Abstract
Programmed cell death (PCD) is a ubiquitous genetically regulated process consisting in an activation of finely controlled signaling pathways that lead to cellular suicide. Although some aspects of PCD control appear evolutionary conserved between plants, animals and fungi, the extent of conservation remains controversial. Over the last decades, identification and characterization of several lesion mimic mutants (LMM) has been a powerful tool in the quest to unravel PCD pathways in plants. Thanks to progress in molecular genetics, mutations causing the phenotype of a large number of LMM and their related suppressors were mapped, and the identification of the mutated genes shed light on major pathways in the onset of plant PCD such as (i) the involvements of chloroplasts and light energy, (ii) the roles of sphingolipids and fatty acids, (iii) a signal perception at the plasma membrane that requires efficient membrane trafficking, (iv) secondary messengers such as ion fluxes and ROS and (v) the control of gene expression as the last integrator of the signaling pathways.
Keywords: plant, programmed cell death, lesion mimic mutants, genetics approaches, immunity responses
Introduction
The decision whether a cell should live or die is fundamental to the survival of multicellular organisms. Programmed Cell Death (PCD), a genetically regulated cellular suicide, has been described in all multicellular organisms and recent studies even suggest that apoptosis-like processes could occur in bacteria (Hakansson et al., 2011; Dwyer et al., 2012). In plants, PCD is involved both in processes required for normal development and to face biotic or abiotic stress (Pennell and Lamb, 1997; Williams and Dickman, 2008). From a cellular perspective, plant PCD can be classified as (i) autolytic PCD, characterized by the formation of large lytic vacuoles and rapid clearance of the cytoplasm due to tonoplast rupture and the release of hydrolases, and (ii) non-autolytic PCD which lacks rapid clearance of the cytoplasm (Van Doorn, 2011). Autolytic PCD occurs mainly during plant development whereas non-autolytic PCD is generally observed in response to stress, for example during the hypersensitive response (HR) induced by pathogens (Van Doorn, 2011). HR is one of the best characterized PCD (Coll et al., 2011) and phytohormones such as salicylic acid (SA), jasmonic acid (JA) or ethylene (ETH), as well as reactive oxygen species such as hydrogen peroxyde (H2O2), singlet oxygen (1O2) or superoxide anion (O−2), are key players for HR development and regulation (Coll et al., 2011). However, our understanding of the cellular events leading to cell death in plants remains largely incomplete.
By contrast, in mammalian cells, the molecular bases of PCD are well described. The main regulators of PCD in animals belong to the Bcl-2 family of proteins composed of pro (Bid, Bad, Bak, and Bax) and anti-apoptotic proteins (Bcl-2 and Bcl-xL) (Youle and Strasser, 2008). Under normal conditions, mitochondrial outer membrane integrity is maintained through a balance between these pro and anti-apoptotic proteins. Perception of a pro-death signal can activate factors such as BAX and BAK (for Bcl2-ASSOCIATED X and Bcl2-HOMOLOGS ANTAGONIST/KILLER) that allow the release of cytochrome c (cyt. c) from mitochondria. Once in the cytoplasm, cyt. c triggers a chain reaction by activating caspases (cysteine-aspartyl proteases), which target various proteins, leading to the activation of several hydrolases and thus to the degradation of most cellular macromolecules (Marino et al., 2014). Based on sequence homology, some caspase-like proteins with protease activities have been identified in plants, such as metacaspases (Tsiatsiani et al., 2011), but this approach was not very fruitful, due to the absence of clear sequence conservation between animals and plants, and it thus remains unclear whether the pathways described in animal cells are conserved in plants. In particular, plant genomes appear to lack Bcl-2 family homologs. Nevertheless, expression of the murine BAX protein in Tobacco or Arabidopsis is sufficient to promote cell death (Lacomme and Santa Cruz, 1999; Kawai-Yamada et al., 2001), and release of cyt. c during plant PCD has been documented (Colombatti et al., 2014), indicating that mitochondria also play a central role during plant PCD. In addition, the BAX-INHIBITOR 1 (BI-1) PCD antagonist is conserved in plants where it can prevent cell death induced by ectopic expression of BAX (Kawai-Yamada et al., 2001), and control cell death progression during pathogen attacks (Watanabe and Lam, 2006). Hence, BI-1 regulates cell death in plants, but data available so far suggest that the molecular mechanisms underlying its action and its partners are not conserved between plants and animals and that sequence similarity searches are thus probably not the best suited to decipher PCD control in plants. Others strong candidates that may function as caspase-like executioners of plant PCD are vacuolar processing enzymes (VPE). Even though they display little sequence homology to animal caspases, they were found to have YVADase (caspase-1 like) activity, and VPE deficiency was shown to prevent virus-induced HR cell death in tobacco (Hatsugai et al., 2004). δVPE was also reported to be involved in the cell death associated with early seed development (Nakaune et al., 2005).
Extensive progress in our understanding of plant PCD in response to stress came from forward genetic approaches and the identification of many mutants displaying spontaneous HR-like cell death, on leaves: the so called lesion-mimic mutants (LMM).
Since about 30 years, at least 60 of these mutants have been isolated in maize (Hoisington et al., 1982), rice (Takahashi et al., 1999), barley (Wolter et al., 1993) and Arabidopsis (Lorrain et al., 2003; Moeder and Yoshioka, 2008). The combined efforts of laboratories and advances in genome annotation of model species allowed mapping of the corresponding mutations in a large number of LMM and the identification of the mutated genes. Table 1 lists the 49 LMM for which the mutated genes have been identified, and summarizes the phenotypes of the mutants, as well as the putative underlying mechanisms causing PCD. Because most mutations present in these LMM are recessive, causing a loss of protein function, this first approach led mainly to the identification of inhibitors of cell death. In order to find positive regulators of PCD, second site mutageneses have been performed by several laboratories. These suppressors are listed in Table 2, indicating which LMM is suppressed, the molecular function of the gene product, and the possible mechanism involved in the suppression of PCD.
Table 1.
Lesion mimic mutants.
| Mutant name | Gene ID | Gene product | PCD-related phenotypes | Possible mechanisms involved | References |
|---|---|---|---|---|---|
| acd1 | At3g44880 | Pheide a oxygenase involved in chlorophyll catabolic process | Lesions on older leaves, or when plants are submitted to a dark-light transfer | Accumulation of photoreactive pheide a | Greenberg and Ausubel, 1993; Gray et al., 1997; Tanaka et al., 2003 |
| lls1 (Zm) | |||||
| acd2 | At4g37000 | Red chlrorophyll catabolite reductase involved in chlorophyll catabolic process | Lesions on older leaves | Accumulation of red chlorophyll catabolites that causes singlet oxygen release | Mach et al., 2001; Yao and Greenberg, 2006; Pruzinska et al., 2007 |
| acd5 | At5g51290 | Ceramide kinase | Small restricted spontaneous lesions on leaves 5 weeks after planting | Accumulation of non-phosphorylated C2 ceramide | Greenberg et al., 2000; Liang et al., 2003; Bi et al., 2011 |
| acd6 (gof) | At4g14400 | Transmembrane protein with ankyrin domain | Yellowing leaves with cell death patches 2 weeks after planting | Constitutive activation of defense responses | Rate et al., 1999; Lu et al., 2003 |
| acd11 | At2g34690 | Protein that transfers Ceramide-1-Phosphate between membrane | Chlorosis that can engulfed the rosette | Ceramide and phytoceramide accumulation | Brodersen et al., 2002; Petersen et al., 2008; Simanshu et al., 2014 |
| agd2 | At4g33680 | Chloroplastic aminotransferase | Spotted necrotic lesions | Unknown | Rate and Greenberg, 2001; Song et al., 2004 |
| bir1 | At5g48380 | Receptor-like kinase belonging to the LRRX group | Lesions in cotyledons and true leaves | Activation of multiple defense signaling pathways | Gao et al., 2009 |
| cad1 | At1g29690 | Protein containing a MACPF domain | HR-like lesions on leaves | Unknown | Morita-Yamamuro et al., 2005 |
| camta3/sr1 | At2g22300 | Calmodulin-binding transcription factor | Chlorotic lesions on rosette leaves | Overexpression of immune regulator such as EDS1 and EIN3 | Yang and Poovaiah, 2002; Galon et al., 2008; Du et al., 2009 |
| er1 (Nt) | |||||
| cat2 | At4g35090 | Dismutation of H2O2 | Necrotic lesions in Long-Days conditions | Intracellular oxidative stress coupled with decreased of Myo-inositol content | Vandenabeele et al., 2004; Queval et al., 2007; Chaouch and Noctor, 2010 |
| cea62 (Os) | Os02g0110200 | hydroperoxyde lyase | Brown lesion spots over the entire leaf surface | Probably due to constitutive induction of JA signaling | Liu et al., 2012 |
| chs2 (gof) | At4g16860 | Encodes the TIR-NBS-LRR protein named RPP4 | Lesions and bleaching when transfer in low temperature conditions | Conditional activation of defense responses | Huang et al., 2010 |
| cpn1/bon1 | At5g61900 | Calcium-dependent phospholipid binding protein | Lesions appear on leaves in low humidity and/or low temperature conditions | Overexpression of SNC1 is necessary to induce lesions | Jambunathan et al., 2001; Jambunathan and McNellis, 2003; Yang and Hua, 2004 |
| cpr5 | At5g64930 | Transmembrane protein with unknown molecular function | Spontaneous lesions and precocious senescence | Unknown | Bowling et al., 1997; Kirik et al., 2001; Yoshida et al., 2002 |
| cpr22 (gof) | At2g46450 and At2g46440 | Cyclic-gated ion channel for Ca2+ | Chlorotic lesions, then plants die within 14 days after planting | The chimeric protein CNGC11/12 created by cpr22 deletion constitutively activate defense signaling | Yoshioka et al., 2001, 2006; Urquhart et al., 2007 |
| cslf6 (Os) | Os08g06380 | Cellulose synthase-like involved in mixed-linkage glucan biosynthesis | Spontaneous, discrete, necrotic lesions in flag and old leaves | Probably due to high decrease in mixed-linkage glucan content | Vega-Sanchez et al., 2012 |
| dnd1 | At5g15410 | Cyclic-gated ion channel for Ca2+ | Small necrotic lesions but also a decrease in HR induction | Unknown | Yu et al., 1998; Clough et al., 2000; Ali et al., 2007 |
| dnd2/hlm1 | At5g54250 | Cyclic-gated ion channel for K+, Na+ and/or Ca2+ | Spontaneus lesions but also a decrease in HR induction | Unknown | Balague et al., 2003; Jurkowski et al., 2004; Rostoks et al., 2006 |
| nec1 (Hv) | |||||
| edr1 | At1g08720 | Map Kinase Kinase Kinase | Necrotic lesions in the case of infection with the powdery mildew | Derepression of a MAP kinase cascade in edr1 seems to be responsible of the cell death phenotype | Frye and Innes, 1998; Frye et al., 2001; Zhao et al., 2014 |
| edr2 | At4g19040 | a PH and STAR domain containing protein | Necrotic lesions in the case of infection with the powdery mildew | Unknown | Tang et al., 2005 |
| edr3 | At3g60190 | Dynamin-related protein with a putative role in Golgi traffic | Necrotic lesions in the case of infection with the powdery mildew | Unknown | Hong et al., 2003; Tang et al., 2006 |
| erh1 | At2g37940 | Inositol phosphorylceramide synthase | Spontaneous lesions in a Col-0 genetic background expressing the resistance gene RPW8 | Ceramide accumulation | Wang et al., 2008 |
| erh1 (Os) | |||||
| exo70b1 | At5g58430 | Exocyst subunit EXO70 family protein B1 with a role in autophagy | HR-like lesions | Unknown | Kulich et al., 2013; Stegmann et al., 2013 |
| flu | At3g14110 | Protein with Coiled-coil TPR domain regulating chlorophyll biosynthesis | Lesions on mature leaves, complete bleaching of seedling after a dark to light transfer | Accumulation of Pchlide in the dark which then generate singlet oxygen in the light condition | Meskauskiene et al., 2001; Op Den Camp et al., 2003; Przybyla et al., 2008; Khandal et al., 2009 |
| tigrina-d.12 (Hv) | |||||
| fzl | At1g03160 | Membrane remodeling GTPase localized in chloroplast enveloppe | Chlorotic lesions on rosettes leaves in Ler ecotype but not in Col-0 | Probably caused by multiple chloroplasts anomalies observed in fzl | Gao et al., 2006; Landoni et al., 2013 |
| len1 | At1g55490 | Chaperonin 60β localized in chloroplast | Spontaneous lesions only in Short-days but not in Long-days condtions | Unknown | Ishikawa et al., 2003; Ishikawa, 2005 |
| les22 (Zm) | GRMZm2G044074 | Uroporphyrynogen decarboxylase (Chlorophyll biosynthesis) | HR-like necrotic lesions | Probably due to accumulation of uroporphyrine | Hu et al., 1998 |
| loh1 | At3g25540 | Ceramide synthase | lesions after a long growth in Short-days condition | Accumulation of free trihydroxy sphingoid bases or ceramide species with C16 fatty acids | Ternes et al., 2011 |
| lrgB | At1g32080 | Intermembrane protein localized in chloroplast envelop inner membrane | Chlorotic lesions on true leaves of juveniles plants | Unknown | Yamaguchi et al., 2012; Yang et al., 2012 |
| lin2 | At1g03475 | Coproporphyrinogen III oxidase involved in chlorophyll biosynthesis | Spontaneous lesions formation on young leaves | Probable accumulation of photosensitizing tetrapyrrole intermediates | Ishikawa et al., 2001; Sun et al., 2011; Guo et al., 2013 |
| Rlin1 (Os) | |||||
| lms (Os) | Os02g0639000 | Protein with a CTD phosphatase domain and two dsRBM motifs | Reddish-brown lesions on leaves and rapid senescence after flowering | Unknown | Undan et al., 2012 |
| lsd1 | At4g20380 | Protein containing three zinc finger domains | Spontaneous and runaway cell death on leaves when plants are transferred from Short-days to Long-days conditions or submitted to a highlight stress | Uncontrolled oxidative stress (anion superoxide and oxydized plastoquinone pool) that is the consequence of misregulation of numerous genes | Jabs et al., 1996; Dietrich et al., 1997; Kliebenstein et al., 1999; Muhlenbock et al., 2008 |
| lsd1 (Os) | |||||
| mips1 | At4g39800 | Myo-inositol-1-phosphate synthase which catalyze the first reaction of Myo-inositol biosynthesis | Necrotic lesions appear on rosette leaves after a transfer from Short-days to Long-days conditions | Not clearly determined but a ceramide accumulation can be observed | Meng et al., 2009; Donahue et al., 2010; Luo et al., 2011 |
| mkk1/mkk2 | At4g26070, At4g29810 | Two Map kinase kinase which act to negatively regulate plant defenses | Reduced growth, lesions that engulfed the whole seedling and lead to lethality | Constitutive activation of defense responses | Ichimura et al., 1998; Gao et al., 2008; Qiu et al., 2008 |
| mpk4 | At4g01370 | Map kinase which acts downstream to MKK1/MKK2 to negatively regulate plant defenses | Reduced growth, small spotted lesions | Constitutive activation of defense responses | Petersen et al., 2000; Su et al., 2007; Gao et al., 2008; Liu et al., 2011 |
| mpk4 (Gm) | |||||
| nsl1 | At1g28380 | Protein containing a MACPF domain | Spotted necrotic lesions | Unknown | Noutoshi et al., 2006 |
| pub13 | At3g46510, Os12g0570000 | Protein with E3 ubiquitin ligase activity | Chlorosis and spotted lesions on leaves in LD conditions | Constitutive activation of defense responses | Zeng et al., 2004; Li et al., 2012 |
| spl11 (Os) | |||||
| rug1 | At5g08280 | Porphobilinogen deaminase involved in chlorophyll biosynthesis | Spontaneous lesions on leaves | Probably due to accumulation of porphobilinogen | Quesada et al., 2013 |
| siz1 | At5g60410 | SUMO E3 ligase | Spontaneous lesions on leaves and dwarfism | Constitutive activation of defense responses | Lee et al., 2007a; Miura et al., 2010 |
| slh1 | At5g45260 | TIR-NBS-LRR with a WRKY domain | Necrotic lesions on rosette and cauline leaves in low humidity | Defect in DNA binding of the protein that conduces to Constitutive activation of defense responses | Noutoshi et al., 2005 |
| smg7 | At5g19400 | Involved in Nonsense-Mediated RNA decay process | Necrotic lesions on rosette leaves and lethality (depending on allele) | Constitutive activation of defense responses | Riehs-Kearnan et al., 2012 |
| spl7 (Os) | Os05g0530400 | Heat stress transcription factor | Small, reddish brown lesions over the whole surface of leaves | Unknown | Yamanouchi et al., 2002 |
| spl28 (Os) | Os01g0703600 | Clathrin adaptator subunit (AP1M1) involved in post-golgi traffic | Spotted lesions on leaves | Unknown | Qiao et al., 2010 |
| ssi2 | At2g43710 | Steroyl-ACP desaturase involved in fatty acid desaturation | HR-like lesions | Increase in 18:0 and decrease in 18:1 fatty acids, inducing, among other things, NO accumulation | Kachroo et al., 2001, 2008; Shah et al., 2001; Mandal et al., 2012 |
| ssi2 (Gm, Os) | |||||
| ssi4 (gof) | Putative TIR-NBS-LRR protein | Chlorotic lesions suppressed by high humidity | Probably caused by constitutive activation of defense signaling | Shirano et al., 2002; Zhou et al., 2004 | |
| syp121/syp122 | At3g11820, At3g52400 | Syntaxins involved in vesicular trafficking | Dwarf phenotype and necrotic lesions | Constitutive activation of defense responses | Zhang et al., 2008 |
| upf1 | At5g47010 | Involved in Nonsense-Mediated RNA decay process | Necrotic lesions on rosette leaves and lethality (depending on allele) | Constitutive activation of defense responses | Riehs-Kearnan et al., 2012 |
| vad1 | At1g02120 | Membrane-associated protein with a GRAM domain | Vascular necrotic lesions | Unknown | Lorrain et al., 2004 |
| xth27 | At2g01850 | Xyloglucan endotransglucosylase/hydrolase | Yellow necrotic spots associated with leaf maturation | Unknown | Matsui et al., 2005 |
Bo, Brassica oleracea; Gm, Glycine max; Hv, Hordeum vulgare (barley); Ta, Triticum aestivum; Zm, Zea mays; Nt, Nicotiana tabbacum; gof, the mutation in correspondings lmm is a Gain of function. In all other lmm, the mutation is recessive.
