Abstract
Background
Stoned olive pomace (SOP), which represents approximately 50% of the conversion process of olives to olive oil, is largely not utilised and creates costs for its disposal and has negative environmental impacts. In vitro trial experiments were employed to study the effect of feeds integrated with this bio-waste, which is rich in polyphenols, on rumen biohydrogenation, using sheep rumen liquor as inoculum.
Results
Fatty acid (FA) analysis and a polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) approach aimed at characterising the microbial community indicated that including SOP in feeds at the level of 50 g/kg and 90 g/kg induced changes in the FA profile and microbial populations. The simultaneous decrease of Butyrivibrio proteoclasticus and accumulation of vaccenic acid was observed. A depression in the populations of Neisseria weaveri, Ruminobacter amylophilus and other unclassified bacteria related to members of the Lachnospiraceae and Pasteurellaceae families was detected, suggesting that these microbial groups may be involved in rumen biohydrogenation.
Conclusions
Supplementation of feeds with SOP alters the rumen bacterial community, including bacteria responsible for the hydrogenation of vaccenic acid to stearic acid, thereby modifying the FA profile of the rumen liquor. Hence, a use of SOP aimed to produce meat or dairy products enriched in functional lipids can be hypothesised.
Keywords: Stoned olive oil pomace, Sheep rumen microbiota, Fatty acid biohydrogenation, PCR-DGGE
Background
The primary factor affecting ruminal biohydrogenation (BH) is the quality of the animal diet, the composition of which can affect the content of healthy fatty acids (FA) in milk and meat. In particular, the concentrate to forage ratio plays an important role in the accumulation of several BH intermediates, especially if the diet is rich in polyunsaturated fatty acids (PUFA) [1]. The inclusion of polyphenols in ruminant feeds has an inhibitory effect on the BH of dietary PUFA, as consequence of their influence on microbial activity and diversity [2]. This has been shown to increase the duodenal flow of bioactive FA, such as vaccenic acid (trans-11 C18:1, VA) and, as consequence, improve the nutritional value of milk fat from large and small dairy ruminants because this FA can be ∆9-desaturated to rumenic acid (cis-9,cis-12 C18:2, RA) in the mammary gland and other tissues. However, from the literature it is well known that the availability of VA in ruminant products is limited at the rumen level by its hydrogenation to stearic acid (C18:0, SA) or by the isomerisation to other C18:1 isomers by microbial activity taking place in the rumen [3].
The rumen microbial community is comprised of an enormous number of microbial species belonging to the Bacteria, Archaea and Eucarya domains. However, only a limited number of rumen microorganisms have been isolated and physiologically characterised thus far [4]. Among the different microbial species already identified, those belonging to the Butyrivibrio group appear of particular interest because they are known to be directly involved in BH [5]. Recent studies carried out on different species of ruminants have reported that diets enriched with polyphenols caused a decrease in SA and a simultaneous decline in key species of Butyrivibrio [2,6].
In the Mediterranean region, during the period of olive oil production, there is a high production of moist virgin olive pomace that presents a problem for its disposal. The most recent stoning virgin olive pomace techniques result in a residual product (stoned olive oil pomace, SOP) after the mechanical extraction of oil from olives followed by the drying of the cake on a fluid bed dryer. SOP is characterised by a high level of polyphenols (approximately 13 g/kg of dry matter) and low lignin content, which is considered to be the main factor that reduces the digestibility of olive pomace or olive cake when these by-products are utilised as animal feed [7-10]. These chemical characteristics render SOP potentially able to interfere with rumen fermentation [7]. However, little information is available in the literature on the effect of the SOP inclusion in ruminant diets on microbial strains involved in BH processes. The aim of the present study was to verify whether SOP supplementation in the sheep diet is able to affect the overall rumen microbial profile and, in particular, the Butyrivibrio group that influences the PUFA profile of rumen liquor (RL).
Results
Rumen liquor fatty acid composition
During the fermentation of the three feeds, the concentration of acetic acid (C2:0) did not exhibit significant differences with the exception of 12 h, when the percentage of this volatile fatty acid was higher in RL fermenting S5 and S9 (Table 1). Moreover, the presence of SOP in feeds significantly increased the concentration of propionic (C3:0), butyric (C4:0) and isovaleric (iso C5:0) acids compared to the content of these FA in RL with C. As consequence, the C2:0/C3:0 ratio in S5 and S9 was significantly lower than that in RL fermented with C at 12 and 24 h (Table 1).
Table 1.
Effect of stoned olive pomace (SOP) concentration (mM) on volatile fatty acid (VFA) production in ruminal fluid at 6, 12 and 24 h of incubation 1
| VFA | Feed | Time (h) | SEM | P 2 | ||
|---|---|---|---|---|---|---|
| 6 | 12 | 24 | FXT | |||
| C2:0 | C | 5.690 cβ | 7.360 bβ | 9.370 a | 0.240 | 0.021 |
| S5 | 6.700 cα | 8.020 bα | 9.190 a | |||
| S9 | 3.690 cγ | 8.530 bα | 9.200 a | |||
| C3:0 | C | 2.450 β | 2.670 β | 3.010 β | 0.340 | 0.047 |
| S5 | 3.090 αβ | 3.250 αβ | 3.790 αβ | |||
| S9 | 3.150 α | 3.990 α | 4.230 α | |||
| C4:0 | C | 3.050 cγ | 3.490 bγ | 3.840 aβ | 0.060 | 0.042 |
| S5 | 3.450 cβ | 3.750 bβ | 4.020 aα | |||
| S9 | 3.670 cα | 3.980 bα | 4.150 aα | |||
| iso C4:0 | C | 0.156 | 0.189 | 0.193 | 0.075 | 0.083 |
| S5 | 0.135 | 0.147 | 0.153 | |||
| S9 | 0.114 | 0.113 | 0.112 | |||
| C5:0 | C | 0.165 | 0.196 | 0.264 | 0.081 | 0.079 |
| S5 | 0.194 | 0.217 | 0.210 | |||
| S9 | 0.105 | 0.233 | 0.235 | |||
| iso C5:0 | C | 0.350 bβ | 0.360 bγ | 0.500 aγ | 0.030 | 0.037 |
| S5 | 0.460 cα | 0.600 bβ | 0.680 aβ | |||
| S9 | 0.490 bα | 0.800 aα | 0.820 aα | |||
| C2/C3 | C | 2.322 bα | 2.756 abα | 3.113 aα | 0.292 | 0.048 |
| S5 | 2.168 α | 2.680αβ | 2.424 β | |||
| S9 | 1.171 bβ | 2.138aβ | 2.175 aβ |
α, β, γ Within a column, means with different Greek superscripts are significantly different (P < 0.05); a, b, c within a row, means with different Latin superscripts are significantly different (P < 0.05). C = control feed; S5 = treatment with 50 g/kg of SOP; S9 = treatment with 90 g/kg of SOP.
1Mean values with their standard errors (SEM); number of samples for each treatment at any time = 3.
2Probability of significant effects due to the interaction of the experimental factors of Feed and Time (FXT).
C14:0 and C16:0 increased in rumen fluid incubated with S5, while with C and S9 the concentration of these FA significantly decreased (Table 2). S5 and S9 significantly increased the C13:0 concentration within 12 h, but at 24 h the percentage of this FA was significantly lower than that found in fermenters containing C (Table 2). C17:0 production was significantly depressed by S9, but not by S5, which was similar to C (Table 2).
Table 2.
