Significance
Enhancers are regions of regulatory DNA that control gene expression and cell fate decisions during development. Enhancers compute the expression pattern of their target gene by reading the concentrations of input regulatory proteins. Many developmental genes contain multiple enhancers that control the same output pattern, but it is unclear whether these enhancers all compute the pattern in the same way. We use measurements in single cells and computational models in Drosophila embryos to demonstrate that two enhancers that encode the same gene expression pattern compute differently: the same regulatory protein represses one enhancer and activates the other. Pairs of enhancers that generate the same pattern by performing different computations may impart special properties to developmental systems.
Keywords: enhancer, computational model, bifunctional transcription factor, Drosophila development, Hunchback
Abstract
Hunchback (Hb) is a bifunctional transcription factor that activates and represses distinct enhancers. Here, we investigate the hypothesis that Hb can activate and repress the same enhancer. Computational models predicted that Hb bifunctionally regulates the even-skipped (eve) stripe 3+7 enhancer (eve3+7) in Drosophila blastoderm embryos. We measured and modeled eve expression at cellular resolution under multiple genetic perturbations and found that the eve3+7 enhancer could not explain endogenous eve stripe 7 behavior. Instead, we found that eve stripe 7 is controlled by two enhancers: the canonical eve3+7 and a sequence encompassing the minimal eve stripe 2 enhancer (eve2+7). Hb bifunctionally regulates eve stripe 7, but it executes these two activities on different pieces of regulatory DNA—it activates the eve2+7 enhancer and represses the eve3+7 enhancer. These two “shadow enhancers” use different regulatory logic to create the same pattern.
Transcription factors (TFs) are typically categorized as activators or repressors, but many TFs can act bifunctionally by both activating and repressing target genes (1–4). Changes in TF activity can result from posttranslational modifications, protein cleavage, or translocation of cofactors into the nucleus (5–7). However, in cases where a TF activates and represses genes in the same cells, bifunctionality is controlled by enhancer sequences, which are responsible for tissue-specific gene expression (8). For example, in Drosophila, Dorsal activates genes when it binds to enhancers alone or near Twist (9, 10) but represses genes when it binds near other TFs (11–13). The DNA sequence of a TF's binding site can also alter TF activity [e.g., the glucocorticoid receptor (14, 15)]. Identifying how the activity of bifunctional TFs is controlled will be critical for inferring accurate gene regulatory networks from genomic data (16).
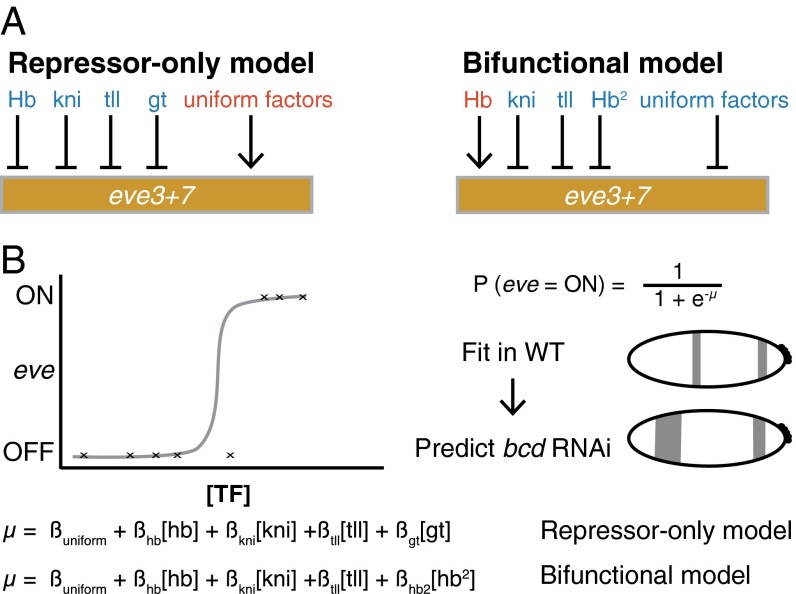
Here, we investigate how TF bifunctionality is controlled using a classic example: the Drosophila gene hunchback (hb) (1, 17, 18). Hb both activates and represses even-skipped (eve) by acting on multiple enhancers. Hb activates eve stripes 1 and 2 and represses stripes 4, 5, and 6 (19–22). Computational models from us and others support the hypothesis that Hb both activates and represses the enhancer that controls eve stripes 3 and 7 (eve3+7) (Fig. 1) (22–25).
Fig. 1.
The repressor-only and bifunctional models formalize two alternative regulator sets for eve stripes 3 and 7. (A) The repressor-only model includes repression (blue) by Hb, knirps (kni), giant (gt), and tailless (tll) and activation (red) by a constant term that represents spatially uniform factors. The bifunctional model includes activation by a linear Hb term and repression by a quadratic Hb term, kni, tll, and uniform factors. (B) A schematic of the logistic regression framework. Logistic regression calculates the probability the target will be ON based on a linear combination of the concentrations of regulators (µ). We fit models in WT and use the perturbed regulator gene expression patterns to predict the perturbed eve patterns in bcd RNAi embryos.
In contrast to others, our computational models of eve3+7 activity do not include regulatory DNA sequence (26–30). Instead, our modeling approach uses regression to identify the activators and repressors that control a given pattern; we refer to the identity and role of the regulators as “regulatory logic.” Modeling regulatory logic without including DNA sequence enables a powerful strategy to dissect gene regulation in a complex locus. We can compare the regulatory logic of an enhancer reporter pattern to that of the corresponding portion of the endogenous pattern to determine whether the annotated enhancer contains all relevant regulatory DNA.
