Abstract
The Cancer Genome Atlas profiled 279 head and neck squamous cell carcinomas (HNSCCs) to provide a comprehensive landscape of somatic genomic alterations. Here we show that human-papillomavirus-associated tumours are dominated by helical domain mutations of the oncogene PIK3CA, novel alterations involving loss of TRAF3, and amplification of the cell cycle gene E2F1. Smoking-related HNSCCs demonstrate near universal loss-of-function TP53 mutations and CDKN2A inactivation with frequent copy number alterations including amplification of 3q26/28 and 11q13/22. A subgroup of oral cavity tumours with favourable clinical outcomes displayed infrequent copy number alterations in conjunction with activating mutations of HRAS or PIK3CA, coupled with inactivating mutations of CASP8, NOTCH1 and TP53. Other distinct subgroups contained loss-of-function alterations of the chromatin modifier NSD1, WNT pathway genes AJUBA and FAT1, and activation of oxidative stress factor NFE2L2, mainly in laryngeal tumours. Therapeutic candidate alterations were identified in most HNSCCs.
Supplementary information
The online version of this article (doi:10.1038/nature14129) contains supplementary material, which is available to authorized users.
Subject terms: Diseases, Head and neck cancer
The Cancer Genome Atlas presents an integrative genome-wide analysis of genetic alterations in 279 head and neck squamous cell carcinomas (HNSCCs), which are classified by human papillomavirus (HPV) status; alterations in EGFR, FGFR, PIK3CA and cyclin-dependent kinases are shown to represent candidate targets for therapeutic intervention in most HNSCCs.
Supplementary information
The online version of this article (doi:10.1038/nature14129) contains supplementary material, which is available to authorized users.
Large-scale analysis of head and neck cancers
Squamous cell head and neck cancer is one of the most common and deadly cancers. Despite initial responses to combinations of surgery, radiation and chemotherapy, approximately half of all tumours recur, usually within two years of initial diagnosis. Molecular markers and targeted therapies have had little impact on this disease to date. Here, The Cancer Genome Atlas team presents a detailed genome-wide overview of alterations and highlights critical genetic events of potential biological and clinical significance in head and neck squamous cell carcinomas (HNSCCs) with different human papillomavirus status. Mutational profiles reveal distinct subgroups of HNSCCs. Mutations in EGFR, FGFRs, PIK3CA and cyclin-dependent kinases represent candidate targets for therapeutic intervention in the majority of HNSCCs.
Supplementary information
The online version of this article (doi:10.1038/nature14129) contains supplementary material, which is available to authorized users.
Main
HNSCCs affect ∼600,000 patients per year worldwide1. They are characterized by phenotypic, aetiological, biological and clinical heterogeneity. Smoking is implicated in the rise of HNSCC in developing countries, and the role of human papillomavirus (HPV) is emerging as an important factor in the rise of oropharyngeal tumours affecting non-smokers in developed countries2. Despite surgery, radiation and chemotherapy, approximately half of all patients will die of the disease. Risk stratification for HNSCC is by anatomic site, stage and histological characteristics of the tumour. Except for HPV status, numerous molecular and clinical risk factors that have been investigated have limited clinical utility.
Published genome-wide profiling studies of HNSCC3,4 are limited to single platforms. To generate an integrated genomic annotation of molecular alterations in HNSCC, The Cancer Genome Atlas (TCGA) has undertaken a comprehensive multi-platform characterization of 500 tumours with the a priori hypothesis of detecting somatic variants present in at least 5% of samples. Here, we report the results for analyses from the first 279 patients with complete data.
Samples and clinical data
The cohort consists primarily of tumours from the oral cavity (n = 172 out of 279, 62%), oropharynx (n = 33 out of 279, 12%), and laryngeal sites (n = 72 out of 279, 26%) (Supplementary Information section 1, Supplementary Table 1.1 and Supplementary Data 1.1). Most patients were male (n = 203 out of 279, 73%) and heavy smokers (mean pack years = 51). Samples were classified as HPV-positive using an empiric definition of >1,000 mapped RNA sequencing (RNA-Seq) reads, primarily aligning to viral genes E6 and E7 (Supplementary Information section 1.2 and Supplementary Fig. 1.1). The HPV status by mapping of RNA-Seq reads was concordant with the genomic, sequencing and molecular data, and indicated that 36 tumours were HPV(+) and 243 were HPV(−) (Supplementary Information section 1.2, Supplementary Fig. 1.1 and Supplementary Data 1.2). Of 33 oropharyngeal tumours, 64% were positive for HPV, compared to 6% of 246 non-oropharyngeal tumours. Molecular HPV signatures were identified using microRNA (miRNA), DNA methylation, gene expression and somatic nucleotide substitutions (Supplementary Information section 1.2 and Supplementary Figs 1.1–1.3). HPV(+) tumours exhibited infrequent mutations in TP53 or genetic alterations in CDKN2A. We evaluated outcome by site, stage, HPV status, molecular subtypes and putative biomarkers (Supplementary Information section 1.3 and Supplementary Figs 1.4 and 1.5). HPV(+) and interestingly patients with HPV(−), TP53 wild-type tumours demonstrated favourable outcomes compared to TP53 mutants and 11q13/CCND1-amplified tumours.
DNA and RNA structural alterations
Most tumours demonstrated copy number alterations (CNAs) including losses of 3p and 8p, and gains of 3q, 5p and 8q chromosomal regions (Fig. 1a, Supplementary Fig. 2.1 and Supplementary Information section 2) resembling lung squamous cell carcinomas (LUSCs)5 (Fig. 1a and Supplementary Figs 2.1 and 2.2). HNSCC genomes showed high instability with a mean of 141 CNAs (amplifications or deletions) from microarray data and 62 structural aberrations (chromosomal fusions) per tumour by ‘high coverage’ whole-genome sequencing (n = 29) (Supplementary Information section 2.2). We observed 39 regions of recurrent copy number loss and 23 regions of recurrent copy number gain (q < 0.1, Supplementary Data 2.1 and 2.2). Both HPV(+) and (−) tumours contained recurrent focal amplifications for 3q26/28, a region involving squamous lineage transcription factors TP63 and SOX2 and the oncogene PIK3CA (Fig. 1b and Supplementary Fig. 2.3).
Figure 1. DNA copy number alterations.
