Abstract
Objectives
To investigate the impact of HAART-induced HIV suppression on levels of 24 serological biomarkers of inflammation and immune activation.
Design
Prospective cohort study.
Methods
Biomarkers were measured with multiplex assays in centralized laboratories using stored serum samples contributed by 1,697 men during 8,903 person-visits in the Multicenter AIDS Cohort Study (MACS) from 1984–2009. Using generalized gamma models, we compared biomarker values across three groups, adjusting for possible confounders: HIV-uninfected (NEG); HIV+, HAART-naïve (NAI); and HAART-exposed with HIV RNA suppressed to <50 copies/mL plasma (SUP). We also estimated changes in biomarker levels associated with duration of HIV suppression, using splined generalized gamma regression with a knot at one year.
Results
Most biomarkers were relatively normalized in the SUP group relative to the NAI group; however, 12 biomarkers in the SUP group were distinct (p<0.002) from NEG values: CXCL10, CRP, sCD14, sTNFR2, TNF-α, sCD27, sGP130, IL-8, CCL13, BAFF, GM-CSF, and IL-12p70. Thirteen biomarkers exhibited significant changes in the first year after viral suppression, but none changed significantly after that time.
Conclusions
Biomarkers of inflammation and immune activation moved toward HIV-negative levels within the first year after HAART-induced HIV suppression. Although several markers of T cell activation returned to levels present in HIV-negative men, residual immune activation, particularly monocyte/macrophage activation, was present. This residual immune activation may represent a therapeutic target to improve the prognosis of HIV-infected individuals receiving HAART.
Keywords: Acquired Immunodeficiency Syndrome; Antiretroviral Therapy, Highly Active; Biological Markers; Inflammation; Prospective Studies; Male
Introduction
The introduction of highly active antiretroviral therapy (HAART) led to greatly increased survival of HIV-infected individuals [1, 2]. Although HAART dramatically reduces HIV viral load, individuals receiving HAART face higher risks than HIV-uninfected individuals of serious non-AIDS-related morbidities, AIDS-defining cancers, and all-cause mortality, depending upon the stage of HIV disease at which HAART was initiated [3]. Though the etiology of these risks remains unclear, persistent dysregulation of inflammatory processes causing chronic immune activation is likely important [4, 5]. For example, T cell activation among HIV-suppressed individuals receiving HAART is negatively associated with immune reconstitution [6, 7]. HIV-induced dysregulation of inflammatory processes occurs via multiple pathways [8], possibly including microbial translocation [9].
The effect of HAART on immune activation and inflammation is incompletely understood. There is evidence that HAART-induced HIV suppression reduces some measures of HIV-induced immune activity, but not necessarily to levels present in HIV-uninfected individuals [10]. In the Multicenter AIDS Cohort Study (MACS), elevated levels of B cell stimulatory cytokines and other immune activation markers did not normalize following HAART initiation [11, 12]. In the Women’s Interagency HIV Study (WIHS), however, serum levels of both inflammatory and anti-inflammatory biomarkers normalized following HAART initiation, though tumor necrosis factor (TNF)-α remained elevated in women despite undetectable HIV RNA [13]. In the Strategies for Management of Antiretroviral Treatment trial, participants with HIV suppression still had higher levels of interleukin (IL)-6, D-dimer, and C-reactive protein (CRP) than HIV-negative individuals from other cohorts [14].
Most studies examining changes in inflammatory biomarker levels in HIV-infected individuals have been limited by small study populations, cross-sectional designs, and/or small numbers of biomarkers. Recent technical advances, especially the development of multiplexed assays, have enabled the more efficient measurement of multiple inflammatory biomarkers. In this study, we compared levels of 24 such biomarkers in stored serum samples from HIV-infected participants in the MACS before and after HAART initiation, and from HIV-uninfected MACS participants who had similar ages and racial profiles to the HIV-infected men studied.
Methods
Study cohort and exposure definitions
The MACS is a prospective cohort study of HIV infection at four sites (Baltimore, MD / Washington, DC; Chicago, IL; Los Angeles, CA; Pittsburgh, PA), comprised of men who have sex with men. Enrollment began in 1984; details of the cohort have been described [15]. Briefly, participants are followed at semiannual study visits with standardized interviews, physical examinations, and phlebotomy for concurrent laboratory testing and storage of plasma and serum (at −80° C), and of peripheral blood mononuclear cells (at −135° C).
MACS person-visits were chosen for inclusion in a large study of biomarkers of inflammation and immune activation. Serum samples were selected at one-year intervals for men with known HIV seroconversion dates, and from visits immediately before and after HAART initiation (and at two-year intervals thereafter) for all HAART users. Four visits from each of 250 HIV-uninfected men from 1984–2009 were chosen as controls, including all HIV-uninfected men with HCV infection.
CD4+ T cell counts were measured with flow cytometry [16] and plasma HIV RNA levels were measured with the Roche ultrasensitive assay sensitive to 50 copies of HIV RNA/mL plasma [17]. We defined person-visits contributed by HIV-infected men on HAART as HIV-suppressed (“SUP”) if plasma HIV RNA was < 50 copies/mL. Measurements from SUP person-visits were compared to those from two reference groups: 1) HIV-uninfected (“NEG”) person-visits and 2) HIV-positive, HAART-naïve (“NAI”) person-visits. Men could contribute visits to multiple categories. We considered age (continuous), race (white/non-white), current smoking (yes/no), chronic hepatitis C infection (defined by detectable HCV RNA), obesity (body mass index >30 kg/m2), uncontrolled diabetes (hemoglobin A1C ≥6.5% or fasting glucose ≥126 mg/dL), and MACS site as possible confounders. We used the most recent covariate values within two years of the serum collection dates.