Effect of the stoned olive pomace (SOP) concentration (g/100 g total fatty acids, FA) on medium chain fatty acid production in ruminal fluid at 6, 12 and 24 h of incubation 1 ; number of samples for each treatment at any time = 3
| FA | Feed | Time (h) | SEM | P 2 | FA | Feed | Time (h) | SEM | P 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 12 | 24 | F | FXT | 6 | 12 | 24 | F | FXT | ||||||
| C12:0 | C | 0.507 aαβ | 0.447 ab | 0.357 bαβ | 0.048 | 0.069 | 0.027 | anteiso C15 | C | 0.510 β | 0.570 α | 0.523 β | 0.020 | 0.996 | 0.049 |
| S5 | 0.403 abβ | 0.363 b | 0.443 aα | S5 | 0.477 bβ | 0.490 aβ | 0.626 aα | ||||||||
| S9 | 0.553 aα | 0.403 b | 0.320 cβ | S9 | 0.700 aα | 0.463 bβ | 0.443 bγ | ||||||||
| C13:0 | C | 0.973 bα | 1.133 bγ | 1.676 aα | 0.041 | 0.791 | 0.034 | iso C16 | C | 0.143 α | 0.147β | 0.123 β | 0.020 | 0.199 | 0.027 |
| S5 | 1.117 bα | 1.360 aβ | 1.340 aγ | S5 | 0.106 bβ | 0.147 abβ | 0.173 aα | ||||||||
| S9 | 0.640 bβ | 1.526 aα | 1.463 aβ | S9 | 0.146 αβ | 0.169 bα | 0.186 aα | ||||||||
| C14:0 | C | 0.847 aβ | 0.823 aα | 0.753 bβ | 0.030 | 0.874 | 0.012 | iso C17 | C | 0.110 | 0.136 β | 0.133 β | 0.030 | 0.002 | 0.044 |
| S5 | 0.663 bγ | 0.730 bβ | 0.913 aα | S5 | 0.123 b | 0.183 aα | 0.176 aαβ | ||||||||
| S9 | 0.930 aα | 0.796 bαβ | 0.593 cγ | S9 | 0.107 b | 0.183 aα | 0.193 aα | ||||||||
| C16:0 | C | 5.440 aα | 5.063 bα | 4.957 bβ | 0.058 | 0.059 | 0.039 | anteiso C17 | C | 0.143 bβ | 0.173 a | 0.174 aα | 0.007 | 0.041 | 0.042 |
| S5 | 4.570 cβ | 4.740 bβ | 5.537 aα | S5 | 0.116 bγ | 0.133 b | 0.201 aα | ||||||||
| S9 | 5.780 aα | 4.327 bγ | 3.860 cγ | S9 | 0.177 aα | 0.133 b | 0.101 cβ | ||||||||
| C17:0 | C | 0.080 bβ | 0.093 b | 0.127 aα | 0.020 | 0.943 | 0.048 | cis-9 C12:1 | C | 0.040 aα | 0.047 aα | 0.013 bγ | 0.030 | 0.061 | 0.002 |
| S5 | 0.073 bβ | 0.103 a | 0.120 aα | S5 | 0.017 cβ | 0.036 bα | 0.050 aα | ||||||||
| S9 | 0.113 aα | 0.089 b | 0.088 bβ | S9 | 0.037 aα | 0.020 bβ | 0.030 abβ | ||||||||
| iso C13 | C | 0.081 | 0.111 β | 0.103 β | 0.020 | 0.610 | 0.048 | cis-9 C14:1 | C | 0.277 bβ | 0.353 aα | 0.287 bαβ | 0.016 | 0.077 | 0.047 |
| S5 | 0.086 | 0.081 β | 0.087 β | S5 | 0.233 bγ | 0.260 bβ | 0.363 aα | ||||||||
| S9 | 0.091b | 0.173 aα | 0.167 aα | S9 | 0.333 aα | 0.273 abβ | 0.250 bβ | ||||||||
| iso C14 | C | 0.087 β | 0.087 | 0.097 β | 0.010 | 0.497 | 0.015 | trans-9 C15:1 | C | 0.077 αβ | 0.076 | 0.053 β | 0.030 | 0.046 | 0.046 |
| S5 | 0.077 bβ | 0.091 b | 0.130 aα | S5 | 0.053 bβ | 0.056 b | 0.093 aα | ||||||||
| S9 | 0.110 aα | 0.081 b | 0.081 bβ | S9 | 0.100 aα | 0.057 b | 0.093 aα | ||||||||
| iso C15 | C | 0.076 aα | 0.073 aαβ | 0.037 bβ | 0.010 | 0.275 | 0.047 | cis-9 C16:1 | C | 0.087 bγ | 0.116 aβ | 0.103 abβ | 0.016 | 0.051 | 0.049 |
| S5 | 0.053 bβ | 0.080 aα | 0.076 aα | S5 | 0.167 aα | 0.149 abα | 0.133 bα | ||||||||
| S9 | 0.073 α | 0.057 β | 0.060 α | S9 | 0.123 β | 0.100 β | 0.093 β |
α, β, γ Within a column, means with different Greek superscripts are significantly different (P < 0.05); a,b,c within a row, means with different Latin superscripts are significantly different (P < 0.05). C = control feed; S5 = treatment with 50 g/kg of SOP; S9 = treatment with 90 g/kg of SOP. 1Mean values with their standard error (SEM); 2Probability of significant effect due to the interaction of the experimental factors Feed and Time (FXT).
At the last point of sampling, the concentration of iso C15, iso C16 and iso C17 was significantly higher in S5 and S9 fermenters than in C (Table 2). With respect to C, anteiso C15 content was depressed during the fermentation of S9 and enhanced when S5 was fermented (Table 2). Moreover, the content of C17 ante increased during the fermentation of S5, whereas S9 exhibited the opposite trend (Table 2). The concentrations of cis-9 C12:1, cis-9 C14:1 and trans-9 C15:1 were characterised by an increasing trend in fermenters containing S5 (Table 2). When RL was incubated with S5, cis-11 C18:1 and cis-13 C18:1 increased significantly after 12 h compared to the fermenters containing C and S9 (Table 3). Moreover, S5 significantly decreased the BH rate of cis-9 C18:1, which exhibited the highest concentration at 24 h (Table 3). VA progressively accumulated during the entire fermentation period when SOP was added to feeds regardless of the percentage of inclusion, as consequence of a decrease in the extent of BH (Table 3). No significant differences among feeds were found for the other trans monoenes (Table 3). RA accumulated at 12 h in all cases but, when S5 and S9 were fermented, its percentage in RL was the highest according to a decrease in the BH rate (Table 3). In contrast, trans-10,cis-12 C18:2 was only detected at 12 h in S5 fermenters (Table 3). The BH rates of linoleic (cis-9,cis-12 C18:2, LA) and α-LNA (cis-9,cis-12,cis-15 C18:3) acids were similar in C and S9 (Table 3). It was simply lowered in S5, leading to a higher accumulation of LA and α-LNA at 24 h. Conjugated linolenic acid (cis-9,trans-11,cis-15 C18:3) and vaccelenic acid (trans-11,cis-15 C18:2) were detected at 24 h only in S9 fermenters (Table 3).
Table 3.
Effect of the stoned olive pomace (SOP) concentration on C18 fatty acid (g/100 g FA) production in ruminal fluid at 6, 12 and 24 h of incubation 1 ; number of samples for each treatment at any time = 3
| FA | Feed | Time (h) | SEM | P 2 | FA | Feed | Time (h) | SEM | P 2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 12 | 24 | FXT | 6 | 12 | 24 | FXT | ||||||
| C18:0 | C | 2.377 cα | 2.590 bγ | 4.167 aα | 0.047 | 0.006 | trans-11 C18:1 | C | 0.580 a | 0.350 bβ | 0.120 cβ | 0.021 | 0.005 |
| S5 | 2.203 cβ | 3.080 bα | 3.446 aβ | S5 | 0.483 c | 0.677 bα | 0.850 aα | ||||||
| S9 | 1.760 cγ | 2.806 bβ | 2.999 aγ | S9 | 0.473 c | 0.653 bα | 0.830 aα | ||||||
| cis-9 C18:1 | C | 2.063 aβ | 1.610 bβ | 1.290 cβ | 0.023 | 0.032 | trans-12 C18:1 | C | 0.047 | 0.047 | 0.053 | 0.024 | 0.485 |
| S5 | 1.950 aγ | 1.526 bγ | 1.567 bα | S5 | 0.056 | 0.050 | 0.043 | ||||||
| S9 | 2.563 aα | 1.663 bα | 1.163 cγ | S9 | 0.053 | 0.036 | 0.029 | ||||||
| cis-11 C18:1 | C | 0.437 | 0.420 | 0.473 | 0.068 | 0.046 | cis-9,cis-12 C18:2 | C | 4.527 aβ | 3.180 bα | 1.733 cβ | 0.043 | 0.031 |
| S5 | 0.340 b | 0.360 b | 0.527 a | S5 | 3.750 aγ | 2.653 bβ | 2.060 cα | ||||||
| S9 | 0.487 | 0.350 | 0.363 | S9 | 4.780 aα | 2.523 bγ | 1.680 cβ | ||||||
| cis-12 C18:1 | C | 0.033 c | 0.150 aα | 0.110 bα | 0.010 | 0.006 | cis-9,trans-11 C18:2 | C | 0.000 b | 0.021 aβ | 0.000 b | 0.020 | 0.033 |
| S5 | 0.050 b | 0.060 bβ | 0.117 aα | S5 | 0.000 b | 0.112 aα | 0.000 b | ||||||
| S9 | 0.040 b | 0.040 bβ | 0.073 aβ | S9 | 0.000 b | 0.113 aα | 0.000 b | ||||||
| cis-13 C18:1 | C | 0.040 bβ | 0.070 aα | 0.050 bβ | 0.013 | 0.046 | trans-10,cis-12 C18:2 | C | 0.000 | 0.000 β | 0.000 | 0.001 | 0.001 |
| S5 | 0.036 bβ | 0.040 bβ | 0.073 aα | S5 | 0.000 b | 0.067 aα | 0.000 b | ||||||
| S9 | 0.080 aα | 0.036 bβ | 0.053 bβ | S9 | 0.000 | 0.000 β | 0.000 | ||||||
| cis-15 C18:1 | C | 0.020 β | 0.020 | 0.037 | 0.009 | 0.045 | cis-9,cis-12,cis-15 C18:3 | C | 0.530 aβ | 0.393 bα | 0.283 cβ | 0.006 | 0.044 |
| S5 | 0.036 β | 0.036 | 0.033 | S5 | 0.400 aγ | 0.357 bβ | 0.370 bα | ||||||
| S9 | 0.053 aα | 0.023 b | 0.020 b | S9 | 0.597 aα | 0.337 bγ | 0.283 cβ | ||||||
| trans-9 C18:1 | C | 0.037 | 0.040 | 0.040 | 0.008 | 0.045 | cis-9,trans-11,cis-15 C18:3 | C | 0.000 | 0.000 | 0.000 β | 0.001 | 0.002 |
| S5 | 0.037 | 0.047 | 0.030 | S5 | 0.000 | 0.000 | 0.000 β | ||||||
| S9 | 0.037 | 0.050 | 0.027 | S9 | 0.000 b | 0.000 b | 0.056 aα | ||||||
| trans-10 C18:1 | C | 0.047 | 0.040 | 0.053 | 0.011 | 0.910 | trans-11,cis-15 C18:2 | C | 0.000 | 0.000 | 0.000 β | 0.001 | 0.004 |
| S5 | 0.060 | 0.063 | 0.057 | S5 | 0.000 | 0.000 | 0.000 β | ||||||
| S9 | 0.043 | 0.050 | 0.047 | S9 | 0.000 b | 0.000 b | 0.143 aα |
α, β, γ Within a column, means with different Greek superscripts are significantly different (P < 0.05); a, b, c within a row, means with different Latin superscripts are significantly different (P < 0.05). C = control feed; S5 = treatment with 50 g/kg of SOP; S9 = treatment with 90 g/kg of SOP. 1Mean values with their standard error (SEM); 2Probability of significant effect due to the interaction of the experimental factors Feed and Time (FXT).