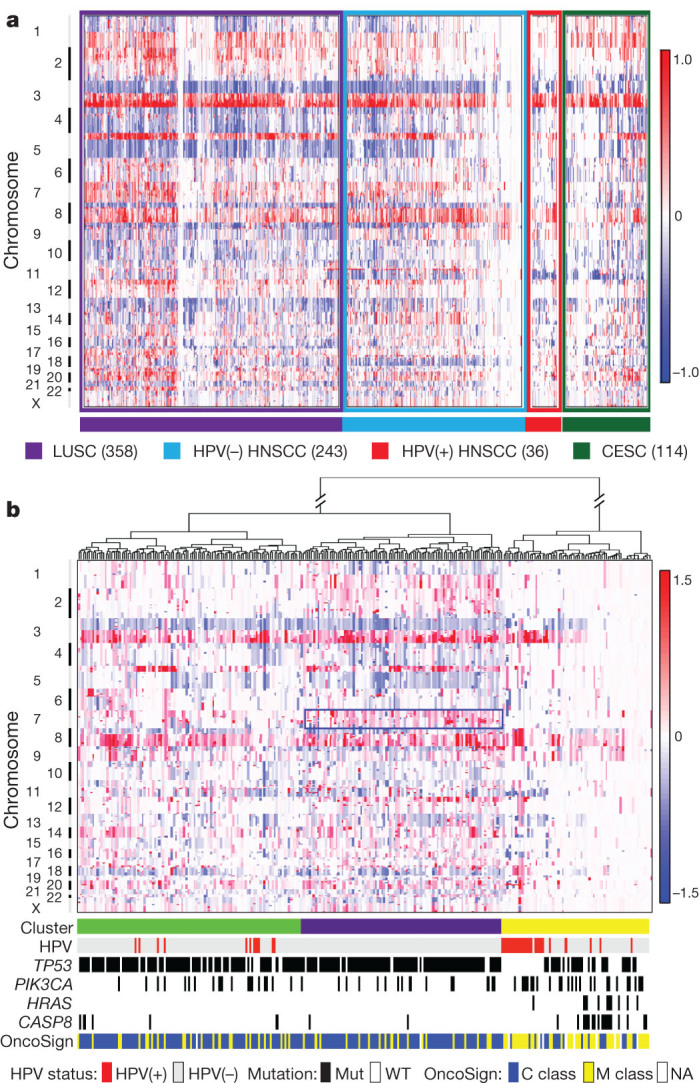
a, Copy number alterations by anatomic site and HPV status for squamous cancers. Lung squamous cell carcinoma (LUSC, n = 358) and cervical squamous cell carcinoma (CESC, n = 114). b, Unsupervised analysis of copy number alteration of HNSCC (n = 279) with associated characteristics. The rectangle indicates chromosome 7 amplifications in the purple cluster. NA, not available.
HPV(+) tumours were distinguished by novel recurrent deletions (n = 5 out of 36, 14%) and truncating mutations (n = 3 out of 36, 8%) of TNF receptor-associated factor 3 (TRAF3) (Supplementary Figs 2.3 and 2.4, and Supplementary Data 2.1). TRAF3 is implicated in innate and acquired anti-viral responses6 including Epstein–Barr, HPV and human immunodeficiency virus (HIV)7,8,9, while loss promotes aberrant NF-κB signalling10. Although TRAF3 inactivation has been reported in haematological malignancies and nasopharyngeal carcinoma11,12, to our knowledge this is the first evidence linking TRAF3 to HPV-associated carcinomas. HPV(+) tumours were also notable for focal amplification of E2F1 and an intact 9p21.3 region containing the CDKN2A gene commonly deleted in HPV(−) tumours.
HPV(−) tumours featured novel co-amplifications of 11q13 (CCND1, FADD and CTTN) and 11q22 (BIRC2 and YAP1), which also contain genes implicated in cell death/NF-κB and Hippo pathways. HPV(−) tumours featured novel focal deletions in the nuclear set domain gene (NSD1) and tumour suppressor genes (for example, FAT1, NOTCH1, SMAD4 and CDKN2A; Supplementary Fig. 2.3). Recurrent focal amplifications in receptor tyrosine kinases (for example, EGFR, ERBB2 and FGFR1) also predominated in HPV(−) tumours. Notably, unsupervised clustering analysis of CNAs identified a mutually exclusive subset of predominantly oral cavity tumours with reduced CNAs, a pattern recently described in cancer as ‘M’ class (tumours driven by mutation rather than CNA)13 (Fig. 1b). This subset in particular contained a new three-gene pattern of activating mutations in HRAS, frequently with inactivating CASP8 mutations, and wild-type TP53. We confirmed a previously reported favourable clinical outcome in tumours with few CNAs14. The three-gene constellation of wild-type TP53 with mutant HRAS and CASP8 suggested an alternative tumorigenesis pathway involving RAS and/or alterations in cell death/NF-κB15. Unsupervised analysis also suggested that clustering was a function of chromosome 7 amplification (including the EGFR locus) in a manner that largely excluded HPV(+) tumours.
To detect additional structural alterations, we interrogated whole-genome and RNA-Seq data (Supplementary Information section 3, Supplementary Data 3.1 and Supplementary Fig. 3.1). Known fusion oncogenes reported in solid tumours including those involving the ALK, ROS or RET genes were not observed in HNSCC. Previously reported FGFR3-TACC3 fusions were present in two HPV(+) tumours (Supplementary Fig. 3.2). Only 1 out of 279 patients showed evidence of the type III isoform of EGFR (vIII), previously described in HNSCC16 (Supplementary Fig. 3.3). Although our investigation did not identify additional novel oncogenic fusions, several tumours demonstrated exon 1 of EGFR or FGFR3 fused to non-recurrent partners, suggesting potential promoter swaps for the partner genes (Supplementary Data 3.1). A low prevalence of an alternative MET transcript with skipped exon 14 was identified in two HPV(−) tumours (Supplementary Fig. 3.4); this finding was reported to be an activating event in non-small cell lung cancer17. Structural alterations (homozygous deletions, intra- and inter-chromosomal fusions) were more commonly associated with loss of function in tumour suppressor genes, most prominently CDKN2A (Supplementary Figs 3.5 and 3.6), followed by TP53, RB1, NOTCH1 and FAT1 (Supplementary Figs 3.7–3.9), than with protein-coding fusion events. RNA-Seq data (Supplementary Data 3.3) demonstrated evidence of alternative splicing in genes not previously described in HNSCC including kallikrein 12 (KLK12) (Supplementary Fig. 3.11), as well as genes such as TP63 with known importance in HNSCC (Supplementary Fig. 3.12).