Biomarker assays
Two multiplex assay platforms were used to quantify 23 serologic markers of immune activation and inflammation. The Meso Scale Discovery (MSD, Gaithersburg, MD) system was used to measure the cytokines IL-1β, IL-2, IL-6, IL-8, IL-10, IL-12p70, TNF-α, granular-macrophage colony-stimulating factor (GM-CSF), and interferon (IFN)-γ (Ultra-Sensitive Human Pro-Inflammatory 9-Plex Kit); and chemokine (C-C motif) ligand (CCL)2, CCL4, CCL11, CCL13, CCL17, chemokine (C-X-C motif) ligand (CXCL)10, and IL-8 (Ultra-Sensitive Human Chemokine 7-Plex Kit), according to the manufacturer’s protocols. The MSD platform is a solid-phase electrochemiluminescence-based assay; MSD plates were analyzed on the SECTOR Imager 2400 (MSD, Gaithersburg, MD).
The Luminex platform (Luminex, Austin, TX) was used according to the manufacturer’s protocol (R&D Systems, Minneapolis, MN) to measure soluble (s)CD14, sCD27, sgp130, sIL-2Rα, sIL-6R, sTNF-R2, B-cell activating factor (BAFF), and CXCL13 using a single lot of assay kits, to eliminate lot-to-lot variability. Serum samples were diluted 1:50. The Luminex platform is a fluorescent bead-based assay; Luminex assay data were collected and analyzed using a BioPlex 200 apparatus and BioPlex Manager software (Bio-Rad, Hercules, CA). With both platforms, all samples from an individual were tested on one plate to minimize variability. Each plate contained samples from both HIV-infected and HIV-uninfected men. One additional marker, CRP, was measured by a reference laboratory (Quest Diagnostics) using a high-sensitivity immunonephelometric assay.
Statistical methods
In the first (across-group) analysis, we initially fit separate generalized gamma models for each biomarker stratified by the three exposure categories described above (SUP, NEG, NAI). Few of the biomarkers exhibited lognormal distributions, which rendered inappropriate the common method of using linear regression on log-transformed biomarker values. Therefore, we employed the generalized gamma (GG) distribution, a highly flexible 3-parameter distribution encompassing several distributional forms such as the exponential, Weibull, and lognormal [18]. We chose the GG model to avoid imposing strong assumptions regarding biomarker distributions.
Location (β), scale (σ), and shape (λ) parameters were allowed to vary by exposure category in unadjusted models. Models were appropriately adjusted for repeated measurements. The possible confounding variables of age, nonwhite race, smoking, HCV infection, obesity, diabetes, and MACS site were chosen for multivariable models via inspection of univariate associations with all biomarkers.
In adjusted models, only the location (β) parameter was allowed to vary by exposure, while σ and λ were held constant for the sake of parsimony. Relative percentiles were calculated comparing SUP values to NAI and NEG values, and comparing NAI to NEG values. With σ and λ held constant, the relative percentile (comparing one group to another) is constant across percentiles of each biomarker. In other words, a constant percent shift in the distribution applies to the median and all other percentiles.
In the second (longitudinal) analysis, we examined individual trajectories of biomarker levels while participants were virally suppressed. Men in this analysis contributed at least one biomarker measurement before HIV suppression; i.e., when HIV RNA was ≥ 50 copies/mL irrespective of HAART use, followed by at least one SUP biomarker person-visit. We defined a baseline value for each biomarker by averaging the last HIV-positive values prior to viral suppression, up to a maximum of three values if available. The date of viral suppression was defined as the midpoint between the latest post-HAART date with detectable plasma HIV RNA and the date of the first visit with undetectable HIV RNA. Only men whose date of viral suppression could be established within ± 1 year were included in this analysis. Men contributed visits until virologic rebound (detectable HIV RNA).
We fit biomarker-specific multivariable generalized gamma models with duration of suppression as the exposure of interest, testing for effect modification by covariates from the first analysis. Standard errors were adjusted for repeated measurements. Again, β could vary by exposure category while σ and λ were held constant. Because many biomarker trajectories exhibited strong discontinuities after about one year of HIV suppression, we employed linear splines with a knot at one year. We then estimated differences in biomarker levels over time as compared to baseline values. We tested for effect modification by including covariate-time interaction terms.
All biomarker values were utilized for each analysis; measurements below the lower limits of detection were handled by modeling the inverse of values. This allowed models to fit distributions using standard methods for right-censoring, avoiding the need for explicit imputation. We subsequently converted predicted values back into the original scale.
In each analysis, we adjusted for multiple tests performed across biomarkers by employing a Bonferroni correction [19, 20] to control the family-wise error rate at an α of 0.05: (0.05/24) = 0.002. Analyses were conducted using SAS v 9.3, Stata 11, and R software.
Results
Across-group analysis
Table 1a displays characteristics of the study population for the across-group analysis. 1,697 men contributed a total of 8,903 person-visits from 1984–2009; 786 men contributed visits to more than one analytical group, and 123 men contributed visits to all three groups. Men contributed a median of 4 person-visits each (interquartile range [IQR] 3–6). SUP person-visits were contributed at an older median age (48) relative to NAI (38) and NEG (42) person-visits. Among SUP person-visits, time since HAART initiation (defined as the midpoint between the last HAART-unexposed visit and the first HAART-exposed visit) ranged from 0 to 14 years with a median of 6.1 years (IQR 3.2–9.4).
Tables 1.
