Abstract
Anti-angiogenic therapy, which suppresses tumor growth by disrupting oxygen and nutrient supply from blood to the tumor, is now widely accepted as a treatment for cancer. To investigate the mechanisms of action of these anti-angiogenesis drugs, new three dimensional (3D) cell culture-based drug screening models are increasingly employed. However, there is no in vitro high-throughput screening (HTS) angiogenesis assay that can provide uniform culture conditions for quantitative assessment of physiological responses to chemoattractant reagents under various concentrations of anti-angiogenesis drugs. Here we describe a method for screening and quantifying the vascular endothelial growth factor (VEGF)-induced chemotactic response on human umbilical vein endothelial cells (HUVECs) cultured under different concentrations of bortezomib, a selective 26S proteasome inhibitor. With this quantitative microfluidic angiogenesis screen (QMAS), we demonstrate that bortezomib-induced endothelial cell death was preceded by a series of morphological changes that develop over several days. We also explore the mechanisms by which bortezomib can inhibit angiogenesis.
Introduction
Drug screening technologies in two dimensions (2D) (i.e. as a flat layer of cells) have been widely adopted in the pharmaceutical industry for drug discovery. These approaches are still in use; however considerable challenges remain in identifying new drugs for cancer treatment in particular due to the critical role of multiple cell types in disease progression and the inability to accurately mimic the cellular environment in vivo [1–5]. Consequentially, a wide gap exists between 2D cell culture assays and animal testing [6]. Moreover, pressures are mounting to reduce animal testing due to its expense, the long times required to obtain results, ethical considerations, and the limitations of an animal model for predicting human responses [7, 8]. In order to overcome the drawbacks of 2D cell culture assays and potentially reduce the need for animal testing, new analytical screening assays in three dimensions (3D) and employing human cells are needed. Cells in vivo are subject to multiple cues that vary in time and space, including gradients of cytokines and secreted proteins from neighboring cells of similar or different type, and their behavior is strongly influenced by mechanical and biochemical interactions with the extracellular matrix (ECM). To meet these challenges, microfluidic Lab-on-a-Chip (LOC) technologies offer promising strategies for addressing the inherent complexity of cellular systems with spatio-temporal multiple cues [9, 10]. In addition, microfluidic approaches have been adopted to create 3D tissue scaffolds with realistic physical, mechanical, and biological properties.
3D cell culture-based microfluidic devices have been previously reported, which enable the formation of cell spheroids in 3D for mimicking the complexity of heterogeneous tumor tissue, for cytotoxicity tests of anticancer drugs in the devices [11–15]. These systems could be used as a assay for screening anticancer drugs in cancer treatment, but, currently, anti-angiogenic therapies (vascular-targeted therapies), which suppress tumor growth by cutting off supply of nutrients and oxygen in the blood to tumor cells, has emerged as an important advance in cancer treatment with a significant reduction in the adverse effects caused by other anti-cancer drugs [16–19]. The potential now exists to develop new approaches to anti-angiogenesis drug screening, which enable monitoring and quantification of cell responses to the anti-angiogenesis drug in addition to screening for cell viability. Recent efforts have produced in vitro microfluidic cell culture models that apply physical and biochemical stimuli within a 3D hydrogel scaffold integrated in the channels and offer a viable solution for monitoring cellular behaviors in response to drug [17–31]. For example, these models have been employed to investigate heterotypic cell–cell interactions in 3D [22], to study neurite responses to growth factor gradients [23], to evaluate and quantify capillary sprouting and angiogenesis from an intact cell monolayer [24, 26], and to examine the effects of interstitial flow on cancer cell morphology and migration [26].
However, the previously developed microfluidic based-platforms are ill-suited to the purpose of screening the effects of various anti-angiogenesis drugs over a range of concentrations. For example, preparing and handling many previously developed microfluidic based-platforms can be time-consuming and tedious. Moreover, larger variability may occur between different chips due to slight variations in protocol caused by testing a large number of chips manually.
Here, we propose a new microfluidic platform (termed the quantitative microfluidic angiogenesis screen (QMAS)), that can monitor and quantify cellular behaviors, such as morphological changes, endothelial cell viability, and formation of angiogenic sprouts, depending on the various concentrations of drug applied (i.e., bortezomib, a selective 26S proteasome inhibitor). This model incorporates a perfusion culture-based system for generating stable concentration gradients in multiple 3D collagen matrices by generating a pressure-driven uniform flow rate through each of the microfluidic channels. We demonstrate these capabilities through a study of a drug, bortezomib, which is known to inhibit angiogenesis in vivo. To gain insight into the anti-angiogenic effects of bortezomib on human umbilical vein endothelial cells (HUVECs), we monitored cell behaviors daily, and investigated the changes of morphological shape and viability of bortezomib-treated HUVEC under various concentrations of bortezomib. Capillary growth and the formation of circular lumen-like structures from an intact endothelial cell monolayer in response to vascular endothelial growth factor (VEGF) are quantified showing that the size and number of lumen-like structures are directly affected by the concentrations of bortezomib. We demonstrate that bortezomib inhibits angiogenesis in a dose-dependent manner and also that it causes cell death of HUVECs at sufficiently high concentrations.