By DNA analysis, most HPV(+) tumours demonstrated clear evidence of host genome integration, usually in a single genomic location per sample and almost always in association with amplifications of the host genome (Supplementary Fig. 3.10 and Supplementary Data 3.2). Interrogation of RNA transcripts confirmed transcription across the viral–human integration locus. However, none of the genes involved were recurrent, suggesting no single driver mechanism related to HPV integration. Similarly, none of the integration sites involved the MYC gene as reported in HPV(+) cell lines18.
Somatic mutations
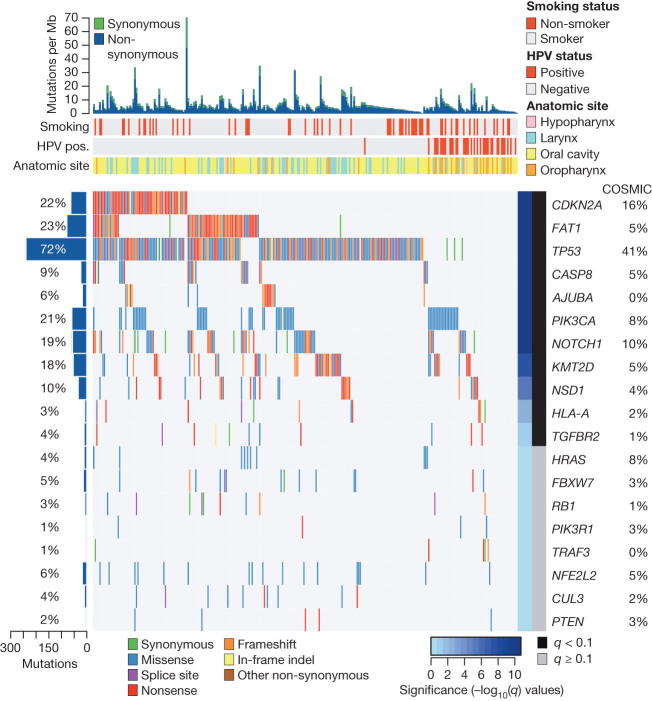
Whole-exome sequencing identified somatically mutated genes, many located in regions of CNAs and annotated in the COSMIC database19 (Fig. 2). The mean sequencing coverage across targeted bases was 95×, with 82% of target bases above 30× coverage. In 279 samples, 12,159 synonymous somatic variants, 37,061 non-synonymous somatic variants, and 2,579 germline single base substitutions from the single nucleotide polymorphism database (dbSNP)20 were detected (Supplementary Information section 4). Targeted re-sequencing of 394 unique regions (Supplementary Fig. 4.1) validated 99% of mutations. Interrogation of RNA for expression of the mutated alleles confirmed the variant in 86% of cases (Supplementary Information section 3.2 and Supplementary Fig. 3.1). In contrast to previous reports, the mutation rates did not differ by HPV status, although transversions at CpG sites were more frequent in HPV(−) tumours and a predominance of TpC mutations were noted in HPV(+) cases3 (Supplementary Fig. 1.1). Mutations were statistically enriched in 11 genes (Fig. 2). Among inactivating mutations (premature termination of the protein by nonsense, frameshift or splice-site mutations), four genes segregated exclusively or predominantly in HPV(−) tumours. Two were associated with cell cycle and survival (CDKN2A (P < 0.01) and TP53 (P < 0.01)) and two were linked to Wnt/β-catenin signalling (FAT1 (P < 0.01) and AJUBA (P = 0.14))21,22. We observed TP53 mutation among HPV(−) samples at higher rates (86%) than have been previously reported19, while only 1 out of 36 HPV(+) cases had a non-synonymous TP53 mutation. Previously unreported somatic mutations and deletions of AJUBA were primarily 5′ inactivating events and clustered missense mutations in the functional LIM domain (Supplementary Fig. 4.2). AJUBA is a centrosomal protein that regulates cell division, vertebrate ciliogenesis and left–right axis determination23. Additionally, AJUBA is subject to EGFR-RAS-MAPK-dependent phosphorylation and implicated in Hippo growth and regeneration pathways conserved from Drosophila to mammals24,25, in ataxia-telangiectasia mutated (ATM) and ATM and Rad-3-related (ATR)-mediated DNA damage response26, and tumour invasion and migration27.
Figure 2. Significantly mutated genes in HNSCC.
Genes (rows) with significantly mutated genes (identified using the MutSigCV algorithim; q < 0.1) ordered by q value; additional genes with trends towards significance are also shown. Samples (columns, n = 279) are arranged to emphasize mutual exclusivity among mutations. Left, mutation percentage in TCGA. Right, mutation percentage in COSMIC (‘upper aerodigestive tract’ tissue). Top, overall number of mutations per megabase. Colour coding indicates mutation type.
A frequently mutated novel gene, the nuclear receptor binding SET domain protein 1 (NSD1), was identified in 33 HNSCCs. Alterations included inactivating mutations (n = 29) and focal homozygous deletions (n = 4). NSD1 is a histone 3 Lys 36 (H3K36) methyltransferase, similar to SETD2, which is frequently mutated in the clear cell variant of renal cell carcinoma, and associated with DNA hypomethylation28. Germline carriers of inactivating mutations in NSD1 are associated with craniofacial abnormalities (Sotos syndrome), and malignancies including squamous carcinoma, implicating NSD1 as a tumour suppressor gene29. Interestingly, NSD1 functions as an oncogene when fused to nucleoporin-98 (NUP98) t(5;11)(q35;p15.5) in haematological cancers with increased H3K36 trimethylation levels at HOXA genes and accompanying transcriptional activation30. Translocations involving other dedicated H3K36 methyltransferase genes including WHSC1 (also known as MMSET and NSD2) are reported in 20% of multiple myelomas. By contrast, NSD1 loss has been associated with sporadic non-melanoma skin cancers31. Significant inactivating mutations were found in genes linked to squamous differentiation including in NOTCH1 (19%), and other non-significant family members (NOTCH2 9%, and NOTCH3 5%, q > 0.1, non-significant), and the TP63 target gene ZNF750 (4%, q > 0.1, non-significant), which falls in a significantly deleted peak at 17q25.3. The analysis identified additional mutations including TRAF3, RB1 and NFE2L2, among others with q values < 1 (non-significant). The frequently mutated apoptosis gene CASP8 displayed clustered missense and other inactivating mutations in the first death effector, intron and caspase peptidase domains. Statistically significant mutations in KMT2D (also known as MLL2) and HLA-A could contribute to defective immunosurveillance. Of known oncogenes, only PIK3CA achieved statistical significance (q < 0.01). Approximately one-quarter of the mutated PIK3CA cases displayed concurrent amplification, with an additional 20% of tumours containing focal amplification without evidence of mutation. Seventy-three per cent of PIK3CA mutations localized to Glu542Lys, Glu545Lys and His1047Arg/Leu hotspots that promote activation, with the remaining mutations of uncertain function. Recurrent activating mutations of HRAS in the GTPase domain in residues 11–13 approached statistical significance (q = 0.2).
