Transcriptome profiling of M. truncatula root hairs during the initial stages of rhizobial infection helps to interpret two decades of research on Medicago and provides a foundation for future studies on host-symbiont interactions in the rhizosphere.
Abstract
Nitrogen-fixing rhizobia colonize legume roots via plant-made intracellular infection threads. Genetics has identified some genes involved but has not provided sufficient detail to understand requirements for infection thread development. Therefore, we transcriptionally profiled Medicago truncatula root hairs prior to and during the initial stages of infection. This revealed changes in the responses to plant hormones, most notably auxin, strigolactone, gibberellic acid, and brassinosteroids. Several auxin responsive genes, including the ortholog of Arabidopsis thaliana Auxin Response Factor 16, were induced at infection sites and in nodule primordia, and mutation of ARF16a reduced rhizobial infection. Associated with the induction of auxin signaling genes, there was increased expression of cell cycle genes including an A-type cyclin and a subunit of the anaphase promoting complex. There was also induction of several chalcone O-methyltransferases involved in the synthesis of an inducer of Sinorhizobium meliloti nod genes, as well as a gene associated with Nod factor degradation, suggesting both positive and negative feedback loops that control Nod factor levels during rhizobial infection. We conclude that the onset of infection is associated with reactivation of the cell cycle as well as increased expression of genes required for hormone and flavonoid biosynthesis and that the regulation of auxin signaling is necessary for initiation of rhizobial infection threads.
INTRODUCTION
Nodulation in legumes requires two coordinated programs, rhizobial infection and nodule organogenesis, that converge to release rhizobia into developing nodule cells where they fix nitrogen. In most legumes, rhizobia enter roots through plant-made infection threads, tubular invaginations, which initiate on growing root hairs. Following rhizobial entrapment within root hairs, infection threads provide a conduit through which rhizobia can colonize the root cortex (Oldroyd et al., 2011). These structures are unique to nitrogen-fixing symbioses and represent a significant innovation in cellular growth and differentiation, but we know very little about how they are formed.
Rhizobia attach to the flank of the root hairs, and subsequent anisotropic growth of the root hair forms a tight curl enclosing rhizobia in an infection pocket formed by the apposed cell walls. The nucleus then doubles in size and moves to a central position in the cell (Dart, 1974) and a broad cytoplasmic bridge forms between the site where the infection thread will initiate and the nucleus (Timmers et al., 1999; Fournier et al., 2008). Once initiated, the infection thread grows intermittently and is colonized by dividing rhizobia (Fournier et al., 2008). As the growing infection thread nears the base of the root hair, the cell wall starts to weaken at the junction of the cytoplasmic bridge. The underlying outer cortical cell then undergoes a similar series of events whereby the nucleus occupies a central position within a large anticlinal cytoplasmic bridge that is aligned with the incoming infection thread (van Brussel et al., 1992). The infection thread progresses through this cell-cell junction and continues growing in this manner through the outer cortical cell layers. When it reaches the cells of the nascent nodule, the infection thread ramifies, extending into the nodule cells, where rhizobia are taken up from the tips of the infection threads by endocytosis to form organelle-like structures called symbiosomes.
Rhizobial infection and nodule organogenesis both require the production of lipochitooligosaccharide Nod factors that are produced by rhizobia in response to plant flavonoids and related compounds (Peters et al., 1986; Subramanian et al., 2006). Infection pocket formation requires localized release of Nod factors (van Batenburg, et al., 1986; Esseling et al., 2003) and the changes in cell architecture that precede infection thread formation result from Nod factor-induced rearrangements in actin filaments (Crdenas et al., 1998; de Ruijter et al., 1999). Perception of Nod factors in Medicago truncatula and Lotus japonicus is mediated by the LysM receptor-like kinases NFP and LYK3 (Radutoiu et al., 2003; Geurts et al., 2005; Smit et al., 2007). Activation of the nodulation signaling pathway to induce gene expression requires several transcription factors, including Nodule Inception (NIN), ERF Required for Nodulation1 (ERN1), and the GRAS transcription factors Nodulation Signaling Pathway1 (NSP1) and NSP2 (Schauser et al., 1999; Oldroyd and Long, 2003; Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Marsh et al., 2007; Middleton et al., 2007), all of which are involved in the formation of infection threads but not root hair deformation. Some components induced have roles in actin nucleation (Yokota et al., 2009; Miyahara et al., 2010; Hossain et al., 2012). NIN, NSP1, and NSP2 act in a signaling pathway required for both rhizobial colonization and nodule organogenesis in L. japonicus (Madsen et al., 2010). NIN is required for the expression of the flotillin encoding genes FLOT2 and FLOT4 in M. truncatula (Haney and Long, 2010) as well as NODULATION PECTATE LYASE1 (NPL1) in L. japonicus (Xie et al., 2012), which are necessary for bacterial infection. NIN is also required for the induction of two CCAAT-box transcription factors Lj-NFYA1 and Lj-NFYB1 in L. japonicus, which are required for nodule organogenesis (Soyano et al., 2013). Mt-NFYA1 (the ortholog of Lj-NFYA1, previously known as HAP2.1) also has a role in infection in Medicago (Combier et al., 2006, 2008; Laporte et al., 2014).
In Medicago, LUMPY INFECTIONS, encoding an E3 ubiquitin ligase is required for infection thread development and is needed for the expression of the infection genes RHIZOBIUM-DIRECTED POLAR GROWTH (RPG; Arrighi et al., 2008; Kiss et al., 2009) and VAPYRIN, a protein of unknown function also required for mycorrhization (Pumplin et al., 2010; Murray et al., 2011). In soybean (Glycine max), knockdown of Nucleolar/Mitochondrial protein involved in Nodulation a impaired infection (Libault et al., 2011). Other legume mutants that are impaired in the sensing of ethylene (sickle) or cytokinin (hyperinfected1) have hyperinfection phenotypes (Penmetsa and Cook, 1997; Murray et al., 2007). However, the relationship of these downstream components required for infection and the transcription factors that control them is not known.
From the first efforts at isolation of proteins and RNA from purified root hairs (Röhm and Werner, 1987; Covitz et al., 1998; Ramos and Bisseling, 2003; Sauviac et al., 2005) to the recent application of different -omics technologies in soybean (Brechenmacher et al., 2009, 2010; Libault et al., 2010), the advantages of isolating root hairs have been recognized. We analyzed the root hair transcriptome of M. truncatula during rhizobial infection, studying gene expression over a time course. By comparing gene induction by Nod factors and Sinorhizobium meliloti in the wild type and the hyperinfected mutant sickle (skl), we identified infection-related genes and processes previously undetected in whole-root studies or in forward genetic analyses. Medicago spp have served as models for phenylpropanoid metabolism for over two decades, and we extend these studies providing new insights into rhizobial infection. In addition, we identified other processes that are regulated in root hairs undergoing rhizobial colonization. Among them are changes in the cell cycle and in the plant hormones strigolactone (SL), gibberellic acid (GA), brassinosteroids (BRs), and auxin, and we reveal a novel role for auxin signaling in rhizobial infection.
RESULTS
The Infectome
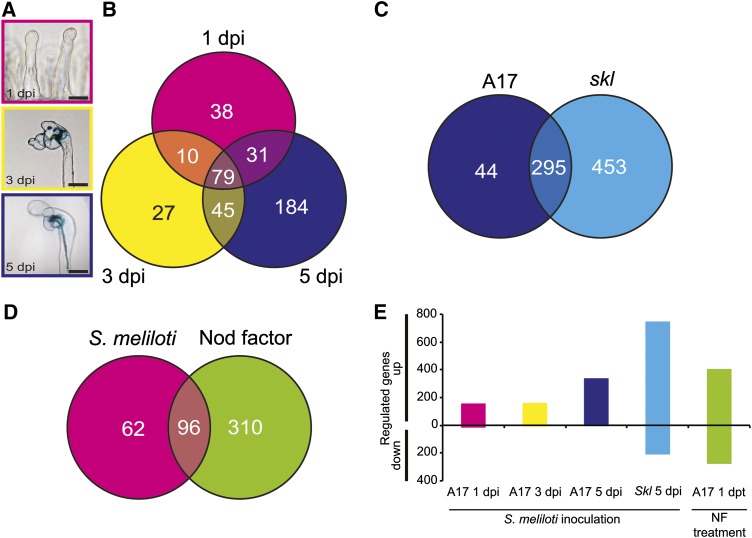
Our main goal was the identification of genes responsible for the developmental changes occurring during rhizobial infection. To accomplish this, wild-type roots were inoculated with S. meliloti Sm1021 or an isogenic mutant (SL44; S. meliloti nodΔD1ABC) that is completely defective for Nod factor signaling, ensuring that the effects we detect are specific for Nod factor signaling rather than other rhizobial-root interactions. We first monitored the progression of rhizobial infection under our selected growth conditions. One day postinoculation (dpi), no curled root hairs or infections were evident; 3 dpi, root hair curling and microcolony formation was observed but no infection threads were evident; 5 dpi, many infection threads had initiated but few had extended into the outer cortex (Figure 1A).
Figure 1.
Experimental Overview of Transcriptome Analysis Performed and Numbers of Significantly Regulated Genes Identified.
(A) Infection time course of wild-type (A17) M. truncatula root hairs from seedlings inoculated with S. meliloti expressing lacZ. Major developmental milestones of infection: 1 dpi represents preinfection before visible root hair curling, at 3 dpi microcolonies had formed within curled root hairs, and at 5 dpi elongating infection threads were seen. Bar = 20 μm.
(B) Genes upregulated in the wild type 1, 3, and 5 dpi with Sm1021.
(C) Genes upregulated 5 dpi with Sm1021 in the wild type and in the hyperinfected skl mutant.
(D) Genes upregulated in wild-type 1 dpi with Sm1021 and 1 d post-treatment with Nod factor.
(E) Summary of up- and downregulated genes for each treatment. dpt, days post-treatment.
Genes with significant regulation relative to control experiments are shown (>2-fold change; P < 0.05).
To enhance identification of infection genes, we also profiled the skl mutant, which, due to a defect in ethylene signaling, had at 5 dpi a 6-fold increase in rhizobial infections relative to the wild type (P value = 0.00004, Student’s t test). We also analyzed wild-type root hairs 24 h after addition of 10 nM Nod factor, a concentration sufficient to induce both calcium spiking and a calcium influx in root hairs that is associated with rhizobial infection (Morieri et al., 2013). A file with all data from individual replicates normalized across treatments is provided in Supplemental Data Set 1.
RNA extracted from root hairs from the different treatment groups at specific time points was analyzed using microarrays (GeneChip Medicago Genome Array, first version). GeneChip hybridizations were performed on three biological replicates per treatment (response of wild-type seedlings to S. meliloti strain Sm1021 (wild type) versus strain SL44 (nodΔD1ABC). All data are available through the Medicago Gene Expression Atlas server (http://mtgea.noble.org/v3/). Comparing the responses of wild-type seedlings to S. meliloti wild type and SL44, we observed a significant and at least 2-fold increase in expression of 158, 161, and 339 genes 1, 3, and 5 dpi, respectively; 79 of these genes were induced at all three time points (Figure 1B; see Methods for description of normalization and statistical analysis). Comparison of the data from root hairs (5 dpi) to data from emerging nodule primordia (Benedito et al., 2008) detected 230 genes that could not be identified in excised root tissue containing nodule primordia (Supplemental Figure 1).
In wild-type seedlings, 93% of the genes that responded to rhizobial infection had increased expression (404 induced, 30 repressed). A larger proportion of the responding genes were repressed in skl (Figure 1E). Responses to Nod factors were more balanced with almost as many genes being repressed as induced (Figure 1E). All data showing significant changes following inoculation or Nod factor treatment are provided in Supplemental Data Set 2.
About 61% (96/158) of the genes induced 1 dpi with S. meliloti 1021 were also induced by Nod factors (Figure 1D), indicating that most of the gene expression changes that occur during preinfection can be accounted for by Nod factors alone. Even more Nod factor-induced genes were also induced at the onset of infection (5 dpi), but at this time point, about twice as many genes were induced compared with the preinfection time points, so the proportion of Nod factor-responsive genes was reduced (41%, 140/339).
The skl Mutation Increases Detection of Infection-Related Genes
At 5 dpi, 87% (295/339) of the genes upregulated in the wild type by strain 1021 were induced in the skl mutant, but an additional 453 upregulated genes were identified (Figure 1C). Genes known to be required for, or associated with infection, revealed that they were on average 2.2-fold more strongly induced in skl than in the wild type (Figure 2) (e.g., NIN expression was induced 26- and 117-fold in the wild type and skl, respectively). Of the 414 genes induced by S. meliloti in the wild type (Figure 1B), 96% had enhanced expression in the skl mutant, including the cytokinin receptor CRE1 (Figure 2). Most of the 410 additional genes discovered in skl also appeared to be slightly increased in the wild type relative to the controls, although they did not show statistically significant differences. Several (47) genes were induced by S. meliloti 1021 in skl and in the wild type by Nod factor but were not significantly induced by S. meliloti in the wild type at any stage; this may be a consequence of a stronger nodulation signaling response in skl compared with the wild type. This global effect of ethylene perception on infection-related gene expression is consistent with the ability of this hormone to suppress Nod factor-induced calcium oscillations (Oldroyd et al., 2001).
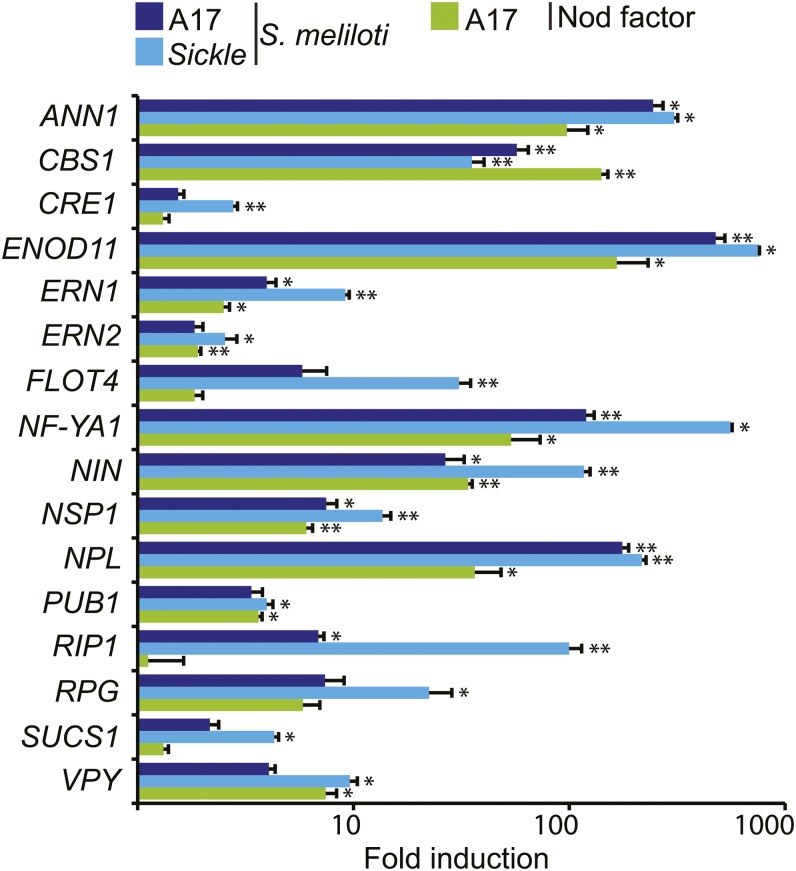
Figure 2.
