Genetic analyses of maize genes in DNA methylation pathways reveal differences between maize and Arabidopsis, including evidence that DNA methylation is required for growth and development in maize.
Abstract
DNA methylation can play important roles in the regulation of transposable elements and genes. A collection of mutant alleles for 11 maize (Zea mays) genes predicted to play roles in controlling DNA methylation were isolated through forward- or reverse-genetic approaches. Low-coverage whole-genome bisulfite sequencing and high-coverage sequence-capture bisulfite sequencing were applied to mutant lines to determine context- and locus-specific effects of these mutations on DNA methylation profiles. Plants containing mutant alleles for components of the RNA-directed DNA methylation pathway exhibit loss of CHH methylation at many loci as well as CG and CHG methylation at a small number of loci. Plants containing loss-of-function alleles for chromomethylase (CMT) genes exhibit strong genome-wide reductions in CHG methylation and some locus-specific loss of CHH methylation. In an attempt to identify stocks with stronger reductions in DNA methylation levels than provided by single gene mutations, we performed crosses to create double mutants for the maize CMT3 orthologs, Zmet2 and Zmet5, and for the maize DDM1 orthologs, Chr101 and Chr106. While loss-of-function alleles are viable as single gene mutants, the double mutants were not recovered, suggesting that severe perturbations of the maize methylome may have stronger deleterious phenotypic effects than in Arabidopsis thaliana.
INTRODUCTION
DNA methylation involves covalent modification of cytosine base in genomic DNA with the addition of a methyl group (Razin and Riggs, 1980). While DNA methylation can be a reversible process with passive loss following DNA replication or active loss catalyzed by a family of DNA glycosylases (Zhang and Zhu, 2012), there is also evidence that DNA methylation patterns can be stably inherited through mitosis and meiosis (Becker et al., 2011; Schmitz et al., 2011; Eichten et al., 2013; Schmitz et al., 2013; Li et al., 2014). DNA methylation is associated with regulation of transposable element expression and transposition and may also play important roles in gene regulation (Ronemus et al., 1996; Bucher et al., 2012; Eichten et al., 2014; Kim and Zilberman 2014). Numerous studies have documented the molecular mechanisms that control DNA methylation patterns and the biological roles of DNA methylation using the model plant Arabidopsis thaliana (Law and Jacobsen, 2010). A recent study (Stroud et al., 2013) documented the genome-wide DNA methylation patterns in a panel of 86 Arabidopsis mutant stocks implicated in gene silencing and histone modifications, providing a high-resolution view of the contributions of many different genes to DNA methylation. While studies in Arabidopsis have highlighted important genes that contribute to shaping the plant methylome, there are many unresolved questions about the roles for homologs of these genes in other plant species with larger genomes, which are dominated by transposable elements.
In plant genomes, DNA methylation occurs in three sequence contexts, CG, CHG, and CHH (H = A, C, or T). There is evidence to suggest that partially distinct genetic pathways involving different methyltransferases regulate DNA methylation in these contexts in Arabidopsis (Stroud et al., 2013). CG methylation is largely maintained by METHYLTRANSFERASE1 (MET1), and CHG methylation is maintained by CHROMOMETHYLASE3 (CMT3). CHH methylation is maintained by the RNA-directed DNA methylation (RdDM) pathway involving DOMAINS REARRANGED METHYLTRANSFERASES1/2 (DRM1/2) and small interfering RNA (Law and Jacobsen, 2010; Matzke and Mosher, 2014). Recent studies in Arabidopsis suggest that the RdDM pathway mainly mediates CHH methylation at short transposons and euchromatic regions, as well as the edges of long heterochromatic transposons (Zemach et al., 2013). CHH methylation within the body of long heterochromatic transposons is maintained separately and independently by the CMT2 enzyme (Zemach et al., 2013; Stroud et al., 2014). Together, RdDM and CMT2 mediate nearly all CHH methylation in the Arabidopsis genome (Zemach et al., 2013). Although specific contexts of DNA methylation are largely attributed to distinct enzymes/pathways, there is evidence of interplay between different pathways. For example, in the double mutant of DRM1 and DRM2, both CHG and CHH methylation were disrupted (Stroud et al., 2013; Zhong et al., 2014). Similarly, CHH methylation at some loci was reduced in plants lacking functional CMT3, the major CHG methylation maintaining enzyme (Bartee et al., 2001; Lindroth et al., 2001; Cao and Jacobsen, 2002; Cao et al., 2003; Stroud et al., 2013). In addition to the above-mentioned DNA methyltransferase enzymes, proper establishment and maintenance of DNA methylation is dependent upon many other genes. These include chromatin remodelers such as Decrease in DNA methylation 1 (DDM1) and Variant in Methylation 1 (VIM1) (Vongs et al., 1993; Woo et al., 2007, 2008; Jeddeloh et al., 1999). There are also genes in the RdDM pathway that either are involved in producing small RNAs or target small RNA to genomic regions for de novo methylation. These include RNA-dependent RNA polymerase 2 (RDR2) and plant-specific RNA polymerases Pol IV and Pol V (reviewed in Law and Jacobsen, 2010; Matzke and Mosher, 2014).
Our understanding of the molecular mechanisms that target DNA methylation patterns in plants is largely based on studies in Arabidopsis, but many differences between Arabidopsis and other plants, including maize (Zea mays), exist. The maize genome is larger than that of Arabidopsis (2500 Mb versus 125 Mb) and contains substantially more transposable elements (Baucom et al., 2009; Schnable et al., 2009). While most Arabidopsis genes are adjacent to other genes, many maize genes are flanked by heavily methylated transposons (West et al., 2014). There are also several differences in the genome-wide distribution of DNA methylation in maize relative to Arabidopsis. Maize contains higher levels of CG and CHG methylation but lower levels of CHH methylation than Arabidopsis (Jeddeloh and Richards, 1996; Gent et al., 2013; Regulski et al., 2013; West et al., 2014). CG and CHG methylation is commonly found in maize transposons but is relatively low near the start and stop of genes. On the contrary, CHH methylation tends to be found upstream of the transcription start site of maize genes and is only associated with certain types of transposons (Gent et al., 2013).
Maize is a model system for the study of epigenetic phenomena including imprinting (Alleman and Doctor, 2000), paramutation (Chandler, 2007; Hollick, 2010), and transposable element inactivation (Fedoroff et al., 1995). Genetic screens have successfully identified mutants that are defective in the establishment and/or maintenance of paramutation (Dorweiler et al., 2000; Hollick and Chandler, 2001). The molecular characterization of these mutants has revealed that most affect components of the RdDM pathway (Alleman et al., 2006; Erhard et al., 2009; Sidorenko et al., 2009; Sloan et al., 2014), while others are novel proteins (Barbour et al., 2012). In some cases, these mutants have been shown to have locus-specific effects on DNA methylation (Lisch et al., 2002; McGinnis et al., 2006; Sloan et al., 2014), but the genome-wide effect of these mutations on DNA methylation has not been characterized. Several mutants that are defective for the transcriptional silencing of a transgene have also been recovered, although the specific causal genes have not been determined (Madzima et al., 2011). In addition, reverse-genetics approaches have been used to isolate mutations in one of the maize chromomethylase genes, Zmet2 (Papa et al., 2001). Plants that are homozygous mutant for zmet2-m1 have reduced levels of CHG methylation (Papa et al., 2001), resulting in altered expression of some genes (Makarevitch et al., 2007).
In this study, we sought to investigate the effects of perturbation of the maize methylome using mutant alleles for at least 11 genes isolated using forward- or reverse-genetic approaches. A combination of whole-genome and sequence-capture bisulfite sequencing was used to study overall changes as well as locus specific effects. We found that in most cases, the loss-of-function alleles for specific genes had limited effects on genome-wide DNA methylation levels. The most substantial effects were observed in lines containing mutations in the chromomethylase genes and in genes that are predicted to function in the RdDM pathway. Notably, double mutants for paralogous genes were not recovered, suggesting severe phenotypic consequences of strong reductions in genomic DNA methylation levels.