Results and Discussion
Characterization of the QMAS for screening anti-angiogenic therapeutic drugs
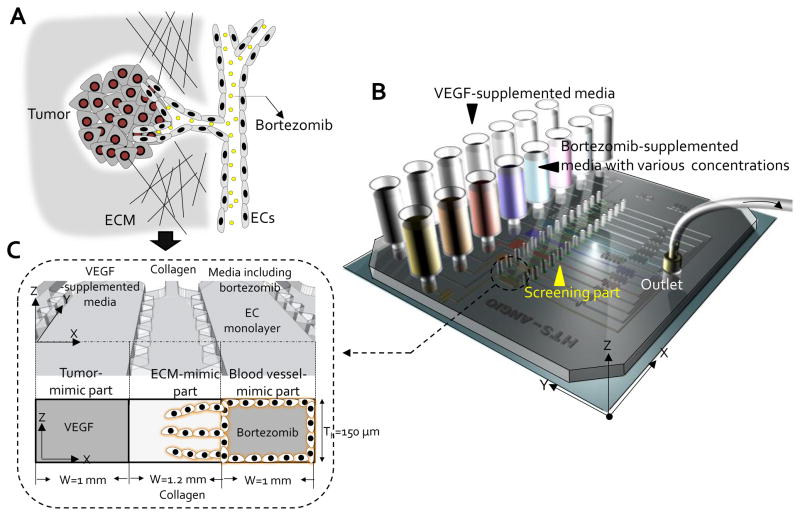
Vascular sprouts grow from existing blood vessels into the interstitial matrix guided by a concentration gradient of factors (e.g., vascular endothelial growth factor (VEGF)) secreted from the tumor during tumor angiogenesis. When an anti-angiogenesis drug is employed in vivo for the treatment of cancer, it inhibits the formation of tumor-associated blood vessels; thereby disrupting the supporting perivascular niche for cancer stem cells [16–18] (Fig. 1A). This in vivo anti-angiogenic therapeutic process was modeled in the QMAS (Fig.1B & C) incorporating the following significant features: (1) 3D hydrogel scaffolds in gel cages (fourteen) mimic the extracellular matrix (ECM) in vivo, (2) confluent endothelial cell (ECs) monolayers in the cell culture channels (eight) mimic pre-existing blood vessels in vivo, (3) VEGF-supplemented media in conditioned media channels (seven) mimic the presence of growth factors (VEGF) secreted by tumor in vivo, and (4) bortezomib-supplemented media with various concentrations in the cell culture channels (eight) mimic the introduction of anti-angiogenesis drugs (i.e. bortezomib, a selective 26S proteasome inhibitor, marketed by Millennium Phamaceuticals as Velcade) in vivo via the circulation (Fig. 1B & Fig. 1C). The QMAS makes it possible for us to monitor, screen, and quantify the angiogenic responses in the 3D hydrogel scaffolds due to various concentrations of bortezomib.
Figure 1. In vivo anti-angiogenesis application and microfluidic assay.
(A) Potential anti-angiogenic therapeutic application in cancer treatment in vivo. (B) Schematic view of the QMAS for screening anti-angiogenic therapeutic drugs. (C) Significant parts of the QMAS are: (1) 3D hydrogel scaffolds in gel cages (fourteen) mimic the extracellular matrix (ECM) in vivo, (2) confluent endothelial cell (ECs) monolayers in the cell culture channels (eight) mimic pre-existing blood vessels in vivo, (3) VEGF-supplemented media in conditioned media channels (seven) mimic the presence of growth factors secreted by tumor in vivo, and (4) bortezomib-supplemented media with various concentrations in the cell culture channels (eight) mimic the introduction of anti-angiogenesis drugs in vivo via the circulation.
The QMAS was fabricated using soft lithography and PDMS replica molding (Fig. S1, see Materials and Methods), and consists of: (1) reservoirs used for providing fresh conditioned media (VEGF-supplemented or Bortezomib-supplemented medias) into all the channels, made by cutting the end of commercial pipette tips (i.e. MBP (Molecular BioProducts)), and media in the reservoirs were replaced daily during cell culture using a multi-tip pipette, (2) a 1st PDMS layer including microchannels for the bortezomib-supplemented media with various concentrations, endothelial cell monolayers and collagen gel-cages, (3) 2nd PDMS layer used only during filling as a manifold layer for filling collagen gel, and (4) a large cover slip at the bottom (TED PELLA, INC. 3 × 3 1/4″, Thick: 0.19 ~0.25mm) (Fig. S1A & S1B).
To mimic the presence of pro-angiogenic factors secreted from the tumor, VEGF (40ng/ml) was introduced into the QMAS, which then diffused into the 3D hydrogel scaffold regions (fourteen) simultaneously. However, establishing stable concentration profiles within the QMAS with integrated porous matrices required that we eliminate any interstitial flow across the matrix resulting from pressure imbalances and this proved challenging. To achieve stability of concentration profiles, the QMAS was designed as a perfusion-based system. The stable concentration profiles could be created in the fourteen-3D hydrogel scaffold matrices by minimizing interstitial flow across them; this was accomplished by a combination of introducing identical volumetric flow rates into the fifteen-independent flow channels and connecting the ends of all flow channels to a single syringe pump at the common outlet-junction (Fig. S2A). The distribution of volumetric flow rates was controlled via design of the hydrodynamic resistance of the microchannels (Fig. S2B). Hydrodynamic simulation using a commercial finite element solver (COMSOL, Burlington, MA) was performed to confirm equal flows in each of the fifteen microchannels, and we confirmed that uniform flow rates were generated during perfusion (Fig. S2C).
Numerical simulations based on a transient solution of the Brinkman equation for porous medium flow and the convection-diffusion equation for a growth factor having the diffusivity of VEGF (D=5 ×10−11 m2/s) demonstrated the generation of a nearly linear concentration gradient of growth factor; details of numerical simulations are provided in SI Materials and Methods (Fig. S3). In order to maintain this gradient over the entire length of the gel regions, the media should be introduced at a flow rate corresponding to a Peclet number (Pe) > 50. To satisfy this condition, the withdrawal flow rate in the syringe pump (Infuse/Withdraw syringe pump, HARVARD) was set to 1.33 μl/min; we used this flow rate for all experiments.