Table 2.
Suppressors of LMM.
| Mutant | lmm supressed | Gene ID | Gene Product | Possible mechanism involved | References |
|---|---|---|---|---|---|
| act1 | ssi2 | At1g32200 | Chloroplastic glycerol-3-phosphate acyltransferase | act1 mutant accumulates 18:1 fatty acid within the chloroplast, this could explain the suppression of ssi2 cell death | Kachroo et al., 2003 |
| adr1 | lsd1 | At1g33560 | CC-NBS-LRR immune receptor | ADR1 is a positive regulator of SA signaling | Bonardi et al., 2011; Roberts et al., 2013 |
| adr1-l1 | lsd1 | At4g33300 | CC-NBS-LRR immune receptor | ADR1-L1 is a positive regulator of SA signaling | Bonardi et al., 2011; Roberts et al., 2013 |
| adr1-l2 | lsd1 | At5g04720 | CC-NBS-LRR immune receptor | ADR1-L2 is a positive regulator of SA signaling | Bonardi et al., 2011; Roberts et al., 2013 |
| edts5/ald1 | edr2 agd2 | At2g13810 | Aminotransferase homologs to AGD2 | Unknown | Nie et al., 2011 |
| bzip10 | lsd1 | At4g02640 | BZIP transcription factor | In wild-type plants, LSD1 retain BZIP10 in cytoplasm. In lsd1 mutants, BZIP10 translocate into nucleus to regulate expression of genes involved in lsd1 cell death | Kaminaka et al., 2006 |
| caa39 | flu | At5g02820 | Subunit of the topoisomerase VI complex (TOP6A) | TOP6A binds to promoter of genes regulated by ROS signaling. | Simkova et al., 2012 |
| cao | lsd1 | At5g50920 | Chaperone necessary for the assembly of the photosystem II antenna | A fully functional photosystem II is necessary for lesion formation in lsd1 | |
| crt1 | ssi4 | At4g36290 | ATPase that interacts with diverse resistance proteins | Unknown | Kang et al., 2008 |
| crt3 | bir1 | At1g08450 | Calreticulin involved in the ER quality control | crt3 and erdj3b are characterized by a decrease of SOBIR1 accumulation | Sun et al., 2014 |
| erdj3b | bir1 | At3g62600 | J domain protein involved in the ER quality control | Sun et al., 2014 | |
| ex1 | flu | At4g33630 | Chloroplastic protein with unknown molecular fucntion | EX1 and EX2 act together to integrate 1O2signal from chloroplasts and subsequently induce signaling responses that induce cell death in flu | Wagner et al., 2004; Lee et al., 2007b; Kim et al., 2012 |
| ex2 | flu | At1g27510 | Chloroplastic protein with unknown molecular fucntion | Lee et al., 2007b; Kim et al., 2012 | |
| hpr1 | edr1 | At5g09860 | Component of the THO/transcrtiption export complex required for mRNA export | A fully functional mRNA export machinery is necessary for cell death in edr1 | Pan et al., 2012 |
| laz1 | acd11 | At4g38360 | Protein with a domain of unknown function (DUF300) | Unknown | Malinovsky et al., 2010 |
| laz2 | acd11 | At1g77300 | Histone lysine 36 methyl transferase | LAZ2 is necessary for the proper expression of LAZ5 which is then a positive mediator of acd11-dependent cell death | Palma et al., 2010 |
| laz5 | acd11 | At5g44870 | Putative TIR-NB-LRR R-protein | LAZ5 is A R-protein which could triggers cell death in the absence of ACD11 | Palma et al., 2010 |
| lol1 | lsd1 | At1g32540 | Protein homologs to LSD1, with 3 zinc finger domains | LOL1 is and LSD1 have antagonistic effects on ROS homeostasis | Epple et al., 2003 |
| mc1 | lsd1 | At1g02170 | Type 1 metacaspase | Unknown | Coll et al., 2010 |
| oxt6 | cat2 | At1g30460 | Subunit of the CPSF complex involved in mRNA polyadenylation (CPSF30) | CPSF30 regulates positively the expression of a large number of genes involved immune responses | Bruggeman et al., 2014 |
| cpr5 | |||||
| lsd1 | |||||
| mips1 | |||||
| plp2 | vad1 | At2g26560 | Lipid acyl hydrolase with wide substrate specificities | PLP2 could provide fatty acid precursors for the biosynthesis of oxylipins | La Camera et al., 2009 |
| prl1 | flu | At4g15900 | Nuclear WD40 protein with unknown molecular function | PRL1 has a role in modeling 1O2-dependent signaling | Baruah et al., 2009 |
| rar1 | ssi4 | At5g51700 | Small protein with two zinc binding domains, involved in defense signaling | A disruption of RAR1 affects a multiple R gene-triggered responses | Zhou et al., 2008 |
| rdc8 | ssi2 | At2g40690 | Glycerol-3-phosphate deshydrogenase (GLY1) | Decrease in glycerol-3-phosphate pool | Kachroo et al., 2004 |
| sdf2 | bir1 | At2g25110 | Protein involved in the ER quality control | Same as crt3 suppressor | Sun et al., 2014 |
| sfd4 | ssi2 | At4g30950 | Fatty acid desaturase involved in the synthesis of polyunsaturated lipids | Decrease in plastid complex lipid species containing hexadecatrienoic acids | Nandi et al., 2003 |
| sobir1 | bir1 | At2g31880 | Receptor-like kinase | In absence of BIR1, SOBIR1 activates resistance signaling pathways | Gao et al., 2009 |
| soldat8 | flu | At2g36990 | SIGMA6 (SIG6) factor of the plastid RNA polymerase | Disruption of SIG6 disturbs chloroplasts homeostasis that could acclimate plants and then suppress flu 1O2-mediated cell death | Coll et al., 2009 |
| soldat10 | flu | At2g03050 | Plastidial protein related to the human mTERF | Same as soldat8 | Meskauskiene et al., 2009 |
| sr1-4D (gof) | edr2 | At2g22300 | Calmodulin-binding transcription factor | Probalby due to enhanced repression of genes involved in edr2 cell death | Nie et al., 2012 |
| summ1 | mkk1/mkk2 | At4g08480 | MAP kinase kinase kinase | In absence of MKK1/MKK2 or MPK4, SUMM1 triggers SUMM2-mediated immune responses | Kong et al., 2012 |
| mpk4 | |||||
| summ2 | mkk1/mkk2 | At1g12280 | Putative NB-LRR R protein | In absence of MKK1/MKK2 or MPK4, SUMM2 activates resistance signaling pathways | Zhang et al., 2012 |
| mpk4 | |||||
| sup6 | acd6 | At5g20660 | Putative Metalloprotease | Unknown | Lu et al., 2009 |
| ulf3/hy1 | flu | At2g26670 | Heme oxygenase involved in biosynthesis of chromophore | Accumulation of heme in hy1 inhibits the Glu-tRNA enzyme conducing to a decrease in Pchlide content | Goslings et al., 2004 |
Together these two types of approaches have greatly contributed to our understanding of the cellular mechanisms governing PCD in plants. This review is mainly focused on the cellular functions or processes that can be grouped in several categories (i) cellular activities linked to chloroplasts activity and light energy capture controlling the onset of PCD, (ii) sphingolipids and fatty acids as regulators of cell death, (iii), signal perception at the plasma membrane that required efficient membrane trafficking (iv) secondary messengers such as ion fluxes and ROS and (v) changes in gene expression as the last integrator of the signaling pathways.
Chloroplasts and light are hubs of stress leading to PCD
Plants need light as the source of energy for photosynthesis, but excess energy can lead to cell damage, cell death and ultimately, plant death. The severity of the symptoms of several LMM, such as lesion simulating disease1 (lsd1) (Mateo et al., 2004), myo-inositol-1-phosphate synthase1 (mips1) (Meng et al., 2009) and catalase2 (cat2) (Queval et al., 2007), is light-dependent, suggesting that chloroplast activity can function as a signal triggering cell death, possibly via ROS production or changes in their redox state. In line with this hypothesis, several LMM mutants are deficient for chlorophyll biosynthesis or degradation, leading to abnormal accumulation of photoreactive molecules and ROS production in the light.
Tight regulation of chlorophyll biosynthesis and degradation is essential to cell survival
The tetrapyrrole biosynthesis pathway is well characterized. It takes place in chloroplasts and through branching leads to production of chlorophyll a/b, hemes, siroheme and phytochromobilin molecules (Mochizuki et al., 2010). Disruption of tetrapyrroles biosynthesis at different stages can lead to lesion-mimic phenotypes due to the abnormal accumulation of photoreactive molecules (Figure 1). For example, the mutant rugosa1 (rug1) is affected in a porphibilinogen deaminase (PGBD), and accumulates porphibilinogen (Quesada et al., 2013). Likewise, accumulation of coproporphyrinogen III in lesion initiation 2/rice lesion initiation 1 (lin2/rlin1) mutants (Ishikawa et al., 2001; Sun et al., 2011), uroporphyrynogen III in lesion22 (les22) mutant (Hu et al., 1998), and protochlorophyllide (Pchlide) in oep16 and flu mutants (Meskauskiene et al., 2001; Samol et al., 2011b) leads to cell death phenotypes.
Figure 1.
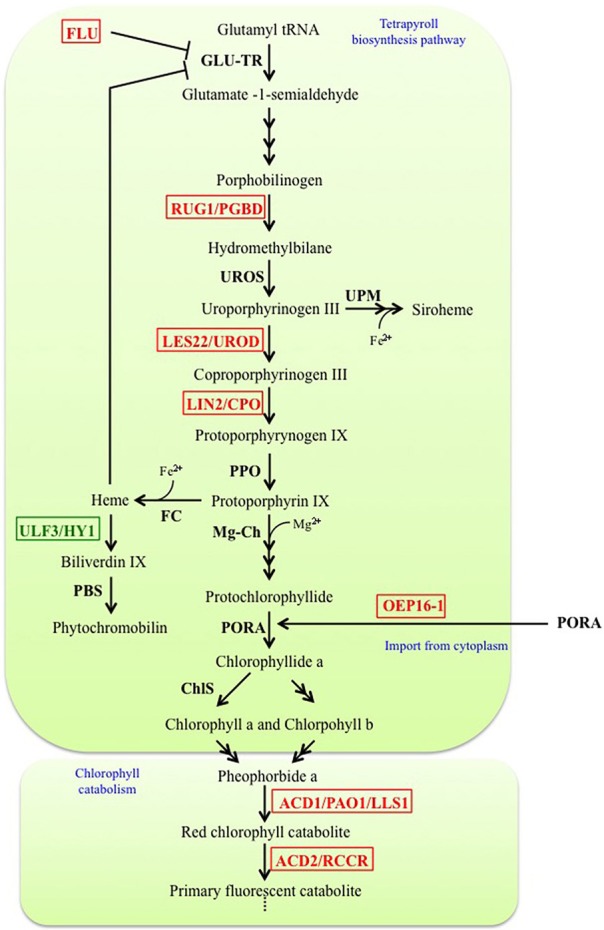
Simplified representation of the tetrapyrrole biosynthesis pathway and chlorophyll catabolism into the chloroplast. Factors disrupted in LMM are indicated in red whereas factor disrupted in suppressors of LMM are in green. ACD1/PAO1/LLS1, Accelerated cell death 1/Pheophorbide a oxygenase/Lethal leaf spot1; ACD2/RCCR, Accelerated cell death 2/Red chlorophyll catabolite reductase; ChlS, Chlorophyll synthase; FC, Fe chelatase; FLU, Fluorescent; GLU-TR, Glutamyl-tRNA reductase; LES22/UROD, Lesion 22/Uroporphyrinogen III decarboxylase; LIN2/CPO, Lesion initiation 2/Coproporphyrinogen III oxidase; Mg-Ch, Mg chelatase; OEP16-1, Outer plastid envelope protein 16-1; PBS, Phytochromobilin synthase; PORA, NADPH-protochlorophyllide oxidoreductase; PPO, Protoporphyrinogen IX oxidase; RUG1/PGBD, Rugosa 1/Porphobilinogen deaminase; ULF3/HY1, FLU3 written backwards/Heme oxygenase 1; UPM, Uroporphyrinogen III methylase; UROS, Uroporphyrinogen III synthase.