Here, we tested the hypothesis that Hb bifunctionally regulates eve3+7. We measured the endogenous eve expression pattern and the expression pattern driven by an eve3+7 enhancer reporter at cellular resolution under multiple genetic perturbations. We then used these data to challenge two computational models of eve3+7 activity. Hb acts only as a repressor in one model, but acts as both an activator and a repressor in the other (Fig. 1) (24). Our modeling indicated that eve3+7 and the endogenous locus use different regulatory logic to position stripe 7. Specifically, eve3+7 is only repressed by Hb, whereas the endogenous stripe 7 is both activated and repressed. We show that an additional sequence is activated by Hb and contributes to the regulation of eve stripe 7 (19, 23, 28, 31–33). Thus, eve stripe 7 is controlled by a pair of shadow enhancers, separate sequences in a locus that drive overlapping spatiotemporal patterns (34). These shadow enhancers respond to Hb in opposite ways and use different regulatory logic.
Results
eve Enhancer Reporter Patterns Do Not Match the Endogenous eve Pattern.
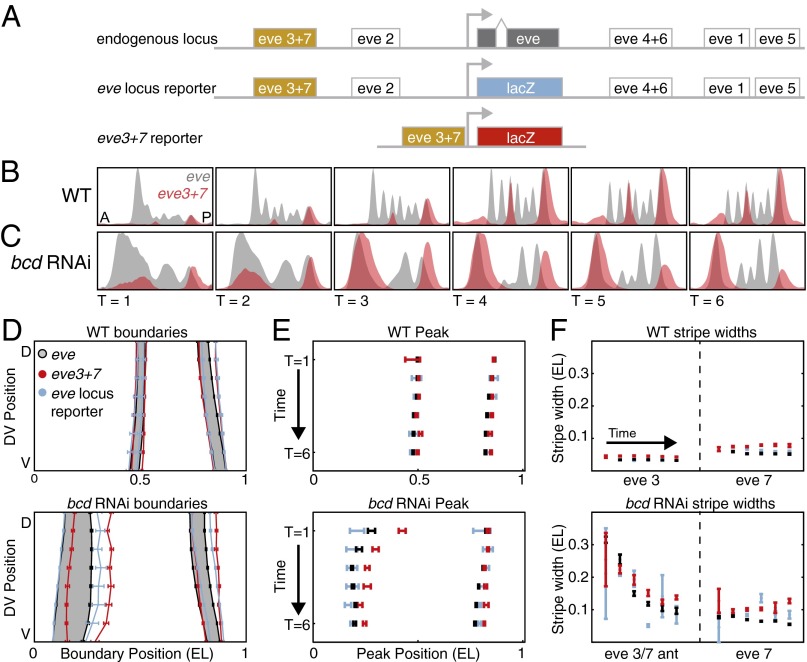
To determine whether Hb bifunctionally regulates eve3+7 we compared the endogenous eve pattern to the pattern driven by a β-galactosidase (lacZ) reporter construct in two genetic backgrounds (Fig. 2A and Figs. S1 and S2). We refer to these data throughout the manuscript as “the eve3+7 reporter pattern” and “the endogenous pattern.” We examined both WT embryos and embryos laid by females expressing short hairpin RNAs against bicoid (bcd RNAi embryos), where expression of all of the regulators, especially Hb, is perturbed (Figs. S3 and S4 and ref. 35). We measured expression patterns quantitatively at cellular resolution using in situ hybridization, two-photon microscopy, and an automated image processing toolkit (Materials and Methods and refs. 36 and 37). We averaged data from many embryos into gene expression atlases (38). Importantly, the eve3+7 reporter pattern results from the activity of eve3+7 alone whereas the endogenous pattern integrates the whole locus.
Fig. 2.
The eve3+7 reporter pattern differs from the endogenous pattern. (A) The eve locus contains five annotated primary stripe enhancers. The endogenous pattern integrates the whole locus. The eve locus reporter pattern also integrates the whole locus, but the transcript is the same as the eve3+7 reporter construct. The eve3+7 reporter construct isolates the activity of the annotated enhancer sequence. (B) WT expression patterns are represented as line traces for a lateral strip of the embryo where anterior–posterior (A-P) position is plotted on the x axis with expression level on the y axis. Endogenous eve pattern (gray), eve3+7 reporter pattern (red). The reporter pattern was manually scaled to match the level of the endogenous pattern; this scaling highlights differences in the position of expression. (C) Line traces in bcd RNAi embryos. Data presented as in B. (D) The boundaries of the endogenous pattern (gray), the eve3+7 reporter pattern (red), and the eve locus reporter pattern (blue) at T = 3. All error bars are the standard error of the mean. The eve locus reporter pattern is more faithful to the endogenous pattern than the eve3+7 reporter pattern, especially in the anterior of bcd RNAi embryos. The endogenous pattern is shaded for visual clarity. (E) Peak positions of stripes 3 and 7, calculated from the line traces in B and C. The eve3+7 reporter pattern shows better agreement to the endogenous pattern in WT than in bcd RNAi embryos. (F) Stripe widths, calculated from the inflection point of the line traces in B and C. The eve3+7 reporter pattern is wider than the corresponding endogenous pattern.