A and B. Characteristics of Multicenter AIDS Cohort Study (MACS) biomarker study population
| (A) Across-group analysis | (B) Longitudinal analysis | ||||||
|---|---|---|---|---|---|---|---|
| HIV-negative (NEG) |
HIV+, HAART- naïve (NAI) |
HIV suppressed (SUP) |
Total | Pre-HIV suppression |
HIV virally suppressed |
Total | |
| Number of individuals* | 625 | 975 | 1,006 | 1,697 | 456 | 456 | 456 |
| Number of person-visits | 1,547 | 3,840 | 3,516 | 8,903 | 456 | 1349 | 1805 |
| Year of person-visits | |||||||
| Median | 2003 | 1991 | 2006 | 2000 | 1999 | 2004 | 2003 |
| IQR | (1998, 2006) | (1988, 1996) | (2003, 2008) | (1990, 2006) | (1997, 2004) | (2001, 2007) | (1999, 2007) |
| Age, years | |||||||
| Median | 42.1 | 38.4 | 48.2 | 43.1 | 44.1 | 48.6 | 47.5 |
| IQR | (33.9, 49.5) | (32.9, 44.8) | (42.6, 53.9) | (35.9, 50.1) | (38.9, 48.9) | (43.3, 54.5) | (42.0, 53.5) |
| Years after HIV suppression | |||||||
| Median | n/a | n/a | 4.2 | 4.2 | n/a | 2.1 | 2.1 |
| IQR | (1.8, 7.5) | (1.8, 7.5) | (0.6, 5.6) | (0.6, 5.6) | |||
| CD4+ cell count, cells/µL | |||||||
| Median | 962 | 507 | 575 | 541 | 365 | 358 | 361 |
| IQR | (746, 1191) | (330, 726) | (415, 767) | (370, 748) | (233, 543) | (240, 536) | (239, 538) |
| % Nonwhite | 32 | 11 | 26 | 20 | 21 | 13 | 15 |
| % Smoking | 40 | 35 | 28 | 33 | 30 | 25 | 26 |
| % Hepatitis C infection | 17 | 4 | 8 | 8 | 7 | 6 | 7 |
| % Obese | 18 | 12 | 20 | 16 | 20 | 19 | 19 |
| % Diabetes | 7 | 1 | 10 | 6 | 4 | 3 | 3 |
| MACS site | |||||||
| % Baltimore | 24 | 29 | 23 | 26 | 23 | 20 | 21 |
| % Chicago | 24 | 22 | 22 | 22 | 21 | 19 | 19 |
| % Pittsburgh | 32 | 22 | 24 | 25 | 22 | 29 | 27 |
| % Los Angeles | 20 | 27 | 30 | 27 | 34 | 33 | 33 |
HAART: highly active antiretroviral therapy, IQR: interquartile range.
Individuals could contribute to more than one group; number given represents unique individuals in each category.
In the SUP group, the median CD4 cell count at HAART initiation was 330 cells/µl (IQR 197–520). The median time between HAART initiation and HIV suppression for those with a known date of suppression (n=683) was 0.5 years (IQR 0.1–1.3). An AIDS-defining illness was diagnosed prior to 4% of NAI person-visits and 15% of SUP person-visits.
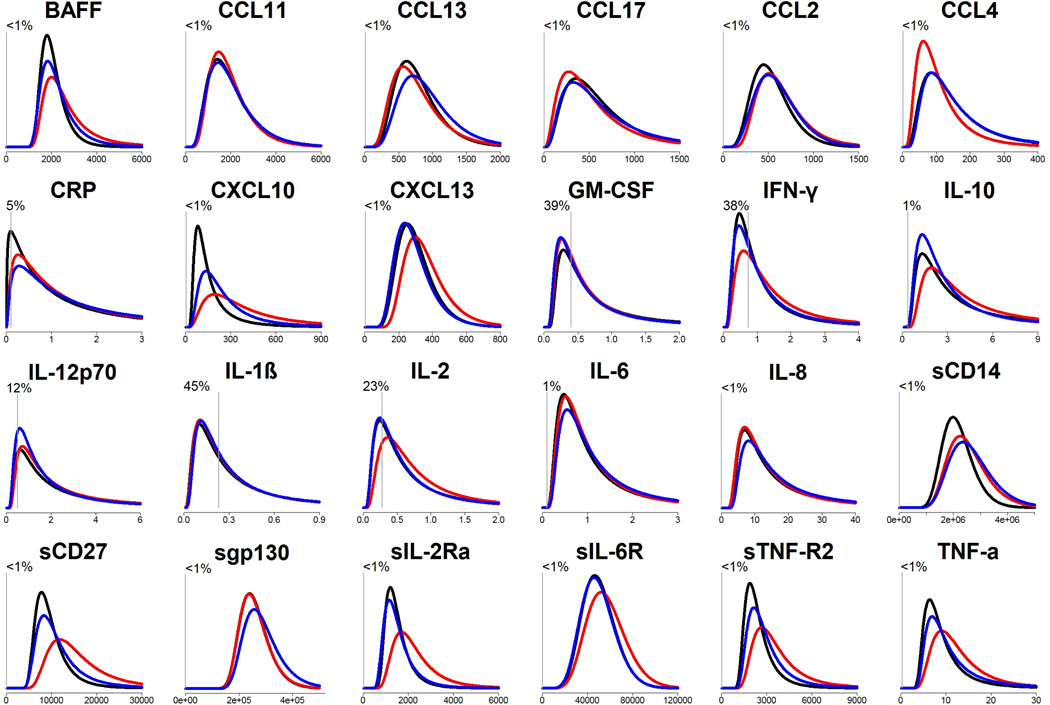
Figure 1 shows estimated distributions of each biomarker for each group, unadjusted for covariates (detailed estimates in Supplemental Table 1). Clear differences across groups are observable for most biomarkers. For example, CXCL10 values were lower and more tightly distributed among the NEG group than values among the SUP group, which in turn were lower than those among the NAI group. As another example, values of CCL4 also differed across groups, but in this case the NAI group was lower than the other two groups. The proportion of undetectable values was ≤1% for most biomarkers, the exceptions being CRP (5%) and 5 cytokines (12–45%).
Figure 1.
Estimated distributions of biomarker levels for HIV-negative, HIV+ HAART-naïve, and HIV+ virally suppressed person-visits.