Induction of Known Symbiotic Genes.
Fold induction of known symbiotic genes 5 dpi with Sm1021 or 1 d post-treatment with Nod factor. References for genes not cited elsewhere in the text: ANN1 (De Carvalho-Niebel et al., 2002), ERN2 (Andriankaja et al., 2007; Cerri et al., 2012), and ENOD11 (Journet et al., 2001). Significant inductions relative to control experiments are shown (*P < 0.05, **P < 0.01). Error bars = se (n = 3).
Highlights from the Infectome
In addition to the known infection-related genes mentioned above (Figure 2), several other symbiosis-related genes were seen to be regulated by S. meliloti 1021 and/or Nod factor. The LysM receptor kinase genes LYK10 and LYE2 were induced by S. meliloti 1021 (Supplemental Figure 2B), whereas three LysM-RLKs (including the Nod factor receptors NFP and LYK3) and two LYK-related (LYR) genes were repressed. Extracellular peroxidases that produce reactive oxygen species in the apoplast are induced early during rhizobial colonization (Cook et al., 1995; Ramu et al., 2002) and are predicted to be secreted to the lumen of the developing infection thread where they may promote hardening of the matrix (Wisniewski et al., 2000; Passardi et al., 2004). We found 10 peroxidase genes, including Rhizobium-Induced Peroxidase1 (RIP1; Cook et al., 1995; Ramu et al., 2002) that were induced in root hairs by both S. meliloti and Nod factors (Supplemental Figure 2A). Eight of these had predicted secretion signal peptides, suggesting increased production of reactive oxygen species in the lumen of the infection thread and/or around root hairs. Several other genes that had not been previously linked to infection were identified. The sucrose synthase gene SUCS1 (Mtr.22018.1.S1_s_at: Medtr4g124660), which is expressed in nodules and required for nitrogen fixation (Baier et al., 2007) and is required for colonization by arbuscular mycorrhizal fungi (Hohnjec et al., 1999; Baier et al., 2010), had increased expression in skl after S. meliloti inoculation. SWEET13, which encodes a sugar transporter (Chen et al., 2012), and a gene involved in actin nucleation ABIL1 (Mtr.20281.1.S1_at: Medtr7g116710), which had very low expression in root hairs in the absence of Nod factor-producing rhizobia, were also induced by S. meliloti. Some evidence for transient defense responses was seen. Another gene encoding a subtilase (Medtr4g102400) was also transiently induced at 1 dpi. This gene does not respond to pathogens and instead is highly expressed in mycorrhized roots. These genes provide potentially useful markers for defense responses and common symbiotic signaling.
Comparison with Soybean
The Medicago lineage diverged from that of Glycine ∼54 million years ago (Lavin et al., 2005). To identify genes that are conserved in host responses to rhizobia, we compared our data with a similar study that monitored gene expression in root hairs from soybean 6 to 48 h after inoculation with Bradyrhizobium japonicum (Libault et al., 2010). Putative M. truncatula orthologs for all G. max genes reported to be increased or decreased by rhizobial inoculation were identified (see Methods), revealing 370 genes with conserved regulation. Of these, 51% were orthologs based on their placement in collinear synteny blocks identified by Li et al. (2012) (Supplemental Data Set 3, Conserved Gm). Among the genes induced in both species were virtually all known genes required for infection, including NIN, PUB1, VPY, RPG, NSP1, NSP2, NPL1, FLOT4, RPG, ERN1, ERN2, NFYA1, and NMN1. As discussed below, induced genes involved in early auxin responses and SL and GA biosynthesis were all conserved between soybean and M. truncatula. Also conserved was the induction of the SWEET13 sugar transporter and the Nod factor hydrolase1 (NFH1), the latter encoding an enzyme involved in specifically inactivating Nod factors in the rhizosphere (Tian et al., 2013). Other genes identified as being induced in both species include those encoding an expansin, peroxidases, proteases, pectinesterases, and pectinesterase inhibitors. Another conserved gene strongly induced by Nod factors and by S. meliloti at all time points tested is the ortholog of Arabidopsis thaliana DOWNY MILDEW RESISTANCE6 (DMR6), which encodes a 2-oxoglutarate-Fe(II) oxygenase of unknown function. This is interesting because DMR6 expression is induced specifically at infection sites, and dmr6 mutants can no longer support growth of the biotrophic oomycete pathogen Hyaloperonospora arabidopsidis (nee H. parasitica; van Damme et al., 2008). The conserved induction of this gene in compatible legumes suggests it has a positive role in biotrophic interactions.
Among the conserved genes suppressed during infection were the JA receptor JAZ2 and several genes involved in polyamine biosynthesis and transport (SPERMIDINE HYDROXYCINNAMOYL TRANSFERASE, POLYAMINE OXIDASE2, and POLYAMINE UPTAKE TRANSPORTER4). In addition, the soybean orthologs of Mt-LYK3, Gm-NFR1a and Gm-NFR1b, were repressed 3 dpi by Bradyrhizobium japonicum (Libault et al., 2010), suggesting negative regulation of the Nod factor receptors is widespread in legumes.
The conservation in transcriptional regulation observed between soybean and Medicago suggests that many of the key genes for symbiosis were recruited prior to the divergence of these lineages (Supplemental Data Set 3, Conserved Gm).
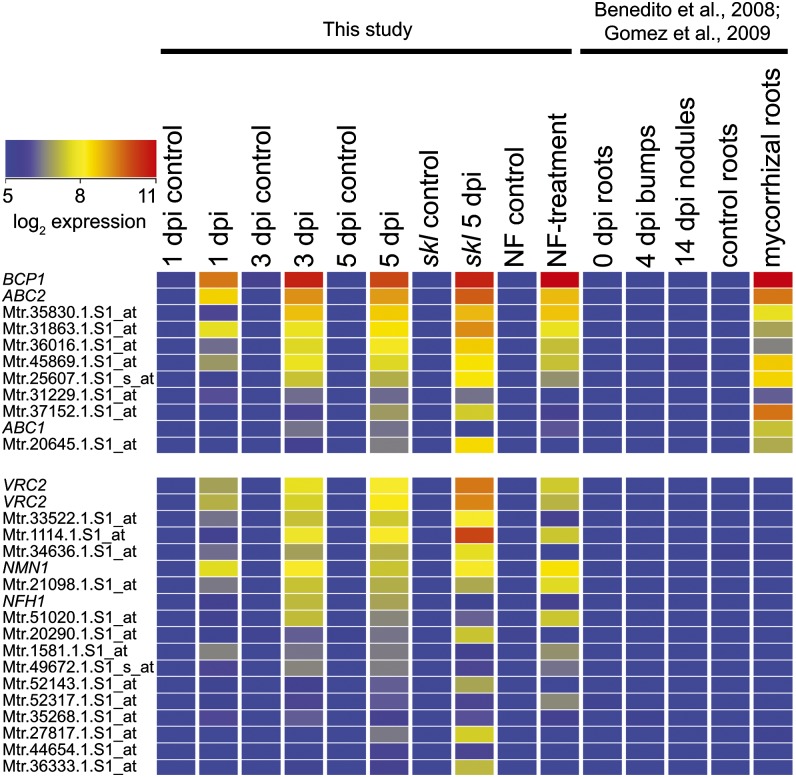
Identification of Root Hair-Specific Genes Induced by Infection
To identify genes that are specific to root hairs during symbiotic infection, we subtracted all nodule-expressed genes (Benedito et al., 2008) from the genes significantly induced to a level >2-fold by S. meliloti. This identified 17 genes induced in the wild type by S. meliloti or Nod factors (Figure 3). Eleven genes meeting these same criteria were also induced in mycorrhizal roots (Figure 3; data from Gomez et al., 2009), including a probe set that corresponded to two blue copper binding proteins, BCP1 (Medtr7g86190) and its close homolog (Medtr7g086160). The BCP1 promoter induces expression in cells containing arbuscules (Hohnjec et al., 2005), but our data suggest that BCP1 may not be specific to mycorrhizal infection. Other genes induced were NFH1 (Tian et al., 2013), a vestitone reductase homolog (designated Vestitone Reductase Cluster2 [VRC2]), two ABC transporters (ABC1 and ABC2), and the ortholog of Nucleolar/Mitochondrial protein involved in Nodulation (NMN1; Libault et al., 2011). Further details on these genes are provided in Supplemental Data Set 4 (Infection Genes).
Figure 3.
Transcriptome Analysis of Purified Root Hairs Reveals Genes with Common Symbiotic and Nodulation Specific Expression.
Heat map displays probe sets that are significantly (P < 0.05) induced at least 2-fold in root hairs 5 dpi with S. meliloti but remain at low levels in a nodule time course (Benedito et al., 2008; Gomez et al., 2009). Names are provided for genes mentioned in the text; otherwise, probe sets are given. Further details including probe sets and corresponding gene models are provided in Supplemental Data Set 4 (Infection genes).
Genes for Legume-Specific Flavonoids Are Increased during Infection
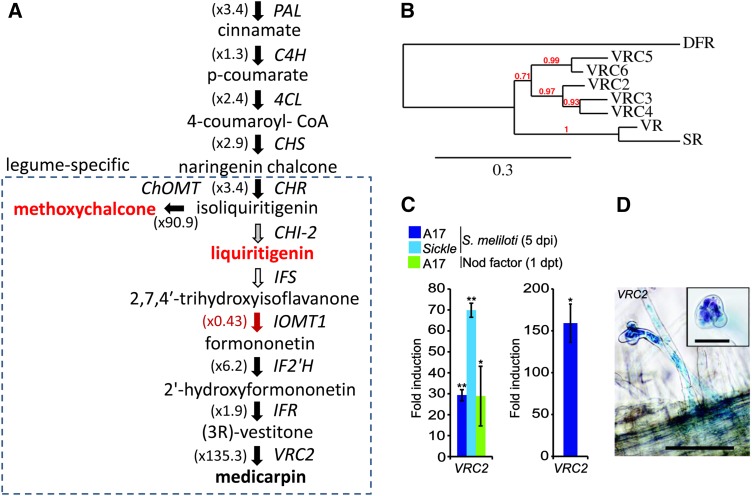
Flavonoids have important roles at different stages in nodulation: They act as rhizobial chemoattractants and nod gene (Nod factor) inducers (Redmond et al., 1986). They have also been implicated in changes in auxin transport in roots during nodule formation (Mathesius et al., 1998; Wasson et al., 2006). As expected from these previous studies, we found that S. meliloti and Nod factors increased expression of two genes encoding phenylammonia lyase isoforms 1 and 2, which catalyze the first committed step in the phenylpropanoid pathway. In parallel, there was increased expression of glutamine synthetase, which catalyzes assimilation of ammonia produced as a by-product of phenylammonia lyase activity, further indicating increased carbon flow into the phenylpropanoid pathway. There was also increased expression of genes encoding a cinnamic acid 4-hydroxylase, two 4-coumarate ligases, a chalcone synthase, and two chalcone reductases (CHRs), the latter being legume specific and acting downstream of chalcone synthase to produce the chalcone isoliquiritigenin (Figure 4; Supplemental Data Set 5, Phenylpropanoids). Notably, two genes involved in lignin biosynthesis from p-coumarate, caffeoyl-CoA O-methyltransferase and caffeic acid-O-methyltransferase (Gowri et al., 1991; Do et al., 2007), were repressed (data not shown), suggesting carbon flow within the pathway is directed mainly toward flavonoid biosynthesis.
Figure 4.
Expression of Genes Involved in Production of Phenylpropanoid Compounds with Activity as S. meliloti nod Gene Inducers and Phytoalexins Are Increased Following Rhizobial Inoculation.
(A) Genes encoding enzymes involved in biosynthesis of the S. meliloti nod gene-inducing compounds liquiritigenin and methoxychalcone and the phytoalexin medicarpin are shown. Genes with expression altered by S. meliloti rhizobia and/or Nod factor are indicated using filled arrows (upregulated in solid black and downregulated are in solid red). Maximum fold increase expression (all treatments) compared with controls is indicated in brackets.
(B) A phylogenetic tree showing vestitone reductase (VR) and five close homologs (Vestitone Reductase Cluster2-6) located within a 71-kb region on M. truncatula chromosome 7. Pea sophorol reductase (SR), which produces the closely related pterocarpin pisatin, is included as a reference. PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; C3H, coumarate 3-hydroxylase; 4CL, 4-coumarate:coA ligase; CHS, chalcone synthase; ChOMT, chalcone O-methyltransferase; CHI, chalcone isomerase; IFS, isoflavone synthase; IF2'H, isoflavone 2'-hydroxylase; IFR, isoflavone reductase.
(C) Analysis of VRC2 expression in root hairs following inoculation with S. meliloti or addition of purified Nod factors as determined by microarray analysis (left). Induction of VRC2 by S. meliloti in wild-type root hairs was confirmed by qPCR (right).
(D) Analysis of pVRC2:GUS expression in hairy roots transformed by A. rhizogenes. Inset shows curled root hairs; rhizobia are stained using magenta-gal to indicate rhizobia within a curled root hair tip. Bars in main image = 100 μm and in inset = 20 μm.
Also induced were a known chalcone O-methyl transferase (ChOMT1) and three additional ChOMT1 homologs (designated ChOMT2-4) that have not been previously identified as being induced (Supplemental Data Set 5, Phenylpropanoids). ChOMT1 acts after CHR to catalyze the conversion of isoliquiritigenin to methoxychalcone (4,4'-dihydroxy-2'-methoxychalcone; Maxwell et al., 1992), which is a potent inducer of S. meliloti nod genes (Maxwell et al., 1989). Isoliquiritigenin can also be converted to another nod gene inducer, liquiritigenin, by Type II chalcone isomerases (CHI-2). A CHI-2 has been characterized in M. sativa but its ortholog in M. truncatula (Medtr1g115820; Jez et al., 2002) is not represented on the microarray used in this study. Liquiritigenin can be converted to dihydroxyflavone, another nod gene inducer by flavone synthase II, which is important for nodulation (Zhang et al., 2007, 2009). We did not see any induction of FSII-1 (which synthesizes flavanones) during infection, while FSII-2, which was found to be induced by S. meliloti (Zhang et al., 2007) was not represented on our microarray.
Genes encoding enzymes active in the branch of the isoflavonoid biosynthetic pathway that produces phytoalexins were also induced, including three genes acting downstream of CHR that are needed for the synthesis of medicarpin (Figure 4A). Medicarpin is produced during nodulation and is induced by Nod factors and rhizobia (Dakora et al., 1993; Phillips et al., 1994; Savouré et al., 1997). S. meliloti also induced isoflavone 2'-hydroxylase (IF2’H), isoflavone reductase (IFR), and two vestitone reductases (VR), which catalyze the penultimate step of the pathway for medicarpin biosynthesis. Two members of a cluster of six VR homologs (Figure 4B) on chromosome 7 were induced. The gene encoding the clear ortholog of the VR gene originally characterized in alfalfa (Medicago sativa; Guo et al., 1994; Guo and Paiva, 1995; Shao et al., 2007) was induced by Nod factors only, while a second isoform, which we designate VRC2, was induced strongly at all three time points following rhizobial inoculation and by Nod factors (Supplemental Data Set 5, Phenylpropanoids). The induction of VRC2 in root hairs of the wild type and skl was confirmed using quantitative PCR (qPCR; Figure 4C). Examination of the Medicago Gene Atlas (MtGEA) revealed that the expression of VRC2 was almost entirely specific to S. meliloti inoculation with the exception of its induction by the cotton (Gossypium hirsutum) root rot pathogen. No other members of this gene cluster (VRC3-6) were induced by either rhizobia or Nod factors. We investigated VRC2 expression further using a promoter-GUS (β-glucuronidase) fusion. This showed that VRC2 expression was specifically associated with the sites of infection and was highest in root hairs forming infection threads (Figure 4D). An isoflavonoid malonyl transferase MAT3 (Medtr7g014160) was also induced by rhizobia and Nod factors. This enzyme catalyzes the malonylation of 7-O-glycosidic (iso) flavones such as formononetin, which may promote their storage in vacuoles, perhaps preventing toxic buildup of intermediates when the pathway is active (Yu et al., 2008).
