; Supplemental Data Set 1I) (Chen et al., 2007). Of these, 483 (95.8%) overlapped annotated protein-coding genes. The primary class of PHAS loci (208 or 41.0% of the total) corresponded to NB-LRR genes, which encoded 79 Toll interleukin 1 receptor (TIR)-NB-LRRs, five coiled-coil (CC)-NB-LRR, and 89 other NB-LRRs (Figure 2A). These phasi-NB-LRRs (pNLs) accounted for 65% (208/319) of all NB-LRRs identified in the soybean genome, including those identified by Kang et al. (2012), plus another 35 phasi-NB-LRR genes identified using the Greenphyl DB (http://www.greenphyl.org) (Supplemental Data Set 1I). The majority of pNL loci were clustered on chromosomes 3, 6, 13, 15, and 16, which contained 30, 21, 15, 14, and 40 pNLs (Figure 2C). The level of phasiRNAs differed among pNLs, with some showing high levels of siRNAs across all analyzed tissues, but others accumulating in a particular tissue, like nodules (Figure 2B). Many receptor-like kinase-encoding genes also generate phasiRNAs, but these were only a small fraction (25 loci) of the ∼600 receptor-like kinase genes known in soybean (Liu et al., 2009). In Arabidopsis, the major group of protein-coding PHAS genes is pentatricopeptide repeat-containing proteins (Howell et al., 2007), but in soybean we found only 15 pentatricopeptide repeat-encoding PHAS loci. Several diverse families of transcription factors accounted for 15% of the PHAS loci (Figure 2A), including 18 PHAS loci from the Aux/IAA and auxin response factor families (AUX-IAA-ARF), 10 PHAS loci in the APETALA2 and ethylene-responsive element binding protein (AP2-EREBPs) gene families, and another 10 PHAS loci from genes encoding MYB/HD-like proteins (Figure 2A). Genes involved in small RNA biogenesis, i.e., DCLs (five loci), SUPPRESSOR OF GENE SILENCING3 (three loci), and AGO2 (one locus), were also as among the soybean PHAS loci, suggesting feedback regulation may occur. Finally, a large number (126) of PHAS loci overlapped with genes of unknown function, many of which are single copy in the genome, suggesting cis- rather than trans-activity (Figure 2A; Supplemental Data Set 1I). As a set, the 504 soybean PHAS loci was significantly larger than but included all of those that we previously identified (41 loci) in soybean (Zhai et al., 2011) due to our much broader and deeper set of new data.
Figure 2.
Protein-Coding PHAS Genes.
The soybean genome contains more protein-coding loci generating phasiRNAs than has been described for any other plant genome.
(A) Categories and numbers of coding PHAS loci.
(B) Expression profiling and hierarchical clustering of PHAS genes in the NB-LRR family.
(C) Distributions and clusters of phasi-NB-LRR genes in the soybean genome.
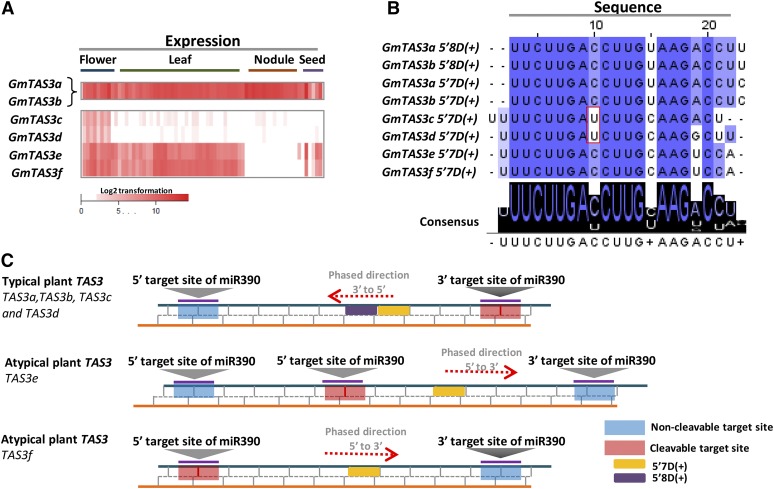
Distinct from protein-coding genes, one group of 21 PHAS loci was predicted to be noncoding. This included six TAS3-like loci and an unnamed TAS-like locus previously reported (Xia et al., 2013). Two TAS3 loci (TAS3a and TAS3b) were highly abundant and in most respects quite similar to those of Arabidopsis, whereas the additional four TAS3 paralogs (TAS3c-f) varied in phasiRNA abundance, sequence conservation, or trigger arrangements (Figure 3). TAS3c and TAS3d produced few phasiRNAs with the exception of floral tissues (Figure 3A); TAS3a and TAS3b accumulated robustly in most tissues, with an abundance gradient over the progression of nodule development (Figure 3A). TAS3e- and TAS3f-derived phasiRNAs were undetectable in nodules (Figure 3A). In addition, we also found a noncoding PHAS locus that produced phasiRNAs exclusively in anthers, as described below.
Figure 3.
Triggers and Processing Mechanisms of Soybean TAS3 TasiRNAs.
(A) The patterns of abundance in flower, leaf, nodule, and seed tissues for the summed total of tasiRNAs from each of the six TAS3 loci present in the soybean genome. TAS3a and TAS3b are identical and thus cannot be measured separately.
(B) TasiARFs derived from TAS3a/b/c/d/e/f. The validated targets for all of the TAS3 5′8D(+) and 5′7D(+) siRNAs were all in the family of auxin response factors (ARFs), consistent with their relatively well conserved sequences (data not shown).
(C) Two or three miR390 target sites exist at soybean TAS3 loci, and the direction of phasing relative to these targets sites suggests a noncanonical processing direction for siRNAs triggered by a 21-nucleotide miRNA at TAS3e and TAS3f.
PhasiRNA biogenesis is typically initiated by target cleavage via an AGO-loaded miRNA trigger, recruiting RDR6 to synthesize dsRNA, the substrate that DCL4 processes into phased 21-nucleotide sRNAs. To identify the miRNA triggers of the PHAS loci, we integrated the soybean miRNAs and PARE data (Xia et al., 2013). This identified 20 miRNA triggers of 127 PHAS loci, with each trigger targeting one to 20 loci (Supplemental Data Set 1I). Three miRNAs triggered more than 10 PHAS loci, including gma-miR167e (triggering 10 PHAS loci), gma-miR2109 (11 loci), and gma-miR1510b-3p (20 loci); the former targets the ARF6 and ARF8 transcription factors and the latter two primarily trigger pNLs. Finally, we observed that a feature observed in Arabidopsis miRNAs that trigger phasiRNA biogenesis, precursors with an asymmetric bulge stem-loop (Manavella et al., 2012), was absent for many of the miRNA triggers that we identified (Supplemental Figure 6).
The Novel Loci and Phasing Patterns of TAS3
In plants, many phased loci are triggered by a one-hit 22 nucleotide miRNA (122), with the production of phasiRNAs downstream of the cleavage site (Chen et al., 2010; Cuperus et al., 2010); this was also true for the phased loci in soybean that we identified (Supplemental Data Set 1I). TAS3 loci are conventionally triggered by miR390 binding at two sites via the two-hit (221) pathway, triggering tasiARF production (Axtell et al., 2006). Conserved tasiARFs were produced from all six soybean TAS3 loci: two produced from TAS3a/b [5′7D(+) and 5′8D(+)] and only one (5′7D(+)) from TAS3c/d/e/f (Figure 3B). There was a single nucleotide variant (C-to-U) found at positions 9 and 10 of the tasiARF GmTAS3c-5′7D(+) and GmTAS3d-5′7D(+) (Figure 3B). Four of the six soybean TAS3 loci, TAS3a/b/c/d, had target sites consistent with the canonical two-hit model (Figure 3C); the other two, TAS3e and f, were atypical. TAS3e has three gma-miR390 binding sites, essentially a three-hit locus, with the middle site cleaved to initiate downstream processing and 5′8D(+) production (Figure 3C). Relative to Arabidopsis TAS3, soybean TAS3e has a noncanonical phasing direction, downstream rather than upstream of the site cleaved by the 21-nucleotide gma-miR390. Similarly, the phasing in TAS3f is downstream of the 5′ miR390 target site, but the position and number of gma-miR390 binding sites is typical of TAS3 loci (Figure 3C).
