Abstract
Background.
Control of C. difficile infection (CDI) is an increasingly difficult problem for healthcare institutions. There are commonly recommended strategies to combat CDI transmission such as oral vancomycin for CDI treatment, increased hand hygiene with soap and water for healthcare workers, daily environmental disinfection of infected patient rooms, and contact isolation of diseased patients. However, the efficacy of these strategies, particularly for endemic CDI, has not been well studied. The objective of this research is to develop a valid agent-based simulation model (ABM) to study C. difficile transmission and control in a mid-sized hospital.
Methods.
We develop an ABM of a mid-sized hospital with agents such as patients, healthcare workers, and visitors. We model the natural progression of CDI in a patient using a Markov chain and the transmission of CDI through agent and environmental interactions. We derive input parameters from aggregate patient data from the 2007-2010 Wisconsin Hospital Association and published medical literature. We define a calibration process, which we use to estimate transition probabilities of the Markov model by comparing simulation results to benchmark values found in published literature.
Results.
Comparing CDI control strategies implemented individually, routine bleach disinfection of CDI+ patient rooms provides the largest reduction in nosocomial asymptomatic colonizations (21.8%) and nosocomial CDIs (42.8%). Additionally, vancomycin treatment provides the largest reduction in relapse CDIs (41.9%), CDI-related mortalities (68.5%), and total patient LOS (21.6%).
Conclusion.
We develop a generalized ABM for CDI control that can be customized and further expanded to specific institutions and/or scenarios. Additionally, we estimate transition probabilities for a Markov model of natural CDI progression in a patient through calibration.
Keywords: clostridium difficile, simulation methods, agent-based simulation, infectious disease control, hospital-acquired infections
INTRODUCTION
Clostridium difficile Infection (CDI) is the leading infectious cause of healthcare-associated diarrhea. CDI affects more than 500,000 Americans every year and is responsible for nearly 20,000 deaths annually. There is strong evidence that the incidence and severity of CDI are increasing over time.(1)(2)(3) Mortality rates from CDI increased more than 400% between 2000 and 2004.(1) In some studies, CDI has surpassed Methicillin-Resistant Staphylococcus aureus (MRSA) as the most common hospital acquired infection.(4)
Recent guidelines from the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA) discuss several strategies for reducing the transmission of C. difficile in a healthcare institution.(5) These include: treatment with antibiotics for patients with CDI, environmental decontamination of rooms with bleach, increased healthcare worker (HCW) hand-hygiene using soap and water, and contact isolation of diseased patients. However, control of CDI continues to be problematic. Adherence to contact isolation and hand hygiene proves to be challenging to achieve.(6) While several studies investigate how these strategies individually affect C. difficile spread in small epidemic settings,(3)(7)(8)(9) to the best of our knowledge, there is no systematic method to evaluate the combined effects of these control measures in endemic and large epidemic situations. Furthermore, many existing models often consider transmission of C. difficile without modeling the interactions among patients, visitors, and HCWs thus ignoring important vectors of transmission and sources of variability.(10)
The complexity of CDI outbreaks and the interactions among patients, visitors, and HCWs present a problem that is too difficult to analytically compute how recommended control strategies affect performance metrics such as total patient length of stay (LOS), prevalence of asymptomatic C. difficile colonization, CDI incidence, and CDI-related mortality. In this study, we develop a discrete-event agent-based simulation model (ABM) that can be used to determine the best CDI control strategies for a particular situation. An ABM is ideal for studying such a problem, because it can model the effects on transmission of C. difficile through agent interactions and the effect of CDI control strategies on agent behavior inside the system. The main objective of this paper is to develop a core framework to model CDI transmission and control in a hospital. This simulation can then be improved upon to build more advanced and complex future models. Additionally, we develop a Markov model of the natural progression of CDI in a patient and estimate the transition probabilities through calibration.
METHODS
Overview of the ABM
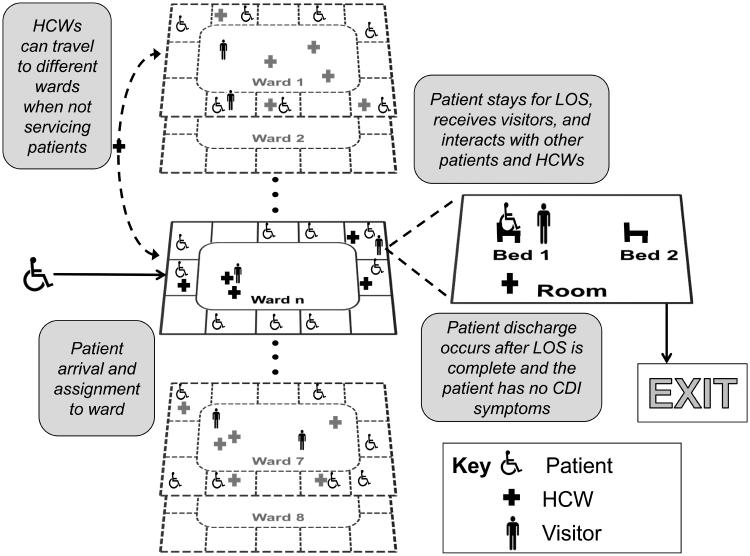
We develop a discrete-event ABM that simulates C. difficile transmission and control in a general mid-sized hospital. An ABM is an extension of traditional discrete-event simulation where the agents in the model have unique attributes and interact with each other. These interactions affect the simulation output and performance of the overall system. The ABM updates occur in discrete-time units each equivalent to 5 simulated minutes. Our model hospital is compartmentalized into 10 wards with 10 rooms per ward, and 2 beds per room. Diagrams of agent flow and environment construction are shown in Figure 1. There are three agent types in our model: patients, HCWs, and visitors. Agents interact with each other and the environment, and these interactions serve as possible transmission routes for C. difficile.
Figure 1.
Agent flow and interactions in the simulated mid-sized hospital.
We assume HCWs and visitors are not susceptible to colonization or infection. However, they may become exposed to C. difficile through interactions and therefore spread C. difficile through the hospital. Hence, HCW and visitor agents have a binary descriptor indicating if they are exposed to C. difficile. Describing C. difficile in patients is more complex, since we consider the possibility of asymptomatic colonization, CDI, and relapse CDI. Therefore, we use a Markov chain to model C. difficile related condition of each patient.
We use this ABM to evaluate four strategies that are commonly used to control the spread of CDI: A standard regimen of vancomycin, hereafter referred to as strategy (V), for treatment of CDI+ patients, increased hand-hygiene with soap and water for HCWs (H), contact isolation of diseased patients (I), and bleach disinfection of rooms that contain diseased patients (B). Additionally we look at a mixed strategy (M) of all four individual strategies at the same time. These strategies affect the behavior of agents, the parameters relevant to C. difficile exposure and contamination, and ultimately, the propagation of CDI in patients. Our ABM can be used to evaluate the impact of applying each strategy individually or a combination of strategies.
Agents & The Environment
Our model consists of three agent types: patients, HCWs, and visitors. Agents can interact with each other and the hospital environment, which is divided into wards consisting of patient rooms and a ward common area. The rooms and the common areas of each ward have a binary descriptor of C. difficile exposure: they are either free of C. difficile or contaminated and thus risk exposing agents.
Patients arrive to the hospital following a Poisson process with arrival rate specified by historical data and are then assigned to a room. Each patient has several descriptive attributes including a unique patient ID, age, length of stay (LOS), remaining stay, designated ward, room assignment, bed assignment, CDI-related state, and exposure level. A patient is provided service for a lognormal distributed period of time and then discharged.
The HCW agents are assigned to specific wards and work in 12-hour shifts. There are 15 HCWs per ward and each HCW services a set of patient rooms. Each HCW makes rounds visiting patients in the ward. Once a service round is completed, an HCW can either return to the ward common area to wait until the next time to he/she tends to patients, or he/she temporarily visits another ward then returns at a later time. HCWs have a binary attribute that indicates if the HCW has been exposed to C. difficile.
At the beginning of each day, a Bernoulli trial determines whether or not a patient receives visitors on that day. A patient may receive between 0 and 3 visitors at a time. Visitor agents arrive to a ward and wait in the common areas or spend time with patients. C. difficile exposure for visitors works in the same fashion as HCWs. Visitors exposed to C. difficile can contribute to the transmission of C. difficile between patient rooms, common areas, and through interactions with the patient they are visiting. While in the hospital, a visitor can either visit a patient in his/her room, or wait in the ward’s common area. We assume patients in shared rooms can receive visitors and HCWs concurrently, and visitors and HCWs can be in a patient room at the same time.
Interactions
Interactions among the agents and interactions with the environment, such as rooms and ward common areas, are the primary modes in which C. difficile spreads. During an interaction, a Bernoulli trial is used to determine whether exposure to C. difficile occurs. The probability of exposure depends on the interaction type, and is estimated from C. difficile contamination studies and expert opinion (Table 1). A patient can become exposed to C. difficile after interacting with an exposed HCW, visitor, or contagious patient with probability peh, pev, or pep respectively. After remaining in a contaminated room for 6 hours, a patient can become exposed to C. difficile with probability penv. Similarly, an HCW can become exposed to C. difficile by interacting with an exposed patient, with probability peh, or being in a contaminated room, with probability penv. Finally, a visitor can become exposed to C. difficile after being in a contaminated room with probability penv, or interacting with a contagious patient, with probability pep.