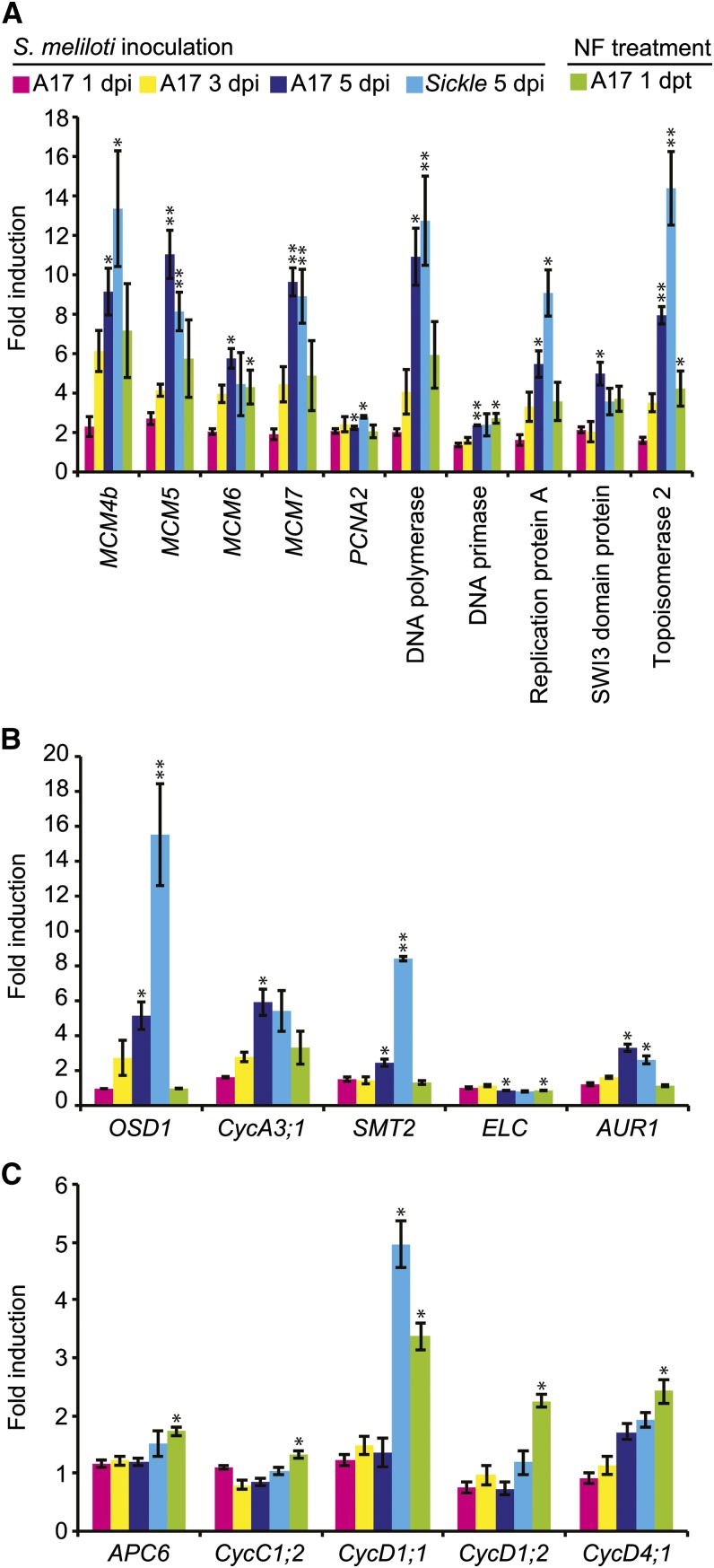
Rhizobial Infection Is Coincident with Increased Expression of Cell Cycle-Related Genes
The greatest number of genes with changed expression was found at the onset of infection (5 dpi). This included the induction of a large set of genes encoding virtually every component of the DNA replication complex and an A-type Cyclin homologous to Arabidopsis CYCA3;1 (Figures 5A and 5B). The expression of this cyclin is very high in meristematic tissues (the shoot apex, root apex, and the internodes; MtGEA), consistent with the proposed role for CYCA3 proteins in mitosis (Takahashi et al., 2010). Increased expression of these genes was statistically significant at 5 dpi (Figure 5B). Several other genes coding for enzymes involved in DNA metabolism were also induced as were several histone-encoding genes (H2A, H2B, and H3 families) and a histone deacetylase. A histone methyltransferase responsible for H3K4 trimethylation was repressed. The details are summarized in Supplemental Data Set 6 (Cell Cycle). In addition, S. meliloti significantly altered expression of several genes involved in the regulation of endoreduplication.
Figure 5.
Cell Cycle-Related Genes Are Regulated during Infection.
(A) Fold induction of replication fork components including mini chromosome maintenance (MCM) genes.
(B) Fold induction of cell cycle regulation genes.
(C) Nod factor treatments induce the expression of the anaphase promoting complex (APC6), a C-type cyclin, and genes encoding three D-type cyclins.
Significant inductions relative to control experiments are shown (*P < 0.05, **P < 0.01). Error bars = se (n = 3).
Based on close homology with Arabidopsis, we identified two genes involved in repressing endoreduplication with increased expression during infection (Figure 5B). The first is OSD1, which is expressed in rapidly dividing cells and has been proposed to repress endoreduplication during mitosis by inhibiting the anaphase-promoting complex/cyclosome (APC/C; Iwata et al., 2011). The second is STEROL METHYLTRANSFERASE2 (SMT2); the Arabidopsis smt2 mutant has increased in ploidy (Hase et al., 2005). A third gene, ELC, which encodes a member of the plant ESCRT I complex that is involved in the switch from mitosis to endoreduplication, has decreased expression (Spitzer et al., 2006). ELC appears to promote endoreduplication by preventing mitosis in cells that have completed S phase; elc mutants feature multiple nuclei in cell that would normally undergo endoreduplication. Interestingly, the lowest expression of the M. truncatula ELC homolog was with S. meliloti 1021 in the skl mutant. This suggests a possible role for ethylene in ELC regulation and is of interest due to a recent report that ethylene-insensitive mutants in L. japonicus have occasional examples where root hair cells infected by rhizobia undergo nuclear division (Gresshoff et al., 2009). A homolog of Arabidopsis MEI-2 Like (AML2) was also repressed. AML2 is an RNA binding protein that promotes meiosis (Kaur et al., 2006).
Nod factors did not significantly alter the expression of the A-type cyclin or endoreduplication-related genes except for ELC, which it repressed, but they did induce three different D-type cyclins and the APC6 subunit of the APC/C (Figure 5C). These data suggest that infection involves reactivation of the cell cycle and repression of endoreduplication.
Infection Alters Expression of Genes Involved in Hormone Biosynthesis and Signaling
GA Biosynthesis
Nod factor signaling appears to increase the production of GA in root hairs cells because the GA biosynthesis genes GIBBERELLIN 3 BETA-HYDROXYLASE1 (GA3OX1), GIBBERELLIN 2-OXIDASE6 (GA2OX6), ENT-COPALYL DIPHOSPHATE SYNTHETASE1 (CPS1), ENT-KAURENE OXIDASE1 (KO1), and ENT-KAURENOIC ACID OXIDASE2 (KAO2) were induced by S. meliloti and by purified Nod factors. Two genes involved in GA regulation and signaling, GAST1 PROTEIN HOMOLOG1 (GASA1) and ZINC FINGER PROTEIN6 (ZFP6), were also induced by Nod factors as was KO1, whereas the gene encoding the GA receptor GA INSENSITIVE DWARF1B (GID1B) was repressed (Supplemental Data Set 7, GA). Many of these genes were also increased after B. japonicum inoculation in soybean.
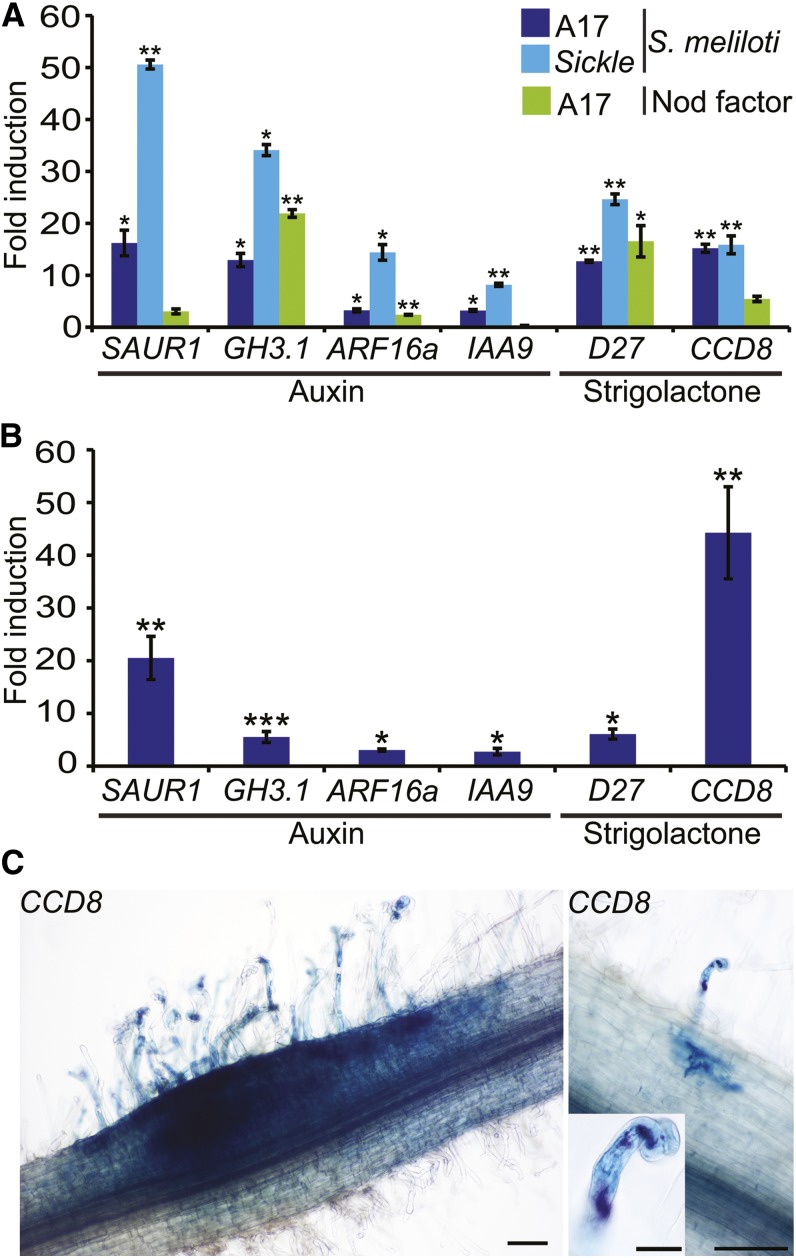
SL Biosynthesis
SLs are carotenoid-derived hormones that act as signaling molecules for arbuscular mycorrhiza (Akiyama et al., 2005). DWARF27 (D27) and CAROTENOID CLEAVAGE DIOXYGENASE8 (CCD8) were induced by S. meliloti, suggesting SLs were being produced (Figures 6A and 6B; Supplemental Data Set 8, SLs). In addition, the carotenoid biosynthesis genes encoding ζ-carotene desaturase (ZDS) and 15-cis-ζ-carotene isomerase (Z-ISO) were induced, as was the ortholog of the cauliflower (Brassica oleracea) Orange gene, which promotes the formation of carotenoid-producing chromoplasts (Lu et al., 2006). These data support increased synthesis of SLs during infection. To assess the potential site of SL production, we fused the promoter of CCD8 to the GUS gene and introduced it into M. truncatula using Agrobacterium rhizogenes-based hairy root transformation. Staining was evident at the sites of S. meliloti infection (Figure 6C) and was initially restricted to colonized root hair cells and was later seen in developing nodule primordia, implying a role for SLs during rhizobial infection.
Figure 6.
Auxin Signaling and SL Biosynthesis Genes Are Induced by S. meliloti and by Nod Factors.
(A) Microarray-based quantification of expression of auxin signaling genes SAUR1, GH3.1, ARF16a, and IAA9 and SL biosynthetic genes D27 and CCD8 at 5 dpi with S. meliloti or 1 d post-treatment with Nod factors in the wild type and skl.
(B) qPCR confirmation of the induction of auxin signaling and SL biosynthetic genes at 5 dpi with S. meliloti.
(C) Analysis of expression of the SL biosynthesis gene CCD8 using a promoter-GUS fusion in A. rhizogenes-induced transgenic roots stained for GUS activity (image on left). The image on the right shows staining of both CCD8:GUS (blue) and S. meliloti (lacZ) (magenta/purple). Bars in main images = 100 μm; insets = 20 μm. Significant induction relative to control experiments are shown (*P < 0.05, **P < 0.01, and ***P < 0.001). Error bars = se (n = 3).
BR Biosynthesis and Signaling
The BR biosynthesis gene DWARF1 (Mtr.10571.1.S1_at: Medtr4g074350) was induced by S. meliloti but not Nod factors. Nod factors did affect BR-related gene expression, repressing the expression of BRASSINAZOLE-RESISTANT1 (BZR1; Mtr.43192.1.S1_at: Medtr5g019550) and SHAGGY-LIKE PROTEIN KINASE23 (SK23; Mtr.17959.1.S1_s_at: Medtr2g083940). BZR1 encodes a transcription factor that is required for normal responses to BR and acts directly to repress the expression of BR biosynthesis genes; SK23 encodes a kinase that phosphorylates BZR1, which excludes it from the nucleus (Ryu et al., 2007). Together, these data suggest that BR synthesis and signaling are triggered during rhizobial infection.
Jasmonic Acid Biosynthesis and Signaling Genes Are Repressed by Nod Factors and by Rhizobial Inoculation in skl
The production of the defense hormone jasmonic acid (JA) appeared to be repressed by S. meliloti in the skl mutant because there was reduced expression of the JA-responsive gene JASMONATE-ZIM-DOMAIN PROTEIN2 (JAZ2), the JA receptor CORONATINE-INSENSITIVE1 (COI1), and the Allene Oxide Synthase1 (AOS1) gene encoding an enzyme required for JA biosynthesis (Supplemental Data Set 9, JA). The marked repression of JA-related responses in skl during infection is presumably in part due to interactions between JA and ethylene signaling. Notably, ethylene is known to induce AOS activity and accumulation of JA (Laudert and Weiler, 1998). However, Nod factors also repressed JA-related responses. Infection of the orthologous Arabidopsis mutant ein2 by the oomycete pathogen Pythium irregular induces much higher levels of JA production than in wild-type plants (Adie et al., 2007). Together these data suggest that repression of JA is a specific feature of rhizobial interactions and not simply a consequence of altered ethylene signaling in skl.