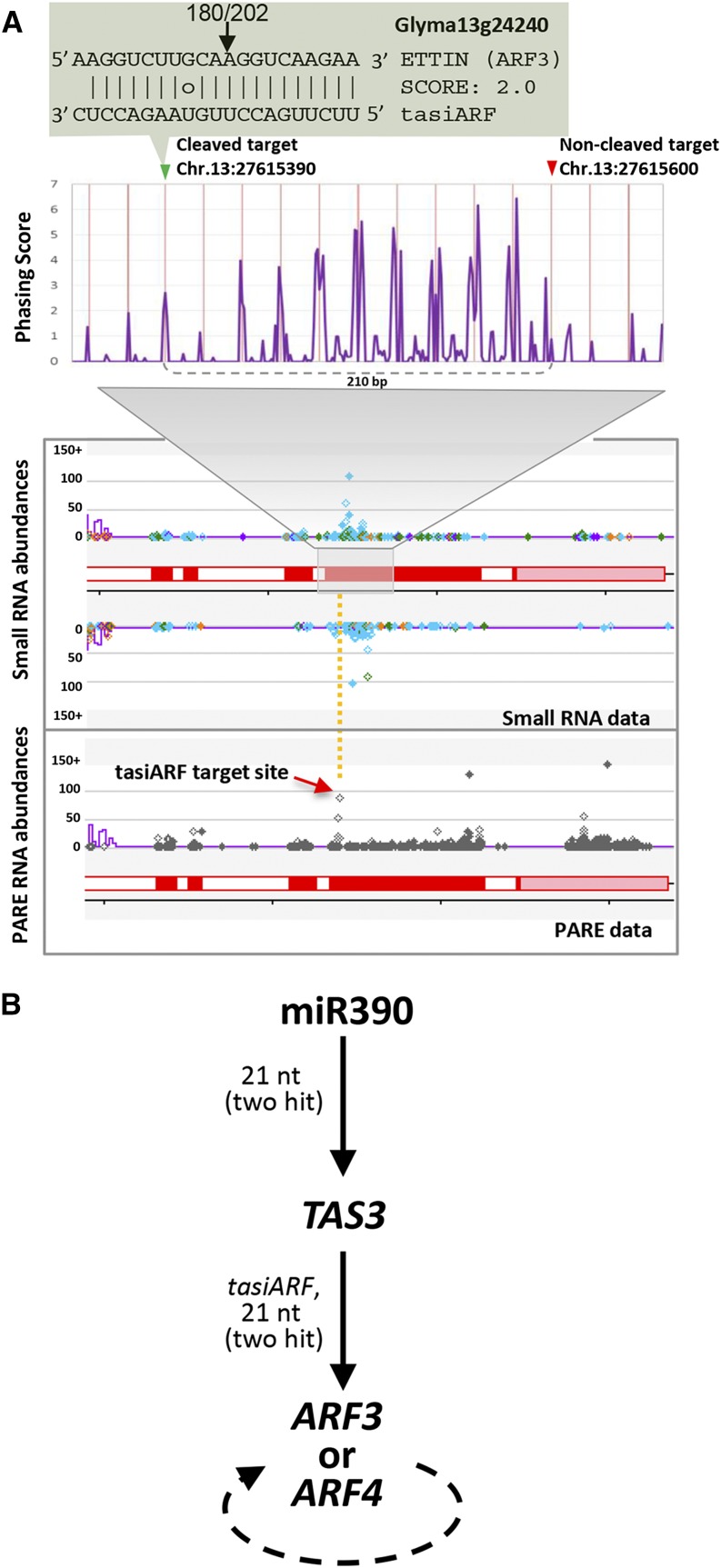
Our data also demonstrate that tasiRNAs can function in two-hit biogenesis to trigger additional secondary siRNAs. The tasiARFs from TAS3 target and cleave transcripts from the ARF3/ETT and ARF4 genes (Allen et al., 2005; Williams et al., 2005; Fahlgren et al., 2006; Hunter et al., 2006; Marin et al., 2010). In soybean, transcripts of ARF3/ETT (Glyma13g24240) and ARF4 (Glyma12g07560) were not only cleaved by the tasiARFs GmTAS3a,b 5′7D(+) and GmTAS3a,b 5′8D(+), but the ARF targets also produced phasiRNAs (Figure 4A; Supplemental Figure 7). Thus, both tasiARFs are phasiRNA triggers, as evidenced by processing downstream from the cleavage site using the 221 pathway. More significantly, this demonstrates that siRNAs can also function as phasiRNA triggers via a two-hit mechanism of biogenesis (Figure 4B).
Figure 4.
The ARF3 PHAS-Locus Triggered by the TasiARF.
(A) The soybean TAS3-derived tasiARFs target ARF3 at two identical sites, a 5′ site for which cleavage was validated by PARE (bottom panel) and a 3′ site for which no cleavage was observed. This two-hit activity of the tasiARFs generated phased siRNAs (middle panel). The y axis is a phasing “score” that is an estimated P value for the significance of phasing (see Methods). The lower two images are our Web browser showing the small RNA (middle) or PARE data (lower), with the orange dashed line indicating the tasiARF cleavage site. Colored spots are small RNAs with abundances indicated on the y axis; light blue spots indicate 21-nucleotide sRNAs, green are 22-nucleotide sRNAs, orange are 24-nucleotide sRNAs, and other colors are other sizes. Red boxes are annotated exons (pink are untranslated regions). Purple lines indicate a k-mer frequency for repeats.
(B) The data from panel A suggest a cascade of two-hit phased siRNA biogenesis in which the 21-nucleotide (nt) miR390 triggers 21-nucleotide tasiARF biogenesis, and via a two-hit mechanism, the tasiARFs trigger the production of additional secondary siRNAs from ARF3 and ARF4 (see Supplemental Figure 7 online). The ARF siRNAs may function in cis or trans.
