Abstract
The replacement of nucleosomal H2A with the histone variant H2A.Z is critical for regulating DNA-mediated processes across eukaryotes and for early development of multicellular organisms. How this variant performs these seemingly diverse roles has remained largely enigmatic. Here, we discuss recent mechanistic insights that have begun to reveal how H2A.Z functions as a molecular rheostat for gene control. We focus on specific examples in metazoans as a model for understanding how H2A.Z integrates information from histone post-translational modifications, other histone variants, and transcription factors (TFs) to regulate proper induction of gene expression programs in response to cellular cues. Finally, we propose a general model of how H2A.Z incorporation regulates chromatin states in diverse processes.
Introduction
Regulation of chromatin dynamics and accessibility is critical for proper control of all DNA-mediated processes, particularly in response to cellular cues. The replication-independent incorporation of histone variants by specific ATP-dependent chromatin remodeling complexes has emerged as a critical mechanism for regulating DNA repair, chromosome segregation, genomic stability, and gene expression [1–3]. The highly conserved H2A-type variant H2A.Z has garnered particular interest over the last several years because it has essential but unknown roles in early metazoan development [4,5]. Recent work demonstrating that H2A.Z is necessary for the proper execution of developmental programs during embryonic stem cell (ESC) differentiation [6–8] as well as for regulating the activation of gene expression patterns during muscle differentiation, T cell activation, and hormone-mediated gene activation [9–12] has led to a model whereby H2A.Z functions to mediate gene induction in response to diverse cellular cues.
Both in vitro and in vivo studies have shown that H2A.Z incorporation can alter nucleosome structure and dynamics, the activity of chromatin remodeling enzymes, and histone modification patterns [2,13]. H2A.Z is enriched at the majority of transcription start sites (TSSs) of RNA polymerase II (RNAPII)-regulated genes across eukaryotes, including mammals as well as regulatory regions, such as enhancers and boundary elements [7,8,14]. Intriguingly, H2A.Z is enriched at both active and silent genes, yet how H2A.Z influences gene expression in a context-dependent manner has remained elusive. In this review, we highlight recent work that reveals new mechanistic insights into how H2A.Z acts as a molecular gatekeeper for RNAPII at promoters and how H2A.Z integrates information from histone post-translational modifications (PTMs), other histone variants, and TFs to affect specific transcriptional outcomes during development, tissue specification, and immune- and hormone-mediated responses. Collectively, these studies suggest that H2A.Z acts as a molecular rheostat for gene control and serve as a paradigm for understanding its broader roles to regulate chromatin states in response to environmental and genetic perturbations.
Regulation of +1 nucleosome and RNA polymerase II progression
H2A.Z is highly enriched at the promoter regions of a large subset of genes across cell types, consistent with a role in transcriptional regulation. Although the precise patterns of enrichment differ slightly among eukaryotes, genome-wide studies from yeast to human indicate that H2A.Z-containing nucleosomes flank the nucleosome-depleted region (NDR) at TSSs [9,15,16]. Why is H2A.Z incorporation at the TSS important for transcriptional regulation? In vitro studies have demonstrated that the nucleosome immediately downstream of the TSS (denoted +1 nucleosome) poses a sizeable barrier to transcription and can direct the orientation of the pre-initiation complex (PICs) and subsequent transcriptional elongation [17,18]. In Drosophila, the +1 nucleosome obstructs RNAPII transit, resulting in the increased stalling and backtracking of the polymerase [19]. Notably, H2A.Z enrichment at the +1 nucleosome correlates with decreased RNAPII stalling, suggesting that its incorporation reduces the high-energy barrier to RNAPII progression. Consistent with this idea, H2A.Z levels anti-correlate with nucleosome turnover, indicating that H2A.Z incorporation at the +1 nucleosome regulates productive elongation by facilitating H2A.Z/H2B dimer loss without depletion of (H3-H4)2 tetramers. These findings suggest that H2A.Z incorporation at the +1 nucleosome regulates transcriptional output by modulating RNAPII kinetics and transcriptional elongation (Figure 1). Given that Drosophila H2A.Z has features of both H2A.Z and H2A.X (known as H2AvD) [20,21], whether H2A.Z incorporation at the +1 nucleosome has similar roles in other organisms remains an open question. Moreover, whereas a large number of genes are regulated by polymerase pausing in Drosophila, not all H2A.Z target genes in other organisms are regulated in this manner [22]. In mouse ESCs, H2A.Z is present at a large set of genes marked by histone 3 lysine 4 trimethylation (H3K4me3) nucleosomes that include both active genes as well as poised, silent developmental genes [6–8,14]. The poised genes harbor H3K27me3, a repressive mark catalyzed by polycomb repressive complex 2 (PRC2) and dramatically less paused RNAPII [7,8,14,22]. Thus, how H2A.Z functions at the +1 nucleosome to mediate these contrasting transcriptional outcomes will likely be influenced by other factors.