In the previously developed systems as well as the QMAS, the collagen hydrogel was confined to the gel regions by a series of equally spaced posts (110 μm, Fig. 1C) that prevent liquid from leaking into adjacent channels. Generally, collagen gel filling by a pipette has been done manually; this is a simple method, but leakage between adjacent channels was sometimes difficult to avoid. In order to improve the consistency of filling, we modified the collagen gel filling method using the manifold layer that can insert the gel to the fourteen-gel regions simultaneously (Fig. S4). When a collagen solution is injected into an inlet of the manifold layer by the syringe pump, the injected solution is distributed among the fourteen microchannels in the manifold layer (2nd PDMS layer), and then simultaneously enters into the fourteen collagen gel regions (1st PDMS layer) (Fig. S4A). Volumetric flow rates in the microchannel network of the manifold layer were controlled by specifying the dimensions, and thereby the hydrodynamic resistance, of the microchannels (Fig. S4B). We again demonstrated that the same volumetric flow rates were generated in the 14 collagen gel regions using hydrodynamic simulation (COMSOL Inc., MA, USA) (Fig. S4C). Experimentally, we observed that the consistency of collagen gel filling into the gel cages was influenced by the surface properties of the bottom layer; for example, a glass surface and PDMS surface behaved differently. When a PDMS surface was used as the bottom layer, collagen filling was more uniform than when glass was used due to its low wettability (data not shown). All reported experiments, however, were conducted with glass as the bottom layer to provide optimal imaging quality. Chip preparation details are provided in Materials & Methods and SI Movie 1.
High throughput angiogenesis assay
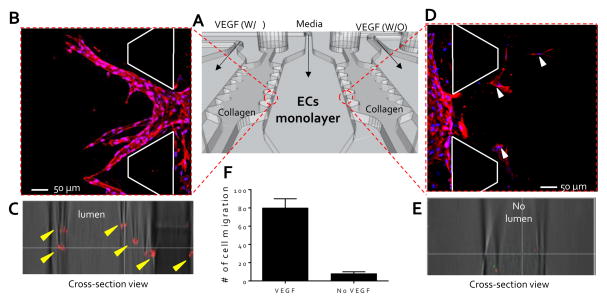
To demonstrate the capabilities of the QMAS as an angiogenesis assay in response to chemoattractant gradients, VEGF-supplemented media (40 ng/ml) were introduced into the VEGF injection channels (#1, #3, #5, & #7) (Fig. S5 & Fig. 2A) and the control mediums without VEGF were injected into the VEGF injection channels (#2, #4, #6, & #7) as reference conditions (Fig. S5 & Fig. 2A). Response of HUVECs was monitored daily for 5 days by phase-contrast microscopy (OLYMPUS, CKK 41, Tokyo, Japan). On day 5, the HUVECs were fixed and stained to examine cell morphologies. Cells rapidly and actively migrated from the endothelial channel into the 3D hydrogel scaffolds (condition-side) toward the VEGF-supplemented media channel (Fig. 2B); while significantly less migration was observed into the opposite scaffold (control-side) toward the control channel without VEGF (Fig. 2D). Confocal microscopy revealed the growth of circular lumen-like structures from the HUVEC monolayer into the condition-side hydrogel scaffolds in response to VEGF (Fig. 2C). No morphological change was evident in the control-side hydrogel scaffolds in immunostained and confocal microscopy images, except that several cells had migrated. These cells were not connected to the endothelial monolayer by vascular stalks presumably due to the absence of a VEGF concentration gradient across the collagen gel (Fig. 2E). These qualitative observations were quantified by counting the number of cells that migrated from each opening (total openings: #6, Fig. 2A) into collagen gel scaffolds (by ImageJ (http://rsbweb.nih.gov/ij/). We found that 79±9.8 cells migrated and proliferated toward the condition-side hydrogel scaffolds per; in contrast, 8±2.3 cells migrated toward the control-side hydrogel scaffolds (Fig. 2F). Using this migration assay, we could assess the response of an endothelial monolayer to various biochemical stimuli simultaneously via achieving the stable concentration gradient toward the fourteen 3D hydrogel scaffolds. The QMAS therefore allows us to monitor and quantify angiogenesis under the in vivo-like situation with uniform experimental conditions (i.e. uniform 3D hydrogel structure conditions & hydrostatic conditions between multiple microchannels) simultaneously and with little variability due to the operator’s technique.
Figure 2. High throughput angiogenesis assay.
(A) Schematic view of a set of experiments to demonstrate the capabilities of the QMAS as an angiogenesis assay in response to chemoattractant gradients; VEGF-supplemented media (40 ng/ml) introduced into the VEGF injection channels (#1, #3, #5, & #7, (Fig. S5)) and control media without VEGF injected into the VEGF injection channels (#2, #4, #6, & #7,(Fig. S5)) as reference conditions. (B) Migration of ECs from the endothelial channel into the 3D hydrogel scaffolds toward the VEGF-supplemented media channel (condition-side). (C) Growth of circular lumen-like structures into the condition-side hydrogel scaffolds in response to VEGF (yellow arrowheads, cross-section view). (D) Minimal migration of the ECs (white arrowheads) into the opposite scaffold toward the control channel without VEGF (control-side). (E) Confocal microscopy confirming the absence of circular lumen-like structures toward the control channel without VEGF (cross-section view). (F) Qualitative analysis showing the number of cells that migrated from each opening into collagen gel scaffolds.
Quantitative analysis of HUVEC morphology by bortezomib
Bortezomib (Millennium Pharmaceuticals, Inc., Cambridge, MA) was chosen to demonstrate the utility of the QMAS for drug screening. Bortezomib, one of the 14 anti-angiogenic therapeutic drugs have been approved by the US Food and Drug Administration (FDA), exerts its anti-angiogenic effect by decreasing the secretion of vascular endothelial growth factor (VEGF) from myeloma cells [32, 33]. We wanted to explore the possibility that bortezomib could act directly on the endothelium, independent of the myeloma cells. In order to model the in vivo situation (Fig. 1A), we first established endothelial cell monolayers on the walls of the channels to mimic in vivo blood vessels through the injection of endothelial cells (HUVECs) by the multiple-pipette (drug injection channels (#1 ~ #8).. Second, VEGF (40 ng/ml)-supplemented media mimicking the growth factors (i.e. VEGF) secreted from the tumor were filled in the VEGF injection channels, and finally, we added media containing a range of concentrations of bortezomib in each reservoir all with the multiple-pipette (drug injection channels (#1 ~ #8), over a logarithmic range (Fig. S6) & a linear range (Fig. S7)). The response of the HUVECs was monitored daily by phase-contrast microscopy for 5 days. Subsequent steps are shown in Supplementary Movie 1.