PhasiRNAs with Differential Expression in Different Tissues and Treatments
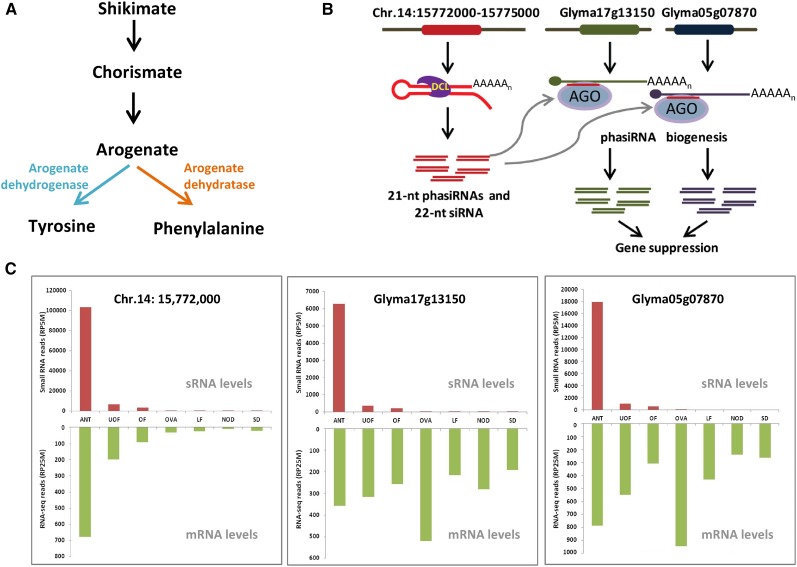
A number of phasiRNAs were expressed preferentially or exclusively in a particular tissue or treatment (Table 1). These analyses were based on our data from leaf, floral parts, and nodule and seed tissues. The phasiRNAs preferentially expressed in flower tissues identified six protein-coding PHAS loci and five non-protein-coding PHAS loci, including TAS3c and TAS3d, and three other novel noncoding loci (Table 1). Two of the six flower-enriched, protein-coding PHAS loci, Glyma05g07870 and Glyma17g13150, encode arogenate dehydrogenase, an enzyme responsible for the synthesis of tyrosine (Figure 5A). PhasiRNAs from these loci also matched to a long-stemmed, hairpin-forming, unannotated region of the genome (Figure 5B). A BLASTX similarity search of this transcript against soybean proteins revealed high similarity to a portion of the arogenate dehydrogenase-encoding genes, but with an open reading frame interrupted by stop codons suggesting that the hairpin transcript is a gene fragment. This locus and the two closely related arogenate dehydrogenase genes (a subset of seven soybean arogenate dehydrogenase-encoding loci), are robust sources of phasiRNAs (Supplemental Figure 8). While the phasiRNAs from these two arogenate dehydrogenase genes were anther specific, the two protein-coding homologs were expressed in other tissues without the accompanying production of phasiRNAs, indicating the phasiRNA trigger is absent from those tissues (Figure 5C). Thus, one model for the biogenesis of these phasiRNAs that is consistent with these data is that the pseudogene hairpin is directly processed by a Dicer protein to generate siRNAs that could function in trans as triggers of secondary siRNAs from the protein-coding genes, with the hairpin mRNA expression restricted to the anthers (Figure 5B).
Table 1. List of PHAS Genes Specific to Tissues or Treatments.
| Tissue/Treatment | Gene ID or Locus Coordinates | Annotation |
|---|---|---|
| Flower | Glyma08g04910 | ATP binding |
| Glyma08g06400 | Armadillo/β-catenin repeat family protein | |
| Glyma05g07870 | Arogenate dehydrogenase | |
| Glyma17g13150 | Arogenate dehydrogenase | |
| Glyma14g04800 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | |
| Glyma13g00630 | Cadmium ion transmembrane transporter | |
| Chr17:5607065:5607900 | TAS3c | |
| Chr13:1198656:1199379 | TAS3d | |
| Chr2:9271806:9271913 | Noncoding RNA | |
| Chr20:158383:158404 | Noncoding RNA | |
| Chr9:45231347:45231516 | Noncoding RNA | |
| Seed | Glyma02g13520 | Heat shock protein binding |
| Glyma05g04900 | MYB DOMAIN PROTEIN5 | |
| Glyma17g15270 | MYB DOMAIN PROTEIN5 | |
| Glyma11g33110 | ACYL ACTIVATING ENZYME12 | |
| Glyma18g05110 | BENZOYLOXYGLUCOSINOLATE1; benzoate-CoA ligase | |
| Chr18:60823864:60824073 | Noncoding RNA | |
| Nodule | Glyma10g01140 | DNA binding protein-related |
| Glyma12g03040 | Disease resistance protein (TIR-NB-LRR class), putative | |
| Glyma15g39460 | Disease resistance protein (CC-NB-LRR class), putative | |
| Glyma15g39620 | Disease resistance protein (NB-LRR class), putative | |
| Glyma15g39660 | Disease resistance protein (CC-NB-LRR class), putative | |
| Drought | Glyma11g13070 | ALLENE OXIDE SYNTHASE; allene oxide synthase |
Figure 5.
Anther-Specific PhasiRNAs Derived from Arogenate Dehydrogenase Loci.
(A) The biochemical pathway in which arogenate dehydrogenase is involved.
(B) Schematic pathway of phasiRNA production from arogenate dehydrogenase-related loci. At left, a gene fragment that forms a hairpin is processed into phasiRNAs.
(C) The levels of read abundances for miRNA triggers and phasiRNAs derived from the two arogenate dehydrogenase PHAS genes in different tissues (red bars) and the levels of gene expression (green bars) are shown as values normalized into RP5M (read per five million) and RP25M (read per 25 million), respectively.
Numerous other PHAS loci showed tissue enrichment or specificity. A set of phasiRNAs preferentially expressed in nodules was produced from NB-LRR genes, genes known for their role in disease resistance (Table 1). Such phasiRNAs could suppress disease resistance in nodules, promoting symbiotic interactions. Seed-preferential phasiRNAs were derived from five protein-coding and a noncoding locus; two of the former, Glyma05g04900 and Glyma17g15270, encode MYB DOMAIN PROTEIN5 (MYB5) and are triggered by miR828, exclusively expressed in seed tissues (Supplemental Figure 9). MYB5 has been reported previously to be involved in differentiation of the outer seed coat (Gonzalez et al., 2009) and anthocyanin biosynthesis (Dubos et al., 2010; Abeynayake et al., 2012), whereas miR828 in Arabidopsis functions to generate MYB-targeting tasiRNAs from TAS4 (Rajagopalan et al., 2006). Thus, relative to soybean, the Arabidopsis TAS4 locus may have evolved to separate the miR828-dependent phasiRNA generation and MYB suppression activities.
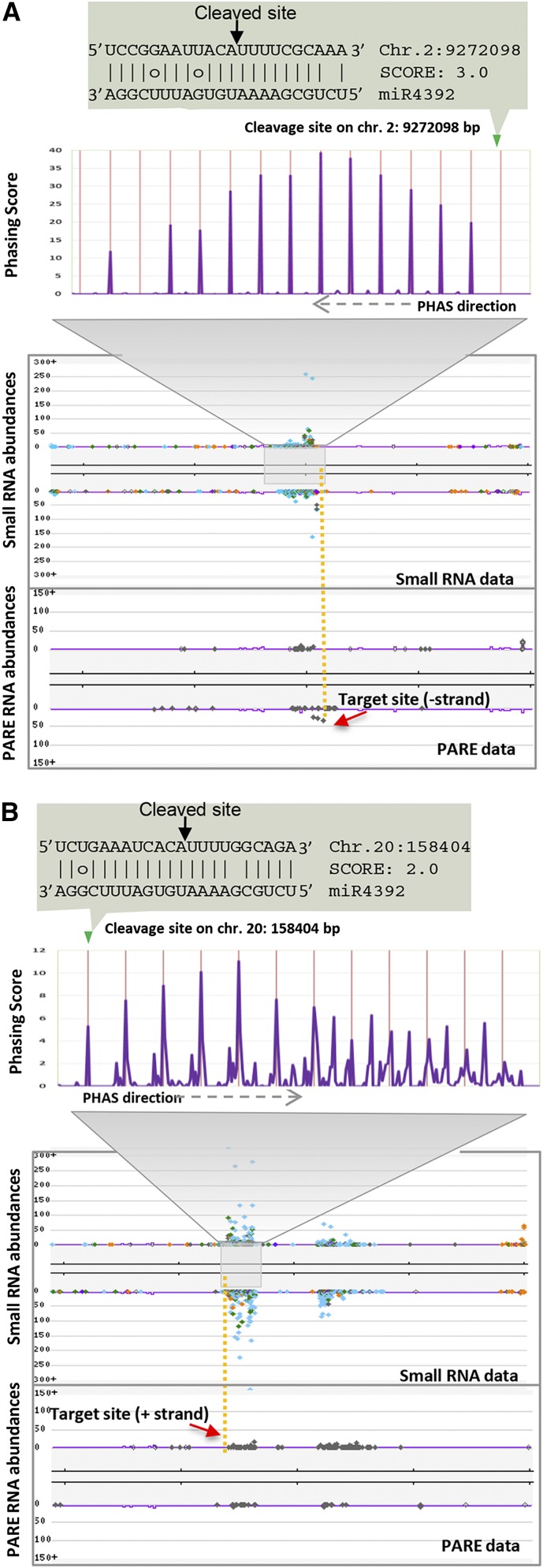
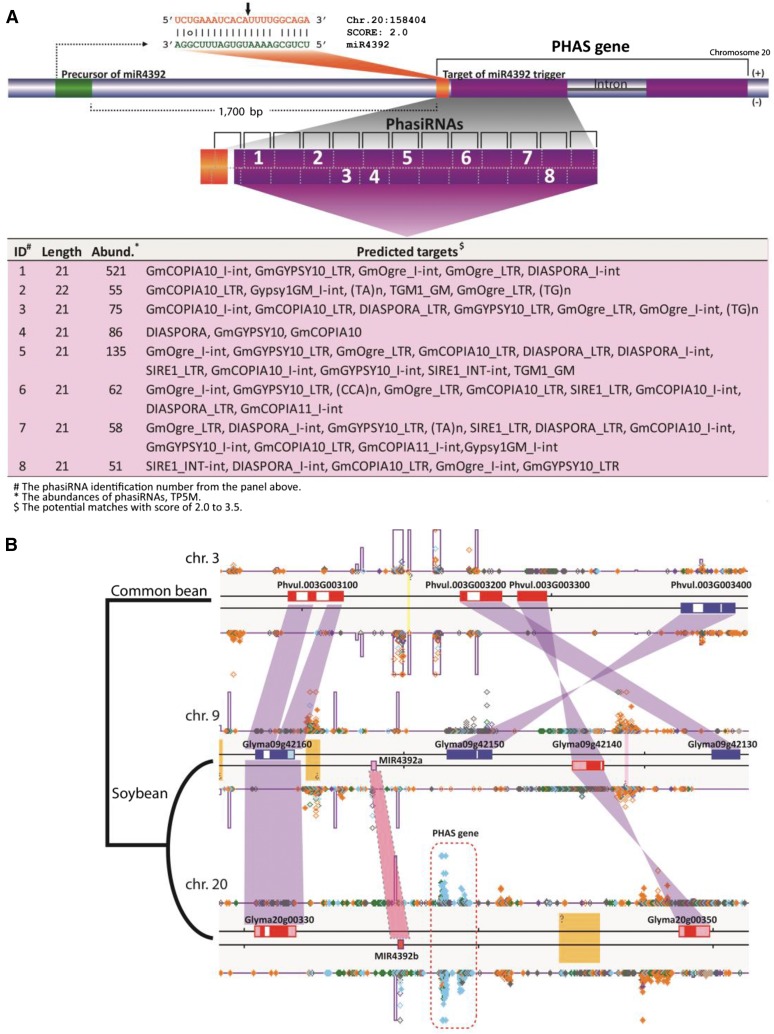