RESULTS
Collection of Mutant Stocks That May Influence DNA Methylation Patterns in Maize
To investigate regulation of DNA methylation in maize, we collected a set of 17 stocks representing up to 11 different genes (Table 1; Supplemental Table 1). Reverse-genetics approaches were used to search for mutant alleles in maize DNA methyltransferase genes that are similar to Arabidopsis MET1, CMT, and DRM genes (Supplemental Figure 1). The maize genome encodes two MET1-like genes (GRMZM2G334041/Zmet1 and GRMZM2G333916) that are present as tandem duplicates. There are two full-length chromomethylase genes in the maize genome, Zmet2 (GRMZM2G025592/Dmt102) and Zmet5 (GRMZM2G005310/Dmt105), that are both more closely related to the Arabidopsis CMT3 than to CMT2. Based on their localization in colinear regions of the maize genome, these two genes are likely retained duplicates derived from the most recent whole-genome duplication event in maize (Swigonová et al., 2004). As noted previously (Zemach et al., 2013), there is no evidence for a CMT2-like gene in the reference genome of B73 (Schnable et al., 2009), and screening of a Mo17 genomic library using chromomethylase-domain probes did not recover any CMT2-like genes. The maize genome also contains several DRM-like genes: Zmet3 (GRMZM2G092497/Dmt103), Zmet6 (GRMZM2G065599/Dmt106), and Zmet7 (GRMZM2G137366/Dmt107). Zmet3 and Zmet7 are retained duplicates from the recent whole-genome duplication event and are both related to the DRM1/2 genes of Arabidopsis. The other gene, Zmet6 (Dmt106), is most similar to DRM3. While the DRM3 genes have domain structures similar to other DRM genes, they are likely catalytically inactive due to altered amino acid sequence at the critical residues in the active site (Henderson et al., 2010). In addition, we also used reverse-genetics approaches to screen for mutant alleles for orthologs of other genes known to play important roles in maintaining DNA methylation, such as DDM1 (Vongs et al., 1993) and VIM1 (Woo et al., 2007, 2008). The maize genome encodes two genes, Chr101 (GRMZM2G177165) and Chr106 (GRMZM2G071025), that are similar to Arabidopsis DDM1 and are likely retained duplicates from the most recent whole-genome duplication event. Three VIM1-like genes (GRMZM2G339151, GRMZM2G461447, and AC191534.3_FGP003) were identified in the maize genome. Two of them are likely retained duplicates (GRMZM2G339151 and GRMZM2G461447).
Table 1. Mutant Stocks Used in This Study.
| Gene Allele | Maize Gene Name | Arabidopsis Ortholog | Gene Function | Maize Gene ID | WGBS Data | SeqCap Data |
|---|---|---|---|---|---|---|
| zmet2-m1 | Zmet2/Dmt102 | CMT3 | Methyltransferase | GRMZM2G025592 | Yes | Yes |
| zmet2-m2 | Zmet2/Dmt102 | CMT3 | Methyltransferase | GRMZM2G025592 | Yes | Yes |
| zmet5-m1 | Zmet5/Dmt105 | CMT3 | Methyltransferase | GRMZM2G005310 | Yes | yes |
| zmet5-m2 | Zmet5/Dmt105 | CMT3 | Methyltransferase | GRMZM2G005310 | No | yes |
| zmet7-m1 | Zmet7/Dmt107 | DRM1/2 | Methyltransferase | GRMZM2G137366 | Yes | Yes |
| zmet7-m2 | Zmet7/Dmt107 | DRM1/2 | Methyltransferase | GRMZM2G137366 | Yes | No |
| zmet7-T03 | Zmet7/Dmt107 | DRM1/2 | Methyltransferase | GRMZM2G137366 | Yes | No |
| chr101-m1 | Chr101 | DDM1 | Chromatin remodeler | GRMZM2G177165 | Yes | Yes |
| chr101-m3 | Chr101 | DDM1 | Chromatin remodeler | GRMZM2G177165 | Yes | Yes |
| chr106-m1 | Chr106 | DDM1 | Chromatin remodeler | GRMZM2G071025 | Yes | Yes |
| chr106-T11 | Chr106 | DDM1 | Chromatin remodeler | GRMZM2G071025 | Yes | Yes |
| vim102-1 | Vim102 | VIM1 | Methylcytosine binding protein | GRMZM2G339151 | Yes | Yes |
| mop1 | Mop1 | RDR2 | RNA-dependent RNA polymerase | GRMZM2G042443 | Yes | Yes |
| mop2 | Mop2 | NRPD2/E2 | RNA polymerase D2/E2 | GRMZM2G054225 | No | Yes |
| mop3 | Mop3 | NRPD1 | RNA polymerase D1 | GRMZM2G007681 | No | Yes |
| tgr1 | Tgr1 | Unknown | Unknown | Unknown | Yes | Yes |
| tgr9 | Tgr9 | Unknown | Unknown | Unknown | Yes | Yes |
We screened TILLING (Till et al., 2004) and transposon insertion populations (Settles et al., 2007; McCarty and Meeley, 2009) to identify loss-of-function alleles for maize methyltransferase, DDM1-like or VIM1-like genes. Characterization efforts focused on alleles with insertion in exons or nonsense mutations. At least two alleles with transposon insertions within exons or nonsense mutations were recovered for Chr101, Chr106, Zmet2, Zmet5, and Zmet7 (Table 1; Supplemental Table 1). No loss-of-function alleles were identified for the MET1-like, Zmet3, or Zmet6 genes. We also obtained a single mutant allele in Vim102 (GRMZM2G339151), which encodes a gene related to Arabidopsis VIM1. No mutant alleles were isolated for the other two VIM1-like genes.
Several mutant lines identified by forward-genetic screens were also included in our analyses (Table 1). The mediator of paramutation1 (mop1), mop2, and mop3 mutants were isolated by screening for lines defective in b1 paramutation (Dorweiler et al., 2000). Mop1 (GRMZM2G042443) encodes an RNA-dependent RNA polymerase that is similar to RDR2 in Arabidopsis and is required for the accumulation of the majority of 24-nucleotide small RNA in maize (Alleman et al., 2006; Nobuta et al., 2008). Mop2 (GRMZM2G054225) encodes a protein that is similar to the second largest subunit of Arabidopsis RNA polymerases IV and V (Sidorenko et al., 2009). Mop3 (GRMZM2G007681) is predicted to encode the largest subunit of RNA Pol IV (Sloan et al., 2014). All these three genes are predicted to play a role in the RdDM pathway. The transgene reactivated1 (tgr1) and tgr9 (Madzima et al., 2011) mutations were isolated by screening for plants defective in transgene silencing (Madzima et al., 2011), and the specific molecular lesions for these mutations have not been determined.
Assessing the Effect of Single Mutants on the Maize Methylome
The relatively large size of the maize genome makes deep coverage DNA methylation profiles for a large panel of mutants expensive and inefficient. Therefore, we used an alternative approach to assess genome-wide and/or locus-specific DNA methylation levels in this panel of mutant stocks. This approach involved low-coverage whole-genome bisulfite sequencing (WGBS) for a subset of the mutant lines and high-coverage sequencing of specific regions that are targeted by sequence capture for most of the stocks in WGBS as well as several additional mutants (Table1; Supplemental Table 1). For WGBS, ∼20 million 100-bp read pairs were generated for each mutant stock, with ∼60% of reads being mapped uniquely, providing ∼1X coverage for the maize genome (Supplemental Table 2). While the low-coverage data are not useful for assessing locus specific effects of the mutations on DNA methylation, it can provide an accurate assessment of total DNA methylation levels and assessment of DNA methylation in categorical genomic regions such as genes or transposable elements. To illustrate this, we performed profiles of DNA methylation over the length of maize genes by subsampling the deeper coverage data for B73 (∼10X coverage). The use of even 5 to 10 million reads in our data set provides very similar DNA methylation profiles as those observed for the deeper coverage data set with relatively little variance among different subsamples (Supplemental Figure 2).
The mutant stocks were obtained from a diverse set of genetic backgrounds (Supplemental Table 1). Many of these alleles have been backcrossed into a common genetic background (B73), but there are likely introgressions from other genotypes, and in some cases, such as the vim102-1 or chr106-T11 alleles, the stocks are in a different genetic background (W22). To control for natural variation in DNA methylation levels or for the influence of DNA sequence polymorphisms, we performed whole-genome bisulfite sequencing for five different maize inbreds (B73, Mo17, Oh43, Tx303, and CML322). The average value for these genotypes is used to represent DNA methylation levels in wild-type plants. By assessing DNA methylation profiles or locus-specific DNA methylation levels in these lines, we can assess how much variation might be contributed by diverse genetic backgrounds.