Figure 1. H2A.Z nucleosome composition at promoters influences nucleosome stability and transcriptional state.
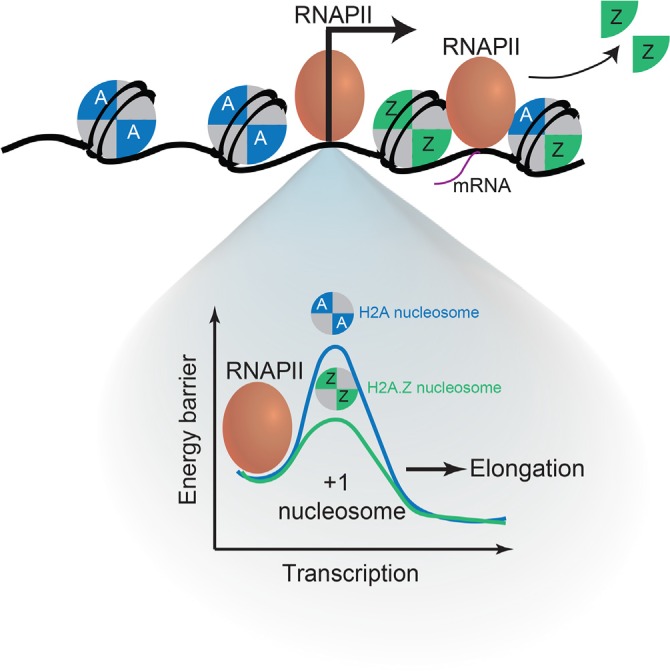
(Top) The transition from homotypic to heterotypic H2A.Z nucleosomes can regulate gene activation through regulation of transcription elongation. (Bottom) H2A.Z nucleosomes can impact the stability of the +1 nucleosome and RNA polymerase II progression.
H2A.Z-associated post-translational modifications correlate with transcriptional outcome
H2A.Z is highly conserved among eukaryotes and ubiquitously expressed across cell types and its replication-independent incorporation is critical for regulating diverse transcriptional outcomes at a large number of RNAPII-regulated promoters. Promoter nucleosome stability and dynamics can be regulated by histone PTMs. Similar to major type histones, variants are also subject to diverse PTMs which are thought to function to recruit downstream effectors, such as histone writers, readers, or erasers to target genes [23,24]. For example, amino-terminal acetylation of H2A.Z (acH2A.Z) is strongly enriched at the 5’ end of active gene promoters [8,14,25,26]. Whereas in Saccharomyces cerevisiae, H2A.Z is acetylated by the ESA1 and NuA4 complexes [27,28], their mammalian counterparts have not been clearly defined. Recent studies showed that the bromodomain-containing protein Brd2 preferentially associates with H2A.Z-containing nucleosomes at active genes and that Brd2 recruitment is necessary for androgen receptor-regulated gene activation [29] (Figure 2A). Brd2 has a higher affinity for H2A.Z nucleosomes compared to canonical H2A, particularly in conjunction with nucleosomes that also harbor H4 amino-terminal acetylation in vitro. H4 acetylation has been implicated in decondensing higher-order chromatin structures and in mediating an active chromatin state [30], suggesting that H4 acetylation marks the transition from a repressed state to an active chromatin state. However, whether H4ac is sufficient or whether additional mechanisms are necessary to drive Brd2 recruitment to target promoters in vivo is not clear. Genome-wide localization studies revealed that acH2A.Z is enriched at activated prostate cancer genes in androgen-sensitive human prostate cancer cells (LNCaP) [26]. Given that bromodomains are recruited to sites of action by recognizing acetylated lysines [31], acH2A.Z may be an additional determinant for Brd2 recruitment at these regions. More broadly, Brd2 is a member of the BET (bromodomain and extra terminal domain) family of proteins that also includes Brd4, which has critical roles in gene activation in ESCs and in cancer [32–36]. Thus, it will be of interest to determine whether acH2A.Z facilitates recruitment of BET proteins during gene activation in vivo.
Figure 2. H2A.Z regulates gene induction.
(A) In response to androgen receptor (AR) signaling, H2A.Z allows binding of AR to its response elements, at both promoters and enhancers, and along with acH4 recruits Brd2 and RNA polymerase II to mediate gene activation. (B) H2A.Z is enriched at both active and silent, poised genes in embryonic stem cells. The effect of H2A.Z on transcription depends on the balance of both activating and repressive histone post-translational modifications as well as H2A.Z-specific deposition and removal complexes in response to cellular cues.