We identified one interesting pair of novel noncoding PHAS loci that were specific to the reproductive tissues and triggered by gma-miR4392, a previously poorly characterized miRNA (Figure 6). gma-miR4392 was particularly enriched in anthers (Figure 1B). Unusually, MIR4392, the locus generating the trigger, is arranged immediately adjacent to one of the PHAS loci that it targets, on chromosome 20 (Figure 7A). We found a homoeologous region on soybean chromosome 9 that also contains MIR4392; however, this copy is likely a pseudogene, since it was not expressed in any tissue analyzed in our study, suggesting a divergence in the ∼13 million years since these regions were duplicated. To assess the conservation of this arrangement in legumes, we identified the syntenic region between soybean and common bean (Phaseolus vulgaris; Figure 7B). MIR4392 was not present within this syntenic region or throughout the genome of common bean. This suggests that the adjacent arrangement of MIR4392 and the PHAS locus emerged in soybean in the 20 million years since the divergence of these species. We next searched for the putative targets of 39 of the most abundant phasiRNAs to infer their function. The best matches were to hundreds of LTR transposable elements (TEs; Figure 7A; Supplemental Data Set 1J). However, the locus itself is single copy, as are the phasiRNAs, despite their TE-targeting capability. Thus, these two PHAS loci may have a specialized function to regulate TE activity during plant reproduction.
Figure 6.
Anther Specificity of Two PHAS Loci.
sRNA and PARE reads mapped onto soybean genome indicating noncoding PHAS loci and their phasing patterns. The features of each panel is as described in previous figures. At the top of each panel, the “cleaved site” indicates weak PARE data supporting the specificity of cleavage but strong correspondence to the initiation site of the phasiRNAs.
(A) A phased locus on chromosome 2 at ∼9,272,000 bp.
(B) A phased locus on chromosome 20 at ∼158,400 bp.
Figure 7.
Pollen-Specific PhasiRNAs Targeting Transposons from the PHAS Locus on Chromosome 20.
(A) A locus on chromosome 20 produces phasiRNAs that are specifically expressed in pollen. These phasiRNAs are triggered by miR4392, the precursor of which located immediately upstream, and also accumulates specifically in pollen. The targets of the most abundant phasiRNAs from the locus were predicted using conventional rules for miRNA target identification. As indicated in the table below, the predicted targets for all of the abundant phasiRNAs belong to a wide variety of transposable elements. The lower table lists phasiRNAs that are abundant and target 10 or more transposon loci; the target data were selected from Supplemental Data Set 1J, which contains more details about these phasiRNAs. The abundance data are the sum of abundance for all the libraries described in this article.
(B) The syntenic region between soybean and common bean. Both MIR4392 and the PHAS locus are located on the chromosome 20 of soybean. The phasiRNA signals detected at the PHAS locus are indicated in the dotted rectangle. The MIR4392 pseudogene located in the duplicated region on the chromosome 9 of soybean is shown. No phasiRNA signal was detected in duplicated region on chromosome 9. The MIR4392 locus is absent in the synthetic region on the chromosome 3 of common bean. No phasiRNA signal was detected in the syntenic region of common bean. Genes located on the sense and antisense strands of the chromosomes are presented in red and blue, respectively.
Finally, we examined PHAS loci showing an abundance graduated over developmental stages or treatment time points. Developmentally varied PHAS loci included the tasiARFs from TAS3a and TAS3b that were highly abundant in the early stage (10 d) of nodule development and gradually reduced in later stages (Supplemental Figure 10), consistent with the abundance of the trigger, miR390. The only PHAS locus showing a gradient over the treatments was the drought-responsive locus Glyma11g13070 encoding allene oxide synthase (AOS). PhasiRNAs from this locus accumulated in the leaf tissues subjected to drought stress in both tolerant and susceptible genotypes. AOS is a member of the large CYP74 cytochrome P450 family and converts lipoxygenase-derived fatty acid hydroperoxide to allene epoxide, the first dedicated step in jasmonate synthesis. AOS is upregulated in drought stress in barley (Hordeum vulgare) (Talamè et al., 2007).
DISCUSSION
We analyzed an extremely large set of small RNAs (>1.9 billion sequences or >1 billion after filtering structural RNAs) generated from various tissues, developmental stages, and treatments of soybean. With these data, we generated an “atlas” of soybean small RNAs. Applying three criteria, abundance, the abundance and strand ratios to the current set of miRBase-annotated miRNAs (miRBase v20), we found that 312 MIRNA loci produce typical miRNAs, while another 203 produce small RNAs more reminiscent of siRNAs with a 24-nucleotide length. An additional 191 annotated miRNAs were weakly abundant and poorly conserved, comparing soybean and Arabidopsis. These poorly accumulating miRNAs were also poorly conserved among legume species (miRBase v20), suggesting that they are soybean specific (Supplemental Data Set 1K). Our analysis of variation in sizes and abundances identified many miRNAs of different sizes than those registered in miRBase, and different variants of the same miRNA accumulated divergently. Thus, we believe that these results can substantially improve the miRBase data for soybean.