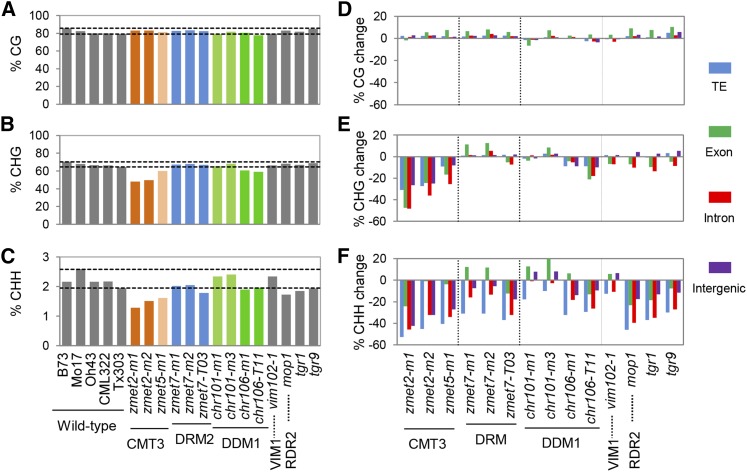
The average levels of CG, CHG, and CHH methylation were determined for each of the mutant and wild-type genotypes using whole-genome bisulfite sequencing data (Figure 1). Methylation levels were defined as the average of 100-bp tiles with data (Methods). None of the genotypes exhibit significantly reduced (>10%) levels of CG methylation (Figure 1A). The biggest reduction was seen in chr106-T11 mutant, but it was <5%. Significant reductions of CHG methylation were observed for zmet2 (27% reduction), zmet5 (10% reduction), and chr106 (11% reduction) and for all the alleles representing these genes (Figure 1B). CHH methylation was influenced in plants with a loss-of-function allele for zmet2 (37% reduction), zmet5 (30% reduction), mop1 (21% reduction), and chr106 (13% reduction) (Figure 1C). These genome-wide DNA methylation levels are likely to be highly influenced by the DNA methylation levels in transposable elements since ∼80% of the maize genome is derived from these elements, and they tend to be highly methylated in CG and CHG contexts (Gent et al., 2013; Regulski et al., 2013; West et al., 2014). In order to assess the effects of these mutants on various genomic features, we separately analyzed transposable elements (TEs), exons, introns, and non-TE intergenic DNA (Figures 1D to 1F). None of the mutants display >10% alterations in CG methylation in any subset of genomic regions, and the variation among the different wild-type genotypes was as large as the variation observed in the mutant lines (Figure 1D). There are similarities and differences in the effects of zmet2 and zmet5 on different genomic regions. Both mutants show a similar significant effect on CHG methylation in exonic and intronic regions, but the effect of zmet5 in transposons and intergenic regions is not as strong as that observed in zmet2 (Figure 1E). The CHG levels were also slightly reduced in chr106 in all four types of genomic features (Figure 1E). For CHH methylation, zmet2, zmet5, and mop1 showed the strongest reduction in all regions. CHH methylation for TEs was also reduced in zmet7, chr106, tgr1, and tgr9, but to a lesser extent than that in zmet2, zmet5, and mop1 (Figure 1F).
Figure 1.
DNA Methylation Levels and Changes in Mutant Genotypes.
(A) to (C) Genome-wide DNA methylation levels in three sequence contexts in the wild type and mutants used in this study. Genes with more than one mutant allele are shown as bars with same color. Gray bars are used to indicate the wild-type genotypes and genes with a single mutant allele. The horizontal dashed lines indicate the highest and lowest DNA methylation levels among the wild-type genotypes. The homologous Arabidopsis genes are indicated at the bottom of the plot.
(D) to (F) Percentage of change in methylation level between the mutant and wild type in the three sequence contexts (CG [D], CHG [E], and CHH [F]) relative to the average of wild-type inbred lines. Methylation change was calculated as the methylation level of (mutant − wild type)/wild type. The level of DNA methylation change was determined for four different types of regions: TE, exon, intron, and intergenic (non-TE) regions. The vertical dashed lines were used to separate classes of mutants for better visualization.
There are many different types of transposable elements in the maize genome with varied profiles of DNA methylation (Eichten et al., 2012; Gent et al., 2013; West et al., 2014). We investigated DNA methylation changes for specific types of transposons in each of the mutant backgrounds (Figure 2). Using available genome-wide annotation (Baucom et al., 2009; Schnable et al., 2009) and previous analyses of DNA methylation levels in flanking regions (Eichten et al., 2012), the maize transposable elements were divided into 15 different classes (www.maizetedb.org). These include two subtypes of LINE elements, three subtypes of LTR retrotransposons (RLG-gypsy, RLC-copia, and RLX-unknown), and five subtypes of DNA transposons (two of which were split into coding and noncoding groups). The LTR retrotransposons were further divided into two classes based on whether the DNA methylation spreads to flanking regions. Although CG methylation remains largely unaffected in most of the mutants, there is evidence for ∼10% CG methylation reduction in the Tc1/Mariner DTT DNA transposon class in mop1 and tgr1 mutants (Figure 2A). The mop1 and tgr1 mutants also have detectable methylation reduction in the CHG context at the DTT transposons (Figure 2B). The zmet2 mutants also exhibit reduced CHG methylation in the DTT family as well as other transposon families. Lower reductions in CHG methylation for many transposon subtypes were also observed in zmet5 and chr106 (Figure 2B). For CHH methylation, all transposon types show significant reduction in the zmet2 and zmet5 mutants (Figure 2C). In contrast, while the zmet7, chr106, mop1, tgr1, and tgr9 mutants influenced CHH methylation for many transposon types, they have limited influence on the LTR transposons classified as “spreading” elements and on the CACTA DNA transposon family (Figure 2C).
Figure 2.
Context-Specific Changes in DNA Methylation Levels at Each Subfamily of TEs.
Only the TE body was used. Heat maps are used to visualize differences in DNA methylation levels relative to the wild type in CG (A), CHG (B), or CHH (C) sequence context. Methylation change was calculated as (mutant – wild type)/wild type, and the wild-type values are the average of the five analyzed inbred lines. Each row represents one transposon family, including two LINE families (LINE_RIL and LINE_RIX), three types of retrotransposons (LTR_RLC, LTR_RLG, and LTR_RLX), which were further classified into spreading and nonspreading five DNA transposons (TIR_DTC, TIR_DTA, TIR_DTH, TIR_DTM, and TIR_DTT) with DTA and DTC families further classified into noncoding type (www.maizetedb.org). Each column represents one mutant, with the genotype shown in the bottom right table. The color scheme for heat map was displayed in the bottom left. Red indicates methylation loss in mutants and green shows methylation gain.
Locus-Specific Changes in CG DNA Methylation in mop and tgr1 Mutants
In order to study the locus-specific effects of these mutations on DNA methylation, a second DNA methylation profiling method was employed. Sequence-capture bisulfite sequencing was used to perform targeted bisulfite sequencing of ∼5 Mb of the maize genome, resulting in consistent, deep coverage of the same regions for all samples tested (Supplemental Table 3 and Supplemental Data Set 1). For genomic regions with coverage in both the sequence capture and low-coverage WGBS, there is quite strong agreement in the DNA methylation levels from both methods (Supplemental Figure 3). The loci targeted by the capture are not representative of the whole genome. Instead, loci with specific methylation states were selected for this capture based on B73 and Mo17 WGBS data (Eichten et al., 2013). This includes 2182 loci that have high methylation in all sequence contexts, 1164 regions with CG/CHG methylation, 330 regions with only CG methylation, 64 regions with only CHG methylation, and 140 regions with particularly high CHH methylation. By focusing on these regions of high methylation, we can assess whether the mutants included in this study affect methylation in the selected regions. We also performed targeted bisulfite sequencing in different maize inbred lines (B73, Mo17, Oh43, W22, and CML322) to provide a control for natural variation in DNA methylation levels. To assess the effects of the mutants upon CG, CHG, or CHH methylation, we identified the subset of targeted loci that exhibit consistently high levels of DNA methylation and coverage for each DNA methylation context in all five wild-type genotypes that were subjected to sequence capture bisulfite sequencing and then determined the level of DNA methylation for these regions in each of the mutant samples (Figures 3 to 5).
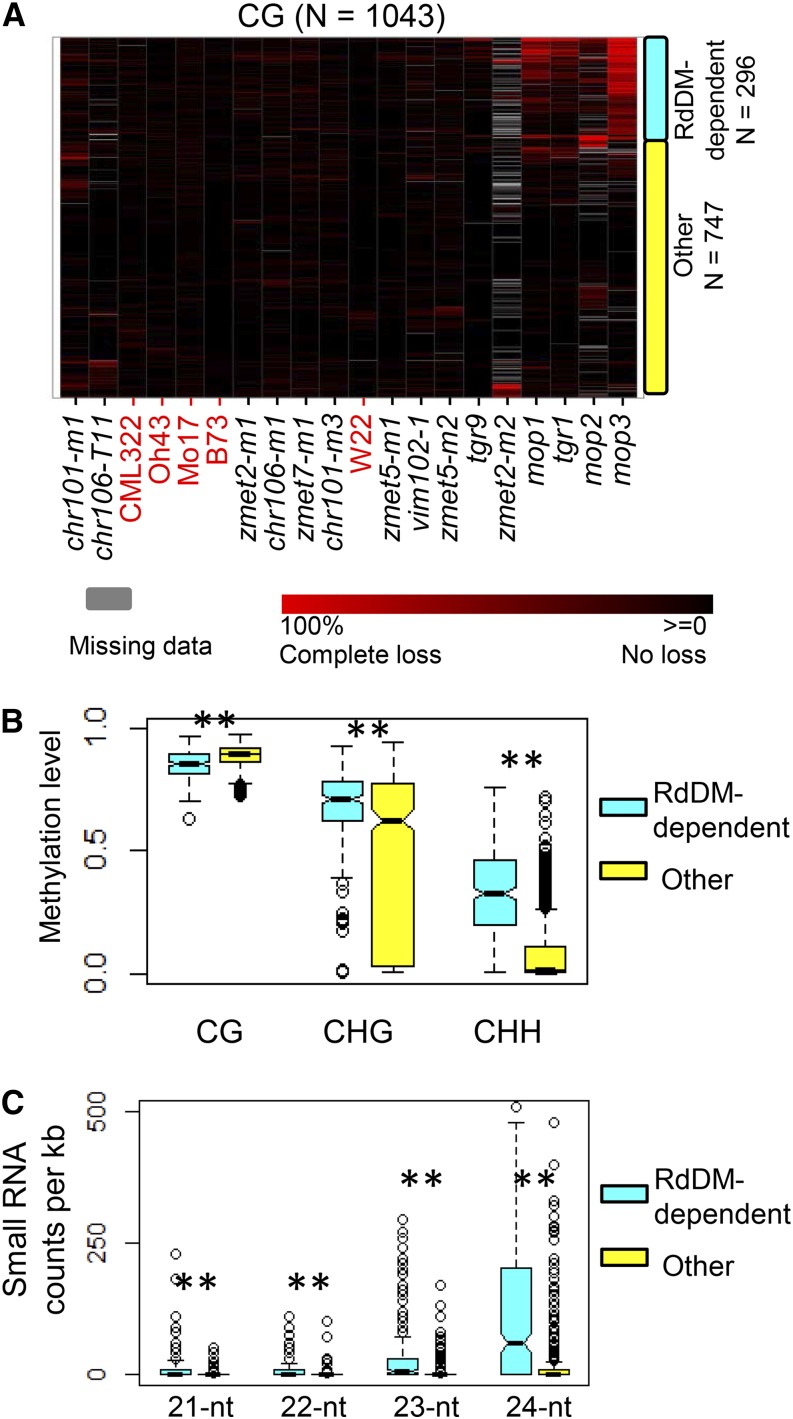
Figure 3.