In addition to active genes, H2A.Z is enriched at silent genes that are poised for activation in ESCs. In mammals, H2A.Z can be ubiquitinated at K120, K121, and K125 residues by the E3 ligase activity of the PRC1 component Ring1b, suggesting a repressive role of this H2A.Z PTM [14,37]. Consistent with this idea, H2A.Z monoubiquitination (H2A.Zub) appears to demarcate facultative heterochromatin [37]. Interestingly, loss of the H2A.Z deubiquitinating enzyme USP10 results in a failure of androgen-receptor target gene activation, suggesting that the removal of this modification is necessary for gene induction [38]. Recent reports showed that PRC1-mediated ubiquitination of core H2A (H2Aub) impedes RNAPII recruitment at bivalent genes [39] and may facilitate PRC2 targeting and the establishment of polycomb domain formation [40,41]. Moreover, H2Aub stimulates PRC2 recruitment to chromatinized templates and catalysis of H3K27me3 in vitro, suggesting that H2Aub is a critical downstream effector of polycomb silencing [42]. Given that PRC1 catalyzes the ubiquitination of both H2A and H2A.Z [14,39], we speculate that H2A.Zub is also a key determinant for regulating RNAPII recruitment and polycomb domain formation.
In mouse ESCs, acH2A.Z and H2A.Zub are co-enriched with H3K27me3 nucleosomes, indicating that dually modified H2A.Z (acH2A.Zub) is present at poised developmental genes [14] (Figure 2B). One attractive model is that H2A.Zub contributes to the recruitment of PRC2 and reduces nucleosome accessibility (e.g. dimer loss) at the +1 nucleosome, whereas acH2A.Z promotes RNAPII progression perhaps through recruitment of BET family members, known regulators of polymerase pause release [43,44]. Notably, H2A.Z facilitates access to both active and repressive complexes to chromatin in ESCs to regulate the balance between self-renewal and differentiation [8]. Thus, the balance of acH2A.Z and H2A.Zub at promoters may be critical for regulating the induction of gene expression programs in response to developmental cues.
Regulation of H2A.Z incorporation is critical for gene regulation
Although histone PTMs provide context-dependent signals to regulate H2A.Z function at promoters, dissecting how this variant is targeted to discrete genomic sites is also important for understanding its regulatory roles. The exchange of H2A for H2A.Z is catalyzed by ATP-dependent complexes in a replication-independent manner in all eukaryotes, namely the SWR1 complex (SRCAP in mouse and human) as well as by p400/Tip60 in higher eukaryotes [45,46]. Notably, INO80, an ATP-dependent remodeler known for its role in DNA repair, has been implicated in transcription-dependent removal of H2A.Z in yeast [47,48]. Whether INO80 functions similarly in higher eukaryotes is yet to be determined. Histone chaperones are additional factors that regulate histone variant incorporation [49]. For example, the metazoan-specific histone chaperone, ANP32E (acidic nuclear phosphoprotein 32 kDa E), which is a member of the p400 complex, regulates variant incorporation by facilitating the removal of H2A.Z dimers as evidenced by increased accumulation of H2A.Z at promoters and enhancers in ANP32E knockout cells [50,51]. Together, these studies suggest that the interplay between H2A.Z-specific ATP-dependent remodelers and histone chaperones is crucial for regulating H2A.Z incorporation.
Until recently, how H2A.Z exchange complexes are targeted to specific sites remained largely unknown. Evidence now suggests that SWR1 and INO80 complexes may recognize NDRs to remodel H2A.Z at the +1 nucleosome by selective positioning of distinct components of the remodeling complex around the NDR [52]. Biochemical analyses suggest that SRCAP and ANP32E recognize specific features of H2A.Z that are divergent from core H2A, such as the C-terminal α-helix to facilitate the exchange reaction [50,53], providing an additional mechanism for how remodelers distinguish between the two histones. Recent work also highlights roles for histone PTMs in proper histone exchange [46,48]. For example, in contrast to its well-documented role in H2A.Z deposition, yeast SRCAP (SWR-C) displays altered substrate specificity in the presence of H3K56ac, leading to the removal of H2A.Z dimers from chromatin [48]. In human ESCs, H3K56ac is enriched at both active and inactive genes that largely overlap targets of the core pluripotency TFs OCT4, SOX2, and NANOG [54]. Notably, H3K56ac is redistributed to developmental genes that are activated in response to retinoic acid during ESC differentiation. Thus, H3K56ac and H2A.Z may cross-talk to regulate H2A.Z dynamics and gene expression across eukaryotes.