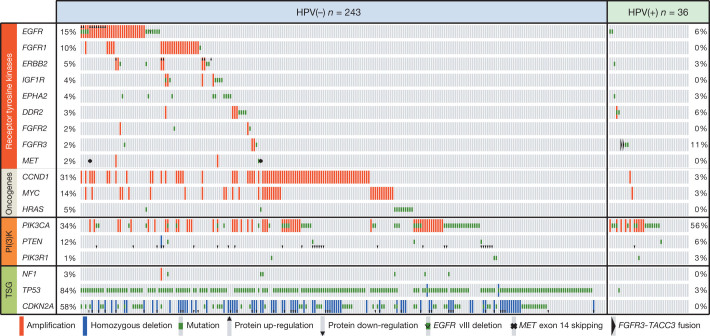
We extended our unsupervised genome-wide analysis of significantly mutated genes as well as genes reported in COSMIC to a subgroup analysis by anatomic sites, tumour versus normal status, HPV status and four previously validated gene expression subtypes32,33 (Supplementary Information section 5, Supplementary Figs 5.1–5.4 and Supplementary Data 4.1 and 5.1–5.4). Additional mutations included TRAF3, RB1 and NFE2L2, among others with q values < 1, and we observed statistical evidence for mutations of HRAS (q = 0 in COSMIC subset) and other genes. Sporadic inactivating mutations and deletions of TGFBR2 were identified primarily in oral cavity tumours, consistent with its role in promoting squamous tumorigenesis in mouse models34. Investigating COSMIC database mutations focused attention on the significant deletion peak at 4q31.3 containing the gene FBXW7, a ubiquitin ligase targeting cyclin E and NOTCH genes, in which we identified mutations that included recurrent Arg505Gly/Leu substitutions (n = 14). Genes with at least one identical mutation previously reported in COSMIC include SCN9A, CHEK2, PTCH1 and PIK3R1. We further focused on somatic alterations and protein expression that represent plausible therapeutic targets (Fig. 3, Supplementary Information sections 6 and 7, Supplementary Figs 6.1 and 6.2 and Supplementary Data 6.1 and 6.2).
Figure 3. Candidate therapeutic targets and driver oncogenic events.
Alteration events for key genes are displayed by sample (n = 279). TSG, tumour suppressor gene.
Integrated genome analysis and pathways
Correlative genetic alteration analysis identified numerous pairwise significant findings (Supplementary Information section 7 and Supplementary Fig. 7.1). In particular, co-amplification of 11q13 containing CCND1, FADD and CTTN and a narrow segment of 11q22 containing the genes with equal evidence for YAP1 and BIRC2 was further characterized (Supplementary Fig. 7.2). Chromosome 11q22 was focally but rarely amplified in the absence of co-amplification of 11q13. This novel finding suggests that the selection pressure for this co-amplification stems from the interaction of BIRC2 with FADD and the caspase cascade that inhibits cell death. Notably, the vast majority of tumours with the 11q13 amplification had large deletions in the telomeric region of 11q22, including other genes known to be important in cell death in cancer such as ATM and CASP1, 4, 5 and 12. Amplification of 11q13 was anti-correlated with CASP8 mutations, suggesting an alternative function of CASP8 and FADD in cell death/NF-κB activation35.
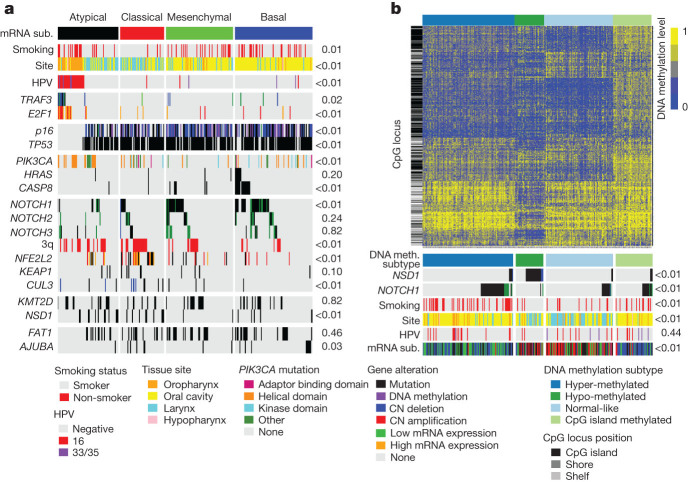
We investigated whether clinical factors, single gene alterations and statistically significant pairwise gene correlations (Supplementary Fig. 7.1 and Supplementary Data 7.1) might segregate previously defined molecular subtypes and/or anatomic sub-sites (Supplementary Data 5.1–5.4). We confirmed reported gene expression subtypes (atypical (24%), mesenchymal (27%), basal (31%) and classical (18%)), and assessed the subtypes for enrichment of somatic alterations32,33 (Supplementary Data 7.1). Notably, TP53 mutation, CDKN2A loss of function, chromosome 3q amplification, alteration of oxidative stress genes (KEAP1, NFE2L2 or CUL3), heavy smoking history (Supplementary Table 1.1) and larynx sub-site co-occurred in most classical subtype tumours (Fig. 4a and Supplementary Information section 7.2), similar to LUSC5 (Supplementary Figs 5.1 and 5.2). Collectively, these findings suggest that the NFE2L2 oxidative stress pathway is a tobacco-related signature across anatomic tumour sites. By contrast, the basal subtype demonstrated inactivation of NOTCH1 with intact oxidative stress signalling and fewer alterations of chromosome 3q. Analysis of the 3q locus highlighted a marked relative decrease of SOX2 expression in basal tumours relative to all other HNSCC and tumour adjacent normal samples (Supplementary Fig. 5.4), supporting the interaction of transcription factors SOX2, TP63, NFE2L2 and NOTCH1 as driving differences between expression subtypes. Additionally, the basal subtype included most tumours with the HRAS–CASP8 co-mutation and most co-amplified 11q13/q22 tumours. These findings along with HRAS mutations implicate disrupted cell death as a major alteration in this subtype36 (Supplementary Fig. 7.2). The atypical subtype was characterized by a lack of chromosome 7 amplifications (Supplementary Fig. 5.3), enrichment of HPV(+) tumours with activating mutations in exon 9 that contains the PIK3CA helical domain. By contrast, the mesenchymal subtype showed high levels of alteration in innate immunity genes, in particular high expression of natural killer cell marker CD56 and a low frequency of HLA class I mutations (Supplementary Fig. 7.3). Among the significantly mutated genes, TP53 (P < 0.001), CASP8 (P = 0.01), NSD1 (P = 0.01) and CDKN2A (0.06) were the most differentially mutated across anatomic sites (Supplementary Data 4.1). Most CASP8 mutations (22 out of 24, 92%) were in oral cavity tumours, whereas TP53, NSD1 and CDKN2A demonstrated decreased mutation rates in oropharyngeal tumours relative to other sites.