Locus-Specific Changes in CG Methylation.
(A) The changes in CG methylation levels in each genotype are plotted for genomic regions that are consistently captured (coverage >10 in all wild-type plants) and have high levels of CG methylation (>60% in all wild-type plants). The level of DNA methylation change in each mutant relative to the wild-type average was calculated and used to perform clustering (Ward’s method) of all mutant and wild-type genotypes with red color indicating loss of DNA methylation. The regions were divided into two groups (indicated on the right side) according to whether they exhibit reduced DNA methylation levels in the mop and tgr1 mutations or not. The “RdDM-dependent” group shows loss of DNA methylation in mop or tgr1 mutant that has a putative function in the RNA-directed DNA methylation pathway, while the “Other” group does not show reduction in these mutants.
(B) Wild-type DNA methylation levels for each sequence context for the two subsets of regions.
(C) Small RNA counts for the two types of regions. Small RNAs with a size between 21 and 24 nucleotides were used. **P < 0.01.
Figure 5.
Clustering of Methylation Change at the Sequence Capture Regions for CHH.
Methylation change was calculated as (genotype – wild type)/wild type, where the wild type is the average of five diverse inbred lines B73, Mo17, Oh43, CML322, and W22 and used for clustering (Ward’s method). The inbred lines were also included in the clustering (highlighted in red) to show variance of methylation levels in these genotypes. Regions with consistent high coverage (>10×) and vary levels of CHH were plotted.
(A) Regions with at least 1% CHH in each of the five wild-type plants and <5% CHH in the average of the wild type.
(B) Regions with at least 5% CHH in each of the five wild-type plants and <20% CHH in the average of the wild type.
(C) Regions with at least 20% CHH in each of the five wild-type plants. The open black boxes are used to highlight mutants of the homologs of CMT3 and genes potentially involved in the RdDM pathway.
(D) Counts of small RNA in the three groups of regions from (A) to (C). Small RNAs with a size between 21 and 24 nucleotides were used. **P < 0.01.
While there was not strong evidence for genome-wide alterations in CG methylation levels (Figures 1A and 1D), there are a subset (296/1043) of loci that exhibit reductions in CG methylation in mop1, mop2, mop3, and tgr1 plants (Figure 3A). These same regions do not show CG methylation change in the tgr9 mutant, which has a similar genetic background as tgr1, or in zmet7 mutants, which may show redundant function with its paralog Zmet3. The regions that exhibit loss of CG methylation in mop1, mop2, mop3, and tgr1 mutants tend to have consistently higher levels of CHH and CHG methylation in wild-type plants compared with regions that do not require functional Mop1, Mop2, Mop3, and Tgr1 genes (Figure 3B). In addition, the genomic loci that exhibit reduced CG methylation in mop1, mop2, mop3, and tgr1 mutants are enriched for small RNAs (Figure 3C), suggesting that these regions might be active targets of the RdDM pathway.
Distinct Control of CHG Methylation by RdDM Components and Zmet2
Locus-specific changes in CHG methylation were assessed for 767 regions that exhibit high (>30%) CHG methylation and coverage (>10 reads per region) in all wild-type genotypes (Figure 4A). One subset of 372 regions requires the RdDM components (Mop1, Mop2, and Mop3), while a distinct set of 86 regions requires Zmet2 (Figure 4A). The remaining 309 regions did not exhibit substantially altered CHG methylation levels in any of the single gene mutations. Genomic regions near longer TEs are more likely to require Zmet2 to maintain proper CHG (Figure 4B), so does regions that are depleted of 21- to 24-nucleotide small interfering RNAs (Figure 4C). On the contrary, the regions that require RdDM components for CHG methylation are enriched for 21- to 24-nucleotide small interfering RNAs (Figure 4C). The analysis of DNA methylation levels in all three sequence contexts for the different subsets of loci reveals that the regions that require RdDM components for proper CHG methylation tend to have elevated CHH levels in wild-type plants, while the regions that require Zmet2 tend to have very low levels of CHH methylation (Figure 4D). The analysis of the low-coverage genome-wide data provided further support for two different pathways for CHG methylation depending on the level of CHH methylation (Figure 4E). Regions with elevated CHH methylation depend upon RdDM components for proper CHG methylation, while regions with low CHH methylation only require Zmet2 (Figure 4E).
Figure 4.
Independent Pathways Contribute to CHG Methylation in Maize.
(A) There are 767 genomic regions (100 bp to 1.1 kb) that have consistent coverage (>10 coverage) and elevated CHG methylation levels (>30%) in all wild-type genotypes (shown in red color). The percentage of change in CHG methylation level relative to wild-type average for each region was calculated for each genotype and used to perform clustering using Ward’s method. The genomic regions were divided into three groups (indicated on right side of plot) based on their loss of methylation in zmet2 (blue, Zmet2-dependent), mop1, mop2, mop3, and tgr1 (cyan, RdDM-dependent) or regions that did not lose methylation in all mutants (yellow, Other).
(B) The size of the nearest TE for each of these three groups of regions from (A).
(C) The number of small RNA reads per kilobase of the three groups of regions. Small RNA data are from Regulski et al. (2013). Small RNAs with a size between 21 and 24 nucleotides were used.
(D) The wild-type level of DNA methylation in each sequence context is shown for the three types of regions using a box plot.
(E) Genome-wide analysis of the change for CHG levels in mop1 and zmet2-m1 at regions with varying levels of CHH methylation. CHH levels were grouped into eight bins: <5%, 5 to 10%, 10 to 20%, 20 to 30%, 30 to 40%, 40 to 50%, 50 to 60%, and 60 to 100%. **P < 0.01.
Overlapping Control of CHH Methylation by RdDM Components and CMT Genes
The sequence capture data included 394 genomic regions with detectable CHH methylation (≥1%) and at least 10 reads in all wild-type genotypes (Figures 5A to 5C). These include 68 regions with 1 to 5% CHH methylation, 102 regions with 5 to 20% CHH methylation, and 224 regions with >20% CHH methylation. These three groups of regions show distinct levels of small interfering RNAs (Figure 5D). Methylation of loci with >5% CHH methylation tends to require the RdDM components Mop1, Mop2, Mop3, as well as Tgr1 (Figures 5B and 5C). However, the loci with lower levels of CHH (1 to 5%) are variable in their requirement for RdDM activity (Figure 5A). CHH methylation changes in lines with mutated CMT3 genes show the opposite trend with greater dependency upon CMT genes for regions with low CHH levels than for regions with elevated CHH levels (Figures 5A to 5C). Consistent with the WGBS data (Figure 2), chr106 and zmet7 also show mild CHH loss at many of the sampled regions.
To further investigate the contribution of RdDM components and CMT genes to CHH methylation, we studied genome-wide CHH profiles at exons, introns, and specific transposon families using WGBS data. Our results suggest multiple pathways are required for the control of CHH methylation levels in maize. The “CHH islands” observed near the 5′ and 3′ ends of many genes (Gent et al., 2013) are most sensitive to the mop1 mutation. However, other mutations, including tgr1, tgr9, zmet2, and zmet5, exhibit partial reductions in the magnitude of this peak and also show reduced CHH methylation levels in introns (Figures 6A and 6B). The analysis of CHH methylation in maize transposons revealed variation in the effect for different subtypes of transposons (Figures 2C and 6C to 6E). Nonspreading class I retrotransposons and class II DNA transposons have high levels of CHH methylation in wild-type plants (Figures 6D and 6E), while class I retrotransposons that result in spreading of CG/CHG methylation to flanking regions tend to have lower CHH methylation (Figure 6C). The low levels of CHH methylation observed within the spreading retrotransposons are reduced in zmet2 but are less altered in mop1 plants (Figure 6C). The elevated CHH methylation in nonspreading retrotransposons and DNA transposons is strongly reduced in mop1, zmet2, and zmet5 mutant plants (Figures 6D and 6E). The 24-nucleotide small RNA levels at each transposon type correspond well with CHH methylation levels of that transposon and the putative genes that mediate the CHH methylation (Supplemental Figure 4). The spreading retrotransposons, which have low CHH methylation levels, tend to be depleted of 24-nucleotide small RNA; thus, the RdDM components whose function depends on 24-nucleotide small RNAs seldom show an effect for this transposon type. Short DNA transposons are particularly enriched for 24-nucleotide small RNAs and show the highest levels of CHH methylation (Supplemental Figure 4). The CHH methylation at this transposon type was significantly reduced in mutants of RdDM components.