Our high-resolution measurements revealed discrepancies between the endogenous pattern and the eve3+7 reporter pattern. In WT embryos, the eve3+7 reporter pattern overlaps the corresponding endogenous eve stripes, but these stripes are broader, have uneven levels, and the peaks lie posterior to the endogenous peaks (Fig. 2). These discrepancies were more pronounced in bcd RNAi embryos than in WT embryos, especially for the anterior stripe (Fig. 2 D–F). When we tested reporters for other eve enhancers we also found discrepancies between reporter patterns and the endogenous pattern (Figs. S1 and S2).
To test whether the discrepancies between the eve3+7 reporter pattern and the endogenous pattern resulted from differences in the eve and lacZ transcripts, we measured the expression driven by a reporter encompassing the entire eve locus where the coding sequence had been replaced with lacZ (eve locus reporter, a gift from Miki Fujioka, Thomas Jefferson University, Philadelphia). In both WT and bcd RNAi embryos the locus reporter pattern was more faithful to the endogenous pattern in terms of stripe peak positions and widths (Fig. 2 and Figs. S1 and S2). Remaining differences between the endogenous and locus reporter patterns arise from differences in the transcripts. Differences between the locus reporter and the eve3+7 reporter patterns may arise from regulatory DNA outside of eve3+7. Together, these data suggest that the eve3+7 reporter construct may not contain all of the regulatory DNA that controls the expression of eve stripes 3 and 7.
Different Computational Models Capture the Behavior of the Endogenous Locus and the Enhancer Reporter After Hb Perturbation.
We used computational models to dissect discrepancies between the eve3+7 reporter pattern and the endogenous pattern. With our collaborators, we previously modeled the regulation of the endogenous eve stripes 3 and 7 in WT embryos and simulated genetic perturbations that mimicked published experimental data (24). These models use logistic regression to directly relate the concentrations of input regulators to output expression in single cells. We constructed two models that together test the hypothesis that Hb both activates and represses eve stripes 3 and 7. In the “repressor-only” model [the linear logistic model in Ilsley et al. (24)], Hb has one parameter and only represses. In the “bifunctional” model [the quadratic logistic model in Ilsley et al. (24)] Hb has two parameters that allow it to both activate and repress (Fig. 1). Both models performed equally well in WT embryos, but we favored the bifunctional model because it predicted the effect of a genetic perturbation. At that time, cellular resolution data for the eve3+7 reporter pattern were not available, so we used a standard assumption to interpret the models: the endogenous expression of eve stripes 3 and 7 could be attributed to the activity of the annotated eve3+7 enhancer.
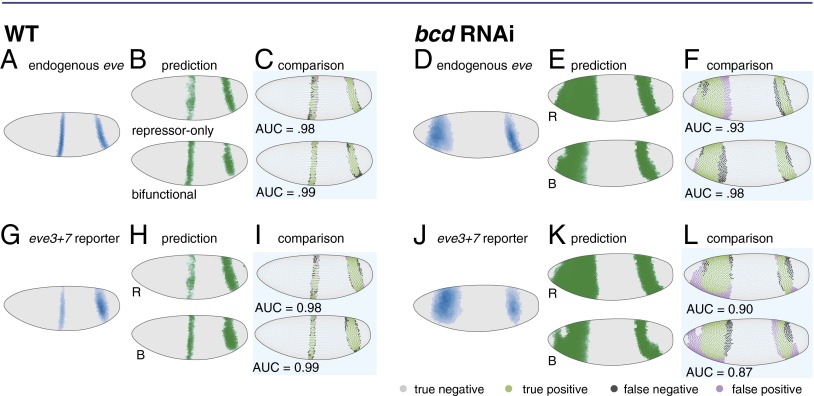
Here, we test this assumption explicitly by modeling the eve3+7 reporter pattern and the endogenous pattern separately. Importantly, it is difficult to interpret the success or failure of a single model. It is much more powerful to compare the performance of two models that together formalize a hypothesis. We compared the performance of the repressor-only and bifunctional models in WT and bcd RNAi embryos. We used Hb protein and giant (gt), tailless (tll), and knirps (kni) mRNA as input regulators and thresholded the endogenous pattern and the reporter pattern for model fitting (Fig. 1 and Materials and Methods). We report our modeling of the third time point, which is representative of results for other time points (Figs. S5 and S6) and evaluated model performance by computing the area under the receiver operating characteristic curve (AUC) (39).
We first analyzed the endogenous pattern: we fit our models in WT embryos and used the resulting parameters to predict expression in bcd RNAi embryos. Both models correctly predicted the positional shifts of stripe 7 and a wide anterior stripe, but the bifunctional model performed better than the repressor-only model (AUCrepressor = 0.93, AUCbifunctional = 0.98; Fig. 3F and Fig. S4).
Fig. 3.
In bcd RNAi embryos the bifunctional model more accurately predicts the endogenous pattern and the repressor-only model more accurately predicts the eve3+7 reporter pattern. (A) The endogenous eve pattern in WT embryos is shown as a rendering of a gene expression atlas. Cells with expression below an ON/OFF threshold (Materials and Methods) are plotted in gray. For cells above this threshold, darker color indicates higher relative amounts. (B) The predictions of the repressor-only (R) and bifunctional (B) models in WT embryos. (C) Comparison of model predictions to the endogenous pattern in WT embryos. Green cells are true positives, purple cells are false positives, dark gray cells are false negatives, and light gray cells are true negatives. For visualization the threshold is set to 80% sensitivity, but the AUC metric quantifies performance over all thresholds. (D) The endogenous eve pattern in bcd RNAi embryos. (E) The predictions of the repressor-only (R) and bifunctional (B) models in bcd RNAi embryos. (F) Comparison of model predictions to the endogenous pattern in bcd RNAi embryos. The bifunctional model more accurately predicts the endogenous pattern in bcd RNAi embryos. (G–L) Same as A–F, respectively, for the eve3+7 reporter pattern. The repressor-only model predicts the eve3+7 reporter pattern more accurately in bcd RNAi embryos. Model parameters and AUC scores are in Tables S1 and S2.