Table 1.
A list of input parameters, values, distribution information, and sources.
| Symbol | Description | Mean Value |
Distribution | Range | Source |
|---|---|---|---|---|---|
| μ LOS | Patient LOS | 4.254 | Lognormal | 1.497 (SD) | WHA Data |
| λ | Patient arrival rate per day | 17.55 | Exponential | -- | AHA |
| μ h | HCW service time in minutes | 10 | Exponential | -- | Expert Opinion |
| nh | Number of HCWs per ward | 15 | Triangular | [13, 17] | Expert Opinion |
| phh | Hand Hygiene Compliance | 0.48 | Triangular | [0.36, 0.6] | Pittet et al.(27) |
| piso | Adherence to Contact Isolation | 0.62 | Triangular | [0.47, 0.78] | Muto et al. (28) |
| μ v | Visitor service time in minutes | 20 | Exponential | -- | Expert Opinion |
| pvis | Probability a patient will receive visitors during on a given day |
0.6 | Triangular | [0.45, 0.75] | Expert Opinion |
| nv | Number of visitors | 2 | Triangular | [1, 3] | Expert Opinion |
| α s | Proportion of patient arrivals in Susceptible state |
0.3018 | Triangular | [0.256, 0.347] | WHA Data |
| α c | Proportion of patient arrivals in Colonized state |
0.076 | Triangular | [0.067, 0.097] | (31,34,48) |
| α d | Proportion of patient arrivals in Diseased state |
0.00242 | Triangular | [0.002, 0.006] | (19,20,31-34,48) |
| α n | Proportion of patients in Not- Susceptible state |
0.6176 | Triangular | [0.549, 0.673] | WHA Data, Extrapolation |
| pvanc | Probability of recovering from CDI with 2.0g/day vancomycin treatment |
0.7981 | Triangular | [0.5985, 0.9975] | (18,24,32,33,49,50) |
| μ vanc | Vancomycin treatment time | 10 | Triangular | [5, 15] | (15) |
| penv | Probability of exposure from interaction with environment |
0.435 | Triangular | [0.3262, 0.5437] | (51) & Expert Opinion |
| ppe | Probability of patient exposure from interacting with contagious patient |
0.24 | Triangular | [0.18, 0.3] | (51,52) & Expert Opinion |
| peh | Probability of HCW exposure given an interaction with contagious patient occurred |
0.48 | Triangular | [0.4, 0.6] | (51-53) & Expert Opinion |
| pep | Probability of patient exposure given an interaction with exposed HCW occurred |
0.5 | Triangular | [0375, 0.625] | (51-53) & Expert Opinion |
| pev | Probability of a visitor exposure given an interaction with exposed patient |
0.5 | Triangular | [0.425, 0.625] | (51,52) & Expert Opinion |
Markov Model
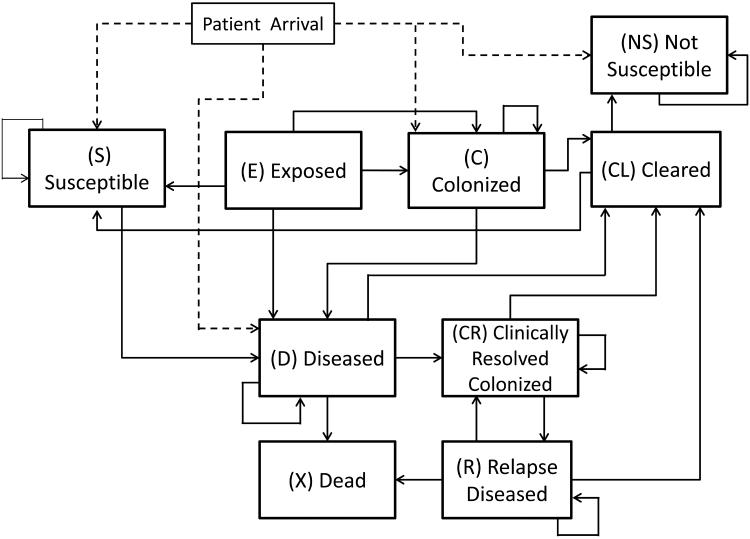
Each patient agent has a dynamic C. difficile related status that is modeled as a discrete-time Markov chain (DTMC). For simplicity and ease of computation, we assume this Markov chain is updated every 6 hours of simulation time. The states of the Markov chain are described by a modified version of the natural history model proposed by Otten et al. (11). Namely, we use the following states to represent the C. difficile related status of a patient: Susceptible (S), Exposed (E), Colonized (C), Diseased (D), Cleared (CL), Clinically Resolved Colonized (CR), Relapse Diseased (R), Dead (X), and Not Susceptible (NS). A patient in the Susceptible (S) state can develop CDI or asymptomatic colonization after exposure to C. difficile. If a patient is exposed to C. difficile after an interaction with a contagious patient, or another exposed agent, he/she will transition to the Exposed state. The Exposed (E) state represents a patient who is exposed to C. difficile through interactions within the past 6 hours. Such a state is particularly useful for examining the effects of interactions among agents. A patient in the Colonized (C) state is asymptomatically colonized with C. difficile and can put others at risk for exposure. A patient in the Diseased (D) state has been diagnosed with active CDI and can expose other patients, HCWs, and visitors to C. difficile. A patient in the Cleared (CL) state once had an infection or colonization, but is now recovered and the patient is no longer contagious. A patient in the Clinically Resolved and Colonized (CR) state once had CDI, but the symptoms have subsided despite the patient still having colonization of C. difficile. Similarly, a patient in the Relapse Diseased (R) state once had CDI, but is experiencing a recurrence of the previous CDI. A patient in the Dead (X) state has died due to CDI-related complications. Finally, a patient in the Not-susceptible (NS) state cannot develop colonization or CDI during his/her stay, i.e. they do not fall into risk categories for CDI or colonization. Figure 3 shows the states and possible transitions of the Markov model.
Figure 3.
State transition diagram of the Markov model for the C. difficile related status of a patient agent.
CDI Control Strategies
Using several studies we estimate Strategy (V) as being effective in resolving CDI in 79.3% of CDI cases without relapse.(11)(12)(13)(14)(15)(16)(17)(18)(19)(20) Due to the increased severity of CDI and the increased number of patients that meet requirements for vancomycin rather than other antibiotics, such as metronidazole, we consider vancomycin as the primary treatment. Other treatment methods such as metronidazole or fidaxomycin can be included in future iterations of the model, and we may estimate the proportion of CDI+ patients who receive each treatment. Metronidazole is shown to be as less effective than vancoymcin at treating CDI.(15,21,22) While fidaxomycin has been shown to be less effective than vancomycin at treating the hyper virulent NAP1 strain, it has been shown to be more effective at treating non NAP1 CDI.(23–25)
We assume 100% of CDI patients will receive oral vancomycin treatment when the strategy is used and the diagnosis of CDI is 100% accurate. Hence, we do not consider any consequences of incorrectly treating patients without CDI. We make this assumption based on the current availability of highly accurate algorithms to detect CDI. Sharp et al. (2010)(26) demonstrate that the use of C.Diff Quik Chek Complete test, which tests for both GDH and Toxin A/B, followed by random-access PCR test for discrepant results from the C.Diff Quik Chek, result in high sensitivity (100%; 95% confidence interval [CI], 89.6 to 100%) and high specificity (99.6%; 95% CI, 97.3 to 99.9%).
Strategy (H) requires all HCWs to thoroughly wash their hands for 25 seconds with soap and water. We use a hand hygiene adherence of 48%, as reported by Pittet et al. who conducted one of the largest studies on hand hygiene in a hospital.(27) We assume each hand hygiene action removes C. difficile from the HCW’s hands between taking care of patients.(3)(9) If increased hand-hygiene measures are not enacted, HCWs may use alternative hand sanitation methods such as alcohol-based hand gel, which are shown to not effectively eliminate C. difficile contamination.(8) Strategy (B) involves the disinfection of rooms with diseased patients every 24 hours and assumes 100% adherence to bleach disinfection when used. Lastly, strategy (I) is the isolation of diseased patients where he/she is placed in his/her own room and do not interact with other patients, and HCWs must wear protective gowns and gloves before entering the room. We assume 62% adherence to proper HCW gowning as indicated by Muto et al.(28) If there is insufficient room capacity, the patient is not isolated. We run 6 different strategies for comparison: no intervention strategy (N), the 4 individual strategies (V), (H), (I), (B), and the mixed strategy (M).
Input Parameters
There are three main sources for our data. The first is aggregate patient data from the Wisconsin Hospital Association (WHA). The second is published papers from medical and epidemiological literature pertaining to CDI. The third data source is published statistics and guidelines from organizations such as the Centers for Disease Control and Prevention (CDC) and the American Hospital Association (AHA).
The WHA dataset consists of patient admissions to all healthcare institutions across the state of Wisconsin from 2007 to 2010. We use this dataset to estimate patient LOS, proportion of arrivals, patient age distribution, and susceptibility to CDI. We use published studies on C. difficile spread and control to estimate essential model parameters of C. difficile control strategies such as the duration of vancomycin treatment and the efficacy of bleach disinfection. A full list of input parameters is presented in Table 1. Additionally, we use published literature to estimate outcome measures such as percentage of colonized patient population, mortality rates, and infection rates; all of which are used as calibration constraints for generating the transition probabilities of the Markov model as explained below. We use the AHA 2010 Fast Fact report to estimate the arrival rate of 17.55 patients per day. We initialize our model with a patient population equal to the average number of patients in the hospital at a given time.
