Abstract
Dental plaque is a dynamic microbial biofilm ecosystem that comprises hundreds of species including difficult-to-cultivate bacteria. We observed the assembly of a plaque bacterial community through 16S rRNA gene analysis. Plaque samples that accumulated on a hydroxyapatite disk for 1, 2, 3, 4, 5, and 7 days with saliva on day 0 were collected from 19 young adults using a removable resin splint. Quantitative PCR analysis showed that the total bacterial amount gradually increased and reached a plateau on day 4. Barcoded pyrosequencing analysis revealed that the microbial richness and diversity particularly increased between days 5 and 7. A principal coordinate analysis plot based on unweighted UniFrac showed the community assembly in a time-related manner, which became increasingly similar to the salivary microbiota. Facultative anaerobic bacteria such as Streptococcus, Neisseria, Abiotrophia, Gemella, and Rothia were predominant in the plaque bacterial community in the earlier days, whereas obligate anaerobes, such as Porphyromonas, Fusobacterium, Prevotella, and Capnocytophaga showed increased dominance on later days. UniFrac analysis also demonstrated that dental caries experience had a significant effect on the assembly process. Our results reveal the development pattern of the plaque bacterial community as well as the inter-individual differences associated with dental caries experience.
Dental plaque is a dynamic microbial biofilm ecosystem that comprises hundreds of species1. The development of biofilm begins with adherence of selected bacteria, including Streptococcus species, to saliva-bathed surfaces. As bacteria cover the substratum, later colonizers attach to the initial colonizers. The biofilm shows increasing diversity over time2. Multiplication of the attached organisms results in an increase in biomass, as well as a population shift due to altered environmental conditions within the biofilm. Marsh et al.3 stated that changes in key environmental factors, such as atmosphere, pH, and nutrients, drive a compositional shift in the microbiota toward a disease-associated community, leading to a greater risk of dental caries or periodontitis.
Microbial succession in dental plaque development was summarized by Ritz4. It is considered to be an ordered sequence of events, with facultative and aerobic bacteria, such as Streptococcus, Neisseria, and Rothia, predominantly colonizing the tooth surface in the early stages, and the proportions of facultative and anaerobic genera, such as Actinomyces, Corynebacterium, Fusobacterium, and Veillonella, increasing gradually with plaque maturation. Furthermore, a spatiotemporal model of oral bacterial colonization was proposed by Kolenbrander et al.5 based on the recognition of salivary pellicle receptors by initial colonizing species and coaggregation by specific bacterial combinations. Conversely, Langfeldt et al.6, using a 16S rRNA gene sequencing approach, recently reported that no systematic time-related pattern was detected in the compositional shift of bacterial biofilm formed on a membrane installed in the oral cavity. Although it is unclear which architecture in the oral cavity was mimicked by the artificial membrane, this result is inconsistent with the conventional concept of oral biofilm establishment. Therefore, it is reasonable that the development of plaque microbiota on tooth surfaces should also be reconsidered using a comprehensive molecular approach, including difficult-to-cultivate bacteria.
In the present study, we observed the assembly process of dental plaque microbiota on a hydroxyapatite disk in 19 young adults through molecular approaches using the 16S rRNA gene. We collected plaque samples for 1, 2, 3, 4, 5, and 7 days using a removable resin splint (Figure 1), and the bacterial community structure was evaluated using a barcoded pyrosequencing approach. In addition, the maturation process of the plaque bacterial community was compared between 9 caries-free (CF) subjects and 10 caries-experienced (CE) subjects.
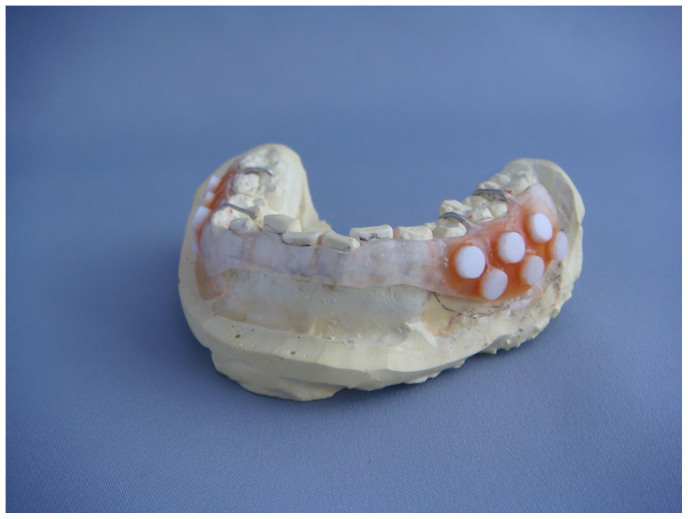
Figure 1. Removable resin splint used in this study.
This photograph was taken by Toru Takeshita (the first author).