Auxin Signaling
Auxin plays an important role during nodule organogenesis (Mathesius et al., 1998), but no role for auxin during rhizobial infection has been demonstrated. Although there was no significant change in expression of genes for auxin biosynthesis of auxin transport (PIN-FORMED [PIN]), S. meliloti induced expression of the auxin-responsive genes AUXIN RESPONSE FACTOR 16a (ARF16a), Gretchen Hagen3.1 (GH3.1), INDOLE-3-ACETIC ACID INDUCIBLE9 (IAA9), and SMALL AUXIN UP RNA1 (SAUR1) (Figures 6A and 6B; Supplemental Data Set 10, Auxin). All of these belong to relatively large gene families but were the only members that responded to infection.
To investigate the location of auxin signaling during infection, we used M. truncatula stably transformed with the auxin reporter DR5:GUS (Zhou et al., 2011) and observed that S. meliloti induced DR5:GUS in both infected and uninfected root hairs over the entire infection zone (root hair differentiation zone) 3 and 5 dpi (Figures 7A and 7B). This increase in auxin signaling is consistent with the observation that auxin signaling is initially downregulated and then increases at the site of inoculation (Mathesius et al., 1998).
Figure 7.
DR5 Is Expressed throughout the Infection Zone after Rhizobial Inoculation.
DR5:GUS expression in stable transgenic M. truncatula seedlings inoculated with S. meliloti. The blue color in (A) and (B) indicates staining of GUS activity. Arrows (A) indicate infection pockets containing microcolonies, and the arrowhead (B) indicates a root hair containing an infection thread. Bars = 50 μm.
(A) 3 dpi. The inset is a magnification of the boxed region.
(B) 5 dpi.
We compared the expression patterns of GH3.1, SAUR1, and ARF16a with that of the DR5 using promoter-GUS fusions in A. rhizogenes-induced hairy roots. GH3.1, SAUR1, and ARF16a were strongly induced in infected root hairs (Figures 8A, 8B, and 9A to 9C), and ARF16a was strongly expressed around infection foci (Figure 10A) and throughout infection thread growth (Figures 10B to 10D). This contrasts with the results obtained using DR5:GUS, which extended to all root hairs in the infection zone, suggesting that infection sites have differential regulation of auxin responses compared with their neighboring cells.
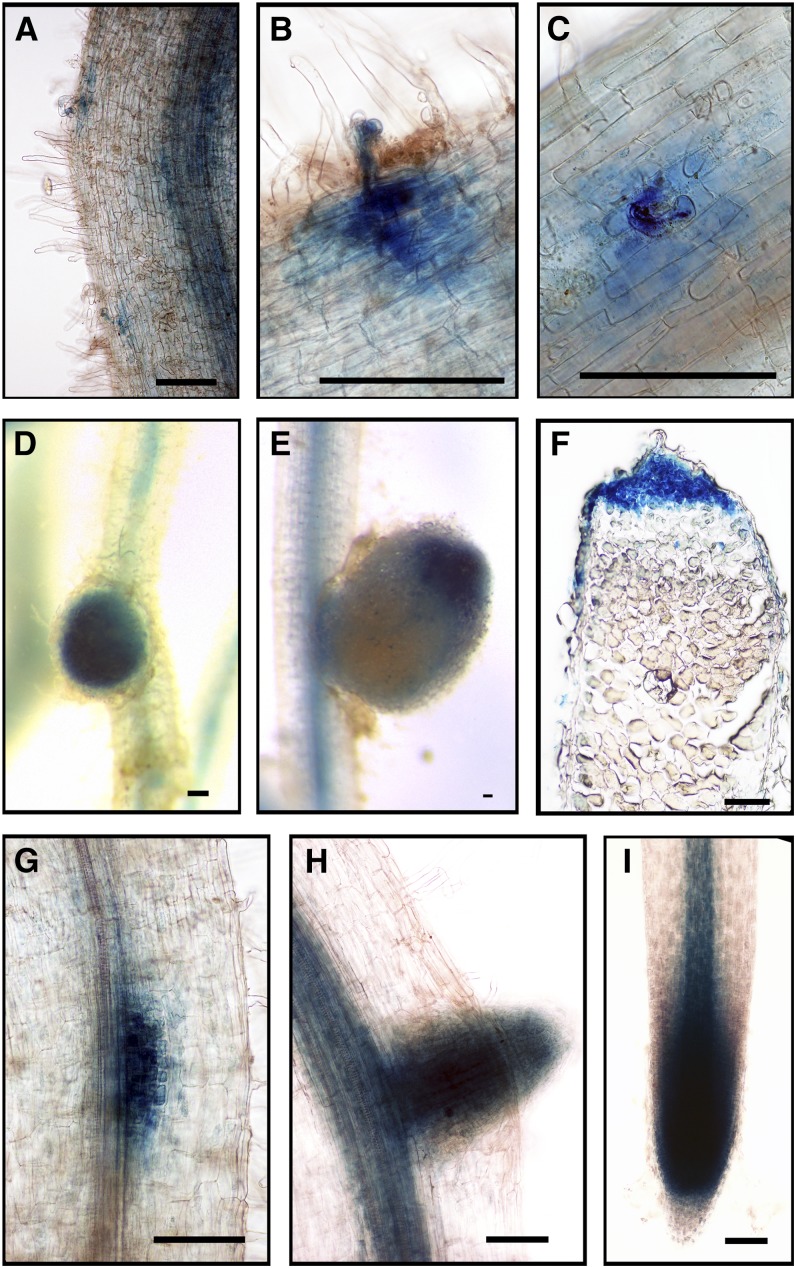
Figure 8.
Expression of GH3.1-GUS Promoter Fusion during Rhizobial Infection and Development of Nodules and Lateral Roots.
(A) to (D) ProGH3.1:GUS expression in A. rhizogenes-induced transgenic hairy roots showing expression at infection sites ([A] and [B]) and in nodules ([C] and [D]), 3 weeks after inoculation with S. meliloti carrying pXLGD4 (lacZ).
(E) to (G) ProGH3.1:GUS expression in a lateral root tip (E), a lateral root primordium (F), and an emerging lateral root (G).
(H) ProGH3.1:GUS expression in zone I and zone II in a mature nodule (3 weeks after inoculation with S. meliloti).
The blue color ([A] to [H]) indicates GUS staining, and the magenta color indicates LacZ staining of S. meliloti. Bars = 100 μm.
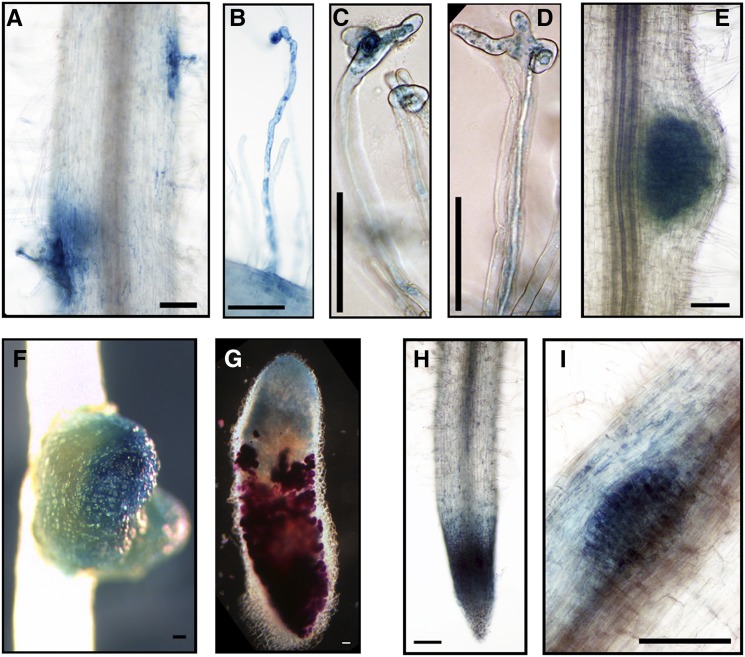
Figure 9.
SAUR1-GUS Promoter Fusion Expression during Rhizobial Infection, and Development of Nodules and Lateral Roots.
A. rhizogenes-induced hairy roots were used to analyze ProSAUR1:GUS expression: (A) to (C) during rhizobial infection; (D) to (F) during nodule development; (G) during the formation of a lateral root primordium; (H) in an emerging lateral root; and (I) in a root tip. The blue color ([A] to [I]) indicates GUS staining of ProSAUR1:GUS activity, and the magenta color (C) indicates LacZ staining of S. meliloti. Bars = 100 μm.
Figure 10.
ARF16a-GUS Promoter Fusion Expression during Rhizobial Infection and in Nodules and Lateral Roots.
A. rhizogenes-induced hairy roots were used to analyze ProARF16a:GUS expression: (A) to (D) during rhizobial infection ([A] and [B], 3 weeks after inoculation; [C] and [D], 3 and 5 dpi); (E) in a young nodule; (F) in an elongated nodule; (G) in a mature nodule (median longitudinal section); (H) in a lateral root tip; and (I) and in a lateral root primordium. The blue color ([A] to [I]) indicates staining of GUS activity, and the magenta color (G) indicates LacZ activity of S. meliloti carrying pXLGD4. Bars = 100 μm.
GH3.1, SAUR1, and ARF16a were expressed uniformly in young nodules, with no apparent polarity (Figures 8C, 9D, and 10E). However, after the nodules matured and their zones were apparent, the expression of these genes was limited to the apex of the elongated nodules (Figures 8D, 9E, and 10F). Longitudinal nodule sections showed that GH3.1 was expressed in the meristem and the infection zone but not in the interzone or the nitrogen fixation zone (Figure 8H). SAUR1 was expressed in a relatively narrow region including the nodule meristem and the distal part of the infection zone (Figure 9F). ARF16a was expressed in the meristem and the infection zone (Figure 10G), but more weakly than GH3.1 and SAUR1 based on the intensity of GUS staining in comparable experiments.
GH3.1, SAUR1, and ARF16a were expressed in lateral root primordia and incipient lateral roots (Figures 8F, 8G, 9G, 9H, and 10I) and in the meristems of mature lateral roots (Figures 9I and 10H) as would be expected because auxin is involved in lateral root initiation and development. The expression pattern of GH3.1 in the root tip was more restricted than that of ARF16a and SAUR1 and was localized mainly to the columella cells of the root cap (Figure 8E). SAUR1 was also expressed in the stele of the root (Figure 9E).
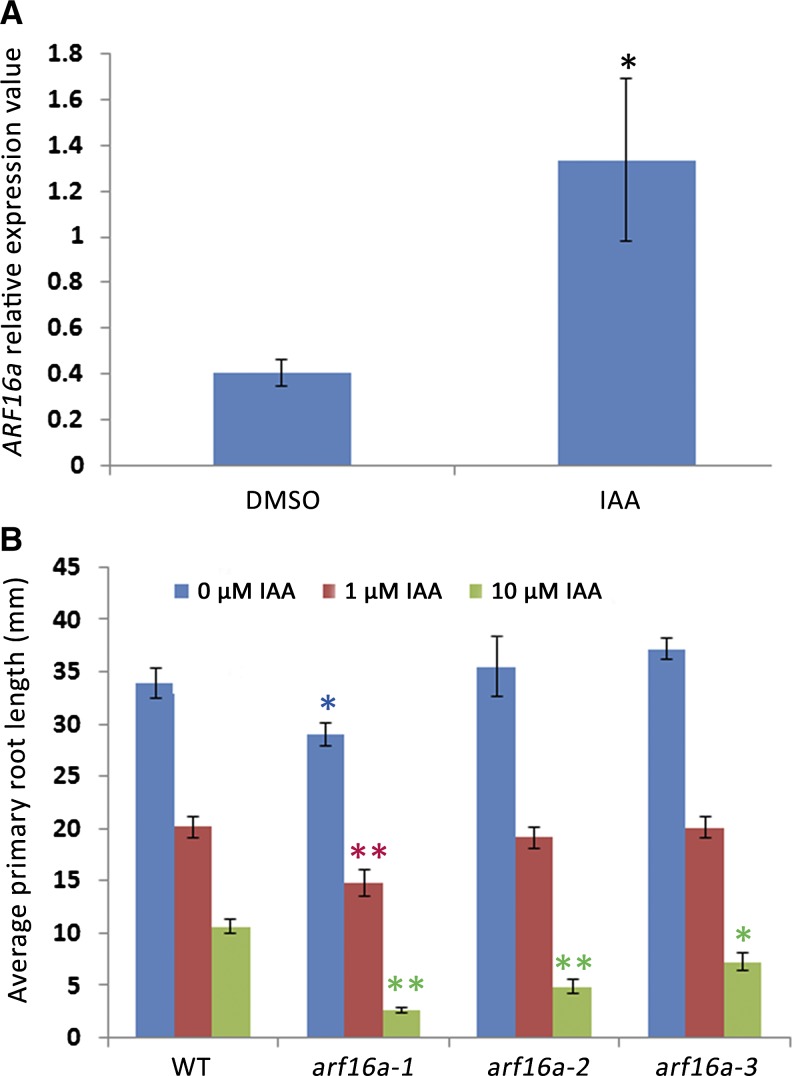
To test if ARF16 acts like its Arabidopsis counterpart, we measured (by quantitative RT-PCR) the auxin responsiveness of ARF16a in roots and saw about a 3-fold increase in expression (Figure 11A). This is consistent with M. truncatula transcriptomics data after NAA treatment in embryogenic leaf explant cultures (Imin et al., 2008) and induction of the Arabidopsis ARF16 ortholog by indole-3-acetic acid (IAA) (Wang et al., 2005). We identified three arf16a:Tnt1 insertional mutants (Figure 12A) through PCR screening. The root growth of these mutants was found to be more sensitive to auxin inhibition than the wild type (Figure 11B), particularly at 10 μM IAA. This is consistent with work in soybean that showed that overexpression of miR160, which targets ARF16 and its close homolog ARF10, resulted in auxin hypersensitivity (Turner et al., 2013).
Figure 11.
ARF16a Responds to Auxin Treatment.
(A) qPCR of relative expression of ARF16a upon treatment with DMSO or 1 μM IAA.
(B) The inhibition of primary root growth by 1 or 10 μM IAA in the wild type (R108), arf16a-1 (NF12634), arf16a-2 (NF13450), and arf16a-3 (NF16027). The histograms show averages (±se) of primary root length 7 d after germination, based on 10 plants for each treatment of each genotype.
Bar = se. Significant (Student’s t test) differences between the wild type and mutants are marked with asterisks (*P ≤ 0.05 and **P ≤ 0.01).