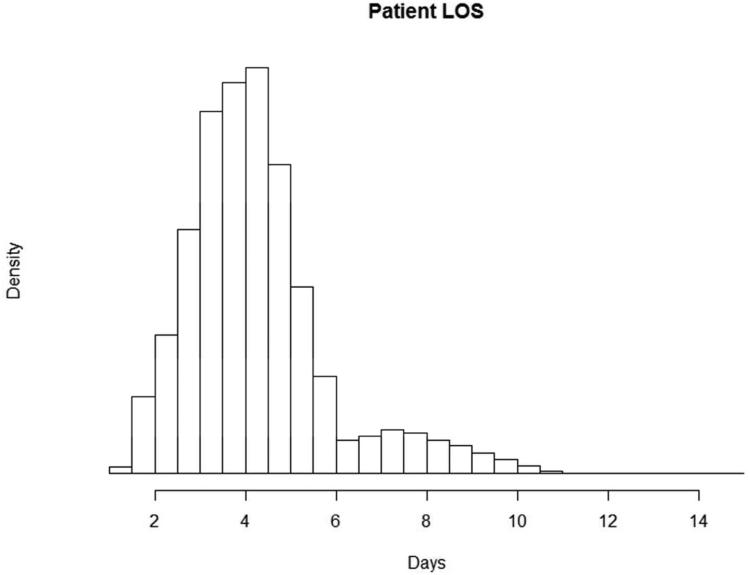
Based on the WHA data, we assume patient LOS to be lognormally distributed (Figure 4), and age to be normally distributed. We calculate the percentage of elderly patients, that is, patients over the age of 65, and considered all these patients to be susceptible to CDI. Therefore, we estimate the proportion of all patients who are susceptible to CDI during his/her stay to be 30.18%. We assume HCW service times and visitor services times are exponentially distributed. All other parameters where distribution data was not readily available are modeled as triangularly distributed with minimums and maximums of 75% and 125% of the mean (mode) values.
Figure 4.
Histogram for patient length of stay from the Wisconsin Hospital Association 2007-2010 dataset.
Calibration of the Markov Model
We use a DTMC to represent the natural progression of CDI in a patient. There is no available data to estimate the state transition probabilities directly, so for this purpose we use a method known as calibration, which is implemented as follows. First, we partition each transition probability pi,j into discretized values using an initial step size. This creates many different instances of P, denoted by Px, whose entries, denoted pi,j,x, are defined as the probability of a patient in state i transitioning to state j in 6 hours . For each Px, we impose constraints on the entries of n-step transition probability matrix, Pxn, which are derived from the WHA dataset and long-term probabilities from published literature. We classify instances of Px that satisfy the constraints listed in Table 2 as feasible and instances of Px that fail to satisfy the constraints as infeasible. This generates a large set of feasible matrices denoted by Π. Then, we randomly sample 2000 matrices from Π into a new set Π’ and run 100 independent replications of the simulation for every Px in Π’. Next, we examine the ABM output and accept the instances of Px whose benchmark values are within the lower and upper bound values obtained from the literature. These four benchmark values are the proportion of patients who experience asymptomatic colonization [0.076,0.127, 0.177](29)(30)(31) (i.e. [lower bound, mean, upper bound]), CDI [0.004 , 0.019, 0.072 ](30)(32)(20)(33)(19)(34), relapse CDI [0.184 , 0.232, 0.273](32)(35)(33), and CDI-related mortality [0.057 , 0.0855, 0.138](20)(19)(32)(34)(33)(36). We reject an instance Px if any of the four benchmark values lies outside of the lower or upper bounds. We then calculate the mean percentage error (MPE) for each benchmark statistic t and for each Px by comparing simulated results in each replication m, denoted ŷt,x,m, with the benchmark values, denoted by yt that are derived from published literature. We define the MPE of benchmark t pertaining to a probability matrix Px as:
and the average mean percentage error (AMPE) as:
We use AMPE to avoid biasing the calibration on the benchmark values with the largest magnitude. We define ΠTOP10 as the set of 10 instances of Px with the smallest AMPE value. ΠTOP10 is then used to run the simulation scenarios for the final results, and the larger set of feasible matrices Π’ is used to explore the effects of error in the estimates for the Markov model transition probabilities.
Table 2.
Markov chain calibration constraints derived from the WHA dataset or published literature. The notation Pi,jn denotes (i,j)th entry in the n-step transition probability matrix P where i and j correspond to states in the Markov model.
| Constraint | Description | Source |
|---|---|---|
| 0 < PS,D20 ≤ 0.00683 | Probability of developing CDI | WHA Data |
| 0 < PS.D28 ≤ 0.0093 | Probability of developing CDI in 1 week | Clabots et al.(30) |
| 0 < PS,D56 ≤ 0.125 | Probability of developing CDI in 1-2 weeks | Clabots et al. (30) |
| 0 < PS,D84 ≤ 0.2787 | Probability of developing CDI in 2-3 weeks | Clabots et al. (30) |
| 0 < PS,D112 ≤ 0.32 | Probability of developing CDI in 3-4 weeks | Glabots et al. (30) |
| 0 < PS,D140 ≤ .5 | Probability of developing CDI in >4 weeks | Clabots et al. (30) |
| 0 < PC,D20 ≤ 0.0227 | Probability of a colonized patient becoming diseased within his/her stay. | Redehngs, et al. (1) |
| 0 < PS,C20 ≤ 0.1937 | Probability of susceptible patient becoming colonized within his/her stay. | McFarland, et al.(54) |
| 0 < PE,D ≤ PS.D < PE,D | Colonized patients are not more likely to develop CDI than susceptible patients |
Johnson, et al.(55) |
| 0.026 ≤ PD,X83 ≤ 0.30 | Probability a diseased patient will die from CDI | Loo, et al.(34) |
| 0 < PD,L12 ≤ 0.23 | Probability of a quick recovery (2.5 days mean time) | CDC estimates Olson, et al.(16) |
| 0 < PD,L88 ≤ 0.595 | Probability of a long recovery (22 days mean time) | |
| 0 < PCL,D ≤ 0.0255 | Probability a patient will develop CDI after being cleared | Olson, et al.(16)& Teasley et al.(15) |
| PR,X > PD,X | Probability of death when relapse diseased is greater than the probability of death in diseased state |
O’Neill, et al.(56) |
Verification and Validation
We use the definitions of verification and validation as presented by the renowned simulation textbook, Simulation Modeling and Analysis by Law (2007, p. 299)(37) in which he describes verification as “determining that a simulation computer program performs as intended, i.e. debugging the computer program.” and validation as, “concerned with determining whether the conceptual simulation model (as opposed to the computer program) is an accurate representation of the system under study.” In another paper on verification and validation, Kleijnen outlines commonly used techniques for verification and validation.(38)
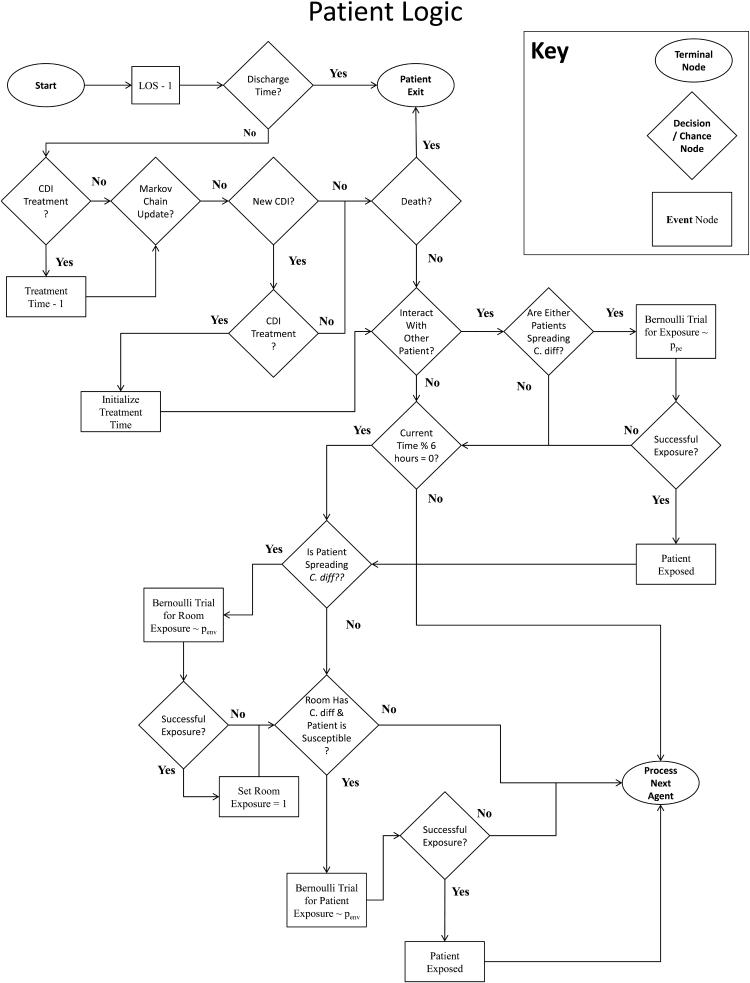
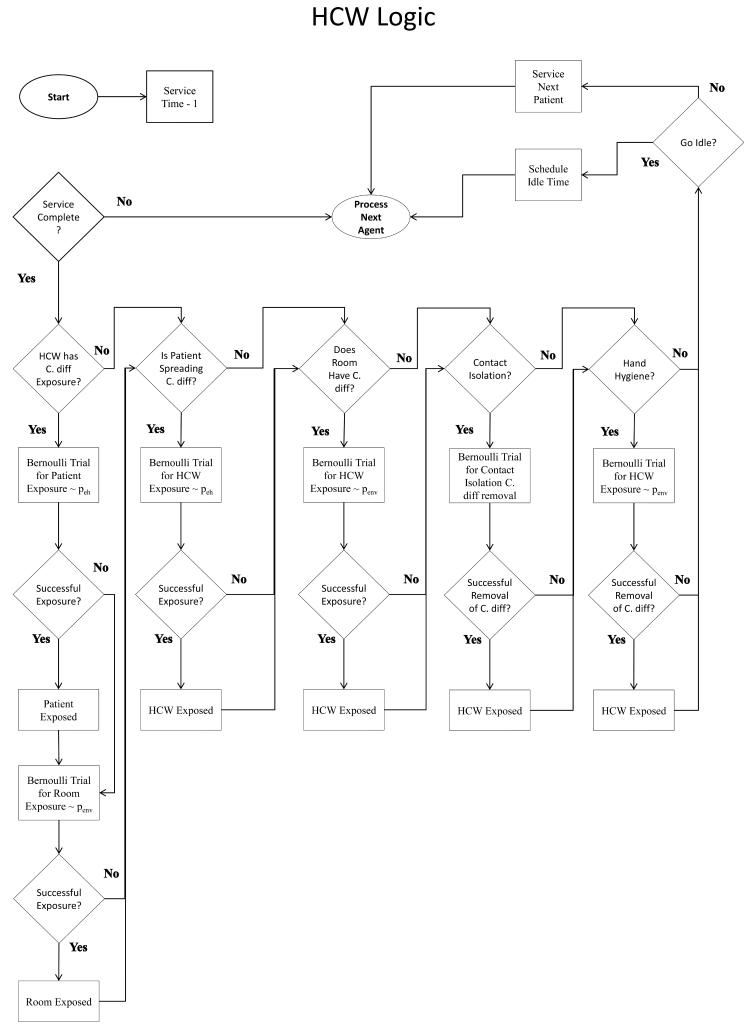