Microbial population profiling
DGGE analysis of PCR-amplified partial 16S rRNA genes was performed on the total bacteria, and Butyrivibrio populations of RL incubated with the three diets. Microbial profiles obtained using universal primers for bacteria displayed a complex band pattern in all samples (Figure 1A). A UPGMA dendrogram separated samples incubated with S5 and S9 diets and collected at 24 h from all the other samples, with 82.8% similarity (Figure 1A). Within the cluster containing S5 and S9 and collected at 24 h two subclusters (86.2% similarity) were evident, based on the percentage of SOP (Figure 1A). Samples collected at 0 and 6 h formed a different group when compared with samples collected at 12 h and with control samples collected at 24 h, with a similarity of 87.6%. A similarity higher than 92% was found in RL samples inoculated with C, S5 and S9 collected at 0 and 6 h (Figure 1A). According to the AMOVA of DGGE banding patterns, there was a significant effect of sampling time on bacterial communities (percentage of variation = 31.29; P < 0.01) and a significant effect of the diet within samples collected at the same time point (percentage of variation = 22.29; P < 0.001).
Figure 1.
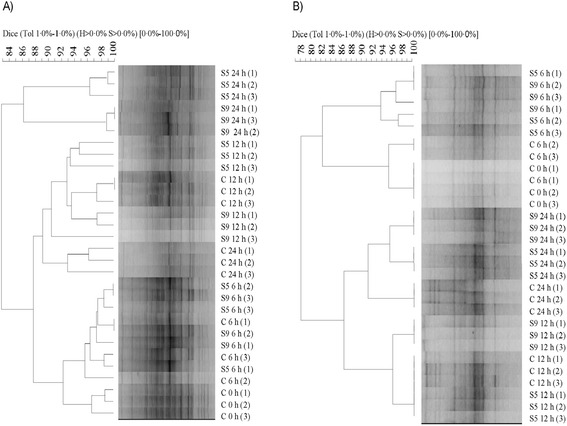
Cluster analysis based on the unweighted pair group method with the arithmetic means of polymerase chain reaction denaturing gradient gel electrophoresis profiles demonstrating the effect of C, S5 and S9 diets on the total bacteria (A) and Butyrivibrio group (B) in rumen liquor collected at 0, 6, 12 and 24 h. C = control feed; S5 = treatment with 50 g/kg of stoned olive pomace; S9 = treatment with 90 g/kg of stoned olive pomace. Scale relates to percent similarity.
PCR-DGGE analysis of members of the Butyrivibrio group exhibited a less complex pattern than the total bacteria (Figure 1B). Two main clusters were evident, separating all samples collected at 0 and 6 h from those collected at 12 and 24 h, with 77.0% similarity (Figure 1B). Subclusters once again clearly reflected the percentage of the amount of SOP added and the collection time (Figure 1B). Control samples collected at 0 and 6 h grouped differently from samples incubated with S5 and S9 diets (Figure 1B), with 81.2% similarity. Moreover, all samples collected at 12 h grouped separately from those collected at 24 h, with 85.6% similarity (Figure 1B). Along within the latter group, samples to which the S5 and S9 diets were added grouped together, separately from control samples, with 90.6% similarity (Figure 1B). AMOVA analysis indicated a significant effect of sampling time on Butyrivibrio DGGE banding patterns (percentage of variation = 58.67; P < 0.01) and of the diet within samples collected at the same time point (percentage of variation = 24.49; P < 0.001).
Sequence analysis of bacterial and Butyrivibrio-specific PCR-DGGE bands
PCR-DGGE bands exhibiting remarkable changes in response to SOP in total bacterial or Butyrivibrio populations (bands 1, 5, 7, 8, 9, 11, 12, 17, 18, 20, 21, 22, 23 and 24) were excised, re-amplified and sequenced (Figure 2). Moreover, to gain more information on the composition of the rumen bacterial community of sheep, ten bands obtained with primers F968/R1401 for total bacteria (bands 2, 3, 4, 6, 10, 13, 14, 15, 16 and 19) were selected and sequenced, even if their intensity was not affected by SOP (Figure 2A). Putative taxonomic identification for each band subjected to sequencing is reported in Table 4.
Figure 2.
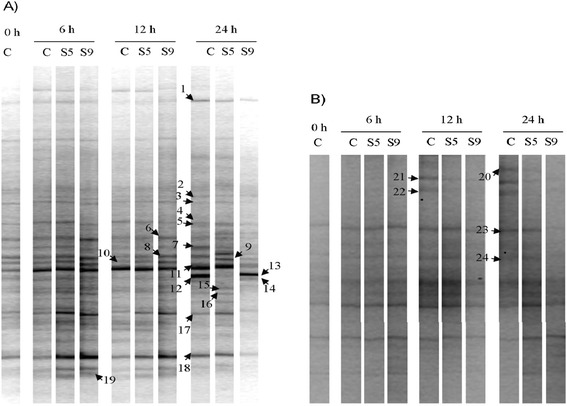
PCR-DGGE profiles of the total bacterial community (A) and Butyrivibrio members (B) in rumen liquor inoculated with the C, S5 and S9 diets and collected at 0, 6, 12 and 24 h. C = control feed; S5 = treatment with 50 g/kg of stoned olive pomace; S9 = treatment with 90 g/kg of stoned olive pomace. Bands indicated by numbers were selected for sequencing.
Table 4.
Identification of selected polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) fragments
| PCR-DGGE band | Nearest match (GenBank accession no.; % sequence similarity) | Taxonomic classification |
|---|---|---|
| Total bacterial community analysis | ||
| 1 | Pasteurella testudinis (NR_042889; 90%) | Unclassified Pasteurellaceae |
| 2 | Bergeriella denitrificans (NR_040933; 99%) | Bergeriella denitrificans |
| 3 | Bergeriella denitrificans (NR_040933; 99%) | Bergeriella denitrificans |
| 4 | Clostridium lavalense (NR_044289; 93%) | Unclassified Clostridiaceae |
| 5 | Neisseria weaveri (NR_025902; 99%) | Neisseria weaveri |
| 6 | Neisseria weaveri (NR_025902; 98%) | Neisseria weaveri |
| 7 | Neisseria weaveri (NR_025902; 99%) | Neisseria weaveri |
| 8 | Neisseria weaveri (NR_025902; 98%) | Neisseria weaveri |
| 9 | Neisseria flavescens (KF030235; 100%) | Neisseria flavescens |
| 10 | Clostridium citroniae (NR_043681; 90%) | Unclassified Clostridiaceae |
| 11 | Ruminobacter amylophilus (NR_026450; 99%) | Ruminobacter amylophilus |
| 12 | Neisseria flavescens (KF030235; 100%) | Neisseria flavescens |
| 13 | Neisseria flavescens (KF030235; 100%) | Neisseria flavescens |
| 14 | Neisseria weaveri (NR_025902; 98%) | Neisseria weaveri |
| 15 | Howardella ureilytica (NR_044022; 94%) | Unclassified Clostridiaceae |
| 16 | Roseburia faecis (NR_042832; 90%) | Unclassified Lachnospiraceae |
| 17 | Butyrivibrio hungatei (NR_025525; 90%) | Unclassified Lachnospiraceae |
| 18 | Butyrivibrio hungatei (NR_025525; 93%) | Unclassified Lachnospiraceae |
| 19 | Ruminococcus torques (NR_036777; 90%) | Unclassified Lachnospiraceae |
| Butyrivibrio-specific analysis | ||
| 20 | Butyrivibrio proteoclasticus (NR_102893; 92%) | Unclassified Lachnospiraceae |
| 21 | Butyrivibrio proteoclasticus (NR_102893; 98%) | Butyrivibrio proteoclasticus |
| 22 | Butyrivibrio proteoclasticus (NR_102893; 99%) | Butyrivibrio proteoclasticus |
| 23 | Robinsoniella peoriensis (NR_041882; 94%) | Unclassified Lachnospiraceae |
| 24 | Eubacterium ruminantium (NR_024661; 92%) | Unclassified Lachnospiraceae |
Phylogenetic analysis of the nineteen sequences of the PCR-DGGE fragments obtained with primers F968/R1401 (total bacteria) and sequences from rumen bacteria of equivalent length retrieved from the GenBank database was performed. The results indicated that seven sequences (bands 4, 10, 15, 16, 17, 18 and 19) were related to known species of Clostridiales (Figure 3), ten sequences (bands 2, 3, 5, 6, 7, 8, 9, 12, 13 and 14) were related to Neisseriales and the remaining two sequences were related to Pasteurellales (band 1) and Aeromonadales (band 11) (Figure 3, Table 4).
Figure 3.
