Significance
Transitions between asymmetric (self-renewal) and symmetric (proliferative) divisions for stem cells are precisely regulated during development and tissue regeneration. Caenorhabditis elegans heterochronic genes encode evolutionarily conserved developmental regulators, including lin-4 (lineage defect) and let-7 (lethal defects) microRNAs and the stem cell factor LIN-28 that control patterns of stem cell self-renewal and proliferation. It is not known how these developmental regulators interface with the machinery of division asymmetry. We report that, in C. elegans, the timing of transitions between asymmetric and symmetric stem cell divisions reflects developmental modulation of the LIT-1/POP-1/APR-1 (loss of intestine/posterior pharynx defect/APC related) asymmetry machinery by the heterochronic genes. These findings illuminate how evolutionarily conserved cellular asymmetry machinery can be coupled to microRNA-regulated developmental pathways for robust stem cell maintenance and proliferation.
Keywords: heterochronic, lit-1/Nlk, stem cell, pop-1/Tcf, asymmetric division
Abstract
Transitions between asymmetric (self-renewing) and symmetric (proliferative) cell divisions are robustly regulated in the context of normal development and tissue homeostasis. To genetically assess the regulation of these transitions, we used the postembryonic epithelial stem (seam) cell lineages of Caenorhabditis elegans. In these lineages, the timing of these transitions is regulated by the evolutionarily conserved heterochronic pathway, whereas cell division asymmetry is conferred by a pathway consisting of Wnt (Wingless) pathway components, including posterior pharynx defect (POP-1)/TCF, APC related/adenomatosis polyposis coli (APR-1)/APC, and LIT-1/NLK (loss of intestine/Nemo-like kinase). Here we explore the genetic regulatory mechanisms underlying stage-specific transitions between self-renewing and proliferative behavior in the seam cell lineages. We show that mutations of genes in the heterochronic developmental timing pathway, including lin-14 (lineage defect), lin-28, lin-46, and the lin-4 and let-7 (lethal defects)-family microRNAs, affect the activity of LIT-1/POP-1 cellular asymmetry machinery and APR-1 polarity during larval development. Surprisingly, heterochronic mutations that enhance LIT-1 activity in seam cells can simultaneously also enhance the opposing, POP-1 activity, suggesting a role in modulating the potency of the cellular polarizing activity of the LIT-1/POP-1 system as development proceeds. These findings illuminate how the evolutionarily conserved cellular asymmetry machinery can be coupled to microRNA-regulated developmental pathways for robust regulation of stem cell maintenance and proliferation during the course of development. Such genetic interactions between developmental timing regulators and cell polarity regulators could underlie transitions between asymmetric and symmetric stem cell fates in other systems and could be deregulated in the context of developmental disorders and cancer.
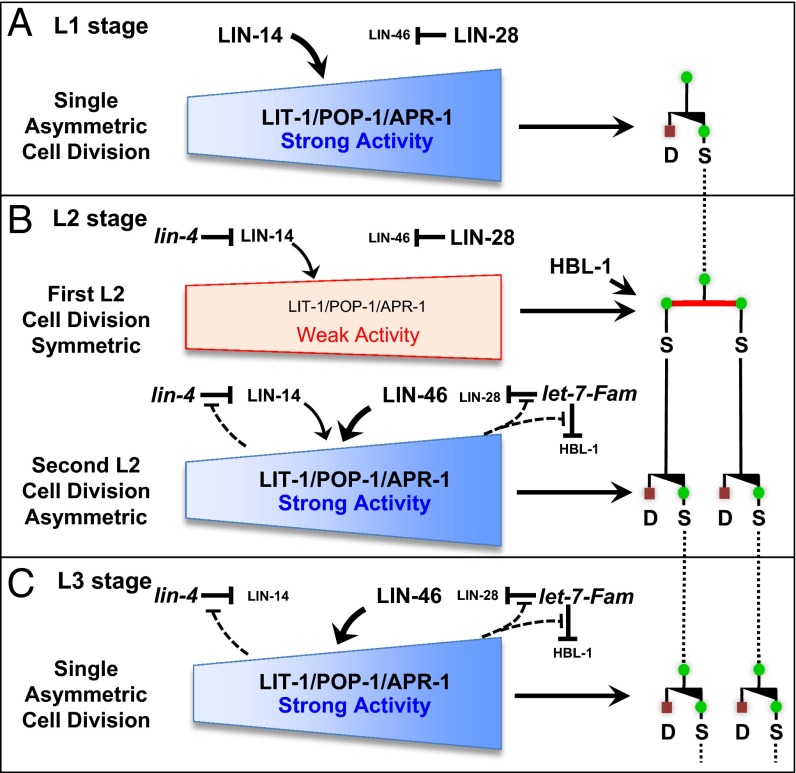
During development and tissue regeneration, stem cells generate cellular diversity through asymmetric divisions that produce a stem cell and a differentiated cell, or alternatively, through symmetric divisions that produce either two stem cells or two differentiated cells (Fig. 1A). The proper regulation of asymmetric vs. symmetric cell division patterns for stem cells is fundamental to the elaboration of developmental patterns of cell fate and for tissue growth and homeostasis (1, 2). Many of the genes that are involved in the regulation of stem cell division asymmetry are evolutionarily conserved and have been found to be associated with carcinogenesis, underlining the biomedical significance of understanding this process (3–5).
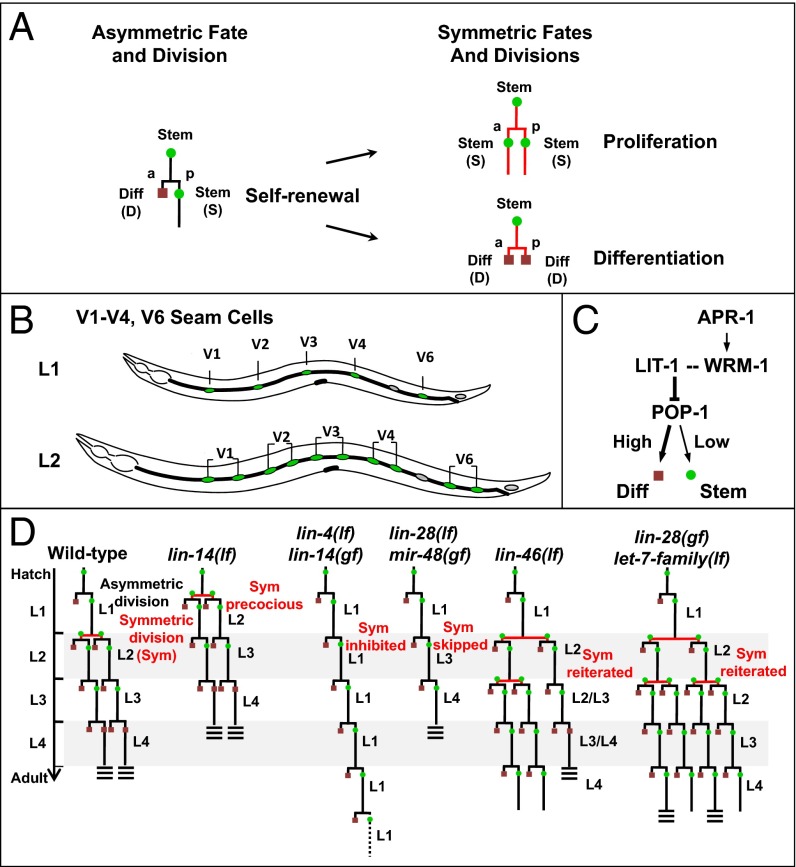
Fig. 1.
Stage-specific asymmetric and symmetric divisions in postembryonic seam cell divisions in the heterochronic mutants. (A) Asymmetric cell division yields one anterior (a) differentiated cell (D, red square) and one posterior (p) stem (seam) cell (S, green circle); the asymmetric division can transit to symmetric division generating either two stem cells or two differentiating cells. (B) Arrangement of lateral hypodermal seam stem cells along one side of an L1 and L2 larva in C. elegans. (C) Genetic relationship of lit-1, wrm-1, and apr-1 on the pop-1 regulation and cell fate of seam cells in C. elegans. (D) Normal stage-specific division pattern of V1–V4/V6 cell lineages during postembryonic development of WT C. elegans; seam cells divide once at the beginning of each larval stage (or twice in the L2) until the final molt, when they terminally differentiate and secrete adult specific cuticular alae (triple bars). These cell lineage patterns include stage-specific asymmetric cell divisions [yielding one differentiated cell (red square) and one stem (seam) cell (green circle) and (specifically in the L2 stage) symmetric divisions (generating two stem cells)]. The normally L2-specific symmetric division occurs precociously (promoted) in L1 stage in the lin-14(lf) mutant, is prevented in the lin-14(gf) and lin-4(lf) mutants, is skipped in lin-28(lf), and is reiterated in lin-46(lf), lin-28(gf), or let-7-family(lf) larvae.
In Caenorhabditis elegans, the lateral hypodermal cell lineages display developmentally regulated transitions between asymmetric and symmetric divisions, providing a model to study the control of stem cell division behavior in vivo (6–8). These stem cells (called “seam” cells) are aligned along each side of the animal (Fig. 1B). Seam cells undergo a single round of asymmetric self-renewing division at each larval stage (L1–L4), producing an anterior daughter that terminally differentiates (fuses to the hyp7 syncytium) and a posterior daughter that maintains stem cell properties and remains a seam cell (Fig. 1D). In addition, at the L2 stage, five of these seam cells, V1–V4 and V6 (V1–V4/V6), undergo a symmetric proliferative division that doubles the number of seam cells to ten (Fig. 1 B and D). In the L4 larval stage, each seam cell exits the cell cycle, fuses with adjacent seam cells, differentiates into a skin cell, and generates a specialized cuticle structure, called alae (9).
