Abstract
Vibrio parahaemolyticus is a marine and estuarine bacterium that has been the leading cause of foodborne outbreaks which leads to a significant threat to human health worldwide. Consumption of seafood contaminated with V. parahaemolyticus causes acute gastroenteritis in individuals. The bacterium poses two main virulence factor including the thermostable direct hemolysin (tdh) which is a pore-forming protein that contributes to the invasiveness of the bacterium in humans and TDH-related hemolysin (trh), which plays a similar role as tdh in the disease pathogenesis. This study aimed to investigate the antimicrobial resistance V. parahaemolyticus strains in shrimps purchased from wetmarkets and supermarkets. The toxR-based PCR assay indicated that a total of 57.8% (185/320) isolates were positive for V. parahaemolyticus. Only 10% (19/185) toxR-positive isolate exhibit the trh gene and none of the isolates were tested positive for tdh. The MAR index was measured for 14 common antimicrobial agents. The results indicated 98% of the isolates were highly susceptible to imipenem, ampicillin sulbactam (96%), chloramphenicol (95%), trimethoprim-sulfamethoxazole (93%), gentamicin (85%), levofloxacin (83%), and tetracycline (82%). The chloramphenicol (catA2) and kanamycin (aphA-3) resistance genes were detected in the resistant V. parahaemolyticus isolates. Our results demonstrate that shrimps are contaminated with V. parahaemolyticus, some of which carry the trh-gene thus being potential to cause food borne illness. The occurrence of multidrug resistance strains in the environment could be an indication of excessive usage of antibiotics in agriculture and aquaculture fields.
Keywords: Vibrio parahaemolyticus, food borne, virulence factors, antimicrobial, MAR index
INTRODUCTION
Vibrio parahaemolyticus is a Gram-negative halophilic, non-spore forming, curved rod-shaped bacterium that naturally lives in estuarine and marine environments worldwide (Su and Liu, 2007; Gode-Potratz et al., 2011; Ceccarelli et al., 2013; Zhang and Orth, 2013; Velazquez-Roman et al., 2014; Wu et al., 2014). While the majority of environmental strains are harmless members of the marine microbiota, some strains are opportunistic pathogens of humans (Johnson et al., 2008). Potential virulent V. parahaemolyticus strains are usually differentiated from likely avirulent strains by the presence of thermostable direct hemoylsin (tdh) and/or TDH related hemolysin (trh) genes (Bej et al., 1999). The tdh and trh genes are the two virulence factors associated with V. parahaemolyticus hemolysis and cytotoxicity activity in the host cell (Broberg et al., 2011; Zhang and Orth, 2013; Letchumanan et al., 2014). This halophile causes acute gastroenteritis in humans following the consumption of raw, undercooked or mishandled seafood (Zarei et al., 2012).
Although V. parahaemolyticus is often found in seafood, not all the strains are considered pathogenic (Velazquez-Roman et al., 2012). The strains isolated from environmental samples usually lack the pathogenic genes tdh and/or trh genes which cause illnesses to humans and marine animals (Deepanjali et al., 2005; Canizalez-Roman et al., 2011; Gutierrez West et al., 2013). Nevertheless, studies from US, Europe and Asia have reported around 0–6% of the environmental samples analyzed to be positive for the presences of V. parahaemolyticus strains with tdh gene and/or trh genes (DePaola et al., 2000; Vuddhakul et al., 2000; Wong et al., 2000; Alam et al., 2002; Hervio-Heath et al., 2002; Haley et al., 2014). In Malaysia, V. parahaemolyticus is naturally distributed in the marine coastal region of Malaysia. Its widespread incidence in the seawater allows the bacteria to use seafood as a vehicle of transmission and cause seafood borne gastroenteritis. V. parahaemolyticus is well known as one of the causative agent in regular institutional food poisoning cases in Malaysia (Al-Othrubi et al., 2014). Lately this bacterium has gain great attention from public due to the frequent rejection of seafood exported to EU countries (Abdul-Rahim et al., 2007; Al-Othrubi et al., 2014).
Conventional phenotyping and biochemical identification techniques of V. parahaemolyticus are often complicated when the strains are isolated from seafood and marine environments (Nishibuchi, 2006). These laborious protocols are mainly useful to estimate total load of V. parahaemolyticus in a sample as a potential risk estimation of the presence of pathogenic strains (Malcolm et al., 2015). Hence, this has raised concerns of many researchers, therefore suggesting the employ of molecular techniques to directly identify virulent markers (tdh+ and trh+), since the pathogenic strains are cause of human illness. The polymerase chain reaction (PCR) assays is one of the molecular techniques that is widely used to detect the presences of pathogenic V. parahaemolyticus strain in food and environment (Panicker et al., 2004; Yamamoto et al., 2008; Paydar et al., 2013; Malcolm et al., 2015). PCR primers can be multiplexed in a single reaction to increase the detection limit or tailored as real-time PCR analysis to provide more rapid results (Grant et al., 2006; Zhang et al., 2014).
Vibrio spp. usually are said to be highly susceptible to most clinically used antibiotics (Mala et al., 2014; Shaw et al., 2014). However, over the years, antibiotic resistance strains has emerged into the environment due the excessive use of antibiotics and other chemotherapeutic agents in human, agriculture, and aquaculture fields (Cabello et al., 2013). In the aquaculture field, antimicrobials are used not to promote growth but rather to prevent (prophylactic use) and treat (therapeutic use) bacterial infections on fish and other invertebrates (Cabello et al., 2013). Oxytetracycline, tetracycline, quinolone, sulphonamides, and trimethoprim are among the antibiotics allowed and used in the Asian aquaculture industry to ensure continuous production of seafood (Rico et al., 2012; Yano et al., 2014). Antimicrobial resistant V. parahaemolyticus strains has been isolated and detected from shrimps in Thailand (Yano et al., 2014), Malaysia (Al-Othrubi et al., 2011; Sani et al., 2013), and China (Peng et al., 2010; Xu et al., 2014). This phenomenon has raised concern due to the increase number of resistance pathogenic V. parahaemolyticus strains in the environment toward clinically used antibiotics. There are many clinically used antibiotics as a choice of treatment for Vibrio spp. infections including cephalothin (first generation cephalosporins), cefuroxime (second generation cephalosporin), cefotaxime and ceftazidime (third generation cephalosporins), tetracycline, doxycycline, or fluoroquinolone (Tang et al., 2002; Al-Othrubi et al., 2014).
The use of antimicrobials in the aquaculture has caused the development of antibiotic resistant bacteria and antibiotic resistant genes. There are numerous antibiotic resistant genes can be found in bacteria and environments. For example, β-lactam and penicillin resistant genes penA and blaTEM-1 (Srinivasan et al., 2005; Zhang et al., 2009), chloramphenicol resistant genes catI, catII, catIII, catIV, and floR (Dang et al., 2007, 2008), tetracycline resistant genes tatA, tatB, tatC, tatD, tatE, tatG, tatH, tatJ, tatY, tatZ, and many more (Macauley et al., 2007; Zhang et al., 2009; Kim et al., 2013). These antibiotic resistant genes can be transfer among different bacteria via conjugation, transduction, or transformation (Manjusha and Sarita, 2011).
Plasmid is one of the important mediators that facilitate in the transfer of antibiotic resistant genes and it can be transmitted to the next generation via vertical gene transfer or exchanged with other bacteria via horizontal gene transfer (Okamoto et al., 2009; Manjusha and Sarita, 2011). Therefore, plasmids are cured in bacteria as a way to eliminate antibiotic resistance. There are various chemical and physical agents have been developed for plasmid curing. The conventional curing agents include chemical agents such as ethidium bromide (EB), acridine, and sodium dodecyl sulfate (SDS), and physical agents such as treatment with ultraviolet and growth at elevated temperature (Liu et al., 2012). A study conducted by Molina-Aja et al. (2002) reported that acridine orange and EB successfully cured the plasmids of Vibrios, whereas SDS did not cure any of the plasmid.
Shrimps are popular seafood with a high market demand in the aquaculture industry in South East Asia (Yano et al., 2014). Usually, shrimps are cooked prior to consumption though with the growing popularity of sushi, this crustacean is often eaten raw. The increase consumption of shrimps has encouraged the expansion of shrimp farming in many countries including Thailand (Yano et al., 2014), China (Xu et al., 2014), and Malaysia. The high contaminations of pathogenic Vibrio spp. in retail seafood in Malaysia recommend that there is a need for adequate consumer protection measures. Hence, monitoring the incidence of V. parahaemolyticus in Malaysian shrimp provides important information to consumers on the food safety. In this study, we evaluated the prevalence, antimicrobial susceptibility, and characterization of antibiotic resistant genes of V. parahaemolyticus in shrimps from retails in Malaysia.