In addition to histone PTMs, H2A.Z incorporation appears to be influenced by DNA methylation levels. Studies in both plants and mammals suggest that H2A.Z and DNA methylation are mutually antagonistic at promoters and within gene bodies [55,56]. Consistent with this idea, 5-aza 2′-deoxycytidine-induced DNA demethylation stimulates SRCAP-mediated H2A.Z incorporation, facilitating nucleosome depletion and gene activation in cancer cells [57]. Although these studies point to a model whereby H2A.Z incorporation is targeted to discrete regions of the genome by a variety of mechanisms, evidence also suggests that H2A.Z can be randomly incorporated at low levels and that the removal of H2A.Z is a key event for regulating chromatin states [58,59]. Thus, a balance between the targeted deposition and removal of H2A.Z is likely critical for maintaining proper chromatin states.
H2A.Z nucleosome composition affects chromatin dynamics
Modulating the number of H2A.Z copies in a nucleosome can also have consequences on nucleosome structure and function. Early structural studies suggested that, owing to steric clashes between the L1 loops of H2A.Z and H2A, formation of a heterotypic H2A.Z nucleosome is unlikely [60], but in vitro and in vivo evidence now indicates that H2A.Z can form both heterotypic and homotypic nucleosomes [16,61]. In vitro biochemical analyses showed that two copies of H2A.Z (homotypic) result in a more stable nucleosome that is refractory to RNAPII progression [62,63]. However, homotypic H2A.Z is enriched at the +1 nucleosome in Drosophila, which is thought to decrease the energy barrier to RNAPII progression, and is depleted downstream of paused polymerase [16]. In mouse trophoblast cells, H2A.Z appears to be redistributed to heterochromatin (for example, telomeres) during G2/M, resulting in a shift from homotypic to heterotypic H2A.Z nucleosomes and to an expanded NDR at the TSS of active H2A.Z genes [64]. Surprisingly, this transition from homotypic to heterotypic H2A.Z nucleosomes during the cell cycle does not appear to correlate with cell cycle-dependent transcriptional changes. Thus, the consequence of H2A.Z composition on transcription is highly complex and likely depends on its levels at the TSS relative to gene bodies as well as the presence of histone modifications or other histone variants or both.
The incorporation of other histone variants with H2A.Z can alter the functional properties of nucleosomes. H3.3 differs from major type H3 by only four or five amino acids in metazoans yet displays distinct regulation and biochemical properties. In mammalian cells, double-variant nucleosomes containing H2A.Z and H3.3 mark regions of dynamic chromatin regulation and are highly salt-labile [65–67]. Moreover, in yeast, where non-centromeric H3 is most similar to vertebrate H3.3, in vitro reconstituted nucleosomes display release of H2A.Z dimers in low salt [68,69]. Together, these findings suggest a conserved mechanism in which H3.3 further regulates the stability of the H2A.Z nucleosome. Notably, whereas H2A.Z facilitates intranucleosomal folding, H3.3 counteracts the H2A.Z-mediated compaction of nucleosomal arrays in vitro [70,71] and can relieve the transcription repression caused by H2A.Z-containing chromatin [71]. In ESCs, H3.3 depletion results in reduced nucleosome turnover and an increase in PRC2 enrichment [6,7,72,73]. Conversely, mutation of the C-terminal H2A.Z acidic patch to resemble core H2A leads to an increase in H2A.Z dynamics, de-repression of poised developmental genes, and increased H3.3 enrichment in ESCs [7]. Together, these studies indicate that H3.3 cooperates with H2A.Z to regulate nucleosome stability, chromatin accessibility, and transcriptional output. Future investigations to determine how the balance between H3.3 and H2A.Z is regulated at specific genomic locations are needed to understand how these variants regulate global gene expression programs.