Figure 6.
Alterations in CHH Methylation Profiles over Genes and Transposons.
(A) CHH levels in exonic and intronic regions in the wild type, mop1, tgr1, and tgr9. The methylation levels at exons and introns were plotted separately. The filled black boxes at the x axis represent either exons or introns. Since the average mRNA size is ∼1.7 kb, we plotted one kilobase from both 5′ and 3′ ends into exons. For introns, three kilobases from both 5′ and 3′ ends were plotted. The exon/intron sizes are not from one single exon/intron; they are concatenated from multiple individual exons or introns. Untranslated regions were considered as exon regions. Also included in the exon plot were 2-kb upstream and downstream regions. The black line indicates average wild-type levels, while the gray shading indicates the variance observed among the wild-type genotypes. The colored lines show the profile for different mutant genotypes. The vertical dashed lines show gene transcriptional start and stop sites.
(B) Similar plots are shown for zmet2 and zmet5, and in both cases these are for the average of the different alleles that were tested.
(C) to (E) The profile of DNA methylation over three major types of transposons is shown for the wild type, zmet2, zmet5, and mop1 using the same color scheme used in (A) and (B). Separate plots are shown for retrotransposon elements that exhibit spreading of CG and CHG methylation to flanking regions (C), retrotransposons that do not exhibit spreading of CG or CHG methylation (D), or DNA transposons (E).
(F) to (K) Similar plots but only including the subset of transposable elements in each class that are <1 kb in length (F) to (H) and show similar plots for elements that are longer than 3 kb ([I] to [K]).
In (C) to (K), the filled black box at the bottom of each plot represent the transposable element, and the two dashed vertical lines in each plot represent the start and end of the transposable elements. Methylation levels in the 1-kb regions that are upstream or downstream of the transposable elements are also shown.
Recent findings in Arabidopsis suggest that mechanisms controlling CHH methylation in transposons vary for long and short TEs (Zemach et al., 2013). Whereas DNA methylation within the bodies of long TEs is maintained by the CMT2 pathway, methylation of short TEs and the edges of long TEs is mediated by the RdDM pathway. We compared the effects of zmet2 and zmet5 with mop1 on CHH methylation in subsets of TEs that are short (<1 kb) or long (>3 kb) (Figures 6F to 6K). The effects for zmet2 and zmet5 on CHH methylation levels in spreading retrotransposons of all sizes are greater than that observed for mop1 (Figures 6F and 6I). For the nonspreading retrotransposons and class II DNA transposons, the effects of mop1 on CHH methylation are greatest for short elements (Figures 6G, 6H, 6J, and 6K). In contrast, the CHH methylation in longer TEs of these types is more dependent upon Zmet2 and Zmet5. It is worth noting that the CHH levels in the wild type are much greater for short TEs (6 to 8%) than for the longer elements (2 to 4%). The levels of CHH methylation in short TEs is even higher than the levels of CHH methylation at the edges of longer TEs. While TE size influenced the requirement for different genes for proper CHH methylation levels, we did not observe differences in the effects of mutations on CHG methylation for different sizes of TEs (Supplemental Figure 5).
Inability to Recover zmet2 zmet5 and chr101 chr106 Double Mutants
Paralogous genes can exhibit redundant function such that abolishing the full functionality of maize DDM1 (Chr101/Chr106), CMT3 (Zmet2/Zmet5), or DRM (Zmet3/Zmet7) genes may require generation of double mutant stocks. These gene pairs show substantially overlapping expression patterns across a collection of ∼60 tissues (Sekhon et al., 2013), with one transcript accumulating more abundantly than the other in most tissues (Zmet2>Zmet5, Chr101>Chr106, and Zmet7>Zmet3) (Supplemental Figure 6). For the Zmet2/Zmet5 and Chr101/Chr106 gene pairs, we had obtained mutant alleles for both genes and made crosses in an attempt to isolate double mutant individuals. Parental plants that were heterozygous for two different alleles or parents that were homozygous mutant for one gene and heterozygous for the other were self-pollinated to create offspring that are expected to segregate for double mutant individuals (Table 2). Despite screening a substantial number of offspring, we were not able to identify any homozygous double mutant individuals for the CMT3- or DDM1-like paralogs in maize (Table 2).
Table 2. Recovery of Double Mutants.
| Parental Genotype | No. of Offspring | Expected Double Mutants | Observed Double Mutants | Observed A:a Gametea | Observed B:b Gameteb |
|---|---|---|---|---|---|
| zmet2-m1/+; zmet5-m1/zmet5-m1 | 13 | 3 | 0 | 17:9 | 0:26 |
| zmet2-m1/zmet2-m1; zmet5-m1/+ | 10 | 3 | 0 | 0:20 | 17:3** |
| zmet2-m1/+; zmet5-m1/+ | 169 | 11 | 0 | 203:135** | 188:150* |
| zmet2-m2/+; zmet5-m1/+ | 79 | 5 | 0 | 93:65* | 90:68 |
| chr101-m3/chr101-m3; chr106-m1/+ | 128 | 32 | 0 | 0:256 | 164:92** |
Deviates from 1:1 at 0.05 level (χ2); **deviates from 1:1 at 0.01 level (χ2).
A and a indicate the wild-type and the mutant gamete of the first gene in the “Parental Genotype” column, respectively.
B and b indicate the wild-type and the mutant gamete of the second gene in “Parental Genotype” column, respectively.
The failure to recover double mutants could be either due to reduced viability or transmission of double mutant gametes through one parent or reduction of viability for the fertilized double mutant zygote. These possibilities were tested by looking at the observed segregation ratio in the offspring of plants from parents with the chr101-m3/chr101-m3; chr106-m1/+ genotype. If the double mutant gamete can only be transmitted through one parent, we would expect a 1:1 segregation ratio for the wild type and heterozygotes at the chr106 locus; otherwise, 1:2 segregation ratio would be expected (Supplemental Figure 7). Genotyping of 128 offspring revealed a segregation ratio of 36:92 (Supplemental Table 4), which is close to 1:2 (P = 0.2332, χ2) and deviates from 1:1 (P = 1.058e-06). A similar analysis was performed on the offspring of zmet2-m1/+; zmet5-m1/zmet5-m1 or zmet2-m1/zmet2-m1; zmet5-m1/+ plants. In both cases, the segregation ratio was consistent with successful transmission of the segregating mutant allele through both parents (Supplemental Table 4). We did not observe substantially decreased germination rates for the offspring of these plants, which suggests that the double mutant individuals do not complete seed development. Our search for double mutant individuals also revealed reduced transmission of the single mutant alleles (Table 2). In most of the screened populations, we found that while the mutant allele could be transmitted, it was found at lower than expected rates. This bias was significant for many of the offspring populations, and the recovery of the mutant gamete is ∼70 to 80% of that of the wild-type gamete for all the alleles we used to create double mutants (Table 2).
DISCUSSION
DNA methylation can contribute to chromatin-based regulation of gene expression and is an important component of some epigenetic phenomena. Understanding the mechanisms that control DNA methylation can help to understand the basis for some examples of epigenetic regulation. The low cost of sequencing and use of bisulfite modification have provided the ability to obtain high-resolution profiles of methylomes. Stroud et al. (2013) provide a seminal example of how WGBS can be used to better understand the role of many different genes to genome-wide DNA methylation patterns by profiling 86 different Arabidopsis mutants. While maize has provided a model system for studying several epigenetic phenomena, such as imprinting, paramutation, and transposable element inactivation, we have a limited understanding of the molecular mechanisms and targets of DNA methylation in the maize genome. In part this is due to the limited access to mutant stocks with documented effects on genome-wide DNA methylation patterns, but it may also be due to the large size and complex organization of the maize genome.