Results
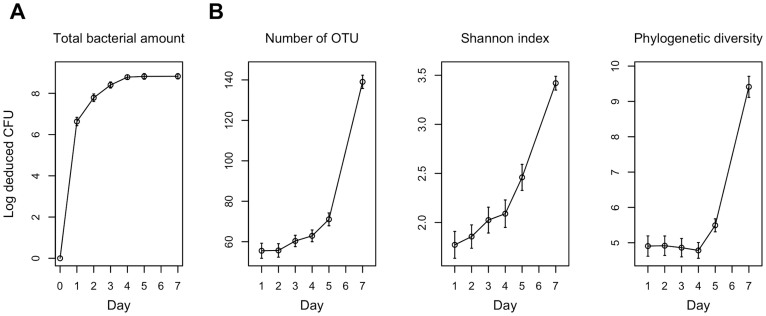
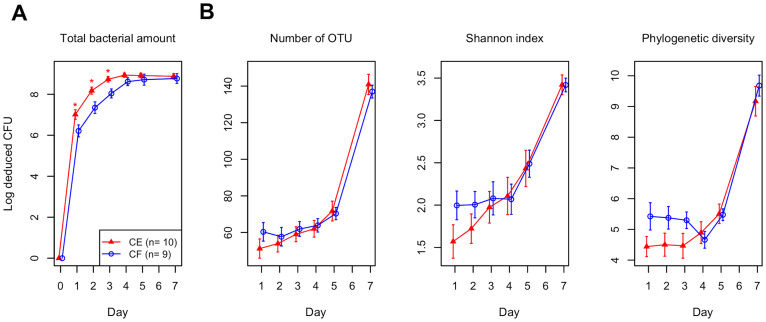
Dental plaque samples that accumulated on a hydroxyapatite disk (5-mm diameter) for 1, 2, 3, 4, 5, and 7 days were retrieved from 19 healthy young adults (Table 1). The total amounts of bacteria on the disks were examined by quantitative PCR analysis of the 16S rRNA gene (Figure 2A). They gradually increased and had almost reached a plateau at about 109 CFU by day 4.
Table 1. Characteristics of study subjects.
| Subject | Age (yr) | Sex | Number of teeth | Number of DT | Number of DMFT | Salivary flow rate (ml/min) | Salivary buffering capacitya |
|---|---|---|---|---|---|---|---|
| E1 | 24 | Male | 29 | 0 | 9 | 1.2 | Intermediate |
| E2 | 26 | Male | 28 | 0 | 16 | 1.1 | High |
| E3 | 22 | Female | 26 | 0 | 10 | 0.8 | High |
| E4 | 22 | Female | 28 | 1 | 11 | 1.2 | High |
| E5 | 22 | Male | 32 | 0 | 12 | 1.8 | High |
| E6 | 24 | Male | 31 | 0 | 10 | 2.0 | High |
| E7 | 28 | Male | 32 | 0 | 14 | 0.5 | Intermediate |
| E8 | 28 | Male | 32 | 0 | 13 | 1.4 | High |
| E9 | 23 | Female | 27 | 2 | 19 | 0.5 | Intermediate |
| E10 | 22 | Female | 31 | 3 | 11 | 1.8 | High |
| F1 | 24 | Male | 29 | 0 | 0 | 1.5 | High |
| F2 | 24 | Male | 28 | 0 | 0 | 1.2 | High |
| F3 | 21 | Female | 29 | 0 | 0 | 0.8 | Intermediate |
| F4 | 22 | Male | 28 | 0 | 0 | 2.4 | High |
| F5 | 20 | Male | 27 | 0 | 0 | 0.8 | High |
| F6 | 25 | Male | 32 | 0 | 0 | 1.3 | High |
| F7 | 25 | Male | 27 | 0 | 0 | 1.3 | High |
| F8 | 22 | Female | 32 | 0 | 0 | 1.0 | Intermediate |
| F9 | 26 | Male | 32 | 0 | 0 | 2.5 | High |
DT, decayed teeth; DMFT, decayed, missing and filled (caries experienced) teeth.
aSalivary buffering capacity was evaluated using Dentbuff strip (Orion Diagnostica, Helsinki).
Figure 2. Temporal change in the total amount of bacteria, microbial richness and diversity.
(A) Temporal change in the total amount of bacteria on a hydroxyapatite disk. (B) Temporal change in the number of operational taxonomic units (OTUs), Shannon diversity index and phylogenetic diversity in the plaque microbiota on a hydroxyapatite disk. To correct for the unequal number of sequences, we evaluated 800 randomly selected sequences. The error bars indicate the standard error.
