Abstract
Although DNA encodes the molecular instructions that underlie control of cell function, it is the proteins that are primarily responsible for implementing those instructions. Therefore, quantitative analyses of the proteome would be expected to yield insights into important candidates for the detection and treatment of disease. We present an iTRAQ (Isobaric Tagging for Relative and Absolute Quantification)-based proteomic analysis of 10 ovarian cancer cell lines and 2 normal ovarian surface epithelial cell lines. We profiled the abundance of 2659 cellular proteins, of which 1273 were common to all 12 cell lines. Of the 1273, 75 proteins exhibited elevated expression, and 164 proteins had diminished expression in the cancerous cells compared to the normal cell lines. The iTRAQ expression profiles allowed us to segregate cell lines based upon sensitivity and resistance to carboplatin. Importantly, we observed no substantial correlation between protein abundance and RNA expression or epigenetic, DNA methylation data. Furthermore, we could not discriminate between sensitivity and resistance to carboplatin on the basis of RNA expression and DNA methylation data alone. This study illustrates the importance of proteomics-based discovery for defining the basis for the carboplatin response in ovarian cancer and highlights candidate proteins, particularly involved in cellular redox regulation, homologous recombination and DNA damage repair, that otherwise could not have been predicted from whole genome and expression data sources alone.
Keywords: iTRAQ, mRNA expression, DNA methylation, carboplatin, resistance
INTRODUCTION
The American Cancer Society projects there will be approximately 21,980 new cases of ovarian cancer in the United States in 2014. Of those, an estimated 14,270 will succumb to the disease. One of the critical steps for treatment of ovarian cancer is identifying those patients that will respond positively to therapy. A majority of the patients that respond initially to a primary treatment of surgery and chemotherapy typically suffer recurrence with a drug-resistant phenotype [1]. Consequently, there is a diagnostic need for both clinical and molecular features, or biomarkers, that can be associated with early detection, survival and disease recurrence. Although significant progress has been made in developing technologies that can simultaneously measure thousands of molecular features in a given patient sample, statistical methods to analyze this flood of data need to be carefully chosen in order to identify biomarkers that are more likely to be validated and thus achieve clinical utility [2]. Additionally, experimental and biological limitations in interpreting transcriptional activity in the context of abundance of cellular proteins have produced few functional genomic and prognostic commonalties from an abundance of ovarian cancer gene expression studies [3, 4].
Proteomic analysis, to define the complement and abundance of cellular proteins, facilitates the identification of pertinent biological processes and the definition of molecular targets that contribute to a disease state. Presently, proteomic analysis of ovarian cancer is in a challenging but promising developmental phase that is aimed at identifying successful strategies for early detection and discovering potential biomarkers of chemosensitivity [5]. In general, proteomic approaches have become an invaluable tool for the discovery of functional associations with oncogenic signaling pathways and changes in protein abundance and modification in the disease state [6, 7]. In ovarian cancer, measurements of cellular protein levels by mass spectrometry have been employed on cell line, tumor and serum samples to identify candidate biomarkers for disease progression, histological subtypes and proteins associated with carboplatin resistance [8-12]. Nevertheless, very few of these studies have been able to quantify protein expression in multiple cancer cell lines or tissue samples. Furthermore, the critical issue of the extent to which changes in protein expression coincide with RNA expression or epigenetic variation within ovarian cancer remains to be defined.
We have employed the Isobaric Tagging for Relative and Absolute Quantification (iTRAQ) approach to examine chemosensitivity in cell models of ovarian cancer. The iTRAQ isobaric chemical labeling technology is often used in discovery MS-based proteomics because it allows multiple points of comparison, initially four, but more recently eight different conditions [13]. The iTRAQ reagents are chemical tags that modify the N-terminus (and the ε-amino group of lysine) of peptides generated by proteolytic digestion of the protein sample. These tags comprise a reporter group of variable Mr (113-119 and 121 in the 8-plex system) and a balancer group that ensures the same total mass (305 Da for the 8-plex set) for each tag. Tryptic peptide samples from various experimental conditions, as many as eight, are labeled separately with an iTRAQ tag and the samples are then combined. The mixture is fractionated by ion exchange chromatography or isoelectric focusing to reduce the complexity of the samples, and each fraction is then subjected to reverse phase nano LC-MS/MS. In MS mode, the multiplexed labeling of any given peptide results in an identical precursor mass; however, following fragmentation in MS/MS mode, each tag releases a unique reporter ion, the intensities of which are used for relative/absolute peptide quantitation. An advantage of this approach is that the multiple samples are analyzed simultaneously. Furthermore, the b- and y-ions derived from the peptides labeled with the iTRAQ tags remain isobaric, resulting in greater signal intensity in the MS/MS spectra.
Here, we performed iTRAQ analysis to quantify protein abundance in 10 ovarian cancer cell lines and 2 normal ovarian surface epithelial cell lines. This was integrated with analysis of DNA methylation and RNA expression data for each gene product identified by iTRAQ. There are two major conclusions of the study. First, we observed no significant correlation between cellular protein content and mRNA expression or methylation data. Therefore, the extent of mRNA expression was not a direct indicator of the extent of protein expression in these ovarian cancer cell models. This illustrates that reliance exclusively on global genomic measurements may miss important features of the disease phenotype. Second, although we observed no correlation between carboplatin resistance and either RNA expression or DNA methylation, we did observe a protein signature that directly correlated with carboplatin resistance. Furthermore, our analysis by iTRAQ revealed a potential role of several redox regulatory proteins and homologous recombination repair (HRR) pathway components in carboplatin resistance.
EXPERIMENTAL METHODS
Cell Culture and RNAi
Ovarian carcinoma-derived cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 μg/ml). Normal human ovarian surface epithelium (HOSE) 11-12 and 6-3 control cell lines were grown in 199: MCDB 105 (1:1) medium containing 10% FBS. Cells were maintained at 37°C in an atmosphere of 5% CO2. Additional information about all of the ovarian cell lines used in this project is presented in Supplementary Table S1.
siRNAs targeting catalase and TXNDC5 were purchased from Sigma: catalase siRNA1-3: SASI_Hs02_00092507, 00092508, 00332471; TXNDC5 siRNA1-3: SASI_Hs02_00171689, 00171690, 00355463. siRNA was delivered into CAOV1 ovarian cancer cells by AMAXA electroporation (Kit T, Program T-016, Lonza).