We next analyzed the eve3+7 reporter pattern: again, we fit both models in WT embryos and used the resulting parameters to predict expression in bcd RNAi embryos. In this case, the repressor-only model was more accurate than the bifunctional model (AUCrepressor = 0.90, AUCbifunctional = 0.87; Fig. 3L). We controlled for several factors that may confound prediction accuracy: we assessed sensitivity to changes in regulator concentrations, refit the models with bcd RNAi data, and refit the models on all of the data, none of which changed our conclusions (Figs. S5 and S6 and Supplemental Note 1).
These results suggest that Hb bifunctionally regulates the endogenous pattern but only represses the reporter pattern. Although the differences in relative model performance are subtle, the results support our hypothesis that the eve3+7 reporter pattern is regulated differently from the endogenous pattern. However, these differences in model performance were not conclusive of their own accord and prompted us to return to the perturbation that previously distinguished the repressor-only and bifunctional models, ventral misexpression of hb (24, 25).
hb Misexpression Confirms That the Endogenous eve Pattern and the eve3+7 Reporter Pattern Respond to Hb Differently.
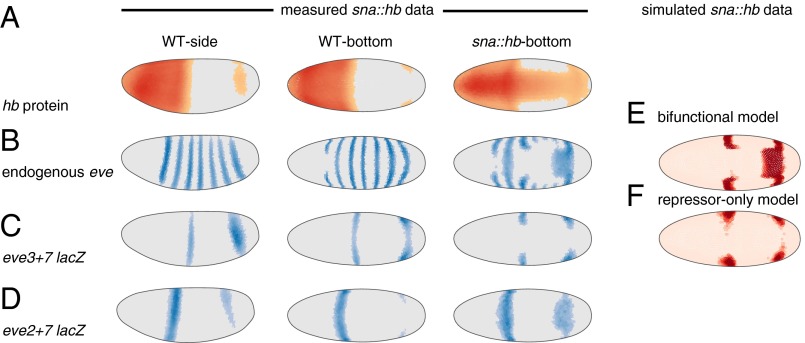
In Ilsley et al. (24) we preferred the bifunctional model because it qualitatively predicted the behavior of a classic genetic perturbation. Misexpressing hb along the ventral surface of the embryo (sna::hb embryos) causes eve stripe 3 to retreat and bend and stripe 7 to bend and bulge (ref. 22 and Fig. 4 A and B). In simulations of this perturbation the bifunctional model predicted this behavior, whereas the repressor-only model predicted retreat of both stripes (Fig. 4 E and F, adapted with permission from ref. 24). We hypothesized that the endogenous and reporter patterns would respond differently to hb misexpression if Hb bifunctionally regulates the endogenous pattern but only represses the reporter pattern.
Fig. 4.
In hb ventral misexpression (sna::hb) embryos the bifunctional model predicts the endogenous pattern whereas the repressor-only model predicts the eve3+7 reporter pattern. (A) Hb protein in WT and sna::hb embryos (Left, lateral view; Right, ventral view). These data are computational renderings of gene expression atlases that average together data from multiple embryos (see Fig. S8 for number of embryos per time point). The relative expression level of each gene is shown in individual cells; cells with expression below an ON/OFF threshold (Materials and Methods) are plotted in gray. For cells above this threshold darker colors indicate higher relative amounts. (B) Endogenous eve pattern. (C) The eve3+7 reporter pattern. Both stripes retreat from ectopic Hb. (D) The eve2+7 reporter pattern. Stripe 7 expands into the ectopic Hb domain. (E and F) Bottom (ventral) view of predictions of the bifunctional model (E) and repressor-only model (F) based on simulated sna::hb data (adapted with permission from ref. 24). OFF cells are light pink and ON cells are red. All data and modeling are from cohort 3 (other time points are shown in Fig. S8).
We measured both patterns quantitatively at cellular resolution in sna::hb embryos (Fig. 4). As previously observed, the endogenous eve stripe 3 retreated from the ventral Hb domain and bent posteriorly, whereas the endogenous stripe 7 expanded and bent anteriorly, consistent with the bifunctional model (Fig. 4 B and E). By contrast, in the eve3+7 reporter pattern both stripes retreated from the ventral Hb domain, consistent with the repressor-only model (Fig. 4 C and F).
Two Shadow Enhancers Enable Bifunctional Hb Regulation of eve Stripe 7.
We hypothesized that additional regulatory DNA in the locus is activated by Hb to produce the eve stripe 7 bulge in sna::hb embryos. We tested an extended version of the minimal eve2 enhancer for this activity based on several previous observations. Hb is known to activate the eve2 enhancer (19, 20, 40, 41); longer versions of eve2 drive stripe 7 in some embryos (28, 31, 32, 40); orthologous eve2 enhancers from other species sometimes drive stripe 7 expression (33, 42); and, finally, in sna::hb embryos the border of the expanded stripe 7 seems to be set by Krüppel (Kr), a known regulator of eve2 (Fig. S7 and refs. 19 and 41). The fragment we chose drives both stripes 2 and 7 (Fig. S8 and Table S3); we call this enhancer reporter construct eve2+7.