H2A.Z is a critical binding platform for pioneer TFs
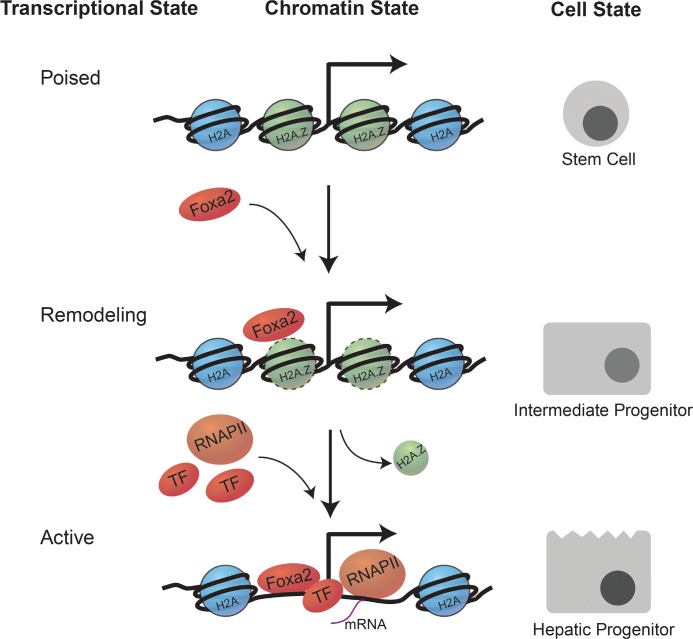
Given that TF binding is influenced by nucleosome density and histone PTMs [69,74] and that H2A.Z exhibits strong genetic interactions with TFs in yeast [74], it is likely that H2A.Z also coordinates transcriptional responses by modulating TF accessibility. Several recent studies in mammals demonstrated that H2A.Z nucleosomes are necessary for the recruitment of pioneer TFs (Figure 3). Unlike classic TFs, pioneer TFs bind nucleosomal DNA and remain bound to their sites through mitosis, providing a level of epigenetic memory [75]. Foxa2 is an example of a pioneer TF that has key roles in development and transcriptional activation. In ESCs, H2A.Z is required for the recruitment of Foxa2 to promoters of genes activated during early endoderm differentiation [76]. Moreover, the binding of Foxa2 to H2A.Z nucleosomes promotes the recruitment of SWI/SNF and INO80 complexes, resulting in H2A.Z removal and nucleosome depletion, establishing a platform for the binding of other TFs. On the other hand, loss of either H2A.Z or Foxa2 impairs gene induction and differentiation. This work adds another layer to how H2A.Z mechanistically regulates gene expression.
Figure 3. H2A.Z acts a scaffold for binding of chromatin remodelers and pioneer transcription factors.
During endodermal differentiation, the pioneer transcription factor Foxa2 binds to H2A.Z nucleosomes, leading to the recruitment of chromatin remodelers, nucleosome depletion, transcription factor binding, and subsequent gene activation.
In addition to its requirement for regulating developmental transitions, H2A.Z is necessary for the expression of circadian-regulated genes by modulating TF binding. CLOCK and BMAL1 are pioneer TFs that bind to circadian clock-regulated genes to induce their expression in a temporally regulated manner [77]. Like binding of Foxa2, binding of these TFs promotes gene activation and the recruitment of RNAPII. Interestingly, CLOCK:BMAL1 binding strongly correlates with cyclical changes in H2A.Z occupancy and nucleosome depletion [78]. Using mouse livers at different time points in the light/dark cycle, investigators demonstrated that H2A.Z-enriched nucleosomes flank CLOCK-binding sites at both promoters and intergenic regulatory regions. Notably, H2A.Z levels oscillate on the basis of CLOCK:BMAL1 binding. The cyclical binding of CLOCK:BMAL1 induces nucleosome depletion at promoters, allowing additional TFs to gain access to DNA. Loss of BMAL1 in mice results in a decreased H2A.Z levels and a higher nucleosome occupancy at CLOCK-binding sites as well as loss of circadian regulation, suggesting that H2A.Z is necessary to maintain these sites in a highly plastic state. Collectively, these studies reveal that H2A.Z provides a crucial binding platform for pioneer TFs to facilitate precise regulation of gene expression programs.
Concluding remarks
Since H2A.Z was first identified over 30 years ago, studies have begun to reveal the many layers of regulation and complex functions of H2A.Z in modulating chromatin dynamics and gene expression. Recent studies have uncovered many players involved in H2A.Z-mediated transcriptional regulation, but how H2A.Z translates upstream signals into diverse transcriptional outcomes is still largely unknown. Although we have focused largely on the roles of H2A.Z at promoters, H2A.Z also appears to be enriched at a small subset of enhancers; however, its roles in enhancer regulation are poorly understood [7,8,14]. Overall levels of H2A.Z are lower at enhancers compared with promoters, whereas H3.3 displays the opposite trend [79]. Thus, the dynamic interplay between H2A.Z and H3.3 at enhancers and promoters may facilitate gene regulation. Consistent with this idea, recent evidence supports the role of H2A.Z in mediating higher-order chromatin interactions between promoters and enhancers [80]. Additional mechanistic studies are needed to understand how histone variants contribute to these critical regulatory interactions.
Insights into the function of H2A.Z in gene induction are critical for understanding how H2A.Z functions in other processes, such as DNA repair and genomic stability. For example, p400-mediated H2A.Z incorporation during double-strand break (DSB) repair stimulates an open chromatin conformation, resulting in the formation of an efficient chromatin template for DSB repair, albeit through different effectors [81]. Interestingly, H2A.Z acetylation and ubiquitylation are also hallmarks of chromatin at DSBs, suggesting a general model by which H2A.Z modulates chromatin in diverse DNA-mediated processes. Given that H2A.Z has been implicated in the activation of estrogen- and androgen-responsive genes in models of breast and prostate cancer [11,38,82,83], continued mechanistic studies into H2A.Z function will be critical to fully understand how H2A.Z contributes to development and disease.