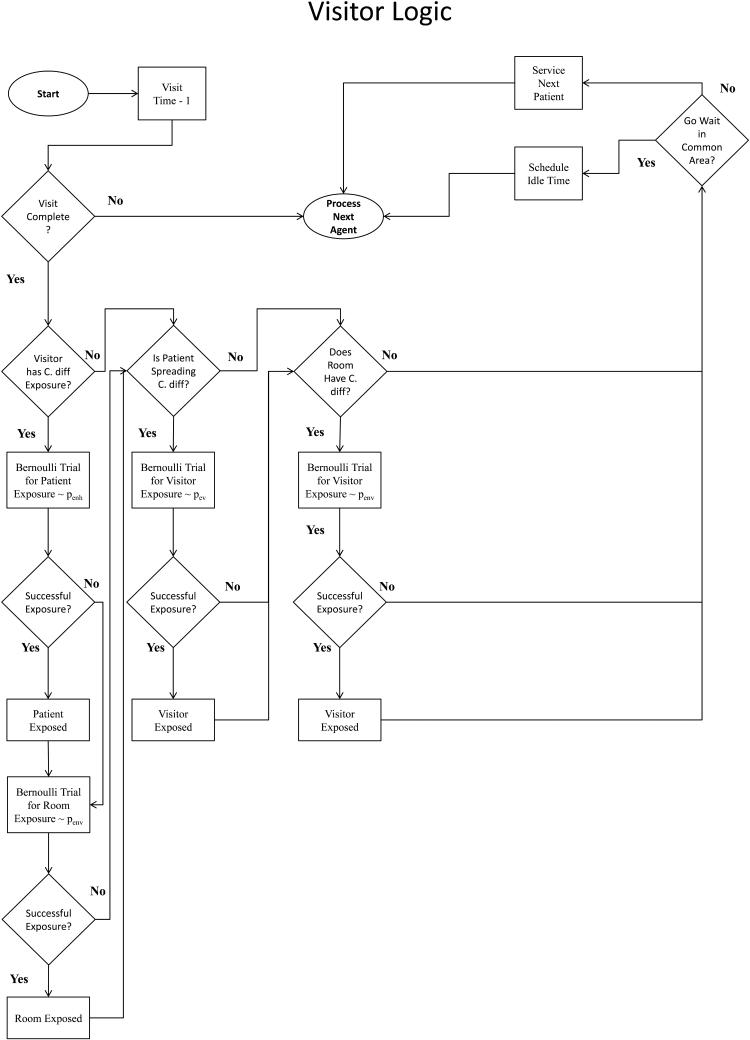
Our simulation model is written in Java using the Eclipse IDE. For verification, agent logic is well documented and compartmentalized by agent-type. Additionally, we perform tracing of agents via simulation output and using the Eclipse IDE debugger. Our tracing method follows randomly selected agents of each type through his/her stay in the hospital and checking whether the behavior of simulated agents match the intended agent logic, which is mapped in Figure 2.
Figure 2.
Flowcharts depicting Agent logic and behavior for (a) Patients, (b) HCWs, and (c) Visitors.
Validation of the simulation model occurs in 5 segments: face validation, Markov model calibration results, calibration robustness, cross validation, and sensitivity analysis. In face validation, one of the authors, Dr. Nasia Safdar, a healthcare epidemiologist and infectious diseases trained physician, provided expertise on C. difficile transmission and control. The agents, environment, methods of interaction, Markov model states, and parameters are also developed in conjunction with Dr. Safdar to ensure the model is a sufficiently accurate representation of a real hospital, while making the necessary simplifying assumptions to create such a model. Calibration of the Markov model is a validation of the natural history model by minimizing the error between the model predictions and the four performance benchmarks. We also study the effects of error in the performance target values by performing simulation runs using randomly sampled Pxs in the set of matrices accepted by the calibration bounds, Π’.
Cross validation is performed by running various scenarios found in published literature on CDI control and comparing our simulation results to those reported in these studies. These performance metrics are not used in the development of the model or calibration of the natural disease progression Markov model. Sensitivity analysis is used to confirm changes in the input parameters correspond with logical changes in the simulation results.
Cross-Validation to Published Literature
We compare the simulation results to expected total patient LOS using our assumptions about patient LOS from the WHA data and published literature on CDI. Estimates are used from Louie et al. (18) who report the mean vancomycin treatment duration as 10 days, and Teasley et al. (15) who find 15%-23% of patients recover naturally in 2-3 days without the use of antibiotics, and Barbut et al.(39) who find CDI is responsible for an 8 to 21 day increase in diseased patient LOS. Using the incidence of CDI and relapse CDI from our simulation results, we can use the parameters from these aforementioned publications and calculate the expected LOS of the 6 strategies discussed previously, and then compare these estimates with results from our simulation. The efficacy of CDI control strategies depends strongly on the susceptibility of patient populations, adherence to CDI control strategies, and treatment of patients. Unfortunately, many of these data are not available from the literature. Hence, validation through comparison to findings in published literature is, at best, limited. For this reason, we refrain from performing statistical tests for validation; however, we demonstrate the feasibility of our results by comparing simulation results of similar scenarios to published studies. For a comparison metric, we use the rate ratio, which is defined as the ratio of CDI incidence after intervention over the CDI incidence before intervention.
Sensitivity Analysis
We perform a deterministic one-way sensitivity analysis where we examine the output and look for a significant change (i.e. >5%) in asymptomatic colonizations, CDIs, relapse CDIs, mortalities, and total patient LOS. We also perform probabilistic sensitivity analysis, where we use a Monte Carlo approach such that parameters that are triangular or uniformly distributed random will have their min/max ranges extended by +/− 50% of their original values, and other parameters will have their attributes changed by +/− 50%.
RESULTS
Markov Model Calibration
The matrices in ΠTOP10 produced the following calibration results: average colonization incidence of 9.17% (MPECOL = 0.576 %), CDI incidence of 1.44% (MPECDI = 3.64%), proportion of relapse CDI as 22.43% (MPERD = 11.091%), CDI-related mortality at 8.53% (MPEMOR = 11.782%), and an AMPE of 6.77%. We use this ΠTOP10 in the simulation for output validation, performance of individual control strategies, and sensitivity analysis. We evaluate the robustness of our selection of Pxs by randomly sampling 10 unique Pxs from Π’ and evaluating the simulation output generated by using these matrices to represent the parameters that dictate the natural progression of C. difficile in a patient. We perform these runs for each of the 6 strategy scenarios (N), (V), (H), (I), (B), and (M) and summarize the results as the maximum deviations from the results across all strategy scenarios. We observe a maximum deviation of 13.3% for colonizations, 11.61% for CDIs, 3.03% for relapse CDIs, 8.46% for mortalities, and 1.73 days for total patient LOS. Table 3 contains the full results of this analysis and the difference range of each non-zero transition probability, pi,j that has been accepted by the calibration. The results demonstrate that the choice of Px in Π’ provides a similar magnitude and rank for the efficacy of each individual CDI control strategy, despite the differences in the values of the pi,js. Therefore, we conclude that our calibration method is stable against error in the choice of calibration targets, and can easily be modified to match new and dynamic strains of C. difficile.