In this study, we demonstrate how the combined use of low-coverage WGBS and high-coverage targeted capture of specific loci following bisulfite conversion can be used to characterize the influence of specific mutant alleles on the maize methylome. This approach could easily be applied to other plant species and will provide lower cost alternatives for understanding how a panel of mutants perturbs the methylome. It is worth noting that the use of either technique alone can miss important observations that are revealed by combining both approaches. For example, zmet2-m1 and zmet2-m2 appear to have the strongest reduction in CHH in the low-coverage WGBS data (Figure 1C), while mop1 has a greater effect in the sequence capture data set (Figure 5). This is likely due to the ascertainment bias for the regions included in the sequence capture. The regions selected for capture often included genomic loci that exhibited unusually high DNA methylation levels in all sequence contexts or rare regions with context specific DNA methylation. The greatest effect of a loss of Mop1 was apparent in regions with high CHH methylation, while the zmet2 mutations had stronger phenotypes in regions with low, but detectable, CHH methylation (Figure 6). Since the majority of the maize genome has quite low CHH levels (West et al., 2014), the cumulative effect of zmet2 mutations may be greater than that of the mop1 mutation even though specific loci with high CHH methylation are impacted more strongly by the mop1 mutation. The combination of these two approaches provides an efficient strategy to study perturbations of DNA methylation in organisms with large and complex genomes.
RdDM Targets in the Maize Genome
The RdDM pathway provides a mechanism to target de novo methylation at specific genomic regions and can induce CHH methylation as well as methylation in other sequence contexts. The mutations in mop1, mop2, and mop3 are all predicted to impair the RdDM pathway based on homology to Arabidopsis genes with roles in RdDM (Alleman et al., 2006; Sidorenko et al., 2009; Sloan et al., 2014). The low-coverage WGBS data provide evidence that CHH levels are reduced in mop1, and the sequence capture data reveal a strong loss of CHH methylation in mop1, mop2, and mop3 for nearly all regions with CHH methylation >5% in the wild type. In regions of elevated CHH methylation, both CHH islands 5′ of genes and certain families of DNA transposons, methylation is strongly reduced in the mop1 mutant. The reduction in CHH methylation observed in mop1 mutants is greater at short DNA transposons and nonspreading retrotransposons than at longer elements of these families. There is only a minor role for Mop1 in CHH methylation of the retrotransposons that exhibit spreading of methylation to surrounding regions. This suggests that similar to RdDM in Arabidopsis (Zemach et al., 2013), maize RdDM machinery is targeted to regions that are near euchromatin and helps to maintain silencing of these regions. We expect that maize DRM-like genes are also important for RdDM in maize. However, the lack of an effect in zmet7 mutants may reflect redundancy of the paralogs Zmet3 and Zmet7 in providing de novo methylation targeting by RdDM.
While the low-coverage WGBS analysis did not reveal a significant role for Mop1 in CG or CHG methylation in most genomic regions, we did find that Mop1 is required for CG and CHG methylation at some transposon types, especially the DTT transposons that has high level of CHH methylation (Gent et al., 2013). The DTT transposon also shows the lowest average size (123 bp) compared with other DNA transposons, making it a more prone to be targeted by the RdDM pathway. Similarly, we also find some genomic regions that require Mop1 for CG or CHG methylation using the sequence capture data. These regions had elevated CHH methylation and tend to be enriched for small interfering RNAs. It may be that active targeting of de novo methylation is required to maintain high levels of methylation in all contexts for these regions. Similarly, in Arabidopsis, the drm1 drm2 double mutant displays loss of methylation not only at CHH sites but also at CHG sites (Zhong et al., 2014), and some of the CHH DMRs also exhibit reduced CG methylation (Stroud et al., 2013).
Although the specific molecular lesion of tgr1 has not been characterized, the DNA methylation changes in this mutant line are quite similar to those observed for the mop1, mop2, and mop3 mutants, suggesting that Tgr1 is a component of the RdDM machinery. The tgr9 mutant individuals do not show the same level of change in CHG or CHH methylation as that observed for the mop1, mop2, and mop3 mutations. A small subset of regions loses CHG or CHH methylation in tgr9, suggesting that this gene has a more locus-specific effect on methylation.
Role of Maize CMT Genes in CHG and CHH Methylation
Chromomethylase genes were originally identified as CHG maintenance methyltransferases (Lindroth et al., 2001; Papa et al., 2001). Recent studies in Arabidopsis on two chromomethylases (CMT2 and CMT3) suggest that different chromomethylase proteins may play distinct roles in DNA methylation (Stroud et al., 2013, 2014; Zemach et al., 2013). The CMT2 gene plays an important role in CHH methylation, particularly in long heterochromatic elements (Zemach et al., 2013), while CMT3 is more important for CHG methylation in a self-reinforcing loop with H3K9me2 (Du et al., 2012; Stroud et al., 2013, 2014) and plays a limited role in CHH methylation (Stroud et al., 2013). While the maize genome encodes two orthologs of CMT3 (Zmet2 and Zmet5), there is no evidence for a CMT2-like gene in the reference maize genome (Zemach et al., 2013). It is possible that maize lacks the functions provided by a CMT2-like gene. Alternatively, the Zmet2 and Zmet5 genes may provide both the CMT2 and CMT3 functions in maize. This latter idea was partly supported by the observation that zmet2-m1 mutant plants have reduced levels of CHH methylation, particularly in the central region of long transposable elements. However, the level of CHH methylation present in the center of long transposable elements in maize is much lower than that observed in Arabidopsis (West et al., 2014). One interpretation of our results is that Zmet2 is recruited to heterochromatic regions by H3K9me2 (Du et al., 2012) and primarily performs CHG methylation, but at a low rate can also catalyze some CHH methylation. Consistent with this are the findings that the Arabidopsis CMT3 can play a role in CHH methylation at some loci (Cao and Jacobsen, 2002) and that CMT3 CHH DMRs tend to be found in LTR/Copia elements (Stroud et al., 2013). It is interesting that maize retrotransposons tend to be quite effectively silenced despite the reduced levels of CHH methylation relative to Arabidopsis.
Requirement for DNA Methylation for Embryo Development in Maize
One of the most interesting findings in this study is the lack of ability to recover specific double mutants that would be predicted to more strongly affect the maize methylome. Despite several genetic screens for mutations affecting epigenetic regulation (Dorweiler et al., 2000; Hollick and Chandler, 2001; Madzima et al., 2011), there are no maize mutants that have been recovered that have lost nearly all DNA methylation. Based on findings in Arabidopsis, we might expect that mutations in the maize orthologs of MET1, DDM1, or CMT genes would have the strongest effect on CG and/or CHG methylation. In maize, there are two MET1 orthologs that are present as a tandem duplication of nearly identical sequences (∼>99% identical), and we have not been able to recover Mu insertions or nonsense TILLING alleles for these genes. Concerted attempts at generating RNA interference lines using the protocols described by McGinnis et al. (2007) could successfully reduce Zmet1 mRNA levels in callus but could not be recovered as plantlets (H. Kaeppler and S.M. Kaeppler, unpublished data). Our attempts to create double mutants for Zmet2/Zmet5 (maize paralogs of CMT3) or Chr101/Chr106 (maize paralogs of DDM1) in this study were not successful. The analysis of the segregation ratios suggest that this lethality most likely happens following fertilization during seed growth and development as opposed to at gamete development. However, we did notice some reductions in transmission of the mutant alleles. Similar observations for reduced transmission of the zmet2-m1 mutant allele were reported by Garcia-Aguilar et al. (2010). While this mutant can be transmitted through the male or female gametes, there is evidence that ∼40% of the pollen grains produced by zmet2-m1 plants are abnormally large and may represent unreduced polyploid gametes. The full seed set for female zmet2-m1 plants suggests that female gametogenesis is not impaired by this mutation (Garcia-Aguilar et al., 2010). In this study, it was difficult to assess the seed set on plants that were homozygous for one mutant allele and segregating for the other. These plants were not vigorous, potentially due to inbreeding depression or due to effects of the mutant alleles, and we could not determine the specific timing of potential seed abortion for double mutant individuals.
Although we cannot rule out the possibility that the maize orthologs of CMT or DDM are required for other functions, our observations are consistent with a suggestion that strong reductions in CG/CHG methylation may have more phenotypic consequences in maize than in Arabidopsis. There is evidence that this may also be true for rice (Oryza sativa) as homozygous met1 mutant individuals were not recovered (Hu et al., 2014). The increased requirement for DNA methylation to successfully complete embryo development in maize but not Arabidopsis could be due to the increased transposon content and higher frequency of genes being located very near to transposons in maize (Schnable et al., 2009). The loss of DNA methylation for these regions may result in misregulation of many genes and impair proper seed development. The difficulty in recovering maize lines with strong reductions in DNA methylation levels has several important implications for our ability to probe the role of DNA methylation in maize. The inability to strongly affect DNA methylation complicates the ability to generate epiRILs (Johannes et al., 2009; Reinders et al., 2009) or similar populations in maize. These epiRILs would be very useful in studying how proper DNA methylation patterns influence cryptic information in the maize genome and for studying the phenotypic role of DNA methylation. The inability to recover maize lines with strongly reduced DNA methylation also greatly complicates our ability to study the role of DNA methylation in epigenetic phenomena, such as paramutation, imprinting, or transposon regulation. Alternative approaches may be necessary to probe the role of DNA methylation in maize.