We first assessed whether the morphological changes were shown in bortezomib treated HUVECs (logarithmic concentrations: 0, 10−5, 10−4, 10−3, 10−2, 10−1, and 1 μM (Fig. S6)). Typical cobblestone-like morphology of HUVECs was observed at the low concentrations of bortezomib (0, 10−5, and 10−4 μM) by day 5, whereas the HUVECs showed some elongated morphological shapes without spindle-like shapes when conditioned with a dose of 10−3 μM (Fig. S8A). Almost all cells died when exposed to bortezomib at higher concentrations (10−2, 10−1, and 1 μM). We investigated whether or not bortezomib-induced endothelial cell death was preceded by observable morphological changes [34] by monitoring cell behaviors daily with the dose of 10−2 μM (Fig. S9). The morphological transition to a spindle-like shape was discernable from day 1, and dead cells, which were detached from the cell surface, were observed from days 2 to 5. Consequently, we determined that bortezomib-induced endothelial cell death was preceded by a series of morphological changes that develop over an extended period. We then screened for the morphological changes in finer detail, over a reduced range of bortezomib concentrations between 10−3 μM (1 nM) and 10−2 μM (10 nM) (Fig. S7). Using a linear variation in concentrations we were able to identify that the morphological changes in HUVECs occurred in the range of 4 nM to 6 nM bortezomib (Fig. S8B). Within this narrow range, HUVECs neither died nor detached from the surface and only showed slight changes in their structures. Almost all cells died at the dose of 8 nM and 10nM, became detached from the surface and were washed away during media changes.
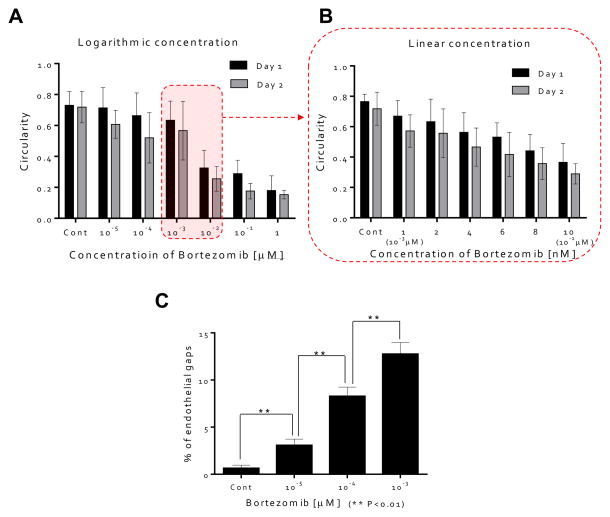
We next studied the morphological changes of HUVECs quantitatively, but limited either the time period of study or the concentration of bortezomib since HUVECs were unable to survive at the higher concentrations (10−2, 10−1, and 1 μM) after 3 days. To analyze the morphological changes of HUVECs quantitatively, circularity was calculated from the area and perimeter of the traced EC (see SI Materials and Methods). The former equals 4π (Area/Perimeter2); a circularity of 1.0 indicates a perfect circle, and as the circularity approaches 0.0, the shape becomes an increasingly elongated ellipse. At the low concentration of bortezomib (0, 10−5, 10−4, and 10−3 μM), circularities were > 0.6, whereas circularities were < 0.4 when treated with the dose of 10−2 μM (Fig.3A). From these results, we observed that HUVEC morphology changed significantly in the concentration range between 10−3 μM and 10−2 μM, so we performed a second set of experiments over a linear concentration range (0, 1 (10−3 μM), 2, 4, 6, 8, and 10 nM (10−2 μM)). We found circularity of the HUVECs to be a sensitive precursor to cell death, which decreased as the concentration and treatment time increased (Fig. 3B). These show that we can demonstrate an effect of bortezomib on the morphogenesis of HUVEC in a dose and time-dependent manner by using the QMAS.
Figure 3. Quantitative analysis of the influence on HUVEC morphology of bortezomib.
(A) Circularities of ECs as a function of bortezomib concentration (0, 10−5, 10−4, 10−3, 10−2, 10−1, and 1 μM). Circularities were > 0.6 for < 10−3 μM), whereas circularities were < 0.4 @ 10−2 μM. (B) Circularities of ECs as a function of bortezomib concentration (0, 1 (10−3 μM), 2, 4, 6, 8, and 10 nM (10−2 μM)). Circularity of the ECs, a sensitive measure of cell health, decreased as the concentration and treatment time increased. (C) HUVECSs were stained with anti-VE-cadherin followed by Alexa 488. Breakdown of VE-cadherin at the doses of bortezomib (10−5, 10−4, & 10−3 μM) was evident through the discontinuity of the green fluorescent line labeling the VE-cadherin junctions compared with intact VE-cadherin junctions in the case of HUVECs cultured in control medium (0 μM). (D) Qualitative analysis of cell junction integrity by measuring the number of gaps observed in the VE-cadherin staining; endothelial gap number increased with increasing concentration of bortezomib at 5 days (0, 10−5, 10−4, and 10−3 μM).
Fluorescence imaging of HUVECs stained for VE-cadherin allowed us to quantitatively assess disruption of VE-cadherin junctions when cells were cultured over 5 days. Breakdown of VE-cadherin was evident through the discontinuity of the green fluorescent line labeling the VE-cadherin junctions compared with intact VE-cadherin junctions in the case of HUVECs cultured in control medium, with no bortezomib (Fig. 3C). We quantified cell junction integrity by measuring the number of gaps observed in the VE-cadherin staining (see Materials and Methods). Endothelial gap formation increased as concentration of bortezomib increased for 5 days (0, 10−5, 10−4, and 10−3 μM) (Fig. 3D), demonstrating significant but sub-lethal effects at these lower concentrations. No data are presented for concentrations of 10−2, 10−1, and 1 μM due to cell death and detachment from the surface. These results show that the disruption of adherent junctions contributes to the increase in vasculature permeability.