Results are from saturated generalized gamma models. X-axis: pg/mL. Y-axis: probability density. Black=HIV-negative (NEG), red=HIV+ HAART-naïve (NAI), blue=HIV+ virally suppressed (SUP). Vertical gray lines represent the median lower limit of detection for assays. The percentage represents % of biomarker measurements that were below the lower limit of detection (right-censored by models by inverting values). Details of model estimates are shown in Supplemental Table 1.
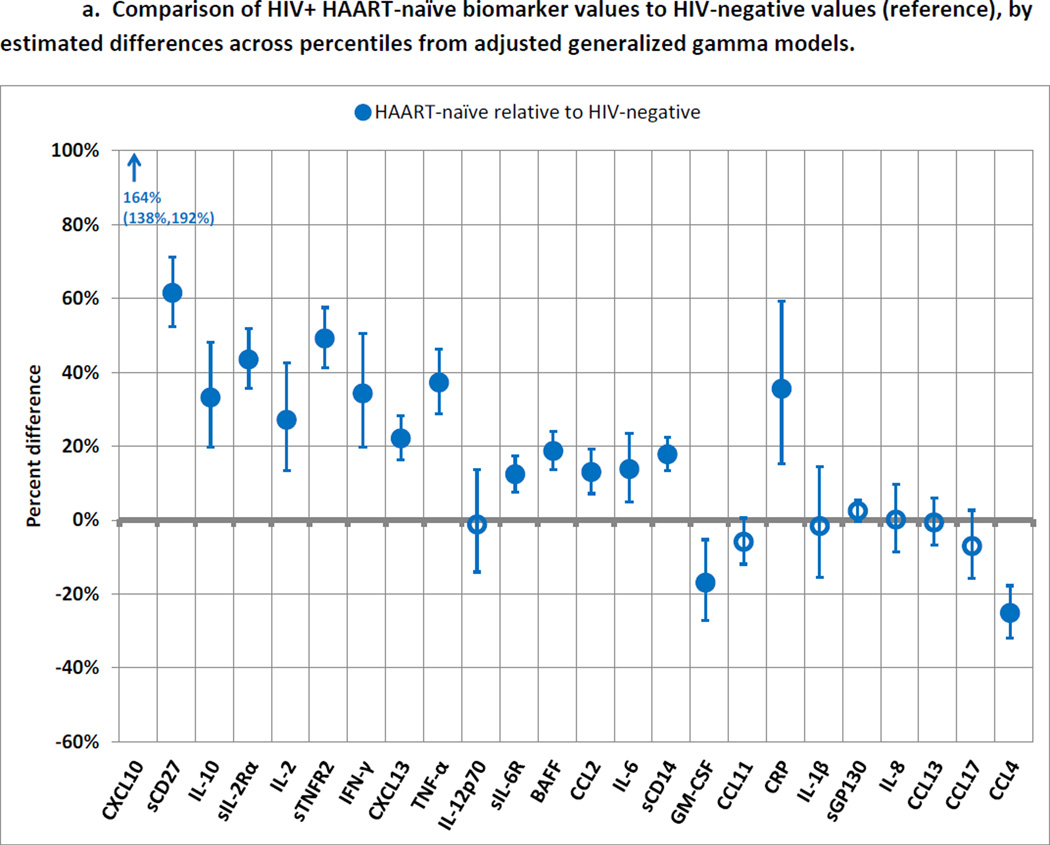
Figure 2a illustrates differences in each percentile of biomarker levels in the NAI group relative to the NEG group, after adjustment for age, race, smoking, HCV infection, obesity, diabetes, and MACS site (detailed estimates in Supplemental Table 2). These results represent the association between untreated HIV infection and biomarker levels. Almost all of the biomarkers were higher in the NAI group, and most of these differences were significant. Only two biomarkers (CCL4 and GM-CSF) were significantly lower in the NAI group.
Figure 2.
a. Comparison of HIV+ HAART-naïve biomarker values to HIV-negative values (reference), by estimated differences across percentiles from adjusted generalized gamma models.
Blue circles represent values of biomarkers among HIV+ HAART-naïve men compared to values among HIV-negative men as the reference category. Error bars represent 99.7% confidence intervals, calculated with Bonferroni adjustment to maintain a family-wise error rate of 0.05. Filled markers represent statistical significance. Results are displayed in the same order as in Figure 2b (sorted by magnitude of difference between HIV-suppressed and HIV+ HAART-naïve groups) to facilitate visual comparisons. Models were adjusted for age, race, smoking, hepatitis C infection, obesity, diabetes, and MACS site. Location parameters of generalized gamma models were allowed to differ by exposure category, while scale and shape parameters were held constant. Details of model estimates are shown in Supplemental Table 2.