The oep16 (Samol et al., 2011b) and flu (Meskauskiene et al., 2001) mutants are both characterized by bleaching of seedlings when they are transferred from dark to light conditions. Unlike other LMM cited above, outer enveloppe protein16-1 (oep16-1) and fluorescent in blue light (flu) are not mutated in genes encoding enzymes involved in the biosynthesis of tetrapyrroles, but are both characterized by over-accumulation of Pchlide in the dark. Indeed, one of the last steps of chlorophyll biosynthesis, namely the conversion of protochlorophyllide (Pchlide) to chlorophyllide (Chlide), is catalyzed by a photoenzyme and can thus not occur in the dark. Down-regulation of chlorophyll biosynthesis in the dark is therefore crucial to avoid the cytotoxic effects of the highly photo-reactive Pchlide when plants reach the light. OEP16 encodes an amino acid-selective channel protein involved in the chloroplast import of the enzyme NADPH:Pchlide oxydoreductase A which converts Pchlide to chlorophyllide a (Samol et al., 2011b). The FLU gene encodes a protein with a coiled-coil TPR domain localized in the chloroplast. FLU acts negatively on the biosynthesis of tetrapyrroles in the dark by inhibiting the activity of Glutamyl-tRNA reductase (GLU-TR) (Meskauskiene and Apel, 2002; Goslings et al., 2004; Apitz et al., 2014), which catalyzes the first step of the tetrapyrrole biosynthetic pathway, thereby preventing Pchlide accumulation. Inhibition of the GLU-TR activity is also observed in the suppressor ulf3/heme oxygenase 1 ulf3/hy1 because of an accumulation of heme, which then prevents Pchlide increase and suppresses cell death in the flu mutant (Goslings et al., 2004). Therefore, FLU appears as a key negative regulator of tetrapyrrole biosynthesis pathway in the dark, and it has been further confirmed by the interaction of FLU with several enzymes of Mg2+ branch of the tetrapyrroles biosynthesis pathway (Kauss et al., 2012). When flu and oep16 mutants are transferred in the light after a prolonged dark period, Pchlide rapidly generates singlet oxygen (1O2) (Meskauskiene et al., 2001; Op Den Camp et al., 2003; Samol et al., 2011a). In flu, the accumulation of this ROS causes the loss of chloroplast integrity followed by a rupture of the central vacuole and finally cell death (Op Den Camp et al., 2003).
Surprisingly, defects in chlorophyll catabolism can also lead to cell death. Indeed, disruption of two enzymes involved in the degradation of chlorophyll generates spontaneous lesions in the accelerated cell death1/lethal-leaf spot1 (acd1/lls1) and accelerated cell death2 (acd2) LMM (Greenberg and Ausubel, 1993; Mach et al., 2001; Tanaka et al., 2003). acd1 is characterized by an accumulation of pheophorbide (Pheide a) due to disruption of ACD1/LLS1/PHEIDE A OXYGENASE1 (PAO1) gene, a Pheide a oxygenase converting Pheide a into red chlorophyll catabolite (RCC) (Gray et al., 1997; Pruzinska et al., 2003; Tanaka et al., 2003). The accumulation of the photoreactive Pheide a induces cell death in an age-dependent (Pruzinska et al., 2005), and, surprisingly, also in a light-independent manner (Hirashima et al., 2009). The ACD2/RED CHLOROPHYLL CATABOLITE REDUCTASE (RCCR) gene encodes a RCC reductase, catalyzing the next reaction that converts RCC to the primary “fluorescent” chlorophyll catabolite (Mach et al., 2001). The spontaneous cell death observed in acd2 is correlated to an accumulation of RCC and 1O2(Pruzinska et al., 2007); moreover, ACD2/RCCR may bind to porphyrin-related molecules in mitochondria to prevent PCD (Yao and Greenberg, 2006). Interestingly, during pathogen infection, ACD2 localizes dynamically between the chloroplast and mitochondria, apparently as a protective response against chloroplast-derived ACD2 substrate molecules that can target mitochondria and induce death (Pattanayak et al., 2012).
Hence, tetrapyrrole biosynthesis and destruction need to be highly controlled to avoid accumulation of photoreactive tetrapyrroles and generation of ROS, which are damaging and/or signaling molecules capable to promote cell death. Although many LMM are affected for chlorophyll biosynthesis or degradation, it remains unclear whether changes in the accumulation of photoreactive chlorophyll precursors or degradation products actually contribute to PCD control during plant development or in response to stress. However, one major finding associated with the description of this class of LMM is the identification of PCD regulatory pathways that respond to ROS production in the chloroplast. Indeed, the triple mutant executer1 executer2 flu (ex1 ex2 flu) still accumulates Pchlide and 1O2, whereas the cell death phenotype is suppressed (Wagner et al., 2004; Lee et al., 2007b). Importantly, EX proteins have been shown to operate in the wild-type under high light stress (Kim et al., 2012). This demonstrates that PCD observed in the flu mutant is not a consequence of cellular damage caused by 1O2 accumulation, and that the plastidial proteins EX1 and EX2 are involved in the retrograde control of nuclear 1O2-responsive genes and are both necessary to integrate the 1O2 signal from chloroplasts to nucleus.
Alteration of chloroplast integrity or activity can modulate PCD
Alteration of chloroplast function or integrity can lead to two opposite effects: promotion or suppression of cell death. For example, disruption of LESION INITIATION1 (LEN1) and LRGB which encode respectively a chloroplastic chaperonin 60 β (Ishikawa et al., 2003) and an intermembrane protein localized in chloroplast envelope (Yamaguchi et al., 2012; Yang et al., 2012), leads to spontaneous cell death, but the underlying mechanisms are still unknown. The FZO-like (FZL) protein, a membrane remodeling GTPase, has a crucial role in thylakoid organization (Gao et al., 2006), and its disruption induces alteration in chloroplast number, size and shape and also the development of spontaneous cell death in fzl mutants, suggesting that damages to the chloroplast membranes could trigger ROS accumulation and induce cell death (Landoni et al., 2013).
Thorough analysis of the lsd1 mutant brought further evidence for the role of chloroplast function in the control of PCD. Indeed this mutant has a conditional phenotype, and displays spontaneous lesions only in long-day condition (Jabs et al., 1996) or high illumination (Mateo et al., 2004). Lesions formation in lsd1 has been shown to depend on the redox status of the plastoquinone pool: this mutant is incapable to confine cell death induced by over-reduction of plastoquinones due to excess excitation energy (Mateo et al., 2004; Muhlenbock et al., 2008). Consistently, a mutation in the CHLOROPHYLL A/B BINDING HARVESTING-ORGANELLE SPECIFIC (CAO) gene encoding a chaperone required for the assembly photosystem II antenna leads to a reduction of lesion formation in lsd1 (Mateo et al., 2004). Likewise, inhibition of chlorophyll biosynthesis abolished cell death in another LMM: the mips1 mutant (Meng et al., 2009) and several mutations affecting chlorophyll metabolism have been reported to suppress lesion formation in len1 (Ishikawa, 2005). Together, these studies support the notion that perturbation of the photosynthetic metabolism can trigger PCD.
Interestingly, alteration of other chloroplast functions can also suppress cell death in some LMM. Indeed, the two suppressors of flu, soldat8 and soldat10 are mutated in genes involved in plastid RNA transcription (Coll et al., 2009; Meskauskiene et al., 2009). Both mutants display disturbance of chloroplast homeostasis that could acclimate plants to stress, thereby preventing flu mediated cell death. The last example of the bipolar function of chloroplasts in PCD is given by the mutant aberrant growth and death2 (agd2), which is deficient for a plastidial aminotransferase (Rate and Greenberg, 2001; Song et al., 2004). Surprisingly, this LMM is suppressed by disruption of AGD2-LIKE DEFENSE RESPONSE PROTEIN 1 (ALD1), another plastidial aminotransferase (Song et al., 2004). The authors suppose that AGD2 synthesizes an amino acid–derived molecule that suppresses defenses, whereas ALD1 generates a related amino acid–derived molecule important for activating defense signaling.
Therefore, chloroplasts are the source of opposite signals able to prevent or to promote cell death, and complex retrograde signaling pathways from plastids to the nucleus are necessary to integrate them and induce appropriate cellular responses (Galvez-Valdivieso and Mullineaux, 2010; Karpinski et al., 2013).
Hence, forward genetic approaches have identified chloroplasts as likely sources of pro-death signals. Indeed, because metabolic pathways occurring in chloroplasts and in particular photosynthesis are exquisitely sensitive to changes in environmental conditions, these organelles appear ideally suited to play a prominent role in stress perception. However, stress perception can occur in various other cellular compartments, notably in cellular membranes which are exposed at early steps of pathogen infections.
Several classes of lipids are key molecules controlling plant PCD
Sphingolipids and oxylipins both possess Fatty Acids (FA) in the composition of their skeleton. Because of their chemical diversity they have the potential to fine-tune many cellular processes. In this section, we will address how the characterization of some specific LMM highlighted the fundamental role of these molecules in the control of plants PCD.
Sphingolipids as inducers of PCD
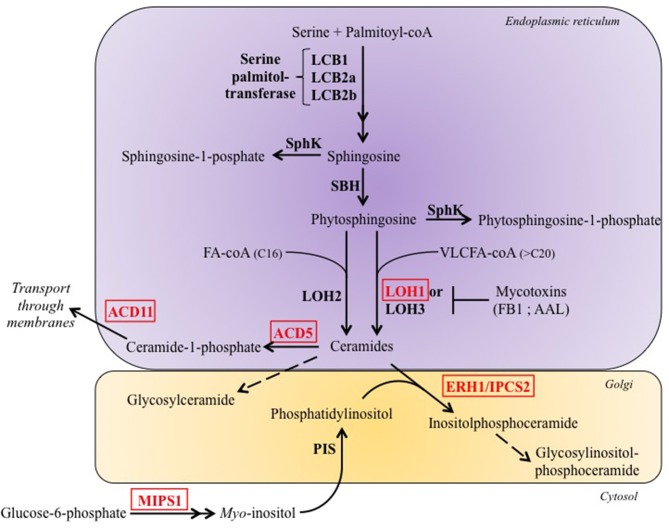
Sphingolipids are a class of lipids present ubiquitously in a large variety of organisms including eukaryotes and bacteria. All complex sphingolipids are composed of a polar head group and an N-acetyl FA linked to a sphingoid long chain base (LCB), via the amine bond, to form the basic unit of all sphingolipids: the ceramide (Pata et al., 2010). Sphingolipids display a high structural diversity caused by variability in the length of the sphingoid LCB and the N-acetyl FA, in the number of saturations, in the degree of hydroxylation, and in the diversity of the head group (Pata et al., 2010). Their biosynthesis starts in the endoplasmic reticulum (ER) and ends in the Golgi apparatus (Figure 2).
Figure 2.
Simplified representation of sphingolipid metabolism in plants. The de novo biosynthesis of ceramides occurs in the endoplasmic reticulum and synthesis of more complex sphingolipids occurs in the Golgi. Sphingosine and phytosphingosine are both referred in the text as Long-chain Basis (LCB). Factors disrupted in LMM are indicated in red. AAL, Alternaria alternata f. sp. lycopersici; ACD5/11, Accelerated cell death 5/11, are a ceramide kinase and a ceramide-1-phosphate transporter, respectively; ERH1/IPCS2, Enhancing RPW8-mediated HR-like cell death 1/Inositolphosphoceramide synthase 2; FB1, Fumosin B1; LCB1/2a/2b, Long-chain base 1/2a/2b, are subunits which form the Serine palmitol-transferase; LOH1/2/2 Lag one homolog1/2/3, are ceramide synthases; MIPS1, Myo-inositol-1-phosphate synthase; PIS, Phosphatidylinositol synthase; SBH, Sphingoid base hydroxylase; SphK, Sphingosin kinase.
Several LMM are characterized by over-accumulation of ceramides due to the disruption of enzymes that convert them in other molecules. It is the case of the accelerated cell death 5 (acd5) mutant (Greenberg et al., 2000), which is deficient for a ceramide kinase (Liang et al., 2003) and of the enhancing RPW8-mediated HR-lihe cell death 1 (erh1) which lacks the inositol-phosphorylceramide synthase 2 (IPCS2). Quantitative sphingolipid profiling indicated that ceramide accumulation in acd5 paralleled the appearance of spontaneous cell death, and it was accompanied by autophagy and mitochondrial ROS accumulation (Bi et al., 2014). IPCS2 catalyzes the production of inositolphosphorylceramide (IPC) and diacylglycerol from ceramide and phosphatidylinositol (Wang et al., 2008). In the absence of this enzyme, synthesis of RESITANCE TO POWDERY MILDEW 8 (RPW8), a protein that confers resistance to powdery mildew, induces an accumulation of ceramides and massive HR-like cell death (Wang et al., 2008). Moreover, the acd5 erh1 double mutant showed an additive effect with more severe cell death (Wang et al., 2008), likely as a consequence of higher levels of ceramides.
The relative accumulation of ceramides and their derivatives thus appear as key elements for the control of plant PCD. Indeed, ceramide-phosphate partially blocks cell death induced by ceramides (Liang et al., 2003), suggesting that ceramides are inducers of PCD whereas their phosphorylated forms block it. This is further supported by the observation that Arabidopsis mutant of the MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1 (MIPS1) forms spontaneous lesions (Meng et al., 2009; Donahue et al., 2010; Luo et al., 2011). Indeed, myo-inositol is used to synthesize numerous compounds in the cell, including phosphatidylinositol, and inhibition of myo-inositol production could thus mimic erh1 PCD (Wang et al., 2008) by preventing the conversion of ceramides in IPC. In agreement with this hypothesis, elevated levels of ceramides and hydroxyceramides were observed in mips1 (Donahue et al., 2010).
Interestingly, membrane transfer of sphingolipid also seems to play a crucial role in regulating plant PCD. Indeed, it has been recently shown that acd11, which displays chlorosis that can lead to plant death (Brodersen et al., 2002), is mutated in a ceramide-1-phosphate (C1P) transfer protein (Simanshu et al., 2014). ACD11 selectively transfers C1P between membranes, and disruption of this activity in acd11 leads to constitutive accumulation of C1P but an even stronger elevation of non-phosphorylated ceramides is observed (Simanshu et al., 2014), which may be responsible for the induction of cell death.