Temporal control of seam cell lineage development relies on a cascade of heterochronic genes including developmentally expressed lin-4 (lineage defect) and let-7 (lethal defects) family microRNAs that regulate the production of key downstream proteins, LIN-14, LIN-28, and HBL-1 (HunchBack Like), and thereby control the timing of stage-specific developmental programs (10, 11). Mutations of these microRNA genes or of downstream heterochronic genes alter the temporal transitions between asymmetric and symmetric seam cell divisions, such that seam cell lineages can either skip the L2 symmetric divisions, resulting in decreased number of seam cells, or reiterate the L2 symmetric divisions, resulting in overproliferation (Fig. 1D). For example, the lin-4 microRNA progressively down-regulates LIN-14 through the L1 and L2 larval stages. LIN-14 is a transcription factor whose developmental expression controls the L2-specific execution of symmetric cell division (12). A high level of LIN-14 in the L1 inhibits symmetric seam cell division and hence specifies an asymmetric division program, whereas down-regulation of LIN-14 by lin-4 causes a switch to symmetric division in the L2 (13). lin-14 loss of function (lf) mutations cause precocious L2 symmetric divisions in the L1 stage (Fig. 1D). By contrast, persistent LIN-14 expression, which occurs in lin-14 gain of function (gf) or lin-4(lf) mutants, prevents symmetric divisions and causes reiteration of the L1 asymmetric cell division pattern at all stages (14) (Fig. 1D). let-7 family microRNAs also contribute to the timing of the L2 symmetric divisions by progressively down-regulating LIN-28 and HBL-1 through the L2 and L3 stages (15). LIN-28 is an evolutionary conserved RNA-binding protein (16) with roles in promoting cell proliferation and pluripotency (17). C. elegans lin-28(lf) mutants skip the L2 symmetric division, resulting in decreased seam cell number and premature adult epidermal differentiation (Fig. 1D). lin-46 encodes a putative scaffolding protein that was identified by mutations that suppress lin-28(lf) phenotypes indicating that lin-46 functions downstream of lin-28 in the regulation of seam cell fate and division asymmetry (18).
How the heterochronic gene pathway regulates the timing of symmetric and asymmetric divisions of seam cells is not well understood. Although, the Wnt (wingless) ligands, including the products of lin-44, cwn-1, egl-20, and cwn-2, are redundantly required to coordinate the orientation and extrinsic polarity of certain cells, they do not affect the intrinsic polarity and asymmetric division of seam cells (19). A noncanonical Wnt asymmetry pathway (Wnt asymmetry) underlies asymmetric cell divisions and cell polarity in most lineages of C. elegans (7, 20–22). For example, reduction of POP-1 (posterior pharynx defect), the C. elegans homolog of the vertebrate TCF transcription factor, affected asymmetric seam cell divisions such that instead of dividing to produce one seam cell and a differentiated cell, pop-1–depleted seam cells divided to produce two seam cells, resulting in an increase in the number of seam cells (23, 24). Reduction of the C. elegans β-catenin homolog, wrm-1 (worm armadillo), was observed to cause both daughters of these divisions to adopt the differentiation fate, resulting in an overall decrease in the number of seam cells (24). APR-1(APC Related) is the worm homolog of mammalian APC (adenomatosis polyposis coli), a conserved cytoplasmic protein with roles in cell polarity and Wnt signaling. APR-1 has been implicated in the regulation of the Wnt pathway in seam cells and is expressed asymmetrically to the anterior cortex of seam cells (25, 26). On activation by Wnt signaling, LIT-1/NLK (loss of intestine/Nemo-like kinase) (27, 28) forms a complex with WRM-1 to phosphorylate POP-1, enhancing POP-1 nuclear export and lowering its level (Fig. 1C) (29, 30). Consistent with this role for LIT-1 and WRM-1 in opposing POP-1, reduction of LIT-1 was reported to decrease the number of seam cells (23, 24), just as was the case for reduction of WRM-1. Despite extensive studies on WRM-1, the nature of decreasing the number of seam cells by lit-1 reduction is not well studied.
Because the heterochronic genes regulate the temporal transitions between asymmetric and symmetric divisions in V1–V4/V6 stem cells and because the noncanonical Wnt asymmetry pathway underlies the polarity of these cells, we postulated that the stage-specific execution of symmetric or asymmetric divisions by seam cells could result from stage-specific modulation of the Wnt asymmetry pathway by the heterochronic genes. Here we report genetic evidence that seam cell transitions between asymmetric and symmetric stem cell fates reflects the action of heterochronic genes in modulating LIT-1/POP-1/APR-1 cellular asymmetry. We show that LIT-1 functions in opposition to POP-1 to promote the asymmetric, self-renewing divisions of V1–V4/V6 seam cells. For seam cells that divide asymmetrically, lit-1 is required for the posterior daughter to express the stem cell fate instead of differentiating fate. We further show that the heterochronic genes either enhance or repress the LIT-1–POP-1 asymmetry axis at specific stages of development. These results suggest a model wherein the timing of stem cell division asymmetry is specified by dynamic genetic interactions between components of heterochronic gene and Wnt cellular asymmetry pathways.
Results
LIT-1 Promotes Seam Cell Division Asymmetry and Opposes Differentiation.
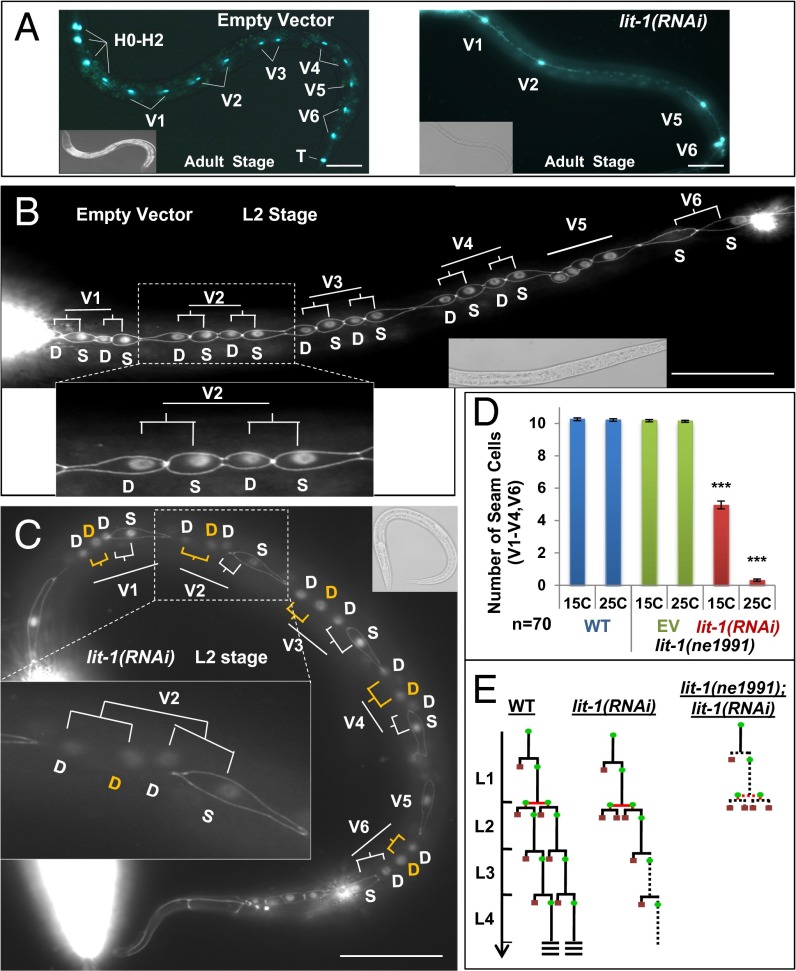
lit-1(RNAi) resulted in a reduced number of cells expressing the seam cell reporter scmp::gfp in L4 and young adult (YAD) animals (Fig. 2A and Fig. S1A) (23, 24). A similar reduction in seam cell number was observed after lit-1(RNAi) of animals expressing a different seam cell specific GFP reporter, bro-1::gfp (31) (Fig. S1B). Normally, each anterior daughter of a V lineage seam cell differentiates and fuses to the hypodermal syncytium (hyp7) (9). This fusion/differentiation involves the loss of junctions between the differentiating cell and its neighbors, as visualized using the cellular junction GFP reporter ajm-1::gfp (32). We found that lit-1(RNAi) of scmp::gfp;ajm-1::gfp animals caused a portion of seam cell daughters at each larval stage to differentiate instead of adopting the stem cell fate, resulting in progressive depletion of the seam cell population (Fig. 2 B and C and Fig. S1A). Cell lineage analysis confirmed that lit-1(RNAi) converts these asymmetric (normally self-renewing) seam cell divisions into symmetric divisions that generate two differentiating cells (Fig. 2E and Fig. S2).
Fig. 2.
lit-1/Nlk promote seam cell division asymmetry and oppose differentiation. (A) lit-1(RNAi) (Right) causes decrease in seam cell number, assayed in young adults (YA) using scmp::gfp, compared with empty vector RNAi controls (Left). (B) Animals doubly marked with scmp::gfp and ajm-1::gfp were used to monitor fusion (differentiation) of seam cells to syncytium during the L2 developmental stage. In the WT, four sisters for each V seam cell lineage are evident just after the L2 asymmetric divisions. The posterior sisters remain seam cells, whereas the anterior sisters differentiate by fusing to the syncytium. (C) lit-1(RNAi) caused premature fusion (differentiation) of seam cells (yellow D) in L2 stage asymmetric division. S, seam cell; D, differentiated cell. (D) lit-1(RNAi) seam cell number phenotype is enhanced in combination with the lit-1(ne1991) hypomorphic mutation. (E) Schematic representation of seam cell division pattern in WT, lit-1(RNAi) and lit-1(RNAi);lit-1(n1991) animals. Seam cell fate: green circles; differentiation fate: red squares. Dotted line, variable fates (seam or differentiation). Anterior is to the left. Error bars indicate ±SEM. *P < 0.05, **P < 0.01, ***P < 0.005, two-tailed t test. (Scale bar, 50 μm.)