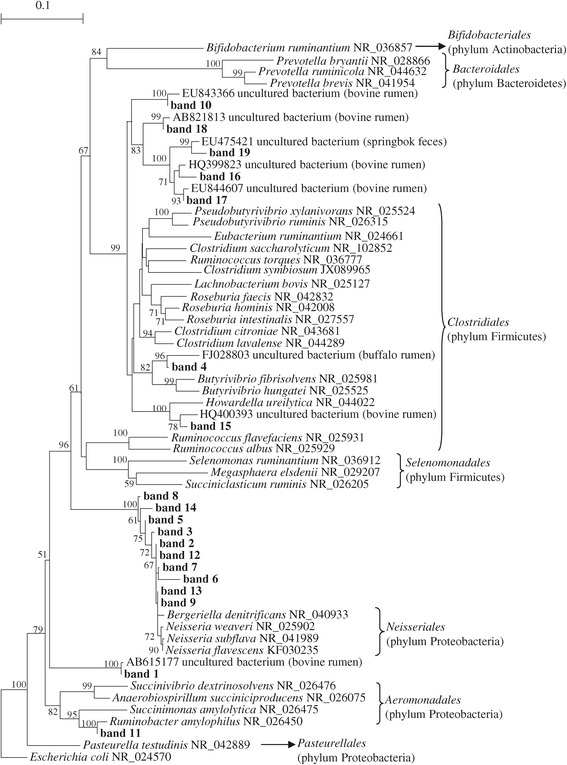
Neighbour-joining tree built using all 16S rRNA sequences obtained from the total bacteria PCR-DGGE gels and sequences of rumen bacteria of equivalent length, retrieved from the GenBank database. Sequences obtained in this study are indicated in boldface. Bootstrap values >50% based on 1000 replications are indicated at the nodes. The 16S rRNA gene sequence of Escherichia coli (NR_024570) was selected as the outgroup.
The analysis of total bacterial PCR-DGGE profiles evidenced that the intensities of seven bands, corresponding to Neisseria weaveri (bands 5, 7 and 8), Ruminobacter amylophilus (band 11), unclassified Pasteurellaceae (band 1) and Lachnospiraceae (bands 17 and 18) were reduced at 24 h in samples receiving the S9 diet in comparison to controls, whereas one band, identified as Neisseria flavescens (band 9), increased in S9 samples at the same sampling time (Figure 2A). On the contrary, minor differences were observed in presence of the S5 diet at 24 h in comparison to controls because only disappearance of band 12 (Neisseria flavescens) and the appearance of band 9 (Neisseria flavescens) were detected (Figure 2A).
A phylogenetic tree was also constructed with the five sequences obtained with the Butyrivibrio-specific primers F968/Bfib and other sequences of equivalent length, representative of bacterial species related to the Lachnospiraceae family. As displayed in Figure 4, two sequences (bands 21 and 22) grouped with sequences representative of Butyrivibrio proteoclasticus, whereas three sequences (band 20, 23 and 24) displayed a very low level of similarity with other known bacterial species belonging to the Lachnospiraceae family.
Figure 4.
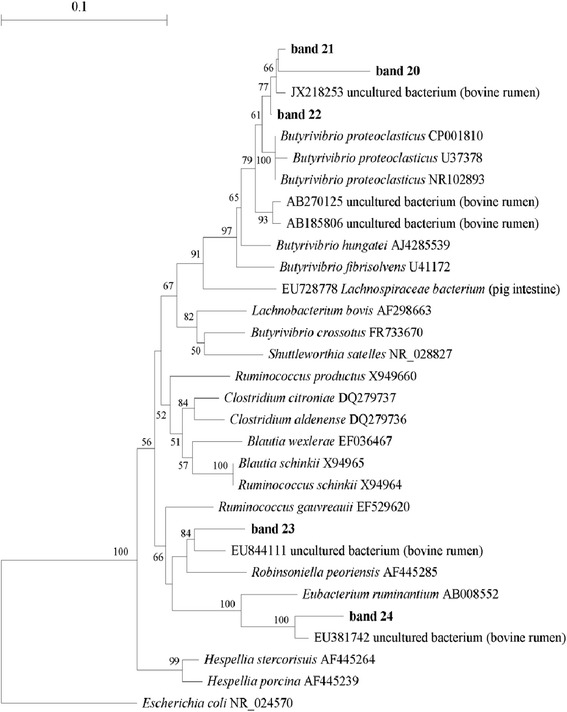
Neighbour-joining tree built using all 16S rRNA sequences obtained from Butyrivibrio -specific PCR-DGGE gels and sequences of rumen bacteria of equivalent length, retrieved from the GenBank database. Sequences obtained in this study are indicated in boldface. Bootstrap values of >50% based on 1000 replications are indicated at the nodes. The 16S rRNA gene sequence of Escherichia coli (NR_024570) was selected as the outgroup.
PCR-DGGE profiles obtained using Butyrivibrio-specific primers exhibited weak changes in the Butyrivibrio community in relation to diets. In S5 and S9 samples, the intensities of bands 21 and 22, identified as Butyrivibrio proteoclasticus, were reduced at 12 h when compared to C (Figure 2B). Moreover, considering samples collected at 24 h, band 20, (unclassified Lachnospiraceae), exhibited a lower intensity than the controls after incubation with the S5 and S9 diets, whereas the intensities of bands 23 and 24 (unclassified Lachnospiraceae) decreased only slightly in the presence of S9 diet (Figure 2B).
Discussion
In the literature it is well known that the inclusion of polyphenols in ruminant diets affects rumen metabolism, decreasing dietary protein degradation and fatty acid BH by means of targeting specific groups of microorganisms [2,6,10,11]. The in vitro degradability of organic matter, which is strongly related to microbial activity in the fore-stomach, is typically low for olive oil cakes on the basis of their polyphenol content. In fact, when dietary polyphenols are inactivated by using poly ethylene glycol (PEG) no detrimental effect has been observed in rumen microorganisms [10,12,13]. SOP contains a high level of polyphenols, and it is hypothesised that this supplement might be useful for improving the content of PUFA in products derived from ruminant livestock, at the same time contributing to the environmental sustainability of animal productions [7,10]. However, until now, there has been a lack of knowledge on the effect of SOP on lipid metabolism and rumen microbial communities involved in fatty acid BH processes. This information is essential to optimise the employment of SOP in ruminant feeding.
Our findings indicated that the inclusion of SOP in feeds stimulated the production of volatile fatty acids (VFA), suggesting that microbial activity was modified by the presence of SOP in feeds: the highest increase of C3:0 in the fermented RL inoculated with S5 and S9 can be related with a good level of amylolytic bacteria activity, while the constant production of C2:0 and the increase of iso C5, arising from microbial degradation of dietary amino acids, can be an indication of stimulated cellulolytic bacteria activity [10]. Iso C5 is the precursor of iso C15 and iso C17, which arise from rumen cellulolytic bacteria metabolism [14]. In our experiment, iso C15 and iso C17 production was stimulated by SOP, confirming that cellulolytic activity was not perturbed. The literature contains controversial results regarding the stimulating or depressing effects of olive oil by-products on rumen VFA production [12,15,16]. In their in vitro study Martin et al. [12] reported low levels of VFA when olive cake was fermented with rumen sheep fluid, while Yanetz-Ruitz et al. [16] obtained values higher than those reported in the former study. These differences are most likely strongly related to the olive variety, different oil extraction procedures used to obtain olive cakes and to the associative effects of the olive by-product with other dietary components.
SOP supplementation in feeds did not protect the double bond cis-9 from saturation, as demonstrated by the BH of OA and RA, which decreased during the fermentation period. A decrease in the isomerisation of OA to other trans C18:1 isomers [17] was hypothesised because no significant variations in the concentrations of these monoenes were detected. The temporary RA accumulation at 12 h in RL fermented with S5 and S9 can be related to a negative feedback effect caused by VA accumulation in these fermenters. This hypothesis can also be extended to conjugated linolenic acid and vaccelenic acid, further precursors of VA from α-LNA biohydrogenation, which appeared only at the end of fermentation in fermenters containing the highest content of SOP. VA accumulation in RL fermented with S5 and S9 is closely related to a decrease in Butyrivibrio proteoclasticum growth, as revealed by PCR-DGGE analysis. SOP did not contribute to the protection of LA and α-LNA from isomerisation to their cis-9,trans-11 isomers, indicating that LA-Isomerase activity is not influenced by SOP inclusion in feeds. Moreover, the shift of LA and α-LNA BH toward the trans-10 isomer was not enhanced. This trend agrees with several studies that demonstrated polyphenols do not favour the increase in the concentrations of trans-10 monoenes, whose synthesis is strongly related to the starch content of the diet [2,6,7].
In this study we employed a PCR-DGGE approach to evaluate the effect of SOP on rumen bacterial and Butyrivibrio communities. PCR-DGGE provides information on the predominant populations in the community and, using this technique it is possible to detect the sequences of the prevalent bacterial populations without the need for large clone libraries [18]. Nevertheless, PCR-DGGE presents some limitations: a single DGGE band may represent several species with identical partial DNA sequences [19], or several bands could be generated from a single organism because of multiple, heterogeneous operons [20]. However PCR-DGGE widely used in microbial ecology investigations.
Cluster analysis of PCR-DGGE profiles obtained with universal primers for the 16S rRNA gene clearly indicated a shift in the total bacterial community in the presence of SOP-enriched diets in comparison to controls, as confirmed by AMOVA. Buccioni et al. [2] demonstrated that polyphenols, such as tannins, affected the FA composition of the sheep rumen bacterial community, suggesting changes in its composition and/or activity in relation to the BH process. In our study, the effect of SOP on rumen bacterial communities appeared to depend on the level of its supplementation in the diet and the incubation time. Indeed, after 24 h of incubation with 90 g/kg of SOP, some bands in the PCR-DGGE profiles exhibited a decreased intensity. Thus the changes observed in the PCR-DGGE banding patterns may reflect the reduced abundance of the most sensitive species of ruminal bacteria to the antimicrobial action of SOP. Our observation is in agreement with previous in vitro studies, supporting the idea that polyphenols from different plants can reduce the activity and proliferation of different ruminal microorganisms [21]. The inhibitory effect exerted by these compounds has been explained by their ability to form complexes with the bacterial wall and to inactivate many extracellular enzymes [22].