It is worth noting that the phosphatidylinositol derivatives phosphoinositides are known to be involved in the transport of sphingolipids. For example, the mammalian ceramide transport protein CERT requires direct binding to phosphatidylinositol-4-phosphate for its function (Hanada et al., 2003). Thus, defects in phosphatidylinositol-4-phosphate accumulation in mips1 may impact ceramide transport as well as their metabolism. Nevertheless, because myo-inositol is involved in a large number and diverse cellular processes (Valluru and Van Den Ende, 2011), it is possible that spontaneous cell death observed in mips1 is not directly the consequence of sphingolipids accumulation.
An additional layer of complexity comes from the fact that disruption of LAG ONE HOMOLOG 1 (LOH1) gene, encoding the ceramide synthase involved in the production of very long chain fatty acids (VLCFA) containing ceramides conduces to spontaneous cell death (Ternes et al., 2011). This finding is consistent with the early observation that the bacterial toxins secreted by necrotrophic pathogens: AAL (named from initials of the producing pathogen, Alternaria alternata f. sp. lycopersici) and Fumonisin B1 (FB1) (Merrill et al., 1993; Wang et al., 1996) target LOHs, and suggests that different families of ceramides may play distinct roles in the control of PCD. Indeed, AAL and FB1 do not target LOH2 which is involved in the synthesis of ceramides containing C16 fatty acids (Markham et al., 2011) and loh1 is characterized by increased levels of LCB, as well as of C16 containing ceramides, but not of VLCFA containing ceramides, indicating that in this case, spontaneous cell death is triggered either by the accumulation of LCB or C16 containing ceramides (Ternes et al., 2011).
Taken together, these studies on LMM indicate that LCB and ceramides are sphingolipids promoting PCD, not only in Arabidopsis but also in monocots (Bi et al., 2011). However, the underlying mechanisms are not well understood. It has been shown that pathogen-specific sphingolipids induced Ca2+ signaling, Mitogen Associated Protein Kinases (MAPK) activation and ROS production in cultured rice cells (Kurusu et al., 2011). Moreover, the suppressor of acd11, lazarus5 (laz5) is deficient for a putative Toll-Interleukin-1 Receptor-Nucleotide Binding Site-Leucine Rich Repeat factor (TIR-NBS-LRR) (Palma et al., 2010). The TIR-NBS-LRRs are generally known to be involved in the recognition of avirulence factors (Rafiqi et al., 2009). Another link between sphingolipids and activation of cell death, is that the MPK6 protein, a positive regulator of plant defenses, has been identified as a potential target of LCBs (Saucedo-Garcia et al., 2011).
Fatty acids and oxylipins
There is major evidence that simple FA can also regulate plant-pathogens interactions and cell death (Walley et al., 2013). The LMM suppressor of SA insensitivity2/fatty acid biosynthesis2 (ssi2/fab2) is mutated in a gene encoding one of the steroyl-ACP desaturase, involved in fatty acid desaturation (Kachroo et al., 2001). In ssi2/fab2, a decrease in 18:1 FA leads to a stabilization of NITRIC OXIDE ASSOCIATED1 (NOA1), an enzyme that regulates NO levels, and thus to an increase in endogenous NO levels (Mandal et al., 2012). This triggers transcriptional up-regulation of NO responsive nuclear genes, thereby activating disease resistance and cell death. A mutation in ACT1, encoding a glycerol-3-phosphate acyltransferase, conduces to accumulation of 18:1 FA and suppresses lesions in the ssi2 act1 double mutant, confirming the role of 18:1 in the regulation of cell death (Kachroo et al., 2003).
One of the key processes in early plant defense signaling is enhanced lipid peroxidation and production of a vast array of FA-derived compounds called oxylipins. After stimuli which activate lipases, FA such as linoleic acid are released from the chloroplastic membrane (Walley et al., 2013). Oxylipins are then produced through parallel and competing branches of the allene oxide synthase (AOS) and hydroperoxide lyase (HPL) pathways (Creelman and Mullet, 1997; Matsui, 2006). The AOS pathway is responsible for stress-inducible production of jasmonates such as jasmonic acid (JA) and methyl jasmonate (MeJA) (Creelman and Mullet, 1997). The HPL pathway produces C6-aldehydes and corresponding derivatives (Matsui, 2006). These two competing pathways are both important in the elicitation of plant defense responses against biotic agents (Walley et al., 2013): impairment of the HPL pathway in the rice LMM constitutive expression of allene oxide synthase gene62 (cea62) causes lesions due to overproduction of JA (Liu et al., 2012). Moreover, the PATATIN-LIKE PROTEIN 2 (PLP2), a pathogen-induced patatin-like lipid acyl hydrolase with a wide range of substrates, seems to be necessary to provide FA precursors for the biosynthesis of oxylipins which are then necessary to spontaneous vascular necrotic lesions observed in the LMM vascular associated death 1 (vad1) (Lorrain et al., 2004; La Camera et al., 2009).
Taken together, these studies demonstrate that FA and oxylipins are other molecules involved in the control of cell death in plants, adding an additional layer of complexity to the control of the plant PCD by cellular metabolites.
From immunity response and cellular trafficking to cell death
Immunity component
In addition of PAMP-triggered immunity, another type of innate immune responses in plants is effector-triggered immunity (ETI), which evolved in plants to detect pathogen effectors and initiate defense response. ETI is mediated by the host resistance (R) genes and recognition of pathogen effectors by R proteins can be either direct or indirect (Chen et al., 2010). In Arabidopsis, R proteins are encoded by approximately 150 genes and are classified according to the domains they are built of, and a number of LMM are deficient for such proteins. The suppressor of SA insensitivity4 (ssi4) LMM is a gain of function (gof) mutant affected in a TIR-NBS-LRR factor (Shirano et al., 2002). Constitutive activation of the SSI4 protein activates SA signaling pathways and induces the formation of chlorotic lesions (Zhou et al., 2004). As previously mentioned, laz5, a suppressor of acd11, is mutated in a gene encoding a putative TIR-NBS-LRR which is required to trigger cell death (Palma et al., 2010). Three other R proteins, ACTIVATED DISEASE RESISTANCE 1 (ADR1), ADR1-LIKE 1 (ADR1-L1) and ADR1-LIKE 2 (ADR1-L2), belonging to the Coiled-Coil-NBS-LRR group are also activators of defense responses and cell death via regulation of SA signaling (Bonardi et al., 2011; Roberts et al., 2013): loss of these three factors allows the suppression of the lsd1 runaway cell death.
Analysis of LMM also allowed the identification of signaling components activated downstream of receptors involved in PTI or ETI. Upon recognition of bacterial flagellin, the plant receptor FLAGELLIN SENSING 2 (FLS2) heterodimerizes with BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) and activates plant defense responses via two parallel MAPK cascades, one involving MPK3 and MPK6 that positively regulate defense responses and one involving MPK4, a negative regulator of R protein signaling (Colcombet and Hirt, 2008; Rasmussen et al., 2012). Indeed, both the mpk4 and the mkk1/mkk2 mutant, which is deficient for the upstream MAPKKs, are LMM that show drastically reduced growth and spontaneous cell death on leaves. These two LMM are suppressed by mutations affecting a MAP kinase kinase kinase (suppressor of mkk1 mkk2 1) and a putative NB-LRR R protein (summ 2) (Kong et al., 2012; Zhang et al., 2012). The authors assumed that the absence of the MKK1/MKK2 or MPK4 protein activates SUMM1, which then triggers SUMM2-mediated immune responses.
Disruption of BIR1, a BAK1-interacting receptor-like kinase, leads to extensive cell death, activation of constitutive defense responses and impairment in the activation of MPK4 (Gao et al., 2009). It likely functions by antagonizing the activity of another RLK: SUPPRESSOR OF BIR1 (SOBIR1). Indeed, disruption of SOBIR1 suppresses lesion formation in bir1 whereas its over-expression in a wild-type context triggers cell death and immune responses signaling pathways (Gao et al., 2009).
Others examples highlight the crucial role of R proteins in regulating cell death in plants. Indeed, constitutive activation of the protein RECOGNITION OF PERONOSPORA PARASITICA 4 (RPP4), a TIR-NBS-LRR, in the chilling-sensitive 2 mutant (chs2), leads to the development of lesions in low temperature condition (Huang et al., 2010). On the contrary, the disruption of a TIR-NBS-LRR protein with a WRKY domain, in the mutant sensitive to low humidity 1 (slh1) induces the apparition of spontaneous lesions (Noutoshi et al., 2005).
One important mechanism controlling the activity of these membrane bound receptors appears to be vesicular trafficking. Indeed, it has been demonstrated that systemic acquired resistance (SAR) requires the regulation of the secretory pathway by the NONEXPRESSER of PR1 (NPR1) protein that directly binds the promoters of genes involved in this pathway (Wang et al., 2005).
Intracellular trafficking
Additional evidence for an involvement of subcellular trafficking in the immune responses and cell death came from the isolation of the exocyst subunit EXO70 family protein B1 (exo70b1), enhanced disease resistance3 (edr3) and spotted lesion28 (spl28) LMM mutants which are all impaired in vesicular trafficking. The EX70B1 gene encodes a subunit of the exocyst complex (Kulich et al., 2013; Stegmann et al., 2013), whereas edr3 is mutated in the dynamin related protein 1E (DRP1E) which could serve to attach vesicles to microtubules (Hong et al., 2003) and SPL28 encodes a clathrin adaptator subunit which may be involved in vesicular trafficking in the post-golgi (Qiao et al., 2010). However, the mechanisms connecting vesicular traffic to the observed spontaneous lesions remain to be elucidated. EXO70B1 may have a positive role for internalization of autophagic bodies in the vacuole (Kulich et al., 2013). Autophagy is a well-documented process with crucial roles in development, immune responses and PCD. For thorough information of this subject, the reader is referred to other reviews (Patel et al., 2006; Yoshimoto, 2010; Hofius et al., 2011). Nevertheless, the mutant exo70b1 is the only autophagic mutant with a known lesion-mimic phenotype, suggesting that mechanisms underlying lesion formation in other mutants deficient for vesicular trafficking may be unrelated to autophagy.
Interestingly, some LMM suppressors are also involved in intracellular trafficking: calreticulin3 (crt3), ER-localized dnaJ-like protein 3b (erdj3b) and stromal-derived factor 2 (sdf2) that have been described as suppressors of bir1 are all mutated in genes involved in the ER quality control (Sun et al., 2014). Disruption of CRT3, ERDJ3B or SDF2 conduces to a reduced accumulation of SOBIR1 (Sun et al., 2014), indicating that the secretory pathway of ER quality control plays important roles in the biogenesis of SOBIR1 and is required for cell death in bir1. Moreover, the acd6 LMM (Rate et al., 1999) harbors a gof mutation in a gene encoding a membrane protein with an ankyrin domain (Lu et al., 2003). ACD6 is localized in the ER and the plasma membrane where it forms complexes with FLS2 and BAK1 (Zhang et al., 2014b). SA signaling increases the plasma membrane pools of ACD6, FLS2, and BAK1, thus, authors suppose that SA improves the efficiency of productive folding and/or complex formation in the ER, such that ACD6, FLS2, and BAK1 reach the cell surface to more effectively promote immune responses (Zhang et al., 2014b). Therefore, ER quality control is an emerging regulatory process of plant defenses via regulation of the abundance and quality of transmembrane receptors and modulation of signal downstream of the receptors (Tintor and Saijo, 2014).
Simultaneous disruption of SYNTAXIN OF PLANTS 121 and 122 (SYP121 and 122), members of the SNARE machinery, in the syp121/syp122 double mutant, leads to constitutive activation of defense responses (Zhang et al., 2008). This activation is dependent of numerous R proteins and results in a dwarf phenotype and the development of lesions.
Together, these studies demonstrate that polarized trafficking and secretory pathways are crucial in the immune and cell death processes. This is supported by the fact that one of the first subcellular response of plants upon detection of pathogens is the reorganization and polarization of cytoskeleton to the site of attack (Hardham et al., 2007), and that RPW8.2, is specifically targeted to the extrashaustrial membrane, the host-pathogen interface (Wang et al., 2009).
Ion fluxes and reactive oxygen species contribute to signal transduction leading to cell death
As seen above, stress can be perceived by plants in a variety of ways depending on its nature. Then, mechanisms such as ionic flux, ROS production and kinase signaling pathways conduce to activation of plant defenses (Muthamilarasan and Prasad, 2013). This part describes how LMMs proved to be powerful tools to decipher all these defense mechanisms.
Signal transduction via ion fluxes
One of the first responses after pathogen recognition is the modification of ionic fluxes. In particular, a high influx of the calcium ion (Ca2+) in the cytoplasm plays a pivotal role in mediating plant immune processes, including stomatal closure, SA production and control of ROS accumulation (Ogasawara et al., 2008; Bashir et al., 2013; Baxter et al., 2014). This influx is then detected by sensors called calmodulins, that directly regulate the activity of target proteins.
Some LMM, such as constitutive expressor of PR genes22 (cpr22), death no death1 (dnd1) or dnd2/ HR-like lesion mimic1 (hlm1) are mutated in genes encoding ion channels allowing the influx or efflux of ions. In cpr22, a deletion of 3 kb between two adjacent genes, named CYCLIC NUCLEOTIDE-GATED ION CHANNEL 11 and 12 (CNGC11 and 12), fuses them and creates a chimeric and constitutively active protein CNGC11/12 responsible for lesions (Yoshioka et al., 2001, 2006). CNGC11 and 12 may act as ion channels for Ca2+, and the phenotype of cpr22 might be due to activation of Ca2+ dependent signaling because channel blockers for this cation inhibit lesions formation (Urquhart et al., 2007). Interestingly, several suppressors of cpr22 are in fact mutated in the chimeric protein CNGC11/12, underlying the importance of some residues for the functionality of the ion channel (Baxter et al., 2008; Chin et al., 2010; Abdel-Hamid et al., 2013). Surprisingly, in dnd1, disruption of CNGC2, encoding another calcic channel (Yu et al., 1998; Clough et al., 2000; Ali et al., 2007) causes opposite phenotypes, namely constitutive activation of broad-spectrum defenses and rare necrotic lesions on the one hand, and disruption of HR cell death on the other hand (Yu et al., 1998; Ahn, 2007). To account for this apparent contradiction, Clough et al. (2000) have postulated that unbalanced ionic homeostasis in dnd1 could mimic a pathogen attack and induce defense response genes, among which catalase and ascorbate peroxidase, that could in turn block HR-related cellular processes (Clough et al., 2000). Furthermore, the lack of HR in dnd1 could be explained by the fact that a Ca2+ influx generated by CNGC2 is necessary to the accumulation of NO and subsequent lesion formation (Ali et al., 2007). CNGC2 physically interacts with another ion channel, CNGC4/HLM1, and they act together to regulate immune responses and floral transition (Chin et al., 2013). Disruption of CNGC4/HLM1, in the dnd2/hlm1 mutant, causes similar phenotypes to dnd1 (Balague et al., 2003; Jurkowski et al., 2004), but unlike CNGC2 the HLM1/CNGC4 protein, which has one ortholog identified in barley (Rostoks et al., 2006; Keisa et al., 2011), has specificity for potassium (K+) and sodium (Na+) (Balague et al., 2003).