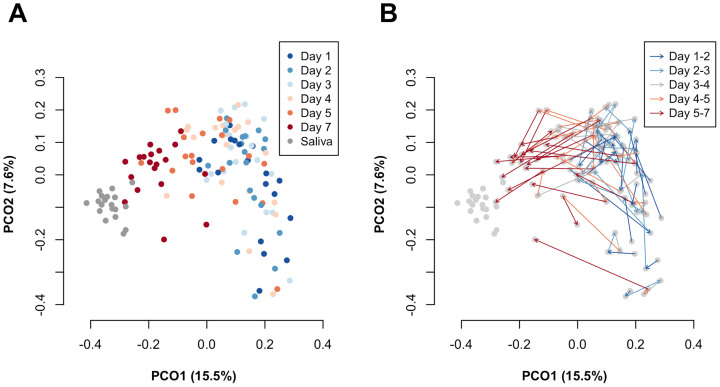
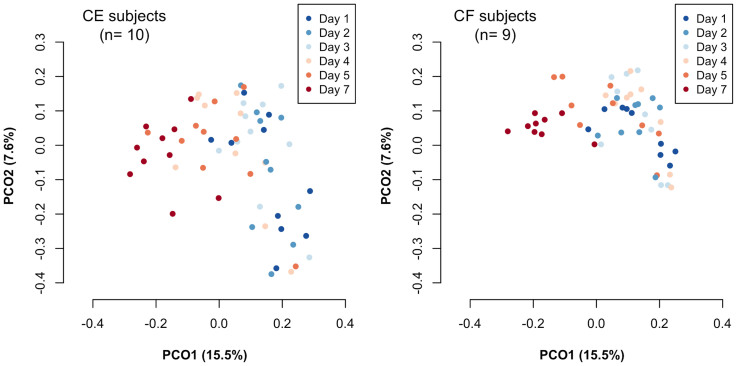
The composition of the bacterial community in accumulating plaque, together with saliva collected just before installing a mouth splint, were investigated using barcoded pyrosequencing analysis of the 16S rRNA gene. We determined 692,400 bacterial 16S rRNA gene sequences (containing the V1–V2 region), of which 470,423 passed quality-control steps (822–7,904 reads per sample, Table S1; average length, 346 ± 18 bases). The sequences were assigned to 4,936 species-level operational taxonomic units (OTUs) using a cutoff distance of 0.03. Alpha diversity metrics including the number of detected OTUs, Shannon diversity index and phylogenetic diversity increased over time, showing an especially large increase between days 5 and 7 (Figure 2B). A principal coordinate analysis (PCoA) plot based on unweighted UniFrac showed a gradual temporal shift of the plaque microbiota to the negative direction of principal coordinate 1 (Figure 3), suggesting that the community assembly of plaque biofilm occurred in a time-related manner. Permutational multivariate analysis of variance (PERMANOVA) confirmed that the plaque accumulation time had a significant effect on the composition of the microbiota (P < 0.001).
Figure 3. A principal coordinate analysis (PCoA) plot showing similar relations among 133 bacterial community samples (7 each from 19 subjects) using the unweighted UniFrac distance metric.
The two components explained 15.5% and 7.6% of the variance, respectively. To correct for the unequal number of sequences, we evaluated 800 randomly selected sequences per sample. (A) Samples collected on different days are depicted using different colors. (B) Plaque samples collected from the same individuals are connected by arrows.
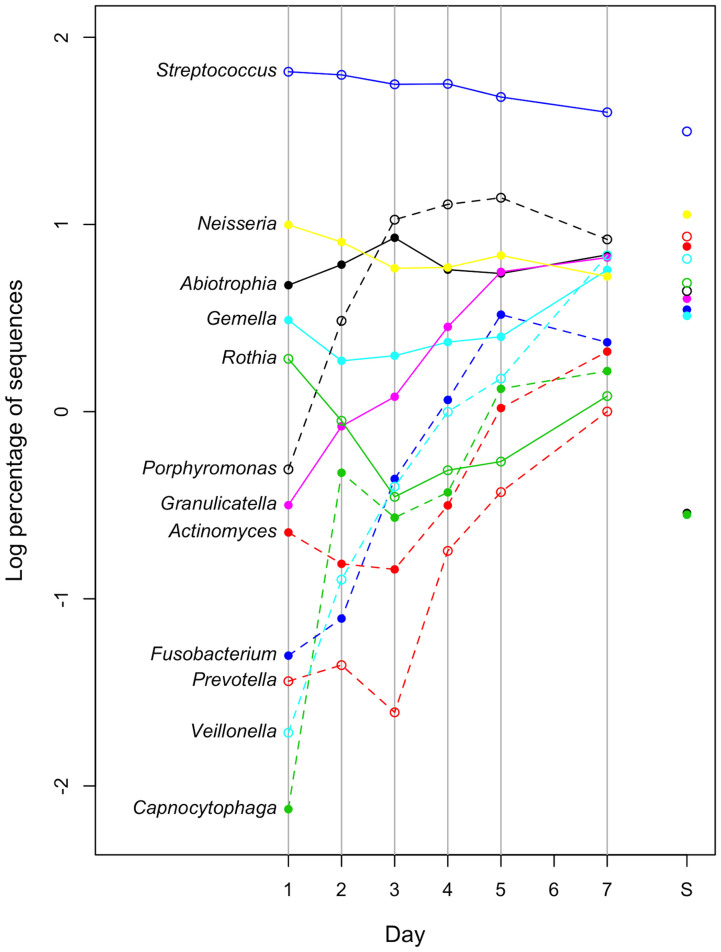
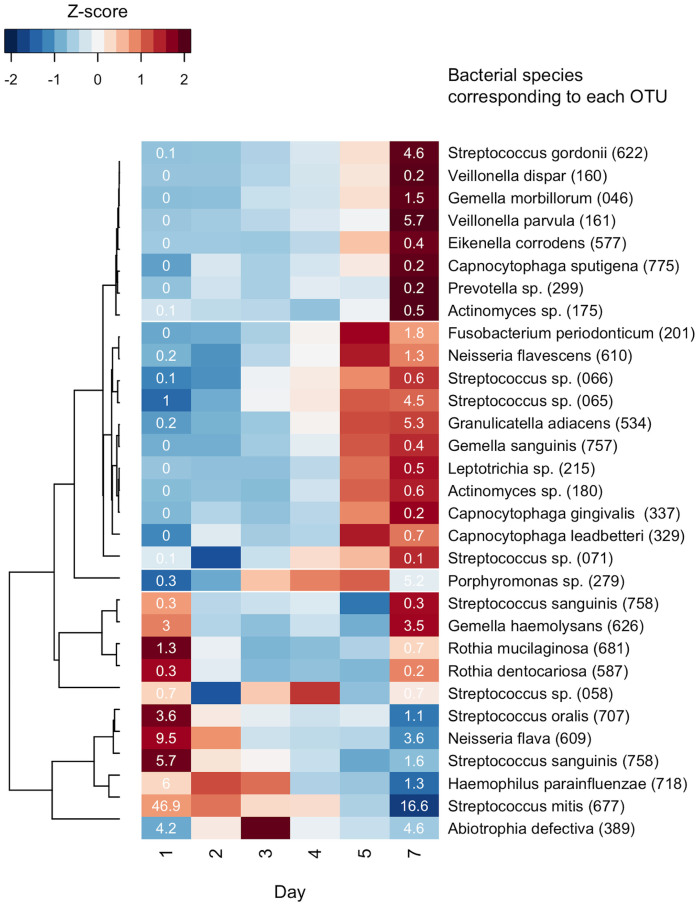
The temporal shift in plaque microbiota composition was also characterized by changes in the relative abundances of bacterial taxa. Plaque samples on the later days exhibited greater microbial richness and diversity (Figure 2B), and were plotted closer to the saliva samples in the PCoA UniFrac diagram (Figure 3), suggesting that a greater variety of salivary bacterial taxa was able to grow in the later stages of plaque development. Of the 53 genera identified in the plaque microbiota, Streptococcus species made up most of the microbiota in each subject on all days (Figure 4). Facultative anaerobic bacteria such as