Until now few studies have been carried out on sheep rumen microbiota using PCR-DGGE analysis followed by sequencing and identification of the dominant bacterial groups. In this study, only eleven PCR-DGGE bands obtained from the total bacterial community analysis were highly related to the 16S rRNA genes of known species, whereas the other bands corresponded to yet unclassified bacteria. This result is not surprising because the use of different culture-independent methods has demonstrated that the rumen microbiota is more diverse than previously hypothesised by considering the number of cultivated species [4]. On the whole, the sequenced bands were related mainly to species belonging to the Clostridiaceae family and the genus Neisseria. The first taxonomic group includes many cellulolytic and amylolytic species, which are often found in the rumen [4]. In contrast, only a Gram-negative carbohydrate-fermenting bacterium similar to Neisseria has been isolated from sheep rumen [23]. However, because members of the Neisseriaceae family are mammalian commensals [24], their presence in the rumen is plausible. In the analysed samples, we also detected the presence of Ruminobacter amylophilus, a typical rumen bacterium that may occur in reasonably large numbers in high grain or high roughage diets [25].
The most interesting changes in PCR-DGGE profiles were observed for the Neisseria flavescens, Neisseria weaveri, Ruminobacter amylophilus species and for members of the Lachnospiraceae and Pasteurellaceae families at 24 h in RL inoculated with S9 diet. Previous in vitro and in vivo studies have indicated that some members of the Lachnospiraceae family, such as Butyrivibrio species, are the main known bacteria involved in rumen BH [26]. Nevertheless, Huws et al. [27] have recently suggested that other yet not known bacterial species may play an important role in the BH process, via analysis of the RL of dairy cows using T-RFLP and DGGE approaches. Thus, the findings thus far from studies on pure cultures may not be sufficient to explain the bacterial contribution to rumen BH in vivo, which appears more complex than previously thought. For example, a previous study by Hudson et al. [28] has indicated that some bacterial species, such as Staphylococcus spp. and Streptococcus bovis, have the capacity to hydrate specific CLA intermediates, diverting them from the BH pathway. Our results strengthen the hypothesis of Huws et al. [27], despite the fact that additional investigations carried out with advanced techniques such as metagenomics and metatranscriptomics could better clarify the potential role of the different bacterial groups in the FA metabolism of sheep rumen.
Previous in vitro experiments have demonstrated that members of the Butyrivibrio group are able to biohydrogenate unsaturated FAs more rapidly than other species, and that only B. proteoclasticus has been recognised to reduce C18:1 to C18:0 [5]. Therefore we performed a Butyrivibrio-specific PCR-DGGE analysis to investigate the effect of SOP supplementation on this taxonomic group in detail, although the Butyrivibrio group comprises only a minor part of ruminal bacteria [29]. Cluster analysis indicated that both diets supplemented with SOP affected the composition of the Butyrivibrio population. Indeed, at both 12 and 24 h of incubation we observed a reduced intensity in specific PCR-DGGE bands. Sequence analysis revealed that two bacterial groups responding negatively to SOP after 12 h of incubation were closely related to B. proteoclasticus (levels of 16S rDNA similarity above 98.0%), which is the only cultivable SA producer. A significant increase in VA was only observed in relation to the incubation time in samples to which SOP had been added. Therefore, we hypothesise that SOP is able to decrease the hydrogenation of trans C18:1 and trans C18:2 intermediates by negatively affecting the growth of B. proteoclasticus or other species of Butyrivibrio not identified here. Our findings agree with the results obtained by Vasta et al. [6], who found a correlation between the reduced abundance of B. proteoclasticus and the simultaneous increase in VA in lamb rumen fluid, following the addition of quebracho tannins to the diet. However, the other few studies available in the literature on the effect of plant extracts rich in polyphenols on ruminal microorganisms have presented contrasting results. According to Ghaffari et al. [30], phenolic compounds from pistachio by-products used in the diet of sheep did not affect the abundance of B. proteoclasticus in rumen fluid. Moreover, extracts from 37 Australian plants containing polyphenols selectively inhibited B. proteoclasticus, and only some of them affected B. fibrisolves [31]. The variable effect of plant extracts on the members of Butyrivibrio group could be related to the type of polyphenols they contain and the supplementation level in the diet, as previously suggested [11].
Conclusions
Supplementation of feeds with SOP inhibited the rumen BH of C18 unsaturated FAs in a dose dependent manner, resulting in a decrease in the SA concentration and in an increase in VA. In particular, changes in rumen fatty acid profiles were associated with changes in the bacterial community, including bacteria responsible for the hydrogenation of VA to SA.
Methods
Feed composition
Feeds used as the substrates of the fermentation were: a control diet (C) in which the SOP was not included and other two diets (S5 and S9) in which the integration with SOP was 50 g/kg of DM and 90 g/kg of DM, respectively. The amount of SOP used in this experiment was chosen with the criterion of practicality under farm conditions (the diets used in this trial were formulated on the basis of previous in vivo trials with Chianina bulls and dairy Comisana sheep; data unpublished). The diets were formulated to be isoproteic and isoenergetic. The ingredients and chemical compositions of the feeds are displayed in Table 5. SOP was obtained after mechanical extraction of virgin olive oil using the following operating conditions [7]: the olives were stoned and malaxed for 40 min at 25°C, and the oil extraction was performed using a three phase decanter (mod. 400 ECO, RCM Rapanelli Costruzioni Meccaniche s.r.l., Bevagna, PG, Italy). After storage at room temperature for 36 hours, stoned olive cake was dried using a fluid bed dryer; the initial temperature of the drying air flow was 120°C and the maximum temperature of olive cake during the drying process was 45°C. The dried stoned olive cake was stored at room temperature. The proximate composition (according to A.O.A.C procedures [32]) of SOP was: DM (873.80 g/kg), crude protein (118.31 g/kg of DM), neutral detergent fibre (490.51 g/kg of DM), acid detergent fibre (347.40 g/kg of DM), acid detergent lignin (85.61 g/kg of DM) and 63.43 g/kg of DM of crude fat in which the main FA contained were C16:0 (12.81 g/100 g of total FA), cis-9 C18:1 (76.43 g/100 g of total FA) and cis-9,cis-12 C18:2 (6.82 g/100 g of total FA). The polyphenol composition of SOP was determined according to Servili et al. [9]: 3,4-dihydroxyphenolethanol (1.16 g/kg DM), 4-hydroxyphenolethanol (0.11 g/kg DM); p-coumaric acid (0.04 g/kg DM), verbascoside (1.33 g/kg DM), 2-(3,4-hydroxyphenyl)ethyl(3S,4E)-4-formyl-3-(2-oxoethyl)hex-4-enoate (1.16 g/kg DM). The total polyphenol content in SOP was 3.80 g/kg DM.
Table 5.
Composition of feeds used as substrates for the fermentation and the main fatty acids (FA) in rumen liquor (RL) at the start of fermentation
| Feed composition | C | S5 | S9 |
|---|---|---|---|
| Ingredients (g/kg DM) | |||
| Grass hay | 103.45 | 103.45 | 98.04 |
| Wheat straw | 103.45 | 103.45 | 98.04 |
| Mais meal | 545.52 | 510.00 | 504.80 |
| Soybean meal | 42.76 | 42.76 | 40.52 |
| Wheat bran | 33.10 | 33.10 | 31.37 |
| Bean flakes | 20.69 | 20.69 | 19.61 |
| Soybean flakes | 12.41 | 12.41 | 11.76 |
| Horsebean flakes | 11.03 | 11.03 | 10.46 |
| Barley | 109.66 | 95.17 | 78.43 |
| Stoned olive oil cake | --- | 50.00 | 90.00 |
| Maize germ meal | 17.93 | 17.93 | 16.99 |
| Chemical composition | |||
| Dry Matter kg | 724.70 | 719.00 | 713.0 |
| expressed as g/kg of DM | |||
| Crude protein (6.25 x N) | 115.91 | 116.23 | 116.40 |
| Crude fat | 23.42 | 24.51 | 25.63 |
| Neutral detergent fibre | 366.00 | 379.40 | 391.81 |
| Acid detergent fibre | 194.73 | 205.63 | 215.82 |
| Lignin | 81.64 | 81.81 | 82.53 |
| Ash | 58.76 | 61.25 | 63.56 |
| Non protein nitrogen | 2.48 | 4.43 | 5.27 |
| Soluble protein | 8.57 | 9.72 | 10.39 |
| Neutral detergent insoluble protein | 2.50 | 5.60 | 7.99 |
| Acid detergent insoluble protein | 1.11 | 2.30 | 3.21 |
| Main fatty acids in RL at the start | |||
| of fermentation (g/100 g of total FA) | |||
| C16:0 | 18.05 | 17.23 | 16.33 |
| C18:0 | 1.90 | 1.79 | 1.79 |
| cis-9 C18:1 | 23.92 | 25.19 | 26.81 |
| cis-9,cis-12 C18:2 | 52.96 | 52.98 | 52.38 |
| cis-9,cis-12,cis-15 C18:3 | 2.67 | 2.44 | 2.32 |
C = control feed; S5 = treatment with 50 g/kg of stoned olive pomace (SOP); S9 = treatment with 90 g/kg of SOP.