LMMs also allowed the identification of factors acting downstream of ion signaling. The COPINE/BONZAI (CPN/BON) gene family encodes Ca2+-dependent, phospholipid-binding proteins that repress cell death (Yang et al., 2006). cpn1/bon1 mutant displays lesions due to the over-accumulation of the TIR-NBS-LRR protein SUPPRESSOR OF NPR1-1 CONSTITUTIVE 1 (SNC1) (Jambunathan et al., 2001; Yang and Hua, 2004). Thanks to its C2 and Willerbrand A domains, involved in Ca2+-dependent phospholipid binding and protein binding, respectively, CPN1/BON1 is localized at plasma membrane (Li et al., 2010) and could recruit partners to the membrane to suppress SNC1 activation (Liu et al., 2005). In addition, cpn1/bon1 is suppressed by mutations in putative R genes (Li et al., 2009), and CPN1/BON1 interacts with the RLKs BAK1 and BIR1 and is phosphorylated by BAK1 (Wang et al., 2011). Given these findings, CPN1/BON1 is probably a signaling protein that integrates calcium and RLKs in the signal transduction pathways involved in cell death and defense responses.
ROS signaling
As mentioned above, ROS production caused by adverse environmental conditions likely functions to trigger PCD. In addition, ROS play a complex role as secondary messengers in the signaling pathways leading to PCD. Their role has been studied in detail in the lsd1 mutant, and these studies led to the conclusion that ROS can have opposing effects on PCD depending on the cellular compartment in which they are produced. LSD1 encodes a protein with three zinc finger domains (Dietrich et al., 1997) and can interact with the three Arabidopsis catalases (Li et al., 2013) that are responsible for the detoxification of the peroxysomal H2O2. These interactions are necessary for a proper catalase activity, and disruption of LSD1 conduces to a decrease in catalase activity (Mateo et al., 2004; Li et al., 2013) thereby triggering cell death. This mechanism is further supported by the observation that mutants deficient for the main leaf catalase CAT2 show high increase in H2O2 cellular content and the apparition of lesions only in long-day conditions (Vandenabeele et al., 2004; Queval et al., 2007). Consistently, inhibition of photorespiration under high CO2 or low O2 conditions decreases the number and the intensity of lesions in lsd1 (Mateo et al., 2004).
H2O2 production in the peroxisome thus appears to trigger PCD. By contrast, apoplastic ROS production may inhibit PCD. In the apoplast, superoxide is produced by transmembrane NADPH oxydases using dioxygen as substrate, and then it is converted in H2O2 by Superoxide dismutases (SOD) (Apel and Hirt, 2004). The lsd1 mutant is affected in the induction of SOD genes which may conduce to high accumulation of superoxide and the runaway cell death (Kliebenstein et al., 1999). However, contrary to what expected, disruption of the two NADPH oxydases, RESPIRATORY BURST OXIDASE HOMOLOG D and F (RbohD and F), does not suppress lesions in lsd1. Indeed, cell death is enhanced in the double mutants lsd1 rbohd and lsd1 rbohf and the triple mutant lsd1 rbohd rbohf is lethal (Torres et al., 2005). Likewise, disruption of the RBOHf gene aggravates lesion formation in the cat2 rbohf double mutant (Chaouch et al., 2012). RbohD and F thus appear as negative regulators of cell death propagation although they are likely to be positive regulators of PCD initiation (Chaouch et al., 2012). How ROS production connects with downstream mechanisms leading to cell death remains to be fully elucidated. Interestingly, a decrease in myo-inositol accumulation is necessary for the development of lesions in the cat2 mutant (Chaouch and Noctor, 2010), providing evidence for a potential link between ROS- and ceramide-dependent signaling.
Therefore, Ca2+ and ROS are key signal molecules for the initiation and the spreading of plant PCD. The LSD1 protein is a crucial hub regulating at once extracellular and intracellular ROS production that allows plants to integrate and coordinate the perception of different biotic and abiotic stresses via profound changes in the expression of the genome.
Integrating signals in the nucleus to modulate gene expression
PCD is an active process that requires changes in nuclear gene expression. Isolation of various LMM or suppressors of LMM has shed light on the role of transcription factors, chromatin modifications, and post-transcriptional factors in the control of PCD.
Modulating gene transcription
The CALMODULIN-BINDING TRANSCRIPTION FACTOR 3/sIGNAL RESPONSIVE 1 (CAMTA3/SR1) transcription factor can integrate several PCD signaling pathways. Its tobacco ortholog, ETHYLENe-UP-REGULATED 1 (ER1), was firstly characterized as a protein involved in plant senescence controlled by ETH (Yang and Poovaiah, 2000), and the Arabidopsis CAMTA3/SR1 was subsequently shown to interact with calmodulin in a Ca2+ dependent manner and to bind DNA CGCG cis-elements (Yang and Poovaiah, 2002). The involvement of CAMTA3/SR1 in the regulation of plant cell death came later with the characterization of camta3/sr1 mutants which display spontaneous chlorotic lesions on rosette leaves (Galon et al., 2008). Further studies demonstrated that CAMTA3/SR1 directly represses the expression of ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) and ETHYLENE-INSENSITIVE 3 (EIN3), encoding two crucial positive regulators of SA and ETH signaling pathways (Du et al., 2009; Nie et al., 2012). Moreover, during pathogen attack, CAMTA3/SR1 interacts with the SR1 INTERACTION PROTEIN 1 (SR1IP1), a ubiquitin E3 ligase, which mediates its degradation, thus removing the repression on EDS1 (Zhang et al., 2014a). Therefore, CAMTA3/SR1 is a Ca2+/calmodulin-dependent transcription factor with an inhibitory function in plant defense and cell death (Du et al., 2009).
Although the LSD1 protein displays three zinc finger domains potentially involved in DNA binding, there is no clear evidence for its role as a transcription factor. Nevertheless, LSD1 could act indirectly on gene expression by modulating the activity of other transcription factors, such as the basic leucine zipper 10 transcription factor (bZIP10). bZIP10 binds on C- and G-box DNA sequences and its disruption partially suppresses the lsd1 phenotype (Kaminaka et al., 2006). LSD1 interacts with bZIP10 and retains it outside of the nucleus. After perception of an appropriate reactive oxygen-derived signal, or in the lsd1 mutant, bZIP10 is dissociated from LSD1 and translocates into the nucleus (Kaminaka et al., 2006), where it may induce the expression of HR- and defense-related genes. Moreover, the LSD1-LIKE 1 (LOL1) gene, acts antagonistically to LSD1 to promote cell death, and its mutation suppresses lsd1 runaway cell death (Epple et al., 2003). These antagonism mechanisms between LSD1, bZIP10, and LOL1, enable fine-tuning of immune responses and cell death mediated by ROS.
The rice LMM lesion mimic and senescence (lms), displaying reddish-brown lesions on leaves and rapid senescence after flowering, is also mutated in a transcriptional regulator (Undan et al., 2012). LMS encodes a protein with a CTD phosphatase domain involved in the dephosphorylation of the CTD of the largest subunit of the RNA polymerase II, thereby regulating transcription of target genes.
The caa39 mutant, a suppressor of flu (Simkova et al., 2012), corresponds to a weak mutant allele of the TOPOISOMERASE VI A-SUBUNIT (TOP6A). TOP6A binds to the promoters of 1O2-responsive genes, and hence could directly regulate their expression (Simkova et al., 2012). TOP6A could therefore act as an integrator of signals generated by ROS formed under adverse environmental conditions.
There is also accumulating evidence for the role of chromatin modifications in the control of plant immunity (Alvarez et al., 2010), some of which come from the characterization of LMM. Mutation in the histone methyl transferase LAZARUS 2/SET DOMAIN GROUP 8 (LAZ2/SDG8), suppresses the cell death occurring in acd11 (Palma et al., 2010). The LAZ2/SDG8 protein binds to the LAZ5 gene, encoding a putative TIR-NBS-LRR protein, and methylates histone H3 on lysine 36. This is necessary for the proper expression of LAZ5, the development of cell death in acd11, and for basal and R protein-mediated resistance in Arabidopsis (Palma et al., 2010). Moreover, MIPS1 has a dual function: it is involved in primary metabolism via synthesis of in myo-inositol but can also be imported into the nucleus where it regulates its own expression (Latrasse et al., 2013). Indeed, MIPS1 binds directly to its own promoter to stimulate transcription by locally inhibiting the spreading of Arabidopsis THRITORAX RELATED 5 and 6 (ATXR5 and 6)-dependent heterochromatin marks coming from a transposable element. Furthermore, upon activation of pathogen response, the inhibitory action of MIPS1 on ATXR5/6 is alleviated and expression of MIPS1 decreases (Latrasse et al., 2013).
Post-transcriptional factors
The final amount of proteins can also be regulated by mRNA processing and export and several suppressors of LMM are affected in these processes.
The pleitropic locus1 (prl1) mutant, a suppressor of flu, is mutated in a gene encoding a protein with a putative function in pre-mRNA splicing (Baruah et al., 2009). Therefore, the PRL1 protein may control the expressions of 1O2-responsive genes to promote cell death occurring in flu.
Recently, the polyadenylation factor subunit Cleavage and polyadenylation specificity factor 30 (CPSF30) has been proposed as a key component of plant PCD (Bruggeman et al., 2014). Indeed, the oxidative stress tolerance6 (oxt6) mutant, with a T-DNA disrupting the gene CPSF30, initially identified by the authors as a suppressor of mips1 also fully suppresses the phenotype of four other LMM: constitutive expressor of PR genes5 (cpr5), cat2, lsd1 and mpk4. Transcriptomic analyses showed that CPSF30, while it is involved in a general process, is specifically required for the proper expression of a large number of genes involved in mechanisms such as defense responses, cell death and abiotic stress responses (Bruggeman et al., 2014) and appears to be required for a correct polyadenylation site choice of the pre-mRNA transcripted from these genes (Thomas et al., 2012; Bruggeman et al., 2014).
Nonsense-mediated RNA decay (NMD) is an important surveillance mechanism that eliminates transcripts with nonsense mutations (Isken and Maquat, 2008). The UPF1 and SMG7 proteins are core components involved in NMD. The analysis of several mutants deficient for these two proteins shows that impaired NMD elicits defense responses and LMM phenotypes which appear to correlate with the extent of NMD deficiency (Riehs-Kearnan et al., 2012).
The LMM enhanced disease resistance1 (edr1) which displays spontaneous cell death in the presence of the biotrophic powdery mildew pathogen, is suppressed by a mutation of the gene HEPARANASE 1 (HPR1), a homolog of human HPR1, which is a component of the THO/TREX complex (Pan et al., 2012). The THO/TREX complex functions in mRNA processing and export. Confirming this, the mRNA export is compromised in the Arabidopsis hpr1 mutant (Pan et al., 2012).
All together, these studies on suppressors of LMMs, and others studies (Gong et al., 2005; Germain et al., 2010; Lee et al., 2012; Vi et al., 2013), clearly highlight the requirement of a functional mRNA processing and export machinery to induce cell death and proper defense responses. Although such a mechanism has never been described to date, it is tempting to speculate that plant pathogens could secret effectors to inhibit this processes, as shown for the human CPSF30 that is targeted by the Nonstructural Protein1 (NS1) of the Influenza A virus, leading to a blockage of the host innate immune response (Das et al., 2008).
Conclusions and perspectives
Genetic approaches through the identification and characterization of LMMs and their related suppressors have enabled to better understand several intra- and extracellular mechanisms involved in fine-tuning plant PCD. Whereas several of these mutants have putative roles in ceramide signaling or chloroplastic function, R proteins are a significant type associated with LMM. Knockout mutants that eliminate host guards mimic the effects of pathogen effectors, and have been found to exhibit R-gene-dependent cell death. Therefore, it is possible that many LMM may correspond to gene functions that are guarded by R-proteins. If so, the diverse functions of these genes may be not directly related to PCD but only implicated in this process due to their targeting by pathogen effectors. It is also interesting to point out that despite extensive analysis of molecular events occurring in some LMMs, whether these deaths are due to PCD or are mechanistically equivalent to HR require further investigations. Indeed in many LMM, deeper biochemical or cytological studies are missing and further characterization would be required to classify LMM according to the different types of PCD involved in their phenotypes.
Altogether, proteins altered in LMM and suppressors of LMM, with other numerous factors, build an extremely complex network regulating the decision to die or not. It is also possible to assume the involvement of a core complex as a central hub integrating several PCD pathways leading to the cell suicide. Deeper genetic characterization, with the analysis of epistatic relationships between LMM and their suppressors will certainly help to decipher the links between the factors of this network and possible cross-talks between different pathways.
As mentioned several times, phytohormones such as SA, JA, and ETH have prominent roles in regulation of HR, and thus PCD, but also more generally in immune responses, senescence, abiotic stress response, plant growth and development (Rivas-San Vicente and Plasencia, 2011; Santino et al., 2013; Zhang and Zhou, 2013; Miura and Tada, 2014). Therefore, phytohormones involvement can be crucial in cell death, and LMM displaying constitutive or conditional activation in their signaling pathways are powerful tools which have also helped to highlight crosstalk between these pathways (Greenberg et al., 2000; Devadas et al., 2002; Derksen et al., 2013; Satoh et al., 2013).
Another open question is which cellular factors are involved in the execution of PCD itself. Indeed, no close orthologs of animal caspases were identified in plants, however, there is extensive evidence for the involvement of caspase-like activities during plant PCD (Reape and McCabe, 2010). To date, only two caspase-like proteins have been identified in plants via LMM approaches. Surprisingly, METACASPASE 1 and 2 (MC1 and MC2) appear to play antagonistic roles in the control of PCD (Coll et al., 2010). Further work will be required to elucidate the specific roles of these caspase-like activities at distinct stages during PCD, the significance of their subcellular localization, their substrate specificities, and the sequence in which they operate and interact with one another.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank C. Bergounioux (IBP, Orsay) for constructive discussions and helpful corrections regarding the manuscript. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Université Paris-Sud (UPSud), the Agence Nationale de la Recherche (MAPK-IPS ANR-2010-BLAN-1613-02) and the Labex Saclay Plant Sciences (SPS).