Figure 4. Integrated analysis of genomic alterations.
a, b, Samples (n = 279) are displayed in columns and grouped by gene expression (a) or methylation (b) subtype (sub.). Unadjusted two-sided Fisher’s exact test P values assess the association of each genomic alteration. Methylation probe location of CpG islands, shores and shelves are shown on the left of b. Annotation shows HPV status and subtype (16, 33 and 35). CN, copy number.
Unsupervised analysis of gene expression by HPV status and of reverse-phase protein arrays (Supplementary Information section 6), DNA methylation (Supplementary Information section 8), and miRNA platforms (Supplementary Information section 9, Supplementary Table 9.1, Supplementary Figs 7.4–7.9 and Supplementary Data 7.1) showed high correlation across platforms (P < 0.01; Fig. 4, Supplementary Information section 7.9 and Supplementary Data 7.2) and coordinated alterations of genes including the epithelial–mesenchymal transition signature37 (Supplementary Figs 7.4, 7.7 and 7.8). However, within the broader cross-platform agreement, individual unsupervised clustering of miRNA, reverse-phase protein arrays and DNA methylation data provided insight into the association of molecular subtypes with single gene alterations. The most notable example was the detection of hypomethylation and loss-of-function mutations of NSD1, and wild-type NOTCH1 in atypical and classical gene expression subtypes (Fig. 4b and Supplementary Table 7.1).
Supervised analyses detected genomic features (miRNA, gene expression and DNA methylation) associated with anatomic site (Supplementary Figs 5.5–5.8 and Supplementary Data 5.1–5.4). A supervised integrated analysis identified target genes that are inversely regulated by miRNAs in HNSCC (Supplementary Information section 7.4). Among these miRNA–messenger RNA networks, let-7c-5p and miR-100-5p exhibited a correlation between low copy number and expression. Let-7c-5p and miR-100-5p were decreased in tumours compared to normal (Supplementary Fig. 7.10). For these miRNAs, deletion was highly associated with increased expression of target genes, including the cell cycle regulator CDK6, transcription factor E2F1 (ref. 38), mitosis regulator PLK1 (ref. 39), and transcription factor HMGA2 (ref. 40; Supplementary Figs 7.10, Supplementary Tables 7.2 and 7.3).
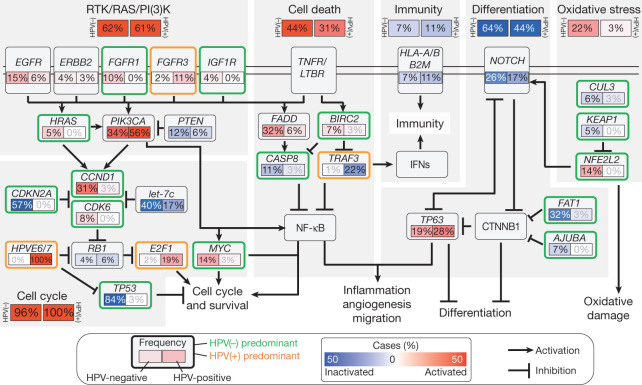
Integrative bioinformatics analysis identified a limited number of pathways targeted by frequent genome alterations (Fig. 5, Supplementary Information section 7, Supplementary Figs 7.11–7.15 and Supplementary Data 7.3). Among receptor tyrosine kinases, EGFR/ERBB2 or FGFR1/3 alterations are the most frequent. Among downstream targets of the receptor tyrosine kinase (RTK)/RAS/phosphatidylinositol-3-OH kinase (PI(3)K) pathway, PIK3CA dominates with occasional HRAS and PTEN alterations. Further downstream, nearly every tumour has alteration of genes governing the cell cycle. The tumour suppressors TP53 and CDKN2A, oncogenes CCND1 and MYC, and the newly identified miRNA let-7c, are most often altered in HPV(−) tumours, whereas viral genes E6, E7 and E2F1 predominate in HPV(+) cases. In addition, we report frequent alterations in genes involved in cell death, NF-κB-mediated survival, or immunity pathways15,35. Co-amplification of FADD ± BIRC2, or CASP8 ± HRAS mutations define exclusive HPV(−) subsets, whereas TRAF3 loss characterizes an HPV(+) subset. These alterations along with PIK3CA and TP63 converge on NF-κB transcription factors that promote cell survival, migration, inflammation and angiogenesis41,42. Furthermore, TRAF3 and/or HLA loss are implicated in deregulation of innate antiviral and adaptive anti-tumour immunity43,44. Further alterations of NOTCH, TP63 and other genes in HPV(−) tumours (FAT1 and AJUBA) recently linked functionally to β-catenin (CTNNB1) are also detected21,22,45. Finally, we highlight a previously underappreciated role for a key transcription factor regulator of oxidative stress, NFE2L2, and its protein complex partners CUL3 and KEAP1 in HPV(−) HNSCCs.
Figure 5. Deregulation of signalling pathways and transcription factors.
Key affected pathways, components and inferred functions, are summarized in the main text and Supplementary Information section 7 for n = 279 samples. The frequency (%) of genetic alterations for HPV(−) and HPV(+) tumours are shown separately within sub-panels and highlighted. Also see Supplementary Fig. 7.15. Pathway alterations include homozygous deletions, focal amplifications and somatic mutations. Activated and inactivated pathways/genes, and activating or inhibitory symbols are based on predicted effects of genome alterations and/or pathway functions.