The seam cell phenotypes of lit-1(RNAi) were not completely penetrant and were somewhat larval stage specific. Seam cells at the L2 and L3 larval stages were more sensitive to lit-1(RNAi) than cells at the L1 stage (Figs. S1A and S2), and the V1–V3 lineages were more sensitive than the V4 and V6 lineages (Fig. S2). Moreover, lit-1(RNAi) did not affect equally all four of the seam cell granddaughters at the L2 stage. One explanation for this incomplete phenotype could be insufficient knockdown of lit-1. To achieve maximal reduction in lit-1 activity, we used lit-1(RNAi) in the background of a temperature-sensitive lit-1 hypomorphic mutation, lit-1(ne1991) (33). lit-1(ne1991) is embryonic lethal at 25 °C but viable at 15 °C. Seam cell numbers are normal for lit-1(ne1991) larvae grown at 15 °C or shifted to 25 °C after embryogenesis. However, lit-1(RNAi) caused a dramatic increase in the frequency of seam cell differentiation for lit-1(ne1991) mutant larvae at 25 °C compared with lit-1(RNAi) alone (Fig. 2D). This enhanced lit-1(RNAi);lit-1(ne1991) phenotype extended to all V lineages and all stages (Fig. 2E).
In embryonic blastomeres and many larval asymmetric cell divisions, POP-1 is asymmetrically localized in the nuclei of sister cells at each asymmetric division, and POP-1 has been proposed to function as a transcriptional repressor at higher nuclear level in anterior sisters and as a transcriptional activator at lower nuclear level in the posterior cells (34, 35). Using a pop-1::gfp transgene (36), we confirmed that nuclear level of GFP-tagged POP-1 was expressed asymmetrically in the L2 asymmetric division, with a higher level in the anterior sister (differentiating fate, syncytium) than the posterior sister (seam fate) (Fig. S3A). Nuclear POP-1 was significantly reduced in pop-1(RNAi) animals where anterior sisters become seam cells instead of syncytium (Fig. S3B). Consistent with LIT-1 acting upstream of POP-1, we observed that lit-1(RNAi) resulted in a significant increase of POP-1 expression in posterior sister cell nuclei (Fig. S3C). These data demonstrate that LIT-1 inhibits the nuclear level of POP-1, which determines the fate of V1-V4/V6 seam cells.
Previous studies have indicated the polarization of seam cell divisions may be largely independent of the known C. elegans Wnt ligands (19, 24) and hence could result from signals other than conventional Wnt ligands. To identify other contributors to seam cell polarity, we conducted a candidate gene screen by RNAi, with emphasis on genes previously implicated in cell polarity (Table S1). Noteworthy genes from this screen include ect-2 (RhoGEF) and cdc-42 (RhoGTPase). ect-2(RNAi) or cdc-42(RNAi) resulted in reduced numbers of seam cells, similar to lit-1(RNAi) (Fig. S4). ect-2 has been implicated in the control of mitotic spindle assembly in Xenopus through activation of cdc-42 (37), and ECT-2 mislocalization is associated with malignant transformation (38). ECT-2 has also been reported to interact with the polarity complex and bind to PAR-6 (partitioning abnormal) (39). This result implies a potential role for PAR-related polarity factors in seam cell asymmetry, perhaps together with Rho family and Wnt components (5). However, we did not observe any effects on seam cell numbers after knockdown of other PAR pathway genes (Table S1).
The Heterochronic Gene lin-14 Promotes Asymmetric and Inhibits Symmetric Cell Division.
It was intriguing that depletion of LIT-1 or POP-1 by RNAi in WT larvae had no significant effects on L1 seam cells, whereas at later larval stages, seam cells were affected. Although this could reflect a relatively inefficient RNAi knockdown in L1 larvae, it is also possible that during normal development the activities of LIT-1 and POP-1 are relatively enhanced in the L1 stage compared with later stages, such that reduction of LIT-1 or POP-1 levels by RNAi may be insufficient to reduce their activities below a threshold in the L1. Because mutants in the heterochronic pathway exhibit temporal changes in the stage specificity of seam cell division asymmetry (Fig. 1D), we hypothesized that certain heterochronic genes, particularly lin-14, may exert an L1-specific potentiation of LIT-1/POP-1 activity to reinforce seam cell asymmetry. The level of LIN-14 is high in the L1 stage of the WT, promoting the expression of L1 cell lineage patterns, including the asymmetric seam cell division characteristic of the L1. LIN-14 decreases after the L1, resulting in the transition from asymmetric seam cell division in the L1 to symmetric division in the L2 (12). We therefore tested whether lin-14 activity potentiates POP-1/LIT-1 activity in seam cells during the L1 stage.
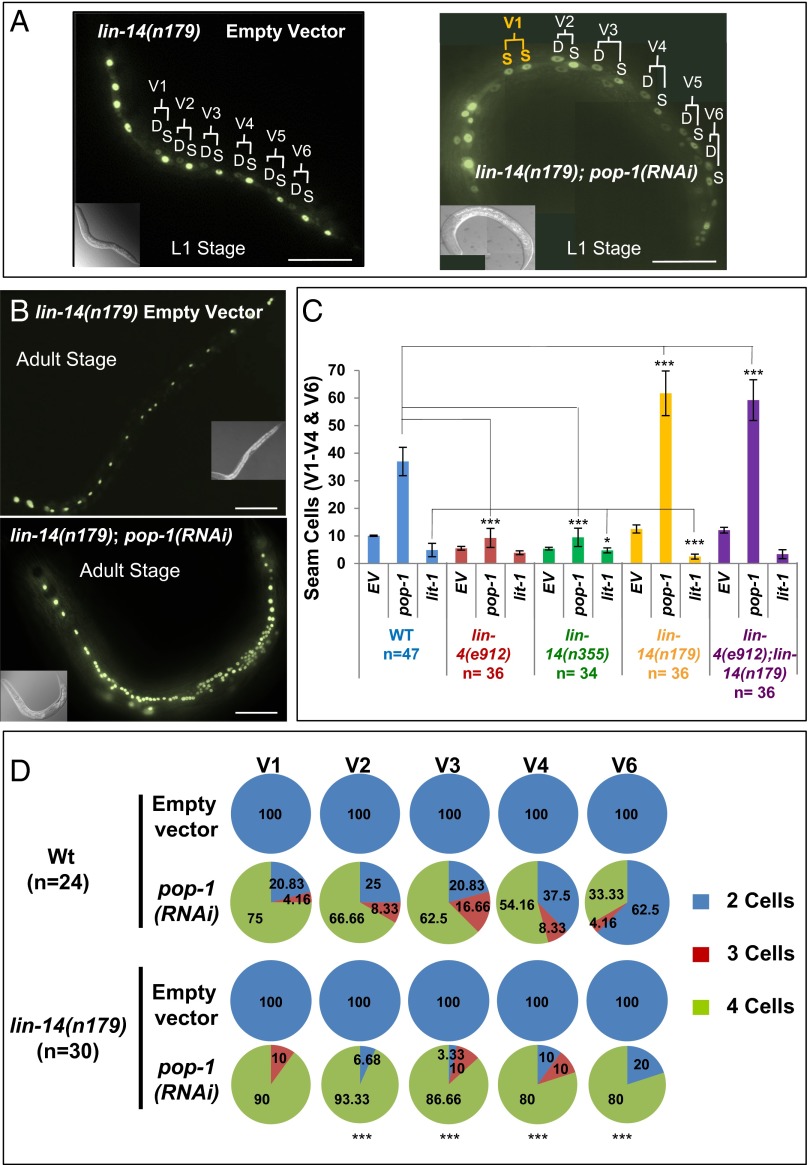
lin-14(n179) is a temperature-sensitive loss-of-function lin-14 mutation. In lin-14(n179) animals at nonpermissive temperature (25 °C), the L1 asymmetric division does not occur, and instead, seam cell lineages express the L2-specific symmetric division precociously in the first larval stage (Fig. 1D) (12, 13). At more permissive temperature (15 °C), this lin-14(n179) precocious L2 phenotype is expressed more weakly. We used the lin-14(n179) mutant at 15 °C as a sensitized background to determine whether lin-14 activity collaborates with pop-1 in the L1. Indeed, we found that pop-1(RNAi) of lin-14(n179) animals at 15 °C caused a 13.3% (n = 45) conversion of asymmetric to symmetric division in the L1 (Fig. 3A). This finding, that reduced lin-14 activity synergizes with pop-1(RNAi) for conversion of asymmetric seam cell divisions into symmetric divisions, suggests that LIN-14 may promote asymmetric division of seam cells in the L1 of WT animals by (directly or indirectly) reinforcing POP-1 activity.
Fig. 3.
lin-14 modulate asymmetric and symmetric cell fates. (A) Effect of pop-1(RNAi) on lin-14(n179) L1 larvae at permissive temperature 15 °C showing GFP-marked seam cells after control RNAi (Left) or pop-1(RNAi) (Right). S indicates a seam cell and D indicates a differentiated cell based on the location, size, and shape of the nucleus, using differential interference contrast (DIC) microscopy (9). (B) Representative of lin-14(n179) young adults (YA) stage at 20 °C showing GFP-marked seam cells after control RNAi (Upper) or pop-1(RNAi) (Lower). (C) Effect of pop-1(RNAi) and lit-1(RNAi) compare with empty vector (EV) control on seam cells numbers in lin-4(e912), lin-14(n355), lin-14(n179), and lin-4(e912);lin-14(n179) backgrounds at 20 °C. (D) Quantitative analysis of the effects of lin-14(n179) on the sensitivity of L2 V1-V4/V6 seam lineages to pop-1(RNAi) at 15 °C. In the second cell division in L2 stage, each V1–V4/V6 seam cell undergoes two rounds of division producing two seam cells and two differentiated cells per lineage. (Upper) Effect of pop-1(RNAi) on individual V1-V4/V6 seam cells in WT by changing the asymmetric to symmetric division producing three or four seam cells. The production of four cells is pronounced as the result of hypersensitivity of lin-14(n179) to pop-1(RNAi) (Lower). Anterior is to the left. Error bars indicate ±SEM. *P < 0.05, **P < 0.01, ***P < 0.005, two-tailed t test. (Scale bar, 50 μm.)