References
- Abdel-Hamid H., Chin K., Moeder W., Shahinas D., Gupta D., Yoshioka K. (2013). A suppressor screen of the chimeric AtCNGC11/12 reveals residues important for intersubunit interactions of cyclic nucleotide-gated ion channels. Plant Physiol. 162, 1681–1693. 10.1104/pp.113.217539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn I. P. (2007). Disturbance of the Ca(2+)/calmodulin-dependent signalling pathway is responsible for the resistance of Arabidopsis dnd1 against Pectobacterium carotovorum infection. Mol. Plant Pathol. 8, 747–759. 10.1111/j.1364-3703.2007.00428.x [DOI] [PubMed] [Google Scholar]
- Ali R., Ma W., Lemtiri-Chlieh F., Tsaltas D., Leng Q., Von Bodman S., et al. (2007). Death don't have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19, 1081–1095. 10.1105/tpc.106.045096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M. E., Nota F., Cambiagno D. A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11, 563–576. 10.1111/j.1364-3703.2010.00621.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K., Hirt H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. 10.1146/annurev.arplant.55.031903.141701 [DOI] [PubMed] [Google Scholar]
- Apitz J., Schmied J., Lehmann M. J., Hedtke B., Grimm B. (2014). GluTR2 complements a hema1 mutant lacking glutamyl-tRNA reductase 1, but is differently regulated at the post-translational level. Plant Cell Physiol. 55, 645–657. 10.1093/pcp/pcu016 [DOI] [PubMed] [Google Scholar]
- Balague C., Lin B., Alcon C., Flottes G., Malmstrom S., Kohler C., et al. (2003). HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15, 365–379. 10.1105/tpc.006999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruah A., Simkova K., Hincha D. K., Apel K., Laloi C. (2009). Modulation of O-mediated retrograde signaling by the PLEIOTROPIC RESPONSE LOCUS 1 (PRL1) protein, a central integrator of stress and energy signaling. Plant J. 60, 22–32. 10.1111/j.1365-313X.2009.03935.x [DOI] [PubMed] [Google Scholar]
- Bashir Z., Ahmad A., Shafique S., Anjum T., Shafique S., Akram W. (2013). Hypersensitive response - A biophysical phenomenon of producers. Eur. J. Microbiol. Immunol. 3, 105–110. 10.1556/EuJMI.3.2013.2.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A., Mittler R., Suzuki N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. 10.1093/jxb/ert375 [DOI] [PubMed] [Google Scholar]
- Baxter J., Moeder W., Urquhart W., Shahinas D., Chin K., Christendat D., et al. (2008). Identification of a functionally essential amino acid for Arabidopsis cyclic nucleotide gated ion channels using the chimeric AtCNGC11/12 gene. Plant J. 56, 457–469. 10.1111/j.1365-313X.2008.03619.x [DOI] [PubMed] [Google Scholar]
- Bi F. C., Liu Z., Wu J. X., Liang H., Xi X. L., Fang C., et al. (2014). Loss of ceramide kinase in arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell. 26, 3449–3467. 10.1105/tpc.114.127050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi F. C., Zhang Q. F., Liu Z., Fang C., Li J., Su J. B., et al. (2011). A conserved cysteine motif is critical for rice ceramide kinase activity and function. PLoS ONE 6:e18079. 10.1371/journal.pone.0018079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V., Tang S., Stallmann A., Roberts M., Cherkis K., Dangl J. L. (2011). Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc. Natl. Acad. Sci. U.S.A. 108, 16463–16468. 10.1073/pnas.1113726108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling S. A., Clarke J. D., Liu Y., Klessig D. F., Dong X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. 10.1105/tpc.9.9.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H. M., Olszak B., Skov S., Odum N., et al. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16, 490–502. 10.1101/gad.218202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q., Garmier M., De Bont L., Soubigou-Taconnat L., Mazubert C., Benhamed M., et al. (2014). The polyadenylation factor subunit CPSF30: a key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 165, 732–746. 10.1104/pp.114.236083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S., Noctor G. (2010). Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 188, 711–718. 10.1111/j.1469-8137.2010.03453.x [DOI] [PubMed] [Google Scholar]
- Chaouch S., Queval G., Noctor G. (2012). AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69, 613–627. 10.1111/j.1365-313X.2011.04816.x [DOI] [PubMed] [Google Scholar]
- Chen Q., Han Z., Jiang H., Tian D., Yang S. (2010). Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J. Mol. Evol. 70, 137–148. 10.1007/s00239-009-9316-4 [DOI] [PubMed] [Google Scholar]
- Chin K., Defalco T. A., Moeder W., Yoshioka K. (2013). The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol. 163, 611–624. 10.1104/pp.113.225680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K., Moeder W., Abdel-Hamid H., Shahinas D., Gupta D., Yoshioka K. (2010). Importance of the alphaC-helix in the cyclic nucleotide binding domain for the stable channel regulation and function of cyclic nucleotide gated ion channels in Arabidopsis. J. Exp. Bot. 61, 2383–2393. 10.1093/jxb/erq072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Fengler K. A., Yu I. C., Lippok B., Smith R. K., Jr., Bent A. F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 97, 9323–9328. 10.1073/pnas.150005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Hirt H. (2008). Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226. 10.1042/BJ20080625 [DOI] [PubMed] [Google Scholar]
- Coll N. S., Danon A., Meurer J., Cho W. K., Apel K. (2009). Characterization of soldat8, a suppressor of singlet oxygen-induced cell death in Arabidopsis seedlings. Plant Cell Physiol. 50, 707–718. 10.1093/pcp/pcp036 [DOI] [PubMed] [Google Scholar]
- Coll N. S., Epple P., Dangl J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. 10.1038/cdd.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N. S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J. L., et al. (2010). Arabidopsis type I metacaspases control cell death. Science 330, 1393–1397. 10.1126/science.1194980 [DOI] [PubMed] [Google Scholar]
- Colombatti F., Gonzalez D. H., Welchen E. (2014). Plant mitochondria under pathogen attack: a sigh of relief or a last breath? Mitochondrion 19, 238–244. 10.1016/j.mito.2014.03.006 [DOI] [PubMed] [Google Scholar]
- Creelman R. A., Mullet J. E. (1997). Biosynthesis and Action of Jasmonates in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. 10.1146/annurev.arplant.48.1.355 [DOI] [PubMed] [Google Scholar]
- Das K., Ma L. C., Xiao R., Radvansky B., Aramini J., Zhao L., et al. (2008). Structural basis for suppression of a host antiviral response by influenza A virus. Proc. Natl. Acad. Sci. U.S.A. 105, 13093–13098. 10.1073/pnas.0805213105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen H., Rampitsch C., Daayf F. (2013). Signaling cross-talk in plant disease resistance. Plant Sci. 207, 79–87. 10.1016/j.plantsci.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Devadas S. K., Enyedi A., Raina R. (2002). The Arabidopsis hrl1 mutation reveals novel overlapping roles for salicylic acid, jasmonic acid and ethylene signalling in cell death and defence against pathogens. Plant J. 30, 467–480. 10.1046/j.1365-313X.2002.01300.x [DOI] [PubMed] [Google Scholar]
- Dietrich R. A., Richberg M. H., Schmidt R., Dean C., Dangl J. L. (1997). A novel zinc finger protein is encoded by the arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. 10.1016/S0092-8674(00)81911-X [DOI] [PubMed] [Google Scholar]
- Donahue J. L., Alford S. R., Torabinejad J., Kerwin R. E., Nourbakhsh A., Ray W. K., et al. (2010). The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 22, 888–903. 10.1105/tpc.109.071779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., et al. (2009). Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Dwyer D. J., Camacho D. M., Kohanski M. A., Callura J. M., Collins J. J. (2012). Antibiotic-induced bacterial cell death exhibits physiological and biochemical hallmarks of apoptosis. Mol. Cell 46, 561–572. 10.1016/j.molcel.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P., Mack A. A., Morris V. R., Dangl J. L. (2003). Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 6831–6836. 10.1073/pnas.1130421100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C. A., Innes R. W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. 10.1105/tpc.10.6.947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C. A., Tang D., Innes R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U.S.A. 98, 373–378. 10.1073/pnas.011405198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y., Nave R., Boyce J. M., Nachmias D., Knight M. R., Fromm H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948. 10.1016/j.febslet.2008.02.037 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G., Mullineaux P. M. (2010). The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiol. Plant. 138, 430–439. 10.1111/j.1399-3054.2009.01331.x [DOI] [PubMed] [Google Scholar]
- Gao H., Sage T. L., Osteryoung K. W. (2006). FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proc. Natl. Acad. Sci. U.S.A. 103, 6759–6764. 10.1073/pnas.0507287103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., et al. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198. 10.1038/cr.2008.300 [DOI] [PubMed] [Google Scholar]
- Gao M., Wang X., Wang D., Xu F., Ding X., Zhang Z., et al. (2009). Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44. 10.1016/j.chom.2009.05.019 [DOI] [PubMed] [Google Scholar]
- Germain H., Qu N., Cheng Y. T., Lee E., Huang Y., Dong O. X., et al. (2010). MOS11: a new component in the mRNA export pathway. PLoS Genet. 6:e1001250. 10.1371/journal.pgen.1001250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Dong C. H., Lee H., Zhu J., Xiong L., Gong D., et al. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17, 256–267. 10.1105/tpc.104.027557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goslings D., Meskauskiene R., Kim C., Lee K. P., Nater M., Apel K. (2004). Concurrent interactions of heme and FLU with Glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 40, 957–967. 10.1111/j.1365-313X.2004.02262.x [DOI] [PubMed] [Google Scholar]
- Gray J., Close P. S., Briggs S. P., Johal G. S. (1997). A novel suppressor of cell death in plants encoded by the Lls1 gene of maize. Cell 89, 25–31. 10.1016/S0092-8674(00)80179-8 [DOI] [PubMed] [Google Scholar]
- Greenberg J. T., Ausubel F. M. (1993). Arabidopsis mutants compromised for the control of cellular-damage during pathogenesis and aging. Plant J. 4, 327–341. 10.1046/j.1365-313X.1993.04020327.x [DOI] [PubMed] [Google Scholar]
- Greenberg J. T., Silverman F. P., Liang H. (2000). Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics 156, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C. Y., Wu G. H., Xing J., Li W. Q., Tang D. Z., Cui B. M. (2013). A mutation in a coproporphyrinogen III oxidase gene confers growth inhibition, enhanced powdery mildew resistance and powdery mildew-induced cell death in Arabidopsis. Plant Cell Rep. 32, 687–702. 10.1007/s00299-013-1403-8 [DOI] [PubMed] [Google Scholar]
- Hakansson A. P., Roche-Hakansson H., Mossberg A. K., Svanborg C. (2011). Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLoS ONE 6:e17717. 10.1371/journal.pone.0017717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., et al. (2003). Molecular machinery for non-vesicular trafficking of ceramide. Nature 426, 803–809. 10.1038/nature02188 [DOI] [PubMed] [Google Scholar]
- Hardham A. R., Jones D. A., Takemoto D. (2007). Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 10, 342–348. 10.1016/j.pbi.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., et al. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305, 855–858. 10.1126/science.1099859 [DOI] [PubMed] [Google Scholar]
- Hirashima M., Tanaka R., Tanaka A. (2009). Light-independent cell death induced by accumulation of pheophorbide a in Arabidopsis thaliana. Plant Cell Physiol. 50, 719–729. 10.1093/pcp/pcp035 [DOI] [PubMed] [Google Scholar]
- Hofius D., Munch D., Bressendorff S., Mundy J., Petersen M. (2011). Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 18, 1257–1262. 10.1038/cdd.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoisington D. A., Neuffer M. G., Walbot V. (1982). Disease lesion mimics in maize. I. Effect of genetic background, temperature, developmental age, and wounding on necrotic spot formation with Les1. Dev. Biol. 93, 381–388. 10.1016/0012-1606(82)90125-7 [DOI] [PubMed] [Google Scholar]
- Hong Z., Geisler-Lee C. J., Zhang Z., Verma D. P. (2003). Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol. Biol. 53, 297–312. 10.1023/B:PLAN.0000006936.50532.3a [DOI] [PubMed] [Google Scholar]
- Hu G., Yalpani N., Briggs S. P., Johal G. S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. 10.1105/tpc.10.7.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li J., Bao F., Zhang X., Yang S. (2010). A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 154, 796–809. 10.1104/pp.110.157610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K., Mizoguchi T., Irie K., Morris P., Giraudat J., Matsumoto K., et al. (1998). Isolation of ATMEKK1 (a MAP kinase kinase kinase)-interacting proteins and analysis of a MAP kinase cascade in Arabidopsis. Biochem. Biophys. Res. Commun. 253, 532–543. 10.1006/bbrc.1998.9796 [DOI] [PubMed] [Google Scholar]
- Ishikawa A. (2005). Tetrapyrrole metabolism is involved in lesion formation, cell death, in the Arabidopsis lesion initiation 1 mutant. Biosci. Biotech. Biochem. 69, 1929–1934. 10.1271/bbb.69.1929 [DOI] [PubMed] [Google Scholar]
- Ishikawa A., Okamoto H., Iwasaki Y., Asahi T. (2001). A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 27, 89–99. 10.1046/j.1365-313x.2001.01058.x [DOI] [PubMed] [Google Scholar]
- Ishikawa A., Tanaka H., Nakai M., Asahi T. (2003). Deletion of a chaperonin 60 beta gene leads to cell death in the Arabidopsis lesion initiation 1 mutant. Plant Cell Physiol. 44, 255–261. 10.1093/pcp/pcg031 [DOI] [PubMed] [Google Scholar]
- Isken O., Maquat L. E. (2008). The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat. Rev. Genet. 9, 699–712. 10.1038/nrg2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabs T., Dietrich R. A., Dangl J. L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273, 1853–1856. 10.1126/science.273.5283.1853 [DOI] [PubMed] [Google Scholar]
- Jambunathan N., McNellis T. W. (2003). Regulation of Arabidopsis COPINE 1 gene expression in response to pathogens and abiotic stimuli. Plant Physiol. 132, 1370–1381. 10.1104/pp.103.022970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambunathan N., Siani J. M., McNellis T. W. (2001). A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13, 2225–2240. 10.1105/tpc.13.10.2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowski G. I., Smith R. K., Jr., Yu I. C., Ham J. H., Sharma S. B., Klessig D. F., et al. (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17, 511–520. 10.1094/MPMI.2004.17.5.511 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Fu D. Q., Havens W., Navarre D., Kachroo P., Ghabrial S. A. (2008). An oleic acid-mediated pathway induces constitutive defense signaling and enhanced resistance to multiple pathogens in soybean. Mol. Plant Microbe Interact. 21, 564–575. 10.1094/MPMI-21-5-0564 [DOI] [PubMed] [Google Scholar]
- Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003). Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15, 2952–2965. 10.1105/tpc.017301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo A., Venugopal S. C., Lapchyk L., Falcone D., Hildebrand D., Kachroo P. (2004). Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 5152–5157. 10.1073/pnas.0401315101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo P., Shanklin J., Shah J., Whittle E. J., Klessig D. F. (2001). A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proc. Natl. Acad. Sci. U.S.A. 98, 9448–9453. 10.1073/pnas.151258398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminaka H., Nake C., Epple P., Dittgen J., Schutze K., Chaban C., et al. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25, 4400–4411. 10.1038/sj.emboj.7601312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. G., Kuhl J. C., Kachroo P., Klessig D. F. (2008). CRT1, an Arabidopsis ATPase that interacts with diverse resistance proteins and modulates disease resistance to turnip crinkle virus. Cell Host Microbe 3, 48–57. 10.1016/j.chom.2007.11.006 [DOI] [PubMed] [Google Scholar]