We used a statistically rigorous method to identify hundreds of PHAS loci in soybean, an approach used with increasing frequency to discover large numbers of such loci (Fei et al., 2013). While soybean PHAS loci have been described previously (Zhai et al., 2011), we had ∼50-fold deeper data (>1 billion small RNAs), generating the greatest number of PHAS loci (504) described for any dicot. All previously reported soybean PHAS loci overlapped with those reported here. Twenty different miRNA triggers were identified for 127 PHAS loci, leaving the triggers unverified for the majority, perhaps due to low precursor expression and a corresponding lack of abundant PARE reads. Alternatively, the triggers might be other phasiRNAs, not miRNAs (such as the case of tasiARFs that we described), and comprehensive target assessment of the many thousands of phasiRNAs is at present computationally challenging. Many PHAS loci in the same gene family shared miRNA triggers, and in many of those cases, this was because the binding sites are in conserved domains. For example, gma-miR1510b-p targets 20 NB-LRR genes at the core P-loop-encoding motif (Zhai et al., 2011).
Among the most intriguing of observations we made is the large number of 21-nucleotide triggers of phasiRNAs. Previous reports show that a 22-nucleotide length plus an asymmetric bulge in the precursor shunts a target mRNA into the 122 phasing pathway (Chen et al., 2010; Cuperus et al., 2010; Zhai et al., 2011; Manavella et al., 2012). Of 20 miRNA triggers, we found 14 of them are a conventional 22-nucleotide in length; the remainder are 21 nucleotides, an unconventional length for PHAS triggers. Manavella et al. (2012) demonstrated that 21-nucleotide miRNAs are competent to trigger secondary siRNA production when derived from an asymmetrically bulged miRNA/miRNA* duplex. However, 10 of the miRNAs that trigger phasiRNAs had no bulge in the precursor stem-loop (Supplemental Figure 6B). One possibility is that these miRNAs trigger phasiRNAs via the two-hit pathway, a pathway almost exclusively represented by miR390-TAS3 (Axtell et al., 2006) and, more recently, an AP2 gene triggered by miR172/miR156 (Zhai et al., 2011). It remains to be determined whether all of these soybean two-hit loci would involve the canonical AGO protein for miRNA binding (AGO1) or the only AGO known to function in the two-hit pathway (AGO7). The 5′ nucleotide has a role in sorting of individual sRNAs into specific AGOs in plants (Mi et al., 2008; Takeda et al., 2008), with 22-nucleotide, 5′U PHAS triggers associated with AGO1 (Cuperus et al., 2010). Most of the triggers identified in this study, including three 21-nucleotide triggers, have a “U” in the 5′-terminus, potentially loaded into AGO1. It is possible that of the 21-nucleotide 5′U miRNAs triggering phasiRNAs, a small portion of 22-nucleotide variants are also produced and act as the triggers of phasiRNA production. Another possibility is that the 21-nucleotide 5′U miRNAs are modified after processing (i.e., tailed) to 22 nucleotides to function as a trigger as recently reported by Zhai et al. (2013). In summary, our study suggests that the number and diversity of phasiRNAs and the diversity of miRNA triggers as well as their mode of action varies substantially between soybean and the model dicot Arabidopsis.
We identified 208 soybean PHAS loci that overlapped with NB-LRR genes, accounting for 41% of the total number of PHAS loci. NB-LRRs are the most numerous disease resistance gene (R gene) class in plants (Dangl and Jones, 2001), with as many as 653 NB-LRR genes in a genome (in rice) (Shang et al., 2009). A high number of phasi-NB-LRR genes has been observed in a number of dicots and as far back as gymnosperms, yet this mode of gene regulation is absent or largely diminished in several species, including Arabidopsis and rice (Källman et al., 2013). In soybean, 319 NB-LRR genes have been characterized (Kang et al., 2012); of these, and like the other well-studied legume M. truncatula (Zhai et al., 2011), the great majority produce 21-nucleotide secondary siRNAs. It was proposed that in the absence of a pathogen, phasiRNAs reduce NB-LRR expression and correspondingly the energetic cost to the plant (Shivaprasad et al., 2012). Alternatively, secondary siRNAs might limit runaway transcription of NB-LRRs in species with a rapidly expanding gene family (Källman et al., 2013). Additional hypotheses for the highly redundant suppression of NB-LRRs by small RNAs have been proposed (Fei et al., 2013). Perhaps the most relevant of these hypotheses in general terms is that phasiRNAs might function to buffer NB-LRR transcript levels, with increased biogenesis and suppression activity when the precursors are more abundant and vice versa when precursor transcript levels are reduced. Since most NB-LRRs in legumes are apparently subject to posttranscriptional control by phasiRNAs, while the two model plants rice and Arabidopsis largely lack phasi-NB-LRRs, those models will likely prove useful systems functional analysis of secondary siRNAs targeting NB-LRRs.
We characterized novel variants of the TAS3 locus in soybean from among its six TAS3 loci. While four TAS3 loci (TAS3a-d) display conventional trigger sites and phasing patterns (Axtell et al., 2006), we discovered two unconventional TAS3 loci (TAS3e and TAS3f). The TAS3e contained three miR390 binding sites instead of two, the phasing indicates downstream instead of upstream processed (relative to miR390 cleavage). By contrast, TAS3f contained the typical two miR390 binding sites, but with TAS3e-like downstream production of phasiRNAs. The phasiRNAs derived from these six TAS3 loci also varied in length and sequence, as well as their accumulation patterns, with the TAS3a and TAS3b tasiARFs present in all tissues at varying levels, whereas the TAS3c and TAS3d tasiARFs were abundant in flower tissues but low or absent in other tissues. The TAS3e and TAS3f phasiRNAs were not expressed in nodules. Thus, the ARF target mRNA abundance in different tissues may be determined or fine-tuned by these diverse different tasiARFs, with particularly diverse tasiARF accumulation in nodule development.
An integration of our work on phasiRNAs with prior publications on miRNA activity suggests that small RNAs play important roles in nodule development. Many miRNAs that we described as PHAS triggers were previously predicted to have a role in nodule development (Subramanian, 2012). For example, miR482, miR1507, and miR1510, which all target NB-LRR PHAS loci (Table 2), were proposed to function during rhizobium colonization in the modulation of defense response/host specificity (Subramanian, 2012), and miR160, which targets the ARF10 PHAS locus, in enhanced auxin responses (Subramanian et al., 2008). During nodule differentiation, miR167, which targets ARF6 and ARF8, has a role in auxin signaling/vascular differentiation (Yang et al., 2006) and miR169, which targets transcripts encoding NF-Y transcription factors, in nodule zone differentiation (Simon et al., 2009). miR172, which targets transcripts for AP2 and RAP2 transcription factors (Table 2), has also been reported to regulate nodulation in soybean (Yan et al., 2013), and miR396, for which one target is a PHAS locus encoding a long-chain fatty acid CoA ligase (Table 2), functions in ROS scavenging (Bazin et al., 2013). The activity of these miRNAs to trigger phasiRNAs is consistent with complex posttranscriptional regulation in nodule development.