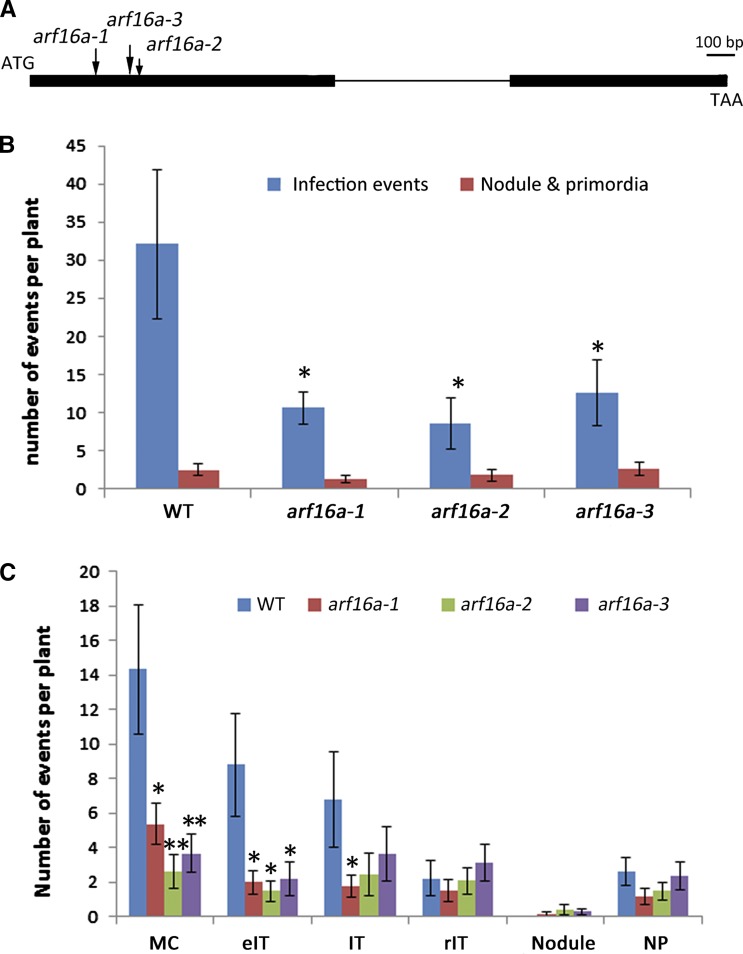
Figure 12.
Nodulation Phenotype of M. truncatula arf16a Mutants.
(A) Gene structure of ARF16a gene. The bold line indicates the exons and thin line indicates an intron. The locations of Tnt1 insertions for the mutant alleles are indicated by arrows. Bar = 100 bp.
(B) Rhizobial infections and formation of nodule primordia in the wild type (R108), arf16a-1(NF12634), arf16a-2 (NF13450), and arf16a-3 (NF16027).
(C) Quantification of different stages of infection and development of nodule primordia in the wild type and arf16a mutants 7 dpi with S. meliloti. Infection events and nodule primordia were scored 7 dpi with S. meliloti 1021 carrying pXLGD4 (lacZ) after LacZ staining. The histograms show averages (±se) based on 14 plants for each line. Significant (Student’s t test) differences between the wild type and mutants are marked with asterisks (*P ≤ 0.05 and **P ≤ 0.01). IT, fully elongated infection thread in root hair; eIT, elongating infection thread in root hair; MC, microcolony; rIT, ramified infection thread in cortex; NP, nodule primordium.
The arf16a Mutant Is Resistant to Infection by S. meliloti
The localized expression patterns of GH3.1, SAUR1, and ARF16a in infected tissues suggested a role for auxin in infection and so the arf16a mutants were examined for S. meliloti-induced infection and nodulation. The number of infection events observed 7 dpi was reduced in the mutants compared with the wild type (Figure 12B). A few normal-looking infection threads were formed (Supplemental Figure 4) and these led to successful nodule colonization (Supplemental Figures 5 and 6). Infection events were classified into four stages of infection thread progression (scoring key provided in Supplemental Figure 3), showing that the reduction in infections could be attributed to a decreased numbers of infection pockets containing microcolonies and elongating threads (Figure 12C). This suggests that the earliest stages of infection were impaired in the mutants. Nodules and nodule primordia were found to be normal in morphology and number (Supplemental Figures 5 and 6). These results indicate a role for ARF16a in the initiation of infection (i.e., formation of the infection pocket and thread initiation) rather than in infection thread extension.
DISCUSSION
Our work has provided insights into the molecular crosstalk that occurs between the rhizobia and legume roots during initiation of infection and has identified some of the biological processes associated with infection thread development.
A key finding is a role for auxin signaling in infection. Four auxin-responsive genes, GH3.1, SAUR1, ARF16a, and IAA9, were more strongly induced by S. meliloti in skl relative to the wild type (Figure 5), consistent with enhanced auxin transport in skl (Prayitno et al., 2006), and reveals an unexpected role for auxin in root-hair infection. Although S. meliloti induced DR5-GUS in all root hairs in the infection zone, expression of GH3.1, ARF16a, and SAUR1 was restricted to infected cells, indicating that auxin signaling in infected and noninfected root hairs is distinct. This could result from an infected cell-specific modification of auxin signaling caused by Nod factors because GH3.1 and ARF16a were induced by purified Nod factors. A local alteration in auxin response must be important because mutation of ARF16a reduced the frequency of infection. Although the function of the putative repressor ARF16a has not been studied directly in legumes, the effect of overexpressing miR160 (which targets ARF16 as well as other closely related ARFs) has been examined in soybean and M. truncatula during nodulation. Overexpression of miR160 in soybean did not affect infections in root hairs; in a similar M. truncatula study, infections were not scored (Bustos-Sanmamed et al., 2013; Turner et al., 2013). The apparent discrepancy between our work and the soybean study could be due to inefficient knockdown of ARF16 by miR160 or the differential regulation of auxin in determinate versus indeterminate nodulation (Subramanian et al., 2006, 2007; Wasson et al. 2006).
GH3, SAUR, and AUX/IAAs regulate auxin responses. Auxin binds to the Transport Inhibitor Response 1 (TIR1) and AUX/IAA coreceptor complex (Calderón Villalobos et al., 2012). TIR1, an F-box protein, promotes degradation of the AUX/IAA proteins, leading to the derepression of ARF transcriptional activators, which induce auxin responses (Tiwari et al., 2001). The TIR1 and IAA regulators act in discrete modules to control different developmental outcomes (De Smet et al., 2010). The localized induction of early auxin response genes in our data suggests that auxin is increased in cells undergoing infection. Rhizobial induction of GH3.1 and SAUR1 is conserved in soybean, indicating a conserved role in phaseoloid and galegoid legumes (Supplemental Data Set 3, Conserved Gm). GH3.1 and SAUR1 are members of large gene families that typically have promoters with auxin response elements that confer sensitivity to repression by ARFs and AUX/IAAs (Li et al., 1994; Ulmasov et al., 1995, 1997; Tiwari et al., 2003). The similar expression patterns of GH3.1, SAUR1, and ARF16a suggest they act as part of a specific auxin regulatory module, possibly along with IAA9. Possibly auxin accumulation in infected root hairs induces expression of SAUR1, GH3.1, and IAA9, but ARF16a (possibly with additional negative feedback from IAA9) may limit their induction. Increased auxin signaling in infected cells is consistent with the increase in auxin reporter gene expression 30 h after rhizobial inoculation of clover (Trifolium repens; Mathesius et al., 1998). Upregulation of an auxin influx transporter has also been reported in Frankia-infected root hair cells in Casuarina glauca, suggesting that an increase in auxin may also occur during infection of non-legume nodules (Péret et al., 2007).
A principal role for auxin is in cell expansion; the long-standing acid growth theory proposes that auxin stimulates acidification of the cell wall, thereby increasing its extensibility. For instance, growth of root hairs can be induced by auxin (Pitts et al., 1998) where it might promote cell wall loosening through localized decreases in pH of the apoplast, a requirement for tip growth (Monshausen et al., 2007). Such cell wall remodeling is likely to occur during the formation of rhizobially induced infection pockets and initiation and growth of infection threads (Oldroyd et al., 2001; Esseling et al., 2003).
The auxin-induced SAUR proteins have a positive role in cell expansion (Spartz et al., 2012) possibly regulating expansion via acid-induced growth. Several SAUR proteins bound to protein phosphatase 2C-D (PP2C-D) proteins inhibiting their phosphatase activity (Spartz et al., 2014). These PP2C-D proteins can regulate the activity of plasma membrane H+-ATPases, suggesting that SAUR proteins may induce cell growth through indirect activation of proton pumps. One such H+-ATPase, HA1, was induced by both S. meliloti and Nod factors. Previously, HA1 was thought to be expressed only in arbusculated cells (Krajinski et al., 2002) where it is required for phosphate uptake during mycorrhization (Krajinski et al., 2014; Wang et al., 2014). Its expression during infection suggests that HA1 may also have a role in auxin-mediated acid-induced cell wall growth during rhizobial infection and arbuscule formation. This hypothesis is consistent with studies showing that Nod factors induce proton efflux from root hairs as well as root hair growth (Allen et al., 1994; Felle et al., 1996; Oldroyd et al., 2001).
The hormones GA, BR, and SL have not been extensively studied in nodulation but appear to be induced during infection. Pea (Pisum sativum) mutants deficient in BRs or GA had reduced nodulation (Ferguson et al., 2005), and SL levels were positively correlated with nodulation in pea and L. japonicus (Soto et al., 2010; Foo and Davies, 2011; Foo and Reid, 2013; Liu et al., 2013), but infection was not characterized in these studies. Our observation that CCD8 is induced in root hairs by S. meliloti taken together with the identification of SLs in M. truncatula root exudates (Liu et al., 2011) raises the possibility that SLs are secreted during infection. Therefore, SLs may either act as an additional cue for rhizobia in the rhizosphere, as seen with mycorrhization (Akiyama et al., 2005), or they may induce developmental changes in root hairs during infection, perhaps analogous to their role in induction of lateral root formation. This could be mediated via effects on root hair growth or by affecting auxin (Crawford et al., 2010). GA and BRs also promote root hair and root growth, respectively (Müssig et al., 2003; Jiang et al., 2007) and might act together with auxin and SLs to promote cell expansion during infection.
Infection Involves Engagement of the Cell Cycle
Nod factors can act as strong mitogens, inducing preinfection threads (PITs) in the outer cortex and cortical cell divisions that induce nodule primordia (van Brussel et al., 1992). The outer cortical cells that make PITs do not divide; they appear to enter the cell cycle and then arrest (Yang et al., 1994). During root hair infection, structures resembling PITs are formed, but this aspect of root hair infection is not well understood. Our findings suggest some interesting leads. Nod factors increased expression of genes encoding three D-type cyclins, a C-type cyclin, and a subunit of the APC/C. D-type cyclins in plants are predicted to control G1 progression in response to extrinsic mitogenic signals (Dahl et al., 1995), C-type cyclins have been implicated in G0-to-G1 entry (Meskiene et al., 1995; Ren and Rollins, 2004), and the APC/C targets mitotic cyclins and aurora kinases for degradation to regulate cell cycle transitions (Peters, 2006). The induction of these cell cycle components along with DNA replication-related components suggests that Nod factors stimulate root hair cells to reenter the cell cycle, presumably completing G1 and then entering S phase. Interestingly, these type-C and D cyclins appear not to be induced in response to S. meliloti, whereas a type-A cyclin, typically associated with G2-M transition was induced. Also induced by rhizobial inoculation was a homolog of AURORA KINASE1, which has been linked to mitosis (Petrovská et al., 2012). There was also increased expression of genes for DNA synthesis, modification, and repair enzymes, reaching the highest expression at 5 dpi, which corresponds to the initiation of infection threads. This suggests that infected root hair cells do not only enter the cell cycle, but may partly enter into the M phase. This difference in cell cycle-related responses between Nod factor treatments and rhizobial inoculation is not surprising, as it has been noted that rhizobia, but not Nod factors, can induce preinfection thread formation in M. sativa (Timmers et al., 1999) and therefore the comparison of these two responses may be particularly informative.
Events that occur in root hairs cells prior to infection thread formation share features with premitotic cells. In the outer cortical cells, the nucleus moves to a central position within a thickened anticlinal cytoplasmic bridge (the preinfection thread), which predicts the route the infection thread will take as it grows through the cell (Timmers et al., 1999). A similar sequence of events gives rise to the phragmosome, a premitotic structure that is visible in highly vacuolated cells prior to the formation of the phragmoplast and cell plate in the same location (Venverloo and Libbenga, 1987; Lloyd, 1991). The events in root hair cells before infection thread formation are quite similar; the nucleus occupies a central position, and a thickened cytoplasmic bridge forms between the nucleus and root hair tip (Fournier et al., 2008). The strong resemblance of the preinfection threads in the outer cortical cells to these premitotic structures has led to the suggestion that they employ the same cytoskeletal machinery (Brewin, 1991). Our data support a model in which Nod factors trigger the entry of root hair cells into the cell cycle, and then, if Nod factor producing rhizobia are present, the cell completes S phase and enters partially into mitosis which may be a prerequisite for preinfection thread formation.
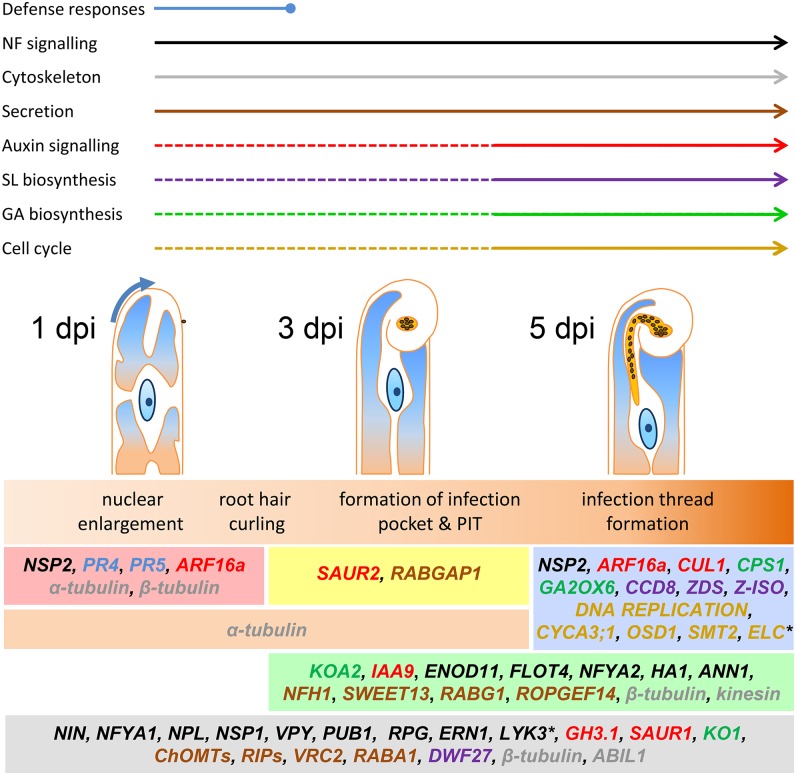
The increased expression of genes for DNA synthesis might suggest an increase in ploidy levels in root hairs. The root hair nucleus practically doubles in size during the preinfection phase (Dart, 1974). Increased ploidy is also a feature of nodule cells; cells in nodule zone II undergo rounds of endoreduplication resulting in cells in the nitrogen fixation zone having ploidy levels up to 64C (Truchet, 1978; Cebolla et al., 1999). The observed expression of homologs of PYM/OSD1, SMT2, and ELC indicate that in root hairs endoreduplication is being supressed. A model for root hair infection suggests an initial increase in ploidy and partial entry into mitosis (as indicated by the induction of the A-type cyclin and the aurora kinase and the suppression of ELC) followed by the induction of PYM/OSD1 and SMT2 to prevent further rounds of endoreduplication. To help relate changes in gene expression with the developmental changes occurring at different stages of infection, we created a timeline showing when specific processes underlying infection occur (Figure 13). From this, it is evident that cell cycle responses primarily occurred when infection threads were forming, coincident with auxin responses and the biosynthesis of SL and GA (Figure 13). Auxin, as the main hormonal player in cell division processes, can be predicted to play a key role in in these events in addition to the role in cell expansion proposed above.