In sna::hb embryos the stripe 7 region of the eve2+7 reporter pattern expanded, recapitulating the bulge observed in the endogenous eve pattern (Fig. 4 B and D). We conclude that Hb activates endogenous eve stripe 7 through the eve2+7 enhancer. Taken together, our results indicate eve stripe 7 expression is controlled by at least two enhancers with different regulatory logic.
Discussion
To test whether Hb can activate and repress the same enhancer we used quantitative data to challenge two computational models that formalize different roles for Hb. We measured expression of endogenous eve and transgenic reporter constructs at cellular resolution under two genetic perturbations. By comparing the regulatory logic of the endogenous and eve3+7 reporter patterns we uncovered two enhancers that both direct expression of eve stripe 7. These shadow enhancers direct the same pattern in different ways: one is activated by Hb whereas the other is repressed. This form of regulatory redundancy enables Hb to “drive with the brakes on” to control eve stripe 7.
Two Shadow Enhancers Control eve Stripe 7 Expression.
Early studies suggested control of eve stripe 7 expression was distributed over DNA encompassing both the minimal eve3+7 and eve2 enhancers (19, 23, 31, 32, 40). We find that there are at least two pieces of regulatory DNA in this region that position stripe 7. The minimal eve3+7 enhancer is repressed by Hb (22, 23, 43), whereas the eve2+7 enhancer, which encompasses the minimal eve2 enhancer, is activated by Hb. This activation may be direct or indirect. Based on the results presented here we cannot rule out the possibility that the bulge of eve2+7 in sna::hb embryos is due to indirect Hb activity, and the result of activation by other TFs and retreat of Gt and Kni (22, 44). However, we hypothesize that Hb activation of eve2+7 is direct. If Hb activation of eve2+7 is indirect, Hb binding to eve2+7 in these cells would have to have little or no effect on stripe 7 expression (45). Moreover, Hb binds to and activates the minimal eve2 enhancer (19, 20, 40, 45, 46).
In addition to responding to Hb in opposite ways, the eve2+7 and eve3+7 enhancers are likely differentially sensitive to additional TFs. eve3+7 is activated by Stat92E and Zelda (43, 47). The anterior border of stripe 7 is set by Kni repression, and the posterior border is set by Hb repression (22, 23, 43). The minimal eve2 enhancer is activated by Bcd and Hb, its anterior boundary is set by Gt, and its posterior boundary is set by Kr (19, 20, 40, 41). In agreement with others, we speculate that the anterior boundary of eve stripe 7 in eve2+7 may be set by Gt (28). Taken together, this evidence argues that eve3+7 and eve2+7 position stripe 7 using different regulatory logic.
The molecular mechanism by which Hb represses and activates remains unclear. One hypothesis is that other TFs bound nearby convert Hb from a repressor into an activator, as is the case for Dorsal (9–13). There is genetic evidence for activator synergy between Bcd and Hb (19, 48), and activator synergy between Hb and Caudal has been proposed by computational work (30). Another hypothesis is that Hb monomers are activators but DNA-bound Hb dimers are repressors (25, 49). Testing these hypotheses will require quantitative data in additional genetic backgrounds and mutagenesis of individual binding sites in the two enhancers.
Comparing the Regulatory Logic of Reporter and Endogenous Patterns May Be Helpful for Mapping Regulatory DNA.
“Veteran enhancer-bashers, and those who carefully read the papers, know that ‘minimal’ enhancer fragments do not always perfectly replicate the precise spatial boundaries of expression of the native gene…” (34). Our data clearly support this often neglected aspect of enhancer reporter constructs. One explanation offered for such discrepancies is different transcript properties. We controlled for this possibility and conclude that transcript properties contribute to the differences between reporter and endogenous patterns but are not the only source. Here, we find that additional regulatory DNA in the locus also plays a role.
Finding all of the active regulatory DNA in a locus is challenging. Enhancer reporter constructs are powerful but can only determine whether a piece of DNA is sufficient to drive a particular pattern in isolation when placed next to the promoter. By comparing the regulatory logic of the eve3+7 reporter pattern and the endogenous pattern we uncovered an additional feature of eve regulation. However, eve3+7 and eve2+7 may not contain all of the DNA that contributes to stripe 7 expression in vivo. Emerging technologies for manipulating the endogenous locus and larger reporter constructs will be helpful for comprehensively mapping regulatory DNA (50–52).
The Bifunctional Model Is a Superposition of Two Computations.
Models are not ends, but merely means to formalize assumptions and develop falsifiable hypotheses (53, 54). The bifunctional model accurately predicts the behavior of the endogenous eve stripe 7 pattern in WT and perturbed embryos, but it does not predict the behavior of either eve3+7 or eve2+7. The interpretation in Ilsley et al. (24) that Hb bifunctionally regulated eve3+7 was based on a common assumption: that the endogenous pattern could be attributed to the annotated enhancer. Here we show that Hb bifunctionality is due to separate enhancers. Ilsley et al. (24) interpreted the success of the bifunctional model as evidence for concentration-dependent control of Hb activation and repression, as has been proposed for Hb and other TFs (17, 25, 55). This interpretation cannot be true because Hb activates and represses in the same cells. Our favored hypothesis is that Hb bifunctionality is controlled by sequence features in each enhancer.