iTRAQ (isobaric tag for relative and absolute quantitation) Analysis
Ovarian cancer cells were lysed in a buffer containing 8M urea, 50mM Triethylammonium bicarbonate (TEAB, pH 8.5) and 0.05% w/v ProteaseMax (Promega) supplemented with phosphatase and protease inhibitor cocktails (P2850, P5726, P8340, Sigma). Protein concentrations were determined by the BCA assay (Pierce). A universal standard was created by pooling 100 μg protein samples from each of the 12 cell lines. This universal standard (100 μg) was incorporated in each 8-plex iTRAQ experiments to enable robust quantitation between experiments. Aliquots of 100 μg total protein from each sample were reduced, alkylated, and then precipitated by a methanol/chloroform precipitation method. Protein pellets were reconstituted in 50 μl of 6M urea/50mM TEAB with sonication [14] and digested by 2% trypsin overnight. iTRAQ reagents (8-plex iTRAQ kit, Sciex) were reconstituted in ~80 μl of isopropanol and incubated with designated tryptic peptide samples at 37°C for 2 hrs with mild agitation. Peptides were desalted on a C18 Sep-Pak cartridge (Waters) and fractionated by high resolution (24-fractions) OFFGEL fractionation [15]. Peptide samples from each fraction were analyzed on a Proxeon Nano LC system coupled to a LTQ-Orbitrap XL mass spectrometer (Thermo) with an 80 minute gradient at a flow rate of 300 nl/min. Survey full-scan spectra were acquired with resolution of 15,000 and mass range from 400-1800 m/z. The top 4 most intense ions were selected for alternated HCD and CID scans. Protein identification and quantification was carried out with Mascot 2.3 (Matrix science) against the human IPI database (87,040 sequences). Methylthiolation of cysteine, N-terminal and lysine 8-plex iTRAQ modifications were set as fixed modifications, methionine oxidation as a variable modification and one missed cleavage allowance. Peptide mass tolerance was set at 20 ppm, with 0.6 Da for fragment ions (decoy database false discovery rate < 2%). iTRAQ ratios were calculated as intensity weighted, using only peptides with expectation values < 0.05 [15].
Analysis of gene expression and detection of methylation in human ovarian cancer cells
We used the Affymetrix Human Genome U133A array: GEO platform identifier GPL96. RNA was isolated using the trizol protocol, converted into cDNA and the double-stranded cDNA was used as the template in an in-vitro transcription reaction containing biotinylated CTP and UTP in addition to the four unmodified ribonucleoside triphosphates. The standard Affymetrix protocol was applied. Final signal intensities were processed using the RMA normalization method in the Affy package of R Bioconductor 2.5.
The protocol for Methylation detection representational Oligonucleotide Microarray Analysis (MOMA) was performed as previously described [16, 17]. Annotated genomic CpG island locations were obtained from the UCSC genome browser. At the time of the experiment the genome contained 26,219 CpG islands in the range of 200–2000 bp. These CpG island locations were covered by MspI restriction fragmentation. Arrays were manufactured by Nimblegen Systems Inc. using the 390,000 probes format. The CpG island annotation from the human genome build 33 (hg17) was used to design a 50-mer tiling array. The primary restriction endonuclease used was MspI. After the digestion linkers were ligated and the material cleaned by phenol chloroform, precipitated, centrifuged, and resuspended. The material was divided in two, half being digested by the endonuclease McrBc according to specification by New England Biolabs and the other half being mock digested. Procedures for hybridization and washing were reported previously [16]. The procedure was performed in duplicate with a dye-swap for the second experiment. The labels were swapped between the McrBc treated and mock samples. For each probe, the geometric mean of the ratios (GeoMeanRatio) of McrBc treated and control samples were then calculated per experiment and its associated dye swap. Microarray images were scanned on GenePix 4000B scanner and data extracted using Nimblescan software (Nimblegen Systems Inc). The GeoMeanRatios of all the samples in a data set were then normalized using a quantile normalization method [18]. All general analysis and statistics were computed using S-plus, R packages and individual Perl/Python scripts.
Cell Viability Assay
Each cell line (7 epithelial ovarian cancer cell lines and 1 control cell line) was seeded at 103 cells/well in a 96-well plate and treated with carboplatin at a concentration range of 0-100 μg/ml in 100 μl of fully supplemented DMEM. After a period of 72 hrs proliferation was assessed by MTT (3-(4.5-dimethylthiazol-2-yl)-2.5-diphenyltetrazolium bromide) assay and absorbance measured at 595nm using the Wallac microplate reader (Perkin Elmer). Absorbance was converted to the percentage of cells surviving and plotted against the concentration to calculate the IC50.
CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used to evaluate the role of Catalase and TXNDC5 in carboplatin resistance. In summary, siRNA was delivered into CAOV1 ovarian cancer cells by AMAXA electroporation (Kit T, Program T-016, Lonza). Twenty-four hours later, cells were trypsinized, re-seeded in a 96-well plate at 2 × 103 cells/well and grown for another 24 hrs. Cells then were treated with carboplatin at a concentration range of 0-100 μg/ml in 200 μl of fully supplemented DMEM. After 24 and 48 hrs, 20 μl of 1/10 diluted CellTiter-Glo reagent was added to each well, mixed for ~15 minutes on an orbital shaker to induce cell lysis, and followed by luminescence reading.
Immunoblotting Analysis
Ovarian cancer cell extracts were prepared in RIPA lysis buffer (50 mM Tris HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing 50 mM sodium fluoride, 1 mM sodium orthovanadate and 1X complete protease inhibitor cocktail (Roche). Total protein concentration was determined by the method of Bradford. Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membrane. Membranes were blocked in 5% BSA in TBST (20mM Tris-HCl pH 7.6, 136 mM NaCl, 0.1% Tween-20) and incubated at 4°C overnight with primary antibodies against RPA1, RPA2, MCM2 (kind gifts from Bruce Stillman Lab), MCM5 (Bethyl Laboratory), catalase (Cell Signaling), TXNDC5 (R&D Systems) and β-Tubulin (Sigma). Proteins were detected with HRP-conjugated secondary antibodies (Jackson Lab) and enhanced chemiluminescence (ECL; Pierce).
Bioinformatic Analysis
All analysis was performed using a combination of Perl/Python scripts and R statistical functions. We used either iTRAQ raw values or iTRAQ log(2) values where applicable. A normal distribution of iTRAQ log(2) data per cell line was applied for the determination of abundance threshold values based on estimated variance. Pearson correlation calculations and unsupervised cluster analysis was performed via R and the gplots library was implemented. Pathway enrichment analysis was performed as described previously [19]. Large datasets are provided in Supplementary Tables 2-7.