Interestingly, we found that lin-14 activity impacts pop-1 activity after the L1 stage also. The overall number of seam cells detected in adult animals is dramatically enhanced after pop-1(RNAi) of lin-14(n179) animals compared with pop-1(RNAi) of WT (Fig. 3 B–D, Table 1, and Fig. S5). Most of these extra seam cells can be attributed to the effects on the division asymmetry of the second round of L2 seam cell divisions. This division is normally asymmetric, where each anterior sister differentiates and fuses to the syncytium and each posterior sister remains a seam cell (Fig. 1D). In the WT animals, POP-1 promotes this second round of L2 asymmetric division; pop-1(RNAi) results in a certain frequency of conversion of the L2 asymmetric division to a symmetric division producing two seam cells (Fig. 3D), and this frequency is highly increased in lin-14(n179) (Figs. 3 B–D and 4 and Table 1). Consistent with this hypersensitivity to pop-1 loss of function being a consequence of reduced LIN-14 activity, lin-14(n179) animals displayed an even stronger hypersensitivity to pop-1(RNAi) and lit-1(RNAi) at nonpermissive temperatures of 20 °C and 25 °C compared with 15 °C. (Fig. S5).
Table 1.
Analysis of genetic interactions between heterochronic genes and lit-1 and pop-1 on average number of V1–V4 and V6 seam cells in young adult animals
| Strains | Empty vector | pop-1(RNAi) | lit-1(RNAi) | n |
| WT (N2)† | 10 | 35.5 ± 7.2* | 3.61 ± 2.2* | 50 |
| lin-4(e912)† | 5 ± 0.6 | 9.2 ± 2.3 | 3.8 ± 0.7 | 34 |
| lin-14(n179)† | 12.5 ± 1.3 | 61.7 ± 8.0* | 2.5 ± 0.8* | 36 |
| lin-14(n355)† | 5 ± 0.4 | 9.5 ± 3.3 | 4.7 ± 0.9 | 34 |
| lin-4(e912);lin14(n179)† | 12.0 ± 1.0 | 59.2 ± 7.3* | 3.3 ± 1.6* | 39 |
| lin-28(n719)† | 5.2 ± 0.4 | 5.2 ± 0.4 | 4.9 ± 0.6 | 34 |
| lin-4(e912);lin-28(n719) | 5.3 ± 0.4 | 5.4 ± 0.5 | 5.2 ± 0.4 | 40 |
| lin-4(e912);lin-28(n947) | 5.3 ± 0.5 | 5.37 ± 0.5 | 5.5 ± 0.5 | 40 |
| lin-28 3′UTR (ΔLCE)‡ | 20.9 ± 2.2 | 42.2 ± 3.4** | 6.7 ± 1.2* | 22 |
| lin-66(ok3326)† | 22.3 ± 2.9 | 40.6 ± 10.6** | 6 ± 1.0* | 23 |
| lin-28(n719);lin-14(n179)† | 5.2 ± 0.4 | 5.5 ± 0.7 | 5.1 ± 0.3 | 44 |
| lin-46(ma146)† | 12.4 ± 0.9 | 52 ± 6.5* | 2.18 ± 1.0* | 35 |
| lin-46(bp312)† | 14.5 ± 1.5 | 56.0 ± 6.0* | 1.7 ± 1.0* | 34 |
| lin-28(n719);lin-46(ma146)† | 13.2 ± 1.5 | 47.7 ± 8.9* | 0.35 ± 0.6* | 34 |
| mir-48 mir-241(nDf51);mir-84(n4037)† | 15.7 ± 1.6 | 49.5 ± 11.5* | 10.4 ± 2.6 | 42 |
| lin-28(n719);mir-48mir-241(nDf51);mir-84(n4037)§ | 5.4 ± 0.5 | 7.0 ± 0.8 | 4.5 ± 0.5 | 36 |
| mir-48(veIs48)¶ | 5.2 ± 0.4 | 5.3 ± 0.5 | 5.2 ± 0.4 | 35 |
| lin-46(ma164);mir-48(veIs48)¶ | 5.3 ± 0.5 | 11.9 ± 2.2*** | — | 36 |
| lin-4(e912);lin-46(ma164)†,|| | 6.2 ± 0.4 | 9.5 ± 1.8 | 4.1 ± 1.0 | 32 |
| hbl-1(ve18)† | 12.8 ± 1.9 | 42.3 ± 5.5* | 5.5 ± 1.6*** | 25 |
P values are based on two-sample equivalence test of means: *P < 0.001, **P < 0.05, ***P < 0.01. n, number of animals.
Animals contain wIs51, which expresses a seam specific GFP, scm::GFP reporter integrated on chromosome V.
Animals contain bpIs145(lin-28::gfp::lin-28).
Animals contain maIs105, which expresses an adult-specific, col-19::GFP reported integrated on chromosome V.
Animals contain veIs48 [mir-48(ve33 +++) sur-5::GFP].
Animals contain unc-76(e911) linked to lin-46.
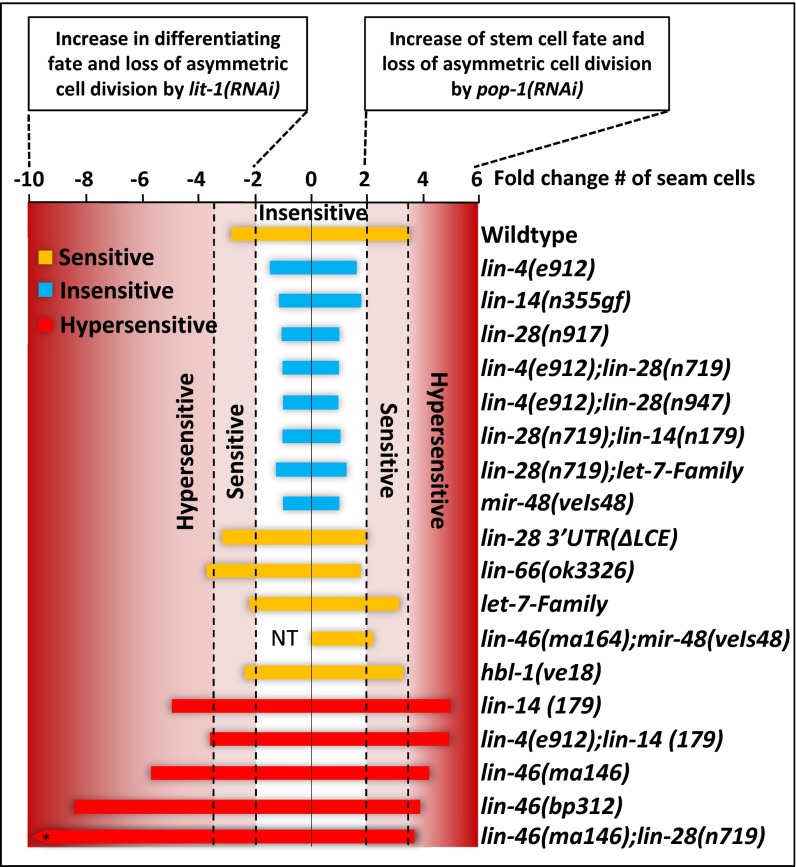
Fig. 4.
Heterochronic genes control the stem cell fate. The graph represents the level of sensitivity or insensitivity of the heterochronic mutants compare with WT based on the data reported in Table 1. The x axis shows the fold change of seam cell number decrease by lit-1(RNAi) or increase by or pop-1(RNAi) (Table 1). Strains are classified by the fold change (either decrease or increase, compared with the WT) in seam cell number that they displayed after pop-1(RNAi) or lit-1(RNAi) as follows: insensitive (blue bars) for less than twofold change, sensitive (yellow bars) for fold change between 2-fold and 3.5-fold, and hypersensitive (red bars) for fold change greater than 3.5. NT, not tested. *The fold decrease of seam cell number by lit-1(RNAi) for lin46(ma146);lin-28(n719) strain is −37.4.
Remarkably, lin-14(n179) animals also showed hypersensitivity to lit-1(RNAi) (Figs. 3C and 4, Fig. S5, and Table 1). It is noteworthy that reduction of lin-14 increases the strength of the lit-1(RNAi) phenotype and the pop-1(RNAi) phenotype, given that pop-1(RNAi) and lit-1(RNAi) result in opposite phenotypes, increased seam cell numbers vs. reduced seam cell numbers, respectively. We interpret this result to indicate that in the WT, LIN-14 promotes seam cell division asymmetry/polarity by effectively reinforcing the activities of opposing determinants for each daughter cell: that is, LIN-14 appears to reinforce LIT-1 activity for the posterior (stem) daughter and to reinforce POP-1 activity for the anterior (differentiating) daughter.
To test whether continuous overexpression of LIN-14 would have the opposite effect on pop-1 and lit-1 function as lin-14(lf), we examined a lin-4 loss of function mutant, lin-4(e912). lin-4 encodes a microRNA that controls the transition from L1 to L2 seam cell fates by progressively down-regulating LIN-14 after the L1 (40). lin-4(e912) (loss of function) animals overexpress LIN-14 continuously, and consequently, lateral hypodermal cell lineages express only asymmetric seam cell divisions at all stages (Fig. 1D). The lateral hypodermal seam cell lineages of lin-4(lf) exhibited insensitivity to both lit-1(RNAi) and pop-1(RNAi) throughout larval development and continued to express asymmetric cell divisions at each stage despite depletion of lit-1 or pop-1 (Figs. 3C and 4 and Table 1). We confirmed that lin-4(lf) mutant is competent for RNAi, by verifying that it is as sensitive as the WT to gfp(RNAi), and hmr-1(RNAi) (41) (Fig. S6).