MATERIALS AND METHODS
SAMPLING
The study mainly focused on two type of Malaysian shrimp, the banana prawn (Penaeus indicus), and red prawn (Solenocera subnuda). A total of 320 shrimp samples of both type were purchased from the local wetmarket and supermarket from January 2014–June 2014. All the samples were sealed and transported in an ice box to the laboratory for analysis on the same day.
ENUMERATION AND ISOLATION OF Vibrio spp. IN SHRIMP SAMPLES
The enumeration and isolation of V. parahaemolyticus was carried out according to the method adapted from Zarei et al. (2012) with some minor modification. Twenty-five grams of samples were weighted and placed into a sterile homogenizer beg containing 225 mL of alkaline peptone water with 2% NaCl, pH 8.5, giving a first 10-1 dilution. The samples were homogenized for 60 s using a stomacher (Bagmixer 400W, Interscience, St Nom, France). Spread plate technique was employed in the enumeration of total presumptive Vibrio spp. in respective samples as described by Beneduce et al. (2010). Hundred microliter of each pre-enriched homogenates with appropriate sample dilution (1:10, 1:100, and 1:1000) were spread in duplicates onto the thiosulphate citrate bile salt sucrose (TCBS) agar (HiMedia, India) and incubated at 37∘C for 18–24 h. After incubation, the total colony count is determined and their concentrations in the original shrimp in cfu/mL were calculated.
For the isolation step, the remaining homogenate is incubated at 37∘C under aerobic conditions for 18 h. After 18 h of incubation and enrichment, a loopful of enriched mixture were streaked onto selective media, TCBS agar (HiMedia, India) and incubated at 37∘C for 18 h. The non-sucrose forming colony appeared green color on TCBS agar were picked and purified by streaking onto Tryptic Soy Agar (TSA; HiMedia, India) plates supplemented with 2% w/v sodium chloride (NaCl; Vivantis, USA). The TSA agar plates are incubated at 37∘C under aerobic conditions for 18–24 h. A loopful of pure isolate are inoculated into respective semi-solid nutrient agar and TSB with 30% glycerol, incubated at 37∘C for 18 h and then stored until further analysis.
DNA EXTRACTION
The DNA from presumptive Vibrio spp. colonies was extracted using direct boiled cell lysate method (Suzita et al., 2010; Vengadesh et al., 2012). The colonies from semisolid nutrient agar are revived in Tryptic Soy Broth (TSB; HiMedia) with 2% w/v NaCl (Vivantis, USA) and incubated in a shaker incubator at 220 rpm for 37∘C for 18 h. Then, 1.5 mL of overnight culture suspension is pipette into a sterile 1.5 mL Eppendorf tube and centrifuged at 10,000 rpm for 5 min. The supernatant were carefully discarded and 1 ml of sterile ultrapure water were added, vortexed, and boiled at 100∘C for 7 min. After 7 min, the tubes are swiftly immerged onto ice for 5 min and then centrifuged at 13,000 rpm for 1 min to separate the debris and DNA contained supernatant. The supernatant were carefully pipetted and transferred into a new 1.5 mL microcentrifuge tube. The crude DNA was used as a template for PCR analysis.
IDENTIFICATION OF Vibrio parahaemolyticus USING toxR-BASED PCR ASSAY
The toxR-based PCR assay was preformed to identify V. parahaemolyticus from all the presumptive isolates. Detection of toxR gene was carried out using primer toxR-F (5′-ATA CGA GTG GTT GCT GTC ATG-3′) and toxR-R (5′-GTC TTC TGA CGC AAT CGT TG-3′) with the expected amplicon size of 368 bp (Kim et al., 1999). The reaction mixture for this PCR assay was performed in a final volume of 20 μL, containing 2 μL of DNA template, 10 μL of 2x Taq PLUS PCR Smart mix 1 (SolGentTM, Korea), 6 μL of sterile distilled water and 1 μL of each primer. toxR-based PCR amplification were performed using PCR thermocycler (Kyratec, SuperCycler Thermal Cycler, Australia) with the following cycling conditions: initial denaturation at 95∘C for 4 min, 35 cycles of 94∘C for 1 min, 68∘C for 1 min and 72∘C for 30 s, and a final elongation at 72∘C for 5 min. PCR products were visualized by 1.5% agarose gel.
DETECTION OF VIRULENCE GENE
The detection of V. parahaemolyticus virulence genes, tdh and trh was performed in a duplex PCR using specific primer adapted from Bej et al. (1999). PCRs were performed in a final volume of 20 μL, containing 2 μL of DNA template, 10 μL of 2x Taq PLUS PCR Smart mix 1 (SolGentTM, Korea), 4 μL of sterile distilled water and 1 μL of each primer. The PCR amplifications were performed using a Thermocycler (Kyratec, SuperCycler Thermal Cycler, Australia) with the following cycling conditions: initial denaturation at 94∘C for 3 min, 30 cycles of 94∘C for 1 min, 58∘C for 1 min and 72∘C for 1 min, and a final elongation at 72∘C for 5 min. All PCR products were visualized by 1.5% agarose gel.
ANTIBIOTIC SUSCEPTIBILITY TEST
Fourteen antibiotic disks (Oxoid, UK) infused with amplicin (10 μg), ampicillin/sulbactam (30 μg), amikacin (30 μg), cefotaxime (30 μg), ceftazidime (30 μg), chloramphenicol (30 μg), gentamicin (30 μg), imipenem (10 μg), kanamycin (30 μg), levofloxacin (5 μg), nalidixic acid (30 μg), oxytetracycline (30 μg), sulfamethoxazole/trimethoprim (25 μg), and tetracycline (30 μg) were used in this study. The antibiotic susceptibility of Vibrio spp. isolates were studied using the disk diffusion method (Yano et al., 2014). The antibiotic disks were dispensed on Mueller Hilton agar (HiMedia, India) supplemented with 2% w/v NaCl (Vivantis, USA) plates with bacterial lawn. After incubation at 37∘C for 18 h, the inhibition zone is measured and interpreted guidelines of the Clinical and Laboratory Standard Institute (CLSI) (2010) M45-A2.
PCR AMPLIFICATION OF ANTIBIOTIC RESISTANCE GENES AND PLASMID CURING
Isolates that showed an antimicrobial resistance phenotype were screened for the presence of genes coding for resistance determinants. The genes associated with resistance to B-lactams (blaTEM, blaSHV, blaOXA), tetracyclines (tetA, tetB, tetC, tetG), chloramphenicols (catA1, catA2, catA3, catB3), and kanamycin (aphA-3) were analyzed by PCR (Kim et al., 2013). PCR primers for each antibiotic gene were performed in a final volume of 20 μL, containing 2 μL of DNA template, 10 μL of 2x Taq PLUS PCR Smart mix 1 (SolGentTM, Korea), 6 μL of sterile distilled water and 1 μL of each primer. All primers are listed in Table 1. The PCR amplifications were performed in a Thermocycler (Kyratec, SuperCycler Thermal Cycler, Australia) with the following conditions: initial denaturation at 95∘C for 3 min, 35 cycles of 94∘C for 30 s, 52∘C for 1 min and 72∘C for 1.5 min, and a final elongation at 72∘C for 6 min. PCR products were visualized by 1.5% agarose gel electrophoresis. The isolates that exhibit presence of antibiotic resistant genes were subjected to plasmid curing.
Table 1.
Polymerase chain reaction (PCR) primers targeting antibiotic resistant gene.
| Antibiotics | Targetgene | Sequence 5′-3′ | Amplicon size (bp) |
|---|---|---|---|
| Ampicillin | blaSHV | FW- TTATCTCCCTGTTAGCCACC RV- GATTTGCTGATTTCGCTCGG |
796 |
| blaOXA | FW- ACCAGATTCAACTTTCAA RV- TCTTGGCTTTTATGCTTG |
589 | |
| blaTEM | FW- ATAAAATTCTTGAAGAC RV- TTACCAATGCTTAATCA |
1073 | |
| Chloram phenicol | catA1 | FW- CGCCTGATGAATGCTCATCCG RV- CCTGCCACTCATCGCAGTAC |
456 |
| catA2 | FW- ATGAATTTTACCAGAATTGATCTGAA RV- ATTTCAGTATGTTATCACACATCATCT |
639 | |
| catA3 | FW- AAATTGGGTTCGCCGTGA RV- ATTTACTGTTACACAACTCTTGTAGCC |
1863 | |
| catB3 | FW- TCAAAGGCAAGCTGCTTTCTGAGC RV- TATTAGACGAGCACAGCATGGGCA |
566 | |
| Kanamycin | aphA-3 | FW- GGGACCACCTATGATGTGGAACG RV- CAGGCTTGATCCCCAGTAAGTC |
600 |
| Tetracycline | tetA | FW- GTAATTCTGAGCACTGTCGC FV- CTGCCTGGACAACATTGCTT |
956 |
| tetB | FW- ACGTTACTCGATGCCAT RV- AGCACTTGTCTCCTGTT |
1169 | |
| tetC | FW- AACAATGCGCTCATCGT RV- GGAGGCAGACAAGGTAT |
1138 | |
| tetG | FW- CCGGTCTTATGGGTGCTCTA RV- CCAGAAGAACGAAGCCAGTC |
603 |
All antibiotic resistant isolates were subjected to a curing treatment using EB (Bio Basic, Canada). The isolates were grown in fresh TSB (HiMedia, India) and TSB supplemented with 0.2 mg/mL EB (Bio Basic, Canada), then incubated at 37∘C for 24 hours under constant agitation. After treatment with the curing agent, the profiles of resistance phenotypes and the related genes were examined for the antibiotic susceptibility profile and presence of resistance gene using PCR.