Table 2. Secondary siRNA-Triggering miRNAs in Soybean.
| miRNA Trigger | Sequence | Length (nt)a | Targeted Gene Family |
|---|---|---|---|
| miR1507a | UCUCAUUCCAUACAUCGUCUGA | 22-nt miRNA | Disease resistance protein (NB-LRR class), putative |
| EPR1 (EARLY-PHYTOCHROME-RESPONSIVE1) | |||
| miR1508a | ACUGCUAUUCCCAUUUCUAAAC | 22-nt miRNA | Pentatricopeptide (PPR) repeat-containing protein |
| miR1509a | UUAAUCAAGGAAAUCACGGUCG | 22-nt miRNA | Noncoding gene |
| miR1510bb | AGGGAUAGGUAAAACAACUACU | 22-nt miRNA | Disease resistance protein (TIR-NB-LRR class), putative |
| miR1514a | UUCAUUUUUAAAAUAGGCAUUG | 22-nt miRNA | No apical meristem (NAM) family protein |
| NTL9 (NAC transcription factor-like 9); transcription factor | |||
| miR1515b | UCAUUUUGCGUGCAAUGAUCUG | 22-nt miRNA | DCL2 (DICER-LIKE2) |
| miR160bb | UGCCUGGCUCCCUGUAUGCCA | 21-nt miRNA | ARF10 (AUXIN RESPONSE FACTOR10) |
| miR167e | UGAAGCUGCCAGCAUGAUCUUA | 22-nt miRNA | ARF6 (AUXIN RESPONSE FACTOR6); transcription factor |
| ARF8 (AUXIN RESPONSE FACTOR8); transcription factor | |||
| Transcription elongation factor-related | |||
| miR169ab | CAGCCAAGGAUGACUUGCCGG | 21-nt miRNA | NF-YA1 (NUCLEAR FACTOR Y, SUBUNIT A1); transcription factor |
| NF-YA10 (NUCLEAR FACTOR Y, SUBUNIT A10); transcription factor | |||
| miR172ab | AGAAUCUUGAUGAUGCUGCAU | 21-nt miRNA | RAP2 (RELATED TO AP2); DNA binding / transcription factor |
| AP2 (APETALA2); transcription factor | |||
| Transducin family protein/WD-40 repeat family protein | |||
| miR2109b | UGCGAGUGUCUUCGCCUCUGA | 21-nt miRNA | Disease resistance protein (TIR-NB-LRR class), putative |
| miR2118a | GGAGATGGGAGGGTCGGTAAAG | 22-nt miRNA | SGS3 (SUPPRESSOR OF GENE SILENCING3) |
| miR319gb | UUGGACUGAAGGGAGCUCCUUC | 22-nt miRNA | TCP family transcription factor, putative |
| TCP3; transcription factor | |||
| TCP4 (TCP family transcription factor 4); transcription factor | |||
| miR390gb | AAGCUCAGGAGGGAUAGCGCC | 21-nt miRNA | TAS3 |
| miR393 | UCCAAAGGGAUCGCAUUGAUCC | 22-nt miRNA | ATCSLE1; cellulose synthase/transferase, transferring glycosyl groups |
| AFB2 (AUXIN SIGNALING F-BOX2); auxin binding | |||
| TIR1 (TRANSPORT INHIBITOR RESPONSE1) | |||
| miR396eb | UUCCACAGCUUUCUUGAACUGU | 22-nt miRNA | Long-chain fatty acid–CoA ligase family protein |
| HSL1 (HAESA-Like 1); ATP binding | |||
| Calcium-transporting ATPase | |||
| miR403ab | UUAGAUUCACGCACAAACUUG | 21-nt miRNA | AGO2 (Argonaute 2); nucleic acid binding |
| miR482b | UCUUCCCUACACCUCCCAUACC | 22-nt miRNA | ATP binding |
| Disease resistance protein (CC-NB-LRR class), putative | |||
| Disease resistance protein (TIR-NB-LRR class), putative | |||
| MEE32 (MATERNAL EFFECT EMBRYO ARREST32) | |||
| RPS2 (RESISTANT TO PSYRINGAE2); protein binding | |||
| miR4392 | UCUGCGAAAAUGUGAUUUCGGA | 22-nt miRNA | Noncoding gene |
| miR828ab | UCUUGCUCAAAUGAGUAUUCCA | 22-nt miRNA | ATMYB5 (MYB DOMAIN PROTEIN5); DNA binding |
nt, nucleotide.
No bulge in the stem-loop.
Two noncoding PHAS loci accumulated to very high levels in soybean anthers, the same tissue in which the trigger of both PHAS loci, miR4392, was highly enriched. While we were unable to validate the result with PARE analysis, target prediction suggests that these phasiRNAs may target hundreds of LTR retrotransposons (Figure 7A). It is possible that they might have a similar role as recently described vegetative nucleus-derived siRNAs that regulate TEs in pollen (Slotkin et al., 2009; Creasey et al., 2014). In M. truncatula, we previously identified an unusual flower-enriched PHAS locus (Zhai et al., 2011). Despite numerous similarities including expression levels as measured by RNA-seq of the miRNA trigger and PHAS locus (Supplemental Figures 11A to 11C), the PHAS genes in M. truncatula and soybean are not orthologous, as evidenced in part by the different miRNA triggers. However, the adjacent arrangement of the trigger and the anther-specific PHAS locus observed in soybean (Supplemental Figure 11B) was entirely reminiscent of that found in M. truncatula, in which there are two adjacent PHAS genes (Supplemental Figure 11C). It is unclear whether the two PHAS genes in M. truncatula are pollen specific because the available data were based on the whole flowers (Zhai et al., 2011). However, the predicted targets of phasiRNAs generated from the M. truncatula PHAS genes are also TEs (Supplemental Data Set 1L), suggesting a mechanism conserved in legumes for the regulation of TEs during reproduction (see the model in Supplemental Figure 12).
Unlike Arabidopsis, and due to challenges such as genetic redundancy, soybean has limited resources of genetic mutants that would facilitate the study of miRNAs and phasiRNAs. However, approaches for targeted genome editing using methods such as TALENs (transcription-activator like effector nucleases) (Christian et al., 2010) or CRISPRs (clustered, regularly interspaced, short palindromic repeats) (Cong et al., 2013) are a promising option for the analysis of small RNAs in soybean. In addition, the reduction of specific miRNAs by short tandem target mimic (Yan et al., 2012) or other target-mimic approaches would be useful as a tool to study the role of miRNA triggers.
Conclusions
We present an analysis of a large small RNA data set for soybean, as well as PARE data, to identify PHAS loci, to evaluate miRBase-annotated miRNAs, and to discover novel miRNAs. Hundreds of PHAS loci were identified, many of which overlapped with protein-encoding genes. The largest class of the protein-coding PHAS loci was NB-LRRs, some of which were abundant specifically in nodules. Two additional TAS3 loci and atypical phasing patterns were also revealed. We demonstrated that phasiRNAs and both known and novel miRNAs were expressed preferentially in particular tissues and treatments. Among these, we identified a pair of novel noncoding PHAS loci and their trigger, both specific to anthers. A specialized function of the phasiRNAs generated from this locus may be to regulate TE activity during plant reproduction.
METHODS
Plant Materials
For reproductive tissues, soybean (Glycine max) plants from the cultivar Williams 82 were grown in the greenhouse under 16 h light/8 h dark at 25°C. Floral tissues were collected from unopened flowers and flowers open for 1 d. Anther and ovary tissues were dissected from unopened flowers. For the nodule tissues, developing nodules were collected from soybean 10, 15, 20, 25, and 30 d after inoculation with Bradyrhizobium japonicum strain USDA110. For water-stressed samples, the inbred lines IA3023 and LD00-3309 were sown in two pots, one as a control and another for the stress treatment. The plants were grown up to V1 stage, and all pots were irrigated once in 2 d to the field capacity (1600 mL water). At V1 stage, the irrigation was withheld from stress pots alone, and the control pots were irrigated until the end of the experiment. Once the 50% of the plants under stress reached a permanent wilting point (leaf water potential ranging from −8 to −10 bars), the leaf samples were collected from both the control and stress pots. For the pathogen-mimic treatments, the leaf samples from three genotypes of soybean, Williams 82, Dassel, and Vinton 81, were treated with chitin octamer and water control for 30 min. The leaf samples from the same genotypes were also treated with conserved 22-amino acid peptide from bacterial flagellin 22 and water control for 30 min. Samples collected from all tissues were immediately frozen in liquid nitrogen before RNA extraction.