Table 3.
Results (min/max) of each strategy scenario using randomly sampled transition probability matrices Px in Π’.
| Strategy | Colonizations | CDIs | Relapse CDIs | CDI-Related Mortalities |
Total Patient LOS |
|---|---|---|---|---|---|
| N | [13.09%,26.39%] | [8.92%,20.53%] | [23.57%,26.44%] | [14.59%,22.62%] | [5.29,7.01] |
| V | [11.29%,21.09%] | [6.58%,13.7%] | [19.4%,20.53%] | [5.21%,9.66%] | [4.26,4.28] |
| I | [12.51%,23.71%] | [7.6%,18.46%] | [23.55%,25.94%] | [14.42%,22.88%] | [5.13,6.71] |
| H | [12.72%,24.29%] | [7.79%,19.14%] | [23.16%,26.19%] | [14.68%,22.42%] | [5.15,6.8] |
| B | [10.62%,19.53%] | [5.83%,12.03%] | [23.27%,25.74%] | [14.15%,22.42%] | [4.93,5.78] |
| M | [8.8%,12.47%] | [2.3%,4.02%] | [18.96%,20.66%] | [5.46%,10.15%] | [4.25,4.26] |
Performance of CDI Control Strategies
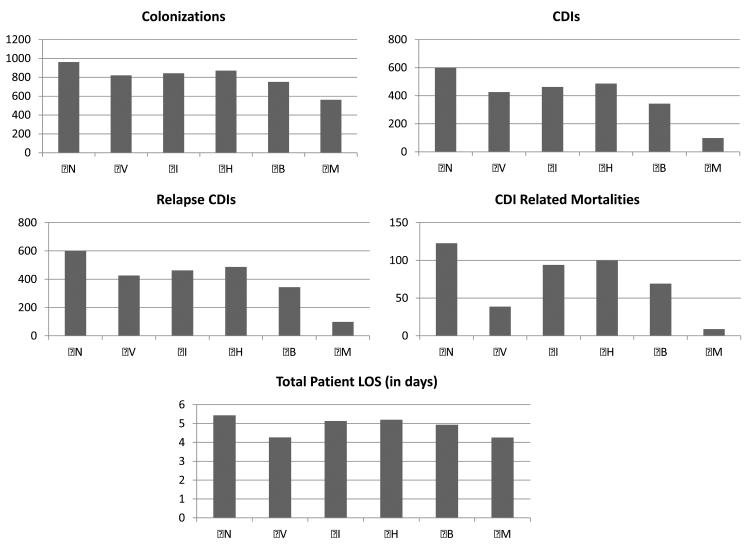
Figure 5 showcases the effects of each scenario, with 95% confidence intervals, on the incidence of asymptotic colonization, CDI, relapse CDI, CDI-related mortality, and total patient LOS. As expected, the mixed strategy (M) results in the largest reduction for all performance metrics: 41.5%, 83.7%, 86.3%, 92.8%, and 21.7% respectively. However, strategy (B) results in the largest reduction in asymptomatic colonizations and CDIs, while strategy (V) results in the largest reduction in total patient LOS, relapse CDIs, and CDI-related mortalities. The results for individual strategies on the aforementioned performance metrics in order of efficacy on CDI reduction: Strategy (B) reduces colonizations by 21.83%, CDIs by 42.84%, relapse CDIs by 41.90%, CDI-related mortalities by 43.68%, and LOS by 9.18%; strategy (V) reduces colonizations by 14.61%, CDIs by 29.09%, relapse CDIs by 41.9 3%, CDI-related mortalities by 68.50%, and LOS by 21.63%.
Figure 5.
Simulation results comparing the number of colonizations, CDIs, relapse CDIs, CDI related mortalities, and total patient LOS over the individual CDI control strategies (V), (I), (H), (B), and mixed strategy (M) to the base case of no interventions (N).
We also compare our simulation results to those found in the literature. First, we compare the results of our model using environmental bleach disinfection of CDI incidence. Safdar et al. find the bleach disinfection rate ratio to be 0.21 (95% CI 0.11 - 0.39). (40) Mayfield et al. report a bleach disinfection rate ratio to be 0.37 (95% CI 0.14 - 0.74)(3) for bone marrow transplant patients, but there was no change in CDI incidence in the neurosurgical intensive care unit and no change in CDI incidence (i.e., rate ratio=1) for general medicine patients. In another study, Wilcox et al.(41) find the bleach disinfection rate ratio to be 0.595 in one ward and notice an increase in CDI (i.e. rate ratio>1) in another ward. Using the same adherence rate of 88% for hand hygiene as Safdar et al.,(40) which was the only study that reported hand hygiene adherence, our simulation results in a bleach-disinfection rate ratio of 0.428 (95% CI 0.401 - 0.455). There are many factors that can be responsible for the wide range of results from these studies such as adherence to other interventions, which was not always present in these studies. Dettonkoffer et al. call for more detailed studies study of disinfection routines to identify the dependent variables of bleach disinfection efficacy.(42)
Next, we compare our simulation results with Apisarnthanarak et al. who report a rate ratio of 0.5 through implementation of contact isolation, increased hand-hygiene, and environmental bleach disinfection.(43) Adherence rates were not available from the study. However, using average adherence rates from published literature,(27)(28) our model reports a rate ratio of 0.546 (95% CI 0.540 – 0.552) with these strategies enacted. To summarize, the outputs from our simulation appear to be within reasonable ranges found in published literature on CDI control. However, due to the complexity of CDI control strategies and the precise data required for validation, such comparisons should be taken as demonstrative.
Finally, we can compare published study estimates of patient LOS with output from our simulation. The expected LOS from published studies for our six strategy scenarios is as follows: strategy (N) 5.41, strategy (V) 4.25, strategy (I) 5.14, strategy (H) 5.19, strategy (B) 4.91, and strategy (M) 4.26 days. Comparatively, our simulation attains the following LOS values: strategy (N) 5.44(95% CI 5.37 – 5.51), strategy (V) 4.26 (95% CI 4.23 - 4.29), strategy (I) 5.13 (95% CI 5.07 – 5.20), strategy (H) 5.20 (95% CI 5.13- 5.27), strategy (B) 4.94 (95% CI 4.88 - 4.99), and strategy (M) 4.26 (95% CI 4.23 - 4.29). The proximity of the LOS results indicates valid flow logic for patients with and without CDI. These results suggest additional validity of the Markov model, namely in the transition probabilities that dictate the duration of CDI.
Results for the Sensitivity Analysis
Changes in parameters that are correlated with a significant increase as compared to the average scenario are mapped in Table 5. As expected, an increased proportion of patients susceptible or colonized to CDI results in an increased incidence of colonizations, CDI, relapse CDI, and mortality. Increasing the probability of environmental contamination and increasing the probability of patient-to-HCW exposure leads to increases in nosocomial colonization and CDI. However, increased exposure probability with visitors did not necessarily lead to significantly more CDIs. These results imply that environmental contamination and HCWs may serve as significant sources of C. difficile transmission in the hospital, however, visitors may play only a minor role. Additionally, longer LOS for patients produced more CDIs, which is likely due to extended opportunities for C. difficile exposure.
Table 5.
Results for one-way sensitivity analysis. The listed parameters resulted in a significant change in output values for the given strategies compared to the average parameter value scenario. A single +/− indicates a change of [5 - 15]%, and a double ++/−− indicates a change of >15%.
