Abstract
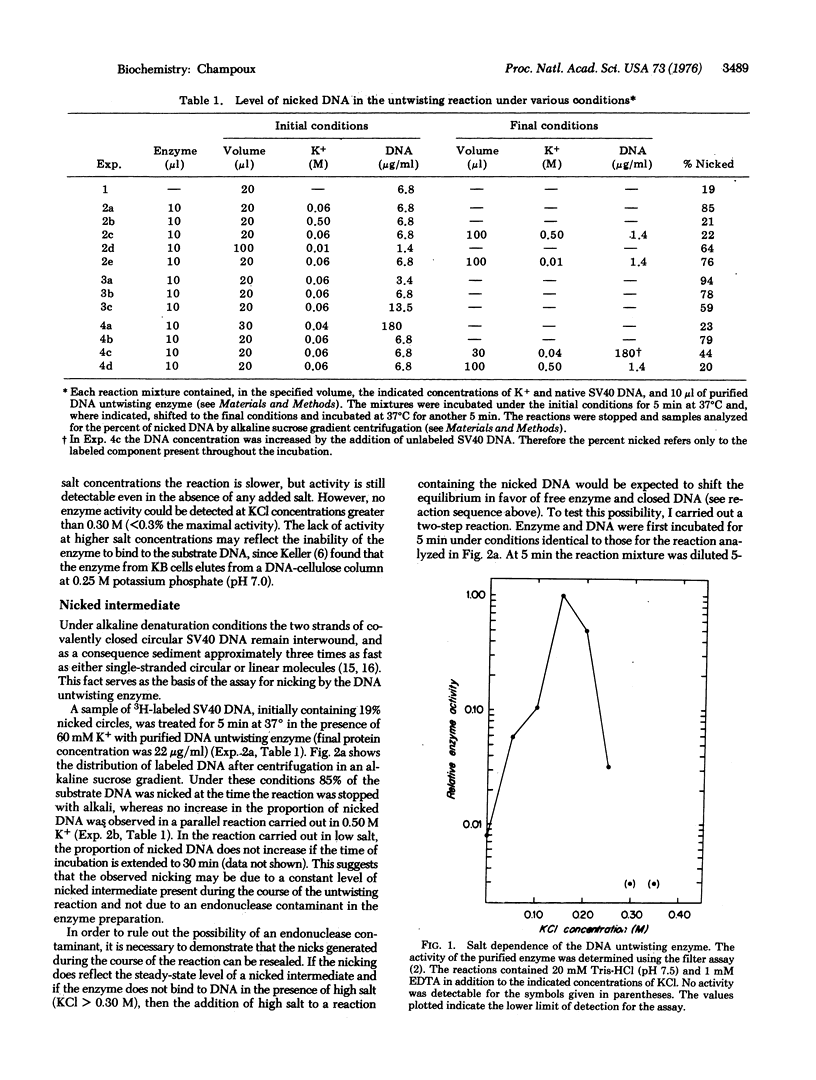
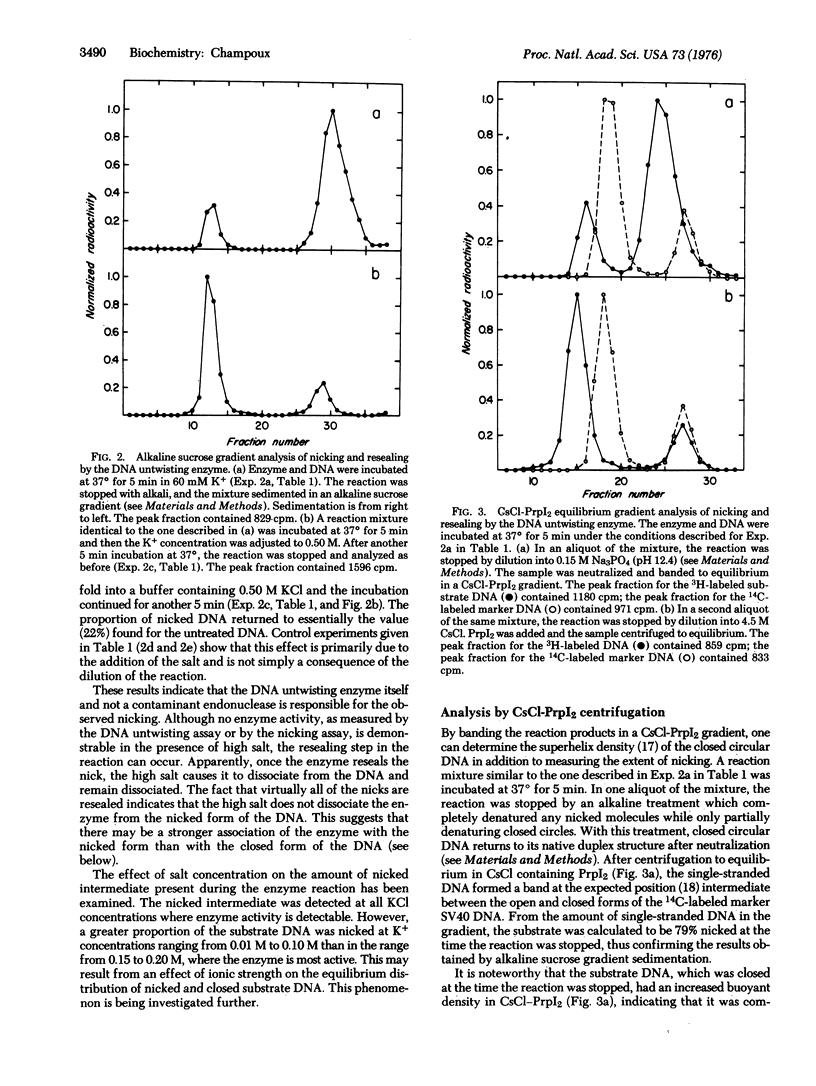
The DNA untwisting enzyme relaxes covalently closed circylar DNAs by the sequential breaking (nicking) and closure of one strand of the duplex. The use of highly purified enzyme from rat liver nuclei at very high protein concentrations has permitted the detection of the nicked intermediate in the reaction. The nicking of closed circular simian virus 40 DNA was measured by alkaline sucrose gradient sedimentation or by equilibrium centrifugation in CsCl gradients containing propidium diiodide. The following observations support the hypothesis that the nicked DNA represents an intermediate in the untwisting reaction. The extent of nicking does not increase with time. Nicking is observed in the range of salt concentrations where the enzyme is active (0.01-0.25 M KCl), but is not observed at 0.50 Mkdl, where enzyme activity is undetectable. The nicked DNA that is generated during the reaction carried out in low salt rapidly disappears if the KCl concentration is raised to 0.50 M. At constant enzyme concentration, the number of nicks in the reaction mixture is independent of DNA concentration in the range from 3 to 14 mug/ml. The addition of an excess of unlabeled DNA to a reaction initially containing labeled nicked DNA partially chases the label from the nicked intermediate into covalently closed circular DNA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baase W. A., Wang J. C. An omega protein from Drosophila melanogaster. Biochemistry. 1974 Oct 8;13(21):4299–4303. doi: 10.1021/bi00718a009. [DOI] [PubMed] [Google Scholar]
- Bauer W., Vinograd J. Interaction of closed circular DNA with intercalative dyes. II. The free energy of superhelix formation in SV40 DNA. J Mol Biol. 1970 Feb 14;47(3):419–435. doi: 10.1016/0022-2836(70)90312-8. [DOI] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., McConaughy B. L. Priming of superhelical SV40 DNA by Escherichia coli RNA polymerase for in vitro DNA synthesis. Biochemistry. 1975 Jan 28;14(2):307–316. doi: 10.1021/bi00673a017. [DOI] [PubMed] [Google Scholar]