To confirm that the insensitivity of lin-4(lf) seam cells to lit-1(RNAi) or pop-1(RNAi) is the result of hyperactivity of lin-14 (the principle target of lin-4), we tested a lin-14 gain-of-function (gf) mutant, lin-14(n355gf) and a lin-4(lf);lin-14(lf) double mutant for sensitivity to lit-1(RNAi) and pop-1(RNAi). lin-14(n355gf) animals exhibited insensitivity to both lit-1(RNAi) and pop-1(RNAi) very similar to lin-4(lf) (Figs. 3C and 4 and Table 1). Because lin-14(gf) animals overexpress lin-14 despite being WT for lin-4, we conclude that the insensitivity of lin-14(gf) animals to lit-1(RNAi) or pop-1(RNAi) likely results directly from hyperactivity of lin-14 and further indicates that the insensitivity of lin-4(lf) to lit-1(RNAi) or pop-1(RNAi) is also a consequence of lin-14 hyperactivity. Consistent with this conclusion, we found that the lin-4(lf);lin-14(lf) double mutant is hypersensitive to lit-1(RNAi) or pop-1(RNAi) much like the single mutant lin-14(lf) (Figs. 3C and 4 and Table 1).
The above findings indicate that lin-14 promotes the activities of both pop-1 and lit-1 in seam cell division asymmetry. We conclude that in WT larval development, lin-14 activity functions in the L1 to reinforce seam cell asymmetric division by enhancing lit-1 and pop-1 activities and that the down-regulation of LIN-14 after the L1 (by the lin-4 microRNA) promotes the transition to symmetric division in the first L2 division. Interestingly, we find that lin-14(lf) results in hypersensitivity to lit-1 or pop-1 RNAi for the second L2 divisions, which are asymmetric in the WT. This result suggests that L2 levels of lin-14 are reduced compared with the L1, such that at the first L2 divisions, LIN-14 level is insufficient to promote asymmetric division of seam cells, but for the second L2 seam cell divisions, a medium level of LIN-14 contributes (along with heterochronic gene lin-46, see below) to promoting strong seam cell asymmetry. A medium level of LIN-14 in the L2 is consistent with the genetic analysis of the stage specificity of lin-14 function in specifying seam cell fates (12).
Let-7 Family microRNAs Act via lin-28 and lin-46 to Modulate Asymmetric and Symmetric Cell Fates.
The let-7-family microRNAs (let-7-Fam), including mir-48, mir-84, and mir-241, function together semi-redundantly to control L2 to L3 fate transitions (15). Animals that were triply mutant for mir-48, mir-84, and mir-241 exhibit reiterations of the L2-specific symmetric seam cell division (15). Conversely, a transgene overexpressing (gain of function) a single member of the family, mir-48(veIs48), results in skipping of the L2-specific symmetric seam cell division (42) (Fig. 1D). We found that loss of the let-7-Fam microRNAs has an opposite effect on pop-1 and lit-1 activity compared with mir-48(veIs48) overexpression: animals that were triply mutant for mir-48, mir-84, and mir-241 loss-of-function mutations were sensitive to pop-1(RNAi) and lit-1(RNAi), whereas mir-48(veIs48) animals were insensitive (Table 1 and Fig. 4). mir-48(veIs48) animals are fully competent for RNAi in general (Fig. S6).
These results indicate that the let-7-Fam microRNAs promote the asymmetry activity of pop-1 and lit-1 in seam cells. Therefore, one or more let-7-Fam target genes would be expected to oppose pop-1 and lit-1 activity. Two major targets of the let-7-Fam microRNAs in the control of seam cell fate progression are the heterochronic genes hbl-1 and lin-28 (15).
In addition to functioning downstream of let-7-Fam microRNAs in the control of L2 and L3 seam cell fates (15), hbl-1 also has essential functions in embryonic development (43). Because the hbl-1 null phenotype is embryonic lethal, the only larva-viable hbl-1 mutants are hypomorphs that do not fully deplete hbl-1 activity. Therefore, we could not critically test the role of hbl-1 in seam cell asymmetry downstream of let-7-Fam. However, we observed that pop-1(RNAi) and lit-1(RNAi) affected seam cell production in hbl-1(ve18 lf) larvae similarly to WT (Table 1 and Fig. 4), suggesting that hbl-1 may function to control symmetric vs. asymmetric seam cell fates independently of pop-1 and lit-1, or this is due to partial loss of hbl-1 activity (Fig. 5).
Fig. 5.
A model of genetic interactions between heterochronic genes and LIT-1/POP-1/APR-1 in the developmental regulation of asymmetric and symmetric seam cell divisions. The stage-specific pattern of asymmetric and symmetric division of seam cells is orchestrated by parallel microRNA-regulated developmental timing pathways. (A) lin-14 acts in the L1 to enhance the activity of the LIT-1/POP-1/APR-1 polarity machinery and hence prevents early expression of the L2 symmetric seam cell division. (B) LIN-14 level is then partially down-regulated in the late L1 stage by the lin-4 microRNA, so that the early L2 symmetric division is permitted because of a below threshold level of LIN-14 together with a parallel activity of lin-28 that inhibits premature activation of lin-46. Our data tentatively place HBL-1 as acting independently of LIT-1/POP-1/APR-1 to promote the symmetric stem cell proliferative division. After the first L2 symmetric division, seam cells immediately switch back to asymmetric division because of the down-regulation of HBL-1 by the let-7-Fam microRNAs, as well as down-regulation of LIN-28, which permits the up-regulation of lin-46, and hence restoration of full LIT-1/POP-1/APR activity. Note that LIN-14 is understood to be partially down-regulated to a medium level in the L2 stage and to a lower level in the L3 (12), and our data from this study confirm that lin-14 activity contributes partially to regulating the activity of LIT-1/POP-1 in the second-L2 asymmetric division (Fig. 3D). The results of Ren and Zhang are incorporated into the model here by invoking feedback circuits wherein LIT-1/POP-1 asymmetry machinery confers robustness to the temporal dynamics of lin-4 and let-7-Fam microRNA activities by negatively modulating those microRNAs during the L2 and L3 stages (45).
The heterochronic gene lin-28 is a key regulator of the timing of L2 asymmetric seam cell fates, downstream of the let-7-Fam microRNAs (15). In lin-28(lf) larvae, the L2-specific symmetric seam cell division is skipped, resulting in decreased seam cell number and precocious execution of L3 adult fates (Fig. 1D) (16, 44). This lin-28(lf) cell lineage phenotype implies that lin-28 activity promotes symmetric seam cell division and/or opposes asymmetric division. To determine whether lin-28 affects seam cell division asymmetry by affecting pop-1 and lit-1 activities, we assayed for changes in seam cell number after lit-1(RNAi) or pop-1(RNAi) of lin-28(lf) animals. lin-28(n719lf) animals are fully competent for RNAi (Fig. S6), yet seam cell number was not changed significantly by lit-1(RNAi) or pop-1(RNAi) (Table 1 and Fig. 4). We interpret these results to indicate that loss of lin-28 increases the asymmetric activity of both lit-1 and pop-1, perhaps raising the threshold for reduction of pop-1 or lit-1 by RNAi. Therefore, we propose that in the WT, lin-28 promotes the L2 symmetric division by inhibiting the asymmetry/polarizing activities of LIT-1 and POP-1. Consistent with this conclusion, strains that overexpress lin-28, either lin-28(gf) (45) or lin-66(lf) (46), are sensitive to lit-1(RNAi) or pop-1(RNAi) (Table 1 and Fig. 4).
Consistent with lin-28 functioning downstream of let-7-Fam microRNAs (15) to regulate pop-1 and lit-1 seam cell polarity, multiply mutant lin-28(lf); mir-48 mir-241; mir-84 worms display insensitivity to both pop-1(RNAi) and lit-1(RNAi), just like lin-28(lf) animals (Table 1 and Fig. 4). Interestingly, although the lin-28 3′UTR is predicted to be targeted by lin-4 as well as the let-7-Fam microRNAs (16), we did not detect a role for lin-28 downstream of lin-4(lf) in the regulation of seam cell asymmetry. Specifically, lin-4(lf);lin-28(lf) doubly mutant animals were insensitive to pop-1(RNAi), like the lin-4(lf) single mutant and like the lin-28(lf) mutant (Table 1 and Fig. 4). These results suggest that the let-7-Fam-lin-28 pathway regulates seam cell division asymmetry downstream or in parallel to the lin-4-lin-14 pathway (Fig. 5). Consistent with this conclusion is that we observed the lin-28(lf);lin-14(lf) double mutant to be insensitive to pop-1(RNAi) or lit-1(RNAi) (Table 1 and Fig. 4), just like the lin-28(lf) mutant.