Neisseria, Abiotrophia, Gemella, and Rothia were predominant in the plaque bacterial community during the earlier days, whereas obligate anaerobes, such as Porphyromonas, Fusobacterium, Prevotella, Veillonella, and Capnocytophaga showed increased dominance on later days, with decreases in facultative members, such as Streptococcus (Figure 4). Of 4,936 species-level OTUs, the representative sequences of 343 OTUs corresponded to the sequences deposited in the Human Oral Microbiome Database (HOMD) with ≥98.5% identity. A total of 31 of these 343 OTUs were commonly detected (15 of 19 subjects) in the plaque bacterial community on day 7, and they made up large proportions of the community (83.6 ± 9.1% and 68.6 ± 8.2% on days 1 and 7, respectively). A temporal shift in their relative abundances is displayed as a heatmap in Figure 5. The OTUs corresponding to Streptococcus species, such as S. mitis, S. sanguinis and S. oralis were more predominant on earlier days. Other than Streptococcus, N. flava, H. parainfluenzae, A. defectiva, R. mucilaginosa, and G. haemolysans could also be considered early colonizers. The relative abundance of the OTU corresponding to Porphyromonas sp. OT-279 peaked on days 4 and 5. Subsequently, several OTUs corresponding to bacterial species such as G. adiacens, Streptococcus sp. OT-65, F. periodonticum and N. flavescens appeared, and bacterial species, including V. parvula, S. gordonii and G. morbillorum were flourishing on day 7.
Figure 4. Temporal shift in the relative abundance of bacterial genera.
Only the 12 predominant genera commonly detected in the day 7 samples (18 of 19 subjects) are shown. S indicates saliva.
Figure 5. Temporal shift in the relative abundance of species-level OTUs.
Only 31 OTUs corresponding to the bacterial species deposited in the Human Oral Microbiome Database25 (Oral taxon IDs were given in parenthesis) with a high (≥98.5%) identity and that were commonly detected (15 of 19 subjects) in the day 7 samples are shown. To show the temporal change in bacterial genera with lower abundance, the relative abundance of each genus was normalized to a mean of 0 and standard deviation of 1 (z-score normalization), and is represented by the color intensity of each grid (blue, low abundance; red, high abundance). The OTUs are ordered according to the result of hierarchical cluster analysis using the correlation distance with average linkage (shown as a dendrogram on the left). The relative abundance (%) of each OTU on day 1 or 7 is displayed in each grid.