Figure 13.
Dynamic Trends in Gene Expression Prior to and during Infection.
Rhizobial preinfection and infection involves transient defense responses and sustained Nod factor signaling, while infection specifically involves hormone biosynthesis and signaling and activation of the cell cycle. Preinfection stage was marked by the induction of two pathogenesis-related genes, PR4 and PR5. Most nodulation pathway genes were induced, and LYK3 was repressed at all time points, while NSP2 was only induced during early preinfection and upon infection. Flavonoid and peroxidase genes were induced at all stages (VRC2, ChOMTs, and RIPs). ENOD11, FLOT4, NFYA2, ANN1, HA1, the Nod factor hydrolase NFH1, and sugar transporter SWEET13 were most strongly induced during formation of the infection pocket and at the onset of infection. RAB GTPases with potential roles in secretion were induced at every stage, while ROPGEF14 was mainly induced at the latter two stages. Tubulins and the cytoskeletal regulator ABIL1 were expressed at every time point, while kinesin was only induced at the latter stages. The initiation of the infection thread coincided with the induction of genes for GA and SL biosynthesis, auxin signaling, and many genes encoding cell cycle components, including most members of the DNA replication complex and an A-type cyclin (CYCA3;1). Genes for repression of endoreduplication (OSD1 and SMT2) were induced, while ELC, which is involved in the switch from mitosis to endoreduplication, is repressed during infection. The color key indicates the processes in which each gene is involved. Genes are grouped based on their trends of induction or repression (the latter indicated by an asterisk) relative to the controls. See Supplemental Table 2 for further details on specific genes.
While hormones involved in growth and development were increased, JA biosynthesis and signaling was repressed by Nod factors and during infection of the skl mutant. The repression of JA-related gene expression in skl may be due to the fact that JA and ethylene responses are partially interdependent (Ballaré, 2011). It is possible that the repression of JA responses is localized to those cells closest to the Nod factor producing rhizobia and could be part of the localized downregulation of plant defenses that are presumably needed to permit rhizobial colonization.
Although JA signaling was repressed by Nod factors, some defense pathway genes were increased. PATHOGENESIS-RELATED genes PR4 (Medtr1g080800) and PR5 (Medtr5g010635) were highly induced at 1, but not at 3 or 5 dpi (Figure 13), and are highly induced by pathogens (MtGEA public gene expression database), indicating a transient defense response. In contrast, Nod factor and S. meliloti increased expression of VRC2 (encoding the second last step in the biosynthesis of the phytoalexin medicarpin) specifically at the sites of infection. This indicates that M. truncatula may produce antimicrobials in during infection and is consistent with the detection of medicarpin during nodulation (Savouré et al., 1997). This could limit opportunistic and/or pathogenic infections (Pandya et al., 2013). S. meliloti is resistant to medicarpin, while several other incompatible rhizobia (Bradyrhizobium and Mesorhizobium spp) are not (Pankhurst and Biggs, 1980), suggesting limitation of infection by inappropriate rhizobia. The identification of what appears to be a nodulation-specific isoform of VR in M. truncatula will allow direct testing of this hypothesis with mutants. Other legumes produce species-specific pterocarpins, such as glyceollin in soybean (Parniske et al., 1991), suggesting that antimicrobials may be used widely to restrict host specificity of rhizobia in legumes.
Increased expression of four different ChOMT genes suggest increased production of the S. meliloti nod gene inducer methoxychalcone in response to S. meliloti; such inducers are needed to initiate and maintain infection (Redmond et al., 1986). The production of these flavonoids is restricted to the root hair differentiation zone (Djordjevic et al., 1987; Peters and Long, 1988), and infective rhizobia can increase the diversity of flavonoids produced (van Brussel et al., 1990). We show here that Nod factors can induce the ChOMT genes and predict that a positive feedback loop is established via the production of methoxychalcone. Nod factor signaling is sustained throughout rhizobial infection thread development (Marie et al., 1992) and is required for appropriate development of the infection thread (Den Herder et al., 2007). Our data reveal that phenylpropanoid metabolism is coordinated during infection, with some pathways such as lignin biosynthesis being repressed, whereas others, such as biosynthesis of both nod gene-inducing and antimicrobial isoflavonoids, are induced (Figure 13).
Nod factor degradation is induced by S. meliloti and by Nod factors (Staehelin et al., 1995), and both S. meliloti and Nod factors induced NFH1 in M. truncatula (Salzer et al., 2004; our data). NFH1 is predicted to be secreted and has been shown to specifically degrade Nod factors (Tian et al., 2013) and so NFH1 induction could limit or interrupt the flavonoid-Nod factor positive feedback loop described above. Similar induction of NFH1 occurs in soybean (Supplemental Data Set 3, Conserved Gm).
Other evidence for inhibition of Nod factor signaling was that the Nod factor receptors NFP and LYK3 were repressed. The Arabidopsis flagellin receptor, FLS2, is targeted for degradation by the ubiquitin E3 ligases PUB12 and PUB13 upon treatment with flagellin (Lu et al., 2011), and LYK3 similarly is targeted by the E3 ligase PUB1 (Mbengue et al., 2010). Taken together, the observations imply that the host regulates Nod factor production, turnover, and perception.
In this analysis, the expression of 1500 genes was altered, and we can discuss only a few in this article. We highlighted genes involved in secondary metabolism and hormone regulation, but many other genes of interest were noted. For example, ABIL1 has been proposed to mediate NAP1 and SCAR/WAVE binding (Basu et al., 2005) so probably acts together with NAP1, PIR1, and ARPC1 to nucleate actin for the development of infection threads (Yokota et al., 2009; Miyahara et al., 2010; Hossain et al., 2012). Unlike other components of the SCAR/WAVE complex, ABIL1 expression in root hairs in the absence of Nod factors was very low, and its increased transcription indicates that regulation of ABIL expression is a potential control for the changes in actin that occur during infection. Also of interest were the SWEET sugar transporter and SUCS1 that are also expressed in mycorrhized roots (Gomez et al., 2009); SWEET may provide sugars to support bacterial growth in the infection thread, and SUCS has a potential role in uptake of sucrose into infected cells. Two RAB GTPases (RABA1 and RABG1), a RAB GTPase activator (RABGAP1), and a ROP-Guanine nucleotide exchange factor (ROFGEF14) were induced by infection; small GTPases feature play central roles in endomembrane trafficking.
A key finding from this temporal analysis is that the onset of infection in root hairs appears to involve reactivation of the cell cycle and that this coincides with the localized activation of an auxin signaling module at infection sites which is required for the initiation of infection. The data further suggest that SLs, BRs, and GAs are produced in root hairs undergoing rhizobial infection. Previous work implicated the role of some of these hormones in nodule organogenesis, but a function for plant hormones during infection thread development had not been proposed. Altogether this work provides a transcriptional framework for infection: a tightly regulated flavonoid-Nod factor signaling loop supported by continuous secretion of proteins and small molecules, including phytoalexins and nutrients (Figure 13). Along with these insights, our study helps interpret two decades of biochemical research in Medicago and provides a foundation for future genetic studies on host-symbiont interactions in the rhizosphere.
METHODS
Isolation of Root Hairs from Roots
Wild-type Medicago truncatula (Jemalong A17) or skl-1 (Penmetsa and Cook, 1997) seeds were scarified with glasspaper and surface sterilized in 10% sodium hypochlorite for 3 min. Seeds were washed five times with sterile distilled water and left to imbibe for 2 h. Seeds were then left on inverted agar plates for 72 h at 4°C before being transferred to 22°C for 16 h. The following day, seedlings with a radical length of ∼1 cm were transferred to 100 × 80-mm pieces of Whatman Envelop Strip (VWR) placed on square (120 × 120 mm) Petri dishes (Fisher Scientific) containing 1.5% agar (10 plants per dish) and 100 nM aminoethoxyvinylglycine. An upper layer of Whatman paper was then inoculated with 600 μL of either wild-type Sinorhizobium meliloti (Sm1021) or S. meliloti nodΔD1ABC (SL44): (OD600 = 0.03) or 500 μL of 1 μM Nod factors using a mucosal atomization device (Intavent Direct). Plates containing 50 mL agar were used providing a final Nod factor concentration of 10 nM. Plates were placed in black bags with only aerial parts exposed to light. Plants were grown in a controlled environment room (22°C; 16 h light, 8 h dark) for the appropriate period of time.
A root hair harvesting protocol was adapted from Ramos and Bisseling (2003). Briefly, root tips were removed and discarded, and the roots were plunged into liquid nitrogen contained in a Teflon-coated loaf tin (Dunelm Mill). A Daler Rowney number 2 filbert paint brush (Dunelm Mill) was then used to break off root hairs that accumulated in the loaf tin. After the seedlings were processed, the tin was placed at a 25° angle until ∼40 mL nitrogen remained. The remaining nitrogen was then poured into a 45-mL PTFE-coated conical centrifuge tube (VWR) and the nitrogen was left to boil off. The purified root hair sample was transferred immediately to −80°C.
RNA Purification from Root Hairs
RNA was extracted using the Qiagen RNeasy micro kit according to the manufactures instructions with the exception of an additional 80% ethanol wash prior to elution in 14 μL RNase free water. Contaminating DNA was removed using the TURBO DNA-free Kit (Ambion). Quantity was measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and quality assessed using a Bioanalyzer 2100 (Agilent).
Microarray Analysis
GeneChip hybridizations were performed on cRNA samples derived from the root hair of 150 M. truncatula seedlings. Three biological replicates were performed for each treatment using a total of 30 GeneChips. M. truncatula A17 was profiled at 1, 3, and 5 dpi following inoculation with S. meliloti Sm1021 or SL44 (S. meliloti nodΔD1ABC). A17 was also profiled at 1 d post-treatment following addition of Nod factors purified from Sm1021 or as control, an equivalent preparation from SL44. The skl mutant was profiled at 5 dpi following inoculation with Sm1021 or SL44.
Labeling was performed using the GeneChip 3′ IVT Express Kit (Affymetrix P/N 901229) according to the manufacturer’s instructions using 150 ng total RNA. Hybridization was done using the GeneChip Hybridization, Wash, and Stain Kit (Affymetrix P/N 900720) with a GeneChip Hybridization Oven 640 (Affymetrix) and a GeneChip Fluidics Station 450 (Affymetrix; protocol FS450_0001). Finally, GeneChips were scanned using a GeneChip Scanner 3000 (Affymetrix).
Normalization and statistical analysis were performed using Genespring 12.0 GX. Background correction, normalization, and probe summarization were performed using robust multichip averaging. Genes with significant changes in expression were identified for each comparison using unpaired t tests. Briefly, probe sets were first filtered by expression, retaining only those with a value of >50 for all three biological replicates in at least one of the two conditions being compared. P values were derived asymptotically and multiple test corrected using Benjamini Hochberg false discovery rate. Normalized data were then exported and further analyzed in Microsoft Excel.
Assessment of Sample Purity
Several of the isolated root hair pellets were visually inspected under a microscope and were found to be free of cells originating from root tips, this being the most likely source of contamination. As a further measure of sample purity, we examined the microarray data for evidence of contaminating transcripts from the root tip or border cells. To do this, we identified genes highly expressed in root tips and border cells and of root hair-specific genes using the differential expression analysis tool available on MtGEAv3. To identify root hair-specific genes, all probe sets with 5-fold higher signal in root hair than in root that had an expression level of <40 in root (370 probe sets) were examined across 74 wild-type root and shoot tissues (no chemical and biotic treatments were included). Expression in border cells was considered separately since they are another type of epidermal cell. A total of 49 root hair specific genes were identified and an additional five genes that also had some expression in border cells (Supplemental Data Set 11, root hair-specific genes). We identified many probe sets with very high levels of expression in root tips and/or border cells with only background levels in root hairs (such as Mtr.26895.1.S1_s_at and Mtr.14964.1.S1_at), indicating that our material was essentially free of contamination from these tissues.
Identification of Soybean Homologs
A similar data set obtained from root hairs of soybean and its symbiont Bradyrhizobium japonicum (Libault et al., 2010) provides an opportunity to identify genes with conserved roles in infection. Putative M. truncatula orthologs for all G. max genes reported to be increased or decreased by rhizobial inoculation were identified using BLASTP. To allow for gene duplication in the M. truncatula lineage post-divergence, the two highest scoring hits for each gene model were retained. Genes that we determined as having significantly changed expression (data for all wild-type rhizobial time-points as well as the skl mutant and Nod factor treatment were used) that have homologs with changed expression as determined by Libault et al. (2010) are listed in Supplemental Data Set 3 (Conserved Gm). Syntenic genes were identified using previously identified synteny blocks (Legume Information Portal web server, LegumeIP; Li et al. 2012).
qPCR
First-strand cDNA was synthesized from root hair RNA using SuperScript II and oligo(dT)12-18 primer (Invitrogen) with 1 μg total RNA in a 20 μL reaction volume. Resulting cDNA was then diluted with 380 μL water and stored at −80°C prior to qPCR. qPCR was performed in 20-μL reactions using SYBR Green JumpStart Taq ReadyMix without MgCl2 (Sigma-Aldrich) and a Bio-Rad CFX96 real-time system. Initial denaturation was at 94°C for 4 min and then 40 cycles of 94°C, 30 s; 60°C, 30 s; and 72°C, 30 s. The melt curves were analyzed between 65 and 90°C at 0.5°C intervals.
Primer3 was used to design primers to generate amplicons of 80 to 120 bp using the default settings. All primers generated a single product of the correct size. The efficiency of all primer pairs was calculated using a dilution series and linear regression of the resulting Ct data points. The stability of five housekeeping genes (protein phosphatase 2A, actin, histone H3, ubiquitin, and TIP41-like protein) was first assessed using the geNorm algorithm (Vandesompele et al., 2002) contained within qbasePLUS (Biogazelle) (Hellemans et al., 2007). Ubiquitin and TIP41-like protein were determined to be the most stable references across all root hair samples and used for subsequent normalization. Normalized relative quantities were calculated using the qBase model (Hellemans et al., 2007), which allows for multiple housekeeping genes and primer specific efficiencies. A two-tailed, type 3 t test was used to test for significant differences in expression using in Microsoft Excel. Primers, primer efficiencies, Ct values, normalized relative quantities, and t tests are detailed in Supplemental Table 1 (Primers).
Agrobacterium rhizogenes Hairy Root Transformation
Plant roots transformed with promoter-GUS constructs were generated in M. truncatula A17 background by hairy root transformation mediated by A. rhizogenes Arqua1 (Boisson-Dernier et al., 2001). The composite plants were transferred to plates containing water agar (with 100 nM aminoethoxyvinylglycine) or a mixture of equal amounts of sand and terra green 4 weeks after transformation and inoculated with Sm1021 pXLGD4 (lacZ). The roots were harvested at different time points for GUS staining, and some samples were then stained with Magental-gal (Melford) for visualization of lacZ-tagged rhizobia as previously reported (Pichon et al., 1994).
Promoter-GUS Analysis
A 2028-bp upstream fragment of CCD8 was amplified from M. truncatula A17 genomic DNA using Phusion High-Fidelity DNA Polymerase (NEB). The fragment was cloned into pDONR207 using Gateway BP Clonase II enzyme mix (Invitrogen). Then the fragment was introduced into a destination vector pKGWFS7 by a LR reaction to make the construct pCCD8:GUS. The other promoter constructs (VRC2, 2197 bp; ARF16a, 1883 bp; GH3.1, 2172 bp; SAUR1, 2269 bp) were made by the same method. The primers used are listed in Supplemental Table 1 (Primers). The DR5:GUS analysis was performed in the R108 background using a stably transformed line (Zhou et al., 2011; Guan et al., 2013).