Conclusion
The TCGA study represents the most comprehensive integrative genomic analysis of HNSCC. Loss of TRAF3, activating mutations of PIK3CA, and amplification of E2F1 in HPV(+) oropharyngeal cancers point to aberrant activation of NF-κB, other oncogenic pathways, and cell cycle, as critical in the pathogenesis and development of new targeted therapies for these tumours. In HPV(−) HNSCCs, mutually exclusive subsets containing amplicons on 11q with CCND1, FADD, BIRC2 and YAP1, or concurrent mutations of CASP8 with HRAS, also target cell cycle, death, NF-κB and other oncogenic pathways. Recent studies predict that the inactivation of AJUBA, as well as FAT1 and NOTCH1, may converge to uncheck Wnt/β-catenin signalling, implicated in deregulation of cell polarity and differentiation. The 3q amplicon found in both HPV(+) and (−) HNSCCs includes transcription factors TP63, SOX2 and signal molecule PIK3CA, which are also implicated in homeostasis of epithelial stem cells and differentiation. Among these, the biological function and agents targeting BIRCs, PI(3)K, Wnt/β-catenin and NOTCH are under investigation. Collectively, these findings provide new insights into HNSCC and suggest that shared and unique alterations might be leveraged to accelerate progress in prevention and therapy across tumour types.
Supplementary information
This file contains Supplementary Text and Data, Supplementary Figures, Supplementary Tables, and additional references. See the Table of Contents for details. (PDF 25182 kb)
This file contains Supplementary Data files and a Supplementary Data Guide. (ZIP 27635 kb)
Acknowledgements
This study was supported by National Institutes of Health (NIH) grants: P50CA097190, P50CA16672, U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845, U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025 and RO1 CA 095419. Supported by The Bobby F. Garrett Fund for Head and Neck Cancer Research and NIDCD Intramural Projects ZIA-DC-000016, 73 and 74.
PowerPoint slides
Author Contributions
The TCGA research network contributed collectively to this study. Biospecimens were provided by the tissue source sites and processed by the biospecimen core resource. Data generation and analyses were performed by the genome sequencing centres, cancer genome characterization centres and genome data analysis centres. All data were released through the data coordinating centre. Project activities were coordinated by the National Cancer Institute and National Human Genome Research Institute project teams. We also acknowledge the following TCGA investigators who made substantial contributions to the project: C.V.W. (manuscript coordinator); V.W. (data coordinator); P.S.H. and D.N.H. (analysis coordinators); Y.W. (data coordinating centre representative); R.A., J.B., T.E.C., J.J.L., M.Parfenov, A.D.T., V.W. and M.D.W. (clinical and HPV analysis); A.D.C. (DNA copy number analysis); J.Cho, A.H., M.Parfenov and M.D.W. (mutation calling); L.C. and L.Danilova (DNA methylation analysis); R.Bowlby, J.P.B., Z.C., H.Cheng, A.H., F.F.L., A.G.R., A.D.S. and C.V.W. (miRNA sequence analysis); L.A.B., L.Diao, P.K., W.L., S.N. and J.W. (reverse-phase protein arrays analysis); L.A.B., L.Diao, S.R.J., P.K.K., V.W., M.D.W. and N.Z. (mRNA sequence analysis); S.Benz, Z.C., R.L.F., S.N., C.R.P., N.Schultz, T.Y.S., C.V.W. and V.W. (pathway analysis); A.E.N., J.R.G. and D.N.H. (project chairs).
Competing interests
The author declare no competing financial interests.
Footnotes
Participants are arranged by area of contribution and then by institution.
The primary and processed data used to generate the analyses can be downloaded by registered users from The Cancer Genome Atlas (https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp, https://cghub.ucsc.edu/ and https://tcga-data.nci.nih.gov/docs/publications/hnsc_2014/).
anaggar@mdanderson.org
Contributor Information
The Cancer Genome Atlas Network:
Michael S. Lawrence, Carrie Sougnez, Lee Lichtenstein, Kristian Cibulskis, Eric Lander, Stacey B. Gabriel, Gad Getz, Adrian Ally, Miruna Balasundaram, Inanc Birol, Reanne Bowlby, Denise Brooks, Yaron S. N. Butterfield, Rebecca Carlsen, Dean Cheng, Andy Chu, Noreen Dhalla, Ranabir Guin, Robert A. Holt, Steven J. M. Jones, Darlene Lee, Haiyan I. Li, Marco A. Marra, Michael Mayo, Richard A. Moore, Andrew J. Mungall, A. Gordon Robertson, Jacqueline E. Schein, Payal Sipahimalani, Angela Tam, Nina Thiessen, Tina Wong, Alexei Protopopov, Netty Santoso, Semin Lee, Michael Parfenov, Jianhua Zhang, Harshad S. Mahadeshwar, Jiabin Tang, Xiaojia Ren, Sahil Seth, Psalm Haseley, Dong Zeng, Lixing Yang, Andrew W. Xu, Xingzhi Song, Angeliki Pantazi, Christopher A. Bristow, Angela Hadjipanayis, Jonathan Seidman, Lynda Chin, Peter J. Park, Raju Kucherlapati, Rehan Akbani, Tod Casasent, Wenbin Liu, Yiling Lu, Gordon Mills, Thomas Motter, John Weinstein, Lixia Diao, Jing Wang, You Hong Fan, Jinze Liu, Kai Wang, J. Todd Auman, Saianand Balu, Thomas Bodenheimer, Elizabeth Buda, D. Neil Hayes, Katherine A. Hoadley, Alan P. Hoyle, Stuart R. Jefferys, Corbin D. Jones, Patrick K. Kimes, Yufeng Liu, J. S. Marron, Shaowu Meng, Piotr A. Mieczkowski, Lisle E. Mose, Joel S. Parker, Charles M. Perou, Jan F. Prins, Jeffrey Roach, Yan Shi, Janae V. Simons, Darshan Singh, Matthew G. Soloway, Donghui Tan, Umadevi Veluvolu, Vonn Walter, Scot Waring, Matthew D. Wilkerson, Junyuan Wu, Ni Zhao, Andrew D. Cherniack, Peter S. Hammerman, Aaron D. Tward, Chandra Sekhar Pedamallu, Gordon Saksena, Joonil Jung, Akinyemi I. Ojesina, Scott L. Carter, Travis I. Zack, Steven E. Schumacher, Rameen Beroukhim, Samuel S. Freeman, Matthew Meyerson, Juok