The total amount of bacteria on the disks, and microbial richness and diversity, increased over time in both CF and CE subjects (Figure 6). In contrast, significantly larger amounts of bacteria were present on the disks of CE subjects than CF subjects on days 1 to 3 (P < 0.05, Student's t-test, respectively), suggesting that the amount of bacteria reached a plateau later in CF subjects than in CE subjects. A PCoA plot based on UniFrac showed that the developing plaque microbiota of CF subjects localized in the positive direction of principal coordinate 2, suggesting that the community assembly of their plaque biofilm occurred by a characteristic process (Figure 7). PERMANOVA confirmed that the dental caries experience also had a significant effect on the microbiota composition (P < 0.001).
Figure 6. Temporal change in the total amount of bacteria, microbial richness and diversity in subjects with caries experience (CE) and caries-free (CF) subjects (10 and 9 subjects, respectively).
(A) Temporal change in the total amount of bacteria on a hydroxyapatite disk in CE and CF subjects. *P < 0.05, using a Student's t-test analyzed per sampling time. The error bars indicate standard error. (B) Temporal change in the number of OTUs, Shannon diversity index and phylogenetic diversity in the plaque microbiota in CF and CE subjects. To correct for the unequal number of sequences, we evaluated 800 randomly selected sequences. The error bars indicate standard error.
Figure 7. A principal coordinate analysis (PCoA) plot showing similar relations among 133 bacterial community samples using the unweighted UniFrac distance metric.
Only plaque samples of subjects with caries experience (CE) and caries-free (CF) subjects were shown in the left and right side of diagrams, respectively. The two components explained 15.5% and 7.6% of the variance, respectively. To correct for the unequal number of sequences, we evaluated 800 randomly selected sequences per sample.
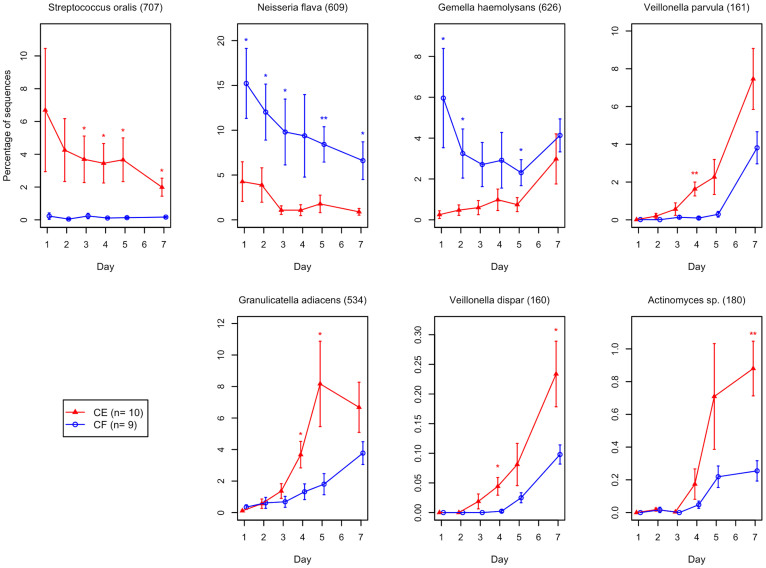
We performed two-way analysis of variance (ANOVA) with repeated measures for the 343 OTUs to explore the important OTUs responsible for the differences in the assembly process of plaque microbiota between CE and CF subjects (Figure 8). Significant effects of dental caries experience were observed in the relative abundance shifts of 7 of 343 OTUs, corresponding to the bacterial species deposited in the HOMD with ≥98.5% identity (P < 0.05, Figure 8). The OTU corresponding to S. oralis OT-707 was observed in a lower proportion of CF subjects than in CE subjects regarding plaque development, particularly on days 3–7. The OTUs corresponding to N. flava OT-609 and G. haemolysans OT-626 were more predominant in CF subjects than in CE subjects, particularly during the earlier days. The OTUs corresponding to V. dispar, V. parvula, and G. adiacens showed significantly lower proportions in CF subjects than in CE subjects on day 4. The OTU corresponding to G. adiacens showed significantly lower proportions in CF subjects than in CE subjects on day 5, and the OTUs corresponding to V. dispar and Actinomyces sp. OT-180 were significantly lower on day 7.
Figure 8. Temporal shift in the relative abundance of species-level OTUs in subjects with and without dental caries experience.
Only seven OTUs in which dental caries experience had a significant effect on their relative abundance (P < 0.05, two-way ANOVA) are shown. **P < 0.01, *P < 0.05, using post-hoc Student's t-test analyzed per sampling time. The error bars indicate standard error. CE, caries-experienced subjects; CF, caries-free subjects.
Discussion
Our results using the 16S rRNA gene sequencing approach revealed microbial succession in dental plaque biofilm established on a hydroxyapatite disk. A plaque microbiota shift was observed in a time-related manner during maturation (Figures 2, 3, 4 and 5), although inter-individual differences, especially associated with dental caries experience were also observed regarding the composition of developing plaque microbiota at each stage (Figures 6, 7 and 8).