In vitro incubation with sheep ruminal fluid
The in vitro incubation was performed according to Tedeschi et al. [33] with several modifications. Four sheep, conditioned with a basal diet formulated to shape rumen microflora and composed of grass hay (770 g/kg DM), soybean meal (55 g/kg DM), barley meal (175 g/kg DM), were used to provide rumen contents. Animals had continuous access to water and mineral blocks. After a 4 week adaptation period, approximately 1 litre of rumen contents was collected from each sheep using a rumen fluid sampling pump on the same day before the morning meal. The handling of the animals was performed according to the Institutional Animal Care and Use Committee of Florence University (IACUC, 2004). The RL was immediately mixed with CO2 to avoid O2 contamination and transferred to the laboratory in a thermostatic box (39°C) under anaerobic conditions. The RL was then filtered through four layers of cheesecloth into a flask under a continuous flow of CO2. An aliquot of the RL was buffered (1:3, v/v) by adding an artificial saliva solution [34]. Feeds (2 g of DM) were incubated in triplicate with 200 ml of inoculum. The incubator consisted of a thermostatic chamber (39°C) equipped with twenty-seven 300 ml glass fermentation vessels provided with two inlets (one to release gas through a valve and one for the pH probe) and connected to an electronic pressure transducer (pre-set at 65 kPa) and an electronic gas valve. When the inside gas pressure reached the pre-set value, the valve was opened, releasing approximately 2 ml of gas. The fermentation pattern was monitored with PC software (Labview 5.0, National Instr., Austin, TX). Each vessel containing substrate inoculated with rumen fluid saturated with CO72 to guarantee anaerobic conditions was continuously stirred. Samples of RL were collected at 6, 12 and 24 h of incubation. At each sampling time, three vessels per treatment were used for the microbial characterisation and FA analysis as follows: the entire contents of each vessel were separated into four aliquots; three aliquots of 1 ml and one aliquot of 150 ml were stored at −80°C for DNA extraction and FA profile determination, respectively, as described below. Each vessel was considered to be a single experimental unit according to Buccioni et al. [2,35]
Samples of RL immediately after the addition of buffer solution (200 ml) and before feed inoculation (as blank to control the quality) and samples of RL (200 ml) inoculated with feeds, at the start of fermentation (t = 0 min), were collected in triplicate for FA profile analysis. The fat content of the RL blank was very low (0.01 g/l), as a consequence of the procedure adopted for the preparation of the inoculum; hence, the initial contribution of RL to FA composition of the inoculum was negligible (data not shown). Table 5 displays the FA composition of RL inoculated with the three diets at the beginning of fermentation. In the feeds the concentration of oleic acid (cis-9 C18:1, OA) increased according to the percentage of SOP inclusion in the diet.
Feed proximate analysis
Samples of feeds were oven-dried at 60°C for 24 h. The dry samples were analysed for crude protein, ash and crude fat according to the 954.01, 954.05 and 920.39 procedures of AOAC (1990) [32], respectively. Neutral detergent fibre, acid detergent fibre and acid detergent lignin were determined using sequential analysis, with sodium sulphite, with heat-stable amylase, and expressed inclusive of residual ash. The carbohydrate and protein degradable fractions (non-protein nitrogen; soluble protein; neutral detergent insoluble protein; acid detergent insoluble protein) were estimated according to the Cornell Net Carbohydrates and Protein System CNCPS [36].
Rumen fatty acid analysis
To determine the FA, each sample (approximately 150 mg) was extracted according to Folch method [37] without drying the final solution containing the lipid extract which was directly methylated using a combination of methods according to Buccioni et al. [2] with the aim to avoid volatile fatty acid (VFA) loss. The first step consisted of an alkaline methylation with sodium methylate/methanol (1 ml of 0.5 M sodium methoxide) to esterify glycerides. The second step involved an acidic methylation using HCl/methanol (1.5 ml of 5% methanolic HCl, 10 min at 50°C) as catalyst to esterify NEFA. Fatty acid methyl esters (FAME) were extracted using n-hexane with C9:0 and C23:0 methyl ester (Sigma Chemical Co., St. Louis, MO) as internal standards for quantification, and maintained in vials with hermetic closure to avoid the loss of volatile components. FAMEs were separated and identified by gas chromatography on a GC equipped with a capillary column (CP-select CB for FAME Varian, Middelburg, The Netherlands: 100 m × 0.25 mm i.d, film thickness 0.20 μm) according to Buccioni et al. [38]. The injector and flame ionisation detector temperatures were 270°C and 300°C, respectively. The programmed temperature was 40°C for 4 min, increased to 120°C at a rate of 10°C/min, maintained at 120°C for 1 min, increased to 180°C at a rate of 5°C/min, maintained at 180°C for 18 min, increased to 200°C at a rate of 2°C/min, maintained at 200°C for 1 min, increased to 230°C at a rate of 2°C/min and maintained at this temperature for 19 min. The split ratio was 1:100, and helium was the carrier gas with a flow rate of 1 ml/min. Individual FAMEs were identified by comparison of the relative retention times of FAME peaks from samples with those from the standard mixture 37 Component FAME Mix C4:0-C24:0 (cod 18919-1AMP, Supelco, Bellefonte, PA, USA), individual trans9 C18:1 and trans11 C18:1 (cod 46903 and v1381, respectively, Sigma-Aldrich, St. Louis, MO, USA), individual cis9, trans11 (cod 1255, Matreya Inc. Pleasant GAP, PA, USA), CLA mix standard (cod 05632, Sigma-Aldrich, St. Louis, MO, USA) and published isomeric profile [39,40]. Determination of the elution sequence of the C18:1 isomers was performed according to Kramer et al. [41]. Moreover, standard mixtures of α-linolenic acid (α-LNA) isomers (47792, Supelco, Chemical Co., St. Louis, MO) and LA isomers (47791, Supelco, Bellefonte, PA, USA) and published isomeric profiles [42] were used to identify the isomers of interest. Two bacterial acid methyl ester mixtures (47080-U Supelco, Chemical Co., Bellefonte, PA, USA; GLC110, Matreya, Pleasant Gap, PA) and individual standards for the methyl esters of iso C14:0, anteiso C14:0, iso C15:0 and anteiso C17:0 (21-1211-11, 21-1210-11, 21-1312-11 and 21-1415-11, Larodan Malmo, SW) were used to identify the branched FA profile. Inter- and intra-assay coefficients of variation were calculated using a reference standard (CRM 164, Community Bureau of Reference, Bruxelles, Belgium), and the detection threshold of FA was 0.01 g/100 g of FA [43]. All FA composition results are expressed as g/100 g of FA.
DNA extraction from rumen microbial samples
Genomic DNA was extracted from 1 ml of rumen microbial suspension using the Fast DNA SPIN kit for soil (Qbiogene, Carlsbad, CA, USA) with some modifications. Briefly, each sample was thawed and transferred to a 15 ml tube containing 4.5 ml of lysis buffer (500 mM-NaCl; 50 mM-Tris–HCl, pH 8.0; 50 mM-EDTA and 4% SDS) and incubated for 15 min at 70°C with gentle shaking by hand every 5 min. After centrifuging at 200 × g at 4°C for 5 min, 1 ml of the supernatant was transferred to a 2 ml centrifuge tube and centrifuged at 14,600 × g at 4°C for 5 min. The supernatant was removed and the pellet was dissolved in 978 μl of sodium phosphate buffer and 122 μl of MT buffer (both solutions are supplied by the Fast DNA SPIN kit for soil). Each sample was homogenised with a FastPrep cell disrupter instrument (Bio101, ThermoSavant, Qbiogene, Carlsbad, CA, USA) for 2 × 40 s at speed 6.0 and then processed according to the manufacturer’s guidelines. This combination of methods was used to maximise the recovery of DNA from ruminal digests. DNA was eluted in sterile water and its integrity was verified by agarose gel electrophoresis. The amount and purity of DNA was measured at 260 and 280 nm using a ND-1000 Spectrophotometer (NanoDrop Technologies, Labtech, Ringmer, UK).
PCR-DGGE analysis of the total bacterial community and Butyrivibrio group
Individual total DNA extracted from rumen samples was diluted to a concentration of 5 ng/μl and 2 μl of diluted DNA was used as template in PCR reactions. Amplification of the V6-V8 region of the 16S rRNA gene was carried out with the primer pair F968GC (5’-CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA CGG GGG GAA CGC GAA GAA CCT TAC-3’) and R1401 (5’-CGG TGT GTA CAA GAC CC-3’) [44] for total bacterial PCR (fragment size ~470 bp) and with F968GC and Bfib (5’-TTC GGG CAT TYC CRA CT-3’) [45] for Butyrivibrio group-specific PCR (fragment size ~470 bp). Reactions were carried out using an iCycler Thermal Cycler (BioRad Laboratories, Hertfordshire, UK) in 25 μl volumes containing 1X PCR buffer (67 mM Tris–HCl, pH 8.8; 1.66 mM (NH4)2SO4; 0.1% Tween-20), 1.5 mM MgCl2, 250 μM deoxynucleotide triphosphates (dNTPs), 400 nM each primer, and 1 U of Polytaq (Polymed, Florence, Italy). Amplifications were performed under the following conditions: an initial denaturation at 94°C for 5 min followed by 35 cycles of 94°C for 20 s, 56°C for 30 s and 72°C for 45 s, and a final extension at 72°C for 10 min. After PCR, amplified products were verified by agarose gel electrophoresis. Subsequently, to perform polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) analysis, amplicons were loaded on a 6% polyacrylamide gel (acrylamide/bis 37.5:1), with a 50-60% denaturing gradient (100% denaturant consisting of 40% v/v deionised formamide, 7 M urea) and electrophoresis was performed in a Phor-U system (Ingeny International, Goes, NL). The gel was run for 17 h at 60°C and 75 V and, after electrophoresis, stained with SYBR® Gold (Molecular Probes, Eugene, OR) and scanned using ChemiDoc XRS (BioRad Laboratories, Hertfordshire, UK).