The bifunctional model effectively behaves as a superposition of the eve3+7 and eve2+7 enhancer activities to accurately predict the behavior of the endogenous locus. It is currently unclear how multiple active enhancers impinge on the same promoter, which makes it challenging to predict their combined behavior. The promoter may integrate information from multiple enhancers in various ways, ranging from independent addition to dominance of one enhancer owing to a long-range repressor (34, 56, 57). The behavior of stripe 7 is not consistent with dominant repression by Hb, but we cannot rule out any other mechanisms. Elucidating how promoters integrate information will be critical for predicting the behavior of complex developmental loci where shadow enhancers are prevalent.
Conclusion
By combining computational modeling and directed experiments we uncovered a previously unidentified feature of a highly studied locus, long held up as a textbook example of modular enhancer organization (58). We tested the hypothesis that Hb bifunctionally regulates the eve3+7 enhancer and discovered that bifunctionality is due to two enhancers that respond to Hb in opposite ways. This example provides an opportunity to uncover how Hb bifunctionality is controlled, which will improve our ability to interpret regulatory DNA and infer connections in gene regulatory networks.
Regulatory redundancy in control of eve stripe 7 expression may have functional consequences. Shadow enhancers may arise from genetic drift (59); however, shadow enhancers in other developmental loci confer robustness to genetic or environmental stresses (56, 60, 61), facilitate temporal refinement of patterns (62), and/or increase expression synchrony and precision (63). This example demonstrates that shadow enhancers can use different regulatory logic to position the same pattern, which may have useful properties for the embryo.
Materials and Methods
Fly Work.
The bcd RNAi gene expression atlas is described in Staller et al. (35) and available at dx.doi.org/10.6084/m9.figshare.1270915 and https://depace.med.harvard.edu. Briefly, we combined short hairpin RNA knockdown of bcd with in situ hybridization and two-photon imaging and automated image segmentation (38, 64–66). Hb protein stains used a guinea pig anti-Hb from John Reinitz, University of Chicago, Chicago, IL. Embryos were partitioned into six time points using the degree of membrane invagination (0–3, 4–8, 9–25, 26–50, 51–75, and 76–100%), which evenly divide the ∼60-min blastoderm stage (37). All enhancer reporters are in pBOY and integrated at attP2 (33, 67) (Table S3). The eve locus lacZ reporter was a gift from Miki Fujioka, Thomas Jefferson University, Philadelphia. hb ventral misexpression was performed as described in Clyde et al. (22) using two copies of the sna::hb transgene on chromosome 2.
Building the Coarsely Aligned sna::hb Gene Expression Atlas.
We determined the genotype of the sna::hb embryos by examining the eve or fushi-tarazu (ftz) mRNA patterns. Embryos were aligned morphologically to create a coarsely registered gene expression atlas (38). Data are available at dx.doi.org/10.6084/m9.figshare.1270915 and https://depace.med.harvard.edu.
Logistic Modeling of Enhancer Gene Regulatory Functions.
The logistic modeling framework was developed and described in detail previously (24). All modeling was performed in MATLAB (MathWorks) using the DIP image toolbox (www.diplib.org) and the PointCloudToolBox (bdtnp.lbl.gov). Ilsley et al. (24) used protein data for Gt, whereas we used mRNA data. For genes where we used mRNA data the mRNA and protein patterns are correlated (38, 68). For the enhancer lacZ reporters we thresholded cells to be ON or OFF by creating a histogram of the expression data (50 bins), identifying the bin with the most counts and adding one SD. Our ON set included all cells expressing the reporter, and our OFF set includes all other cells. All regulators are maintained as continuous values.
To threshold the endogenous WT eve pattern into ON and OFF cells we used 0.2 for all time points (24). To threshold the endogenous eve patterns in the bcd RNAi atlas we used the lowest threshold that would separate the stripes: 0.1, 0.15, 0.15, 0.2, and 0.21 for T = 2 through T = 6, respectively. To compare the modeling of the reporter and the endogenous patterns, the ON set included all cells in the endogenous eve stripes 3 and 7 and the OFF set included all other cells. This OFF set is different from that in Ilsley et al. (24), but this change does not have a large effect on the AUC scores in bcd RNAi embryos (Tables S1 and S2).
Sensitivity Analysis.
For the sensitivity analysis (Fig. S5), for each TF we scaled the concentration of the bcd RNAi atlas in silico and recomputed the model AUC scores.
Binding Site Predictions.
For the Kr binding site analysis in Fig. S7 we predicted binding sites using PATSER (stormo.wustl.edu) with a position weight matrix derived from bacterial one-hybrid data (69). Binding sites were visualized using InSite (cs.utah.edu/∼miriah/projects).
Quantifying Concordance Between Reporters and Endogenous Patterns.
For each embryo we used the PointCloudToolBox in MATLAB to find pattern boundaries by creating 16 anterior–posterior line traces and finding the inflection point of each trace (adapted from DEMO_EVESTRIPES in the PointCloudToolBox, bdtnp.lbl.gov). Finding the boundary by using half the maximum value of the stripe peak identifies a very similar boundary to the inflection point. To find the peaks of the endogenous and reporter stripes we took one lateral line trace (extractpattern function) and found the local maxima.