Initially, we expected that the results of this study would differ from the plaque assembly process reported by Ritz4 due to the difference between their cultivation-based method and our molecular approach, which allowed determination of the composition of the microbiota, including difficult-to-cultivate species. However, the plaque-development process suggested by our results is unexpectedly consistent with that study. The bacterial communities were assembled in a time-related manner, from an early biofilm dominated by Streptococcus species to a complex community with high levels of Gram-negative anaerobes (Figure 4). This process demonstrates the effectiveness of a classic cultivation-based approach for providing an overview of oral bacterial communities, as well as the reliability of our results. In addition, it included more information regarding candidate early, middle, and late colonizer genera (Figures 4 and 5), adding to those reported previously7. Of the bacterial genera missed in Ritz's study4, Porphyromonas species are noteworthy as a dominant middle colonizer because they displayed similar behavior to Fusobacterium in developing plaque microbiota (Figure 4). Fusobacterium species are considered important “bridge” organisms that coaggregate initial, early, and late colonizers, and contribute to an increase in microbial diversity in plaque development8. Our results suggest that Porphyromonas species, particularly Porphyromonas sp. OT-279, may be a new target for the examination of plaque biofilm formation mechanisms.
Total amounts of bacteria had almost plateaued by day 4 (Figure 2A). The multiplication and shedding of bacteria presumably kept occurring after the plateau was reached, but about 109 CFU would represent a threshold amount of dental plaque that could form on the hydroxyapatite disk of 5-mm diameter. Conversely, the alpha diversity metrics increased rapidly between days 5 and 7 (Figure 2B). In particular, phylogenetic diversity which accounts for species evolutionary relationships did not become elevated until day 4, suggesting that bacteria taxonomically distinct from the early colonizers begin to participate in the plaque microbiota development after day 4. This is confirmed in Figure 5. These findings suggest that accumulation of a threshold amount of bacteria in plaque biofilm triggers major shifts in plaque microbiota composition. They also imply that saturation of plaque biomass does not indicate the ultimate state of the plaque microbiota succession, and the plaque bacterial communities at day 7 might not yet have reached their final composition because the complexity of the plaque microbiota at this timepoint was increasing (Figure 2B).
When the subjects were classified into two groups according to dental caries status, notable differences in the plaque bacterial community between CF and CE subjects were observed from day 1. The OTUs corresponding to N. flava and G. haemolysans made up a higher proportion of the microbiota in CF than in CE subjects, particularly during the early stages (Figure 8). When considering the lower total amount of bacteria in CF subjects (Figure 6A), the marked growth of Streptococcus during the early stages was inhibited in the more complex plaque microbiota of the CF subjects. These bacteria were implicated as candidate species that might protect against caries onset and progression in a previous clinical study with a longitudinal design9. Determining their interaction with acid producers in initial biofilm formation might facilitate the development of novel approaches that might enable assembly of healthy plaque that is less prone to a shift toward a pathogenic community.
Although the differences in the amount of bacteria between CF and CE subjects lessened over time (Figure 6A), the OTU corresponding to S. oralis was more predominant in CE subjects than in CF subjects (Figure 8). In addition, the growth of non-Streptococcus acid producers, such as Actinomyces and Granulicatella species, during the later stages was slower in CF subjects (Figure 8). Veillonella species, which metabolize lactate, were also less predominant in later plaque (Figure 8), in accordance with several previous studies, indicating that elevated levels of Veillonella were associated with higher acid production within plaque9,10,11,12. The developing plaque in CF subjects likely had a lower acidogenic potential than that in CE subjects during the later stage of the current study period. Because formation, succession, and removal of dental plaque on the tooth surface occur repeatedly, the differences observed in the assembly process may be involved in the susceptibility to dental caries. Further evaluation of the microbiota shift upon carbohydrate ingestion and pH measurement within dental plaque would be helpful to clarify their contribution to the progression of dental caries.
We performed multiple statistical tests for 343 OTUs to identify those responsible for the differences in the assembly process of plaque microbiota between CE and CF subjects (Figure 8). We did not adjust for multiple testing, because such adjustments are not strictly required in exploratory analyses13. As the choice and number of tested hypotheses is data dependent in exploratory analyses and a clear structure in the multiple tests is missing, an appropriate multiple test adjustment is difficult or even impossible13. Additionally, the number of tests was too large to use Bonferroni adjustment, which is the simplest and best-known multiple test procedure. With as many as 300 tests, this procedure has insufficient power to detect any true effect13. Actually, no OTU was selected after the adjustment, although PERMANOVA analysis on UniFrac results confirmed differences in the assembly process between CE and CF subjects (Figure 7, P < 0.001). On the other hand, interpretation of the significant results in the exploratory analyses needs careful consideration, as the significant relationships shown in this study were only exploratory results. Further, confirmatory studies with larger sample sizes are required to identify important bacterial species in the plaque assembly process of CF and CE subjects.