RESULTS
Quantitative proteomic analysis of control and ovarian cancer cell lines by iTRAQ
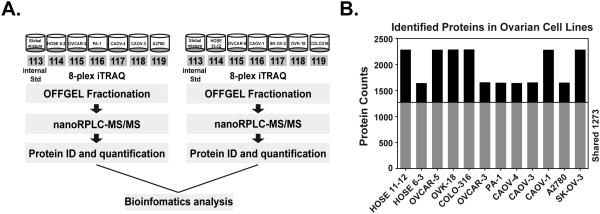
We performed an 8-plex iTRAQ analysis to evaluate systematically and quantitatively the change in protein expression profile between normal ovarian epithelial control cells and carcinoma-derived cell lines. To allow comparison between multiple runs, an internal control was developed, which was composed of an equal mixture of all the samples. The results of this sample were used to normalize the multiple runs. Protein samples from each cell line and from the internal control were analyzed on a Proxeon Nano LC system coupled to a LTQ-Orbitrap XL mass spectrometer. The flow chart of the analysis was illustrated in Fig 1A. To optimize the experimental conditions and test data reproducibility, a pilot experiment was performed involving HOSE 6-3, OVCAR-3, PA-1, CAOV-4, CAOV-3 and A2780 cell lines. Among 881 proteins identified in common between the pilot and final experiment for these 6 cell lines, we observed consistently high correlation (Fig S1: Spearman Correlation > 0.71; Pearson correlation > 0.76). In the final study of 12 cell lines, we identified 87,040 peptides, which represented 3099 proteins. Further protein identification and quantitation with Mascot identified 2657 proteins, of which 1273 were found in all 10 ovarian carcinoma-derived cell lines and the 2 control lines that were examined (Fig 1B and Table S2&3). The number of proteins captured by iTRAQ per cell line ranged from 1642 to 2289.
Figure 1. Quantitative proteomic analysis of ovarian control and cancer cell lines.
(A) Experimental strategy for iTRAQ-based quantitative proteomics analysis. (B) Identification of shared ovarian proteome and distinct proteins from each cell line.
Analysis of variability in iTRAQ values among ovarian epithelial cell lines
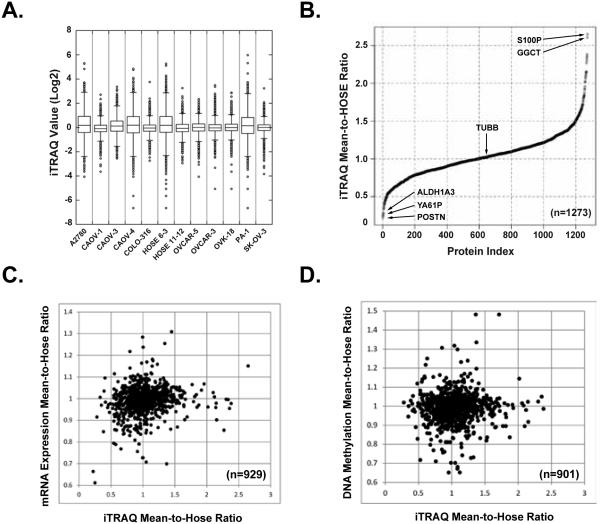
We examined the distribution of protein expression among all the cell lines to define the scope of the changes we observed and to demonstrate the legitimacy of the dataset. In Fig 2A, we present a box plot of the raw iTRAQ values to illustrate the variation between the internal control and each cell line and the range of iTRAQ measurements. The iTRAQ log(2) values of 50% of the proteins identified per cell line were between −1 and 1 (within the box, Fig 2A), and the median varied little among all cell lines. This indicated that there was no substantial change in ~50% of the proteins detected in each cell line. When this was extended from 1 to 2 standard deviations away from the mean (within 25-75 percentile, Fig 2A), we captured >98% of the total protein detected. In each cell line, there was a small number of proteins that displayed a greater magnitude of change; however, these were not consistent across cell lines.
Figure 2. Variability analysis of iTRAQ values among ovarian cell lines.
(A) A box plot of the raw iTRAQ values to illustrate the variation between cell lines and the range of iTRAQ measurements. The distribution range of iTRAQ measurement per protein identified is presented for 10 ovarian cancer cell lines and 2 normal ovarian surface epithelial cell lines. The log2 value of each iTRAQ measurement was used to determine protein abundance per cell line. (B) Variability comparison in expression levels of individual protein between 10 ovarian cancer cell lines and 2 normal control cell lines. Waterfall plot was employed to summarize the “mean-hose-ratio” for each of the 1273 common proteins. (C) Correlations of iTRAQ “mean-hose-ratio” with mRNA expression. (D) Correlations of iTRAQ “mean-hose-ratio” with DNA methylation.
We compared the variability in expression of individual proteins between 10 ovarian cancer cell lines and 2 normal controls (HOSE 6-3 & Hose11-12). We identified 1273 proteins in common among all the cell lines and determined the “mean-hose-ratio” for each individual protein, which was the average iTRAQ value from the 10 ovarian cancer cell lines divided by the average iTRAQ value of the 2 normal control cell lines (Table S4). TUBB (tubulin beta chain) served as a control in this analysis, with a mean-hose-ratio of 1.01. Among the 1273 proteins, a total of 75 proteins displayed a mean-hose-ratio greater than 1.5, whereas there were 164 proteins with a mean-hose-ratio less than 0.75. The distribution of the ratios is summarized in Fig. 2B and all the data are presented in Table S3-4. Furthermore, we applied the same “mean-hose-ratio” algorithm to mRNA expression and DNA methylation data for these cell lines (Table S4), and calculated the correlation between iTRAQ and mRNA expression, or between iTRAQ and DNA methylation. Interestingly, there was no overall correlation of iTRAQ protein expression with either mRNA expression (Fig 2C) or DNA methylation (Fig 2D)
Use of unsupervised clustering of iTRAQ ratios to segregate cells according to sensitivity to carboplatin.
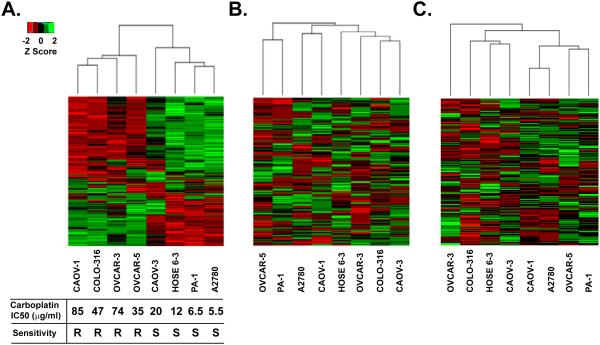
We chose 8 out of the 12 ovarian cell lines, 7 cancer and 1 control, for which we had mRNA expression and DNA methylation data in addition to the results of the iTRAQ analysis. Using median absolute deviation (MAD) filtering analysis, we calculated the extent to which expression of a particular protein in a given cell, measured by the iTRAQ score, varied relative to the median level of expression of that same protein in all of the cell lines tested. We performed this analysis on Z-scores obtained from the iTRAQ log(2) ratio data for 1279 proteins among each of the 8 cell lines (Table S5). The 300 most variable proteins (Table S6) were selected for unsupervised hierarchical clustering analysis using a Euclidean distance complete method. Interestingly, using a heat map to illustrate the expression level of each protein for each cell line (Fig 3A), we were able to place these proteins into two classes. Accordingly, we were able to segregate the 8 cell lines into two major clusters, which, unexpectedly, coincided with the cellular response to carboplatin. Cluster 1 comprised those cell lines with an IC50 in the range 35 - 85 μg/ml, which we categorized as the “carboplatin-resistant group”. This included cell lines CAOV-1, COLO-316, OVCAR-3, and OVCAR-5. Cluster 2, the “carboplatin-sensitive group”, comprised cell lines with IC50 lower than 20 μg/ml and contained A2780, PA-1, CAOV-3 and the normal ovarian surface epithelial cell line Hose 6-3. Consistent with the outcome of this clustering analysis, candidates associated with DNA damage repair and redox regulation segregated according to the iTRAQ data as would have been expected between chemoresistant and chemosensitive cell lines (Table S7).