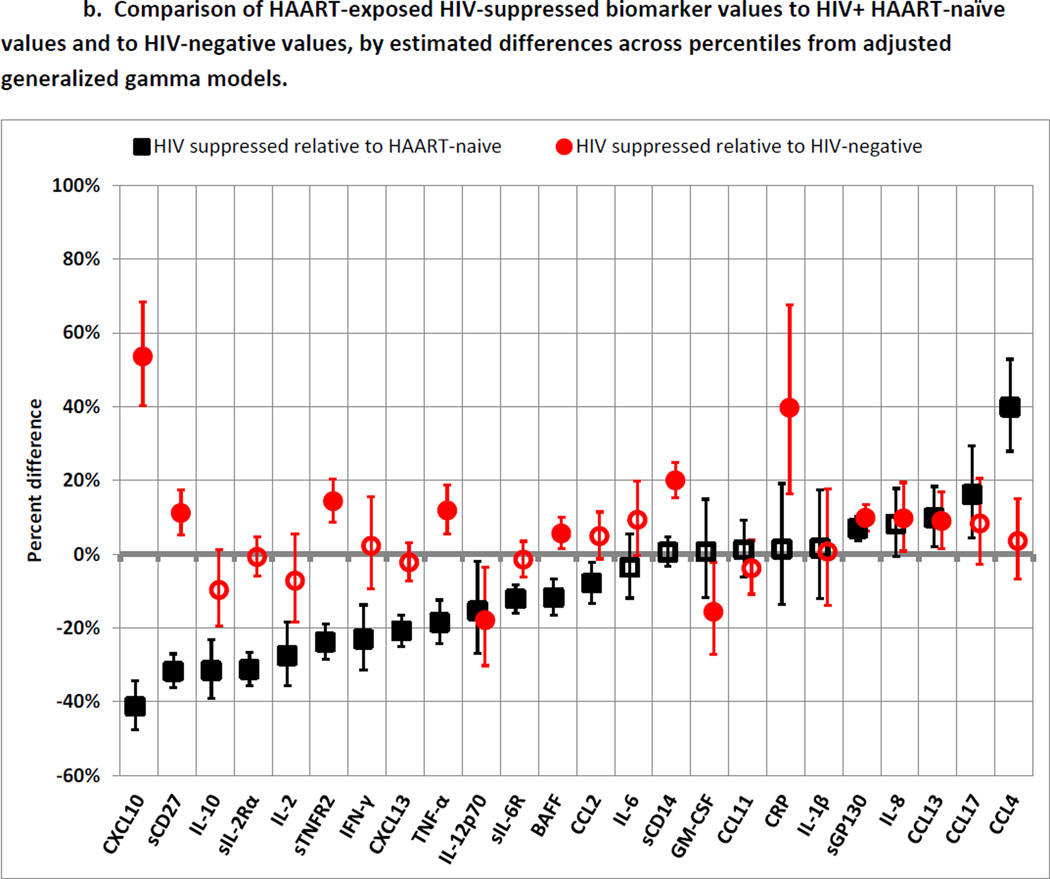
b. Comparison of HAART-exposed HIV-suppressed biomarker values to HIV+ HAART-naïve values and to HIV-negative values, by estimated differences across percentiles from adjusted generalized gamma models.
Black squares represent values of biomarkers among HIV-suppressed men compared to values among HIV+ HAART-naïve men as the reference category. Red circles represent values of biomarkers among HIV-suppressed men compared to values among HIV-negative men as the reference category. Error bars represent 99.7% confidence intervals, calculated with Bonferroni adjustment to maintain a family-wise error rate of 0.05. Filled markers represent statistical significance. Models were adjusted for age, race, smoking, hepatitis C infection, obesity, diabetes, and MACS site. Location parameters of generalized gamma models were allowed to differ by exposure category, while scale and shape parameters were held constant. Details of model estimates are shown in Supplemental Table 2.
Figure 2b illustrates differences across percentiles in biomarker levels for the SUP group compared to the NAI and NEG groups. These results represent the association between HAART-induced HIV suppression and biomarker levels. Fifteen of the 17 significant differences between the NAI and NEG groups (Figure 2a) were smaller when comparing the SUP group to the NEG group. Thirteen biomarkers were significantly (p<0.002) lower in the SUP group than the NAI group: CXCL10 (−41%), sCD27 (−32%), IL-10 (−32%), sIL-2Rα (−31%), IL-2(−28%), sTNFR2 (−24%), IFN-γ (−23%), CXCL13 (−21%), TNF-α (−19%), IL-12p70 (−15%), sIL-6R (−12%), BAFF (−12%), and CCL2 (−8%). All but one (IL-12p70) of these differences represented normalization, since NAI values were higher than NEG values (Figure 2a). In fact, 7 of these (IL-10, sIL-2Rα, IL-2, IFN-γ, CXCL13, sIL-6R, and CCL2) were statistically indistinguishable between the SUP and NEG groups, while CXCL10 (+54%), sCD27 (+11%), sTNFR2 (+14%), TNF-α (+12%), and BAFF (+6%) were still significantly higher in the SUP group than the NEG group.
Seven biomarkers were statistically indistinguishable between the NAI and SUP groups and thus appeared unaffected by HAART-induced HIV suppression: IL-6, sCD14, GM-CSF, CCL11, CRP, IL-1β, and IL-8. Finally, four biomarkers were higher in the SUP group than in the NAI group: sGP130 (+7%), CCL13 (+10%), CCL17 (+16%), and CCL4 (+40%). For CCL4, which was lower in the NAI group than the NEG group, this difference represented normalization after HAART initiation.
Longitudinal analysis
The goal of the longitudinal analysis was to define how biomarkers changed with time after HAART-induced viral suppression. 550 men from the SUP group were excluded from the analysis because either their dates of HIV suppression were not known within ± 1 year (n=323), or they lacked biomarker measurements prior to suppression or during the period of suppression (n=227). Thus, 456 men contributing 1,805 person-visits were eligible for the longitudinal analysis. Characteristics of these men are shown in Table 1b; in general, this study population closely resembled the population in the SUP group. Men contributed a median of 2 (IQR 1–4) post-suppression visits; visits occurred at a median of 2.14 (IQR 0.6–5.6) years after onset of suppression.