Insertional Mutant Screening, Genotyping, and Phenotyping
Tnt1 retrotransposon insertions in ARF16a were screened using a nested PCR approach (Cheng et al., 2011, 2014). Subsequent genotyping in the R2 generation progeny was performed using primers ARF16_geno_F2 and Tnt1F for the arf16a-1 allele and ARF16_geno_F2 and Tnt1R for the arf16a-2 and arf16a-3 alleles (Supplemental Table 1, Primers). Methods for nodulation assay and rhizobial infection assay were described by Guan et al. (2013).
Phylogenetic Tree
Alignments were made with MUSCLE (Edgar, 2004), the phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v3.0; Guindon and Gascuel, 2003), and reliability for internal branches was assessed using the aLRT test (SH-Like; Anisimova and Gascuel, 2006).
Auxin Treatments
To study the responses of ARF16a to exogenous auxin, M. truncatula A17 seedlings germinated overnight were grown vertically on agarose medium for 3 d at 23°C. The seedlings were then immersed in either 1 μM IAA dissolved in 10% DMSO or 10% DMSO alone for 24 h in the dark. Finally, root tips of the treated seedlings were removed, and only the root zone containing root hairs was collected for extraction of RNA. Eight seedlings were used per replicate and three biological replicates were used for qPCR analysis.
To test auxin sensitivity, wild-type M. truncatula (R108) and arf16a mutants were germinated overnight in the dark at 22°C and were then transferred to BNM media plates containing 0, 1, or 10 μM IAA. Then the plants were then grown at 22°C in a controlled environment chamber with 16 h/8 h (light/dark) photoperiod for 7 d, at which point the length of the primary root was measured.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Differentially Expressed Genes (P < 0.05) in Wild-Type Plants Infected by Sm1021.
Supplemental Figure 2. Regulation of Genes Encoding Peroxidases, and LYSM Proteins during Infection.
Supplemental Figure 3. Depiction of the Different Categories of Infection Events Used to Characterize the Infection Phenotype of mtarf16 Mutants.
Supplemental Figure 4. Infection Threads and Nodule Primordia from the Wild Type (R108) and arf16a Mutants.
Supplemental Figure 5. Nodule Numbers of arf16a Mutants Are Not Different from the Wild Type.
Supplemental Figure 6. Nodules of arf16a Mutants Have Normal Morphology.
Supplemental Table 1. Primers Used in This Study.
Supplemental Table 2. Details for Genes in Figure 13.
Supplemental Data Set 1. Complete Data Set for All Treatments and Controls.
Supplemental Data Set 2. Genes Induced or Repressed after Rhizobial Inoculation or Nod Factor Treatment.
Supplemental Data Set 3. Genes Induced or Repressed in G. max and M. truncatula Root Hairs by Rhizobia or Nod Factors.
Supplemental Data Set 4. Genes Expressed Only in Root Hairs of Seedlings Inoculated with Rhizobia or by Rhizobia and in Mycorrhized Roots
Supplemental Data Set 5. Phenylpropanoid-Related Genes Regulated by Rhizobial Inoculation or Nod Factors.
Supplemental Data Set 6. Cell Cycle-Related Genes Regulated by Rhizobial Inoculation or Nod Factors.
Supplemental Data Set 7. GA-Related Genes Regulated by Rhizobial Inoculation or Nod Factors.
Supplemental Data Set 8. SL-Related Genes Regulated by Rhizobial Inoculation or Nod Factors
Supplemental Data Set 9. JA-Related Genes Regulated by Rhizobial Inoculation or Nod Factors.
Supplemental Data Set 10. Auxin-Related Genes Regulated by Rhizobial Inoculation or Nod Factors.
Supplemental Data Set 11. Genes Expressed Only in Root Hair Cells.
Supplementary Material
Acknowledgments
We thank Michael Schultze for critical reading of the article, Julie Ellwood for help in formatting the article, and Graham McGrann, Grant Calder, and Ali Pendle for technical advice. This work was supported by the Biotechnology and Biological Sciences Research Council (Grants BB/G023832/1 and BB/L010305/1 [David Phillips Fellowship]) and the John Innes Foundation (to S.R. and J.A.D.). G.M. was funded by a Marie Curie European Union grant (MRTN-CT-2006-035546) within the “Nodperception” Network to the John Innes Centre and CNRS.
AUTHOR CONTRIBUTIONS
A.B., C.L., J.A.D., and J.D.M. designed the research. A.B., C.L., G.M., N.S., S.R., and C.R. performed the research. A.B., M.T., and J.D.M. analyzed the data. A.B., C.L., G.E.D.O., J.A.D., and J.D.M. wrote the article.
Glossary
- SL
strigolactone
- GA
gibberellic acid
- BR
brassinosteroid
- dpi
days postinoculation
- CHR
chalcone reductase
- MtGEA
Medicago Gene Atlas
- JA
jasmonic acid
- IAA
indole-3-acetic acid
- PIT
preinfection thread
- qPCR
quantitative PCR
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adie B.A., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Allen N.S., Bennett M.N., Cox D.N., Shipley A., Ehrhardt D.W., Long S.R. (1994). Effects of Nod factors on alfalfa root hair Ca++ and H+ currents and on cytoskeletal behaviour. In Advances in Molecular Genetics of Plant-Microbe Interactions, Vol. 3. M.J. Daniels, J.A. Downie, and A.E. Osbourn, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 107–113. [Google Scholar]
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Gascuel O. (2006). Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55: 539–552. [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., Godfroy O., de Billy F., Saurat O., Jauneau A., Gough C. (2008). The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. USA 105: 9817–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M.C., Barsch A., Küster H., Hohnjec N. (2007). Antisense repression of the Medicago truncatula nodule-enhanced sucrose synthase leads to a handicapped nitrogen fixation mirrored by specific alterations in the symbiotic transcriptome and metabolome. Plant Physiol. 145: 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M.C., Keck M., Gödde V., Niehaus K., Küster H., Hohnjec N. (2010). Knockdown of the symbiotic sucrose synthase MtSucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol. 152: 1000–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré C.L. (2011). Jasmonate-induced defenses: a tale of intelligence, collaborators and rascals. Trends Plant Sci. 16: 249–257. [DOI] [PubMed] [Google Scholar]
- Basu D., Le J., El-Essal Sel.-D., Huang S., Zhang C., Mallery E.L., Koliantz G., Staiger C.J., Szymanski D.B. (2005). DISTORTED3/SCAR2 is a putative Arabidopsis WAVE complex subunit that activates the Arp2/3 complex and is required for epidermal morphogenesis. Plant Cell 17: 502–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito V.A., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55: 504–513. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700. [DOI] [PubMed] [Google Scholar]
- Brechenmacher L., Lee J., Sachdev S., Song Z., Nguyen T.H., Joshi T., Oehrle N., Libault M., Mooney B., Xu D., Cooper B., Stacey G. (2009). Establishment of a protein reference map for soybean root hair cells. Plant Physiol. 149: 670–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brechenmacher L., Lei Z., Libault M., Findley S., Sugawara M., Sadowsky M.J., Sumner L.W., Stacey G. (2010). Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 153: 1808–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin N.J. (1991). Development of the legume root nodule. Annu. Rev. Cell Biol. 7: 191–226. [DOI] [PubMed] [Google Scholar]
- Bustos-Sanmamed P., Mao G., Deng Y., Elouet M., Abbas Khan G., Bazin J., Turner M., Subramanian S., Yu O., Crespi M., Lelandais-Brière C. (2013). Overexpression of miR160 affects root growth and nitrogen-fixing nodule number in Medicago truncatula. Funct. Plant Biol. 40: 1208–1220. [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L.I., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A., Vinardell J.M., Kiss E., Oláh B., Roudier F., Kondorosi A., Kondorosi E. (1999). The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 18: 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crdenas L., Vidali L., Domnguez J., Prez H., Snchez F., Hepler P.K., Quinto C. (1998). Rearrangement of actin microfilaments in plant root hairs responding to rhizobium etli nodulation signals. Plant Physiol. 116: 871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri M.R., Frances L., Laloum T., Auriac M.C., Niebel A., Oldroyd G.E.D., Barker D.G., Fournier J., de Carvalho-Niebel F. (2012). Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 160: 2155–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Q., Qu X.Q., Hou B.H., Sosso D., Osorio S., Fernie A.R., Frommer W.B. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211. [DOI] [PubMed] [Google Scholar]
- Cheng X., Wang M., Lee H.K., Tadege M., Ratet P., Udvardi M., Mysore K.S., Wen J. (2014). An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 201: 1065–1076. [DOI] [PubMed] [Google Scholar]
- Cheng X., Wen J., Tadege M., Ratet P., Mysore K.S. (2011). Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol. Biol. 678: 179–190. [DOI] [PubMed] [Google Scholar]
- Combier J.P., de Billy F., Gamas P., Niebel A., Rivas S. (2008). Trans-regulation of the expression of the transcription factor MtHAP2-1 by a uORF controls root nodule development. Genes Dev. 22: 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., Vernié T., Ott T., Gamas P., Crespi M., Niebel A. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20: 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D., Dreyer D., Bonnet D., Howell M., Nony E., VandenBosch K. (1995). Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7: 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covitz P.A., Smith L.S., Long S.R. (1998). Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA library. Plant Physiol. 117: 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S., Shinohara N., Sieberer T., Williamson L., George G., Hepworth J., Müller D., Domagalska M.A., Leyser O. (2010). Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913. [DOI] [PubMed] [Google Scholar]
- Dahl M., Meskiene I., Bögre L., Ha D.T., Swoboda I., Hubmann R., Hirt H., Heberle-Bors E. (1995). The D-type alfalfa cyclin gene cycMs4 complements G1 cyclin-deficient yeast and is induced in the G1 phase of the cell cycle. Plant Cell 7: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakora F.D., Joseph C.M., Phillips D.A. (1993). Alfalfa (Medicago sativa L.) root exudates contain isoflavonoids in the presence of Rhizobium meliloti. Plant Physiol. 101: 819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart P.J. (1974). The infection process. In The Biology of Nitrogen Fixation, Quispel A., ed (Amsterdam: North-Holland Publishing, pp. 382–420. [Google Scholar]
- De Carvalho-Niebel F., Timmers A.C., Chabaud M., Defaux-Petras A., Barker D.G. (2002). The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. Plant J. 32: 343–352. [DOI] [PubMed] [Google Scholar]
- Den Herder J., Vanhee C., De Rycke R., Corich V., Holsters M., Goormachtig S. (2007). Nod factor perception during infection thread growth fine-tunes nodulation. Mol. Plant Microbe Interact. 20: 129–137. [DOI] [PubMed] [Google Scholar]
- de Ruijter N., Bisseling T., Emons A. (1999). Rhizobium Nod factors induce an increase in sub-apical fine bundles of actin filaments in Vicia sativa root hairs within minutes. Mol. Plant Microbe Interact. 12: 829–832. [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I., et al. (2010). Bimodular auxin response controls organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 2705–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic M.A., Redmond J.W., Batley M., Rolfe B.G. (1987). Clovers secrete specific phenolic compounds which either stimulate or repress nod gene expression in Rhizobium trifolii. EMBO J. 6: 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do C.T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., Lapierre C., Jouanin L. (2007). Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129. [DOI] [PubMed] [Google Scholar]
- Esseling J.J., Lhuissier F.G., Emons A.M. (2003). Nod factor-induced root hair curling: continuous polar growth towards the point of nod factor application. Plant Physiol. 132: 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle H.H., Kondorosi E., Kondorosi A., Schultze M. (1996). Rapid alkalinization in alfalfa root hairs in response to rhizobial lipochitooligosaccharide signals. Plant J. 10: 295–301. [Google Scholar]
- Ferguson B.J., Ross J.J., Reid J.B. (2005). Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 138: 2396–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo E., Davies N.W. (2011). Strigolactones promote nodulation in pea. Planta 234: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Foo E., Reid J.B. (2013). Strigolactones: New physiological roles for an ancient signal. J. Plant Growth Regul. 32: 429–442. [Google Scholar]
- Fournier J., Timmers A.C., Sieberer B.J., Jauneau A., Chabaud M., Barker D.G. (2008). Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148: 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts R., Fedorova E., Bisseling T. (2005). Nod factor signaling genes and their function in the early stages of Rhizobium infection. Curr. Opin. Plant Biol. 8: 346–352. [DOI] [PubMed] [Google Scholar]
- Gomez S.K., Javot H., Deewatthanawong P., Torres-Jerez I., Tang Y., Blancaflor E.B., Udvardi M.K., Harrison M.J. (2009). Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 9: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowri G., Bugos R.C., Campbell W.H., Maxwell C.A., Dixon R.A. (1991). Stress responses in alfalfa (Medicago sativa L.): X. Molecular cloning and expression of S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase, a key enzyme of lignin biosynthesis. Plant Physiol. 97: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresshoff P.M., Lohar D., Chan P.K., Biswas B., Jiang Q., Reid D., Ferguson B., Stacey G. (2009). Genetic analysis of ethylene regulation of legume nodulation. Plant Signal. Behav. 4: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D., et al. (2013). Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol. 162: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704. [DOI] [PubMed] [Google Scholar]
- Guo L., Dixon R.A., Paiva N.L. (1994). Conversion of vestitone to medicarpin in alfalfa (Medicago sativa L.) is catalyzed by two independent enzymes. Identification, purification, and characterization of vestitone reductase and 7,2′-dihydroxy-4′-methoxyisoflavanol dehydratase. J. Biol. Chem. 269: 22372–22378. [PubMed] [Google Scholar]
- Guo L., Paiva N.L. (1995). Molecular cloning and expression of alfalfa (Medicago sativa L.) vestitone reductase, the penultimate enzyme in medicarpin biosynthesis. Arch. Biochem. Biophys. 320: 353–360. [DOI] [PubMed] [Google Scholar]
- Haney C.H., Long S.R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. USA 107: 478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase Y., Fujioka S., Yoshida S., Sun G., Umeda M., Tanaka A. (2005). Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J. Exp. Bot. 56: 1263–1268. [DOI] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec N., Becker J.D., Pühler A., Perlick A.M., Küster H. (1999). Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol. Gen. Genet. 261: 514–522. [DOI] [PubMed] [Google Scholar]
- Hohnjec N., Vieweg M.F., Pühler A., Becker A., Küster H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 137: 1283–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.S., Liao J., James E.K., Sato S., Tabata S., Jurkiewicz A., Madsen L.H., Stougaard J., Ross L., Szczyglowski K. (2012). Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol. 160: 917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imin N., Goffard N., Nizamidin M., Rolfe B.G. (2008). Genome-wide transcriptional analysis of super-embryogenic Medicago truncatula explant cultures. BMC Plant Biol. 8: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata E., Ikeda S., Matsunaga S., Kurata M., Yoshioka Y., Criqui M.C., Genschik P., Ito M. (2011). GIGAS CELL1, a novel negative regulator of the anaphase-promoting complex/cyclosome, is required for proper mitotic progression and cell fate determination in Arabidopsis. Plant Cell 23: 4382–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jez J.M., Bowman M.E., Noel J.P. (2002). Role of hydrogen bonds in the reaction mechanism of chalcone isomerase. Biochemistry 41: 5168–5176. [DOI] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145: 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet E.P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D.G., Gianinazzi-Pearson V. (2001). Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14: 737–748. [DOI] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kaur J., Sebastian J., Siddiqi I. (2006). The Arabidopsis-mei2-like genes play a role in meiosis and vegetative growth in Arabidopsis. Plant Cell 18: 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E., Oláh B., Kaló P., Morales M., Heckmann A.B., Borbola A., Lózsa A., Kontár K., Middleton P., Downie J.A., Oldroyd G.E.D., Endre G. (2009). LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 151: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinski F., Courty P.E., Sieh D., Franken P., Zhang H., Bucher M., Gerlach N., Kryvoruchko I., Zoeller D., Udvardi M., Hause B. (2014). The H+-ATPase HA1 of Medicago truncatula is essential for phosphate transport and plant growth during arbuscular mycorrhizal symbiosis. Plant Cell 26: 1808–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajinski F., Hause B., Gianinazzi-Pearson V., Franken P. (2002). Mtha1, a plasma membrane H+-ATPase gene from Medicago truncatula, shows arbuscule-specific induced expression in mycorrhizal roots. Plant Biol. 4: 754–761. [Google Scholar]
- Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., Jardinaud M.F., Mun J.H., Larrainzar E., Cook D.R., Gamas P., Niebel A. (2014). The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 65: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert D., Weiler E.W. (1998). Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 15: 675–684. [DOI] [PubMed] [Google Scholar]
- Lavin M., Herendeen P.S., Wojciechowski M.F. (2005). Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 54: 575–594. [DOI] [PubMed] [Google Scholar]
- Li J., Dai X., Liu T., Zhao P.X. (2012). LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Res. 40: D1221–D1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu Z.B., Shi X., Hagen G., Guilfoyle T.J. (1994). An auxin-inducible element in soybean SAUR promoters. Plant Physiol. 106: 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Farmer A., Brechenmacher L., Drnevich J., Langley R.J., Bilgin D.D., Radwan O., Neece D.J., Clough S.J., May G.D., Stacey G. (2010). Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152: 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Govindarajulu M., Berg R.H., Ong Y.T., Puricelli K., Taylor C.G., Xu D., Stacey G. (2011). A dual-targeted soybean protein is involved in Bradyrhizobium japonicum infection of soybean root hair and cortical cells. Mol. Plant Microbe Interact. 24: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Liu J., Novero M., Charnikhova T., Ferrandino A., Schubert A., Ruyter-Spira C., Bonfante P., Lovisolo C., Bouwmeester H.J., Cardinale F. (2013). Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 64: 1967–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., et al. (2011). Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23: 3853–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd C.W. (1991). Cytoskeletal elements of the phragmosome establish the division plane in vacuolated higher plant cells. In The Cytoskeletal Basis of Plant Growth and Form, Lloyd C.W., ed (San Diego, CA: Academic Press; ), pp. 245–257. [Google Scholar]
- Lu S., et al. (2006). The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of β-carotene accumulation. Plant Cell 18: 3594–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie C., Barny M.A., Downie J.A. (1992). Rhizobium leguminosarum has two glucosamine synthases, GlmS and NodM, required for nodulation and development of nitrogen-fixing nodules. Mol. Microbiol. 6: 843–851. [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E.D. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Schlaman H.R., Spaink H.P., Of Sautter C., Rolfe B.G., Djordjevic M.A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14: 23–34. [DOI] [PubMed] [Google Scholar]
- Maxwell C.A., Edwards R., Dixon R.A. (1992). Identification, purification, and characterization of S-adenosyl-L-methionine: isoliquiritigenin 2′-O-methyltransferase from alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 293: 158–166. [DOI] [PubMed] [Google Scholar]
- Maxwell C.A., Hartwig U.A., Joseph C.M., Phillips D.A. (1989). A chalcone and two related flavonoids released from alfalfa roots induce nod genes of Rhizobium meliloti. Plant Physiol. 91: 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C., Cullimore J., Lefebvre B. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskiene I., Bögre L., Dahl M., Pirck M., Ha D.T., Swoboda I., Heberle-Bors E., Ammerer G., Hirt H. (1995). cycMs3, a novel B-type alfalfa cyclin gene, is induced in the G0-to-G1 transition of the cell cycle. Plant Cell 7: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahara A., Richens J., Starker C., Morieri G., Smith L., Long S., Downie J.A., Oldroyd G.E.D. (2010). Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol. Plant Microbe Interact. 23: 1553–1562. [DOI] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Messerli M.A., Shi C., Gilroy S. (2007). Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA 104: 20996–21001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morieri G., Martinez E.A., Jarynowski A., Driguez H., Morris R., Oldroyd G.E.D., Downie J.A. (2013). Host-specific Nod-factors associated with Medicago truncatula nodule infection differentially induce calcium influx and calcium spiking in root hairs. New Phytol. 200: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252. [DOI] [PubMed] [Google Scholar]
- Müssig C., Shin G.H., Altmann T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133: 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E.D., Engstrom E.M., Long S.R. (2001). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13: 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E.D., Long S.R. (2003). Identification and characterization of Nodulation-Signaling Pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd G.E.D., Murray J.D., Poole P.S., Downie J.A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45: 119–144. [DOI] [PubMed] [Google Scholar]
- Pandya M., Kumar G.N., Rajkumar S. (2013). Invasion of rhizobial infection thread by non-rhizobia for colonization of Vigna radiata root nodules. FEMS Microbiol. Lett. 348: 58–65. [DOI] [PubMed] [Google Scholar]
- Pankhurst C.E., Biggs D.R. (1980). Sensitivity of Rhizobium to selected isoflavonoids. Can. J. Microbiol. 26: 542–545. [DOI] [PubMed] [Google Scholar]
- Parniske M., Ahlborn B., Werner D. (1991). Isoflavonoid-inducible resistance to the phytoalexin glyceollin in soybean rhizobia. J. Bacteriol. 173: 3432–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F., Penel C., Dunand C. (2004). Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 9: 534–540. [DOI] [PubMed] [Google Scholar]
- Penmetsa R.V., Cook D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530. [DOI] [PubMed] [Google Scholar]
- Péret B., Swarup R., Jansen L., Devos G., Auguy F., Collin M., Santi C., Hocher V., Franche C., Bogusz D., Bennett M., Laplaze L. (2007). Auxin influx activity is associated with Frankia infection during actinorhizal nodule formation in Casuarina glauca. Plant Physiol. 144: 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Peters N.K., Frost J.W., Long S.R. (1986). A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233: 977–980. [DOI] [PubMed] [Google Scholar]
- Peters N.K., Long S.R. (1988). Alfalfa root exudates and compounds which promote or inhibit induction of Rhizobium meliloti nodulation genes. Plant Physiol. 88: 396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovská B., Cenklová V., Pochylová Z., Kourová H., Doskočilová A., Plíhal O., Binarová L., Binarová P. (2012). Plant Aurora kinases play a role in maintenance of primary meristems and control of endoreduplication. New Phytol. 193: 590–604. [DOI] [PubMed] [Google Scholar]
- Phillips D.A., Dakora F.D., Sande E., Joseph C.M., Zoń J. (1994). Synthesis, release, and transmission of alfalfa signals to rhizobial symbionts. Plant Soil 161: 69–80. [Google Scholar]
- Pichon M., Journet E.P., de Billy F., Dedieu A., Truchet G., Barker D.G. (1994). ENOD12 gene expression as a molecular marker for comparing rhizobium-dependent and independent nodulation in alfalfa. Mol. Plant Microbe Interact. 7: 740–747. [Google Scholar]
- Pitts R.J., Cernac A., Estelle M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16: 553–560. [DOI] [PubMed] [Google Scholar]
- Prayitno J., Rolfe B.G., Mathesius U. (2006). The Ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol. 142: 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumplin N., Mondo S.J., Topp S., Starker C.G., Gantt J.S., Harrison M.J. (2010). Medicago truncatula Vapyrin is a novel protein required for arbuscular mycorrhizal symbiosis. Plant J. 61: 482–494. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Ramos J., Bisseling T. (2003). A method for the isolation of root hairs from the model legume Medicago truncatula. J. Exp. Bot. 54: 2245–2250. [DOI] [PubMed] [Google Scholar]
- Ramu S.K., Peng H.M., Cook D.R. (2002). Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol. Plant Microbe Interact. 15: 522–528. [DOI] [PubMed] [Google Scholar]
- Redmond J.R., Batley M., Djordjevic M.A., Innes R.W., Keumpel P.L., Rolfe B.G. (1986). Flavones induce expression of nodulation genes in Rhizobium. Nature 323: 632–635. [Google Scholar]
- Ren S., Rollins B.J. (2004). Cyclin C/cdk3 promotes Rb-dependent G0 exit. Cell 117: 239–251. [DOI] [PubMed] [Google Scholar]
- Röhm M., Werner D. (1987). Isolation of root hairs from seedlings of Pisum sativum. Identification of root hair specific proteins by in situ labelling. Physiol. Plant. 69: 129–136. [Google Scholar]
- Ryu H., Kim K., Cho H., Park J., Choe S., Hwang I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer P., Feddermann N., Wiemken A., Boller T., Staehelin C. (2004). Sinorhizobium meliloti-induced chitinase gene expression in Medicago truncatula ecotype R108-1: a comparison between symbiosis-specific class V and defence-related class IV chitinases. Planta 219: 626–638. [DOI] [PubMed] [Google Scholar]
- Sauviac L., Niebel A., Boisson-Dernier A., Barker D.G., de Carvalho-Niebel F. (2005). Transcript enrichment of Nod factor-elicited early nodulin genes in purified root hair fractions of the model legume Medicago truncatula. J. Exp. Bot. 56: 2507–2513. [DOI] [PubMed] [Google Scholar]
- Savouré A., Sallaud C., El-Turk J., Zuanazzi J., Ratet P., Schultze M., Kondorosi A., Esnault R., Kondorosi E. (1997). Distinct response of Medicago suspension cultures and roots to Nod factors and chitin oligomers in the elicitation of defense-related responses. Plant J. 11: 277–287. [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Shao H., Dixon R.A., Wang X. (2007). Crystal structure of vestitone reductase from alfalfa (Medicago sativa L.). J. Mol. Biol. 369: 265–276. [DOI] [PubMed] [Google Scholar]
- Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145: 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Soto M.J., Fernandez-Aparicio M., Castellanos-Morales V., Garcia-Garrido J.M., Ocampo J.A., Delgado M.J., Vierheilig H. (2010). First indications for the involvement of strigolactones on nodule formation in alfalfa (Medicago sativa). Soil Biol. Biochem. 42: 383–385. [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. (2013). Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Lee S.H., Wenger J.P., Gonzalez N., Itoh H., Inzé D., Peer W.A., Murphy A.S., Overvoorde P.J., Gray W.M. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Ren H., Park M.Y., Grandt K.N., Lee S.H., Murphy A.S., Sussman M.R., Overvoorde P.J., Gray W.M. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer C., Schellmann S., Sabovljevic A., Shahriari M., Keshavaiah C., Bechtold N., Herzog M., Müller S., Hanisch F.G., Hülskamp M. (2006). The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133: 4679–4689. [DOI] [PubMed] [Google Scholar]
- Staehelin C., Schultze M., Kondorosi E., Kondorosi A. (1995). Lipo-chitooligosaccharide nodulation signals from Rhizobium meliloti induce their rapid degradation by the host plant alfalfa. Plant Physiol. 108: 1607–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Stacey G., Yu O. (2006). Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 48: 261–273. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Stacey G., Yu O. (2007). Distinct, crucial roles of flavonoids during legume nodulation. Trends Plant Sci. 12: 282–285. [DOI] [PubMed] [Google Scholar]
- Takahashi I., Kojima S., Sakaguchi N., Umeda-Hara C., Umeda M. (2010). Two Arabidopsis cyclin A3s possess G1 cyclin-like features. Plant Cell Rep. 29: 307–315. [DOI] [PubMed] [Google Scholar]
- Tian Y., Liu W., Cai J., Zhang L.Y., Wong K.B., Feddermann N., Boller T., Xie Z.P., Staehelin C. (2013). The nodulation factor hydrolase of Medicago truncatula: characterization of an enzyme specifically cleaving rhizobial nodulation signals. Plant Physiol. 163: 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., Truchet G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Wang X.J., Hagen G., Guilfoyle T.J. (2001). AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchet G. (1978). Sur l'état diploide des cellules du méritème des nodules radiculaires des légumineuses. Ann. Sci. Nat. Bot. Paris 19: 3–38. [Google Scholar]
- Turner M., Nizampatnam N.R., Baron M., Coppin S., Damodaran S., Adhikari S., Arunachalam S.P., Yu O., Subramanian S. (2013). Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 162: 2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Liu Z.B., Hagen G., Guilfoyle T.J. (1995). Composite structure of auxin response elements. Plant Cell 7: 1611–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme M., Huibers R.P., Elberse J., Van den Ackerveken G. (2008). Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 54: 785–793. [DOI] [PubMed] [Google Scholar]
- van Brussel A.A., Bakhuizen R., van Spronsen P.C., Spaink H.P., Tak T., Lugtenberg B.J., Kijne J.W. (1992). Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257: 70–72. [DOI] [PubMed] [Google Scholar]
- van Brussel A.A., Recourt K., Pees E., Spaink H.P., Tak T., Wijffelman C.A., Kijne J.W., Lugtenberg B.J. (1990). A biovar-specific signal of Rhizobium leguminosarum bv. viciae induces increased nodulation gene-inducing activity in root exudate of Vicia sativa subsp. nigra. J. Bacteriol. 172: 5394–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Batenburg F.H.D., Jonker R., Kijne J. (1986). Rhizobium induces marked root hair curling by redirection of tip growth - a computer-simulation. Physiol. Plant. 66: 476–480. [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: 0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venverloo C., Libbenga K. (1987). Regulation of the plane of cell-division in vacuolated cells. 1. The function of nuclear positioning and phragmosome formation. J. Plant Physiol. 131: 267–284. [Google Scholar]
- Wang J.-W., Wang L.-J., Mao Y.-B., Cai W.-J., Xue H.-W., Chen X.-Y. (2005). Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., et al. (2014). A H+-ATPase that energizes nutrient uptake during mycorrhizal symbioses in rice and Medicago truncatula. Plant Cell 26: 1818–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson A.P., Pellerone F.I., Mathesius U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18: 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J.P., Rathbun E.A., Knox J.P., Brewin N.J. (2000). Involvement of diamine oxidase and peroxidase in insolubilization of the extracellular matrix: implications for pea nodule initiation by Rhizobium leguminosarum. Mol. Plant Microbe Interact. 13: 413–420. [DOI] [PubMed] [Google Scholar]
- Xie F., Murray J.D., Kim J., Heckmann A.B., Edwards A., Oldroyd G.E., Downie J.A. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. USA 109: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.C., de Blank C., Meskiene I., Hirt H., Bakker J., van Kammen A., Franssen H., Bisseling T. (1994). Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6: 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K., et al. (2009). Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell 21: 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.H., Chen M.H., Liu C.J. (2008). Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J. 55: 382–396. [DOI] [PubMed] [Google Scholar]
- Zhang J., Subramanian S., Stacey G., Yu O. (2009). Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 57: 171–183. [DOI] [PubMed] [Google Scholar]
- Zhang J., Subramanian S., Zhang Y., Yu O. (2007). Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol. 144: 741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Han L., Hou C., Metelli A., Qi L., Tadege M., Mysore K.S., Wang Z.Y. (2011). Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell 23: 2106–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.