STATISTICAL ANALYSIS
The experimental data was analyzed by using SPSS software version 20. Statistical analysis was performed in order to determine whether there were any significant difference in the species of crustaceans and the MAR index of resistant isolates using the independent t-test. The significance level was set at p-value of <0.05.
RESULTS
PREVALENCE OF Vibrio parahaemolyticus IN SHRIMP
Vibrio parahaemolyticus is a foodborne pathogen with a worldwide distribution but its densities in the environment and seafood vary depending on the season, location, sample type, and analytical methodology employed (Martinez-Urtaza et al., 2008; Zarei et al., 2012). In the present study, banana prawn (P. indicus; n = 160) and red prawn (S. subnuda; n = 160) samples were collected from three wetmarket and three supermarket. V. parahaemolyticus was found in all the shrimp samples using the conventional plating method. A total of 320 presumptive V. parahaemolyticus colonies that appeared green or bluish green on TCBS were picked. The results from the PCR showed positive amplification of toxR gene in 57.8% (185/320) of the V. parahaemolyticus isolates. Out of the 185 positive V. parahaemolyticus isolates, 52% (97/185) of the isolates are from red prawn (S. subnuda), and 48% (88/185) of the isolates are from banana prawn (P. indicus).
Table 2 summarizes the total Vibrio densities (log cfu/mL) in shrimp. The banana prawn and red prawn collected from all the wetmarket and supermarket had a mean total Vibrio count range of 4.36 log cfu/mL to 6.34 log cfu/mL. Among the shrimp samples collected from wetmarket, the red prawn sample from wetmarket A had the highest mean total Vibrio counts of 6.34 log cfu/mL. The shrimps from wetmarket B had mean total Vibrio counts of 5.04 log cfu/mL, which is the lowest compared to the samples from wetmarket A and wetmarket C. Based on the results, the supermarket samples was least contaminated compared to wetmarket samples with mean total Vibrio counts of 4.35 log cfu/mL to 4.43 log cfu/mL.
Table 2.
Total Vibrio densities (log cfu/mL) in shrimp.
| Type of sample | Total Vibrio densities (log cfu/mL) |
|||||
|---|---|---|---|---|---|---|
| Wetmarket A | Wetmarket B | Wetmarket C | Supermarket A | Supermarket B | Supermarket C | |
| Banana Prawn | 6.24 | 5.04 | 5.16 | 4.36 | 4.40 | 4.21 |
| Red Prawn | 6.34 | 5.04 | 5.19 | 4.43 | 4.35 | 4.36 |
DETECTION OF tdh and trh
To detect pathogenic isolates, tdh and trh genes were amplified using a duplex PCR. No tdh-positive V. parahaemolyticus isolates were detected among the 185 toxR-positive isolates. However, 19 (10%) trh-positive isolates were identified among the V. parahaemolyticus isolates. Table 3 shows the trh-positive strains from the both shrimp species.
Table 3.
List of trh-positive Vibrio parahaemolyticus isolates.
| Strains | Shrimp species | Location | toxR-positive | trh-positive |
|---|---|---|---|---|
| VP89 | Red Prawn | Supermarket C | + | + |
| VP90 | Red prawn | Supermarket C | + | + |
| VP91 | Red prawn | Supermarket C | + | + |
| VP92 | Red prawn | Supermarket C | + | + |
| VP93 | Banana prawn | Wetmarket A | + | + |
| VP94 | Banana prawn | Wetmarket A | + | + |
| VP95 | Banana prawn | Wetmarket A | + | + |
| VP96 | Banana prawn | Wetmarket A | + | + |
| VP97 | Banana prawn | Wetmarket A | + | + |
| VP98 | Banana prawn | Wetmarket A | + | + |
| VP99 | Banana prawn | Wetmarket A | + | + |
| VP100 | Banana prawn | Wetmarket A | + | + |
| VP101 | Banana prawn | Wetmarket A | + | + |
| VP102 | Banana prawn | Wetmarket A | + | + |
| VP103 | Banana prawn | Wetmarket A | + | + |
| VP175 | Banana prawn | Supermarket C | + | + |
| VP176 | Banana prawn | Supermarket C | + | + |
| VP177 | Banana prawn | Supermarket C | + | + |
| VP178 | Banana prawn | Supermarket C | + | + |
ANTIMICROBIAL SUSCEPTIBILITIES OF Vibrio parahaemolyticus ISOLATES
All 14 antibiotics used in this study are among the antibiotics recommended by Centre for Disease Control and Prevention (CDC) for the treatment of Vibrio spp. infections that includes fluoroquinolones (levofloxacin), cephalosporins (cefotaxime and ceftazidime), aminoglycosides (amikacin and gentamicin), and folate pathway inhibitors (trimethoprim-sulfamethoxazole; Daniels et al., 2000; Shaw et al., 2014). Table 4 summarizes the percentage of antibiotic resistant of V. parahaemolyticus isolated from shrimp. The results indicated 82% of the isolates were resistant to ampicillin. Besides ampicillin, this study found that isolates exhibited resistant profile to aminoglycosides antimicrobial agents. A total of 95 isolates (51%) were resistant to amikacin, 28% resistant to kanamycin and 11% were gentamicin-resistant isolates. A high percentage of antibiotic resistant profile was also detected among the V. parahaemolyticus isolates toward the third generation cephalosporins (cefotaxime 37% and ceftazidime 15%). In the present study, high susceptibility to antibiotics including imipenem (98%), ampicillin sulbactam (96%), chloramphenicol (95%), trimethoprim-sulfamethoxazole (93%), gentamicin (85%), levofloxacin (83%), and tetracycline (82%) were observed among the V. parahaemolyticus isolates.
Table 4.
The percentage of antibiotic resistant of V. parahaemolyticus isolated from shrimp.
| Antimicrobial agent | No of Isolates (* %) |
|||||
|---|---|---|---|---|---|---|
| Red prawn |
Banana prawn |
Total isolates for each antimicrobial (** %) | ||||
| Wetmarket (n = 65) | Supermarket (n = 32) | Wetmarket (n = 37) | Supermarket (n = 51) | |||
| Ampicillin | 45 (69) | 32 (100) | 35 (95) | 39 (76) | 151 (82) | |
| Ampicillin/sulbactam | 0 (0) | 0 (0) | 0 (0) | 3 (6) | 3 (2) | |
| Amikacin | 20 (31) | 17 (53) | 22 (6) | 36 (71) | 95 (51) | |
| Cefotaxime | 13 (20) | 12 (38) | 9 (24) | 35 (68) | 69 (37) | |
| Ceftazidime | 0 (0) | 1 (3) | 2 (5) | 25 (49) | 28 (15) | |
| Chloramphenicol | 0 (0) | 5 (16) | 1 (3) | 2 (4) | 8 (4) | |
| Gentamicin | 0 (0) | 1 (3) | 0 (0) | 20 (39) | 21 (11) | |
| Imipenem | 0 (0) | 1 (3) | 1 (3) | 2 (4) | 4 (2) | |
| Kanamycin | 3 (5) | 5 (16) | 12 (32) | 32 (63) | 52 (28) | |
| Levofloxacin | 0 (0) | 0 (0) | 0 (0) | 17 (33) | 17 (9) | |
| Nalidixic acid | 0 (0) | 0 (0) | 0 (0) | 35 (68) | 35 (19) | |
| Oxytetracycline | 3 (5) | 7 (22) | 3 (8) | 22 (43) | 35 (19) | |
| Sulfamethoxazole /trimethoprim | 0 (0) | 0 (0) | 1 (3) | 6 (12) | 7 (4) | |
| Tetracycline | 3 (5) | 3 (9) | 5 (14) | 20 (39) | 31 (17) | |
( * % ) – No of isolates/Total isolates per source; (** %) – No of isolates/Total isolates.
In the current study, a high percentage (83%) of isolates have MAR index more than 0.2. The range of MAR index was from 0.00 to 0.79, with the highest MAR index attributed from an isolate (VP152; Table 5) from supermarket banana prawn which exhibited resistant to 11 antibiotics. Gwendelynne et al. (2005) stated that MAR indices higher that 0.2 could be due to contamination from high risk sources, thus leading to human health risk. A total of 49/185 isolates exhibited MAR index of 0.07, indicating the isolates were resistant to at least one type of antibiotic. About 28% of the isolates were resistant to three different antibiotics and have a MAR index of 0.21. The study also noted shrimp samples from wetmarket and supermarket had difference MAR indices.