Cho, Lynda Chin, Gad Getz, Michael S. Noble, Daniel DiCara, Hailei Zhang, David I. Heiman, Nils Gehlenborg, Doug Voet, Pei Lin, Scott Frazer, Petar Stojanov, Yingchun Liu, Lihua Zou, Jaegil Kim, Carrie Sougnez, Stacey B. Gabriel, Michael S. Lawrence, Donna Muzny, HarshaVardhan Doddapaneni, Christie Kovar, Jeff Reid, Donna Morton, Yi Han, Walker Hale, Hsu Chao, Kyle Chang, Jennifer A. Drummond, Richard A. Gibbs, Nipun Kakkar, David Wheeler, Liu Xi, Giovanni Ciriello, Marc Ladanyi, William Lee, Ricardo Ramirez, Chris Sander, Ronglai Shen, Rileen Sinha, Nils Weinhold, Barry S. Taylor, B. Arman Aksoy, Gideon Dresdner, Jianjiong Gao, Benjamin Gross, Anders Jacobsen, Boris Reva, Nikolaus Schultz, S. Onur Sumer, Yichao Sun, Timothy A. Chan, Luc G. Morris, Joshua Stuart, Stephen Benz, Sam Ng, Christopher Benz, Christina Yau, Stephen B. Baylin, Leslie Cope, Ludmila Danilova, James G. Herman, Moiz Bootwalla, Dennis T. Maglinte, Peter W. Laird, Timothy Triche, Jr, Daniel J. Weisenberger, David J. Van Den Berg, Nishant Agrawal, Justin Bishop, Paul C. Boutros, Jeff P. Bruce, Lauren Averett Byers, Joseph Califano, Thomas E. Carey, Zhong Chen, Hui Cheng, Simion I. Chiosea, Ezra Cohen, Brenda Diergaarde, Ann Marie Egloff, Adel K. El-Naggar, Robert L. Ferris, Mitchell J. Frederick, Jennifer R. Grandis, Yan Guo, Robert I. Haddad, Peter S. Hammerman, Thomas Harris, D. Neil Hayes, Angela B. Y. Hui, J. Jack Lee, Scott M. Lippman, Fei-Fei Liu, Jonathan B. McHugh, Jeff Myers, Patrick Kwok Shing Ng, Bayardo Perez-Ordonez, Curtis R. Pickering, Michael Prystowsky, Marjorie Romkes, Anthony D. Saleh, Maureen A. Sartor, Raja Seethala, Tanguy Y. Seiwert, Han Si, Aaron D. Tward, Carter Van Waes, Daryl M. Waggott, Maciej Wiznerowicz, Wendell G. Yarbrough, Jiexin Zhang, Zhixiang Zuo, Ken Burnett, Daniel Crain, Johanna Gardner, Kevin Lau, David Mallery, Scott Morris, Joseph Paulauskis, Robert Penny, Candace Shelton, Troy Shelton, Mark Sherman, Peggy Yena, Aaron D. Black, Jay Bowen, Jessica Frick, Julie M. Gastier-Foster, Hollie A. Harper, Kristen Leraas, Tara M. Lichtenberg, Nilsa C. Ramirez, Lisa Wise, Erik Zmuda, Julien Baboud, Mark A. Jensen, Ari B. Kahn, Todd D. Pihl, David A. Pot, Deepak Srinivasan, Jessica S. Walton, Yunhu Wan, Robert A. Burton, Tanja Davidsen, John A. Demchok, Greg Eley, Martin L. Ferguson, Kenna R. Mills Shaw, Bradley A. Ozenberger, Margi Sheth, Heidi J. Sofia, Roy Tarnuzzer, Zhining Wang, Liming Yang, Jean Claude Zenklusen, Charles Saller, Katherine Tarvin, Chu Chen, Roni Bollag, Paul Weinberger, Wojciech Golusiński, Paweł Golusiński, Matthew Ibbs, Konstanty Korski, Andrzej Mackiewicz, Wiktoria Suchorska, Bartosz Szybiak, Maciej Wiznerowicz, Ken Burnett, Erin Curley, Johanna Gardner, David Mallery, Robert Penny, Troy Shelton, Peggy Yena, Christina Beard, Colleen Mitchell, George Sandusky, Nishant Agrawal, Julie Ahn, Justin Bishop, Joseph Califano, Zubair Khan, Jeff P. Bruce, Angela B. Y. Hui, Jonathan Irish, Fei-Fei Liu, Bayardo Perez-Ordonez, John Waldron, Paul C. Boutros, Daryl M. Waggott, Jeff Myers, William N. William, Jr, Scott M. Lippman, Sophie Egea, Carmen Gomez-Fernandez, Lynn Herbert, Carol R. Bradford, Thomas E. Carey, Douglas B. Chepeha, Andrea S. Haddad, Tamara R. Jones, Christine M. Komarck, Mayya Malakh, Jonathan B. McHugh, Jeffrey S. Moyer, Ariane Nguyen, Lisa A. Peterson, Mark E. Prince, Laura S. Rozek, Maureen A. Sartor, Evan G. Taylor, Heather M. Walline, Gregory T. Wolf, Lori Boice, Bhishamjit S. Chera, William K. Funkhouser, Margaret L. Gulley, Trevor G. Hackman, D. Neil Hayes, Michele C. Hayward, Mei Huang, W. Kimryn Rathmell, Ashley H. Salazar, William W. Shockley, Carol G. Shores, Leigh Thorne, Mark C. Weissler, Sylvia Wrenn, Adam M. Zanation, Simion I. Chiosea, Brenda Diergaarde, Ann Marie Egloff, Robert L. Ferris, Marjorie Romkes, Raja Seethala, Brandee T. Brown, Yan Guo, Michelle Pham, and Wendell G. Yarbrough
References
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oganesyan G, et al. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 7.Karim R, et al. Human papillomavirus (HPV) upregulates the cellular deubiquitinase UCHL1 to suppress the keratinocyte’s innate immune response. PLoS Pathog. 2013;9:e1003384. doi: 10.1371/journal.ppat.1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eliopoulos AG, et al. CD40-induced growth inhibition in epithelial cells is mimicked by Epstein–Barr Virus-encoded LMP1: involvement of TRAF3 as a common mediator. Oncogene. 1996;13:2243–2254. [PubMed] [Google Scholar]
- 9.Imbeault M, et al. Acquisition of host-derived CD40L by HIV-1 in vivo and its functional consequences in the B-cell compartment. J. Virol. 2011;85:2189–2200. doi: 10.1128/JVI.01993-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ni CZ, et al. Molecular basis for CD40 signaling mediated by TRAF3. Proc. Natl Acad. Sci. USA. 2000;97:10395–10399. doi: 10.1073/pnas.97.19.10395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung GT, et al. Constitutive activation of distinct NF-κB signals in EBV-associated nasopharyngeal carcinoma. J. Pathol. 2013;231:311–322. doi: 10.1002/path.4239. [DOI] [PubMed] [Google Scholar]
- 12.Annunziata CM, et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nature Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smeets SJ, et al. Genetic classification of oral and oropharyngeal carcinomas identifies subgroups with a different prognosis. Cellular Oncol. 2009;31:291–300. doi: 10.3233/CLO-2009-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayo MW, et al. Requirement of NF-κB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 16.Sok JC, et al. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin. Cancer Res. 2006;12:5064–5073. doi: 10.1158/1078-0432.CCR-06-0913. [DOI] [PubMed] [Google Scholar]