| Name | Δ | Colonizations | CDIs | Relapse CDIs | Deaths | Total Patient LOS |
|---|---|---|---|---|---|---|
| − | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M− | N− | |
| α s | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | |||
| + | N++ V++ I++ H++ B++ M+ | N+ | ||||
| M++ | M++ | M++ | ||||
|
| ||||||
| − | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N− V− I− H− B− M− | N− | |
| α c | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | |||
| + | N+ V+ I+ H+ B+ M+ | N+ | ||||
| M++ | M++ | M++ | ||||
|
| ||||||
| − | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N− V− I− H− B− M− | N− | |
| α d | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | |||
| + | N+ V+ I+ H+ B+ M+ | N+ | ||||
| M++ | M++ | M++ | ||||
|
| ||||||
| N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | ||||
| − | N++ V++ I++ H++ B++ M+ | N+ | ||||
| α ns | M++ | M++ | M++ | |||
| + | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M− | N− | |
|
| ||||||
| − | V+ M+ | V++ M+ | V++ M++ | V++ M++ | V+ M+ | |
| Pvanc | ||||||
| + | V− M− | V− M− | V− M− | V− M− | V− M− | |
|
| ||||||
| − | V− M− | V− M− | ||||
| μ vanc | ||||||
| + | V+ M+ | V+ M+ | ||||
|
| ||||||
| − | N− V− I− H− M− | N− V− I− H− B− M− | N− V− I− H− B− M− | N− V− I− H− B− M− | ||
| Penv | ||||||
| + | N+ V+ H+ I+ M+ | N+ V+ I+ H+ B+ M+ | N+ I+ H+ B+ M+ | N+ V+ I+ H+ B+ M+ | ||
|
| ||||||
| − | N− V− H− B− M− | H− I− M− | H− I− M− | H− I− M− | ||
| Ppe | ||||||
| + | N+ I+ H+ B+ M+ | H+ I+ M+ | H+ I+ M+ | H+ I+ M+ | ||
|
| ||||||
| − | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N−− V−− I−− H−− B−− M−− | N− V− I− H− B− M− | N−− V− I− H− B− M−− | |
| Peh | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | N++ V++ I++ H++ B++ | |||
| + | N+ V+ I+ H+ B+ M+ | N+ V+ I+ H+ B+ M+ | ||||
| M++ | M++ | M++ | ||||
|
| ||||||
| − | N− V− H− B− M− | N− H− B− M− | N− H− B+ | N− H− B− M− | ||
| Pev | ||||||
| + | N+ I+ H+ B+ M+ | N+ I+ H+ B+ M+ | N+ H− B+ | N+ I+ H+ B+ M+ | ||
|
| ||||||
| − | N− V− I− H− B− M− | N− V− I− H− B− M− | N− V− I− H− B− M− | N− V− I− H− B− M− | N− V− I− H− B− M− | |
| μ LOS | ||||||
| + | N+ V+ I+ H+ B+ M+ | N+ V+ I+ H+ B+ M+ | N+ V+ I+ H+ B+ M+ | N+ V+ I+ H+ B+ M+ | N+ V+ I+ H+ B+ M+ | |
|
| ||||||
| − | N− H− I− B− M− | N− H− I− B− M− | N− H− I− B− M− | N− H− I− B− M− | ||
| λ | ||||||
| + | N+ H+ I+ B+ M+ | N+ H+ I+ B+ M+ | N+ H+ I+ B+ M+ | N+ H+ I+ B+ M+ | ||
|
| ||||||
| − | N− H− I− B− M− | N− H− I− B− M− | N− H− I− B− M− | |||
| nhcws | ||||||
| + | N+ H+ I+ B+ M+ | N+ H+ I+ B+ M+ | N+ H+ I+ B+ M+ | |||
|
| ||||||
| − | H++ M+ | H++ M+ | H++ M+ | H+ M+ | H+ | |
| Phh | ||||||
| + | H−− M− | H−− M− | H−− M− | H− M− | H− | |
|
| ||||||
| − | I++ M+ | I++ M+ | I++ M+ | I+ M+ | I+ | |
| Piso | ||||||
| + | I−− M− | I−− M− | I−− M− | I− M− | I− | |
|
| ||||||
| − | I− B− M− | I− B− M− | I− | I− B− M− | ||
| Pv | ||||||
| + | I+ B+ M+ | I+ B+ | I+ | I+ B+ M+ | ||
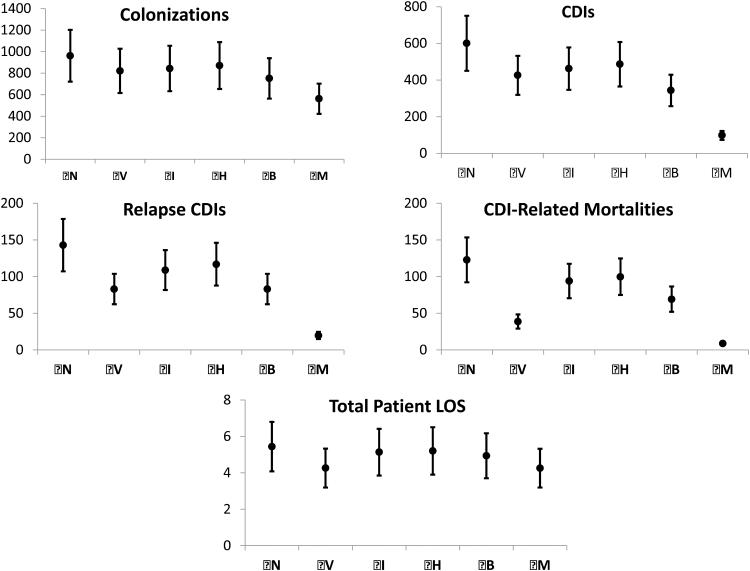
Figure 6 shows the results of the probabilistic sensitivity analysis including the 95% CIs for colonization, CDI, mortality, relapse CDI, and total patient LOS for each scenario. Overall, while increased variability in input parameters leads to increases in CDI related outcomes; the trend of strategy efficacy remains consistent with the average results. Additionally, increased uncertainty in input parameters did not result in large variability in the number of CDIs and relapse CDIs when strategy (M) is implemented. This is likely due to strategy (M) eliminating most nosocomial CDI due to the effectiveness of combined strategies.
Figure 6.
Results for the Monte Carlo sensitivity analysis. We compared individual CDI control strategies (V), (I), (H), (B), and mixed strategy (M) to the base case of no interventions (N).
DISCUSSION
In this study, we achieve two goals. First, we develop an ABM that models C. difficile spread and control in a hospital. Second, we use parameter estimates from clinical studies on CDI and our ABM to estimate transition probabilities for a Markov model that describes the natural progression of CDI in a patient. Together, these provide a framework for future development of more complex models that may help clinicians and infectious disease experts understand the dynamics of C. difficile transmission and the efficacy of individual CDI control strategies.
There have been few studies using agent-based modeling for nosocomial infectious diseases. Ruben et al.(44) also use an agent-based approach to study isolation and treatment policies for CDI. However, they use a simpler stochastic model for CDI progression in a patient, and details on any calibration or validation methods used in their model were not publicly available at the time this article was submitted. Barnes et al.(45) present an agent-based model for MRSA transmission and control that also considers interactions between patients, visitors, and HCWs, common MRSA control strategies, and a stochastic model for MRSA acquisition. We use this model as a starting point in the development of our ABM and expand these methods with additional complexities. For example, Barnes et al. do not evaluate the effectiveness of environmental disinfection or consider HCW interactions with other HCWs. Several other models study nosocomial infections using differential equations with an estimated basic reproduction number value for CDI transmission. However, an ABM can analyze the system dynamics and complexity of interactions in a hospital and give a detailed account of C. difficile spread, without uniform generalizations on transmissibility. When modeling the natural history of CDI in a patient, Lanzas et al.(10) use a more compact disease state transition model and exclude distinct states for relapse colonized and diseased patients. We consider low-probability events and relapse CDI patients as possible sources for CDI outbreaks, and hence we use a more complete transition model, which include the aforementioned relapse states. Furthermore, modeling interactions among agents allows us to evaluate the effects of individual infection control strategies, whereas evaluating adjustments to the parameters of closed-form expressions may not capture a sufficient level of detail for small-scale disease transmission dynamics.
Our model is not without its limitations. Several of our parameters are approximated from limited data. The admission rates for Colonized, Infected, or Not-susceptible patients are not explicitly available from our data or the literature; hence we rely on age as the only determining factor for CDI susceptibility. The inclusion of relative-risk for different patient conditions would provide insight to CDI spread in at-risk patients. However, limited data on disease progression for specific patient types make it difficult to calibrate the state transition probabilities of the Markov model for many different patient types. We also assume the probability of exposure to C. difficile after interacting with a contagious patient is the same for both HCWs and visitors. The exact relationship between these probabilities is not known at this time since the available data on the transmission of C. difficile spores to HCWs from patients is limited. That is, we are unaware of any studies on C. difficile transmission to visitors.
Additionally, since there may be different ways that the ABM matches observed data, our calibration method may not be very precise due to the unavailability of explicit data, which may also explain the variation given in Table 3. Furthermore, only a generalized strain of C. difficile. That is, we do not consider the possibility of a patient with the hyper virulent NAP1 strain, which has been shown to result in increased severity and transmissibility.(46) We make this assumption due to the lack of control studies on the NAP1 strain for CDI, and the availability of data on C. difficile over other strains. The incidence of relapse CDI for patients who do not receive antibiotic treatment is not well documented. Therefore, we assume the chance of relapse for CDI patients without vancomycin is the same as those who receive vancomycin. Moreover, the behavior of agents and hospital design in our model is simplistic. However, additional detail without explicit information would add uncertainty and increase the difficulty of validation. Cost of CDI control is not included in our model at this time, as the evaluation of costs would require additional analysis to determine accuracy of such cost estimates. In one study, the cost of vancomycin ranges from $71 to $143 per day depending on regimen.(47) However, many CDI control costs require further study of indirect costs, which require more complexities and detail in the ABM. For example, the cost of increased hand hygiene measures depends heavily on HCW type (i.e. physician, nurse, or ancillary) and time. Using 2012 average nurse and physician salary information from the US Bureau of Labor Statistics (USBLS), we estimate the cost of increased hand hygiene measures between $0.23 and $0.62 per hand washing episode. Bleach disinfection costs associated with environmental services personnel time are estimated between $3.08 and $4.11 per room using USBLS information. Furthermore, the cost of contact isolation ranges from $0.46 to $0.66 per protective gown, but the cost of reduced patient capacity due to contact isolation is unknown. Hence, we leave the topic of cost analysis to be addressed in future work.