Table 5.
Antibiograms and multiple antimicrobial resistance (MAR) indices of V. parahaemolyticus strains.
| Antibiograms | Strains | Total antibiotic resistance | MAR index |
|---|---|---|---|
| AK/AMP/CAZ/CN/CTX/K/NA/OT/SAM/SXT/TE | VP152 | 11 | 0.79 |
| AK/AMP/CAZ/CN/CTX/K/LEV/ NA/OT/TE | VP134, VP135, VP139 | 10 | 0.71 |
| AK/AMP/CAZ/CN/CTX/K/NA/OT/SXT/TE | VP158 | 10 | 0.71 |
| AK/CAZ/CN/CTX/K/LEV/NA/OT/TE | VP136 | 9 | 0.64 |
| AK/C/CAZ/CN/CTX/K/NA/OT/TE | VP160 | 9 | 0.64 |
| AK/AMP/CAZ/CN/CTX/K/NA/ SAM | VP142 | 8 | 0.57 |
| AK/AMP/CAZ/CTX/CN/K/OT/TE | VP153 | 8 | 0.57 |
| AK/AMP/CAZ/CN/CTX/K/LEV/ NA | VP165 | 8 | 0.57 |
| AK/CAZ/CN/CTX/K/LEV/NA/SXT | VP174 | 8 | 0.57 |
| AK/CAZ/CN/CTX/K/NA/OT/TE | VP162 | 8 | 0.57 |
| AK/AMP/C/CAZ/CN/K/NA/ SXT | VP158 | 8 | 0.57 |
| AK/AMP/K/LEV/NA/OT/TE | VP130 | 7 | 0.5 |
| AK/AMP/CAZ/CN/CTX/K/NA | VP163 | 7 | 0.5 |
| AK/AMP/CTX/LEV/NA/OT/TE | VP137, VP167 | 7 | 0.5 |
| AK/AMP/CAZ/CN/K/NA/OT | VP138 | 7 | 0.5 |
| AK/AMP/CTX/K/IMP/OT | VP71 | 6 | 0.43 |
| AK/CAZ/CN/CTX/K/NA | VP132, VP133, VP140, VP141, VP143 | 6 | 0.43 |
| AMP/IPM/LEV/NA/OT/TE | VP145,VP146 | 6 | 0.43 |
| AK/AMP/CAZ/CTX/K/NA | VP151 | 6 | 0.43 |
| AK/AMP/CAZ/CTX/K/SAM | VP157 | 6 | 0.43 |
| AK/CAZ/CTX/K/NA/SXT | VP161 | 6 | 0.43 |
| AK/AMP/CTX/K/LEV/NA | VP166, VP169 | 6 | 0.43 |
| AMP/CTX/NA/OT/SXT/TE | VP170 | 6 | 0.43 |
| AK/AMP/CAZ/CTX/K | VP102, VP103, VP148 | 5 | 0.36 |
| AMP/LEV/NA/OT/TE | VP131 | 5 | 0.36 |
| CAZ/CTX/K/NA/OT | VP168 | 5 | 0.36 |
| AMP/CTX/NA/OT/TE | VP173 | 5 | 0.36 |
| AK/AMP/C/OT/TE | VP183 | 5 | 0.36 |
| AMP/CTX/OT/TE | VP159 | 4 | 0.29 |
| AK/AMP/CAZ/CTX | VP90 | 4 | 0.29 |
| AK/AMP/K/SXT | VP119 | 4 | 0.29 |
| AK/AMP/OT/TE | VP125 | 4 | 0.29 |
| AMP/NA/OT/TE | VP129 | 4 | 0.29 |
| LEV/NA/OT/TE | VP144 | 4 | 0.29 |
| AK/AMP/LEV/NA | VP171 | 4 | 0.29 |
| AK/AMP/CTX/K | VP73, VP77, VP82, VP95, VP104, VP117, VP149, VP154, VP156 | 4 | 0.29 |
| AK/AMP/K/CN | VP84 | 4 | 0.29 |
| AK/AMP/CTX | VP21, VP30, VP52, VP54, VP64, VP70, VP78, VP87, VP88, VP89, VP91, VP111, VP126 | 3 | 0.21 |
| AK/AMP/K | VP55, VP93, VP101, VP110, VP118, VP123, VP128, VP150 | 3 | 0.21 |
| AMP/OT/TE | VP59, VP72, VP74, VP108, VP120 | 3 | 0.21 |
| AMP/CTX/K | VP43 | 3 | 0.21 |
| AK/AMP/IMP | VP114 | 3 | 0.21 |
| AMP/C/CTX | VP182 | 3 | 0.21 |
| AMP/C/OT | VP179, VP180, VP181 | 3 | 0.21 |
| AMP/LEV/NA | VP172 | 3 | 0.21 |
| C/OT/TE | VP184 | 3 | 0.21 |
| AMP/TE | VP2, VP105 | 2 | 0.14 |
| OT/TE | VP5 | 2 | 0.14 |
| AMP/K | VP12 | 2 | 0.14 |
| AMP/CTX | VP28, VP29, VP46, VP50, VP99, VP127, VP147, VP175, | 2 | 0.14 |
| AK/AMP | VP31, VP34, VP41, VP45, VP46, VP57, VP60, VP61, VP63, VP66, VP79, VP83, VP85, VP107, VP109, VP112, VP113, VP121, VP124, VP178 | 2 | 0.14 |
| AK/CTX | VP42, VP44 | 2 | 0.14 |
| AMP | VP6, VP7, VP8, VP9, VP10, VP11, VP16, VP18, VP19, VP20, VP22, VP25, VP27, VP32, VP33, VP35, VP37, VP38, VP48, VP51, VP53, VP58, VP62, VP67, VP68, VP69, VP75, VP76, VP80, VP81, VP86, VP92, VP94, VP96, VP97, VP100, VP106, VP115, VP116, VP122, VP155, VP164, VP176, VP177 | 1 | 0.07 |
| AK | VP47, VP49 | 1 | 0.07 |
| CTX | VP23, VP26, VP40 | 1 | 0.07 |
AMP, Amplicin; OT, Oxytetracycline; NA, Nalidixic acid; C, Chloramphenicol; CTX, Cefotaxime; SXT, Suphamethox/Trithoprim; IMP, Imipenem; AK, Amikacin; SAM, Ampicillin/Sulbactam; LEV, Levofloxacin; CAZ, Ceftazidime; K, Kanamycin; CN, Gentamicin; TE, Tetracycline.
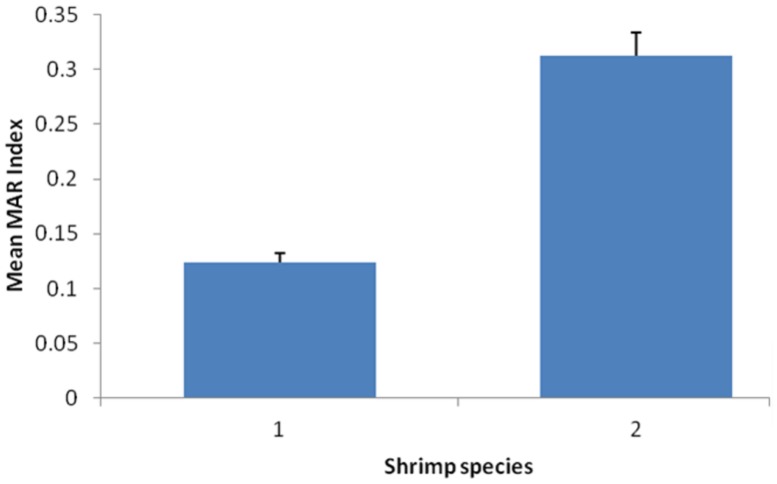
Furthermore, the mean of MAR index for V. parahaemolyticus isolates of different shrimp species were compared and the results showed there was significantly difference with p < 0.05 (Figure 1). The mean MAR index of the V. parahaemolyticus isolates recovered from banana prawn species were significantly higher than mean MAR index of V. parahaemolyticus isolates from the red prawn species. Results in this study highlighted that the banana prawn isolates are possible exposed to various antimicrobials that lead to emerging of multiple antibiotic resistant strains.
FIGURE 1.
Comparison of the mean of MAR index for Vibrio parahaemolyticus isolates of different shrimp species. Each bar represents the mean of the MAR index for V. parahaemolyticus strains from different shrimp species. Label 1, red prawn; Label 2, banana prawn.