Figure 3. Hierarchical clustering of iTraq z-scores for ovarian cancer cell lines.
(A) The top 300 most variable iTRAQ proteins determined by median absolute deviation were clustered using the Eculidean complete method for the indicated ovarian cancer cell lines. Minimum z-scores are depicted in red and maximum z-scores are in green. Each row represents an individual protein, each column is the sum of the proteins identified in the indicated cell line. Carboplatin survival curves were determined by performing MTT cell viability assay and plotted for the 7 epithelial ovarian cancer cell lines and one control cell line. Absorbance was converted to the percentage of cells surviving and plotted against the concentration to calculate the IC50. Carboplatin-resistant cell lines were labeled “R” and cell lines sensitive to carboplatin treatment were labeled “S”. (B) Unsupervised hierarchical clustering of the same 300 most variable iTRAQ proteins using mRNA expression data. (C) Unsupervised hierarchical clustering of the same 300 most variable iTRAQ proteins using DNA methylation data.
We also examined data from standard Affymetrix arrays to quantify mRNA expression and MOMA methylation arrays to assess DNA methylation for the MAD-filtered iTRAQ proteins. It is important to note that in contrast to data on protein expression, we could not generate a similar segregation pattern when using either MAD-filtered mRNA expression data (Fig 3B) or DNA methylation data (Fig 3C) for the same top 300 protein candidates. Furthermore, when a similar unsupervised clustering analysis was performed solely on mRNA expression and DNA methylation data for all of the candidate proteins identified in the iTRAQ experiment in these cell lines, neither data set produced significant clusters nor did they define chemosensitive cellular subtypes.
Protein abundance did not correlate with mRNA expression or DNA methylation.
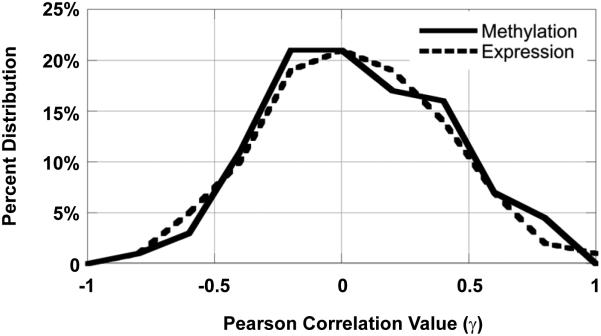
In light of the fact that we were able to classify cell lines as either carboplatin-sensitive or carboplatin-resistant only on the basis of the protein signature, we examined further whether there were correlations between protein expression and mRNA expression, as well as protein expression and DNA methylation. We assigned a positive correlation to decreased DNA methylation and increased iTRAQ values since decreased promoter methylation corresponds biologically to an increase in gene expression and therefore presumably increased protein abundance. The normal distributions for correlation of either iTRAQ-DNA methylation or iTRAQ-RNA expression show that protein level was poorly correlated with either mRNA expression or DNA methylation (Fig. 4). A positive correlation of r ≥ 0.5 between mRNA expression and cellular protein concentration was observed only in 11% of the proteins; conversely even an anti-correlation r ≤ −0.5 was observed in 7.5% of the proteins sampled. These results show that there were roughly equal numbers of genes for which gene expression correlated with protein level (high gene expression and high protein level or low gene expression and low protein expression) as there were genes for which expression did not correlate with protein level (high gene expression and low protein or low gene expression and high protein level). Similar results were observed for DNA methylation.
Figure 4. Correlation of iTRAQ Data with RNA Expression and DNA Methylation.
The correlation distribution of RNA expression (dashed line) and DNA methylation (solid line) with iTraq data is shown at r = 0.2 binned increments. Note: a positive correlation for DNA methylation and iTraq values signifies hypomethylation with increased iTraq protein value.
Validation of differentially expressed proteins
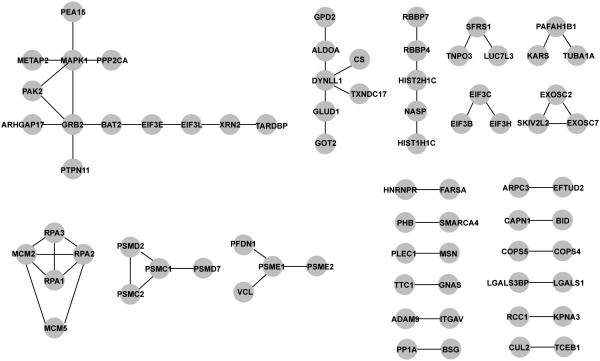
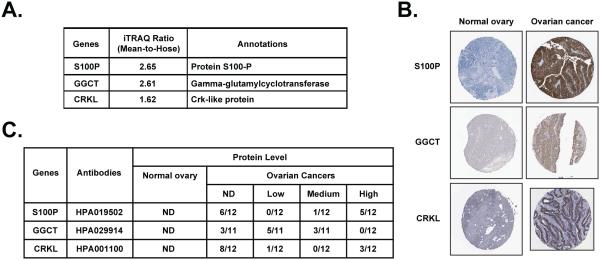
The 300 most variable proteins, through which we generated signatures that distinguished between carboplatin-resistant and -sensitive cell lines, were filtered through the Human Protein Reference Database (HPRD, Release 9) to illustrate potential protein-protein interactions. This analysis highlighted 74 of the 300 proteins, with 59 interactions among them, including 12 single edge (pairs) interactions, 8 multi-protein complex interactions and two larger protein interaction networks of 6 and 13 edges (Fig 5). Of particular note, we identified the RPA-MCM complex; correlation between the expression of RPA (replication protein A), a single-stranded DNA binding protein required for recombination and DNA repair, and the MCM2 and MCM5 mini-chromosome maintenance complex components has been reported and implicated as a predictor of survival in serous ovarian carcinomas [20]. In addition, we performed pathway enrichment analysis for these 300 proteins using the 169 pathways available at the time of analysis in the KEGG database. The most significant of these were hsa04540-Gap Junction (p=0.0068) and hsa03440-Homologous recombination (p=0.0146), both of which have been implicated in sensitivity to chemotherapeutic agents, such as carboplatin. Finally, we searched the Human Protein Atlas to compare the protein expression levels of the top candidates from our analysis between normal ovary and malignant ovarian carcinomas. We observed higher expression of S100P, GGCT and CRKL in human ovarian cancer samples compared to normal ovary, in which these proteins were undetectable (Fig 6).
Figure 5. Proteomics-derived ovarian cancer modules.