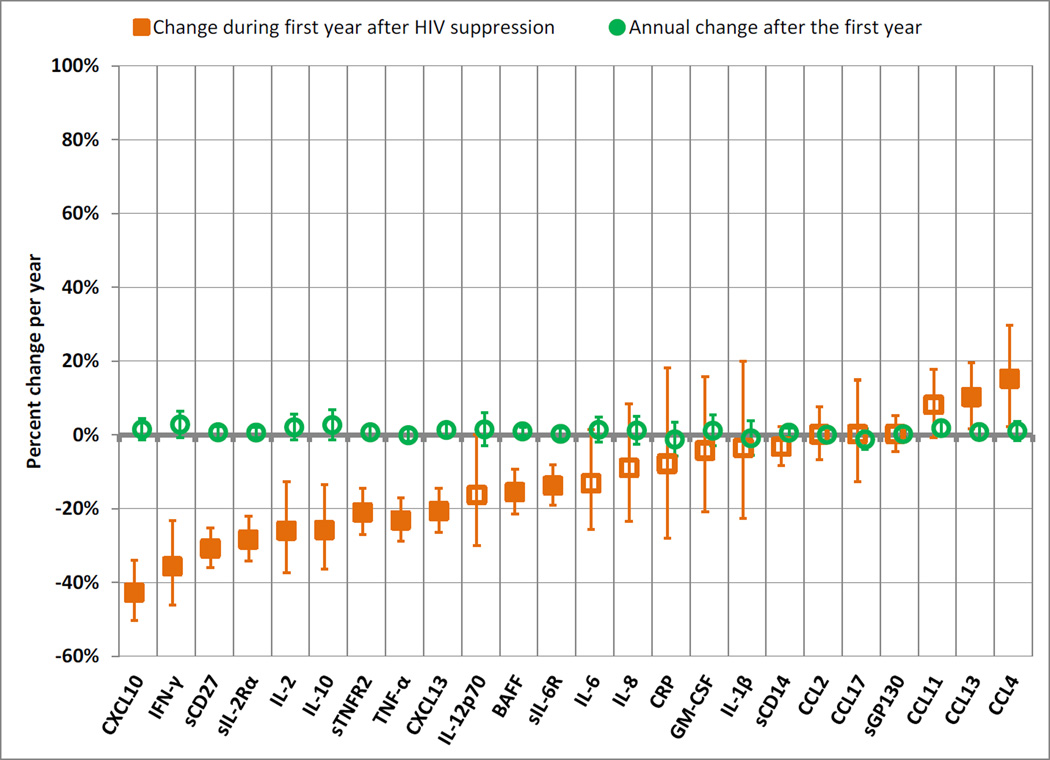
Figure 3 displays two sets of relative percentiles, the first representing the change during the first year of HIV suppression and the second the annual change thereafter (detailed model estimates in Supplemental Table 3). In the first year, 18 of the 24 biomarkers declined, and 11 of these declines were significant: CXCL10 (change of −43% during one year of viral suppression), IFN-γ (−36%), sCD27 (−31%), sIL-2Rα (−28%), IL-2 (−26%), IL-10 (−26%), sTNFR2 (−23%), TNF-α (−21%), CXCL13 (−21%), BAFF (−16%), and sIL-6R (−14%). The chemokines CCL4 (+15%) and CCL13 (+10%) showed significant increases during the first year of viral suppression. These results were congruent with those from the first analysis: of the biomarkers that were lower among the SUP group than the NAI group in the across-group analysis, all but CCL2 and IL12p70 exhibited significant declines during the first year in the longitudinal analysis, and the decline in IL12p70 in the longitudinal analysis was nearly significant. After year 1, biomarker trajectories were uniformly flat, with no biomarker changing significantly.
Figure 3.
Changes inbiomarker values during the first year after HIV viral suppression, and annual changes for each subsequent year, by estimated differences across percentiles from generalized gamma models.
Orange squares represent the percent change in biomarker values during the first year after HIV suppression. Green circles represent the annual percent change in biomarker values after the first year of HIV suppression. Error bars represent 99.7% confidence intervals, calculated with Bonferroni adjustment to maintain a family-wise error rate of 0.05. Filled markers represent differences that were statistically significant after Bonferroni adjustment (p<0.002). Location parameters of generalized gamma models were allowed to differ by exposure category, while scale and shape parameters were held constant. Details of model estimates are shown in Supplemental Table 3.
To test for effect modification by the measured covariates described in the Methods, we allowed changes during the first year and during subsequent years to differ by covariate values. No covariates exhibited evidence of interaction with time across multiple biomarkers.
We performed several sensitivity analyses: restriction of the SUP group to men who were ART-naïve before HAART initiation; changing the time origin from the date of viral suppression to the date of HAART initiation; using a less strict definition of HIV suppression (<1000 copies/mL); permitting intermittent virologic rebound in the longitudinal analysis; using different splines or modeling nonlinearity using polynomial terms; and testing for confounding/effect modification by CD4+ cell count, statin use, and calendar period. None of these variations substantially altered the results.
Discussion
To our knowledge, this is the largest study yet reported to examine the effect of HAART-induced HIV suppression on inflammatory and immune biomarkers, both in sample size and in the number of biomarkers assessed. The MACS was well-suited for addressing these questions, because serum samples were available for a long follow-up time, and because the MACS recruited HIV-uninfected men from the same population as the men with untreated HIV infection and with HAART-induced HIV suppression. Moreover, the timing of viral suppression was well-characterized, as were values of possible confounding covariates.
These aspects of the study design allowed us to identify a constellation of biomarkers of immune activation and inflammation that were: a) abnormal in untreated HIV infection (17 markers) and b) showed at least some restoration toward HIV-negative levels with HIV suppression (15 markers). Among those showing restoration toward HIV-negative levels, 7 markers were indistinguishable from HIV-negative levels among the HIV-suppressed group: the cytokines IL-10, IL-2, and IFN-γ, the cytokine receptors sIL-2Rα and sIL-6R, and the chemokines CCL2 and CXCL13. All of these markers are associated with immune activation. For example, CXCL13 is produced by CD4+ T follicular helper cells and drives B cell migration to germinal centers in secondary lymphoid organs [21]. Therefore, it appears that HAART may be resolving ongoing immune activation, perhaps by removing antigenic stimulation by HIV.
The 7 markers that were still abnormally high among the HIV-suppressed were the chemokine CXCL10 (also known as interferon gamma-induced protein 10 or IP-10), the soluble cytokine receptors sCD27 and sTNFR2, the proinflammatory cytokines TNF-α and BAFF, the soluble scavenger receptor sCD14, and the acute phase reactant CRP. Interestingly, these biomarkers include members of the TNF (TNF-α, BAFF) or the TNF-receptor (sCD27, sTNFR2) superfamilies [22], and a receptor associated with microbial translocation and stimulation by LPS (sCD14) [23]. Meanwhile, several important markers of T-cell activation (IL-2, sIL-2Ra, IFN-γ) were essentially normalized among the HIV-suppressed. These data strongly suggest that some of the systemic inflammation that remains after HIV suppression may be related to ongoing monocyte/macrophage activation.