Acknowledgments
We thank members of the Boyer lab, especially Lauren Surface and Joe Wamstad for insightful discussions and critical evaluation of the manuscript.
Abbreviations
- ANP32E
acidic nuclear phosphoprotein 32 kDa E
- BET
bromodomain and extra terminal domain
- DSB
double-strand break
- ESC
embryonic stem cell
- H2Aub
H2A ubiquitination
- H2A.Zub
H2A.Z ubiquitination
- acH2A.Z
acetylated H2A.Z
- NDR
nucleosome-depleted region
- PRC
polycomb repressive complex
- PTM
post-translational modification
- RNAPII
RNA polymerase II
- TF
transcription factor
- TSS
transcription start site
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at: http://f1000.com/prime/reports/b/7/1
*The contribution of these two authors is equal.
References
- 1.Sarma K, Reinberg D. Histone variants meet their match. Nature reviews. Molecular cell biology. 2005;6:139–49. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 2.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes & development. 2014;28:672–82. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skene PJ, Henikoff S. Histone variants in pluripotency and disease. Development (Cambridge, England) 2013;140:2513–24. doi: 10.1242/dev.091439. [DOI] [PubMed] [Google Scholar]
- 4.Maze I, Noh K, Soshnev AA, Allis CD. Every amino acid matters: essential contributions of histone variants to mammalian development and disease. Nature reviews Genetics. 2014;15:259–71. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bönisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic acids research. 2012;40:10719–41. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–61. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1147365
- 7.Subramanian V, Mazumder A, Surface LE, Butty VL, Fields PA, Alwan A, Torrey L, Thai KK, Levine SS, Bathe M, Boyer LA. H2A.Z acidic patch couples chromatin dynamics to regulation of gene expression programs during ESC differentiation. PLoS genetics. 2013;9:e1003725. doi: 10.1371/journal.pgen.1003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell stem cell. 2013;12:180–92. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutcliffe EL, Parish IA, He YQ, Juelich T, Tierney ML, Rangasamy D, Milburn PJ, Parish CR, Tremethick DJ, Rao S. Dynamic histone variant exchange accompanies gene induction in T cells. Molecular and cellular biology. 2009;29:1972–86. doi: 10.1128/MCB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schones DE, Cui K, Cuddapah S, Roh T, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1107027
- 11.Dryhurst D, McMullen B, Fazli L, Rennie PS, Ausió J. Histone H2A.Z prepares the prostate specific antigen (PSA) gene for androgen receptor-mediated transcription and is upregulated in a model of prostate cancer progression. Cancer letters. 2012;315:38–47. doi: 10.1016/j.canlet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Cuadrado A, Corrado N, Perdiguero E, Lafarga V, Muñoz-Canoves P, Nebreda AR. Essential role of p18Hamlet/SRCAP-mediated histone H2A.Z chromatin incorporation in muscle differentiation. The EMBO journal. 2010;29:2014–25. doi: 10.1038/emboj.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure (London, England:1993) 2008;16:166–79. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome biology. 2012;13:R85. doi: 10.1186/gb-2012-13-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS biology. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nature structural & molecular biology. 2010;17:1500–7. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13491999
- 18.Nock A, Ascano JM, Barrero MJ, Malik S. Mediator-regulated transcription through the +1 nucleosome. Molecular cell. 2012;48:837–48. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Molecular cell. 2014;53:819–30. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718304563
- 20.Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, Glaser RL. Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. The Journal of biological chemistry. 2000;275:23267–72. doi: 10.1074/jbc.M910206199. [DOI] [PubMed] [Google Scholar]
- 21.Madigan JP, Chotkowski HL, Glaser RL. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic acids research. 2002;30:3698–705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes & development. 2011;25:742–54. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/9653956
- 23.Sevilla A, Binda O. Post-translational modifications of the histone variant H2AZ. Stem cell research. 2014;12:289–95. doi: 10.1016/j.scr.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell research. 2011;21:564–78. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruce K, Myers FA, Mantouvalou E, Lefevre P, Greaves I, Bonifer C, Tremethick DJ, Thorne AW, Crane-Robinson C. The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic acids research. 2005;33:5633–9. doi: 10.1093/nar/gki874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdés-Mora F, Song JZ, Statham AL, Strbenac D, Robinson MD, Nair SS, Patterson KI, Tremethick DJ, Stirzaker C, Clark SJ. Acetylation of H2A.Z is a key epigenetic modification associated with gene deregulation and epigenetic remodeling in cancer. Genome research. 2012;22:307–21. doi: 10.1101/gr.118919.110. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717957008