DETECTION OF ANTIMICROBIAL RESISTANCE GENE AND PLASMID CURING
Plasmid curing was performed to detect the location of antibiotic resistance genes in the resistant V. parahaemolyticus isolates. All the chloramphenicol-resistant V. parahaemolyticus isolates (n = 8) were screened for chloramphenicol catA1, catA2, catA3, and catB3 genes. Only catA2 gene was detected and present in all the eight chloramphenicol-resistant V. parahaemolyticus isolates. Two V. parahaemolyticus isolates (VP160 and VP185) from banana prawn Supermarket B lost their resistance toward chloramphenicol as a result of plasmid curing. The PCR result showed catA2 gene was not present after plasmid curing suggesting the gene is located in the plasmid of the isolate. The plasmid curing result for another six chloramphenicol-resistant V. parahaemolyticus isolates (VP179, VP180, VP181, VP182, VP183, VP184) showed possibility that is chromosomal-mediated since the isolates exhibit positive amplification with catA2 gene and demonstrated phenotypic resistance toward chloramphenicol on the disk diffusion test after plasmid curing.
The kanamycin aphA-3 gene was detected in 15 out of 52 kanamycin-resistant V. parahaemolyticus isolates (VP84, VP133, VP134, VP136, VP138, VP140, VP142, VP143, VP151, VP153, VP160, VP161, VP162, VP163, VP174). This study did not detect any β-lactam resistance genes (blaSHV, blaOXA, and blaTEM) which are normally found as plasmid-encoded β-lactamase and tetracycline resistant genes (tetA, tetB, tetC, and tetG) among the V. parahaemolyticus isolates.
DISCUSSION
The presence of V. parahaemolyticus in shrimp samples suggests a public health concern to those who consume shrimp raw or undercooked shrimps. The risk is substantial regardless of pathogenicity island presence in the genomes of circulating V. parahaemolyticus, since some illnesses are caused by isolates lacking of tdh, trh, or T3SS2 (García et al., 2009; Harth et al., 2009; Chao et al., 2010; Haley et al., 2014). A total of 185 isolates recovered in this study were toxR-positive with a mean total Vibrio count range of 4.36 log cfu/mL to 6.34 log cfu/mL. Results of this study demonstrated a high level of V. parahaemolyticus contamination in shrimp samples from wetmarket compared to supermarket samples.
Since there is limited access to the main source origin of the samples, we scrutinized the environment at the site whereby samples are collected. The occurrence of Vibrio spp. in both shrimp species samples are probably a reflection of the atmosphere at the wetmarket and supermarket. The contamination occurs due to way of handling shrimps by wetmarket retailers is less hygienic compared to supermarkets retailers. The wetmarket retailers display the shrimps on ice rack left on the normal room temperature which allows the ice to melt faster. Where else, at the supermarket, retailers display their shrimps on ice in air conditional room temperature. The difference of temperature in the environment contributes to the high counts of V. parahaemolyticus in wetmarket as seen in our study. Previous study has indicated that seafood such as fish should be kept in cold condition during transit and storage to reduce the risk and level of Vibrios (Elhadi et al., 2004). Sudha et al. (2014) have also reported higher contamination of shellfishes with pathogenic Vibrios isolated from samples collected from the roadside stalls compared to markets in Cochin, India.
The study results are in line with previous studies that stated the occurrence of V. parahaemolyticus in supermarket samples is possibly due to lack of hygiene, improper handling, cross contamination, or difference in storage temperature during the capture to the supermarket (Yang et al., 2008; Tunung et al., 2010; Sudha et al., 2014). V. parahaemolyticus cells multiple rapidly with increase in ambient temperature, hence non-refrigerated post harvest storage could possible lead consumers to the exposure of potentially pathogenic V. parahaemolyticus strains (Sudha et al., 2014). This pathogen cell viability could be reduced if the samples are maintained on ice (Su and Liu, 2007). Therefore, in effort to reduce the risk of V. parahaemolyticus in seafood, retailers from wetmarket, and supermarket should be well educated on seafood handling techniques, storage temperature conditions, and proper hygiene.
Seafood is known as a vehicle of transmission of food borne bacteria. The virulence of these bacteria plays an important role in causing human illness. In order to assess the real risk factor of V. parahaemolyticus in the food sample, their identification should be followed with the detection of the virulence genes. Only 10% (19/185) of V. parahaemolyticus isolates in the study was trh-positive and none was tdh-positive. These results are not surprising since it is reported that environmental V. parahemolyticus strains are considered to be non pathogenic due to lacking tdh or trh, but a small percentage of these environmental strains harbor either one or both of the virulence factors (Wong et al., 2000; Alam et al., 2002; Hervio-Heath et al., 2002; Velazquez-Roman et al., 2012; Haley et al., 2014).
The trh gene is considered as a virulence factor of V. parahaemolyticus and plays a similar role as tdh gene in the pathogenesis of V. parahaemolyticus (Nelapati et al., 2012). The study findings are in agreement with another study that reported the detection of 12% (6/50) isolates to harbor trh gene in the food samples in Malaysia (Paydar et al., 2013). A recent study reported high occurrence of tdh+ and trh+ isolates in shrimp and cockles in Malaysia. In that study, 26 isolates were positive for trh virulence gene and only eight isolates positive for tdh virulence gene isolated from cockles and shrimp (Al-Othrubi et al., 2014). The variation in the occurrence of pathogenic V. parahaemolyticus between the current study and previous study may be associated with differences in sampling techniques, sample sources, and the detection techniques employed. Wilson and Salyers (2003) stated that environmental factor including interaction with other hosts plays a huge effect in the evolution of certain pathogens.
The study results demonstrated 82% of the isolates were resistance to ampicillin. It is in agreement with other studies that reported V. parahaemolyticus isolated from seafood samples are commonly resistance to ampicillin (Okuda et al., 1997; Han et al., 2007; Al-Othrubi et al., 2014). The ampicillin-resistant pattern could be due to the fact that first generation antibiotics including ampicillin is misused in the environment thus reducing the susceptibility and low efficiency of ampicillin in treatment of Vibrio infection (Sudha et al., 2014). Furthermore, high percentage of antibiotic resistance profile was also detected among the V. parahaemolyticus isolates toward the third generation cephalosporins (cefotaxime 37% and ceftazidime 15%). The results are in agreement with Sahilah et al. (2014) that the presence of cefuroxime (second generation cephalosporin) and ceftazidime-resistant V. parahaemolyticus isolates were evidenced in shellfish samples from Terengganu, Malaysia. A study in Korea also demonstrated high percentage (70–80%) of the V. parahaemolyticus isolates from seafood are resistant to both cefotaxime and ceftazidime (Jun et al., 2012). In contrast, a study in U.S. reported low percentage (3%) of V. parahaemolyticus isolates are resistant to cefotaxime (Shaw et al., 2014). The discrepancies in the literature regarding the susceptibility of V. parahaemolyticus to cefotaxime could be related to test methodology or geographical variation of samples.
In the present study, high susceptibility to antibiotics including imipenem (98%), ampicillin sulbactam (96%), chloramphenicol (95%), trimethoprim-sulfamethoxazole (93%), gentamicin (85%), levofloxacin (83%), and tetracycline (82%) were observed among the V. parahaemolyticus isolates. These findings were in agreement with literature stating most V. parahaemolyticus isolates were susceptible to chloramphenicol and tetracycline (Han et al., 2007; Sahilah et al., 2014; Sudha et al., 2014). V. parahaemolyticus strains isolated from fish were also reported to be highly susceptible to imipenem (Noorlis et al., 2011). Based on these findings, besides the widely use tetracycline, imipenem could be prescribed by doctors as a treatment for bacterial infection.
The shrimp collected from wetmarket and supermarket had different MAR indices with a range from 0.00 to 0.79. The highest MAR index attributed from an isolate (VP152) from supermarket banana prawn which exhibited resistance to 11 antibiotics. Gwendelynne et al. (2005) stated that MAR indices higher that 0.2 could be due to contamination from high risk sources, thus leading to human health risk. The findings were in agreement with researchers in Malaysia which reported that the antimicrobial susceptibility of V. parahaemolyticus varied and influenced the resistance level depending on the source of sample obtained (Khan et al., 2007; Tunung et al., 2012) and the differences in the geographical location (Lesley et al., 2011). Moreover, this was further supported by the findings in this study showing that the mean MAR index of V. parahaemolyticus isolates from the two shrimps species varied significantly with p < 0.05. A higher mean MAR index was observed in the V. parahaemolyticus isolates from banana prawn collected from both wetmarket and supermarket. The banana prawn is more exposed to various antimicrobial in the environment compared to the red prawn. The high occurrence of multiple antibiotic resistance strains in this study could be due to intense usage of antibiotics to fight against bacterial infections in the aquaculture sector and maintaining a continuous production and supply of shrimp. A study in Thailand have highlighted that shrimp farming in the inland environments might increase the opportunity for dissemination of resistance genes among bacteria (Yano et al., 2014). Literature have also stated resistant V. parahaemolyticus strains could be isolated from samples collected from location with frequent usage of antibiotics. This is due to mutation that has modified the target site or transport mechanism which causes the antibiotics to become inactive on cell (Zulkifli et al., 2009). Such widespread usage of antimicrobial has increased antibiotic resistance among environmental bacteria including potential Vibrio spp. (Tendencia and de la Peña, 2001; Yano et al., 2014).