Proliferation rate and cell viability in bortezomib treated HUVECs
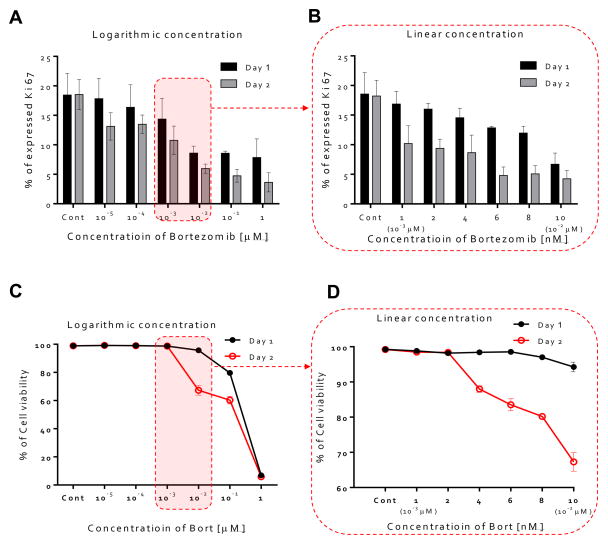
We next studied whether or not HUVEC proliferation was affected by the treatment of bortezomib using Ki67 expression, a marker of proliferation also used for drug discovery [35–37]. Studies were conducted, in HUVECs cultured over 2 days under the logarithmic (Fig. S6) and linear concentrations (Fig. S7) of bortezomib. We found that Ki67 was expressed at higher levels in the absence of bortezomib (0 μM, average=18.2%), and its expression ratio gradually decreased with increasing bortezomib concentration at day 1 (from 18.2% at 0 μM to 7.8% at 1 μM), but declined more by day 2 (from 18.2% at 0 μM to 3.6% at 1 μM, Fig.4A). Ki67 expression ranged from 18.5% at 0 nM to 6.70% at 10 nM on day 1 and 18.34% at 0 nM to 4.24% at 10 nM on day 2 (Fig. 4B). Ki67 was expressed for less than 5% in HUVECs when conditioned with 6 nM of bortezomib, which indicates that the proliferation rate varies in a time and dose-dependent manner.
Figure 4. Proliferation rate and cell viability in bortezomib treated HUVECs.
(A) Percentages of proliferation rate over a logarithmic range of bortezomib concentrations (0, 10−5, 10−4, 10−3, 10−2, 10−1, and 1 μM). Ki67 was expressed at higher levels in the absence of bortezomib (0 μM, average=18.2%), and its expression ratio gradually decreased with increasing bortezomib concentration at day 1 (from 18.2% at 0 μM to 7.8% at 1 μM), but declined more by day 2 (from 18.2% at 0 μM to 3.6% at 1 μM). (B) Percentages of proliferation rate over a linear range of bortezomib concentrations (0, 1 (10−3 μM), 2, 4, 6, 8, and 10 nM (10−2 μM)). Ki67 expression ranged from 18.5% at 0 nM to 6.70% at 10 nM on day 1 and 18.34% at 0 nM to 4.24% at 10 nM on day 2. (C) Cell viability percentages over a logarithmic range of bortezomib concentrations. Cell viability did not decrease with the addition of bortezomib < 10−2 μM for 1 day, but significantly decreased at 10−2 μM (cell viability < 67 %, day 2). (D) Percentages of cell viability on the dose of linear concentration of bortezomib. Cell viability was still mostly maintained for doses < 4 to 6 nM (cell viability > 80%, day 2).
We also hypothesized that the potent anti-angiogenic capacity of bortezomib was in part a consequence of regression of blood vessels, so we next evaluated the HUVEC’s cell viability on the exposure of the bortezomib. Cell viability did not decrease with the addition of bortezomib on day 1 (Fig. 4C & D). On day 2, cell viability was still mostly maintained for doses < 4 to 6 nM (cell viability > 80%, Fig. 4D), but was significantly decreased at concentrations of 10−2 μM (cell viability < 67 %, Fig. 4D). According to these results, we have also shown that the cell viability varies with the time and dose-dependent manner, and that bortezomib is capable of inducing the death of HUVECs.
Quantitative analysis of anti-angiogenesis in bortezomib-treated endothelial cells
Vascular endothelial growth factor (VEGF)–induced endothelial cell migration is an important component of tumor angiogenesis. VEGF regulates key steps in the angiogenic process, including endothelial cell migration and tube formation [38]. To find a treatment condition that did not result in significant toxicity for HUVECs and inhibit the growth of new blood vessels from pre-existing ones, we studied the effects of bortezomib over a logarithmic concentration range of bortezomib on the VEGF induced migration of HUVECs using the QMAS.
We found that bortezomib treatment significantly suppressed VEGF-induced migration at the higher doses of bortezomib (10−2, 10−1, & 1 μM). Confocal microscopy revealed the growth of circular lumen-like structures from the HUVEC monolayer into the collagen scaffold in response to VEGF at lower bortezomib concentrations (0, 10−5, 10−4, & 10−3 μM), but no morphogenesis for doses > 10−2 μM was evident in immunostained and confocal microscopy images (Fig. S10A). The formation of circular lumen-like structures was preceded by a period of VEGF-induced migration, and was also reduced with increasing dose of bortezomib. We also monitored daily the cellular behaviors at the dose of 10−2 μM to assess defects in cell migration due to bortezomib. HUVECs migrated toward the 3D hydrogel scaffolds in response to VEGF for 2 days, but those migrated cells regressed after 3 days (Fig. S10B).