- 27.Keogh M, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes & development. 2006;20:660–5. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta M, Braberg H, Wang S, Lozsa A, Shales M, Solache A, Krogan NJ, Keogh M. Individual lysine acetylations on the N terminus of Saccharomyces cerevisiae H2A.Z are highly but not differentially regulated. The Journal of biological chemistry. 2010;285:39855–65. doi: 10.1074/jbc.M110.185967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Draker R, Ng MK, Sarcinella E, Ignatchenko V, Kislinger T, Cheung P. A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS genetics. 2012;8:e1003047. doi: 10.1371/journal.pgen.1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/720224432
- 30.Shia W, Pattenden SG, Workman JL. Histone H4 lysine 16 acetylation breaks the genome's silence. Genome biology. 2006;7:217. doi: 10.1186/gb-2006-7-5-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denis GV. Bromodomain coactivators in cancer, obesity, type 2 diabetes, and inflammation. Discovery medicine. 2010;10:489–99. [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Walker SR, Nelson EA, Cerulli R, Xiang M, Toniolo PA, Qi J, Stone RM, Wadleigh M, Bradner JE, Frank DA. Targeting STAT5 in hematologic malignancies through inhibition of the bromodomain and extra-terminal (BET) bromodomain protein BRD2. Molecular cancer therapeutics. 2014;13:1194–205. doi: 10.1158/1535-7163.MCT-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Molecular cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1103930
- 34.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nature reviews. Cancer. 2012;12:465–77. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718140971
- 36.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718000122
- 37.Sarcinella E, Zuzarte PC, Lau Priscilla NI, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Molecular and cellular biology. 2007;27:6457–68. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1089782
- 38.Draker R, Sarcinella E, Cheung P. USP10 deubiquitylates the histone variant H2A.Z and both are required for androgen receptor-mediated gene activation. Nucleic acids research. 2011;39:3529–42. doi: 10.1093/nar/gkq1352. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/719377177
- 39.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nature cell biology. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1098169
- 40.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen Lars LP, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–59. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718415419
- 41.Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, Marco V de, Elderkin S, Koseki H, Klose R, Heger A, Brockdorff N. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell reports. 2014;7:1456–70. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718415386
- 42.Kalb R, Latwiel S, Baymaz HI, Jansen Pascal WTC, Müller CW, Vermeulen M, Müller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nature structural & molecular biology. 2014;21:569–71. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718397247
- 43.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Molecular and cellular biology. 2013;33:2497–507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718000706
- 44.Anand P, Brown JD, Lin CY, Qi J, Zhang R, Artero PC, Alaiti MA, Bullard J, Alazem K, Margulies KB, Cappola TP, Lemieux M, Plutzky J, Bradner JE, Haldar SM. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–82. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718058296
- 45.Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–7. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 46.Choi J, Heo K, An W. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic acids research. 2009;37:5993–6007. doi: 10.1093/nar/gkp660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–13. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/10351956
- 48.Watanabe S, Radman-Livaja M, Rando OJ, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science (New York, N.Y.) 2013;340:195–9. doi: 10.1126/science.1229758. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717999660
- 49.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nature structural & molecular biology. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Obri A, Ouararhni K, Papin C, Diebold M, Padmanabhan K, Marek M, Stoll I, Roy L, Reilly PT, Mak TW, Dimitrov S, Romier C, Hamiche A. ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature. 2014;505:648–53. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718252262
- 51.Mao Z, Pan L, Wang W, Sun J, Shan S, Dong Q, Liang X, Dai L, Ding X, Chen S, Zhang Z, Zhu B, Zhou Z. Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z. Cell research. 2014;24:389–99. doi: 10.1038/cr.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718310243
- 52.Yen K, Vinayachandran V, Pugh BF. SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell. 2013;154:1246–56. doi: 10.1016/j.cell.2013.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718110119
- 53.Hong J, Feng H, Wang F, Ranjan A, Chen J, Jiang J, Ghirlando R, Xiao TS, Wu C, Bai Y. The catalytic subunit of the SWR1 remodeler is a histone chaperone for the H2A.Z-H2B dimer. Molecular cell. 2014;53:498–505. doi: 10.1016/j.molcel.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718270679
- 54.Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, Grunstein M. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Molecular cell. 2009;33:417–27. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/722242022
- 55.Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–9. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1123983
- 56.Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome research. 2010;20:1383–90. doi: 10.1101/gr.106542.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang X, Noushmehr H, Han H, Andreu-Vieyra C, Liang G, Jones PA. Gene reactivation by 5-aza-2’-deoxycytidine-induced demethylation requires SRCAP-mediated H2A.Z insertion to establish nucleosome depleted regions. PLoS genetics. 2012;8:e1002604. doi: 10.1371/journal.pgen.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/720224769