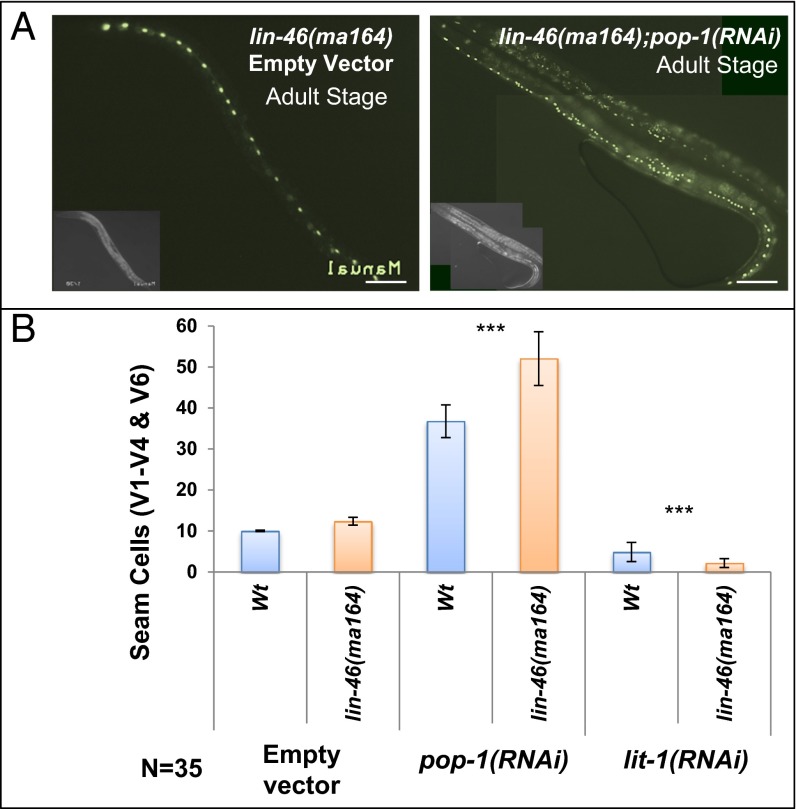
lin-46 was identified by mutations that suppress the developmental timing defects of lin-28(lf) and encodes a conserved protein with roles in mediating the action of LIN-28 in controlling the stage specificity of symmetric seam cell division (18). We used a putative null allele, lin-46(ma164) (18) (a premature stop codon in exon 4), to test whether lin-46 acts downstream of lin-28 in modulating seam cell division asymmetry. Interestingly, lin-28(lf);lin-46(lf) double mutant animals displayed a hypersensitivity to pop-1(RNAi) or lit-1(RNAi) (Table 1 and Figs. 4 and 6), opposite to the insensitivity of the lin-28(lf) single mutant. Moreover, singly mutant lin-46(lf) (18) and lin-46(bp312) (45) animals also exhibited hypersensitivity to pop-1(RNAi) or lit-1(RNAi) (Figs. 4 and 6 and Table 1). Therefore, lin-46 acts downstream of lin-28 and the let-7-Fam microRNAs to reinforce seam cell division asymmetry. This conclusion is supported by the finding that lin-46(lf) also partially suppresses the insensitivity of mir-48 overexpression to pop-1(RNAi); we observed that lin-46(ma164);mir-48(veIs48) animals exhibited an increase in seam cell number after pop-1(RNAi) (Fig. 6 and Table 1).
Fig. 6.
lin-46 modulates asymmetric and symmetric cell fates. (A) Representative lin-46(ma164) adults showing GFP-marked seam cells after control RNAi (Left) or pop-1(RNAi) (Right). Anterior is to the left. (B) Effect of pop-1(RNAi) and lit-1(RNAi) on seam cells numbers in lin-46(ma164) and WT control. Error bars indicate ±SEM. *P < 0.05, **P < 0.01, ***P < 0.005, two-tailed t test. (Scale bar, 50 μm.)
APR-1/APC Depolarization in Seam Cells Promoted by lit-1/pop-1 and Heterochronic Genes.
APR-1 is the worm homolog of mammalian APC, a conserved cytoplasmic protein with roles in cell polarity and Wnt signaling. C. elegans APR-1 has been implicated in the regulation of the Wnt/MAPK pathway in seam cells (26). We observed APR-1::GFP asymmetrically concentrated to the anterior cortex of V1–V4/V6 lineage seam cells before and after asymmetric divisions in the L2 stage (Fig. S7 B and C) similar to the previous report (25). To test whether the asymmetric cortical localization of APR-1 reflects the cell polarizing activity of POP-1/LIT-1, we examined APR-1::GFP localization after lit-1(RNAi) and pop-1(RNAi). lit-1(RNAi) or pop-1(RNAi) disrupted APR-1 polarity (Fig. S7 E and F). APR-1 is known to promote the asymmetrical nuclear localization of POP-1 (47), and therefore, these findings support a novel reciprocal regulatory interaction between cortical APR-1 and the LIT-1/POP-1 axis.
To test whether the L2-specific symmetric division is associated with a concomitant loss of APR-1 cortical asymmetry, we monitored APR-1::GFP before and during the early L2 symmetric division of WT V1–V4/V6 cell lineages. Surprisingly, at the end of the L1 stage, just before execution of the symmetric division of V1–V4/V6 in the early L2, APR-1::GFP appears asymmetrically expressed, with enriched APR-1::GFP on the anterior cortex of seam cells (Fig. S7A). Apparently, although APR polarized, the early L2 seam cells are nevertheless specified to divide symmetrically to produce two stem cells (Fig. S7 B and D).
Our analysis of genetic interactions between heterochronic genes and POP-1/LIT-1 [evidenced by effects of heterochronic gene mutations on the response of seam cells to pop-1(RNAi) or lit-1(RNAi)] suggests that heterochronic genes act to regulate POP-1/LIT-1 asymmetry/polarizing activity in the L1–L3 stages. To test whether the developmental modulation of LIT-1/POP-1 asymmetry activity by the heterochronic genes (described above) is reflected by changes in cortical APR-1 asymmetry, we examined the developmental localization of APR-1::GFP expression in seam cells of WT larvae and heterochronic mutants. lin-14(RNAi) or lin-46(RNAi) abolished APR-1 polarity (Fig. S7 G and H). This result is consistent with our findings that lin-14(lf) or lin-46(lf) mutations sensitized seam cells to pop-1(RNAi) or lit-1(RNAi) and supports the model that in WT development, lin-14 and lin-46 exert stage-specific reinforcement of seam cell asymmetry/polarity. By contrast, lin-28(lf) had no evident effect on APR-1 polarity (Fig. S8), consistent with the proposed asymmetry/polarity-opposing role for lin-28 (Fig. 5).
These findings indicate that lin-14 and lin-46 contribute to the proper localization of Wnt-asymmetry pathway components and cell fate determination in seam cells. Although mutations in heterochronic genes can affect APR-1 polarity, those effects could be indirect, and in the WT, the specification of L2-specific symmetric seam cell divisions may involve a transient effect of the heterochronic pathway on the polarity machinery downstream of APR-1.
Discussion
The heterochronic gene pathway controls the timing of stage-specific transitions between asymmetric and symmetric divisions in lateral hypodermal cell lineages (6, 11). lin-28 and hbl-1 promote expression of the symmetric division in the L2, whereas lin-14 acts in the L1 to prevent expression of the symmetric division. lin-4 and let-7 family microRNAs temporally regulate lin-14, lin-28 and hbl-1 to restrict the symmetric division to only the L2 stage. Here we show that in lateral hypodermal cells, microRNA-regulated developmental timing pathways genetically interact with LIT-1/POP-1/APR-1 cellular asymmetry machinery to regulate the stage-specific execution of asymmetric vs. symmetric stem cell divisions and cell fates. Our findings that the heterochronic genes stage specifically modulate pop-1 and lit-1 activities provides a framework for understanding the control of transitions from asymmetric to symmetric fates in C. elegans development and perhaps in animal development and disease more generally (Fig. 1).
Regulation of Cell Division Asymmetry and Cell Fate in Seam Cells by LIT-1 Through Negative Regulation of POP-1.
The asymmetric stem cell divisions in the seam cell lineages are associated with asymmetric patterns of nuclear localization of LIT-1, WRM-1, and POP-1 between daughter cells that are similar to the patterns of these proteins in the C. elegans embryo (25, 26, 32, 34). In the worm embryo, Wnt signaling results in LIT-1 becoming phosphorylated and hence forming a complex with WRM-1(β-catenin) to reduce POP-1 activity through phosphorylation and nuclear export (29, 30). LIT-1 appears to play a similar role in asymmetric postembryonic lineages by reducing nuclear POP-1 levels in the seam cell daughter. Knockdown of WRM-1 activity causes a decrease in seam cell number, similar to the previously reported decrease in seam cells after experimental reduction of LIT-1 (23, 24). However, previous reports did not characterize the precise cell lineage transformation associated with reduction of seam cell number on lit-1 knockdown. Here, we show that lit-1(RNAi) reduces the number of seam cells by conversion of asymmetric divisions to symmetric divisions that produce two differentiated cells. It is striking that loss of function of either POP-1 or LIT-1 converts asymmetric seam cell divisions to symmetric, but importantly, with opposite consequences to cell fates: loss of POP-1 results in production of two stem cell daughters, whereas loss of LIT-1 results in two differentiated daughters. These findings emphasize how the coordinated activity of both POP-1 and LIT-1 are required to generate the asymmetric seam cell divisions that produce one stem cell sister and one differentiated cell sister.
Stage-Specific Heterochronic Gene Activities Promoting Seam Cell Division Asymmetry.
Our results suggest that two parallel microRNA-regulated LIN-14 and LIN-46 pathways promote the stage-specific execution of asymmetric seam cell divisions in the L1 stage, mid-L2 stage, and L3 stage (Fig. 5). According to our model, microRNA-mediated developmental regulation of the LIN-14 and LIN-46 asymmetry-promoting pathways helps to specify the stage specific expression of asymmetric seam cell division; the single symmetric division in the early L2 stage is proposed to result from a transient phase of appropriately low activities of lin-14 and lin-46 (Fig. 5).
One of the key findings underlying our conclusions is that lin-14(lf) mutants exhibited more severe pop-1(RNAi) or lit-1(RNAi) phenotypes in the L1 than did the WT. By contrast, lin-4(lf) mutants, which overexpress lin-14 at all stages, exhibited insensitivity to pop-1(RNAi) or lit-1(RNAi). These results suggest that LIN-14 prevents symmetric seam cell division in the L1 by (directly or indirectly) enhancing the activity of the fate determinant POP-1 and also its negative regulator, LIT-1. We also observed that reduction of lin-14 function resulted in reduced polarity of APR-1 cortical localization in L1 stage seam cells, which we posit may reflect an indirect effect of lin-14 on APR-1 polarity via LIT-1/POP-1.