Chloramphenicol (catA2) gene was detected in eight chloramphenicol-resistance V. parahaemolyticus isolates. Two of the isolates had the gene present in their plasmid where else another six isolates showed possibility of chromosomal-mediated since the isolates exhibit positive amplification with catA2 gene and demonstrated phenotypic resistance to chloramphenicol on the disk diffusion test after plasmid curing. These results are in agreement with previous study stating the presence of V. parahaemolyticus isolates that demonstrated chromosomal-mediated resistance against chloramphenicol (Devi et al., 2009). Researchers have also reported the presence of antibiotic resistance gene located in the chromosome of Vibrio spp. (Son et al., 1998; Manjusha and Sarita, 2011). The result of plasmid curing revealed that kanamycin-resistant V. parahaemolyticus isolates were potentially chromosomal-mediated since the isolates exhibit positive amplification with aphA-3 gene and demonstrated phenotypic resistance or intermediate to kanamycin on the disk diffusion test after plasmid curing.
This study did not detect any β-lactam resistance genes (blaSHV, blaOXA, and blaTEM) which are normally found as plasmid-encoded β-lactamase and tetracycline resistance (tetA, tetB, tetC, and tetG) gene among the V. parahaemolyticus isolates. It is well known that the ampicillin-resistance genes are very diverse. Therefore, the negative results of all the tested β-lactamase genes and tetracycline genes could possibly due to possession of other encoding genes in all the ampicillin-resistant or tetracycline resistance V. parahaemolyticus isolates of the present study. For instance, a class A extended-spectrum- β-lactamase gene, blaPER-1, which is mostly associated with Gram-negative clinical pathogens such as Pseudomonas aeruginosa (Qing et al., 2014) was also detected in V. parahaemolyticus (Wong et al., 2012; Liu et al., 2013). Another literature has also stated that ampicillin resistance of V. parahaemolyticus was not conferred by the bla gene but was mediated by an efflux system (Pazhani et al., 2014). A study also has reported the occurrence of tetracycline genes tetM and tetS in Vibrio spp. from seawater in Japan and Korea which could be an important reservoir of tetracycline resistance genes in the marine environment (Kim et al., 2004).
To the best of our knowledge, our findings represents the first comprehensive report about the prevalence, antibiotic resistance profile, antibiotic resistance genes detection, and plasmid curing of V. parahaemolyticus isolates from shrimps in Malaysia. Most gastroenteritis cases are attributed by consumption of seafood including shrimps. Shrimps may act as a vehicle to disseminate potential pathogens to the consumers. The occurrence of pathogenic V. parahaemolyticus in banana prawn (P. indicus) and red prawn (S. subnuda) in this study requires extended surveillance in the region and across the country. Hence, continuous monitoring of V. parahaemolyticus strains and their susceptibility to antibiotics is necessary to ensure the best treatment for patients with gastroenteritis and ensure seafood safety.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a University of Malaya for High Impact Research Grant (UM-MOHE HIR Nature Microbiome Grant No. H-50001-A000027) awarded to Kok-Gan Chan and External Industry Grants from Biotek Abadi Sdn Bhd (vote no. GBA-808138 and GBA-808813) awarded to Learn-Han Lee.
REFERENCES
- Abdul-Rahim M., Jamal K. H., Son R. (2007). Joint Food Safety and Quality Division, Ministry of Health Malaysia/ National Food Safety Research Center, Faculty of Food Science and Technology, University Putra Malaysia Expert Consultation on Risk Assessment of Vibrio parahaemolyticus in Black Tiger Prawn (Penaeus monodon). [Google Scholar]
- Alam M. J., Tomochika K. I., Miyoshi S. I., Shinoda S. (2002). Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208 83–87 10.1111/j.1574-6968.2002.tb11064.x [DOI] [PubMed] [Google Scholar]
- Al-Othrubi S. M., Alfizah H., Son R., Humin N., Rahaman J. (2011). Rapid detection and E-test antimicrobial susceptibility testing of Vibrio parahaemolyticus isolated from seafood and environmental sources in Malaysia. Saudi Medic. J. 32 400–406. [PubMed] [Google Scholar]
- Al-Othrubi S. M., Kqueen C. Y., Mirhosseini H., Hadi Y. A., Radu S. (2014). Antibiotic resistance of Vibrio parahaemolyticus Isolated from cockles and shrimp sea food marketed in Selangor, Malaysia. Clin. Microbiol. 3 148–154 10.4172/2327-5073.1000148 [DOI] [Google Scholar]
- Bej A. K., Patterson D. P., Brasher C. W., Vickery M. C., Jones D. D., Kaysner C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shell fish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Meth. 36 215–225 10.1016/S0167-7012(99)00037-8 [DOI] [PubMed] [Google Scholar]
- Beneduce L., Vernile A., Spano G., Massa S., Lamacchia F., Oliver, J D.(2010). Occurrence of Vibrio vulnificus in mussel farms from the Varano lagoon environment. Lett. Appl. Microb. 51 443–449 10.1111/j.1472-765X.2010.02917.x [DOI] [PubMed] [Google Scholar]
- Broberg C. A., Calder T. J.,, Orth K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13 992–1001 10.1016/j.micinf.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello F. C., Godfrey H. P., Tomova A., Ivanova L., Dölz H., Millanao A., et al. (2013). Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Appl. Eviron. Microbiol. 15 1917–1942 10.1111/1462-2920.12134 [DOI] [PubMed] [Google Scholar]
- Canizalez-Roman A., Flores-Villasenor H., Zazueta-Beltran J., Muro-Amador S., Leon-Sicairos N. (2011). Comparative evaluation of a chromogenic agar medium-PCR protocol with a conventional method for isolation of Vibrio parahaemolyticus strains from environmental and clinical samples. Can. J. Microbiol. 57 136–142 10.1139/W10-108 [DOI] [PubMed] [Google Scholar]
- Ceccarelli D., Hasan N. A., Hug A., Colwell R. R. (2013). Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 3:97 10.3389/fcimb.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G., Jiao X., Zhou X., Wang F., Yang Z., Huang J., et al. (2010). Distribution of genes encoding four pathogenicity islands (VPaIs), T6SS, biofilm, and type I pilus in food and clinical strains of Vibrio parahaemolyticus in China. Foodborne Pathog. Dis. 7 649–658 10.1089/fpd.2009.0441 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standard Institute (CLSI). (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, Approved Guideline, 2nd Edn. Wayne, PA: Clinical Laboratory Standard Institute. [Google Scholar]
- Dang H. Y., Ren J., Song L. S., Sun S., An L. G. (2008). Dominant chloramphenicol-resistant bacteria and resistance genes in coastal marine waters of Jiazhou Bay, China. World J. Microbiol. Biotech. 24 209–217 10.1007/s11274-007-9458-8 [DOI] [Google Scholar]
- Dang H. Y., Zhang X. X., Song L. S., Chang Y. Q., Yang G. P. (2007). Molecular determination of oxytetracycline-resistant bacteria and their resistance genes from mariculture environments of China. J. Appl. Microbiol. 103 2580–2592 10.1111/j.1365-2672.2007.03494.x [DOI] [PubMed] [Google Scholar]
- Daniels N. A., MacKinnon L., Bishop R., Altekruse S., Ray B., Hammond R. M., et al. (2000). Vibrio parahaemolyticus infection in the United States, 1973-1998. J. Infect. Dis. 181 1661–1666 10.1086/315459 [DOI] [PubMed] [Google Scholar]
- Deepanjali A., Kumar H. S., Karunasagar I., Karunasagar I. (2005). Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 71 3575–3580 10.1128/AEM.71.7.3575-3580.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaola A., Kaysner C. A., Bowers J., Cook D. W. (2000). Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and NewYork (1997 and 1998). Appl. Environ. Microbiol. 66 4649–4654 10.1128/AEM.66.11.4649-4654.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi R., Surendran P. K., Chakraborty K. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the coast of India. World J. Microbiol. Biotech. 25 2005–2012 10.1007/s11274-009-0101-8 [DOI] [Google Scholar]
- Elhadi N., Radu S., Chen C. H., Nishibuchi M. (2004). Prevalence of potentially pathogenic Vibrio species in the seafood marketed in Malaysia. J. Food Protect. 67 1469–1475. [DOI] [PubMed] [Google Scholar]