Protein-protein interactions of the 300 MAD filtered iTRAQ proteins examined through the HPRD (Release 9) data base.
Figure 6. Expression of S100P, GGCT and CRKL in normal ovary and ovarian carcinoma tissue samples.
(A) “Mean-hose-ratio” of S100P, GGCT and CRKL from iTRAQ. (B) Representative tissue microarray cores showing high expression of S100P, GGCT and CRKL in malignant ovarian carcinoma sections compared to normal ovary. Data were adapted from The Human Protein Atlas. (C) Summary of the immunohistochemistry result for all three proteins.
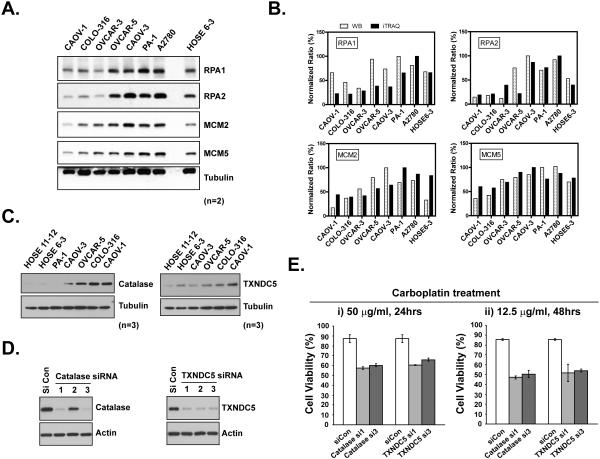
We examined the DNA damage response proteins RPA1, RPA2, MCM2 and MCM5 to validate further the differential protein expression between carboplatin-sensitive and carboplatin-resistant ovarian cancer cell lines. Carboplatin-resistant cell lines (CAOV-1, COLO316 and OVCAR-3) displayed low expression level of all the proteins tested compared to the carboplatin-sensitive cell lines (A2780, PA-1, CAOV-3 and normal control HOSE 6-3) (Fig 7A). An exception is OVCAR-5 cell line, which, based on protein expression analysis, would have been predicted to behave like the sensitive cell lines; however, it displayed a low carboplatin IC50 (35 μg/μl) compared to the other cells in the resistant class. We quantified the expression of these proteins in all the cells tested, by immunoblotting using β-tubulin as an internal normalization control, and as shown in Fig 7B, we observed a similar expression trend to that revealed by iTRAQ analysis.
Figure 7. Biochemical validation of the role of iTRAQ candidate proteins in carboplatin chemosensitivity.
(A) Cells were lysed in SDS-containing RIPA buffer and protein expression levels were measured by immunoblotting with the indicated antibodies against RPA1, RPA2, MCM2, MCM5 and β-Tubulin, used as a normalization control. Results have been verified in duplicate (n=2). (B) Results from (A) were quantitated with program ImageJ and the relative ratio after normalization was compared in parallel with iTRAQ value per gene per cell line. (C) Analysis of the expression of catalase and TXNDC5 by immunoblotting with indicated antibodies. Results have been verified in triplicate (n=3). (D) Control and target gene siRNA were electroporated into CAOV1 cells. After 48hrs, knockdown efficiency was demonstrated by immunoblotting. (E) 24hrs after siRNA delivery, CAOV1 cells were re-seeded into 96-well plates. Carboplatin was then added and incubated for indicated time. CellTiter-Glo Luminescent Cell Viability Assay was performed to illustrate the carboplatin response. Results represent mean ± S.D. from three independent experiments.
A mechanism that has been proposed to contribute to carboplatin resistance involves a more reductive environment in the resistant cell, which can detoxify carboplatin-induced reactive oxygen species (ROS) production. Consistently, we detected higher expression of three reductase proteins, namely catalase, TXNDC5 and TXNDC17 in carboplatin-resistant ovarian cancer cell lines (Table S7). The upregulation of catalase and TXNDC5 in ovarian cancer cells compared to normal control, especially the most resistant to carboplatin, was confirmed by immunoblotting (Fig 7C). To evaluate further the importance of these genes in carboplatin resistance, we applied RNAi and selected 3 siRNAs for each gene. As shown in Fig 7D, 2 of the 3 siRNAs against catalase and all 3 siRNAs against TXNDC5 resulted in robust knockdown of the targeted proteins. Furthermore, suppression of these genes in the most resistant CAOV1 cells, with two different siRNAs, consistently increased the sensitivity to carboplatin (Fig 7E), highlighting their potential importance in carboplatin resistance.
DISCUSSION
In order to understand the functional differences between a normal and disease state, it is important to define the identity and quantity of the cellular proteins, the machines that implement the function of the cell. In the context of cancer, such insights have the potential to reveal more precisely the mechanistic determinants of oncogenesis, cancer progression and therapy response within cancer cells than an analysis of the cancer genome alone. We have applied iTRAQ, which is one method of quantitative proteomics that can provide insight into the functional significance of changes encoded in the cancer genome. Here, we have presented a proteomic profiling of 10 ovarian cancer cell lines and 2 normal surface epithelial cell lines. As has been observed in other proteomic studies [21, 22], the patterns of mRNA expression did not correlate with the levels of the cognate proteins. Our analysis extends this further to demonstrate that DNA methylation, as another representation of gene regulation, also did not correlate with cellular protein expression.
Our study already illustrates how proteomic analysis has the potential to contribute insights into cancer etiology. When we compared the variability in expression levels of individual proteins between the panel of ovarian cancer cell lines and the normal controls, we observed that among 1273 proteins in common, 75 were over-expressed >1.5 fold, whereas 164 proteins under-expressed <0.75 fold. The protein that was overexpressed to the greatest extent was S100P, a member of the S100 family of proteins that contains 2 EF-hand calcium-binding motifs (Fig 2A). Consistent with our findings, increased levels of S100P have been reported in multiple tumor cell lines and different tumor types. It has been proposed that S100P may function in tumor growth and metastasis by Ca2+-dependent interaction with several critical regulatory proteins [23]. Interestingly, we observed relatively high protein expression of S100P in our carboplatin-sensitive cell lines, including PA-1 (iTRAQ: 1.974), A2780 (1.462), HOSE 6-3 (1.159), and relatively low expression in carboplatin-resistant cell lines, including COLO-316 (0.181), OVCAR-5 (0.225), CAOV-1 (0.797), which is also consistent with a contribution of S100P to chemosensitivity to carboplatin and paclitaxel in ovarian cancer cells [24]. Additional candidates identified as overexpressed in the ovarian cancer cells in our analysis include GGCT (gamma-glutamycyclotransferase), which has been shown to be a potential biomarker in esophageal squamous tumors [25] and CRKL (Crk-like protein), a known oncogene [26].