RNA Extraction and Sequencing of sRNAs and PARE
Total RNA was isolated from the plant materials using Concert Plant RNA Reagent (Invitrogen/Life Technologies). Small RNA libraries were constructed using the TruSeq Small RNA Sample Preparation Kits (Illumina). PARE libraries were constructed as previously described (Zhai et al., 2014). The libraries were sequenced on an Illumina HiSequation 2000 at the Delaware Biotechnology Institute (Newark, DE).
Computational Analysis of Sequencing Data
The raw sequencing data were trimmed to remove adaptor sequences and then mapped to the soybean genome (DOE-JGI Community Sequencing Program v1.1) using Bowtie (Langmead et al., 2009). Reads that perfectly matched the soybean genome, excluding those matching tRNAs, rRNAs, snRNA, and snoRNAs, were used for further study. Soybean mature miRNAs and their precursors were retrieved from miRBase (version 20; http://www.mirbase.org/).
miRNA Prediction Pipeline
The miRNA prediction pipeline is outlined in Supplemental Figure 4. Individual steps in the process were performed with Perl scripts as described (Jeong et al., 2011) combined with miREAP (https://sourceforge.net/projects/mireap/) and CentroidFold (Sato et al., 2009). miREAP was used to evaluate the pairing of the miRNA and miRNA* with the parameters set to allow a maximal distance of 400 nucleotides between miRNA and miRNA* (-d 400), extending 25 nucleotides at the end of the precursor (-f 25), with filters optimized for animal miRNAs turned off and including minor tuning for plant miRNA characteristics (our modified version of miREAP is available upon request). In addition, two miRNA features were requested: single-strand bias ≥0.9 and an abundance bias ≥0.7 based on the features of conserved miRNAs. CentroidFold was used with default settings to visualize the overall miRNA precursor structure for manual evaluation.
miRNA Target Prediction and PARE Validation
Genome-wide targets of 394 microRNAs were identified and validated; this represents 312 typical miRNAs, 15 marginal miRNAs, 44 new miRNA variants, and 23 novel miRNAs. Validation was performed using the using the sPARTA package (Kakrana et al., 2015). Target prediction was performed using sPARTA’s built-in target prediction module miRferno with standard scoring schema and score cutoff ≤7, followed by PARE-based validation of predicted targets. The validated miRNA-target interactions that passed a threshold of a corrected P value cutoff of 0.05 and with PARE read abundance ≥5 at the cleavage site were considered for further interpretation.
Phasing Analysis
After mapping sRNA reads to soybean genome, individual sRNAs were denoted with their matching coordinates. A two-nucleotide positive offset was added for sRNA matching to the antisense strand because of the existence of two-nucleotide overhang at the 3′ end of sRNA duplex. A genome-wide search was performed using a nine-cycle sliding window (189 bp) with each shift of three cycles (63 bp), and windows were reported when ≥10 unique reads fell into a nine-cycle window, ≥50% of matched unique reads were 21 nucleotides in length, and with ≥3 unique reads fell into a certain register. Next reported windows with overlapping region were combined into a single longer window. Then, a P value was calculated for each window based on the mapping results using an algorithm of Xia et al. (2013). As a final check of loci with phasing P value ≤ 0.001, P value and abundances of small RNAs from each locus were graphed and checked visually to remove false positives such as miRNA loci with numerous low abundance peaks could incorrectly pass our filters. Unannotated tRNA and rRNA-like loci were also manually removed.
Analysis of Differential Abundance of miRNAs
Differential expression for all miRNAs was analyzed in pairwise (i.e., control versus stress treatment) for water-stressed and pathogen-mimic-treated samples based on the read abundance data and using Bioconductor’s R package “baySeq”(Hardcastle and Kelly, 2010). miRNAs accumulating to significantly different levels were identified based on the estimated posterior likelihoods of ≥0.95.
Accession Numbers
The soybean small RNA and PARE sequencing data were submitted to the NCBI Gene Expression Omnibus under accession number GSE58779.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Small RNA Size Distribution from Different Soybean Tissues.
Supplemental Figure 2. Leaf Small RNA Size Distributions in Arabidopsis and Soybean.
Supplemental Figure 3. Examples of miRBase-Annotated miRNAs in Our Four Categories of Assessment.
Supplemental Figure 4. The Pipeline for the Identification of Novel miRNA and miRNA Variants.
Supplemental Figure 5. Abundance Profiles of the miR171 Family.
Supplemental Figure 6. Secondary Structures for the Stem-Loop Region of Precursors of the Soybean miRNAs That are Triggers of PHAS Loci.
Supplemental Figure 7. PhasiRNAs from the ARF4 Locus Are Triggered by TasiARFs.
Supplemental Figure 8. Several Soybean Loci Related to Arogenate Dehydrogenase-Encoding Transcripts Are Abundant Sources of 21-Nucleotide PhasiRNAs.
Supplemental Figure 9. Seed-Specific PHAS Loci from MYB5 Genes.
Supplemental Figure 10. Decrease of miR390 and TAS3a and TAS3b Read Abundances during Nodule Development.
Supplemental Figure 11. Colocalization of Legume Anther- or Flower-Specific PHAS Loci and Their miRNA Triggers.
Supplemental Figure 12. A Model of Transposable Element Regulation in Legume Pollen by Pollen-Specific PhasiRNAs.
Supplemental Data Set 1A. Number of Small RNAs from Soybean Libraries, from SBS Sequencing.
Supplemental Data Set 1B. Evaluation of Annotated Soybean miRNAs from miRBase (Version 20).
Supplemental Data Set 1C. Novel Soybean miRNAs.
Supplemental Data Set 1D. Soybean miRNA Variants.
Supplemental Data Set 1E. Bayseq Results for Biotic and Abiotic Stressed Soybean.
Supplemental Data Set 1F. Number of PARE Reads from Soybean Libraries, by SBS Sequencing.
Supplemental Data Set 1G. PARE Validation of miRNA Targets from miRBase Version 20.
Supplemental Data Set 1H. PARE Validation of Targets of Novel miRNAs and Variants.
Supplemental Data Set 1I. PHAS Loci in Soybean.
Supplemental Data Set 1J. Repeat Targets of PhasiRNAs from Soybean Anthers.
Supplemental Data Set 1K. Species Specificity of Soybean Low Abundance miRNAs.
Supplemental Data Set 1L. Repeat Targets of PhasiRNAs from Medicago Flower.
Supplementary Material
Acknowledgments
We thank Moaine Elbaidouri and Scott A. Jackson (University of Georgia) for helpful discussions about the synteny analysis. We also thank George Graef (University of Nebraska) for providing Vinton81 and Dassel seeds. This work was supported by funding from the United Soybean Board and the North Central Soybean Research Program. Research in the G.S. laboratory was funded by a grant from the U.S. National Science Foundation Plant Genome Program (Grant DBI-0421620).
AUTHOR CONTRIBUTIONS
S.A. generated the sequencing data. S.A., R.X., A.K., and J.X. carried out bioinformatic analyses. S.A., K.H., Z.Y., O.V.-L., T.A.M., S.P., H.T.N., and G.S. generated materials and data. S.A., R.X., J.Z., and B.C.M. conceived of the study, participated in its design and coordination, and wrote the article. All authors read and approved the final article.
Glossary
- miRNA
microRNA
- siRNA
small interfering RNA
- dsRNA
double-stranded RNA
- phasiRNA
phased siRNA
- PARE
parallel analysis of RNA end
- TP5M
transcripts per five million
- tasiARF
trans-acting short interfering RNA-auxin response factor
- TE
transposable element
Footnotes
Online version contains Web-only data.