METHODS
Plant Materials
Supplemental Table 1 lists the 17 maize (Zea mays) mutants used in this study. For each mutant, the source, genetic background, and type of mutation (if known) as well as other details about these mutants are indicated. All the plants, except tgr1, which was grown in the field, were grown in a greenhouse condition with a cycle of 15 h of light and 9 h of dark. For mop1 and tgr1, the immature ear shoot was used for DNA isolation, and for the rest of the genotypes, the fully expanded 3rd leaf of seedlings was harvested for DNA extraction.
Genotyping of Double Mutants
We tried to create double mutants for two pairs of genes, Zmet2/Zmet5, both of which are homologs of Arabidopsis thaliana CMT3 gene, and Chr101/Chr106, both of which are homologs of Arabidopsis chromatin remodeler DDM1. Each of the four genes has two different mutant alleles (Supplemental Table 1). Combinations of multiple alleles were tested. Table 2 and Supplemental Table 4 list the genotypes for which we self-pollinated plants and used the derived ears to search for double mutants. We grew the kernels from each self-pollinated ear in the greenhouse, and DNA was isolated from the leaf tissues using standard CTAB method. We performed PCR on the isolated DNA using primers that can differentiate mutant from wild-type allele. Primer sequences and representative band pattern for the wild-type and mutant alleles are listed in Supplemental Table 5.
Whole-Genome Bisulfite Sequencing and Analysis
One microgram of genomic DNA was used for library construction. DNA was sheared to fragments between 200 and 300 bp. The fragments were subject to end repair, dA tailing, and ligating to methylated adapters, followed by bisulfite conversion. The bisulfite converted libraries were then PCR amplified and purified, and sequenced if they passed quality control (size range between 200 to ∼400 bp and concentration greater than 2 nM), which was checked by analyzing the library on an Agilent DNA 1000 chip. The 100-bp paired end reads were obtained for each library using Illumina HiSeq2000. Reads were trimmed to remove adapters and bases with poor sequencing quality using FASTQC. The aligner Bismark (version 0.10.0; Krueger and Andrews, 2011) was used to map quality-passed reads to maize B73 reference genome version 2, allowing one mismatch in the 25-nucleotide seed sequence (-N 1 -L 25). Methylation status at each cytosine was then determined using the methylation extractor command in Bismark. Due to the large size of the maize genome, the methylation level at each sequence context (CG, CHG, and CHH) was calculated for nonoverlapping 100-bp sliding tiles across the whole genome instead of individual cytosine using the weighted DNA methylation computing method (#C/(#C+#T)) (Schultz et al., 2012).
We computed genome-wide methylation levels by averaging all the tiles with coverage. Each 100-bp tile was then associated with the nearest TEs (ZmB73_5b_MTEC) or genes (ZmB73_5b_FGS) using the closestBed command from BEDTools (Quinlan and Hall, 2010). The tiles that overlap each genomic feature were flagged by assigning a distance of 0 between the 100-bp tile and the genomic feature. We filtered the files overlapping four genomic features: exons, introns, transposons, and nontransposon intergenic regions. Nontransposon intergenic regions were defined as regions that didn’t overlap any genes or transposons. We used the average of all 100-bp tiles overlapping a specific genomic feature to represent methylation levels of that genomic feature. The methylation levels at different transposon subfamilies were calculated in a similar way: 100-bp tiles overlapping each subfamily were identified using BEDTools, and methylation levels at those tiles were then averaged to represent methylation level at each transposon subfamily. The above methylation levels were calculated for each mutant as well as the wild-type inbred lines (B73, Mo17, Oh43, CML322, W22, and Tx303). The relative DNA methylation loss at each genomic feature was calculated as (mutant − wild type)/wild type, whereas DNA methylation level in the wild type was calculated as the average of the wild-type inbreds.
To generate line plots for methylation levels over the transposon and its flanking regions, we first identified 100-bp tiles in or close to each transposon type. The distance between the tiles and transposon was determined as the distance between the midpoint of each tile and the start position of the transposon. Tiles within, upstream of, and downstream of a transposon were assigned 0, a negative, and a positive value respectively. Tiles within a transposon were further positioned at a scale of 1 to 1000 bp by normalizing the distance between tile and transposon start site to the size of the transposon, multiplied by 1000. We divided each of the three 1000-bp bins (upstream, within, and downstream) into 20 equal bins each with 50 bp. Methylation at CG, CHG, and CHH was calculated for each bin for each transposon and were then averaged over transposons within the same subfamily. The averaged methylation level was then plotted against the position of each bin and displayed in a line plot (example in Figure 6C). Similar plots were generated for transposons with different sizes (examples in Figures 6F to 6K).
For line plots at genes, a total of 10-kb genomic region, including the regions 3 kb into genes from both the 5′ end and 3′ end, along with 2 kb surrounding regions on each side were plotted. For genes that are less than 3 kb, only the actual gene region was used; thus, those genes will have no contribution to methylation level of regions that are beyond their size. The 10-kb region was divided into 200 bins of 50 bp each. The methylation level for each sequence context was calculated for each bin for each gene and was then averaged over all genes for a particular bin. The derived methylation level was used to plot against the center position of each bin (see example in Figure 6A).
Sequence Capture, Sequencing, and Analysis
The sequence capture bisulfite sequencing was performed using the Roche NimbleGen SeqCap Epi protocol (http://www.nimblegen.com/products/lit/07162839001_NG_SQC_UG_SeqCapEpi_0114.pdf). In short, a standard bisulfite-treated library was constructed, followed by hybridization to probes designed to target a set of genomic regions selected for analysis (Supplemental Data Set 1). After hybridization, the captured library was recovered, amplified using 16 PCR cycles, and sequenced on MiSeq using 150 cycles from both ends (see Supplemental Table 3 for read numbers and mapping efficiency for each sample). The probe sequences used to perform the capture were designed to target all possible sequences that might result from different DNA methylation states.
Reads were aligned to maize genome version 2 using the aligner Bismark allowing one mismatch in the 25-nucleotide seed sequence (-N 1 -L 25). Only uniquely mapped reads were kept for subsequent analysis. Conversion rate was determined using the cytosines mapped to the unmethylated chloroplast genome. The output alignment file from Bismark was used to derive methylation level (i.e., number of methylated and unmethylated reads) for each cytosine in all three sequence contexts (CG, CHG, and CHH) using methylation extractor from Bismark. The methylation level of a specific target region was calculated based on the cytosines within the region using the weighted DNA methylation method (#C/(#C+#T)). Read coverage per target regions was obtained by counting the number of reads overlapping with the target regions, which was determined using BEDTools. The DNA methylation levels for each captured region are available in Supplemental Data Set 1, and accession numbers for the raw data are available in Supplemental Table 3.
Analysis of Small RNA
We used a small RNA data set from the study by Regulski et al. (2013) (Gene Expression Omnibus accession number GSE39232). This data set was generated from sequencing small RNAs that were isolated in the B73 genotype from shoot tissue at the coleoptile stage 5 d after planting. The FASTX toolkit was used to remove 3′ adapters, control sequencing quality, and select small RNAs with a size between 21 and 24 nucleotides. We removed the types of small RNA that were mapped to tRNA, rRNA, and microRNA. The remaining small RNA was mapped to maize genome version 2, allowing only one mapping location. Small RNA abundance was then computed for the target regions that were included in sequence capture. To make small RNA levels comparable across different target regions, we normalized small RNA counts to the size of target regions. The normalized small RNA counts were then plotted using boxplot for regions where DNA methylation was mediated by different pathways.
Accession Numbers
Sequencing data from this study can be found in the NCBI Sequence Read Archive (SRA) under the accession number SRP048795. The SRA number for each experiment can be found in Supplemental Tables 2 and 3.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of DNA Methyltransferases in Maize and Arabidopsis.
Supplemental Figure 2. Methylation Profiles over Genes and Surrounding Regions in Subsets of the B73 WGBS Data Set.
Supplemental Figure 3. Comparison of Methylation Levels Measured by Low-Coverage WGBS and SeqCap for Each Sequence Context.
Supplemental Figure 4. Small RNA Profiles at Different Transposon Types and Sizes.
Supplemental Figure 5. Regulation of CHG Methylation by chr106, zmet2, and zmet5 across Transposons with Different Sizes.
Supplemental Figure 6. Expression Patterns of Three Paralogous Pairs in Different Tissues of B73.
Supplemental Figure 7. Two Scenarios Leading to Reduced Recovery of Double Mutant.
Supplemental Table 1. Summary of Mutants.
Supplemental Table 2. Mapping Summary of Whole-Genome Bisulfite Sequencing Data Set.
Supplemental Table 3. Mapping Summary of Targeted Bisulfite Sequencing Data Set.
Supplemental Table 4. Isolation of Double Mutants.
Supplemental Table 5. Primers Used in This Study.
Supplemental Data Set 1. Data for Sequence Capture Libraries.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Tree in Supplemental Figure 1.