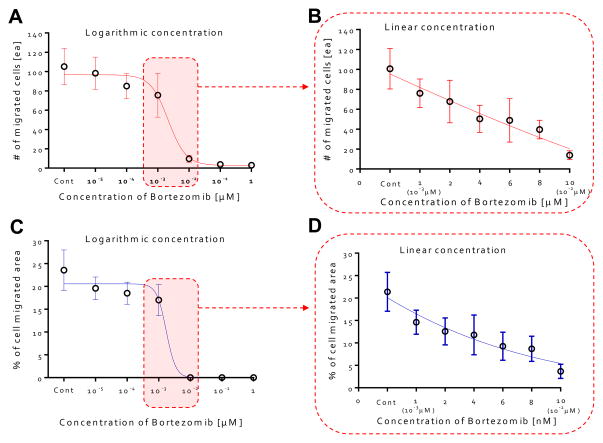
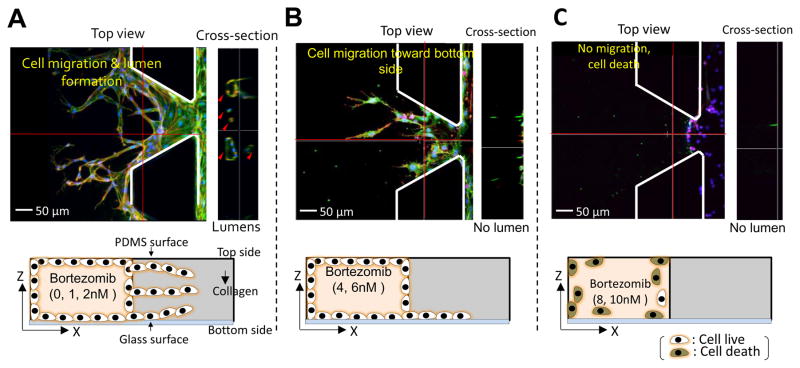
The number of migrated cells generally decreased at doses of < 10−3 μM, but the number drastically declined when 10−2 μM was applied (Fig. 5A). The percentage of image area occupied by the migrated cells fell dramatically at ~ 10−2 μM, in agreement with our results on the number of migrated cells (Fig. 5C). For finer discrimination, we screened for the cellular behaviors in the linear concentration range of bortezomib (1, 2, 4, 6, 8, & 10 nM). The number of migrated cells and % of area occupied by the migrated cells exhibited a nearly linear dependence along the linear dose range region (Fig. 5B & D). As expected, cells actively migrated at all heights inside the collagen gel scaffolds at the concentration of 0 nM, 1nM, and 2nM (Fig. 6A) with lumen-like structures, but at concentrations of 4nM and 6nM, although endothelial cells started out in the gel, they wound up at the bottom with little or no lumen-like structures (Fig. 6B). Neither cell migration nor lumen-like structures formed at doses of 8nM and 10nM (Fig. 6C). From these results, we determined that bortezomib by itself inhibited VEGF-induced angiogenesis, and were able to identify the proper dosage of bortezomib (4 nM & 6 nM) that did not prove fatal toxicity to HUVECs while being able to inhibit the growth of new blood vessels from a pre-existing monolayer.
Figure 5. Quantitative analysis of anti-angiogenesis in bortezomib-treated endothelial cells.
(A) The number of migrated cells as a function of dose with logarithmic variations in concentration of bortezomib (0, 10−5, 10−4, 10−3, 10−2, 10−1, and 1 μM). # of migrated cells generally decreased at doses < 10−3 μM, but the number drastically declined when 10−2 μM was applied. (B) The number of migrated cells as a function of dose with linear variations in concentration of bortezomib (0, 1 (10−3 μM), 2, 4, 6, 8, and 10 nM (10−2 μM)). # of migrated cells exhibited nearly a linear dependency along the linear dose range region. (C) Percentages of cell migrated area as a function of dose of bortezomib (0, 10−5, 10−4, 10−3, 10−2, 10−1, and 1 μM). % of area occupied by the migrated cells fell dramatically at the dose of 10−2 μM, in agreement with our results on the number of migrated cells. (D) Percentages of cell migrated area as a function of dose with linear variations in concentration of bortezomib. % of area occupied by the migrated cells exhibited nearly a linear dependency along the linear dose range region.
Figure 6. Projected images showing the effect of bortezomib dose;
(A) Cells actively migrated towards upper, middle, and bottom region of collagen gel scaffolds and formed lumen-like structures at concentrations of 0 nM, 1nM, and 2nM. (B) At concentrations of 4nM and 6nM, although endothelial cells started out throughout the gel, they wound up migrating to the bottom and failed to create lumen-like structures. (C) Neither cell migration nor lumen-like structures formed at doses of 8nM and 10nM.
Conclusions
In the current study, we have proposed and described the QMAS as a quantitative anti-angiogenesis screening assay. Its utility was demonstrated using a known anti-angiogenic agent, bortezomib. An important attribute of the QMAS is its ability to screen multiple concentrations simultaneously, allowing a side-by-side comparison of the dose- and time-dependent effects of drug in an in vivo-like situation. Minimizing the need for repeated experiments on multiple devices will likely reduce experiment-to-experiment variability, and lead to assays that are less sensitive to user experience. With this microfluidic-based in vitro assay, we explored the possibility that bortezomib, which has been reported to act decreasing the VEGF secretion from myeloma cells [32, 33]. Our results suggest an additional mechanism, that bortezomib can inhibit angiogenesis by influencing the endothelium directly. In addition, we successfully identified a dosage of bortezomib that did not result in fatal toxicity for HUVECs while inhibiting the growth of lumen like structures from the intact HUVECs monolayer. The proposed QMAS allows experiments in a more biologically relevant microenvironment that represents a compromise between mimicking all the complexity of the in vivo condition and current in vitro 2D-based screening systems. Thus, QMAS provides quantitative information on the intricacies of cellular behaviors using a less labor intensive, time-saving method as compared to previously developed in vitro angiogenesis assays. Recently, much effort is being directed towards the design of cancer treatment by the combination of anti-angiogenesis drugs and conventional anti-cancer drugs. The present QMAS lends itself to assays for combination therapies to search for synergistic effects, and studies are underway with combination therapies in our group.