Untreated HIV infection has been reported to be associated with changes in macrophage phenotype from M1 activation in early infection toward M2 activation later, and eventually toward dysfunctional macrophage activity [24, 25]. In this study, HAART-naïve men exhibited evidence for both M1 activation (high levels of CXCL10, sIL-2Rα, TNF-α, IL-6, and CCL2) and M2 activation (high levels of IL-10 and low levels of CCL4). HIV suppression appeared to reduce levels of several M1-associated biomarkers and to increase levels of the M2-associated chemokines CCL13 and CCL17. However, some M1 biomarkers remained higher among HIV-suppressed men than among HIV-negative men (CXCL10, TNF-α, sTNFR2, and CRP).
This residual inflammation persisted even after many years of HIV suppression. Nearly all of the (partial or complete) restoration occurred during the first year after HAART-induced viral suppression, and the annual change in the markers after the first year was essentially zero. Identifying the underlying processes that cause this residual inflammation among individuals responding well to HAART may aid in setting therapy targets. For example, in the general population, elevated CRP levels are associated with kidney disease mortality [26], cardiovascular disease [27], and colon cancer [28]. Moreover, residual immune activation itself may promote the persistence of HIV despite HAART [29]. The finding that biomarkers were stable after one year of viral suppression suggests that repeated assessments of these biomarkers may not be needed for studies of long-term clinical outcomes in people with stable viral suppression. Further study will be needed to confirm this point.
This study had several limitations. First, although applying a standardized analytical approach across all biomarkers facilitated interpretation of results, it also meant that we did not tailor individual models with unique covariates for each biomarker. Instead, we examined results across all biomarkers and chose models that balanced parsimony with capturing potential confounders. It is reassuring that 55% of the men in the NAI group also contributed to the SUP group, because uncontrolled potential confounders may vary less within individuals. Second, because we performed multiple tests without individualized hypotheses, we adjusted the alpha level for each test using a Bonferroni correction. This approach is known to be highly conservative and to result in low power [30]. When the tests are correlated, as they were here, the conservatism of the Bonferroni approach is particularly acute. Nonetheless, our results still demonstrated strong exposure-outcome associations. Third, the study was limited to men.
This study also had important strengths. First, as mentioned above, the nature of the MACS permitted long-term assessment of biomarker trajectories following the precisely defined date of HIV suppression. Second, the inclusion of HIV-uninfected men from the same population provided an optimal comparison group, allowing us to identify biomarkers that returned to normal levels or remained elevated or suppressed after HIV suppression [20]. Third, this study benefited from the multiplex assays that measured biomarker levels even at low concentrations, as the broad patterns discussed above would not have been identifiable had a smaller set of biomarkers been assessed. Laboratory variability was diminished by the use of centralized laboratories and by the quality control measures described in the Methods. Finally, using generalized gamma models allowed flexibility and minimized assumptions about the shape of the biomarker distributions.
Using serum markers as a proxy for inflammatory and/or immune hyperactivity states holds real utility, especially as levels of these markers may be ascertained via highly sensitive, reproducible, and multiplexed assays. Next steps include identifying associations with morbidity and mortality among these biomarkers, and inferring biological mechanisms for these associations from clusters of biomarkers that behave similarly [31]. Developing reliable surrogates of harmful inflammation or immune activation will facilitate development of effective interventions, and we believe this study is a useful contribution to that process.
Supplementary Material
Acknowledgment
Funding: The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute (UO1-AI-35042, UL1-RR025005, UM1-AI-35043, UO1-AI-35039, UO1-AI-35040 and UO1-AI-35041). Additional funding is provided by the UCLA Older Americans Independence Center Inflammatory Biology Core (5P30 AG028748).
Additional acknowledgements: The authors wish to thank Larry Magpantay, Guadalupe Peña, and Jose Leon Merino (UCLA AIDS Institute), who did the Luminex assays, and Joseph Lopez who did the MSD assays, for their expert technical support and assistance. We also thank the Becton Dickenson Immune Function Laboratory at the Johns Hopkins Bloomberg School of Public Health for providing technical and analytical assistance with all MSD assays.
Footnotes
Conflicts of interest: none declared.
Author contributions: N.I.W. was responsible for conceptualizing the study, carried out the analysis in close collaboration with L.P.J. and had primary responsibility for preparing the manuscript. L.P.J., J.B.M., E.C.B., B.M., S.P., O.M.M., and J.H.B. were responsible for the design and conduct of the cohort studies and of the biomarker sub-study, which serve as the basis for the work presented here. L.P.J. helped design the initial stages of the analysis and contributed to the drafting of the manuscript. J.B.M., E.C.B., and J.H.B. implemented critical revisions of the manuscript and provided important intellectual content.
Ethics committee approval: Studies were approved by the Committees of Human Research of participating institutions.