References
- Abeynayake S.W., Panter S., Chapman R., Webster T., Rochfort S., Mouradov A., Spangenberg G. (2012). Biosynthesis of proanthocyanidins in white clover flowers: cross talk within the flavonoid pathway. Plant Physiol. 158: 666–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18: 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., Vaucheret H. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16: 927–932. [DOI] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A.M., Carrington J.C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221. [DOI] [PubMed] [Google Scholar]
- Axtell M.J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64: 137–159. [DOI] [PubMed] [Google Scholar]
- Axtell M.J., Jan C., Rajagopalan R., Bartel D.P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127: 565–577. [DOI] [PubMed] [Google Scholar]
- Bazin J., Khan G.A., Combier J.P., Bustos-Sanmamed P., Debernardi J.M., Rodriguez R., Sorin C., Palatnik J., Hartmann C., Crespi M., Lelandais-Brière C. (2013). miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 74: 920–934. [DOI] [PubMed] [Google Scholar]
- Carthew R.W., Sontheimer E.J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Li Y.H., Wu S.H. (2007). Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3318–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Chen L.T., Patel K., Li Y.H., Baulcombe D.C., Wu S.H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. USA 107: 15269–15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25: 21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186: 757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey K.M., Zhai J., Borges F., Van Ex F., Regulski M., Meyers B.C., Martienssen R.A. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508: 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J.T., Carbonell A., Fahlgren N., Garcia-Ruiz H., Burke R.T., Takeda A., Sullivan C.M., Gilbert S.D., Montgomery T.A., Carrington J.C. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17: 997–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581. [DOI] [PubMed] [Google Scholar]
- Dunoyer P., Schott G., Himber C., Meyer D., Takeda A., Carrington J.C., Voinnet O. (2010). Small RNA duplexes function as mobile silencing signals between plant cells. Science 328: 912–916. [DOI] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95: 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944. [DOI] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M.A., Collado M., Muñoz-Fontela C., Matheu A., Marcos-Villar L., Arroyo J., Esteban M., Serrano M., Rivas C. (2006). Antiviral action of the tumor suppressor ARF. EMBO J. 25: 4284–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German M.A., Luo S., Schroth G., Meyers B.C., Green P.J. (2009). Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc. 4: 356–362. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Mendenhall J., Huo Y., Lloyd A. (2009). TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 325: 412–421. [DOI] [PubMed] [Google Scholar]
- Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866. [DOI] [PubMed] [Google Scholar]
- Hardcastle T.J., Kelly K.A. (2010). baySeq: empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. (2007). Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 19: 926–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutiérrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., Park S., Zhai J., Gurazada S.G., De Paoli E., tMeyers B.C., Green P.J. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., et al. (2013). Parallel analysis of RNA ends enhances global investigation of microRNAs and target RNAs of Brachypodium distachyon. Genome Biol. 14: R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C., Kasprzewska A., Tennessen K., Fernandes J., Nan G.L., Walbot V., Sundaresan V., Vance V., Bowman L.H. (2009). Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 19: 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakrana A., Hammond R., Patel P., Nakano M., Meyers B.C. (2015). sPARTA: a parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets, including new miRNA target-identification software. Nucleic Acids Res. 42: e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källman T., Chen J., Gyllenstrand N., Lagercrantz U. (2013). A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 162: 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.J., Kim K.H., Shim S., Yoon M.Y., Sun S., Kim M.Y., Van K., Lee S.H. (2012). Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol. 12: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Habu Y. (2011). siRNA-mediated chromatin maintenance and its function in Arabidopsis thaliana. Biochim. Biophys. Acta 1809: 444–451. [DOI] [PubMed] [Google Scholar]
- Kim V.N. (2005). MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 6: 376–385. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wang X., Zhang S., Liu D., Duan Y., Dong W. (2012). Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE 7: e39650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wei W., Ouyang S., Zhang J.S., Chen S.Y., Zhang W.K. (2009). Analysis of expressed receptor-like kinases (RLKs) in soybean. J. Genet. Genomics 36: 611–619. [DOI] [PubMed] [Google Scholar]
- Liu Y., Wang Y., Zhu Q.H., Fan L. (2013). Identification of phasiRNAs in wild rice (Oryza rufipogon). Plant Signal. Behav. 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Bouché N. (2008). MicroRNA-directed regulation: to cleave or not to cleave. Trends Plant Sci. 13: 359–367. [DOI] [PubMed] [Google Scholar]
- Manavella P.A., Koenig D., Weigel D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. USA 109: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin E., Jouannet V., Herz A., Lokerse A.S., Weijers D., Vaucheret H., Nussaume L., Crespi M.D., Maizel A. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S., Voinnet O., Mette M.F., Matzke M., Vaucheret H., Ding S.W., Pruss G., Vance V.B. (2002). RNA silencing and the mobile silencing signal. Plant Cell 14 (suppl.): S289–S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Hamada M., Asai K., Mituyama T. (2009). CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 37: W277–W280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H., Tsugane K., Kanno T., Nishimura T. (2012). DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol. 53: 766–784. [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Shang J., et al. (2009). Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site—leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes. Genetics 182: 1303–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.M., Patel K., Bond D.M., Santos B.A., Baulcombe D.C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S.A., Meyers B.C., Sherrier D.J. (2009). MicroRNAs in the rhizobia legume symbiosis. Plant Physiol. 151: 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Vaughn M., Borges F., Tanurdzić M., Becker J.D., Feijó J.A., Martienssen R.A. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q.X., Liu Y.F., Hu X.Y., Zhang W.K., Ma B., Chen S.Y., Zhang J.S. (2011). Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol. 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S. (2012). MicroRNA regulation of symbiotic nodule development in legumes. In MicroRNAs in Plant Development and Stress Responses, R. Sunkar, ed (Berlin, Heidelberg: Springer), pp. 177–195. [Google Scholar]
- Subramanian S., Fu Y., Sunkar R., Barbazuk W.B., Zhu J.K., Yu O. (2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49: 493–500. [DOI] [PubMed] [Google Scholar]
- Talamè V., Ozturk N.Z., Bohnert H.J., Tuberosa R. (2007). Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J. Exp. Bot. 58: 229–240. [DOI] [PubMed] [Google Scholar]
- Tuteja J.H., Zabala G., Varala K., Hudson M., Vodkin L.O. (2009). Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell 21: 3063–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L., Carles C.C., Osmont K.S., Fletcher J.C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. USA 102: 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R., Meyers B.C., Liu Z., Beers E.P., Ye S., Liu Z. (2013). MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in Eudicots. Plant Cell 25: 1555–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Gu Y., Jia X., Kang W., Pan S., Tang X., Chen X., Tang G. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Hossain M.S., Wang J., Valdés-López O., Liang Y., Libault M., Qiu L., Stacey G. (2013). miR172 regulates soybean nodulation. Mol. Plant Microbe Interact. 26: 1371–1377. [DOI] [PubMed] [Google Scholar]
- Yang J.H., Han S.J., Yoon E.K., Lee W.S. (2006). Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 34: 1892–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M., Peragine A., Park M.Y., Poethig R.S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19: 2164–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala G., Campos E., Varala K.K., Bloomfield S., Jones S.I., Win H., Tuteja J.H., Calla B., Clough S.J., Hudson M., Vodkin L.O. (2012). Divergent patterns of endogenous small RNA populations from seed and vegetative tissues of Glycine max. BMC Plant Biol. 12: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Arikit S., Simon S.A., Kingham B.F., Meyers B.C. (2014). Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods 67: 84–90. [DOI] [PubMed] [Google Scholar]
- Zhai J., et al. (2013). Plant microRNAs display differential 3′ truncation and tailing modifications that are ARGONAUTE1 dependent and conserved across species. Plant Cell 25: 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25: 2540–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Li G., Wang J., Fang J. (2012). Identification of trans-acting siRNAs and their regulatory cascades in grapevine. Bioinformatics 28: 2561–2568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.