Among those proteins shown to be down-regulated, periostin (POSTN) is a ligand for integrins that has been reported to induce attachment and spreading, consistent with a role in cell adhesion [27]. ALDH1A3, a member of the aldehyde dehydrogenase family of detoxifying enzymes, has previously been linked to breast cancer. A lack of expression of ALDH1A3 protein is thought to underlie the impaired production of retinoic acid in several breast cancer cell lines relative to normal breast tissue [28]. Expression of secernin-1 (SCRN1), first characterized in mast cells with a unique function in exocytosis [29], is a novel prognostic biomarker of synovial sarcoma, expression of which is associated with metasis-free survival [30]. We detected expression of secernin-1 in normal control ovarian cell line HOSE6-3 (iTRAQ: 7.624), whereas we observed consistently lower levels in the cancer cell lines. Furthermore, we observed that the levels of secernin-1 were lowest in “carboplatin-resistant” cell lines, including CAOV-1 (1.083) and OVCAR-5 (0.866), compared with the “carboplatin-sensitive” cell lines including A2780 (2.195) and PA-1 (2.547). Consequently, our data suggest that secernin-1 may have potential as a prognostic biomarker in ovarian cancer. Of note, we reported previously that the protein tyrosine phosphatase PTP1B, encoded by the PTPN1 gene, served to inhibit IGF-1R signaling that was essential for ovarian cancer cell proliferation, migration, invasion and anchorage independent growth [31]. In fact, we observed down-regulation of PTP1B (mean-hose-ratio: 0.82) among all the ovarian carcinoma-derived cell lines tested, compared with HOSE 11-12 & 6-3, consistent with enhanced IGF-1R signaling in the cancer cells.
Using an ICAT-based quantitative proteomic technique, Stewart and co-workers published an analysis of proteins associated with cisplatin resistance in ovarian cancer cells [21]. Compared to their work, our studies have extended the scope of the cell lines analyzed and increased the number of proteins captured in our iTRAQ-based quantitative analysis. However, perhaps the most striking results of our analysis were the observation that quantitative proteomics can be used to distinguish cells on the basis of their sensitivity to carboplatin treatment and the fact that such a “protein signature” could not be discerned from analysis of either DNA methylation of mRNA expression. This reinforces our position that quantitative analyses of the proteome have the potential to provide crucial insights that will help us to understand the molecular mechanism of chemoresistance.
The active form of cis-platin is a “bi-aquated” species, in which chloro groups are substituted with water molecules. These activated cis-platin derivatives bind DNA and generate inter-strand crosslinks, which trigger the cytotoxic response. In addition, the hydrated cis-platin interacts with endogenous nucleophiles, including cysteines in such molecules as reduced glutathione, GSH. This has the potential to trigger an oxidative stress condition, which also favors DNA damage [32]. Interestingly, it has been reported that cis-platin resistance correlates with elevated protein expression of GSH, together with the enzyme that catalyzes GSH synthesis, γ-glutamylcysteine synthetase, and the enzyme that mediates the conjugation of carboplatin and GSH, glutathione S-transferase in patient-derived cells [33]. In our studies, we also observed high protein expression levels of glutathione S-transferase (especially glutathione S-transferase omega 1, GSTO1) and glutathione reductase in ovarian cancer cell lines that are resistant to carboplatin (Table S6). Furthermore, we observed that proteins related to thioredoxin, namely thioredoxin domain containing 5 & 17, also displayed elevated expression in carboplatin-resistance ovarian cancer cell lines (Table S7 and Fig 7C). A similar pattern was observed for catalase (Table S7 and Fig 7C). Importantly, we demonstrated that our panel of carboplatin-resistant cells could be sensitized to the effects of the chemotherapeutic agent by RNAi-induced downregulation of either TXNDC5 or catalase (Fig 7D-E), suggesting that an enhanced anti-oxidant environment within the cell may contribute to chemotherapy resistance. The use of cytotoxic gold compounds, which display similar structural features to platinum (II) complexes, has been proposed as one approach to overcoming resistance to platinum-based therapies. It is interesting to note that proteomic profiling of the effects of such gold compounds on A2780 ovarian cancer cells also revealed changes in proteins involved in control of redox homeostasis, for example, thioredoxin-like protein 1 [34] and GSH and thioredoxin reductase 1 (TXNRD1) [35].
Our quantitative analysis also revealed the potential importance of DNA damage repair proteins, including elevated levels of single strand DNA-binding proteins RPA1-3 and MCM2/MCM5 in carboplatin-sensitive cells. Furthermore, RPA plays an essential role in the recruitment, binding and stabilization of BRCA2/RAD51 in DNA damage response [36]. Perhaps an increase in RPA concentration may compete with RAD51 for single strand DNA localization and BRCA2 interaction thereby stalling DNA repair and making the cells sensitive to carboplatin? In addition, our iTRAQ data also highlight other signaling components/pathways that may be crucial to carboplatin resistance. Jab1/CSN5, which is a highly conserved protein complex that is implicated in cell cycle control, apoptosis as well as the DNA damage response and DNA repair, is overexpressed in various cancers including nasopharyngeal carcinoma, where it coincides with poor prognosis. It has been reported that suppression of Jab1/CSN5 induced chemosensitivity in nasopharyngeal carcinoma [37], consistent with our observation that it was elevated in the carboplatin-resistant ovarian cancer cell lines. In addition, we observed that the levels of intergrin-linked kinase (ILK) were also increased in the carboplatin-resistant cancer ovarian cells, consistent with reports that KP-392, a small molecule inhibitor of ILK, exerts a beneficial therapeutic effect in combination with cisplatin in lung cancer [38]. Galectin-1/LGALS1, another protein whose function is involved in cell-cell and cell-matrix interactions, was also elevated in resistant cell groups. Interestingly, aberrant expression of Galectin-1 could promote lung cancer progression and chemoresistance (to cisplatin) by upreguating p38MAPK, ERK and cyclooxygenase-2 [39]. We also detected high expression of calcium-dependent phospholipid binding protein annexin A4 in resistant cells, down-regulation of which by RNAi could sensitize mesothelioma cells to cisplatin [40]. In contrast, we demonstrated that expression of phospholipase A2-activating protein (PLAA), RNA-binding protein RBM3 and Bid, which have been reported to enhance cisplatin-induced apoptosis [41, 42], were lower in the resistant compared to sensitive ovarian cancer cells in our study.
In summary, although current high-throughput genomic strategies focus on the discovery of gene mutations and altered transcription to help identify novel drug targets, here we demonstrate the importance of proteomic analysis to reveal changes in the level of expression of proteins that are critical for therapy response and treatment outcome, changes that would not be revealed by analysis of mRNA expression or methylation data alone. Our study to date highlights the potential significance of proteins that regulate cellular redox status and DNA damage repair, suggesting additional approaches to enhance chemotherapeutic strategies for treatment of patients with ovarian cancer. Overall, our results reinforce the importance of developing further appropriate technologies for quantitative analyses of the proteome to reveal novel candidates for the detection and treatment of disease.
Supplementary Material
Figure S1. Comparison of two sets of iTRAQ measurements among 6 ovarian cancer cell lines.