Supplementary Material
Acknowledgments
We thank Miki Fujioka for sharing the eve locus reporter flies ahead of publication; Kelly Eckenrode for staining the WT reporter embryos; Steve Small for the sna::hb flies; Garth Ilsley for developing the initial models, help with figures, and stimulating discussions; John Reinitz for the Hb antibody; and Steve Small, Becky Ward, Garth Ilsley, Peter Combs, Alistair Boettiger, two anonymous reviewers, and members of the A.H.D. laboratory for comments on the manuscript. This work was supported by the Harvard Herchel Smith Graduate Student Fellowship (to M.V.S.), Jane Coffin Childs Memorial Fund for Medical Research (to Z.W.), National Institutes of Health (NIH) Grant K99HD073191 (to Z.W.), and NIH Grant U01 GM103804-01A1 (to A.H.D.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413877112/-/DCSupplemental.
References
- 1.Struhl G, Johnston P, Lawrence PA. Control of Drosophila body pattern by the hunchback morphogen gradient. Cell. 1992;69(2):237–249. doi: 10.1016/0092-8674(92)90405-2. [DOI] [PubMed] [Google Scholar]
- 2.Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: A multifaceted protein beyond a transcription factor. Transcription. 2010;1(2):81–84. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Stefano B, et al. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature. 2014;506(7487):235–239. doi: 10.1038/nature12885. [DOI] [PubMed] [Google Scholar]
- 4.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 5.Lynch VJ, May G, Wagner GP. Regulatory evolution through divergence of a phosphoswitch in the transcription factor CEBPB. Nature. 2011;480(7377):383–386. doi: 10.1038/nature10595. [DOI] [PubMed] [Google Scholar]
- 6.Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 7.Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126(Pt 10):2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine M, Cattoglio C, Tjian R. Looping back to leap forward: Transcription enters a new era. Cell. 2014;157(1):13–25. doi: 10.1016/j.cell.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirokawa JM, Courey AJ. A direct contact between the dorsal rel homology domain and Twist may mediate transcriptional synergy. Mol Cell Biol. 1997;17(6):3345–3355. doi: 10.1128/mcb.17.6.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72(5):741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 11.Dubnicoff T, et al. Conversion of dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev. 1997;11(22):2952–2957. doi: 10.1101/gad.11.22.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores-Saaib RD, Jia S, Courey AJ. Activation and repression by the C-terminal domain of Dorsal. Development. 2001;128(10):1869–1879. doi: 10.1242/dev.128.10.1869. [DOI] [PubMed] [Google Scholar]
- 13.Ratnaparkhi GS, Jia S, Courey AJ. Uncoupling dorsal-mediated activation from dorsal-mediated repression in the Drosophila embryo. Development. 2006;133(22):4409–4414. doi: 10.1242/dev.02643. [DOI] [PubMed] [Google Scholar]
- 14.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Latchman DS. Transcription factors: Bound to activate or repress. Trends Biochem Sci. 2001;26(4):211–213. doi: 10.1016/s0968-0004(01)01812-6. [DOI] [PubMed] [Google Scholar]
- 16.Kim HD, Shay T, O’Shea EK, Regev A. Transcriptional regulatory circuits: Predicting numbers from alphabets. Science. 2009;325(5939):429–432. doi: 10.1126/science.1171347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz C, Tautz D. Autonomous concentration-dependent activation and repression of Krüppel by hunchback in the Drosophila embryo. Development. 1994;120(10):3043–3049. doi: 10.1242/dev.120.10.3043. [DOI] [PubMed] [Google Scholar]
- 18.Zuo P, et al. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991;5(2):254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]
- 19.Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev. 1991;5(5):827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- 20.Arnosti DN, Barolo S, Levine M, Small S. The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development. 1996;122(1):205–214. doi: 10.1242/dev.122.1.205. [DOI] [PubMed] [Google Scholar]
- 21.Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126(11):2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clyde DE, et al. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426(6968):849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 23.Small S, Blair A, Levine M. Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev Biol. 1996;175(2):314–324. doi: 10.1006/dbio.1996.0117. [DOI] [PubMed] [Google Scholar]
- 24.Ilsley GR, Fisher J, Apweiler R, DePace AH, Luscombe NM. Cellular resolution models for even skipped regulation in the entire Drosophila embryo. eLife. 2013;2:e00522. doi: 10.7554/eLife.00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papatsenko D, Levine MS. Dual regulation by the Hunchback gradient in the Drosophila embryo. Proc Natl Acad Sci USA. 2008;105(8):2901–2906. doi: 10.1073/pnas.0711941105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Samee MAH, Blatti C, Sinha S. Thermodynamics-based models of transcriptional regulation by enhancers: the roles of synergistic activation, cooperative binding and short-range repression. PLOS Comput Biol. 2010;6(9):e1000935. doi: 10.1371/journal.pcbi.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazemian M, et al. Quantitative analysis of the Drosophila segmentation regulatory network using pattern generating potentials. PLoS Biol. 2010;8(8):e1000456. doi: 10.1371/journal.pbio.1000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssens H, et al. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat Genet. 2006;38(10):1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- 29.Sherman MS, Cohen BA. Thermodynamic state ensemble models of cis-regulation. PLOS Comput Biol. 2012;8(3):e1002407. doi: 10.1371/journal.pcbi.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim A-R, et al. Rearrangements of 2.5 kilobases of noncoding DNA from the Drosophila even-skipped locus define predictive rules of genomic cis-regulatory logic. PLoS Genet. 2013;9(2):e1003243. doi: 10.1371/journal.pgen.1003243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goto T, Macdonald P, Maniatis T. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell. 1989;57(3):413–422. doi: 10.1016/0092-8674(89)90916-1. [DOI] [PubMed] [Google Scholar]
- 32.Harding K, Hoey T, Warrior R, Levine M. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J. 1989;8(4):1205–1212. doi: 10.1002/j.1460-2075.1989.tb03493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4(6):e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barolo S. Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. BioEssays. 2012;34(2):135–141. doi: 10.1002/bies.201100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staller MV, et al. 2015. A gene expression atlas of a bicoid-depleted Drosophila embryo reveals early canalization of cell fate. Development, 10.1242/dev.117796.