We acknowledge that there could be several outstanding issues, which must be overcome in subsequent designs. These include (1) establishing a longer-term culture in the model; (2) conducting a detailed study comparing the QMAS and animal experimental results; (3) increasing the number of screening locations; and (4) testing various cell types. Nevertheless, this assay can provide a representative tissue specific responses; it can be used throughout early drug discovery, from target identification and validation to primary screening, lead identification and optimization, and safety and toxicology screening.
Materials and Methods
Cell Seeding & perfusion culture
Human umbilical vein endothelial cells (HUVECs) (Lonza, NJ, USA) or RFP-expressing HUVECs (Angio-Proteomie; Boston, MA, USA) were seeded and cultured in the cell culture channels in direct contact with collagen hydrogel scaffolds, prepared by diluting a 4.0-mg/ml solution of type I collagen (BD Biosciences, MA, USA) to 2.5 mg/ml and polymerizing at an initial pH of 7.4. The cell culture channels were filled with HUVEC suspensions prepared in medium at a density of 2 × 106 cells/ml. After forming an endothelial cell monolayer, the reservoirs were added and each conditioned medium was injected using a multi-tipped pipette. Flow was then generated through the microfluidic channels by an external pump (the withdrawal flow rate=1.33 μl/min, Microsyringe pump, HARVARD, USA) and continued throughout the experiment in order to maintain a stable concentration profile across the 3D hydrogel scaffolds. The device was then placed in a 37°C incubator for 5 days. Subsequent steps are shown in Supplementary Movie 1.
Preparation of the microfluidic device
Chips were fabricated using an SU-8 fabrication method and replica molding. An SU-8 (Microchem, Newton, MA, USA) photoresist was used for the development of SU-8 microstructures. Both the fabricated PDMS device and glass cover slip were autoclaved and dried at 80°C overnight. They were then plasma treated (PDC-001, Harrick, CA, USA) in air, and bonded together to form a closed microfluidic channel. After recovering hydrophobic surface characteristics in the gel filling region, scaffold material was introduced into fourteen gel regions to form a gel scaffold using the manifold layer. In these experiments, type I collagen (BD Biosciences, MA, USA) was used as the scaffold material and was gelled by incubating for 30 minutes at 37°C. Gel stiffness was varied by adjusting the solution pH; we used pre-polymerized collagen solutions (2.5mg/ml) of pH 7.4. Following gelation, fibronectin (dilution ratio=1:10, Sigma–Aldrich) was introduced into all microfluidic channels to improve the cell attachment, and after treatment for 1 day, fresh cell culture medium was injected and the device was placed in an incubator (37°C) for cell seeding.
Fixation, imaging, and quantification of cell migration
Image J software (http://rsbweb.nih.gov/ij/) was used to measure % of projected areas and number (#) of migrated cells. The F-actin distribution and number of cells involved in “capillary -like” networks or tube structures were assessed after 5 days culture in the device. Cells were fixed with 4% paraformaldehyde (Sigma–Aldrich) for 15 min at room temperature. During fixation, the pressure head and flow were maintained in experimental devices to preserve protein expression and activation. After fixation with 4.0% PFA, actin filaments and nuclei were then stained with rhodamine-phalloidin (Sigma–Aldrich) and 4′6-diamidino-2-phenylindole (DAPI; Sigma–Aldrich), respectively. Fluorescent images were obtained using a confocal microscope ((OLYMPUS, FLUOVIEW FV 1000, Tokyo, Japan).
Analysis of gaps and disruption of VE-cadherin
Disruption of VE-cadherin was identified from the analysis of discontinuity of green fluorescence at VE-cadherin junctions between HUVECs. Gap area within the disrupted VE-cadherin junctions was determined from six images. Gap area was quantified as the ratio of pixels within all the gaps and total number of pixels in one image. The average percentage of endothelial gap was calculated using Image J software.
HUVEC proliferation and cell viability assays
HUVECs were treated with various concentrations of bortezomib in the model for the indicated periods of time. Ki-67, a common proliferation marker, was labeled with rabbit polyclonal antibody (Abcam) at 1:500 dilutions to analyze HUVEC proliferation ratio. Cell viability was tested using the LIVE/DEAD reduced-biohazard viability/cytotoxicity test (Molecular Probes, Invitrogen, Cergy Pontoise, France). Staining and analysis were performed as recommended by the manufacturer. Cell number was determined using ImageJ. All experiments were repeated at least twice at different times.
Statistical analysis
Numerical values were expressed as means ± SDs of more than three independent experiments. Statistical significance was determined via one-way analysis of variance (ANOVA). Paired Student’s t-tests were conducted to compare means when the ANOVAs indicated a significant difference. Statements of significance were based on P-values < 0.01.
Supplementary Material
Acknowledgments
This research was supported in part by the US National Institutes of Health (R33 CA174550) and the National Science Foundation (CBET-0939511); Seok Chung was supported by the Seoul R&BD Program (PA090930). This work was also supported by the 2013 Kyungil University Research Grant.