- Karpinski S., Szechynska-Hebda M., Wituszynska W., Burdiak P. (2013). Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 36, 736–744. 10.1111/pce.12018 [DOI] [PubMed] [Google Scholar]
- Kauss D., Bischof S., Steiner S., Apel K., Meskauskiene R. (2012). FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg(++)-branch of this pathway. FEBS Lett. 586, 211–216. 10.1016/j.febslet.2011.12.029 [DOI] [PubMed] [Google Scholar]
- Kawai-Yamada M., Jin L., Yoshinaga K., Hirata A., Uchimiya H. (2001). Mammalian Bax-induced plant cell death can be down-regulated by overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. U.S.A. 98, 12295–12300. 10.1073/pnas.211423998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keisa A., Kanberga-Silina K., Nakurte I., Kunga L., Rostoks N. (2011). Differential disease resistance response in the barley necrotic mutant nec1. BMC Plant Biol. 11:66. 10.1186/1471-2229-11-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandal D., Samol I., Buhr F., Pollmann S., Schmidt H., Clemens S., et al. (2009). Singlet oxygen-dependent translational control in the tigrina-d.12 mutant of barley. Proc. Natl. Acad. Sci. U.S.A. 106, 13112–13117. 10.1073/pnas.0903522106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Meskauskiene R., Zhang S., Lee K. P., Lakshmanan Ashok M., Blajecka K., et al. (2012). Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039. 10.1105/tpc.112.100479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Bouyer D., Schobinger U., Bechtold N., Herzog M., Bonneville J. M., et al. (2001). CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr. Biol. 11, 1891–1895. 10.1016/S0960-9822(01)00590-5 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. J., Dietrich R. A., Martin A. C., Last R. L., Dangl J. L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12, 1022–1026. 10.1094/MPMI.1999.12.11.1022 [DOI] [PubMed] [Google Scholar]
- Kong Q., Qu N., Gao M., Zhang Z., Ding X., Yang F., et al. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24, 2225–2236. 10.1105/tpc.112.097253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich I., Pecenkova T., Sekeres J., Smetana O., Fendrych M., Foissner I., et al. (2013). Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic 14, 1155–1165. 10.1111/tra.12101 [DOI] [PubMed] [Google Scholar]
- Kurusu T., Hamada H., Sugiyama Y., Yagala T., Kadota Y., Furuichi T., et al. (2011). Negative feedback regulation of microbe-associated molecular pattern-induced cytosolic Ca2+ transients by protein phosphorylation. J. Plant Res. 124, 415–424. 10.1007/s10265-010-0388-4 [DOI] [PubMed] [Google Scholar]
- La Camera S., Balague C., Gobel C., Geoffroy P., Legrand M., Feussner I., et al. (2009). The Arabidopsis patatin-like protein 2 (PLP2) plays an essential role in cell death execution and differentially affects biosynthesis of oxylipins and resistance to pathogens. Mol. Plant Microbe Interact. 22, 469–481. 10.1094/MPMI-22-4-0469 [DOI] [PubMed] [Google Scholar]
- Lacomme C., Santa Cruz S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. U.S.A. 96, 7956–7961. 10.1073/pnas.96.14.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landoni M., De Francesco A., Bellatti S., Delledonne M., Ferrarini A., Venturini L., et al. (2013). A mutation in the FZL gene of Arabidopsis causing alteration in chloroplast morphology results in a lesion mimic phenotype. J. Exp. Bot. 64, 4313–4328. 10.1093/jxb/ert237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrasse D., Jegu T., Meng P. H., Mazubert C., Hudik E., Delarue M., et al. (2013). Dual function of MIPS1 as a metabolic enzyme and transcriptional regulator. Nucleic Acids Res. 41, 2907–2917. 10.1093/nar/gks1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Kim D. S., Hwang B. K. (2012). The pepper RNA-binding protein CaRBP1 functions in hypersensitive cell death and defense signaling in the cytoplasm. Plant J. 72, 235–248. 10.1111/j.1365-313X.2012.05063.x [DOI] [PubMed] [Google Scholar]
- Lee J., Nam J., Park H. C., Na G., Miura K., Jin J. B., et al. (2007a). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49, 79–90. 10.1111/j.1365-313X.2006.02947.x [DOI] [PubMed] [Google Scholar]
- Lee K. P., Kim C., Landgraf F., Apel K. (2007b). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 10270–10275. 10.1073/pnas.0702061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Ahn I. P., Ning Y., Park C. H., Zeng L., Whitehill J. G., et al. (2012). The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159, 239–250. 10.1104/pp.111.192617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen L., Mu J., Zuo J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. 10.1104/pp.113.225805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gou M., Sun Q., Hua J. (2010). Requirement of calcium binding, myristoylation, and protein-protein interaction for the Copine BON1 function in Arabidopsis. J. Biol. Chem. 285, 29884–29891. 10.1074/jbc.M109.066100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Pennington B. O., Hua J. (2009). Multiple R-like genes are negatively regulated by BON1 and BON3 in arabidopsis. Mol. Plant Microbe Interact. 22, 840–848. 10.1094/MPMI-22-7-0840 [DOI] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J. T., Luo S., Lu H., Greenberg J. T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17, 2636–2641. 10.1101/gad.1140503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Jambunathan N., McNellis T. W. (2005). Transgenic expression of the von Willebrand A domain of the BONZAI 1/COPINE 1 protein triggers a lesion-mimic phenotype in Arabidopsis. Planta 221, 85–94. 10.1007/s00425-004-1413-4 [DOI] [PubMed] [Google Scholar]
- Liu J. Z., Horstman H. D., Braun E., Graham M. A., Zhang C., Navarre D., et al. (2011). Soybean homologs of MPK4 negatively regulate defense responses and positively regulate growth and development. Plant Physiol. 157, 1363–1378. 10.1104/pp.111.185686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li F., Tang J., Wang W., Zhang F., Wang G., et al. (2012). Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS ONE 7:e50089. 10.1371/journal.pone.0050089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Lin B., Auriac M. C., Kroj T., Saindrenan P., Nicole M., et al. (2004). Vascular associated death1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. Plant Cell 16, 2217–2232. 10.1105/tpc.104.022038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S., Vailleau F., Balagué C., Roby D. (2003). Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 8, 263–271. 10.1016/s1360-1385(03)00108-0 [DOI] [PubMed] [Google Scholar]
- Lu H., Rate D. N., Song J. T., Greenberg J. T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15, 2408–2420. 10.1105/tpc.015412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Salimian S., Gamelin E., Wang G., Fedorowski J., Lacourse W., et al. (2009). Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J. 58, 401–412. 10.1111/j.1365-313X.2009.03791.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Qin G., Zhang J., Liang Y., Song Y., Zhao M., et al. (2011). D-myo-inositol-3-phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell 23, 1352–1372. 10.1105/tpc.111.083337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach J. M., Castillo A. R., Hoogstraten R., Greenberg J. T. (2001). The Arabidopsis-accelerated cell death gene ACD2 encodes red chlorophyll catabolite reductase and suppresses the spread of disease symptoms. Proc. Natl. Acad. Sci. U.S.A. 98, 771–776. 10.1073/pnas.021465298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky F. G., Brodersen P., Fiil B. K., McKinney L. V., Thorgrimsen S., Beck M., et al. (2010). Lazarus1, a DUF300 protein, contributes to programmed cell death associated with Arabidopsis acd11 and the hypersensitive response. PLoS ONE 5:e12586. 10.1371/journal.pone.0012586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M. K., Chandra-Shekara A. C., Jeong R. D., Yu K., Zhu S., Chanda B., et al. (2012). Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 24, 1654–1674. 10.1105/tpc.112.096768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G., Niso-Santano M., Baehrecke E. H., Kroemer G. (2014). Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 15, 81–94. 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham J. E., Molino D., Gissot L., Bellec Y., Hematy K., Marion J., et al. (2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23, 2362–2378. 10.1105/tpc.110.080473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A., Muhlenbock P., Rusterucci C., Chang C. C., Miszalski Z., Karpinska B., et al. (2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136, 2818–2830. 10.1104/pp.104.043646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A., Yokoyama R., Seki M., Ito T., Shinozaki K., Takahashi T., et al. (2005). AtXTH27 plays an essential role in cell wall modification during the development of tracheary elements. Plant J. 42, 525–534. 10.1111/j.1365-313X.2005.02395.x [DOI] [PubMed] [Google Scholar]
- Matsui K. (2006). Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin. Plant Biol. 9, 274–280. 10.1016/j.pbi.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Meng P. H., Raynaud C., Tcherkez G., Blanchet S., Massoud K., Domenichini S., et al. (2009). Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS ONE 4:e7364. 10.1371/journal.pone.0007364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill A. H., Jr., Wang E., Gilchrist D. G., Riley R. T. (1993). Fumonisins and other inhibitors of de novo sphingolipid biosynthesis. Adv. Lipid Res. 26, 215–234. [PubMed] [Google Scholar]
- Meskauskiene R., Apel K. (2002). Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett. 532, 27–30. 10.1016/S0014-5793(02)03617-7 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R., Nater M., Goslings D., Kessler F., Op Den Camp R., Apel K. (2001). FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831. 10.1073/pnas.221252798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R., Wursch M., Laloi C., Vidi P. A., Coll N. S., Kessler F., et al. (2009). A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses (1)O(2)-induced cell death. Plant J. 60, 399–410. 10.1111/j.1365-313X.2009.03965.x [DOI] [PubMed] [Google Scholar]
- Miura K., Lee J., Miura T., Hasegawa P. M. (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51, 103–113. 10.1093/pcp/pcp171 [DOI] [PubMed] [Google Scholar]
- Miura K., Tada Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 5:4. 10.3389/fpls.2014.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Grimm B., Masuda T., Moulin M., Smith A. G., et al. (2010). The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci. 15, 488–498. 10.1016/j.tplants.2010.05.012 [DOI] [PubMed] [Google Scholar]
- Moeder W., Yoshioka K. (2008). Lesion mimic mutants: a classical, yet still fundamental approach to study programmed cell death. Plant Signal. Behav. 3, 764–767. 10.4161/psb.3.10.6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita-Yamamuro C., Tsutsui T., Sato M., Yoshioka H., Tamaoki M., Ogawa D., et al. (2005). The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol. 46, 902–912. 10.1093/pcp/pci095 [DOI] [PubMed] [Google Scholar]
- Muhlenbock P., Szechynska-Hebda M., Plaszczyca M., Baudo M., Mateo A., Mullineaux P. M., et al. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356. 10.1105/tpc.108.059618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamilarasan M., Prasad M. (2013). Plant innate immunity: an updated insight into defense mechanism. J. Biosci. 38, 433–449. 10.1007/s12038-013-9302-2 [DOI] [PubMed] [Google Scholar]
- Nakaune S., Yamada K., Kondo M., Kato T., Tabata S., Nishimura M., et al. (2005). A vacuolar processing enzyme, deltaVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell 17, 876–887. 10.1105/tpc.104.026872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A., Krothapalli K., Buseman C. M., Li M., Welti R., Enyedi A., et al. (2003). Arabidopsis sfd mutants affect plastidic lipid composition and suppress dwarfing, cell death, and the enhanced disease resistance phenotypes resulting from the deficiency of a fatty acid desaturase. Plant Cell 15, 2383–2398. 10.1105/tpc.015529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie H., Wu Y., Yao C., Tang D. (2011). Suppression of edr2-mediated powdery mildew resistance, cell death and ethylene-induced senescence by mutations in ALD1 in Arabidopsis. J. Genet. Genomics 38, 137–148. 10.1016/j.jgg.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Nie H., Zhao C., Wu G., Wu Y., Chen Y., Tang D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158, 1847–1859. 10.1104/pp.111.192310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutoshi Y., Ito T., Seki M., Nakashita H., Yoshida S., Marco Y., et al. (2005). A single amino acid insertion in the WRKY domain of the Arabidopsis TIR-NBS-LRR-WRKY-type disease resistance protein SLH1 (sensitive to low humidity 1) causes activation of defense responses and hypersensitive cell death. Plant J. 43, 873–888. 10.1111/j.1365-313X.2005.02500.x [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Kuromori T., Wada T., Hirayama T., Kamiya A., Imura Y., et al. (2006). Loss of Necrotic Spotted Lesions 1 associates with cell death and defense responses in Arabidopsis thaliana. Plant Mol. Biol. 62, 29–42. 10.1007/s11103-006-9001-6 [DOI] [PubMed] [Google Scholar]
- Ogasawara Y., Kaya H., Hiraoka G., Yumoto F., Kimura S., Kadota Y., et al. (2008). Synergistic activation of the Arabidopsis NADPH oxidase AtrbohD by Ca2+ and phosphorylation. J. Biol. Chem. 283, 8885–8892. 10.1074/jbc.M708106200 [DOI] [PubMed] [Google Scholar]
- Op Den Camp R. G., Przybyla D., Ochsenbein C., Laloi C., Kim C., Danon A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332. 10.1105/tpc.014662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F. G., Fiil B. K., Nielsen H. B., Brodersen P., et al. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6:e1001137. 10.1371/journal.ppat.1001137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Liu S., Tang D. (2012). HPR1, a component of the THO/TREX complex, plays an important role in disease resistance and senescence in Arabidopsis. Plant J. 69, 831–843. 10.1111/j.1365-313X.2011.04835.x [DOI] [PubMed] [Google Scholar]
- Pata M. O., Hannun Y. A., Ng C. K. (2010). Plant sphingolipids: decoding the enigma of the Sphinx. New Phytol. 185, 611–630. 10.1111/j.1469-8137.2009.03123.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Caplan J., Dinesh-Kumar S. P. (2006). Autophagy in the control of programmed cell death. Curr. Opin. Plant Biol. 9, 391–396. 10.1016/j.pbi.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Pattanayak G. K., Venkataramani S., Hortensteiner S., Kunz L., Christ B., Moulin M., et al. (2012). Accelerated cell death 2 suppresses mitochondrial oxidative bursts and modulates cell death in Arabidopsis. Plant J. 69, 589–600. 10.1111/j.1365-313X.2011.04814.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell R. I., Lamb C. (1997). Programmed cell death in plants. Plant Cell 9, 1157–1168. 10.1105/tpc.9.7.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M., Brodersen P., Naested H., Andreasson E., Lindhart U., Johansen B., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. 10.1016/S0092-8674(00)00213-0 [DOI] [PubMed] [Google Scholar]
- Petersen N. H., McKinney L. V., Pike H., Hofius D., Zakaria A., Brodersen P., et al. (2008). Human GLTP and mutant forms of ACD11 suppress cell death in the Arabidopsis acd11 mutant. FEBS J. 275, 4378–4388. 10.1111/j.1742-4658.2008.06584.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska A., Anders I., Aubry S., Schenk N., Tapernoux-Luthi E., Muller T., et al. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19, 369–387. 10.1105/tpc.106.044404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska A., Tanner G., Anders I., Roca M., Hortensteiner S. (2003). Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. U.S.A. 100, 15259–15264. 10.1073/pnas.2036571100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinska A., Tanner G., Aubry S., Anders I., Moser S., Muller T., et al. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139, 52–63. 10.1104/pp.105.065870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyla D., Gobel C., Imboden A., Hamberg M., Feussner I., Apel K. (2008). Enzymatic, but not non-enzymatic, 1O2-mediated peroxidation of polyunsaturated fatty acids forms part of the EXECUTER1-dependent stress response program in the flu mutant of Arabidopsis thaliana. Plant J. 54, 236–248 10.1111/j.1365-313X.2008.03409.x [DOI] [PubMed] [Google Scholar]