We develop the model with a level of detail that we believe appropriately captures interactions among agents, while being generalized to avoid any major flaws in our assumptions or limitations from scarce data. Furthermore, we do not consider modeling environmental disinfection strategies beyond bleach since clinical data on the efficacy of newer technology such as hydrogen peroxide vapor and UV light disinfection are limited and these tools are not extensively used at this time. Since our model's primary focus is on horizontal transmission of C. difficile, we choose not to introduce additional complexity by adding antibiotic stewardship constraints.
This model serves as a framework for more complex models. Future iterations of the model will include multiple strains of CDI, additional antibiotic treatments (i.e. metronidazole and fidaxomycin), relative-risk of CDI due to a patient’s underlying condition, antibiotic stewardship, and the possibility of treating false positive CDIs. Additionally, due to the large number of parameters required for such a model, we plan to conduct a large observational study, which is necessary to obtain the data needed to perform a more rigorous calibration and validation of the model.
Table 4.
Spread of pi,j Values Across all Pxs in Π’ Accepted by the Calibration.
| CL | S | E | C | D | CR | R | X | NS | |
|---|---|---|---|---|---|---|---|---|---|
| CL | 0 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0.02 |
| S | 0 | <.001 | <.001 | <.001 | <.001 | 0 | 0 | 0 | 0 |
| E | 0 | 0.862 | 0 | 0.480 | 0.382 | 0 | 0 | 0 | 0 |
| C | 0.012 | 0 | 0 | <.001 | <.001 | 0 | 0 | 0 | 0 |
| D | 0.010 | 0 | 0 | 0 | 0.011 | 0.016 | 0 | 0.009 | 0 |
| CR | 0.019 | 0 | 0 | 0 | <.001 | 0.003 | 0 | ||
| R | 0.011 | 0 | 0 | 0 | 0 | 0 | 0.008 | 0.012 | 0 |
| X | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Acknowledgment
The authors thank Dr. Ajay Sethi and three anonymous reviewers for their suggestions and insights, which improved this paper.
Support: Supported through grant 2UL1TR000427 from the CTSA program of NCRR NIH and grant CMII-0844423 from the National Science Foundation.
Footnotes
Parts of this paper were presented in abstract form at the 2011 and 2013 annual meetings of the Society for Medical Decision Making.
REFERENCES
- 1.Redelings MD, Sorvillo F, Mascola L. Increase in Clostridium difficile–related Mortality Rates, United States, 1999–2004. Emerg Infect Dis. 2007 Sep;13(9):1417–9. doi: 10.3201/eid1309.061116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly CP, LaMont JT. Clostridium difficile — More Difficult Than Ever. N Engl J Med. 2008;359(18):1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 3.Mayfield JL, Leet T, Miller J, Mundy LM. Environmental control to reduce transmission of Clostridium difficile. Clin Infect Dis Off Publ Infect Dis Soc Am. 2000 Oct;31(4):995–1000. doi: 10.1086/318149. [DOI] [PubMed] [Google Scholar]
- 4.Miller B, Chen L, Sexton D, Anderson D. The impact of hospital-onset healthcare facility associated (HO-HCFA) Clostridium difficile infection (CDI) in community hospitals: Surpassing methicillin-resistant Staphylococcus aureus (MRSA) as the new superbug; Fifth Decennial International Conference on Healthcare, Associated Infections, Society for Healthcare Epidemiology of America; Atlanta. [Google Scholar]
- 5.Erik R, Dubberke MD, Dale N, Gerding MD, David Classen MM, Kathleen M, Arias MC, Kelly Podgorny RMC, Deverick J, Anderson MM, et al. Strategies to Prevent Clostridium difficile Infections in Acute Care Hospitals. Infect Control Hosp Epidemiol. 2008;29(S1):S81–S92. doi: 10.1086/591065. [DOI] [PubMed] [Google Scholar]
- 6.Novoa AM, Pi-Sunyer T, Sala M, Molins E, Castells X. Evaluation of hand hygiene adherence in a tertiary hospital. Am J Infect Control. 2007;35(10):676–83. doi: 10.1016/j.ajic.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Goldmann D, Huskins W. Control of nosocomial antimicrobial-resistant bacteria: a strategic priority for hospitals worldwide. Clin Infect Dis Off Publ Infect Dis Soc Am. 1997 Jan;24(Suppl 1):S139–45. doi: 10.1093/clinids/24.supplement_1.s139. [DOI] [PubMed] [Google Scholar]
- 8.Oughton MT, Loo VG, Dendukuri N, Fenn S, Libman MD. Hand hygiene with soap and water is superior to alcohol rub and antiseptic wipes for removal of Clostridium difficile. Infect Control Hosp Epidemiol Off J Soc Hosp Epidemiol Am. 2009 Oct;30(10):939–44. doi: 10.1086/605322. [DOI] [PubMed] [Google Scholar]
- 9.Eckstein BC, Adams D a, Eckstein EC, Rao A, Sethi AK, Yadavalli GK, et al. Reduction of Clostridium Difficile and vancomycin-resistant Enterococcus contamination of environmental surfaces after an intervention to improve cleaning methods. BMC Infect Dis. 2007 Jan;7:61. doi: 10.1186/1471-2334-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanzas C, Dubberke ER, Lu Z, Reske KA, Gröhn YT. Epidemiological Model forClostridium difficileTransmission in Healthcare Settings. Infect Control Hosp Epidemiol. 2011;32(6):553–61. doi: 10.1086/660013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva J, Jr, Batts DH, Fekety R, Plouffe JF, Rifkin GD, Baird I. Treatment of Clostridium difficile colitis and diarrhea with vancomycin. Am J Med. 1981 Nov;71(5):815–22. doi: 10.1016/0002-9343(81)90369-7. [DOI] [PubMed] [Google Scholar]
- 12.Fekety R, Silva J, Kauffman C, Buggy B, Deery HG. Treatment of antibiotic-associated Clostridium difficile colitis with oral vancomycin: comparison of two dosage regimens. Am J Med. 1989 Jan;86(1):15–9. doi: 10.1016/0002-9343(89)90223-4. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett JG, Tedesco FJ, Shull S, Lowe B, Chang T. Symptomatic relapse after oral vancomycin therapy of antibiotic-associated pseudomembranous colitis. Gastroenterology. 1980 Mar;78(3):431–4. [PubMed] [Google Scholar]
- 14.Bartlett JG. Treatment of antibiotic-associated pseudomembranous colitis. Rev Infect Dis. 1984 Apr;6(Suppl 1):S235–241. doi: 10.1093/clinids/6.supplement_1.s235. [DOI] [PubMed] [Google Scholar]
- 15.Teasley D, Olson M, Gebhard R, Gerding D, Peterson L, Schwartz M, et al. PROSPECTIVE RANDOMISED TRIAL OF METRONIDAZOLE VERSUS VANCOMYCIN FOR CLOSTRIDIUM-DIFFICILE-ASSOCIATED DIARRHOEA AND COLITIS. The Lancet. 1983 Nov 5;322(8358):1043–6. doi: 10.1016/s0140-6736(83)91036-x. [DOI] [PubMed] [Google Scholar]
- 16.Olson MM, Shanholtzer CJ, Lee JT, Gerding DN. Ten years of prospective Clostridium difficile-associated disease surveillance and treatment at the Minneapolis VA Medical Center, 1982-1991. Infect Control Hosp Epidemiol Off J Soc Hosp Epidemiol Am. 1994 Jun;15(6):371–81. doi: 10.1086/646934. [DOI] [PubMed] [Google Scholar]
- 17.De Lalla F, Nicolin R, Rinaldi E, Scarpellini P, Rigoli R, Manfrin V, et al. Prospective study of oral teicoplanin versus oral vancomycin for therapy of pseudomembranous colitis and Clostridium difficile-associated diarrhea. Antimicrob Agents Chemother. 1992 Oct;36(10):2192–6. doi: 10.1128/aac.36.10.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, et al. Fidaxomicin versus Vancomycin for Clostridium difficile Infection. N Engl J Med. 2011;364(5):422–31. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 19.Wenisch JM, Schmid D, Tucek G, Kuo H-W, Allerberger F, Michl V, et al. A prospective cohort study on hospital mortality due to Clostridium difficile. Infection. 2012 Oct 1;40(5):479–84. doi: 10.1007/s15010-012-0258-1. [DOI] [PubMed] [Google Scholar]
- 20.Pepin J, Valiquette L, Alary ME, Villemure P, Pelletier A, Forget K, et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Cmaj. 2004;171(5):466–72. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musher DM, Aslam S, Logan N, Nallacheru S, Bhaila I, Borchert F, et al. Relatively poor outcome after treatment of Clostridium difficile colitis with metronidazole. Clin Infect Dis Off Publ Infect Dis Soc Am. 2005 Jun 1;40(11):1586–90. doi: 10.1086/430311. [DOI] [PubMed] [Google Scholar]
- 22.Zar FA, Bakkanagari SR, Moorthi KMLST, Davis MB. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin Infect Dis Off Publ Infect Dis Soc Am. 2007 Aug 1;45(3):302–7. doi: 10.1086/519265. [DOI] [PubMed] [Google Scholar]