- DULBECCO R., VOGT M. EVIDENCE FOR A RING STRUCTURE OF POLYOMA VIRUS DNA. Proc Natl Acad Sci U S A. 1963 Aug;50:236–243. doi: 10.1073/pnas.50.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Upholt W. B., Vinograd J. A buoyant method for the determination of the superhelix density of closed circular DNA. J Mol Biol. 1971 Nov 28;62(1):1–19. doi: 10.1016/0022-2836(71)90127-6. [DOI] [PubMed] [Google Scholar]
- Grossman L. I., Watson R., Vinograd J. Restricted uptake of ethidium bromide and propidium diiodide by denatured closed circular DNA in buoyant cesium chloride. J Mol Biol. 1974 Jun 25;86(2):271–283. doi: 10.1016/0022-2836(74)90018-7. [DOI] [PubMed] [Google Scholar]
- Keller W. Characterization of purified DNA-relaxing enzyme from human tissue culture cells. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2550–2554. doi: 10.1073/pnas.72.7.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Morgan A. R. Partial purification of "omega" protein from calf thymus. Biochemistry. 1975 Nov 18;14(23):5205–5209. doi: 10.1021/bi00694a029. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman A., Champoux J. J., Dulbecco R. Characterization of the replicative intermediates of polyoma virus. Virology. 1974 Jan;57(1):147–160. doi: 10.1016/0042-6822(74)90116-0. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Vosberg H. P., Grossman L. I., Vinograd J. Isolation and partial characterisation of the relaxation protein from nuclei of cultured mouse and human cells. Eur J Biochem. 1975 Jun 16;55(1):79–93. doi: 10.1111/j.1432-1033.1975.tb02140.x. [DOI] [PubMed] [Google Scholar]
- WEIL R., VINOGRAD J. THE CYCLIC HELIX AND CYCLIC COIL FORMS OF POLYOMA VIRAL DNA. Proc Natl Acad Sci U S A. 1963 Oct;50:730–738. doi: 10.1073/pnas.50.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]