Supplementary Material
Acknowledgments
Library sequencing was performed at the University of Minnesota Genomics Center. Data analysis was performed using the tools and resources provided by the iPlant Collaborative. Computational support and data storage was provided by the Texas Advanced Computing Center at the University of Texas at Austin. The Minnesota Supercomputing Institute provided access to software and user support for data analyses. We thank Steve Jacobsen and Robert Schmitz for helpful discussions regarding this project. This work was supported by a grant from the National Science Foundation to N.M.S. and M.W.V. (DBI-1237931).
AUTHOR CONTRIBUTIONS
M.W.V. and N.M.S. designed the research. Q.L., S.R.E., P.J.H., V.M.Z., J.W., H.R., D.L.B., T.A.R., and J.A.J. performed research. T.F.M., A.E.S., J.H., K.M.M., R.B.M., and O.N.D. contributed new materials. Q.L., S.R.E., J.S., and N.M.S. analyzed data. Q.L., S.M.K., and N.M.S. wrote the article.
Glossary
- RdDM
RNA-directed DNA methylation
- WGBS
whole-genome bisulfite sequencing
- TE
transposable element
Footnotes
Online version contains Web-only data.
References
- Alleman M., Doctor J. (2000). Genomic imprinting in plants: observations and evolutionary implications. Plant Mol. Biol. 43: 147–161. [DOI] [PubMed] [Google Scholar]
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J.E., White J., Sikkink K., Chandler V.L. (2006). An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442: 295–298. [DOI] [PubMed] [Google Scholar]
- Barbour J.E., Liao I.T., Stonaker J.L., Lim J.P., Lee C.C., Parkinson S.E., Kermicle J., Simon S.A., Meyers B.C., Williams-Carrier R., Barkan A., Hollick J.B. (2012). required to maintain repression2 is a novel protein that facilitates locus-specific paramutation in maize. Plant Cell 24: 1761–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L., Malagnac F., Bender J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15: 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucom R.S., Estill J.C., Chaparro C., Upshaw N., Jogi A., Deragon J.M., Westerman R.P., Sanmiguel P.J., Bennetzen J.L. (2009). Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5: e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker C., Hagmann J., Müller J., Koenig D., Stegle O., Borgwardt K., Weigel D. (2011). Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480: 245–249. [DOI] [PubMed] [Google Scholar]
- Bucher E., Reinders J., Mirouze M. (2012). Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr. Opin. Plant Biol. 15: 503–510. [DOI] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13: 2212–2217. [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. (2002). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. USA 99 (suppl. 4): 16491–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V.L. (2007). Paramutation: from maize to mice. Cell 128: 641–645. [DOI] [PubMed] [Google Scholar]
- Dorweiler J.E., Carey C.C., Kubo K.M., Hollick J.B., Kermicle J.L., Chandler V.L. (2000). mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12: 2101–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., Ellis N.A., Makarevitch I., Yeh C.T., Gent J.I., Guo L., McGinnis K.M., Zhang X., Schnable P.S., Vaughn M.W., Dawe R.K., Springer N.M. (2012). Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 8: e1003127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., et al. (2013). Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S.R., Schmitz R.J., Springer N.M. (2014). Epigenetics: beyond chromatin modifications and complex genetic regulation. Plant Physiol. 165: 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhard K.F. Jr., Stonaker J.L., Parkinson S.E., Lim J.P., Hale C.J., Hollick J.B. (2009). RNA polymerase IV functions in paramutation in Zea mays. Science 323: 1201–1205. [DOI] [PubMed] [Google Scholar]
- Fedoroff N., Schläppi M., Raina R. (1995). Epigenetic regulation of the maize Spm transposon. BioEssays 17: 291–297. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M., Michaud C., Leblanc O., Grimanelli D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J.I., Ellis N.A., Guo L., Harkess A.E., Yao Y., Zhang X., Dawe R.K. (2013). CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Deleris A., Wong W., Zhong X., Chin H.G., Horwitz G.A., Kelly K.A., Pradhan S., Jacobsen S.E. (2010). The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 6: e1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick J.B. (2010). Paramutation and development. Annu. Rev. Cell Dev. Biol. 26: 557–579. [DOI] [PubMed] [Google Scholar]
- Hollick J.B., Chandler V.L. (2001). Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., et al. (2014). Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. USA 111: 10642–10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A., Richards E.J. (1996). mCCG methylation in angiosperms. Plant J. 9: 579–586. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes T.L., Richards E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22: 94–97. [DOI] [PubMed] [Google Scholar]
- Johannes F., et al. (2009). Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5: e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.Y., Zilberman D. (2014). DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 19: 320–326. [DOI] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Eichten S.R., Hermanson P.J., Springer N.M. (2014). Inheritance patterns and stability of DNA methylation variation in maize near-isogenic lines. Genetics 196: 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A.M., Cao X., Jackson J.P., Zilberman D., McCallum C.M., Henikoff S., Jacobsen S.E. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080. [DOI] [PubMed] [Google Scholar]
- Lisch D., Carey C.C., Dorweiler J.E., Chandler V.L. (2002). A mutation that prevents paramutation in maize also reverses Mutator transposon methylation and silencing. Proc. Natl. Acad. Sci. USA 99: 6130–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzima T.F., Mills E.S., Gardiner J.M., McGinnis K.M. (2011). Identification of epigenetic regulators of a transcriptionally silenced transgene in maize. G3 (Bethesda) 1: 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevitch I., Stupar R.M., Iniguez A.L., Haun W.J., Barbazuk W.B., Kaeppler S.M., Springer N.M. (2007). Natural variation for alleles under epigenetic control by the maize chromomethylase zmet2. Genetics 177: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408. [DOI] [PubMed] [Google Scholar]
- McCarty D.R.andMeeley R.B. (2009). Transposon resources for forward and reverse genetics in maize. In Handbook of Maize: Genetics and Genomics, Bennetzen J.L., Hake S.C., eds (Berlin: Springer), pp. 561–584. [Google Scholar]
- McGinnis K., et al. (2007). Assessing the efficiency of RNA interference for maize functional genomics. Plant Physiol. 143: 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Springer C., Lin Y., Carey C.C., Chandler V. (2006). Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173: 1637–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuta K., et al. (2008). Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. USA 105: 14958–14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa C.M., Springer N.M., Muszynski M.G., Meeley R., Kaeppler S.M. (2001). Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A.D. (1980). DNA methylation and gene function. Science 210: 604–610. [DOI] [PubMed] [Google Scholar]
- Regulski M., et al. (2013). The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J., Wulff B.B., Mirouze M., Marí-Ordóñez A., Dapp M., Rozhon W., Bucher E., Theiler G., Paszkowski J. (2009). Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. 23: 939–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M.J., Galbiati M., Ticknor C., Chen J., Dellaporta S.L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273: 654–657. [DOI] [PubMed] [Google Scholar]
- Schmitz R.J., He Y., Valdés-López O., Khan S.M., Joshi T., Urich M.A., Nery J.R., Diers B., Xu D., Stacey G., Ecker J.R. (2013). Epigenome-wide inheritance of cytosine methylation variants in a recombinant inbred population. Genome Res. 23: 1663–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R.J., Schultz M.D., Lewsey M.G., O’Malley R.C., Urich M.A., Libiger O., Schork N.J., Ecker J.R. (2011). Transgenerational epigenetic instability is a source of novel methylation variants. Science 334: 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P.S., et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schultz M.D., Schmitz R.J., Ecker J.R. (2012). ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28: 583–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Briskine R., Hirsch C.N., Myers C.L., Springer N.M., Buell C.R., de Leon N., Kaeppler S.M. (2013). Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS ONE 8: e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settles A.M., et al. (2007). Sequence-indexed mutations in maize using the UniformMu transposon-tagging population. BMC Genomics 8: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko L., Dorweiler J.E., Cigan A.M., Arteaga-Vazquez M., Vyas M., Kermicle J., Jurcin D., Brzeski J., Cai Y., Chandler V.L. (2009). A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 5: e1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, A.E., Sidorenko, L., and McGinnis, K.M. (2014). Diverse gene silencing mechanisms with distinct requirements for RNA polymerase subunits in Zea mays Genetics 198: 1031–1042. [DOI] [PMC free article] [PubMed]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., Patel D.J., Jacobsen S.E. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Greenberg M.V., Feng S., Bernatavichute Y.V., Jacobsen S.E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonová Z., Lai J., Ma J., Ramakrishna W., Llaca V., Bennetzen J.L., Messing J. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14 (10A): 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B.J., et al. (2004). Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol. 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongs A., Kakutani T., Martienssen R.A., Richards E.J. (1993). Arabidopsis thaliana DNA methylation mutants. Science 260: 1926–1928. [DOI] [PubMed] [Google Scholar]
- West P.T., Li Q., Ji L., Eichten S.R., Song J., Vaughn M.W., Schmitz R.J., Springer N.M. (2014). Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS ONE 9: e105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Dittmer T.A., Richards E.J. (2008). Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 4: e1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Pontes O., Pikaard C.S., Richards E.J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu J.K. (2012). Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 77: 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Du J., Hale C.J., Gallego-Bartolome J., Feng S., Vashisht A.A., Chory J., Wohlschlegel J.A., Patel D.J., Jacobsen S.E. (2014). Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.