- 23.Cornely OA, Crook DW, Esposito R, Poirier A, Somero MS, Weiss K, et al. Fidaxomicin versus vancomycin for infection with Clostridium difficile in Europe, Canada, and the USA: a double-blind, non-inferiority, randomised controlled trial. Lancet Infect Dis. 2012 Apr;12(4):281–9. doi: 10.1016/S1473-3099(11)70374-7. [DOI] [PubMed] [Google Scholar]
- 24.Golan Y, Epstein L. Safety and efficacy of fidaxomicin in the treatment of Clostridium difficile-associated diarrhea. Ther Adv Gastroenterol. 2012 Nov;5(6):395–402. doi: 10.1177/1756283X12461294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, et al. Fidaxomicin versus Vancomycin for Clostridium difficile Infection. N Engl J Med. 2011;364(5):422–31. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 26.Sharp SE, Ruden LO, Pohl JC, Hatcher PA, Jayne LM, Ivie WM. Evaluation of the C.Diff Quik Chek Complete Assay, a New Glutamate Dehydrogenase and A/B Toxin Combination Lateral Flow Assay for Use in Rapid, Simple Diagnosis of Clostridium difficile Disease. J Clin Microbiol. 2010 Jun 1;48(6):2082–6. doi: 10.1128/JCM.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittet D, Mourouga P, Perneger TV. Compliance with Handwashing in a Teaching Hospital. Ann Intern Med. 1999;130(2):126–30. doi: 10.7326/0003-4819-130-2-199901190-00006. [DOI] [PubMed] [Google Scholar]
- 28.Muto C, Pokrywka M, Shutt K, Mendelsohn A, Nouri K, Posey K, et al. A Large Outbreak of Clostridium difficile, Associated Disease With an Unexpected Proportion of Deaths and Colectomies at a Teaching Hospital Following Increased Fluoroquinolone Use. Infect Control Hosp Epidemiol. 2005;26(3):273–80. doi: 10.1086/502539. [DOI] [PubMed] [Google Scholar]
- 29.Leekha S, Aronhalt KC, Sloan LM, Patel R, Orenstein R. Asymptomatic Clostridium difficile colonization in a tertiary care hospital: Admission prevalence and risk factors. Am J Infect Control. 2013 May;41(5):390–3. doi: 10.1016/j.ajic.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. C. difficile by Hospitalized Patients : Evidence for Acquisition of Clostridium New Admissions as a Source of Infection Colonized. Analysis. 2011;166(3):561–7. doi: 10.1093/infdis/166.3.561. N D. [DOI] [PubMed] [Google Scholar]
- 31.McFarland LV, Surawicz CM, Stamm WE. Risk Factors for Clostridium difficile Carriage and C. difficile-Associated Diarrhea in a Cohort of Hospitalized Patients. J Infect Dis. 1990;162(3):678–684. doi: 10.1093/infdis/162.3.678. [DOI] [PubMed] [Google Scholar]
- 32.Norén T, Åkerlund T, Bäck E, Sjöberg L, Persson I, Alriksson I, et al. Molecular Epidemiology of Hospital-Associated and Community-Acquired Clostridium difficile Infection in a Swedish County. J Clin Microbiol. 2004 Aug;42(8):3635–43. doi: 10.1128/JCM.42.8.3635-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dubberke ER, Butler AM, Reske KA, Agniel D, Olsen MA, D’Angelo G, et al. Attributable Outcomes of Endemic Clostridium difficile-associated Disease in Nonsurgical Patients. Emerg Infect Dis. 2008 Jul;14(7):1031–8. doi: 10.3201/eid1407.070867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, et al. A Predominantly Clonal Multi-Institutional Outbreak of Clostridium difficile–Associated Diarrhea with High Morbidity and Mortality. N Engl J Med. 2005;353(23):2442–9. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 35.Ghantoji SS, Sail K, Lairson DR, DuPont HL, Garey KW. Economic healthcare costs of Clostridium difficile infection: a systematic review. J Hosp Infect. 2010 Apr;74(4):309–18. doi: 10.1016/j.jhin.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Kenneally C, Rosini JM, Skrupky LP, Doherty JA, Hollands JM, Martinez E, et al. Analysis of 30-day mortality for clostridium difficile-associated disease in the icu setting*. CHEST J. 2007 Aug 1;132(2):418–24. doi: 10.1378/chest.07-0202. [DOI] [PubMed] [Google Scholar]
- 37.Law AM. Simulation Modeling & Analysis. 4th McGraw-Hill; New York: 2007. p. 768. [Google Scholar]
- 38.Kleijnen JPC. Verification and validation of simulation models. Eur J Oper Res. 1995;82(1):145–62. [Google Scholar]
- 39.Barbut F, Petit JC. Epidemiology of Clostridium difficile-associated infections. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2001 Aug;7(8):405–10. doi: 10.1046/j.1198-743x.2001.00289.x. [DOI] [PubMed] [Google Scholar]
- 40.Nasia Safdar, Sandra Olson, Linda McKinley. Controlling an Outbreak of Clostridium Difficile - Associated Diarrhea Using Sodium Hypochlorite Solution for Environmental Decontamination. Geriatr Nur (Lond) (Pending Revision) [Google Scholar]
- 41.Wilcox M, Fawley W, Wigglesworth N, Parnell P, Verity P, Freeman J. Comparison of the effect of detergent versus hypochlorite cleaning on environmental contamination and incidence of Clostridium difficile infection. J Hosp Infect. 2003 Jun;54(2):109–14. doi: 10.1016/s0195-6701(02)00400-0. [DOI] [PubMed] [Google Scholar]
- 42.Dettenkofer M, Wenzler S, Amthor S, Antes G, Motschall E, Daschner FD. Does disinfection of environmental surfaces influence nosocomial infection rates? a systematic review. Am J Infect Control. 2004;32(2):84–9. doi: 10.1016/j.ajic.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Apisarnthanarak A, Zack J, Mayfield J. Effectiveness of environmental and infection control programs to reduce transmission of Clostridium difficile. Clin Infect Dis. 2004;39:601–2. doi: 10.1086/422523. al. et. [DOI] [PubMed] [Google Scholar]
- 44.Rubin MA, Jones M, Leecaster M, Khader K, Ray W, Huttner A, et al. A simulation-based assessment of strategies to control clostridium difficile transmission and infection. PloS One. 2013;8(11):e80671. doi: 10.1371/journal.pone.0080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barnes S, Golden B, Wasil E. MRSA Transmission Reduction Using Agent-Based Modeling and Simulation. Inf J Comput. 2010;22(4):635–46. [Google Scholar]
- 46.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366(9491):1079–84. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 47.Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. 2013 Apr;108(4):478–498. doi: 10.1038/ajg.2013.4. quiz 499. [DOI] [PubMed] [Google Scholar]
- 48.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. Acquisition of Clostridium difficile by Hospitalized Patients: Evidence for Colonized New Admissions as a Source of Infection. J Infect Dis. 1992;166(3):561–561. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 49.Musher DM, Logan N, Bressler AM, Johnson DP, Rossignol J-F. Nitazoxanide versus vancomycin in Clostridium difficile infection: a randomized, double-blind study. Clin Infect Dis Off Publ Infect Dis Soc Am. 2009 Feb 15;48(4):e41–46. doi: 10.1086/596552. [DOI] [PubMed] [Google Scholar]
- 50.Shah D, Dang M-D, Hasbun R, Koo HL, Jiang Z-D, DuPont HL, et al. Clostridium difficile infection: update on emerging antibiotic treatment options and antibiotic resistance. Expert Rev Anti Infect Ther. 2010 May;8(5):555–64. doi: 10.1586/eri.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sethi AK, Al-Nassir WN, Nerandzic MM, Bobulsky GS, Donskey CJ. Persistence of skin contamination and environmental shedding of Clostridium difficile during and after treatment of C. difficile infection. Infect Control Hosp Epidemiol Off J Soc Hosp Epidemiol Am. 2010 Jan;31(1):21–7. doi: 10.1086/649016. [DOI] [PubMed] [Google Scholar]
- 52.Bobulsky GS, Al-Nassir WN, Riggs MM, Sethi AK, Donskey CJ. Clostridium difficile skin contamination in patients with C. difficile-associated disease. Clin Infect Dis Off Publ Infect Dis Soc Am. 2008 Feb 1;46(3):447–50. doi: 10.1086/525267. [DOI] [PubMed] [Google Scholar]
- 53.Landelle C, Verachten M, Legrand P, Girou E, Barbut F, Buisson CB. Contamination of Healthcare Workers’ Hands with Clostridium difficile Spores after Caring for Patients with C. difficile Infection. Infect Control Hosp Epidemiol. 2014 Jan;35(1):10–5. doi: 10.1086/674396. [DOI] [PubMed] [Google Scholar]
- 54.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989 Jan 26;320(4):204–10. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 55.Johnson S, Gerding DN. Clostridium difficile-Associated Diarrhea. Clin Infect Dis. 2011;26(5):1027–34. doi: 10.1086/520276. [DOI] [PubMed] [Google Scholar]
- 56.O’Neill GL, Beaman MH, Riley TV. Relapse Versus Reinfection with Clostridium Difficile. Epidemiol Infect. 1991;107(03):627–35. doi: 10.1017/s0950268800049323. [DOI] [PMC free article] [PubMed] [Google Scholar]