References
- 1.Takei J. J AATEX. 2006;11:170–176. [Google Scholar]
- 2.Fang C, Avis I, Salomon D, Cuttitta F. J Cancer. 2013;4:402–415. doi: 10.7150/jca.6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horning JL, Sahoo SK, Vijayaraghavalu S, Dimitrijevic S, Vasir JK, Jain TK. Mol Pharm. 2008;5:849–862. doi: 10.1021/mp800047v. [DOI] [PubMed] [Google Scholar]
- 4.Takara K, Sakaeda T, Yagami T, Kobayashi H, Ohmoto N, Horinouchi M, Nishiguchi K, Okumura K. Biol Pharm Bull. 2002;25:771–778. doi: 10.1248/bpb.25.771. [DOI] [PubMed] [Google Scholar]
- 5.Yamori T, Matsunaga A, Sato S, Yamazaki K, Komi A, Ishizu K, Mita I, Edatsugi H, Matsuba Y, Takezawa K, Nakanishi O, Kohno H, Nakajima Y, Komatsu H, Andoh T, Tsuruo T. Cancer Res. 1999;59:4042–4049. [PubMed] [Google Scholar]
- 6.Pampaloni F, Reynaud EG, Stelzer EH. Nat Rev Mol Cell Biol. 2007;8:839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 7.Kobuke K, Piccolo F, Garringer KW, Moore SA, Sweezer E, Yang B, Campbell KP. Human Molecular Genetics. 2008;17:1201–1213. doi: 10.1093/hmg/ddn009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackam DG, Redelmeier DA. Journals JAMA. 2006;296:1731–1732. doi: 10.1001/jama.296.14.1731. [DOI] [PubMed] [Google Scholar]
- 9.Wlodkowic D, Darzynkiewicz Z. World J Clin Onclo. 2010;1:18–23. doi: 10.5306/wjco.v1.i1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huh D, Hamilton GA, Ingber DE. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen MC, Gupta M, Cheung KC. Biomed Microdevices. 2010;12:647–654. doi: 10.1007/s10544-010-9417-2. [DOI] [PubMed] [Google Scholar]
- 12.Zang R, Li D, Tang IC, Wang J, Yang ST. IJBWI. 2012;1:31–51. [Google Scholar]
- 13.Zhao L, Caot JT, Wu ZQ, Li JX, Zhu JJ. J Biomed Nanotechnol. 2013;9(3):348–356. doi: 10.1166/jbn.2013.1546. [DOI] [PubMed] [Google Scholar]
- 14.Wu LY, Di Carlo D, Lee LP. Biomed Microdevices. 2008;10:197–202. doi: 10.1007/s10544-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 15.Kim C, Bang JH, Kim YE, Lee SH, Kang JY. Lab Chip. 2012;12:4135–4142. doi: 10.1039/c2lc40570a. [DOI] [PubMed] [Google Scholar]
- 16.Kubota Y. J Med. 2012;61:47–56. doi: 10.2302/kjm.61.47. [DOI] [PubMed] [Google Scholar]
- 17.Wu HC, Huang CT, Chang DK. J Cancer Mol. 2008;4:37–45. [Google Scholar]
- 18.Samant RS, Shevde LA. Oncotarget. 2011;2:122–134. doi: 10.18632/oncotarget.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tandle A, Blazer DG, Libutti SK. J Transl Med. 2004;2:1–20. doi: 10.1186/1479-5876-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon N, Baskaran H, Dertinger S, Whitesides G, Van De Water L, Toner M. Nature Biotechnol. 2002;20:826–830. doi: 10.1038/nbt712. [DOI] [PubMed] [Google Scholar]
- 21.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Nature materials. 2007;6:908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 22.Sudo R, Chung S, Zervantonakis IK, Vicherman V, Toshimitsu Y, Griffith LG, Kamm RD. FASEB J. 2009;23:2155–2164. doi: 10.1096/fj.08-122820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kothapalli CR, van Veen E, de Valence S, Chung S, Zervantonakis IK, Gertler FB, Kamm RD. Lab Chip. 2011;11:497–507. doi: 10.1039/c0lc00240b. [DOI] [PubMed] [Google Scholar]
- 24.Vickerman V, Blundo J, Chung S, Kamm RD. Lab Chip. 2008;8:1468–1477. doi: 10.1039/b802395f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung S, Sudo R, Mack PJ, Wan CR, Vickerman V, Kamm RD. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 26.Polacheck WJ, Charest JL, Kamm RD. Proc Natl Acad Sci. 2011;108:11115–11120. doi: 10.1073/pnas.1103581108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung S, Sudo R, Zervantonakis IK, Rimchala T, Kamm RD. Adv Matter. 2009;21:4863–4867. doi: 10.1002/adma.200901727. [DOI] [PubMed] [Google Scholar]
- 28.Kim C, Chung S, Yuchun L, Kim MC, Chan JK, Asada HH, Kamm RD. Lab Chip. 2012;12:2942–2950. doi: 10.1039/c2lc40182g. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen DH, Stapleton SC, Yang MT, Cha SS, Choi CK, Galie PA, Chen CS. Proc Natl Acad Sci. 2013;110:6712–6717. doi: 10.1073/pnas.1221526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, Kermani P, Hempstead B, Fischbach-Teschl C, López JA, Stroock AD. Proc Natl Acad Sci. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen MB, Srigunapalan S, Wheeler AR, Simmons CA. Lab Chip. 2013;13:2591–2598. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi T, Hideshima T, Anderson KC. Br J Haematol. 2003;120:10–17. doi: 10.1046/j.1365-2141.2003.03929.x. [DOI] [PubMed] [Google Scholar]
- 33.Hideshima T, Chauhan D, Podar K, Schlossman RL, Richardson P, Anderson KC. Semin Oncol. 2001;28:607–612. doi: 10.1016/s0093-7754(01)90033-8. [DOI] [PubMed] [Google Scholar]
- 34.Belloni D, Veschini L, Foglieni C, Dell’Antonio G, Caligaris-Cappio F, Ferrarini M, Ferrero E. Exp Cell Res. 2010;316:1010–1018. doi: 10.1016/j.yexcr.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Scholzen T, Gerdes J. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 36.Urruticoechea A, Smith IE, Dowsett M. J Clin Oncol. 2005;23:7212–7220. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- 37.Johannessen AL, Torp SH. Pathol Oncol Res. 2006;12:143–147. doi: 10.1007/BF02893360. [DOI] [PubMed] [Google Scholar]
- 38.Lamalice L, Boeuf FL, Jacques H. Circulation Research. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.