- 58.Hardy S, Robert F. Random deposition of histone variants: A cellular mistake or a novel regulatory mechanism? Epigenetics: official journal of the DNA Methylation Society. 2010;5:368–72. doi: 10.4161/epi.5.5.11787. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/720580194
- 59.Hardy S, Jacques P, Gévry N, Forest A, Fortin M, Laflamme L, Gaudreau L, Robert F. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS genetics. 2009;5:e1000687. doi: 10.1371/journal.pgen.1000687. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1165439
- 60.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nature structural biology. 2000;7:1121–4. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 61.Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell. 2010;143:725–36. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/7830956
- 62.Park Y, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. The Journal of biological chemistry. 2004;279:24274–82. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 63.Thakar A, Gupta P, McAllister WT, Zlatanova J. Histone variant H2A.Z inhibits transcription in reconstituted nucleosomes. Biochemistry. 2010;49:4018–26. doi: 10.1021/bi1001618. [DOI] [PubMed] [Google Scholar]
- 64.Nekrasov M, Amrichova J, Parker BJ, Soboleva TA, Jack C, Williams R, Huttley GA, Tremethick DJ. Histone H2A.Z inheritance during the cell cycle and its impact on promoter organization and dynamics. Nature structural & molecular biology. 2012;19:1076–83. doi: 10.1038/nsmb.2424. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/722615061
- 65.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes & development. 2007;21:1519–29. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1088008
- 66.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nature genetics. 2009;41:941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shu H, Nakamura M, Siretskiy A, Borghi L, Moraes I, Wildhaber T, Gruissem W, Hennig L. Arabidopsis replacement histone variant H3.3 occupies promoters of regulated genes. Genome biology. 2014;15:R62. doi: 10.1186/gb-2014-15-4-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Molecular cell. 2002;9:1191–200. doi: 10.1016/S1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1007380
- 69.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–31. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1029195
- 70.Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nature structural biology. 2002;9:172–6. doi: 10.1038/nsb0402-316b. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1004759
- 71.Chen P, Zhao J, Wang Y, Wang M, Long H, Liang D, Huang L, Wen Z, Li W, Li X, Feng H, Zhao H, Zhu P, Li M, Wang Q, Li G. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes & development. 2013;27:2109–24. doi: 10.1101/gad.222174.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718118332
- 72.Goldberg AD, Banaszynski LA, Noh K, Lewis PW, Elsaesser SJ, Stadler S, Dewell S, Law M, Guo X, Li X, Wen D, Chapgier A, DeKelver RC, Miller JC, Lee Y, Boydston EA, Holmes MC, Gregory PD, Greally JM, Rafii S, Yang C, Scambler PJ, Garrick D, Gibbons RJ, Higgs DR, Cristea IM, Urnov FD, Zheng D, Allis CD. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsässer SJ, Chapgier A, Goldberg AD, Canaani E, Rafii S, Zheng D, Allis CD. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–20. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wan Y, Saleem RA, Ratushny AV, Roda O, Smith JJ, Lin C, Chiang J, Aitchison JD. Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Molecular and cellular biology. 2009;29:2346–58. doi: 10.1128/MCB.01233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/719781868
- 75.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes & development. 2011;25:2227–41. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–16. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718685256
- 77.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annual review of neuroscience. 2012;35:445–62. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Menet JS, Pescatore S, Rosbash M. CLOCK:BMAL1 is a pioneer-like transcription factor. Genes & development. 2014;28:8–13. doi: 10.1101/gad.228536.113. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718229803
- 79.Chen P, Wang Y, Li G. Dynamics of histone variant H3.3 and its coregulation with H2A.Z at enhancers and promoters. Nucleus (Austin, Tex.) 2014;5:21–7. doi: 10.4161/nucl.28067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dalvai M, Bellucci L, Fleury L, Lavigne A, Moutahir F, Bystricky K. H2A.Z-dependent crosstalk between enhancer and promoter regulates cyclin D1 expression. Oncogene. 2013;32:4243–51. doi: 10.1038/onc.2012.442. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718099455
- 81.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Molecular cell. 2012;48:723–33. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717988297
- 82.Dryhurst D, Ausió J. Histone H2A.Z deregulation in prostate cancer. Cause or effect? Cancer metastasis reviews. 2014;33:429–39. doi: 10.1007/s10555-013-9486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gévry N, Hardy S, Jacques P, Laflamme L, Svotelis A, Robert F, Gaudreau L. Histone H2A.Z is essential for estrogen receptor signaling. Genes & development. 2009;23:1522–33. doi: 10.1101/gad.1787109. [DOI] [PMC free article] [PubMed] [Google Scholar]