- 36.Luengo Hendriks CL, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: Data acquisition pipeline. Genome Biol. 2006;7(12):R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keränen SVE, et al. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution II: Dynamics. Genome Biol. 2006;7(12):R124. doi: 10.1186/gb-2006-7-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fowlkes CC, et al. A quantitative spatiotemporal atlas of gene expression in the Drosophila blastoderm. Cell. 2008;133(2):364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 39.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 40.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11(11):4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanojevic D, Small S, Levine M. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science. 1991;254(5036):1385–1387. doi: 10.1126/science.1683715. [DOI] [PubMed] [Google Scholar]
- 42.Peterson BK, et al. Big genomes facilitate the comparative identification of regulatory elements. PLoS ONE. 2009;4(3):e4688. doi: 10.1371/journal.pone.0004688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struffi P, et al. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development. 2011;138(19):4291–4299. doi: 10.1242/dev.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu D, Small S. Precise registration of gene expression boundaries by a repressive morphogen in Drosophila. Curr Biol. 2008;18(12):868–876. doi: 10.1016/j.cub.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X-Y, et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 2008;6(2):e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanojević D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. 1989;341(6240):331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- 47.Yan R, Small S, Desplan C, Dearolf CR, Darnell JE., Jr Identification of a Stat gene that functions in Drosophila development. Cell. 1996;84(3):421–430. doi: 10.1016/s0092-8674(00)81287-8. [DOI] [PubMed] [Google Scholar]
- 48.Simpson-Brose M, Treisman J, Desplan C. Synergy between the hunchback and bicoid morphogens is required for anterior patterning in Drosophila. Cell. 1994;78(5):855–865. doi: 10.1016/s0092-8674(94)90622-x. [DOI] [PubMed] [Google Scholar]
- 49.Bieler J, Pozzorini C, Naef F. Whole-embryo modeling of early segmentation in Drosophila identifies robust and fragile expression domains. Biophys J. 2011;101(2):287–296. doi: 10.1016/j.bpj.2011.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venken KJ, Bellen HJ. Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and ΦC31 integrase. Methods Mol Biol. 2012;859:203–228. doi: 10.1007/978-1-61779-603-6_12. [DOI] [PubMed] [Google Scholar]
- 51.Ren X, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA. 2013;110(47):19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crocker J, Stern DL. TALE-mediated modulation of transcriptional enhancers in vivo. Nat Methods. 2013;10(8):762–767. doi: 10.1038/nmeth.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gunawardena J. Models in biology: ‘Accurate descriptions of our pathetic thinking’. BMC Biol. 2014;12:29. doi: 10.1186/1741-7007-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wunderlich Z, DePace AH. Modeling transcriptional networks in Drosophila development at multiple scales. Curr Opin Genet Dev. 2011;21(6):711–718. doi: 10.1016/j.gde.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Kelley KM, Wang H, Ratnam M. Dual regulation of ets-activated gene expression by SP1. Gene. 2003;307:87–97. doi: 10.1016/s0378-1119(03)00445-1. [DOI] [PubMed] [Google Scholar]
- 56.Dunipace L, Ozdemir A, Stathopoulos A. Complex interactions between cis-regulatory modules in native conformation are critical for Drosophila snail expression. Development. 2011;138(18):4075–4084. doi: 10.1242/dev.069146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci USA. 2011;108(33):13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maeda RK, Karch F. Gene expression in time and space: Additive vs hierarchical organization of cis-regulatory regions. Curr Opin Genet Dev. 2011;21(2):187–193. doi: 10.1016/j.gde.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 59.Lynch M. The frailty of adaptive hypotheses for the origins of organismal complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8597–8604. doi: 10.1073/pnas.0702207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20(17):1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466(7305):490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunipace L, Saunders A, Ashe HL, Stathopoulos A. Autoregulatory feedback controls sequential action of cis-regulatory modules at the brinker locus. Dev Cell. 2013;26(5):536–543. doi: 10.1016/j.devcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325(5939):471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fowlkes CC, et al. A conserved developmental patterning network produces quantitatively different output in multiple species of Drosophila. PLoS Genet. 2011;7(10):e1002346. doi: 10.1371/journal.pgen.1002346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wunderlich Z, et al. Dissecting sources of quantitative gene expression pattern divergence between Drosophila species. Mol Syst Biol. 2012;8:604. doi: 10.1038/msb.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staller MV, et al. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 2013;193(1):51–61. doi: 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pisarev A, Poustelnikova E, Samsonova M, Reinitz J. FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution. Nucleic Acids Res. 2009;37(Database issue):D560–D566. doi: 10.1093/nar/gkn717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Noyes MB, et al. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36(8):2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.