- Qiao Y., Jiang W., Lee J., Park B., Choi M. S., Piao R., et al. (2010). SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol. 185, 258–274. 10.1111/j.1469-8137.2009.03047.x [DOI] [PubMed] [Google Scholar]
- Qiu J. L., Zhou L., Yun B. W., Nielsen H. B., Fiil B. K., Petersen K., et al. (2008). Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 148, 212–222. 10.1104/pp.108.120006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V., Sarmiento-Manus R., Gonzalez-Bayon R., Hricova A., Ponce M. R., Micol J. L. (2013). PORPHOBILINOGEN DEAMINASE deficiency alters vegetative and reproductive development and causes lesions in Arabidopsis. PLoS ONE 8:e53378. 10.1371/journal.pone.0053378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G., Issakidis-Bourguet E., Hoeberichts F. A., Vandorpe M., Gakiere B., Vanacker H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. 10.1111/j.1365-313X.2007.03263.x [DOI] [PubMed] [Google Scholar]
- Rafiqi M., Bernoux M., Ellis J. G., Dodds P. N. (2009). In the trenches of plant pathogen recognition: role of NB-LRR proteins. Semin. Cell Dev. Biol. 20, 1017–1024. 10.1016/j.semcdb.2009.04.010 [DOI] [PubMed] [Google Scholar]
- Rasmussen M. W., Roux M., Petersen M., Mundy J. (2012). MAP Kinase Cascades in Arabidopsis Innate Immunity. Front. Plant Sci. 3:169. 10.3389/fpls.2012.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D. N., Cuenca J. V., Bowman G. R., Guttman D. S., Greenberg J. T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11, 1695–1708. 10.1105/tpc.11.9.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D. N., Greenberg J. T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27, 203–211. 10.1046/j.0960-7412.2001.1075umedoc.x [DOI] [PubMed] [Google Scholar]
- Reape T. J., McCabe P. F. (2010). Apoptotic-like regulation of programmed cell death in plants. Apoptosis 15, 249–256. 10.1007/s10495-009-0447-2 [DOI] [PubMed] [Google Scholar]
- Riehs-Kearnan N., Gloggnitzer J., Dekrout B., Jonak C., Riha K. (2012). Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res. 40, 5615–5624. 10.1093/nar/gks195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. 10.1093/jxb/err031 [DOI] [PubMed] [Google Scholar]
- Roberts M., Tang S., Stallmann A., Dangl J. L., Bonardi V. (2013). Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 9:e1003465. 10.1371/journal.pgen.1003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks N., Schmierer D., Mudie S., Drader T., Brueggeman R., Caldwell D. G., et al. (2006). Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Mol. Genet. Genomics 275, 159–168. 10.1007/s00438-005-0073-9 [DOI] [PubMed] [Google Scholar]
- Samol I., Buhr F., Springer A., Pollmann S., Lahroussi A., Rossig C., et al. (2011a). Implication of the oep16-1 mutation in a flu-independent, singlet oxygen-regulated cell death pathway in Arabidopsis thaliana. Plant Cell Physiol. 52, 84–95. 10.1093/pcp/pcq176 [DOI] [PubMed] [Google Scholar]
- Samol I., Rossig C., Buhr F., Springer A., Pollmann S., Lahroussi A., et al. (2011b). The outer chloroplast envelope protein OEP16-1 for plastid import of NADPH:protochlorophyllide oxidoreductase A in Arabidopsis thaliana. Plant Cell Physiol. 52, 96–111. 10.1093/pcp/pcq177 [DOI] [PubMed] [Google Scholar]
- Santino A., Taurino M., De Domenico S., Bonsegna S., Poltronieri P., Pastor V., et al. (2013). Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 32, 1085–1098. 10.1007/s00299-013-1441-2 [DOI] [PubMed] [Google Scholar]
- Satoh M., Tokaji Y., Nagano A. J., Hara-Nishimura I., Hayashi M., Nishimura M., et al. (2013). Arabidopsis mutants affecting oxylipin signaling in photo-oxidative stress responses. Plant Physiol. Biochem. 81, 90–95. 10.1016/j.plaphy.2013.11.023 [DOI] [PubMed] [Google Scholar]
- Saucedo-Garcia M., Guevara-Garcia A., Gonzalez-Solis A., Cruz-Garcia F., Vazquez-Santana S., Markham J. E., et al. (2011). MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 191, 943–957. 10.1111/j.1469-8137.2011.03727.x [DOI] [PubMed] [Google Scholar]
- Shah J., Kachroo P., Nandi A., Klessig D. F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. 10.1046/j.1365-313x.2001.00992.x [DOI] [PubMed] [Google Scholar]
- Shirano Y., Kachroo P., Shah J., Klessig D. F. (2002). A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 14, 3149–3162. 10.1105/tpc.005348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu D. K., Zhai X., Munch D., Hofius D., Markham J. E., Bielawski J., et al. (2014). Arabidopsis accelerated cell death 11, ACD11, is a ceramide-1-phosphate transfer protein and intermediary regulator of phytoceramide levels. Cell Rep. 6, 388–399. 10.1016/j.celrep.2013.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkova K., Moreau F., Pawlak P., Vriet C., Baruah A., Alexandre C., et al. (2012). Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 16360–16365. 10.1073/pnas.1202041109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. T., Lu H., Greenberg J. T. (2004). Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, aberrant growth and death2 and AGD2-LIKE DEFENSE RESPONSE PROTEIN1, encoding novel aminotransferases. Plant Cell 16, 353–366. 10.1105/tpc.019372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann M., Anderson R. G., Westphal L., Rosahl S., McDowell J. M., Trujillo M. (2013). The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal. Behav. 8:e27421. 10.4161/psb.27421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S. H., Suarez-Rodriguez M. C., Krysan P. (2007). Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity. FEBS Lett. 581, 3171–3177. 10.1016/j.febslet.2007.05.083 [DOI] [PubMed] [Google Scholar]
- Sun C., Liu L., Tang J., Lin A., Zhang F., Fang J., et al. (2011). RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J. Genet. Genomics 38, 29–37. 10.1016/j.jcg.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Sun T., Zhang Q., Gao M., Zhang Y. (2014). Regulation of SOBIR1 accumulation and activation of defense responses in bir1-1 by specific components of ER quality control. Plant J. 77, 748–756. 10.1111/tpj.12425 [DOI] [PubMed] [Google Scholar]
- Takahashi A., Kawasaki T., Henmi K., Shi I. K., Kodama O., Satoh H., et al. (1999). Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 17, 535–545. 10.1046/j.1365-313X.1999.00405.x [DOI] [PubMed] [Google Scholar]
- Tanaka R., Hirashima M., Satoh S., Tanaka A. (2003). The Arabidopsis-accelerated cell death gene ACD1 is involved in oxygenation of pheophorbide a: inhibition of the pheophorbide a oxygenase activity does not lead to the “stay-green” phenotype in Arabidopsis. Plant Cell Physiol. 44, 1266–1274. 10.1093/pcp/pcg172 [DOI] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C. A., Innes R. W. (2005). Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein. Plant J. 44, 245–257. 10.1111/j.1365-313X.2005.02523.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Ade J., Frye C. A., Innes R. W. (2006). A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 47, 75–84. 10.1111/j.1365-313X.2006.02769.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., et al. (2011). Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 192, 841–854. 10.1111/j.1469-8137.2011.03852.x [DOI] [PubMed] [Google Scholar]
- Thomas P. E., Wu X., Liu M., Gaffney B., Ji G., Li Q. Q., et al. (2012). Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24, 4376–4388. 10.1105/tpc.112.096107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintor N., Saijo Y. (2014). ER-mediated control for abundance, quality, and signaling of transmembrane immune receptors in plants. Front. Plant Sci. 5:65. 10.3389/fpls.2014.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. A., Jones J. D., Dangl J. L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37, 1130–1134. 10.1038/ng1639 [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P. V. (2011). Metacaspases. Cell Death Differ. 18, 1279–1288. 10.1038/cdd.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undan J. R., Tamiru M., Abe A., Yoshida K., Kosugi S., Takagi H., et al. (2012). Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice (Oryza sativa L.). Genes Genet. Syst. 87, 169–179. [DOI] [PubMed] [Google Scholar]
- Urquhart W., Gunawardena A. H., Moeder W., Ali R., Berkowitz G. A., Yoshioka K. (2007). The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol. Biol. 65, 747–761. 10.1007/s11103-007-9239-7 [DOI] [PubMed] [Google Scholar]
- Valluru R., Van Den Ende W. (2011). Myo-inositol and beyond–emerging networks under stress. Plant Sci. 181, 387–400. 10.1016/j.plantsci.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S., Vanderauwera S., Vuylsteke M., Rombauts S., Langebartels C., Seidlitz H. K., et al. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58. 10.1111/j.1365-313X.2004.02105.x [DOI] [PubMed] [Google Scholar]
- Van Doorn W. G. (2011). Classes of programmed cell death in plants, compared to those in animals. J. Exp. Bot. 62, 4749–4761. 10.1093/jxb/err196 [DOI] [PubMed] [Google Scholar]
- Vega-Sanchez M. E., Verhertbruggen Y., Christensen U., Chen X., Sharma V., Varanasi P., et al. (2012). Loss of Cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 159, 56–69. 10.1104/pp.112.195495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vi S. L., Trost G., Lange P., Czesnick H., Rao N., Lieber D., et al. (2013). Target specificity among canonical nuclear poly(A) polymerases in plants modulates organ growth and pathogen response. Proc. Natl. Acad. Sci. U.S.A. 110, 13994–13999. 10.1073/pnas.1303967110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Przybyla D., Op Den Camp R., Kim C., Landgraf F., Lee K. P., et al. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185. 10.1126/science.1103178 [DOI] [PubMed] [Google Scholar]
- Walley J. W., Kliebenstein D. J., Bostock R. M., Dehesh K. (2013). Fatty acids and early detection of pathogens. Curr. Opin. Plant Biol. 16, 520–526. 10.1016/j.pbi.2013.06.011 [DOI] [PubMed] [Google Scholar]
- Wang D., Weaver N. D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. 10.1126/science.1108791 [DOI] [PubMed] [Google Scholar]
- Wang H., Li J., Bostock R. M., Gilchrist D. G. (1996). Apoptosis: a functional paradigm for programmed plant cell death induced by a host-selective phytotoxin and invoked during development. Plant Cell 8, 375–391. 10.1105/tpc.8.3.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wen Y., Berkey R., Xiao S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21, 2898–2913. 10.1105/tpc.109.067587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang X., Tangchaiburana S., Ndeh R., Markham J. E., Tsegaye Y., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20, 3163–3179. 10.1105/tpc.108.060053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Meng P., Zhang X., Ren D., Yang S. (2011). BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 67, 1081–1093. 10.1111/j.1365-313X.2011.04659.x [DOI] [PubMed] [Google Scholar]
- Watanabe N., Lam E. (2006). Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 45, 884–894. 10.1111/j.1365-313X.2006.02654.x [DOI] [PubMed] [Google Scholar]
- Williams B., Dickman M. (2008). Plant programmed cell death: can't live with it; can't live without it. Mol. Plant Pathol. 9, 531–544. 10.1111/j.1364-3703.2008.00473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter M., Hollricher K., Salamini F., Schulze-Lefert P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Takechi K., Myouga F., Imura S., Sato H., Takio S., et al. (2012). Loss of the plastid envelope protein AtLrgB causes spontaneous chlorotic cell death in Arabidopsis thaliana. Plant Cell Physiol. 53, 125–134. 10.1093/pcp/pcr180 [DOI] [PubMed] [Google Scholar]
- Yamanouchi U., Yano M., Lin H., Ashikari M., Yamada K. (2002). A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7530–7535. 10.1073/pnas.112209199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Hua J. (2004). A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16, 1060–1071. 10.1105/tpc.020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Yang H., Grisafi P., Sanchatjate S., Fink G. R., Sun Q., et al. (2006). The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J. 45, 166–179. 10.1111/j.1365-313X.2005.02585.x [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B. W. (2000). An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 275, 38467–38473. 10.1074/jbc.M003566200 [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B. W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. 10.1074/jbc.M207941200 [DOI] [PubMed] [Google Scholar]
- Yang Y., Jin H., Chen Y., Lin W., Wang C., Chen Z., et al. (2012). A chloroplast envelope membrane protein containing a putative LrgB domain related to the control of bacterial death and lysis is required for chloroplast development in Arabidopsis thaliana. New Phytol. 193, 81–95. 10.1111/j.1469-8137.2011.03867.x [DOI] [PubMed] [Google Scholar]
- Yao N., Greenberg J. T. (2006). Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18, 397–411. 10.1105/tpc.105.036251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Ito M., Nishida I., Watanabe A. (2002). Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 29, 427–437. 10.1046/j.0960-7412.2001.01228.x [DOI] [PubMed] [Google Scholar]
- Yoshimoto K. (2010). Plant autophagy puts the brakes on cell death by controlling salicylic acid signaling. Autophagy 6, 192–193. 10.4161/auto.6.1.10843 [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Kachroo P., Tsui F., Sharma S. B., Shah J., Klessig D. F. (2001). Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J. 26, 447–459. 10.1046/j.1365-313X.2001.2641039.x [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H. G., Kachroo P., Masmoudi K., Berkowitz G., et al. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18, 747–763. 10.1105/tpc.105.038786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R. J., Strasser A. (2008). The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59. 10.1038/nrm2308 [DOI] [PubMed] [Google Scholar]
- Yu I. C., Parker J., Bent A. F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. U.S.A. 95, 7819–7824. 10.1073/pnas.95.13.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L. R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., et al. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. 10.1105/tpc.104.025171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhou C. (2013). Signal transduction in leaf senescence. Plant Mol. Biol. 82, 539–545. 10.1007/s11103-012-9980-4 [DOI] [PubMed] [Google Scholar]
- Zhang L., Du L., Shen C., Yang Y., Poovaiah B. W. (2014a). Regulation of plant immunity through ubiquitin-mediated modulation of Ca(2+) -calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. 10.1111/tpj.12473 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lenk A., Andersson M. X., Gjetting T., Pedersen C., Nielsen M. E., et al. (2008). A lesion-mimic syntaxin double mutant in Arabidopsis reveals novel complexity of pathogen defense signaling. Mol. Plant 1, 510–527. 10.1093/mp/ssn011 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shrestha J., Tateda C., Greenberg J. T. (2014b). Salicylic acid signaling controls the maturation and localization of the Arabidopsis defense protein ACCELERATED CELL DEATH6. Mol. Plant. 8, 1365–1383. 10.1093/mp/ssu072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wu Y., Gao M., Zhang J., Kong Q., Liu Y., et al. (2012). Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11, 253–263. 10.1016/j.chom.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Zhao C., Nie H., Shen Q., Zhang S., Lukowitz W., Tang D. (2014). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10:e1004389. 10.1371/journal.pgen.1004389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Menke F. L., Yoshioka K., Moder W., Shirano Y., Klessig D. F. (2004). High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J. 39, 920–932. 10.1111/j.1365-313X.2004.02180.x [DOI] [PubMed] [Google Scholar]
- Zhou F., Mosher S., Tian M., Sassi G., Parker J., Klessig D. F. (2008). The Arabidopsis gain-of-function mutant ssi4 requires RAR1 and SGT1b differentially for defense activation and morphological alterations. Mol. Plant Microbe Interact. 21, 40–49. 10.1094/MPMI-21-1-0040 [DOI] [PubMed] [Google Scholar]