Both Spearman correlation (S-cor) and Pearson correlation (P-cor) were calculated to evaluate the correlation between two replicates. To minimize clustering of the data points and to facilitate visual examination, iTRAQ measurements were log2-transformed.
Table S1: Background information for 12 ovarian cell lines
Table S2: List of 2657 proteins with iTRAQ value among 2 normal control epithelial ovarian cell lines and 10 epithelial ovarian cancer cell lines.
Table S3: List of 1273 proteins with iTRAQ value found in common among 2 normal control epithelial ovarian cell lines and 10 epithelial ovarian cancer cell lines.
Table S4: Mean-to-Hose Ratio of iTRAQ, mRNA and methylation data
Table S5: List of 1279 common proteins with iTRAQ value among 8 cell lines subjected to Median absolute deviation (MAD) filtering analysis.
Table S6: iTRAQ clustering data list of 300 MAD filtered genES
ACKNOWLEDGMENTS/FUNDING
This research was supported by grants from the National Institutes of Health [CA53840 and GM55989] to N.K.T., and the National Institutes of Health, National Cancer Institute CSHL Cancer Centre Support Grant [CA45508]. N.K.T. is also grateful for support from the following foundations; The Gladowsky Breast Cancer Foundation, The Don Monti Memorial Research Foundation, Hansen Memorial Foundation, West Islip Breast Cancer Coalition for Long Island, Glen Cove CARES, Find a Cure Today (FACT), Constance Silveri, Robertson Research Fund and the Masthead Cove Yacht Club Carol Marcincuk Fund.
Footnotes
The authors have no conflicts of interest to report.
REFERENCES
- 1.Romero I, Bast RC., Jr. Minireview: human ovarian cancer: biology, current management, and paths to personalizing therapy. Endocrinology. 2012;153:1593–1602. doi: 10.1210/en.2011-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J. Clin. Oncol. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinopoulos PA, Spentzos D, Cannistra SA. Gene-expression profiling in epithelial ovarian cancer. Nat. Clin. Pract. Oncol. 2008;5:577–587. doi: 10.1038/ncponc1178. [DOI] [PubMed] [Google Scholar]
- 4.Cox B, Kislinger T, Emili A. Integrating gene and protein expression data: pattern analysis and profile mining. Methods. 2005;35:303–314. doi: 10.1016/j.ymeth.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Meani F, Pecorelli S, Liotta L, Petricoin EF. Clinical application of proteomics in ovarian cancer prevention and treatment. Mol. Diagn. Ther. 2009;13:297–311. doi: 10.1007/BF03256335. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 7.Jorgensen C, Locard-Paulet M. Analysing signalling networks by mass spectrometry. Amino Acids. 2012;43:1061–1074. doi: 10.1007/s00726-012-1293-z. [DOI] [PubMed] [Google Scholar]
- 8.Boylan KL, Andersen JD, Anderson LB, Higgins L, Skubitz AP. Quantitative proteomic analysis by iTRAQ(R) for the identification of candidate biomarkers in ovarian cancer serum. Proteome Sci. 2010;8:31. doi: 10.1186/1477-5956-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagne JP, Ethier C, Gagne P, Mercier G, Bonicalzi ME, Mes-Masson AM, Droit A, Winstall E, Isabelle M, Poirier GG. Comparative proteome analysis of human epithelial ovarian cancer. Proteome Sci. 2007;5:16. doi: 10.1186/1477-5956-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian Y, Yao Z, Roden RB, Zhang H. Identification of glycoproteins associated with different histological subtypes of ovarian tumors using quantitative glycoproteomics. Proteomics. 2011;11:4677–4687. doi: 10.1002/pmic.201000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldemarson S, Krogh M, Alaiya A, Kirik U, Schedvins K, Auer G, Hansson KM, Ossola R, Aebersold R, Lee H, Malmstrom J, James P. Protein expression changes in ovarian cancer during the transition from benign to malignant. J. Proteome Res. 2012;11:2876–2889. doi: 10.1021/pr201258q. [DOI] [PubMed] [Google Scholar]
- 12.Wang LN, Tong SW, Hu HD, Ye F, Li SL, Ren H, Zhang DZ, Xiang R, Yang YX. Quantitative proteome analysis of ovarian cancer tissues using a iTRAQ approach. J. Cell. Biochem. 2012;113:3762–3772. doi: 10.1002/jcb.24250. [DOI] [PubMed] [Google Scholar]
- 13.Beck F, Burkhart JM, Geiger J, Zahedi RP, Sickmann A. Robust workflow for iTRAQ-based peptide and protein quantification. Methods Mol. Biol. 2012;893:101–113. doi: 10.1007/978-1-61779-885-6_8. [DOI] [PubMed] [Google Scholar]
- 14.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ, Kauppinen S. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A, Riggs M, Banerjee N, Synnestvedt M, Schlichting E, Karesen R, Shama Prasada K, Rotti H, Rao R, Rao L, Eric Tang MH, Satyamoorthy K, Lucito R, Wigler M, Dimitrova N, Naume B, Borresen-Dale AL, Hicks JB. DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol. Oncol. 2011;5:77–92. doi: 10.1016/j.molonc.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamalakaran S, Kendall J, Zhao X, Tang C, Khan S, Ravi K, Auletta T, Riggs M, Wang Y, Helland A, Naume B, Dimitrova N, Borresen-Dale AL, Hicks J, Lucito R. Methylation detection oligonucleotide microarray analysis: a high-resolution method for detection of CpG island methylation. Nucleic Acids Res. 2009;37:e89. doi: 10.1093/nar/gkp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Wrzeszczynski KO, Varadan V, Byrnes J, Lum E, Kamalakaran S, Levine DA, Dimitrova N, Zhang MQ, Lucito R. Identification of tumor suppressors and oncogenes from genomic and epigenetic features in ovarian cancer. PLoS One. 2011;6:e28503. doi: 10.1371/journal.pone.0028503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levidou G, Ventouri K, Nonni A, Gakiopoulou H, Bamias A, Sotiropoulou M, Papaspirou I, Dimopoulos MA, Patsouris E, Korkolopoulou P. Replication protein A in nonearly ovarian adenocarcinomas: correlation with MCM-2, MCM-5, Ki-67 index and prognostic significance. Int. J. Gynecol. Pathol. 2012;31:319–327. doi: 10.1097/PGP.0b013e31823ef92e. [DOI] [PubMed] [Google Scholar]