- 17.Kong-Beltran M, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–289. doi: 10.1158/0008-5472.CAN-05-2749. [DOI] [PubMed] [Google Scholar]
- 18.Popescu NC, DiPaolo JA, Amsbaugh SC. Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet. Cell Genet. 1987;44:58–62. doi: 10.1159/000132342. [DOI] [PubMed] [Google Scholar]
- 19.Forbes, S. A. et al. The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr. Protoc. Hum. Genet. Chapter 10, Unit 10.11 (2008) [DOI] [PMC free article] [PubMed]
- 20.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris LG, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nature Genet. 2013;45:253–261. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haraguchi K, et al. Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3β-mediated phosphorylation of β-catenin. Oncogene. 2008;27:274–284. doi: 10.1038/sj.onc.1210644. [DOI] [PubMed] [Google Scholar]
- 23.Nagai Y, et al. The LIM protein Ajuba is required for ciliogenesis and left-right axis determination in medaka. Biochem. Biophys. Res. Commun. 2010;396:887–893. doi: 10.1016/j.bbrc.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Sun G, Irvine KD. Ajuba family proteins link JNK to Hippo signaling. Sci. Signal. 2013;6:ra81. doi: 10.1126/scisignal.2004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy BV, Irvine KD. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell. 2013;24:459–471. doi: 10.1016/j.devcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalan S, Matveyenko A, Loayza D. LIM protein Ajuba participates in the repression of the ATR-mediated DNA damage response. Front. Genet. 2013;4:95. doi: 10.3389/fgene.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nola S, et al. Ajuba is required for Rac activation and maintenance of E-cadherin adhesion. J. Cell Biol. 2011;195:855–871. doi: 10.1083/jcb.201107162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature499, 43–49 (2013) [DOI] [PMC free article] [PubMed]
- 29.Fickie MR, et al. Adults with Sotos syndrome: review of 21 adults with molecularly confirmed NSD1 alterations, including a detailed case report of the oldest person. Am. J. Med. Genet. A. 2011;155:2105–2111. doi: 10.1002/ajmg.a.34156. [DOI] [PubMed] [Google Scholar]
- 30.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nature Cell Biol. 2007;9:804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 31.Quintana RM, et al. A transposon-based analysis of gene mutations related to skin cancer development. J. Invest. Dermatol. 2013;133:239–248. doi: 10.1038/jid.2012.245. [DOI] [PubMed] [Google Scholar]
- 32.Chung CH, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 33.Walter V, et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE. 2013;8:e56823. doi: 10.1371/journal.pone.0056823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu SL, et al. Loss of transforming growth factor-β type II receptor promotes metastatic head-and-neck squamous cell carcinoma. Genes Dev. 2006;20:1331–1342. doi: 10.1101/gad.1413306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberst A, Green DR. It cuts both ways: reconciling the dual roles of caspase 8 in cell death and survival. Nature Rev. Mol. Cell Biol. 2011;12:757–763. doi: 10.1038/nrm3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SJ, et al. Opposite role of Ras in tumor necrosis factor-α-induced cell cycle regulation: competition for Raf kinase. Biochem. Biophys. Res. Commun. 2001;287:1140–1147. doi: 10.1006/bbrc.2001.5713. [DOI] [PubMed] [Google Scholar]
- 37.Byers LA, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong JV, Dong P, Nevins JR, Mathey-Prevot B, You L. Network calisthenics: control of E2F dynamics in cell cycle entry. Cell Cycle. 2011;10:3086–3094. doi: 10.4161/cc.10.18.17350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanhaji M, et al. Polo-like kinase 1 inhibitors, mitotic stress and the tumor suppressor p53. Cell Cycle. 2013;12:1340–1351. doi: 10.4161/cc.24573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morishita A, et al. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013;73:4289–4299. doi: 10.1158/0008-5472.CAN-12-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bancroft CC, et al. Effects of pharmacologic antagonists of epidermal growth factor receptor, PI3K and MEK signal kinases on NF-κB and AP-1 activation and IL-8 and VEGF expression in human head and neck squamous cell carcinoma lines. Int. J. Cancer. 2002;99:538–548. doi: 10.1002/ijc.10398. [DOI] [PubMed] [Google Scholar]
- 42.Yang X, et al. ΔNp63 versatilely regulates a broad NF-κB gene program and promotes squamous epithelial proliferation, migration, and inflammation. Cancer Res. 2011;71:3688–3700. doi: 10.1158/0008-5472.CAN-10-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keating PJ, et al. Frequency of down-regulation of individual HLA-A and -B alleles in cervical carcinomas in relation to TAP-1 expression. Br. J. Cancer. 1995;72:405–411. doi: 10.1038/bjc.1995.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esteban F, et al. Lack of MHC class I antigens and tumour aggressiveness of the squamous cell carcinoma of the larynx. Br. J. Cancer. 1990;62:1047–1051. doi: 10.1038/bjc.1990.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nature Rev. Cancer. 2009;9:587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Text and Data, Supplementary Figures, Supplementary Tables, and additional references. See the Table of Contents for details. (PDF 25182 kb)
This file contains Supplementary Data files and a Supplementary Data Guide. (ZIP 27635 kb)