The PCR-DGGE banding patterns obtained were analysed using the GelCompar II Software v 4.6 software package (Applied Maths, Saint-Martens-Latem, Belgium). Normalisation of bands within and between gels was performed by defining an active reference system. To summarise the species number of rumen bacterial communities, each band was considered as corresponding to a single microbial species. Bands with a minimum area below 1% were discarded. The banding patterns of PCR-DGGEs were further analysed by hierarchical cluster analysis based on the positions and presence/absence of bands in different profiles. The resultant binary matrices were translated into distance matrices using the Dice similarity coefficient and utilised to construct dendrogram using the unweighted pair group method using arithmetic average (UPGMA) algorithm. Analysis of Molecular Variance (AMOVA) was conducted using Arlequin 3.0 software [46], to compare the DGGE patterns and detect statistically significant differences between bacterial community structures in relation to sampling time and diet.
Sequence analysis of PCR-DGGE fragments
A total of 24 bands were excised from DGGE gels and were placed in 20 μl of distilled water. The PCR products were eluted through freezing and thawing according to Throbäck et al. [18] and reamplified using the F968/R1401 or F968/Bfib primers without a GC clamp, as described above. The fresh PCR products were then sequenced using the dideoxy chain termination method at BMR Genomics sequencing service (BMR Genomics srl, Padova, Italy). Chromatograms were visualised using the Finch TV computer software (ver. 1.4.0, Geospiza, Seattle, WA, USA) and, to obtain reliable results, we carefully verified the absence of many ambiguous peaks in each sequence. Nucleotide sequences were compared against all sequences in GenBank using the BLASTN program [47] with the aim of identifying the microorganisms corresponding to each selected band. Taxonomic identification was achieved by using different sequence similarity thresholds: a similarity ≥97% for a species level identification and 95%, 90%, 85%, 80% and 75% for assignment at the genus, family, order, class and phylum levels, respectively [48].
For phylogenetic analysis, sequences were aligned together with other sequences of equivalent length retrieved from the GenBank database, using the ClustalX 2.0.11 multiple sequence alignment software [49]. Distance calculation was performed according to Jukes and Cantor [50] followed by phylogenetic tree construction using the neighbour-joining algorithm [51] by means of TREECON 1.3b [52]. The robustness of each node was evaluated by bootstrap analysis with 1000 replicates.
Statistical analysis of fatty acid data
FA concentration data were processed with the General Linear Model of SAS [53] using the following linear model with fixed factors of diet and incubation time as well as their interaction:
where yij is the observation; μ is the overall mean; Di is the diet (i = 1 to 3); Tj is the incubation time (j = 1 to 3); Di × Tj is the interaction between diet and incubation time and eij is the residual error. Multiple comparisons of means were made using Tukey’s test. Main effects and differences were considered significant when P < 0.05.
Availability of supporting data
Nucleotide sequences from this study have been deposited in the GenBank database. Those from DGGE bands obtained with universal primer pair F968GC/R1401 targeting bacterial 16SrRNA gene have been deposited in the GenBank database under the accession numbers KF976364–KF976382. Those from DGGE bands obtained with primer pair F968GC/Bfib specific for the Butyrivibrio group have been deposited in GenBank under accession numbers KF976383-KF976387.
Band matching tables of Bacteria and Butyrivibrio DGGE profiles according to diet and time of sampling have been deposited to LabArchives, LLC (http://www.labarchives.com/) at https://dx.doi.org/10.6070/H4HH6H16.
Aknowledgments
This work was supported by the Agricultural Department or Regione Toscana- Italy (NUTRIFOROIL Project DD 6107/2013), and by University of Florence (Fondi di Ricerca di Ateneo 2012–13).
Abbreviations
- α-LNA
α-linolenic acid
- BH
Biohydrogenation
- CALNA
Conjugated linolenic acid
- CLA
Conjugated linoleic acids
- FA
Fatty acids
- FAME
Fatty acid methylesters
- LA
Linoleic acid
- OA
Oleic acid
- PCR-DGGE
Polymerase chain reaction denaturing gradient gel electrophoresis
- RA
Rumenic acid
- RL
Rumen liquor
- SA
Stearic acid
- SOP
Stoned olive pomace
- VA
Vaccenic acid
- VFA
Volatile fatty acids
- VLA
Vaccelenic acid
Footnotes
Grazia Pallara, Arianna Buccioni, Roberta Pastorelli, Marcello Mele, Luciana Giovannetti and Carlo Viti contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GP, AB and CV conceived the study; AB and SM performed fatty acid analysis; GP and RP performed DGGE experiments and analysis; GP, AB, RP, LG and CV prepared tables and figures; SM, MM, SR, AM, MP and MS gave technical support and conceptual advice. All authors discussed the results and implications and commented on the manuscript at all stages. All authors read and approved the final manuscript.
Contributor Information
Grazia Pallara, Email: grazia.pallara@unifi.it.
Arianna Buccioni, Email: arianna.buccioni@unifi.it.
Roberta Pastorelli, Email: roberta.pastorelli@entecra.it.
Sara Minieri, Email: sara.minieri@unifi.it.
Marcello Mele, Email: marcello.mele@unipi.it.
Stefano Rapaccini, Email: stefano.rapaccini@unifi.it.
Anna Messini, Email: anna.messini@unifi.it.
Mariano Pauselli, Email: mariano.pauselli@unipg.it.
Maurizio Servili, Email: maurizio.servili@unipg.it.
Luciana Giovannetti, Email: luciana.giovannetti@unifi.it.
Carlo Viti, Email: carlo.viti@unifi.it.
References
- 1.Mele M, Buccioni A, Petacchi F, Serra A, Banni S, Antongiovanni M, Secchiari P. Effect of forage/concentrate ratio and soy bean oil supplementation on milk yield and composition from Sarda ewes. Anim Res. 2006;55:273–285. doi: 10.1051/animres:2006019. [DOI] [Google Scholar]
- 2.Buccioni A, Minieri S, Rapaccini S, Antongiovanni M, Mele M. Effect of chestnut and quebracho tannins on fatty acid profile in rumen liquid- and solid-associated bacteria: an in vitro study. Animal. 2011;5:1521–1530. doi: 10.1017/S1751731111000759. [DOI] [PubMed] [Google Scholar]
- 3.Griinari JM, Bauman DE. Biosynthesis of Conjugated Linoleic Acid and its Incorporation into Meat and Milk in Ruminants. In: Yurawecz MP, Mossoba MM, Kramer JKG, Pariza MW, Nelson GJ, editors. Advances in Conjugated Linoleic Acid Research. Champaign: AOCS Press; 1999. pp. 180–200. [Google Scholar]
- 4.Kong YH, Teather R, Forster R. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol Ecol. 2010;74:612–622. doi: 10.1111/j.1574-6941.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- 5.Wallace RJ, Chaudhary LC, McKain N, Mcewan NR, Richardson AJ, Vercoe PE, Walker ND, Paillard D. Clostridium proteoclasticum: a ruminal bacterium that forms stearic acid from linoleic acid. FEMS Microbiol Lett. 2006;265:195–201. doi: 10.1111/j.1574-6968.2006.00487.x. [DOI] [PubMed] [Google Scholar]
- 6.Vasta V, Yanez-Ruiz DR, Mele M, Serra A, Luciano G, Lanza M, Biondi L, Priolo A. Bacterial and protozoal communities and fatty acid profile in the rumen of sheep fed a diet containing added tannins. Appl Environ Microbiol. 2010;76:2549–2555. doi: 10.1128/AEM.02583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luciano G, Pauselli M, Servili M, Mourvaki E, Serra A, Monahan FJ, Lanza M, Priolo A, Zinnai A, Mele M. Dietary olive cake reduces the oxidation of lipids, including cholesterol, in lamb meat enriched in polyunsaturated fatty acids. Meat Sci. 2013;93:703–714. doi: 10.1016/j.meatsci.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 8.Molina Alcaide E, Nefzaoui A. Recycling of olive oil by-products: possibilities of utilization in animal nutrition. Int Biodet Biodeg. 1996;38:227–235. doi: 10.1016/S0964-8305(96)00055-8. [DOI] [Google Scholar]
- 9.Servili M, Esposto S, Fabiani R, Urbani S, Taticchi A, Mariucci F, Selvaggini R, Montedoro GF. Phenolic compounds in olive oil: Antioxidant, health and sensoryactivities according to their chemical structures. Inflammopharmacology. 2009;17:1–9. doi: 10.1007/s10787-008-8014-y. [DOI] [PubMed] [Google Scholar]
- 10.Molina-Alcaide E, Yanez-Ruiz DR. Potential use of olive by-products in ruminant feeding: a review. Anim Feed Sci Technol. 2008;147:247–264. doi: 10.1016/j.anifeedsci.2007.09.021. [DOI] [Google Scholar]
- 11.Patra AK, Saxena J. Exploration of dietary tannins to improve rumen metabolism and ruminant nutrition. J Sci Food Agric. 2011;91:24–37. doi: 10.1002/jsfa.4152. [DOI] [PubMed] [Google Scholar]
- 12.Martin Garcia AI, Yanez-Ruitz D, Moumen A, Molina-Alcaide E. Effect of poly ethylene glycol, urea and sufflower meal supply on two stage olive cake fermentation. Anim Res. 2004;53:245–257. doi: 10.1051/animres:2004017. [DOI] [Google Scholar]