LIN-14 protein is abundantly expressed in L1 seam cells (48), consistent with our finding that lin-14 acts in the L1 to enhance the activity of the LIT-1/POP-1/APR-1 polarity machinery and reinforce the asymmetry of the L1 seam cell division. lin-14 activity is known to be down-regulated by lin-4 microRNA (48, 49): first to a medium level for seam cells undergoing L2 divisions and then further reduced for L3 seam cells (12). Accordingly, we propose that the first L2 seam cell division is symmetric on account of a below-threshold level of LIN-14, in combination with and a parallel activity of lin-28 that inhibits premature activation of lin-46 (see below).
In contrast to lin-14, lin-28 appears to oppose lit-1 and pop-1 asymmetry activity. In lin-28(lf) mutant larvae, all seam cell divisions are asymmetric (16), and unlike the WT, lin-28(lf) animals do not express symmetric cell divisions after knockdown of pop-1 or lit-1. These results suggest that in the WT, lin-28 functions in the first L2 cell division to blunt the activities of both POP-1 and LIT-1 and thereby promote symmetric seam cell division. Our results indicate that lin-28(lf) enhances the asymmetry/polarization of seam cells via inappropriate activity of lin-46, which was previously identified as a downstream mediator for the temporal control of lateral hypodermal cell fates by lin-28 (18). We observed that lin-46(lf) restores the sensitivity of a lin-28(lf) mutant to lit-1(RNAi) or pop-1(RNAi), and that lin-46(lf) animals are hypersensitive to pop-1 or lit-1 knockdown.
The independence of the parallel lin-14 and lin-46 pathways is emphasized by our finding that elevated activity of either lin-14 alone, or lin-46 alone, can reinforce pop-1 and lit-1 asymmetry activities. Specifically, we observed that lin-28(lf) (which results in lin-46 hyperactivity) caused insensitivity to lit-1(RNAi) or pop-1(RNAi) even in a lin-14(lf) background (Table 1 and Fig. 4) and that lin-4(lf) (which results in lin-14 hyperactivity) resulted in insensitivity to lit-1(RNAi) or pop-1(RNAi), even in a lin-46(lf) background (Table 1 and Fig. 4).
After the early L2 symmetric division, seam cells immediately switch back to asymmetric division, and we propose that this switch is promoted by the down-regulation of LIN-28 by the let-7-Fam microRNAs, which permits the up-regulation of lin-46 and hence restoration of full LIT-1/POP-1/APR activity. Interestingly, it has been reported that LIN-46 is expressed in the L2 stage primarily in pairs of adjacent seam cells (18), suggesting that lin-46 is specifically up-regulated immediately following the first L2 symmetric cell division, which would support a major role for lin-46 in the switch back to asymmetry for the second L2 seam cell division.
The expression of the L2 symmetric division is also promoted by HBL-1, which like LIN-28, is down-regulated by the let-7-Fam microRNAs after the L2 (50, 51). In addition to these larval developmental timing roles, HBL-1 also has essential roles in embryonic development, so we were not able to test the effects of complete loss of hbl-1 activity on seam cell polarity or sensitivity to pop-1 or lit-1 knockdown. Our observation that a hypomorphic hbl-1 mutant exhibited no differences in pop-1(RNAi) or lit-1(RNAi) phenotypes compared with the WT could be attributable to residual hbl-1 activity in the mutant. Alternatively, it is possible that hbl-1 may act to promote symmetric seam cell division downstream or parallel of pop-1 and lit-1 (Fig. 5).
We do not know whether these heterochronic gene activities affect seam cell polarity mechanisms directly or indirectly. It is possible that the LIN-14 transcription factor may regulate a gene expression program that facilitates stabilization of the asymmetry/polarity framework for seam cells, for example, by (directly or indirectly) promoting expression of both lit-1 and pop-1. LIN-46 is similar to mammalian gephyrin (18), a multifunctional scaffolding protein, suggesting that LIN-46 could act as a scaffold for a multiprotein assemblies that impact asymmetric cell division and polarity. It remains to be determined whether the effects of LIN-46 on pop-1 and lit-1 activity in seam cells reflect direct interactions between LIN-46 and cell polarity proteins.
Regulatory Networks Underlying the Developmentally Robust Expression of Stem Cell Division Asymmetry.
Our findings that the heterochronic genes modulate the LIT-1/POP-1/APR-1 asymmetry machinery present a particularly interesting complement to previous reports showing the reciprocal: that the asymmetry machinery can affect the developmental timing phenotypes of heterochronic mutants. Parry et al. identified pop-1 as a gene that functionally opposes let-7 microRNA activity in the context of vulva development (52). Ren and Zhang reported that lit-1 activity can modulate the developmental timing phenotypes of heterochronic mutants, perhaps by modulating the activities of the lin-4 and let-7-Fam microRNAs (45). They demonstrated that the lit-1(bp239) mutation can suppress the retarded heterochronic defects of mutations in dcr-1, one of the core players in the microRNA and RNAi biogenesis pathways (45). Ren and Zhang proposed that the POP-1/LIT-1 asymmetry pathway negatively regulates lin-4 and let-7-Fam microRNAs in the context of the control of symmetric and asymmetric seam cell divisions in the L2 stage (Fig. 5). Therefore, our results, taken together with those previous findings, suggest a model wherein the LIT-1/POP-1/APR-1 asymmetry machinery and the heterochronic genes are engaged in developmentally dynamic mutual regulatory interactions that result in stage-specific transitions between symmetric and asymmetric cell divisions in the seam cell lineages (Fig. 5).
It is possible that the proposed complex functional interactions between developmental timing regulators and components of the LIT-1/POP-1 asymmetry pathway (Fig. 5) contribute to the exceedingly robust temporal patterns of asymmetric and symmetric cell divisions and cell fates in WT C. elegans development. We observed that seam cell APR-1 polarity can be compromised by reduction of LIT-1 or POP-1 or by reducing the activity of heterochronic genes that promote asymmetric division (lin-14 or lin-46), suggesting that these heterochronic genes contribute to polarized APR-1 localization perhaps by enhancing LIT-1/POP-1 activity.
Interestingly, we observed that APR-1 appeared polarized even at the early L2 symmetric division (Fig. S7). This finding suggests that these stem cells may continuously maintain a measure of intrinsic asymmetry (albeit, as we propose, weakened at the early L2) and that the heterochronic genes may also regulate the coupling of stem and differentiated fates to LIT-1/POP-1/APR-1 polarity. Perhaps the maintenance of APR-1 asymmetry (and hence self-renewal divisions) is the default state of stem cells more generally, and symmetric stem cell division in other systems can also result from a transient uncoupling of polarity machinery from cell fate.
Our proposed model for the regulation of stem cell division asymmetry/polarity in C. elegans may represent a general strategy for regulated robustness of stem cell asymmetry and cell fates during normal development (Fig. 5). It is possible that evolutionarily conserved components of the C. elegans heterochronic and Wnt asymmetry pathways could play similar functions in mammalian stem cell lineages. For example, LIT-1/Nlk is a serine-threonine kinase with conserved functions in MAPK and Wnt signaling pathways, and dysregulation of LIT-1/Nlk has been reported to correlate with tumorigenesis in mammals (53–56). It is possible that the conserved tumor suppressor function of the let-7 microRNA (57, 58) may in part reflect conserved roles in regulating LIT-1/POP-1/APR-1 asymmetry and hence promoting asymmetric (nonproliferative) cell division. Similarly, the conserved role for LIN-28 in promoting stem cell fate in diverse contexts, including cancer (17, 59, 60), may reflect a general activity for LIN-28 in opposing LIT-1/POP-1/APR-1 stem cell division asymmetry.
Materials and Methods
Genetic Methods and Nematode Growth Condition.
C. elegans culture and genetic manipulation was as previously described (61). WT animals were C. elegans N2 Bristol. All experiments were carried out at 20 °C except where otherwise indicated. The putative null allele of lin-46(ma164) was sequenced, and a premature stop codon in exon 4 was identified as a point mutation guanine to adenine in lin-46(ma164) exon-4 causing a stop code in the sense strand [CGA (Arg.) to UGA (Stop)].
Seam Cell Division and Phenotypic Analysis.
Lateral hypodermal seam cells in anesthetized (1 mM levamisole) living worms were identified using Nomarski differential interference contrast (DIC) microscopy by scoring their characteristic anatomical locations and nuclear morphology (9) and/or using fluorescence microscopy to detect the expression of the scmp::gfp transgene. For newborn sister nuclei from asymmetric divisions, the GFP reporter intensity in the D cell is initially similar to the S cell but then fades in the D cell over the course of 1 h or so, as the D nucleus fuses with the hyp7 syncytium. Therefore, the fates of young sister nuclei from seam cell divisions are scored primarily by DIC microscopy using their distinct nuclear locations, sizes, and shapes (9). Specifically, soon after birth, S cell nuclei are relatively elongated compared with D cell nuclei, which are relatively rounded. Also, S cell nuclei remain precisely positioned along the lateral midline of the animal, whereas D nuclei adopt ventro-lateral and dorso-lateral positions. Developmental stage was assessed by the extent of gonad and germ-line development using DIC microscopy. Seam cell divisions were monitored using a Zeiss Axio Imager D1 dissecting stereomicroscope with an AxioCam MRm camera and an X-Cite 120Q light source. Cell fusion was visualized using the junction marker ajm-1::gfp (62).
RNA Interference.
Bacterial RNAi feeding strains were from the Ahringer RNAi library (63). dsRNA-expressing bacteria were induced in culture and seeded on NGM plates containing 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), 50 µg/mL ampicillin, and 12.5 µg/mL tetracycline. Empty vector was used as a negative control. Unless otherwise indicated, animals were fed RNAi food throughout larval development.
Temperature Shift Experiments.
Temperature-sensitive mutants were cultured at permissive temperature (15 °C). Embryos were isolated by bleaching of gravid adults and were transferred to empty vector control or RNAi plates and maintained at the restrictive temperature (25 °C or 26.5 °C) during postembryonic development.
Confocal Microscopy.