- 21.Stewart JJ, White JT, Yan X, Collins S, Drescher CW, Urban ND, Hood L, Lin B. Proteins associated with Cisplatin resistance in ovarian cancer cells identified by quantitative proteomic technology and integrated with mRNA expression levels. Mol. Cell. Proteomics. 2006;5:433–443. doi: 10.1074/mcp.M500140-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Hornshoj H, Bendixen E, Conley LN, Andersen PK, Hedegaard J, Panitz F, Bendixen C. Transcriptomic and proteomic profiling of two porcine tissues using high-throughput technologies. BMC Genomics. 2009;10:30. doi: 10.1186/1471-2164-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Hu H, Tong X, Jiang Q, Zhu H, Zhang S. Calcium-binding protein S100P and cancer: mechanisms and clinical relevance. J. Cancer Res. Clin. Oncol. 2012;138:1–9. doi: 10.1007/s00432-011-1062-5. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, He Z, Gao J, Hu S, Huang M, Liu M, Zheng J, Tang H. S100P sensitizes ovarian cancer cells to carboplatin and paclitaxel in vitro. Cancer Lett. 2008;272:277–284. doi: 10.1016/j.canlet.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 25.Takemura K, Kawachi H, Eishi Y, Kitagaki K, Negi M, Kobayashi M, Uchida K, Inoue J, Inazawa J, Kawano T, Board PG. gamma-Glutamylcyclotransferase as a novel immunohistochemical biomarker for the malignancy of esophageal squamous tumors. Hum. Pathol. 2014;45:331–341. doi: 10.1016/j.humpath.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 26.Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene. 2001;20:6348–6371. doi: 10.1038/sj.onc.1204779. [DOI] [PubMed] [Google Scholar]
- 27.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 28.Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF-7 cells. Cancer Res. 2001;61:7065–7070. [PubMed] [Google Scholar]
- 29.Way G, Morrice N, Smythe C, O'Sullivan AJ. Purification and identification of secernin, a novel cytosolic protein that regulates exocytosis in mast cells. Mol. Biol. Cell. 2002;13:3344–3354. doi: 10.1091/mbc.E01-10-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suehara Y, Tochigi N, Kubota D, Kikuta K, Nakayama R, Seki K, Yoshida A, Ichikawa H, Hasegawa T, Kaneko K, Chuman H, Beppu Y, Kawai A, Kondo T. Secernin-1 as a novel prognostic biomarker candidate of synovial sarcoma revealed by proteomics. J. Proteomics. 2011;74:829–842. doi: 10.1016/j.jprot.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 31.Fan G, Lin G, Lucito R, Tonks NK. Protein-tyrosine Phosphatase 1B Antagonized Signaling by Insulin-like Growth Factor-1 Receptor and Kinase BRK/PTK6 in Ovarian Cancer Cells. J. Biol. Chem. 2013;288:24923–24934. doi: 10.1074/jbc.M113.482737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, Castedo M, Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 33.Lewis AD, Hayes JD, Wolf CR. Glutathione and glutathione-dependent enzymes in ovarian adenocarcinoma cell lines derived from a patient before and after the onset of drug resistance: intrinsic differences and cell cycle effects. Carcinogenesis. 1988;9:1283–1287. doi: 10.1093/carcin/9.7.1283. [DOI] [PubMed] [Google Scholar]
- 34.Guidi F, Landini I, Puglia M, Magherini F, Gabbiani C, Cinellu MA, Nobili S, Fiaschi T, Bini L, Mini E, Messori L, Modesti A. Proteomic analysis of ovarian cancer cell responses to cytotoxic gold compounds. Metallomics : integrated biometal science. 2012;4:307–314. doi: 10.1039/c2mt00083k. [DOI] [PubMed] [Google Scholar]
- 35.Gamberi T, Massai L, Magherini F, Landini I, Fiaschi T, Scaletti F, Gabbiani C, Bianchi L, Bini L, Nobili S, Perrone G, Mini E, Messori L, Modesti A. Proteomic analysis of A2780/S ovarian cancer cell response to the cytotoxic organogold(III) compound Aubipy(c) J. Proteomics. 2014;103:103–120. doi: 10.1016/j.jprot.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Cousineau I, Belmaaza A. EMSY overexpression disrupts the BRCA2/RAD51 pathway in the DNA-damage response: implications for chromosomal instability/recombination syndromes as checkpoint diseases. Mol Genet Genomics. 2011;285:325–340. doi: 10.1007/s00438-011-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y, Zhang Q, Atsaves V, Yang H, Claret FX. Suppression of Jab1/CSN5 induces radio- and chemo-sensitivity in nasopharyngeal carcinoma through changes to the DNA damage and repair pathways. Oncogene. 2013;32:2756–2766. doi: 10.1038/onc.2012.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Costello PC, Pham NA, Pintillie M, Jabali M, Sanghera J, Tsao MS, Johnston MR. Integrin-linked kinase inhibitor KP-392 demonstrates clinical benefits in an orthotopic human non-small cell lung cancer model. J. Thorac. Oncol. 2006;1:771–779. [PubMed] [Google Scholar]
- 39.Chung LY, Tang SJ, Sun GH, Chou TY, Yeh TS, Yu SL, Sun KH. Galectin-1 promotes lung cancer progression and chemoresistance by upregulating p38 MAPK, ERK, and cyclooxygenase-2. Clin. Cancer Res. 2012;18:4037–4047. doi: 10.1158/1078-0432.CCR-11-3348. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita T, Nagano K, Kanasaki S, Maeda Y, Furuya T, Inoue M, Nabeshi H, Yoshikawa T, Yoshioka Y, Itoh N, Abe Y, Kamada H, Tsutsumi Y, Tsunoda S. Annexin A4 is a possible biomarker for cisplatin susceptibility of malignant mesothelioma cells. Biochem. Biophys. Res. Commun. 2012;421:140–144. doi: 10.1016/j.bbrc.2012.03.144. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, Suarez G, Sha J, Sierra JC, Peterson JW, Chopra AK. Phospholipase A2-activating protein (PLAA) enhances cisplatin-induced apoptosis in HeLa cells. Cell. Signal. 2009;21:1085–1099. doi: 10.1016/j.cellsig.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 42.Ehlen A, Brennan DJ, Nodin B, O'Connor DP, Eberhard J, Alvarado-Kristensson M, Jeffrey IB, Manjer J, Brandstedt J, Uhlen M, Ponten F, Jirstrom K. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J. Transl. Med. 2010;8:78. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of two sets of iTRAQ measurements among 6 ovarian cancer cell lines.
Both Spearman correlation (S-cor) and Pearson correlation (P-cor) were calculated to evaluate the correlation between two replicates. To minimize clustering of the data points and to facilitate visual examination, iTRAQ measurements were log2-transformed.
Table S1: Background information for 12 ovarian cell lines
Table S2: List of 2657 proteins with iTRAQ value among 2 normal control epithelial ovarian cell lines and 10 epithelial ovarian cancer cell lines.
Table S3: List of 1273 proteins with iTRAQ value found in common among 2 normal control epithelial ovarian cell lines and 10 epithelial ovarian cancer cell lines.
Table S4: Mean-to-Hose Ratio of iTRAQ, mRNA and methylation data
Table S5: List of 1279 common proteins with iTRAQ value among 8 cell lines subjected to Median absolute deviation (MAD) filtering analysis.
Table S6: iTRAQ clustering data list of 300 MAD filtered genES