- García K., Torres R., Uribe P., Hernández C., Rioseco M., Romero J., et al. (2009). Dynamics of clinical and environmental Vibrio parahaemolyticus strains during seafood-related summer diarrheaoutbreaks in Southern Chile. Appl. Environ. Microbiol. 75 7482–7487 10.1128/AEM.01662-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gode-Potratz C. J., Kustusch R. J., Breheny P. J., Weiss D. S., McCarter L. L. (2011). Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol. Microbiol. 79 240–263 10.1111/j.1365-2958.2010.07445.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M. A., Hu J., Jinneman K. C. (2006). Multiplex real-time PCR detection of heatlabile and heat-stable toxin genes in enterotoxigenic Escherichia coli. J. Food Protect. 69 412–416. [DOI] [PubMed] [Google Scholar]
- Gutierrez West C. K., Klein S. L., Lovell C. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79 2247–2252 10.1128/AEM.03792-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwendelynne B. T., Son R., Nishibuchi M., Raha A. R., Suhaimi N., Lesley M., et al. (2005). Characterization of Vibrio parahaemolyticus isolated from coastal seawater in Peninsular Malaysia. Southeast Asian J. Trop. Med. Pub. Health 36 940–945. [PubMed] [Google Scholar]
- Haley B. J., Kokashvili T., Tskshvediani A., Janelidze N., Mitaishvili N., Grim C. J., et al. (2014). Molecular diversity and predictability of Vibrio parahaemolyticus along the Georgian coastal zone of the Black Sea. Front. Microbiol. 5:45 10.3389/fmicb.2014.00045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F., Walker R. D., Janes M. E., Prinyawinwatkul W., Ge B. (2007). Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73 7096–7098 10.1128/AEM.01116-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth E., Matsuda L., Hernández C., Rioseco M., Romero J., González-Escalona N., et al. (2009). Epidemiology of Vibrio parahaemolyticus outbreaks Southern Chile. Emerg. Infect. Dis. 15 163–168 10.3201/eid1502.071269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervio-Heath D., Colwell R. R., Derrien J. M., Robert-Pillot A., Fournier J. M., Pommepuy M. (2002). Occurrence of pathogenic vibrios in coastal areas of France. J. Appl. Microbiol. 92 1123–1135 10.1046/j.1365-2672.2002.01663.x [DOI] [PubMed] [Google Scholar]
- Johnson C. N., Flowers A. R., Young V. C., Gonzalez-Escalona N., DePaola A., Norie N. F., III, et al. (2008). Genetic relatedness among tdh+ and trh+ Vibrio parahamolyticus cultured from Gulf of Mexico oysters (Crassostrea virginica) and surrounding water and sediment. Microb. Ecol. 57 437–443 10.1007/s00248-008-9418-3 [DOI] [PubMed] [Google Scholar]
- Jun J. W., Kim J. H., Choresca C. H., Shin S. P., Han J. E., Han S. Y., et al. (2012). Isolation, molecular characterization and antibiotic susceptibility of Vibrio parahaemolyticus in Korean seafood. Foodborne Pathog. Dis. 9 224–231 10.1089/fpd.2011.1018 [DOI] [PubMed] [Google Scholar]
- Khan A. W., Hossain S. J., Uddin S. N. (2007). Isolation, identification and determination of antibiotics susceptibility of Vibrio parahaemolyticus from shrimps at Khulna Region of Bangladesh. Res. J. Microbiol. 2 216–227 10.3923/jm.2007.216.227 [DOI] [Google Scholar]
- Kim M., Kwon T. H., Jung S. M., Cho S. H., Jin S. Y., Park N. H., et al. (2013). Antibiotic resistance of bacteria isolated from the internal organs of edible snow crabs. PLoS ONE 8:70887 10.1371/journal.pone.0070887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. R., Nonaka L., Suzuki S. (2004). Occurrence of tetracycline resistance genes tet(M) and tet(S) in bacteria from marine aquaculture sites. FEMS Microbiol. Lett. 237 147–156 10.1111/j.1574-6968.2004.tb09690.x [DOI] [PubMed] [Google Scholar]
- Kim Y. B., Okuda J., Matsumoto C., Takahashi N., Hashimoto S., Nishibuchi M. (1999). Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37 1173–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesley M. B., Velnetti L., Cheah Y. K., Son R., Kasing A., Samuel L., et al. (2011). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from cockles (Anadara granosa) at Tanjung Karang, Kuala Selangor. Int. Food Res. J. 18 1183–1188. [Google Scholar]
- Letchumanan V., Chan K., Lee L. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence and advance molecular identification techniques. Front. Microbiol. 5:705 10.3389/fmicb.2014.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Wong M. H. Y., Chen S. (2013). Molecular characterisation of a multidrug resistance conjugative plasmid from Vibrio parahaemolyticus. Int. J. Antimicrob. Agents 42 575–579 10.1016/j.ijantimicag.2013.08.014 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang D., Wang H., Feng E., Zhu L., Wang H. (2012). Curing of plasmid pX01 from Bacillus anthracis using plasmid incompatibility. PLoS ONE 7:e29875 10.1371/journal.pone.0029875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley J. J., Adams C. D., Mormile M. R. (2007). Diversity of tet resistance genes in tetracycline resistant bacteria isolated from a swine lagoon with low antibiotic impact. Can. J. Microbiol. 53 1307–1315 10.1139/W07-104 [DOI] [PubMed] [Google Scholar]
- Mala E., Oberoi A., Alexander V. S. (2014). Vibrio isolates from cases of acute diarrhea and their antimicrobial susceptibility pattern in a tertiary care hospital. Int. J. Basic Appl. Sci. 3 35–37. [Google Scholar]
- Malcolm T. T. H., Cheah Y. K., Radzi C. W. J. W. M., Kasim F. A., Kantilal H. K., John T. Y. H., et al. (2015). Detection and quantification of pathogenic Vibrio parahaemolyticus in shellfish by using multiplex PCR and loop-mediated isothermal amplification assay. Food Control 47 664–671 10.1016/j.foodcont.2014.08.010 [DOI] [Google Scholar]
- Manjusha S., Sarita G. B. (2011). Plasmid associated antibiotic resistance in Vibrio isolated from coastal waters of Kerala. Int. Food Res. J. 18 1171–1181. [Google Scholar]
- Martinez-Urtaza J., Huapaya B., Gavilan R. G., Blanc-Abad V., Ansede-Bermejo J., Cadarso-Suarez C., et al. (2008). Emergence of Asiatic Vibrio disease in South America in phase with El Nino. Epidemiology 19 829–837 10.1097/EDE.0b013e3181883d43 [DOI] [PubMed] [Google Scholar]
- Molina-Aja A., Garcia-Gasca A., Abreu-Grobois A., Bolan-Mejia C., Roque A., Gomez-Gil B. (2002). Plasmid profiling and antibiotic resistance of Vibrio strains isolated from cultured penaeid shrimp. FEMS Microbiol. Lett. 213 7–12 10.1111/j.1574-6968.2002.tb11278.x [DOI] [PubMed] [Google Scholar]
- Nelapati S., Nelapati K., Chinnam B. K. (2012). Vibrio parahaemolyticus-an emerging foodborne pathogen-a Review. Vet. World. 5 48–62 10.5455/vetworld.2012.48-63 [DOI] [Google Scholar]
- Nishibuchi M. (2006). The biology of vibrios: molecular identification. ASM 4 44–64. [Google Scholar]
- Noorlis A., Ghazali F. M., Cheah Y. K., Tuan Zainazor T. C., Ponniah J., Tunung R., et al. (2011). Prevalence and quantification of Vibrio species and V. parahaemolyticus in freshwater fish at hypermarket level. Int. Food Res. J. 18 673–679. [Google Scholar]
- Okamoto A. S., Andreatti Filho R. L., Rocha T. S., Menconi A., Marietto-Goncalves G. A. (2009). Detection and transfer of antimicrobial resistance gene integron in Salmonella Enteritidis derived from avian material. Rev. Brasil. Ciên. Avíc. 11 195–201 10.1590/S1516-635X200900030009 [DOI] [Google Scholar]
- Okuda J., Ishibashi M., Hayakawa E., Nishino T., Takeda Y., Mukhopadhyay A. K., et al. (1997). Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35 3150–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker G., Myers M. L., Bej A. K. (2004). Rapid detection of Vibrio vulnificus in shellfish and Gulf of Mexico water by real-time PCR. Appl. Environ. Microbiol. 70 498–507 10.1128/AEM.70.1.498-507.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paydar M., Teh C. S. J., Thong K. L. (2013). Prevalence and characterisation of potentially virulent Vibrio parahaemolyticus in seafood in Malaysia using conventional methods, PCR and REP-PCR. Food Control 32 13–18 10.1016/j.foodcont.2012.11.034 [DOI] [Google Scholar]