References
- 1.Egger M, Hirschel B, Francioli P, Sudre P, Wirz M, Flepp M, et al. Impact of new antiretroviral combination therapies in HIV infected patients in Switzerland: prospective multicentre study. BMJ. 1997;315:1194–1199. doi: 10.1136/bmj.315.7117.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Detels R, Muñoz A, McFarlane G, Kingsley LA, Margolick JB, Giorgi J, et al. Effectiveness of potent antiretroviral therapy on time to AIDS and death in men with known HIV infection duration. Multicenter AIDS Cohort Study Investigators. JAMA. 1998;280:1497–1503. doi: 10.1001/jama.280.17.1497. [DOI] [PubMed] [Google Scholar]
- 3.Wada N, Jacobson L, Cohen M, French AL, Phair JP, Muñoz A. Cause-specific life expectancies after age 35 for HIV-infected and HIV-negative individuals followed simultaneously in long-term cohort studies: 1984–2008. Am J Epidemiol. 2013;177:116–125. doi: 10.1093/aje/kws321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lederman MM, Funderburg NT, Sekaly RP, Klatt NR, Hunt PW. Residual immune dysregulation syndrome in treated HIV infection. Advances in immunology. 2013;119:51–83. doi: 10.1016/B978-0-12-407707-2.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plaeger SF, Collins BS, Musib R, Deeks SG, Read S, Embry A. Immune activation in the pathogenesis of treated chronic HIV disease: a workshop summary. AIDS research and human retroviruses. 2012;28:469–477. doi: 10.1089/aid.2011.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. The Journal of infectious diseases. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 7.Lederman MM, Calabrese L, Funderburg NT, Clagett B, Medvik K, Bonilla H, et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. The Journal of infectious diseases. 2011;204:1217–1226. doi: 10.1093/infdis/jir507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazenberg MD, Otto SA, van Benthem BH, Roos MT, Coutinho RA, Lange JM, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 9.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nature medicine. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 10.Hatano H. Immune activation and HIV persistence: considerations for novel therapeutic interventions. Current opinion in HIV and AIDS. 2013;8:211–216. doi: 10.1097/COH.0b013e32835f9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Regidor DL, Detels R, Breen EC, Widney DP, Jacobson LP, Palella FJ, Jr, et al. Effect of highly active antiretroviral therapy on biomarkers of B-lymphocyte activation and inflammation. AIDS. 2011;25:303–314. doi: 10.1097/QAD.0b013e32834273ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Widney DP, Breen EC, Boscardin WJ, Kitchen SG, Alcantar JM, Smith JB, et al. Serum levels of the homeostatic B cell chemokine, CXCL13, are elevated during HIV infection. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2005;25:702–706. doi: 10.1089/jir.2005.25.702. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan RC, Landay AL, Hodis HN, Gange SJ, Norris PJ, Young M, et al. Potential cardiovascular disease risk markers among HIV-infected women initiating antiretroviral treatment. Journal of acquired immune deficiency syndromes. 2012;60:359–368. doi: 10.1097/QAI.0b013e31825b03be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuhaus J, Jacobs DR, Jr, Baker JV, Calmy A, Duprez D, La Rosa A, et al. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. The Journal of infectious diseases. 2010;201:1788–1795. doi: 10.1086/652749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 16.Hultin LE, Menendez FA, Hultin PM, Jamieson BD, O'Gorman MR, Borowski L, et al. Assessing immunophenotyping performance: proficiency-validation for adopting improved flow cytometry methods. Cytometry. Part B, Clinical cytometry. 2007;72:249–255. doi: 10.1002/cyto.b.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider MF, Margolick JB, Jacobson LP, Reddy S, Martinez-Maza O, Munoz A. Improved estimation of the distribution of suppressed plasma HIV-1 RNA in men receiving effective antiretroviral therapy. Journal of acquired immune deficiency syndromes. 2012;59:389–392. doi: 10.1097/QAI.0b013e318246bfce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox C, Chu H, Schneider MF, Muñoz A. Parametric survival analysis and taxonomy of hazard functions for the generalized gamma distribution. Stat Med. 2007;26:4352–4374. doi: 10.1002/sim.2836. [DOI] [PubMed] [Google Scholar]
- 19.Dunn OJ. Multiple comparisons among means. Journal of the American Statistical Association. 1961;50:1096–1121. [Google Scholar]
- 20.Bosch RJ, Zhang X, Sandler NG. Study design issues in evaluating immune biomarkers. Current opinion in HIV and AIDS. 2013;8:147–154. doi: 10.1097/COH.0b013e32835d3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crotty S. Follicular helper CD4 T cells (TFH) Annual review of immunology. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 22.Kumar A, Abbas W, Herbein G. TNF and TNF receptor superfamily members in HIV infection: new cellular targets for therapy? Mediators of inflammation. 2013;2013:484378. doi: 10.1155/2013/484378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. The Journal of infectious diseases. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassol E, Cassetta L, Rizzi C, Alfano M, Poli G. M1 and M2a polarization of human monocyte-derived macrophages inhibits HIV-1 replication by distinct mechanisms. Journal of immunology. 2009;182:6237–6246. doi: 10.4049/jimmunol.0803447. [DOI] [PubMed] [Google Scholar]
- 25.Herbein G, Varin A. The macrophage in HIV-1 infection: from activation to deactivation? Retrovirology. 2010;7:33. doi: 10.1186/1742-4690-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, He J, Zhang F, Huang C, Wu Y, Han Y, et al. Prognostic role of C-reactive protein and interleukin-6 in dialysis patients: a systematic review and meta-analysis. Journal of nephrology. 2013;26:243–253. doi: 10.5301/jn.5000169. [DOI] [PubMed] [Google Scholar]
- 27.Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. The New England journal of medicine. 1997;336:973–979. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- 28.Swede H, Hajduk AM, Sharma J, Rawal S, Rasool H, Vella AT, et al. Baseline serum C-reactive protein and death from colorectal cancer in the NHANES III cohort. International journal of cancer. Journal international du cancer. 2014;134:1862–1870. doi: 10.1002/ijc.28504. [DOI] [PubMed] [Google Scholar]
- 29.Klatt NR, Chomont N, Douek DC, Deeks SG. Immune activation and HIV persistence: implications for curative approaches to HIV infection. Immunological reviews. 2013;254:326–342. doi: 10.1111/imr.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995;57:289–300. [Google Scholar]
- 31.Kamat A, Misra V, Cassol E, Ancuta P, Yan Z, Li C, et al. A plasma biomarker signature of immune activation in HIV patients on antiretroviral therapy. PloS one. 2012;7:e30881. doi: 10.1371/journal.pone.0030881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.