- 13.Martin AI, Yanez-Ruitz DR, Moumen A, Molina-Alcaide E. Effect of polyethylene glycol, urea and sunflower meal on olive (olea europea var europea) leaf fermentation in continuous fermenters. Small Rum Res. 2006;61:53–61. doi: 10.1016/j.smallrumres.2005.01.005. [DOI] [Google Scholar]
- 14.Vlaeminck B, Fievez V, Tamminga S, Dewhurst RJ, van Vuuren A, De Brabander D, Demeyer D. Milk odd- and branched-chain fatty acids in relation to the rumen fermentation pattern. J Dairy Sci. 2006;89:3954–3964. doi: 10.3168/jds.S0022-0302(06)72437-7. [DOI] [PubMed] [Google Scholar]
- 15.Ramos-Cormenzana A, Juárez-Jiménez B, Garcia-Pareja MP. Antimicrobial activity of olive mill waste-waters (alpechin) and biotransformed olive oil mill wastewater. Int Biodeter Biodegrad. 1996;38:283–290. doi: 10.1016/S0964-8305(96)00061-3. [DOI] [Google Scholar]
- 16.Yanez-Ruitz DR, Moumen A, Martin Garcia AI, Molina-Alcaide E. Ruminal fermentation and degradation pattern, protozoa population and urinary purine derivatives escretion in goats and wethers fed diets based on two-stage olive oil cake: effect of PEG supply. J Anim Sci. 2004;82:2023–2032. doi: 10.2527/2004.8272023x. [DOI] [PubMed] [Google Scholar]
- 17.Mosley EE, Powell GL, Riley MB, Jenkins TC. Microbial biohydrogenation of oleic acid to trans isomers in vitro. J Lipid Res. 2002;43:290–296. [PubMed] [Google Scholar]
- 18.Throback IN, Enwall K, Jarvis A, Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol Ecol. 2004;49:401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Vallaeys T, Topp E, Muyzer G, Macheret V, Laguerre G, Rigaud A, Soulas G. Evaluation of denaturing gradient gel electrophoresis in the detection of 16S rDNA sequence variation in rhizobia and methanotrophs. FEMS Microbiol Ecol. 1997;24:279–285. doi: 10.1111/j.1574-6941.1997.tb00445.x. [DOI] [Google Scholar]
- 20.Rainey FA, Wardrainey NL, Janssen PH, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308(T) contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 21.Min BR, Barry TN, Attwood GT, Mcnabb WC. The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review. Anim Feed Sci Tech. 2003;106:3–19. doi: 10.1016/S0377-8401(03)00041-5. [DOI] [Google Scholar]
- 22.Jones GA, Mcallister TA, Muir AD, Cheng KJ. Effects of sainfoin (Onobrychis-viciifolia Scop) condensed tannins on growth and proteolysis by 4 strains of ruminal bacteria. Appl Environ Microbiol. 1994;60:1374–1378. doi: 10.1128/aem.60.4.1374-1378.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsden SR, Volcani BE, Gilchrist FMC, Lewis D. Properties of a fatty acid forming organism isolated from the rumen of sheep. J Bacteriol. 1956;72:681–689. doi: 10.1128/jb.72.5.681-689.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sørum H, Sunde M. Resistance to antibiotics in the normal flora of animals. Vet Res. 2001;32:227–241. doi: 10.1051/vetres:2001121. [DOI] [PubMed] [Google Scholar]
- 25.Caldwell DR, Bryant MP. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Microbiol. 1966;14:794–801. doi: 10.1128/am.14.5.794-801.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boeckaert C, Vlaeminck B, Fievez V, Maignien L, Dijkstra J, Boon N. Accumulation of trans C-18:1 fatty acids in the rumen after dietary algal supplementation is associated with changes in the Butyrivibrio community. Appl Environ Microbiol. 2008;74:6923–6930. doi: 10.1128/AEM.01473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huws SA, Kim EJ, Lee MRF, Scott MB, Tweed JKS, Pinloche E, Wallace RJ, Scollan ND. As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environ Microbiol. 2011;13:1500–1512. doi: 10.1111/j.1462-2920.2011.02452.x. [DOI] [PubMed] [Google Scholar]
- 28.Hudson JA, Cai Y, Corner RJ, Morvan B, Joblin KN. Identification and enumeration of oleic acid and linoleic acid hydrating bacteria in the rumen of sheep and cows. J Appl Microbiol. 2000;88:286–292. doi: 10.1046/j.1365-2672.2000.00968.x. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson DM, Weimer PJ. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol. 2007;75:165–174. doi: 10.1007/s00253-006-0802-y. [DOI] [PubMed] [Google Scholar]
- 30.Ghaffari MH, Tahmasbi AM, Khorvash M, Naserian AA, Ghaffari AH, Valizadeh H. Effect of pistachio by-products in replacement of alfalfa hay on populations of rumen bacteria involved in biohydrogenation and fermentative parameters in the rumen of sheep. J Anim Physiol An N. 2014;98:578–586. doi: 10.1111/jpn.12120. [DOI] [PubMed] [Google Scholar]
- 31.Durmic Z, McSweeney CS, Kemp GW, Hutton P, Wallace RJ, Vercoe PE. Australian plants with potential to inhibit bacteria and processes involved in ruminal biohydrogenation of fatty acids. Anim Feed Sci Tech. 2008;145:271–284. doi: 10.1016/j.anifeedsci.2007.05.052. [DOI] [Google Scholar]
- 32.Association of Official Analytical Chemists . Official Methods of Analysis. 16. Arlington, VA: AOAC; 1995. [Google Scholar]
- 33.Tedeschi LO, Konohoff PJ, Karges K, Gibson ML. Effects of chemical composition variation on the dynamics of ruminal fermentation and biological value of corn milling (co) products. J Dairy Sci. 2009;92:401–413. doi: 10.3168/jds.2008-1141. [DOI] [PubMed] [Google Scholar]
- 34.McDougall EI. Studies on ruminant saliva. I The composition and output of sheep’s saliva. Biochem. 1948;43:99–109. [PMC free article] [PubMed] [Google Scholar]
- 35.Buccioni A, Antongiovanni M, Petacchi F, Mele M, Serra A, Serra P, Benvenuti D. Effect of dietary fat quality on C18:1 fatty acids and conjugated linoleic acid production: an in vitro rumen fermentation study. Anim Feed Sci Technol. 2006;127:268–282. doi: 10.1016/j.anifeedsci.2005.09.007. [DOI] [Google Scholar]
- 36.Licitra G, Hernandez TM, VanSoest PJ. Standardization of procedures for nitrogen fractionation of ruminant feeds. Anim Feed Sci Tech. 1996;57:347–358. doi: 10.1016/0377-8401(95)00837-3. [DOI] [Google Scholar]
- 37.Folch J, Lees M, Sloane-Stanely GH. A simple method for the isolation and purification of total lipids from animal tissues. J Clin Microbiol. 1957;226:497–507. [PubMed] [Google Scholar]
- 38.Buccioni A, Rapaccini S, Antongiovanni M, Minieri S, Conte G, Mele M. Conjugated linoleic acid and C18:1 isomers content in milk fat of sheep and their transfer to Pecorino Toscano cheese. Int Dairy J. 2010;20:190–194. doi: 10.1016/j.idairyj.2009.10.001. [DOI] [Google Scholar]
- 39.Griinari JM, Dwyer MA, McGiure MA, Bauman DE, Palmquist DL, Nurmela KV. Trans octa decenoic acids and milk fat depression in lactating dairy cows. J Dairy Sci. 1988;81:1251–1261. doi: 10.3168/jds.S0022-0302(98)75686-3. [DOI] [PubMed] [Google Scholar]
- 40.Kramer JKG, Cruz-Hernandez C, Deng ZY, Zhou JQ, Jahreis G, Dugan MER. Analysis of conjugated linoleic acid and trans 18:1 isomers in synthetic and animal products. Am J Clin Nutr. 2004;79:1137S–1145S. doi: 10.1093/ajcn/79.6.1137S. [DOI] [PubMed] [Google Scholar]
- 41.Kramer KG, Hernandez M, Cruz-Hernandez C, Kraft J, Dugan MER. Combining Results of Two GC Separations Partly Achieves Determination of All cis and trans 16:1, 18:1, 18:2 and 18:3 Except CLA Isomers of Milk Fat as Demonstrated Using Ag-Ion SPE Fractionation. Lipids. 2008;43:259–273. doi: 10.1007/s11745-007-3143-4. [DOI] [PubMed] [Google Scholar]
- 42.Destaillats F, Trottier JP, Galvez JMG, Angers P. Analysis of alpha-linolenic acid biohydrogenation intermediates in milk fat with emphasis on conjugated linolenic acids. J Dairy Sci. 2005;88:3231–3239. doi: 10.3168/jds.S0022-0302(05)73006-X. [DOI] [PubMed] [Google Scholar]
- 43.Contarini G, Povolo M, Pelizzola V, Monti L, Lercker G. Interlaboratory evaluation of milk fatty acid composition by using different GC operating conditions. J Food Compos Anal. 2003;32:131–140. doi: 10.1016/j.jfca.2013.08.008. [DOI] [Google Scholar]
- 44.Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim EJ, Huws SA, Lee MRF, Wood JD, Muetzel SM, Wallace RJ, Scollan ND. Fish oil increases the duodenal flow of long chain polyunsaturated fatty acids and trans-11 18:1 and decreases 18:0 in steers via changes in the rumen bacterial community. J Nutr. 2008;138:889–896. doi: 10.3945/jn.108.089912. [DOI] [PubMed] [Google Scholar]
- 46.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinf Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 48.Webster NS, Taylor MW, Behnam F, Lucker S, Rattei T, Whalan S, Horn M, Wagner M. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettingan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Jukes TH, Cantor CR. Evolution of Protein Molecules. In: Munro HN, editor. Mammalian Protein Metabolism. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 51.Saitou N, Nei M. The neighbor-joining method - a New method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 52.Van de Peer Y, De Wachter R. Treecon for Windows - a software package for the construction and drawing of evolutionary trees for the Microsoft Windows Environment. Comput Applic Biosci. 1994;10:569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 53.SAS Institute . SAS User’s Guide: Statistics. Version 8.0. Cary, NC: SAS Institute Inc; 1999. [Google Scholar]