We observed plaque accumulation on hydroxyapatite disks instead of natural tooth surfaces. Tooth enamel is mostly (96%) composed of hydroxyapatite, and the remainder is organic substances and fluid. The pristine artificial surfaces of the disks were bathed in saliva immediately after installing a mouth splint, and were coated with salivary proteins in the same way as natural teeth in the oral cavity. In addition, considering the consistency of our results with those of a previous study using natural teeth4, our model is considered to mimic the bacterial community assembly on intact enamel surfaces. However, the variability in microbial colonization patterns occurring at various ecological niches in the oral cavity must also be considered14. Different processes of plaque development might be observed in other dental sites, such as gingival sulcus and dental fissures in molars.
Our results indicate the bacterial communities were assembled in a time-related manner consistent with a previous study4, and reconfirm that dental plaque biofilm that forms on tooth enamel is a dynamic microbial community. The microbial composition gradually shifts with accumulation time on tooth surfaces; therefore, the sampling of dental plaque microbiota as a representative of an individual oral microbiome should be performed with caution.
Methods
Study population
Subjects were recruited from among undergraduate students at Kyushu University, Faculty of Dental Science, and were aged 20 to 28 years. After an oral examination by a dentist, 9 CF subjects and 10 CE subjects (who had experienced ≥9 teeth with caries) were enrolled. Although three CE subjects had active caries regions as well as previously filled teeth, all of them were not obvious cavity but were localized only in the enamel or were secondary caries along the inlay margin. The ethics committee of Kyushu University Faculty of Dental Science approved the study design and the procedure for obtaining informed consent (Reference number, 20-11 and 26-130). The subjects reported no antibiotic intake in the study period and the preceding 1 month. No subjects with severe periodontitis or any systemic disease were included in the study population. All experiments were performed in accordance with the approved guidelines.
Oral device
Removable resin splints were fabricated for each subject to cover the mandibular buccal gingiva and were retained with a wire clasp. On the left and right sides of the buccal surface, six 5-mm-diameter hydroxyapatite disks (Cellyard, Hoya, Japan) were attached to the splint using wax (Figure 1). After paraffin-stimulated saliva collection, subjects wore the splint for 7 days, except while eating, drinking liquids other than water, or tooth-brushing, when the splint was immersed in phosphate-buffered saline (PBS; pH 7.4). At 1, 2, 3, 4, 5, and 7 days later, one of the disks was retrieved from each side of the splint after rinsing with PBS. We initially focused on the plaque development process especially in an early stage, therefore we omitted day-6 sample collection due to the limited space on the splint.
DNA extraction
Disks covered with plaque biofilm were placed in microcentrifuge tubes containing 500-µl lysis buffer (10 mM Tris, 1 mM EDTA, 1% SDS). After ultrasonic vibration to completely detach the biofilm, bacterial DNA extraction was performed as described previously15. The left side samples were used for the later analysis, except when the corresponding disk compromised. DNA extraction from saliva samples was performed according to a protocol described previously15.
Quantification of total bacteria by real-time PCR
Quantitative PCR was performed using a QuantiFast SYBR green PCR kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The universal bacterial primers 806F (5′-TTA GAT ACC CYG GTA GTG G-3′) and 926R (5′-CCG TCA ATT YCT YYG AGT TT-3′) were used and details of the procedure have been reported previously16. The amounts of total bacteria were calculated using the comparative CT method, and DNA extracted from Streptococcus mutans Xc was used as the real-time PCR control.
Barcoded pyrosequencing analysis
In total, 133 samples (7 each from 19 subjects) were assessed by barcoded pyrosequencing analysis of the 16S rRNA gene. The samples were separated into three groups: plaque samples taken on days 1 and 2, and those of CE subjects taken on day 3 (totaling 48 samples); plaque samples taken on days 3 and 4, and those of CF subjects taken on day 3 (totaling 47 samples); and plaque samples taken on day 7 and saliva samples (totaling 38 samples). The 16S rRNA genes of each sample in each group were amplified using the following primers: 338R with the 454 Life Sciences (Roche, Basel, Switzerland) adaptor B sequence (5′-CCT ATC CCC TGT GTG CCT TGG CAG TCT CAG TGC TGC CTC CCG TAG GAG T-3′) and 8F with the 454 Life Sciences adaptor A and 48 different sample-specific six-base barcodes (5′-CCA TCT CAT CCC TGC GTG TCT CCG ACT CAG NNN NNN AGA GTT TGA TYM TGG CTC AG-3′). PCR amplification was performed as previously described17. The amplicons were gel-purified using a Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI) according to the manufacturer's instructions. DNA concentration and quality were assessed using a spectrophotometer (NanoDrop Technologies, Wilmington, DE), and equal amounts of DNA from each group were pooled together. Pyrosequencing was conducted for each mixture using a 454 Life Sciences Genome sequencer FLX instrument (Roche) at Hokkaido System Science Co., Ltd. (Sapporo, Japan).