Anesthetized animals were imaged with a Nikon TE-2000E2 inverted confocal microscope with a motorized Z drive equipped with optics for bright field, DIC, fluorescence, phase contrast, and 100× Plan Apo objective microscopy. MetaMorph software (version 2.5) was used for image acquisition and analysis. Approximately 40-mm Z-stacks at 4-mm intervals were captured with a 100× Plan Apo objective lens. For further analysis and quantification of GFP intensity of combined Z-stacks, the ImageJ program (version 1.46) was used by subtracting the background fluorescence, which was defined as the autofluorescence signal from an adjacent region of similar size. For polarity analysis, the localization of GFP::APR-1 protein was measured for anterior and posterior sides of the cells by drawing a line across the middle of the cell to separate the anterior and posterior sides for V cells in L1 asymmetric and L2 symmetric division. The intensity of GFP::APR-1 is measured for the whole anterior and posterior daughters in L2 asymmetric division. About 10 stacks for each single image and 6–28 images were used for each APR-1 polarity analysis.
Statistical Analysis.
The data for all figures were analyzed by two-tail unpaired t test except for cortical expression of APR-1, which was analyzed by paired t test. Error bars indicate ±SEM. The statistical analysis for the Table 1 is examined by the equivalence test of the means (64).
Supplementary Material
Acknowledgments
We thank Drs. Craig Mello, Allison Woollard, and Eric Moss and Caenorhabditis Genetics Center [funded by National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440)] for worm strains; Drs. Parisa Kalantari and Nick Willis for critical reading of the manuscript; Dr. Paul Furcinitti for advice and help with confocal microscopy; members of the V.R.A. laboratory for discussions; and Dr. Hamid Ashrafi for help with statistical analysis. This work was supported by the Sass Foundation for Medical Research and Leukemia and Lymphoma Society awards to O.F.H. and NIH Award R01-GM34028 to V.R.A., who is also a member of the UMass Diabetes Endocrinology Research Center (Grant DK32520).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422852112/-/DCSupplemental.
References
- 1.Clevers H. Stem cells, asymmetric division and cancer. Nat Genet. 2005;37(10):1027–1028. doi: 10.1038/ng1005-1027. [DOI] [PubMed] [Google Scholar]
- 2.Neumüller RA, Knoblich JA. Dividing cellular asymmetry: Asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23(23):2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441(7097):1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 4.Grifoni D, et al. aPKCzeta cortical loading is associated with Lgl cytoplasmic release and tumor growth in Drosophila and human epithelia. Oncogene. 2007;26(40):5960–5965. doi: 10.1038/sj.onc.1210389. [DOI] [PubMed] [Google Scholar]
- 5.Knoblich JA. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11(12):849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nimmo RA, Slack FJ. An elegant miRror: MicroRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118(4):405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizumoto K, Sawa H. Two betas or not two betas: Regulation of asymmetric division by beta-catenin. Trends Cell Biol. 2007;17(10):465–473. doi: 10.1016/j.tcb.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Joshi PM, Riddle MR, Djabrayan NJ, Rothman JH. Caenorhabditis elegans as a model for stem cell biology. Dev Dyn. 2010;239(5):1539–1554. doi: 10.1002/dvdy.22296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 10.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17(11):R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 11.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21(4):511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambros V, Horvitz HR. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1987;1(4):398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- 13.Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes Dev. 1996;10(23):3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- 14.Chalfie M, Horvitz HR, Sulston JE. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24(1):59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 15.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9(3):403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88(5):637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Pepper AS, et al. The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development. 2004;131(9):2049–2059. doi: 10.1242/dev.01098. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto Y, Takeshita H, Sawa H. Multiple Wnts redundantly control polarity orientation in Caenorhabditis elegans epithelial stem cells. PLoS Genet. 2011;7(10):e1002308. doi: 10.1371/journal.pgen.1002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips BT, Kimble J. A new look at TCF and beta-catenin through the lens of a divergent C. elegans Wnt pathway. Dev Cell. 2009;17(1):27–34. doi: 10.1016/j.devcel.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herman M. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. 2001;128(4):581–590. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- 22.Whangbo J, Harris J, Kenyon C. Multiple levels of regulation specify the polarity of an asymmetric cell division in C. elegans. Development. 2000;127(21):4587–4598. doi: 10.1242/dev.127.21.4587. [DOI] [PubMed] [Google Scholar]
- 23.Banerjee D, Chen X, Lin SY, Slack FJ. kin-19/casein kinase Iα has dual functions in regulating asymmetric division and terminal differentiation in C. elegans epidermal stem cells. Cell Cycle. 2010;9(23):4748–4765. doi: 10.4161/cc.9.23.14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gleason JE, Eisenmann DM. Wnt signaling controls the stem cell-like asymmetric division of the epithelial seam cells during C. elegans larval development. Dev Biol. 2010;348(1):58–66. doi: 10.1016/j.ydbio.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wildwater M, Sander N, de Vreede G, van den Heuvel S. Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development. 2011;138(20):4375–4385. doi: 10.1242/dev.066431. [DOI] [PubMed] [Google Scholar]
- 26.Mizumoto K, Sawa H. Cortical beta-catenin and APC regulate asymmetric nuclear beta-catenin localization during asymmetric cell division in C. elegans. Dev Cell. 2007;12(2):287–299. doi: 10.1016/j.devcel.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Ishitani T, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399(6738):798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 28.Meneghini MD, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399(6738):793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- 29.Rocheleau CE, et al. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97(6):717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 30.Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117(1):95–106. doi: 10.1016/s0092-8674(04)00203-x. [DOI] [PubMed] [Google Scholar]
- 31.Kagoshima H, et al. The C. elegans CBFbeta homologue BRO-1 interacts with the Runx factor, RNT-1, to promote stem cell proliferation and self-renewal. Development. 2007;134(21):3905–3915. doi: 10.1242/dev.008276. [DOI] [PubMed] [Google Scholar]
- 32.Takeshita H, Sawa H. Asymmetric cortical and nuclear localizations of WRM-1/beta-catenin during asymmetric cell division in C. elegans. Genes Dev. 2005;19(15):1743–1748. doi: 10.1101/gad.1322805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura K, et al. Wnt signaling drives WRM-1/beta-catenin asymmetries in early C. elegans embryos. Genes Dev. 2005;19(15):1749–1754. doi: 10.1101/gad.1323705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin R, Hill RJ, Priess JR. POP-1 and anterior-posterior fate decisions in C. elegans embryos. Cell. 1998;92(2):229–239. doi: 10.1016/s0092-8674(00)80917-4. [DOI] [PubMed] [Google Scholar]
- 35.Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev Biol. 2005;285(2):584–592. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Siegfried KR, Kidd AR, 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004;166(1):171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatsumoto T, Sakata H, Dasso M, Miki T. Potential roles of the nucleotide exchange factor ECT2 and Cdc42 GTPase in spindle assembly in Xenopus egg cell-free extracts. J Cell Biochem. 2003;90(5):892–900. doi: 10.1002/jcb.10750. [DOI] [PubMed] [Google Scholar]
- 38.Saito S, et al. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279(8):7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- 39.Liu XF, Ishida H, Raziuddin R, Miki T. Nucleotide exchange factor ECT2 interacts with the polarity protein complex Par6/Par3/protein kinase Czeta (PKCzeta) and regulates PKCzeta activity. Mol Cell Biol. 2004;24(15):6665–6675. doi: 10.1128/MCB.24.15.6665-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 41.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427(6975):645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 42.Li M, Jones-Rhoades MW, Lau NC, Bartel DP, Rougvie AE. Regulatory mutations of mir-48, a C. elegans let-7 family MicroRNA, cause developmental timing defects. Dev Cell. 2005;9(3):415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205(2):240–253. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- 44.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226(4673):409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 45.Ren H, Zhang H. Wnt signaling controls temporal identities of seam cells in Caenorhabditis elegans. Dev Biol. 2010;345(2):144–155. doi: 10.1016/j.ydbio.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Morita K, Han M. Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J. 2006;25(24):5794–5804. doi: 10.1038/sj.emboj.7601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugioka K, Mizumoto K, Sawa H. Wnt regulates spindle asymmetry to generate asymmetric nuclear β-catenin in C. elegans. Cell. 2011;146(6):942–954. doi: 10.1016/j.cell.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 48.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57(1):49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 49.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 50.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4(5):625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 51.Lin SY, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4(5):639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 52.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17(23):2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, Ma Z, Chen X, Zhang J. Prognostic significance of nemo-like kinase in nasopharyngeal carcinoma. Mol Med Rep. 2014;10(1):131–136. doi: 10.3892/mmr.2014.2190. [DOI] [PubMed] [Google Scholar]
- 54.Li M, et al. Prognostic significance of nemo-like kinase (NLK) expression in patients with gallbladder cancer. Tumour Biol. 2013;34(6):3995–4000. doi: 10.1007/s13277-013-0988-4. [DOI] [PubMed] [Google Scholar]
- 55.Mendes-Pereira AM, Lord CJ, Ashworth A. NLK is a novel therapeutic target for PTEN deficient tumour cells. PLoS ONE. 2012;7(10):e47249. doi: 10.1371/journal.pone.0047249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan Z, et al. NLK is a key regulator of proliferation and migration in gallbladder carcinoma cells. Mol Cell Biochem. 2012;369(1-2):27–33. doi: 10.1007/s11010-012-1365-0. [DOI] [PubMed] [Google Scholar]
- 57.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Copley MR, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15(8):916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 60.Cai WY, et al. The Wnt-β-catenin pathway represses let-7 microRNA expression through transactivation of Lin28 to augment breast cancer stem cell expansion. J Cell Sci. 2013;126(Pt 13):2877–2889. doi: 10.1242/jcs.123810. [DOI] [PubMed] [Google Scholar]
- 61.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128(15):2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- 63.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2(1):RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schuirmann DJ. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J Pharmacokinet Biopharm. 1987;15(6):657–680. doi: 10.1007/BF01068419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.