- Pazhani G. P., Bhowmik S. K., Ghosh S., Guin S., Dutta S., Rajendran K., et al. (2014). Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS ONE 8:2815 10.1371/journal.pntd.0002815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng F. M., Jiang D. Y., Ruan H. H., Liu H. Q., Zhou L. P. (2010). Pathogenic investigation on a food poisoning induced by Vibrio parahaemolyticus. Prev. Med. Trib. 16 746–747. [Google Scholar]
- Qing Y., Cao K. Y., Fang Z. L., Huang Y. M., Zhang X. F., Tian G. B., et al. (2014). Outbreak of PER-1 and diversity of β-lactamases among ceftazidime-resistant Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 63 386–392 10.1099/jmm.0.069427-0 [DOI] [PubMed] [Google Scholar]
- Rico A., Satapornvanit K., Haque M. M., Min J., Nguyen P. T., Telfer T., et al. (2012). Use of chemicals and biological products in Asian aquaculture and their potential environmental risks: a critical review. Rev. Aquac. 4 75–93 10.1111/j.1753-5131.2012.01062.x [DOI] [Google Scholar]
- Sahilah A. M., Laila R. A., Sallehuddin H. M., Osman H., Aminah A., Ahmah Azuhairi A. (2014). Antibiotic resistance and molecular typing among cockle (Anadara granosa) strains of Vibrio parahaemolyticus by polymerase chain reaction (PCR)-based analysis. World J. Microbiol. Biotech. 30 649–659 10.1007/s11274-013-1494-y [DOI] [PubMed] [Google Scholar]
- Sani N. A., Ariyawansa S., Babji A. S., Hashim J. K. (2013). The risk assessment of Vibrio parahaemolyticus in cooked black tiger shrimps (Penaeus monodon) in Malaysia. Food Control 31 546–552 10.1016/j.foodcont.2012.10.018 [DOI] [Google Scholar]
- Shaw K. S., Rosenberg Goldstein R. E., He X., Jacobs J. M., Crump B. C., Sapkota A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Cheaspeake Bay and Maryland coastal bay. PLoS ONE 9:e89616 10.1371/journal.pone.0089616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V., Nam H. M., Nguyen L. T., Tamilselvam B., Murinda S. E., Oliver S. P. (2005). Prevalence of antimicrobial resistance genes in Listeria monocytogenes isolated from dairy farms. Foodborne Pathog. Dis. 2 201–211 10.1089/fpd.2005.2.201 [DOI] [PubMed] [Google Scholar]
- Son R., Elhadi N., Hassan Z., Rusul G., Lihan S., Fifadara N., et al. (1998). Characterization of Vibrio vulnificus isolated from cockles (Anadara granosa): antimicrobial resistance, plasmid profiles and random amplification of polymorphic DNA analysis. FEMS Microbiol. Lett. 165 139–143 10.1111/j.1574-6968.1998.tb13138.x [DOI] [PubMed] [Google Scholar]
- Su C. Y., Liu C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24 549–558 10.1016/j.fm.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Sudha S., Mridula C., Silvester R., Hatha A. A. M. (2014). Prevalence and antibiotic resistance of pathogenic Vibrios in shellfishes from Cochin market. Indian J. Mar. Sci. 43 815–824. [Google Scholar]
- Suzita R., Abu Bakar F., Son R., Abdul Amir A. S. (2010). Detection of Vibrio cholerae in raw cockles (Anadara granosa) by polymerase chain reaction. Int. Food Res. J. 3 675–680. [Google Scholar]
- Tang H. J., Chang M. C., Ko W. C., Huang K. Y., Lee C. L., Chuang Y. C. (2002) In vitro and in vivo activities of newer fluoroquinolones against Vibrio vulnificus. Antimicrob. Agents Chemother. 46 3580–3584 10.1128/AAC.46.11.3580-3584.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendencia E. A., de la Peña L. D. (2001). Antibiotic resistance of bacteria from shrimp ponds. Aquaculture 195 193–204 10.1016/S0044-8486(00)00570-6 [DOI] [Google Scholar]
- Tunung R., Jeyaletchumi P., Noorlis A., Tang Y. H., Sandra A., Ghazali F. M., et al. (2012). Biosafety of Vibrio parahaemolyticus from vegetables based on antimicrobial sensitivity and RAPD profiling. Int. Food Res. J. 19 467–474. [Google Scholar]
- Tunung R., Margaret S. P., Jeyaletchumi P., Chai L. C., Zainazor T. C., Ghazali F. M., et al. (2010). Prevalence and quantification of Vibrio in raw salad vegetables at retail level. J. Microbiol. Biotech. 20 391–396. [PubMed] [Google Scholar]
- Velazquez-Roman J., León-Sicairos N., de Jesus Hernández-Díaz L., Canizalez-Roman A. (2014). Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 3:110 10.3389/fcimb.2013.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Roman J., Leon-Sicairos N., Flores-Villasenor H., Villafana-Rauda S., Canizalez-Roman A. (2012). Association of pandemic Vibrio parahaemolyticus O3: K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl. Environ. Microbiol. 78 1794–1803 10.1128/AEM.06953-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vengadesh L., Son R., Yoke-Kqueen C. (2012). Molecular quantitation and characterization of Vibrio cholera from different seafood obtained from wetmarket and supermarket. Int. Food Res. J. 19 45–50. [Google Scholar]
- Vuddhakul V., Chowdhury A., Laohaprertthisan V., Pungrasamee P., Patararungrong N., Thianmontri P. (2000). Isolation of a pandemic O3:K6 clone of a Vibrio parahaemolyticus strain from environmental and clinical sources in Thailand. Appl. Environ. Microbiol. 66 2685–2689 10.1128/AEM.66.6.2685-2689.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B. A., Salyers A. A. (2003). Is the evolution of bacterial pathogens an out-of-body experience? Trends Microbiol. 11 347–350 10.1016/S0966-842X(03)00179-3 [DOI] [PubMed] [Google Scholar]
- Wong H. C., Liu S. H., Ku L. W., Lee I. Y., Wang T. K., Lee Y. S., et al. (2000). Characterization of Vibrio parahaemolyticus isolates obtained from foodborne illness outbreaks during 1992 through 1995 in Taiwan. J. Food Protect. 63 900–906. [DOI] [PubMed] [Google Scholar]
- Wong M. H., Liu M., Wan H. Y., Chen S. (2012). Characterization of extended-spectrum-β-lactamase-producing Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 56 4026–4028 10.1128/AAC.00385-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wen J., Ma Y., Ma X., Chen Y. (2014). Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus, China, 2003-2008. Food Control 46 197–202 10.1016/j.foodcont.2014.05.023 [DOI] [Google Scholar]
- Xu X., Wu Q., Zhang J., Cheng J., Zhang S., Wu K. (2014). Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Control 46 81–85 10.1016/j.foodcont.2014.04.042 [DOI] [Google Scholar]
- Yamamoto A., Iwahori J., Vuddhakul V., Charernjiratrag W., Vose D., Osaka K., et al. (2008). Quantitative modeling for risk assessment of Vibrio parahaemolyticus in bloody clams in southern Thailand. Int. J. Food Microbiol. 124 70–78 10.1016/j.ijfoodmicro.2008.02.021 [DOI] [PubMed] [Google Scholar]
- Yang Z., Jiao X., Zhou X., Cao G., Fang W., Gu R. (2008). Isolation and molecular characterization of Vibrio parahaemolyticus from fresh, low-temperature preserved, dried and salted seafood products in two coastal areas of eastern China. Int. J. Food Microbiol. 125 279–285 10.1016/j.ijfoodmicro.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Yano Y., Hamano K., Satomi M., Tsutsui I., Ban M., Aue-umneoy D. (2014). Prevalence and antimicrobial susceptibility of Vibrio species related to food safety isolated from shrimp cultured at inland ponds in Thailand. Food Control 38 30–45 10.1016/j.foodcont.2013.09.019 [DOI] [Google Scholar]
- Zarei M., Borujeni M. P., Jamnejad A., Khezrzadeh M. (2012). Seasonal prevalence of Vibrio species in retail shrimps with an emphasis on Vibrio parahaemolyticus. Food Control 25 107–109 10.1016/j.foodcont.2011.10.024 [DOI] [Google Scholar]
- Zhang L., Orth K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 16 70–77 10.1016/j.mib.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang X. X., Zhang T., Fang H. H. P. (2009). Antibiotic resistance genes in water environment. Appl. Microbiol. Biotech. 82 397–414 10.1007/s00253-008-1829-z [DOI] [PubMed] [Google Scholar]
- Zhang Z., Xiao L., Lou Y., Jin M., Liao C., Malakar P. K., et al. (2014). Development of a multiplex real-time PCR method for simultaneous detection of Vibrio parahaemolyticus, Listeria monocytogenes and Salmonella spp. in raw shrimp. Food Cont. 51 31–36 10.1016/j.foodcont.2014.11.007 [DOI] [Google Scholar]
- Zulkifli Y., Alitheen N. B., Raha A. R., Yeap S. K., Marlina, Son R., et al. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from cockles in Padang, Indonesia. Int. Food Res. J. 16 53–58. [Google Scholar]