Data analysis and taxonomy assignment
Sequences obtained from all samples were analyzed together. Sequences were excluded from the analysis using a script written in PHP if they were shorter than 240 bases or had an average quality score <25, and subsequently removed using a script written in R if they did not include the correct primer sequence, had a homopolymer run >6 nt, or contained ambiguous characters. The remaining sequences were assigned to the appropriate sample by examining the six-base barcode sequence. Similar sequences were clustered into operational taxonomic units (OTUs) using UCLUST18 in QIIME19, with a minimum pairwise identity of 97%. The most abundant sequence in each OTU was chosen to represent that OTU. The representative sequences were aligned using PyNAST20 and the Greengenes database21 using a minimum identity of 75%. Chimeras were removed from the representative set on the basis of identification as being chimeric using Chimera Slayer22. After chimera elimination, a relaxed neighbor-joining tree was built using FastTree23. The UniFrac metric24 was used to determine the dissimilarity between any pair of bacterial communities. The similarity relationship, assessed using the UniFrac metric, was presented in a PCoA plot, drawn using R 3.0.1. The taxonomy of representative sequences was determined using the RDP classifier with a minimum support threshold of 80% and the RDP taxonomic nomenclature (to the genus level). For each representative sequence of the OTUs, nearest-neighbor species with ≥98.5% identity were selected as candidates using BLAST searches against 831 oral bacterial 16S rRNA gene sequences (HOMD 16S rRNA RefSeq version 13.2) in the Human Oral Microbiome Database25.
Statistical analysis
All of the statistical analyses were conducted using R 3.0.1 (available from URL: http://www.r-project.org/). Student's t-test was used for comparison of the total bacterial amount per sampling time. Number of OTU and Shannon index were calculated using vegan library. Phylogenetic diversity was calculated using pd function in picante library. Permutational multivariate analysis of variance (PERMANOVA) was used to assess the effects of plaque accumulation time and dental caries experience on plaque microbiota composition using adonis function in vegan library based on 9,999 permutations. Two-way analysis of variance (ANOVA) with repeated measures was used to assess the effects of plaque accumulation time and dental caries experience on the relative abundance of each OTU, followed by post-hoc Student's t-test.
Author Contributions
T.T., M.Y., Y.S. and Y.Y. designed and supervised the research project. T.T. wrote the paper. M.Y., T.T. and Y.S. conducted oral examination and sample collection. M.Y. conducted molecular analysis. T.T. conducted data processing. T.T., M.F. and N.E. conducted statistical analysis. All authors read and approved the manuscript.
Supplementary Material
Supplemental Material
Acknowledgments
This study was supported in part by Grants-in Aid for Scientific Research 25463249 (T.T.), 22592337 (Y.S.) and 25293428 (Y.Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Filoche S., Wong L. & Sissons C. H. Oral biofilms: emerging concepts in microbial ecology. J Dent Res 89, 8–18 (2010). [DOI] [PubMed] [Google Scholar]
- Rickard A. H., Gilbert P., High N. J., Kolenbrander P. E. & Handley P. S. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol 11, 94–100 (2003). [DOI] [PubMed] [Google Scholar]
- Marsh P. D. Dental plaque as a biofilm and a microbial community - implications for health and disease. BMC Oral Health 6 Suppl 1S14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz H. L. Microbial population shifts in developing human dental plaque. Arch Oral Biol 12, 1561–1568 (1967). [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E. et al. Communication among oral bacteria. Microbiol Mol Biol Rev 66, 486–505 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfeldt D. et al. Composition of microbial oral biofilms during maturation in young healthy adults. PLoS One 9, e87449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander P. E. et al. Bacterial interactions and successions during plaque development. Periodontol 2000 42, 47–79 (2006). [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J. Jr, Periasamy S. & Jakubovics N. S. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8, 471–480 (2010). [DOI] [PubMed] [Google Scholar]
- Gross E. L. et al. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7, e47722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aas J. A. et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46, 1407–1417 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner A. C. et al. Cultivable anaerobic microbiota of severe early childhood caries. J Clin Microbiol 49, 1464–1474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E. L. et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol 48, 4121–4128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender R. & Lange S. Adjusting for multiple testing--when and how? J Clin Epidemiol 54, 343–349 (2001). [DOI] [PubMed] [Google Scholar]
- Simon-Soro A. et al. Microbial geography of the oral cavity. J Dent Res 92, 616–621 (2013). [DOI] [PubMed] [Google Scholar]
- Takeshita T., Nakano Y. & Yamashita Y. Improved accuracy in terminal restriction fragment length polymorphism phylogenetic analysis using a novel internal size standard definition. Oral Microbiol Immunol 22, 419–428 (2007). [DOI] [PubMed] [Google Scholar]
- Takeshita T. et al. The ecological proportion of indigenous bacterial populations in saliva is correlated with oral health status. ISME J 3, 65–78 (2009). [DOI] [PubMed] [Google Scholar]
- Takeshita T. et al. Enteral tube feeding alters the oral indigenous microbiota in elderly adults. Appl Environ Microbiol 77, 6739–6745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72, 5069–5072 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21, 494–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C. & Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–8235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010; 10.1093/database/baq013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material